Abstract
A reduction in trabecular bone mass is often associated with an increase in marrow fat in osteoporotic bones. The molecular mechanisms underlying this inverse correlation are incompletely understood. Here, we report that mice lacking tuberous sclerosis 1 (Tsc1) in Osterix-expressing cells had a significant decrease in trabecular bone mass characterized by decreased osteoblastogenesis, increased osteoclastogenesis, and increased bone marrow adiposity in vivo. In vitro study showed that Tsc1-deficient bone marrow stromal cells (BMSCs) had decreased proliferation, decreased osteogenic differentiation, and increased adipogenic differentiation in association with the downregulation of Wnt/β-catenin signaling. Mechanistically, TSC1 deficiency led to autophagy suppression and consequent Notch1 protei n increase, which mediated the GSK3β-independent β-catenin degradation. Together, our results indicate that Tsc1 controls the balance between osteoblast and adipocyte differentiation of BMSCs.
Keywords: OSTEOBLAST, ADIPOCYTE, TSC1, WNT, B-CATENIN, BONE MARROW, NOTCH1, AUTOPHAGY
Introduction
An increase in marrow adipose tissue often accompanies a decrease in bone mass in osteoporosis(1) and other conditions that induce bone loss.(2–4) This inverse relationship is also reported in mouse models, which provide important tools to determine underlying molecular mechanisms. Multipotent bone marrow stromal cells (BMSCs) can differentiate into both adipocytes and osteoblasts upon specific stimulation for cell differentiation. The osteoblast-adipocyte lineage commitment of BMSCs has direct relevance to the regulation of bone development and the maintenance of bone homeostasis. The insight into the molecular mechanisms governing the balance between osteoblast and adipocyte differentiation in bone marrow can facilitate developing new therapies to treat osteoporosis and other bone loss conditions.
Wnt/β-catenin signaling regulates both osteoblastogenesis and adipogenesis.(5,6) β-catenin conditional knockout in osteo-blast progenitor cells decreases bone mass and increases bone marrow adiposity.(7,8) β-catenin deficient BMSCs have decreased osteogenic but enhanced adipogenic ability.(7) It has been demonstrated that core-binding factor subunit beta controls the balance between osteoblast and adipocyte lineage through promoting Wnt/β-catenin signaling.(9) Similarly, we showed that focal adhesion kinase inhibits the adipogenic differentiation potential and enhances the osteogenic differentiation potential of BMSCs through enhancing β-catenin signaling.(10)
Mechanistic target of rapamycin complex 1 (mTORC1) plays critical roles in cell growth, cellular metabolism, and autophagy.(11,12) Tuberous sclerosis complex 1 (TSC1) and TSC2 are tumor suppressor genes and TSC1/TSC2 complex is an upstream inhibitor of mTORC1.(13) Germline mutations in either TSC1 or TSC2 leads to the activation of mTORC1 signaling and causes a human disease called tuberous sclerosis with a birth rate of 1 in 5800, characterized by the presence of benign congenital tumors in multiple organs.(14,15) Osseous manifestations of tuberous sclerosis are very diverse and complex, including calvaria hyperostosis, periosteal new bone formation, osteoporotic lesion in hand bones, and cyst-like radiolucent lesions.(16,17) Recently, Tsc1 or Tsc2 mutation mouse models have been used to determine the impact and mechanisms of upregulated mTORC1 signaling in bone homeostasis. Tsc2 deletion in mature osteoblasts(18) and Tsc1 deletion in osteoblast progenitors(19) both lead to increased bone mass. Hypomineralization, decreased bone mineral density, and/or greatly increased cortical porosity are reported in these two models. In contrast, we found that Tsc1 deletion in neural crest–derived cells results in an increase in craniofacial bone mass in association with a hypermineralization phenotype.(20,21) The phenotypic difference among these mouse models highlighted the complexity of mTORC1 regulation on bone homeostasis. To further determine the role of TSC1/mTORC1 signaling in bone homeostasis, we generated a Tsc1 conditional knockout mouse model by deleting Tsc1 in Osterix-expressing cells. We showed that Tsc1 deletion led to a decrease in trabecular bone mass in association with an increase in bone marrow adiposity and a decrease in Wnt/β-catenin signaling. Our data indicate that Tsc1 is a novel regulator governing the osteoblast-adipocyte differentiation in bone marrow.
Materials and Methods
Mice
The generation of floxed Tsc1 (Tsc1F/F), floxed Fip200 (Fip200F/F), Osterix-Cre (Osx-Cre), and GFP-LC3 transgenic mice has been reported.(22–25) All mice were in a C57BL/6 background after at least eight-generation backcrossing. Tsc1F/F mice were bred with Osx-Cre/+ mice to generate male Tsc1F/+;Osx-Cre/+ (CHet) mice. Male CHet mice were then bred with female Tsc1F/+ mice to generate Tsc1F/F;Osx-Cre/+ (conditional knockout [CKO]), CHet, and Osx-Cre/+ (Osx-Cre) mice. In some experiments, CHet mice were bred with Tsc1F/F mice to generate Tsc1F/F, CHet, and CKO mice. Tsc1F/F mice were bred with GFP-LC3 mice to generate female Tsc1F/F;GFP-LC3/GFP-LC3 mice, which were used in the breeding with male CHet mice in GFP-LC3 puncta experiments. Mice were housed at 22 ± 2°C on a 12-hour:12-hour light/dark cycle under pathogen-free conditions. Local, state, and federal regulations were followed for mouse handling. Mice were euthanized by overdose of carbon dioxide. The investigators who analyzed the various bone parameters were blind to the identity of the mice. All experimental procedures were approved by the Institutional Animal Care and Use Committee at the University of Michigan.
Skeleton preparation
Skeleton preparation was performed as reported.(26)
Micro–computed tomography analysis
Male Tsc1F/F, Osx-Cre, CHet, and CKO mice were euthanized at the age of 1 month (n = 9 for each group). Femurs were analyzed by micro–computed tomography (μCT) as reported.(10,27,28) In brief, the trabecular region of interest (ROI) from the distal femora was identified as 10% of the bone length starting at the interface between the growth plate and metaphyseal trabecular bone extending into the metaphysis.
Histomorphometry
Histomorphometry measurements were conducted as described.(10,27,29,30) All measurements were performed following the published guidelines.(31) The trabecular ROI from the distal femora was identified as a 1-mm × 0.8-mm rectangle, in the center of the bone, which was 0.5 mm below the growth plate.
RNA extraction and qRT-PCR
RNA extraction and qRT-PCR analyses were performed as described.(27) For qRT-PCR analyses, equal amounts of cDNA template were used for SYBR Green PCR Core reagents system (Qiagen, Valencia, CA, USA). Primer sequences used for qRT-PCR are provided in Supporting Table S1.
Bone marrow culture
BMSCs were collected from the femur and tibia of 24-day-old to 28-day-old control, Osx-Cre, CHet, or CKO mice. Briefly, bone marrow were flushed and cultured in α-MEM with 10% FBS for 3 days and rinsed with medium to remove nonadherent cells. Adherent cells were cultured for 7 to 8 days as described.(10)
Osteoclast differentiation
Bone marrow was flushed from femur and tibia of 1-month-old control or CKO mice and bone marrow cells were cultured in α-MEM with 10% FBS for 1 day and nonadherent cells were cultured with 20 ng/mL M-CSF (#416-ML-010; R&D Systems, Minneapolis, MN, USA) for 3 days. The medium was replaced every 48 hours. Bone marrow–derived macrophages (BMMs) were harvested to induce osteoclast formation. BMMs were plated (1 × 104 cells/well in 48-well plates) and treated with or without 30 ng/mL of RANKL (#462-TEC-010; R&D Systems, Minneapolis, MN, USA) in the presence of M-CSF (20 ng/mL) for 4 to 6 days. The cells were fixed and stained using a tartrate resistant acid phosphatase (TRAP) staining kit (#387A-1KT; Sigma-Aldrich, St. Louis, MO, USA). Osteoclast-like cells were defined as pink-colored TRAP-positive multinucleated (>5 nuclei) cells (MNCs).
Osteoblast and osteoclast co-culture
Bone marrow was flushed from femur and tibia of 1-month-old control, CHet, or CKO mice and bone marrow cells were cultured in α-MEM with 10% FBS for 1 day. For bone marrow-derived osteoblasts, adherent cells were cultured for 10 days with medium replacement every 48 hours. Nonadherent cells were cultured in the presence of 20 ng/mL M-CSF for 3 days to differentiate to BMMs. The medium was replaced every 48 hours. After 3 days, BMMs were plated (0.5 × 105 cells/well or 1 × 105 cells/well in 48-well plates) with bone marrow-derived osteo-blast cells (1 × 104 cells/well or 2 × 104 cells/well in 48-well plates) in the presence of 20 nM 1α, 25-dyhydroxy vitamin D3 (D1530, Sigma-Aldrich, St. Louis, MO, USA) and 1mM prostaglandin E2 (P0409, Sigma-Aldrich, St. Louis, MO, USA) to induce the expression of osteoclastogenic M-CSF and RANKL, and to permit the osteoclast differentiation for 7 days. The cells were fixed and stained using a TRAP staining kit. Osteoclast-like cells were defined as pink-colored TRAP-positive MNCs (>3 nuclei).
Osteoblast differentiation assay
BMSCs were seeded at 100,000 cells per well in a 12-well plate. Cells were cultured in α-MEM with 10% FBS containing 50 mg/mL of ascorbic acid and 10 mM of beta-glycerophosphate. The medium was changed every other day. The alkaline phosphatase (ALP) staining and Alizarin Red (AR) staining was performed at day 7 and day 21, respectively, as described.(10)
Proliferation assay
BMSCs were seeded at 80,000 cells per well in a 12-well plate with osteogenic differentiation medium. Cells were trypsinized on day 3 or day 7 and cell numbers were counted. For immunostaining using anti-Ki67 antibody (#9129; Cell Signaling Technology, Beverly, MA, USA), cells were fixed with 4% paraformaldehyde and immunostained on day 3 or day 7 as described.(10)
Western blot
Western blot was performed as described.(32) Antibody information is described as follows. Anti-TSC1: #4906, Cell Signaling Technology, Beverly, MA, USA; anti-β-catenin: #8480, Cell Signaling Technology, Beverly, MA, USA; anti-active β-catenin: #8814, Cell Signaling Technology, Beverly, MA, USA; anti-p-GSK-3b (S9A): #5558, Cell Signaling Technology, Beverly, MA, USA; anti-GSK-3b: #9315, Cell Signaling Technology, Beverly, MA, USA; anti-p-S6: #5364, Cell Signaling Technology, Beverly, MA, USA; anti-S6: #2217, Cell Signaling Technology, Beverly, MA, USA; anti-p62: #5114, Cell Signaling Technology, Beverly, MA, USA; anti-LC3: #2775, Cell Signaling Technology, Beverly, MA, USA; anti-Vinculin: #V4505, Sigma-Aldrich, St. Louis, MO, USA; anti-a-Tubulin: #T6199, Sigma-Aldrich, St. Louis, MO, USA; and anti-Notch1: #ab52627; Abcam, Cambridge, MA, USA.
BMSC adipocyte differentiation
BMSCs were seeded at 400,000 cells/well in a 12-well plate with adipogenic differentiation medium. Briefly, cells were cultured in α-MEM with 10% FBS containing 1 μM dexamethasone, 5 μg/mL troglitazone, 50 μM 3-isobutyl-1-methylxanthine, and 1 μg/mL insulin for 2 days and then cultured in a-MEM with 10% FBS containing 1 μg/mL insulin for 3 days. Oil-red O staining was performed as described.(10)
Starvation
Amino acid-free Earle’s balanced salt solution (EBSS, #SH30029.02; Hyclone Laboratories, Logan, UT, USA) without FBS was used as starvation medium.
siRNA knockdown
Lipofectamine RNAi MAX (Invitrogen, Carlsbad, CA, USA) was used to transfect BMSCs or mouse embryonic fibroblasts (MEFs) with siRNA in Opti-MEM medium (Invitrogen). The efficiency of transfection was monitored by Western blot. The sequences of siRNA were as follows: Notch1: sense: 5′-AACAGCAUCAUCC AAAUCCAACUGA-3′, and anti-sense: 5′-UCAGUUGGAUUUGG AUGAUGCUGUUUG-3′. Nonspecific siRNA sequences were used as negative controls: sense: 5′-CGUUAAUCGCGUAUAAUACGCG UAT-3′, and antisense: 5′-AUACGCGUAUUAUACGCGAUUAACG AC-3′. (TriFECTa DsiRNA Duplex; Integrated DNA Technologies [IDT], Coralville, IA, USA).
Statistical analysis
For two-test group comparison, Student’s t test was used. For the comparison in multiple groups, one-way ANOVA with Tukey’s multiple comparison tests were performed using GraphPad Prism 7.0 (GraphPad Software, Inc., La Jolla, CA, USA). For both tests, *p < 0.05 was considered to be significant.
Results
Deletion of Tsc1 in Osterix+ cells leads to decreased trabecular bone mass in mice
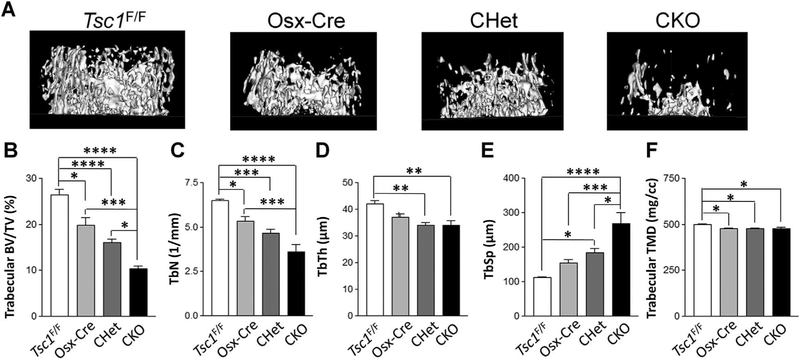
To determine the function of Tsc1 in early osteoblast lineage cells, we generated Tsc1 CKO mice with Osterix-Cre (Osx-Cre), which has Cre recombinase activity in osteoprogenitor cells.(22) Floxed Tsc1 (Tsc1F/F), Osx-Cre/+ (Osx-Cre), Tsc1F/+;Osx-Cre mice (CHet), and Tsc1F/F;Osx-Cre (CKO) mice were generated as described in Materials and Methods. To determine the effects of Tsc1 deletion on early skeleton development, whole-mount skeleton staining was performed on newborn mice. Major skeletal elements appeared normal in CKO mice at birth (Supporting Fig. S1). CKO mice had similar body weight compared to other littermate mice at the neonatal stage (Supporting Fig. S2), but grew slower after birth (Supporting Fig. S3). At 1 month old, CKO mice were lighter than Tsc1F/F, Osx-Cre, and CHet mice (Supporting Fig. S3). The CKO mice died around 1 month of age due to an unidentified reason. Next, μCT analysis was performed to determine the effects of Tsc1 deletion on bone structure. CKO mice had a 35% to 60% decrease in femoral trabecular bone volume fraction (BV/TV) compared to Tsc1F/F, Osx-Cre, and CHet mice (Fig. 1A, B), indicating Tsc1 deletion led to decreased femoral trabecular bone mass. Trabecular bone mass decrease in CKO was contributed by the decrease in both trabecular number (Tb.N) (Fig. 1C) and trabecular thickness (Tb.Th) (Fig. 1D). Correspondingly, CKO had a 45% to 139% increase in trabecular spacing (Tb.Sp) compared to Tsc1F/F, Osx-Cre, and CHet mice (Fig. 1E). There was a small decrease in trabecular TMD in CKO compared to Tsc1F/F but not to Osx-Cre and CHet mice (Fig. 1F). In contrast to the decrease in trabecular bone mass, CKO mice had a small increase in femoral cortical bone mass as well as calvarial bone mass compared to Tsc1F/F, Osx-Cre, and CHet mice (data not shown). The increase in cortical bone mass was consistent with a previous report using similar mouse model, which ascribed the cortical bone mass increase to enhanced proliferation in TSC1-deficient osteoblasts.(19) Altogether, our data showed that Tsc1 deletion in osterix-expressing cells led to a significant decrease in femoral trabecular bone mass.
Fig. 1.
Tsc1 deletion leads to decreased femoral trabecular bone mass. μCT analysis in femurs of 1-month-old male Tsc1F/F, Osterix-Cre/+ (Osx-Cre), Tsc1F/+;Osx-Cre/+ (CHet), and Tsc1F/F;Osx-Cre/+ (CKO) mice. (A) Representative μCT image of femoral trabecular bone. (B) BV/TV. (C) Tb.N. (D) Tb.Th. (E) Tb.Sp, trabecular spacing. (F) TMD. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, #0.05 < p < 0.1, n = 9. Values are shown as mean + SE. BV/TV = bone volume per tissue volume; Tb.N = trabecular number; Tb.Th = trabecular thickness; Tb.Sp = trabecular spacing; TMD = tissue mineral density.
Decreased femoral trabecular bone mass in Tsc1-deficient mice is due to decreased osteoblastogenesis in association with increased osteoclastogenesis and increased bone marrow adipogenesis
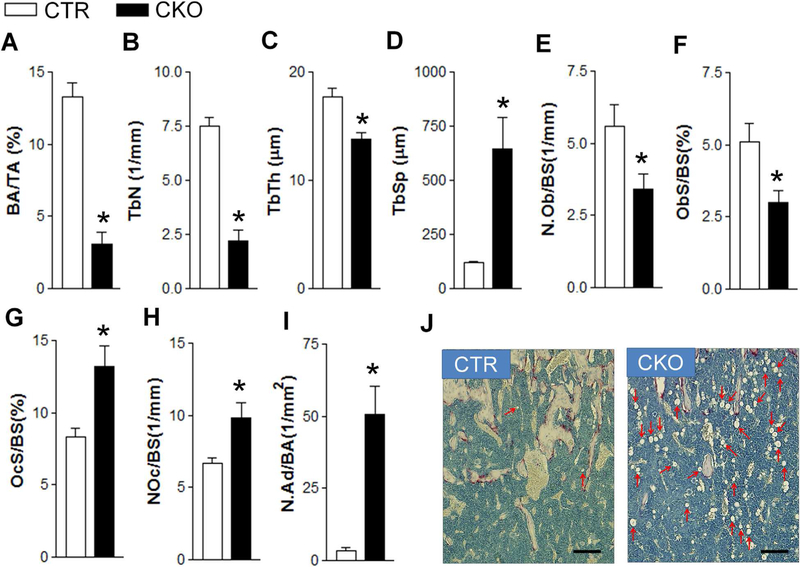
Consistent with μCT measurements, histomorphometry analysis on femurs showed a 77% decrease in BV/TV (Fig. 2A), a 71% decrease in Tb.N (Fig. 2B), a 22% decrease in Tb.Th (Fig. 2C), and a 441% increase in Tb.Sp (Fig. 2D) in CKO mice. At the cellular level, there was a 40% decrease in both osteoblast number (N.Ob) (Fig. 2E) and surface (Ob.S/BS) (Fig. 2F), and a 46% and 60% increase in osteoclast number (N.Oc) (Fig. 2G) and surface(Oc.S/BS) (Fig. 2H), respectively. Notably, there was a 13-fold increase in bone marrow adipocyte number in CKO mice (Fig. 2I, J). In addition, there was a slight increase in the length of hypertrophic zone of femoral growth plate in CKO mice (Supporting Fig. S4). Collectively, compromised osteoblasto-genesis in association with increased adipogenesis and increased osteoclastogenesis contributed to decreased femoral trabecular bone mass in CKO mice in vivo.
Fig. 2.
Histomorphometric analysis of control and Tsc1 CKO mice. (A) BV/TV, bone volume per tissue volume. (B) Tb.N. (C) Tb.Th. (D) Tb.Sp. (E) N.Ob/BS. (F) Ob.S/BS. (G) Oc.S/BS. (H) N.Oc/BS. (I) N.Ad/BA. (J) Histological images showing bone marrow adipocytes (indicated by arrows). *p < 0.05, n = 9. Scale bar = 50 μm. Values are shown as mean + SE. Tb.N = trabecular bone number; TbTh = trabecular thickness; Tb.Sp = trabecular spacing; N.Ob/BS = osteoblast number per bone surface; Ob.S/BS = osteoblast surface per bone surface; Oc.S/BS = osteoclast surface per bone surface; N.Oc/BS = osteoclast number per bone surface; N.Ad/BA = adipocyte number per bone area; CTR = control (Tsc1F/F).
Increased osteoclastogenesis in Tsc1-deficient mice is associated with increased Mcsf expression
To understand the mechanism of increased osteoclastogenesis in CKO bone, we first determined the effect of Tsc1 deletion in Osterix-expressing cells on the osteoclastogenic potential of bone marrow cells. Data showed that bone marrow cells of CKO mice had the same osteoclastogenic potential as that of control (CTR) mice (Fig. 3A), which supported a cell nonautonomous effect of Tsc1 deletion in osteoblastic lineage on osteoclastic lineage. Next, we determined the effect of Tsc1 deletion on the expression of Opg, Rankl, and Mcsf, three important osteoblast-secreted mediators of osteoclastogenesis in both BMSCs (Fig. 3B) and femur (Fig. 3C). Because the expression of both Opg and Rankl was increased similarly, their changes were unlikely responsible for the increased osteoclastogenesis. However, there was a significant increase in Mcsf expression in TSC1-deficient BMSCs and femur. Mcsf is a positive regulator for the proliferation, differentiation, and survival of monocytic lineage cells.(33) Thus, increased Mcsf expression may be at least partially responsible for the enhanced osteoclastogenesis in CKO bone. To further determine whether osteoblasts/BMSCs in CKO mice were responsible for the increased osteoclastogenesis, we performed a co-culture experiment using both BMSCs and BMMs. Our data showed that compared to the BMSCs isolated from control and CHet mice, BMSCs isolated from CKO mice could better support the osteoclastogenesis of BMMs isolated from control mice (Fig. 3D) or CKO mice (Supporting Fig. S5).
Fig. 3.
Tsc1 deletion in Osterix-expressing cells does not affect the osteoclastogenic potential of bone marrow cells but increases Mcsf expression. (A) BMMs were cultured in the presence of 20 ng/mL of M-CSF and 30 ng/mL of RANKL for indicated days and stained for TRAP. Numbers of TRAP+ MNCs (>5 nuclei) were counted (right panel). Scale bar = 200 μm. (B, C) Quantitative PCR analyses of the mRNA levels of Opg, Rankl, Mcsf in BMSCs (B) and femur (C). (D) A total of 100,000 BMMs isolated from CTR mice were cultured with 20,000 BMSCs isolated from CTR, CHet, and CKO mice in the presence of 20 nM 1α, 25-dyhydroxy vitamin D3 and 1 μM prostaglandin E2 for 7 days. Numbers of TRAP+ MNCs (>3 nuclei) were counted (right panel). Scale bar = 100 μm. *p < 0.05, n = 3. Values are shown as mean + SE. TRAP = tartrate resistant acid phosphatase; TRAP+ = TRAP-positive; MNC = multinucleated cell; BMM = bone marrow–derived macrophage; CTR = control (Tsc1F/F).
Tsc1 deletion leads to decreased proliferation, compromised osteogenic differentiation, and enhanced adipogenic differentiation of BMSCs
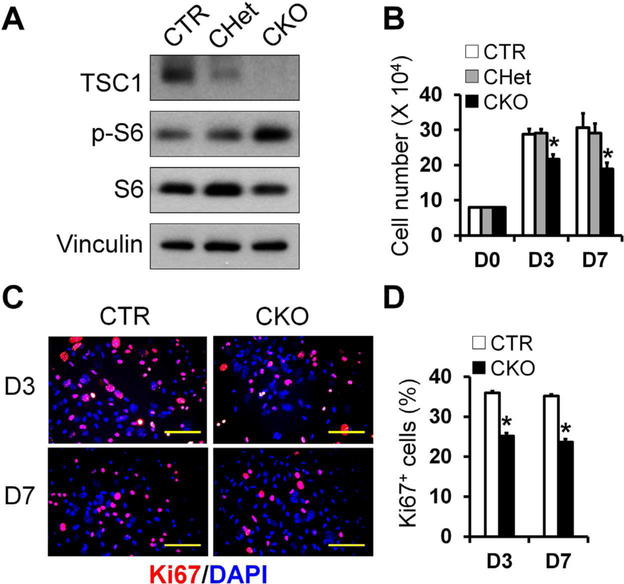
To further determine the mechanisms responsible for the decreased osteoblastogenesis observed in the femoral trabecular bone of CKO mice, we first investigated the effect of Tsc1 deletion on BMSC proliferation. BMSCs from CKO mice had efficient Tsc1 deletion and expected increase in the level of phosphorylated S6, an mTORC1 activation marker (Fig. 4A). TSC1-deficient BMSCs had decreased proliferation ability shown by decreased cell number and decreased percentage of Ki67-positive cells (Fig. 4B–D). Of note, BMSCs isolated from CHet mice (Fig. 4B) or Osx-Cre mice (Supporting Fig. S6) had comparable proliferation compared to controls.
Fig. 4.
TSC1-deficient BMSCs have decreased proliferation. BMSCs were isolated from femur and tibia of CTR, CHet, and CKO mice as described in Materials and Methods. (A) TSC1 was efficiently deleted in BMSCs in association with increased phosphor-S6 level in CKO group as shown by Western blot analysis. (B) Quantitation of cell number cultured for 3 and 7 days in osteogenic medium. n = 3 per group. (C) Representative merged fluorescent images of Ki67 (red) and DAPI (blue) staining in CTR and CKO osteoblasts at 3rd and 7th day, in the same cultures system as B. Scale bar = 50 μm. (D) Quantification of Ki67-positive (Ki67+) in the cultures shown in C. n = 3 per group, *p < 0.05. Values are presented as mean + SE. DAPI = 4,6-diamidino-2-phenylindole; CTR = control (Tsc1F/F).
Next, we determined the effect of Tsc1 deletion on the osteogenic differentiation of BMSCs.
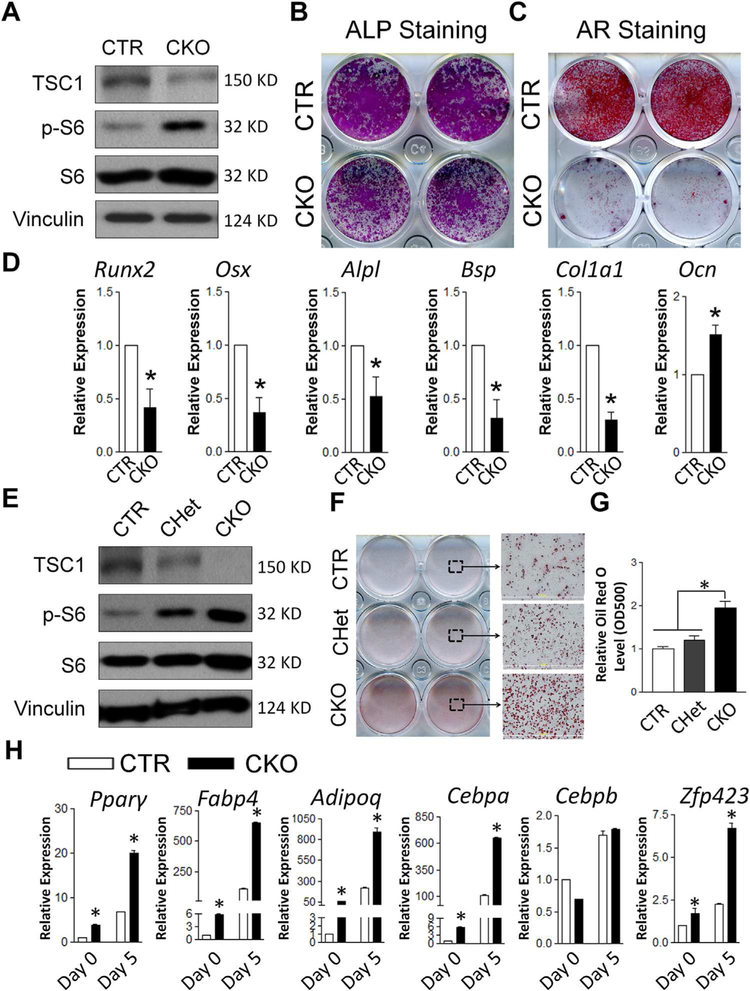
Deletion of TSC1 in BMSCs (Fig. 5A) resulted in significantly compromised early and terminal differentiation (Fig. 5B, C). TSC1-deficient BMSCs had decreased mRNA expression of master osteoblastic transcription factors Runx2 and Osx, as well as most osteoblast differentiation marker genes, including Alpl, Bsp, and ColIa1 (Fig. 5D). Notably, Ocn expression was increased in TSC1-deficient BMSCs, which was opposite to other marker genes. The mechanism of increased Ocn expression in TSC1-deficient osteoblasts is unknown. Similarly, others reported an increase of Ocn expression in TSC2-deficient osteoblasts, which had compromised osteogenic differentiation in vitro.(18) Altered FoxO1 activity due to dysregulated mTOR was suggested to be the mechanism responsible for this osteoblast differentiation-independent increase of Ocn expression.(18) Thus, our data suggested that compromised osteoblast differentiation likely contributed to the decreased bone mass in CKO mice in vivo.
Fig. 5.
Tsc1 deletion leads to compromised osteogenic and enhanced adipogenic differentiation in BMSCs. (A) TSC1 was effectively deleted in CKO BMSCs as shown by Western blot. (B, C) Osteogenic differentiation of BMSCs were analyzed by ALP staining at day 7 (B) and AR staining at day 21 (C). Images shown were the representatives of five independent experiments. (D) qPCR analysis of the mRNA expression of osteoblast differentiation markers. *p < 0.05, n = 3. Values are shown as mean + SE. (E) TSC1 was effectively deleted in CKO BMSCs and partially deleted in CHet BMSCs as shown by Western blot. (F) Oil red O staining showed enhanced adipogenic differentiation of TSC1-deficient BMSCs, in adipogenic medium for 5 days. (G) The quantitation of Oil red O staining described in F. (H) qPCR analysis of the mRNA expression of adipogenic differentiation markers and transcription factors, in the cultures described in F. *p < 0.05, n = 3. Values are shown as mean + SE. ALP = alkaline phosphatase; AR = Alizarin Red; Runx2 = Runt-related transcription factor 2; Osx = osterix; Alpl = alkaline phosphatase; Bsp = bone sialoprotein protein; ColIa1 = collagen, type I, alpha 1; Ocn = osteocalcin; CTR = control (Tsc1F/F).
To determine the mechanism of increased bone marrow adipocytes in Tsc1 CKO mice, we first assessed the effect of Tsc1 deletion on the adipogenic potential of BMSCs. As expected, TSC1 was effectively deleted in CKO culture and decreased in CHet culture with corresponding increase in p-S6 level (Fig. 5E). In response to adipogenic stimuli, the TSC1-deficient BMSCs exhibited significantly enhanced adipogenic differentiation compared to CTR and CHet BMSCs (Fig. 5F, G). mRNA levels of adipogenic differentiation marker genes and master adipocyte transcription factors including peroxisome proliferator-activated receptor gamma (Pparg), fatty acid binding protein 4 (Fabp4, also known as Ap2), adiponectin (Adipoq), CCAAT/enhancer-binding protein alpha (Cebpa, also known as C/EBPα), and zinc finger protein 423 (Zfp423) were increased in TSC1-deficient BMSCs (Fig. 5H). Notably, even before the adipogenic induction at the start of the culture (day 0), the mRNA expression levels of these adipogenic genes were significantly increased. This indicated that TSC1-deficient BMSCs had acquired some extent of intrinsic adipocyte phenotype.
Tsc1 deletion leads to mTORC1-dependent downregulation of Wnt/β-catenin signaling
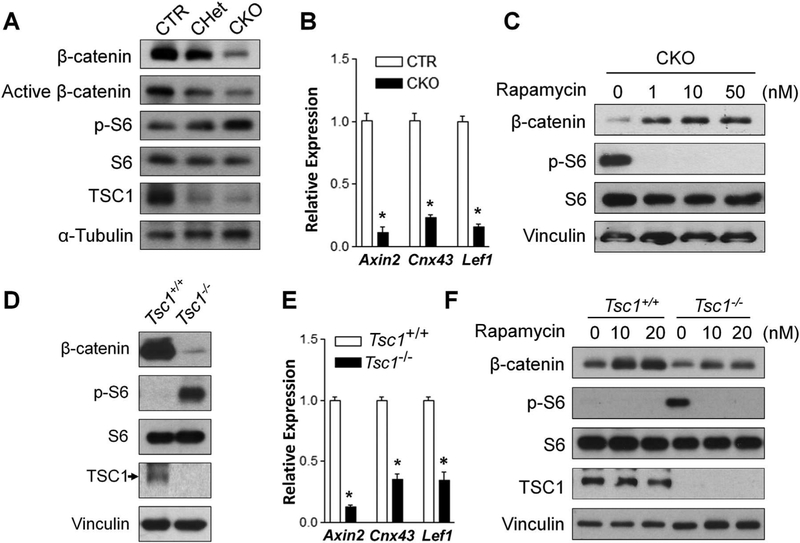
Wnt/β-catenin signaling plays important role in both osteo-blastogenesis and adipogenesis.(5,6) It has been reported that β-catenin deletion in Osterix-expressing cells results in decreased femoral trabecular bone mass in association with increased bone marrow adiposity.(7,8) Thus, we examined the effect of Tsc1 deletion on β-catenin level. Our data showed significantly reduced total and active β-catenin protein levels in TSC1-deficient BMSCs (Fig. 6A). BMSCs isolated from CHet mice (Fig. 6A) or Osx-Cre mice (Supporting Fig. S7) had comparable β-catenin protein levels compared to controls. The mRNA expression level of Wnt/β-catenin target genes including Axin2, Cnx43, and Lef1 was decreased in TSC1-deficient BMSCs (Fig. 6B). Thus, Tsc1 deletion in BMSCs downregulates Wnt/β-catenin signaling by decreasing β-catenin protein level. Treatment with rapamycin, an mTORC1 inhibitor, increased the β-catenin protein level in TSC1-deficient BMSCs (Fig. 6C), suggesting that the downregulation of β-catenin protein in TSC1-deficient BMSCs was mTORC1-dependent. To elucidate whether the regulation of Wnt/β-catenin signaling pathway by TSC1 is specific to BMSCs, we determined the effect of Tsc1 deletion on Wnt/β-catenin signaling pathway in MEFs. Similar to BMSCs, Tsc1–/– MEFs had decreased β-catenin protein level (Fig. 6D) and decreased mRNA expression level of Wnt/β-catenin target genes (Fig. 6E). Rapamycin treatment increased β-catenin protein level in Tsc1–/– MEFs (Fig. 6F). Altogether, our data demonstrated that Tsc1 deletion led to mTORC1-dependent reduction of β-catenin protein level and downregulation of the Wnt/β-catenin signaling pathway in both BMSCs and MEFs.
Fig. 6.
Tsc1 deletion leads to mTORC1-dependent downregulation of Wnt/β-catenin signaling. (A) BMSCs were analyzed by Western blotting with indicated antibodies. (B) qPCR analysis of the mRNA expression of β-catenin target genes Axin2, Cnx43, and Lef1 in BMSCs. n = 3. (C) TSC1 CKO BMSCs were treated with rapamycin at indicated concentration for 24 hours and analyzed by Western blotting with indicated antibodies. (D) MEF TSC1+/+ and TSC1–/– cells were analyzed by Western blotting with indicated antibodies. (E) qPCR analysis of the mRNA expression of β-catenin target genes Axin2, Cnx43, and Lef1 in MEFs. n = 3. (F) MEFs were treated with rapamycin at 10 nM or 20 nM for 24 hours and analyzed by Western blotting with indicated antibodies. For B and E, *p < 0.05, values are shown as mean + SE. CTR = control (Tsc1F/F).
β-catenin downregulation in TSC1-deficient cells is non-canonically regulated but Notch1-dependent
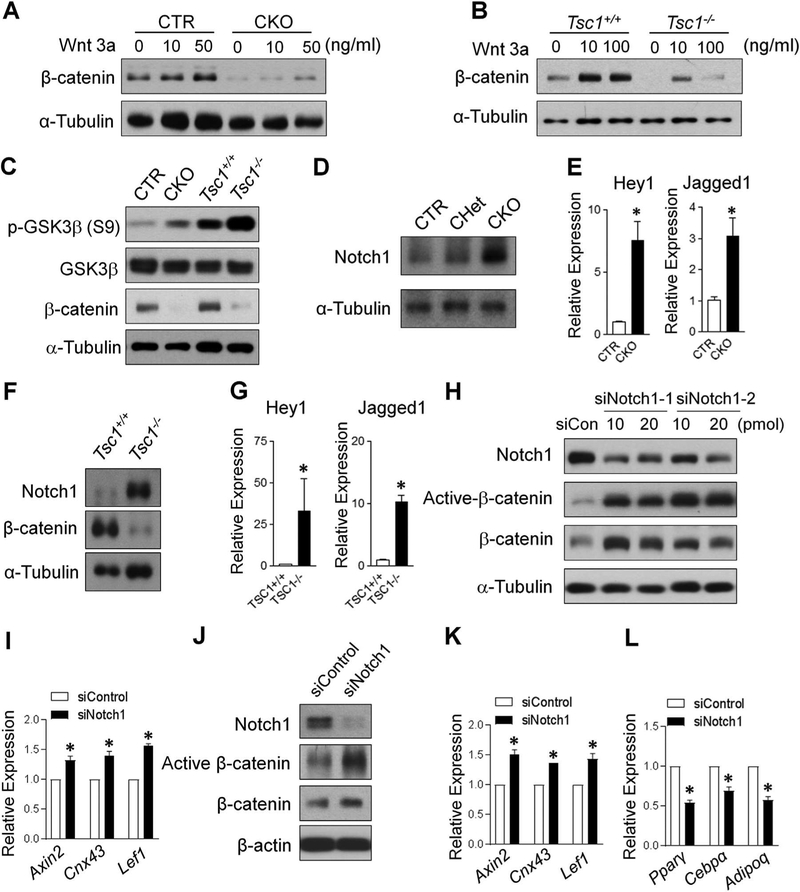
To elucidate the mechanisms underlying the β-catenin down-regulation in TSC1-deficient cells, we determined the effect of Tsc1 deletion on the response to exogenous canonical Wnt ligand in both BMSCs (Fig. 7A) and MEFs (Fig. 7B). As expected, the control cells had increased β-catenin level upon Wnt3a treatment. However, TSC1-deficient BMSCs and MEFs had a blunted increase in β-catenin protein level upon Wnt3a treatment, indicating that the β-catenin downregulation in TSC1-deficient cells was likely not due to the deficiency in Wnt ligands. Downstream of the Wnt ligands and receptors, β-catenin is regulated by a destruction complex in which GSK3 is an important component.(5) To determine whether the down-regulation of β-catenin in TSC1-deficient cells was mediated by increased activity of GSK3 destruction complex, phosphor-GSK3β (S9) level was analyzed. Phosphorylation of the N terminal Ser 9 of GSK3β leads to the inhibition of GSK3 function. Thus the increase in the phosphor-GSK3β (S9) level is expected to have lower β-catenin degradation activity, which would result in higher β-catenin level.(34) Intriguingly, phosphor-GSK3β (S9) was significantly increased in TSC1-deficient BMSCs and MEFs (Fig. 7C), suggesting that the downregulation of β-catenin in TSC1-deficient cells was not mediated through canonical GSK destruction pathway.
Fig. 7.
β-catenin downregulation in TSC1-deficient cells is non-canonically regulated and Notch1-dependent. (A) BMSCs were treated with Wnt3a at indicated concentration for 24 hours and (B) MEFs were treated with Wnt3a at indicated concentration for 48 hours and analyzed by Western blotting with indicated antibodies. (C) The cell lysates from BMSCs (CTR and CKO) and MEFs (Tsc1+/+ and Tsc1–/–) were subjected to immunoblot analysis with indicated antibodies. (D) BMSCs or (F) MEFs were analyzed by Western blotting with indicated antibodies. (E, G) qPCR analysis of the mRNA expression of Notch1 target genes Hey1 and Jagged1 in BMSCs (E) or MEFs (G). n = 3, *p < 0.05. (H, I) Control siRNA (siCon or siControl) and siNotch1 (1 and 2) (10 or 20 pmol) were transduced to Tsc1–/– MEFs for 24 hours and analyzed by Western blotting with indicated antibodies (H) and qPCR analysis of the mRNA expression of β-catenin target genes using 10 pmol siControl and siNotch1–1 (I). (J–L) siControl and siNotch1–1 at 10 pmol were transduced to TSC1-deficient BMSCs for 24 hours and analyzed by Western blotting with indicated antibodies (J) and qPCR analysis of the mRNA expression of β-catenin target genes (K) and adipocyte marker genes (L). For E, G, I, K, and L, n = 3, *p < 0.05. Values are shown as mean + SE. CTR = control (Tsc1F/F).
mTORC1 signaling can enhance Notch signaling and increase Notch1 expression.(35) Overexpression of Notch intracellular domain can impair osteoblast differentiation and inhibit Wnt/β-catenin signaling.(36) Thus, we determined to what extent that the Notch1 signaling was altered in TSC1-deficient cells. In contrary to the decrease in β-catenin, TSC1-deficient BMSCs had significantly increased Notch1 protein level (Fig. 7D) in association with an increase in mRNA expression level of Hey1 and Jagged1 (Fig. 7E), two downstream target genes of Notch1 signaling. In MEFs, a similar inverse relationship between the protein level of β-catenin and Notch1 as well as the increase of Notch signaling due to Tsc1 deletion was observed (Fig. 7F, G). Notch1 protein can mediate GSK3-independent active β-catenin degradation in stem and colon cancer cells through lysosomal degradation.(37) Thus, we next determined whether TSC1 regulated β-catenin through Notch1. Using siRNA approach, we deleted Notch1 in Tsc1–/– MEFs cells and found that Notch1 knockdown increased the active and total β-catenin level (Fig. 7H). In association with the increase in active β-catenin level, there was increased mRNA expression level of Wnt/bcatenin target genes (Fig. 7I). In TSC1-deificent BMSCs, Notch1 knockdown led to an increase in active β-catenin protein level (Fig. 7J), an increase in the mRNA expression level of Wnt/β-catenin target genes (Fig. 7K) and an inverse decrease in the mRNA expression level of adipogenic factors (Fig. 7L). Altogether, our data showed that β-catenin downregulation in TSC1-deficient cells was mediated by increased Notch1 protein level.
Tsc1 deletion leads to autophagy inhibition and Notch1 is an autophagy target
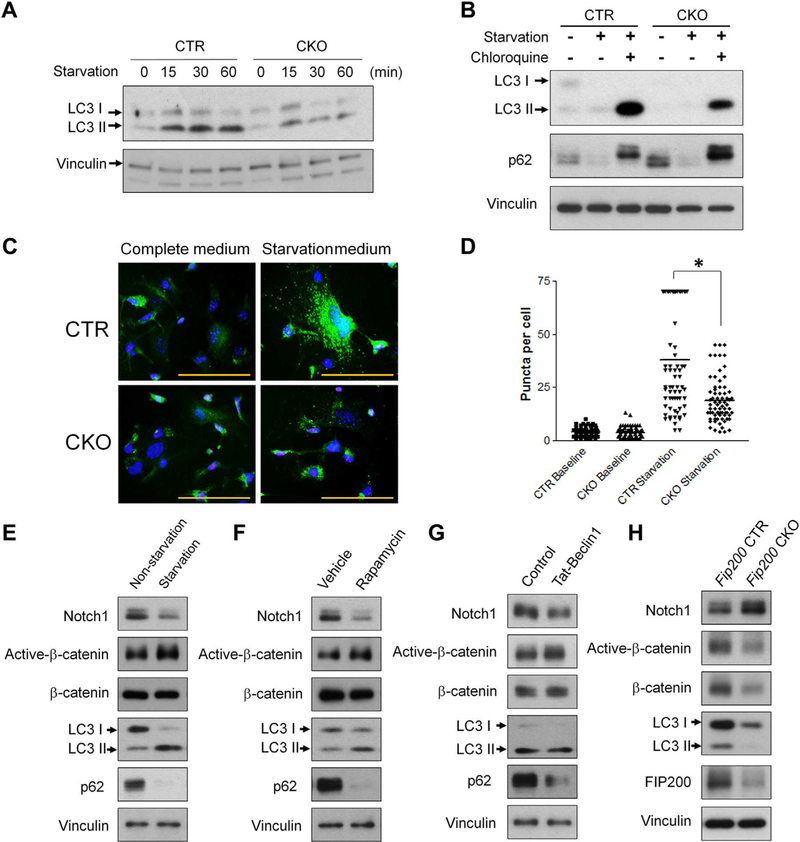
Last, we determined the mechanism of Notch1 regulation by Tsc1 deletion. mTORC1 is a master regulator of autophagy(38–40) and Tsc1 deletion can suppress autophagy as a result of mTORC1 activation.(41–43) It is reported that plasma membrane-resident Notch1 can be degraded by autophagy and autophagy inhibition can increase Notch1 level on the plasma membrane in human embryonic kidney cells.(44) In addition, autophagy deficiency can trigger precocious Notch signaling activation during Drosophila oogenesis.(45) Thus, we hypothesized that TSC1 regulates Notch1 through the autophagy pathway in BMSCs. To test this hypothesis, we first determined the effect of Tsc1 deletion on BMSC autophagy. In response to starvation stimulation, there was increased LC3-II level in both control and TSC1-deficient BMSCs, however, the increase of LC3-II in TSC1-deficient cells was milder, indicating a defect in autophagy response (Fig. 8A). Our data also showed a defective autophagy flux in CKO cells (Fig. 8B). Furthermore, we examined GFP-LC3 puncta formation in response to starvation in isolated BMSCs from CKO and control mice containing transgenic GFP-LC3. As expected, robustly increased GFP-LC3 puncta in BMSCs was observed after starvation treatment, but this increase was significantly blunted in BMSCs isolated from CKO;GFP-LC3 mice compared to that from CTR;GFP-LC3 mice (Fig. 8C, D). Thus, the autophagy function was suppressed in TSC1-deficient BMSCs.
Fig. 8.
TSC1 deletion leads to decreased autophagy in BMSCs. (A) BMSCs were cultured in complete medium or starved for up to 60 min and analyzed by Western blotting with indicated antibodies. (B) Control and TSC1-deficient BMSCs were cultured in the complete medium or starved for 2 hours with or without 100 μM chloroquine. Immunoblot analysis was performed with indicated antibodies. (C) BMSCs isolated from GFP-LC3 transgenic CTR or CKO mice were cultured in complete medium or starved for 2 hours. The cells were subject to fluorescent microscope analysis after fixation and DAPI staining. Scale bar = 50 μm. (D) Numbers of GFP+ puncta per cells were counted, *p < 0.05. (E) TSC1-deficient BMSCs were starved for 1 hour and analyzed by Western blotting with indicated antibodies. (F) TSC1-deficient BMSCs were treated with rapamycin (200 nM) for 1 hour and analyzed by Western blotting with indicated antibodies. (G) TSC1-deficient BMSCs were treated with control peptide or Tat-Beclin1 peptide (10 mM) for 1 hour and analyzed by Western blotting with indicated antibodies. (H) BMSCs isolated from control (Fip200F/F) and Fip200 CKO (Fip200F/F;Osx-Cre/+) mice were analyzed by Western blotting with indicated antibodies. DAPI = 4,6-diamidino-2-phenylindole; CTR = control (Tsc1F/F).
To determine whether the suppressed autophagy in TSC1-deficient BMSCs is responsible for the upregulation of Notch1 and the inverse downregulation of active β-catenin, the TSC1-deficient BMSCs were subject to autophagy induction either by starvation (Fig. 8E) or being treated with rapamycin (Fig. 8F) or Tat-Beclin1, a novel autophagy-inducing peptide(46) (Fig. 8G). In response to the autophagy stimuli, there was a significant decrease in Notch1 protein in association with autophagy activations as indicated by the increase in LC3-II level and decrease in p62 level, indicating Notch1 is an autophagy target in BMSCs. In contrast to the significant decrease in Notch1 level, there was a moderate increase in the active β-catenin level in response to the autophagy activation. Thus, autophagy activation in TSC1-deficient BMSCs led to increased autophagic degradation of Notch1, which in turn restored the decreased active β-catenin level by damping the Notch1-mediated GSK3β-independent degradation of active β-catenin, which is shown to occur in lysosomes.(37) Due to the nonselective strong autophagy activation by starvation, rapamycin, or Tat-Beclin1treatment, β-catenin could be directly subject to autophagic degradation. Thus, the β-catenin increase mediated by the downregulation of Notch1 was counterbalanced by the autophagic degradation of β-catenin. Hence, the increase of active β-catenin level in TSC1-deficient BMSCs in response to these strong autophagy stimuli appears very mild. To further support the notion that autophagy could regulate β-catenin level through Notch1, we determined the effect of autophagy suppression by deleting Fip200, an essential autophagy gene, in osterix+ cells on the β-catenin and Notch 1 level. As expected, Fip200 deletion in BMSCs led to decrease LC3-II protein level (Fig. 8H) and increased p62 (data not shown). Importantly, autophagy deficiency led to increased Notch1 level but decreased β-catenin level (Fig. 8H). This further supports the regulatory axis of autophagy/Notch1/β-catenin in BMSCs.
Discussion
In this work, we identified a novel signaling cascade regulating the commitment of BMSCs into osteoblasts versus adipocytes. Ablation of Tsc1 in Osterix-expressing cells leads to a decrease in trabecular bone mass and an increase in bone marrow adipogenesis in association with decreased β-catenin level, which is mediated by increased Notch1 protein level via autophagy suppression. It is known that Wnt/β-catenin signaling favors osteoblast over adipocyte lineage commitment.(6–8,22) Thus, we propose a model (Fig. 9A, B) in which the decreased Wnt/β-catenin signaling due to the increased Notch1 protein level via autophagy suppression in TSC1-deficient BMSCs accounts for the dysregulated osteoblast-adipocyte lineage commitment and subsequent osteoporotic phenotype in association with increased marrow adipose tissue. It is known that Osterix-Cre transgene causes some skeletal development defects.(26,47,48) Our data showed that Osterix-Cre does not affect in vitro BMSC proliferation (Supporting Fig. S6) and osteogenic differentiation(26); neither did it affect the Notch1 and β-catenin level in BMSCs (Supporting Fig. S7). Thus, we concluded that the Tsc1 deletion itself was the major contributor of both the in vivo bone phenotype of Tsc1-deficient mice and the in vitro differentiation defect of Tsc1-deficient BMSCs. However, because the CHet mice had similar decrease in trabecular bone mass and body weight to Osterix-Cre mice, the trabecular bone mass decrease in CKO mice was likely due to the combined effects of Tsc1 deletion and the presence of Osterix-Cre transgene.
Fig. 9.
Proposed model of TSC1 regulation on the balance between osteoblast and adipocyte differentiation in BMSCs. (A) When TSC1 is present, TSC1-TSC2 complex inhibits mTORC1 activity; PAS develops into autophagosome, which degrades Notch1. In the presence of sufficient β-catenin/Wnt signaling, BMSCs preferentially differentiate into osteoblasts. (B) When TSC1 is deleted, mTORC1 activity is upregulated, which in turn inhibits the autophagosome formation and autophagy-mediated Notch1 degradation. Increased Notch1 can mediate the β-catenin degradation through lysosomal degradation.(37) In the absence of sufficient β-catenin/Wnt signaling, BMSCs preferentially differentiate into adipocytes. PAS = pre-autophagosomal structure.
mTORC1 and Wnt/β-catenin mediate two important signaling pathways that have significant impacts on major cellular functions. The connection between mTORC1 signaling and Wnt signaling is not completely new. It has been shown that Wnt signaling increases mTORC1 activity through the inhibition of GSK3 kinase that promotes TSC1/TSC2 activity by directly phosphorylating TSC2.(49) Our data revealed a novel interaction between these two types of signaling in which mTORC1 activation downregulates Wnt/β-catenin signaling, thus forming a negative feedback loop. Furthermore, our data showed that this downregulation of Wnt/β-catenin signaling by mTORC1 activation is mediated through a GSK3-independent but Notch1-dependent mechanism via autophagy suppression.
The mammalian skeleton contains cortical and trabecular bone. Even though they are generated by the same cell types, there are significant differences between them in both structure(50,51) and response to external factors such as athletic training(52) and parathyroid hormone deficiency and excess.(53) In addition, cortical and trabecular bone have different microenvironments and modeling/remodeling mechanisms. Cortical bone growth mainly occurs through periosteal expansion at periosteum. On the other hand, trabecular bone growth mainly occurs in the bone marrow, which contains multipotent BMSCs that can differentiate into osteoblasts and adipocytes. Despite these significant differences, very little is known about the cellular and molecular mechanisms that are responsible for the differential regulation of cortical versus trabecular bone. In this study, we found that Tsc1 deletion in Osterix-expressing cells decreased trabecular bone mass but increased cortical bone mass. Mechanistically, our data showed that TSC1-deficient BMSCs had decreased proliferation, decreased osteogenic differentiation, and increased adipogenic differentiation. In addition, CKO bone had increased osteoclasts, which further contributed to the decrease in trabecular bone mass. The molecular mechanisms that differentially regulate cortical and trabecular bone remain to be determined.
A recent study also investigated the effect of Tsc1 deletion in Osterix-expressing cells on bone homeostasis.(19) It is interesting to note that this study showed that both cortical and trabecular bone mass is increased in mice at 10 weeks of age when Tsc1 is deleted in Osterix-expressing cells.(19) The detailed bone analysis at earlier age such as 1 month old was not reported in that research. Our study showed the differential effect of Tsc1 deletion on cortical and trabecular bone at 1 month of age. One limitation of our work is the early death of the Tsc1 CKO mice after 1 month of age. This prevented us from determining whether Tsc1 differentially regulates cortical versus trabecular bone in a later development stage. The timing of the sudden death of these otherwise normal Tsc1 CKO mice may suggest a likely hematopoietic failure. The reason of the early death of Tsc1 CKO mice remains to be determined. The mechanisms underlying the likely difference between our study and the aforementioned study(19) is currently unknown but it may be related to the difference in genetic background of the mice (C57BL/6Ncrl versus 129S4/SvJae) as well as the difference in development stages (1 month versus 10 weeks). If the difference in trabecular bone phenotype was indeed due to the difference in genetic background, it highlights the importance of back-crossing to known genetic background in bone research. On the other hand, whenever possible, thorough analysis at both earlier and later developmental stages is desirable to ensure the complete understanding of bone phenotype. Of note, another group similarly reported that Tsc1 deletion in Osterix-expression cells leads to decreased trabecular bone mass in association with increased bone marrow adipogenesis and decreased β-catenin protein level in osteoblasts.(54)
It has been reported that the Osx-Cre transgene is also expressed in the hypertrophic or late proliferating chondrocytes.(55,56) Hypertrophic chondrocytes can transdifferentiate to become osteoblasts contributing to endochondral bone formation.(57) This raised the possibility that decreased femoral trabecular bone mass may also be partially contributed by Tsc1 deletion in chondrocytes. Indeed, our data showed that CKO mice had slightly elongated hypertrophic zone without significant change in proliferative zone (Supporting Fig. S4). The extent of contribution by Tsc1 deletion in chondrocytes to decreased femoral trabecular bone in our mouse model remains to be determined.
Supplementary Material
Acknowledgments
This work was supported by NIH grant (AR062030 to FL, CA211066 and NS094144 to JG). μCT work was partly supported by a P30 Core Center award to University of Michigan from NIAMS (AR 69620). We thank Linford Williams for histomorphometry analysis.
Footnotes
Disclosures
All authors state that they have no conflicts of interest.
Additional Supporting Information may be found in the online version of this article.
References
- 1.Justesen J, Stenderup K, Ebbesen EN, Mosekilde L, Steiniche T, Kassem M. Adipocyte tissue volume in bone marrow is increased with aging and in patients with osteoporosis. Biogerontology. 2001;2(3):165–71. [DOI] [PubMed] [Google Scholar]
- 2.Minaire P, Neunier P, Edouard C, Bernard J, Courpron P, Bourret J. Quantitative histological data on disuse osteoporosis: comparison with biological data. Calcif Tissue Res. 1974;17(1):57–73. [DOI] [PubMed] [Google Scholar]
- 3.Wronski TJ, Morey-Holton E, Jee WS. Skeletal alterations in rats during space flight. Adv Space Res. 1981;1(14):135–40. [DOI] [PubMed] [Google Scholar]
- 4.Nuttall ME, Gimble JM. Is there a therapeutic opportunity to either prevent or treat osteopenic disorders by inhibiting marrow adipo-genesis? Bone. 2000;27(2):177–84. [DOI] [PubMed] [Google Scholar]
- 5.Liu F, Kohlmeier S, Wang CY. Wnt signaling and skeletal development. Cell Signal. 2008;20(6):999–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prestwich TC, Macdougald OA. Wnt/beta-catenin signaling in adipogenesis and metabolism. Curr Opin Cell Biol. 2007;19(6): 612–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Song L, Liu M, Ono N, Bringhurst FR, Kronenberg HM, Guo J. Loss of wnt/beta-catenin signaling causes cell fate shift of preosteoblasts from osteoblasts to adipocytes. J Bone Miner Res. 2012;27(11): 2344–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen J, Long F. beta-catenin promotes bone formation and suppresses bone resorption in postnatal growing mice. J Bone Miner Res. 2013;28(5):1160–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu M, Wang Y, Shao JZ, Wang J, Chen W, Li YP. Cbfbeta governs osteoblast-adipocyte lineage commitment through enhancing beta-catenin signaling and suppressing adipogenesis gene expression. Proc Natl Acad Sci U S A. 2017;114(38):10119–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun C, Yuan H, Wang L, et al. FAK promotes osteoblast progenitor cell proliferation and differentiation by enhancing Wnt signaling. J Bone Miner Res. 2016;31(12):2227–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149(2):274–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Russell RC, Fang C, Guan KL. An emerging role for TOR signaling in mammalian tissue and stem cell physiology. Development. 2011;138(16):3343–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18(16):1926–45. [DOI] [PubMed] [Google Scholar]
- 14.Osborne JP, Fryer A, Webb D. Epidemiology of tuberous sclerosis. Ann N Y Acad Sci. 1991;615:125–7. [DOI] [PubMed] [Google Scholar]
- 15.Curatolo P, Bombardieri R, Jozwiak S. Tuberous sclerosis. Lancet. 2008;372(9639):657–68. [DOI] [PubMed] [Google Scholar]
- 16.Holt JF, Dickerson WW. The osseous lesions of tuberous sclerosis. Radiology. 1952;58(1):1–8. [DOI] [PubMed] [Google Scholar]
- 17.Umeoka S, Koyama T, Miki Y, Akai M, Tsutsui K, Togashi K. Pictorial review of tuberous sclerosis in various organs. Radiographics. 2008;28(7):e32. [DOI] [PubMed] [Google Scholar]
- 18.Riddle RC, Frey JL, Tomlinson RE, et al. Tsc2 is a molecular checkpoint controlling osteoblast development and glucose homeostasis. Mol Cell Biol. 2014;34(10):1850–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang B, Wang Y, Wang W, et al. mTORC1 prevents preosteoblast differentiation through the notch signaling pathway. PLoS Genet. 2015;11(8):e1005426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fang F, Sun S, Wang L, et al. Neural crest-specific TSC1 deletion in mice leads to sclerotic craniofacial bone lesion. J Bone Miner Res. 2015;30(7):1195–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei X, Hu M, Cvanaugh B, et al. The impact and mechanism of Tsc1 deletion in craniofacial bone development. J Bone Miner Res. 2016;31 Suppl 1 [Presented orally at: Annual Meeting American Society for Bone and Mineral Research (ASBMR); 2016 Sep 16–19; Atlanta, GA, USA; Presentation Number: 1050]. Available from: http://www.asbmr.org/education/AbstractDetail?aid=ca7a3c3ea839-4db1-bbf3-0a8d15f871b7. [Google Scholar]
- 22.Rodda SJ, McMahon AP. Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development. 2006; 133(16):3231–44. [DOI] [PubMed] [Google Scholar]
- 23.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15(3):1101–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwiatkowski DJ, Zhang H, Bandura JL, et al. A mouse model of TSC1 reveals sex-dependent lethality from liver hemangiomas, and up-regulation of p70S6 kinase activity in Tsc1 null cells. Hum Mol Genet. 2002;11(5):525–34. [DOI] [PubMed] [Google Scholar]
- 25.Gan B, Peng X, Nagy T, Alcaraz A, Gu H, Guan JL. Role of FIP200 in cardiac and liver development and its regulation of TNFalpha and TSC-mTOR signaling pathways. J Cell Biol. 2006;175(1):121–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang L, Mishina Y, Liu F. Osterix-Cre transgene causes craniofacial bone development defect. Calcif Tissue Int. 2015;96(2):129–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu F, Fang F, Yuan H, et al. Suppression of autophagy by FIP200 deletion leads to osteopenia in mice through the inhibition of osteoblast terminal differentiation. J Bone Miner Res. 2013;28(11): 2414–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei X, Hu M, Mishina Y, Liu F. Developmental regulation of the growth plate and cranial synchondrosis. J Dent Res. 2016;95(11): 1221–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chandhoke TK, Huang YF, Liu F, et al. Osteopenia in transgenic mice with osteoblast-targeted expression of the inducible cAMP early repressor. Bone. 2008;43(1):101–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu F, Lee SK, Adams DJ, Gronowicz GA, Kream BE. CREM deficiency in mice alters the response of bone to intermittent parathyroid hormone treatment. Bone. 2007;40(4):1135–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dempster DW, Compston JE, Drezner MK, et al. Standardized nomenclature, symbols, and units for bone histomorphometry: a 2012 update of the report of the ASBMR Histomorphometry Nomenclature Committee. J Bone Miner Res. 2013;28(1):2–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu F, Lee JY, Wei H, et al. FIP200 is required for the cell-autonomous maintenance of fetal hematopoietic stem cells. Blood. 2010;116(23): 4806–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao Q, Shao J, Chen W, Li YP. Osteoclast differentiation and gene regulation. Front Biosci. 2007;12:2519–29. [DOI] [PubMed] [Google Scholar]
- 34.Rayasam GV, Tulasi VK, Sodhi R, Davis JA, Ray A. Glycogen synthase kinase 3: more than a namesake. Br J Pharmacol. 2009;156(6): 885–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma J, Meng Y, Kwiatkowski DJ, et al. Mammalian target of rapamycin regulates murine and human cell differentiation through STAT3/p63/Jagged/Notch cascade. J Clin Invest. 2010;120(1):103–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zanotti S, Smerdel-Ramoya A, Stadmeyer L, Durant D, Radtke F, Canalis E. Notch inhibits osteoblast differentiation and causes osteopenia. Endocrinology. 2008;149(8):3890–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kwon C, Cheng P, King IN, et al. Notch post-translationally regulates beta-catenin protein in stem and progenitor cells. Nat Cell Biol. 2011;13(10):1244–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22(2): 124–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jung CH, Ro SH, Cao J, Otto NM, Kim DH. mTOR regulation of autophagy. FEBS Lett. 2010;584(7):1287–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chan EY. mTORC1 phosphorylates the ULK1-mAtg13-FIP200 autophagy regulatory complex. Sci Signal. 2010;2(84):pe51. [DOI] [PubMed] [Google Scholar]
- 41.Papadakis M, Hadley G, Xilouri M, et al. Tsc1 (hamartin) confers neuroprotection against ischemia by inducing autophagy. Nat Med. 2013;19(3):351–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Castets P, Lin S, Rion N, et al. Sustained activation of mTORC1 in skeletal muscle inhibits constitutive and starvation-induced autophagy and causes a severe, late-onset myopathy. Cell Metab. 2013;17(5):731–44. [DOI] [PubMed] [Google Scholar]
- 43.Castets P, Ruegg MA. MTORC1 determines autophagy through ULK1 regulation in skeletal muscle. Autophagy. 2013;9(9):1435–7. [DOI] [PubMed] [Google Scholar]
- 44.Wu X, Fleming A, Ricketts T, et al. Autophagy regulates Notch degradation and modulates stem cell development and neuro-genesis. Nat Commun. 2016;7:10533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barth JM, Hafen E, Kohler K. The lack of autophagy triggers precocious activation of Notch signaling during Drosophila oogenesis. BMC Dev Biol. 2012;12:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shoji-Kawata S, Sumpter R, Leveno M, et al. Identification of a candidate therapeutic autophagy-inducing peptide. Nature. 2013; 494(7436):201–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davey RA, Clarke MV, Sastra S, et al. Decreased body weight in young Osterix-Cre transgenic mice results in delayed cortical bone expansion and accrual. Transgenic Res. 2012;21(4):885–93. [DOI] [PubMed] [Google Scholar]
- 48.Huang W, Olsen BR. Skeletal defects in Osterix-Cre transgenic mice. Transgenic Res. 2015;24(1):167–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Inoki K, Ouyang H, Zhu T, et al. TSC2 integrates Wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth. Cell. 2006;126(5):955–68. [DOI] [PubMed] [Google Scholar]
- 50.Goodyear SR, Gibson IR, Skakle JM, Wells RP, Aspden RM. A comparison of cortical and trabecular bone from C57 Black 6 mice using Raman spectroscopy. Bone. 2009;44(5):899–907. [DOI] [PubMed] [Google Scholar]
- 51.Gong JK, Arnold JS, Cohn SH. Composition of trabecular and cortical bone. Anat Rec. 1964;149:325–31. [DOI] [PubMed] [Google Scholar]
- 52.Ducher G, Tournaire N, Meddahi-Pelle A, Benhamou CL, Courteix D. Short-term and long-term site-specific effects of tennis playing on trabecular and cortical bone at the distal radius. J Bone Miner Metab. 2006;24(6):484–90. [DOI] [PubMed] [Google Scholar]
- 53.Duan Y, De Luca V, Seeman E. Parathyroid hormone deficiency and excess: similar effects on trabecular bone but differing effects on cortical bone. J Clin Endocrinol Metab. 1999;84(2): 718–22. [DOI] [PubMed] [Google Scholar]
- 54.Han Q, LIu K, Zhu Y, Chen Q, Ouyang H. Osteoblast-specific deletion of Tsc1 leads to reduced osteoblastogenesis and enhanced bone marrow adipogenesis in vivo. J Bone Miner Res. 2016;31 Suppl 1 [Poster presented at: Annual Meeting American Society for Bone and Mineral Research (ASBMR); 2016 Sep 16–19; Atlanta, GA, USA; Presentation Number: LB-SU0357]. Available from: http://www.asbmr.org/education/AbstractDetail?aid=6b3536c2-5f59-47d6-a89d-9a785aa9a5a2. [Google Scholar]
- 55.Xiong J, Onal M, Jilka RL, Weinstein RS, Manolagas SC, O’Brien CA. Matrix-embedded cells control osteoclast formation. Nat Med. 2011;17(10):1235–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kobayashi T, Lu J, Cobb BS, et al. Dicer-dependent pathways regulate chondrocyte proliferation and differentiation. Proc Natl Acad Sci U S A. 2008;105(6):1949–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou X, von der Mark K, Henry S, Norton W, Adams H, de Crombrugghe B. Chondrocytes transdifferentiate into osteoblasts in endochondral bone during development, postnatal growth and fracture healing in mice. PLoS Genet. 2014;10(12):e1004820. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.