Abstract
Background
Detection of circulating tumor DNA can be limited due to their relative scarcity in circulation, particularly while patients are actively undergoing therapy. Exosomes provide a vehicle through which cancer-specific material can be enriched from the compendium of circulating non-neoplastic tissue-derived nucleic acids. We carried out a comprehensive profiling of the pancreatic ductal adenocarcinoma (PDAC) exosomal ‘surfaceome’ in order to identify surface proteins that will render liquid biopsies amenable to cancer-derived exosome enrichment for downstream molecular profiling.
Patients and methods
Surface exosomal proteins were profiled in 13 human PDAC and 2 non-neoplastic cell lines by liquid chromatography–mass spectrometry. A total of 173 prospectively collected blood samples from 103 PDAC patients underwent exosome isolation. Droplet digital PCR was used on 74 patients (136 total exosome samples) to determine baseline KRAS mutation call rates while patients were on therapy. PDAC-specific exosome capture was then carried out on additional 29 patients (37 samples) using an antibody cocktail directed against selected proteins, followed by droplet digital PCR analysis. Exosomal DNA in a PDAC patient resistant to therapy were profiled using a molecular barcoded, targeted sequencing panel to determine the utility of enriched nucleic acid material for comprehensive molecular analysis.
Results
Proteomic analysis of the exosome ‘surfaceome’ revealed multiple PDAC-specific biomarker candidates: CLDN4, EPCAM, CD151, LGALS3BP, HIST2H2BE, and HIST2H2BF. KRAS mutations in total exosomes were detected in 44.1% of patients undergoing active therapy compared with 73.0% following exosome capture using the selected biomarkers. Enrichment of exosomal cargo was amenable to molecular profiling, elucidating a putative mechanism of resistance to PARP inhibitor therapy in a patient harboring a BRCA2 mutation.
Conclusion
Exosomes provide unique opportunities in the context of liquid biopsies for enrichment of tumor-specific material in circulation. We present a comprehensive surfaceome characterization of PDAC exosomes which allows for capture and molecular profiling of tumor-derived DNA.
Keywords: exosomes, liquid biopsies, pancreatic cancer, proteomics, next-generation sequencing
Introduction
An emerging body of literature demonstrates that comprehensive characterization of cancer genomes has both diagnostic and prognostic utility, and may provide insights into formulating individualized treatment strategies [1, 2]. However, even with large-scale sequencing efforts, accessibility of tumor tissue is often limited by both patient- and/or system-centered factors. Tissue sampling of pancreatic ductal adenocarcinoma (PDAC) may be limited to an initial diagnostic fine needle aspiration (FNA), while risk of biopsy, locally destructive therapies, cost, or facility capabilities may limit further sampling efforts. ‘Liquid biopsy’ is a less invasive strategy for tumor sampling, which may circumvent the need for tissue biopsy while still affording high-resolution profiling of the genomic landscapes of visceral cancers. Within the field of liquid biopsy, tumor-derived extracellular vesicles (EVs), such as exosomes, are a source of high-quality nucleic acids for molecular characterization with inherent utility for diagnostic and therapeutic purposes [3].
Exosomes are nanometer-sized membrane-limited EVs that arise from endosomal biogenesis pathways and serve as critical means of cell–cell communication [4]. Tumor-derived exosomes contain membrane-tethered proteins, microRNAs, and as recently demonstrated, entire genomic complements of DNA (exoDNA) [5–8]. Exosomes are shed from both tumor and non-neoplastic cells into the peripheral circulation. Therefore, one of the potential pitfalls of utilizing exosomes, and essentially any liquid biopsy component including circulating tumor DNA (ctDNA), as a surrogate for the tumor genome is that genetic information obtained from such exosomes will be diluted in large part with the DNA of non–cancer cell-derived exosomes. Exosomal surface biomarkers provide a means to separate cancer from non-cancer-derived exosomes through the use of bead-based selection of such markers. While exosomes are known to express tetraspanins such as CD63, CD9, and CD81, these biomarkers are not specific to cancer-derived exosomes. Specific markers to distinguish normal and cancer tissue-derived exosomes is an area of active research, particularly in PDAC where the use of such biomarkers has great potential in early disease detection [9, 10]. Here, we identify a panel of PDAC-specific exosomal surface proteins, demonstrate the ability to enrich for PDAC-derived exosomes in circulation using these identified proteins, and then show the feasibility of mutation profiling of enriched exoDNA samples through next-generation sequencing (NGS).
Patients and methods
Exosomes were isolated from a total of 13 human PDAC cell lines, MIAPaCa-2, Pa01C, Pa02C, Pa03C, Pa04C, Pa07C, Pa08C, Pa09C, Pa021C, Pa028C, PATC43, PATC66, and PATC92, and two non-neoplastic cell lines, CAF19 and HPNE. Proteins from exosome surface were tagged with biotin and separated from the cargo compartment after being affinity captured to monomeric avidin and subject to liquid chromatography–mass spectrometry (MS) (supplementary Figure S1, available at Annals of Oncology online). Identified exosomal surface proteins were curated based on their tumor-specific expression when compared with non-neoplastic sources. Biomarker candidates based on this initial screen were then validated through western blot where protein expression was evaluated between PDAC and non-neoplastic cell lines. The selected list of protein biomarker candidates was then evaluated in prospectively collected clinical samples from early stage (resectable) and late stage (locally advanced and metastatic) PDAC patients. First, a baseline exoDNA mutant KRAS detection rate using droplet digital PCR (ddPCR) was determined for 74 patients undergoing active therapy (supplementary Table S1, available at Annals of Oncology online). Tumor-specific exosome enrichment was then carried out on a separate cohort of 29 patients through an immunocapture-based methodology for exoDNA KRAS mutation detection. Finally, tumor-enriched exoDNA from a metastatic PDAC patient was subject to a molecular barcoded targeted cancer panel for comprehensive molecular profiling. Detailed methods can be found in the supplementary Materials and methods, available at Annals of Oncology online.
Results
‘Surfaceome’ profiling of exosomes
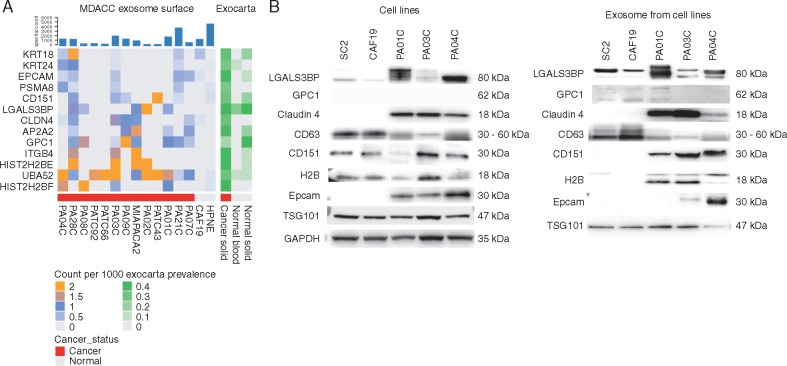
Surface and cargo exosomal proteins were profiled in 13 human PDAC cell lines and 2 non-neoplastic cell lines (HPNE and CAF19) through liquid chromatography–MS. Proteomes from exosome surface and cargo were fractionated at an intact protein level and then subjected to trypsin digestion and MS-based analysis. A total of 7086 proteins (corresponding to 3663 gene symbols) were identified (supplementary Table S2, available at Annals of Oncology online). Requiring expression on the surface of at least three samples (i.e. the proposed exosomal ‘surfaceome’) demonstrated the presence of canonical proteins universally expressed in exosomal populations including CD81, CD9, and TSG101 resulting in 1057 proteins (corresponding to 482 genes; supplementary Table S2, available at Annals of Oncology online). In order to identify a panel of PDAC-specific surface exosomal markers, resulting ‘surfaceome’ proteins that were found to be expressed in at least three PDAC cell lines with a maximum of 1 spectral count being expressed in non-neoplastic cell lines were considered candidate PDAC-specific exosomal surface markers. In addition, we annotated these candidates using the EV database ExoCarta (database of exosomes proteomics, including data from 160 exosome experiments and 166 samples based on MS analyses), which contains human exosome protein profiles from normal and cancer tissue sources to effectively assess the absence of our candidate proteins from vesicles of non-neoplastic origin. Further curation and validation of these biomarkers was prioritized based on biological rationale and availability of targeting antibodies (supplementary Materials and methods, available at Annals of Oncology online) (Figure 1).
Figure 1.
Cancer-specific exosomal biomarker selection and validation. (A) Heat map representation of proteins that are expressed on the surface of pancreatic ductal adenocarcinoma exosomes that are not expressed (or expressed at very low levels) on the surface of HPNE and CAF19 exosomes. (B) Western blot validation of identified candidate biomarkers: protein expression analysis of cell lysates (left) compared with exosomes (right) of neoplastic (Pa01C, Pa03C, and Pa04C) and non-neoplastic (CAF19 and SC2) cell lines. CD63 and TSG101 are used as antibody controls for identification of exosome populations. Most selected biomarkers show enriched specificity towards being present in cancer exosomes versus normal exosomes.
Biomarker validation
Candidate proteins were validated through western blot analysis of PDAC cell line-derived exosomes from Pa01C, Pa03C, and Pa04C (Figure 1). Non-neoplastic cell lines CAF19 and SC2 were used as controls. Candidate biomarkers were detected within protein lysates of cell lines with varying degrees of sensitivity and specificity, but were effectively enriched within the exosome protein fractions. In other words, protein biomarkers such as CD151 and HistoneH2B (H2B) are found in the protein lysates from all cell lines, including non-neoplastic cell lines, but are only found within exosomes derived from PDAC cell lines. On the other hand, LGALS3BP is present in all exosomal populations but is overexpressed in tumor-derived exosomes when compared with non-neoplastic sources. In contrast, the recently published PDAC exosomal biomarker glypican-1 (GPC1) did not demonstrate significant expression in tumor-derived exosomes and in fact appeared to be selectively expressed in non-neoplastic sources when four separate GPC1 antibodies were tested, including the originally reported clone (ThermoFisher, PA5-28055, Waltham, MA) (supplementary Figure S2, available at Annals of Oncology online) [9]. This profiling analysis led to a final antibody cocktail targeting the following candidate biomarkers: anti-CLDN4, EPCAM, CD151, LGALS3BP, HIST2H2BE and HIST2H2BF, respectively, used for subsequent enrichment studies.
Validation of capture assay in clinical samples
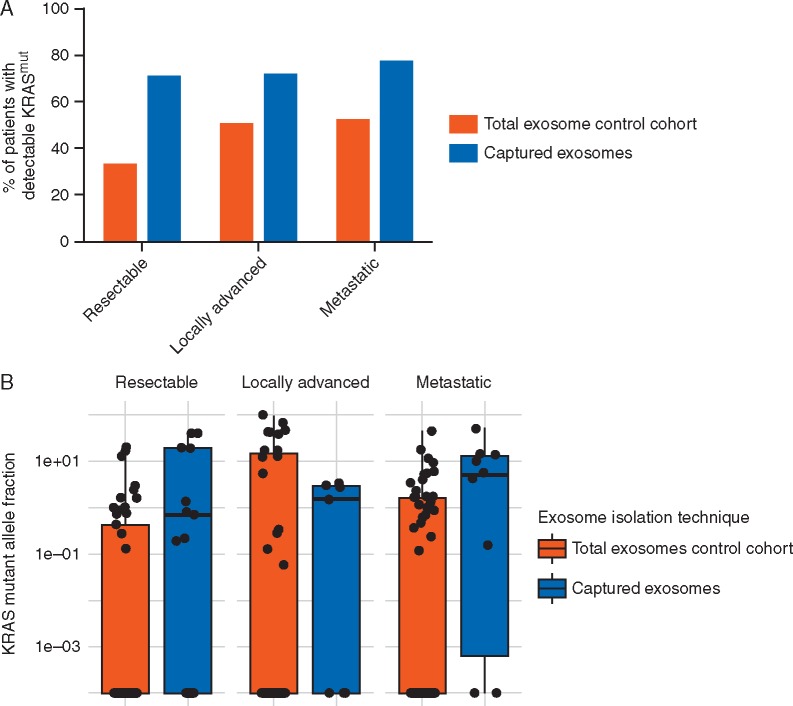
Following the selection of our candidate biomarkers, we designed an immunocapture pulldown assay to specifically capture enriched populations of cancer-derived exosomes (see supplementary Materials and methods, available at Annals of Oncology online). We next aimed to implement our enrichment methodology for PDAC-derived exosomes on patient plasma samples to determine its effectiveness at detecting tumor-derived DNA during therapy (supplementary Table S2, available at Annals of Oncology online). Detection rates for mutant KRAS exoDNA in a control cohort of 136 prospective samples that did not undergo capture enrichment (total exosomes) taken during active chemotherapeutic intervention was 32.7% (17/52), 50% (15/30), and 51.8% (28/54) in resectable, locally advanced, and metastatic disease, respectively, as defined by American Joint Committee on Cancer guidelines (Figure 2A). In 37 samples that underwent exosome capture as previously described, our mutation detection rate increased to 70.6% (12/17), 71.4% (5/7), and 76.9% (10/13) in resectable, locally advanced, and metastatic disease, respectively. Of note, these detection rates reach the theoretical upper limit of detection of our ddPCR multiplex assay which can detect up to 80% of known KRAS mutations found in PDAC [11]. This suggests that most patients undergoing therapy have tumor-derived material in circulation that is typically overwhelmed by non-neoplastic tissue-derived exosomes. Harvested exoDNA from both protocols yielded an average of 19.17 ng (0.11–125.72 ng) and 24.13 ng (0.12–636.00 ng) for captured exosomes and total exosomes, respectively. Overall positive call rate among all combined patients is associated with the pulldown cohort (P = 0.002) where a pulldown sample is 3.28 (95% CI: 1.41–8.19) times more likely to have KRAS detected. Importantly, exosome capture not only increases the proportion of cases with detectable mutant alleles but also leads to a statistically significant increase in KRAS MAF within each category, serving as a surrogate for tumor enrichment capability (Figure 2B).
Figure 2.
exoDNA KRAS mutant detection in circulation. (A) Percent of patients with detectable mutant KRAS in exoDNA among those patient samples that did and did not undergo capture enrichment. When comparing the percentages of patients with detectable KRAS in the pulldown-cohort versus the total exosome cohort, the pulldown-cohort consistently detects KRAS in a higher proportion of patients across stages. This increase in call-rate was statistically significant in resectable patients (P = 0.024) where pulldown samples were 4.11 (95% CI: 1.14–17.19) more likely to have KRAS detected. (B) KRAS mutant allele frequency (MAF) comparisons of captured exosomes versus total exosomes, there was a statistically significant difference showing increased KRAS MAFs from the captured exosomes for resectable and metastatic patients (P = 0.003 and 0.015, respectively, using one-sided Wilcoxon Rank Sum tests).
Enriched cancer-specific exosomal cargo is amenable to comprehensive molecular profiling by NGS
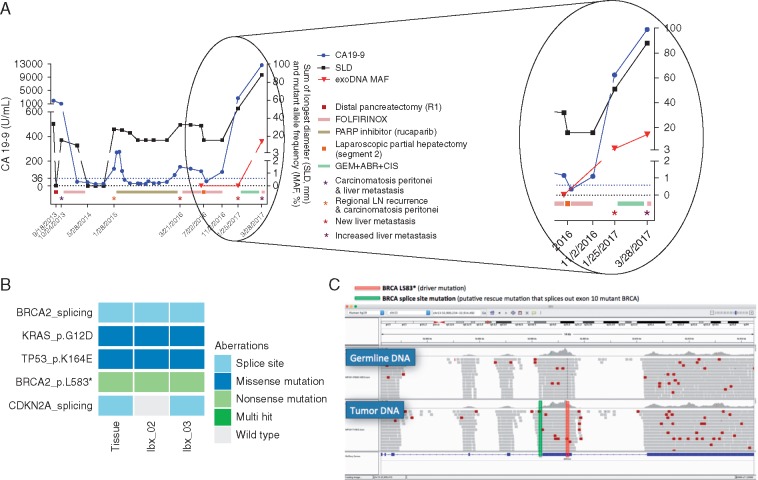
A metastatic PDAC patient who underwent prior tumor resection and subsequently developed liver metastasis underwent liquid biopsies for exosome isolation. The emergence of metastasis corresponded with clinically detectable resistance to a Rucaparib (PARP1 inhibitor) clinical trial in which the patient was stratified into due to a somatic frameshift BRCA2 (L583*) mutation with associated loss of heterozygosity. Plasma-derived exosomes were isolated and profiled for KRAS mutant detection revealing an increase in KRAS mutant burden during disease progression (Figure 3). In an on-treatment blood draw where no exoDNA mutant KRAS was detected based on ddPCR, we subsequently attempted exoDNA enrichment resulting in an increase in mutational KRAS allelic fraction from 0% to 3.2%. More importantly, the amount of DNA material was sufficient for subsequent NGS using a molecular barcoding approach. This resulted in the detection of the known driver mutations that were present in the patient’s original primary tumor, and subsequently detected in the metastatic liver tissue, including mutations in KRAS, TP53, and BRCA2. Notably a secondary mutation in BRCA2 was also detected in liquid biopsies, which was not present in the original primary tissue, likely arising during PARP inhibitor therapy. This mutation resided immediately before exon 10 where the BRCA2 (L583*) mutation was present allowing for the entire exon to be spliced out and leading to transcription of a full mRNA molecule. Tumor exosomal DNA enrichment thus allowed us to detect this putative mechanism of resistance to PARP inhibitor, underscoring the utility of liquid biopsies in facilitating therapeutic stratification.
Figure 3.
(A) Clinical course of a pancreatic ductal adenocarcinoma patient who underwent prior pancreatic tumor resection, and subsequently progressed while on Parp-1 inhibitor therapy. ExoDNA enrichment led to capture of tumor derived material which was not previously detectable. (B) Relevant mutations found in the metastatic tissue and liquid biopsies 6 months (lbx_02) and 9 months (lbx_03) after tissue biopsy. Of note is the presence of a stopgain BRCA mutation (L583*) that was correlated to her prolonged response to PARP1 inhibitor therapy. (C) A subsequent mechanism of resistance was detected in the liquid biopsies and confirmed in the tissue in the form of a BRCA2 splice site variant which splices out the aberrant stopgain mutation. SLD sum of the largest dimension; exoDNA MAF represent the KRAS mutant allele fraction. ABR, abraxane (nab-paclitaxel); CIS, cisplatin; GEM, gemcitabine.
Discussion
We have carried out proteomic profiling of exosomes isolated from a panel of PDAC cells in order to identify a candidate list of cancer-specific surface exosomal proteins (the PDAC exosomal ‘surfaceome’). We validated the cancer specificity of these exosomal proteins by performing the same proteomic profiling in non-neoplastic pancreatic cell types and examining which candidate proteins were differentially and preferentially expressed by the collective PDAC exosomal ‘surfaceome’. The resultant PDAC-exosome-specific markers can be exploited using an immunocapture assay for enrichment of tumor-specific material in liquid biopsies. This allows for subsequent molecular analysis of tumor material with implications for early detection, longitudinal disease monitoring (especially in low tumor volume settings), and therapeutic stratification during targeted therapy.
As it is possible that the exosome ‘surfaceome’ may evolve throughout disease progression and may, in fact, be a product of the intrinsic heterogeneity found in PDAC, we opted to pursue a multiplexed panel of antibodies against six candidate biomarkers for validation. These included CLDN4, EPCAM, CD151, LGALS3BP, HIST2H2BE, and HIST2H2BF. As evidenced by our data, these biomarkers appear to greatly enhance not only the fraction of patients at each PDAC stage with detectable mutant molecules but also the mutant allelic fraction per se at each stage, suggesting enrichment for the tumor-derived nucleic acid component. The latter has direct implications for downstream molecular assessment using NGS that can be pursued in liquid biopsy samples.
Mechanisms of DNA packaging within exosomes remain largely unknown as opposed to the apoptotic/necrotic pathways that are mostly recognized as sources of ctDNA in circulation. In the nucleus, histones are essential for chromatin structure and play a crucial role during gene transcription and silencing. Interestingly, histones have also been found outside the nucleus, in the cytosol, mitochondria, and cell membrane [12]. Extrachromosomal Histone-H2B has been identified as a cytosolic DNA sensor for aberrant self and non-self double-stranded DNA, which mediates an innate immune response and co-localizes within the mitochondrial membrane [13, 14]. Upon detection of cytosolic DNA, H2B has been described to partially associate with mitochondria and co-localize with the late endosome marker CD63 [15]. Both mitochondria and endosomes are known to generate multivesicular bodies that can fuse with the cell membrane and generate exosomes [16]. Therefore, the relative enrichment of H2B within the exosome compartment of cancer cells suggests that this protein may be interacting with mutant DNA that originated in the nucleus and which subsequently becomes packaged within exosomes for transport.
Not unexpectedly, the other candidate exosomal ‘surfaceome’ proteins identified in our analysis have been independently implicated in cancer initiation and progression of PDAC. For example, the extracellular matrix glycoprotein LGALS3BP is overexpressed by neoplastic cells with a role in promoting cell viability, migration, and metastasis, resulting in its role as a potential biomarker associated with prognosis and response to therapy [16, 17]. Other identified biomarkers such as the tetraspanin family member CD151 have also been implicated in cancer initiation and metastasis; in fact, exosomal CD151 per se has previously been shown to facilitate metastasis through induction of epithelial to mesenchymal transition in PDAC cell lines [18]. The family of claudin proteins is involved in the formation of tight junctions, with overexpression of CLDN4 previously described in the context of PDAC [19]. Notably, this overexpression was present in both human archival material and genetically engineered mouse models at the stage of PDAC precursor lesions (pancreatic intraepithelial neoplasia), with implications for early detection [20]. Finally, expression of epithelial markers in circulation has been best characterized in the context of circulating tumor cells (CTCs). Specifically, the use of EPCAM to isolate and quantify CTCs has led to FDA-approved prognostic tests in colorectal, breast, and prostate cancers [21]. As the majority of content in circulation is derived from blood components such as peripheral blood mononuclear cells, the presence of circulating material expressing epithelial proteins such as EPCAM are postulated to represent tumor-derived origins. This is further supported by our own data, which suggest that EPCAM in circulation may represent a cancer-specific exosomal biomarker [22].
Previous work has demonstrated the utility of the biomarker GPC1 as a highly sensitive and specific exosomal biomarker for detection of PDAC [9]. While our proteomics data does confirm that GPC1 is expressed on the PDAC-derived exosomal ‘surfaceome’, upon incorporation of public EV databases, this protein appears to be also enriched in exosomes originating from normal tissues. Furthermore, our experimental data confirm the presence of GPC1 in non-neoplastic cell lines including CAF19 and SC2, while not being expressed within the exosomes of three representative PDAC cell lines following attempted validation using multiple commercially available antibodies. A recent study by Yang et al. also found that GPC1 as a single exosome marker was not optimal in PDAC plasma samples, although it could potentially be used as a component of a multi-analyte panel [10]. Thus, the significance of GPC1 in PDAC liquid biopsies will require future clarification.
Among limitations of this study is the fact that we were unable to obtain matched captured and non-captured total exosome samples from the same patient due to the volumes of plasma required to pursue both protocols. The purpose of utilizing these volumes (∼11.7 ml of plasma) was to have sufficient nucleic acid material for downstream NGS analysis. Additionally, our relatively small sample size which underwent exosome capture may limit the generalization of our conclusions and would require further validation in larger cohorts. It would also be prudent to perform this analysis on a cohort of healthy controls in order to effectively validate the specificity of our cancer-derived exosome capture approach for KRAS mutation detection. Finally, it is important to note the feasibility of implementing such a protocol in the clinics. Whereas plasma processing and DNA isolation for ctDNA can be carried out within a day, the need to isolate exosomes using a bead immunocapture-based approach followed by DNA isolation would require 4 days in addition to the required infrastructure needed for ultracentrifugation. Although it is not a significant processing time difference, new exosome isolation approaches are being developed to decrease cost and increase efficiency of specific exosome capture without the need for ultracentrifugation. This includes the use of antibody-coated chips and microfluidic-based approaches which can capture specific exosome populations of interest [23, 24]
The need to enrich for tumor-derived material in circulation is underlined by the difficulties in detecting rare circulating mutant molecules in a heterogeneous milieu that is typically overwhelmed by non-neoplastic tissue-derived DNA. This is particularly compounded in the context of patients undergoing therapy where mutant DNA might be at levels that are undetectable with conventional ultra-sensitive digital PCR techniques. The ability to detect latent mutant molecules has implications in uncovering emerging mechanisms of resistance or vulnerability nodes before these become clinically evident, thus allowing for more effective therapeutic stratification. As typical circulating biomarkers such as ctDNA are not amenable to enrichment methodologies, we present exosomes as a viable alternative to capture tumor-specific material. This can come in the form of not only DNA but also mRNA and proteins that are sourced from the originating tumor cell. Indeed, we have demonstrated how a tumor enrichment platform can lead to detectable tumor material in those patients initially thought to be free of circulating mutant molecules. But more importantly, specific tumor exosome enrichment leads to an augmentation of mutant genomic equivalents that are subsequently amenable to NGS. For example, in our cohort of resectable patients, 44% of patient samples from total exosomes had sufficient quantity and quality of DNA to undergo downstream molecular barcoding (as defined by >1% KRAS mutant AF and >1 ng of isolated DNA), compared with 67% of patient samples that were subject to exosome capture. This enrichment then permits elucidation of emerging mechanisms of resistance, such as a secondary BRCA2 mutation that reverts PARP sensitivity, as we have demonstrated in our study.
Funding
MD Anderson Moonshot Program and the Khalifa Bin Zayed Al-Nahyan Foundation (no grant numbers apply); the National Institute of Health (U01CA196403 and U01CA200468 to AM); the Cancer Prevention Research Institute of Texas (RP160517 to AM and RP140106 to VB); a faculty fellowship from The University of Texas MD Anderson Cancer Center Duncan Family Institute for Cancer Prevention and Risk Assessment (MC) (no grant number applies); a foreign postdoctoral fellowship grant, Chile – CONICYT (JC) (no grant number applies).
Disclosure
The authors have declared no conflicts of interest.
Supplementary Material
References
- 1. Bailey P, Chang DK, Nones K. et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016; 531: 47–52. http://dx.doi.org/10.1038/nature16965 [DOI] [PubMed] [Google Scholar]
- 2. Waddell N, Pajic M, Patch AM. et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 2015; 518: 495–501. http://dx.doi.org/10.1038/nature14169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. San Lucas FA, Allenson K, Bernard V. et al. Minimally invasive genomic and transcriptomic profiling of visceral cancers by next-generation sequencing of circulating exosomes. Ann Oncol 2016; 27: 635–664. http://dx.doi.org/10.1093/annonc/mdv604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kalluri R. The biology and function of fibroblasts in cancer. Nat Rev Cancer 2016; 16: 582–598. http://dx.doi.org/10.1038/nrc.2016.73 [DOI] [PubMed] [Google Scholar]
- 5. Costa-Silva B, Aiello NM, Ocean AJ. et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat Cell Biol 2015; 17: 816–826. http://dx.doi.org/10.1038/ncb3169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kahlert C, Melo SA, Protopopov A. et al. Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J Biol Chem 2014; 289: 3869–3875. http://dx.doi.org/10.1074/jbc.C113.532267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Melo SA, Sugimoto H, O'Connell JT. et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell 2014; 26: 707–721. http://dx.doi.org/10.1016/j.ccell.2014.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Thakur BK, Zhang H, Becker A. et al. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res 2014; 24: 766–769. http://dx.doi.org/10.1038/cr.2014.44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Melo SA, Luecke LB, Kahlert C. et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 2015; 523: 177–182. http://dx.doi.org/10.1038/nature14581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yang KS, Im H, Hong S. et al. Multiparametric plasma EV profiling facilitates diagnosis of pancreatic malignancy. Sci Transl Med 2017; 9(391): eaal3226 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Biankin AV, Waddell N, Kassahn KS. et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 2012; 491: 399–405. http://dx.doi.org/10.1038/nature11547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kobiyama K, Takeshita F, Jounai N. et al. Extrachromosomal histone H2B mediates innate antiviral immune responses induced by intracellular double-stranded DNA. J Virol 2010; 84: 822–832. http://dx.doi.org/10.1128/JVI.01339-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Choi YS, Hoon Jeong J, Min HK. et al. Shot-gun proteomic analysis of mitochondrial D-loop DNA binding proteins: identification of mitochondrial histones. Mol Biosyst 2011; 7: 1523–1536. http://dx.doi.org/10.1039/c0mb00277a [DOI] [PubMed] [Google Scholar]
- 14. Kobiyama K, Kawashima A, Jounai N. et al. Role of Extrachromosomal Histone H2B on Recognition of DNA Viruses and Cell Damage. Front Genet 2013; 4: 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shen YJ, Le Bert N, Chitre AA. et al. Genome-derived cytosolic DNA mediates type I interferon-dependent rejection of B cell lymphoma cells. Cell Rep 2015; 11: 460–473. http://dx.doi.org/10.1016/j.celrep.2015.03.041 [DOI] [PubMed] [Google Scholar]
- 16. Sugiura A, McLelland GL, Fon EA, McBride HM.. A new pathway for mitochondrial quality control: mitochondrial-derived vesicles. Embo j 2014; 33: 2142–2156. http://dx.doi.org/10.15252/embj.201488104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nigjeh EN, Chen R, Allen-Tamura Y. et al. Spectral library-based glycopeptide analysis-detection of circulating galectin-3 binding protein in pancreatic cancer. Proteomics Clin Appl 2017; 11: 9-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yue S, Mu W, Erb U, Zoller M.. The tetraspanins CD151 and Tspan8 are essential exosome components for the crosstalk between cancer initiating cells and their surrounding. Oncotarget 2015; 6: 2366–2384. http://dx.doi.org/10.18632/oncotarget.2958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nichols LS, Ashfaq R, Iacobuzio-Donahue CA.. Claudin 4 protein expression in primary and metastatic pancreatic cancer: support for use as a therapeutic target. Am J Clin Pathol 2004; 121: 226–230. http://dx.doi.org/10.1309/K144PHVDDUPDD401 [DOI] [PubMed] [Google Scholar]
- 20. Neesse A, Hahnenkamp A, Griesmann H. et al. Claudin-4-targeted optical imaging detects pancreatic cancer and its precursor lesions. Gut 2013; 62: 1034–1043. http://dx.doi.org/10.1136/gutjnl-2012-302577 [DOI] [PubMed] [Google Scholar]
- 21. Ligthart ST, Coumans FA, Bidard FC. et al. Circulating Tumor Cells Count and Morphological Features in Breast, Colorectal and Prostate Cancer. PLoS One 2013; 8: e67148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kalra H, Simpson RJ, Ji H. et al. Vesiclepedia: a compendium for extracellular vesicles with continuous community annotation. PLoS Biol 2012; 10: e1001450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liang K, Liu F, Fan J. et al. Nanoplasmonic Quantification of Tumor-derived Extracellular Vesicles in Plasma Microsamples for Diagnosis and Treatment Monitoring. Nat Biomed Eng 2017; 1: 0021 http://dx.doi.org/10.1038/s41551-016-0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kanwar SS, Dunlay CJ, Simeone DM, Nagrath S.. Microfluidic device (ExoChip) for on-chip isolation, quantification and characterization of circulating exosomes. Lab Chip 2014; 14: 1891–1900. http://dx.doi.org/10.1039/C4LC00136B [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.