Abstract
In each somatic cell of a female mammal one X chromosome is transcriptionally silenced via X-chromosome inactivation (XCI), initiating early in development. Although XCI events are conserved in mouse and human post-implantation development, regulation of X-chromosome dosage in pre-implantation development occurs differently. In pre-implantation development, mouse embryos undergo imprinted form of XCI, yet humans lack imprinted XCI and instead regulate gene expression of both X-chromosomes by dampening transcription. The long non-coding RNA Xist/XIST is expressed in mouse and human pre- and post-implantation development to orchestrate XCI, but its role in dampening is unclear. In this review, we discuss recent advances in our understanding of the role of Xist in X-chromosome dosage compensation in mouse and human.
Keywords: Xist, X-inactivation, embryonic stem cells, lncRNAs, dosage compensation
X chromosome dosage compensation
Male (XY) and female (XX) eutherian (see Glossary) mammals have equivalent expression levels of most X- chromosome genes despite the presence of an extra X chromosome in females. This X- chromosome dosage compensation is due to the phenomenon of X-chromosome inactivation (XCI), which refers to the transcriptional silencing and heterochromatinization of one of the two X chromosomes in females early in embryonic development [1]. Most of our knowledge of XCI is based on mouse studies, where two types of XCI exist: imprinted and random (Figure 1A).
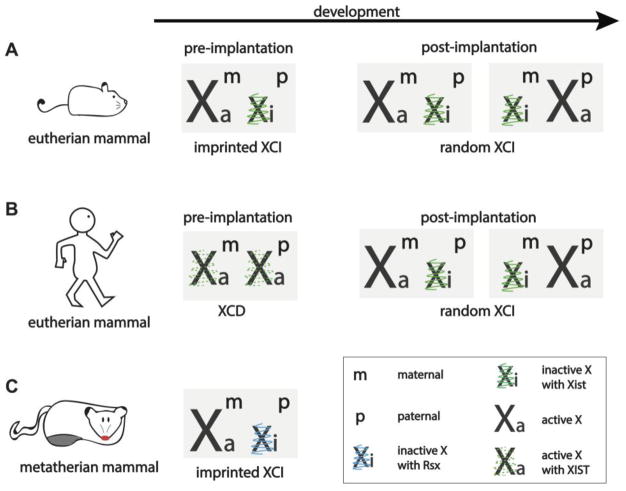
Figure 1. X chromosome inactivation in different mammals.
The X-chromosome states of eutherian (A and B) and metatherian (C) female mammals are shown in embryonic development. A) Imprinted XCI occurs in mouse pre-implantation development, but it is re-set in the cells that develop into the embryo to give way to random XCI, resulting in a mosaic adult female mouse. Both imprinted and random XCI in the mouse are regulated by the lncRNA Xist. B) Humans have evolved away from imprinted XCI as they dosage compensate in pre-implantation development by turning down transcription from both X chromosomes via XCD. Moreover, XIST is expressed on both dampened X chromosomes, where its functional role remains to be determined. In post-implantation development, similar to the mouse, human females display random XCI mediated by XIST. C) Metatherians, such as the marsupial opossum (Monodelphis domestica) dosage-compensate by inactivating the paternally-inherited X chromosome using the lncRNA Rsx. This imprinted dosage-compensation is maintained throughout marsupial development, resulting in a female adult with a transcriptionally inactive paternal X chromosome in all of its cells. XCI = X chromosome inactivation, Xist/XIST = X inactive specific transcript, Xa = active X chromosome, Xi = inactive X chromosome, Xm = maternal X chromosome, Xp = paternal X chromosome, XCD = X chromosome dampening, Rsx = RNA on the silent X.
In imprinted XCI, which initiates in all cells of the female mouse four- to eight-cell stage pre-implantation embryo, the paternally inherited X chromosome (Xp) undergoes inactivation, while the maternally inherited X chromosome (Xm) remains active [2,3]. As pre-implantation development progresses to form the blastocyst, cells of the trophectoderm layer, which give rise to extra-embryonic tissues (e.g. the placenta), maintain their imprinted XCI state. In contrast, epiblast cells of the blastocyst, which give rise to the embryo proper, reactivate the inactive Xp, re-establishing a state with two active X-chromosomes. The biallelic X-linked gene expression of epiblast cells is resolved again via XCI, but in this second wave of XCI either the Xp or the Xm is chosen at random for inactivation. Random XCI is maintained in all descendent somatic cells throughout life, resulting in adult mice that are a mosaic of cells expressing either maternal or paternal alleles of X-linked genes [4,5]. A group of X-linked genes express both the maternal and the paternal allele in each cell since these genes escape XCI and are thus the exception to the rule (reviewed by [6]). The chromosome-wide inactivation of the X chromosome, both in imprinted and random XCI, appears to always be governed by the lncRNA X inactive specific transcript (Xist), which is encoded in the X-inactivation center (XIC) of the X chromosome [7].
The occurrence of both imprinted and random XCI in the same species, as is the case in mouse, may not be very common. Most mammals studied utilize only one form of XCI for X-chromosome dosage compensation. In marsupials, only imprinted XCI is observed where the Xp is exclusively chosen for inactivation [8] (Figure 1C). Contrary to this, imprinted XCI does not occur in rabbit, pig, [9,10] horse, or human development [11,12,13] based on analysis of pre-implantation blastocysts [9,10,13] or placental tissues [11,12]. In human post-implantation development, both extra-embryonic and embryonic lineages dosage-compensate via random XCI [12,14] (Figure 1B). However, in the first week of human development, prior to implantation and XCI, the existence of a novel gene-dosage regulation has recently been uncovered [13]. Here, both X chromosomes remain active from the onset of zygotic gene activation until the blastocyst stage [9,13] (Figure 1B). However, transcription from both X chromosomes is tuned down, or dampened, resulting in a net reduction of X-linked gene expression in female blastocyst cells [13]. X-chromosome dampening (XCD) has not been observed in any other mammal yet, but it has been reported in the nematode Caenorhabditis elegans [15], although the underlying mechanism in human and nematode may differ. In the XX hermaphrodite C. elegans, the 3D conformation of the X chromosomes is remodeled to reduce chromosome-wide gene expression by half in order to achieve gene-expression balance between XX hermaphrodites and XO males [16]. 3D chromosome conformation also differs between the active and inactive X chromosomes in mammals [17], suggesting that chromosome confirmation remodeling might also be at play in human XCD. However, unlike the mammalian inactive X-chromosome (Xi), lncRNAs have not been reported to regulate X-chromosome dose in the C. elegans. Instead, dampening in the nematode is carried out by the dosage compensation complex, a condensin-containing multi-subunit protein assembly that binds at multiple sites along the X chromosome and leads to chromosome-wide compaction and gene repression [18].
Whether XCD observed in human pre-implantation development and C. elegans are related at the molecular level needs further investigation. Moreover, when single cell RNA-sequencing data (Box 1) of pre-implantation human blastocysts are analyzed using different bioinformatics tools and approaches, the dosage compensation observed in human pre-implantation embryos has been interpreted as initiation of XCI rather than dampening of both X chromosomes [19,20]. Fortunately, naïve human embryonic stem cells (hESCs), which are the in vitro counterparts of the pluripotent cells in the human pre-implantation embryo, exhibit XCD and thus can be used as a model system to address XCD and its relationship to the initiation of XCI further [21].
Box 1. Methods.
Sox2 promoter-driven Cre recombinase: the Cre-recombinase enzyme, which recombines DNA at specific DNA sequences, coined loxP sites, is expressed only in cells where the Sox2 gene is actively transcribed. This system is engineered by cloning the Sox2 promoter and enhancer region upstream of the Cre recombinase gene. When the same cell contains DNA sequences flanked with loxP sites of the same orientation, the intervening sequence will be removed – or deleted.
Tetraploid complementation assay is a method for creating mice where all the cells of the embryo proper are derived from mouse pluripotent stem cells (PSCs) upon blastocyst injection of these cells. To accomplish this, cells of two-cell embryos are fused experimentally to form a tetraploid pre-implantation embryo. While the tetraploid cells of the blastocyst can develop into the extraembryonic tissues required for in utero development, they cannot contribute to the embryo proper. Hence, when such tetraploid blastocysts are injected with diploid mouse PSCs, all the tissues of the embryo proper come from these injected diploid cells, creating a non-chimeric mouse.
Single cell RNA-sequencing is a method of measuring global gene expression from individual cells to determine gene expression patterns unique to individual cells that would be otherwise be lost in bulk RNA-sequencing (from a group of cells) due to averaging of the data.
In this review, we discuss the experimental evidence examining the role of Xist in X-chromosome dosage compensation via imprinted and random XCI in mouse. We also consider XIST function in early human development and in human pluripotent stem cells (PSCs), reflecting on potential molecular mechanisms which might regulate context-dependent XIST function.
Long non-coding RNAs are key players in X-chromosome regulation
An intriguing fact about X-chromosome dosage regulation in all mammals is the utilization of long non-coding RNAs (lncRNAs) such as Jpx, Ftx, Tsix, and XACT, most of which are located in the XIC of the X chromosome [20,22,23–28] (Figure 2). When expressed, Xist is exclusively associated with the X-chromosome from which it is expressed, acting only in cis [29]. The lncRNAs Jpx and Ftx exert their function by acting as activators of Xist to fine-tune Xist expression and thus indirectly regulate XCI. The Jpx lncRNA product acts either in cis or in trans [23] and binds the Xist repressor CTCF, taking away repression of Xist transcription [22]. Contrary to this, the Ftx transcript itself is not required for Xist regulation: it is the act of transcription of the Ftx locus that leads to Xist expression in cis [25]. Moreover, in mice, the lncRNA Tsix, which is transcribed in anti-sense orientation to Xist, represses Xist expression, thus ensuring Xist induction on the Xi and protecting the Xa (active X chromosome) from ectopic silencing by Xist [26,27]. Tsix is not expressed in human pre-implantation development [13], and the roles of Jpx and Ftx are yet to be examined in humans. A recently discovered lncRNA called XACT (X active coating transcript) that is unique to human PSCs, but does not reside in the XIC, may aid in maintaining transcriptional activity of the X chromosome from which it is expressed by counteracting XIST [20,28]. Interestingly, correlative studies suggest that XIST RNA, in addition to XCI, might also mediate the dampening of the transcriptional output of both X chromosomes of female human pre-implantation blastocysts [13,21] (discussed below). While Xist is unique to placental mammals, marsupials also use a cis-acting lncRNA encoded on the X chromosome, termed Rsx (RNA-on-the-silent X), which in many ways appears to act like Xist in XCI [8] (Figure 1). Taken together, it is rather interesting that different lncRNAs have evolved to regulate gene expression chromosome-wide in cis. This is perhaps due to the unique ability of lncRNAs to bind distant sites on chromatin while still tethered to their transcription loci. Xist [30] and other lncRNAs such as HOTTIP (HOXA transcript at the distal tip), which is encoded on mouse chromosome 6 and activates genes in its neighborhood [31], reach their target chromatin sites simply by proximity – by being close to these sites in 3D space due to the folding of chromatin within the nucleus (reviewed and illustrated in [32]). Understanding how X-chromosome dosage is regulated via lncRNAs will thus not only shed light onto X-chromosome biology but can serve as a starting point in understanding how lncRNAs localize to and act on chromatin in general.
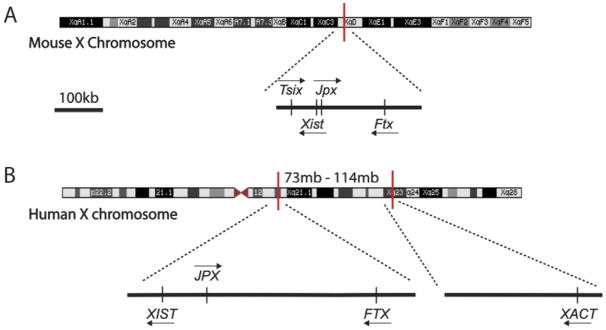
Figure 2. Long non-coding RNAs involved in X chromosome dosage regulation in mouse and human.
The X inactivation center (XIC) is located on the X chromosome and harbors the master regulator of XCI – the long non-coding RNA (lncRNA) Xist. A) In the mouse, Xist itself is positively regulated by the lncRNAs Jpx and Ftx, which are also encoded in the XIC, upstream of the Xist gene. The lncRNA Tsix, which is anti-sense to Xist, has a mutually exclusive expression pattern with Xist: it is expressed bi-allelically from both X chromosomes prior to XCI. Tsix ‘protects’ the active X chromosome in pluripotency from being silenced by Xist upon induction of XCI, and is thought to be a repressor of Xist.
B) The human XIST gene is also encoded in the XIC of the human X chromosome. The lncRNAs JPX and FTX are also upstream of XIST in the human XIC, similar to mouse. The role of JPX and FTX in regulating XIST expression in human is speculated based on mouse studies. Unlike mouse, the human XIC does not contain the XIST anti-sense lncRNA TSIX, since TSIX expression is not detected in pre-implantation blastocysts or human embryonic stem cells. A novel, human-specific lncRNA, X active coating transcript (XACT), however, is encoded about 40Mb upstream of the human XIC and seems to antagonize XIST in naïve pluripotency. Similar to mouse Tsix, expression of XACT is unique to pluripotent cells and not detected in somatic cells.
Xist is required for XCI in mouse
Xist is the best studied lncRNA to date. As the name suggests, XIST was discovered due to its association with the inactive X-chromosome – X inactive specific transcript [33–37]. The requirement of Xist for XCI was first directly implicated using female mouse embryonic stem cells (mESCs) with a mutated Xist gene lacking the first two thirds of exon 1 [38]. When induced to differentiate, both in vitro and in vivo (using aggregation chimeras), the X chromosome bearing the mutant truncated Xist gene was always spared from inactivation while Xist was expressed from the wild type X chromosome, causing non-random silencing [38]. This study demonstrated that Xist is required for choosing the chromosome for inactivation, and suggested, without direct evidence, that cis-expression of Xist is required for XCI.
Since mESCs are derived from the epiblast cells of the blastocyst, the role of Xist in pre-implantation development, from zygote to blastocyst formation, cannot be studied using these cells. Therefore, to further investigate the role of Xist in early development, mESCs were used to generate chimeric mice containing cells with a large deletion of Xist, which were then mated with wild type mice to generate hemizygote males or heterozygote females [39]. When the mutant Xist was inherited from the mother, both normal female and male pups were born. However, female embryos with a paternally-inherited mutant Xist had severe prenatal growth defects and survived until approximately embryonic day 10.5 (E10.5). In these embryos the extraembryonic tissues failed to develop due to the lack of imprinted XCI of the paternally inherited X chromosome in these tissues [39] (Figure 3). Due to the pre-determined choice of the paternal X chromosome for silencing in imprinted XCI, this experiment is the first clear demonstration that XCI, specifically imprinted XCI, cannot be initiated without Xist. Note that there is no paternal inheritance of mutant Xist to be studied in males since males inherit a Y chromosome but no X chromosome from their father. The inability of the extraembryonic tissue to support the growth of the embryo in the absence of X-chromosome dosage compensation was investigated further and this failure was attributed to exhaustion of the extra-embryonic ectoderm due to premature cell differentiation [40].
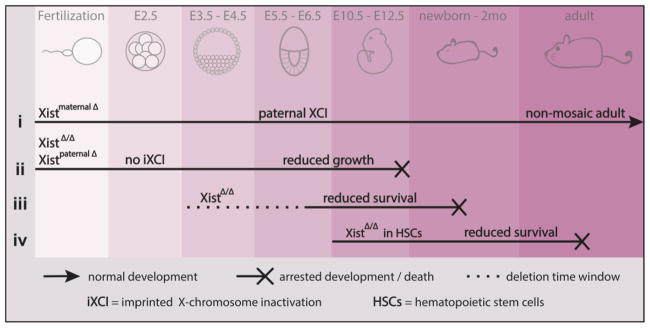
Figure 3. Lack of Xist at various developmental time-points highlights its importance in normal development.
Summary of key studies addressing the role of Xist in mouse development from fertilization to birth and into adulthood. (i) When a zygote is formed with a maternally deleted Xist (inherited from the egg), mouse development progresses normally and results in non-mosaic adults where all cells inactivate the paternally inherited X chromosome (since only that X has the only functional Xist allele). (ii) However, when Xist is deleted from both X chromosomes (from the egg and the sperm), or from only the paternal X chromosome (sperm), extra-embryonic tissues fail to develop in the absence of imprinted XCI since this process requires paternally-inherited Xist, and thus mouse development halts 5–7 days post implantation. (iii) Conditional Xist deletion from both X chromosomes in epiblast cells that give rise to the embryo is often embryonically lethal, and if pups are born, they display partial loss of X-chromosome silencing and do not survive to adulthood. (iv) When Xist is deleted several days post-implantation, specifically in hematopoietic stem cells (HSCs), after the establishment of the Xi, pups are born but succumb to Multilineage Dysplasia as early as 1.5 months after birth. E = embryonic day, Xist = X inactive specific transcript, iXCI = imprinted X-chromosome inactivation, HSCs = hematopoietic stem cells.
A recent study used single cell RNA-sequencing to provide high temporal and chromosome-wide resolution of X-linked gene silencing in mouse pre-implantation development and its dependence on Xist RNA [3]. Comparisons of female wild type to mutant embryos carrying a paternal Xist deletion at the 8-, 16-, 32-cell, and blastocyst stages validated the need for Xist in initiating imprinted XCI [3], which was also demonstrated recently using RNA-sequencing of single embryos [2]. Such high-resolution data were important to clarify the role of Xist in imprinted XCI and to rule out prior arguments for an Xist-independent imprinted XCI [41]. The dynamics of silencing during the initiation of imprinted XCI revealed that genes are silenced with different kinetics. Genes silenced at an earlier stage of pre-implantation development were those near the XIC (where the Xist gene is located) in 3D space [3,30]. Intriguingly, this recapitulates the finding that at the initiation of random XCI, Xist first contacts the sites of the chromosome closest to its site of transcription in 3D, rather than linear space [30]. This correlation between the kinetics of imprinted XCI and the proximity to the Xist locus in 3D space independently supports the notion that imprinted XCI is mediated by Xist, and additionally suggests that the mechanism of Xist spreading in cis during the initiation of imprinted and random XCI is conserved.
Requirement of XCI in the development of the mouse embryo proper
Although the above-mentioned paternal Xist deletion experiment demonstrated that Xist is required for imprinted XCI, the early embryonic lethality due to malfunctioning extraembryonic tissues have made addressing the requirement of Xist in the embryo itself unfeasible with a germline mutation of Xist. A homozygous Xist knockout is required to address this question, which, when using a germline mutation, affects imprinted XCI as well (Figure 3). However, the maternal germline deletion of Xist allele was valuable to demonstrate the requirement of a functional Xist allele for choosing the X chromosome for XCI in vivo [42], extending the prior mESC study [38]. In the meantime, experiments demonstrating the sufficiency of Xist RNA for silencing was demonstrated. Particularly, ectopic expression of Xist cDNA from autosomes or the X chromosome [43,44], or activation of the endogenous Xist allele from the single X chromosome in male mESCs with an inducible promoter [30] demonstrated that Xist expression is sufficient to cause silencing in cis. These gain-of-function experiments also opened the way for dissection of Xist RNA for its functional units [43].
Studying the need for Xist in random XCI requires a unique approach that specifically deletes Xist in embryonic tissues. A recent study took on the challenge to assess the importance of X-chromosome dosage compensation by random XCI in mouse embryonic development. To silence Xist specifically in the embryo while sparing the extra-embryonic tissue, Xist was conditionally deleted in the epiblast lineage [45]. While most mutant mice died in utero, surprisingly some mice survived to term, but exhibited growth retardation with reduced body size, dying within one month after birth (Figure 3). Only one mouse, which was a mosaic of XX and XO cells – cells that had lost one of their two X chromosomes - survived to adulthood. These data indicate that Xist and random XCI are required for normal embryonic development. However, given the survival to term, the phenotype appeared much weaker than expected and was a surprise – since XCI occurs soon after implantation the expectation was that Xist loss should have led to early embryonic lethality. This can perhaps be explained by another unexpected observation, namely that X-chromosome dosage compensation was not completely wiped out upon deletion of Xist. The authors reported partial X-chromosome dosage compensation in the absence of Xist, concluding that an Xist-independent mechanism was responsible [45].
This phenotype of partial X-chromosome dosage compensation in Xist mutant mice was observed due to less-than-expected increase in average expression levels of all genes by RNA-sequencing when compared to wild-type female mice, and due to the presence of mono-allelic gene expression in some, but not all cells at the single cell level of a few X-linked genes, assessed by fluorescent in situ hybridization [45]. However, understanding the Xist mutant mouse model system used [45] can perhaps better explain these above-mentioned observations. Deletion of Xist in the epiblast lineage was accomplished with the Sox2 promoter-driven Cre recombinase [45] (Box 1). However, the efficiency of Xist excision by the Sox2-driven Cre was not measured in the developmental interval around the induction of random XCI [46], as embryos were only harvested at E8.5 to confirm Xist deletion, a time point at which XCI has already occurred [45]. Originally, Sox2-driven Cre recombinase activity has been shown to occur in blastocyst outgrowths in culture and in all cells of the epiblast in E6.5 embryos in vivo, by assessing the removal of a ‘stop’ cassette in front of a beta-galactosidase reporter gene integrated into the ROSA26 locus [47,49]. Together, these findings suggest that the ROSA26 reporter recombined before E6.5 and possibly before XCI would be initiated in vivo, indicating that the Sox2-driven Cre-recombinase may be ideally suited to delete Xist before induction of random XCI. However, since chromatin accessibility may be different for the Xist locus in comparison to ROSA26, a region on mouse chromosome 6 identified because of its high recombination frequency [49], the Cre-mediated excision kinetics may differ for the two loci and thus need to be independently determined for the Xist locus. Additionally, the DNA segment flanked by LoxP sites, which need to come together for Cre-mediated recombination, is significantly longer in the Xist deletion construct [45] compared to the ROSA26 beta-galactosidase reporter system [48], and therefore potentially less favorable for deletion, again arguing for the need to establish in vivo Xist deletion kinetics in this mouse model. Hence, it cannot be ruled out that Xist deletion may have occurred after initiation/completion of random XCI.
Previous studies have shown that deletion of Xist has no dramatic short-term effect on the silent state of genes on the X chromosome when it occurs after the inactive X chromosome is fully established [50,51] (see below). Depending on the proportion of cells undergoing random XCI prior to Xist deletion, the embryos would then survive to term and demonstrate incomplete dosage compensation at organism level due to the mixture of XaXa and XaXi cells (Xa is the active X chromosome, Xi is the inactive X chromosome). Xist deletion post XCI establishment would also explain the low number of pups surviving to term, since if not enough cells per embryo undergo X-chromosome dosage compensation prior to Xist deletion, the embryo would not be viable, arguing for the importance of Xist in XCI and embryonic development. Taken together, additional experiments with a temporally precise Xist deletion (and confirmation of Xist deletion before induction of random XCI) are required to dissect whether a novel embryonic X-chromosome dosage compensation mechanism in the absence of Xist or the delayed deletion of Xist relative to the onset of XCI explain the surprisingly weak consequences of the current Xist deletion in the embryo [45]. For instance, using homozygous Xist knockout mESCs in tetraploid complementation assays [52] (Box 1) could provide wild-type extraembryonic tissues capable of supporting normal development while all cells of the epiblast, derived from mESCs, would lack Xist, ruling out the possibility of random XCI occurring in any fraction of the cells. It would be interesting to see if/when development would fail in this scenario, but our hypothesis is that no viable pups would be obtained from such mice. Regardless, the current data argue that X-chromosome dosage compensation mediated by Xist is critical for embryonic development.
The influence of XCI on the developmental potential of female cells has been shown with mESCs, as the double dose of X-linked genes delays the differentiation of these cells due to its stabilizing effect on the naïve pluripotent state [55]. This stabilization is achieved via inhibition of the MAPK and Gsk3 pathways and stimulation of the Akt pathway, and XCI is needed to properly exit naïve pluripotency [53]. The delayed exit from pluripotency in the presence of two active X chromosomes may also occur in vivo [53], since embryos with a single X chromosome undergo accelerated development [54].
Xist executes XCI by recruiting a diverse set of proteins
During initiation of XCI, Xist recruits numerous silencing factors to the X chromosome to establish facultative heterochromatin, also known as the Barr body [55]. This is accompanied by epigenetic changes including substitution of certain core histones, covalent modifications of histone tails, and promoter CpG methylation (reviewed by [1]). Although some of these Xist-induced epigenetic remodeling steps were discovered years ago, most of the proteins binding Xist directly and indirectly were identified only recently using mass spectrometry-based approaches and genetic screens [56–60]. While some of the Xist-binding proteins influence histone modifications (via the activation of the histone deacetylase HDAC3 through the engagement of SPEN by the 5′ end of Xist) [57] or nuclear positioning of the Xist-coated Xi (through the binding of lamin B receptor (LBR) to Xist) [61], others induce RNA modification of adenosine methylation (m6A) on Xist to influence its silencing ability [62]. Identification of the Xist interactome – i.e. the proteins that directly or indirectly bind to Xist - has created a newfound appreciation of the multiple roles Xist plays in XCI, spanning from orchestrating chromosome-wide silencing, localizing itself to chromatin, altering chromatin state, and remodeling the 3D chromosome architecture, recently reviewed in [55]. While the primary sequence of Xist RNA is not well conserved between mouse and human, the gene structure (exons/introns) as well as the presence of key repeat regions [37] are conserved in the two species. Some of these repeat regions are important for Xist function since they are the sites where proteins that directly interact with Xist bind to [56,57,63]. Therefore, although the mass-spectrometry based unbiased approaches have identified the mouse Xist interactome [56–58], it is safe to predict that the humanXIST interactome will largely overlap with that of mouse [64].
Xist is required for long-term maintenance of random XCI
The Xi with Xist expression remains inactive in all somatic progeny of cells. In short-term in vitro studies (days – weeks), Xist does not seem to play a major role in the maintenance of the silent status of genes in random XCI in somatic cells [44,50]. This appears to also be the case in maintenance of imprinted XCI in the vole Microtus Levis, where ablation of Xist expression via deletion of its promoter region in trophoblast stem cells, which have already undergone imprinted XCI, does not lead to transcriptional reactivation or loss of repressive chromatin marks of the Xi [65]. However, a longer-term in vivo mouse study suggests that the prolonged absence of Xist in mice, initiated in the blood lineage using a tissue-specific Cre-recombinase, induces at least partial reactivation of genes on the X chromosome [51]. Notably, the experimentally induced deletion of Xist in hematopoietic cells in mice results in poor postnatal survival and development of myelodysplasia and various cancers of the blood with 100% penetrance [51] (Figure 3). The inevitable development of cancer in the absence of Xist clearly labels Xist as a potent tumor-suppressor, most likely due to its requirement in the maintenance of gene silencing in somatic cells. In agreement with this in vivo mouse study, abnormal reactivation of the Xi has also been reported in human breast cancer cells, although here an extra dose of X-linked genes is either due to Xi erosion or loss of an Xi combined with an Xa duplication [66]. However, the connection between XIST-dependent maintenance of XCI and cancer formation in humans needs to be further explored. Since the importance of Xist in maintaining XCI only became obvious from mouse in vivo studies, it is critical to address the role of XIST in human cancers with carefully designed experiments.
There are two instances in mouse development that require reactivation of the Xi: once in cells of the inner cell mass (ICM) of the blastocyst, when imprinted XCI needs to be reversed prior to induction of random XCI, and once more in the development of primordial germ cells, prior to meiosis [67]. In both cases, shutdown of Xist expression from the existing Xi precedes the removal of chromosome-wide transcriptional repression [68,69]. Xi reactivation is also observed in vitro, when female mouse somatic cells such as mouse embryonic fibroblasts are reprogrammed to form induced pluripotent stem cells (iPSCs). Whereas the necessity of Xist loss in Xi reactivation is difficult to address in the in vivo scenarios described above, the in vitro reprogramming system has allowed detailed studies of this relationship. Using ectopic maintenance of expression or the deletion of Xist in a reprogramming experiment, it was demonstrated that Xist loss is necessary, but not sufficient, for Xi reactivation in iPSC generation, reviewed in more detail in [67]. During reprogramming to iPSCs, Xi reactivation is one of the last steps of reprogramming, requiring DNA demethylation in addition to Xist RNA loss [70]. Similarly, in somatic cells, DNA demethylating agents, such as 5-aza-dC, induce reactivation of genes of the somatic Xi, albeit in a small proportion of cells, via induction of global DNA demethylation [71,72]. DNA methylation works in synergy with Xist RNA and histone hypoacetylation [71], as well as the H3K9 trimethylation pathway [72,73] in maintaining the inactive state of the somatic Xi. In fact, it takes the synergism of triple-drug combinations targeting DNA methylation, topoisomerase activity (involved in relieving torsional stress during DNA replication and transcription) combined with knockdown of an Xist-interacting protein to obtain dramatic re-activation of the Xi, and even then the re-activation is not for all silenced genes [58]. Complete chromosome-wide reactivation of all silenced X-linked genes in somatic cells has not been reported thus far, highlighting the unbreachable nature of the multiple epigenetic layers protecting the Xi.
Xi reactivation in human pluripotent stem cells
Studies of the relationship between XIST and Xi reactivation in human cells are not as straightforward as in mouse, mainly because 1) there is no imprinted XCI to be reversed in human pre-implantation development [9], 2) reactivation of the Xi in human primordial germ cells is difficult to study due to the hurdles associated with obtaining appropriate tissue samples and the lack of a human germ cell culture system that recapitulates Xi reactivation, and 3) reprogramming of human somatic cells under standard conditions does not lead to Xi reactivation [74] as it results in iPSCs that are in a developmentally advanced – primed – pluripotent state [75]. However, when conventional human iPSCs and ESCs in the primed pluripotent state are expanded in culture, XIST expression becomes gradually lost, which is accompanied by methylation of the XIST promoter [74,76–78]. The XIST loss in these pluripotent cells is usually accompanied by partial reactivation of the Xi, where transcriptional repression of some, but not all genes on the Xi goes away, hence the overall inactive state of the Xi erodes, a phenomenon coined Xi erosion [77–80] (Figure 4). Importantly, erosion differs from escape of XCI as the genes undergoing erosion are initially silent on the Xi in early passage hPSCs and become reactivated upon extended passaging of these cells [80], whereas escape is not passage-dependent and rather includes genes on the Xi in chromosome regions with reduced Xist occupancy [30,81]. The extent of erosion of the Xi, i.e. the number of genes affected by this process, varies between individual human pluripotent stem cell (hPSC) lines; however, XIST loss occurs in nearly all hPSC lines studied over time in culture and often leads to Xi erosion [78,80] (Figure 4). Currently it remains to be tested whether loss of XIST expression causes Xi erosion, but the fact that no Xi erosion is observed while XIST is expressed in newly derived human iPSC lines [74] suggests such a causative relationship. While XIST may have a protective role in preventing Xi erosion, another X-linked lncRNA, XACT (X active coating transcript), has been implicated in driving Xi erosion in primed hPSCs [79]. While the eroded Xi does not interfere with hPSC growth or ability to differentiate, it does modulate these processes [80,82]. Moreover, when primed hPSCs with Xi erosion are differentiated, the reactivated genes on the Xi do not get re-silenced, resulting in somatic cells that at least partially lack dosage compensation of X-linked genes [80] (see [83] and [84] for detailed review) (Figure 4). Methods of repairing or preventing Xi erosion of female hPSCs are needed for their use in disease modeling [77] and regenerative medicine, particularly when considering X-linked diseases. For instance, iPSCs or iPSC-derived neurons from female patients with Lesch-Nyhan syndrome, a devastating disease affecting neurologic, cognitive, and behavioral functions [85], can be used to model the disease only in the presence of a faithfully silenced Xi. This is because the disease phenotype is caused by a heterozygous mutation in the X-linked HPRT1 gene, leading to HPRT1 insufficiency in cells where the non-mutant HPRT1 resides on the Xi. When the region of the Xi harboring the HPRT1 gene undergoes erosion, it results in expression of the non-mutant HPRT1 gene product, overriding HPRT1 insufficiency. Thus, the Lesch-Nyhan diseases phenotypes can no longer be faithfully recapitulated with cultured iPSCs or iPSC-derived neurons in the presence of Xi erosion [77]. Additionally, in regenerative medicine such as cell replacement therapies, introducing cells with an Xi erosion into a patient may be treacherous because these cells lack proper dosage compensation of X-linked genes, a phenomenon observed in cancers [66].
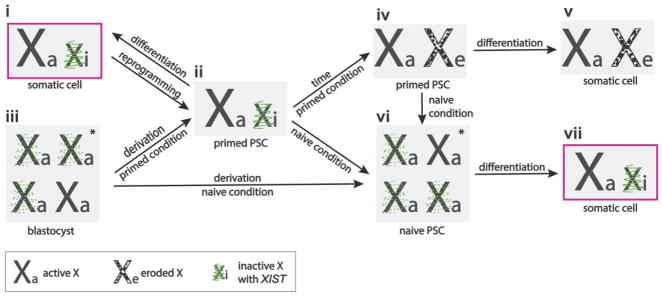
Figure 4. The X-chromosome state of naïve and primed human pluripotent stem cells.
(i) Female human somatic cells have an active and an XIST-expressing inactive X chromosome (Xa and Xi). (ii) Reprogramming of these cells to primed pluripotency does not change the X-chromosome state. (iii) Similarly, derivation of hPSCs from a pre-implantation blastocyst stabilizes the post-XCI state in primed pluripotent culture conditions. (iv) Over time in culture, the Xi loses expression of XIST and undergoes epigenetic erosion, resulting in partial reactivation and thus double-dose of the X-linked genes that fall in these eroded regions in primed hPSCs. (v) Although these cells can differentiate into somatic lineages, the resulting differentiated cells maintain the eroded X (Xe).
Female pre-implantation blastocysts have two active X chromosomes and express XIST, serving as a unique scenario where XIST expression does not cause XCI. (vi) When hESCs are derived under naïve pluripotent culture conditions, or when primed hPSCs are adapted to such naïve conditions, the X-chromosome state of resulting hPSCs resembles that of the pre-implantation blastocyst. (vii) Similar to normal development, differentiation of naïve hPSCs results in XIST-mediated XCI. *denotes the state found in majority of cells. hPSCs = human pluripotent stem cells, XIST = X inactive specific transcript, Xi = inactive X chromosome, Xa = active X chromosome, Xe = eroded X chromosome.
The role of XIST in early human development
It is interesting to note that primed hESCs do not reflect the X-chromosome state of the human pre-implantation embryos from which they are derived: all cells of a female human blastocyst, including those of the epiblast lineage, have two active X chromosomes and simultaneously express XIST [9,13], (Figure 4). The recent discovery of this non-silencing XIST in early human development has intrigued many researchers who study X-chromosome dosage compensation, including us. The two immediate questions regarding this unusual X-chromosome state are, 1) what role, if any, does XIST have, and 2) what is the molecular mechanism disabling XIST from silencing the X chromosome(s). These and many other questions cannot be addressed with in vitro studies of conventional (primed) hESCs since their X-chromosome state is different from the cells of the blastocyst from which they are derived, most plausibly due to suboptimal cell culture composition used (reviewed in more detail in [83]). However, recently devised cell culture conditions, which have been formulated to support cells in a naïve (pre-implantation) pluripotent state, allow growth of hESCs that better resemble the pluripotent state of cells in the pre-implantation blastocyst from which they are derived [86,87]. Most importantly, the X-chromosome state of these naïve hESCs recapitulates many aspects of the human blastocyst, where female cells have two active X chromosome and express XIST [20,21]. While most of the cells in a pre-implantation female human blastocyst express XIST bi-allelically, this pattern is a minority in naïve hESCs which exhibit mostly mono-allelic XIST expression [21]. Hence, naïve hESCs resemble the blastocyst, but not perfectly, as there is still room for improvement in the naïve culture media formulation. The molecular mechanism behind the non-silencing XIST is currently not understood, but investigating XIST-interacting proteins and XIST RNA modifications, which have recently been demonstrated to be crucial for Xist’s silencing role in the mouse [55], warrant further investigation. Current naïve culture conditions will allow such studies since naïve hESCs exhibit non-silencing XIST, albeit mostly mono-allelically [21].
In addition to recapitulating the X-state of the pre-implantation blastocyst, naïve hPSCs allow XIST-mediated induction of XCI upon differentiation [21]. When primed hPSCs with large regions of Xi erosion are adapted to naïve pluripotency and then differentiated, the erosion is, for the first time, reversed and replaced with XCI [21] (Figure 4). Hence, the transition to the naïve state resets the X-chromosome abnormalities of the primed pluripotent state. However, when primed hPSCs are adapted to naïve pluripotency, the memory of the starting Xi does not get lost in the naïve transition, since upon differentiation the starting Xi becomes silenced despite the presence of de novo XCI [21]. Therefore, although naïve hPSCs allow studies of de novo XCI in humans for the first time, they cannot be used to study choice of XCI since the process is non-random. The epigenetic memory of the starting Xi is unlikely due to DNA methylation, since the naïve state results in robust hypo-methylation of DNA [21,88,89], but may be due to the presence of histone modifications. For instance, it is possible that tri-methylation of histone H3 lysine 27 (H3K27me3), which was recently shown to regulate Xist imprinting in mice [90], marks the inactive or the active X chromosome through the transitions from primed to naïve pluripotency and eventually differentiation. It is, however, not clear whether naïve hESCs directly derived from the blastocyst or somatic cells directly reprogrammed to the naïve state can undergo random XCI upon differentiation.
Since there is no imprinted XCI in early human development, it has been unclear how X-chromosome dosage is compensated prior to onset of random XCI. Single cell RNA-sequencing of human pre-implantation embryos demonstrates gradual and time-dependent reduction of X-linked gene expression from both X chromosomes in embryonic days 4 to 7 in development [13]. This gradual dampening of X-linked gene expression correlates with upregulation of XIST [13]. X-chromosome dampening has also been observed in naïve XIST-expressing hESCs, further suggesting a novel role of XIST in human naïve pluripotency [21]. Independent analysis of the sequencing data from the pre-implantation blastocyst [13] and naïve hESC [21] studies has instead suggested the presence of XCI instead of XCD in human pre-implantation development [19]. If XIST is truly initiating XCI in the human blastocyst, given the fact that it is expressed from both X chromosomes in most cells, there must be a critical time-point at which point the cell decides to limit XIST’s silencing function to a single X chromosome, since silencing both X chromosomes is lethal due to phenotypic nullisomy of most X-chromosome genes [44,91]. Interestingly, blastocyst outgrowth studies demonstrated the presence of a XIST-negative transitionary state between the XIST-expressing blastocyst cells and the XIST-expressing XCI cells [80]. In the transition from XCD to XCI in hPSCs an XIST-negative state is also observed [21]. These data suggest that X-chromosome dosage compensation via XCD does not lead to the initiation of XCI.
If XIST is responsible for XCD in naïve pluripotency, it might do so by mediating accumulation of some, but perhaps not all chromatin modifications that are also responsible for XCI. For instance, H3K27me3 [92] accumulates on the XIST -coated active X chromosome in naïve hESCs [21], which might be responsible for dampening of X-linked gene expression. Another hypothesis is that expression of the lncRNA XACT may counteract some but not all functions of XIST, thereby achieving dampening instead of silencing. Indeed, it has recently been shown that XACT prevents accumulation of Xist when ectopically expressed on the mouse X chromosome [20], consistent with the idea that XACT can limit XIST’s activity in naive hPSCs. Interestingly, in a fraction of cells of rabbit blastocysts, Xist gets expressed from both X chromosomes, initiating silencing of both X-chromosomes before resolving to mono-allelic XCI via unknown mechanisms [9]. It is possible that the human scenario derives from such a mode of initiation of XCI and that XACT has evolved in primates to alleviate the detrimental consequences of inactivating both X-chromosomes for too long or in too many cells.
Regardless, naïve hPSCs, for the first time, allow detailed molecular studies of XIST and XCD, as well as the transition to XCI as cells exit pluripotency. Moreover, by studying these cells we can now gain insight into early human pre-implantation development and understand how it compares to what we already know in the mouse model organism.
Concluding Remarks and Future Directions
The biology of Xist unites researchers from multiple disciplines, including but not limited to those studying sex-chromosome dosage regulation, epigenetics (lncRNAs and chromatin remodeling), cell fate changes (reprogramming), cancer biology, disease pathogenesis (X-linked disorders), as well as development. Xist’s ability to recruit such a diverse group of researchers has enabled rapid advancement in understanding how Xist functions at the molecular level. We now understand that Xist acts as a scaffold to bring proteins to their site of action, effectively increasing the concentration of these proteins in a localized manner. Thanks to its very long-lived outcome and ability to be used in an allele-specific manner, Xist’s ability to silence genes can be mined for therapeutic purposes to balance gene expression in trisomic diseases such as Down Syndrome [93]. Furthermore, increased understanding of Xist can help engineer variants of this lncRNA to silence smaller and specific regions of a chromosome, increasing its therapeutic potential to silence mutant genes in an allele-specific manner.
Xist’s unique expression pattern in human pre-implantation development (expression without silencing) is a great marker of human naïve pluripotency [21] which can be used to develop new and improved naïve culture conditions in the future. Lastly, from an evolutionary perspective, Xist is an interesting lncRNA to study since it carries out similar functions in mouse and human despite lack of sequence conservation, but also seems to have evolved extra functions in a context-dependent manner, which requires further investigation (see Outstanding Questions). Both mouse and human Xist serve as a wonderful model for expanding our knowledge on lncRNA function, while learning about development and dosage regulation.
Outstanding Questions.
Requirement of Xist in random X-chromosome inactivation (XCI) in vivo needs to be addressed with mouse models where deletion of Xist can be confirmed prior to induction of X-chromosome inactivation.
Molecular mechanisms regulating X-chromosome dampening (XCD) in human pre-implantation embryos and naïve pluripotent stem cells are yet to be discovered.
What is preventing XIST from silencing X-linked gene expression in human pre-implantation development and naïve pluripotent stem cells?
Our knowledge of Xist biology from mouse studies needs to be extended to human XIST: what are the protein binding partners of human XIST? What are the steps in the initiation of XCI?
Role of XIST and XACT in human pre-implantation development and naïve pluripotent stem cells needs to be examined with deletion studies.
Why do naïve human pluripotent stem cells undergo non-random XCI? What is the epigenetic memory on the X-chromosomes in these cells?
An improved culture condition for naïve human pluripotent stem cells that allows higher rate of bi-allelic XIST cells and random XCI upon differentiation needs to be devised.
Are there other eutherians that exhibit XCD in pre-implantation development similar to humans?
Trends.
X inactive specific transcript (Xist) is a long non-coding RNA that remains associated with the X chromosome from which it is expressed.
Xist is unequivocally required for the imprinted form of X-chromosome inactivation (XCI) in mice in vivo, but demonstration of its indisputable requirement for random XCI in vivo is yet to be shown.
Loss of Xist expression in mice in vivo and in conventional human pluripotent stem cells correlates with partial re-activation of genes residing on the inactive X chromosome, suggesting an important role of Xist in maintenance of the silent state of genes on the inactive X chromosome.
Human pre-implantation embryos have a unique X-chromosome dosage compensation state called X-chromosome dampening (XCD), where transcriptional output is tuned down from both X chromosomes. Correlative observations suggest that XCD might be mediated by XIST.
Glossary
- Erosion
loss of transcriptional silencing and DNA methylation for genes on the inactive X chromosome in cultured primed human pluripotent stem cells. Eroded regions of the inactive X chromosome usually contain more than one gene, and fail to re-inactivate upon exit from pluripotency.
- ESC
embryonic stem cells are derived from the inner cell mass of the blastocyst before it implants in the uterus. ESCs are pluripotent and can self-renew indefinitely.
- Escape
from X-chromosome inactivation happens in both human and mouse development when a small portion of X-linked genes do not undergo transcriptional silencing in early development despite residing on the inactive X-chromosome.
- Eutheria
type of mammals whose fetal development requires a placenta (e.g. mouse and human). This excludes pouched mammals (marsupials, e.g. kangaroo) and egg-laying mammals (monotremes, e.g. platypus).
- Facultative heterochromatin
a subtype of heterochromatin that can also be present as euchromatin (actively transcribed chromatin) in a different context, such as the X chromosome before and after X-chromosome inactivation.
- Heterochromatin
regions of chromatin with low or no transcriptional activity.
- Heterochromatinization
formation of heterochromatin via changes to the epigenome, such as covalent modifications of histones and methylation of DNA, which results in transcriptional inactivity.
- Histone hypoacetylation
removal of the acetyl groups from histones, leading to reduced transcriptional activity.
- LncRNA
long non-coding ribonucleic acid. These RNA molecules are more than 200 nucleotides long and do not encode proteins.
- IPSCs
induced pluripotent stem cells derived from somatic cells by transcription factor overexpression-induced reprogramming.
- Metatheria
also marsupials, these mammals give live birth after a rather short gestation time, after which development of the newborn continues in the mother’s pouch.
- Myelodysplasia
multiple types of cancers of blood progenitor cells in the bone marrow which interferes with normal maturation of blood cells, resulting in reduced platelet, or red or white blood cell count.
- Naïve pluripotency
pluripotency state of cells of epiblast cells in the pre-implantation blastocyst.
- Pluripotency
cell state that can give rise to any of the cell types in the body.
- Primed pluripotency
pluripotency state of cells of epiblast cells in the post-implantation embryo.
- PSC
pluripotent stem cells are obtained either from pre-implantation blastocysts (ESCs) or via reprogramming of somatic cells (iPSCs).
- Reprogramming
forced expression of key transcription factors which remodel the epigenome of differentiated cells (i.e. fibroblasts) to transition these cells into the developmentally early, pluripotent state.
- Xa
active X chromosome.
- XACT
X active coating transcript is a recently discovered long non-coding RNA on the X chromosome, present in humans but not mice and expressed only in pluripotent cells from non-silent X chromosomes.
- XCD
X-chromosome dampening is the tuning down, but not complete silencing of genes on the X chromosome. Transcriptional output from a dampened X chromosome is less than that from an active X but more than that from an inactive X-chromosome.
- XCI
X-chromosome inactivation is the transcriptional silencing of most genes on the X chromosome in female mammalian cells.
- Xi
inactive X chromosome with most genes not transcribed due to X-chromosome inactivation.
- Xi reactivation
of the inactive X-chromosome refers to when all silenced genes become transcribed to resemble the state of the active X-chromosome. This is a chromosome-wide event and occurs in both mouse and human development during germ cell development (cells that give rise to gametes). In mouse development Xi reactivation also occurs in the transition from imprinted to random X-chromosome inactivation.
- Xist
X inactive specific transcript is a lncRNA gene on the X chromosome. It does not leave the nucleus and stays associated with the chromosome from which it is transcribed. In mouse, studies have shown that it is required and sufficient for the induction of X-chromosome inactivation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dixon-McDougall T, Brown C. The making of a Barr body: the mosaic of factors that eXIST on the mammalian inactive X chromosome 1. Biochem Cell Biol. 2016;94:56–70. doi: 10.1139/bcb-2015-0016. [DOI] [PubMed] [Google Scholar]
- 2.Wang F, et al. Regulation of X-linked gene expression during early mouse development by Rlim. eLife. 2016;5 doi: 10.7554/eLife.19127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borensztein M, et al. Xist-dependent imprinted X inactivation and the early developmental consequences of its failure. Nat Struct Mol Biol. 2017;24:226–233. doi: 10.1038/nsmb.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lyon MF. Sex chromatin and gene action in the mammalian X-chromosome. Am J Hum Genet. 1962;14:135–148. [PMC free article] [PubMed] [Google Scholar]
- 5.Wu H, et al. Cellular Resolution Maps of X Chromosome Inactivation: Implications for Neural Development, Function, and Disease. Neuron. 2014;81:103–119. doi: 10.1016/j.neuron.2013.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balaton BP, Brown CJ. Escape Artists of the X Chromosome. Trends Genet. 2016;32:348–359. doi: 10.1016/j.tig.2016.03.007. [DOI] [PubMed] [Google Scholar]
- 7.Gendrel AV, Heard E. Noncoding RNAs and Epigenetic Mechanisms During X-Chromosome Inactivation. Annu Rev Cell Dev Biol. 2014;30:561–580. doi: 10.1146/annurev-cellbio-101512-122415. [DOI] [PubMed] [Google Scholar]
- 8.Grant J, et al. Rsx is a metatherian RNA with Xist-like properties in X-chromosome inactivation. Nature. 2012;487:254–258. doi: 10.1038/nature11171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okamoto I, et al. Eutherian mammals use diverse strategies to initiate X-chromosome inactivation during development. Nature. 2011;472:370–374. doi: 10.1038/nature09872. [DOI] [PubMed] [Google Scholar]
- 10.Hwang JY, Oh JN, Park CH, Lee DK, Lee CK. Dosage compensation of X-chromosome inactivation center-linked genes in porcine preimplantation embryos: Non-chromosome-wide initiation of X-chromosome inactivation in blastocysts. Mech Dev. 2015;138:246–255. doi: 10.1016/j.mod.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 11.Wang X, Miller DC, Clark AG, Antczak DF. Random X inactivation in the mule and horse placenta. Genome Res. 2012;22:1855–1863. doi: 10.1101/gr.138487.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moreira de Mello JC, et al. Random X Inactivation and Extensive Mosaicism in Human Placenta Revealed by Analysis of Allele-Specific Gene Expression along the X Chromosome. PLoS ONE. 2010;5:e10947. doi: 10.1371/journal.pone.0010947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petropoulos S, et al. Single-Cell RNA-Seq Reveals Lineage and X Chromosome Dynamics in Human Preimplantation Embryos. Cell. 2016;165:1012–1026. doi: 10.1016/j.cell.2016.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deng X, Berletch JB, Nguyen DK, Disteche CM. X chromosome regulation: diverse patterns in development, tissues and disease. Nat Rev Genet. 2014;15:367–378. doi: 10.1038/nrg3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meyer BJ. Targeting X chromosomes for repression. Curr Opin Genet Dev. 2010;20:179–189. doi: 10.1016/j.gde.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crane E, et al. Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature. 2015;523:240–244. doi: 10.1038/nature14450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pandya-Jones A, Plath K. The ‘lnc’ between 3D chromatin structure and X chromosome inactivation. Semin Cell Dev Biol. 2016;56:35–47. doi: 10.1016/j.semcdb.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Albritton SE, Ercan S. Caenorhabditis elegans Dosage Compensation: Insights into Condensin-Mediated Gene Regulation. Trends Genet. 2018;34:41–53. doi: 10.1016/j.tig.2017.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moreira de Mello JC, Fernandes GR, Vibranovski MD, Pereira LV. Early X chromosome inactivation during human preimplantation development revealed by single-cell RNA-sequencing. Sci Rep. 2017;7 doi: 10.1038/s41598-017-11044-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vallot C, et al. XACT Noncoding RNA Competes with XIST in the Control of X Chromosome Activity during Human Early Development. Cell Stem Cell. 2017;20:102–111. doi: 10.1016/j.stem.2016.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sahakyan A, et al. Human Naive Pluripotent Stem Cells Model X Chromosome Dampening and X Inactivation. Cell Stem Cell. 2017;20:87–101. doi: 10.1016/j.stem.2016.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun S, et al. Jpx RNA Activates Xist by Evicting CTCF. Cell. 2013;153:1537–1551. doi: 10.1016/j.cell.2013.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carmona S, Lin B, Chou T, Arroyo K, Sun S. LncRNA Jpx induces Xist expression in mice using both trans and cis mechanisms. PLOS Genet. 2018;14:e1007378. doi: 10.1371/journal.pgen.1007378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chureau C, et al. Ftx is a non-coding RNA which affects Xist expression and chromatin structure within the X-inactivation center region. Hum Mol Genet. 2011;20:705–718. doi: 10.1093/hmg/ddq516. [DOI] [PubMed] [Google Scholar]
- 25.Furlan G, et al. The Ftx Noncoding Locus Controls X Chromosome Inactivation Independently of Its RNA Products. Mol Cell. 2018;70:462–472.e8. doi: 10.1016/j.molcel.2018.03.024. [DOI] [PubMed] [Google Scholar]
- 26.Lee JT, Davidow LS, Warshawsky D. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat Genet. 1999;21:400–404. doi: 10.1038/7734. [DOI] [PubMed] [Google Scholar]
- 27.Gayen S, Maclary E, Buttigieg E, Hinten M, Kalantry S. A Primary Role for the Tsix lncRNA in Maintaining Random X-Chromosome Inactivation. Cell Rep. 2015;11:1251–1265. doi: 10.1016/j.celrep.2015.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vallot C, et al. XACT, a long noncoding transcript coating the active X chromosome in human pluripotent cells. Nat Genet. 2013;45:239–241. doi: 10.1038/ng.2530. [DOI] [PubMed] [Google Scholar]
- 29.Jonkers I, et al. Xist RNA Is Confined to the Nuclear Territory of the Silenced X Chromosome throughout the Cell Cycle. Mol Cell Biol. 2008;28:5583–5594. doi: 10.1128/MCB.02269-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Engreitz JM, et al. The Xist lncRNA Exploits Three-Dimensional Genome Architecture to Spread Across the X Chromosome. Science. 2013;341:1237973–1237973. doi: 10.1126/science.1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang KC, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Engreitz JM, Ollikainen N, Guttman M. Long non-coding RNAs: spatial amplifiers that control nuclear structure and gene expression. Nat Rev Mol Cell Biol. 2016;17:756–770. doi: 10.1038/nrm.2016.126. [DOI] [PubMed] [Google Scholar]
- 33.Brown CJ, et al. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- 34.Brockdorff N, et al. Conservation of position and exclusive expression of mouse Xist from the inactive X chromosome. Nature. 1991;351:329–331. doi: 10.1038/351329a0. [DOI] [PubMed] [Google Scholar]
- 35.Borsani G, et al. Characterization of a murine gene expressed from the inactive X chromosome. Nature. 1991;351:325–329. doi: 10.1038/351325a0. [DOI] [PubMed] [Google Scholar]
- 36.Brown CJ, et al. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- 37.Brockdorff N, et al. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71:515–526. doi: 10.1016/0092-8674(92)90519-i. [DOI] [PubMed] [Google Scholar]
- 38.Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379:131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- 39.Marahrens Y, Panning B, Dausman J, Strauss W, Jaenisch R. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev. 1997;11:156–166. doi: 10.1101/gad.11.2.156. [DOI] [PubMed] [Google Scholar]
- 40.Mugford JW, Yee D, Magnuson T. Failure of extra-embryonic progenitor maintenance in the absence of dosage compensation. Dev Camb Engl. 2012;139:2130–2138. doi: 10.1242/dev.076497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kalantry S, Purushothaman S, Bowen RB, Starmer J, Magnuson T. Evidence of Xist RNA-independent initiation of mouse imprinted X-chromosome inactivation. Nature. 2009;460:647–651. doi: 10.1038/nature08161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marahrens Y, Loring J, Jaenisch R. Role of the Xist gene in X chromosome choosing. Cell. 1998;92:657–664. doi: 10.1016/s0092-8674(00)81133-2. [DOI] [PubMed] [Google Scholar]
- 43.Wutz A, Rasmussen TP, Jaenisch R. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat Genet. 2002;30:167–174. doi: 10.1038/ng820. [DOI] [PubMed] [Google Scholar]
- 44.Wutz A, Jaenisch R. A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol Cell. 2000;5:695–705. doi: 10.1016/s1097-2765(00)80248-8. [DOI] [PubMed] [Google Scholar]
- 45.Yang L, Kirby JE, Sunwoo H, Lee JT. Female mice lacking Xist RNA show partial dosage compensation and survive to term. Genes Dev. 2016;30:1747–1760. doi: 10.1101/gad.281162.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rastan S. Timing of X-chromosome inactivation in postimplantation mouse embryos. J Embryol Exp Morphol. 1982;71:11–24. [PubMed] [Google Scholar]
- 47.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 48.Hayashi S, Lewis P, Pevny L, McMahon AP. Efficient gene modulation in mouse epiblast using a Sox2Cre transgenic mouse strain. Mech Dev. 2002;119(Suppl 1):S97–S101. doi: 10.1016/s0925-4773(03)00099-6. [DOI] [PubMed] [Google Scholar]
- 49.Friedrich G, Soriano P. Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev. 1991;5:1513–1523. doi: 10.1101/gad.5.9.1513. [DOI] [PubMed] [Google Scholar]
- 50.Csankovszki G, Panning B, Bates B, Pehrson JR, Jaenisch R. Conditional deletion of Xist disrupts histone macroH2A localization but not maintenance of X inactivation. Nat Genet. 1999;22:323–324. doi: 10.1038/11887. [DOI] [PubMed] [Google Scholar]
- 51.Yildirim E, et al. Xist RNA Is a Potent Suppressor of Hematologic Cancer in Mice. Cell. 2013;152:727–742. doi: 10.1016/j.cell.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nagy A, et al. Embryonic stem cells alone are able to support fetal development in the mouse. Dev Camb Engl. 1990;110:815–821. doi: 10.1242/dev.110.3.815. [DOI] [PubMed] [Google Scholar]
- 53.Schulz EG, et al. The two active X chromosomes in female ESCs block exit from the pluripotent state by modulating the ESC signaling network. Cell Stem Cell. 2014;14:203–216. doi: 10.1016/j.stem.2013.11.022. [DOI] [PubMed] [Google Scholar]
- 54.Thornhill AR, Burgoyne PS. A paternally imprinted X chromosome retards the development of the early mouse embryo. Dev Camb Engl. 1993;118:171–174. doi: 10.1242/dev.118.1.171. [DOI] [PubMed] [Google Scholar]
- 55.da Rocha ST, Heard E. Novel players in X inactivation: insights into Xist-mediated gene silencing and chromosome conformation. Nat Struct Mol Biol. 2017;24:197–204. doi: 10.1038/nsmb.3370. [DOI] [PubMed] [Google Scholar]
- 56.Chu C, et al. Systematic Discovery of Xist RNA Binding Proteins. Cell. 2015;161:404–416. doi: 10.1016/j.cell.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McHugh CA, et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 2015;521:232–236. doi: 10.1038/nature14443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Minajigi A, et al. A comprehensive Xist interactome reveals cohesin repulsion and an RNA-directed chromosome conformation. Science. 2015;349:aab2276–aab2276. doi: 10.1126/science.aab2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moindrot B, et al. A Pooled shRNA Screen Identifies Rbm15, Spen, and Wtap as Factors Required for Xist RNA-Mediated Silencing. Cell Rep. 2015;12:562–572. doi: 10.1016/j.celrep.2015.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Monfort A, et al. Identification of Spen as a Crucial Factor for Xist Function through Forward Genetic Screening in Haploid Embryonic Stem Cells. Cell Rep. 2015;12:554–561. doi: 10.1016/j.celrep.2015.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen CK, et al. Xist recruits the X chromosome to the nuclear lamina to enable chromosome-wide silencing. Science. 2016;354:468–472. doi: 10.1126/science.aae0047. [DOI] [PubMed] [Google Scholar]
- 62.Patil DP, et al. m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016 doi: 10.1038/nature19342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pintacuda G, et al. hnRNPK Recruits PCGF3/5-PRC1 to the Xist RNA B-Repeat to Establish Polycomb-Mediated Chromosomal Silencing. Mol Cell. 2017;68:955–969.e10. doi: 10.1016/j.molcel.2017.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Van Nostrand EL, et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP) Nat Methods. 2016;13:508–514. doi: 10.1038/nmeth.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shevchenko AI, et al. Impact of Xist RNA on chromatin modifications and transcriptional silencing maintenance at different stages of imprinted X chromosome inactivation in vole Microtus levis. Chromosoma. 2018;127:129–139. doi: 10.1007/s00412-017-0650-9. [DOI] [PubMed] [Google Scholar]
- 66.Chaligné R, Heard E. X-chromosome inactivation in development and cancer. FEBS Lett. 2014;588:2514–2522. doi: 10.1016/j.febslet.2014.06.023. [DOI] [PubMed] [Google Scholar]
- 67.Pasque V, Plath K. X chromosome reactivation in reprogramming and in development. Curr Opin Cell Biol. 2015;37:75–83. doi: 10.1016/j.ceb.2015.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mak W, et al. Reactivation of the paternal X chromosome in early mouse embryos. Science. 2004;303:666–669. doi: 10.1126/science.1092674. [DOI] [PubMed] [Google Scholar]
- 69.de Napoles M, Nesterova T, Brockdorff N. Early Loss of Xist RNA Expression and Inactive X Chromosome Associated Chromatin Modification in Developing Primordial Germ Cells. PLoS ONE. 2007;2:e860. doi: 10.1371/journal.pone.0000860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pasque V, et al. X Chromosome Reactivation Dynamics Reveal Stages of Reprogramming to Pluripotency. Cell. 2014;159:1681–1697. doi: 10.1016/j.cell.2014.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Csankovszki G, Nagy A, Jaenisch R. Synergism of Xist RNA, DNA methylation, and histone hypoacetylation in maintaining X chromosome inactivation. J Cell Biol. 2001;153:773–784. doi: 10.1083/jcb.153.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Minkovsky A, et al. The Mbd1-Atf7ip-Setdb1 pathway contributes to the maintenance of X chromosome inactivation. Epigenetics Chromatin. 2014;7:12. doi: 10.1186/1756-8935-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Keniry A, et al. Setdb1-mediated H3K9 methylation is enriched on the inactive X and plays a role in its epigenetic silencing. Epigenetics Chromatin. 2016;9 doi: 10.1186/s13072-016-0064-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tchieu J, et al. Female Human iPSCs Retain an Inactive X Chromosome. Cell Stem Cell. 2010;7:329–342. doi: 10.1016/j.stem.2010.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nichols J, Smith A. Naive and Primed Pluripotent States. Cell Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 76.Shen Y, et al. X-inactivation in female human embryonic stem cells is in a nonrandom pattern and prone to epigenetic alterations. Proc Natl Acad Sci. 2008;105:4709–4714. doi: 10.1073/pnas.0712018105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mekhoubad S, et al. Erosion of Dosage Compensation Impacts Human iPSC Disease Modeling. Cell Stem Cell. 2012;10:595–609. doi: 10.1016/j.stem.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nazor KL, et al. Recurrent Variations in DNA Methylation in Human Pluripotent Stem Cells and Their Differentiated Derivatives. Cell Stem Cell. 2012;10:620–634. doi: 10.1016/j.stem.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vallot C, et al. Erosion of X Chromosome Inactivation in Human Pluripotent Cells Initiates with XACT Coating and Depends on a Specific Heterochromatin Landscape. Cell Stem Cell. 2015;16:533–546. doi: 10.1016/j.stem.2015.03.016. [DOI] [PubMed] [Google Scholar]
- 80.Patel S, et al. Human Embryonic Stem Cells Do Not Change Their X Inactivation Status during Differentiation. Cell Rep. 2017;18:54–67. doi: 10.1016/j.celrep.2016.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Simon MD, et al. High-resolution Xist binding maps reveal two-step spreading during X-chromosome inactivation. Nature. 2013;504:465–469. doi: 10.1038/nature12719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bruck T, Yanuka O, Benvenisty N. Human pluripotent stem cells with distinct X inactivation status show molecular and cellular differences controlled by the X-Linked ELK-1 gene. Cell Rep. 2013;4:262–270. doi: 10.1016/j.celrep.2013.06.026. [DOI] [PubMed] [Google Scholar]
- 83.Sahakyan A, Plath K, Rougeulle C. Regulation of X-chromosome dosage compensation in human: mechanisms and model systems. Philos Trans R Soc B Biol Sci. 2017;372:20160363. doi: 10.1098/rstb.2016.0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Geens M, et al. Female human pluripotent stem cells rapidly lose X chromosome inactivation marks and progress to a skewed methylation pattern during culture. Mol Hum Reprod. 2016;22:285–298. doi: 10.1093/molehr/gaw004. [DOI] [PubMed] [Google Scholar]
- 85.Bell S, Kolobova I, Crapper L, Ernst C. Lesch-Nyhan Syndrome: Models, Theories, and Therapies. Mol Syndromol. 2016;7:302–311. doi: 10.1159/000449296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Theunissen TW, et al. Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell. 2014;15:471–487. doi: 10.1016/j.stem.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Takashima Y, et al. Resetting Transcription Factor Control Circuitry toward Ground-State Pluripotency in Human. Cell. 2014;158:1254–1269. doi: 10.1016/j.cell.2014.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pastor WA, et al. Naive Human Pluripotent Cells Feature a Methylation Landscape Devoid of Blastocyst or Germline Memory. Cell Stem Cell. 2016;18:323–329. doi: 10.1016/j.stem.2016.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Theunissen TW, et al. Molecular Criteria for Defining the Naive Human Pluripotent State. Cell Stem Cell. 2016 doi: 10.1016/j.stem.2016.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Inoue A, Jiang L, Lu F, Zhang Y. Genomic imprinting of Xist by maternal H3K27me3. Genes Dev. 2017;31:1927–1932. doi: 10.1101/gad.304113.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Guyochin A, et al. Live Cell Imaging of the Nascent Inactive X Chromosome during the Early Differentiation Process of Naive ES Cells towards Epiblast Stem Cells. PLoS ONE. 2014;9:e116109. doi: 10.1371/journal.pone.0116109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Plath K, Zhang Y, Panning B, Otte AP. Role of Histone H3 Lysine 27 Methylation in X Inactivation. Science. 2003;300:131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- 93.Jiang J, et al. Translating dosage compensation to trisomy 21. Nature. 2013;500:296–300. doi: 10.1038/nature12394. [DOI] [PMC free article] [PubMed] [Google Scholar]