Abstract
Ferroptosis is considered genetically and biochemically distinct from other forms of cell death. In this study, we examined whether ferroptosis shares cell death pathways with other types of cell death. When human colon cancer HCT116, CX-1, and LS174T cells were treated with ferroptotic agents such as sorafenib (SRF), erastin (ERA), and artesunate (ART), data from immunoblot assay showed that ferroptotic agents induced endoplasmic reticulum (ER) stress and the ER stress response-mediated expression of death receptor 5 (DR5), but not death receptor 4 (DR4). An increase in the level of DR5, which is activated by binding to tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) and initiates apoptosis, was probably responsible for synergistic apoptosis when cells were treated with ferroptotic agent in combination with TRAIL. This collateral effect was suppressed in C/EBP (CCAAT-enhancer-binding protein)-homologous protein (CHOP)-deficient mouse embryonic fibroblasts or DR5 knockdown HCT116 cells, but not in p53-deficient HCT116 cells. The results from in vitro studies suggest the involvement of the p53-independent CHOP/DR5 axis in the synergistic apoptosis during the combinatorial treatment of ferroptotic agent and TRAIL. The synergistic apoptosis and regression of tumor growth were also observed in xenograft tumors when SRF and TRAIL were administered to tumor-bearing mice.
Introduction
Ferroptosis is a unique iron-dependent form of non-apoptotic regulated cell death, identified in 2012 by the lab of Dr. Brent R. Stockwell.1 Ferroptosis results from the integration of two different pathways. In the presence of ferroptosis-inducing agents, the iron storage protein ferritin and/or the ferritinophagy cargo receptor NCOA4 (nuclear receptor coactivator 4) are degraded via ferritinophagy and release ferrous iron, which generates reactive oxygen species through the Fenton reaction and subsequently induces lipid peroxidation.2,3 At the same time, ferroptosis-inducing agents inhibit biosynthesis or utilization of the antioxidant glutathione by inhibiting the Na+-independent cystine–glutamate Xc− antiporter, glutathione (GSH) synthesis, the glutathione-dependent antioxidant enzyme glutathione peroxidase 4 (GPX4), or glutathione S-transferase.1,4–8 Thus, the accumulation of lipid peroxidation and depletion of plasma membrane polyunsaturated fatty acids have been well known to result in the lethal event of ferroptosis.1,9–11
Previous studies have shown that inhibition of cystine-glutamate exchange by ferroptotic agents leads to activation of an endoplasmic reticulum (ER) stress response and upregulation of the CHAC1 (glutathione-specific gamma-glutamylcyclotransferase 1) gene.12,13 Ferroptotic agent-induced ER stress is probably due to glutathione depletion, which results in a decrease in the GSH to GSSG (glutathione disulfide) ratio.14,15 The perturbation in redox status affects the activities of resident ER-folding enzymes and subsequently leads to unfolded protein response (UPR) stress.15 Indeed, data from microassay studies also reveal that ferroptotic agent promotes expression of the ER stress marker ATF4 (activating transcription factor 4)-dependent gene,16,17 which is a basic leucine zipper transcription factor that regulates several UPR target genes.18 Previous studies show that the ATF4-CHOP (C/EBP (CCAAT-enhancer-binding protein)-homologous protein) signal pathway regulates induction of several pro-apoptotic proteins such as PUMA (p53 upregulated modulator of apoptosis), GADD34 (growth arrest and DNA damage-inducible protein), ERO1α (endoplasmic reticulum, oxidoreductin-1α), Bim (Bcl-2 [B-cell lymphoma 2]-like protein 11), and NOXA (Latin for damage).19,20 Recent studies reveal that, unlike apoptotic agent, ferroptotic agent-induced PUMA expression does not induce apoptosis.17 These results suggest that ferroptotic agent-induced PUMA sustains a biochemically inactive state during treatment with ferroptotic agent alone.21
Although ferroptosis is considered genetically and biochemically distinct from other forms of regulated cell death, emerging evidence suggests that ferroptosis often shares common pathways with other types of cell death.22 In this study, we hypothesized that ferroptosis and apoptosis share ER stress pathways. ER stress is sensed by three upstream signaling proteins: PERK (protein kinase RNA (PKR)-like ER kinase), IRE1 (inositol requiring protein 1), and ATF6 (activating transcription factor 6). Previous studies show that CHOP binds to the promoter of the TRAIL-R2 (tumor necrosis factor-related apoptosis-inducing ligand receptor 2) during ER stress and induces TRAIL-R2 expression through the ER stress-mediated PERK-eIF2α (eukaryotic initiation factor 2α)-ATF4-CHOP pathway.23–29 TRAIL-R2, also known death receptor 5 (DR5), is a cell surface receptor which binds to TRAIL and forms trimer. Trimerization of TRAIL-R2 results in activation of pro-caspase-8 through the recruitment of Fas-associating protein with death domain (FADD) and procaspase-8 and then the formation of the death-inducing signaling complex (DISC).30,31 Activated caspase-8 leads to activation of downstream executioners caspase-3, -6, and -7 through two different pathways: mitochondria-dependent and -independent mechanisms.32–37
In this study, we observed that a combined treatment of ferroptotic agents with TRAIL markedly enhanced TRAIL-induced apoptosis. Our studies suggest that ferroptotic agent-induced ER stress response plays an important role in the enhancement of TRAIL-induced apoptosis through upregulation of DR5 expression.
Materials and Methods
Cell Lines and Cell Culture Conditions
Human colon cancer CX1, LS174T, and HCT116 cells were previously obtained from American Type Culture Collection (ATCC, Manassas, VA). Bak-deficient (Bak−/−), Bax-deficient (Bax−/−), Bax and Bak-deficient (Bax−/−/Bak−/−), and p53-deficient (p53−/−) HCT116 cells were provided by Dr. B. Vogelstein (Johns Hopkins University, Baltimore, MD). CHOP-deficient (CHOP−/−) and WT MEF cell lines were provided by Dr. Randal J. Kaufman (Sanford Burnham Medical Research Institute, CA). Each cell line was maintained in a medium as follows: HCT116 cells in McCoy’s 5A; LS174T and MEF cells in Dulbecco’s Modified Eagle Medium; CX1 cells in Roswell Park Memorial Institute medium-1640 supplemented with 2 mM glutamine. All cell lines were maintained with 10% fetal bovine serum and incubated in a humidified atmosphere of 5% CO2 at 37°C.
Chemicals and Reagents
For production of recombinant human TRAIL, a human TRAIL cDNA fragment (amino acids 114–281) obtained by RT-PCR was cloned into a pET-23d plasmid (Novagen, Madison, WI), and His-tagged TRAIL protein was purified using the Qiagen express protein purification system (Qiagen, Valencia, CA). Soluble recombinant murine TRAIL was purchased from R&D systems (Minneapolis, MN). Artesunate, erastin, and sorafenib were purchased from Selleckchem (Houston, TX).
Microarray Assay
HCT116 cells were treated with 50 μM ART for 24 h and total RNA was extracted using the RNeasy mini kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. RNA quality and integrity were verified using the Agilent 2100 Bioanalyzer system (Agilent Technologies, Santa Clara, CA). Biotin-labeled cRNA samples for hybridization on Illumina HumanHT-12 v4 Expression BeadChip (Illumina, Inc., San Diego, CA) were prepared according to Illumina’s recommended sample labeling procedure.
Cell Survival Assay
For quantification of cell survival rate, cells were trypsinized and stained with trypan blue followed by counting with a hemocytometer under microscope.
Western Blotting and Antibodies
Immunoblotting was carried out as previously described.17 The following antibodies were used in this study: anti-PARP-1, anti-cleaved caspase-3, anti-cleaved caspase-8, anti-cleaved caspase-9, anti-DR5, anti-PUMA, and anti-CHOP from Cell signaling Technology (Beverly, MA), and anti-actin, goat anti-rabbit IgG-horseradish peroxidase (HRP), and goat anti-mouse IgG-HRP from Santa Cruz Biotechnology (Santa Cruz, CA).
Combination Index (CI)
CIs were calculated using the CompuSyn software (ComboSyn, Inc., Paramus, NJ, USA). The extent of synergism/antagonism was determined based on CI values. In general, CI values below 1 suggest synergy, whereas CI values above 1 indicate antagonism between the drugs. CI values in the 0.9–1.10 range mainly indicate additive effects; 0.85–0.9, slight synergy; 0.7–0.85, moderate synergy; 0.3–0.7, synergy; 0.1–0.3, strong synergy; <0.1, very strong synergy.
Small Interfering RNA (siRNA)
DR5 siRNA (sc-40237) and negative control siRNA (sc-37007) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Cells were transfected with siRNA oligonucleotides using Lipofectamine RNAiMax (Invitrogen), according to the manufacturer’s instructions. After 24 h of transfection, cells were treated with erastin and TRAIL for further analysis.
Immunofluorescence Staining
The procedures were performed as previously described (1). Tissue sections were incubated with the primary antibodies overnight at 4°C, and then incubated with a secondary Alexa Fluor®-594-conjugated (Molecular Probes, Eugene, OR, USA). The nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) and visualized using fluorescence microscopy.
TUNEL Assay
An in situ cell apoptosis detection kit (Trevigen, Gaithersburg, MD, USA) was used to detect apoptosis in paraffin-embedded tissues. In situ cell death detection kit, TMR red (Roche, Indianapolis, USA) was used to detect apoptotic cell death in adherent cells. The staining was performed according to the manufacturer’s instructions. Briefly, sections of paraffin-embedded tissues were deparaffinized and permeabilized with proteinase K. Adherent cells were fixed before permeabilization. DNA strand breaks were end-labeled with terminal transferase and visualized using fluorescence microscopy.
Animal Model
Four-week-old female BALB/c nude mice were obtained from Orient Bio (Seongnam, Kyonggi-Do, South Korea) and housed in a specific pathogen-free environment. The animals were acclimated for one week prior to the study and were provided free access to food and water. Mice were fed ad libitum and maintained in environments with controlled temperature of 22–24°C and 12 h light and dark cycles. Luciferase-expressing HCT116 (HCT116-Luci) cells (1 × 107) in 100 μL of culture medium were mixed with 100 μL of Matrigel and implanted subcutaneously into five-week-old BALB/c nude female mice. Prior to treatment with SRF and TRAIL, tumor size was measured two to three times per week until the volume reached approximately 100 mm3. Tumor volume was calculated as W2 × L × 0.52, where L is the largest diameter and W is the diameter perpendicular to L. After establishment of these tumor xenografts, mice were randomized into four groups of five mice per group. Animals were treated with saline (control), SRF alone, TRAIL alone, and SRF in combination with TRAIL (S + T). SRF group: 15 mg/kg –three times per week – intratumoral injection —two-week treatment. TRAIL group: 200 μg/kg –twice per week—intratumoral injection—two-week treatment. For imaging assay, animals were anesthetized and subjected to NightOWL LB983 bioluminescence imaging (BLI) system (Berthold Technologies, TN, USA). D-luciferin sodium salt (BioVision Inc. Milpitas, CA) at 50 mg/kg was administered intraperitoneally as a substrate before BLI imaging. The captured images were quantified using the IndiGo™ software package. All animal procedures were carried out in accordance with guidelines approved by the Korea University Institutional Animal Care and Use Committee.
Statistical Analysis
All the values are represented as mean ± S.D. Statistical analysis was performed using one-way analysis of variance (ANOVA) followed by Bonferroni’s test or by the Student’s t-test as indicated using GraphPad Prism 7 software. P value less than 0.05 was defined as statistical significance. Where indicated *p < 0.05; **p < 0.01; ***p < 0.001.
Results
TRAIL- induced apoptosis is promoted by treatment with ER stress-inducing ferroptotic agents in human colon cancer cells
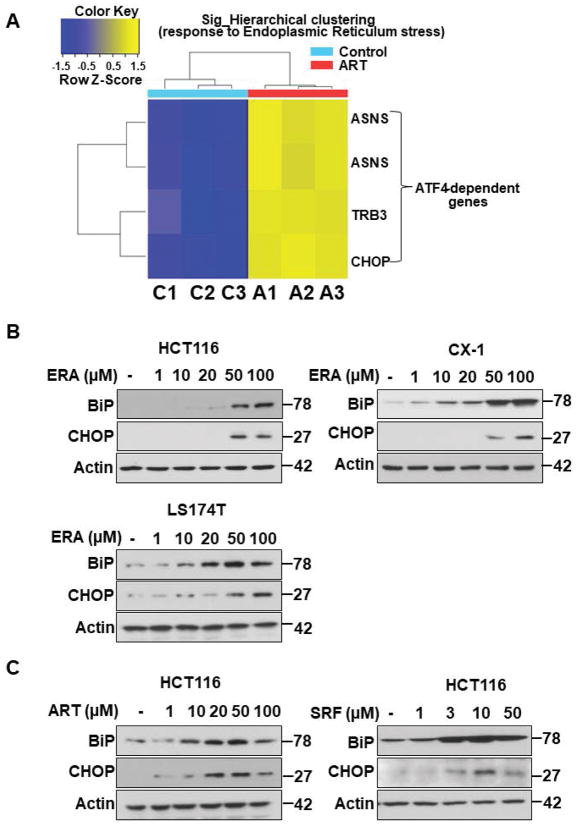
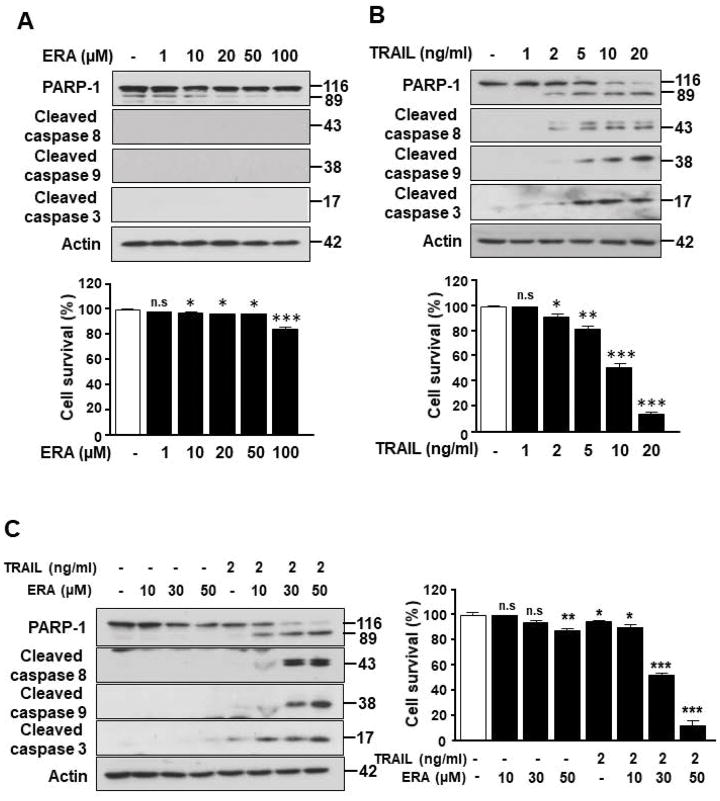
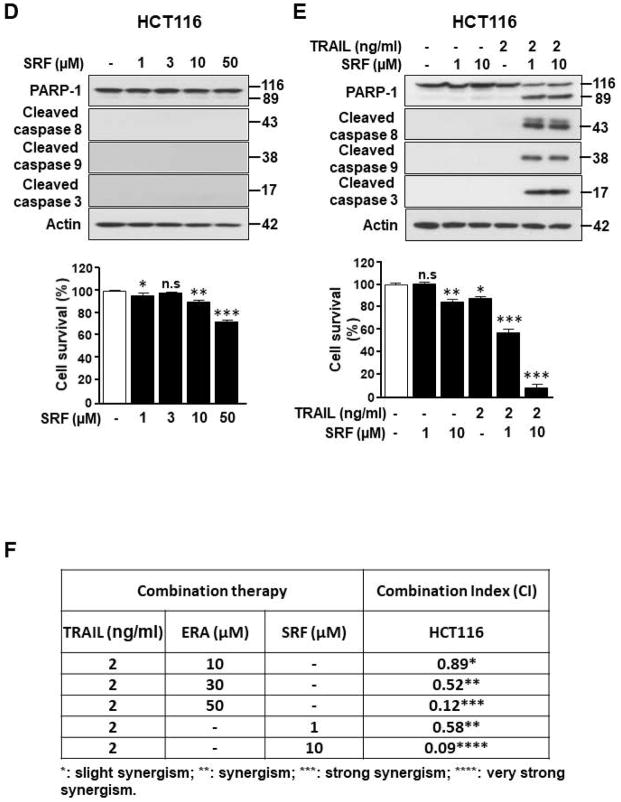
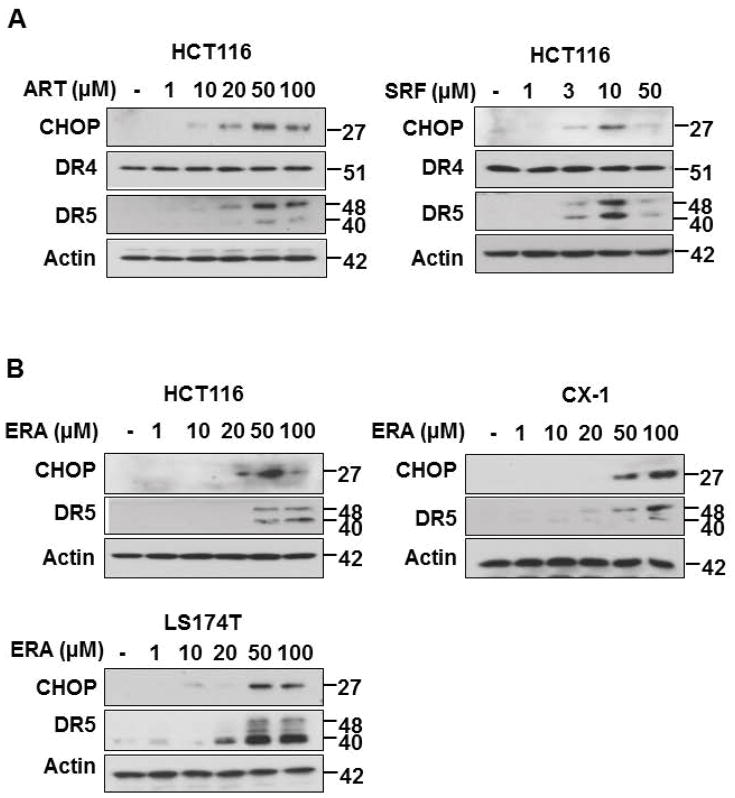
We previously proposed that ferroptosis-induced ER stress response plays an important role in the crosstalk with other types of cell death.17 To examine this possibility, we investigated induction of the ER stress response during treatment with several different ferroptotic agents in a variety of cell lines and then analyzed the process using microarray and immunoblotting assay. Data from microarray assay show that artesunate (ART) promotes ER stress response marker ATF4 (activating transcription factor 4)-dependent gene expression such as ASNS (asparagine synthetase) and CHOP (Fig. 1A). Immunoblotting assay data in Fig. 1B also show an induction of the ER stress response in three different colon cancer cell lines: HCT116, CX-1, and LS174T cells. The expression of the ER stress response-related protein CHOP and the ER chaperone protein BiP (binding immunoglobulin protein) was increased during erastin (ERA) treatment in a dose-dependent manner. We also confirmed the ER stress response using ART and sorafenib (SRF) in HCT116 cells (Fig. 1C). The CHOP and BiP protein expression levels were increased in a dose-dependent manner during treatment with ART and SRF. Next, we examined the crosstalk between ferroptosis and other types of cell death, in particular apoptosis. For this study, HCT116 cells were treated with ERA/SRF alone, recombinant human TRAIL alone, and the combination of ERA/SRF and TRAIL (Fig. 2). We observed a dose-dependent ferroptotic effect of ERA/SRF and apoptotic effect of TRAIL (Figs. 2A, 2B, and 2D). Unlike ferroptotic agent-induced cytotoxicity, TRAIL clearly induced cleavage of PARP (poly (ADP-ribose) polymerase), the hallmark feature of apoptosis, and caspase 8/9/3. As shown in Figs. 2C and 2E, a synergistic apoptotic death was observed with the combinatorial treatment compared with any single treatment. Data from combination index analysis showed that there was slight, moderate, strong, or very strong synergy of ferroptotic agent in combination of TRAIL (Fig. 2F). The synergistic effects were ferroptotic agent- and dose-dependent.
Figure 1. Ferroptotic agents induce ER stress in human colon cancer cells.
(A) Microarray assay for detection of artesunate (ART)-induced ER stress-associated gene expression. HCT116 cells were treated with 50 μM ART for 24 h and triplicate Illumina Gene Expression Microarrays were performed with BeadArray microarray technology. (B, C) Immunoblot assay for detection of ferroptotic agent-induced ER stress in HCT116, CX-1, and LS174T cells. Cells were treated with various doses of erastin (ERA: 1 to 100 μM), ART (1–100 μM), or sorafenib (SRF: 1–50 μM) for 24 h. Whole-cell extracts were then analyzed with immunoblotting assay using indicated antibodies.
Figure 2. Ferroptotic agents enhance TRAIL-induced apoptosis in HCT116 cells.
Cells were treated with various doses of ERA (A) or SRF (D) for 24 h or TRAIL for 4 h (B). Cell lysates were analyzed with immunoblotting assay using indicated antibodies (upper panels). Cell survival was determined using trypan blue exclusion assay (lower panels). (C and E) Cells were preteated with various doses of ERA (C) or SRF (E) for 20 h and then exposed to TRAIL (2 ng/ml) for an additional 4 h. Cell lysates were analyzed with immunoblotting assay using indicated antibodies and cell survival was determined using trypan blue exclusion assay. Error bars represent the mean ± SD from triplicate experiments. For statistical analysis, Student’s t-test (two-sided, paired) was used. p-values: n.s. = not significant; *, 0.05; **, 0.01; ***, 0.001. (F) Combination index (CI) for combination of TRAIL and ERA or SRF in HCT116 cells was calculated using CompuSyn software.
Role of PUMA in the synergistic apoptosis during the combinatorial treatment of ferroptotic agent and TRAIL
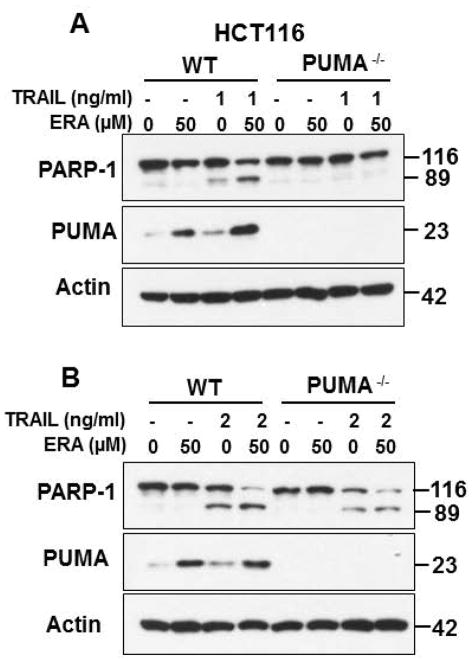
Our previous studies demonstrated that ER stress-induced PUMA plays an important role in the crosstalk between ferroptosis and apoptosis.17 We further examined our previous studies. As shown in Fig. 3, the combinatorial treatment of ERA and TRAIL enhanced apoptosis. Interestingly, the combination treatment-enhanced apoptosis was completely (Fig. 3A) or partially (Fig. 3B) suppressed in PUMA knockout (PUMA−/−) HCT116 cell line. These differential effects were dependent on dose of TRAIL. These results suggest that multiple factors may be involved in the enhancement of apoptosis during treatment of ERA and TRAIL. This possibility was further examined.
Figure 3. Role of PUMA in the synergistic apoptosis during the combinatorial treatment of ferroptotic agent and TRAIL.
HCT116 WT and HCT116 PUMA KO cells were pretreated with 50 μM ERA for 20 h and then treated to 1 ng/ml TRAIL (A) or 2 ng/ml TRAIL (B) for additional 4 h. Whole-cell extracts were analyzed with immunoblotting using indicated antibodies.
Ferroptotic agent induces ER stress response-associated DR5 expression but not DR4 expression
Previous studies reveal that CHOP regulates ER stress-induced apoptosis through promoting death receptor 5 (DR5) expression and subsequently facilitating the DR5-caspase 8 pathway in human cancer cells.23–29 Since data from Fig. 1 show that a variety of ferroptotic agents induce CHOP expression, we hypothesized that the combination of ferroptotic agent and TRAIL enhances apoptosis through overexpression of DR5. This possibility was examined in a variety of colon cancer cell line (HCT116, CX-1, and LS174T cells) and a variety of ferroptotic agents (ART, SRF, ERA). Fig. 4 shows that ferroptotoic agents induced DR5 expression in colon cancer cell lines.
Figure 4. Ferroptotic agents induce ER stress-associated DR5 expression in human colon cancer cells.
(A) HCT116 cells were treated with various doses of ART and SRF for 24h. Whole-cell lysates were analyzed with immunoblotting assay using indicated antibodies. (B) HCT116, CX1, and LS174T cells were treated with various doses of ERA for 24 h. Whole-cell lysates were analyzed with immunoblotting assay using indicated antibodies.
Ferroptotic agent promotes TRAIL-induced apoptosis via the p53-independent CHOP-DR5 pathway
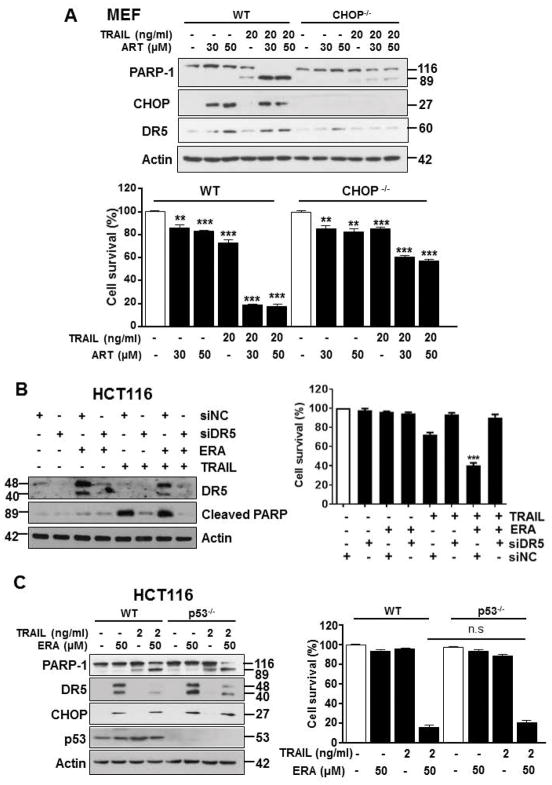
We further investigated the involvement of the CHOP-DR5 pathway in the synergistic apoptosis during the combinatorial treatment of ferroptotic agent and TRAIL. For this study, we employed CHOP knockout (CHOP−/−) mouse embryonic fibroblast (MEF) and DR5 small interfering RNA (siDR5). Data from Fig. 5A shows that a combination of mouse TRAIL and ART induced synergistic apoptosis in MEF wild-type (WT) cells and its synergistic apoptosis as well as DR5 expression were suppressed in CHOP−/− MEFs. Similar results were observed in DR5 knockdown HCT116 cells (Fig. 5B). Suppression of DR5 expression inhibited the combinatorial treatment-induced synergistic apoptosis. We next observed the combinatorial treatment-induced apoptosis in HCT116 WT and p53 knockout (p53−/−) cells (Fig. 5C). The combined treatment of TRAIL and ERA induced synergistic apoptosis in HCT116 WT and its synergistic apoptosis was not inhibited in p53−/− cells. Moreover, the combinatorial treatment-induced CHOP and DR5 expression was not inhibited in p53−/− cells. These results suggest that the p53-indepdent CHOP-DR5 pathway plays an important role in the combinatorial treatment-induced synergistic apoptosis.
Figure 5. Ferroptotic agents promote TRAIL-induced apoptosis via the p53-independent CHOP-DR5 pathway.
(A) Mouse embryonic fibroblasts (MEFs) wild-type (WT) or MEF CHOP knockout (CHOP−/−) cells were treated with ART alone (30 or 50 μM) for 24 h, murine TRAIL (mTRAIL; 20 ng/ml) alone for 4 h, or pretreated with ART (30 or 50 μM) for 20 h and then exposed to mTRAIL (20 ng/ml) for an additional 4 h. Whole-cell lysates were analyzed with immunoblotting assay using indicated antibodies (upper panel). Cell survival was analyzed using trypan blue exclusion assay (lower panel). (B) HCT116 cells were transfected with control siRNA (siNC) or DR5 siRNA (siDR5) for 48 h and then treated with ERA (50 μM) for 20 h and then exposed to TRAIL (1 ng/ml) for an additional 4 h. Whole-cell lysates were analyzed with immunoblotting assay using indicated antibodies (left panel). Cell survival was analyzed using trypan blue exclusion assay (right panel). (C) HCT116 WT or HCT116 p53 −/− cells were pretreated with ERA (50 μM) for 20 h and then exposed to TRAIL (2 ng/ml) for an additional 4 h. Whole-cell lysates were analyzed with immunoblotting assay using indicated antibodies (left panel). Cell survival was analyzed using trypan blue exclusion assay (right panel). Error bars represent the mean ± SD from triplicate experiments. For statistical analysis, Student’s t-test (two-sided, paired) was used. p-values: n.s. = not significant; **P < 0.01, ***P < 0.001.
Role of Bax and Bak in the crosstalk between ferroptosis and apoptosis
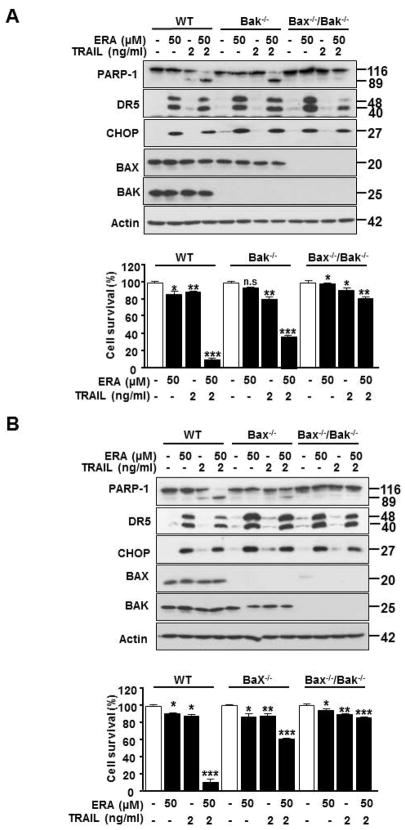
It is well-known that apoptosis occurs through the mitochondria-dependent and -independent pathways. To examine which pathway plays an important role in the combinatorial treatment-induced synergistic apoptosis, we employed HCT116 WT, Bak (Bcl-2 homologous antagonist killer) single knockout (Bak−/−), Bax (Bcl-2–associated X protein) single knockout (Bax−/−), and Bak and Bax double knockout (Bak−/−/Bax−/−) cells. Figure 6 shows that ERA-induced CHOP and DR expression was not affected in Bak−/−, Bax−/−, and Bak−/−/Bax−/− cells. However, the combinatorial treatment-induced synergistic apoptosis was suppressed in Bax−/− and Bak−/−/Bax−/− cells, but not Bak−/− cells. These results suggest that Bax rather than Bak plays an important role in the combinatorial treatment-induced synergistic apoptosis.
Figure 6. The role of Bax and Bak in ERA in combination with TRAIL-induced apoptosis in HCT116 cell.
(A) HCT116 WT, Bak−/− single knockout or Bak−/−/Bax−/− double knockout cells were pretreated with ERA (50 μM) for 20 h and then exposed to TRAIL (5 ng/ml) for an additional 4 h. Whole-cell lysates were analyzed with immunoblotting assay using indicated antibodies (upper panel). Cell survival was analyzed using trypan blue exclusion assay (lower panel). (B) HCT116 WT, Bax−/− or Bak−/−/Bax −/− cells were pretreated with ERA (50 μM) for 20 h and then exposed to TRAIL (5 ng/ml) for an additional 4 h. Whole-cell lysates were analyzed with western blotting assay with indicated antibodies (upper panel). Cell survival (lower panel) was analyzed using trypan blue exclusion. Error bars represent the mean ± SD from triplicate experiments. For statistical analysis, Student’s t-test (two-sided, paired) was used. p-values: n.s. = not significant; **P < 0.01, ***P < 0.001.
Effect of the combined treatment of SRF and TRAIL on the growth of xenograft tumors
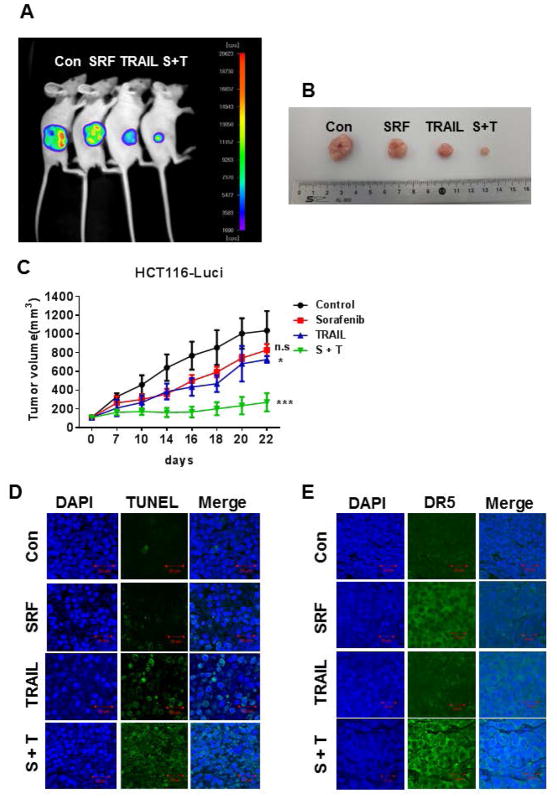
Since we observed a synergistic apoptotic death when human colon cancer cells were treated with ferroptotic agent in combination with TRAIL, we next investigated in vivo studies to assess the effect of the combinatorial treatment with SRF and TRAIL on the growth of luciferase-expressing HCT116 (HCT116-Luci) xenograft tumors (Fig. 7). SRF alone did not significantly affect tumor growth compared with the control group, despite the tendency for decreased tumor volume. TRAIL alone caused a decreased in tumor growth (p < 0.05). The combinatorial treatment of SRF and TRAIL was significantly more effective in inhibiting tumor growth compared with single treatment (p < 0.001) (Fig. 7C). Data from terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL) assay show that the combinatorial treatment effectively induced apoptosis (Fig. 7D). We also observed an increase in the DR5 expression level with treatment of SRF only as well as the combinatorial treatment of SRF and TRAIL (Fig. 7E). These data suggest that the combination of SRF and TRAIL-induced DR5 expression may play an important role in the regression of tumor growth.
Figure 7. A combinatorial treatment of SRF and TRAIL synergistically inhibits tumor growth.
Nude mice were subcutaneously inoculated with 1×106 HCT116-Luci cells. When the tumor volume reached approximately 100 mm3, tumor-bearing mice were treated with sorafenib (SRF or S, 15 mg/kg, three times per week, intratumoral) alone, TRAIL (T, 200 μg/kg, three times per week, intratumoral) alone, or a combination of sorafenib and TRAIL (S + T). (A) Mice were imaged using the NightOWL LB983 bioluminescence imaging (BLI) system. Representative images are shown on day 22. (B) Tumor tissues were harvested on day 22 and displayed. (C) Line graph illustrating the tumor volume (mm3) in HCT116-Luci tumor-bearing mice treated with phosphate-buffered saline alone, sorafenib alone, TRAIL alone, or the combination from day 0 to day 22. Error bars represent the mean ± SD from five mice. For statistical analysis, Student’s t-test (two-sided, paired) was used. p-values: *, 0.05; ***, 0.001. (D) Tumor tissues were harvested on day 22 and subjected to TUNEL assay and DAPI (4′,6-diamidino-2-phenylindole) staining. Cell nuclei were stained with DAPI. Apoptosis was detected using TUNEL assay. (E) Tumor tissues were harvested at day 22 and subjected to immunofluorescence staining with anti-DR5 antibody and DAPI. Representative images are shown (magnification is 200X and scale bar represents 20 μm).
Discussion
Several conclusions can be drawn after considering the data presented in the current study. First, ferroptotic agents (ERA, ART, and SRF) induce ER stress-mediated upregulation of TRAIL receptor DR5. A combined treatment of ferroptotic agents and TRAIL enhances TRAIL-induced apoptosis through upregulation of DR5.
Several researchers have reported that the ER stress response mediated by the PERK-eIF2α-ATF4-CHOP pathway is involved in regulation of the DR5 gene expression.23–29 Our studies confirmed an increase in expression of the ER stress-mediated DR5 gene during ferroptotic agent treatment (Figs. 4–6). Previous studies have shown that apoptotic agent-induced overexpression of DR5 leads to activation of caspase-8 without interaction with TRAIL.24,28 However, an interesting aspect of ferroptotic agent-induced upregulation of DR5 is that, unlike apoptotic agent-induced in a ligand-independent manner apoptotic death of DR5, ferroptotic agent-induced overexpressed DR5 does not induce apoptosis. Nevertheless, a combined treatment of ferroptotic agent and TRAIL enhances TRAIL-induced apoptosis in in vitro (Figs. 2, 3, 5, 6) and in vivo (Fig. 7). These results suggest two different findings: (1) no crosstalk between ferroptosis and apoptosis occurs with single-agent treatment, and (2) a crosstalk between ferroptosis and apoptosis occurs when treatments are combined.
In this study, we observed that ER stress response-mediated expression of DR5 played an important role in synergistic apoptosis when cells were treated with ferroptotic agent in combination with TRAIL. It is well-known that death receptors (DRs) such as DR4 and DR5 bind to procaspase-8 and form the DISC.30 Autoactivation of caspase-8 at the DISC leads to further triggering of signaling molecules downstream, which results in the activation of executioner caspases-3, -6, and -7, which culminates in apoptotic death.32 However, our studies clearly reveal that neither apoptosis nor caspase-8 activation occurs during treatment with ferroptotic agents, even though ferroptotic agents induce upregulation of DR5 expression. How ferroptotic agent-induced DR5 sustains a biochemically inactive state during treatment with ferroptotic agent alone is an enigma that needs clarification. It is possible that ferroptotic agent-induced DR5 remains inactive through binding with anti-apoptotic regulator, cellular FADD-like IL-1β-converting enzyme-inhibitory protein (c-FLIP). c-FLIP binds to FADD and/or procaspase-8 in a TRAIL-dependent and-independent fashion, which in turn prevents DISC formation and subsequent activation of the caspase cascade.39 This possibility needs to be further examined to understand the role of C-FLIP in the antagonistic relationship between ferroptosis and apoptosis.
TRAIL initiates the extrinsic pathway by binding to DRs such as DR4 and DR5 and activates caspase-8 through DISC formation.30,32 Activated caspase-8 induces apoptosis through mitochondrial-dependent and -independent pathways. In the mitochondrial-dependent pathway, Bax (Bcl-2-associated X protein) and Bak (Bcl-2 homologous antagonist killer) play a critical role in regulating cellular commitment to apoptosis. Bax and Bak form homo- and hetero-oligomers and oligomerized Bax and Bak’s insertion into the mitochondrial outer membrane, permeabilization, and depolarization of the mitochondria promote cytochrome c release.33,34,36 Released cytochrome c facilitates the formation of the Apaf1 (apoptosis signal-regulating kinase)/caspase-9 apoptosome, which activates caspase-9 and subsequently, caspase-3. Data from Bax−/−, Bak−/−, and Bax/Bak−/− cell lines (Fig. 6) clearly demonstrate that a combinatorial treatment of ERT and TRAIL-enhanced apoptosis is mediated through the mitochondrial-dependent pathway. Moreover, a recent report suggests that ferroptotic agent-induced ER stress-mediated PUMA also plays an important role in the enhancement of TRAIL-induced apoptosis by ferroptotic agent treatment.17 Thus, our studies suggest that DR5 as well as PUMA are involved in this process. Obviously, these possibilities need to be further examined to clarify the role of DR5 and PUMA in the combinatorial treatment-induced synergistic apoptosis.
Conclusions
We examined a possible mechanism of the combined treatment of ferroptotic agents and TRAIL-enhanced apoptosis. The results from in vitro studies suggest that ferroptotic agents induce upregulation of TRAIL receptor DR5 through the p53-independent CHOP pathway and this upregulation of DR5 plays an important role in the synergistic apoptosis during the combinatorial treatment of ferroptotic agent and TRAIL. The results from in vivo studies demonstrated an enhancement of tumoricidal efficacy when ferroptotic agent SRF and TRAIL were combined.
Acknowledgments
Grant sponsor: NCI R03CA205267, R03CA212125, R01CA215481, P30CA047904, and NRF-2015R1D1A1A01058303
We thank Christine Heiner (Department of Surgery, University of Pittsburgh) for her critical reading of the manuscript. This work was supported by the following grants: National Cancer Institute (NCI) grants R03 CA205267, R03 CA212125 (Y.J.L.), R01CA215481 (J. Y.) and National Research Foundation (NRF) of Korea grant NRF-2015R1D1A1A01058303 (D.H.L.). This project used the University of Pittsburgh Cancer Institute Core Facility and was supported in part by award P30CA047904.
Abbreviations used in this paper
- Apaf1
apoptosis signal-regulating kinase
- ART
artesunate
- ASNS
asparagine synthetase
- ATCC
American Tissue Type Culture Collection
- ATF4
activating transcription factor 4
- ATF6
activating transcription factor-6
- Bak
Bcl-2 homologous antagonist killer
- Bax
Bcl-2–associated X protein
- Bcl-2
B-cell lymphoma 2
- Bim
Bcl-2-like protein 11
- BLI
bioluminescence imaging
- c-FLIP
FADD-like IL-1β-converting enzyme-inhibitory protein
- C/EBP
CCAAT-enhancer-binding protein
- CHAC1
glutathione-specific gamma-glutamylcyclotransferase 1
- CHOP
CCAAT-enhancer-binding protein homologous protein
- CI
combination index
- DAPI
4′,6-diamidino-2-phenylindole
- DISC
death-inducing signaling complex
- DR
death receptor
- DR4
death receptor 4
- DR5
death receptor 5
- eIF2α
eukaryotic initiation factor 2α
- ER
endoplasmic reticulum
- ERA
erastin
- ERO1α
endoplasmic reticulum, oxidoreductin-1α
- GADD34
growth arrest and DNA damage-inducible protein
- GPX4
glutathione peroxidase 4
- GSH
glutathione
- HRP
horseradish peroxidase
- IRE1
inositol requiring protein-1
- MEF
mouse embryonic fibroblast
- NCOA4
nuclear receptor coactivator 4
- PARP
poly (ADP-ribose) polymerase
- PERK
PKR-like ER kinase
- PKR
protein kinase RNA
- PUMA
p53 upregulated modulator of apoptosis
- siDR5
DR5 small interfering RNA
- siRNA
small interfering RNA
- SRF
sorafenib
- TRAIL
tumor necrosis factor-related apoptosis-inducing ligand
- TUNEL
terminal deoxynucleotidyl transferase dUTP nick-end labeling
- UPR
unfolded protein response
- WT
wild-type
Footnotes
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Author Contributions
Y.S.L., D.H.L., S.Y.J. and S.H.P. collected the data and performed the experiments. J.Y. provided materials. S.C.O., Y.S.P., H.A.C. and D.L.B. designed and concepted the work. Y.S.L. and Y.J.L. participated in analyzing data and interpretation. Y.J.L. drafted the manuscript. All authors read and approved the final manuscript.
References
- 1.Dixon SJ, Lemberg KM, Lamprecht MR, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Braughler JM, Duncan LA, Chase RL. The involvement of iron in lipid peroxidation. Importance of ferric to ferrous ratios in initiation. J Biol Chem. 1986;261:10282–10289. [PubMed] [Google Scholar]
- 3.Gao M, Monian P, Pan Q, Zhang W, Xiang J, Jiang X. Ferroptosis is an autophagic cell death process. Cell Res. 2016;26:1021–1032. doi: 10.1038/cr.2016.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Friedmann Angeli JP, Schneider M, Proneth B, et al. Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat Cell Biol. 2014;16:1180–1191. doi: 10.1038/ncb3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang WS, SriRamaratnam R, Welsch ME, et al. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156:317–331. doi: 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conrad M, Friedmann Angeli JP. Glutathione peroxidase 4 (Gpx4) and ferroptosis: what’s so special about it? Mol Cell Oncol. 2015;2:e995047. doi: 10.4161/23723556.2014.995047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xie Y, Hou W, Song X, et al. Ferroptosis: process and function. Cell Death Differ. 2016;23:369–379. doi: 10.1038/cdd.2015.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lisewski AM, Quiros JP, Ng CL, et al. Supergenomic network compression and the discovery of EXP1 as a glutathione transferase inhibited by artesunate. Cell. 2014;158:916–928. doi: 10.1016/j.cell.2014.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Magtanong L, Ko PJ, Dixon SJ. Emerging roles for lipids in non-apoptotic cell death. Cell Death Differ. 2016;23:1099–1109. doi: 10.1038/cdd.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang WS, Stockwell BR. Ferroptosis: Death by Lipid Peroxidation. Trends Cell Biol. 2016;26:165–176. doi: 10.1016/j.tcb.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang WS, Kim KJ, Gaschler MM, Patel M, Shchepinov MS, Stockwell BR. Peroxidation of polyunsaturated fatty acids by lipoxygenases drives ferroptosis. Proc Natl Acad Sci U S A. 2016;113:E4966–4975. doi: 10.1073/pnas.1603244113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dixon SJ, Patel DN, Welsch M, et al. Pharmacological inhibition of cystine-glutamate exchange induces endoplasmic reticulum stress and ferroptosis. Elife. 2014;3:e02523. doi: 10.7554/eLife.02523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rahmani M, Davis EM, Crabtree TR, et al. The kinase inhibitor sorafenib induces cell death through a process involving induction of endoplasmic reticulum stress. Mol Cell Biol. 2007;27:5499–5513. doi: 10.1128/MCB.01080-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao JY, Dixon SJ. Mechanisms of ferroptosis. Cell Mol Life Sci. 2016;73:2195–2209. doi: 10.1007/s00018-016-2194-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dixon BM, Heath SH, Kim R, Suh JH, Hagen TM. Assessment of endoplasmic reticulum glutathione redox status is confounded by extensive ex vivo oxidation. Antioxid Redox Signal. 2008;10:963–972. doi: 10.1089/ars.2007.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ohoka N, Yoshii S, Hattori T, Onozaki K, Hayashi H. TRB3, a novel ER stress-inducible gene, is induced via ATF4-CHOP pathway and is involved in cell death. EMBO J. 2005;24:1243–1255. doi: 10.1038/sj.emboj.7600596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hong SH, Lee DH, Lee YS, et al. Molecular crosstalk between ferroptosis and apoptosis: emerging role of ER stress-induced p53-independent PUMA expression. Oncotarget. 2017;8:115164–115178. doi: 10.18632/oncotarget.23046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oslowski CM, Urano F. Measuring ER stress and the unfolded protein response using mammalian tissue culture system. Methods Enzymol. 2011;490:71–92. doi: 10.1016/B978-0-12-385114-7.00004-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghosh AP, Klocke BJ, Ballestas ME, Roth KA. CHOP potentially co-operates with FOXO3a in neuronal cells to regulate PUMA and BIM expression in response to ER stress. PLoS One. 2012;7:e39586. doi: 10.1371/journal.pone.0039586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Urra H, Dufey E, Lisbona F, Rojas-Rivera D, Hetz C. When ER stress reaches a dead end. Biochim Biophys Acta. 2013;1833:3507–3517. doi: 10.1016/j.bbamcr.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 21.Lee YS, Lee DH, Choudry HA, Bartlett DL, Lee YJ. Ferroptosis-induced endoplasmic reticulum stress: cross-talk between ferroptosis and apoptosis. Mol Cancer Res. 2018;16:1073–1076. doi: 10.1158/1541-7786.MCR-18-0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nikoletopoulou V, Markaki M, Palikaras K, Tavernarakis N. Crosstalk between apoptosis, necrosis and autophagy. Biochim Biophys Acta. 2013;1833:3448–3459. doi: 10.1016/j.bbamcr.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 23.Lu M, Lawrence DA, Marsters S, et al. Opposing unfolded-protein-response signals converge on death receptor 5 to control apoptosis. Science. 2014;345:98–101. doi: 10.1126/science.1254312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamaguchi H, Wang HG. CHOP is involved in endoplasmic reticulum stress-induced apoptosis by enhancing DR5 expression in human carcinoma cells. J Biol Chem. 2004;279:45495–45502. doi: 10.1074/jbc.M406933200. [DOI] [PubMed] [Google Scholar]
- 25.Edagawa M, Kawauchi J, Hirata M, et al. Role of activating transcription factor 3 (ATF3) in endoplasmic reticulum (ER) stress-induced sensitization of p53-deficient human colon cancer cells to tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL)-mediated apoptosis through up-regulation of death receptor 5 (DR5) by zerumbone and celecoxib. J Biol Chem. 2014;289:21544–21561. doi: 10.1074/jbc.M114.558890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin-Perez R, Palacios C, Yerbes R, et al. Activated ERBB2/HER2 licenses sensitivity to apoptosis upon endoplasmic reticulum stress through a PERK-dependent pathway. Cancer Res. 2014;74:1766–1777. doi: 10.1158/0008-5472.CAN-13-1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin-Perez R, Niwa M, Lopez-Rivas A. ER stress sensitizes cells to TRAIL through down-regulation of FLIP and Mcl-1 and PERK-dependent up-regulation of TRAIL-R2. Apoptosis. 2012;17:349–363. doi: 10.1007/s10495-011-0673-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li T, Su L, Lei Y, Liu X, Zhang Y, Liu X. DDIT3 and KAT2A proteins regulate TNFRSF10A and TNFRSF10B expression in endoplasmic reticulum stress-mediated apoptosis in human lung cancer cells. J Biol Chem. 2015;290:11108–11118. doi: 10.1074/jbc.M115.645333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun SY, Liu X, Zou W, Yue P, Marcus AI, Khuri FR. The farnesyltransferase inhibitor lonafarnib induces CCAAT/enhancer-binding protein homologous protein-dependent expression of death receptor 5, leading to induction of apoptosis in human cancer cells. J Biol Chem. 2007;282:18800–18809. doi: 10.1074/jbc.M611438200. [DOI] [PubMed] [Google Scholar]
- 30.Ganten TM, Haas TL, Sykora J, et al. Enhanced caspase-8 recruitment to and activation at the DISC is critical for sensitisation of human hepatocellular carcinoma cells to TRAIL-induced apoptosis by chemotherapeutic drugs. Cell Death Differ. 2004;11(Suppl 1):S86–96. doi: 10.1038/sj.cdd.4401437. [DOI] [PubMed] [Google Scholar]
- 31.Chang DW, Xing Z, Capacio VL, Peter ME, Yang X. Interdimer processing mechanism of procaspase-8 activation. EMBO J. 2003;22:4132–4142. doi: 10.1093/emboj/cdg414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li P, Nijhawan D, Budihardjo I, et al. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- 33.Wei MC, Lindsten T, Mootha VK, et al. tBID, a membrane-targeted death ligand, oligomerizes BAK to release cytochrome c. Genes Dev. 2000;14:2060–2071. [PMC free article] [PubMed] [Google Scholar]
- 34.Grinberg M, Sarig R, Zaltsman Y, et al. tBID Homooligomerizes in the mitochondrial membrane to induce apoptosis. J Biol Chem. 2002;277:12237–12245. doi: 10.1074/jbc.M104893200. [DOI] [PubMed] [Google Scholar]
- 35.Logue SE, Cleary P, Saveljeva S, Samali A. New directions in ER stress-induced cell death. Apoptosis. 2013;18:537–546. doi: 10.1007/s10495-013-0818-6. [DOI] [PubMed] [Google Scholar]
- 36.Eskes R, Desagher S, Antonsson B, Martinou JC. Bid induces the oligomerization and insertion of Bax into the outer mitochondrial membrane. Mol Cell Biol. 2000;20:929–935. doi: 10.1128/mcb.20.3.929-935.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baliga B, Kumar S. Apaf-1/cytochrome c apoptosome: an essential initiator of caspase activation or just a sideshow? Cell Death Differ. 2003;10:16–18. doi: 10.1038/sj.cdd.4401166. [DOI] [PubMed] [Google Scholar]
- 38.Kim BR, Jeong YA, Na YJ, et al. Genipin suppresses colorectal cancer cells by inhibiting the Sonic Hedgehog pathway. Oncotarget. 2017;8:101952–101964. doi: 10.18632/oncotarget.21882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Safa AR. c-FLIP, a master anti-apoptotic regulator. Exp Oncol. 2012;34:176–184. [PMC free article] [PubMed] [Google Scholar]