Abstract
The earlier an activation of a G protein-dependent signalling cascade at a G protein-coupled receptor (GPCR) is probed, the less amplificatory effects contribute to the measured signal. This is especially useful in case of a precise quantification of agonist efficacies, and is of paramount importance, when determining agonist bias in relation to the β-arrestin pathway. As most canonical assays with medium to high throughput rely on the quantification of second messengers, and assays affording more proximal readouts are often limited in throughput, we developed a technique with a proximal readout and sufficiently high throughput that can be used in live cells. Split luciferase complementation (SLC) was applied to assess the interaction of Gαq with its effector phospholipase C-β3. The resulting probe yielded an excellent Z’ value of 0.7 and offers a broad and easy applicability to various Gαq-coupling GPCRs (hH1R, hM1,3,5R, hNTS1R), expressed in HEK293T cells, allowing the functional characterisation of agonists and antagonists. Furthermore, the developed sensor enabled imaging of live cells by luminescence microscopy, as demonstrated for the hM3R. The versatile SLC-based probe is broadly applicable e.g. to the screening and the pharmacological characterisation of GPCR ligands as well as to molecular imaging.
Introduction
G protein-coupled receptors (GPCR) consist of seven transmembrane helices and are responsible for transducing stimuli, e.g. by hormones or neurotransmitters, across the cellular membrane. They represent the largest of all protein superfamilies in the human genome comprising more than 1000 different receptors1, and are the most important drug targets with approximately 34% of all drugs addressing GPCRs2. Agonist binding to a GPCR leads to the activation of heterotrimeric G proteins comprising an α, β and γ subunit. Binding of an agonist to a GPCR leads to a structural rearrangement resulting in an exchange of GDP for GTP within the α subunit.
There are four major subfamilies of Gα proteins of which we focussed on the αq type that upon activation of the receptor, interacts with effector proteins of the phospholipase C (PLC) class and triggers their enzymatic activity. PLCs catalyse the formation of inositol trisphosphate (IP3) and diacylglycerol (DAG) from phosphatidylinositol 4,5-bisphosphate. DAG diffuses in the cell membrane, whereas IP3 activates Ca2+ channels within the membrane of the endoplasmic reticulum and/or the cellular membrane, both leading to a transient increase in the concentration of Ca2+ in the cytosol. The latter is involved in a plethora of physiological processes such as rearrangements of the cytoskeleton and regulation of gene transcription3.
In case of the second messengers IP3 and Ca2+, changes in intracellular levels are usually measured by liquid-scintillation counting or luminometry. When cells are incubated with tritiated myo-inositol, radioactive IP3 (IP1, IP2) levels can be determined4, whereas Förster resonance energy transfer (FRET)5- and split-luciferase complementation (SLC)-based6 assays make use of specific interactions of IP1 or IP3 with various optical probes. Most often intracellular Ca2+ levels are measured either with fluorescent chelators, changing their optical properties upon complexation of Ca2+ ions7–9 or a calcium-dependent luciferase (aequorin)7,10.
However, substantial amplification, taking place with every step in the signalling cascade, potentially masking effects at earlier stages11 can lead to misinterpretation, e.g. partial agonists appear as a full agonists in assays with a distal readout3,12. Furthermore, since there is an increasing interest in the discrimination of biased agonism with respect to G protein-dependent and β-arrestin-dependent pathways13, techniques are needed, allowing a proximal quantification of signalling events14. Such methods comprise [35S]GTPγS-incorporation15 and steady-state [32,33P]-based GTPase assays16 or FRET17- and bioluminescence resonance energy transfer (BRET)-based techniques18.
Most of the aforementioned approaches are compromised e.g. by requiring cell lysis, the preparation of membranes, the availability of radiolabelled chemicals or by low throughput. To overcome these limitations, we decided to use SLC to quantify the interaction of Gαq with PLC-β3. This technology is based on two catalytically inactive complementary fragments of a luciferase, reconstituting a functional enzyme, catalysing the oxidation of a substrate with concomitant emission of light, when brought in close proximity19. The two fragments are fused to two proteins of which a specific interaction is expected, in this case Gαq and PLC-β3. Probes, based on SLC, have become valuable tools for the quantification of protein-protein interactions (PPI) in general20–22, but also in the field of GPCR research. In this context, SLC was successfully applied to probe the interaction of β-arrestins with GPCRs23–25 and to the quantification of second messengers such as cAMP26 and IP36,26. Advantages of SLC involve a high signal-to-background (S/B) ratio, enabling live cell and in vivo imaging27 and the availability of luciferases catalysing chemical reactions, accompanied by the emission of bright light of different wavelengths (broad spectral diversity)28–30.
We applied SLC to probe the Gαq/PLC-β3 interaction (Fig. 1A) by means of a modified luciferase from the click-beetle Pyrophorus plagiophthalamus (λmax = 613 nm). The enzyme was split into two fragments, a larger N-terminal fragment (CBRN) consisting of the amino acids 1–416 and a smaller C-terminal fragment (CBRC) composed of amino acids 395–542. We generated two sets of fusion proteins of which the first one represents CBRN fused either N-, or C-terminally to PLC-β3 and in the second one CBRC was fused terminally to Gαq. As both termini of Gα subunits are known to be crucial not only for interactions with a respective GPCR the βγ-complex but also for the association with the cellular membrane31–34, CBRC was also integrated in three different flexible loop regions of Gαq. The combination of those fusion proteins, giving the highest S/B ratio upon complementation was used as a sensor to probe the activation of different Gαq-coupled receptors. We demonstrate that the new probe is of value for the functional characterisation of GPCR ligands and for imaging receptor activation in live cells.
Figure 1.
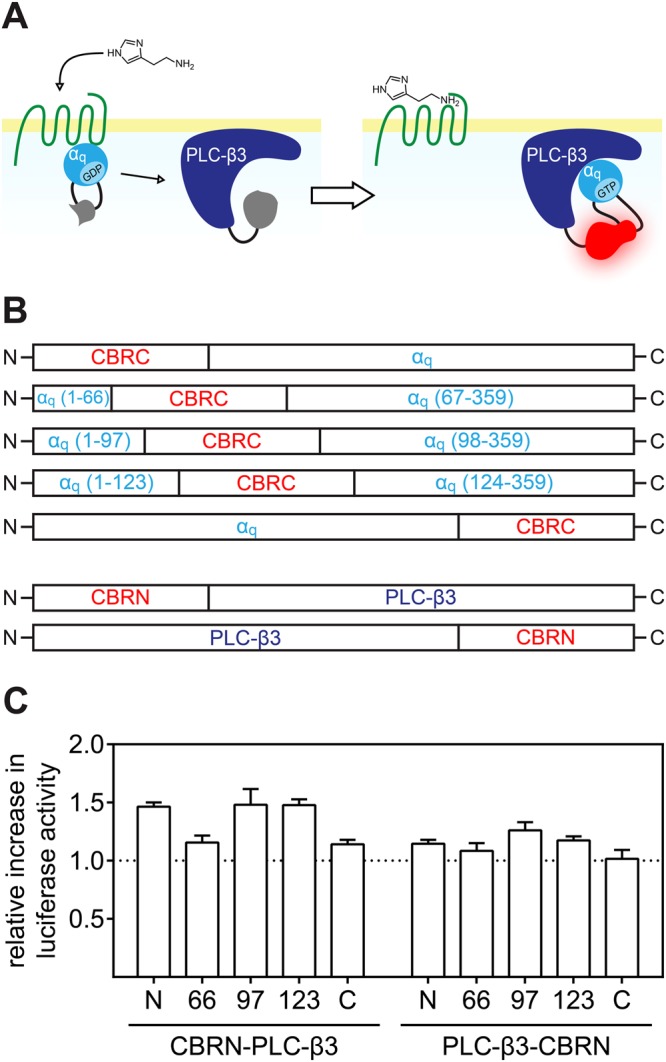
Schematic illustration of the sensor principle and the fusion protein library used to determine the best combination of proteins. The activation of the Gαq pathway was probed by fusing complementary luciferase fragments to Gαq and PLC-β3 (A). A fusion protein library was generated by fusing CBRC to Gαq terminally and in three loop regions (numbers in parentheses denote amino acid positions) and by fusing CBRN either N-, or C-terminally to PLC-β3 (B). The different combinations of Gαq and PLC-β3 fusion proteins were expressed in HEK293T cells, co-expressing the hH1R. The relative increase in luminescence of cells stimulated with 10 µM histamine compared to unstimulated cells is shown for each combination (C). Data are presented as means ± SEM from three independent experiments, each performed in triplicate.
Material and Methods
Materials
Dulbecco’s modified Eagle’s medium (DMEM) with and without phenol red and phosphate-buffered saline (PBS) were from Sigma (Germany). Leibovitz’ L-15 medium (L-15) and Hank’s balanced salt solution (HBSS) were from Gibco (Germany). Fura-2 AM, fetal calf serum (FCS), trypsin and geneticin (G418) were from Merck Biochrom (Germany). D-Luciferin was purchased as potassium salt either from Wako (Japan) or from Pierce (Germany) and was dissolved in HBSS at a concentration of 400 mM. Puromycin was obtained from Invivogen (France). The pCBR-control vector and the Bright-Glo luciferase assay reagent were from Promega (Japan and Germany). The pcDNA4 vector was from Thermo Scientific (Germany), whereas the pIRESpuro3 vector was from Clontech (France). Depending on their physicochemical properties, when possible, ligands were dissolved in H2O; otherwise DMSO (Merck) was used as solvent. Histamine dihydrochloride (his), was from Acros Organics (Belgium), betahistine dihydrochloride (betahis), diphenhydramine hydrochloride (diph), cyproheptadine hydrochloride (cyp), maprotiline hydrochloride (map), carbachol chloride (car), iperoxo iodide (iper), N-methyl scopolamine bromide (NMS), atropine (atr), propantheline bromide (prop) and pirenzepine dihydrochloride (pir) were from Sigma (Germany), whereas mepyramine maleate (mep) was from Tocris Bioscience (MO, USA). UR-KUM530 (KUM530)35 and histaprodifen (histapro) were kindly provided by Prof. Dr. Sigurd Elz (University of Regensburg, Germany). Oxotremorine sesquifumarate (oxo) was from MP Biomedicals (CA, USA), xanomeline (xan) was synthesized in-house according to a standard procedure36. Neurotensin (8–13) (NT(8-13)) was from Synpeptide (China). SR142948A was gift from Dr. Harald Hübner (University of Erlangen, Germany). FR900359 was purchased from the Institute of Pharmaceutical Biology, University of Bonn (Germany).
Cell cultivation
In this study, the HEK293T cell line, obtained from the German collection of microorganisms and cell cultures (DSMZ), was used. Cells were routinely monitored for mycoplasma contamination using the Venor GeM Mycoplasma Detection Kit (Minerva Biolabs, Germany) and were negative. Unless otherwise stated, the cells were cultivated in DMEM containing 10% FCS (full medium) at 37 °C in a water-saturated atmosphere containing 5% CO2.
Generation of plasmids
Plasmids encoding different human GPCRs were obtained from the Missouri cDNA resource center (MO, USA). The other plasmids used were generated by standard PCR and restriction techniques within the pcDNA backbone, unless otherwise stated. A plasmid encoding the red-emitting click-beetle luciferase (pCBR-control) was used as a template in different polymerase chain reactions (PCR) to generate the sequences encoding the two slightly overlapping N-terminal (CBRN, encoding amino acids 1–416) and C-terminal (CBRC, encoding amino acids 394–542) fragments of the luciferase. CBRN was then used to prepare plasmids encoding fusion proteins consisting of PLC-β3 fused either N- or C-terminally to CBRN. CBRC was used to generate plasmids of five different fusion proteins in which CBRC was fused to both termini of Gαq or integrated into the Gαq sequence after amino acids 66, 97 (Gαq(97)) and 123 (Gαq(123)), respectively. The linker sequences used to connect the luciferase fragments to either PLC-β3 or Gαq consisted of flexible Gly and Ser residues. The cDNA encoding the Gαq(123) fusion protein and the N-terminally-tagged PLC-β3 were then subcloned into a pIRESpuro3 vector separated by a P2A autoproteolysis site37, yielding the pIRESpuro3 CBRN-PLC-β3-2A-Gαq(123) vector. Cleavage of the P2A site was controlled by immunoblotting (Fig. S2). All plasmids were quality controlled by means of enzyme restriction analysis and sequencing.
Identification of the best pair of Gαq and PLC-β3 fusion proteins
HEK293T cells were seeded on a 6-well plate (Sarstedt, Germany) at a density of 7 · 105 cell/well. The next day, the cells were transfected with plasmids encoding the different combinations of fusion proteins and the human histamine H1 receptor (hH1R). After 48 h of incubation, the cells were detached by trypsinization, centrifuged and resuspended in DMEM devoid of phenol red, supplemented with FCS (5%). The concentration was adjusted to 1.11 · 106 cells/mL; 90 µL of this suspension were seeded into each well of a white 96-well plate (Greiner Bio One, Germany), and the cells were incubated overnight. To induce interaction of the two fusion proteins, histamine was added at a concentration of 10 µM to the cells. In a control experiment, only DMEM without phenol red, the vehicle of histamine, was applied. After 25 min, 50 µL of medium were aspirated and replaced by 50 µL of Bright-Glo luciferase reagent. The cells were vigorously shaken for 2 min before luminescence was detected using a Genios Pro plate reader (Tecan) for 1 s per well.
Generation of stable expression cell lines
HEK293T cells were transfected with the pIRESpuro3 CBRN-PLC-β3-2A-Gαq(123) vector as described above. After two days of incubation, the cells were detached using trypsin and were seeded into a 75-cm2 cell culture flask. Then, the cells were allowed to attach and puromycin was added at a concentration of 0.75 µg/mL. The cells were cultured upon changing the medium at regular intervals until stable growth was observed again. Subsequently, plasmids encoding cDNAs of GPCRs were transfected in the same way, with the exception that selection was achieved in the presence of geneticin.
Quantification of agonistic potencies and antagonistic activities using the developed probe
Cells, expressing the developed Gαq/PLC-β3 sensor in combination with one of the GPCRs, were detached from a 75-cm2 flask by trypsinization and centrifuged (700 g for 5 min). The pellet was resuspended in assay medium consisting of L-15 with 5% FCS and the density of the suspension was adjusted to 1.25 · 106 cells/mL. Then, 80 µL of this suspension were seeded into each well of a 96-well plate, and the plate was incubated at 37 °C in a humidified atmosphere (without additional CO2) overnight. On the next day, 10 µL of 10 mM D-luciferin (Pierce) were added to the cells, and the plate was transferred into a pre-warmed microplate luminescence reader (either a Genios Pro (Tecan, Austria) or an EnSpire (Perkin-Elmer, Germany)). The cells were allowed to equilibrate inside the reader for 10 min, before the basal luminescence was determined, by recording the luminescence for the entire plate ten times with an integration time of 1 s per well. In the meantime, serial dilutions of agonists were prepared, the resulting solutions were also pre-warmed to 37 °C and subsequently added to the cells. Thereafter, luminescence was recorded for 30 plate repeats amounting to a time period of 50 min. Negative controls (solvent) and positive controls (reference full agonist, histamine (hH1R), carbachol (hM1,5R), oxotremorine (hM3R)) eliciting a maximal response (100%) were included for subsequent normalization of the data. In case of the antagonist mode, antagonists were added 15 min prior to the initial thermal equilibration period to ensure an equilibrium between antagonists and receptors, before agonists were added. The pKb-values of antagonists were determined according to the Cheng-Prusoff equation38. FR900359 was pre-incubated for 20 min, before cells were stimulated with agonists. After acquisition of the data, the peak luminescence intensities obtained after stimulation were used for quantitative analysis using Prism 5 (Graph Pad).
Fura-2 Ca2+ assay
HEK293T cells expressing either the hH1R alone, or co-expressing the Gαq/PLC-β3 sensor, were incubated with Fura-2 AM analysed in cuvettes using a LS50 B luminescence spectrophotometer (Perkin-Elmer, Germany). Fura-2 calcium assays were essentially performed as described previously39.
Live cell luminescence microscopy
HEK293T cells, expressing hM3R and the Gαq/PLC-β3 sensor, were seeded on a 35-mm cell culture dish (Iwaki, Japan) in full medium at a density of 106 cells/dish and were incubated overnight. The next day, 30 µL of 1 M HEPES buffer (pH 7.4) and 7.5 µL of 400 mM D-luciferin (Wako) were added. The cells were transferred to a IX-81 microscope (Olympus, Japan), equipped with a super-cooled EM-CCD camera (Hamamatsu photonics, Japan), with its stage heated to 37 °C, and bioluminescence microscopy was performed essentially as described25. Briefly, the images were acquired with an exposure time of 5 min per frame. After the first frame, oxotremorine was added to a final concentration of 100 nM. In case of the antagonist mode, atropine was added to a final concentration of 100 nM prior to the very first frame.
Results and Discussion
Development of the Gαq activation sensor
In a conventional approach to develop a SLC-based PPI probe, a set of fusion proteins, comprising the luciferase fragments and the two host proteins is engineered and expressed. Usually, both luciferase fragments are fused to both termini of the host proteins and all combinations of the resulting fusion proteins are analysed with respect to their ability to restore the luminescence signal. As our aim was to probe the Gαq/PLC-β3 interaction (Fig. 1A), and because Gαq is a rather small protein of which it is well known that both termini are of major importance for interactions with βγ-subunits, with a GPCR and the association with the cell membrane31–34, we pursued a slightly different strategy: we fused the smaller luciferase fragment (CBRC) to Gαq and incorporated CBRC into three different flexible loop regions, localized within the helical domain of Gαq40 (Fig. 1B). The complementing part of the luciferase (CBRN) was fused either to the N-, or the C-terminus of PLC-β3 (Fig. 1B). To identify the best combination of fusion proteins in terms of luminescence intensity and S/B ratio, we expressed all combinations of Gαq and PLC-β3 fusion proteins shown in Fig. 1B in HEK293T cells co-expressing the human histamine H1 receptor (hH1R). The resulting transfectants were stimulated with 10 µM histamine for 25 min before the cells were lysed and the substrate was added. The detected luminescence was normalized i.e. divided by the luminescence intensity emitted from unstimulated cells. The low normalized luminescence shown in Fig. 1C suggests that the C-terminus of PLC-β3 is rather far away from the interaction site with Gαq. This is supported by the crystal structure of the Gαq/PLC-β3 complex40. The CBRN-PLC-β3 fusion protein gave higher S/B ratios, especially when used in combination with the Gαq variant, in which CBRC was incorporated after amino acid 97 or 123. However, the overall luminescence intensity was higher for Gαq(123) (Fig. S1).
Although the construct, in which CBRC was N-terminally fused to Gαq, showed higher luminescence (Fig. S1), but only an S/B ratio comparable to that of Gαq(97) and Gαq(123), respectively, this fusion protein was not considered, because Yu et al. reported that an N-terminal fusion of green fluorescent protein to Gαs resulted in a lack of association with the cell membrane34. Therefore, we favoured Gαq(123) in combination with CBRN-PLC-β3.
We further optimized the sensor mainly with respect to handling. For this purpose, a vector plasmid, enabling convenient multi-cistronic expression of both fusion proteins in a fixed stoichiometry (1:1), using a self-cleaving P2A peptide sequence37, separating CBRN-PLC-β3 and Gαq(123), was constructed. Furthermore, we fabricated a HEK293T cell line, characterized by stable integration of the aforementioned plasmid into the genome as a versatile platform for the analysis of different GPCRs upon co-transfection. Cleavage of the P2A sequence was proven by immunoblotting using an anti-Gαq antibody (Fig. S2). The western blots also revealed that the expression level of the modified Gαq was similar to that of endogenous Gαq in HEK293T cells (Fig. S2).
Characterization of the new probe
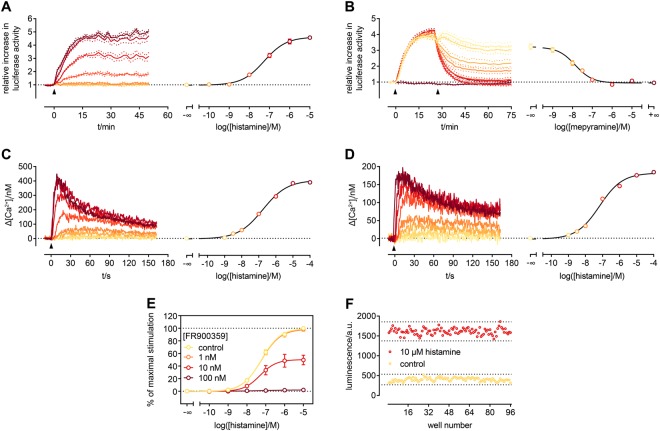
To overcome the low S/B ratio, we added the substrate D-luciferin to live cells (in culture medium) rather than performing endpoint measurements after lysis of the cells. Thereby, we were able to follow the kinetics of the reaction (Fig. 2A). The sensor responded to an activation of the hH1R by increasing the concentration of histamine with a gradual increase in luminescence, which can be converted to a concentration-response-curve (CRC), yielding an EC50 value in very good agreement with data obtained from canonical assays3,16.
Figure 2.
Characterization of the Gαq activation sensor. All experiments were performed with HEK293T cells, expressing the Gαq sensor and the hH1R, except for C were the sensor was not present. Increasing concentrations of histamine (addition indicated by arrow) lead to proportionally increasing luminescence emitted from the cells, which can be converted to a CRC (A). The opposite (a gradual decrease in luminescence) became obvious, when stimulated cells (300 nM histamine, first arrow) were subsequently treated with the selective hH1R agonist mepyramine (second arrow) (B). In case of the highest concentration, luminescence decreased to basal levels, indicating full reversibility of the sensor interaction. Since the observed activation kinetics in A was slower than expected for G protein activation, a kinetic Fura-2 assay for the quantification of [Ca2+]i was performed, to guarantee that the sensor does not negatively influence downstream signalling. In cells, in which the sensor was present, the kinetics was the same (C) when compared to cells devoid of the sensor (D). The concentration-dependent response to histamine (addition indicated by arrow) was similar (hH1R alone: pEC50: 7.1 ± 0.1; Gαq sensor present: pEC50: 6.8 ± 0.1), too. Furthermore, we were able to show that the modified Gαq as part of the sensor was still prone to inhibition by FR900359 (E). The sensor shows an exceptionally good Z’ of 0.7 (F). Data in A-D are representative of at least two independent experiments. Data in E are presented as mean along with their SEM from five independent experiments performed in triplicate. Data in F was obtained from an entire 96-well plate.
Additionally, we could show that the interaction of the two sensor proteins was fully reversible as demonstrated in Fig. 2B. The addition of mepyramine, an hH1R-specific antagonist, after activation of the receptor with histamine (300 nM) led to a concentration-dependent decrease in luminescence, down to the background level. Similar experiments were performed with a sensor in which Gαq(123) was replaced by Gαq(97). The concentration-dependent response to histamine and the reversibility of the interaction of the two sensor proteins was still given, but the S/B ratio was approx. 3-fold lower (Fig. S3).
It turned out that the quality of D-luciferin, the pH of the surrounding medium and the temperature were critical for the validity of the results. D-luciferin from different suppliers was compared (data not shown) as a substrate of the developed probe, and only those products are listed in the method section, which afforded robust results. When experiments were performed at RT instead at 37 °C or in DMEM, in equilibrium with atmospheric CO2 (around 450 ppm), no or only very weak luminescence was detected (data not shown).
As shown in Fig. 2A, onset kinetics of the sensor was slower than expected for the activation of Gα subunits18,41. We hypothesize that the observed slowdown is related to the folding of the full-length luciferase (maturation) upon association of the two fragments22, thereby neither affecting the Gαq/PLC-β3 interaction itself nor downstream signalling. To prove this, we performed a Fura-2 Ca2+ assay, affording kinetic information on the transient increase in cytosolic Ca2+ with a high temporal resolution. Although the amplitudes of the Ca2+ transients were higher in the presence of the sensor (Fig. 2C) – presumably due to differences in cell density and/or by an additional expression of exogenous Gαq and PLC-β3 proteins – the kinetics of intracellular Ca2+ mobilisation observed was nearly identical to that obtained from cells expressing the hH1R alone (Fig. 2D). Accordingly, the histamine concentration-dependent response was not influenced by the presence of the sensor.
The Gαq inhibitor FR900359 is a valuable tool for analysing signalling pathways involving Gαq3,42. Therefore, we examined the susceptibility of the modified Gαq to inhibition by FR900359 (Fig. 2E). We were able to inhibit sensor activation stepwise by applying FR900359 at increasing concentrations until the signal was totally abolished at a concentration of 100 nM.
Furthermore, we were interested in the performance of the sensor, when applied in an assay for ligand characterization. With S/B ratios around 5 (Fig. S3) and a Z’ value of 0.7 (Fig. 2F) the new probe should give excellent robust readouts in functional assays.
Characterization of reference ligands at five different GPCRs
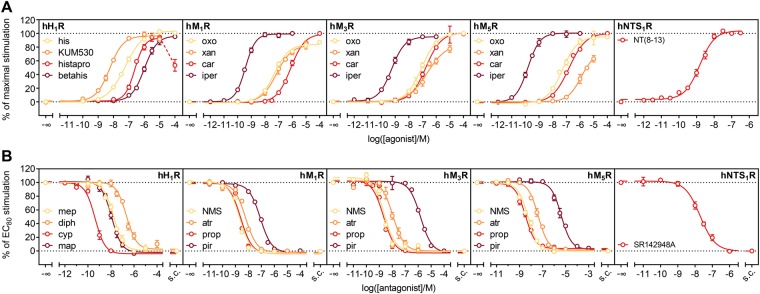
Aiming at a sensor, broadly applicable to diverse Gαq-coupled GPCRs, especially to pharmacologically characterize ligands of the respective receptors, we analysed five different GPCRs of the histamine (hH1R), muscarinic acetylcholine (hM1,3,5R) and neurotensin (hNTS1R) family. Our focus lied on literature-described standard agonists in terms of potencies (pEC50) and efficacies (Emax), as well as on antagonists concerning their antagonistic activity (pKb). The characterization always included a reference agonist that was able to maximally activate the receptor (defined as 100%). This was either the endogenous agonist or a pharmacologically comparable compound at the respective receptor. All other analysed ligands were normalized regarding their efficacy to the particular reference agonist (Table 1).
Table 1.
pEC50, Emax and pKb values of compounds analysed at the hH1R, the hM1,3,5Rs and the hNTS1R.
| compound | pEC50 | %Emax | N | pKb | N | |
|---|---|---|---|---|---|---|
| hH1R | histamine | 7.21 ± 0.07 | 100 | 8 | ||
| UR-KUM530 | 8.22 ± 0.04 | 97.8 ± 3.6 | 7 | |||
| histaprodifen | 6.54 ± 0.07 | 95.6 ± 1.0 | 4 | |||
| betahistine | 5.95 ± 0.07 | 98.3 ± 2.1 | 4 | |||
| mepyramine | 8.48 ± 0.07 | 4 | ||||
| diphenhydramine | 7.36 ± 0.07 | 5 | ||||
| cyproheptadine | 10.12 ± 0.06 | 3 | ||||
| maprotiline | 8.74 ± 0.10 | 4 | ||||
| hM1R | carbachol | 6.12 ± 0.08 | 100 | 4 | ||
| xanomeline | 7.19 ± 0.17 | 80.6 ± 3.2 | 3 | |||
| oxotremorine | 7.32 ± 0.05 | 83.6 ± 1.8 | 3 | |||
| iperoxo | 9.42 ± 0.05 | 99.8 ± 2.3 | 4 | |||
| N′-methyl scopolamine | 9.35 ± 0.07 | 3 | ||||
| atropine | 8.93 ± 0.05 | 3 | ||||
| propantheline | 9.26 ± 0.05 | 3 | ||||
| pirenzepine | 7.76 ± 0.05 | 3 | ||||
| hM3R | oxotremorine | 7.09 ± 0.09 | 100 | 8 | ||
| xanomeline | 6.51 ± 0.11 | 87.2 ± 6.0 | 5 | |||
| carbachol | 6.65 ± 0.06 | 101.0 ± 4.9 | 5 | |||
| iperoxo | 9.24 ± 0.10 | 96.4 ± 1.3 | 4 | |||
| N′-methyl scopolamine | 9.34 ± 0.04 | 5 | ||||
| atropine | 8.69 ± 0.08 | 5 | ||||
| propantheline | 9.37 ± 0.05 | 5 | ||||
| pirenzepine | 6.57 ± 0.03 | 5 | ||||
| hM5R | carbachol | 6.78 ± 0.06 | 100 | 5 | ||
| xanomeline | 5.88 ± 0.14 | 73.3 ± 2.8 | 4 | |||
| oxotremorine | 7.19 ± 0.06 | 101.4 ± 4.3 | 4 | |||
| iperoxo | 9.80 ± 0.07 | 101.4 ± 1.1 | 4 | |||
| N′-methyl scopolamine | 9.52 ± 0.08 | 5 | ||||
| atropine | 8.66 ± 0.05 | 5 | ||||
| propantheline | 9.82 ± 0.08 | 5 | ||||
| pirenzepine | 6.65 ± 0.09 | 5 | ||||
| hNTS1R | neurotensin (8–13) | 8.79 ± 0.09 | 100 | 8 | ||
| SR142948A | 8.20 ± 0.06 | 4 |
Live HEK293T cells, expressing the developed sensor and the indicated receptor, were investigated regarding their response to standard agonists and antagonist. Data is given as mean ± SEM. N denotes the number of biological replicates, each determined in triplicate.
At the hH1R, the endogenous agonist histamine, the slightly more potent phenylhistamine derivative UR-KUM530, histaprodifen and betahistine, a drug approved for the treatment of Ménière’s disease, were analysed (Fig. 3A). Except for histaprodifen, where luminescence decreased after having reached a plateau, compounds gave robust CRCs. For histaprodifen and derivatives thereof, at higher concentrations toxic effects were reported3, which might compromise the luminescence signal. The obtained potencies – with no more than half an order of magnitude difference in EC50 – and efficacies were in good agreement with values obtained from the [32P]GTPase assay35,43 and results from assays addressing alternative signalling pathways, second messengers and holistic methods3. The only exception was histaprodifen, which was not always described as a full agonist as in our case3,16,35,44,45. Antagonistic activities of the reference ligands mepyramine, diphenhydramine, cyproheptadine and maprotiline also aligned well with values described in literature. The corresponding pKb-values differed no more than half a log unit, although especially cyproheptadine was described controversially with at least one and a half orders of magnitude difference in activity across different functional and competition binding assays3,43,46,47. For this compound, our data align best with those from competition binding and functional holistic assays3,46.
Figure 3.
Characterization of standard ligands at the hH1R, the hM1,3,5R and the hNTS1R. Live HEK293T cells, stably expressing the developed sensor and the indicated receptor, were analysed regarding their response to standard agonists (A) and antagonists (B) for the respective receptors. The substrate D-luciferin was added directly to the cells, and the experiment was carried out at 37 °C. Agonist data was normalized to a reference full agonist for each receptor, maximal stimulation of which was defined as 100% (hH1R: histamine, hM1,5R: carbachol, hM3R: oxotremorine). pEC50, Emax and pKb values are listed in Table 1 and were in good accordance with data described in literature. Data are presented as means ± SEM of at least three independent experiments, each performed in triplicate. s.c.: solvent control.
In literature, controversial data were reported for the analysed agonists at the muscarinic receptors, i.e. the ranges of potencies and efficacies for several compounds are very wide (Table S1). The potencies, we determined, fit well into the reported ranges, and, with a few exceptions, the efficacies too. For e.g. xanomeline at the hM3R, or oxotremorine at the hM5R, different Emax values were reported. Although the developed sensor delivers a readout proximal to the receptor, a potential receptor reserve might still influence the observed ligand efficacies48, which might be an explanation for the aforementioned discrepancies. This can, of course, also apply to most of the literature reported data. The determined activities (pKb values) of the hMxR antagonists were comparable to those obtained from other functional assays or pKi values from radioligand competition binding, with differences not greater than half an order of magnitude49–56.
In case of the peptidergic hNTS1R, the potency of NT(8–13) was comparable to that determined by BRET between G protein subunits57, or in a MAPK-dependent reporter gene assay58. The same holds true for the antagonist SR142948A, the pKb of which matches well with values obtained by IP3 quantification or radioligand binding59,60.
Live cell luminescence
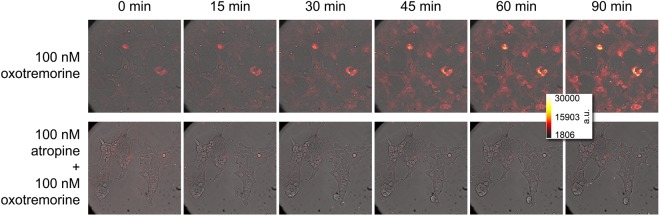
To explore the potential of the developed probe in view of future applications, such as multiparametric measurements, e.g. in combination with impedance-based cell sensing, and imaging in laboratory animals, we decided to do live cell bioluminescence microscopy. For this purpose, cells, expressing the hM3R and the sensor, were investigated under an inverted microscope equipped with a super-cooled EM-CCD camera. Stimulation of the cells resulted in an increase in luminescence over time, whereas pre-incubation with the antagonist atropine abolished sensor activation completely, as shown in Fig. 4 and Videos S1 and S2. Although confocal resolution was not reached, luminescence was predominantly observed on the edges of the cells, indicating sensor activation associated with the cellular membrane. These observations are in agreement with the fact that both sensor proteins are membrane-associated either due to palmytoylation as in case of Gαq or via hydrophobic regions as in the catalytic core of PLC-β340.
Figure 4.
Live cell luminescence imaging of Gαq sensor-expressing HEK293T cells stimulated via the hM3R. Shown are the results of one experiment, performed in the agonist (N = 3) and the antagonist (N = 2) mode, respectively. D-Luciferin was added to the cells before they were transferred to the bioluminescence microscope with its stage warmed to 37 °C. The first frame always shows cells before stimulation. All images were taken with an exposure time of 5 min and are presented as arbitrary light units in false colour. Upon stimulation with 100 nM oxotremorine (approx. EC60), a constant saturable increase in luminescence was observed leading to a plateau after approx. 45 min (cf. Fig. S6). No increase was detectable, when the cells were pre-incubated with atropine (100 nM). The supplementary videos clips S1 and S2 show all acquired frames of which only selected are shown here.
Conclusion
The SLC approach was applied to the interaction of Gαq and PLC-β3 involved in the signalling cascade of GPCRs. This led, to the best of our knowledge, to the first described SLC-based probe, in which one of the two luciferase fragments was incorporated into the protein sequence of one of the host proteins rather than attached to the termini. As a probe for the Gαq/PLC-β3 interaction that makes genetical receptor modifications unnecessary and with its excellent Z’ value of 0.7, the sensor is very suitable for ligand characterization, which was shown for five different GPCRs. Furthermore, the sensor proved to be useful for imaging, as shown for live cell bioluminescence microscopy. Beyond the here described applications the sensor might become a valuable tool for de-orphanization and subsequent determination of signalling pathways of orphan GPCRs, the analysis of Gαq activation in cells endogenously expressing Gαq protein-coupled receptors and imaging in laboratory animals.
Electronic supplementary material
Live cell luminescence microscopy – agonist mode
Live cell luminescence microscopy – antagonist mode
Acknowledgements
A PhD position awarded to T.L. from the International Doctoral Program “Receptor Dynamics” Elite Network of Bavaria is gratefully acknowledged. This study was supported by the Graduate Training Program GRK 1910 (“Medicinal Chemistry of Selective GPCR Ligands”) of the Deutsche Forschungsgemeinschaft (T.L., A.B., G.B.) and the Japan Society for the Promotion of Science (Grants-in-Aid for Scientific Research (S) 26220805) (T.O.).
Author Contributions
T.L. conceived the project with input from T.O., C.H., A.B. and G.B. T.L. performed experiments and data analysis with continuous supervision by T.O., C.H., A.B. and G.B. T.O. provided important plasmids and microscopy equipment. T.L. and G.B. wrote the manuscript with input from T.O. and C.H.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Armin Buschauer is deceased.
Contributor Information
Timo Littmann, Email: timo.littmann@ur.de.
Günther Bernhardt, Email: guenther.bernhardt@ur.de.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-35615-w.
References
- 1.Katritch V, Cherezov V, Stevens RC. Structure-function of the G protein-coupled receptor superfamily. Annu Rev Pharmacol Toxicol. 2013;53:531–556. doi: 10.1146/annurev-pharmtox-032112-135923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hauser AS, Attwood MM, Rask-Andersen M, Schiöth HB, Gloriam DE. Trends in GPCR drug discovery: new agents, targets and indications. Nat Rev Drug Discov. 2017;16:829–842. doi: 10.1038/nrd.2017.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lieb S, et al. Label-free versus conventional cellular assays: Functional investigations on the human histamine H1 receptor. Pharmacol Res. 2016;114:13–26. doi: 10.1016/j.phrs.2016.10.010. [DOI] [PubMed] [Google Scholar]
- 4.Benjamin ER, et al. A miniaturized column chromatography method for measuring receptor-mediated inositol phosphate accumulation. J Biomol Screen. 2004;9:343–353. doi: 10.1177/1087057103262841. [DOI] [PubMed] [Google Scholar]
- 5.Trinquet E, et al. D-myo-inositol 1-phosphate as a surrogate of D-myo-inositol 1,4,5-tris phosphate to monitor G protein-coupled receptor activation. Anal Biochem. 2006;358:126–135. doi: 10.1016/j.ab.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 6.Ataei F, Torkzadeh-Mahani M, Hosseinkhani S. A novel luminescent biosensor for rapid monitoring of IP3 by split-luciferase complementary assay. Biosens Bioelectron. 2013;41:642–648. doi: 10.1016/j.bios.2012.09.037. [DOI] [PubMed] [Google Scholar]
- 7.Hulme, E. C. Receptor-Effector Coupling: A Practical Approach. Vol. 68 (Oxford University Press, 1990).
- 8.Grynkiewicz G, Poenie M, Tsien RY. A new generation of Ca2+ indicators with greatly improved fluorescence properties. J Biol Chem. 1985;260:3440–3450. [PubMed] [Google Scholar]
- 9.Gee KR, et al. Chemical and physiological characterization of fluo-4 Ca2+-indicator dyes. Cell Calcium. 2000;27:97–106. doi: 10.1054/ceca.1999.0095. [DOI] [PubMed] [Google Scholar]
- 10.McCormack, J. G. & Cobbold, P. H. Cellular Calcium: A Practical Approach. Vol. 80 (Oxford University Press, 1991).
- 11.Schrage R, De Min A, Hochheiser K, Kostenis E, Mohr K. Superagonism at G protein-coupled receptors and beyond. Br J Pharmacol. 2016;173:3018–3027. doi: 10.1111/bph.13278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nordemann U, et al. Luciferase reporter gene assay on human, murine and rat histamine H4 receptor orthologs: correlations and discrepancies between distal and proximal readouts. PLoS One. 2013;8:e73961. doi: 10.1371/journal.pone.0073961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manglik A, et al. Structure-based discovery of opioid analgesics with reduced side effects. Nature. 2016;537:185–190. doi: 10.1038/nature19112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kenakin T, Christopoulos A. Signalling bias in new drug discovery: detection, quantification and therapeutic impact. Nat Rev Drug Discov. 2013;12:205–216. doi: 10.1038/nrd3954. [DOI] [PubMed] [Google Scholar]
- 15.Hilf G, Gierschik P, Jakobs KH. Muscarinic acetylcholine receptor-stimulated binding of guanosine 5′-O-(3-thiotriphosphate) to guanine-nucleotide-binding proteins in cardiac membranes. Eur J Biochem. 1989;186:725–731. doi: 10.1111/j.1432-1033.1989.tb15266.x. [DOI] [PubMed] [Google Scholar]
- 16.Strasser A, Striegl B, Wittmann HJ, Seifert R. Pharmacological profile of histaprodifens at four recombinant histamine H1 receptor species isoforms. J Pharmacol Exp Ther. 2008;324:60–71. doi: 10.1124/jpet.107.129601. [DOI] [PubMed] [Google Scholar]
- 17.Bünemann M, Frank M, Lohse MJ. Gi protein activation in intact cells involves subunit rearrangement rather than dissociation. Proc Natl Acad Sci USA. 2003;100:16077–16082. doi: 10.1073/pnas.2536719100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galés C, et al. Real-time monitoring of receptor and G-protein interactions in living cells. Nat Methods. 2005;2:177–184. doi: 10.1038/nmeth743. [DOI] [PubMed] [Google Scholar]
- 19.Yoshimura H, Ozawa T. Methods of split reporter reconstitution for the analysis of biomolecules. Chem Rec. 2014;14:492–501. doi: 10.1002/tcr.201402001. [DOI] [PubMed] [Google Scholar]
- 20.Paulmurugan R, Gambhir SS. Monitoring protein-protein interactions using split synthetic renilla luciferase protein-fragment-assisted complementation. Anal Chem. 2003;75:1584–1589. doi: 10.1021/ac020731c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirohama M, et al. Assay methods for small ubiquitin-like modifier (SUMO)-SUMO-interacting motif (SIM) interactions in vivo and in vitro using a split-luciferase complementation system. Anal Biochem. 2014;448:92–94. doi: 10.1016/j.ab.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 22.Hattori M, Ozawa T. Split luciferase complementation for analysis of intracellular signaling. Anal Sci. 2014;30:539–544. doi: 10.2116/analsci.30.539. [DOI] [PubMed] [Google Scholar]
- 23.Dixon AS, et al. NanoLuc Complementation Reporter Optimized for Accurate Measurement of Protein Interactions in Cells. ACS Chem Biol. 2016;11:400–408. doi: 10.1021/acschembio.5b00753. [DOI] [PubMed] [Google Scholar]
- 24.Littmann T, et al. Recruitment of β-arrestin 1 and 2 to the β2-adrenoceptor: analysis of 65 ligands. J Pharmacol Exp Ther. 2015;355:183–190. doi: 10.1124/jpet.115.227959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Misawa N, et al. Rapid and high-sensitivity cell-based assays of protein-protein interactions using split click beetle luciferase complementation: an approach to the study of G-protein-coupled receptors. Anal Chem. 2010;82:2552–2560. doi: 10.1021/ac100104q. [DOI] [PubMed] [Google Scholar]
- 26.Takeuchi M, Nagaoka Y, Yamada T, Takakura H, Ozawa T. Ratiometric bioluminescence indicators for monitoring cyclic adenosine 3′,5′-monophosphate in live cells based on luciferase-fragment complementation. Anal Chem. 2010;82:9306–9313. doi: 10.1021/ac102692u. [DOI] [PubMed] [Google Scholar]
- 27.Takakura H, Hattori M, Takeuchi M, Ozawa T. Visualization and quantitative analysis of G protein-coupled receptor-β-arrestin interaction in single cells and specific organs of living mice using split luciferase complementation. ACS Chem Biol. 2012;7:901–910. doi: 10.1021/cb200360z. [DOI] [PubMed] [Google Scholar]
- 28.Nakajima Y, et al. Multicolor luciferase assay system: one-step monitoring of multiple gene expressions with a single substrate. Biotechniques. 2005;38:891–894. doi: 10.2144/05386ST03. [DOI] [PubMed] [Google Scholar]
- 29.Hida N, et al. High-sensitivity real-time imaging of dual protein-protein interactions in living subjects using multicolor luciferases. PLoS One. 2009;4:e5868. doi: 10.1371/journal.pone.0005868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Villalobos V, et al. Dual-color click beetle luciferase heteroprotein fragment complementation assays. Chem Biol. 2010;17:1018–1029. doi: 10.1016/j.chembiol.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Conklin BR, Farfel Z, Lustig KD, Julius D, Bourne HR. Substitution of three amino acids switches receptor specificity of Gqα to that of Giα. Nature. 1993;363:274–276. doi: 10.1038/363274a0. [DOI] [PubMed] [Google Scholar]
- 32.Hepler JR, et al. Functional importance of the amino terminus of Gqα. J Biol Chem. 1996;271:496–504. doi: 10.1074/jbc.271.1.496. [DOI] [PubMed] [Google Scholar]
- 33.Rasmussen SG, et al. Crystal structure of the β2 adrenergic receptor-Gs protein complex. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu JZ, Rasenick MM. Real-time visualization of a fluorescent Gαs: Dissociation of the activated G protein from plasma membrane. Mol Pharmacol. 2002;61:352–359. doi: 10.1124/mol.61.2.352. [DOI] [PubMed] [Google Scholar]
- 35.Strasser A, Wittmann HJ, Kunze M, Elz S, Seifert R. Molecular basis for the selective interaction of synthetic agonists with the human histamine H1-receptor compared with the guinea pig H1-receptor. Mol Pharmacol. 2009;75:454–465. doi: 10.1124/mol.108.053009. [DOI] [PubMed] [Google Scholar]
- 36.Kane BE, Grant MK, El-Fakahany EE, Ferguson DM. Synthesis and evaluation of xanomeline analogs - probing the wash-resistant phenomenon at the M1 muscarinic acetylcholine receptor. Bioorg Med Chem. 2008;16:1376–1392. doi: 10.1016/j.bmc.2007.10.058. [DOI] [PubMed] [Google Scholar]
- 37.Kim JH, et al. High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS One. 2011;6:e18556. doi: 10.1371/journal.pone.0018556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheng Y, Prusoff WH. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 39.Müller M, et al. Synthesis and neuropeptide Y Y1 receptor antagonistic activity of N,N-disubstituted ω-guanidino- and ω-aminoalkanoic acid amides. Arch Pharm (Weinheim) 1997;330:333–342. doi: 10.1002/ardp.19973301104. [DOI] [PubMed] [Google Scholar]
- 40.Lyon AM, Dutta S, Boguth CA, Skiniotis G, Tesmer JJ. Full-length Gαq-phospholipase C-β3 structure reveals interfaces of the C-terminal coiled-coil domain. Nat Struct Mol Biol. 2013;20:355–362. doi: 10.1038/nsmb.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Milde M, Rinne A, Wunder F, Engelhardt S, Bünemann M. Dynamics of Gαi1 interaction with type 5 adenylate cyclase reveal the molecular basis for high sensitivity of Gi-mediated inhibition of cAMP production. Biochem J. 2013;454:515–523. doi: 10.1042/BJ20130554. [DOI] [PubMed] [Google Scholar]
- 42.Schrage R, et al. The experimental power of FR900359 to study Gq-regulated biological processes. Nat Commun. 2015;6:10156. doi: 10.1038/ncomms10156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seifert R, et al. Multiple differences in agonist and antagonist pharmacology between human and guinea pig histamine H1-receptor. J Pharmacol Exp Ther. 2003;305:1104–1115. doi: 10.1124/jpet.103.049619. [DOI] [PubMed] [Google Scholar]
- 44.Lim HD, et al. Evaluation of histamine H1-, H2-, and H3-receptor ligands at the human histamine H4 receptor: identification of 4-methylhistamine as the first potent and selective H4 receptor agonist. J Pharmacol Exp Ther. 2005;314:1310–1321. doi: 10.1124/jpet.105.087965. [DOI] [PubMed] [Google Scholar]
- 45.Patil R, Elz S, Reiser O. Side-chain modified analogues of histaprodifen: asymmetric synthesis and histamine H1-receptor activity. Bioorg Med Chem Lett. 2006;16:672–676. doi: 10.1016/j.bmcl.2005.10.030. [DOI] [PubMed] [Google Scholar]
- 46.Moguilevsky N, et al. Stable expression of human H1-histamine-receptor cDNA in Chinese hamster ovary cells. Pharmacological characterisation of the protein, tissue distribution of messenger RNA and chromosomal localisation of the gene. Eur J Biochem. 1994;224:489–495. doi: 10.1111/j.1432-1033.1994.00489.x. [DOI] [PubMed] [Google Scholar]
- 47.Appl H, et al. Interactions of recombinant human histamine H1R, H2R, H3R, and H4R receptors with 34 antidepressants and antipsychotics. Naunyn Schmiedebergs Arch Pharmacol. 2012;385:145–170. doi: 10.1007/s00210-011-0704-0. [DOI] [PubMed] [Google Scholar]
- 48.Bräuner-Osborne H, Ebert B, Brann MR, Falch E, Krogsgaard-Larsen P. Functional partial agonism at cloned human muscarinic acetylcholine receptors. Eur J Pharmacol. 1996;313:145–150. doi: 10.1016/0014-2999(96)00501-8. [DOI] [PubMed] [Google Scholar]
- 49.Bolden C, Cusack B, Richelson E. Antagonism by antimuscarinic and neuroleptic compounds at the five cloned human muscarinic cholinergic receptors expressed in Chinese hamster ovary cells. J Pharmacol Exp Ther. 1992;260:576–580. [PubMed] [Google Scholar]
- 50.Bräuner-Osborne H, Brann MR. Pharmacology of muscarinic acetylcholine receptor subtypes (m1-m5): high throughput assays in mammalian cells. Eur J Pharmacol. 1996;295:93–102. doi: 10.1016/0014-2999(95)00639-7. [DOI] [PubMed] [Google Scholar]
- 51.Burstein ES, Spalding TA, Brann MR. Pharmacology of muscarinic receptor subtypes constitutively activated by G proteins. Mol Pharmacol. 1997;51:312–319. doi: 10.1124/mol.51.2.312. [DOI] [PubMed] [Google Scholar]
- 52.Christopoulos A, Pierce TL, Sorman JL, El-Fakahany EE. On the unique binding and activating properties of xanomeline at the M1 muscarinic acetylcholine receptor. Mol Pharmacol. 1998;53:1120–1130. [PubMed] [Google Scholar]
- 53.Dong GZ, Kameyama K, Rinken A, Haga T. Ligand binding properties of muscarinic acetylcholine receptor subtypes (m1-m5) expressed in baculovirus-infected insect cells. J Pharmacol Exp Ther. 1995;274:378–384. [PubMed] [Google Scholar]
- 54.Huang F, et al. Receptor binding studies of soft anticholinergic agents. AAPS PharmSci. 2001;3:E30. doi: 10.1208/ps030430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Watson N, Daniels DV, Ford AP, Eglen RM, Hegde SS. Comparative pharmacology of recombinant human M3 and M5 muscarinic receptors expressed in CHO-K1 cells. Br J Pharmacol. 1999;127:590–596. doi: 10.1038/sj.bjp.0702551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wess J, Lambrecht G, Mutschler E, Brann MR, Dörje F. Selectivity profile of the novel muscarinic antagonist UH-AH 37 determined by the use of cloned receptors and isolated tissue preparations. Br J Pharmacol. 1991;102:246–250. doi: 10.1111/j.1476-5381.1991.tb12161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Besserer-Offroy E, et al. The signaling signature of the neurotensin type 1 receptor with endogenous ligands. Eur J Pharmacol. 2017;805:1–13. doi: 10.1016/j.ejphar.2017.03.046. [DOI] [PubMed] [Google Scholar]
- 58.Kling RC, et al. Development of Covalent Ligand-Receptor Pairs to Study the Binding Properties of Nonpeptidic Neurotensin Receptor 1 Antagonists. ACS Chem Biol. 2016;11:869–875. doi: 10.1021/acschembio.5b00965. [DOI] [PubMed] [Google Scholar]
- 59.Gully D, et al. Biochemical and pharmacological activities of SR 142948A, a new potent neurotensin receptor antagonist. J Pharmacol Exp Ther. 1997;280:802–812. [PubMed] [Google Scholar]
- 60.Keller M, et al. Mimicking of Arginine by Functionalized Nω-Carbamoylated Arginine As a New Broadly Applicable Approach to Labeled Bioactive Peptides: High Affinity Angiotensin, Neuropeptide Y, Neuropeptide FF, and Neurotensin Receptor Ligands As Examples. J Med Chem. 2016;59:1925–1945. doi: 10.1021/acs.jmedchem.5b01495. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Live cell luminescence microscopy – agonist mode
Live cell luminescence microscopy – antagonist mode