Summary
Lineage-committed αβ and γδ T cells are thought to originate from common intrathymic multipotent progenitors following instructive T cell receptor (TCR) signals. A subset of lymph node and mucosal Vγ2+ γδ T cells is progγδTCR in development of these cells remains controversial. Here we generated reporter mice for the rammed intrathymically to produce IL-17 (Tγδ17 cells), however the role of the Tγδ17 lineage-defining transcription factor SOX13 and identified fetal-origin, intrathymic Sox13+ progenitors. In organ culture developmental assays, Tγδ17 cells derived primarily from Sox13+ progenitors, and not from other known lymphoid progenitors. Single cell transcriptome assays of the progenitors found in TCR-deficient mice demonstrated that Tγδ17 lineage programming was independent of γδTCR. Instead, generation of the lineage committed progenitors and Tγδ17 cells was controlled by TCF1 and SOX13. Thus, T lymphocyte lineage fate can be prewired cell-intrinsically and is not necessarily specified by clonal antigen receptor signals.
Keywords: T cells, thymus, IL-17, development, T cell receptor, ranscription factor, SOX13
eTOC Blurb
The role of TCR in establishing IL-17 producing γδ T (Tγδ17) cell lineage identity during thymic development remains controversial. Spidale et al. identify a Sox13hi progenitor to Vγ2+ Tγδ17 cells and demonstrate that progenitor programming is dependent on SOX13 and TCF7 but independent of γδTCR.
Graphical Abstract
Introduction
Mammalian lymphoid immunity consists of two distinct branches: the fast acting innate lymphocytes and the slower, but expansive and sterilizing, adaptive lymphocytes (Kang and Malhotra, 2015). Innate lymphocytes include γδ T cells, αβ T cells with restricted T cell receptor (TCR) repertoire, B1 cells and innate lymphoid cells (ILCs) that lack clonal antigen receptors, including natural killer (NK) cells. Adaptive B and T lymphocytes express clonal antigen receptors and are central mediators of rapid recall responses to previously encountered pathogens. The developmental origin of the two branches is under intense investigations, historically starting with the lineage commitment processes producing γδ T and αβ T cells from what was assumed to be a common progenitor (Narayan and Kang, 2010). This was logically centered on the role distinct TCR plays in specifying cell fate. However, with the increasing appreciation of ontogenically restricted emergence of the innate responders in the fetuses (Bando et al., 2015; Constantinides et al., 2014; Haas et al., 2012; Havran and Allison, 1990), the possibility that distinct progenitors are lineage-specified molecularly prior to antigen receptor signaling has gained increased traction conceptually (Kang and Malhotra, 2015; Mold et al., 2010; Yuan et al., 2012).
Among T cell subtypes, interleukin-17 (IL-17) producing γδ T cells (Tγδ17 cells) are the prototypic innate T lymphocytes stationed at mucosal barriers, namely the dermis, reproductive organs, and oral cavities of the mouse and humans (Chien et al., 2013). In contrast to adaptive IL-17 producing αβ T (Th17) cells, Tγδ17 cells are programmed during thymic development for the earliest IL-17 response to various pathogens, such as cutaneous Staphylococcus aureus and Candida albicans (Cho et al., 2010; Kashem et al., 2015; Malhotra et al., 2013; Narayan et al., 2012). In these settings, Tγδ17 cells have been shown to respond to cytokines, predominantly IL-23 and IL-1, rather than overt TCR triggering (Cai et al., 2011; Sutton et al., 2009). Similarly, in response to cutaneous application of the toll-like receptor 7 agonist Imiquimod, neonatal-origin Vγ2+ Tγδ17 cells are responsible for an IL-17-dependent psoriasis-like disease (Gray et al., 2013; Malhotra et al., 2013).Furthermore, Tγδ17 cells have been associated with both anti- and pro-tumor functions (Coffelt et al., 2015; Ma et al., 2011), demonstrating the extensive contribution of Tγδ17 cells to immunity and inflammation.
Among the earliest T cells to develop, Tγδ17 cells represent the “second wave” of fetal γδ T cells and are composed of subsets utilizing distinct TCRVγ chains, Vγ4 or Vγ2 TCR [Garman nomenclature, (Garman et al., 1986)]. Mature Vγ4+ Tγδ17 cells develop first (Haas et al., 2012), express an invariant Vγ4Vδ1 TCR without junctional sequence diversity. Mature Vγ2+ Tγδ17 cells emerge in the late fetal and neonatal period, express a TCR junctional diversified repertoire paired with several different Vδ chains (Kashani et al., 2015; Wei et al., 2015), and are the dominant functional subtype in the dermis. The association of specific Vγ/Vδ chains with discrete functional properties of γδ T cells bearing those chains has often been interpreted to imply a role for the TCR in specifying γδ T cell lineage fate and function. Indeed, most recent studies have suggested that strength of γδTCR signaling during development in the thymus dictates functional properties (Lee et al., 2014; Wencker et al., 2014), with Tγδ17 cells having different TCR signaling requirements than IFNγ- producing γδ T cells. However, the data is mixed (Munoz-Ruiz et al., 2016), and critically, whether TCR signaling primarily permits prewired effector lineage committed cells to mature (“permissive”)rather than specifying cell fate (“instructive”) is unresolved.
Developing intrathymic Tγδ17 cells are marked by a unique transcriptome distinct from other lymphocyte subtypes, including ILCs (Narayan et al., 2012; Robinette et al., 2015). Chief among the factors required for Tγδ17 programming is the high-mobility group (HMG) transcription factor (TF) Sry (sex determining region Y)-box 13 (SOX13) (Gray et al., 2013; Malhotra et al., 2013; Melichar et al., 2007). SOX13 is expressed by all immature (CD24hi) γδ thymocytes, but at the highest level in those expressing the TCRVγ2 chain, which includes the immediate precursors to Tγδ17 cells. Concordantly, development of Vγ2+ Tγδ17 cells, but not the Vγ4+ subset, absolutely requires Sox13, and in its absence the core Tγδ17 transcriptome fails to establish, including Rorc, Blk, and Maf. Given that Sox13 is not induced by TCR signaling, it was possible that SOX13 first specifies Tγδ17 lineage differentiation prior to TCR expression or signaling. Here, using Sox13 reporter mice, we identified TCR-negative Tγδ17 progenitors and found, via single-cell transcriptomic analyses, clonal heterogeneity among Sox13 reporter marked cells. Lineage specifying clonal gene signatures were independent of TCR; rather, the generation of Tγδ17 progenitors was controlled by HMG TFs.
Results
Identification of candidate γδ T cell progenitors using SOX13 reporter mice
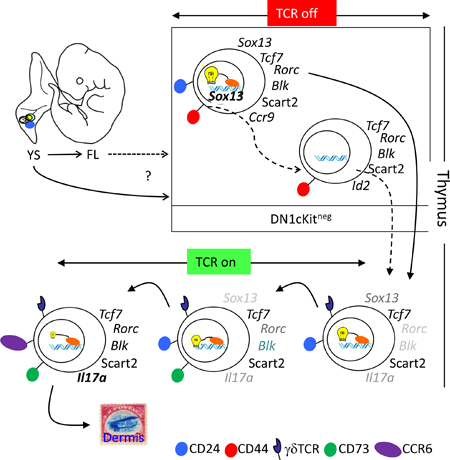
Previous gene expression analyses identified SOX13 as one of only a few TFs highly restricted to the γδ T cell lineage in the hematopoietic system (Jojic et al., 2013), and SOX13 is absolutely required for the intrathymic development of Tγδ17 cells. Thus, we hypothesized that high SOX13 expression may distinguish progenitors pre-specified toward the Tγδ17 lineage. To track SOX13-expressing cells, we developed reporter mice with a nuclear ECFP-Cre fusion protein coding gene under control of the endogenous Sox13 locus (Sox13ecfp/+ mice, Figure S1A), which demonstrated an ECFP expression pattern consistent with Sox13 expression previously determined by a genome wide transcriptome analysis (Narayan et al., 2012) (Figure S1B–C). ECFP was observed in all thymic γδ T cells, with the highest levels found in immature (CD24hi) subsets and subsequent decreased expression upon maturation. ECFP expression was also observed in some invariant Natural Killer T (iNKT) cells, but not in other αβ T lineage cells (e.g. CD4+CD8+ double positive [DP] thymocytes, Malhotra et al., manuscript submitted). Given the expression of Sox13 in the hematopoietic stem cell and its lymphoid progeny (McKinney-Freeman et al., 2012; Melichar et al., 2007) (Figure S1D), Sox13 reporter mice were used to track the origin of Tγδ17 cells, focusing on developmentally staged thymic precursors (traditionally termed CD4 and CD8 double negative [DN]) lacking the expression of TCR, coreceptors for MHC, and additional markers of mature leukocyte lineages (Lin-, see Methods). DN1 precursor thymocytes (CD44+CD25-c-Kit+/−Lin-/lo) are the most undifferentiated, and can be further stratified into subsets (a to e) based on c-Kit and CD24 expression. With the exception of DN1e, DN1 subsets lack expression of RAG proteins and have been described as having Tcr loci in unrearranged state (Porritt et al., 2004). Previous studies showed that adult DN1a/b (also termed early thymic progenitors, ETPs) are the primary generators of conventional αβ T cells but they do not appear to differentiate into DN1d/e cells (Benz et al., 2008), while DN1c cells are CD11c+ and likely represent committed progenitors to intrathymic CD8α+-type dendritic cells (Luche et al., 2011). Among intrathymic precursors, only DN1d cells were uniformly (75% to 95% in postnatal mice) ECFP+ (Figure 1A). ECFP was not detected in other DN thymocytes, including DN2 cells (c-Kit+CD25+CD44+) that express some Sox13 (Melichar et al., 2007), suggesting that ECFP was a proxy for cells with highest Sox13 transcriptional activity. Given the unique high expression and/or retention of SOX13-ECFP by DN1d cells, this population was further investigated. SOX13-ECFP was also observed in fetal (Embryonic day (E) 16), neonatal and young adult DN1d precursors (Figure 1A–B). Consistent with a previous study (Ramond et al., 2014), DN1d cells were not prevalent prior to E16.5 (Figure 1C), and in some E16.5 DN1d-like cells, Sox13 expression was evident (Figure 1B). Additionally, some DN1d cells were already endowed with proteins essential for developing Tγδ17 cells and effector function, including the TFs RORγt and cMAF, the signaling molecule BLK, and the scavenger receptor SCART2 (5830411N06Rik) (Kisielow et al., 2008; Narayan et al., 2012) (Figure 1D). While some DN1e cells also expressed RORγt and BLK (Figure 1D), none of these markers of Tγδ17 cells were expressed in c-Kit+ DN1 or DN2 cells (data not shown). Most DN1 cells, but not all DN1d and DN1e cells, expressed the HMG TF TCF1, marking them as T cell lineage cells that had received Notch signals (Weber et al., 2011) and smaller subsets expressed its close relative LEF1 (Figure 1E) that in adults is associated with cells programmed to produce IFNγ (Malhotra et al., 2013). These results suggested that ECFP+ DN1d cells share molecular features of developing immature thymocytes programmed to become Tγδ17 cells.
Figure 1. Identification and characterization of SOX13-ECFP expressing thymic progenitors.

(A) Expression of ECFP driven by the Sox13 promoter in DN subsets (bold black line) of 10 day (d10) old mice. Light gray plots indicate background fluorescence from DN subsets of non-reporter mice (Sox13-ECFP mice, n = 28). (B) Assessment of ECFP expression by DN1d cells from Sox13ECFP/+mice analyzed at the indicated embryonic day or time post-birth. (C) Representative DN1 subset distributions during ontogeny. (D) Expression of intranuclear RORγt, MAF, and BLK and surface Scart2 by DN1d and DN1e cells in thymuses of E18.5 fetuses. (E) Analysis of TCF1 and LEF1expression in E18.5 fetal thymocyte progenitors.
Tγδ17 gene network is established in SOX13+ DN1 progenitors
SOX13 works in concert with a battery of additional TFs to impose the Tγδ17 effector identity. We hypothesized that a SOX13+ γδ T cell progenitor would also exhibit such molecular hallmarks. To systematically establish precursor cell lineage relationships, global transcriptome analysis of thymic progenitor subsets by deep sequencing was performed. DN1d cells were molecularly distinct from other T precursor subsets: DN1a/b (c-Kit+), DN1e and DN2 (Figure 2A), and principal co-ordinate analysis (PCoA, of all differentially expressed genes detected from pairwise comparisons of DN1ab, DN1d, DN1e, and immature CD24hi Vγ2+ (ImmV2) thymocytes as the input gene list) aligned DN1d cells most closely to ImmV2 thymocytes (Figure 1B), a heterogeneous population that we had previously characterized as most similar to CD4+CD8+ DP cells largely due to similar metabolic gene signatures (Narayan et al., 2012). Overall, ~600 genes were differentially expressed between DN1d and ImmV2 cells, the immediate precursor of Tγδ17 cells (Figure 2C, bottom, and Tables S1–3), representing the smallest divergence when compared to pairwise comparisons of other thymic subsets. DN1d cells also expressed genes active in non-T cell types, in particular myeloid cells (Csf2rb2, Cdh1, and Gp49a for examples). A focused analysis of TFs revealed that the transcriptional regulatory network of DN1d cells overlapped with ImmV2 thymocytes, with all known TFs involved in Tγδ17 development expressed precociously and at relatively high amounts in DN1d cells (Figure 2D and Table S4). These TFs include Bcl11b, Bhlhe40, Etv5, Id3, Notch1, Sox4 and Vdr, which in concert with Sox13, Tcf7, Rorc and Maf specify the effector function of Tγδ17 cells (Jojic et al., 2013; Malhotra et al., 2013; Shibata et al., 2014). Notably, DN1d cells did not express Il17a as assessed by RNA-Seq. Il17a-IRES-egfp (Il17aegfp/egfp) reporter mice were used to confirm that DN1d cells did not transcribe Il17a (Fig. S1F), befitting their status as progenitors. DN1 cell phenotypes were unstable upon culture, especially upon stimulation, precluding IL-17 protein analysis (Fig. S1E). Il17a EGFP was first detected in ImmV2 cells that also lacked expression of CD73 (Fig. S1G–H), a proposed marker of post-γδTCR signaled cells (Coffey et al., 2014), indicating an onset of Il17a transcription prior to known TCR signal-mediated alterations. Together, these findings suggested that a pre-wired genetic program exists in DN1d progenitors to generate innate Tγδ17 cells, without γδTCR assembly.
Figure 2. DN1d thymic progenitors exhibit molecular hallmarks of Tγδ17 cells.
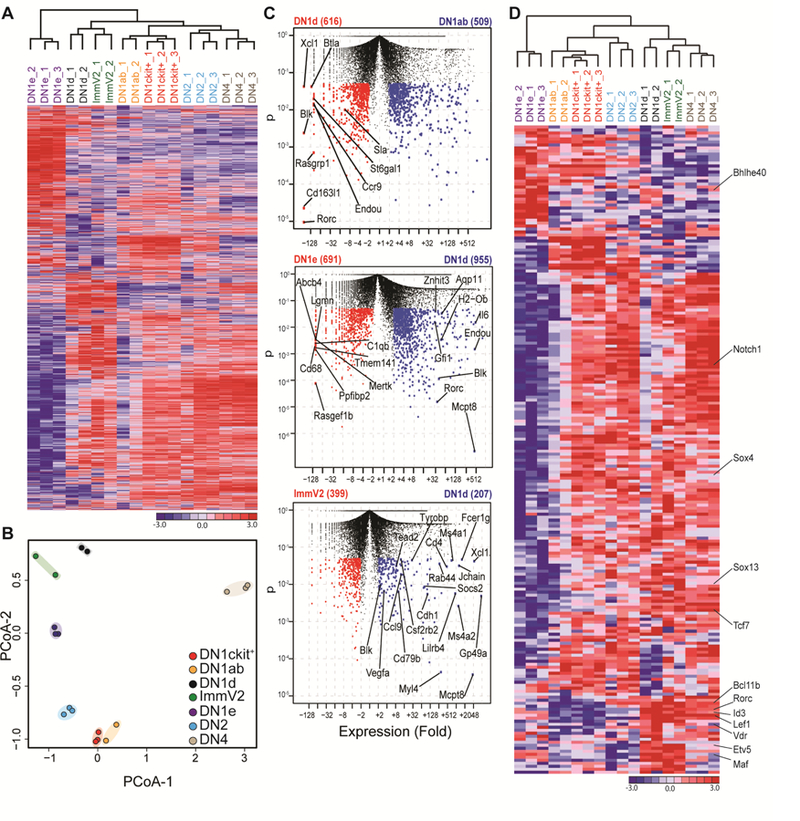
(A) Thymocyte progenitor subsets were sorted from d7–10 old mice and subjected to RNA sequencing analysis. Hierarchical clustering and heat map rendering of differentially expressed genes (DEGs: >2-fold change and p-value < 0.05) from population-level RNA-Seq. (B) Principal coordinate analysis (PCoA) of indicated thymic progenitor subsets and immature Vγ2+ thymocytes (ImmV2), the immediate precursors to Vγ2+ Tγδ17 cells. (C) Pairwise comparisons of genes increased and decreased in expression (red, blue dots) in indicated subsets plotted as expression fold change versus p-value; numbers in brackets indicate transcripts significantly increased in expression by at least 2-fold and p< 0.05. (D) Hierarchical clustering and heat map rendering of differentially expressed transcription factors (p< 0.01) in indicated subsets.
Extensive heterogeneity within DN1d cells
The transcriptome profiles established above suggested that DN1d cells may exhibit characteristics of multipotent progenitors and retain the ability to adopt at least two disparate cell fates, myeloid lineage and Tγδ17 lineage. Alternatively, DN1d cells as originally defined may contain a mixture of cells, some of which possess a myeloid transcriptional profile and others that possess the Tγδ17 profile. To distinguish between these possibilities, we performed high-sensitivity single cell quantitative PCR analysis of select genes identified by RNA-seq (Figure 3A–C). For comparison, DN2 progenitors, which are largely T lineage committed and also known to express low amounts of Sox13 transcripts (Melichar et al., 2007), but not to levels detectable by the SOX13-ECFP reporter (Figure 1A), were also analyzed. Strikingly, Sox13 transcripts were not detectable in over half of DN1d cells, suggesting that the Cre-ECFP composite nuclear reporter can be stably retained in DN1d cells without significant concomitant Sox13 transcription, as shown previously for other reporters that can bind to DNA (Wilson et al., 2008). Given that Sox13 was detected in ~80% of single DN2 thymocytes, at levels on average at least 10-fold lower than in Sox13 transcript+ DN1d cells and not sufficient for reporter marking (Figure 3D), it was unlikely that the assay was underestimating ECFP+ cells that had concurrent Sox13 transcription. Critically, a substantial fraction (~40%) of DN1d cells expressed a near-complete array of genes associated with Tγδ17 functional programming, including Sox13 (SOX13+ progenitors (Soxpro) cluster, Figure 3A, D). Notably, there was a strong association of a thymic-homing receptor CCR9 with the Soxpro cluster.CCR9+ DN1d cells were most prevalent in fetal thymi and they decreased in proportion with increasing age (Figure S2A).
Figure 3. Heterogeneity among DN1d cells identified by single-cell transcriptomics.

(A) Single-cell gene expression analysis of DN1d (four denoted clusters) and DN2 (clustered to far right) thymocytes from d7–10 old mice. Expression of 47 genes are visualized by hierarchical clustering and displayed by heat map rendering of global Z scores. Each column is expression profile of single cells and each row is a target gene, denoted on the right. The blue dot marks the row for Sox13. (B) Principle coordinate analysis (PCoA) of individual DN1d and DN2 cells in two dimensions. Boxed “Soxpro” cluster (n=83/223 total DN1d cells) contain DN1d cells with the Tγδ17 gene signature. (C) PCoA of genes that define cell clusters depicted in Panel B, with genes most strongly associated with the “Soxpro” DN1d cluster (low in PC2) indicated by gene symbol. (D)Violin plots of 47 genes analyzed.
While γδTCR was not detected on DN1d cell surface or in the cytoplasm (Figure S2B), most Soxpro cells expressed at relatively low levels rearranged Trgv4-Trgj1 (Vγ2-Jγ1), but not rearranged Trvd2–2 (Vδ4-Jδ1) or Trdv5 (Vδ5-Jδ1) genes (Figure 3D), suggesting that the TCRVγ repertoire of Tγδ17 cells may be programmed early in differentiation. The estimated frequency of rearranged Vγ2-Jγ1 gene, of which only ~20% are in frame (Kang et al., 1995), at the DNA level in DN1d cells (~25% of Vγ2+ T cells, Figure S2C–D) was largely concordant with the single cell transcriptome data. This rearrangement pattern was intact in Tcrd−/− mice that cannot generate functional γδTCR complex (see below), indicating programmed gene rearrangement rather than TCR selection as the mechanistic basis for the observed pattern. Three additional clusters of DN1d cells were also distinguished (designated A, B, and C; Figure 3A). Cluster A DN1d cells were characterized by a generic T cell lineage gene expression pattern, with variable transcription of core T cell-lineage genes and few cells with Sox13 and/or rearranged Vγ2 gene transcripts. Cluster B DN1d cells also expressed some T-lineage genes, but showed a notable coordinate expression of Il7r and Id2, both markers of early innate lymphoid cell (ILC) precursors and other immature γδ T cell subsets (Figure S2E). Lastly, a separate cluster of cells (Cluster C) expressed some or all of the myeloid lineage genes (Cdh1, Csf2rb2, Gp49a, Spi1) but not T cell lineage defining genes, including Sox13. By comparison, single DN2 cell gene expression patterns indicated relatively uniform T-lineage potential. Consistent with FACS and population RNA-seq analyses, most Tγδ17 genes were not expressed in single DN2 cells (Figure 3B, D). Together, these data indicated that the DN1d subset as currently identified was heterogeneous, with very few if any cells exhibiting a multi-primed lymphoid-myeloid gene expression pattern.
Generation of Tγδ17 cells from DN1d progenitors
Previous studies have shown that DN1d cells can generate αβ lineage (i.e. DP) cells in in vitro cultures on stromal cell lines transduced with Notch ligands (which promote T lineage fate), but lack the robust proliferative burst typical of canonical T cell progenitors such as DN1a/b cells (Porritt et al., 2004). However, gene expression profiling suggested that some DN1d cells were functionally programmed to become γδ T cells, in particular Tγδ17 cells. To directly test whether ECFP+ DN1d cells are developmentally programmed to generate specific T cell types, neonatal thymic precursor cell subsets were sorted by flow cytometry (Figure S3A) and used to reconstitute intact fetal thymic lobes in hanging drop organ cultures (hFTOC), which fosters development of T lineage cells in an environment more physiologically representative of cells developing in vivo. Consistent with previous work, limited DP development and proliferation were detected in total DN1d cell reconstituted hFTOC lobes (Figure S3B); however, ECFP+ DN1d cells generated a higher frequency of γδTCR+ T cells, compared to DN2 cells or ETPs, and the majority of DN1d-derived γδ T cells expressed the Vγ2 TCR associated with neonatal Tγδ17 cells, with a negligible fraction of Vγ1.1 TCR+ cells (Figure 4A). Parallel analyses of total DN1d cells from WT C57BL/6J (B6) non-reporter mice demonstrated that developmental tendencies of total DN1d cells were similar to those of ECFP+ DN1d cells, albeit with somewhat reduced efficiency of overall γδ T cell generation and an increased αβ lineage DP cell development (Figure S3B–C). Crucially, ~20% of Vγ2+ T cells from DN1d cells expressed CCR6 (Figure 4A), and the majority of CCR6+ cells expressed Scart2 (Figure S3B). Conversely, ETPs and DN2 precursors did not generate CCR6+ Tγδ17 cells, even after extended culture (Figure S3D), while DN1e cells did not reliably reconstitute hFTOC lobes (data not shown). As DN1d cells and DN2 cells generated significantly different numbers of total cells per thymic lobe during hFTOC (Figure 4A, bottom left), it was possible that total cellularity and cell composition differences influenced developmental outcomes. To address this possibility, hFTOC were established in which individual thymic lobes were reconstituted with a 1:1 mix of CD45.2+ DN1d cells and CD45.1+ DN2 cells. Even under these conditions only DN1d cells generated CCR6 Vγ2+ cells (Figure S3E), indicating that Tγδ17 developmental potential in the hFTOC setting was primarily progenitor cell intrinsic.
Figure 4. Soxpro cells generate Tγδ17.
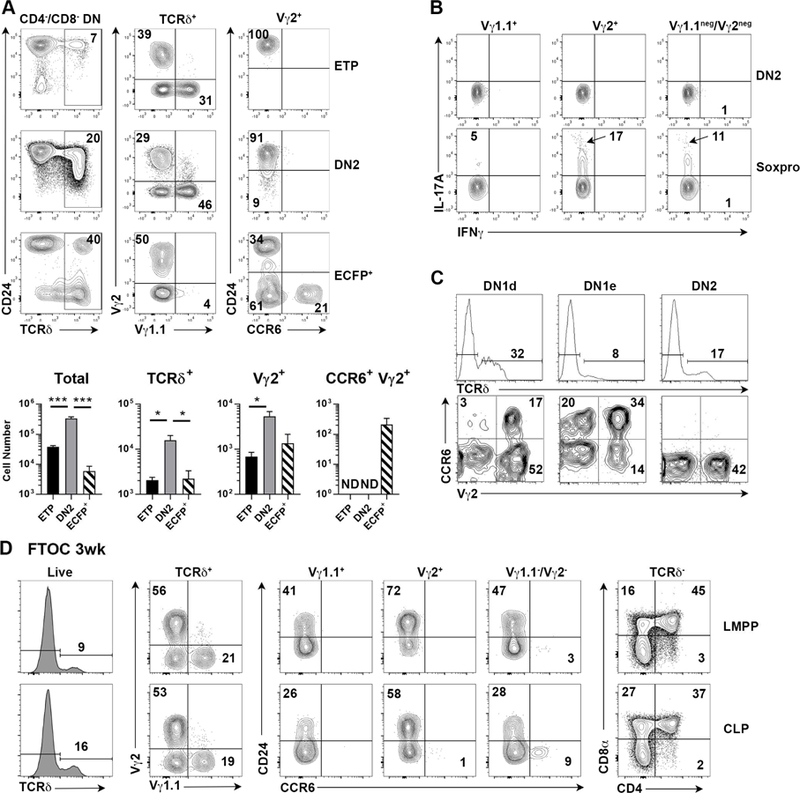
(A) Thymic lobes were seeded with ETP, DN2, or Sox13-ECFP+ DN1d cells sorted from d7–10 old Sox13ecfp/= mice and then cultured for 7 days before analysis. Two to four repopulated lobes were pooled for analyses. Summary data combines three hFTOC experiments analyzed at 7–8 d of culture. *, p<.05; ***, p<.001 by one-way ANOVA with Turkey’s multiple comparisons test. ND, not detected. (B) DN2 or total DN1d progenitor cells were sorted from d7–10 old WT B6 thymi, used to repopulate alymphoid fetal thymic lobes by hanging drop culture, and then cultured under standard hFTOC conditions. After 1 wk of hFTOC culture, cells were stimulated with PMA and Ionomycin to assess production of IL-17A and IFNγ. Gates for cytokine+ cells were based on non-stimulated controls stained with anti-cytokine Abs. Data are pooled from 6 lobes. (C) Developmental potential of indicated precursor subsets sorted from d7–10 old Tcrb−/− mice was assessed on OP9-DLL1 stromal cells. Top row is gated on total live cells and bottom row is gated on TCRδ+ cells. Data are representative of one of three experiments. (D) Fetal liver progenitors from E13.5 embryos were sorted and used to repopulate hFTOC lobes. Analyses shown were performed on d20 of culture. Quadrants without an inset number indicate <1% cells. Data are representative of 2 experiments.
To confirm that DN1d cell progenies were indeed Tγδ17 cells, intracellular IL-17A was analyzed after brief in vitro stimulation of cells recovered from DN1d cell hFTOC lobes. IL-17 production was readily detected in Vγ2+ cells at a frequency strongly correlated with the frequency of CCR6+ cells. IL-17 production was also detected in the Vγ2-/Vγ1.1- subset (presumably Vγ4+ Tγδ17 cells, see below) (Figure. 4B, Figure S3F). In contrast, γδ T cells from DN1d cells did not produce IFNγ. Similar results were obtained when DN1d cells were analyzed in the OP9 stromal cell assay (Figure 4C, Figure S4A–B), even in the absence of Notch signals. Both CCR9+ and CCR9- DN1d cells generated Tγδ17 cells (Fig. S3A), albeit with different kinetics that require further characterization (data not shown). Development of CCR6+ Vγ2+ cells was enhanced when DN1d cells from GS2Tg mice (bearing a productively rearranged Vγ2 transgene) (Kang et al., 1998) were assessed (Figure S4C), suggesting Tγδ17 development may be restrained by poor in-frame Vγ2 rearrangement (Kang et al., 1995). However, DN2 precursor cells from GS2Tg mice did not generate CCR6+ γδ T cells, indicating that Vγ2 expression alone was insufficient to promote Tγδ17 differentiation.
Since DN1e cells did not reliably reconstitute hFTOC lobes, we used OP9 stromal cells to assess DN1e cell developmental potential. DN1e cells also generated CCR6+ Vγ2+ and Vγ2- cells in vitro (Figure S4A). As DN1e cells did not express Sox13 (Figure 1A, 2D) or CD24, the expression of which are decreased upon maturation in γδ thymocytes, we addressed the possibility that DN1e cells may be a product of DN1d cells. After 3d of hFTOC culture, over half of the cells in DN1d cell reconstituted lobes were Lin-, and the majority of these cells were DN1e cells (Figure S5A). These data suggest that some DN1e cells may be a downstream developmental intermediate of DN1d cells.
Unlike DN2 cells and the majority of conventional T cell precursors, DN1d cells do not appear to arise from ETPs (Benz et al., 2008; Porritt et al., 2004). Whether DN1d cells are derived from earlier lymphoid progenitors was unknown. Given the emergence of DN1d cells during fetal thymic development (Figure 1B–C), fetal liver T cell progenitors preceding ETPs, namely lymphoid-primed multipotent progenitors (LMPPs) and common lymphoid progenitors (CLPs, Figure S5B), were assessed for the ability to generate DN1d cells and Tγδ17 cells. While CD24+c-Kit- DN1d-like cells were variably observed from LMPP and CLP at different days of culture (Figure S5C ), neither progenitor types generated Vγ2+ Tγδ17 cells at any point during extended culture (up to 4 weeks, Figure 4D, Figure S5D–E), though CCR6+ Vγ1.1-/Vγ2- cells did develop by 3wk of culture, consistent with the fetal ontogeny of Vγ4+ Tγδ17 cells. Both LMPPs and CLPs robustly generated other γδ T cell subsets and αβ T cells. While we cannot exclude the possibility that some additional extrathymic priming of LMPP and/or CLP is necessary to foster Vγ2+ Tγδ17 developmental potential, these data suggested that fully mature Vγ2+ Tγδ17 and bona fide Soxpro cells may not follow the predominant lymphopoietic developmental pathway.
If not from the conventional lymphopoietic pathway we explored the possibility that Tγδ17 cells may originate from early embryonic hematopoietic tissues. We first tested whether some E9.5–10.5 yolk sac (YS) cells, the principal site of embryonic hematopoiesis (Boiers et al., 2013), had requisite properties of Tγδ17 cell progenitors. YS cells had intracellular SOX13-ECFP reporter and a small subset expressed the T cell lineage HMG TF TCF1 (Figure S6A). SOX13 reporter expression was diminished in E11.5 fetal liver (FL) cells, one day after the time when the classical hematopoietic stem cells (HSCs) are reliably detected. Initial studies using the OP9 culture suggested that YS cells can generate Vγ2+ Tγδ17 cells (data not shown). To verify this in a more physiological setting we performed YS-hFTOCs. E10.5 YS cells variably produced the first wave of fetal γδ T cells that express Vγ3 TCR (in ~ a third of individual hFTOC) but not postnatal Vγ1.1+ γδ T cells. Instead, the γδ T cell progeny in all FTOCs expressed Vγ2+ and/or Vγ4+ TCR. Among these, Tγδ17 cells were observed, although their generation was efficient in ~1/3 of the lobes (Figure S6B and data not shown). These results supported the possibility that Tγδ17 cells may originate from embryonic hematopoietic tissues, rather than the conventional lymphopoietic pathway. A definitive demonstration waits physiological testing of YS developmental potential in utero and/or improved culture system to favor lymphopoiesis from defined YS cell subsets.
DN1d cells are generated and lineage-programmed independently of γδTCR signaling
A prevailing model of T cell lineage diversification posits that specific γδTCR signals initially instruct the development and functional programming of Tγδ17 cells. To definitively test this paradigm we systematically investigated the role of TCR in establishing the DN1d identity. TCR γ, δ, or β were not detected on the cell surface or intracellularly (Figure S2B, S7A), however some Tcrg and Tcrd gene rearrangements were observed in DN1d cells (Figure S2C–D), raising the possibility that DN1d cells had silenced TCR expression, but their generation may have been dependent on TCR. This possibility was systematically tested. DN1d cells were generated in TCR transgenic mice with ectopic early expression of rearranged TCRβ or TCRγ chains (Figure S7B), and Tcrd−/−, Tcrb−/−, or Ptcra−/− mice produced similar frequencies of DN1d precursors as wild-type (WT) mice (Figure 5A, H.J. Felhing, personal communications). Predictably, Sox13 expression was normal in Tcrb−/−DN1d cells, and Vγ2+ IL-17+ cell numbers were normal in the Tcrb−/− thymus (Figure S7C), indicating that αβTCR is dispensable for DN1d generation.
Figure 5. DN1d cell generation is independent of maximal TCR signaling or RAG.

(A) DN1 subsets from d7–10 old mice of the indicated genotypes were analyzed by FACS. Data are from one of three independent experiments. (Tcrd−/− N = 11 mice, Tcrb−/− N = 8). (B) DN1 subset analysis from 6 weeks old WT B6 or 6F/6F mice. (C) Representative intranuclear staining for RORγt and BLK in ETP and DN1d cells from 6wk old WT B6 and 6F/6F mice. (D) Summary of total DN1d cell numbers and RORγt+ BLK+ DN1d cell numbers in 2wk old and 6wk old WT and 6F/6F mice. 6wk n=16 (WT) or 18 (6F/6F); 2wk n=6 each. (E) Analysis of Rag1−/− DN1 subsets from E16.5, 17.5, and 18.5 fetuses and <24h old Neonates (Neo), top to bottom row, respectively. E16.5 n=3 (WT) or 5 (−/−); E17.5 n=10 (WT) or 8 (−/−); E18.5 n=8 (WT) or 7 (−/−); Neo n=4 each. * p<.05, ** p<.01, *** p<.001 by unpaired t-test. (F) FACS analysis of RORγt, BLK, and Scart2 expression by DN1 subsets of E18.5 Rag1−/− mice. Data are representative of 3 independent experiments. (G) Neonatal (<48h old) Rag1−/−c-Kitneg DN1 progenitors were sorted and analyzed by single-cell qPCR. Rag1−/− cells were segregated into Sox13+ (n=25) and Sox13− (n=198) cells and then compared against WT Soxpro from Figure 3 via hierarchical clustering. (H) Violin plot analysis of gene expression of cells from Panel F.
We next assessed TCR signaling requirement for DN1d cell development. For this we used mice with signaling-defective CD3ζ chains, in which the six phosphorylation-targeted tyrosine (Y) residues are replaced by phenylalanine (F), such that T cells from these 6F/6F mice only have approximately 40% signaling capacity of WT T cells (Hwang et al., 2012). Consistent with a requirement for TCR signaling at some stage of Tγδ17 cell generation (Wencker et al., 2014), these mice produced significantly fewer Vγ2+ T cells, especially in the lymph nodes, and no Tγδ17 cells (Figure S7D). Though proportionally overrepresented, the numbers of Vγ1.1+ γδ T cells were normal in 6F/6F mice. The defect in immature (CD24+) Vγ2+ T cells were observed prior to the expression of CD73, a marker of TCR signaled cells (Coffey et al., 2014) with significantly diminished expression of Tγδ17 cell signature antigens, such as Scart2 and BLK, but also reduced expression of the IFNγ-producing γδ T cell associated marker CD27 (Figure S7E). Critically, DN1d cells expressing the Tγδ17 signature proteins RORγt and BLK were generated and maintained in 6F/6F mice (Figure 5B-D). These results indicate that normal TCR signaling was not required for Tγδ17-programmed DN1d cell generation, but it was necessary at the earliest transition of DN1d cells into ImmV2 cells or in immature CD73- Vγ2+ Tγδ17 cells.
Given the requirement for normal cortical thymic epithelial cells (cTECs) for Tγδ17 cell development (Mair et al., 2015; Nitta et al., 2015) and the published defects in cTECs in Rag1−/− mice (Klug et al., 1998) it was possible that postnatal Rag1−/− mice do not support DN1d cell development. DN1d cells were generated normally in E16.5 Rag1−/− thymus, consistent with the lack of TCR requirement for their generation, and were numerically increased at E17.5 and E18.5, but appeared to lose the expression of CD24 at birth (Figure 5E and see below). DN1e cells exhibited a similar trend as DN1d cells, except were further increased at birth, while ETP cell numbers were identical at each age. Given this we predicted that Sox13+ cells would nevertheless be present in total c-Kitneg DN1 (combined DN1d and DN1e) cells in postnatal Rag1−/− thymuses. We analyzed postnatal progenitors to most appropriately permit comparison to the original single-cell profiling performed on post-natal DN1d cells of WT mice (Figure 3). Single-cell transcriptome analysis of Rag1−/− c- Kitneg DN1 progenitors identified ~11% of cells (n=25/223) that expressed Sox13 with concurrent expression of molecular hallmarks of Soxpro/Tγδ17 cells, including expression of Rorc, Blk, and/or Maf, which were confirmed by FACS (Figure 5F–H). A reduced frequency of BLK+ cells was observed (Figure S7F), which may be related to reduced levels of Sox13, which binds to Blk transcriptional control elements and directly regulate its expression (Malhotra et al., 2013). This frequency of Soxpro cells in Rag1−/− was within the range (~15–18%) found in c- Kitneg DN1d+e cells of WT mice. Interestingly, many (15–20%) c- Kitneg DN1 cells (Sox13+ and Sox13-) expressed high levels of Rorc, Id2, Il7r, Zbtb16, and Nfil3 (Figure 5G), suggesting an ILC-like phenotype that may correspond to the IL-17+ ILC3-like effectors in the Rag1−/− thymus (Haas et al., 2012).
Finally, to rigorously test the role of γδTCR on the DN1d precursor gene expression signature we determined single cell molecular properties of Tcrd−/− DN1d cells. Tcrd−/− mice have normal αβ T cell development and cTECs, thus removing the confounding parameter of Rag1−/− mice. Single-cell gene expression analysis demonstrated that Tcrd−/− DN1d cells were nearly identical to WT Soxpro cells, including expression of all core Tγδ17 genes examined (Figure 6A–B). Further, these DN1d cells that cannot produce γδTCR complex rearranged and expressed Tcrg and Tcrd genes even to a greater extent than WT DN1d cells (Figure 6B, third row rightmost column and fifth row second from left column), strongly indicating that the TCR gene rearrangements observed in WT DN1d cells are genomically programmed rather than selected based on functional TCRs. Consistent with single-cell analysis, Tcrd−/− DN1d cells maintained uniform expression of SOX13-ECFP reporter as WT cells (Figure 6C), and expressed the signature Tγδ17 markers Scart2, RORγt, BLK and MAF (Figure 6D-E and data not shown). Additionally, nearly 2-fold more DN1d cells were recovered from Tcrd−/− thymuses compared to B6 controls (Figure 6F), indicating that DN1d cells can accumulate when they are blocked from assembling the γδTCR. Together, these findings prove that lineage programmed DN1d Soxpro cells develop independent of γδTCRs.
Figure 6. γδTCR is dispensable for DN1d cell generation and Tγδ17 gene programming.
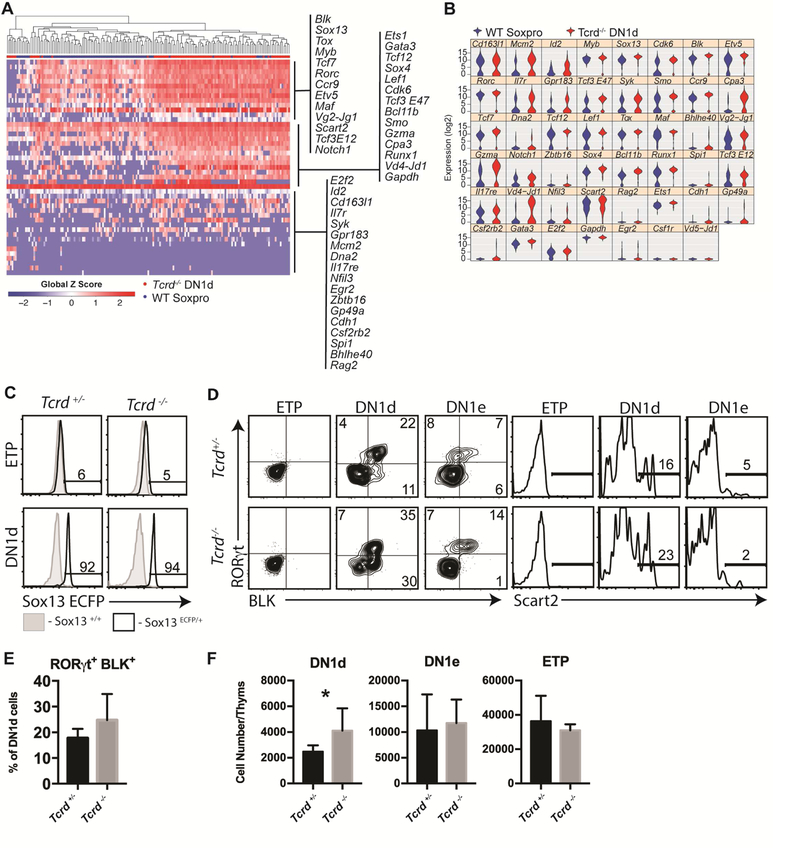
(A) DN1d cells from d7–10 old Tcrd−/− thymi were sorted and analyzed by single-cell qPCR (n=86). Data are visualized by hierarchical clustering and displayed by heat map rendering of global Z scores and compared to WT Soxpro from Figure 3. (B) Violin plot of single-cell qPCR data presented in Panel A. Note that Tcrd−/− mice are deficient in the constant region, but do not exhibit any deficiencies in VDJ rearrangement. (C) Sox13-ECFP reporter signal was assessed in ETP or DN1d cells from Tcrd+/−Sox13 ecfp/+ and Tcrd−/−Sox13ecfp/+ littermates. Histograms are representative of 6 Tcrd+/−Sox13 ecfp/+ mice analyzed across three independent experiments. (D) Analysis of Scart2, RORγt, and BLK expression by DN1 progenitors from Tcrd−/− mice. (E) Summary data of n=4 mice assessed in Panel D. Data are from 1 of 3 representative experiments. (F) Enumeration of total DN1d cells, DN1e cells, and ETP cells isolated per thymus from Tcrd+/− or Tcrd−/− mice. n=5 (+/−) or 7 (−/−) pooled from 2 independent experiments. *, p<.05 by unpaired t test.
SOX13 and TCF1 are required for DN1d cell generation and functional programming
SOX13, a TCR signaling-independent TF, and the HMG TF TCF1 (encoded by Tcf7) coordinately regulate Tγδ17 differentiation (Gray et al., 2013; Malhotra et al., 2013). To determine whether this requirement was at the level of Soxpro cells genetic studies were performed to determine factors necessary for DN1d precursor generation. Persistence of DN1d cells per se was not dependent on Sox13 alone as mice lacking Sox13 had normal DN1d cell frequency and number (Figure 7A). Next, mice with compound deficiencies in Sox13 and Tcf7 were analyzed. Sox13+/+Tcf7−/− mice had a reduced frequency, but not numbers, of DN1d cells. However, both Sox13−/−Tcf7+/− and Sox13+/−Tcf7−/− mice had sharply reduced DN1d cell numbers, and Sox13−/−Tcf7−/− had a further reduction in DN1d cell numbers, though this reduction was not statistically significant by comparison to Sox13−/−Tcf7+/− or Sox13+/−Tcf7−/− mice (Figure 7B–C). These results indicated TCF1 and SOX13 were required for normal DN1d cell generation. We next assessed whether Sox13 itself is required for the differentiation of Soxpro cells to Tγδ17 cells. BLK expression was severely reduced and RORγt was expressed at a similar overall frequency but at reduced levels in Sox13−/− DN1d and DN1e cells (Figure 7D). These data indicated that effector lineage programming was deficient in the absence of Sox13, though some RORγt can be induced, most likely by SOX4 as previously shown for immature γδ thymocytes (Malhotra et al., 2013). Furthermore, Sox13−/− DN1d cells did not generate CCR6+ Tγδ17 cells in hFTOC assay (Figure 7E). These results indicated that TCF1 and SOX13 coordinately regulated Soxpro generation and differentiation of Soxpro cells into Tγδ17 cells.
Figure 7. HMG TFs regulate DN1d and Tγδ17 functional programming.
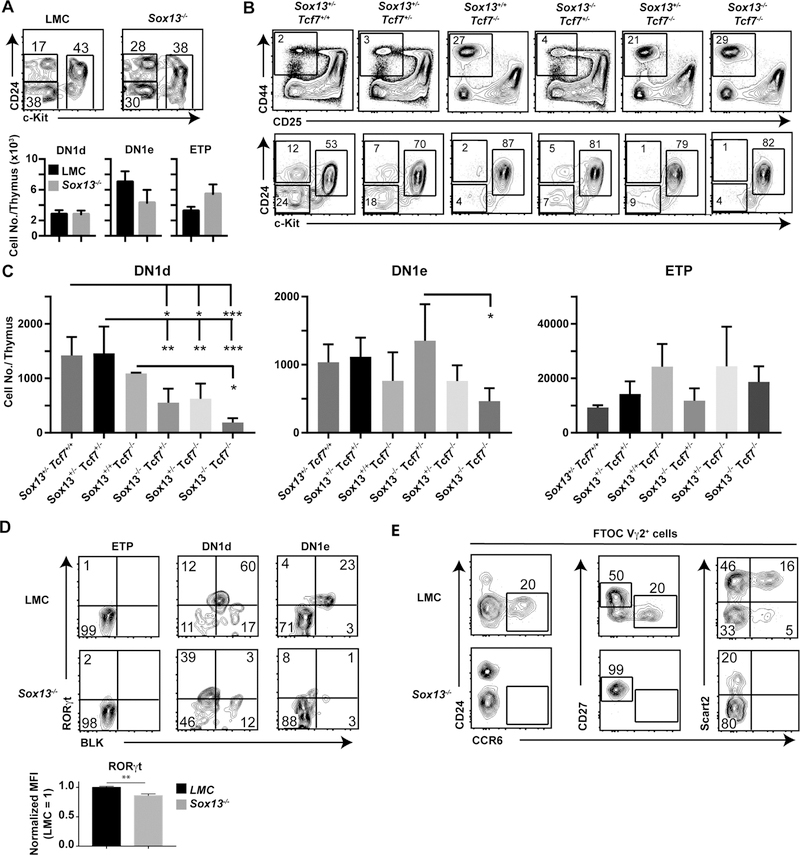
(A) Analysis of DN1 progenitor distribution in d7–10 old littermate control 129.Sox13+/− (LMC) and 129.Sox13−/− mice (N=9 each). (B) Representative DN subset profiles (Top) and DN1 subset distributions (Bottom) in neonatal mice (<48h old) lacking Sox13 and Tcf7. Analysis was performed in neonates prior to ectopic activation of T cells observed in young Tcf7−/− mice. (C) Summary of analysis performed in Panel B. Sox13+/−Tcf7+/+ n=4, Sox13+/−Tcf7+/− n=5, Sox13+/+Tcf7−/− n=2, Sox13−/−Tcf7+/− n=4, Sox13+/−Tcf7−/− n=5, Sox13−/−Tcf7−/− n=4 pooled from 2 independent experiments. * p<.05, ** p<.01, *** p<.001 by one-way ANOVA with Turkey’s multiple comparisons test. (D) Representative RORγt+BLK+ intracellular profiles in mice (d7–10 old) deficient for SOX13. Summary data n=6 (LMC) or7 (−/−) from 2 independent experiments expressed as “normalized MFI” in which LMC mice were normalized to 1 to permit comparison across experiments. **, p<.01 by unpaired t test. (E) Differentiation of DN1d cells sorted from d7–10 old LMC or Sox13−/− mice in hFTOC analyzed at d7 of culture. Shown are phenotypes of Vγ2+ T cells. Data are representative of three independent experiments. Quadrants and gates without an inset number indicate <1% cells in gate.
Discussion
Evolution of animal immune systems involved the emergence of innate, mucosal tissue-tropic and adaptive-like lymphatic lymphocyte lineages in addition to a subset that produces soluble antigen-sensing receptors in the last common ancestor to jawed and jawless fish ~560 million years ago (Hirano et al., 2013). While the immune systems of jawed and jawless fish diverged and became equipped with distinct clonal antigen sensory receptors, genetic programming of lymphoid effector subtypes appears conserved, with the SOX-TCF HMG network already established in the γδ T cell- like VLRC+ lymphocytes (SOXhi) and αβ T cell-like VLRA+ lymphocytes (TCFhi) of the jawless fish lamprey (Hirano et al., 2013). Our data further support the model that a gene network-driven lymphocyte effector lineage diversification preceded acquisition of distinct sensory antigen receptor complexes that have been used to classify lymphocyte types. Interestingly, ILC equivalents have yet to be identified in jawless fish, and their evolutionary relationship to T cell equivalents remains to be determined.
Since the initial identification of a second TCR composed of γδ heterodimeric chains, and the distinct immunological role of T cells bearing the γδTCR, the relative impact of TCR signals versus other developmental cues such as Notch and WNT signaling in directing progenitor cell lineage fate has been challenging to resolve (Narayan and Kang, 2010). This debate was reinvigorated by the discovery that many γδ T cells exit the thymus already programmed for peripheral effector functions (Narayan et al., 2012; Ribot et al., 2009; Shibata et al., 2008). The pattern of distinct intrathymic effector γδ T cell subsets extends to αβTCR+ iNKT and MR1-restricted mucosal- associated invariant T cell lineages (Engel et al., 2016; Lee et al., 2013; Rahimpour et al., 2015). This developmental complexity partly accounts for discrepancies in data interpretations from studies of innate T cell development that rely on the expression of distinct TCR type as the sole cell identifier. Moreover, restricted generation of some innate T cell subsets in ontogeny (Grigoriadou et al., 2003; Haas et al., 2012; Yuan et al., 2012), tailored to their postulated critical function early in life, necessitated reevaluation of developmental requirements for these cells established primarily from studies of adult mice.
Through the use of SOX13 reporter mice, developmental assays, and single-cell transcriptomics, we have demonstrated that the Tγδ17 cell fate was preprogrammed in DN1d progenitor cells that were first detected in E16.5 thymus. Analysis of DN1d cells in Tcrd−/− mice unequivocally demonstrated an intact Tγδ17 molecular signature, providing clear support for a central prediction of the pre-programming (“stochastic”) model of T cell lineage diversification. That Tcrvg and Tcrvd rearrangements remained enriched in Tcrd−/− DN1d cells that cannot be selected by γδTCR signaling also indicated that these rearrangements are molecularly targeted during developmental programming, potentially accounting for some of the restricted TCRVγ usage observed by distinct γδ T cell effectors.
While γδTCR signaling was dispensable for DN1d cell generation or Tγδ17 gene network programming, T cells do not develop without TCR and it was important to pinpoint the γδTCR-dependent developmental checkpoint(s) for Tγδ17 cells. Our results showed that TCR signaling is first required to generate Tγδ17-skewed immature Vγ2+ thymocytes from pre-programmed DN1d cells. Whether this signal was mostly trophic, akin to preTCR for αβ T cells, or it sustained the initial genetic programming has not been determined. γδTCR may need to signal again during the generation of mature Tγδ17 thymocytes from the immature TCR+ state as the second TCR-dependent maturation checkpoint (Coffey et al., 2014), and transcriptome changes associated with Tγδ17 maturation overlap with that of the maturation gene signature of single-positive CD4+ and CD8+ αβ thymocytes that are controlled by αβTCR (Narayan et al., 2012; Park et al., 2010).
We had previously shown that Vγ2+ Tγδ17 cells could be generated from adult c-Kit+ DN thymocytes using the OP9 stroma (Malhotra et al., 2013). But using more physiologically relevant hFTOCs that can provide the requisite thymic epithelial cells no significant Vγ2+ Tγδ17 cell potential was observed from neonatal or adult c-Kit+ DN thymocytes or FL LMPPs or CLPs. This result suggested that either the Vγ2+ Tγδ17 cells generated from adult thymic precursors in vitro represent the “induced” adult Tγδ17 cells (Buus et al., 2017) and/or the OP9 cultures can promote ectopic Tγδ17 cell differentiation from adult thymocytes in the presence of high NOTCH signaling. Among γδ T cells Vγ2+ Tγδ17 cell development in neonates is highly sensitive to undefined properties of thymic epithelial microenvironments (Mair et al., 2015; Nitta et al., 2015) and caution must be exercised ininterpreting results from in vitro stromal cultures.
We identified developmental steps in the generation of innate Tγδ17 cells, but heterogeneity within c-Kit- DN1 cells and its implication for the generation of other lymphoid subtypes in the thymus remain to be established. As most DN1d cells are marked with SOX13-ECFP expression, yet only ~40% actively express Sox13 transcript, it seems likely that most DN1d cells derive from a common SOX13hi state, with some cells retaining the ECFP reporter despite ceasing Sox13 transcription. This pattern is akin to other DNA associated nuclear reporters that are retained without ongoing transcription, a property that has been used to mark stem/progenitor cells (Wilson et al., 2008). These data, combined with the aberrant DN1d cells in Sox13−/− mice, also indicated that, at least at the DN1d stage, ongoing high levels of Sox13 transcription were needed to maintain and fullyenforce Tγδ17 programming.
Extensive molecular heterogeneity was also evident for DN1e cells based on ICS flow cytometry and single cell RNAseq analysis (N.A.S., J.K., and the Immunological Genome Project Consortium, unpublished). Currently, a lack of tools to segregate DN1e subtypes and the inefficiency with which DN1e cells populate hFTOCs severely limited the ability to determine whether most DN1e cells are progeny of DN1d cells or derived from independent developmental pathways. Further, gene expression patterns of Sox13 transcript-negative DN1d and DN1e cells supported the possibility that other innate αβ and γδ T cell subsets and ILCs can also originate from c-Kit− DN1 cells and the enhanced Type 3 cytokine signature in Rag1−/− cKit− DN1 cells suggest that TCR signals limit the generation of previously observed ILC3-like cells in the thymus that may originate from cKit− DN1 cells (Haas et al., 2012).
Collectively, our data demonstrated that the lineage identity of the innate Tγδ17 subset is predetermined at the progenitor level before expression of and input from γδTCR in the thymus. Our findings predict that DN1d cells, which do not express Rag genes, arose from progenitors that at some point transiently expressed RAG. That these innate T cells arise from Rag1/2+ progenitors that have some TCRγ and/or δ gene transcription and rearrangements has parallels in innate NK and ILC development (Robinette et al., 2015; Veinotte et al., 2006; Yang et al., 2011), and early embryonic progenitors with lymphoid potential can express Rag genes (Boiers et al., 2013; de Andres et al., 2002). Identification of fetal preprogrammed Tγδ17 cell progenitors now permits a rational search and identification of prethymic embryonic sources of the T cell progenitors. There were discrete Tcf7+ or Sox13+ YS and aorta-gonad-mesonephros hemogenic endothelial cells at E8.5-E11.5, and initial analysis of E10.5 YS cell developmental potential indicated that these cells were biased to generate Vγ2+ and Vγ4+ Tγδ17 cells. Whether the pre-HSC embryonic hematopoiesis is programmed to generate lymphocytes such as Tγδ17 cells necessary for mucosal immunity, particularly for fetuses and newborns, is a central issue to be solved.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Joonsoo Kang (Joonsoo.kang@umassmed.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
129/Sv.Sox13−/−, 129/Sv.GS2 (Vγ2TCR) Tg, C57BL/6J.Cd2476F/6F, C57BL/6J.Cd2476Y/6Y,C57BL/6J.Tcrb−/−, C57BL/6J.Tcrd−/−, C57BL/6J.Rag1−/−, C57BL/6J.Tcf7−/−, 129.Sox13Ecfp/+, and C57BL/6J.Il17a egfp/egfp (Jackson Laboratories Stock #018472) mice were generated and/or maintained in-house in a specific pathogen-free barrier facility. The Sox13−/− mice used in this study were maintained on a 129/Sv genetic background and are normal in fertility and growth (Malhotra et al., 2013; Melichar et al., 2007). For RNA-seq analyses, C57BL/6 (B6) mice were purchased from Charles River for cell sorting. Males and females (B6 background) were used for experiments, but sex-matched within an experiment. No differences were observed between sexes. Ages of mice used for experiments are indicated in Figure Legends. Animals were randomly allocated to797experimental groups. All experiments performed were approved by the University of Massachusetts Medical School IACUC.
METHOD DETAILS
Antibodies (Abs) and Flow Cytometry
Flow cytometry staining was performed in 96-well microtiter plates. Antibody cocktails were diluted in PBS (Gibco) + 0.5% BSA (Fisher Scientific) or 2% FBS (Sigma-Aldrich), and cells were stained in 50 μl for 20 min at 4°C. Abs to the following cell surface markers were purchased from BD Biosciences, BioLegend, or eBioscience: Lineage markers [anti-CD3ε (145–2C11), anti-CD8α (53–6.7), anti-CD11b (M1/70), anti-CD11c (N418), anti-CD19 (1D3), anti-GR1 (RB6–8CS), anti-NK1.1 (PK136), anti-CD49b (DX5), anti-TER-119 (TER-119), anti-TCRβ (H57–597), and anti-TCRδ (GL3), all of which were used as Biotin conjugates except anti-TCRδ, which was used in a separate channel to permit rigorous exclusion of γδTCR+ cells (see Figure S3A for example gating scheme)] , anti-CD4 (RM4–5), anti-CD16 (2.4G2), anti-CD25 (PC61, IL-2Rα), anti-CD24 (heat-stable antigen, HSA; M1/69), anti-CD27 (LG.3A10), anti-CD44 (IM7), anti-CD45.2 (104), anti-CD49b(DX5), anti-CCR6(140706), anti-c-Kit (CD117, 2B8), anti-Sca1 (D7), anti-TCRδ (GL3), IL-7Rα/CD127 (A7R34),streptavidin PerCp-Cyanine5.5, streptavidin APC-eFluor780, and streptavidin BB515. Anti-Vγ2(UC3–10A6) was purchased from Leinco Technologies or eBioscience. Anti-Vγ1.1 (2.11) was purchased from Biolegend. Intracellular IL-17A (17B7; eBioscience) and IFN-γ (XMG1.2; BD Biosciences) were detected with the Cytofix/Cytoperm kit (BD Biosciences). Intranuclear staining was done with the Foxp3 Staining Kit (eBioscience) with the following antibodies: anti-BLK (3262), anti-TCF1 (C46C7), anti-LEF1 (C18A7; all from Cell Signaling), anti-RORγt (AFKJS-9, eBioscience) and anti-cMAF (sym0F1, eBioscience). Intracellular detection of TCRδ and TCRγ was performed following fixation in 4% paraformaldehyde (Electron Microscopy Sciences) + 0.01% Tween-20 in FACS Buffer (PBS + 0.5% BSA + 2mM EDTA) and permeabilization by 0.1% Triton X-100 (Sigma-Aldrich) in FACS Buffer. Anti-rabbit IgG Ab from Life Technologies was used as the secondary reagent for LEF1, TCF1 and BLK detection. Characterization of anti-Scart2 Ab (25A2) has been reported (Kisielow et al., 2008). Anti-rat IgG (mouse cross-absorbed) from Southern Biotechnology Associates was used as a secondary reagent for Scart2 detection. Dead cells were excluded from all analyses using propidium iodide (PI), fixable LIVE/DEAD cell stains (both Life Technologies), or 7-aminoactinomycin D (7-AAD, BD). For FACS purification of DN progenitors, DN cells were enriched by depletion using anti-CD8 Dynabeads (ThermoFisher) following the manufacturer’s recommendations, and then CD8-depleted thymocytes were first gated strictly on TCRδ- prior to gating on Lin-/lo. For intracellular cytokine staining, cells were stimulated in vitro with 10 ng/mL phorbol 12-myristate 13-acetate (PMA) + 500 ng/mL Ionomycin (both Sigma-Aldrich) in the presence of GolgiStop and GolgiPlug (BD Biosciences) for 3 hours, surface stained, LIVE/DEAD labeled, fixed/permeabilized, and then stained for indicated intracellular cytokines. Data were acquired on a BD LSRII cytometer or FACSAria (BD Biosciences) and analyzed using FlowJo (Treestar). For “optimal ECFP detection” of Sox13 reporter signal on the FACSAria, the ECFP bandpass filter (470/20) was placed in the “A” (i.e. first to receive) detector without a dichroic mirror.
OP9 cell culture
ETP (ckit+CD44+CD25−Lin−), DN1d (ckit−CD44+CD25−CD24+Lin−), DN1e (ckit−CD44+CD25−CD24−Lin−),and DN2 (ckit+CD44+CD25+Lin−) cells were sorted from B6, B6:Tcrb−/− or B6:TCRγ transgenic mice and plated onto OP9-DLL1, OP9-DLL4 or control OP9-GFP vector alone (no DLLs) transduced monolayers (made by J.C. Zuniga-Pflucker and provided by A. Bhandoola). Most assays were performed using precursor subsets sorted from Tcrb−/− mice for optimal cell yield and aside from higher frequencies of TCRδ+ cells generated all other phenotypic parameters of progeny were similar to those tracked with B6 precursors. Test subsets were plated at various numbers (routinely 5 × 103 to 2 ×104) per well in αMEM media (Gibco) containing 20% FBS (Hyclone),1ng/ml IL-7 (eBioscience) and 5ng/ml Flt3L (eBioscience). Varied input cell numbers within the tested range did not significantly impact observed developmental potentials. Cells were analyzed by flow cytometry 7–12 days post culture.
Fetal thymic organ culture (FTOC)
Fetal thymic lobes were isolated from embryos of gestational day 15 timed pregnant B6 dams and cultured for 5–6 d on 0.8 μm isopore membranes (Millipore) atop gelfoam sponges (Pfizer) floating in DMEM-10 [DMEM, high-glucose (Life Technologies) + 10% FBS (Sigma-Aldrich), 10 mM HEPES,4mM L-Glutamine, 1 mM Sodium Pyruvate, 1x non-essential amino acids, 100 U/mL Penicillin, 100 μg/mL Streptomycin, and 55 μM 2-mercaptoethanol (all media supplements purchased from Gibco)] containing 1.35 mM 2’-deoxyguanosine (2-dG, Sigma-Aldrich) to deplete endogenous lymphocytes. The night before cell sorting and repopulation by progenitors, media was replaced with fresh DMEM-10 without 2-dG to wash residual 2-dG that otherwise may negatively affect progenitor repopulation and viability. To repopulate lobes, 20 μl hanging drop cultures containing 5 × 103 to 2 ×104 FACS-purified thymic progenitors and 1 thymic lobe per well were established in Terasaki plates (Greiner Bio-one) and cultured for 24 h. Repopulated lobes were then returned to FTOC (Day 0) as above but without 2-dG, and media was refreshed every 3–4 d. Due to the low yield from DN1d repopulated lobes, FTOC cells were often mixed with 0.5–1×106 CD45.1+ thymocytes to help normalize cell number per well and facilitate gating (FTOC cells identified as CD45.2+). Tests with DN2-derived FTOC cells demonstrated that this strategy did not affect results compared to FTOC-alone wells (data not shown).
YS-hFTOCs
Il17aegfp/egfp mice were used for timed mating. YS was digested with 5mg/mL Type IV collagenase (Worthington Biochemical Corporation) and 10mg/mL DNAse I (Roche) with 10%FBS at 37°C for 20 minutes and a single cell suspension was obtained by mechanical meshing through a 37 μM filter. 2 × 104 YS cells were plated on OP9-DL1 stromal cells with 1ng/ml IL-7 for 7 days. The primed YS cells were then harvested and without further purification used to repopulate alymphoid E14.5 thymic lobes (2×104 per lobe per Terasaki plate well) in hanging drops for 1 day and cultured for further 2–3 weeks under standard hFTOC conditions before analysis.
PCR Analysis
For analysis of genomic DNA (gDNA) TCR gene rearrangement, cells were sorted or isolated as indicated and then lysed overnight at 37°C in 50 mM Tris-Cl pH 8, 1% SDS, 25 mM EDTA, 250 µg/mL Proteinase K. DNA was isolated by phenol:chloroform extraction, chloroform extraction, and isopropanol precipitation and then solubilized in 10 mM Tris + 1mM EDTA buffer at 1000 cell equivalents per µL. PCR was performed using Taq Polymerase (ThermoFisher) under the following cycling conditions: 94°C, 3 min then 94°C, 30s; 55°C 30s; 72°C 60s for 32 cycles. Reaction products were immediately electrophoresed on a 1.5% agarose gel and visualized with ethidium bromide. The DN2.3 cell line is a TCRγδ+ thymoma with 2 Vγ2-Jγ1 rearranged alleles.
For traditional RT-qPCR analysis, cells were sorted directly into 500 μL TRIzol Reagent (Thermo Fisher Scientific) and RNA was isolated following the protocol recommended by the manufacturer. RNA was converted to cDNA using AffinityScript reverse transcriptase (Agilent) and oligo(dT)priming. Equivalent quantities of cDNA were used for qPCR employing the iQ SYBR Green master mix (Bio-Rad) and a CFX96 thermal cycler (Bio-Rad) with thermal melt curve analysis to assess specific amplification. Expression of Sox13 was analyzed by subtracting the Sox13 threshold cycle from the Gapdh threshold cycle, and then exponentiating this difference to the base 2.
RNA-seq
Libraries for RNA-sequencing were prepared using the NuGEN Ovation RNA-seq v2 library preparation kit (NuGEN Technologies Inc.). Briefly, 10,000 cells per subset were sorted (from 10 day old B6 mice) into the NuGEN Prelude Direct Lysis Buffer and total RNA was inputted directly into the first strand synthesis step of the Ovation RNA-seq v2 system. 1ng of SPIA amplified fragmented ds-cDNA was used to prepare libraries using the NuGEN Ovation Ultralow DR Multiplex System. Size distribution and qualitative assessment of libraries was performed using the Bioanalyzer DNA Chip 1000. Libraries were quantified using the KAPA Universal Library quantification kit prior to multiplexing and sequenced on the Illumina HiSeq platform. Sequenced reads were mapped to the mouse NCBI Ref-sequences using Bowtie. Differentially expressed genes (DEGs) were detected using the criteria of having an average RPKM count of at least 2.5 per sample, at least 2-fold change, and a p-value of <0.05 or 0.01 as indicated. The principal co-ordinate analysis plot was produced using edgeR’s plotMDS function using all differentially expressed genes detected from pairwise comparisons of DN1ab, DN1d, DN1e, and Immv2 as the input gene list. Volcano plots showing fold change versus p-value were generated using R. Hierarchical clustering and heat maps of DEGs and differentially expressed annotated transcription factors were generated using the GenePattern program of the Broad Institute.
Single-cell qPCR gene expression
Single-cell capture, RNA preparation, reverse transcription, and preamplification were performed using a C1 auto-prep system (Fluidigm). Thymic progenitors were FACS-purified in bulk and suspended at 6×105 cells/mL in RPMI 1640 + 20% FBS (sort collection media). The cell suspension was mixed at a 60:40 ratio with C1 suspension reagent and loaded into a 5–10 μM IFC for PreAmp.Capture of single cells was visually confirmed by phase-contrast microscopy. Sorted cells were loaded into the IFC and lysed within 1 hour of sort completion. Lysis, reverse transcription, and preamplification reagents were a combination of the C1 single-cell reagent kit for Preamp (Fluidigm) and the Single Cell-to-Ct Kit (ThermoFisher) as recommended by the manufacturer.Gene expression was assessed using a 48×48 Dynamic Array on a BioMark HD (Fluidigm, run by the High Throughput Gene Expression Biomarker Core), employing SsoFast EvaGreen detection chemistry (Bio-Rad). Primers for dynamic array were selected from PrimerBank (https://pga.mgh.harvard.edu/primerbank/), prior publications, or designed using PrimerBlast (optimal Tm 60°C, amplicon size 70–200bp, 1 primer must span an exon junction or be separated by an intron) and synthesized by Integrated DNA Technologies. Primer sequences are compiled in Table S5. Dynamic array quantification cycle (Cq) and pass vs. fail calling were determined automatically by the Fluidigm Real-Time PCR Analysis software using the Auto (Detectors) setting. Amplification and melt curves were manually inspected to confirm specific amplification, and genes hwith spurious melt peaks were recorded as negative. Exported Cq data was analyzed in R using the Fluidigm Singular Toolkit, first eliminating wells containing 0 or >1 cells and wells exhibiting no amplification. Remaining single-cell data was analyzed using the autoAnalysis function to generate hierarchical clustering maps, principal coordinate analysis, and violin plots. For correlation plots, exported Log2 Expression values (Log2Exp) were plotted and statistics calculated using GraphPad Prism. A total of 223 WT B6 DN1d, 78 WT B6 DN2, 86 Tcrd−/− DN1d, and 223 Rag1−/− c-Kitneg DN1 single cells were analyzed.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis of RNA-seq data was performed using R. Principal coordinate analysis and calculation of DEGs was performed in edgeR. Summary data from FACS analyses were analyzed in GraphPad Prism software using statistical tests indicated in Figure Legends. The mean of all samples in a group is used to represent the central tendency of the dataset, and all error bars represent standard deviation of biological replicates. Sample size was not determined prior to experimentation. No randomization of experiments was conducted. Experimenters were not blinded during performance or analysis of the experiments.
DATA AND SOFTWARE AVAILABILITY
The single-cell qPCR datasets have been deposited in Gene Expression Omnibus under accession number GSE118770. The bulk RNA sequencing datasets have been deposited in Gene Expression Omnibus under accession number GSE118924.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Hamster anti-mouse CD3ε Biotin | eBioscience | Cat#13–0033-85, clone 500A2 |
| Rat anti-mouse CD8α Biotin | BD Biosciences | Cat#553028, clone 53–6.7 |
| Rat anti-mouse CD11b Biotin | BD Biosciences | Cat#557395, clone M1/70 |
| Hamster anti-mouse CD11c Biotin | eBioscience | Cat#13–0114-82, clone N418 |
| Rat anti-mouse CD19 Biotin | eBioscience | Cat#13–0193-82, clone 1D3 |
| Rat anti-mouse Gr-1 Biotin | eBioscience | Cat#13–5931-82, clone RB6–8C5 |
| Mouse anti-mouse NK1.1 Biotin | BD Biosciences | Cat#553163, clone PK136 |
| Rat anti-mouse CD49b Biotin | eBioscience | Cat#13–5971-82, clone DX5 |
| Rat anti-mouse Ter-119 Biotin | BD Biosciences | Cat#553672, clone Ter-119 |
| Hamster anti-mouse TCRβ Biotin | eBioscience | Cat#13–5961-85, clone H57–597 |
| Hamster anti-mouse TCRδ BV421 | Biolegend | Cat#118120, clone GL3 |
| Hamster anti-mouse TCRδ PerCP-eFluor 710 | eBioscience | Cat#46–5711-82, clone GL3 |
| Hamster anti-mouse TCRδ APC | eBioscience | Cat#13–5711-82, clone GL3 |
| Hamster anti-mouse Vγ2 Biotin | Leinco Technologies | Cat#T211, clone UC3–10A6 |
| Hamster anti-mouse Vγ2 PE-Cy7 | eBioscience | Cat#25–5828-80, clone UC3–10A6 |
| Hamster anti-mouse Vγ1.1 FITC | eBioscience | Cat#141104, clone 2.11 |
| Rat anti-mouse CD24 BV605 | Biolegend | Cat#101827, clone M1/69 |
| Rat anti-mouse CD24 eFluor 405 | eBioscience | Cat#48–0242-82, clone M1/69 |
| Rat anti-mouse CCR6 Alexa Fluor 647 | BD Biosciences | Cat#557976, clone 140706 |
| Hamster anti-mouse CD27 V450 | BD Biosciences | Cat#561245, clone LG.3A10 |
| Rat anti-mouse CD4 V500 | BD Biosciences | Cat#560782, clone RM4–5 |
| Rat anti-mouse CD8α APC-Cy7 | BD Bioscience | Cat#557654, clone 53–6.7 |
| Rat anti-mouse CD44 PE | eBioscience | Cat#12–0441-81, clone IM7 |
| Rat anti-mouse CD44 FITC | BD Biosciences | Cat#553133, clone IM7 |
| Rat anti-mouse CD44 BV711 | Biolegend | Cat#103057, clone IM7 |
| Rat anti-mouse CD25 PE-Cy7 | BD Biosciences | Cat#552880, clone PC61 |
| Rat anti-mouse CD25 Alexa Fluor 700 | eBioscience | Cat#56–0251-80, Clone PC61 |
| Rat anti-mouse c-Kit APC | eBioscience | Cat#17–1171-82, clone 2B8 |
| Rat anti-mouse c-Kit PE | eBioscience | Cat#12–1172-81, clone ACK2 |
| Mouse anti-mouse CD45.2 APC-eFluor 780 | eBioscience | Cat#47–0454-82, clone 104 |
| Rat anti-mouse Sca-1 PE-Cy7 | eBioscience | Cat#25–5981-81, clone D7 |
| Rat anti-mouse Flt3 PE | eBioscience | Cat#12–1351-81, clone A2F10 |
| Rat anti-mouse CD127 BV421 | Biolegend | Cat#135027, clone A7R34 |
| Rat anti-mouse IL-17A PE | eBioscience | Cat#12–7177-81, clone ebio17B7 |
| Rat anti-mouse IFNγ PE-Cy7 | BD Biosciences | Cat#557649, clone XMG1.2 |
| Rabbit anti-mouse BLK | Cell Signaling Technology |
Cat#3262, Rabbit Polyclonal |
| Rabbit anti-mouse TCF1 | Cell Signaling Technology |
Cat#2206, clone C46C7 |
| Rabbit anti-mouse LEF1 | Cell Signaling Technology |
Cat#2286, clone C18A7 |
| Rat anti-mouse RORγt PE | eBioscience | Cat#12–6988-80, clone AFKJS-9 |
| Mouse anti-mouse c-MAF PerCP-eFluor 710 | eBioscience | Cat#46–9855-41, clone sym0F1 |
| Rat anti-mouse Scart2 | J. Kisielow | Cloned hybridoma 25A2 |
| Streptavidin Brilliant Blue 515 | BD Biosciences | Cat#564453 |
| Streptavidin BV605 | BD Biosciences | Cat#563260 |
| Streptavidin PerCP-eFluor 710 | eBioscience | Cat#46–4317-82 |
| Goat anti-Rabbit IgG Alexa Fluor 647 | ThermoFisher | Cat#A21245, Goat Polyclonal |
| Goat anti-Rabbit IgG Alexa Fluor 488 | ThermoFisher | Cat#A11034, Goat Polyclonal |
| Goat anti-Rat IgG PE | Southern Biotech | Cat#3050–09, Goat Polyclonal |
| Rat anti-mouse Anti-CD16/32 | Biolegend | Cat#101321, clone 93 |
| Bacterial and Virus Strains | ||
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| 2’-deoxyguanosine | Sigma-Aldrich | Cat#D0901 |
| Recombinant murine IL-7 | eBioscience | Cat#14–8071-62 |
| Recombinant murine Flt3L | eBioscience | Cat#14–8513-80 |
| Fetal Bovine Serum | Sigma-Aldrich | Cat#F-4135 |
| Fetal Bovine Serum (Characterized) | Hyclone | Cat#SH0071.03 |
| Bovine Serum Albumin | Fisher Scientific | Cat#BP9703100 |
| Paraformaldehyde (16%) | Electron Microscopy Sciences |
Cat#15710 |
| Propidium Iodide (1mg/mL) | ThermoFisher | Cat#P3566 |
| 7-AAD | BD Biosciences | Cat#559925 |
| Phorbol 12-myristate 13-acetate (PMA) | Sigma-Aldrich | Cat#P8139 |
| Ionomycin Calcium Salt | Sigma-Aldrich | Cat#I3909 |
| GolgiStop (Monensin) | BD Biosciences | Cat#554724 |
| GolgiPlug (Brefeldin A) | BD Biosciences | Cat#555029 |
| Collagenase type IV | Worthington Biochemical Corporation |
Cat#LS004189 |
| DNAse I, grade II | Roche | Cat#10104159001 |
| Phenol:Chloroform:Isoamyl Alcohol | ThermoFisher | Cat#15593031 |
| Trizol | ThermoFisher | Cat#155596026 |
| Critical Commercial Assays | ||
| Cytofix/Cytoperm Kit | BD Biosciences | Cat#554714 |
| Foxp3/Transcription Factor Staining Buffer Set | eBioscience | Cat#00–5523-00 |
| LIVE/DEAD Fixable Aqua Dead Cell Stain Kit | ThermoFisher | Cat#L34966 |
| Dynabeads Mouse CD8 | ThermoFisher | Cat#11447D |
| Taq DNA Polymerase | ThermoFisher | Cat#10342020 |
| AffinityScript Reverse Transcription Kit | Agilent | Cat#200436 |
| iQ SYBR Green SuperMix | Bio-Rad | Cat#1708880 |
| Ovation RNA-seq v2 Library Preparation Kit | NuGEN | Cat#7102–08 |
| C1 Single-Cell Reagent Kit for Preamp | Fluidigm | Cat#100–5319 |
| Single Cell-to-CT qRT-PCR Kit | ThermoFisher | Cat#4458237 |
| SsoFast EvaGreen Supermix | Bio-Rad | Cat#172–5211 |
| Deposited Data | ||
| Single-cell RT-PCR analysis of Tgd17 progenitors | This Paper | GEO: GSE118770 |
| RNA Sequencing of Tgd17 progenitors | This Paper | GEO: GSE118924 |
| Experimental Models: Cell Lines | ||
| OP9-GFP | (Schmitt and Zuniga-Pflucker, 2002) | |
| OP9-DLL1 | (Schmitt and Zuniga-Pflucker, 2002) | |
| OP9-DLL4 | (Schmitt and Zuniga-Pflucker, 2006) | |
| Experimental Models: Organisms/Strains | ||
| C57BL/6J (B6) | Jackson Laboratories | Stock#000664 |
| B6.Tcrb−/− (B6.129P2-Tcrbtm1Mom/J) | Jackson Laboratories | Stock#002118 |
| B6.Tcrd−/− (B6.129P2-Tcrdtm1Mom/J) | Jackson Laboratories | Stock#002120 |
| B6.Rag1−/− (B6.129S7-Rag1tm1Mom/J) | Jackson Laboratories | Stock#002216 |
| B6.Tcf7−/− | (Verbeek et al., 1995) | |
| B6.Il17aegfp/egfp (C57BL/6-Il17atm1Bcgen/J) | Jackson Laboratories | Stock#018472 |
| 129/Sv.Sox13−/− | Malhotra et al | |
| 129/Sv.GS2 | (Kang et al., 1998) | |
| B6.Cd2476Y/6Y | (Hwang et al., 2012) | |
| B6.Cd2476F/6F | (Hwang et al., 2012) | |
| 129/Sv.Sox13ecfp/+ | This Study | |
| Oligonucleotides | ||
| See Table S5 for Primer sequences used in single-cell qPCR, traditional qPCR, and genomic DNA PCR |
Integrated DNA Technologies |
|
| Recombinant DNA | ||
| Software and Algorithms | ||
| FlowJo V10.4.2 | FlowJo, LLC | https://www.flowjo.com/solutions/flowjo/downloads |
| Prism V7.04 | GraphPad Software, Inc. |
https://www.graphpad.com/scientific-software/prism/ |
| R Studio V1.1.447 | RStudio | https://www.rstudio.com/products/rstudio/download/ |
| edgeR | (Robinson et al., 2010) | https://bioconductor.org/packages/release/bioc/html/edgeR.html |
| Singular Analysis Toolset V3.5 | Fluidigm | https://www.fluidigm.com/software |
| GenePattern | Broad Institute | http://software.broadinstitute.org/cancer/software/genepattern/ |
| Bowtie | (Langmead and Salzberg, 2012) | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| Other | ||
Supplementary Material
Highlights.
DN1d thymocyte progenitors are Sox13hi and express Tγδ17 genes
DN1d cells generate Vγ2+ Tγδ17 cells in developmental assays
DN1d cell transcriptome is programmed independently of γδTCR expression/signaling
SOX13 and TCF7 regulate DN1d cell generation and programming
Acknowledgements
We thank A. Bhandoola for reagents, N. Malhotra, S. Kumar and H.J. Felhing for unpublished data, K. Richards-Hrdlicka and K. Tanriverdi for assistance with C1/BioMark assays, P. Wang and H. Wang for assistance with RNA-seq data analysis, Y.M. Morillon II for critical reading of the manuscript, and UMMS FACS Core for cell sorting. Supported by NIH/NCI grants CA100382, AI101301 to J.K.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
References
- Bando JK, Liang HE, and Locksley RM (2015). Identification and distribution of developing innate lymphoid cells in the fetal mouse intestine. Nature immunology 16, 153–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benz C, Martins VC, Radtke F, and Bleul CC (2008). The stream of precursors that colonizes the thymus proceeds selectively through the early T lineage precursor stage of T cell development. J Exp Med 205, 1187–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiers C, Carrelha J, Lutteropp M, Luc S, Green JC, Azzoni E, Woll PS, Mead AJ, Hultquist A,Swiers G, et al. (2013). Lymphomyeloid contribution of an immune-restricted progenitor emerging prior to definitive hematopoietic stem cells. Cell stem cell 13, 535–548. [DOI] [PubMed] [Google Scholar]
- Buus TB, Odum N, Geisler C, and Lauritsen JPH (2017). Three distinct developmental pathways for adaptive and two IFN-gamma-producing gammadelta T subsets in adult thymus. Nat Commun 8, 1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Shen X, Ding C, Qi C, Li K, Li X, Jala VR, Zhang HG, Wang T, Zheng J, and Yan J(2011). Pivotal role of dermal IL-17-producing gammadelta T cells in skin inflammation. Immunity 35, 596–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien YH, Zeng X, and Prinz I (2013). The natural and the inducible: interleukin (IL)-17-producing gammadelta T cells. Trends in immunology 34, 151–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho JS, Pietras EM, Garcia NC, Ramos RI, Farzam DM, Monroe HR, Magorien JE, Blauvelt A, Kolls JK, Cheung AL, et al. (2010). IL-17 is essential for host defense against cutaneous Staphylococcus aureus infection in mice. The Journal of clinical investigation 120, 1762–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffelt SB, Kersten K, Doornebal CW, Weiden J, Vrijland K, Hau CS, Verstegen NJ,Ciampricotti M, Hawinkels LJ, Jonkers J, and de Visser KE (2015). IL-17-producing gammadelta T cells and neutrophils conspire to promote breast cancer metastasis. Nature 522, 345–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffey F, Lee SY, Buus TB, Lauritsen JP, Wong GW, Joachims ML, Thompson LF, Zuniga-Pflucker JC, Kappes DJ, and Wiest DL (2014). The TCR ligand-inducible expression of CD73 marks gammadelta lineage commitment and a metastable intermediate in effector specification. J Exp Med 211, 329–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen NR, Brennan PJ, Shay T, Watts GF, Brigl M, Kang J, Brenner MB, and ImmGen Project C (2013). Shared and distinct transcriptional programs underlie the hybrid nature of iNKT cells. Nature immunology 14, 90–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinides MG, McDonald BD, Verhoef PA, and Bendelac A (2014). A committed precursor to innate lymphoid cells. Nature 508, 397–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Andres B, Gonzalo P, Minguet S, Martinez-Marin JA, Soro PG, Marcos MA, and Gaspar ML (2002). The first 3 days of B-cell development in the mouse embryo. Blood 100, 4074–4081. [DOI] [PubMed] [Google Scholar]
- Engel I, Seumois G, Chavez L, Samaniego-Castruita D, White B, Chawla A, Mock D, Vijayanand P, and Kronenberg M (2016). Innate-like functions of natural killer T cell subsets result from highly divergent gene programs. Nature immunology 17, 728–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garman RD, Doherty PJ, and Raulet DH (1986). Diversity, rearrangement and expression of murine T cell gamma genes. Cell 45, 733–742. [DOI] [PubMed] [Google Scholar]
- Gray EE, Ramirez-Valle F, Xu Y, Wu S, Wu Z, Karjalainen KE, and Cyster JG (2013).Deficiency in IL-17-committed Vgamma4(+) gammadelta T cells in a spontaneous Sox13-mutant CD45.1(+) congenic mouse substrain provides protection from dermatitis. Nature immunology 14, 584–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoriadou K, Boucontet L, and Pereira P (2003). Most IL-4-producing gamma delta thymocytes of adult mice originate from fetal precursors. J Immunol 171, 2413–2420. [DOI] [PubMed] [Google Scholar]
- Haas JD, Ravens S, Duber S, Sandrock I, Oberdorfer L, Kashani E, Chennupati V, Fohse L, Naumann R, Weiss S, et al. (2012). Development of interleukin-17-producing gammadelta T cells is restricted to a functional embryonic wave. Immunity 37, 48–59. [DOI] [PubMed] [Google Scholar]
- Havran WL, and Allison JP (1990). Thy-1+ dendritic epidermal cells of adult mice arise from fetal thymic precursors. Nature 344, 68–70. [DOI] [PubMed] [Google Scholar]
- Hirano M, Guo P, McCurley N, Schorpp M, Das S, Boehm T, and Cooper MD (2013). Evolutionary implications of a third lymphocyte lineage in lampreys. Nature 501, 435–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang S, Song KD, Lesourne R, Lee J, Pinkhasov J, Li L, El-Khoury D, and Love PE (2012). Reduced TCR signaling potential impairs negative selection but does not result in autoimmune disease. J Exp Med 209, 1781–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jojic V, Shay T, Sylvia K, Zuk O, Sun X, Kang J, Regev A, Koller D, Immunological Genome Project C, Best AJ, et al. (2013). Identification of transcriptional regulators in the mouse immune system. Nature immunology 14, 633–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Baker J, and Raulet DR (1995). Evidence that productive rearrangements of TCR γ genes influence the fate of progenitor cells to differentiate into αβ or γδ T cells. Eur. J. Immunol [DOI] [PubMed] [Google Scholar]
- Kang J, Coles M, Cado D, and Raulet DH (1998). The developmental fate of T cells is critically influenced by TCRgammadelta expression. Immunity 8, 427–438. [DOI] [PubMed] [Google Scholar]
- Kang J, and Malhotra N (2015). Transcription factor networks directing the development, function, and evolution of innate lymphoid effectors. Annual review of immunology 33, 505–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashani E, Fohse L, Raha S, Sandrock I, Oberdorfer L, Koenecke C, Suerbaum S, Weiss S, and Prinz I (2015). A clonotypic Vgamma4Jgamma1/Vdelta5Ddelta2Jdelta1 innate gammadelta T-cell population restricted to the CCR6(+)CD27(−) subset. Nat Commun 6, 6477. [DOI] [PubMed] [Google Scholar]
- Kashem SW, Riedl MS, Yao C, Honda CN, Vulchanova L, and Kaplan DH (2015). Nociceptive Sensory Fibers Drive Interleukin-23 Production from CD301b+ Dermal Dendritic Cells and Drive Protective Cutaneous Immunity. Immunity 43, 515–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisielow J, Kopf M, and Karjalainen K (2008). SCART scavenger receptors identify a novel subset of adult gammadelta T cells. J Immunol 181, 1710–1716. [DOI] [PubMed] [Google Scholar]
- Klug DB, Carter C, Crouch E, Roop D, Conti CJ, and Richie ER (1998). Interdependence of cortical thymic epithelial cell differentiation and T-lineage commitment. Proceedings of the National Academy of Sciences of the United States of America 95, 11822–11827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nature methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SY, Coffey F, Fahl SP, Peri S, Rhodes M, Cai KQ, Carleton M, Hedrick SM, Fehling HJ, Zuniga-Pflucker JC, et al. (2014). Noncanonical mode of ERK action controls alternative alphabeta and gammadelta T cell lineage fates. Immunity 41, 934–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, Holzapfel KL, Zhu J, Jameson SC, and Hogquist KA (2013). Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nature immunology 14, 1146–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luche H, Ardouin L, Teo P, See P, Henri S, Merad M, Ginhoux F, and Malissen B (2011). The earliest intrathymic precursors of CD8alpha(+) thymic dendritic cells correspond to myeloid-type double-negative 1c cells. Eur J Immunol 41, 2165–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Aymeric L, Locher C, Mattarollo SR, Delahaye NF, Pereira P, Boucontet L, Apetoh L, Ghiringhelli F, Casares N, et al. (2011). Contribution of IL-17-producing gamma delta T cells to the efficacy of anticancer chemotherapy. J Exp Med 208, 491–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair F, Joller S, Hoeppli R, Onder L, Hahn M, Ludewig B, Waisman A, and Becher B (2015). The NFkappaB-inducing kinase is essential for the developmental programming of skin-resident and IL-17-producing gammadelta T cells. Elife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra N, Narayan K, Cho OH, Sylvia KE, Yin C, Melichar H, Rashighi M, Lefebvre V,Harris JE, Berg LJ, et al. (2013). A network of high-mobility group box transcription factors programs innate interleukin-17 production. Immunity 38, 681–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney-Freeman S, Cahan P, Li H, Lacadie SA, Huang HT, Curran M, Loewer S, Naveiras O, Kathrein KL, Konantz M, et al. (2012). The transcriptional landscape of hematopoietic stem cell ontogeny. Cell stem cell 11, 701–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melichar HJ, Narayan K, Der SD, Hiraoka Y, Gardiol N, Jeannet G, Held W, Chambers CA, and Kang J (2007). Regulation of gammadelta versus alphabeta T lymphocyte differentiation by the transcription factor SOX13. Science 315, 230–233. [DOI] [PubMed] [Google Scholar]
- Mold JE, Venkatasubrahmanyam S, Burt TD, Michaelsson J, Rivera JM, Galkina SA, Weinberg K, Stoddart CA, and McCune JM (2010). Fetal and adult hematopoietic stem cells give rise to distinct T cell lineages in humans. Science 330, 1695–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Ruiz M, Ribot JC, Grosso AR, Goncalves-Sousa N, Pamplona A, Pennington DJ, Regueiro JR, Fernandez-Malave E, and Silva-Santos B (2016). TCR signal strength controls thymic differentiation of discrete proinflammatory gammadelta T cell subsets. Nature immunology 17, 721–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan K, and Kang J (2010). Disorderly conduct in gammadelta versus alphabeta T cell lineage commitment. Seminars in immunology [DOI] [PMC free article] [PubMed]
- Narayan K, Sylvia KE, Malhotra N, Yin CC, Martens G, Vallerskog T, Kornfeld H, Xiong N,Cohen NR, Brenner MB, et al. (2012). Intrathymic programming of effector fates in three molecularly distinct gammadelta T cell subtypes. Nature immunology 13, 511–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitta T, Muro R, Shimizu Y, Nitta S, Oda H, Ohte Y, Goto M, Yanobu-Takanashi R, Narita T,Takayanagi H, et al. (2015). The thymic cortical epithelium determines the TCR repertoire of IL- 7-producing gammadeltaT cells. EMBO reports 16, 638–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park K, He X, Lee HO, Hua X, Li Y, Wiest D, and Kappes DJ (2010). TCR-mediated ThPOK induction promotes development of mature (CD24-) gammadelta thymocytes. The EMBO journal 29, 2329–2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porritt HE, Rumfelt LL, Tabrizifard S, Schmitt TM, Zuniga-Pflucker JC, and Petrie HT (2004). Heterogeneity among DN1 prothymocytes reveals multiple progenitors with different capacities to generate T cell and non-T cell lineages. Immunity 20, 735–745. [DOI] [PubMed] [Google Scholar]
- Rahimpour A, Koay HF, Enders A, Clanchy R, Eckle SB, Meehan B, Chen Z, Whittle B, Liu L, Fairlie DP, et al. (2015). Identification of phenotypically and functionally heterogeneous mouse mucosal-associated invariant T cells using MR1 tetramers. J Exp Med 212, 1095–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramond C, Berthault C, Burlen-Defranoux O, de Sousa AP, Guy-Grand D, Vieira P, Pereira P, and Cumano A (2014). Two waves of distinct hematopoietic progenitor cells colonize the fetal thymus. Nature immunology 15, 27–35. [DOI] [PubMed] [Google Scholar]
- Ribot JC, deBarros A, Pang DJ, Neves JF, Peperzak V, Roberts SJ, Girardi M, Borst J, Hayday AC, Pennington DJ, and Silva-Santos B (2009). CD27 is a thymic determinant of the balance between interferon-gamma- and interleukin 17-producing gammadelta T cell subsets. Nature immunology 10, 427–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinette ML, Fuchs A, Cortez VS, Lee JS, Wang Y, Durum SK, Gilfillan S, Colonna M, and Immunological Genome C (2015). Transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nature immunology 16, 306–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt TM, and Zuniga-Pflucker JC (2002). Induction of T cell development from hematopoietic progenitor cells by delta-like-1 in vitro. Immunity 17, 749–756. [DOI] [PubMed] [Google Scholar]
- Schmitt TM, and Zuniga-Pflucker JC (2006). T-cell development, doing it in a dish. Immunological reviews 209, 95–102. [DOI] [PubMed] [Google Scholar]
- Serwold T, Hochedlinger K, Inlay MA, Jaenisch R, and Weissman IL (2007). Early TCR expression and aberrant T cell development in mice with endogenous prerearranged T cell receptor genes. J Immunol 179, 928–938. [DOI] [PubMed] [Google Scholar]
- Shibata K, Yamada H, Nakamura M, Hatano S, Katsuragi Y, Kominami R, and Yoshikai Y (2014). IFN-gamma-producing and IL-17-producing gammadelta T cells differentiate at distinct developmental stages in murine fetal thymus. J Immunol 192, 2210–2218. [DOI] [PubMed] [Google Scholar]
- Shibata K, Yamada H, Nakamura R, Sun X, Itsumi M, and Yoshikai Y (2008). Identification of CD25+ gamma delta T cells as fetal thymus-derived naturally occurring IL-17 producers. J Immunol 181, 5940–5947. [DOI] [PubMed] [Google Scholar]
- Sutton CE, Lalor SJ, Sweeney CM, Brereton CF, Lavelle EC, and Mills KH (2009). Interleukin-1 and IL-23 induce innate IL-17 production from gammadelta T cells, amplifying Th17 responses and autoimmunity. Immunity 31, 331–341. [DOI] [PubMed] [Google Scholar]
- Veinotte LL, Greenwood CP, Mohammadi N, Parachoniak CA, and Takei F (2006). Expression of rearranged TCRgamma genes in natural killer cells suggests a minor thymus-dependent pathway of lineage commitment. Blood 107, 2673–2679. [DOI] [PubMed] [Google Scholar]
- Verbeek S, Izon D, Hofhuis F, Robanus-Maandag E, te Riele H, van de Wetering M, Oosterwegel M, Wilson A, MacDonald HR, and Clevers H (1995). An HMG-box-containing T-cell factor required for thymocyte differentiation. Nature 374, 70–74. [DOI] [PubMed] [Google Scholar]
- Weber BN, Chi AW, Chavez A, Yashiro-Ohtani Y, Yang Q, Shestova O, and Bhandoola A (2011). A critical role for TCF-1 in T-lineage specification and differentiation. Nature 476, 63–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei YL, Han A, Glanville J, Fang F, Zuniga LA, Lee JS, Cua DJ, and Chien YH (2015). A Highly Focused Antigen Receptor Repertoire Characterizes gammadelta T Cells That are Poised to Make IL-17 Rapidly in Naive Animals. Front Immunol 6, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wencker M, Turchinovich G, Di Marco Barros R, Deban L, Jandke A, Cope A, and Hayday AC(2014). Innate-like T cells straddle innate and adaptive immunity by altering antigen-receptor responsiveness. Nature immunology 15, 80–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A, Laurenti E, Oser G, van der Wath RC, Blanco-Bose W, Jaworski M, Offner S, Dunant CF, Eshkind L, Bockamp E, et al. (2008). Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell 135, 1118–1129. [DOI] [PubMed] [Google Scholar]
- Yang Q, Saenz SA, Zlotoff DA, Artis D, and Bhandoola A (2011). Cutting edge: Natural helper cells derive from lymphoid progenitors. J Immunol 187, 5505–5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Nguyen CK, Liu X, Kanellopoulou C, and Muljo SA (2012). Lin28b reprograms adult bone marrow hematopoietic progenitors to mediate fetal-like lymphopoiesis. Science 335, 1195–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


