Abstract
NBEA is a candidate gene for autism, and de novo variants have been reported in neurodevelopmental disease (NDD) cohorts. However, NBEA has not been rigorously evaluated as a disease gene, and associated phenotypes have not been delineated. We identified 24 de novo NBEA variants in patients with NDD, establishing NBEA as an NDD gene. Most patients had epilepsy with onset in the first few years of life, often characterized by generalized seizure types, including myoclonic and atonic seizures. Our data show a broader phenotypic spectrum than previously described, including a Myoclonic-Astatic Epilepsy (MAE)-like phenotype in a subset of patients.
Introduction:
NBEA encodes Neurobeachin, a brain-specific kinase-anchoring protein implicated in vesicle trafficking and synaptic structure and function 1. It is not currently associated with disease in the Online Mendelian Inheritance in Man (OMIM) database but is a candidate gene for autism based on linkage studies and the identification of microdeletions and a reciprocal balanced translocation involving NBEA in patients with autism 2–6. More recently, two de novo NBEA variants were identified through whole exome sequencing (WES) in large cohorts of patients with neurodevelopmental disease (NDD) phenotypes, implicating NBEA as a candidate gene 7.
Here we report 24 de novo NBEA variants, including 18 identified through clinical and research sequencing and six de novo deletions identified by array or whole genome sequencing (WGS). De novo NBEA mutations were associated with NDD in all patients and epilepsy in the majority. Of those with epilepsy, most had generalized seizure types and seizure onset in the first few years of life. A subset had features of Myoclonic-Astatic Epilepsy (MAE), with toddler age onset of myoclonic, atonic and/or myoclonic-atonic seizures and, in some cases, developmental regression with seizure onset. Epilepsy has not been described in previous reports implicating NBEA as an autism candidate gene, but our data support that it is a major feature in patients with variants in NBEA.
Methods:
NBEA variants were identified by research trio WES for individuals 1, 6, 10, 14 and 18 and research trio WGS for individual 13 and 24. Individuals 11 and 12 and their parents underwent sequencing of a panel of candidate genes based off of a previous sequencing project. Variants identified in individuals 1, 10, 12, 13, 14 and 18 were confirmed by Sanger sequencing. For individual 24, the deletion was confirmed by research GSA array. For individual 19, the deletion was identified through a research protocol using SNP array. NBEA variants were identified in individuals 2–5, 7–9, 15–17 through clinical WES, and were either Sanger confirmed or met laboratory criteria for reporting based on WES only. Individuals 20–23 and their parents underwent clinical microarray. Informed consent was obtained by parents and all research studies received prior approval by the appropriate Institutional Review Board. Groups were connected through GeneMatcher 8.
Results:
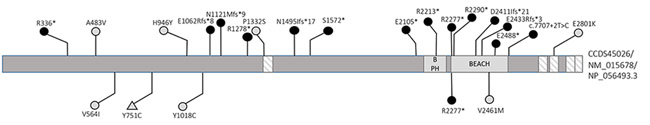
NBEA variants were identified in 22 previously unreported cases and two cases already reported in the literature 7. All variants were confirmed to be de novo with the exception of individual 13. For individual 13, parents were unavailable for Sanger confirmation, but we presumed (based on demographic information and referral site) that this was the same person included in an epileptic encephalopathy cohort, where the variant was confirmed to be de novo 9, 10. Variant types identified included nonsense (8/24), frameshift (5/24), missense (4/24), intragenic deletion (5/24), splice site (1/24), and a multi-gene deletion (1/24). All variants were absent from gnomAD and from 12,325 internal Institute for Genomic Medicine (IGM) controls 11. Each of the missense variants were predicted to be deleterious by at least one computational model; of the four missense variants, three were predicted probably damaging and one possibly damaging (ind. 17) by PolyPhen2 and all had an adjusted CADD score greater than 20. Two were found within WD-40 repeats12 (Figure 1). NBEA encodes two CCDS transcripts—the 2946 amino acid CCDS45026/NM_015678 (NP_056493.3) and a shorter 739 amino acid CCDS55894/NM_001204197 (NP_001191126.1). All variants identified in our cohort affected the longer transcript, and eight affected both. Nonsense and frameshift variants from our cohort occurred throughout the protein (NP_056493.3). All deletions were intragenic, except for the 2.87 Mb deletion identified in individual 19, which contains multiple other genes, none of which are known to be associated with disease based on OMIM.
Figure 1:
Mutational landscape of NBEA
Domain organization of the NBEA protein is based on Uniprot. Boxes indicate functional domains; beach type PH (BPH), BEACH domain, and WD-40 repeat domains (lined) are shown. The positions of variants described in our cohort are shown above the protein and those described in other cohorts are shown below. Black circles represent nonsense, frameshift, and splice site variants, and light circles represent missense variants. All variants identified in a person with a NDD are represented with circle, and the variant identified in a control individual is represented with a triangle.
All patients had NDD, including developmental delay (DD) and/or intellectual disability (ID) (Table 1). Age of ambulation ranged from 11 months to 3.5 years; one person (individual 11) is non-ambulatory as an adult. All patients had some level of speech delay, and two patients are non-verbal at 11 and 19 years (Table 2). Two patients had developmental regression noted at the time of seizure onset. Half of the patients (12/24) had autism or prominent autistic features.
Table 1:
Summary of clinical and epilepsy characteristics by variant type
| Clinical Characteristic | All variants, n = 24 |
LoF variants*, n = 20 |
Missense variants, n = 4 |
| Neurodevelopmental disability | 24 (100 %) | 19 (100 %) | 4 (100 %) |
| Developmental regression | 2 (8.33 %) | 2 (10 %) | 0 |
| Autistic features or Autism | 12 (50 %) | 10 (50 %) | 2 (50 %) |
| Microcephaly or borderline microcephaly | 3 (12.50 %) | 3 (15 %) | 0 |
| Epilepsy | 15 (62.50 %) | 12 (60 %) | 3 (75 %) |
| Epilepsy Characteristic | All variants, n = 15 | LoF variants, n = 12 | Missense variants, n = 3 |
| Age of epilepsy onset | |||
| < 1 year | 2 (13.33 %) | 2 (16.67 %) | 0 |
| 1-4 years | 11 (73.33 %) | 8 (66.67 %) | 3 (100 %) |
| > 4 years | 2 (13.33 %) | 2 (16.67 %) | 0 |
| Generalized seizures | 12 (80 %) | 10 (83.33 %) | 2 (66.67 %) |
| Myoclonic | 7 (46.67 %) | 6 (50 %) | 1 (33.33 %) |
| Atonic and/or myoclonic-atonic | 5 (33.33 %) | 5 (41.67 %) | 0 |
| Absence or atypical absence | 5 (33.33 %) | 5 (41.67 %) | 0 |
| Tonic, clonic and/or tonic-clonic | 10 (66.67 %) | 8 (66.67 %) | 2 (66.67 %) |
| Focal and generalized seizures | 4 (26.67 %) | 2 (16.67 %) | 2 (66.67 %) |
| Focal seizures only | 1 (6.67 %) | 0 | 1 (33.33 %) |
| Unclassified seizures only | 2 (13.33 %) | 2 (16.67 %) | 0 |
| Epileptiform abnormalities on EEG | |||
| Generalized spike/polyspike and wave | 9 (60 %) | 8 (66.67 %) | 1 (33.33 %) |
| Focal and generalized discharges | 1 (6.67 %) | 1 (8.33 %) | 0 |
| Focal discharges | 2 (13.33 %) | 1 (8.33 %) | 1 (33.33 %) |
| Unclassified discharges | 2 (13.33 %) | 2 (16.67 %) | 0 |
| No epileptiform abnormalities | 1 (6.67 %) | 0 | 1 (33.33 %) |
All deletions, frameshift, nonsense and splice-site were considered loss of function (LoF) for this table
Table 2:
Genotype and phenotype details for individuals with NBEA variants
| Variant (NM_015678.4); variant type |
Sex, Age (years) |
Development | Autism | Seizure Types | Age at 1st seizure |
|
|---|---|---|---|---|---|---|
| 1 | c.1006C>T; p.Arg336*; NS | M, 6 | No motor delay; mild speech delay | − | Myoclonic, Astatic, Absence, Myoclonic-Clonic-Tonic, GTC, Tonic, Focal unaware, SE | 3 yr |
| 2 | c.6829C>T; p.Arg2277*; NS | F, 21 | W=18 m.; NV | ++ | Tonic, GTC | 3 yr |
| 3 | c.3994C>T; p.Pro1332Ser; MS | M, 4 | W=30 m.; speaks some two word phrases | + | None | N/A |
| 4 | c.4484del; p.Asn1495Ilefs*17; FS |
M, 19 | W=2.5 yr; speaks words at 16 yr | ++ | unknown | 3.5-4 yr |
| 5 | c.6313G>T; p.Glu2105*; NS | M, 18 | W=15-17 m; slightly delayed speech; regression at 2 yr. | − | Myoclonic, Atonic, Atypical Absence, Clonic, GTC | 2 yr |
| 6 | c.7294_7295dup; p.Glu2433Argfs*3; FS | F, 13 | W=prior to 15 mo.; first word=prior to 2 yr. | − | none | N/A |
| 7 | c.7707+2T>C; SS | F, 18 | W-unavailable, speech delay | − | Febrile, Absence | <1 yr |
| 8 | c.6868C>T; p.Gln2290*; NS |
M, 3 | W=19 m; speaks <10 words at 3 yr | + | none | N/A |
| 9 | c.7462G>T; p.Glu2488*; NS |
F, 19 | W=12 m; speech delay | − | none | N/A |
| 10 | c.7230del; p.Asp2411Ilefs*21; FS |
M, 9 | W=11 m; first word=16 m. | ++ | Myoclonic, Myoclonic-atonic, Atypical Absence, GTC | 19 m |
| 11 | c.3183delA; p.Glu1062Argfs*8; FS |
M, 19 | Non-ambulatory; NV | + | Myoclonic, Myoclonic-atonic, GTC, Focal unaware, SE | 19 m |
| 12 | c.1448C>T; p.Ala483Val; MS |
F, 11 | W=15 m; first word=30 m | − | Febrile, hemi-convulsive, GTC | 1 yr |
| 13 | c.6637C>T; p.Arg2213*; NS | F, 23 | normal until regression at 26 m. | − | Atonic, Tonic, GTC | 26 m |
| 14 | c.2836C>T; p.His946Tyr; MS | F, 20 | W=18 m; words only (no phrases) at 2 yr | − | Nocturnal frontal lobe seizures | 2 yr |
| 15 | c.3832C>T; p.Arg1278*; NS | F, 5 | W=1 yr.; severe speech delay | − | None | N/A |
| 16 | c.3362del; p.Asn1121Metfs*9; FS | F, 24 | W=2 yr; first word=2 yr | ++ | Generalized | 19 yr |
| 17 | c.8401G>A; p.Glu2801Lys; MS | M, 11 | W=15 m; 10 words at 24 m | + | Myoclonic, Tonic, Focal, Focal to GTC, Febrile | 14 m |
| 18 | c.4715C>A; p.Ser1572*; NS | F, 3 | W=14 m; 10-15 words at 2 yr | + | None (paroxysmal spells not confirmed to be seizures) | N/A |
| 19 | chr13:33957317-36828237 ×1; MGD | M, 15 | W=3.5 yr; first word=14 m, stagnation until 4 yr | − | Myoclonic, GTC, epileptic spasms/tonic | 18 m |
| 20 | chr13:35590335-35940429 ×1; IGD | M, 16 | W=15 m; first word=3 yr | − | None | N/A |
| 21 | chr13:35574513-36163037 ×1; IGD | M, 16 | W=2 yr; NV at 11 yr | + | None | N/A |
| 22 | chr13:36038249-36141224 ×1; IGD | M, 11 | W-unavailable, IQ=~60 | + | Myoclonic, GTC, Tonic | 8 m |
| 23 | chr13:35963197-36125577 ×1; IGD | M, 5 | W=18 m; delayed speech | + | Possible Absences | 4 yr |
| 24 | chr13:35700830-35887000 ×1; IGD | F, 9 | W=17 m; first word=18–20 m | − | Non-convulsive SE, generalized | 2 yr |
NS=nonsense, MS=missense, FS=frameshift, SS=splice site, MGD=multigene deletion, IGD=intragenic deletion, M=male, F=female, W=age at walking, NV=non-verbal, m= month(s), yr=year(s), Autism: + indicates autistic features, ++ indicates autism diagnosis, GTC = generalized tonic clonic, SE = status epilepticus, N/A=not applicable
The majority of patients (15/24) had epilepsy, and two additional patients have presumed or possible epilepsy. Of those with classified epilepsy, one had focal epilepsy; the rest have generalized epilepsy or mixed (focal and generalized) epilepsy with multiple seizure types. Myoclonic seizures were common, present in almost half of those with epilepsy (7/15). Of these, four presented with a MAE-like phenotype, with toddler onset atonic and/or myoclonic-atonic seizures. Myoclonic seizures were more common in patients with LoF variants (6/12, 50%) than those with missense variants (1/3, 33.3%), and all cases of atonic and/or myoclonic-atonic seizures (n=5) were found in individuals with a LoF mutation.
About three-quarters of patients with epilepsy had seizure onset between 1–4 years of age. The two individuals with seizure onset at less than 1 year of age both had LoF variants. Response to treatment was variable, some patients had cessation of seizures between the ages of 3 and 19 years old, while others remain refractory to treatment. Some success was reported using valproic acid, ethosuximide (with valproic acid), levetiracetam, lamotrigine, benzodiazepines and dietary therapy, usually in some combination (Table 2). The interictal EEG often showed diffuse slowing and included generalized epileptiform abnormalities in the majority of patients with epilepsy (9/15, 60%). MRIs did not show any distinctive features in most patients.
Seven participants had behavior problems including aggression (4/24) and attention deficits and hyperactivity (4/24). Eight patients had abnormal movements, including wide-based, uncoordinated gait (6/24) and dystonic movements (3/24); one additional patient was noted to be “clumsy”. Other features reported in more than one patient include hypotonia of variable severity (8/24), microcephaly or borderline microcephaly (3/24), recurrent infections (2/24) and eczema or dry skin (3/24). All of those with microcephaly had LoF variants.
Discussion:
We have demonstrated that NBEA is an NDD gene, associated with early childhood epilepsy, which is consistent with what is known about the biology of NBEA. Neurobeachin (NBEA) is a brain-specific multidomain scaffolding protein belonging to the family of BEACH (Beige and Chediak-Higashi) domain-containing proteins, which play a role in vesicle trafficking and dynamics 1, 13. NBEA localizes to vesicular structures at the trans-face of the Golgi and within neuronal dendrites and appears to regulate synaptic structure and function through targeted trafficking of post-synaptic proteins 14, 15. In zebrafish, post-synaptic NBEA is essential for both electrical and chemical synapse formation and maintenance of dendritic complexity 16. Our work adds to a growing body of literature implicating genes encoding synaptic proteins in neurodevelopmental disorders associated with epilepsy, autism and intellectual disability, although the specific mechanism by which these variants give rise to NDD is unknown. 17, 18
NBEA has been considered an autism candidate gene, and de novo NBEA variants have been seen in NDD cohorts. A large genetic linkage study indicated the 19 cM segment on chromosome 13 containing NBEA as a candidate region for autism 2. A patient with sporadic autism was found to have a de novo balanced reciprocal translocation (t(5;13)(q12.1;q13.2)) disrupting NBEA 3. Three patients with autism have been described in the literature with monoallelic deletions that include NBEA 4–6. Two individuals with de novo NBEA single nucleotide variants (individuals 13 and 14) were previously described in a cohort with ID and/or DD 7. While NBEA has been prioritized as a strong candidate epileptic encephalopathy (EE) gene based on high level of co-expression with known EE genes in the adult and developing brain, an association with epilepsy has not previously been reported 19.
In reporting 24 patients with NDD harboring de novo variants absent from population databases, we provide substantial genetic evidence implicating NBEA in disease. The majority of the variants (20/24) predict loss of function and NBEA is extremely intolerant to loss-of-function variation (pLI=1, RVIS (ExAC v2)=1.16%)11, 20, suggesting a haploinsufficiency disease mechanism. Our work complements experimental data demonstrating a role for NBEA at the synapse and autism-like behaviors modeled in the Nbea+/− mouse21. Applying ClinGen’s clinical validity of gene-disease associations framework, we therefore conclude that there is “strong” evidence that pathogenic variants in NBEA cause NDD with and without epilepsy22.
In terms of the overall phenotype of the cohort, language delay was universal, but the vast majority of patients were ultimately verbal, and almost all patients were ambulatory by 4 years of age; half had autism or autistic features and half had epilepsy within the first years of life, typically a generalized epilepsy often with myoclonic seizures. A subset had an MAE-like phenotype.
MAE is rare, accounting for only 1–2% of epilepsy in the first decade of life 23. While a genetic etiology was speculated in the first description by Doose, the vast majority of cases of MAE remain genetically unexplained. MAE-like phenotypes have been described in association with variants in SLC2A1, SCN1A, SCN1B, GABRG2 and GABRB3 24–26. The core NBEA associated epilepsy in our cohort involves toddler-age onset of multiple generalized seizure types, especially myoclonic seizures, with MAE-like phenotypes similar to what has been described for CHD2 and SLC6A1. As with these genes, the phenotypic spectrum of NBEA epilepsy includes patients with features atypical for MAE, such as developmental delay preceding seizure onset and the presence of focal epileptiform discharges and/or focal seizures in some patients 27–29. A limitation of the current study is the lack of standardization in the evaluation of patients. Diagnosis of seizure types and syndromes and EEG interpretation were as determined by the treating neurologists. Future work with centralized review of polygraphic video-EEG for each reported seizure type and a common neuropsychological battery applied universally across the cohort will give a better understanding of the NBEA phenotypic spectrum.
While the small number of, and lack of recurrent, missense variants limit our ability to make definite phenotype-genotype correlations, it is perhaps notable that MAE seizure types, including myoclonic, atonic and/or myoclonic-atonic seizures, were seen almost exclusively with LoF variants. Additionally, epilepsy with onset in infancy and microcephaly or borderline microcephaly were each only seen with LoF variants, possibly reflecting a more severe phenotypic spectrum in this group.
In addition to the cases from our cohort, three other de novo missense variants are reported in denovo-db (http://denovo-db.gs.washington.edu) from various NDD cohorts, although NBEA variants were not significant in any of these cohorts. The cohorts included autism spectrum disorders, schizophrenia, and developmental disability/intellectual disability, but none of these individuals were specifically reported to have epilepsy or seizures 30–32. Individual 4 was previously included in an ID cohort, and as mentioned before, individual 13 was probably previously included in an epileptic encephalopathy cohort9, 10, 33. Notably, the same nonsense variant seen in individual 2 was identified in a (possibly) unique person in an autism spectrum disorder cohort; individual 2 also had a diagnosis of autism 34. One de novo missense variant was identified in a control individual; this was not located in a known functional domain and is predicted benign by PolyPhen-2 (Figure 1). 34
In conclusion, by gathering genetic data from multiple sites connected through GeneMatcher, we identified 24 de novo variants in NBEA identified through either research or clinical genetic testing. This study implicates NBEA as a neurodevelopmental gene with distinctive epilepsy presentations in the first years of life, overlapping MAE phenotypes.
Supplementary Material
Acknowledgements
We are very appreciative of all the families who enrolled in the various research studies involved. Individual 6 was enrolled through the CAUSES study, which is funded by British Columbia Children’s Hospital Foundation (Mining for Miracles) and Genome BC. Investigators in the CAUSES study include Shelin Adam, Christele Du Souich, Alison Elliott, Anna Lehman, Jill Mwenifumbo, Tanya Nelson, Clara Van Karnebeek, and Jan Friedman. We also thank individuals and groups who contributed exome sequence samples for analysis, including the Epi4K Consortium, the Epilepsy Phenome Genome Project (EPGP), EuroEPINOMICS-RES Consortium, and Dr. Vandana Shashi and Dr. Ann Bergin. This publication was supported by the National Center for Advancing Translational Sciences, National Institutes of Health, through Grant Number UL1TR001873. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Potential Conflicts of Interest
There are no potential conflicts of interest relevant to this manuscript.
References
- 1.Wang X, Herberg FW, Laue MM, et al. Neurobeachin: A protein kinase A-anchoring, beige/Chediak-higashi protein homolog implicated in neuronal membrane traffic. J Neurosci 2000;20:8551–8565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrett S, Beck JC, Bernier R, et al. An autosomal genomic screen for autism. Collaborative linkage study of autism. Am J Med Genet 1999;88:609–615. [DOI] [PubMed] [Google Scholar]
- 3.Castermans D, Wilquet V, Parthoens E, et al. The neurobeachin gene is disrupted by a translocation in a patient with idiopathic autism. J Med Genet 2003;40:352–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith M, Woodroffe A, Smith R, et al. Molecular genetic delineation of a deletion of chromosome 13q12-->q13 in a patient with autism and auditory processing deficits. Cytogenet Genome Res 2002;98:233–239. [DOI] [PubMed] [Google Scholar]
- 5.Steele MM, Al-Adeimi M, Siu VM, Fan YS. Brief report: A case of autism with interstitial deletion of chromosome 13. J Autism Dev Disord 2001;31:231–234. [DOI] [PubMed] [Google Scholar]
- 6.Ritvo ER, Mason-Brothers A, Menkes JH, Sparkes RS. Association of autism, retinoblastoma, and reduced esterase D activity. Arch Gen Psychiatry 1988;45:600. [DOI] [PubMed] [Google Scholar]
- 7.Bowling KM, Thompson ML, Amaral MD, et al. Genomic diagnosis for children with intellectual disability and/or developmental delay. Genome Med 2017;9:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sobreira N, Schiettecatte F, Valle D, Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum Mutat 2015;36:928–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Epi4K Consortium. De novo mutations in epileptic encephalopathies. Nature 2013;501:217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Euro E-RESC, Epilepsy Phenome/Genome Project, Epi4K Consortium. De novo mutations in synaptic transmission genes including DNM1 cause epileptic encephalopathies. Am J Hum Genet 2014;95:360–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016;536:285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 2014;46:310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cullinane AR, Schaffer AA, Huizing M. The BEACH is hot: a LYST of emerging roles for BEACH-domain containing proteins in human disease. Traffic 2013;14:749–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nair R, Lauks J, Jung S, et al. Neurobeachin regulates neurotransmitter receptor trafficking to synapses. J Cell Biol 2013;200:61–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Niesmann K, Breuer D, Brockhaus J, et al. Dendritic spine formation and synaptic function require neurobeachin. Nat Commun 2011;2:557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller AC, Voelker LH, Shah AN, Moens CB. Neurobeachin is required postsynaptically for electrical and chemical synapse formation. Curr Biol 2015;25:16–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zoghbi HY, Bear MF. Synaptic dysfunction in neurodevelopmental disorders associated with autism and intellectual disabilities. Cold Spring Harb Perspect Biol 2012;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fukata Y, Fukata M. Epilepsy and synaptic proteins. Curr Opin Neurobiol 2017;45:1–8. [DOI] [PubMed] [Google Scholar]
- 19.Oliver KL, Lukic V, Freytag S, Scheffer IE, Berkovic SF, Bahlo M. In silico prioritization based on coexpression can aid epileptic encephalopathy gene discovery. Neurol Genet 2016;2:e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Samocha KE, Robinson EB, Sanders SJ, et al. A framework for the interpretation of de novo mutation in human disease. Nat Genet 2014;46:944–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nuytens K, Gantois I, Stijnen P, et al. Haploinsufficiency of the autism candidate gene Neurobeachin induces autism-like behaviors and affects cellular and molecular processes of synaptic plasticity in mice. Neurobiol Dis 2013;51:144–151. [DOI] [PubMed] [Google Scholar]
- 22.Strande NT, Riggs ER, Buchanan AH, et al. Evaluating the Clinical Validity of Gene-Disease Associations: An Evidence-Based Framework Developed by the Clinical Genome Resource. Am J Hum Genet 2017;100:895–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guerrini R, Mari F, Dravet C. Idiopathic Myoclonic Epilepsies in Infancy and Early Childhood. Epileptic syndromes in infancy, childhood and adolescence 2012. [Google Scholar]
- 24.Mullen SA, Marini C, Suls A, et al. Glucose transporter 1 deficiency as a treatable cause of myoclonic astatic epilepsy. Arch Neurol 2011;68:1152–1155. [DOI] [PubMed] [Google Scholar]
- 25.Moller RS, Wuttke TV, Helbig I, et al. Mutations in GABRB3: From febrile seizures to epileptic encephalopathies. Neurology 2017;88:483–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sands TT, Choi H. Genetic Testing in Pediatric Epilepsy. Curr Neurol Neurosci Rep 2017;17:45. [DOI] [PubMed] [Google Scholar]
- 27.Chenier S, Yoon G, Argiropoulos B, et al. CHD2 haploinsufficiency is associated with developmental delay, intellectual disability, epilepsy and neurobehavioural problems. J Neurodev Disord 2014;6:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johannesen KM, Gardella E, Linnankivi T, et al. Defining the phenotypic spectrum of SLC6A1 mutations. Epilepsia 2018;59:389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carvill GL, McMahon JM, Schneider A, et al. Mutations in the GABA Transporter SLC6A1 Cause Epilepsy with Myoclonic-Atonic Seizures. Am J Hum Genet 2015;96:808–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Rubeis S, He X, Goldberg AP, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 2014;515:209–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deciphering Developmental Disorders Study. Prevalence and architecture of de novo mutations in developmental disorders. Nature 2017;542:433–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fromer M, Pocklington AJ, Kavanagh DH, et al. De novo mutations in schizophrenia implicate synaptic networks. Nature 2014;506:179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lelieveld SH, Reijnders MR, Pfundt R, et al. Meta-analysis of 2,104 trios provides support for 10 new genes for intellectual disability. Nat Neurosci 2016;19:1194–1196. [DOI] [PubMed] [Google Scholar]
- 34.Iossifov I, O’Roak BJ, Sanders SJ, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature 2014;515:216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.