The germline genome is constantly challenged by retrotransposons, the harmful mobile genetic elements that can copy and paste themselves in the genome to introduce insertional mutations [1]. In humans, the most successful retrotransposons is the long-interspersed element type 1 (LINE-1, or L1) family that comprises ∼17% of the human genome. Mobilization of L1 can generate insertional mutations that drive tumorigenesis and human genetic diseases [2]. Because of the danger for mutations to be passed along to the next generation, retrotransposons such as L1 have to be especially tightly controlled in the germline. To cope with this threat, germ cells have developed the PIWI-interacting RNA (piRNA) pathway as a germ cell-specific small RNA-based immune system to suppress retrotransposon activities [3,4]. The central players in this pathway are PIWI proteins and their associated 24–32 nt piRNAs wherein the PIWI protein uses piRNA as a guide to recognize and suppress retrotransposon expression through transcriptional and posttranscriptional gene silencing.
The timing and frequency of how individual L1s propagate and mobilize within the genome during germ cell development is unknown because the mobilization of individual L1s is extremely difficult to track due to the abundance of preexisting L1 copies in the genome. In a report by Newkirk et al. [5], a research group led by Wenfeng An solved this mystery by elegantly tracking the mobilization of a single copy L1 reporter transgene in a piRNA-deficient background in mice. This allows for the first time the accurate monitoring of L1 transposition in developing germ cells at different stages of spermatogenesis.
To address the above question, Newkirk et al. generated a mouse line with a stable inheritance of a single copy of L1 reporter transgene. This mouse model is advantageous over previous L1 reporter transgenic lines (e.g. CAG-L1 [6]) in that it contains a transgene with an endogenous mouse L1 promoter, codon-optimized L1 ORF1 and ORF2 sequences, and an EGFP-based retrotransposition indicator cassette. The stable single copy of L1 transgene in mice is important in that it faithfully mimics the endogenous L1 promoter DNA methylation dynamics, which is not represented by mouse models with high copy L1 transgenes or human tandem L1 transgenes. These features together ensure the optimal expression and insertion of L1 transgene that recapitulates that of endogenous L1s.
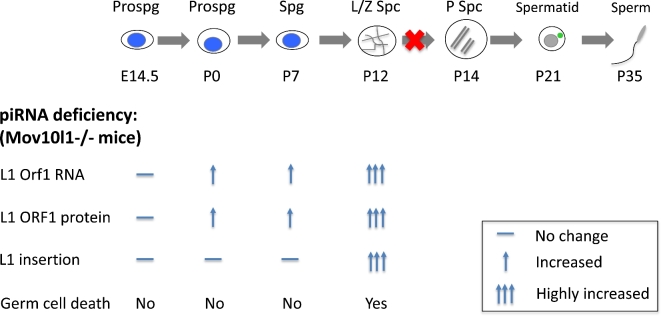
Although piRNAs are essential for retrotransposon silencing and male fertility in mice, a long puzzling question for reproductive biologists in the piRNA field is that whether piRNA deficiency-triggered elevated L1 activity indeed directly drives germ cell demise. It has been noticed in various piRNA mutants that the defective piRNA production and L1 ORF1 protein overexpression start in fetal germ cells, whereas germ cell arrest does not occur until meiosis [1]. It is unclear why there is a delay from L1 expression to germ cell death. Understanding the timing and frequency of L1 mobilization (insertion) in piRNA-deficient background holds the key to solve this puzzle. To do this, the authors crossed the newly developed L1 reporter line (referred to as SN1) to Mov10l1–/– mice that are deficient in piRNAs. MOV10l1 is an RNA helicase critical for piRNA biogenesis and Mov10l1–/– mice show L1 upregulation and germ cell arrest at meiotic prophase I [7,8]. By tracing de novo insertions of SN1 L1 reporter transgene in control and Mov10l1–/– germ cells, the authors showed that at postnatal day 7, despite a 37-fold increase in ORF1 mRNA levels and a significant elevation in ORF1 protein expression, L1 transposition rate is low in Mov10l1–/– spermatogonia. This low L1 insertions rate is similar to that of the control. Strikingly, the dramatic L1 insertions occur days later in Mov10l1–/– spermatocytes at the onset of meiosis. At P14, new SN1 L1 insertions increase by 144-fold in Mov10l1–/– spermatocytes. This delayed onset of L1 retrotransposition correlates perfectly with the timing of germ cell death (Figure 1). Would this increased retrotransposition be powerful enough to drive the meiotic arrest in Mov10l1–/– germ cells? By estimating the frequency of retrotransposition, the number of new insertions per cell was 2.25 for endogenous L1s. This seemingly low numbers of insertions per cell appear unlikely to directly cause germ cell demise. Accordingly, the authors demonstrate that the pharmacological inhibition of L1 retrotransposition in turn did not rescue the meiotic phenotype of Mov10l1–/– mice. Thus, the sheer numbers of new L1 insertional mutations in the germline may not be the decisive factor to drive piRNA-deficient germ cell death. However, one cannot exclude the possibilities that other aspects of elevated L1 activities in mutant germ cells may contribute to germ cell arrest. One possibility is that overexpression of L1 ORF2, encoding both an endonuclease and a reverse transcriptase necessary for L1 transposition, may introduce excessive DNA damage without completing L1 insertions in piRNA-deficient germ cells. Another possibility is that L1 insertions may disrupt transcriptomic integrity, which systematically influences germ cell health [9]. These possibilities need to be further investigated.
Figure 1.
Schematic illustration of the developmental timing of L1 expression and insertion in piRNA-deficient germ cells during spermatogenesis. In piRNA-deficient Mov10l1–/– germ cells, L1 RNA and protein upregulation is first seen in fetal/perinatal prospermatogonia (Prospg), which is, however, not accompanied by obvious increased L1 insertions and germ cell death. Drastic L1 overexpression and mobilization occurs in leptotene/zygotene spermatocytes (L/Z Spc), which correlates with the timing of germ cell arrest before the pachytene spermatocyte (P Spc) stage of meiotic prophase I.
In summary, Newkirk et al. created an L1 transgenic mouse line uniquely suitable to monitor L1 mobilization in vivo. It reveals the stage-specific increases in L1 retrotransposition during spermatogenesis and confirms that an intact piRNA pathway is critical to prevent L1 mobilization. This SN1 L1 mouse model will also be a useful tool for reproductive biologists to study the mechanism of female germ cell retrotransposon regulation and the effect of genome perturbation by L1 in gametes on embryonic development and offspring health and fertility. Lastly, it can serve as a new model for studying a critical aspect of environmental epigenetics [10], i.e. how environmental factors alter retrotransposon regulation and impart genetic changes through insertional mutagenesis.
References
- 1. Bao J, Yan W. Male germline control of transposable elements. Biol Reprod 2012; 86(162):161–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hancks DC, Kazazian HH. Roles for retrotransposon insertions in human disease. Mobile DNA 2016; 7:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol 2011; 12:246–258. [DOI] [PubMed] [Google Scholar]
- 4. Juliano C, Wang J, Lin H. Uniting germline and stem cells: the function of Piwi proteins and the piRNA pathway in diverse organisms. Annu Rev Genet 2011; 45:447–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Newkirk SJ, Lee S, Grandi FC, Gaysinskaya V, Rosser JM, Vanden Berg N, Hogarth CA, Marchetto MCN, Muotri AR, Griswold MD, Ye P, Bortvin A et al. . Intact piRNA pathway prevents L1 mobilization in male meiosis. Proc Natl Acad Sci USA 2017; 114:E5635–E5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. An W, Han JS, Wheelan SJ, Davis ES, Coombes CE, Ye P, Triplett C, Boeke JD. Active retrotransposition by a synthetic L1 element in mice. Proc Natl Acad Sci USA 2006; 103:18662–18667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zheng K, Xiol J, Reuter M, Eckardt S, Leu NA, Mclaughlin KJ, Stark A, Sachidanandam R, Pillai RS, Wang PJ. Mouse MOV10L1 associates with Piwi proteins and is an essential component of the Piwi-interacting RNA (piRNA) pathway. Proc Natl Acad Sci USA 2010; 107:11841–11846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vourekas A, Zheng K, Fu Q, Maragkakis M, Alexiou P, Ma J, Pillai RS, Mourelatos Z, Wang PJ. The RNA helicase MOV10L1 binds piRNA precursors to initiate piRNA processing. Genes Dev 2015; 29:617–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Inoue K, Ichiyanagi K, Fukuda K, Glinka M, Sasaki H. Switching of dominant retrotransposon silencing strategies from posttranscriptional to transcriptional mechanisms during male germ-cell development in mice. PLoS Genet 2017; 13:e1006926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rosenfeld CS. Animal models to study environmental epigenetics1. Biol Reprod 2010; 82:473–488. [DOI] [PubMed] [Google Scholar]