Abstract
Dinoflagellates are a diverse group of unicellular primary producers and grazers that exhibit some of the most remarkable features known among eukaryotes. These include gigabase-sized nuclear genomes, permanently condensed chromosomes and highly reduced organelle DNA. However, the genetic inventory that allows dinoflagellates to thrive in diverse ecological niches is poorly characterised. Here we systematically assess the functional capacity of 3,368,684 predicted proteins from 47 transcriptome datasets spanning eight dinoflagellate orders. We find that 1,232,023 proteins do not share significant sequence similarity to known sequences, i.e. are “dark”. Of these, we consider 441,006 (13.1% of overall proteins) that are found in multiple taxa, or occur as alternative splice variants, to comprise the high-confidence dark proteins. Even with unknown function, 43.3% of these dark proteins can be annotated with conserved structural features using an exhaustive search against available data, validating their existence and importance. Furthermore, these dark proteins and their putative homologs are largely lineage-specific and recovered in multiple taxa. We also identified conserved functions in all dinoflagellates, and those specific to toxin-producing, symbiotic, and cold-adapted lineages. Our results demonstrate the remarkable divergence of gene functions in dinoflagellates, and provide a platform for investigations into the diversification of these ecologically important organisms.
Introduction
Dinoflagellates are a diverse group of phytoplankton that are ubiquitous in marine and fresh waters. About 2300 dinoflagellate species have been described1,2, most of which are photosynthetic. However, mixotrophy3,4 that combines phototrophy and ingestion of prey (heterotrophy) is common. Photosynthetic dinoflagellates form the base of food webs and sustain global aquatic ecosystems via primary production and cycling of organic carbon and nitrogen. Bloom-forming dinoflagellates, predominantly in the orders Gonyaulacales and Gymnodiniales, can cause “red tides” (harmful algal blooms) and produce toxins that pose serious human health risks5. Other dinoflagellates, particularly Symbiodiniaceae6 (Suessiales), are symbionts in corals and other coral reef animals7,8. Dinoflagellates are also found in extreme environments, with multiple cold-adapted (psychrophilic) species described in the polar regions9,10. The capacity of dinoflagellates to thrive in diverse ecological niches, and the remarkable sequence divergence and complexity of their genomes when compared to other eukaryotes, have led researchers to grumble that dinoflagellates are in fact aliens from “outer space”11.
The genetic capacity and features that are common to all dinoflagellate lineages, or those related to niche specialisation (e.g., bloom formation, symbiotic lifestyle and cold adaptation), remain poorly understood. Symbiodiniaceae species are the only dinoflagellates for which genome data are available12–15. However, the functional capacity of dinoflagellate genes is poorly understood when relying on the commonly used annotation approach, whereby predicted proteins are compared against a set of curated proteins of known function that are largely derived from model organisms. The often-overlooked proteins of unknown function (i.e. “dark” proteins), and the corresponding dark genes, may be highly conserved in closely related species and represent unique lineage-specific features. Whereas genome data from dinoflagellates are limited, transcriptome data provide an avenue for the exploration of gene functions that drive niche specialisation in these species16,17.
Here we use available dinoflagellate transcriptome data to systematically investigate gene functions that are common (and unique) to distinct dinoflagellate lineages, and identify the conserved dark proteins. We also investigate gene functions and pathways that are enriched in toxin-producing, symbiotic, and cold-adapted dinoflagellates.
Results and Discussion
We retrieved 64 publicly available dinoflagellate transcriptomes and their predicted proteins18–20 (Supplementary Table S1). To avoid potential biases arising from codon degeneracy, we restricted our analysis to proteins, using the amino acid sequences predicted from these transcriptomes. We filtered the datasets using stringent cri-teria, including the recovery of core conserved eukaryote proteins21 as an indicator of dataset completeness (see Methods). This approach resulted in the final 47 datasets, representing 3,368,684 protein sequences from eight taxonomic orders (Table 1).
Table 1.
The final 47 datasets used in this study.
| Taxon | Order | No. non-redundant protein sequences |
|---|---|---|
| Dinophysis acuminata DAEP01 | Dinophysiales | 83,934 |
| Alexandrium catenella OF101 | Gonyaulacales | 68,889 |
| Alexandrium margalefi AMGDE01CS-322 | Gonyaulacales | 50,502 |
| Alexandrium monilatum CCMP3105 | Gonyaulacales | 87,380 |
| Alexandrium tamarense CCMP1771 | Gonyaulacales | 114,975 |
| Azadinium spinosum 3D9 | Gonyaulacales | 70,040 |
| Ceratium fusus PA161109 | Gonyaulacales | 68,969 |
| Gambierdiscus australes CAWD 149 | Gonyaulacales | 48,770 |
| Gambierdiscus caribaeus | Gonyaulacales | 290,362 |
| Gonyaulax spinifera CCMP409 | Gonyaulacales | 39,652 |
| Lingulodinium polyedra CCMP1738 | Gonyaulacales | 96,319 |
| Protoceratium reticulatum CCCM535-CCMP1889 | Gonyaulacales | 75,595 |
| Pyrodinium bahamense pbaha01 | Gonyaulacales | 99,554 |
| Amphidinium carterae CCMP1314 | Gymnodiniales | 35,832 |
| Amphidinium massartii CS-259 | Gymnodiniales | 49,240 |
| Gymnodinium catenatum GC744 | Gymnodiniales | 82,846 |
| Karenia brevis CCMP2229 | Gymnodiniales | 79,497 |
| Karenia brevis SP1 | Gymnodiniales | 83,816 |
| Karenia brevis SP3 | Gymnodiniales | 69,522 |
| Karenia brevis Wilson | Gymnodiniales | 90,529 |
| Karlodinium micrum CCMP2283 | Gymnodiniales | 57,487 |
| Togula jolla CCCM725 | Gymnodiniales | 42,196 |
| Noctiluca scintillans | Noctilucales | 40,801 |
| Oxyrrhis marina LB1974 | Oxyrrhinales | 34,348 |
| Oxyrrhis marina | Oxyrrhinales | 43,246 |
| Brandtodinium nutricula RCC3387 (“Brandtodinium nutriculum” in MMETSP) | Peridiniales | 66,253 |
| Durinskia baltica CSIRO CS-38 | Peridiniales | 88,656 |
| Glenodinium foliaceum CCAP1116/3 | Peridiniales | 106,311 |
| Heterocapsa arctica CCMP445 | Peridiniales | 45,573 |
| Heterocapsa rotundata SCCAP K-0483 | Peridiniales | 43,925 |
| Heterocapsa triquestra CCMP448 | Peridiniales | 57,688 |
| Kryptoperidinium foliaceum CCMP1326 | Peridiniales | 161,360 |
| Peridinium aciculiferum PAER-2 | Peridiniales | 53,784 |
| Scrippsiella hangoei-like SHHI-4 | Peridiniales | 74,092 |
| Scrippsiella hangoei SHTV5 | Peridiniales | 74,862 |
| Scrippsiella trochoidea CCMP3099 | Peridiniales | 101,032 |
| Prorocentrum minimum CCMP1329 | Prorocentrales | 85,555 |
| Prorocentrum minimum CCMP2233 | Prorocentrales | 79,005 |
| Pelagodinium beii RCC1491 | Suessiales | 47,797 |
| Polarella glacialis CCMP1383 | Suessiales | 58,545 |
| Polarella glacialis CCMP2088 | Suessiales | 33,576 |
| Symbiodinium sp. C15 (Cladocopium) | Suessiales | 37,221 |
| Symbiodinium sp. C1 (Cladocopium) | Suessiales | 45,710 |
| Symbiodinium sp. CCMP2430 (Symbiodinium) | Suessiales | 43,277 |
| Symbiodinium sp. CCMP421 (Effrenium) | Suessiales | 72,087 |
| Symbiodinium sp. D1a (Durusdinium) | Suessiales | 44,936 |
| “Symbiodinium” sp. Mp | Suessiales | 43,138 |
Reference phylogeny and data completeness
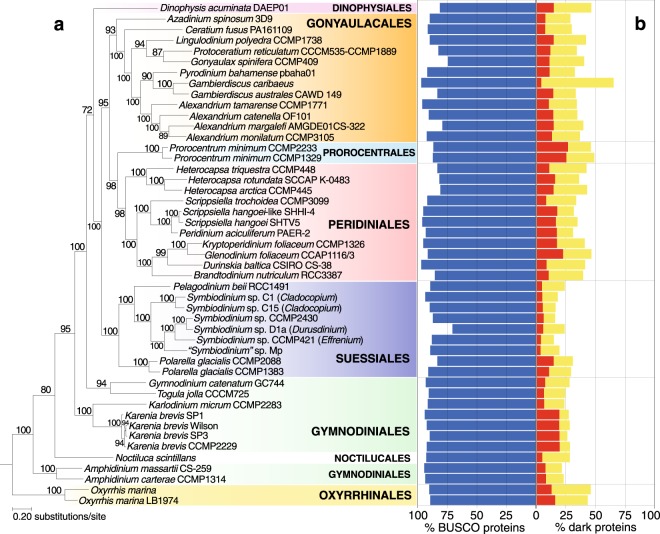
An earlier study by Price and Bhattacharya22 demonstrated the utility of constructing a phylogeny using high-throughput transcriptome data. Following a similar approach22, we inferred a maximum-likelihood tree using these data comprising 1043 single-copy protein sets (Fig. 1a; see Methods). The statistics of the concatenated alignment (209,857 aligned positions) and the associated individual 1043 alignments used for inferring this tree are shown in Supplementary Tables S2 and S3, respectively. On average, each taxon contributes 22.13% of the aligned residues in the concatenated alignment (Supplementary Fig. S1A). The maximum-likelihood tree inferred from these sets (Fig. 1a) is largely topologically congruent to the published phylogeny22 (normalised Robinson-Foulds23 distance = 0.17). The backbone node for each taxonomic order is strongly supported (bootstrap support [BS] > 95% based on ultrafast bootstrap approximation24) in the tree (Fig. 1a) except for the Gonyaulacales and Gymnodiniales, as was also found in the earlier study22. Thus, phylogenetic signal from dinoflagellate transcriptomes is largely consistent in these two independent analyses. The sole member of the Dinophysiales, Dinophysis acuminata DAEP01, is placed as the basal lineage in the clade including Gonyaulacales, Prorocentrales, Peridiniales, and Suessiales (BS 72%; Fig. 1a); this taxon was sister to Prorocentrum minimum in the earlier published trees22,25. The placement of Dinophysiales at the base of this clade of five orders lends support to the earlier phylogeny and the single origin of the theca in dinoflagellates (comparable BS 72% in the tree of Janouškovec et al.25). The differential placement of Gonyaulacales and Suessiales relative to Peridiniales within this clade may be due to more aligned positions used for inferring the tree in Fig. 1a (based on 209,857 positions across 1043 protein sets) than those used in the earlier study25 (based on 29,400 positions across 101 protein sets). We note with caution that the high percentage of undetermined characters (on average 77.87% per taxon; Supplementary Table S2 and Fig. S1A) in our concatenated alignment may have resulted in a reduced information content, but 22.13% of this alignment, based on a larger number of protein sets, still comprises 46,449 amino acid positions. Although we required that each orthologous set contains sequences from ten or more taxa (see Methods), we cannot exclude the possibility that some sequences may have arisen from eukaryote prey of the mixotrophic taxa. However, the strong node support for each dinoflagellate order in the tree suggests that the impact of eukaryote contaminants on our inferred phylogeny is likely to be negligible. The presence of highly diverged homologs originating from non-dinoflagellate eukaryotic contaminants would likely weaken node support in the tree.
Figure 1.
(a) Maximum-likelihood phylogeny inferred using the 1043 orthologous protein sets. Support values, based on 2000 ultrafast bootstrap approximations24, are shown at the internal nodes. The unit of branch length is the number of substitutions per site. (b) The percentage of recovered alveolate + stramenopile BUSCO proteins and of dark proteins in each dataset. High- and low-confidence dark proteins are shown in red and yellow bars, respectively.
On average, 208.6 (89.1%) of the 234 alveolate + stramenopile BUSCO proteins26 were recovered in each of these 47 datasets, indicating their high extent of completeness (Fig. 1b). In an independent assessment at the order level (Supplementary Table S1), we recovered a high proportion of these conserved proteins, e.g. 233 of the 234 (99.6%) among the Peridiniales datasets. The sole dataset (Dinophysis acuminata) from the order Dinophysiales is reasonably complete, with the recovery of 190 (81.2% of 234) alveolate + stramenopile BUSCO proteins. The recovery of multiple homologs in some of the taxa may be due to true gene duplications or alternatively, reflect alternative splicing events.
Prevalence of dark genes in dinoflagellates
Of all 3,368,684 proteins, 1,232,023 (36.57%) do not share significant sequence similarity to UniProt entries. The functions of these proteins are thus unknown, and we consider them as “dark” proteins. The average percentage of dark proteins in each dataset is 33%; the minimum is 15.2% in Symbiodinium sp. CCMP421 (now Effrenium), and the maximum is 63.5% in Gambierdiscus caribaeus (Fig. 1b). Although the number of dark proteins identified here may be somewhat dependent on the amount of data and the sequence length (low regression R2 values < 0.40 in Supplementary Fig. S2), these aspects have minimal impact on our broader interpretation that dark proteins are common in dinoflagellates.
We clustered the 3,368,684 protein sequences into 162,126 homologous sets of two or more sequences (see Methods). Of these sets (containing 2,554,321 proteins), 103,620 (63.9%) containing 441,006 proteins (17.27% of 2,554,321) are dark (hereafter the high-confidence set; see Methods). Within the 103,620 sets, 100,661 (97.14%) contain proteins from multiple taxa, whereas 2959 (2.86%) are taxon-specific; the latter must reflect e.g. alternative splice variants, because our approach excluded identical proteins from each taxon (Methods). The dark protein sets have an average size of 4.26, compared to the average size of 36.12 for the annotated sets, indicating that the dark protein families, although relatively more abundant, are smaller in size and more taxon-specific than annotated proteins. Of the 814,363 (unclustered) singleton proteins, 791,017 (97.13%) are dark (hereafter, the low-confidence set). These results suggest that dark proteins are prevalent in dinoflagellates and comprise an unexplored resource from which we can derive insights into the functional capabilities of these organisms.
Are dark genes in dinoflagellates from outer space?
In the absence of functional annotation based on full-length protein sequences, conserved structural features such as protein domains can be used to illuminate the potential roles dark proteins play in dinoflagellate biology. The amino acid profile of the high-confidence dark proteins is largely similar to that of the annotated proteins; the proportions of four of the 20 amino acids are significantly different between the two sets (at 95% confidence interval of 10,000 comparisons of random subsamples; see Methods and Supplementary Fig. S3).
The putative functions of high-confidence dark proteins were further inferred though annotation of Pfam domains. Of the 441,006 proteins, only 6168 (1.4%) had Pfam annotations. In comparison, 31.38% of all proteins in this study were annotated with Pfam domains, indicating that these dark proteins are so highly diverged that their homologs (if any exist) are poorly represented in the curated databases. Although 202 (3.3%) of the 6168 Pfam-annotated dark proteins share significant similarity (BLASTP, E ≤ 10−5) with sequences in the more-inclusive RefSeq protein database, the majority (78.2%) of recovered top hits (Supplementary Table S4) are “hypothetical”, “uncharacterized”, “predicted”, “X-containing”, “X-like” or putative proteins. We therefore maintain that these proteins are dark. The dark proteins are shorter than the average length in these datasets (234.2 and 109.3 amino acids respectively for high- and low-confidence dark proteins, compared to 291.8 overall). This is likely not in itself sufficient to explain the inability to annotate dark proteins with functions27. It is possible (indeed likely) that some low-confidence dark proteins are artefacts arising from sequencing error or transcriptome mis-assembly. Of the 103,620 dark homologous sets, most (100,661; 97.14%) have proteins from multiple taxa. The recovery of these proteins in multiple datasets suggests that their prominence in dinoflagellates is unlikely to have arisen primarily from artefacts.
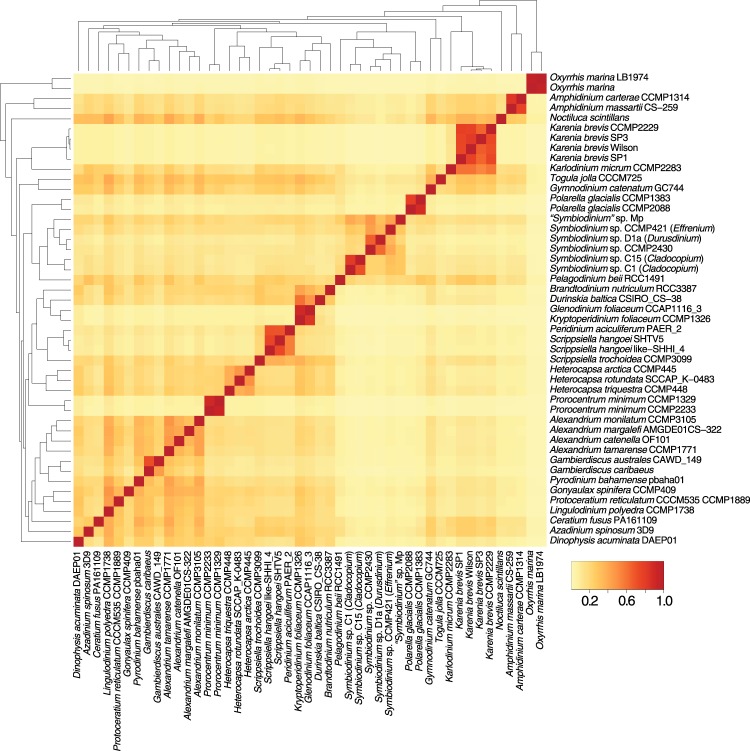
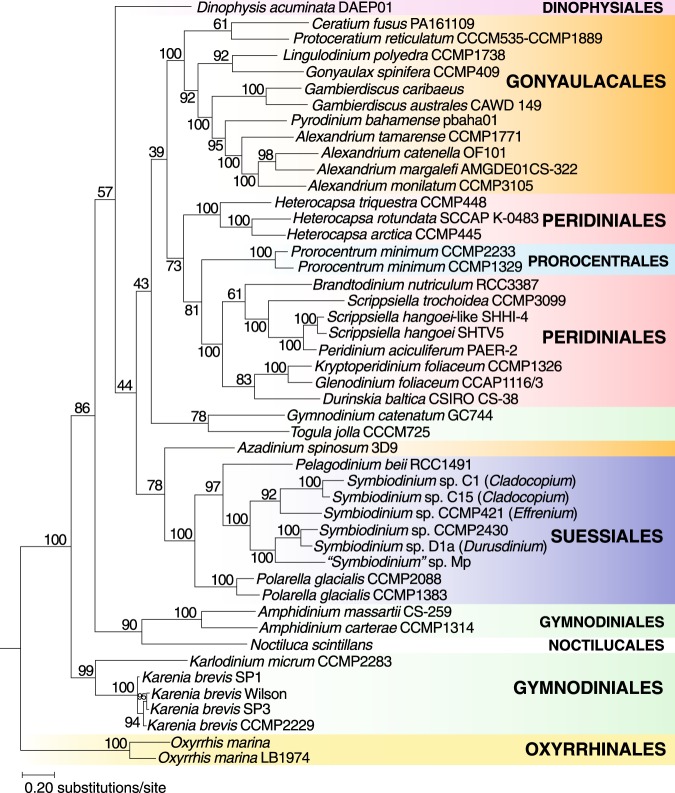
Figure 2 shows the proportion of 100,661 multi-taxon dark protein sets that are shared pairwise between taxa, with reference to the phylogenetic relationship of these taxa (based on Fig. 1a). We observed higher proportions of these sets among closely related taxa, such as among the strains of Karenia, Oxyrrhis, and Polarella, indicating that these dark proteins are lineage- or species-specific innovations. Interestingly, 403/1043 (38.6%) of the single-copy sets used to construct our reference phylogenetic tree (Fig. 1a) are dark. The maximum-likelihood tree inferred from these 403 dark protein sets is shown in Fig. 3. The statistics of the concatenated alignment (71,346 aligned positions) are shown in Supplementary Table S5. Each taxon on average contributes to 18.98% of the aligned residues in the concatenated alignment (Supplementary Fig. S1B). The tree topology is largely congruent with our reference phylogeny in Fig. 1a, indicating that these dark proteins and dark protein sets are indeed dinoflagellate proteins (and unlikely to be artefacts), are predominantly lineage-specific, and are more rarely shared between distantly related lineages. This latter observation suggests a more general insight. Shared phylogenetic information is lost with time and divergence, supporting the adage that adaptive evolution is local28 and its footprints (be it novel gene origin or lateral genetic transfer) are most obvious in recently split taxa. The use of BUSCO proteins is useful for assessing genome completeness or broad patterns of genome growth/reduction but provides little insight into how specialised functions or lineages evolve. This is the realm of dark proteins that still remain poorly characterised.
Figure 2.
Heat map showing the proportion of dark protein sets shared between taxa used in this study. Each row is normalised by the total number of protein sets of which the taxon is a member. The order of the species on both axes and their associated dendrograms follow the phylogeny in Fig. 1a.
Figure 3.
Maximum-likelihood phylogeny reconstructed using the 403 strictly orthologous dark protein sets. Support values, based on 2000 ultrafast bootstrap approximations24, are shown at the internal nodes. The unit of branch length is the number of substitutions per site.
Enrichment analysis comparing the annotated Pfam domains in high-confidence dark proteins and those in all datasets shows that functions related to calcium binding, protein localisation, protein degradation, protein-protein interaction, cell cycle regulation, and photosynthesis are over-represented (Supplementary Table S6). These functions may play a role in the ability of dinoflagellates to adapt to a rapidly changing environment, and may represent the putative functions of dark proteins. Protein degradation could be important for removing misfolded proteins that result from rapid changes in the local environment.
To further explore the conserved structural features of high-confidence dark proteins, we expanded our annotation strategy to include multiple methods and databases available via InterProScan. Using this approach, conserved features were annotated in 190,950 (43.3% of 441,006) high-confidence dark proteins. Of these, 37,270 proteins contain putative transmembrane domains (see Methods). We annotated conserved features in 36,352 proteins using SUPERFAMILY, ProSiteProfile, Pfam, PANTHER and Gene3D (in comparison to 6168 using Pfam alone). The ten most abundant domains identified by each of these in silico approaches are shown in Supplementary Table S7. The EF-hand, ubiquitin, zinc finger and IQ (calmodulin-binding) motifs are among the most abundant domains. The remaining 121,373 dark proteins are annotated with one or more secondary structures. Therefore, even though the functions of most dark proteins remain elusive, a substantial proportion of these proteins contain conserved structural features.
Core functions in dinoflagellate lineages
For all proteins in each dataset, we annotated function based on significant sequence similarity to known proteins in UniProt, protein domains in Pfam29, membrane transporters30, and Gene Ontology terms (Supplementary Table S1). To assess the core protein functions in dinoflagellates, we identified the Pfam domains and membrane transporters that are the most abundant across all taxa (Supplementary Fig. S4). The prevalent domains and transporters that were recovered among the top ten and top 20 in each taxon are shown in Table 2. The prevalence of protein kinase, RNA recognition and ankyrin repeat domains implicates functions in a diverse array of important cellular processes, including proliferation, cell cycle, signal transduction and RNA splicing. The prevalent membrane transporters (Table 2) include those related to transport of ions, metabolites, sugars and lipids is critical to all dinoflagellates (i.e., as in most mixotrophic lineages), potentially for nutrient uptake and osmoregulation.
Table 2.
Prevalent protein domains and membrane transporters annotated in dinoflagellate proteins consistently recovered among the top ten and among the top 20 in each of the 47 taxa.
| Pfam domain (Pfam identifier) | Membrane transporter (family identifier) |
|---|---|
| Among top 10 in each taxon | |
| Protein kinase (PF00069) | Eukaryotic Nuclear Pore Complex (E-NPC) Family (1.I.1) |
| RNA recognition motif (PF00076) | Mitochondrial Carrier (MC) Family (2.A.29) |
| Ankyrin repeats (3 copies) (PF12796) | Ankyrin (Ankyrin) Family (8.A.28) |
| EF-hand domain pair (PF13499) | ATP-binding Cassette (ABC) Superfamily (3.A.1) |
| Drug/Metabolite Transporter (DMT) Superfamily (2.A.7) | |
| Among top 20 in each taxon | |
| WD40 repeat (PF00400) | Voltage-gated Ion Channel (VIC) Superfamily (1.A.1) |
| MORN repeat (PF02493) | The Major Facilitator Superfamily (MFS) (2.A.1) |
| P-type ATPase (P-ATPase) Superfamily (3.A.3) | |
Core functions in toxic dinoflagellates
To identify functions common to toxic dinoflagellates, protein annotations among taxa from Gonyaulacales and Gymnodiniales (hereinafter, the G + G dataset) were contrasted to those of all taxa. We found significant over-representations of the Voltage-gated Ion Channel (VIC) Superfamily (1.A.1) and the Monovalent Cation:Proton Antiporter-1 (CPA1) Family (2.A.36) in the G + G dataset (Supplementary Table S8).
The Voltage-gated Ion Channel (VIC) Superfamily (1.A.1) is the most over-represented membrane transporter family. These ion channels are critical in the maintenance of ion concentrations and gradients across cell membranes. The sodium and calcium voltage-gated ion channels are also the target for the majority of dinoflagellate toxins31. In eukaryotes, these channels are highly glycosylated with sialic acid, which is known to modulate the excitability of voltage-gated ion channels32,33. Pfam domains of Glycosyltransferase family 29 (PF00777) and Kelch motif (PF01344), as well as the GO terms sialylation (GO:0097503) and sialyltransferase activity (GO:0008373) are over-represented in the G + G dataset. This indicates that functions related to the processing and attachment of sialic acids to other macromolecules are prominent in toxic dinoflagellates.
Whereas sialic acid had not been described in dinoflagellates34, it has been reported in other algae35,36. A gene related to sialyltransferase is differentially (more highly) expressed in toxin-producing strains of Alexandrium minutum than in non-toxic species37. Sialic acid was previously reported to be absent from the symbiotic dinoflagellates38. Here we found that the glycosyltransferase domain was almost completely absent from Symbiodiniaceae taxa (and from all lineages of Suessiales, except for three domain matches found in the Symbiodinium sp. CCMP421 [Effrenium] dataset).
Because voltage-gated ion channels are important in toxic dinoflagellates, the function of the channels must be unaffected by the toxins that these dinoflagellates release. In snakes, voltage-gated sodium channels that are resistant to tetrodotoxin (a toxin similar to saxitoxin from dinoflagellates39) have a reduced channel activity compared to those that are susceptible40. We hypothesise that a similar situation may occur in toxic dinoflagellates, i.e. voltage-gated ion channels are resistant to their own toxins and have a reduced activity. The link between sialic acid and these ion channels may represent a functional innovation in toxin-producing dinoflagellates, with the dinoflagellates using sialic acid to modulate (increase or recover) the activity of these toxin-resistant channels.
Known dinoflagellate toxins are polyketides produced by the multi-domain polyketide synthase (PKS) enzyme family5. The Beta-ketoacyl synthase, N-terminal domain (PF00109), one of the main PKS domains, and the Beta-ketoacyl synthase, C-terminal domain (PF02801) that is often associated with the N-terminal domain, are over-represented in the G + G dataset (Supplementary Table S8). The Acyl transferase domain (PF00698), another primary PKS domain, is over-represented with an adjusted p-value of 2.24 × 10−6. The cellular component GO term for polyketide synthase complex (GO:0034081) is also enriched.
Core functions in symbiotic dinoflagellates
Dinoflagellates in the family of Symbiodiniaceae6 form critical symbiotic relationships with marine invertebrates, notably reef-building corals. Disruption of this symbiosis due to environmental stress can lead to bleaching and eventual death of the host animal. A few dinoflagellate lineages also form symbiotic relationships with zooplankton (Brandtodinium nutricula) and foraminifera (Pelagodinium beii). Comparison of annotated Pfam domains in these symbiotic taxa against those in all taxa, shows that functions related to protein-protein interaction (potentially involved in host-symbiont recognition41–43), extracellular matrix, photosynthesis, signal transduction, membrane transport, and cell adhesion are over-represented in the symbiotic lineages (Supplementary Table S9).
Earlier studies of Symbiodiniaceae genomes revealed extensive lineage-specific divergence12–14,16, and genome-wide positive selection of symbiosis-related functions15. Features known to be prevalent in Symbiodiniaceae, including Chlorophyll a-b binding protein (PF00504), Ankyrin repeats (3 copies) (PF12796) and EF-hand domain pair (PF13499)12,14–16, were also significantly over-represented. Carbonic anhydrase (PF00484), involved in carbon dioxide sequestration for photosynthesis, was likewise over-represented (Supplementary Table S9). Nitrogen has been shown to be important for dinoflagellate-coral symbiosis, particularly in nutrient-poor tropical waters. It has even been suggested that the coral host uses ammonium limitation as a means of controlling the symbiont population44. Terms for nitrogen utilisation (such as ammonium, nitrate, and nitrite transport) are over-represented, confirming the importance of these processes to symbiotic dinoflagellates. Analysis of available Symbiodiniaceae genomes has shown a high level of sequence divergence even between closely related lineages15.
Core functions in cold-adapted dinoflagellates
Although most dinoflagellates occur in tropical and subtropical regions, a few psychrophilic species have been described. To identify the functional characteristics of cold-adapted dinoflagellates, we compared the four psychrophilic species (those isolated from either the Arctic or Antarctic circles): two Suessiales (Polarella glacialis CCMP1383 and Polarella glacialis CCMP2088) and two Peridiniales (Heterocapsa arctica CCMP445 and Scrippsiella hangoei-like SHHI-4) against all taxa. Pfam domains related to cold adaptation were over-represented (Supplementary Table S10). The DUF3494 (PF11999) domain (which is shared by type 1 ice-binding proteins45) was the most significantly enriched, and cold-shock (PF00313) domain the third most enriched. DUF347 (repeat of unknown function) (PF03988) is the second most over-represented domain, ATP synthase (E/31 kDa) subunit (PF01991) the fourth-most, and Chlorophyll a-b-binding protein (PF00504) the fifth-most. The enrichment of chlorophyll-binding proteins is likely due to the primarily photosynthetic lifestyle of cold-adapted dinoflagellates compared to the mixotrophic lifestyle of other dinoflagellate taxa.
We further compared the cold-adapted Peridiniales against all Peridiniales taxa (Supplementary Table S11). Mixotrophy was reported in Scrippsiella spp. and Heterocapsa spp46; they comprise six of the 11 Peridiniales taxa in our dataset. Over-represented domains in cold-adapted Peridiniales include DUF347 (PF03988), chlorophyll a-b-binding protein (PF00504), DUF3494 (PF11999), and peridinin-chlorophyll a binding protein (PF02429). Similarly, we compared the cold-adapted Suessiales against all members of this lineage (Supplementary Table S12) and did not observe a significant enrichment of domains related to photosynthetic functions. This observation may not be surprising, because Suessiales lineages are photoautotrophs. A large number of over-represented domains with functions related to RNA processing (e.g. DEAD/DEAH box helicase (PF00270) and multiple [PPR_2 and PPR_3] PPR repeat (PF01535) domains) were recovered in the cold-adapted Suessiales. The Ion transport protein (PF00520) domain is under-represented in these taxa (Supplementary Table S12).
Species that thrive in extreme cold conditions must adapt to slow enzyme kinetics, which results in a decreased rate of catalysis. One postulated mechanism to deal with this issue is the up-regulation of proteins or substrates that might otherwise limit biochemical processes. The quantity of synthesised ribosomal proteins47 and ATP48 has also been shown to increase with decreasing temperature in psychrophilic species. In cold-adapted dinoflagellates (Supplementary Table S10), a number of ATP synthase subunits, ribosomal proteins and photosynthesis-related domains are over-represented. Our results suggest that an increased genetic capacity for these functions in psychrophilic dinoflagellates may compensate for low enzyme kinetics. This hypothesis remains to be tested as additional genomic and functional data from these dinoflagellates become available.
Conclusions
Our study represents the most comprehensive in silico analysis, to date, of dinoflagellate transcriptomes and their functional capacities. We offer the first glimpses into the inventory of dark proteins in dinoflagellates, highlighting putative functions. Dark proteins represent a treasure trove of knowledge into local adaptation, because their functions are directly related to the diversification of lineages. We also identify potential functions that are shared across all analyzed dinoflagellate datasets, thus representing a putative set of defining features for these taxa. Enrichment analysis identifies features that define selective constraints on dinoflagellates to toxin biosynthesis, and to symbiotic and cold-adapted lifestyles. These results provide a foundational platform for further investigations of lineage-specific diversification, and of adaptation of dinoflagellates to their environments. However, most dinoflagellate genes are known to be constitutively expressed irrespective of growth conditions49,50, thus these transcriptome datasets do not allow us to adequately assess niche-specific gene expression and functional features; these questions can be addressed when genome data from the relevant taxa become available. The development and deployment of genetic methods such as CRISPR-Cas9, transposon-based mutagenesis, and RNAi are urgently needed to test hypotheses about genes that putatively define locally adapted dinoflagellate lineages.
Methods
Data
The predicted protein sequences from 62 assembled transcriptomes were retrieved from the Microbial Eukaryote Transcriptome Sequencing Project (MMETSP)18. Transcriptomes of Gambierdiscus caribaeus19 and Alexandrium tamarense CCMP159820 were also acquired to create the initial pool of transcriptomes used in this study (64 in total; Supplementary Table S1). Eight of these transcriptomes (Akashiwo sanguinea CCCM885, Gyrodinium dominans SPMC 103, Lessardia elongata SPMC 104, Oxyrrhis marina CCMP1788, Prorocentrum lima CCMP684, Prorocentrum micans CCCM845, Pyrocystis lunula CCCM517 and Thoracosphaera heimii CCCM670-CCMP1069) were removed because they contained <1000 proteins; Crypthecodinium cohnii Seligo and Symbiodinium sp. Clade A were also removed, as they are potentially mislabelled22.
The Benchmarking Universal Single-Copy Orthologs (BUSCO v3.0.2b)26 program (using the alveolate_stramenophiles_ensembl, eukaryota_odb9 and protists_ensembl datasets; retrieved 22 September 2017), BLASTP searches (v2.3.0, e-value 1e-10) using the same three BUSCO datasets and BLASTP searches (v2.3.0, e-value 1e-10) using the protein orthologs from the Core Eukaryotic Genes (CEGs)21 were used to assess the completeness of each transcriptome. Seven transcriptomes (Alexandrium andersonii CCMP2222, Alexandrium fundyense CCMP1719, Alexandrium minutum CCMP113, Alexandrium tamarense CCMP1598, Amoebophrya sp. Ameob2, Oxyrrhis marina CCMP1795, Symbiodinium [now Fugacium] kawagutii CCMP2468) which all had >80%, >40% and >65% missing genes in the alveolate-stramenophiles, eukaryota and protists datasets, and also had <80% recovery of CEGs, were removed.
The proportion of each transcriptome with similarity to the RefSeq bacterial proteins database (release 80) was assessed using BLASTP (v2.2.28, e-value 1e-10); sequences matching at >90% identity were considered as putative bacterial contaminants. All transcriptomes analysed had <1% of their sequences sharing >90% similarity with bacterial protein sequences; the highest proportion was found in Glenodinium foliaceum CCAP1116_3 (0.67%; 714 sequences) and Symbiodinium sp. D1a (now Durusdinium, 0.45%; 203 sequences). As the putative bacterial sequences in each transcriptome was <1%, all transcriptomes (including the putative bacterial sequences) were retained and no filtering was conducted. To reduce redundancy of protein sequences in each of the 47 transcriptome datasets, each dataset was clustered independently using CD-HIT (v4.6.5, identity 100%, word length 5)51; only the longest ‘representative’ sequences (Table 1) were retained and used in subsequent identification of homologous sets.
Identification of homolog groups and phylogenetic reconstruction
Construction of a maximum-likelihood phylogenetic tree consisting of all samples used in this study was conducted using the method described in Price and Bhattacharya22. Putatively homologous protein sets were constructed using OrthoFinder v1.1.8 (inflation 1.5)52. Similar to the “set B” clusters in Price and Bhattacharya22, we selected sequence sets (represented by ≥ 10 taxa) in which all taxa have only one sequence representation except for one taxon X that has two copies. The two sequences from taxon X were then removed from the sequence set before phylogenetic inference. This approach yielded 1043 single-copy sets for phylogenetic inference. For each of these sets, the sequences were aligned using MAFFT v7.31053 (--localpair --maxiterate 1000). Alignments were trimmed in two stages using trimAL v1.2rev5954: (1) the automated heuristic selection method (-automated1) was first used, then (2) taxa in which 50% of the sequence did not overlap with 50% of the other sequences were removed (-resoverlap 0.5 -seqoverlap 50). A maximum-likelihood tree then was inferred using the partitioned analysis implemented in IQ-TREE v1.5.555; the best evolutionary model for each trimmed alignment was selected using IQ-TREE56, with models considered unlinked. Support of nodes in the inferred consensus tree was determined using 2000 ultrafast bootstraps24. Alignment statistics were generated using AMAS57. The distance between our tree and the one published was calculated using the Robinson-Foulds metric as implemented in PHYLIP58.
Functional annotation of proteins
Each protein was queried using BLASTp (v2.3.0; -evalue 1e-5, -max_target_seqs 20) against separate SwissProt and TrEMBL databases (UniProt release 2017_07). We consider a protein to be “dark” (without a known function) if it, or any protein in the set it is part of, has no significant match to any UniProt entry. Gene Ontology (GO; http://geneontology.org/) terms were assigned using UniProt-GOA mapping (release 2017_09). Membrane transporters were identified by linking SwissProt annotations (release 2016_06), assigned using BLASTp (v2.3.0; -evalue 1e-10, -max_target_seqs 20), with the transporter classifications present in the Transporter Classification Database (retrieved 26 May 2017)30. The transcriptomes were annotated with Pfam domains using pfam_scan.pl (v1.5; database release 30) at E-value < 0.001 following earlier studies16,59,60, and InterProScan (v5.27-66.0) using all analysis packages except SignalP. Proteins were considered to contain a putative transmembrane domain if identified as such by both the Phobius and TMHMM packages.
Enrichment analysis of function
For Pfam domains and transporter classifications, each identifier was assessed for enrichment against a background set using Fisher’s exact test, with correction for multiple testing using the Benjamini and Hochberg method61. GO enrichment was conducted using the topGO R package62, applying the Fisher’s Exact test with the ‘elimination’ methods to correct for the hierarchical structure of GO terms.
Comparison of amino acid profiles between dark versus annotated proteins
We performed a random subsampling test to assess the statistical significance of the difference in proportion we observed for each amino acid between the high-confidence dark and the annotated protein sets. In the subsampling step, for each amino acid, we sampled its proportion from 100 randomly selected individual sequences (in the annotated set versus the dark set), and conducted Student’s t-test to assess the significance of the difference between their means; a Benjamini-Hochberg61 adjusted p-value ≤ 0.05 is considered statistically significant. We carried out this subsampling step 10,000 times, and assessed the number of times that the difference in proportions (of each amino acid in turn) is significant between the two sets. At 95% confidence interval (≥ 9500 tests returned a significant adjusted p-value), the difference in proportions of the amino acid is considered significant.
Electronic supplementary material
Acknowledgements
T.G.S. is supported by an Australian Government Research Training Program (RTP) Scholarship. This project was supported by an Australian Research Council grant (DP150101875) awarded to M.A.R., C.X.C. and D.B, and the computational resources of the National Computational Infrastructure (NCI) National Facility systems through the NCI Merit Allocation Scheme (Project d85) awarded to M.A.R. and C.X.C.
Author Contributions
T.G.S. and C.X.C. conceived the study; T.G.S., C.X.C., M.A.R. and D.B. designed the experiments and interpreted the results; T.G.S. conducted the experiments, prepared all figures and tables, and the first draft of the manuscript; all authors prepared, wrote, reviewed, commented on and approved the final manuscript.
Data Availability
The sources of datasets analysed during the current study are included in this published article and its Supplementary Information files, as detailed in Supplementary Table S1.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-35620-z.
References
- 1.Gómez F. A list of free-living dinoflagellate species in the world’s oceans. Acta Bot. Croat. 2005;64:129–212. [Google Scholar]
- 2.Taylor FJR, Hoppenrath M, Saldarriaga JF. Dinoflagellate diversity and distribution. Biodivers. Conserv. 2008;17:407–418. doi: 10.1007/s10531-007-9258-3. [DOI] [Google Scholar]
- 3.Stoecker DK, Hansen PJ, Caron DA, Mitra A. Mixotrophy in the marine plankton. Ann. Rev. Mar. Sci. 2017;9:311–335. doi: 10.1146/annurev-marine-010816-060617. [DOI] [PubMed] [Google Scholar]
- 4.Caron DA. Mixotrophy stirs up our understanding of marine food webs. Proc. Natl. Acad. Sci. USA. 2016;113:2806–2808. doi: 10.1073/pnas.1600718113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kellmann R, Stüken A, Orr RJ, Svendsen HM, Jakobsen KS. Biosynthesis and molecular genetics of polyketides in marine dinoflagellates. Mar. Drugs. 2010;8:1011–1048. doi: 10.3390/md8041011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.LaJeunesse TC, et al. Systematic revision of Symbiodiniaceae highlights the antiquity and diversity of coral endosymbionts. Curr. Biol. 2018;28:2570–2580. doi: 10.1016/j.cub.2018.07.008. [DOI] [PubMed] [Google Scholar]
- 7.Baker AC. Flexibility and specificity in coral-algal symbiosis: diversity, ecology, and biogeography of Symbiodinium. Annu. Rev. Ecol. Evol. Syst. 2003;34:661–689. doi: 10.1146/annurev.ecolsys.34.011802.132417. [DOI] [Google Scholar]
- 8.Suggett DJ, Warner ME, Leggat W. Symbiotic dinoflagellate functional diversity mediates coral survival under ecological crisis. Trends Ecol. Evol. 2017;32:735–745. doi: 10.1016/j.tree.2017.07.013. [DOI] [PubMed] [Google Scholar]
- 9.Horiguchi T. Heterocapsa arctica sp. nov. (Peridiniales, Dinophyceae), a new marine dinoflagellate from the arctic. Phycologia. 1997;36:488–491. doi: 10.2216/i0031-8884-36-6-488.1. [DOI] [Google Scholar]
- 10.Montresor M, Procaccini G, Stoecker DK. Polarella glacialis, gen. nov., sp. nov. (Dinophyceae): Suessiaceae are still alive! J. Phycol. 1999;35:186–197. doi: 10.1046/j.1529-8817.1999.3510186.x. [DOI] [Google Scholar]
- 11.John, U., Mock, T., Valentin, K., Cembella, A. D. & Medlin, L. Dinoflagellates come from outer space but haptophytes and diatoms do not. In Harmful Algae 2002 (eds Steidinger, K. A., Landsberg, J. H., Tomas, C. R. & Vargo, G. A.) 428–430 (Florida Fish and Wildlife Conservation Commission and Intergovernmental Oceanographic Commission of UNESCO, St Petersburg(FL), 2004).
- 12.Shoguchi E, et al. Draft assembly of the Symbiodinium minutum nuclear genome reveals dinoflagellate gene structure. Curr. Biol. 2013;23:1399–1408. doi: 10.1016/j.cub.2013.05.062. [DOI] [PubMed] [Google Scholar]
- 13.Lin S, et al. The Symbiodinium kawagutii genome illuminates dinoflagellate gene expression and coral symbiosis. Science. 2015;350:691–694. doi: 10.1126/science.aad0408. [DOI] [PubMed] [Google Scholar]
- 14.Aranda M, et al. Genomes of coral dinoflagellate symbionts highlight evolutionary adaptations conducive to a symbiotic lifestyle. Sci. Rep. 2016;6:39734. doi: 10.1038/srep39734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu H, et al. Symbiodinium genomes reveal adaptive evolution of functions related to coral-dinoflagellate symbiosis. Commun. Biol. 2018;1:95. doi: 10.1038/s42003-018-0098-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.González-Pech RA, Ragan MA, Chan CX. Signatures of adaptation and symbiosis in genomes and transcriptomes of Symbiodinium. Sci. Rep. 2017;7:15021. doi: 10.1038/s41598-017-15029-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meng A, et al. Analysis of the genomic basis of functional diversity in dinoflagellates using a transcriptome-based sequence similarity network. Mol. Ecol. 2018;27:2365–2380. doi: 10.1111/mec.14579. [DOI] [PubMed] [Google Scholar]
- 18.Keeling PJ, et al. The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol. 2014;12:e1001889. doi: 10.1371/journal.pbio.1001889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Price DC, et al. Analysis of Gambierdiscus transcriptome data supports ancient origins of mixotrophic pathways in dinoflagellates. Environ. Microbiol. 2016;18:4501–4510. doi: 10.1111/1462-2920.13478. [DOI] [PubMed] [Google Scholar]
- 20.Chan CX, et al. Analysis of Alexandrium tamarense (dinophyceae) genes reveals the complex evolutionary history of a microbial eukaryote. J. Phycol. 2012;48:1130–1142. doi: 10.1111/j.1529-8817.2012.01194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parra G, Bradnam K, Ning Z, Keane T, Korf I. Assessing the gene space in draft genomes. Nucleic Acids Res. 2009;37:289–297. doi: 10.1093/nar/gkn916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Price DC, Bhattacharya D. Robust Dinoflagellata phylogeny inferred from public transcriptome databases. J. Phycol. 2017;53:725–729. doi: 10.1111/jpy.12529. [DOI] [PubMed] [Google Scholar]
- 23.Robinson DF, Foulds LR. Comparison of phylogenetic trees. Math. Biosci. 1981;53:131–147. doi: 10.1016/0025-5564(81)90043-2. [DOI] [Google Scholar]
- 24.Minh BQ, Nguyen MA, von Haeseler A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013;30:1188–1195. doi: 10.1093/molbev/mst024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janouškovec J, et al. Major transitions in dinoflagellate evolution unveiled by phylotranscriptomics. Proc. Natl. Acad. Sci. USA. 2017;114:E171–E180. doi: 10.1073/pnas.1614842114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 27.Frith MC, et al. The abundance of short proteins in the mammalian proteome. PLoS Genet. 2006;2:e52. doi: 10.1371/journal.pgen.0020052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rose MR, et al. The effects of evolution are local: evidence from experimental evolution in Drosophila. Integr. Comp. Biol. 2005;45:486–491. doi: 10.1093/icb/45.3.486. [DOI] [PubMed] [Google Scholar]
- 29.Finn RD, et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saier MH, Jr, Tran CV, Barabote RD. TCDB: the Transporter Classification Database for membrane transport protein analyses and information. Nucleic Acids Res. 2006;34:D181–D186. doi: 10.1093/nar/gkj001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang DZ. Neurotoxins from marine dinoflagellates: a brief review. Mar. Drugs. 2008;6:349–371. doi: 10.3390/md6020349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marban E, Yamagishi T, Tomaselli GF. Structure and function of voltage-gated sodium channels. J. Phycol. 1998;508:647–657. doi: 10.1111/j.1469-7793.1998.647bp.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scott H, Panin VM. N-glycosylation in regulation of the nervous system. Adv. Neurobiol. 2014;9:367–394. doi: 10.1007/978-1-4939-1154-7_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Warren L. The distribution of sialic acids in nature. Comp. Biochem. Physiol. 1963;10:153–171. doi: 10.1016/0010-406X(63)90238-X. [DOI] [PubMed] [Google Scholar]
- 35.Mamedov T, Yusibov V. Green algae Chlamydomonas reinhardtii possess endogenous sialylated N-glycans. FEBS Open Bio. 2011;1:15–22. doi: 10.1016/j.fob.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Preisfeld A, Ruppel HG. Detection of sialic-acid and glycosphingolipids in Euglena gracilis (euglenozoa) Arch. Protistenkd. 1995;145:251–261. doi: 10.1016/S0003-9365(11)80320-9. [DOI] [Google Scholar]
- 37.Yang I, et al. Comparative gene expression in toxic versus non-toxic strains of the marine dinoflagellate Alexandrium minutum. BMC Genomics. 2010;11:248. doi: 10.1186/1471-2164-11-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Markell DA, Trench RK. Macromolecules exuded by symbiotic dinoflagellates in culture: amino acid and sugar composition. J. Phycol. 1993;29:64–68. doi: 10.1111/j.1529-8817.1993.tb00280.x. [DOI] [Google Scholar]
- 39.Jost MC, et al. Toxin-resistant sodium channels: parallel adaptive evolution across a complete gene family. Mol. Biol. Evol. 2008;25:1016–1024. doi: 10.1093/molbev/msn025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brodie ED, Brodie ED. Costs of exploiting poisonous prey: evolutionary trade-offs in a predator-prey arms rage. Evolution. 1999;53:626–631. doi: 10.1111/j.1558-5646.1999.tb03798.x. [DOI] [PubMed] [Google Scholar]
- 41.Schwarz JA, et al. Coral life history and symbiosis: functional genomic resources for two reef building Caribbean corals, Acropora palmata and Montastraea faveolata. BMC Genomics. 2008;9:97. doi: 10.1186/1471-2164-9-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jernigan KK, Bordenstein SR. Ankyrin domains across the Tree of Life. PeerJ. 2014;2:e264. doi: 10.7717/peerj.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nguyen MT, Liu M, Thomas T. Ankyrin-repeat proteins from sponge symbionts modulate amoebal phagocytosis. Mol. Ecol. 2014;23:1635–1645. doi: 10.1111/mec.12384. [DOI] [PubMed] [Google Scholar]
- 44.Gordon BR, Leggat W. Symbiodinium-invertebrate symbioses and the role of metabolomics. Mar. Drugs. 2010;8:2546–2568. doi: 10.3390/md8102546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raymond JA. The ice-binding proteins of a snow alga, Chloromonas brevispina: probable acquisition by horizontal gene transfer. Extremophiles. 2014;18:987–994. doi: 10.1007/s00792-014-0668-3. [DOI] [PubMed] [Google Scholar]
- 46.Stoecker DK. Mixotrophy among dinoflagellates. J. Eukaryot. Microbiol. 1999;46:397–401. doi: 10.1111/j.1550-7408.1999.tb04619.x. [DOI] [Google Scholar]
- 47.Toseland A, et al. The impact of temperature on marine phytoplankton resource allocation and metabolism. Nat. Clim. Change. 2013;3:979–984. doi: 10.1038/nclimate1989. [DOI] [Google Scholar]
- 48.Napolitano MJ, Shain DH. Distinctions in adenylate metabolism among organisms inhabiting temperature extremes. Extremophiles. 2005;9:93–98. doi: 10.1007/s00792-004-0424-1. [DOI] [PubMed] [Google Scholar]
- 49.Liew YJ, Li Y, Baumgarten S, Voolstra CR, Aranda M. Condition-specific RNA editing in the coral symbiont Symbiodinium microadriaticum. PLoS Genet. 2017;13:e1006619. doi: 10.1371/journal.pgen.1006619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moustafa A, et al. Transcriptome profiling of a toxic dinoflagellate reveals a gene-rich protist and a potential impact on gene expression due to bacterial presence. PLoS ONE. 2010;5:e9688. doi: 10.1371/journal.pone.0009688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 52.Emms DM, Kelly S. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015;16:157. doi: 10.1186/s13059-015-0721-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chernomor O, von Haeseler A, Minh BQ. Terrace aware data structure for phylogenomic inference from supermatrices. Syst. Biol. 2016;65:997–1008. doi: 10.1093/sysbio/syw037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Borowiec ML. PeerJ. 2016. AMAS: a fast tool for alignment manipulation and computing of summary statistics; p. e1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Felsenstein J. PHYLIP–Phylogeny inference package (Version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 59.Finn R, Griffiths-Jones S, Bateman A. Identifying protein domains with the Pfam database. Curr Protoc Bioinformatics. 2003;1:2.5.1–2.5.19. doi: 10.1002/0471250953.bi0205s01. [DOI] [PubMed] [Google Scholar]
- 60.Shoguchi E, et al. Two divergent Symbiodinium genomes reveal conservation of a gene cluster for sunscreen biosynthesis and recently lost genes. BMC Genomics. 2018;19:458. doi: 10.1186/s12864-018-4857-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.R Core Team R: a language and environment for statistical computing (2015).
- 62.Alexa, A. & Rahnenführer, J. topGO: enrichment analysis for Gene Ontology. R package version 2.22.0 (2010).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sources of datasets analysed during the current study are included in this published article and its Supplementary Information files, as detailed in Supplementary Table S1.