Abstract
Aims: Rheumatoid arthritis (RA) is characterized by cartilage and bone damage leading to disability. Here, the association between microRNA (miRNA) polymorphisms and susceptibility to RA was evaluated by performing an updated meta-analysis and systematic review.
Main methods: An electronic search of databases including PubMed and Embase was performed from inception to December 8, 2017 to retrieve studies investigating the association between miRNA polymorphisms and RA risk. Two reviewers independently screened literature according to the inclusion and exclusion criteria and extracted data. The meta-analysis was conducted using Stata 14.0 software.
Key findings: Thirteen case-control studies with 2660 cases and 4098 controls were screened out after a systematic search. One study from the miR-146a rs2910164 G > C polymorphism group and two from the miR-499 rs3746444 T > C polymorphism group were excluded because of deviations from Hardy-Weinberg equilibrium. Pooled analysis demonstrated that miR-146a rs2910164 G > C polymorphism was not significantly associated with susceptibility to RA. However, a significant association was observed between miR-499 rs3746444 T > C polymorphism and RA risk (C vs. T: OR = 1.22, 95% CI = 1.05–1.42, P = 0.008; TC vs. TT: OR = 1.26, 95% CI = 1.05–1.50, P = 0.011; TC/CC vs. TT: OR = 1.26, 95% CI = 1.07–1.5, P = 0.007). Subgroup analysis based on ethnicity showed no significant association between miR-499 T > C polymorphism and susceptibility to RA in the Asian population (P > 0.05). However, in Caucasian population, the C allele in the miR-499 T > C polymorphism was a contributor to RA susceptibility in some genetic models (C vs. T: OR = 1.64, 95% CI = 1.28–2.11, P < 0.001; TC vs. TT: OR = 1.95, 95% CI = 1.40–2.71, P < 0.001; TC/CC vs. TT: OR = 1.96, 95% CI = 1.43–2.69, P < 0.001).
Significance: The miR-146a rs2910164 G > C polymorphism was not associated with susceptibility to RA. In the Caucasian population, the C allele in the miR-499 T > C polymorphism contributed to RA susceptibility.
Keywords: microRNA, single nucleotide polymorphism, rheumatoid arthritis, meta-analysis, systematic review
Introduction
Rheumatoid arthritis (RA), one of the most prevalent chronic inflammatory diseases, is characterized by damage to cartilage and bone leading to disability (Smolen et al., 2016). It is a substantial burden for individuals and society, with up to 0.5% of the global population affected (Neogi et al., 2010), and the identification of novel therapeutic strategies to control inflammation and reduce severe consequences is increasingly imperative. The pathogenesis of RA is closely associated with genetic and environmental factors, with the major histocompatibility complex (MHC) region at 6p21 contributing to 30–50% of genetic liability to RA (Yamamoto et al., 2015; Okada et al., 2016; Terao et al., 2016). Cigarette smoking also plays an important role in RA risk (Sokolove et al., 2016).
Rheumatoid arthritis risk was recently shown to be associated with epigenetic regulation including DNA methylation, histone modification, and transcriptional/post-transcriptional regulation (Klein and Gay, 2015). MicroRNAs (miRNAs), which are the most widely studied class of non-coding RNAs, function in the post-transcriptional regulation of gene expression by controlling the translation of mRNA into proteins (He and Hannon, 2004). Moreover, miRNAs play a significant role in regulating biological processes, including proliferation, apoptosis, and development (Esteller, 2011).
A growing body of evidence indicates that single nucleotide polymorphisms (SNPs) in miRNAs may affect RA susceptibility because certain SNPs located in miRNA encoding genes can alter miRNA expression, pri-miRNA and pre-miRNA processing, or miRNA–mRNA interactions (Furer et al., 2010; Dai and Ahmed, 2011).
Ciccacci et al. (2016) reported that the C allele of rs2910164 in miR-146a has a protective role in the pathogenesis of RA. However, Piatek et al. (2016) reported no significant association between miR-146a polymorphisms and RA susceptibility or progression. With regard to miR-499, the effect of C allele of rs3746444 on RA risk is also controversial (El-Shal et al., 2013; Zhang et al., 2013). The association between miRNA polymorphisms and susceptibility to RA remain unclear. Because of increasing number of case-control studies published, an updated meta-analysis is required to evaluate the relationship between miRNA polymorphisms and RA, including subgroup analyses investigating ethnicity-specific effects.
Materials and Methods
Literature Search Strategy
A comprehensive search of the PubMed and Embase databases was performed to retrieve studies investigating the association between miRNA polymorphisms and RA susceptibility published in English up to December 8, 2017. The search terms were as follows: “rheumatoid arthritis” “arthritis” “microRNA” “miRNAs” “polymorphism” and “polymorphisms.” Relevant journals and the references of the included studies were also reviewed.
Inclusion and Exclusion Criteria
The inclusion criteria of our meta-analysis were as follows: (1) case-control studies; (2) investigation of miRNA polymorphisms and RA susceptibility; (3) detailed numbers of alleles and genotypes between cases and controls; (4) full text published in English; (5) control group consistent with Hardy-Weinberg Equilibrium (HWE); and (6) any specific miRNA with at least two case-control studies to evaluate the association between single nuclear polymorphism and RA susceptibility.
The exclusion criteria were as follows: (1) reviews or case reports without controls; (2) no available data reported; and (3) duplicated reports.
Data Extraction
Two investigators (Zhou and Jiang) screened the literature and extracted data independently according to the inclusion and exclusion criteria. Discrepancies and differences were resolved by a third investigator (Zhu) until consensus was reached. The following data were retrieved: author list, year of publication, ethnicity, sample size, alleles, genotype of each gene polymorphism, and HWE.
Data Synthesis and Statistical Analysis
The odds ratios (ORs) with their corresponding 95% confidence intervals (CIs) were used to assess the strength of the association between polymorphisms in miRNAs and RA. For miR-146a rs2910164 G > C, the dominant (GC + CC vs. GG), recessive (CC vs. GC + GG), heterozygous (GC vs. GG), homozygous (CC vs. GG), and allelic (C vs. G) genetic models were used to calculate pooled ORs. These models were also used to evaluate the miR-499 rs3746444 T > C polymorphism, including the dominant (TC + CC vs. TT), recessive (CC vs. TC + TT), heterozygous (TC vs. TT), homozygous (CC vs. TT), and allelic (C vs. T) models.
The Chi square-based Q-test was used to assess the between-study heterogeneity and a P-value of 0.1 was considered to indicate significant heterogeneity among studies (Lau et al., 1997). The I2 metric value was also used to describe the proportion of total variation in study estimates due to heterogeneity, with values of 25, 50, and 75% considered as evidence of low, moderate, and high heterogeneity, respectively (Higgins and Thompson, 2002). The pooled OR estimation of each study was calculated by the fixed-effects model (the Mantel–Haenszel method) when the P-value was >0.1 and I2 was <50% (Mantel and Haenszel, 1959). Otherwise, the random-effects model (the DerSimonian and Laird method) was used (DerSimonian and Laird, 1986). Subgroup analyses according to ethnicity were also performed to calculate ethnic-specific ORs. The HWE was tested for each study by comparing the observed and expected genotype frequencies of the control group (Chi-square test). Publication bias was evaluated with Egger’s linear regression and Begg’s funnel plots (Hayashino et al., 2005). Statistical analyses were performed using the Stata software (version 14.1; Stata Corp., College Station, TX, United States) with a two-sided P-value. P < 0.05 was considered significant.
Results
Study Characteristics
The results of the systematic search and selection performed according to the inclusion and exclusion criteria are shown in Figure 1. A total of 13 articles with 2660 cases and 4098 controls were finally included in the meta-analysis (Chatzikyriakidou et al., 2010; Yang et al., 2011, 2016; Jimenez-Morales et al., 2012; El-Shal et al., 2013; Hashemi et al., 2013; Zhang et al., 2013; Zhou X. et al., 2015; Bogunia-Kubik et al., 2016; Ciccacci et al., 2016; Toraih et al., 2016; Fattah et al., 2017; Hassine et al., 2017). The 13 articles comprised 16 studies, including 9 studies on miR-146a rs2910164 and 7 studies on miR-499 rs3746444. Table 1 shows the characteristics of the retrieved studies. Various genotyping methods were used, including PCR-RFLP, TaqMan, and PCR. The genotype distribution of the controls in all studies was consistent with HWE, except one study from the miR-146a polymorphism group (El-Shal et al., 2013) and two from the miR-499 polymorphism group (Hashemi et al., 2013; Toraih et al., 2016).
FIGURE 1.
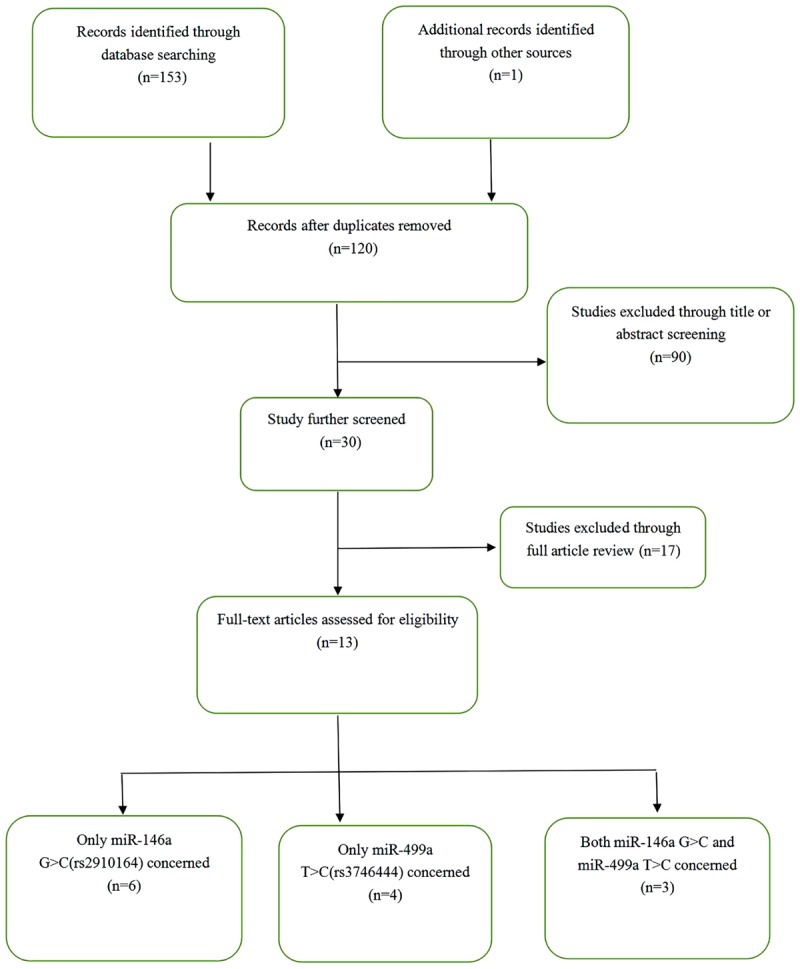
Flow chart showing the detailed steps for literature selection.
Table 1.
The basic characteristics of the retrieved studies.
| First author | Year | Country | Ethnicity | Genotyping method | Sample size |
Genotypes (case/control) |
HWE (P) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | Case | Control | ||||||||||
| GG | GC | CC | GG | GC | CC | ||||||||
| miR-146a G > C | |||||||||||||
| Hasssine | 2017 | Tunisia | Caucasian | PCR-RFLP | 165 | 150 | 94 | 63 | 8 | 68 | 69 | 13 | 0.441 |
| Ciccacci | 2016 | Italia | Caucasian | TaqMan PCR | 192 | 298 | 109 | 69 | 14 | 158 | 117 | 23 | 0.836 |
| Bogunia-Kubik | 2016 | Poland | Caucasian | PCR–RFLP | 111 | 130 | 72 | 32 | 7 | 88 | 36 | 6 | 0.361 |
| Zhou | 2015 | China | Asian | PCR | 598 | 821 | 114 | 283 | 201 | 151 | 385 | 285 | 0.296 |
| El-Shal | 2013 | Egypt | Caucasian | PCR-RFLP | 217 | 245 | 30 | 103 | 84 | 15 | 119 | 111 | 0.021 |
| Hashemi | 2013 | Iran | Caucasian | T-ARMS-PCR | 104 | 110 | 57 | 39 | 8 | 64 | 37 | 9 | 0.280 |
| Jimenez-Morales | 2012 | China | Asian | TaqMan PCR | 210 | 531 | 102 | 80 | 28 | 236 | 229 | 66 | 0.369 |
| Yang | 2011 | China | Asian | PCR-RFLP | 208 | 240 | 28 | 95 | 85 | 30 | 116 | 94 | 0.529 |
| Chatzikyriakidou | 2010 | Greece | Caucasian | PCR-SSCP | 136 | 147 | 73 | 53 | 10 | 80 | 53 | 14 | 0.240 |
| TT | CT | CC | TT | CT | CC | ||||||||
| miR-499a T > C | |||||||||||||
| Fattah | 2017 | Egypt | Caucasian | PCR-RFLP | 100 | 100 | 33 | 53 | 14 | 49 | 41 | 10 | 0.742 |
| Yang | 2016 | China | Asian | TaqMan PCR | 386 | 576 | 282 | 99 | 5 | 443 | 125 | 8 | 0.807 |
| Toraih | 2016 | Egypt | Caucasian | TaqMan PCR | 95 | 200 | 50 | 15 | 30 | 82 | 68 | 50 | <0.001 |
| El-Shal | 2013 | Egypt | Caucasian | PCR-RFLP | 217 | 245 | 113 | 93 | 11 | 167 | 70 | 8 | 0.841 |
| Zhang | 2013 | China | Asian | MALDI-TOF MS | 206 | 466 | 159 | 44 | 3 | 346 | 110 | 10 | 0.719 |
| Hashemi | 2013 | Iran | Caucasian | T-ARMS-PCR | 104 | 110 | 46 | 32 | 26 | 74 | 25 | 11 | <0.001 |
| Yang | 2011 | China | Asian | PCR-RFLP | 208 | 240 | 159 | 42 | 7 | 182 | 53 | 5 | 0.624 |
Meta-Analysis
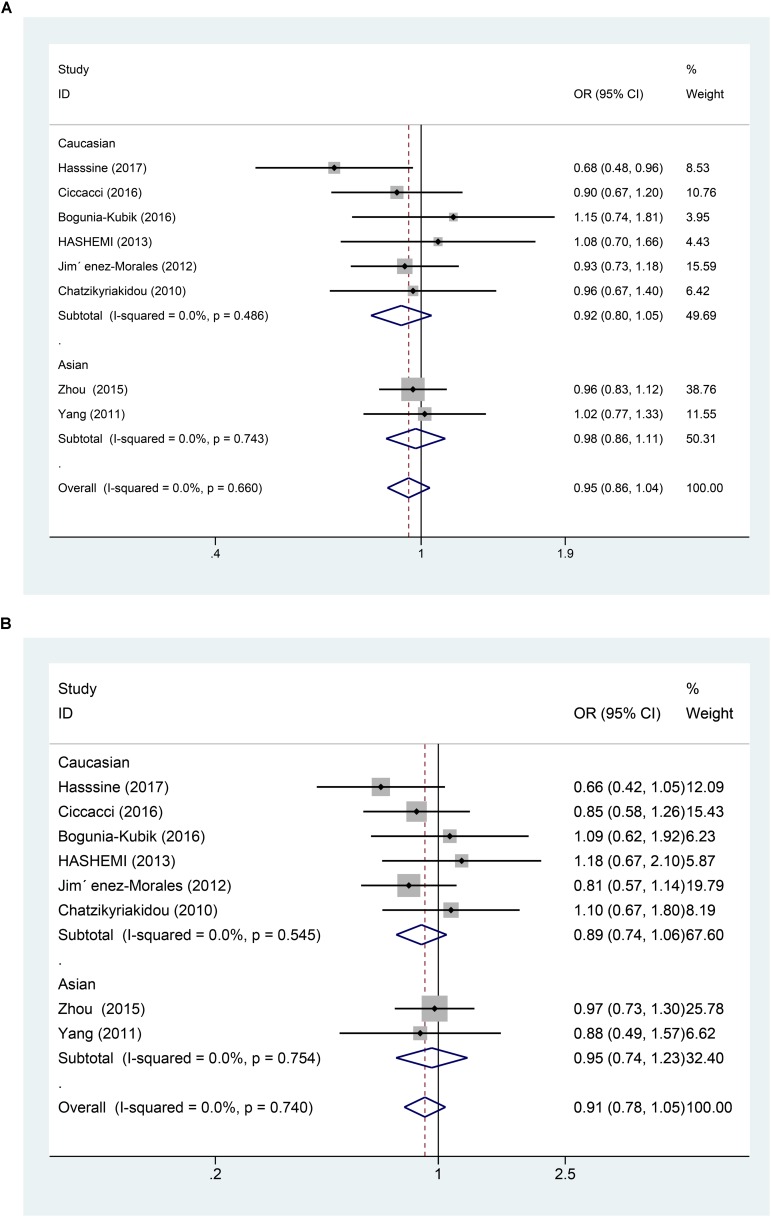
In all the populations, the miR-146a rs2910164 G > C polymorphism was not significantly associated with increased susceptibility to RA (C vs. G: OR = 0.95, 95% CI = 0.86–1.04, Ph = 0.66; CC vs. GG: OR = 0.91, 95% CI = 0.75–1.12, Ph = 0.87; GC vs. GG: OR = 0.91, 95% CI = 0.78–1.05, Ph = 0.74; GC/CC vs. GG: OR = 0.91, 95% CI = 0.79–1.04, Ph = 0.69; CC vs. GC/GG: OR = 0.97, 95% CI = 0.82–1.14, Ph = 0.896). Subgroup analysis based on ethnicity showed no significant association between the miR-146a C/G polymorphism and susceptibility to RA in the Asian and Caucasian populations (P > 0.05) (Table 2 and Figures 2A,B, 3A).
Table 2.
Meta-analysis results of the miR-146a rs3027898 and miR-499a rs3746444 polymorphisms with genetic susceptibility to rheumatoid arthritis.
| Variables | N | C vs. G |
CC vs. GG |
GC vs. GG |
GC/CC vs. GG |
CC vs. GC/GG |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% Cl) | Ph | I2(%) | OR (95% Cl) | Ph | I2(%) | OR (95% Cl) | Ph | I2(%) | OR (95% Cl) | Ph | I2(%) | OR (95% Cl) | Ph | I2(%) | ||
| miR-146a G > C | ||||||||||||||||
| Caucasian | 6 | 0.92 (0.80–1.05) | 0.486 | 0.0 | 0.88 (0.64–1.20) | 0.692 | 0.0 | 0.89 (0.74–1.06) | 0.545 | 0.0 | 0.89 (0.75–1.05) | 0.476 | 0.0 | 0.93 (0.69–1.26) | 0.772 | 0 |
| Asian | 2 | 0.98 (0.86–1.11) | 0.743 | 0.0 | 0.94 (0.72–1.23) | 0.915 | 0.0 | 0.95 (0.74–1.23) | 0.754 | 0.0 | 0.95 (0.75–1.21) | 0.896 | 0.0 | 0.98 (0.81–1.19) | 0.593 | 0 |
| All ethnicity | 8 | 0.95 (0.86–1.04) | 0.660 | 0.0 | 0.91 (0.75–1.12) | 0.870 | 0.0 | 0.91 (0.78–1.05) | 0.740 | 0.0 | 0.91 (0.79–1.04) | 0.690 | 0.0 | 0.97 (0.82–1.14) | 0.896 | 0 |
|
C vs. T |
CC vs. TT |
TC vs. TT |
TC/CC vs. TT |
CC vs. TC/TT |
||||||||||||
| OR (95% Cl) | Ph | I2(%) | OR (95% Cl) | Ph | I2(%) | OR (95% Cl) | Ph | I2(%) | OR (95% Cl) | Ph | I2(%) | OR (95% Cl) | Ph | I2(%) | ||
| miR-499a T > C | ||||||||||||||||
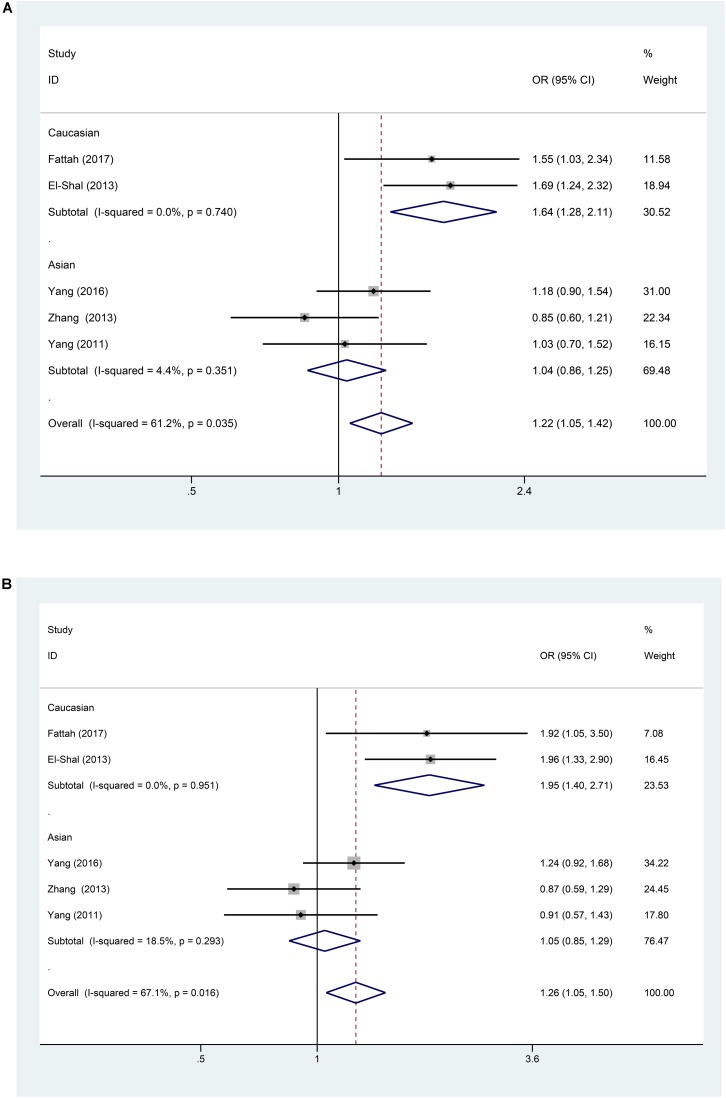
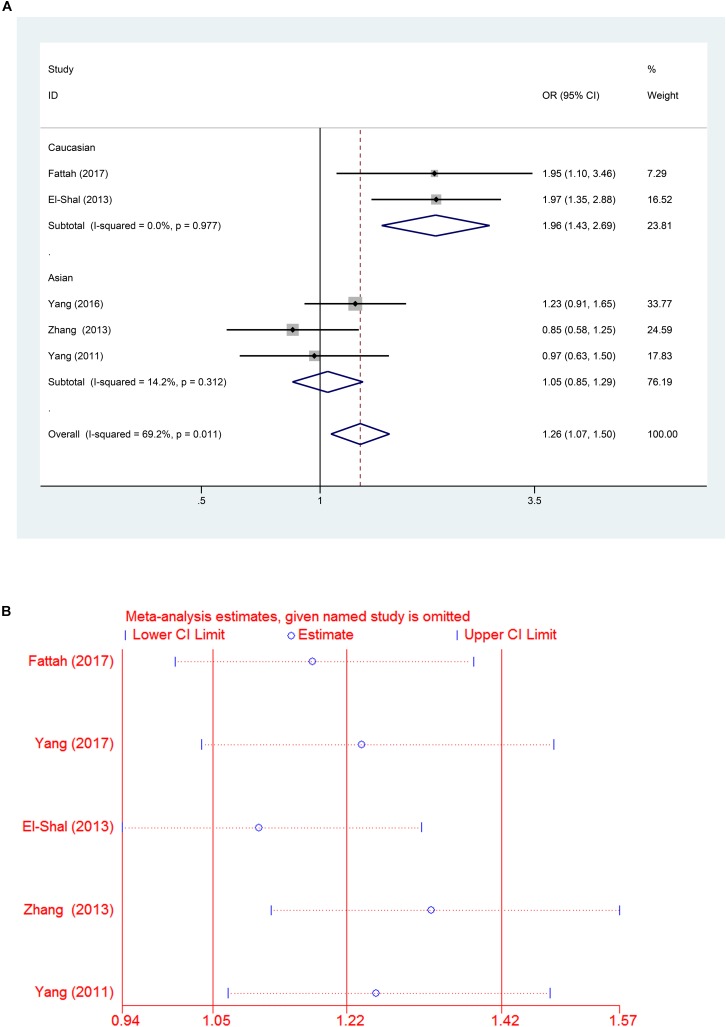
| Caucasian | 2 | 1.64 (1.28–2.11) | 0.740 | 0.0 | 2.06 (1.06–3.97) | 0.973 | 0.0 | 1.95 (1.40–2.71) | 0.951 | 0.0 | 1.96 (1.43–2.69) | 0.977 | 0.0 | 1.52 (0.81–2.86) | 0.906 | 0.0 |
| Asian | 3 | 1.04 (0.86–1.25) | 0.351 | 4.4 | 1.03 (0.53–2.01) | 0.598 | 0.0 | 1.05 (0.85–1.29) | 0.293 | 18.5 | 1.05 (0.85–1.29) | 0.312 | 14.2 | 1.03 (0.53–2.01) | 0.592 | 0 |
| All ethnicity | 5 | 1.22 (1.05–1.42) | 0.035 | 61.2 | 1.46 (0.92–2.32) | 0.558 | 0.0 | 1.26 (1.05–1.50) | 0.016 | 67.1 | 1.26 (1.07–1.50) | 0.011 | 69.2 | 1.26 (0.80–1.99) | 0.790 | 0.0 |
CI, confidence interval, OR, odds ratio, I2 = the variation in OR attributable to heterogeneity.
FIGURE 2.
(A) Forest plot for association between miR-146a rs302789 polymorphisms and risk of rheumatoid arthritis under allele genetic model (C vs. G) after stratification analysis by ethnicity. (B) Forest plot for association between miR–146a rs302789 polymorphisms and risk of rheumatoid arthritis under heterozygote genetic model (GC vs. GG) after stratification analysis by ethnicity.
FIGURE 3.
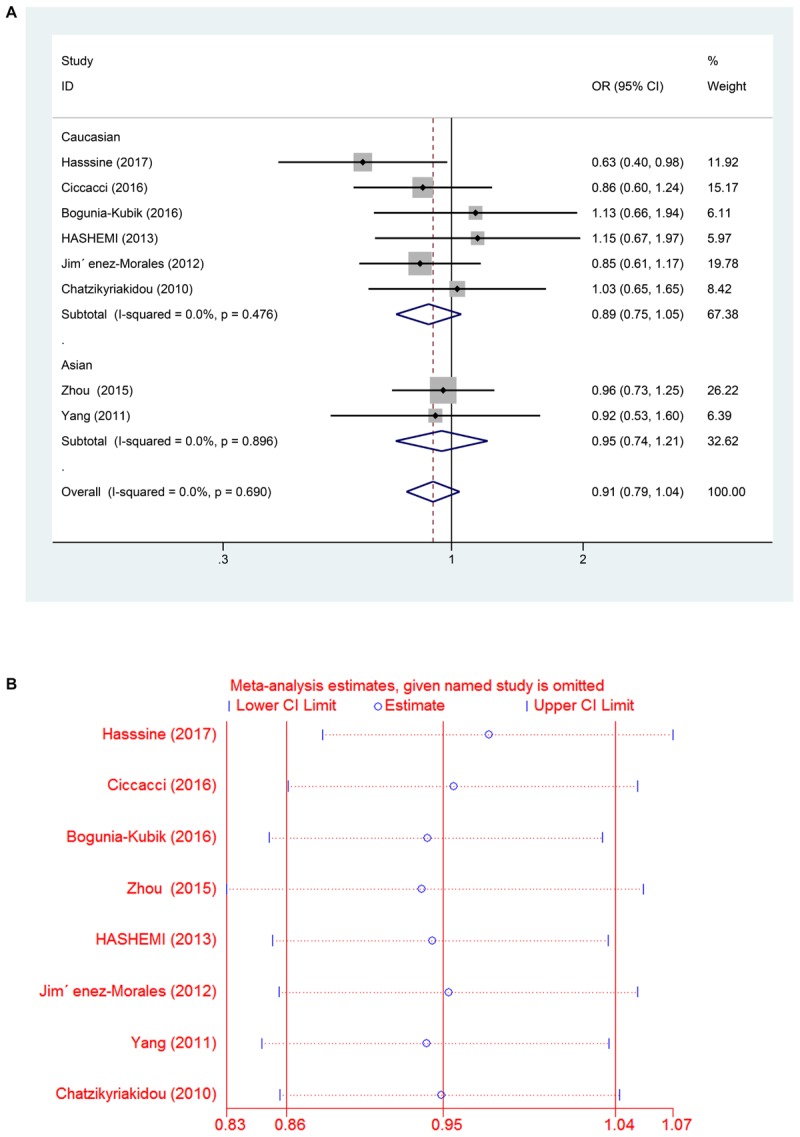
(A) Forest plot for association between miR-146a rs302789 polymorphisms and risk of rheumatoid arthritis under dominant genetic model (GC/CC vs. GG) after stratification analysis by ethnicity. (B) The sensitivity analysis for studies of the association between miR–146a rs302789 polymorphisms and risk of rheumatoid arthritis under allele genetic model (C vs. G).
A significant association between the miR-499 rs3746444 T > C polymorphism and RA risk was observed in all populations, as shown in Table 2 (C vs. T: OR = 1.22, 95% CI = 1.05–1.42, Ph = 0.035; TC vs. TT: OR = 1.26, 95% CI = 1.05–1.50, Ph = 0.016; TC/CC vs. TT: OR = 1.26, 95% CI = 1.07–1.50, Ph = 0.011). However, no significant associations were found in CC vs. TT: OR = 1.46, 95% CI = 0.92–2.32, P = 0.558 and CC vs. TC/TT: OR = 1.26, 95% CI = 0.8–1.99, P = 0.79 (all P > 0.05, Table 2). Subgroup analysis based on ethnicity showed no significant association between the miR-146a C/T polymorphism and susceptibility to RA in the Asian population (P > 0.05) (Table 2). However, in the Caucasian population, the C allele in the miR-499 T > C polymorphism was a contributor to RA susceptibility in some genetic models (C vs. T: OR = 1.64, 95% CI = 1.28–2.11, Ph = 0.74; TC vs. TT: OR = 1.95, 95% CI = 1.40–2.71, Ph = 0.951; TC/CC vs. TT: OR = 1.96, 95% CI = 1.43–2.69, Ph = 0.977) (all P < 0.001, Table 2 and Figures 4A,B, 5A).
FIGURE 4.
(A) Forest plot for association between miR-499a rs3746444 polymorphisms and risk of rheumatoid arthritis under allele genetic model (C vs. T) after stratification analysis by ethnicity. (B) Forest plot for association between miR–499a rs3746444 polymorphisms and risk of rheumatoid arthritis under heterozygote genetic model (TC vs. TT) after stratification analysis by ethnicity.
FIGURE 5.
(A) Forest plot for association between miR-499a rs3746444 polymorphisms and risk of rheumatoid arthritis under dominant genetic model (TC/CC vs. TT) after stratification analysis by ethnicity. (B) The sensitivity analysis for studies of the association between miR–499a rs3746444 polymorphisms and risk of rheumatoid arthritis under allele genetic model (C vs. G).
Sensitivity Analysis and Publication Bias
A leave-one-out analysis was performed to investigate the influence of each individual study on the pooled ORs (Figures 3B, 5B). The results indicated no significant alteration in the pooled ORs after exclusion of any study on miR-146a rs2910164 G > C and miR-499 rs3746444 T > C. Moreover, because of the limited number of studies included (<10), publication bias was not evaluated.
Discussion
Genetic and epigenetic factors play pivotal roles in cancer and autoimmune diseases. MiRNAs are important for the regulation of the cartilage-invading phenotype of RA synovial fibroblasts, leukocyte activation, and cytokine production, and their roles contribute to our understanding of the mechanisms underlying RA and provide novel therapeutic targets for the treatment of RA (Baxter et al., 2012).
MiR-146a, one of the most extensively investigated miRNAs, exerts significant effects on cancer and autoimmune diseases. MiR-146a is widely expressed and involved in the differentiation and activation of the innate and adaptive immune systems (O’Connell et al., 2010). It suppresses NF-κB signaling by downregulating the NF-κB signaling transducers interleukin-1 (IL-1) receptor associated kinase 1 and tumor necrosis factor receptor-associated factor 6. Meanwhile, miR-146a directly activates signal transducer and activator of transcription 1 to promote the activity of Treg cells in RA, which mediate the main mechanisms underlying RA (Zhou Q. et al., 2015). Polymorphisms of miR-146a are associated with various cancers and autoimmune diseases (Wang et al., 2012; Xu et al., 2012).
A single nucleotide polymorphism consisting of the G/C nucleotide substitution in miR-146a rs2910164 could result in the replacement of a G:U pair by a C:U mismatch (Hassine et al., 2017). However, the association between mir146a polymorphisms and RA remains controversial. Because of the increasing number of studies, we set strict inclusion and exclusion criteria to avoid potential bias from inadequate studies. Data from Singh et al. (2014). were not included in this meta-analysis because the patients were diagnosed as juvenile idiopathic arthritis (JIA), which includes other types of arthritis in young patients. Data from El-Shal et al. (2013) were also excluded from the pooled analysis for mir146a because of deviation from HWE in the control group. The results of the updated meta-analysis suggest that mir146a polymorphisms are not associated with RA risk, which is consistent with previous studies (Li et al., 2014; Lee and Bae, 2015).
The mechanism underlying the association between RA and miR-499 was investigated previously. One possible explanation is that mir-499 modulates the expression of regulatory factor X4, thus regulating the human leukocyte antigen DRB1, which is closely associated with RA (van Gaalen et al., 2004; Yang et al., 2011). Moreover, miR-499 targets IL-17 receptor B, IL-6, and several cytokines, which are important factors involved in RA pathogenesis (Ying et al., 2011; El-Shal et al., 2013). Although previous studies demonstrated the association between miR-499 polymorphisms and RA, the limited study number and high heterogeneity decreased the robustness of the conclusions. In the present study, two studies from the miR-499 polymorphism group were excluded from the meta-analysis because the genotype distribution of the controls in all studies was inconsistent with HWE (Hashemi et al., 2013; Toraih et al., 2016). Fattah et al. (2017) found that miR-499 rs3746444 T > C is associated with the susceptibility to RA, and the heterozygote is characterized by higher levels of C-reactive protein, anti-cyclic citrullinated peptide, antibody and disease activity score 28 than those of the homozygote (Fattah et al., 2017). Hashemi et al. (2013) also reported that the C allele of the mir-499 rs3746444 polymorphism is a risk factor for RA. Although the overall results of the pooled analysis indicated that the miR-499 T > C polymorphism was associated with RA risk, the high heterogeneity suggested an insufficient robustness of the conclusion. In the subgroup analysis according to ethnicity, a strong correlation between the miR-499 T > C polymorphism and RA was observed in the Caucasian group, which suggested that the C allele in the miR-499 T > C polymorphism was an obvious contributor to RA susceptibility. However, no relationship was observed in the Asian group. The results were in consistency with previous meta-analysis (Fu et al., 2016), however, Fu et al. (2016) drew a conclusion about miR-499 polymorphism group based on three studies including two studies (El-Shal et al., 2013; Hashemi et al., 2013) from the Caucasian group and one (Yang et al., 2011) from the Asian group. As increasing number of case-control studies published, this updated meta-analysis retrieved five studies including two (El-Shal et al., 2013; Fattah et al., 2017) from the Caucasian group and three from Asian group (Yang et al., 2011, 2016; Zhang et al., 2013). Due to the inconsistency with HWE, two studies (Hashemi et al., 2013; Toraih et al., 2016) were excluded from the miR-499 polymorphism group, which increased the robustness of this meta-analysis.
The present study had several limitations. Firstly, the variation in sample size and in genotyping methods was a limitation of the study. Secondly, all studies included in our meta-analyses were in accordance with HWE, and because of the limited number of included studies, no publication bias could be tested. Because of the limited quantity and quality of the included studies, the above conclusion needs to be further verified in additional high quality studies.
Conclusion
In conclusion, the present meta-analysis showed no significant association between miR-146a and RA risk, which is consistent with previous studies. However, the present study demonstrated the contribution of the C allele in the miR-499 T > C polymorphism to RA susceptibility in the Caucasian population, and this association was not present in the Asian population.
Author Contributions
XZ conceived and designed the study and contributed to statistical analyses. MZ, BJ, and MX contributed to eligible study collection and data extraction. XZ and BJ prepared the tables and figures. XZ, MZ, and MX wrote and revised the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Baxter D., McInnes I. B., Kurowska-Stolarska M. (2012). Novel regulatory mechanisms in inflammatory arthritis: a role for microRNA. Immunol. Cell Biol. 90 288–292. 10.1038/icb.2011.114 [DOI] [PubMed] [Google Scholar]
- Bogunia-Kubik K., Wysoczanska B., Piatek D., Iwaszko M., Ciechomska M., Swierkot J. (2016). Significance of polymorphism and expression of miR-146a and NFkB1 genetic variants in patients with rheumatoid arthritis. Arch. Immunol. Ther. Exp. 64(Suppl. 1), 131–136. 10.1007/s00005-016-0443-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatzikyriakidou A., Voulgari P. V., Georgiou I., Drosos A. A. (2010). A polymorphism in the 3’-UTR of interleukin-1 receptor-associated kinase (IRAK1), a target gene of miR-146a, is associated with rheumatoid arthritis susceptibility. Joint Bone Spine 77 411–413. 10.1016/j.jbspin.2010.05.013 [DOI] [PubMed] [Google Scholar]
- Ciccacci C., Conigliaro P., Perricone C., Rufini S., Triggianese P., Politi C., et al. (2016). Polymorphisms in STAT-4, IL-10, PSORS1C1, PTPN2 and MIR146A genes are associated differently with prognostic factors in Italian patients affected by rheumatoid arthritis. Clin. Exp. Immunol. 186 157–163. 10.1111/cei.12831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai R., Ahmed S. A. (2011). MicroRNA, a new paradigm for understanding immunoregulation, inflammation, and autoimmune diseases. Transl. Res. 157 163–179. 10.1016/j.trsl.2011.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DerSimonian R., Laird N. (1986). Meta-analysis in clinical trials. Control Clin. Trials 7 177–188. 10.1016/0197-2456(86)90046-2 [DOI] [PubMed] [Google Scholar]
- El-Shal A. S., Aly N. M., Galil S. M. A., Moustafa M. A., Kandel W. A. (2013). Association of microRNAs genes polymorphisms with rheumatoid arthritis in Egyptian female patients. Joint Bone Spine 80 626–631. 10.1016/j.jbspin.2013.03.005 [DOI] [PubMed] [Google Scholar]
- Esteller M. (2011). Non-coding RNAs in human disease. Nat. Rev. Genet. 12 861–874. 10.1038/nrg3074 [DOI] [PubMed] [Google Scholar]
- Fattah S. A., Ghattas M. H., Saleh S. M., Abo-Elmatty D. M. (2017). Pre-micro RNA-499 gene polymorphism rs3746444 T/C is associated with susceptibility to rheumatoid arthritis in egyptian population. Indian J. Clin. Biochem. 33 96–101. 10.1007/s12291-017-0652-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L., Jin L., Yan L., Shi J., Wang H., Zhou B., et al. (2016). Comprehensive review of genetic association studies and meta-analysis on miRNA polymorphisms and rheumatoid arthritis and systemic lupus erythematosus susceptibility. Hum. Immunol. 77 1–6. 10.1016/j.humimm.2014.09.002 [DOI] [PubMed] [Google Scholar]
- Furer V., Greenberg J. D., Attur M., Abramson S. B., Pillinger M. H. (2010). The role of microRNA in rheumatoid arthritis and other autoimmune diseases. Clin. Immunol. 136 1–15. 10.1016/j.clim.2010.02.005 [DOI] [PubMed] [Google Scholar]
- Hashemi M., Eskandari-Nasab E., Zakeri Z., Atabaki M., Bahari G., Jahantigh M., et al. (2013). Association of pre-miRNA-146a rs2910164 and premiRNA-499 rs3746444 polymorphisms and susceptibility to rheumatoid arthritis. Mol. Med. Rep. 7 287–291. 10.3892/mmr.2012.1176 [DOI] [PubMed] [Google Scholar]
- Hassine H. B., Boumiza A., Sghiri R., Baccouche K., Boussaid I., Atig A., et al. (2017). Micro RNA-146a but not IRAK1 is associated with rheumatoid arthritis in the tunisian population. Genet. Test. Mol. Biomarkers 21 92–96. 10.1089/gtmb.2016.0270 [DOI] [PubMed] [Google Scholar]
- Hayashino Y., Noguchi Y., Fukui T. (2005). Systematic evaluation and comparison of statistical tests for publication bias. J. Epidemiol. 15 235–243. 10.2188/jea.15.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L., Hannon G. J. (2004). MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 5 522–531. 10.1038/nrg1379 [DOI] [PubMed] [Google Scholar]
- Higgins J. P., Thompson S. G. (2002). Quantifying heterogeneity in a meta-analysis. Stat. Med. 21 1539–1558. 10.1002/sim.1186 [DOI] [PubMed] [Google Scholar]
- Jimenez-Morales S., Gamboa-Becerra R., Baca V., Del Rio-Navarro B. E., Lopez-Ley D. Y., Velazquez-Cruz R., et al. (2012). MiR-146a polymorphism is associated with asthma but not with systemic lupus erythematosus and juvenile rheumatoid arthritis in Mexican patients. Tissue Antigens 80 317–321. 10.1111/j.1399-0039.2012.01929.x [DOI] [PubMed] [Google Scholar]
- Klein K., Gay S. (2015). Epigenetics in rheumatoid arthritis. Curr. Opin. Rheumatol. 27 76–82. 10.1097/bor.0000000000000128 [DOI] [PubMed] [Google Scholar]
- Lau J., Ioannidis J. P., Schmid C. H. (1997). Quantitative synthesis in systematic reviews. Ann. Intern. Med. 127 820–826. 10.7326/0003-4819-127-9-199711010-00008 [DOI] [PubMed] [Google Scholar]
- Lee Y. H., Bae S. C. (2015). The miR-146a polymorphism and susceptibility to systemic lupus erythematosus and rheumatoid arthritis : a meta-analysis. Z. Rheumatol. 74 153–156. 10.1007/s00393-014-1509-6 [DOI] [PubMed] [Google Scholar]
- Li K., Tie H., Hu N., Chen H., Yin X., Peng C., et al. (2014). Association of two polymorphisms rs2910164 in miRNA-146a and rs3746444 in miRNA-499 with rheumatoid arthritis: a meta-analysis. Hum. Immunol. 75 602–608. 10.1016/j.humimm.2014.05.002 [DOI] [PubMed] [Google Scholar]
- Mantel N., Haenszel W. (1959). Statistical aspects of the analysis of data from retrospective studies of disease. J. Natl. Cancer Inst. 22 719–748. [PubMed] [Google Scholar]
- Neogi T., Aletaha D., Silman A. J., Naden R. L., Felson D. T., Aggarwal R., et al. (2010). The 2010 American College of Rheumatology/European League Against Rheumatism classification criteria for rheumatoid arthritis: phase 2 methodological report. Arthritis Rheum. 62 2582–2591. 10.1002/art.27580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell R. M., Rao D. S., Chaudhuri A. A., Baltimore D. (2010). Physiological and pathological roles for microRNAs in the immune system. Nat. Rev. Immunol. 10 111–122. 10.1038/nri2708 [DOI] [PubMed] [Google Scholar]
- Okada Y., Suzuki A., Ikari K., Terao C., Kochi Y., Ohmura K., et al. (2016). Contribution of a Non-classical HLA Gene, HLA-DOA, to the Risk of Rheumatoid Arthritis. Am. J. Hum. Genet. 99 366–374. 10.1016/j.ajhg.2016.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatek D., Swierkot J., Iwaszko M., Ciechomska M., Wiland P., Bogunia-Kubik K. (2016). Relationships between the miRNA-146a polymorphism and expression in patients with rheumatoid arthritis. Arch. Immunol. Ther. Exp. Conf. 64(1 Suppl. 1):S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S., Rai G., Aggarwal A. (2014). Association of microRNA-146a and its target gene IRAK1 polymorphism with enthesitis related arthritis category of juvenile idiopathic arthritis. Rheumatol. Int. 34 1395–1400. 10.1007/s00296-014-3001-7 [DOI] [PubMed] [Google Scholar]
- Smolen J. S., Aletaha D., McInnes I. B. (2016). Rheumatoid arthritis. Lancet 388 2023–2038. 10.1016/s0140-6736(16)30173-8 [DOI] [PubMed] [Google Scholar]
- Sokolove J., Wagner C. A., Lahey L. J., Sayles H., Duryee M. J., Reimold A. M., et al. (2016). Increased inflammation and disease activity among current cigarette smokers with rheumatoid arthritis: a cross-sectional analysis of US veterans. Rheumatology 55 1969–1977. 10.1093/rheumatology/kew285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terao C., Ikari K., Nakayamada S., Takahashi Y., Yamada R., Ohmura K., et al. (2016). A twin study of rheumatoid arthritis in the Japanese population. Mod. Rheumatol. 26 685–689. 10.3109/14397595.2015.1135856 [DOI] [PubMed] [Google Scholar]
- Toraih E. A., Ismail N. M., Toraih A. A., Hussein M. H., Fawzy M. S. (2016). Precursor miR-499a variant but not miR-196a2 is associated with rheumatoid arthritis susceptibility in an egyptian population. Mol. Diagn. Ther. 20 279–295. 10.1007/s40291-016-0194-3 [DOI] [PubMed] [Google Scholar]
- van Gaalen F. A., van Aken J., Huizinga T. W., Schreuder G. M., Breedveld F. C., Zanelli E., et al. (2004). Association between HLA class II genes and autoantibodies to cyclic citrullinated peptides (CCPs) influences the severity of rheumatoid arthritis. Arthritis Rheum. 50 2113–2121. 10.1002/art.20316 [DOI] [PubMed] [Google Scholar]
- Wang J., Bi J., Liu X., Li K., Di J., Wang B. (2012). Has-miR-146a polymorphism (rs2910164) and cancer risk: a meta-analysis of 19 case-control studies. Mol. Biol. Rep. 39 4571–4579. 10.1007/s11033-011-1247-7 [DOI] [PubMed] [Google Scholar]
- Xu W. D., Lu M. M., Pan H. F., Ye D. Q. (2012). Association of MicroRNA-146a with autoimmune diseases. Inflammation 35 1525–1529. 10.1007/s10753-012-9467-0 [DOI] [PubMed] [Google Scholar]
- Yamamoto K., Okada Y., Suzuki A., Kochi Y. (2015). Genetic studies of rheumatoid arthritis. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 91 410–422. 10.2183/pjab.91.410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B., Zhang J. L., Shi Y. Y., Li D. D., Chen J., Huang Z. C., et al. (2011). Association study of single nucleotide polymorphisms in pre-miRNA and rheumatoid arthritis in a Han Chinese population. Mol. Biol. Rep. 38 4913–4919. 10.1007/s11033-010-0633-x [DOI] [PubMed] [Google Scholar]
- Yang X. K., Li P., Zhang C., Leng R. X., Li S., Liu J., et al. (2016). Association between IRAK1 rs3027898 and miRNA-499 rs3746444 polymorphisms and rheumatoid arthritis: a case control study and meta-analysis. Z. Rheumatol. 76 622–629. 10.1007/s00393-016-0169-0 [DOI] [PubMed] [Google Scholar]
- Ying B., Shi Y., Pan X., Song X., Huang Z., Niu Q., et al. (2011). Association of polymorphisms in the human IL-10 and IL-18 genes with rheumatoid arthritis. Mol. Biol. Rep. 38 379–385. 10.1007/s11033-010-0119-x [DOI] [PubMed] [Google Scholar]
- Zhang H., Pu J., Wang X., Shen L., Zhao G., Zhuang C., et al. (2013). IRAK1 rs3027898 C/A polymorphism is associated with risk of rheumatoid arthritis. Rheumatol. Int. 33 369–375. 10.1007/s00296-012-2379-3 [DOI] [PubMed] [Google Scholar]
- Zhou Q., Haupt S., Kreuzer J. T., Hammitzsch A., Proft F., Neumann C., et al. (2015). Decreased expression of miR-146a and miR-155 contributes to an abnormal Treg phenotype in patients with rheumatoid arthritis. Ann. Rheum. Dis. 74 1265–1274. 10.1136/annrheumdis-2013-204377 [DOI] [PubMed] [Google Scholar]
- Zhou X., Zhu J., Zhang H., Zhou G., Huang Y., Liu R. (2015). Is the microRNA-146a (rs2910164) polymorphism associated with rheumatoid arthritis? Association of microRNA-146a (rs2910164) polymorphism and rheumatoid arthritis could depend on gender. Joint Bone Spine 82 166–171. 10.1016/j.jbspin.2014.12.009 [DOI] [PubMed] [Google Scholar]