GNCIA allows a sensitive and selective detection of HIV-1 infections with a sensitivity of 5 pg/ml.
Abstract
We have engineered streptavidin-labeled fluorescent gold nanoclusters to develop a gold nanocluster immunoassay (GNCIA) for the early and sensitive detection of HIV infection. We performed computational simulations on the mechanism of interaction between the nanoclusters and the streptavidin protein via in silico studies and showed that gold nanoclusters enhance the binding to the protein, by enhancing interaction between the Au atoms and the specific active site residues, compared to other metal nanoclusters. We also evaluated the role of glutathione conjugation in binding to gold nanoclusters with streptavidin. As proof of concept, GNCIA achieved a sensitivity limit of detection of HIV-1 p24 antigen in clinical specimens of 5 pg/ml, with a detection range up to1000 pg/ml in a linear dose-dependent manner. GNCIA demonstrated a threefold higher sensitivity and specificity compared to enzyme-linked immunosorbent assay for the detection of HIV p24 antigen. The specificity of the immunoassay was 100% when tested with plasma samples negative for HIV-1 p24 antigen and positive for viruses such as hepatitis B virus, hepatitis C virus, and dengue. GNCIA could be developed into a universal labeling technology using the relevant capture and detector antibodies for the specific detection of antigens of various pathogens in the future.
INTRODUCTION
Metal nanoclusters are an interesting class of materials. They are isolated particles with up to hundreds of metal ions and a size comparable to the Fermi wavelength of electrons (1). Their unique characteristic properties arise as a result of their size being restricted to around 2 nm, which restricts the motion of their free electrons in a confined space that results in discrete electronic states (2). These molecular-like transitions generate intense fluorescence, and thus, nanoclusters have been extensively studied because of their wide range of potential applications in bioimaging, biosensing, catalysis, and various other applications (3–6). Moreover, metal nanoclusters show size-dependent tunable fluorescence from visible to near-infrared regions with high quantum yields (7). Their importance lies in their biocompatibility for conjugation with various other biological molecules. Bioconjugated fluorescent metal nanoclusters have been considered as powerful materials for the detection of low-concentration analytes due to their facile synthesis, low cost, excellent biocompatibility, and multifunctional surface chemistry (8).
Gold is the most extensively studied material due to its stable chemical property, facile synthesis, and nontoxicity. With sizes approaching the nanoscale level, radical changes are observed in their characteristics. Gold nanoparticles (NPs) exhibit distinctive properties that differ from other NPs, such as their surface plasmon resonance effect, size-dependent electronic properties, and their photo-thermal effect in biological therapy (9). Because of these unique characteristics, gold NPs have been considered as one of the key materials for nanoscience and nanotechnology. However, recent advances make it possible to further restrict the size of the gold NP to a few nanometers, permitting the synthesis of gold nanoclusters (AuNCs) with tunable size or emission colors. AuNCs have an extremely high surface-to-volume ratio, which allows for further surface modification and controllable bioconjugation. With all the above properties, AuNCs can be used in a wide range of applications, such as biodetection, biosensing, biological labeling, and bioimaging (10–14). Photoluminescent AuNCs have emerged as an interesting sensing material, mainly because of their ease in preparation, biocompatibility, large Stokes shift, and long lifetime (15).
One of the most popular reactions used in NP-based biosensing is the interaction of biotin and streptavidin. Streptavidin-biotin biochemistry has been exploited for use in many protein and nucleic acid detection technologies (16). Biotin is stable and small and can be easily functionalized onto antibodies, which can react with streptavidin-conjugated labels, allowing sensitive and specific detection. The streptavidin-biotin interaction has been widely in use for the development of robust and highly sensitive assays. Therefore, it is envisaged that the combination of intense fluorescence properties of AuNCs and conjugation with streptavidin will allow for the detection of low-concentration analytes. This may provide attractive multifunctional features for the development of AuNC-based assays.
Thus, we have engineered streptavidin-conjugated AuNCs (AuNC-SA) for applications in biosensing and biodetection. We have evaluated the thermodynamics and the mechanism of interactions between the nanoclusters and the streptavidin protein via in silico studies using computational simulations. Further, on the basis of the results of the simulations, we have experimentally shown the successful conjugation of AuNCs with streptavidin. Our findings confirm that fluorescent-conjugated nanocluster-based immunoassays could increase detection sensitivity and assay stability and have applications for improved diagnosis and point-of-care use. A gold nanocluster immunoassay (GNCIA) could be developed into a universal labeling technology by replacing capture and detection antibodies for the detection of antigens of various pathogens in the future.
RESULTS
Mechanistic rationale of conjugation of AuNC for application in immunoassay
The interaction of biotin and streptavidin has been widely adopted as the sensing interaction for many biomolecular assays for detection. Streptavidin is a protein that is conjugated to the NP label using well-established N-(3-dimethylamino propyl)-N′-ethylcarbodiimide hydrochloride (EDC)–N-hydroxysuccinimide(NHS) chemistry because, by itself, the protein interacts weakly with the NPs. The streptavidin-biotin complex is the strongest known noncovalent interaction between a protein and a ligand (17). Bond formation between biotin and streptavidin is very rapid and, once formed, is unaffected by extremes of pH, temperature, organic solvents, and other denaturing agents. Many proteins, such as antibodies, can be tagged with several biotin tags, each having the possibility of binding to a biotin-binding protein (18). Thus, stable conjugation of streptavidin to the nanoclusters is essential, and each parameter involved in the conjugation needs to be investigated.
To understand the mechanism of streptavidin conjugation with AuNC and visualize the role of EDC-NHS conjugation, we pursued the thermodynamics of this interaction at a molecular level. This required probing into the energetics of different steps in the conjugation that were accordingly simulated. For computational analysis, we considered representative nanocluster systems analogous to the experimental work, building clusters of increasing size in the range of 1 to 57 atoms. We carried out the studies using Gaussian 09, Molecular Operating Environment (MOE), and HEX.
Thermodynamics of formation of the conjugated AuNC systems
First, we analyzed the thermodynamics of EDC-NHS activation step. Gaussian was used to generate structures in three steps, as noted by the reaction below
The energies of the individual reactants and products in the process described above were calculated for their MMFX94-optimized confirmations and tabulated in Table 1. Values from the table indicate that, with an increasing cluster size from 0 to 13, the energy decreases, rendering the most stable conformation in Au13-glutathione–NHS. This demonstrates the driving spontaneity with the growth in AuNC because of the greater negative free energy. The net free energy was found to be positive in the absence of the gold cluster, indicating that the reaction would not be thermodynamically spontaneous in the absence of the AuNC.
Table 1. Energy profiles of the individual steps in the synthesis of the glutathione capped AuNCs.
| Process step | Energy (E0) (kcal/mol) | Energy (E1) | Energy (E6) | Energy (E13) |
| Aux-glutathione + EDC + NHS | 44.47 | 41.333 | 41.974 | 971.502 |
| Aux-glutathione–EDC + NHS | 42.622 | 33.818 | 43.684 | 911.661 |
| Aux-glutathione–NHS | 46.307 | 23.553 | 24.186 | 878.414 |
| Net energy gain | 1.837 | −17.78 | −17.788 | −93.088 |
Role of the Aux clusters in formation of AuNC-SA
Figure 1 shows the binding interaction of the nanocluster with the protein through interactions between the Au and the particular active site residues. The role of AuNCs is very important, as established by the energy calculations performed in the previous step. Further, increasing the size of the cluster enhances the binding affinity.
Fig. 1. Effect of the presence of Au clusters on the glutathione interaction with the streptavidin protein as captured by MOE.
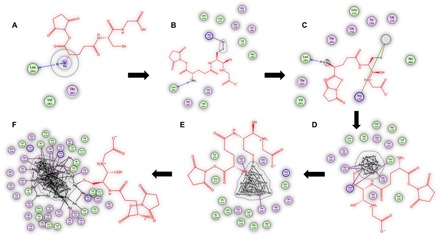
(A) Au0-glutathione, (B) Au1-glutathione, (C) Au6-glutathione, (D) Au13-glutathione, (E) Au20-glutathione, and (F) Au57-glutathione interactions with the streptavidin protein, as determined by MOE docking. Red lines indicate the glutathione structure, while green and purple circles indicate the amino acids involved in the interaction with gold atoms and clusters.
Effect of surface functionalization on streptavidin conjugation to AuNC
EDC-NHS activation is followed by conjugation with streptavidin. Nanocluster interactions with EDC-NHS are mediated through the ligands on the surface of the clusters. We evaluated the role of glutathione in facilitating this process. In this regard, we carried out the simulations by varying the number of glutathione molecules on the nanoclusters. Au13 was taken as a model nanocluster to study the role of increasing the number of glutathione tails. Our studies show that increasing the number of the glutathione-NHS tails on the AuNC enhanced binding to streptavidin, as shown in fig. S1, given that each tail acquires its own binding residues in the active site pocket.
Effect of glutathione conjugation on the stability of AuNC-SA
The driving force for glutathione conjugation is the thermodynamic spontaneity for the reaction with negative total free energy that stabilizes the conjugated moiety. Thus, to determine the stability of the entire structure, we evaluated the binding score of the gold cluster interaction with the streptavidin protein. Further, we calculated the difference in the stability in the presence and absence of glutathione. The difference in the binding score confirms the substantial enhancement in the binding affinity, as shown in Table 2. Thus, we observed that the presence of glutathione is paramount for conjugation while stabilizing the conjugated system.
Table 2. Comparison of the role of the glutathione in the conjugation reaction with streptavidin for increasing cluster size.
| Nanocluster |
Binding score without glutathione-NHS to streptavidin (kcal/mol) |
Binding score with glutathione-NHS to streptavidin (kcal/mol) |
| Au1 | −42.25 | −279.06 |
| Au6 | −116.65 | −282.52 |
| Au13 | −137.72 | −345.92 |
| Au20 | −203.13 | −382.04 |
| Au57 | −318.67 | −388.8 |
Further, with respect to the interaction with streptavidin, our studies indicate that conjugation of glutathione-NHS tails plays a key role. Thus, we studied the change in the binding score for a variety of ligands. The results showed that the best values were obtained with glutathione-NHS tail compared to various other capping tails that are commonly used in the synthesis of AuNCs. This study was performed on Au13 as the common core, and results are presented in table S1.
While AuNCs are the most widely studied nanoclusters, other metal nanoclusters that are being studied for biosensing applications have also been synthesized. Thus, we compared the binding scores of other nanoclusters such as silver and copper in conjugation with streptavidin. Our simulations showed that gold has the best binding score compared to silver and copper nanoclusters. Results are tabulated in table S2.
Characterization of the glutathione-functionalized AuNCs
The morphological characterization of the synthesized nanoclusters is discussed in the Supplementary Materials. The transmission electron microscopy (TEM) image indicated the size of the clusters to be around 2 nm or less (fig. S2). We performed the ultraviolet-visible (UV-Vis) and photoluminiscence characterization and observed that nanoclusters were highly fluorescent and emitting at 615 nm (excitation at 350 nm), as seen in fig. S3.
Fluorescence polarization measurements
The confirmation of the conjugation of nanocluster with streptavidin, as predicted by the computational study, was obtained experimentally from fluorescence polarization (FP) measurements. Increase in the value of FP indicates that the size of the bioconjugated NP has increased after conjugation to the protein (19). The streptavidin binding to the nanoclusters results in an increase in size, which decreases the rotation when compared to the unconjugated nanoclusters. This results in a higher magnitude of FP for the conjugated nanoclusters compared to the unconjugated nanoclusters. We used a SpectraMax M5 plate reader to perform the measurements in the FP mode. The FP values of unconjugated and streptavidin-conjugated nanoclusters from the FP equation were obtained by measuring the P and S polarization values based on the formula (20)
where ∥ is the parallel polarized emission, ⊥ is the perpendicularly polarized emission, and G is the correction factor that is evaluated for the sensitivity of the instrument.
The value of G, as obtained from the instrument, was 1. From the calculations, we observed that the FP value for unconjugated particles was 0.098, while for the conjugated particles, the FP value was 0.131. Data are presented in table S3. As it can be inferred, the smaller moieties have a small FP value, and larger particles have a relatively higher FP value. Thus, comparing the FP values helps in monitoring the functionalization. It was seen that the FP value of the AuNC-SA was definitely higher than pristine AuNCs, and this increase in the value confirmed the conjugation of streptavidin to AuNCs.
To understand the increase in FP upon conjugation, the relationship between FP and size of the fluorophores needs to be comprehended. The unconjugated particles are smaller in size, which causes their movement in solution, making the incident plane-polarized excitation energy to be depolarized. This causes the low polarization value to be lower in magnitude (21, 22). However, when streptavidin is conjugated to AuNCs, there is a decrease in the motion of the particle as the size and volume of the particles increase. This leads to a reduction in the rotational motion of the particle, which results in the excitation energy and the transmitted energy to be in the same state of polarization. This exhibits as a higher magnitude of FP value (23). This is the reason why a higher value of FP is observed for the conjugated particle than for the unconjugated particle.
Application of AuNC-SA in GNCIA for the detection of HIV-1 p24 antigen
We next used the AuNC-SA in gold nanocluster immunoassay(GNCIA). HIV-1 p24 antigen was selected as the target viral protein in this study due to its abundance in the early stages of HIV infection. This is the stage where the antibody concentrations are below the measureable limits. The immunoassay deploys a sandwich immunoassay format, which involves an antibody-antigen-antibody sandwich complex, and the detection is based on chemistry between the biotinylated detector antibody and AuNC-SA, which is illustrated in the schematic (Fig. 2). Strong noncovalent chemistry between biotin and streptavidin is the principal interaction involved in the detection process, which immobilizes the fluorophore to the sandwich complex. Upon excitation, the fluorophore emits, and the emitted signal intensity is proportional to the amount of AuNC-SA. The amount of AuNC-SA is in turn proportional to the amount of p24 present. Thus, the concentration of p24 present in the sample determines the signal intensity. The fluorescence signal intensity was then recorded using a spectrophotometer (SpectraMax M5 microplate reader) (24). The calibration plot for the signal intensity versus the concentration of purified HIV-1 p24 antigen was plotted with the measured data, as depicted in Fig. 3.
Fig. 2. Schematic representation of GNCIA in detection of HIV-1 p24 antigen.
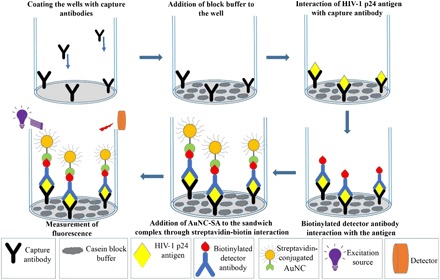
Capture antibodies are coated on the microtiter wells that capture the HIV-1 p24. Subsequently, secondary biotinylated anti-p24 antibodies, which bind to the captured p24 antigen, are added. The immunocomplex then attaches to the AuNC-SA, which recognizes and interacts with the above biotinylated sandwich complex. After multiple wash steps, the signal released from the wells is recorded with a SpectraMax M5 multiplate reader (27).
Fig. 3. Results of GNCIA.
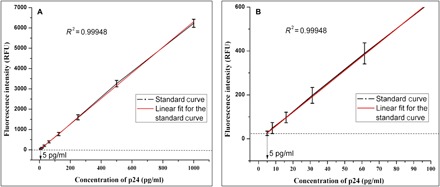
(A) Calibration plot of (GNCIA). The target analyte in this study was the purified HIV-1 p24 antigen [range, 5 to 1000 pg/ml in serial dilution in phosphate-buffered saline (PBS)]. The dashed line represents the cutoff value of negative control. Error bars represent the SD of at least three independently repeated experiments for each assay. RFU, relative fluorescence units. (B) The calibration curve of GNCIA with resolved axis that shows the lowest concentration measured at 5 pg/ml (27).
Demonstration of performance of AuNC-SA through GNCIA
We obtained the signal cutoff value for GNCIA based on the sum of the means methods, which is based on the sum of the means of the signal intensity of negative controls plus double their SD. The cutoff value was calculated to be 10.15 relative fluorescence units (25). For the linear dynamic range of 5 to 1000 pg/ml, the limit of detection was found to be 3.8 pg/ml for a signal-to-cutoff (S:Co) value of 2:1. This ratio is important as it is the criteria for determining whether the sample was HIV positive or HIV negative (26). The equation presented below was the equation of linear correlation obtained on the basis of the calibration curve
where FI is fluorescence intensity and C is the concentration of p24.
It can be seen that there is an excellent linear correlation between the concentrations of HIV-1 p24 and the fluorescence intensity in GNCIA. This is further confirmed by the value of the coefficient of correlation, which was determined to be R2 = 0.99941. This also goes on to suggest that the nature of GNCIA is that of a linear dose-response assay (27). Furthermore, we also report that GNCIA has realized an analytical sensitivity at the picogram level, which is as good as or better than sensitivities achieved using traditional colorimetric enzyme-linked immunosorbent assay (ELISA). Table 3 compares the sensitivity of different detection techniques involving AuNCs, including the present study.
Table 3. Comparison of sensitivity of GNCIA with other NP-based immunoassays and conventional ELISA.
| Serial number | Name of the assay | Sensitivity measured (pg/ml) | Reference |
| 1 | Zinc oxide NP immunoassay | 25.0 | (37) |
| 2 | Carbon dots immunoassay | 20.0 | (20) |
| 3 | Conventional ELISA | 15.0 | (38) |
| 4 | Fluorescent silver NP immunoassay | 10.0 | (27) |
| 5 | Inductively coupled plasma mass spectrometry–based gold NP immunoassay | 7.5 | (39) |
| 6 | GNCIA | 5.0 | Present study |
Effect of interfering protein and coexistent virions on the immunoassay protocol
The samples that are used for testing with the immunoassay contain various other proteins and biomolecules. This further demands the testing of the interference from other coexisting biomolecules on the immunoassay. For purposes of testing, bovine serum albumin (BSA) was taken as a model protein. The results of the assay confirmed that BSA has no effect on the signal intensities in the immunoassay. Figure S4A proves that there is hardly any variation in the signal strength with changing BSA concentration. This further demonstrates the stability of the assay (27).
In addition, to further examine the degree of specificity and cross-reactivity of GNCIA, we spiked hepatitis C virus (HCV)–positive plasma samples with HIV-1 p24 antigen and tested with GNCIA. To perform the test, we spiked a fixed concentration of 250 pg/ml in the samples. The results, as shown in fig. S4B, indicate that there is no visible change in the fluorescence signal from the HCV virions, suggesting that GNCIA is a remarkably specific and stable testing procedure.
Optimization of GNCIA
Capture antibody
The sensitivity of the assay is substantially affected by the concentration of capture antibodies (27). There is an optimum concentration of capture antibody at which the signal strength is maximum; at concentrations higher than the specific optimum concentration, the background signal is also higher, and at concentrations below this, the signal strength shall be weak (28). The concentration of capture antibody requisite for the assay was optimized by testing different concentrations of 0.2, 0.5, 1.0, 1.2, and 1.5 μg/ml, followed by checking the signal response. The signal response was weak for 0.2, 0.5, and 1.0 μg/ml, as shown in fig. S5A. There was no steady correlation between the observed signal and antibody concentration. However, a strong response was observed at 1.2 μg/ml. For concentrations higher than 1.2 μg/ml, there was no change in the response. This behavior can be attributed to the saturation of the detection area with capture antibodies at 1.2 μg/ml (27). As a result, for further studies, 1.2 μg/ml was chosen as the optimum concentration of capture antibody.
AuNC-SA
AuNC-SA play the role of the fluorescent probes in GNCIA. AuNC-SA allow the streptavidin-biotin interaction, which in turn allows AuNCs to indirectly interact with the detector antibody. When the probes are excited with the excitation signal, the emission signal is recorded. The amount of antigen present is calculated from the intensity of the output (fluorescence) signal (27). A sensitive spectrophotometer (SpectraMax M5) was used for recording the emitted signal. The calibration curve is plotted on the basis of the linear correlation observed between the fluorescence intensity and the p24 antigen concentration. The concentration of AuNC-SA greatly affects the performance of the assay. Seven concentrations of AuNC-SA (0.05, 0.1, 0.25, 0.5, 1.0, 1.5, and 2.0 μg/ml) were taken under consideration to study their effects on the signal-to-blank ratio (S:B ratio) in GNCIA. The blank measurements were recorded by performing the immunoassay without the p24 antigen. The concentration of AuNC-SA was optimized by calculating the S:B ratio for fluorescence signals measured for p24 antigen with a fixed concentration of 250 pg/ml. Figure S5B shows an absence of linearity in the relationship between the concentration of AuNC-SA and the S:B ratio. The S:B ratio was detected to be maximum at a concentration of 0.5 μg/ml. This can be attributed to the increase in the signals measured from the blank wells. With an increase in the concentration of AuNC-SA past the optimal concentration, there was a corresponding increase of blank intensity, which is due to the nonspecific absorption of AuNC-SA in the wells.
Clinical evaluation of GNCIA
We evaluated the sensitivity of GNCIA by testing samples from infected individuals (HIV-positive samples). The testing protocol requires the plasma samples to be diluted 100 times before experimentation. Fifty HIV-positive samples (which were confirmed by commercial ELISA kits) were tested using GNCIA, wherein no false negatives were recorded. We evalutated specificity using 10 HBV-positive/HIV-negative and 10 HCV-positive/HIV-negative plasma samples (27). The signal intensity obtained from these samples was minimal, comparable to the fluorescence signal measured from the negative control. Fluorescence signal intensities from different plasma samples are compared in Fig. 4 (A to C). Thus, it was proved that the assay is specific to p24 antigen and is not susceptible to cross-reactivity effects from other virions and biomolecules. Data are summarized in table S4. High specificity and no cross-reactivity are features of GNCIA.
Fig. 4. Application of GNCIA to clinical samples.
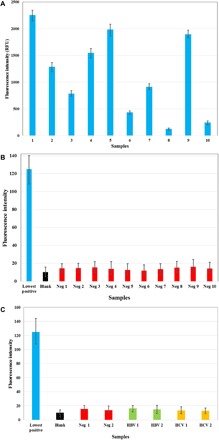
(A) Results of GNCIA for 10 samples tested as HIV positive chosen at random (blue bars). (B) Results of GNCIA for two randomly chosen samples tested as HIV negative in comparison with the lowest positive tested sample and blank. Blue and black bars indicate the intensity of the HIV-positive sample and the blank. (C) Results of GNCIA for two randomly chosen HIV-negative, HBV-positive/HIV-negative, and HCV-positive/HIV-negative samples, respectively, in comparison with the intensity of the lowest positive tested sample and blank. Blue and black bars indicate the intensity of the HIV-positive sample and the blank, while the red, green, and yellow bars indicate HIV-negative, HBV-positive/HIV-negative, and HCV-positive/HIV-negative samples, respectively (27).
An important observation was that some of the samples that were borderline positive by the current assay system were detected by GNCIA. Hence, all the samples tested were positive, and no discrepancies were noted. Similarly, all HIV-negative samples from healthy individuals were negative using GNCIA. Future studies will be dedicated to the performance of GNCIA with clinical samples that contain extremely low quantities of virus.
GNCIA could achieve high-sensitivity levels without any major requirement of sophisticated instruments and costly reagents, unlike nucleic acid testing. GNCIA has been proved to be a highly sensitive immunoassay due to its inherent features of stable signal strength and high signal-to-noise ratio. The assay does not use detection methodologies based on enzymatic reactions, specific high-end instrumentation for performing reactions, and specialized storage conditions for reagents. The high specificity of AuNC-SA is a great strength in this protocol, which shall allow GNCIA to be developed into a complete labeling technology applicable to any detection protocol. GNCIA can be easily modified for the detection of any disease agent by choosing the relevant primary and detector antibodies specific to antigens of disease. Further, GNCIA does not demand technically trained staff to perform the assay, as it is similar to traditional ELISA. GNCIA has the potential to be developed into a rapid and ultrasensitive testing platform for clinical diagnosis and laboratory research in resource-limited settings.
DISCUSSION
In this study, we have demonstrated a computational methodology to verify and test the possibility of using AuNC-SA in biological detections. On the basis of our results, we have validated the feasibility of using AuNCs to detect HIV-1 p24 antigen at picogram levels using an antigen-based sandwich immunoassay (GNCIA). These AuNCs exhibited high photostability, narrow emission widths, and high quantum yield when compared to dye-doped NPs. The characteristic large Stokes shift and very low nonspecific binding have resulted in the reduction of background fluorescence. This has enabled the enhanced detection sensitivity. Biocompatibility of AuNCs is another very vital characteristic that is useful for targeted diagnostic applications.
On the basis of our studies of reaction energetics and binding mechanisms, we could study the feasibility of successful conjugation of nanoclusters with the streptavidin protein. While the results showed that the conjugation will be stable, we tested the predictions using the conjugated nanoclusters in immunoassay for detection of HIV. We observed an analytical sensitivity of 5 pg/ml in GNCIA, which exhibited threefold higher analytical sensitivity when compared to traditional ELISA. A strong linear correlation that was observed between the target concentration and signal intensity shall be useful for quantitative detection of antigen in plasma samples.
Our findings confirm that the application of streptavidin-conjugated fluorescent AuNCs in the immunoassay has markedly increased the detection sensitivity. This shows that GNCIA can have applications in improved diagnosis and point-of-care use. There are multiple assay platforms where these nanoclusters can be used to develop a robust sensing platform. Although many such assays exist, GNCIA holds the capability to detect HIV-1 p24 antigen in the picogram per milliliter range, which makes it a great tool in identifying early HIV infection cases. These research findings are very vital for those working on creating highly sensitive labels and assays for the estimation of HIV incidence and detection of acute cases. As part of the strategy toward an AIDS-free generation, this highly sensitive GNCIA for HIV detection would also assist in improving the blood safety by decreasing the antibody-negative window period in patients in resource-limited settings where nucleic acid testing is not possible.
In GNCIA, the background fluorescence is not sufficiently low enough to allow for higher S:B ratios. This limits its sensitivity and prevents attaining subpicogram-level limits of detection. To circumvent this issue, we need to use signal enhancement methodologies that can specifically enhance the fluorescence signal and suppress the background noise. Metal-enhanced fluorescence (29) or surface plasmon–coupled emission (30) may be incorporated for signal enhancement to achieve higher S:B ratios, thus realizing still better sensitivities with fluorescent reporters. Sandwich immunoassay formats have the limitation that there are multiple incubation steps involved, which increase the assay time substantially. In a clinical setting, a longer assay time can mean a longer wait period for the test results, which can be a problem at the point-of-care use. The assay duration can be reduced by transferring the assay from the microplate platform to a microfluidic platform. The use of paper as a substrate has been widely adopted for rapid assays. Moreover, there will also be a reduction in the consumption of reagents, resulting in reduced cost of the assay. Paper-based microfluidic GNCIA reduces the assay period from 5 to 6 hours to almost 1 hour (31). Addressing these shortcomings can render GNCIA to be an ultrasensitive, highly specific, affordable, user-friendly, rapid, and robust platform for early detection of diseases in point-of-care applications for resource-limited settings.
MATERIALS AND METHODS
Chemicals
Chloroauric acid (HAuCl4), glutathione, EDC (98%), sulfo-NHS, BSA, casein block buffer, and streptavidin were of analytical grade, purchased from Sigma-Aldrich (India), and used without further purification. PBS buffer (pH 7.2) and carbonate-bicarbonate buffer were prepared in laboratory by following the established procedures, and a blocking buffer was purchased from Thermo Fisher Scientific. All experiments were performed with double-distilled water. Sri Sathya Sai Institute of Higher Medical Sciences (Prasanthigram, India) provided the plasma samples from HIV-infected and healthy individuals (study ID SSIHL/IEC/PSN/BS/2012/01). The samples were tested in a blinded manner after their details were coded. Samples were tested with a Microlisa HIV Ag and Ab ELISA kit (J. Mitra & Co. Pvt. Ltd.) and determined whether they were positive or negative.
Computation simulations on streptavidin conjugation of AuNC
Given the minimal mechanistic understanding of the immunoassay in the sensing, we pursued the rationale that drives the appropriate design of the gold assay. In this regard, we evaluated the mechanism of the interaction between the nanoclusters and the streptavidin protein via in silico studies using the computational simulations. First, to explain the thermodynamics of these processes, we sought to understand the energetics of the individual reactants and products at every step. Each of these structures was built and energetically minimized in Gaussian 09 (32). Second, we elucidated the role of the growing Au clusters in the interaction where the various sizes of gold clusters (Aux) were built using Gaussian ranging from Au1 to Au57. For this, we obtained the density functional theory–optimized structures ranging from Ag1 to Ag57 atoms to be treated as consequent ligands (33). The smallest being the neutral Ag atom, to the consequent icosahedral Ag13, tetrahedral Ag20, and the biggest being the icosahedral core–based Ag66 cluster whose energetics were determined by MOE using MMFX94 force field (34). Last, we increased the number of glutathione-NHS tails attached to the individual gold Ag13 nanocluster in G09 and also built assemblies of multiple capping agents. This ascertained the role of glutathione conjugation in the assay through the HEX docking interactions of these conjugated species with streptavidin (35).
Synthesis and characterization of AuNCs
The method that we followed was chemical reduction of gold salt. Chloroauric acid (HAuCl4) is the salt of gold ions that acts as the source in the synthesis. Freshly prepared aqueous solutions of HAuCl4 (20 mM, 0.50 ml) and glutathione (100 mM, 0.15 ml) were mixed with 4.35 ml of ultrapure water at 25°C. The reaction mixture was heated to 70°C under gentle stirring (500 rpm) for 24 hours. An aqueous solution of strongly orange-emitting AuNCs was formed. Synthesized AuNCs were centrifuge dried and stored by refrigeration at 4°C for additional characterization and use in immunoassay.
To understand the structure and morphological characteristics including the size of the AuNC dispersion, a Joel 1400 TEM operated at 80 kV was used. AuNCs were prepared for the imaging by drop-casting them on a copper grid and vacuum drying before imaging. The optical characterization studies were performed using a Shimadzu 2450 UV-Vis spectrophotometer for UV-Vis absorption characterization and a SpectraMax M5 microplate reader for fluorescence emission characterization. Dilution of samples was carried out to obtain the absorbance and fluorescence spectra.
Bioconjugation of streptavidin to glutathione-functionalized AuNCs
Carboxyl groups present on the glutathione-functionalized AuNCs were covalently conjugated with primary amines of streptavidin using EDC/sulfo-NHS chemistry (36). In the initial step, 20 mg of AuNC dispersed in 10 mM PBS was washed in a Nanosep centrifugal ultrafiltration device (molecular weight cutoff, 300 kDa; Pall Life Sciences, Ann Arbor, MI, USA). After washing, carboxyl groups present on AuNCs were activated with 10 mM EDC and 20 mM sulfo-NHS in PBS buffer for 30 min. After washing these activated particles with glycine buffer, 50 μl of streptavidin (1 mg/ml) in the carbonate buffer (pH 9.0) solution was added. The subsequent step involved incubation of the mixture for 24 hours at room temperature, followed by washing five times with glycine buffer. The resultant streptavidin-conjugated nanoclusters were diluted to 0.1 mg/ml in PBS and stored at 4°C for immunoassay experiments.
Fluorescence polarization
Since there was no direct way to determine whether conjugated NPs were produced, FP experiments were used. The FP value was obtained by exciting the sample at 345 nm, recording its emission at 615 nm, and then using the value in the formula
The FP mode was used to obtain I∥ and I⊥ values in the plate reader (SpectraMax M5 plate reader), which were then substituted in the formula to obtain the FP ratio. The higher magnitude of FP of conjugated AuNCs in comparison to unconjugated AuNCs confirms the conjugations of AuNCs.
HIV-1 p24 antigen detection by GNCIA
In this experiment, the chosen format is the sandwich immunoassay format where p24 antigen is sandwiched between the capture and detector antibodies. The fluorophore used here is the AuNCs bound to the detector antibodies.
Protocol for GNCIA
An established protocol for an NP-based immunoassay was followed (27). A capture antibody concentration of 2 μg/ml was prepared by diluting with carbonate-bicarbonate buffer [100 mM (pH 9.6)]. Fifty-five microliters of capture antibody at a concentration of 2 μg/ml was coated onto a 96-well plate and left for overnight incubation at 4°C. After the incubation step, the wells were repeatedly washed five times with wash buffer, following which 300 μl of casein blocking buffer was added per well. The wells were allowed to incubate for 30 min at 37°C so that all the sites are blocked for any nonspecific adsorption. Different concentrations of antigen solution were prepared by diluting a stock p24 solution with block buffer or plasma sample. To each well, 100 μl of the p24 antigen was added at an incubation temperature of 37°C, with shaking for 1 hour. The wells were again repeatedly washed five times with wash buffer, following which a biotinylated detector antibody (100 μl per well) was added and incubated for 30 min at 37°C. To this sandwiched complex, 100 μl of AuNC-SA (0.5 μg/ml) was added to each well, and the mixture was incubated for 30 min at 37°C with shaking. This was followed by final washing five times with PBS with Tween 20 buffer to reduce the background noise and prevent nonspecific interactions. Last, the sandwiched immunocomplex was excited with the excitation signal, and the emitted fluorescent signals from the sandwich immunocomplexes were read directly from the microplate in the end point format with the SpectraMax microplate reader (excitation at 345 nm and emission at 615 nm) (20). All experiments were performed in triplicate. Fluorescence intensity values were plotted against the concentration of p24 antigen. A sample with an S:Co ratio greater than 2 was considered to be positive for HIV-1 p24.
Control experiment
The effect of interference of proteins was tested by performing a control experiment that involved using BSA as an interfering protein and studying the effects of its presence on GNCIA (27). Five various concentrations of BSA were taken (0.1, 0.5, 1.0, 5.0, and 10.0 μg/ml) for a fixed p24 concentration of 500 pg/ml. The effects on the presence of BSA on the fluorescence signal was observed.
Additional tests were also performed to see the interference effect of coexistent virions. HCV was taken as the interfering moiety, as it is a commonly known coinfection in HIV patients (27). Thus, it was necessary to study the effect of the presence of HCV as an interfering moiety in the immunoassay. To that effect, solutions with 10×, 20×, 50×, and 100× dilutions of the HCV-positive plasma sample spiked with p24 antigen (500 pg/ml) were prepared in addition to the undiluted plasma sample. The immunoassay was conducted on the samples spiked with p24 antigen. The signal intensities were recorded and plotted for further analysis.
HIV detection in blood plasma samples
Preparing plasma samples
Before the preparation of the plasma samples for the immunoassay, they were confirmed to be HIV negative or HIV positive using standard HIV detection kits. The preparation of the test sample was carried out by diluting 188 μl of casein block buffer with 2 μl of plasma being tested. Furthermore, 10 μl of 10% aqueous solution of Triton X-100 was added to the above prepared solution (27).
Validation of GNCIA with clinical samples
Testing the performance of the assay with clinical samples is essential to evaluate its deployment in clinical and real world testing. Validation of GNCIA was performed using 50 HIV-positive and 50 HIV-negative samples (27). Sample preparation was carried out on the basis of the protocol described before. The cross-reactivity was also evaluated with 10 HBV-positive/HIV-negative and 10 HCV-positive/HIV-negative plasma samples.
Supplementary Material
Acknowledgments
We thank Bhagawan Sri Sathya Sai Baba for constant motivation and support. A.D.K. thanks DST for support through the DST-INSPIRE Fellowship program, Ministry of Science and Technology, Government of India. We also thank the Sri Sathya Sai Institute of Higher Medical Sciences for providing the plasma samples for clinical analysis. The findings and conclusions in this report are those of us and do not necessarily represent the views of the U.S. Food and Drug Administration and the U.S. Department of Health and Human Services. Funding: No funding was involved in this work. Ethics statement: We confirm that informed consent was obtained after the nature and possible consequences of the studies were explained. The study was performed in compliance with the guidelines of Institutional Ethics Committee of Sri Sathya Sai Institute of Higher Learning. The study was approved under the study ID SSIHL/IEC/PSN/BS/2012/01. Author contributions: K.V. and I.K.H. conceived and planned the study. A.D.K., M.K.H., and L.A.A.C. designed experiments and analyzed data. A.D.K. synthesized and characterized NPs. A.D.K. and C.S.M. performed computational simulations. A.D.K. and L.A.A.C. performed the assay. A.D.K., K.V., and I.K.H. wrote the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/11/eaar6280/DC1
Fig. S1. HEX docking scores of the best poses for the increasing glutathione-NHS tails on the AuNC core with the streptavidin protein.
Fig. S2. TEM image of AuNCs, which indicates the size of the AuNCs to be around 2 nm or less.
Fig. S3. Absorption and emission spectra of AuNCs.
Fig. S4. Effect of the presence of interfering moieties on the GNCIA stability.
Fig. S5. Optimization of concentration of capture antibodies and AuNC-SA for optimal assay performance.
Table S1. Comparison of HEX energies of AuNC-SA for various capping agents on Au13 cluster.
Table S2. Comparison of binding scores for gold, silver, and copper nanoclusters for increasing cluster sizes, where x denotes the number of atoms.
Table S3. Comparison of FP values of unconjugated AuNCs with conjugated AuNCs.
Table S4. Results of clinical serum analysis that shows the high specificity of GNCIA.
REFERENCES AND NOTES
- 1.Sun H.-T., Sakka Y., Luminescent metal nanoclusters: Controlled synthesis and functional applications. Sci. Technol. Adv. Mater. 15, 014205 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu G., Ma K., Cui Q., Wu F., Xu S., Synthesis of DNA-templated fluorescent gold nanoclusters. Gold Bull. 45, 69–74 (2012). [Google Scholar]
- 3.Goswami N., Yao Q., Luo Z., Li J., Chen T., Xie J., Luminescent metal nanoclusters with aggregation-induced emission. J. Phys. Chem. Lett. 7, 962–975 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Jin R., Zeng C., Zhou M., Chen Y., Atomically precise colloidal metal nanoclusters and nanoparticles: Fundamentals and opportunities. Chem. Rev. 116, 10346–10413 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Zheng J., Nicovich P. R., Dickson R. M., Highly fluorescent noble-metal quantum dots. Annu. Rev. Phys. Chem. 58, 409–431 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chatterjee D. K., Gnanasammandhan M. K., Zhang Y., Small upconverting fluorescent nanoparticles for biomedical applications. Small 6, 2781–2795 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Sun Y., Wu J., Wang C., Zhao Y., Lin Q., Tunable near-infrared fluorescent gold nanoclusters: Temperature sensor and targeted bioimaging. New J. Chem. 41, 5412–5419 (2017). [Google Scholar]
- 8.Tan X., Jin R., Ultrasmall metal nanoclusters for bio-related applications. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 5, 569–581 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Kvítek O., Siegel J., Hnatowicz V., Švorčík V., Noble metal nanostructures influence of structure and environment on their optical properties. J. Nanomater. 2013, 743684 (2013). [Google Scholar]
- 10.Cai H., Wang Y., He P., Fang Y., Electrochemical detection of DNA hybridization based on silver-enhanced gold nanoparticle label. Anal. Chim. Acta 469, 165–172 (2002). [Google Scholar]
- 11.Malon A., Vigassy T., Bakker E., Pretsch E., Potentiometry at trace levels in confined samples: Ion-selective electrodes with subfemtomole detection limits. J. Am. Chem. Soc. 128, 8154–8155 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang X., Li D., Xu X., Ying Y., Li Y., Ye Z., Wang J., Immunosensors for detection of pesticide residues. Biosens. Bioelectron. 23, 1577–1587 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Bakalova R., Zhelev Z., Ohba H., Baba Y., Quantum dot-based western blot technology for ultrasensitive detection of tracer proteins. J. Am. Chem. Soc. 127, 9328–9329 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Ornberg R. L., Harper T. F., Liu H., Western blot analysis with quantum dot fluorescence technology: A sensitive and quantitative method for multiplexed proteomics. Nat. Methods 2, 79–81 (2005). [Google Scholar]
- 15.Yuan X., Luo Z., Yu Y., Yao Q., Xie J., Luminescent noble metal nanoclusters as an emerging optical probe for sensor development. Chem. Asian J. 8, 858–871 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Dundas C. M., Demonte D., Park S., Streptavidin–biotin technology: Improvements and innovations in chemical and biological applications. Appl. Microbiol. Biotechnol. 97, 9343–9353 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Chivers C. E., Koner A. L., Lowe E. D., Howarth M., How the biotin–streptavidin interaction was made even stronger: Investigation via crystallography and a chimaeric tetramer. Biochem. J. 435, 55–63 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Avidin-Biotin Interaction (2017); https://www.thermofisher.com/in/en/home/life-science/protein-biology/protein-biology-learning-center/protein-biology-resource-library/pierce-protein-methods/avidin-biotin-interaction.html.
- 19.Jameson D. M., Ross J. A., Fluorescence polarization/anisotropy in diagnostics and imaging. Chem. Rev. 110, 2685–2708 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chunduri L. A. A., Haleyurgirisetty M. K., Patnaik S., Bulagonda P. E., Kurdekar A., Liu J., Hewlett I. K., Kamisetti V., Development of carbon dot based microplate and microfluidic chip immunoassay for rapid and sensitive detection of HIV-1 p24 antigen. Microfluid. Nanofluidics 20, 167 (2016). [Google Scholar]
- 21.Lea W. A., Simeonov A., Fluorescence polarization assays in small molecule screening. Expert Opin. Drug Discov. 6, 17–32 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fluorescence Polarization (2010); https://www.thermofisher.com/in/en/home/references/molecular-probes-the-handbook/technical-notes-and-product-highlights/fluorescence-polarization-fp.html.
- 23.Bruno A., de Lisio C., Minutolo P., Time resolved fluorescence polarization anisotropy of carbonaceous particles produced in combustion systems. Opt. Express 13, 5393–5408 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Chunduri L. A. A., Kurdekar A., Haleyurgirisetty M. K., Bulagonda E. P., Kamisetti V., Hewlett I. K., Femtogram level sensitivity achieved by surface engineered silica nanoparticles in the early detection of HIV infection. Sci. Rep. 7, 7149 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saltzman C. L., Herzenberg J. E., Phillips W. A., What nononcologists need to know about MEN IIb. Oncology (Williston Park). 2, 15 (1988). [PubMed] [Google Scholar]
- 26.Shrivastava A., Gupta V., Methods for the determination of limit of detection and limit of quantitation of the analytical methods. Chron. Young Sci. 2, 21–25 (2011). [Google Scholar]
- 27.Kurdekar A. D., Chunduri L. A. A., Chelli S. M., Haleyurgirisetty M. K., Bulagonda E. P., Zheng J., Hewlett I. K., Kamisetti V., Fluorescent silver nanoparticle based highly sensitive immunoassay for early detection of HIV infection. RSC Adv. 7, 19863–19877 (2017). [Google Scholar]
- 28.Liu J., Du B., Zhang P., Haleyurgirisetty M., Zhao J., Ragupathy V., Lee S., DeVoe D. L., Hewlett I. K., Development of a microchip Europium nanoparticle immunoassay for sensitive point-of-care HIV detection. Biosens. Bioelectron. 61, 177–183 (2014). [DOI] [PubMed] [Google Scholar]
- 29.H. Ju, X. Zhang, J. Wang, Signal amplification for nanobiosensing, in NanoBiosensing (Springer, 2011), pp. 39–84. [Google Scholar]
- 30.Mulpur P., Podila R., Lingam K., Krishna Vemula S., Sathish Ramamurthy S., Kamisetti V., Rao A. M., Amplification of surface plasmon coupled emission from graphene–Ag hybrid films. J. Phys. Chem. C 117, 17205–17210 (2013). [Google Scholar]
- 31.Kurdekar A., Chunduri L. A. A., Bulagonda E. P., Haleyurgirisetty M. K., Kamisetti V., Hewlett I. K.. Comparative performance evaluation of carbon dot-based paper immunoassay on Whatman filter paper and nitrocellulose paper in the detection of HIV infection. Microfluid. Nanofluidics 20, 99 (2016). [Google Scholar]
- 32.M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, G. A. Petersson, H. Nakatsuji, X. Li, M. Caricato, A. Marenich, J. Bloino, B. G. Janesko, R. Gomperts, B. Mennucci, H. P. Hratchian, J. V. Ortiz, A. F. Izmaylov, J. L. Sonnenberg, D. Williams-Young, F. Ding, F. Lipparini, F. Egidi, J. Goings, B. Peng, A. Petrone, T. Henderson, D. Ranasinghe, V. G. Zakrzewski, J. Gao, N. Rega, G. Zheng, W. Liang, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, K. Throssell, J. A. Montgomery Jr., J. E. Peralta, F. Ogliaro, M. Bearpark, J. J. Heyd, E. Brothers, K. N. Kudin, V. N. Staroverov, T. Keith, R. Kobayashi, J. Normand, K. Raghavachari, A. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, J. M. Millam, M. Klene, C. Adamo, R. Cammi, J. W. Ochterski, R. L. Martin, K. Morokuma, O. Farkas, J. B. Foresman, D. J. Fox, Gaussian 09, Revision A.02 (2009); http://gaussian.com/.
- 33.Gieseking R. L., Ratner M. A., Schatz G. C.. Semiempirical modeling of Ag nanoclusters: New parameters for optical property studies enable determination of double excitation contributions to plasmonic excitation. J. Phys. Chem. A 120, 4542–4549 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Molecualr Operating Environment, (2016); www.chemcomp.com/MOE-Molecular_Operating_Environment.htm.
- 35.Hex Protein Docking, (2013); http://hex.loria.fr/.
- 36.G. T. Hermanson, Functional targets for bioconjugation, in Bioconjugate Techniques (Academic Press, ed. 3, 2013), chap. 2, pp. 127–228. [Google Scholar]
- 37.Avinash Chunduri L. A., Kurdekar A., Pradeep B. E., Haleyurgirisetty M. K., Venkataramaniah K., Hewlett I. K., Streptavidin conjugated ZnO nanoparticles for early detection of HIV infection. Adv. Mater. Lett. 8, 472–480 (2017). [Google Scholar]
- 38.Tang S., Hewlett I., Nanoparticle-based immunoassays for sensitive and early detection of HIV-1 capsid (p24) antigen. J. Infect. Dis. 201 (suppl. 1), S59–S64 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.He Q., Zhu Z., Jin L., Peng L., Guo W., Hu S., Detection of HIV-1 p24 antigen using streptavidin–biotin and gold nanoparticles based immunoassay by inductively coupled plasma mass spectrometry. J. Anal. At. Spectrom 29, 1477–1482 (2014). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/11/eaar6280/DC1
Fig. S1. HEX docking scores of the best poses for the increasing glutathione-NHS tails on the AuNC core with the streptavidin protein.
Fig. S2. TEM image of AuNCs, which indicates the size of the AuNCs to be around 2 nm or less.
Fig. S3. Absorption and emission spectra of AuNCs.
Fig. S4. Effect of the presence of interfering moieties on the GNCIA stability.
Fig. S5. Optimization of concentration of capture antibodies and AuNC-SA for optimal assay performance.
Table S1. Comparison of HEX energies of AuNC-SA for various capping agents on Au13 cluster.
Table S2. Comparison of binding scores for gold, silver, and copper nanoclusters for increasing cluster sizes, where x denotes the number of atoms.
Table S3. Comparison of FP values of unconjugated AuNCs with conjugated AuNCs.
Table S4. Results of clinical serum analysis that shows the high specificity of GNCIA.


