Abstract
Autism spectrum disorder (ASD) is a highly prevalent and complex genetic disorder. The complex genetic make-up of ASD has been extensively studied and both common and rare genetic variants in up to 1000 genes have been linked to increased ASD risk. While these studies highlight the genetic complexity and begin to provide a window for delineating pathways at risk in ASD, the pathogenicity and specific contribution of many mutations to the disorder are poorly understood. Defining the convergent pathways disrupted by this large number of ASD-associated genetic variants will help to understand disease pathogenesis and direct future therapeutic efforts for the groups of patients with distinct etiologies. Here, we review some of the common regulatory pathways including chromatin remodeling, transcription, and alternative splicing that have emerged as common features from genetic and transcriptomic profiling of ASD. For each category, we focus on one gene (CHD8, FOXP1, and RBFOX1) that is significantly linked to ASD and functionally characterized in recent years. Finally, we discuss genetic and transcriptomic overlap between ASD and other neurodevelopmental disorders.
Keywords: transcription, FOXP1, splicing, CHD8, RBFOX1, network, autism
Introduction
Autism spectrum disorder (ASD) is a neurodevelopmental condition with a prevalence of 1 in 59 children in the United States per recent estimates1. ASD is characterized by impairments in reciprocal social interactions as well as the presence of repetitive and restricted behaviors and interests2. The increased prevalence of the disease in siblings of ASD patients and greater ASD concordance rates in monozygotic twins compared with dizygotic twins confirmed that ASD has a major heritable component3,4,5-7. Within the last decade, numerous large-scale family-based whole exome and genome sequencing studies have identified a rapidly growing number of genes linked to ASD8-16. These studies, which include family cohorts with sporadic ASD (simplex) or with more than one affected individual (multiplex), resulted in the discovery of rare or common variants with various inheritance patterns. Interestingly, these studies uncovered the involvement of non-inherited genetic variants such as de novo (spontaneous)8,13,14,17 and somatic mutations18-20. De novo mutations could arise in the germ cells of one parent or in the fertilized egg during embryogenesis resulting in an affected child with unaffected parents. Somatic mutations can occur at the later stages of development and yield mosaic individuals with distinct genomic content in subsets of cells21. Recurrence of genetic variants in independent cohorts as well as overlap of genes with inherited, de novo, and somatic mutations substantiates the pathogenicity of these mutations in ASD22 and rank them to a “high-confidence” category. Taken together, these studies underscore the complexity of the genetic landscape of the disease and begin to illuminate the biological pathways at risk in ASD. This complex genetic architecture also raises the possibility that certain combinations of common genetic variants contribute to ASD by modifying the pathogenic effects of rare inherited, de novo or somatic mutations.
Given the progress in gene discovery in large-scale family based studies, the pressing challenge now is to prioritize high-confidence causal genes in ASD for further functional studies validating and defining the pathogenicity. Several approaches have been taken to pinpoint high confidence causative genetic variants. First, recurrent mutations of a given gene found in independent family cohorts and unrelated individuals with ASD can “rank” the gene to a high confidence category23,24. Second, predictions of the damaging potential of a mutation to gene structure and function are also taken into account when prioritizing the loss-of-function (LOF) mutations8. Third, a network-level approach involving functional annotation, gene lists implicated in brain development, neuronal function or monogenetic syndromes is being used to assess the functional relevance of newly identified genes25,26. Fourth, integrating transcriptomic analysis of ASD post-mortem tissue is also providing information on the pathways disrupted in ASD27-34. Knowledge acquired from transcriptome studies can be used as a framework to assess the novel candidate genes for their involvement in particular pathways affected in ASD. These approaches have helped predict pathogenicity of the large catalog of ASD-associated genetic variants; however, the functional impact of each mutation on the developing brain still needs to be determined and validated in experimental models including three-dimensional brain organoids22 and rodent models36.
The polygenic nature of ASD is also supported by the fact that many high confidence ASD mutations reside in regulatory genes encoding chromatin remodelers, transcription factors (TFs) and RNA binding proteins (RBPs) that can further regulate a multitude of developmental programs rather than a single gene function. Thus, loss-of-function mutations within one of these key master regulators can cause ASD by leading to dysregulation of an entire network of genes that coexpress and function together during critical windows of neurodevelopment.
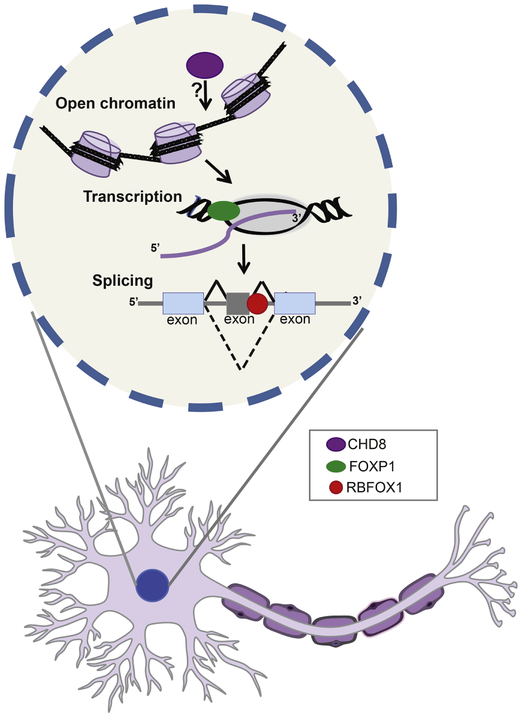
Here, we will review the recent progress on three gene regulatory pathways implicated in ASD as common mechanisms (chromatin remodeling, transcriptional control and alternative splicing) with the focus and example of high-confidence and relatively well-studied ASD genes linked to these respective pathways (CHD8, FOXP1, RBFOX1) (Figure 1). We will describe the mechanistic insights that have emerged from cell and animals models for these high-confidence ASD genes. Finally, we will discuss increasing evidence for shared molecular features of ASD with other neurodevelopmental disorders, in particular schizophrenia (SCZ).
Figure 1: Regulatory genes disrupted in ASD.
High confidence ASD genes regulate multiple levels of gene expression. CHD8 (purple) is a chromatin remodeler and is associated with open chromatin and active promoters. FOXP1 is a transcriptional factor (green) active or repress its transcriptional targets. RBFOX1 (red) is a RNA binding protein and regulates RNA metabolism including splicing.
Chromatin Remodeling
Regulation of gene expression plays a predominant role in cell fate determination and maintenance during human brain development37. The local chromatin state surrounding any given gene is an important determinant for the gene to be “on” or “off” and is regulated by the chromatin remodeling complexes38. Proper regulation of chromatin states is critical for ensuring key genetic programs are in place during developmental stages. The role of gene regulation at the chromatin level in human cortical development and function is further supported by the identification of mutations in chromatin remodeling genes linked to neurodevelopmental and neuropsychiatric disorders39,40.
The gene encoding the chromodomain helicase DNA-binding protein 8, CHD8, has emerged as a high-confidence ASD gene. Recurrence of rare, de novo, LOF mutations in CHD8 among unrelated individuals with ASD points to chromatin remodeling as a converging molecular disease mechanism8,13,14,17,23,41,42. In addition to typical features of ASD, patients harboring CHD8 mutations often are co-morbid for macrocephaly, facial dysmorphisms, and intellectual disability42,43.
CHD8 is an ATP-dependent chromatin remodeling protein, which is a member of the chromodomain-helicase-DNA binding protein family. Regulatory roles for CHD8 in Wnt signaling44 and apoptosis45 have been implicated; however, knowledge of the cellular function of CHD8, particularly in brain, is sparse. The strong genetic link of CHD8 in ASD has fuelled mechanistic studies geared towards understanding the role of CHD8 in brain development and function along with the consequences of reduced CHD8 levels in animal and cell models.
Modeling disease-associated haploinsufficieny of CHD8 through knockdown studies in human neural cell models followed by RNA-sequencing (RNA-seq) facilitated the identification of the subset of genes regulated by CHD846-48. While reduced CHD8 levels lead to altered expression of hundreds of genes, other known ASD risk genes are significantly enriched among the downregulated but not upregulated genes upon CHD8 reduction46-48. The use of chromatin immunoprecipitation-sequencing (ChIP-seq) in cell models46,47 and human midfetal brain tissue47 showed that CHD8 binds to active promoter regions marked with trimethylated histone H3 lysine 4. In agreement with knockdown studies, genes that are identified as direct targets of CHD8 in developing human brain are also enriched for ASD candidate genes47. These data highlight the possibility that the majority of ASD risk genes are co-expressed and subject to co- and crossregulation. CHD8 is likely to have a prominent regulatory role in critical co-expression networks and the loss of CHD8 thereby contributes to ASD pathogenesis by disrupting numerous downstream cellular processes.
Multiple genetically modified mice with decreased expression of CHD8 have been developed to characterize the impact of reduced CHD8 levels on brain development and behavioral outcomes (Table 1). Whereas homozygous deletion of Chd8 is embryonically lethal in mice45, haploinsufficient models of Chd8 have been established through conventional exon targeting49,50, in utero knockdown of CHD851, or introducing a gene-disrupting mutation via CRISPR/Cas9 gene editing52,53. Consistent with the macrocephaly observed in patients harboring CHD8 mutations42, imaging49,50,53 and histological52 examination of heterozygous Chd8 mutant mice show increased brain volume relatively to wild type littermates. CHD8 mutant and in utero knockdown models manifest some degree of altered behavior potentially relevant to clinical features of ASD however; these results are highly variable between studies. While two of the heterozygous knockout49,53 and knockdown models51 show mild deficits in social interaction, two different knockout models are reported to have normal social behavior50,52. Similarly, cognitive deficits are found only in one of the models52. The most recent study also reported a motor deficit in mutant mice50, while former models did not show atypical motor function. Platt et al53 observed an increased acquired motor learning phenotype in Chd8 knock-in mice. The authors linked this behavior to synaptic dysfunction of spiny projection neurons in the nucleus accumbens (NAc) via a region-specific targeting of Chd8 in adult animals in NAc53. These findings implicate region-specific roles for CHD8 and support the role of NAc dysfunction in ASD. Although these studies are able to recapitulate various behavioral aspects of the disease, discrepancies in the behavioral outcomes among studies need to be addressed. While such behavioral assays could be confounded by genetic background, sex, or age of the animals tested as well as the sensitivity of the techniques, these differences might also result from uncharacterized differences in CHD8 dose among different models.
Table 1:
Mouse models of regulatory ASD genes (CHD8, FOXP1, RBFOX1).
| Reference | Strategy Used |
Morphological Phenot |
Behavioral Phenotype |
Physiological Deficits |
Downstream Targets | |
|---|---|---|---|---|---|---|
| CHD8 | Katayama et al49, 2016 | Heterozygous s knockout (Cre-mediated) | macrocephaly | Increased anxiety, deficits in social behaviour, normal learning | n.d. | neuronal development, REST complex |
| Durak et al.51, 2016 | in utero knockdown (in developing cortex) | Defective neural progenitor proliferation | deficits in social behaviour, normal learning | n.d. | cell cycle, Wnt signaling | |
| Platt et al.53, 2017 | heterozygou s knockout (CRISPR-mediated) | macrocephaly | mild social defects, increased anxiety, no repetative behaviour, increased acquired motor learning | decreased inhibitory signaling in SPNs of Nac | chromatin remodelling, mRNA processing, cell cycle, Wnt signaling | |
| Gompers et al.52, 2016 | heterozygou s knockout (CRISPR-mediated) | macrocephaly | normal social behaviour, no repetative behaviour, learning and memory impairment | n.d. | RNA processing, chromatin remodelling, cell cycle | |
| Suetterlin et al.50, 2018 | heterozygou s knockout (Cre-mediated) | macrocephaly | normal social behaviour, delayed motor development, hypoactivity in adults | n.d. | CNS development, cell adhesion, axon guidance | |
| FOXP1 | Bacon et al.65, 2015 | brain-specific knockout (Nestin.Cre-mediated) | enlarged lateral ventricle, abnormalitie s in striatum, decreased neuronal density in hippocampu s | increased repetitive behavior, decreased social interest and impairment s in patial memory | decreased excitability and increased excitatory synaptic transmission in hippocampal pyramidal neurons. | chromatin, nucleosome, cell cyle |
| Araujo et al.66, 2015 | heterozygous knockout | n.d. | defects in neonatal ultrasonic vocalizations | increased excitability of striatal SPNs | striatal development | |
| Li et al.67, 2015 | in utero knockdown (in developing cortex) | defective neuronal migration, defective Neurite development | n.d. | n.d. | n.d. | |
| Araujo et al.68, 2017 | conditional knockout in forebrain pyramidal neurons (Emx.Cre-mediated) | decreased hippocampal volume | hyperactivity, decreased sociability, impaired hippocampal-based spatial learning | decreased late-phase long-term potentiatio n (LTP) response | neurogenesis, neural differantiation, synaptic transmission | |
| Usui et al.69, 2017 | conditional knockout in forebrain pyramidal neurons (Emx.Cre-mediated) | reduced neocortical size and mispositioning of deep layer neurons | impaired postnatal vocal communicatio n | n.d. | neurogenesi s and neuronal migration | |
| RBFOX1 | Gehman et al.77, 2011 | brain-specific knockout (Nestin.Cre-mediated) | normal gross morphology | seizures | hyperexcitbility in hippocampal neurons | SNARE complex, neurotransmitter genes, ion channels |
| Hamada et al.79, 2015 | in utero knockdown (in developing cortex) | defects in neuronal migration, neuronal placement, and dendritic arbor formation | n.d. | n.d. | n.d. |
SPN: spiny projection neuron, Nac: nucleus accumbens, n.d: not determined
Gene expression studies of these mice captured subtle yet widespread changes in gene expression consistent with the studies in ASD human data. Differentially expressed genes are consistently enriched for functional annotations including chromatin and histone modification, and cell-cycle regulation49,51,52. These data suggest involvement of a network of epigenetic modifiers in CHD8-mediated gene regulation. Dysregulation of cell-cycle genes are consistent with the macrocephaly phenotypes. More specifically, mice heterozygous for Chd8 show elevated expression levels of genes involved in early fetal development and downregulation of genes expressed during mid-fetal stages, indicating a developmental delay49. Based on gene expression profiles, each study has identified potential downstream mechanisms of Chd8 haploinsufficiency including activation of the REST complex49 or disrupted Wnt signaling51,53. Remarkably, Gompers et al.52 identified downregulation of a group of genes responsible for RNA processing and widespread alternative splicing changes in Chd8 heterozygous mice. Thus, CHD8 can indirectly regulate alternative splicing, another convergent mechanism implicated in ASD discussed below, by controlling the expression of RNA processing genes.
Genetic evidence for the involvement of CHD8 in ASD is particularly strong. Studies of cell and animal models show that CHD8 is required for neuronal function and regulates a network of genes critical for early neurodevelopment. Moving forward to therapeutic strategies would require addressing a number of remaining questions including: 1) What is the mechanism CHD8 uses to either repress or activate genes? 2) What is the therapeutic window for reversing phenotypes related to dysfunction of CHD8? 3) Can CHD8-regulated events also be dysregulated due to environmental factors in the absence of a mutation?
Transcriptional Control
Transcription factors play a key role in intricate regulation of the spatial and temporal gene expression patterns important for brain development37,54. Work over the past few decades has identified a number of transcription factors that cooperatively and/or hierarchically control proper brain development. Variants in genes encoding transcription factor and dysregulated gene expression have been reported in neurodevelopmental disorders highlighting the need for the identification of gene networks regulated by the transcription factors implicated in both brain development and disease states.
The gene encoding the Forkhead box transcription factor 1, FOXP1, has been implicated in neurodevelopmental disorders such as ASD and ID55. Numerous studies have identified geneinterrupting variants of FOXP1 including heterozygous deletions, duplications, and missense and nonsense mutations in both case reports and recent large-scale profiling of patients with ASD and ID, ranking FOXP1 as one of the high-confidence causal ASD genes17,56-58,40,59-61. Moreover, FOXP2, a paralog of FOXP1 is linked to human speech and language development suggesting a prominent role for FOXP proteins in human cognitive function including language55,62-64. Collectively, these studies have provided strong evidence for FOXP1 mutations underlying specific cognitive phenotypes, and have prompted research on FOXP1 function in brain.
Several groups have begun to elucidate a role for FOXP1 in neurodevelopment and cognitive function (Table 1). Mice with brain-specific deletion of Foxp1 exhibit widespread morphological defects throughout the brain including enlarged lateral ventricles, impaired striatal development, and decreased density of CA1 neurons in hippocampus65. These structural alterations are accompanied by an excitatory/inhibitory (E/I) imbalance in hippocampal CA1 neurons. Behavioral deficits in these mice include increased repetitive behaviors, decreased social interest and impairments in spatial memory. Moreover, heterozygous knockout Foxp1 mice modeling patient-relevant haploinsufficiency show increased excitability of striatal spiny projection neurons (SPNs) and defects in neonatal ultrasonic vocalizations (USVs)66. Gene expression and co-expression module preservation analyses of the heterozygous knockout mice with human neuronal data demonstrate that Foxp1 orchestrates gene expression networks important for striatal development and function that are at risk in ASD. These results from the brain-specific and heterozygous knockout mice models support the functional significance of FOXP1 in neurodevelopment, social and cognitive function, and vocal communication; however, these studies are limited in linking behavioral outcomes to region-specific defects caused by the loss of Foxp1. This is relevant because unlike CHD8, FOXP1 is not widely expressed in the brain.
To address a region-specific role of Foxp1, in utero knockdown of Foxp1 expression in developing neocortex results in defective neuronal migration and neurite development; however, behavioral outcomes of decreased cortical levels of Foxp1 in this model have yet to be reported67. A more complete characterization of the brain region-specific role of Foxp1 comes from studies of conditional knockout mice with loss of Foxp1 in the pyramidal neurons of the neocortex and the hippocampus68. These mice exhibit hyperactivity, decreased sociability, impaired hippocampal-based spatial learning and memory highlighting the role of Foxp1 in the hippocampus68. Consistent with behavioral deficits indicative of impaired hippocampal function, these mice present with a decreased late-phase long-term potentiation (LTP) response. Pathways disturbed due to loss of Foxp1 in the hippocampus that could potentially contribute to the LTP and spatial learning deficits were examined using genomic approaches. Gene ontology categories of differentially expressed genes downstream of Foxp1 in the hippocampus include abnormal synaptic transmission, and abnormal learning/memory/conditioning in agreement with the behavioral and electrophysiological characterization68. In addition, deletion of Foxp1 in pyramidal neurons of the forebrain results in impaired vocal communication in postnatal stages in mice69. Structural changes that occurred from loss of cortical Foxp1 include reduced overall neocortical size and mispositioning of neurons in the deep layers of the mouse neocortex69. Transcriptional networks regulated by Foxp1 in early development include genes that are responsible for neurogenesis and neuronal migration. Both in hippocampus and neocortex, Foxp1 regulated genes are enriched for other ASD genes68,69. Taken together, these studies provide insights into the role of Foxp1 in distinct brain regions and highlights brain-region specific features of a complex disorder.
In summary, Foxp1 regulates distinct sets of transcriptional programs in different brain regions and loss of Foxp1 function yields social and cognitive deficits. Disentangling these diverse functions of Foxp1 in different brain regions and cell-types will be important for understanding region-specific pathophysiology of the disease and guiding future therapeutic efforts. Future studies focused on the role of Foxp1 in striatum will be important for complete understanding of the molecular basis for complex ASD presentation as striatal circuits are affected in ASD and brain-specific Foxp1 knockout mice show striking striatal defects.
Alternative Splicing
There is a growing body of evidence showing the prominent role of alternative post-transcriptional processing events including alternative splicing (AS) and polyadenylation in human brain. Considering the limited number of protein coding genes in the human genome, AS is increasingly recognized as the primary source of transcriptomic and proteomic diversity and complexity driving the species-specific features of humans including brain evolution70-72. AS is coor post-transcriptionally regulated by RNA binding proteins (RBPs) and tightly controlled during normal development stages in a tissue-specific manner73. Consistent with the presumed role of alternative splicing regulation in human brain, erroneous AS regulation has been implicated in many neurologic diseases including frontotemporal dementia and myotonic dystrophy74.
Transcriptomic profiling of ASD has increasingly pointed to dysregulation of AS as a convergent mechanism for disease pathogenesis. ASD-linked copy number variations and chromosomal translocation in one particular neuronal RBP with a role in AS, RBFOX1, have been highlighted in patient cohorts11,75,76. In addition, transcriptomic analyses of ASD postmortem brains have identified dysregulated RBFOX1 function as a common feature of genetically distinct ASD cases, supporting a prominent role for loss and/or dysregulation of RBFOX1 activity in ASD pathogenesis27,28.
Several studies have begun to characterize the role of RBFOX1 and understand the functional impact of defective RBFOX1 function (Table 1). Brain-specific knockout mice show spontaneous and induced seizures and aberrantly increased neuronal activity77. The loss of Rbfox1 resulted in increased excitability in dentate gyrus consistent with the imbalanced E/I activity observed in other ASD models77. A separate study also reported decreased inhibitory synaptic transmission in CA1 neurons of this model78. Whole-transcriptome profiling by RNA-seq identified gene expression and alternative splicing changes in the knockout mice77. In utero knockdown of Rbfox1 caused defects in neuronal migration, neuronal placement, and dendritic arbor formation during corticogenenis; however, behavioral consequences of these defects have yet to be determined79. Specific functional consequences of loss of Rbfox proteins were also investigated in motor neurons differentiated from mouse embryonic stem cells (ESCs) lacking all three members of the Rbfox protein family (Rbfox1, Rbfox, and Rbfox3)80. These triple knockout neurons show immature electrophysiological activity and defective axon initial segment assembly (AIS). Remarkably, defects in AIS have previously been implicated for ASD as high-confidence variants are found in genes involved in this process including SCN2A81. Depletion of Rbfox proteins led to missplicing of genes encoding cytoskeletal, cell membrane and synaptic proteins. Moreover, studies in mice delineated differential roles of cytoplasmic and nuclear isoforms of Rbfox182. In addition to canonical splicing regulation by nuclear Rbfox1, the cytoplasmic isoform of Rbfox1 elicits distinct functions including regulation of RNA stability and translation. Vamp1, a vSNARE protein was identified as one of the downstream targets of cytoplasmic Rbfox1 and shown to be downregulated in Rbfox1 knockout mice as a result of loss of post-transcriptional regulation. Forced expression of Vamp1 using AAV mediated delivery in Rbfox1 knockout mice rescued inhibitory synaptic transmission defects in those mice78. These findings suggest several aspects of RNA metabolism including translation efficiency and stability might be dysregulated in ASD due to the loss of RBFOX1 function.
Studies of human neural progenitor cells suggested that a larger network of RBPs along with RBFOX1 is co-expressed during development, and can potentially function together in post-transcriptional regulation of cortical development83,84. Moreover, the overlapping targets in this network are important for neuronal development and are likely disrupted in ASD. For example, the ELAVL2 binding motif was enriched in alternatively spliced exons in human neurons with decreased levels of RBFOX1 suggesting a coordinated combinatorial regulation of RNA processing by RBFOX1 and ELAVL2. Consistent with this hypothesis, transcripts misspliced in postmortem ASD brains are also enriched for cellular targets of several RBPs including SRRM429 and PTB127.
These data highlight RBP function, including those of RBFOX1, as essential for cortical development and function, and at risk in ASD. Taken together, dysregulation of RNA processing may be a unifying feature of genetically diverse ASD cases, and regulation of these processes might be viable targets for therapeutic strategies.
Overlapping Pathways
Genetic and transcriptomic profiling of ASD has found overlapping molecular underpinning for ASD and other neurodevelopmental disorders, in particular schizophrenia (SCZ). In fact, genetic variants in CHD885, FOXP186, and RBFOX187 have been also reported in patients with SCZ. Additional de novo mutations have been identified in overlapping chromatin and synaptic genes in ASD and SCZ patients8,88. Moreover, examination of microarray and RNA-seq data from postmortem brain tissue across several neuropsychiatric disorders including ASD and SCZ has revealed similar transcriptome signatures for ASD and SCZ with downregulation of synaptic genes and upregulation of astroglial genes31. While these data highlight the possibility of shared pathways at risk in both ASD and SCZ, these results cannot explain how perturbations of similar genes results in strikingly distinct clinical representations. One potential explanation is that distinct combinations of common genetic variants in each individual genome determine the expressivity and penetrance of rare, disease-associated variants. Additionally, the biological impact of different mutations on the same gene might lead to distinct pathogenesis leading to ASD or SCZ. For example, missense variants in the SCN2A gene encoding for a neuronal sodium channel have been linked to both ASD59 and infantile seizures89. The majority of SCN2A variants associated with infantile seizures are predicted to have gain of function effects leading to a hyperexcitability phenotype90,91. In contrast, bioinformatics and electrophysiological characterization of ASD-associated missense SCN2A variants have revealed their LoF effects leading to drastic reduction on channel conductance92. One recent example of differential pathogenesis caused by distinct mutations of the same gene came from patients harboring mutations in the PUM1 gene, a gene encoding an RBP93. Based on the severity of each LoF mutation in this gene, individuals harboring mutations either present with a severe developmental delay or developed an adult-onset ataxia93. A similar dichotomous effect was also reported for genetic variants of the gene RORα, which encodes the RAR-related orphan nuclear receptor alpha94. In this case, individuals with LoF variants presented with ID and ASD, whereas individuals with dominant toxic variants showed ID and ataxia. Finally, it is worth noting that these transcriptome data come from bulk brain tissue, and thus the reported gene expression levels represent the average levels across highly heterogeneous cell types and fail to determine cell-specific differences between unaffected, ASD, and SCZ brain tissue. Integrating data from studies profiling single-cell RNA-sequencing (scRNA-seq) of unaffected human brain95-97 with the list of disease associated genes lists27,28,32,98,99 has begun to provide insights into the cell-specific biology of these disorders. One such study100 showed that high confidence ASD-candidate genes and downstream targets of ASD gene including those of CHD8 are enriched in inhibitory neurons. While this finding is in agreement with the E/I imbalance hypothesis, this study is limited to cortical scRNA-seq and lacks data from other brain regions such as the striatum and hippocampus, which are known to be involved in ASD pathology. A similar approach101 identified the cell types affected in SCZ by integrating both mouse and human scRNA-seq datasets35,95,102-106 with genes linked to SCZ107. Inclusion of datasets spanning more diverse brain regions in this study led to the identification of SPNs, pyramidal cells in hippocampal CA1, pyramidal cells of the somatosensory cortex and cortical interneurons as cell types connected to SCZ101. Ultimately, scRNA-seq of disease-affected brain tissue will be more informative to characterize cell-specific pathways perturbed in ASD.
Conclusion
Genomics has identified altered regulatory processes including chromatin remodeling, transcription and alternative splicing as key contributors to ASD. Functionally relevant disease models have begun to provide insights into the basic function of ASD-associated chromatin remodelers, TFs, and RBPs. Collectively, these data demonstrate the involvement of functionally connected gene regulatory networks in ASD pathogenesis. However, these regulatory networks might be biased or underexplored due to the nature of the genetic studies focusing on LoF mutations and haploinsufficiency as primary disease mechanisms. Future studies should include identifying potential gain-of-function mutations, as neurons are known to be sensitive to overexpression and/or misfolding of certain proteins.
Highlights.
Chromatin remodeling, transcription, and alternative splicing are disrupted in ASD.
CHD8, FOXP1, and RBFOX1 are high confidence ASD genes related to these functions.
Cell and animal models have begun to elucidate the molecular function of these genes.
There are converging molecular pathways between ASD and other neurodevelopmental disorders.
Grant information
This work is supported by grants from the National Institutes of Health (R01DC014702 and R01MH102603 to GK and 5T32DA007290-25 to FA), the Simons Foundation Autism Research Initiative (project 401220 to GK), and a James S. McDonnell Foundation 21st Century Science Initiative in Understanding Human Cognition – Scholar Award to GK.
Abbreviations
- AIS
axon initial segment
- AS
alternative splicing
- ASD
autism spectrum disorder
- ChlP-seq
chromatin immunoprecipitation-sequencing
- E/I
excitatory/inhibitory
- ESCs
embryonic stem cells
- ID
intellectual disability
- LTP
long-term potentiation
- SCZ
schizophrenia
- SPN
spiny projection neurons
- RBP
RNA-binding protein
- scRNA-seq
single-cell RNA-sequencing
- TF
transcription factor
Footnotes
Competing interests
The authors declare that they have no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Baio J et al. Prevalence of Autism Spectrum Disorder Among Children Aged 8 Years - Autism and Developmental Disabilities Monitoring Network, 11 Sites, United States, 2014. MMWR Surveill Summ 67, 1–23, doi:10.15585/mmwr.ss6706a1 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Diagnostic and Statistical Manual of Mental Disorders | DSM Library. [DOI] [PubMed] [Google Scholar]
- 3.Rosenberg RE et al. Characteristics and concordance of autism spectrum disorders among 277 twin pairs. Arch Pediatr Adolesc Med 163,907–914, doi:10.1001/archpediatrics.2009.98 (2009). [DOI] [PubMed] [Google Scholar]
- 4.Bailey A et al. Autism as a strongly genetic disorder: evidence from a British twin study. Psychol Med 25, 63–77 (1995). [DOI] [PubMed] [Google Scholar]
- 5.Ozonoff S et al. Recurrence Risk for Autism Spectrum Disorders: A Baby Siblings Research Consortium Study. Pediatrics, peds.2010–2825, doi:10.1542/peds.2010-2825 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sandin S et al. The familial risk of autism. JAMA 311, 1770–1777, doi:10.1001/jama.2014.4144 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colvert E et al. Heritability of Autism Spectrum Disorder in a UK Population-Based Twin Sample. JAMA Psychiatry 72, 415–423, doi:10.1001/jamapsychiatry.2014.3028 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Rubeis S et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 515, 209–215, doi:10.1038/nature13772 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaugler T et al. Most genetic risk for autism resides with common variation. Nat. Genet 46, 881–885, doi:10.1038/ng.3039 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuen RKC et al. Whole genome sequencing resource identifies 18 new candidate genes for autism spectrum disorder. Nat Neurosci 20, 602–611, doi:10.1038/nn.4524 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sebat J et al. Strong association of de novo copy number mutations with autism. Science 316, 445–449, doi:10.1126/science.1138659 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanders SJ et al. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron 70, 863–885, doi:10.1016/j.neuron.2011.05.002 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanders SJ et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature 485, 237–241, doi:10.1038/nature10945 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neale BM et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature 485, 242–245, doi:10.1038/nature11011 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iossifov I et al. De novo gene disruptions in children on the autistic spectrum. Neuron 74, 285–299, doi:10.1016/j.neuron.2012.04.009 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu TW et al. Using whole-exome sequencing to identify inherited causes of autism. Neuron 77, 259–273, doi:10.1016/j.neuron.2012.11.002 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iossifov I et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature 515, 216–221, doi:10.1038/nature13908 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lim ET et al. Rates, distribution and implications of postzygotic mosaic mutations in autism spectrum disorder. Nat Neurosci 20, 1217–1224, doi:10.1038/nn.4598 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krupp DR et al. Exonic Mosaic Mutations Contribute Risk for Autism Spectrum Disorder. Am J Hum Genet 101, 369–390, doi:10.1016/j.ajhg.2017.07.016 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.D’Gama AM et al. Targeted DNA Sequencing from Autism Spectrum Disorder Brains Implicates Multiple Genetic Mechanisms. Neuro 88, 910–917, doi:10.1016/j.neuron.2015.11.009 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poduri A, Evrony GD, Cai X & Walsh CA Somatic mutation, genomic variation, and neurological disease. Science 341, 1237758, doi:10.1126/science.1237758 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ayhan F & Konopka G Genomics of autism spectrum disorder: approach to therapy. F1000Res 7, doi:10.12688/f1000research.13865.1 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Roak BJ et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science 338, 1619–1622, doi:10.1126/science.1227764 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O’Roak BJ et al. Recurrent de novo mutations implicate novel genes underlying simplex autism risk. Nat Commun 5, 5595, doi:10.1038/ncomms6595 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krishnan A et al. Genome-wide prediction and functional characterization of the genetic basis of autism spectrum disorder. Nat Neurosci 19, 1454–1462, doi:10.1038/nn.4353 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duda M et al. Brain-specific functional relationship networks inform autism spectrum disorder gene prediction. Transl Psychiatry 8, 56, doi:10.1038/s41398-018-0098-6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Voineagu I et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature 474, 380–384, doi:10.1038/nature10110 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parikshak NN et al. Genome-wide changes in lncRNA, splicing, and regional gene expression patterns in autism. Nature 540, 423–427, doi:10.1038/nature20612 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Irimia M et al. A highly conserved program of neuronal microexons is misregulated in autistic brains. Cell 159, 1511–1523, doi:10.1016/j.cell.2014.11.035 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wright C et al. Altered expression of histamine signaling genes in autism spectrum disorder. Transl Psychiatry 7, e1126, doi:10.1038/tp.2017.87 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gandal MJ et al. Shared molecular neuropathology across major psychiatric disorders parallels polygenic overlap. Science 359, 693–697, doi:10.1126/science.aad6469 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu X et al. Disruption of an Evolutionarily Novel Synaptic Expression Pattern in Autism. PLoS Biol 14, e1002558, doi:10.1371/journal.pbio.1002558 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gupta S et al. Transcriptome analysis reveals dysregulation of innate immune response genes and neuronal activity-dependent genes in autism. Nat Commun 5, 5748, doi:10.1038/ncomms6748 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chow ML et al. Age-dependent brain gene expression and copy number anomalies in autism suggest distinct pathological processes at young versus mature ages. PLoS Genet 8, e1002592, doi:10.1371/journal.pgen.1002592 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Habib N et al. Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat Methods 14, 955–958, doi:10.1038/nmeth.4407 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chahrour M et al. Current Perspectives in Autism Spectrum Disorder: From Genes to Therapy. J Neurosci 36, 11402–11410, doi:10.1523/JNEUROSCI.2335-16.2016 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Popovitchenko T & Rasin MR Transcriptional and Post-Transcriptional Mechanisms of the Development of Neocortical Lamination. Front Neuroanat 11,102, doi:10.3389/fnana.2017.00102 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ronan JL, Wu W & Crabtree GR From neural development to cognition: unexpected roles for chromatin. Nat Rev Genet 14, 347–359, doi:10.1038/nrg3413 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCarthy SE et al. De novo mutations in schizophrenia implicate chromatin remodeling and support a genetic overlap with autism and intellectual disability. Mol Psychiatry 19, 652–658, doi:10.1038/mp.2014.29 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vissers LE, Gilissen C & Veltman JA Genetic studies in intellectual disability and related disorders. Nat Rev Genet 17, 9–18, doi:10.1038/nrg3999 (2016). [DOI] [PubMed] [Google Scholar]
- 41.O’Roak BJ et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 485, 246–250, doi:10.1038/nature10989 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bernier R et al. Disruptive CHD8 mutations define a subtype of autism early in development. Cell 158, 263–276, doi:10.1016/j.cell.2014.06.017 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barnard RA, Pomaville MB & O’Roak BJ Mutations and Modeling of the Chromatin Remodeler CHD8 Define an Emerging Autism Etiology. Front Neurosci 9, 477, doi:10.3389/fnins.2015.00477 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thompson BA, Tremblay V, Lin G & Bochar DA CHD8 is an ATP-dependent chromatin remodeling factor that regulates beta-catenin target genes. Mol Cell Biol 28, 3894–3904, doi:10.1128/MCB.00322-08 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nishiyama M et al. CHD8 suppresses p53-mediated apoptosis through histone H1 recruitment during early embryogenesis. Nat Cell Biol 11, 172–182, doi:10.1038/ncb1831 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sugathan A et al. CHD8 regulates neurodevelopmental pathways associated with autism spectrum disorder in neural progenitors. Proc Natl Acad Sci U S A 111, E4468–4477, doi:10.1073/pnas.1405266111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cotney J et al. The autism-associated chromatin modifier CHD8 regulates other autism risk genes during human neurodevelopment. Nat Commun 6, 6404, doi:10.1038/ncomms7404 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilkinson B et al. The autism-associated gene chromodomain helicase DNA-binding protein 8 (CHD8) regulates noncoding RNAs and autism-related genes. Transl Psychiatry 5, e568, doi:10.1038/tp.2015.62 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Katayama Y et al. CHD8 haploinsufficiency results in autistic-like phenotypes in mice. Nature 537, 675–679, doi:10.1038/nature19357 (2016). [DOI] [PubMed] [Google Scholar]
- 50.Suetterlin P et al. Altered Neocortical Gene Expression, Brain Overgrowth and Functional Over-Connectivity in Chd8 Haploinsufficient Mice. Cereb Cortex 28, 2192–2206, doi:10.1093/cercor/bhy058 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Durak O et al. Chd8 mediates cortical neurogenesis via transcriptional regulation of cell cycle and Wnt signaling. Nat Neurosci 19, 1477–1488, doi:10.1038/nn.4400 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gompers AL et al. Germline Chd8 haploinsufficiency alters brain development in mouse. Nat Neurosci 20, 1062–1073, doi:10.1038/nn.4592 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Platt RJ et al. Chd8 Mutation Leads to Autistic-like Behaviors and Impaired Striatal Circuits. Cell Rep 19, 335–350, doi:10.1016/j.celrep.2017.03.052 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nord AS, Pattabiraman K, Visel A & Rubenstein JLR Genomic perspectives of transcriptional regulation in forebrain development. Neuron 85, 27–47, doi:10.1016/j.neuron.2014.11.011 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bowers JM & Konopka G The role of the FOXP family of transcription factors in ASD. Dis Markers 33, 251–260, doi:10.3233/DMA-2012-0919 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stessman HA et al. Targeted sequencing identifies 91 neurodevelopmental-disorder risk genes with autism and developmental-disability biases. Nat Genet 49, 515–526, doi:10.1038/ng.3792 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Le Fevre AK et al. FOXP1 mutations cause intellectual disability and a recognizable phenotype. Am J Med Genet A 161A, 3166–3175, doi:10.1002/ajmg.a.36174 (2013). [DOI] [PubMed] [Google Scholar]
- 58.Lozano R, Vino A, Lozano C, Fisher SE & Deriziotis P A de novo FOXP1 variant in a patient with autism, intellectual disability and severe speech and language impairment. Eur J Hum Genet 23, 1702–1707, doi:10.1038/ejhg.2015.66 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sanders SJ et al. Insights into Autism Spectrum Disorder Genomic Architecture and Biology from 71 Risk Loci. Neuron 87, 1215–1233, doi:10.1016/j.neuron.2015.09.016 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Siper PM et al. Prospective investigation of FOXP1 syndrome. Mol Autism 8, 57, doi: 10.1186/s13229-017-0172-6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meerschaut I et al. FOXP1-related intellectual disability syndrome: a recognisable entity. J Med Genet 54, 613–623, doi:10.1136/jmedgenet-2017-104579 (2017). [DOI] [PubMed] [Google Scholar]
- 62.Fisher SE & Scharff C FOXP2 as a molecular window into speech and language. Trends Genet 25, 166–177, doi:10.1016/j.tig.2009.03.002 (2009). [DOI] [PubMed] [Google Scholar]
- 63.Bacon C & Rappold GA The distinct and overlapping phenotypic spectra of FOXP1 and FOXP2 in cognitive disorders. Hum Genet 131, 1687–1698, doi:10.1007/s00439-012-1193-z (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Konopka G & Roberts TF Insights into the Neural and Genetic Basis of Vocal Communication. Cell 164, 1269–1276, doi:10.1016/j.cell.2016.02.039 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bacon C et al. Brain-specific Foxp1 deletion impairs neuronal development and causes autistic-like behaviour. Mol Psychiatry 20, 632–639, doi:10.1038/mp.2014.116 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Araujo DJ et al. FoxP1 orchestration of ASD-relevant signaling pathways in the striatum. Genes Dev 29, 2081–2096, doi:10.1101/gad.267989.115 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li X et al. Foxp1 regulates cortical radial migration and neuronal morphogenesis in developing cerebral cortex. PLoS One 10, e0127671, doi:10.1371/journal.pone.0127671 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Araujo DJ et al. Foxp1 in Forebrain Pyramidal Neurons Controls Gene Expression Required for Spatial Learning and Synaptic Plasticity. J Neurosci 37, 10917–10931, doi:10.1523/JNEUROSCI.1005-17.2017 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Usui N et al. Foxp1 regulation of neonatal vocalizations via cortical development. Genes Dev 31,2039–2055, doi:10.1101/gad.305037.117 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pan Q, Shai O, Lee LJ, Frey BJ & Blencowe BJ Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 40, 1413–1415, doi:10.1038/ng.259 (2008). [DOI] [PubMed] [Google Scholar]
- 71.Raj B & Blencowe BJ Alternative Splicing in the Mammalian Nervous System: Recent Insights into Mechanisms and Functional Roles. Neuron 87, 14–27, doi:10.1016/j.neuron.2015.05.004 (2015). [DOI] [PubMed] [Google Scholar]
- 72.Wang ET et al. Alternative isoform regulation in human tissue transcriptomes. Nature 456, 470–476, doi:10.1038/nature07509 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nussbacher JK, Batra R, Lagier-Tourenne C & Yeo GW RNA-binding proteins in neurodegeneration: Seq and you shall receive. Trends Neurosci 38, 226–236, doi:10.1016/j.tins.2015.02.003 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Scotti MM & Swanson MS RNA mis-splicing in disease. Nat Rev Genet 17, 19–32, doi: 10.1038/nrg.2015.3 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Martin CL et al. Cytogenetic and molecular characterization of A2BP1/FOX1 as a candidate gene for autism. Am J Med Genet B Neuropsychiatr Genet 144B, 869–876, doi:10.1002/ajmg.b.30530 (2007). [DOI] [PubMed] [Google Scholar]
- 76.Wintle RF et al. A genotype resource for postmortem brain samples from the Autism Tissue Program. Autism Res 4, 89–97, doi:10.1002/aur.173 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gehman LT et al. The splicing regulator Rbfox1 (A2BP1) controls neuronal excitation in the mammalian brain. Nat Genet 43, 706–711, doi:10.1038/ng.841 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vuong CK et al. Rbfox1 Regulates Synaptic Transmission through the Inhibitory Neuron-Specific vSNARE Vamp1. Neuron 98, 127–141e127, doi:10.1016/j.neuron.2018.03.008 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hamada N et al. Role of the cytoplasmic isoform of RBFOX1/A2BP1 in establishing the architecture of the developing cerebral cortex. Mol Autism 6, 56, doi:10.1186/s13229-015-0049-5 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jacko M et al. Rbfox Splicing Factors Promote Neuronal Maturation and Axon Initial Segment Assembly. Neuron 97, 853–868 e856, doi:10.1016/j.neuron.2018.01.020 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sanders SJ et al. Progress in Understanding and Treating SCN2A-Mediated Disorders. Trends Neurosci 41, 442–456, doi:10.1016/j.tins.2018.03.011 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lee JA et al. Cytoplasmic Rbfox1 Regulates the Expression of Synaptic and Autism-Related Genes. Neuron 89, 113–128, doi:10.1016/j.neuron.2015.11.025 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fogel BL et al. RBFOX1 regulates both splicing and transcriptional networks in human neuronal development. Hum Mol Genet 21, 4171–4186, doi:10.1093/hmg/dds240 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Berto S, Usui N, Konopka G & Fogel BL ELAVL2-regulated transcriptional and splicing networks in human neurons link neurodevelopment and autism. Hum Mol Genet 25, 2451–2464, doi:10.1093/hmg/ddw110 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kimura H et al. Identification of a rare variant in CHD8 that contributes to schizophrenia and autism spectrum disorder susceptibility. Schizophr Res 178, 104–106, doi:10.1016/j.schres.2016.08.023 (2016). [DOI] [PubMed] [Google Scholar]
- 86.Autism Spectrum Disorders Working Group of The Psychiatric Genomics, C. Metaanalysis of GWAS of over 16,000 individuals with autism spectrum disorder highlights a novel locus at 10q24.32 and a significant overlap with schizophrenia. Mol Autism 8, 21, doi: 10.1186/s13229-017-0137-9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen J et al. Genetic Relationship between Schizophrenia and Nicotine Dependence. Sci Rep 6, 25671, doi:10.1038/srep25671 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fromer M et al. De novo mutations in schizophrenia implicate synaptic networks. Nature 506, 179–184, doi:10.1038/nature12929 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Howell KB et al. SCN2A encephalopathy: A major cause of epilepsy of infancy with migrating focal seizures. Neurology 85, 958–966, doi:10.1212/WNL.0000000000001926 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Scalmani P et al. Effects in neocortical neurons of mutations of the Na(v)1.2 Na+ channel causing benign familial neonatal-infantile seizures. J Neurosci 26, 10100–10109, doi:10.1523/JNEUROSCI.2476-06.2006 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ogiwara I et al. De novo mutations of voltage-gated sodium channel alphaII gene SCN2A in intractable epilepsies. Neurology 73, 1046–1053, doi:10.1212/WNL.0b013e3181b9cebc (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ben-Shalom R et al. Opposing Effects on NaV1.2 Function Underlie Differences Between SCN2A Variants Observed in Individuals With Autism Spectrum Disorder or Infantile Seizures. Biol Psychiatry 82, 224–232, doi:10.1016/j.biopsych.2017.01.009 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gennarino VA et al. A Mild PUM1 Mutation Is Associated with Adult-Onset Ataxia, whereas Haploinsufficiency Causes Developmental Delay and Seizures. Cell 172, 924–936 e911, doi:10.1016/j.cell.2018.02.006 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Guissart C et al. Dual Molecular Effects of Dominant RORA Mutations Cause Two Variants of Syndromic Intellectual Disability with Either Autism or Cerebellar Ataxia. Am J Hum Genet 102, 744–759, doi:10.1016/j.ajhg.2018.02.021 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Darmanis S et al. A survey of human brain transcriptome diversity at the single cell level. Proc Natl Acad Sci U S A 112, 7285–7290, doi:10.1073/pnas.1507125112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lake BB et al. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science 352, 1586–1590, doi:10.1126/science.aaf1204 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Camp JG et al. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc Natl Acad Sci U S A 112, 15672–15677, doi:10.1073/pnas.1520760112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Abrahams BS et al. SFARI Gene 2.0: a community-driven knowledgebase for the autism spectrum disorders (ASDs). Mol Autism 4, 36, doi:10.1186/2040-2392-4-36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Xu LM et al. AutismKB: an evidence-based knowledgebase of autism genetics. Nucleic Acids Res 40, D1016–1022, doi:10.1093/nar/gkr1145 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang P, Zhao D, Lachman HM & Zheng D Enriched expression of genes associated with autism spectrum disorders in human inhibitory neurons. Transl Psychiatry 8, 13, doi:10.1038/s41398-017-0058-6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Skene NG et al. Genetic identification of brain cell types underlying schizophrenia. Nat Genet 50, 825–833, doi:10.1038/s41588-018-0129-5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zeisel A et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 347, 1138–1142, doi:10.1126/science.aaa1934 (2015). [DOI] [PubMed] [Google Scholar]
- 103.Romanov RA et al. Molecular interrogation of hypothalamic organization reveals distinct dopamine neuronal subtypes. Nat Neurosci 20, 176–188, doi:10.1038/nn.4462 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.La Manno G et al. Molecular Diversity of Midbrain Development in Mouse, Human, and Stem Cells. Cell 167, 566–580 e519, doi:10.1016/j.cell.2016.09.027 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tasic B et al. Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat Neurosci 19, 335–346, doi:10.1038/nn.4216 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gokce O et al. Cellular Taxonomy of the Mouse Striatum as Revealed by Single-Cell RNA-Seq. Cell Rep 16, 1126–1137, doi:10.1016/j.celrep.2016.06.059 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pardinas AF et al. Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat Genet 50, 381–389, doi:10.1038/s41588-018-0059-2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]