Abstract
Friedreich ataxia (FRDA) is a multisystem genetic disorder caused by GAA repeat expansion mutations within the FXN gene, resulting in heterochromatin formation and deficiency of frataxin protein. Elevated levels of the FXN antisense transcript (FAST-1) have previously been detected in FRDA. To investigate the effects of FAST-1 on the FXN gene expression, we first stably overexpressed FAST-1 in non-FRDA cell lines and then we knocked down FAST-1 in FRDA fibroblast cells. We observed decreased FXN expression in each FAST-1 overexpressing cell type compared to control cells. We also found that FAST-1 overexpression is associated with both CCCTC-Binding Factor (CTCF) depletion and heterochromatin formation at the 5′UTR of the FXN gene. We further showed that knocking down FAST-1 in FRDA fibroblast cells significantly increased FXN expression. Our results indicate that FAST-1 can act in trans in a similar manner to the cis-acting FAST-1 overexpression that has previously been identified in FRDA fibroblasts. The effects of stably transfected FAST-1 expression on CTCF occupancy and heterochromatin formation at the FXN locus suggest a direct role for FAST-1 in the FRDA molecular disease mechanism. Our findings also support the hypothesis that inhibition of FAST-1 may be a potential approach for FRDA therapy.
Introduction
Friedreich ataxia (FRDA), the most prevalent inherited ataxia, is an autosomal recessive neurodegenerative disorder, primarily affecting the nervous system and the heart. This progressive disease is characterized by limb and gait ataxia, dysarthria, hypertrophic cardiomyopathy and skeletal abnormalities1. Most patients are homozygous for expanded GAA triplet repeat within the first intron of the frataxin (FXN) gene2. Unaffected individuals have up to ~40 triplets, whereas in FRDA patients the number of GAA repeats can be from 70 to 17003,4. The expanded repeats cause a severe deficiency of transcriptional initiation of the FXN gene that ultimately leads to reduction of the essential mitochondrial protein frataxin5,6. Frataxin (FXN) is a nuclear encoded, highly conserved protein which is involved in iron-sulfur cluster (ISC) biosynthesis and regulating mitochondrial iron transport and respiration7,8. Although the exact molecular mechanism of FXN gene silencing is still unknown, accumulating evidence indicates that epigenetic changes play a crucial role in inhibition of FXN transcription. Work with transgenic mice showed that it is the intrinsic property of the expanded GAA repeat that causes heterochromatin formation to exert its epigenetic gene silencing effect9. FRDA alleles have been shown to be enriched for molecular signatures of heterochromatin including histone H3 and H4 deacetylation, histone trimethylation (H3K9me3 and H3K27me3), CpG methylation and non-coding RNA transcription10–14. Investigating DNA methylation profiles of the FXN gene in FRDA cell models, human and transgenic mouse tissues demonstrated elevated CpG methylation levels upstream of the expanded repeats. The amount of DNA methylation correlates with the extent of GAA expansion, phenotype severity and age of disease onset12,15,16. Interestingly, no changes in DNA methylation have been detected in the 5′ untranslated region (UTR) of the FXN gene. Enrichment of repressive chromatin marks at the FXN promoter, upstream and downstream GAA regions have been reported in lymphoblastoid and fibroblast cells14,17 and in FRDA human and transgenic mouse brain and heart tissues12. A number of studies have demonstrated that reversing epigenetic changes via administration of histone deacetylase inhibitors (HDACi) can restore FXN transcription in FRDA10,18. These results further support the hypothesis that transcriptional silencing is due to epigenetic aberrations. In FRDA, heterochromatin encompasses the FXN transcription start site (FXN-TSS) and silences the promotor activity6. In addition, severe depletion of the chromatin insulator protein CTCF has been identified at the 5′ untranslated region (UTR) of the FXN gene in FRDA patients. An antisense transcript named FAST-1 (FXN Antisense Transcript – 1), whose sequence overlaps with the CTCF binding site, has also been discovered. FAST-1 expression is significantly increased in FRDA and is associated with the severe CTCF depletion and heterochromatin formation in the 5′UTR of the FXN gene14,19. Natural antisense transcripts (NATs) have long been described as ‘junk DNA’ or transcriptional noise due to their low expression and unknown function. However, in recent years, antisense transcripts have emerged as key regulators of gene expression in an epigenetic manner20–23.
Literature supporting the notion that antisense transcripts are involved in heterochromatin formation and the regulation of their partner mRNA expression inspired us to further investigate the characteristics of FAST-1. We first identified a full-length FAST-1 transcript with a total length of 523 bp in size containing a poly (A) tail. Mapping the 3′ and 5′ ends of the FAST-1 transcript onto the genome showed that FAST-1 transcription overlaps with the FXN-TSS and CTCF binding site in the 5′UTR of the FXN gene. Therefore, we decided to investigate potential effects of altered FAST-1 expression on FXN expression in three different types of cell lines. We report that FAST-1 overexpression is consistently associated with reduced CTCF occupancy, heterochromatin formation and decreased FXN expression. We also show that knocking down FAST-1 expression results in increased FXN expression in FRDA fibroblast cells, thereby revealing FAST-1 to be a potential FRDA therapeutic target.
Results
Identification of FAST-1 by rapid amplification of cDNA ends
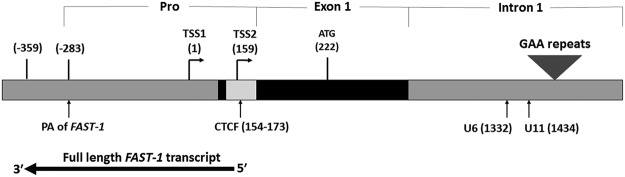
To determine the exact size and location of FAST-1, 5′ and 3′ rapid amplification of cDNA ends (RACE) experiments were performed with total RNA and poly(A) + RNA. Mapping the 5′ and 3′ ends of FAST-1 transcript onto the genome precisely localised them to nucleotides + 164 and −359 of the FXN gene, respectively, and the total length of FAST-1 was found to be 523 bp in size. A poly (A) signal was also identified in the FAST-1 sequence at FXN nucleotide positions −283 to −288 (Fig. 1).
Figure 1.
The 5′ end of FXN gene showing the region corresponding to the full length FAST-1 transcript. It also contains a polyadenylation signal (PA) located between −283 to −288. The 5′-end of FAST-1 coincides with the CTCF binding site in the FXN 5′UTR. Pro = FXN promoter, TSS = transcription start site, U6 and U11 = CpG sites.
Stable overexpression of FAST-1 in non-FRDA cells
To assess the effect of FAST-1 on FXN expression, we generated three non-FRDA cell lines - HeLa, HEK293 and fibroblast - that stably overexpress FAST-1 cDNA under the control of a CMV promoter. Twelve stable FAST-1 overexpressing HeLa cell lines, twelve stable FAST-1 overexpressing HEK293 cell lines and three stable FAST-1 overexpressing fibroblast cell lines were developed. To confirm FAST-1 overexpression in FAST-1 transfected cells, qRT-PCR measurements were performed, using primers designed to detect human-specific frataxin antisense transcript, FAST-1. Analysis of qRT-PCR measurement confirmed a very significant increase of FAST-1 expression in transfected HeLa cells (396%, P < 0.001), transfected HEK293 cells (660%, P < 0.05) and transfected fibroblast cells (332%, P < 0.01) compared with non-transfected control cells (Fig. 2). No such differences were detected with the empty vector-transfected cells.
Figure 2.
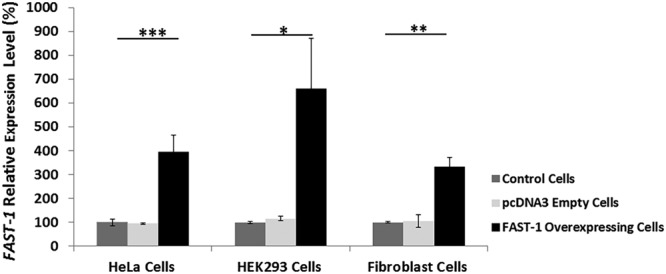
Relative FAST-1 expression in FAST-1 overexpressing cells. FAST-1 overexpression was confirmed in FAST-1 transfected cells compared to non-transfected and empty-vector-transfected control cells. Data were normalized to the mean FAST-1 level of non-transfected control cells taken as 100%. (n = 12, FAST-1 overexpressing HeLa and HEK293 clones), (n = 3, FAST-1 overexpressing fibroblast clones), *p < 0.05, **p < 0.01, ***p < 0.001, Bars represent SEMs.
Overexpression of FAST-1 reduces the FXN mRNA and protein expression
Analysis of FAST-1 overexpressing cells revealed a very significant reduction of FXN mRNA in the FAST-1 overexpressing HeLa cells (45%, P < 0.05), HEK293 cells (47%, P < 0.001) and fibroblast cells (42%, P < 0.001) compared with non-transfected control cells. There was no significant difference detected between the empty vector-transfected and non-transfected control cells (Fig. 3a).
Figure 3.
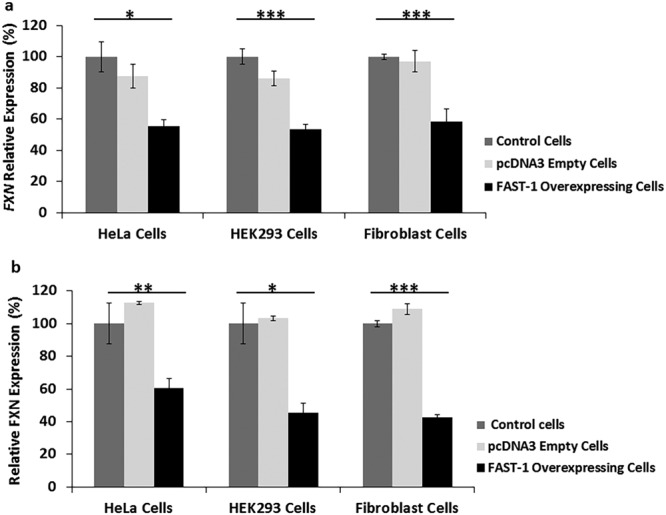
Frataxin expression levels in FAST-1 overexpressing cells. (a) qRT–PCR analysis of FXN mRNA levels in FAST-1 overexpressing cells. Data were normalized to the mean FXN level of non-transfected control cells taken as 100%. (b) Dipstick immunoassay of human frataxin protein. Data were normalized to the mean FXN level of non-transfected control cells taken as 100%. (n = 12, FAST-1 overexpressing HeLa and HEK293 clones), (n = 3, FAST-1 overexpressing fibroblast clones), *p < 0.05, **p < 0.01, ***p < 0.001, Bars represent SEMs.
To determine the levels of human frataxin expression in the FAST-1 overexpressing cells, frataxin protein expression levels were measured by lateral flow immunoassay with the Frataxin Protein Quantity Dipstick assay kit. Analysis of FAST-1 overexpressing cells revealed that frataxin expression levels were significantly decreased to (40%, P < 0.01) in the FAST-1 overexpressing HeLa cells, HEK293 cells (55%, P < 0.05) and fibroblast cells (58%, P < 0.001) compared with non-transfected control cells (Fig. 3b). No such differences were detected with the empty vector-transfected cells.
FAST-1 copy number associated gene expression changes
The FAST-1 copy number was investigated in FAST-1 overexpressing HeLa cell genomic DNA samples using TaqMan real-time PCR, compared to non-transfected HeLa cells that contain two copies of the FXN locus. To assess the reliability of each copy number call, the results were analysed by Applied Biosystems CopyCaller™ Software v.2.0. Seven out of twelve FAST-1 overexpressing HeLa cell lines, together with the endogenous non-transfected HeLa cell control, had acceptable confidence values and absolute zero score to pass the test. The results indicated that FAST-1 copy number is positively correlated with increased FAST-1 expression (R = 0.7, P < 0.05) (Fig. 4a), which in turn is negatively correlated with FXN expression by the analysis of eleven transfected HeLa cell lines and the endogenous non-transfected HeLa cell control (R = −0.58, P < 0.05) (Fig. 4b). This suggests a concentration-dependent molecular mechanism of action for FAST-1, rather than a potential threshold effect.
Figure 4.
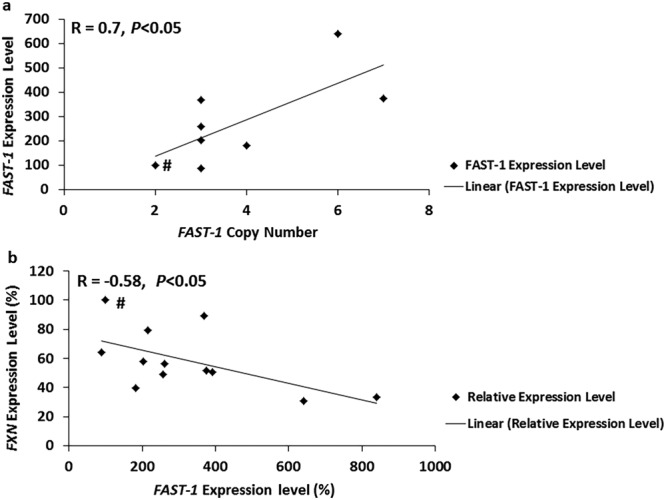
(a) FAST-1 copy number in FAST-1 overexpressing clones. TaqMan copy number assays were applied in duplicates. Non-transfected HeLa cells known to have two copies of FAST-1 were used as the calibrator samples. (n = 7, FAST-1 overexpressing HeLa clones). (b) Scatter plot of FXN mRNA expression level versus FAST-1 expression in FAST-1 overexpressing HeLa cells. There was a significant negative correlation between FAST-1 expression and FXN expression in FAST-1 overexpressing HeLa cells. (n = 11, FAST-1 overexpressing HeLa clones).
FAST-1 overexpressing HeLa cells show reduced occupancy of CTCF and marks of heterochromatin formation at the FXN 5′UTR locus
It has previously been reported that CTCF depletion results in increased FAST-1 transcription14. To elucidate the relationship between CTCF and FAST-1, CTCF occupancy was measured in FAST-1 overexpressing HeLa cells. Three different FAST-1 overexpressing HeLa clones, which showed the lowest level of FXN expression, empty vector-transfected cells and non-transfected control HeLa cells, were utilized in this experiment. CTCF ChIP analysis revealed that CTCF occupancy was decreased sharply at the 5′UTR of the FXN gene. In addition, comparison of CTCF occupancy in empty transfected-vector and non-transfected HeLa control cells did not show a similar reduction (Fig. 5a). We also investigated acetylated and trimethylated histone H3K9 modifications by ChIP analysis at the FXN 5′UTR region. The ChIP results were normalized to the ‘input’, and negative control antibody values were taken into account. ChIP assay results showed significantly decreased acetylation of H3K9 (Fig. 5b) and enrichment of H3K9me3 (Fig. 5c) at the FXN 5′UTR region in FAST-1 overexpressing HeLa cells compared to non-transfected control cells. There were no statistically significant differences in H3K9 acetylation and trimethylation levels between the empty vector-transfected and non-transfected control cells.
Figure 5.
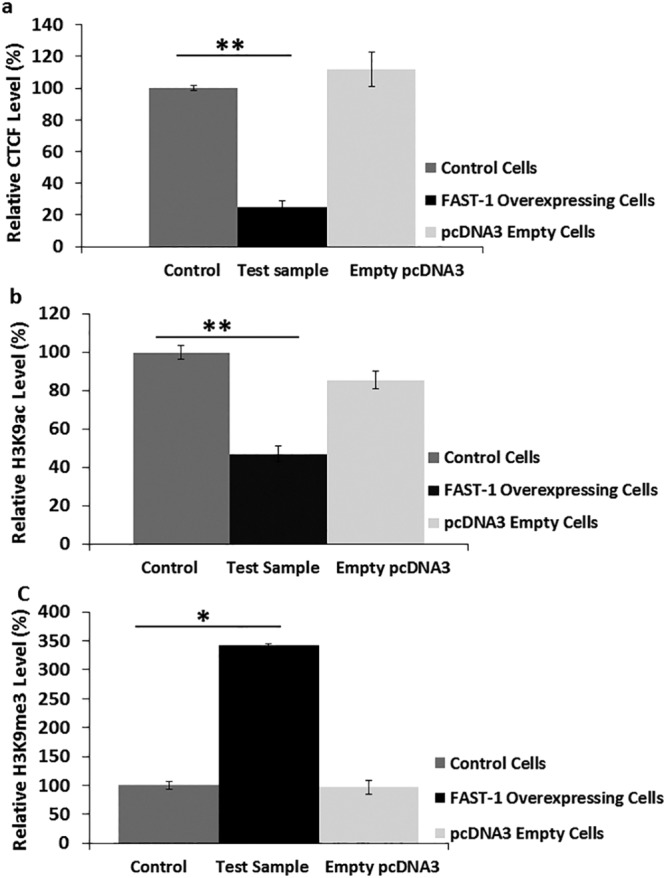
(a) CTCF analysis. ChIP analysis showing the relative CTCF occupancy in the FXN 5′UTR of FAST-1 overexpressing HeLa cells. (b,c) Analysis of histone modifications in FAST-1 overexpressing HeLa cells. ChIP quantitative PCR results for the FXN 5′UTR amplified regions are represented as the relative amount of immunoprecipitated DNA compared with ‘input’ DNA, having taken negligible −Ab control values into account. Mean values from the non-transfected Hela control cells are 100%. The data were from three independent chromatin preparations, with each experiment done in triplicate. (n = 3, FAST-1 overexpressing HeLa clones). *p < 0.05, **p < 0.01, Bars represent SEMs.
FXN gene DNA methylation levels are altered in FAST-1 overexpressing fibroblast cells
It has been reported that CpG sites, designated U6 and U11, at the upstream GAA repeat region show elevated levels of DNA methylation in FRDA patients compared to controls11,19. Therefore, we chose to investigate the DNA methylation status at CpG U6 and U11 in two FAST-1 overexpressing fibroblast cell lines (FAST-1B, FAST-1C), together with FRDA fibroblasts and unaffected control fibroblasts. Results from MethylScreen assays revealed a non-significant small increase in DNA methylation at CpG site U6, with DM values increasing from 14% in unaffected control fibroblasts to 18% and 23% in FAST-1 B and FAST-1 C, respectively (Fig. 6a). At CpG site U11, the proportion of densely methylated templates in FAST-1 overexpressing fibroblasts significantly increased from 8% in unaffected controls to 79% (P < 0.01) and 76% (P < 0.001) in FAST-1 B and FAST-1 C, respectively, similar to the levels found in FRDA fibroblasts (Fig. 6b).
Figure 6.
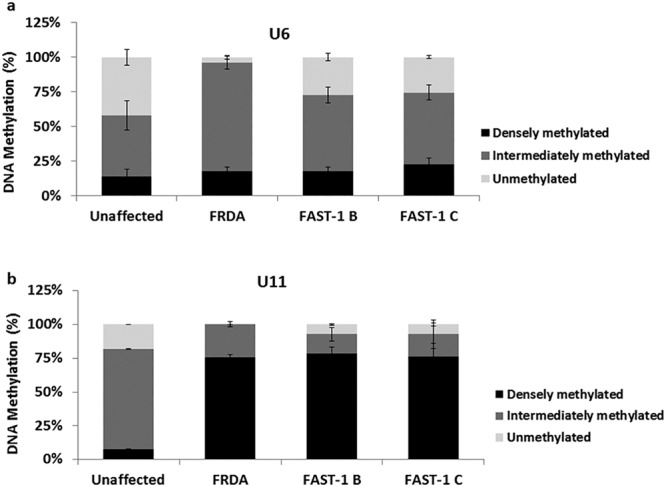
DNA methylation levels in FAST-1 overexpressing fibroblast cells. MethylScreen analysis of CpG site (a) U6 and (b) U11 in the FXN upstream GAA repeat region of DNA from FRDA, unaffected control and FAST-1 overexpressing fibroblast cells. (n = 2, FAST-1 overexpressing fibroblasts).Bars represent SEMs.
Knocking down FAST-1 in FRDA fibroblast cells increases FXN expression
To investigate the effect of FAST-1 silencing on the FXN expression in FRDA cells, FRDA and normal fibroblast cell lines were transduced with lentiviral (LV) vectors expressing shRNAs targeting FAST-1 (pLKO.1-puro-CMV-tGFP). The efficacy of shRNA knockdown was assessed by qRT-PCR. Analysis of FAST-1 expression levels in the FAST-1 knockdown control and FRDA fibroblast cell lines revealed that FAST-1 levels decreased to 63% (P < 0.01) and 43% (P < 0.01), respectively (Fig. 7). After confirming efficient FAST-1 knockdown in FRDA fibroblast cells, FXN mRNA expression was measured in FAST-1 knockdown cells. Our results demonstrated that the levels of FXN expression in FAST-1 knockdown FRDA cells increased by 1.5-fold (P < 0.05) when compared to FRDA fibroblast cells. In contrast, FXN expression in normal fibroblast cells treated with LV FAST-1 showed a non-statistically significant 0.8-fold decrease in the FXN expression. FXN expression remained unchanged in FRDA fibroblast cells treated with scrambled control shRNA (Fig. 8).
Figure 7.
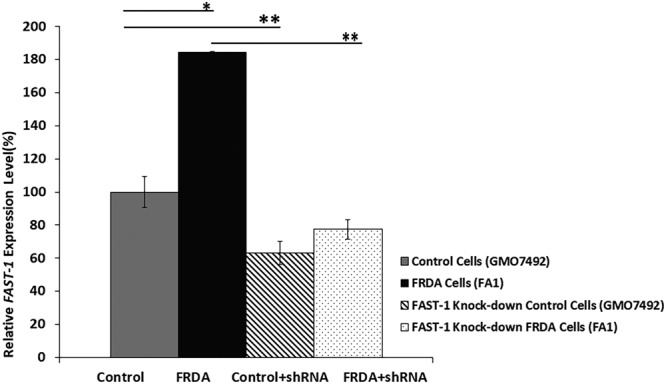
Quantitative RT–PCR analysis of FAST-1 expression levels in FRDA fibroblasts and FAST-1 knockdown fibroblast cells. qRT-PCR showed approximately twice as much FAST-1 expression in FRDA cell lines versus non-FRDA controls. The efficacy of shRNA knockdown was assessed by qRT-PCR, demonstrating a significant reduction in the levels of FAST-1 in both control fibroblast and FAST-1 knockdown FRDA fibroblast cells. (n = 3). *p < 0.05, **p < 0.01, Bars represent SEMs.
Figure 8.
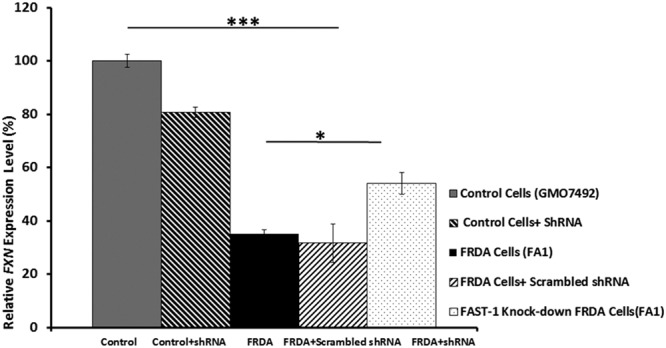
Quantitative RT–PCR analysis of FXN expression levels in control and FAST-1 knockdown cell lines. Knocking down FAST-1 in FRDA fibroblast cells increased FXN expression, but not to the level of control cells; however it did not significantly affect FXN expression in control cells. FXN expression in FRDA cells treated with scrambled shRNA remained unchanged. (n = 3). *p < 0.05, ***p < 0.001, Bars represent SEMs.
Restoring aconitase activity following knockdown of FAST-1
As a biomarker of frataxin function within cells, aconitase activity was measured in FAST-1 knockdown fibroblast cells 1 month after treatment with LV FAST-1 using the Aconitase Assay Kit (Cayman). Normal FRDA fibroblast cells (GMO7492) and FRDA cells treated with scrambled shRNA were used as the controls. All data were calibrated to the mean of the aconitase activity in normal fibroblast cells, which was set to 100%. The aconitase activity in FRDA cells was determined to be 49% (P < 0.05) compared to normal control fibroblast cell levels. However, after knocking down FAST-1 in FRDA fibroblast cells, aconitase activity significantly increased to a value of 195% (P < 0.01). No significant difference was detected in FRDA fibroblast cells treated with scrambled shRNA (Fig. 9).
Figure 9.
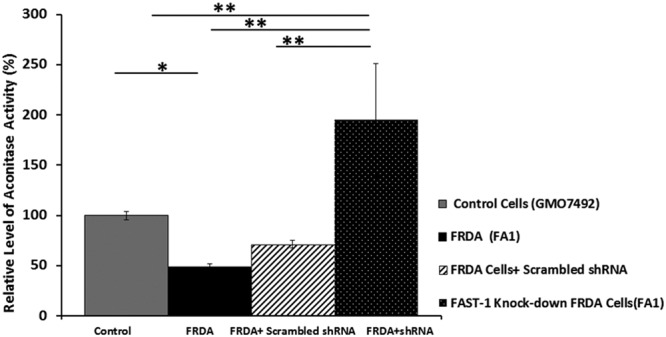
Aconitase activity levels in control and FAST-1 knockdown cell lines. The experiment was performed in triplicate with values being calculated relative to citrate synthase activity. Aconitase activity in normal fibroblast cells was set at 100% all other samples were measured relative to these normal fibroblast cells. *p < 0.05, **p < 0.01, Bars represent SEMs.
Discussion
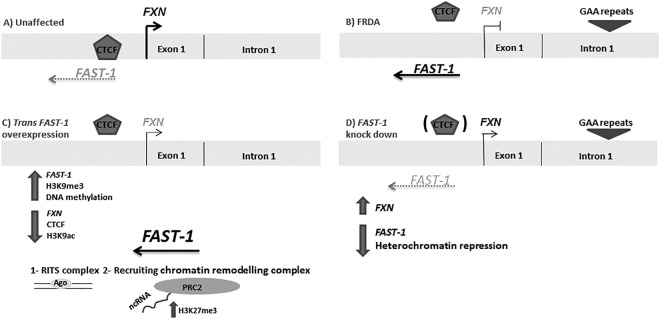
Although it is more than 20 years since the identification of the genetic mutation underlying FRDA, the exact mechanisms by which GAA repeat expansions lead to FXN gene silencing are still not fully understood. However, several potential mechanisms have been proposed based on experimental evidence. For example, it has been suggested that abnormal triplex conformation, such as sticky DNA adopted by GAA repeat expansion and/or RNA•DNA hybrids, might impede the transcription of the FXN gene24,25. In addition, several previous studies of FRDA mouse models, human tissues and cells have highlighted the possibility of epigenetic changes being also either directly or indirectly involved in FXN gene silencing12. Significant enrichment of the repressive histone modifications at the 5′UTR of the FXN gene, together with elevated levels of the non-coding RNA (FAST-1) and hypermethylation of CpG regions upstream of the GAA repeat are the epigenetic changes that have been identified in FRDA10,11,26. Enrichment of these repressive chromatin marks in the FRDA alleles provides evidence of the heterochromatin formation in the promoter region, involving the critical +1 nucleosome and/or the first intron of the FXN gene. Bidichandani and colleagues showed that in FRDA patient-derived fibroblasts, the FXN Antisense Transcript (FAST-1) was expressed at higher levels. Previous in vitro studies in our lab also detected high levels of FAST-1 in the YG8R and YG22R-derived fibroblast cells27. In addition, it has been reported that higher levels of FAST-1 were associated with the severe CTCF depletion and coincidentally heterochromatin formation in the 5′UTR of the FXN gene14. It is important to note that FRDA cells do not have a generalized defect in CTCF binding14. Our results from overexpressing FAST-1 in three different non-FRDA cell lines now demonstrate that FAST-1 can act in trans in a similar manner to the cis-acting FAST-1 overexpression that has previously been identified in FRDA fibroblasts. This mechanism of action, where an antisense RNA acts both in cis and trans and induces epigenetic silencing of its partner sense gene, has been reported for the p15 and huntingtin (HTT) naturally occurring antisense transcripts21,28. We also observed that FAST-1 overexpressing HeLa cells have a reduced occupancy of CTCF, a decreased level of H3K9ac and increased levels of H3K9me3 at the 5′UTR of the FXN gene. This is interesting, because the start of FAST-1 transcription overlaps with the CTCF binding site in the 5′UTR of the FXN gene. The effects of FAST-1 overexpression on CTCF, H3K9ac and H3K9me3 at the FXN 5′UTR locus indicated a potential causative role for FAST-1 in FRDA.
Thus, it is plausible that FAST-1 overexpression may displace CTCF from the 5′UTR of the FXN gene, which would result in secondary changes in chromatin structure that are incompatible with CTCF binding to DNA (Fig. 10)29,30. It is well established that it is the intrinsic property of the expanded GAA repeat that causes heterochromatin formation and exert its gene silencing effect9. It is not clear yet how GAA repeat expansion results in CTCF dislodgement. The expanded GAA repeat drives chromatin changes in intron 1 and this repeat-proximal heterochromatin spreads from the expanded GAA in intron 1 to the upstream regions of the FXN gene encompassing the FXN promoter and CTCF binding site6,10. One possibility is that the heterochromatin emanating from the expanded GAA repeat displaces CTCF from its binding site. Considering the reduced occupancy of CTCF, H3K9me3 enrichment and decreased H3K9ac levels, all hallmarks of heterochromatin formation, at the 5′UTR of the FXN gene in FAST-1 overexpressing HeLa cells, it can be speculated that FAST-1 RNA, via subsequent heterochromatin, may indirectly displace CTCF. Indeed, several studies have reported that association of H3K9me3 with loss of CTCF binding is a common event in epigenetic silencing of cancer-related genes such as p16 and p5331,32. When the chromosomal boundaries are destabilised through dissociation of CTCF and long-range epigenetic organisations are lost, repressive chromatin can spread passively into the promoter region of the FXN gene and inhibit its transcription14,31. In addition, given the enrichment of CTCF at the border of lamina-associated domains (LADs), loss of CTCF can affect the position of genomic loci relative to the nuclear lamina (NL), a location with a generally repressive environment33,34. With this in mind, it has been reported that the majority of expanded FXN alleles are positioned at the NL. Thus, CTCF loss and subsequent heterochromatin formation may contribute to FXN relocation to the NL and its repression35. Recent studies suggest that CTCF has a contradictory function to its classical enhancer-blocker function. It has been proposed that CTCF can tether distant chromatin sites together, create a loop and physically bridge enhancer-promoter interaction36,37. In addition, it has been demonstrated that gene looping plays an important role in restricting divergent transcription of non-coding RNA. It can be speculated that CTCF may facilitate enhancer-promoter interactions at the 5′UTR of the FXN gene to drive the FXN expression. FAST-1 overexpression and resultant local depletion of CTCF could abolish this CTCF-mediated enhancer-promoter interaction and consequently reduce FXN gene expression. Non-coding RNAs have also been found to interact with transcription factors and regulate local gene expression. Two transcription factors, SRF and TFAP2, have been found to regulate expression of the FXN gene. They bind directly to a region that is located close to the start of FAST-1 antisense transcript38. Therefore, it is possible that FAST-1 acts as a decoy, reducing the availability of these transcription factors required for the FXN gene transcription.
Figure 10.
Role of FAST-1 in FRDA. (A) In unaffected individuals, the FXN expression is normal, FAST-1 expression level is low and CTCF is bound to its binding site at 5′UTR of the FXN gene. (B) In FRDA patients, FAST-1 expression is elevated and CTCF is depleted from the 5′UTR. (C) Following FAST-1 overexpression, the FXN gene and protein expression were decreased. FAST-1 overexpressing HeLa cells showed reduced occupancy of CTCF, a decreased level of H3K9ac and an increased level of H3K9me3- a hallmark of silenced chromatin- at the 5′UTR of the FXN gene. It is plausible to speculate that FAST-1 overexpression through yet unknown mechanisms (RITS, PRC2) triggers heterochromatin formation and subsequently displaces CTCF from 5′UTR of the FXN gene. (D) Knocking down FAST-1 in FRDA fibroblast cells resulted in a 1.5-fold increase in the FXN gene expression. Knocking down FAST-1 may possibly reduce the heterochromatin repression leading to increase in frataxin expression.
Antisense transcription is a very common phenomenon in mammalian transcriptomes. Although NATs can show mRNA-like characteristics such as having a poly A tail and a 5′ cap structure, the function of these transcripts is not well understood39. Experimental evidence suggests that non-coding RNA can influence gene expression by recruiting chromatin remodelling complexes to specific alleles and mediating heterochromatin formation and gene silencing. It has been shown that the polycomb repressive complex 2 (PRC2) associates with almost 3000 NATs in mouse embryonic stem cells40,41. Indeed, antisense transcripts can act as scaffolds or directly recruit PRC2 to chromatin in cis and trans manners to induce trimethylation of H3K27 and consequently establish a silent chromatin state. According to this mechanism, even a relatively low abundance of antisense transcript can mediate transcriptional repression42. PRC2 has also been reported to bind to ncRNA molecules in a size-dependent manner, predominantly around TSSs to maintain the repressed chromatin state43,44. In addition, recent studies suggest that there is a cooperative mechanism between H3K27 and H3K9 methylation marks, where H3K9me3 can crosstalk with the Polycomb H3K27me3 modification to cooperate in gene silencing. Indeed, H3K27me3-bound PRC2 stabilizes H3K9me3-anchored HP1 and reinforces heterochromatin formation and ensuing gene repression. Significant enrichment of H3K9 and H3K27 methylation and HP1 has been reported at the FXN locus of FRDA-derived fibroblast cells. Here in FAST-1 overexpressing HeLa cells, we also identified significant enrichment of H3K9me3 at the 5′UTR of the FXN gene. Moreover, knowing that genomic repeats are targets of Polycomb complexes and that repeat sequences might provide a binding platform for polycomb group proteins, it is conceivable to speculate that the expanded GAA repeat and high levels of FAST-1 either directly or indirectly recruit PRC2, and through a cooperative mechanism dictate heterochromatin formation and FXN gene silencing in FRDA19,45. Alternatively, overexpression of FAST-1 could be leading to the generation of endogenous short RNAs (endo-siRNAs) thus leading to promotion of the RNA interference pathway46.
Our results show that knocking down FAST-1 in FRDA fibroblast cells increases FXN gene expression (Fig. 10). Therefore, it can be concluded that, since FAST-1 is associated with epigenetic repression of the FXN gene, inhibition of FAST-1 may be an approach to increase the FXN transcripts and stimulate subsequent protein expression. Indeed, our results demonstrate that knocking down FAST-1 in FRDA results in a significant increase in aconitase enzyme activity, a good indicator of frataxin function within cells. Our data suggest that since FAST-1 is associated with FXN gene silencing, inhibition of FAST-1 may be an approach for FRDA therapy. Considering the nature of NATs and the fact that many currently available drugs would not affect the activity of non-coding RNA molecules, developing new methods to disrupt the function of NATs seems necessary.
Methods
Characterization of antisense FAST-1 transcripts by rapid amplification of cDNA ends (RACE)
RACE experiments were done with DNase I-treated RNA from human FRDA fibroblast cell line (GM04078); 5′- and 3′- RACE were carried out by using the SMARTerTM RACE cDNA amplification kit (Clontech). The gene- specific primers used in each case are:
RACE-FAST-F2 5′-GACCTCCAAGCTTTGCCTCCCTCAAG-3′.
RACE-FAST-R1 5′-GCACCCACTTCCCAGCAAGACAGCAG-3′.
Following the primary FAST-1 RACE PCR, nested PCRs were performed using the NUP primer (nested universal primer A, supplied with the SMARTer RACE cDNA amplification kit) and a gene specific primer:
RACE-N-FAST-R2 5′-GACAGCAGCTCCCAAGTTCCTCCTG-3′ for the 5′nested PCR.
RACE-N-FAST-F1 5′-GACCCAAGGGAGACTGCAGCCTGGTG-3′ for the 3′nested PCR.
The PCR products were gel purified and cloned for the sequencing analysis.
FAST-1 overexpression
Initially, a full-length FAST-1 sequence (523 bp) was cloned into the expression plasmid pcDNA3. One day before transfection, HeLa, HEK293 and fibroblast cells were seeded in 6 well plates and grown in DMEM containing 10% FBS and antibiotics (penicillin/streptomycin). Two micrograms each of pcDNA3 empty vector and pcDNA3-FAST-1 (Addgene) were transfected into cells by using Lipofectamine 3000 (Life Technologies) and Neon™ transfection system. After 48 h, 200, 400 and 100 μg/ml of G418 (Gibco) were added to the medium of HeLa, HEK293 and fibroblast cells, respectively, to select for cells carrying the plasmid. After a further 16 days G418-resistant colonies were picked for individual culture, and were maintained in complete medium (DMEM medium containing 10% fetal calf serum) with appropriate G418 concentration.
Quantitative RT-PCR
RNA was isolated from cells using the Trizol (Invitrogen) method. Following DNase treatment, FAST-1 cDNA was obtained using the Cloned AMV First-Strand cDNA Synthesis Kit (Invitrogen) with the strand specific FAST RT primer (5′-CCAAGCAGCCTCAATTTGTG-3′). For FXN and HPRT quantifications, cDNA was synthesised using the oligo (dT)20 primer.
Levels of FAST-1 and FXN mRNA expression were assessed by quantitative RT–PCR using a QuantStudio 7 Flex Real-Time PCR instrument and SYBR® Green (Applied Biosystems) with the following primers:
N-FAST F2- forward 5′-GACCCAAGGGAGACTGCAG-3′ and
FAST-R1 -reverse 5′-CACTTCCCAGCAAGACAGC-3′
FXN-h–forward 5′-CAGAGGAAACGCTGGACTCT-3′ and
FXN-h-reverse 5′-AGCCAGATTTGCTTGTTTGGC-3′
HPRT-h-forward 5′-GGTGAAAAGGACCCCACGA-3′and
HPRT-h-reverse 5′-TCAAGGGCATATCCTACAACA-3′
The mRNA expression levels of HPRT were also measured in all samples to normalise the gene expression levels to avoid sample-to-sample differences in RNA input, RNA quality and reverse transcription efficiency. Power SYBR® Green Master Mix (Applied Biosystems) was used along with 2 µl of sample cDNA in 10 µl reaction mixture. PCR conditions were set as10 min at 95 °C for enzyme activation followed by 40 two-step cycles (15 sec at 95 °C and 1 min at 60 °C). Reactions were carried out in triplicate for each biological sample and each experiment was repeated at least two times. Values were expressed relative to HPRT and expression levels were calculated by 2−ΔΔCt method and RQ manager software (Applied Biosystems).
Frataxin dipstick assay
Protein concentration was quantified by BCA assay and the levels of frataxin protein were measured by lateral flow immunoassay with the Frataxin Protein Quantity Dipstick Assay Kit (MitoSciences, Eugene, OR, USA) according to the manufacturer’s instructions47. Signal intensity was measured with a Hamamatsu ICA-1000 Immunochromatographic Reader (MitoSciences).
FAST-1 copy number assay
To detect the number of FAST-1 copies in FAST-1 overexpressing cell lines, a custom TaqMan® copy number assay was designed by Life Technologies with a specific TaqMan® FAM™ dye-labeled MGB probe. In brief, 20 ng of genomic DNA was combined with 2 × TaqMan universal master mix, custom designed TaqMan® copy number assay for detecting FAST-1, and TaqMan copy number reference assay for RNase P in a 20 μl reaction volume. The assay was performed using the QuantStudio 7 Flex Real-Time PCR instrument (Applied Biosystems) and the following thermal cycling conditions: 50 °C for 2 minutes, 95 °C for 10 minutes, and 40 cycles of 95 °C for 15 seconds and 60 °C for 1 minute. Samples were assayed using triplicate wells for each gene of interest and copy numbers were estimated by relative quantitation (RQ) normalised to the known copy number of the reference sequence using the comparative Ct (ΔΔCt) method. The Ct data were subsequently compared to a calibrator sample of non-transfected control HeLa cells containing two copies of the target sequence, analysed by Applied Biosystems CopyCaller Software (v.2.0; Applied Biosystems) according to the product instruction.
Chromatin immunoprecipitation-qPCR assay
Histone modifications and CTCF occupancy levels at the 5′UTR of FXN gene were detected by ChIP analysis of FAST-1 overexpressing HeLa cells. This procedure was performed by using ChIP qPCR kit (Chromatrap) with an acetylated H3 (Lys9) (Upstate, 7-352), trimethyl-H3 (Lys9) (Upstate, 07-442) and CTCF (Upstate, 7-729), antibody on formaldehyde cross-linked samples.
DNA was then sheared by sonication, followed by immunoprecipitation. For each experiment, normal rabbit serum (SIGMA) was used as a negative control. After reversal of cross-linking, quantitative RT–PCR amplification of the resultant co-immunoprecipitated DNA was carried out with SYBR® Green in a QuantStudio 7 Flex Real-Time PCR instrument (Applied Biosystems) using the following primers;
FXN-ChIP-forward 5′-TCCTGAGGTCTAACCTCTAGCTGC-3′ and
FXN-ChIP-reverse 5′-CGAGAGTCCACATGCTGCTCC-3′
GAPDH-ChIP-forward 5′-TCGACAGTCAGCCGCATCT-3′ and
GAPDH-ChIP-reverse 5′-CTAGCCTCCCGGGTTTCTCT-3′
FXN-pro-forward 5′- CCCCACATACCCAACTGCTG-3′and
FXN-pro-reverse 5′-GCCCGCCGCTTCTAAAATTC-3′
The data were from three independent chromatin preparations, with each experiment done in triplicate.
MethylScreen assay
DNA methylation analysis was performed using the ‘MethylScreen’ method19, which uses combined restriction digestion of DNA with methylation sensitive and methylation dependent restriction enzymes, MSRE and MDRE respectively. MethylScreen was used to analyse the two CpG sites, CpG6 and CpG1111, at FXN locus upstream of the GAA repeat. 1 µg of genomic DNA was digested with: (1) a MSRE, (2) MDRE, (3) both MSRE and MDRE (double digest, DD), and (4) neither MSRE or MDRE (mock control). The MSREs used for CpGs 6 and 11 were AjiI (Fermentas) and Hpy188III (New England Biolabs), respectively38. The MDRE used for all two CpGs was McrBc (Fermentas). A 50 ng aliquot of digested DNA was then amplified by quantitative PCR using SYBR® Green (Applied Biosystems) and an ABI 7900 Fast Real-Time PCR System (Applied Biosystems) with the following primers: CpG6 F 5′-GAAGATGCCAAGGAAGTGGTAG-3′ and R 5′-GAGCAACACAAATATGGCTTGG-3′; CpG11 F 5′-GATCCGTCTGGGCAAAGGCCAG-3′ and R 5′-ATCCCAAAGTTTCTTCAAACACAATG-3′. PCR quantification was carried out using the ΔCt method (values were calculated as 2ΔCt (mock – digest) with the mock value set at 100%) and RQ Manager Software (Applied Biosystems). Each qRT-PCR reaction was performed in triplicate. MethylScreen DNA methylation values were then calculated as follows: Densely methylated (DM) = (MSRE − DD)/(100 − DD) × 100; unmethylated (UM) = (MDRE − DD)/(100 − DD) × 100; intermediately methylated (IM) = 100–(DM + UM).
Knocking down FAST-1 expression
To knock down FAST-1 expression, Sigma custom cloning MISSION shRNA team designed a short hairpin RNA, cloned it into a pLKO.1-puro-CMV-tGFP vector and provided us with ready to use lentiviral particles for transduction. To achieve a stable FAST-1 knockdown, FRDA and normal fibroblast cell lines were transduced with lentiviral (LV) vectors expressing shRNAs targeting FAST-1 (pLKO.1-puro-CMV-tGFP). Several precautions were taken in order to control off-target effects and specificity, as follows: (i) pLKO.1-puro Non-Target shRNA was used as the negative control; (ii) The normal fibroblast cell line was also transduced with LV FAST-1 to assess the specificity of any FAST-1 knockdown effect on FRDA cells. Briefly, cells were seeded into 6-well plates with culture medium containing 10% FBS until 80% confluence, and the cells were then incubated with FAST-1 and scrambled LV particles (25ul–100ul of 5.7 × 107 to achieve MOI values of 5–20) in serum-free culture medium supplemented with polybrene (8 μg/ml) for 6 h. Then the supernatant was replaced with 1 ml fresh medium and incubated at 37 °C and 5% CO2 for 72 hours to allow time for the expression of the reporter and puromycin resistance genes. After 72 hours, the selective antibiotic, puromycin, was added to the media at a final concentration of 0.2 μg/μl. The selection media was replaced every 3 days and the final cell harvest for RNA analysis was performed at 2 weeks post-transduction.
Aconitase assay
Aconitase activities were determined using the Aconitase Assay Kit (Cayman Chemical Company, 705502). To perform the assays, cultured cells were washed with cold PBS, scraped off the flask and pelleted by centrifugation at 800 g for 10 min. Cell pellets were suspended in cell lytic buffer (Cayman) for 15 min at 4 °C, then centrifuged at 10,000 g for 10 min, followed by BCA protein concentration determination and dilution of cell protein lysates to 0.5ug/µl in 1x assay buffer (Cayman). 50 µl of 0.5 ug/µl cell protein lysates were added to 200 µl of substrate mix (50 mM Tris/HCl pH 7.4, 0.4 mM NADP, 5 mM Na citrate, 0.6 mM MgCl2, 0.1% (v/v) Triton X-100 and 1 U isocitrate dehydrogenase) and the reactions were incubated at 37 °C for 15 min, followed by spectrophotometric 340 nm absorbance measurements every minute for 15 min at 37 °C to determine the reaction slope. Aconitase activities of samples were then normalized to citrate synthase activities, which were determined using a citrate synthase assay kit (Sigma, CS0720).
Statistical analysis
Statistical analyses that compared two groups of data were performed using the Student’s t test, with a P value of <0.05 taken to indicate a statistically significant difference. Correlation and regression analyses of FAST-1 copy number, FAST-1 expression and FXN expression were performed using Microsoft Excel data analysis tools.
Acknowledgements
We would like to thank Sanjay Bidichandani (University of Oklahoma College of Medicine) for his most useful comments on this work. This work was partially supported by funding from the European Union Seventh Framework Programme [FP7/2007–2013] under grant agreement number 242193/EFACTS to MAP. AB was the recipient of a Research Fellowship from Ataxia UK.
Author Contributions
H.M., M.S., A.B. and S.A. performed experiments; H.M., M.S., A.B., S.A. and M.A.P. conceived and designed the study; H.M., S.A. and M.A.P. wrote the manuscript; all authors read and approved the manuscript.
Availability of Data and Material
All data generated or analysed during this study are included in this published article.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Pandolfo M. Friedreich ataxia. Arch. Neurol. 2008;65:1296–1303. doi: 10.1001/archneur.65.10.1296. [DOI] [PubMed] [Google Scholar]
- 2.Campuzano V, et al. Friedreich’s Ataxia: Autosomal Recessive Disease Caused by an Intronic GAA Triplet Repeat Expansion. Science. 1996;271:1423–1427. doi: 10.1126/science.271.5254.1423. [DOI] [PubMed] [Google Scholar]
- 3.Dürr A, et al. Clinical and genetic abnormalities in patients with Friedreich’s ataxia. N. Engl. J. Med.. 1996;335:1169–1175. doi: 10.1056/NEJM199610173351601. [DOI] [PubMed] [Google Scholar]
- 4.Pandolfo M. Iron metabolism and mitochondrial abnormalities in Friedreich ataxia. Blood Cells Mol. Dis. 2002;29:536–547. doi: 10.1006/bcmd.2002.0591. [DOI] [PubMed] [Google Scholar]
- 5.Campuzano V, et al. Frataxin is reduced in Friedreich ataxia patients and is associated with mitochondrial membranes. Hum. Mol. Gen. 1997;6:1771–1780. doi: 10.1093/hmg/6.11.1771. [DOI] [PubMed] [Google Scholar]
- 6.Chutake YK, et al. Altered nucleosome positioning at the transcription start site and deficient transcriptional initiation in Friedreich ataxia. J. Biol. Chem. 2014;289:15194–15202. doi: 10.1074/jbc.M114.566414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rotig A, et al. Aconitase and mitochondrial iron-sulphur protein deficiency in Friedreich ataxia. Nat. Genet. 1997;17:215–217. doi: 10.1038/ng1097-215. [DOI] [PubMed] [Google Scholar]
- 8.Colin F, et al. Mammalian frataxin controls sulfur production and iron entry during de novo Fe4S4 cluster assembly. J. Am. Chem. Soc. 2013;135:733–740. doi: 10.1021/ja308736e. [DOI] [PubMed] [Google Scholar]
- 9.Saveliev A, et al. DNA triplet repeats mediate heterochromatin-protein-1-sensitive variegated gene silencing. Nature. 2003;422:909–913. doi: 10.1038/nature01596. [DOI] [PubMed] [Google Scholar]
- 10.Herman D, et al. Histone deacetylase inhibitors reverse gene silencing in Friedreich’s ataxia. Nat. Chem. Biol. 2006;2:551–558. doi: 10.1038/nchembio815. [DOI] [PubMed] [Google Scholar]
- 11.Greene E, et al. Repeat-induced epigenetic changes in intron 1 of the frataxin gene and its consequences in Friedreich ataxia. Nucleic Acids Res. 2007;35:3383–3390. doi: 10.1093/nar/gkm271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Al-Mahdawi S, et al. The Friedreich ataxia GAA repeat expansion mutation induces comparable epigenetic changes in human and transgenic mouse brain and heart tissues. Hum. Mol. Gen. 2008;17:735–746. doi: 10.1093/hmg/ddm346. [DOI] [PubMed] [Google Scholar]
- 13.Soragni E, et al. Long intronic GAA•TTC repeats induce epigenetic changes and reporter gene silencing in a molecular model of Friedreich ataxia. Nucleic Acids Res. 2008;36:6056–6065. doi: 10.1093/nar/gkn604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Biase I, et al. Epigenetic silencing in Friedreich ataxia is associated with depletion of CTCF (CCCTC-binding factor) and antisense transcription. PLoS One. 2009;4:7914. doi: 10.1371/journal.pone.0007914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Castaldo I, et al. DNA methylation in intron 1 of the frataxin gene is related to GAA repeat length and age of onset in Friedreich ataxia patients. J. Med. Genet. 2008;45:808–812. doi: 10.1136/jmg.2008.058594. [DOI] [PubMed] [Google Scholar]
- 16.Evans‐Galea MV, et al. FXN methylation predicts expression and clinical outcome in Friedreich ataxia. Ann. Neurol. 2012;71:487–497. doi: 10.1002/ana.22671. [DOI] [PubMed] [Google Scholar]
- 17.Kumari D, Biacsi RE, Usdin K. Repeat expansion affects both transcription initiation and elongation in friedreich ataxia cells. J. Biol. Chem. 2011;286:4209–4215. doi: 10.1074/jbc.M110.194035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rai M, et al. HDAC inhibitors correct frataxin deficiency in a Friedreich ataxia mouse model. PloS one. 2008;3:e1958. doi: 10.1371/journal.pone.0001958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Al-Mahdawi S, et al. Friedreich ataxia patient tissues exhibit increased 5-hydroxymethylcytosine modification and decreased CTCF binding at the FXN locus. PLoS One. 2013;8:74956. doi: 10.1371/journal.pone.0074956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohhata T, et al. Crucial role of antisense transcription across the Xist promoter in Tsix-mediated Xist chromatin modification. Development. 2008;135:227–235. doi: 10.1242/dev.008490. [DOI] [PubMed] [Google Scholar]
- 21.Yu Wenqiang, Gius David, Onyango Patrick, Muldoon-Jacobs Kristi, Karp Judith, Feinberg Andrew P., Cui Hengmi. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451(7175):202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khorkova O, et al. Natural antisense transcripts. Hum. Mol. Gen. 2014;23:R54–R63. doi: 10.1093/hmg/ddu207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pelechano V, Steinmetz LM. Gene regulation by antisense transcription. Nat. Rev. Genet. 2013;14:880–893. doi: 10.1038/nrg3594. [DOI] [PubMed] [Google Scholar]
- 24.Sakamoto N, et al. GGA· TCC-interrupted triplets in long GAA· TTC repeats inhibit the formation of triplex and sticky DNA structures, alleviate transcription inhibition, and reduce genetic instabilities. J. Biol. Chem. 2001;276:27178–27187. doi: 10.1074/jbc.M101852200. [DOI] [PubMed] [Google Scholar]
- 25.Grabczyk E, Mancuso M, Sammarco MC. A persistent RNA· DNA hybrid formed by transcription of the Friedreich ataxia triplet repeat in live bacteria, and by T7 RNAP in vitro. Nucleic Acids Res. 2007;35:5351–5359. doi: 10.1093/nar/gkm589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim E, Napierala M, Dent SY. Hyperexpansion of GAA repeats affects post-initiation steps of FXN transcription in Friedreich’s ataxia. Nucleic Acids Res. 2011;39:8366–8377. doi: 10.1093/nar/gkr542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sandi, M. Identification and quantification of FXN antisense transcript 1 (FAST-1) in friedreich ataxia. Brunel University Research Archive (BURA) PhD thesis at, https://bura.brunel.ac.uk/bitstream/2438/12464/1/FulltextThesis.pdf (2015).
- 28.Chung DW, et al. A natural antisense transcript at the Huntington’s disease repeat locus regulates HTT expression. Hum. Mol. Gen. 2011;20:3467–3477. doi: 10.1093/hmg/ddr263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lefevre P, et al. The LPS-induced transcriptional upregulation of the chicken lysozyme locus involves CTCF eviction and noncoding RNA transcription. Mol. Cell. 2008;32:129–139. doi: 10.1016/j.molcel.2008.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Witcher M, Emerson BM. Epigenetic silencing of the p16INK4a tumor suppressor is associated with loss of CTCF binding and a chromatin boundary. Mol. Cell. 2009;34:271–284. doi: 10.1016/j.molcel.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soto-Reyes E, Recillas-Targa F. Epigenetic regulation of the human p53 gene promoter by the CTCF transcription factor in transformed cell lines. Oncogene. 2010;29:2217–2227. doi: 10.1038/onc.2009.509. [DOI] [PubMed] [Google Scholar]
- 33.Guelen L, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 34.Yáñez-Cuna JO, van Steensel B. Genome–nuclear lamina interactions: from cell populations to single cells. Curr. Opin. Genetics Dev. 2017;43:67–72. doi: 10.1016/j.gde.2016.12.005. [DOI] [PubMed] [Google Scholar]
- 35.Silva AM, et al. Expanded GAA repeats impair FXN gene expression and reposition the FXN locus to the nuclear lamina in single cells. Hum. Mol. Gen. 2015;24:3457–3471. doi: 10.1093/hmg/ddv096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oti M, et al. CTCF-mediated chromatin loops enclose inducible gene regulatory domains. BMC Genomics. 2016;17:252. doi: 10.1186/s12864-016-2516-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu Z, Tee W-W. Enhancers and chromatin structures: regulatory hubs in gene expression and diseases. Biosci. Rep. 2017;37:13. doi: 10.1042/BSR20160183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li K, et al. Expression of human frataxin is regulated by transcription factors SRF and TFAP2. PLoS One. 2010;5:12286. doi: 10.1371/journal.pone.0012286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Villegas VE, Zaphiropoulos PG. Neighboring gene regulation by antisense long non-coding RNAs. Int. J. Mol. Sci. 2015;16:3251–3266. doi: 10.3390/ijms16023251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao J, et al. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol. Cell. 2010;40:939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Halley P, Khorkova O, Wahlestedt C. Natural antisense transcripts as therapeutic targets. Drug Discov. Today. 2013;10:119. doi: 10.1016/j.ddmec.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holoch D, Moazed D. RNA-mediated epigenetic regulation of gene expression. Nat. Rev. Genet. 2015;16:71–84. doi: 10.1038/nrg3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davidovich C, et al. Promiscuous RNA binding by Polycomb repressive complex 2. Nat. Struct. Mol. Biol. 2013;20:1250. doi: 10.1038/nsmb.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Kruijsbergen I, Hontelez S, Veenstra GJC. Recruiting polycomb to chromatin. Int. J. Biochem. Cell Biol. 2015;67:177–187. doi: 10.1016/j.biocel.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leeb M, et al. Polycomb complexes act redundantly to repress genomic repeats and genes. Genes Dev. 2010;24:265–276. doi: 10.1101/gad.544410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Werner A, et al. Contribution of natural antisense transcription to an endogenous siRNA signature in human cells. BMC Genomics. 2014;15:19. doi: 10.1186/1471-2164-15-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Willis JH, et al. Lateral-flow immunoassay for the frataxin protein in Friedreich’s ataxia patients and carriers. Mol. Gen. Metab. 2008;94:491–497. doi: 10.1016/j.ymgme.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analysed during this study are included in this published article.