Abstract
Butyrivibrio fibrisolvens forms part of the gastrointestinal microbiome of ruminants and other mammals, including humans. Indeed, it is one of the most common bacteria found in the rumen and plays an important role in ruminal fermentation of polysaccharides, yet, to date, there is no closed reference genome published for this species in any ruminant animal. We successfully assembled the nearly complete genome sequence of B. fibrisolvens strain INBov1 isolated from cow rumen using Illumina paired-end reads, 454 Roche single-end and mate pair sequencing technology. Additionally, we constructed an optical restriction map of this strain to aid in scaffold ordering and positioning, and completed the first genomic structure of this species. Moreover, we identified and assembled the first chromid of this species (pINBov266). The INBov1 genome encodes a large set of genes involved in the cellulolytic process but lacks key genes. This seems to indicate that B. fibrisolvens plays an important role in ruminal cellulolytic processes, but does not have autonomous cellulolytic capacity. When searching for genes involved in the biohydrogenation of unsaturated fatty acids, no linoleate isomerase gene was found in this strain. INBov1 does encode oleate hydratase genes known to participate in the hydrogenation of oleic acids. Furthermore, INBov1 contains an enolase gene, which has been recently determined to participate in the synthesis of conjugated linoleic acids. This work confirms the presence of a novel chromid in B. fibrisolvens and provides a new potential reference genome sequence for this species, providing new insight into its role in biohydrogenation and carbohydrate degradation.
Keywords: Butyrivibrio fibrisolvens, cow rumen, genome sequencing, INBov1
Data Summary
This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under accession GCA_003175155.1. All the sequencing data used in this experiment and assembly details are under NCBI BioProject PRJNA412083. https://www.ncbi.nlm.nih.gov/bioproject/PRJNA412083/
Impact Statement.
Currently, all assembly projects available for Butyrivibrio fibrisolvens species provide the genome sequence information in more than 60 unordered sequences. In the present study, we assembled the complete genomic structure of B. fibrisolvens in one sequence. We identified ~96 % of the bases, and established the number and position of the ~4 % unidentified bases. Furthermore, we identified a chromid, the first sequenced chromid in this bacterial species. The results presented here will contribute to our understanding of the role of the B. fibrisolvens as part of the rumen microbiota.
Introduction
Butyrivibrio fibrisolvens is part of the gastrointestinal microbiome of ruminants and other mammals, including humans. This species belongs to the genus Butyrivibrio (class Clostridia) which comprises non-spore-forming, monotrichous, anaerobic, butyric-acid-producing, curved rod-shaped bacteria [1]. It is ubiquitously present in the gastrointestinal tract of many animals and in high abundance in the bovine rumen, which suggests that this organism plays an important role in the ruminal fermentation of polysaccharides involved in cellulose degradation [2]. Therefore, B. fibrisolvens can provide genetic resources of potential use in the utilization of vegetal biomass for the development of third-generation biofuels. Moreover, this species participates in the biohydrogenation of polyunsaturated fatty acids in ruminants and has been proposed to improve the fatty acid profile of milk and meat from ruminant animals and thus the creation of healthier food products [3]. This species has also been evaluated in mice as a probiotic that prevents enterocolitis [4] and colorectal cancer [5].
As of now, there is still no closed circular genome sequence for B. fibrisolvens in any public databases (see Genome Properties section). There are nine genome assembly projects of B. fibrisolvens deposited in the NCBI genome database, each one in more than 60 unordered sequences. These genome sequences have been provided by the Hungate1000 Consortium [6] and the DOE Joint Genome Institute (JGI). One previous study [7], using DNA isolation, suggests that the B. fibrisolvens genome contains a chromid with an estimated molecular weight of 200 MDa. Nevertheless, this has not been confirmed by sequencing and the structure and function of this putative chromid remain unknown. Thus, the aim of this work is two-fold: to contribute to the production of a nearly complete genome sequence for the B. fibrisolvens INBov1 strain obtained from cow rumen, and to provide the first chromid sequence of this species, contributing new insight into its characteristics.
Methods
Genome sequencing and assembly
This genome assembly project was performed at Instituto Nacional de Tecnología Agropecuaria (INTA) within the bioinformatics unit. The results of this Whole Genome Shotgun project have been deposited at DDBJ/ENA/GenBank under accession GCA_003175155.1. Details of bacterial isolation, growth conditions and species characterization methods and results are available in File S1 (available in the online version of this article).
For genome assembly we obtained a complex dataset of reads from different instruments and from different library preparations, as well as an optical map restriction digest. Overall, we had ~750 000 sequences of Illumina paired-end (PE) reads (2×250 bp) produced by the sequencing services using Miseq (Illumina) performed at INTA, Instituto de Biotecnología (Argentina), Consorcio Argentino de Tecnología Genómica (CATG). Additionally, we had 220 000 single-end (SE) reads (300 bp in length) and 200 000 mate pair (MP) reads (300 bp in length, insert size ~2000 bp) which were obtained through GS 454 FLX technology (Instituto Indear of Rosario, Argentina). In addition, we created an optical restriction map of B. fibrisolvens INBov1 with the enzyme KpnI to aid in scaffold ordering (OpGen Technologies). Quality control was tested using FastQC [8] and the Illumina PE and 454 SE reads were trimmed using Trimmomatic [9]. Almost half of the Illumina PE reads were extended using the FLASh program [10].
A detailed description and discussion of the assembly methods and genome annotation is included in File S1.
Results
Assembly and genome organization
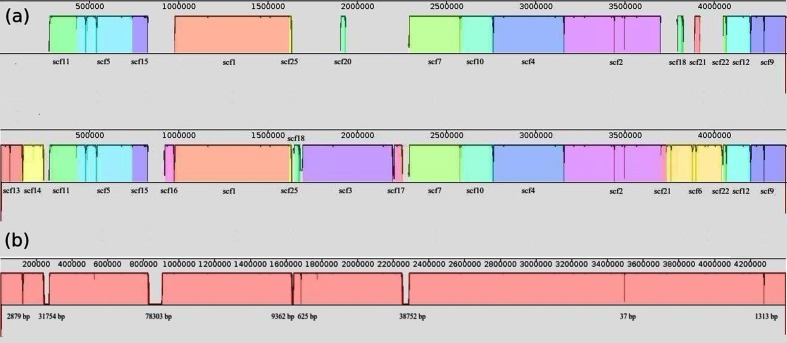
After analysis of the different assembly trials tested (see ‘Assembly discussion’ in File S1), the final workflow chosen to reconstruct the genome was via the Newbler software because fewer scaffolds were produced and higher map coverage values were obtained through this workflow. Map coverage refers to the percentage of sequence assembled which aligns in accordance with the restriction map provided by the optical mapping results. The 25 scaffolds obtained with Newbler with only 454 reads (SE and MP) were used to reconstruct the genome sequence. Soma, a scaffold-restriction map aligner pipeline, placed 15 scaffolds in the optical map alignment. In a following step, after further analysis using NEBcutter [11], six new scaffolds were placed by manual alignment. We also relocated two small scaffolds and removed one (see Fig. 1a). A complete description of the manual alignment process is described in the Supplementary Material.
Fig. 1.
(a) Top: visualization using the program Mauve [18] of scaffolds placed by Soma in the optical restriction map (70 % map coverage). Bottom: structure of the genome sequence after manual placing of scaffolds with NEBcutter (95 % map coverage). (b) Final genomic sequence of B. fibrisolvens (4 398 850 bp, complete genomic structure with 96 % of identified bases) after using GapCloser. Gap length is shown (gap regions smaller than 10 bases are not shown).
In order to reduce the number of gaps in the scaffolds, we performed gap filling with the Illumina PE reads. GapCloser [12] was used with the Illumina PE reads to close 93 % of the gaps present in all of the scaffolds (41 570 total bases). Moreover, the genome size was estimated from the optical map restriction data, which gave a value of 4 327 514 bp. This was similar to the size estimated from the kmer distribution, using the Illumina PE reads, which was 4 407 001 bp (see File S1). Overall, we assembled close to 96 % of the genome sequence in one scaffold of 4 398 850 bp and had only 163 074 unidentified bases (see Fig. 1b). The position and number of the unidentified bases was established by alignment of the MP reads and by positioning the scaffolds in the restriction map. Because the position and number of the unidentified bases could be determined, we were able to complete the full genomic structure of the genome. We also identified one large unplaced scaffold (266 542 bp) as a new putative chromid. This scaffold did not align with any region of the restriction map and, among other plasmidic features, contained a repA gene, which encodes a plasmid replication initiator protein [13, 14] (see ‘Genome insights from the genome sequence’ section).
Genome properties and statistics
The genome of strain INBov1 contains one scaffold of 4 279 765 bp (163 097 gaps), representing 96 % of the estimated complete genome sequence and the complete genomic structure. One chromid in one sequence of 266 542 bp (pINBov266), four scaffolds in a range of ~2–107 kbp (total of 174 701 bp) and 64 small contigs smaller than ~2 kbp (total of 41 201 bp) remained unplaced but all contained at least one annotated gene. The total size of the non-redundant genome data set is 4 721 197 bp, including the chromid sequence (4 457 655 bp without counting the chromid). The RAST server (http://rast.nmpdr.org) [15] annotated, including all the sequences, 4027 coding sequences that correspond to 3947 proteins and 80 RNAs. The G+C content calculated by RAST is 39.9 %. According to COG annotation, 3121 genes (including 155 genes encoded in the chromid) were classified by using the WebMGA server [16] (see Table 1).
Table 1. COG annotation statistics.
| COG code | Genes | Percentage of total genes | Description |
|---|---|---|---|
| J | 174 | 4.4 | Translation, ribosomal structure and biogenesis |
| A | 0 | 0.0 | RNA processing and modification |
| K | 214 | 5.4 | Transcription |
| L | 164 | 4.2 | Replication, recombination and repair |
| B | 0 | 0.0 | Chromatin structure and dynamics |
| D | 52 | 1.3 | Cell cycle control, cell division, chromosome partitioning |
| V | 103 | 2.6 | Defence mechanisms |
| T | 186 | 4.7 | Signal transduction mechanisms |
| M | 223 | 5.7 | Cell wall/membrane biogenesis |
| N | 32 | 0.8 | Cell motility |
| U | 30 | 0.8 | Intracellular trafficking and secretion |
| O | 78 | 2.0 | Post-translational modification, protein turnover, chaperones |
| C | 110 | 2.8 | Energy production and conversion |
| G | 322 | 8.2 | Carbohydrate transport and metabolism |
| E | 172 | 4.3 | Amino acid transport and metabolism |
| F | 84 | 2.1 | Nucleotide transport and metabolism |
| H | 107 | 2.7 | Coenzyme transport and metabolism |
| I | 63 | 1.6 | Lipid transport and metabolism |
| P | 110 | 2.8 | Inorganic ion transport and metabolism |
| Q | 11 | 0.3 | Secondary metabolite biosynthesis, transport and catabolism |
| R | 372 | 9.4 | General function prediction only |
| S | 249 | 6.3 | Function unknown |
| – | 266 | 6.7 | Multiple classes |
| – | 1092 | 20.9 | Not in COGs |
The statistics obtained with the genome of strain INBov1 are very similar to those observed in the other B. fibrisolvens genomes deposited in the NCBI genome database. Median values for these genomes are 4.7 Mb for genome size, 39.7 % for G+C content and 3764 for proteins annotated. The exception is the genome of strain 16/4 (GenBank: GCA_000209815.1), the metrics of which differ considerably and this strain behaves as an outlier; it presents a genome size of 3.16 Mb, G+C content of 38.6 % and 2966 proteins annotated. Therefore, we also evaluated the 16S rRNA gene sequence of strain 16/4 (GenBank: AJ250365.2) to assess its species identity. The results obtained by using the identification service of the EzBioCloud database [17] showed that the closest species to strain 16/4 is Pseudobutyrivibrio ruminis DSM 9787T (GenBank: X95893) with a sequence similarity of 98.18 %. The level of sequence similarity between the 16S rRNA genes of strain 16/4 and B. fibrisolvens NCDO2221T (GenBank: X89970.1) is 88.56 %, considerably lower than the species threshold proposed by several authors [18, 19]. This suggests that strain 16/4 might have been incorrectly classified as a member of B. fibrisolvens by NCBI.
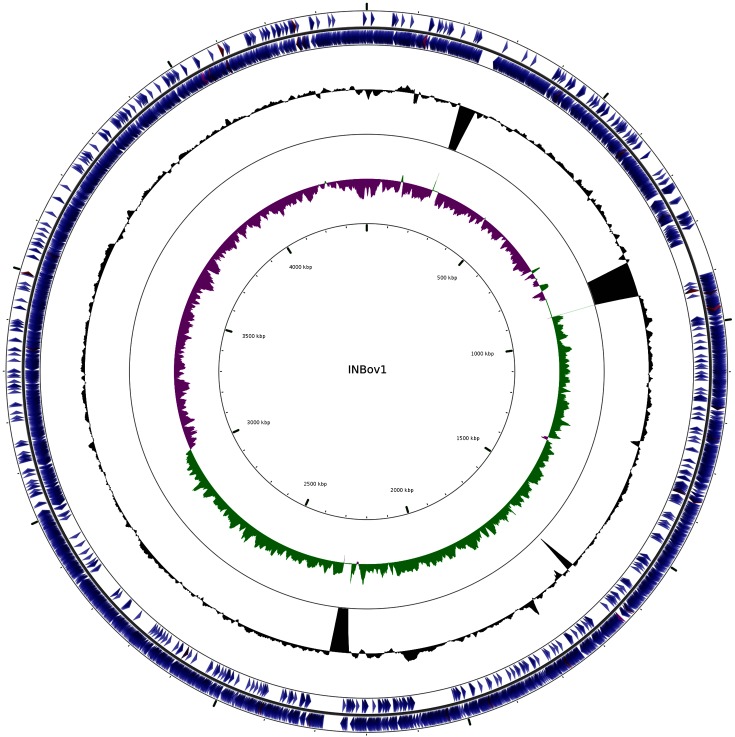
In Fig. 2 the genome sequence of INBov1 is visualized by using CGview [20]. The GC skew shows a characteristic asymmetry in the nucleotide frequency present in most prokaryotes where a higher frequency of guanines is found in the leading strand [21], in accordance with the Theta replication model. Therefore, the origin of replication and the site of termination of the genome are generally located in regions where the skew in the GC content shifts. This GC skew adds further confidence that this is a well-assembled genome.
Fig. 2.
Circular visualization of the B. fibrisolvens INBov1 chromosome. The image shows (from outside to centre): genes on the forward strand, genes on the reverse strand (coding sequences in blue, tRNAs in red, rRNAs in purple). The G+C content is in black with peaks indicating higher or lower values than the average G+C content (peaks out/inside, respectively). There are four noticeable peaks inside that correspond to the largest gap regions, as shown in Fig. 1. The inner circle shows the GC skew. Positive values correspond to green peaks, indicating that the amounts of guanines are enriched in the top strand versus the amount of cytosines in the bottom strand. Purple peaks represent the opposite.
Insights from the genome sequence
Following annotation of the INBov1 genome, we focused on analysis of carbohydrate active enzyme (CAZymes) families due to their potential in many biotechnological applications. We also performed a comparative analysis of the genomes and carbohydrate enzymes of INBov1 and the other species of the genus Butyrivibrio: Butyrivibrio hungatei MB2003, Butyrivibrio proteoclasticus B316 and Butyrivibrio crossotus DSM 2876 (GenBank IDs: GCA_001858005.1, GCA_000145035.1 and GCA_000156015.1). We used the Carbohydrate Active Enzymes database (http://www.cazy.org) [22] and the DBCan server (http://csbl.bmb.uga.edu/dbCAN/) [23] to annotate the INBov1 enzymes.
INBov1, as expected, encodes an extensive repertoire of CAZymes with 114 glycosyl hydrolases (GHs), 33 carbohydrate esterases (CEs), three polysaccharide lyases (PLs) and 86 glycosyl transferases (GTs) encoded in the genome, indicating that INBov1 has a similar distribution of CAZymes to B. proteoclasticus B316. Moreover, INBov1 and B. proteoclasticus B316 are also similar in terms of genome size and number of genes.
INBov1 has a role in the biohydrogenation of unsaturated acids. However, a gene encoding linoleate isomerase (EC 5.2.1.5) was absent from this strain. Interestingly, INBov1 does encode an oleate hydratase (EC 4.2.1.53) involved in the biohydrogenation of oleic acids. No linoleate isomerase genes were found in the other Butyrivibrio species, and oleate hydratase genes were only found in B. crossotus.
The INBov1 genome encodes a full-length enolase (EC 4.2.1.11), which is present on the main chromosome. This glycolytic pathway enzyme, also known as phosphopyruvate hydratase, is responsible for the conversion of 2-phosphoglycerate (2 PG) to phosphoenolpyruvate (PEP). Enolases have recently been linked to the biohydrogenation of linoleic acid in Lactobacillus plantarum [24]. Genes encoding enolase were also present in B. hungatei and B. crossotus.
INBov1 encodes two l-lactate dehydrogenase (EC 1.1.1.27) genes, one on the main chromosome and the other on its chromid. A gene encoding this enzyme was also found in B. hungatei. Genes encoding enolase and l-lactate dehydrogenase are co-localized in the INBov1 genome, an observation that is consistent with a recent study [25] on the rumen microbiome of members of the Hungate1000 Collection.
INBov1 lacks genes encoding two key glucose metabolism enzymes, namely d-glucose phosphotransferase (EC 2.7.1.199), involved in glucose uptake, and phosphoglucomutase (EC 5.4.2.2), required for the inter-conversion of d-glucose 1-phosphate to d-glucose 6-phosphate. Genes encoding phosphoglucomutase were, however, present in B. proteoclasticus and B. hungatei. In contrast, INBov1 does have a phosphomannomutase (EC 5.4.2.8) and genes encoding this enzyme were found in all the Butyrivibrio species with the exception of B. crossotus. An interesting finding is that INBov1 and B. proteoclasticus encode copies of this gene on both their main chromosome and their chromids.
The ubiquitous presence and high abundance of B. fibrisolvens in ruminants suggest that this species plays a significant role in cellulose degradation. Consequently, we characterized the INBov1 genes involved in the cellulolytic process. The glycosyl hydrolases that play a major role in cellulolysis are the endoglucanases (EC 3.2.1.4), β-glucosidases (EC 3.2.1.21) and exoglucanases, which include celodextrinases (EC 3.2.1.91) and cellobiohydrolases (EC 3.2.1.176; EC 3.2.1.74) [26]. Other non-glycosyl hydrolase enzymes have also recently been found to participate in cellulolysis. Laccases (EC 1.10.3.2) and peroxidases (EC 1.11.1) have been shown to participate in the degradation of lignin. Polysaccharide monooxygenases (PMOs) and lytic polysaccharide monooxygenases (LPMOs) also play a role in the cellulose decrystallization process [26].
The INBov1 genome encodes 41 genes related to cellulolytic processes. Among these were several genes encoding endoglucanase and β-glucosidase. The endoglucanase genes were only found on the main chromosome, while genes encoding β-glucosidases were found on both the chromosome and the chromid; a similar situation was observed in B. proteoclasticus. Endoglucanase genes were found in all Butyrivibrio species, while β-glucosidase genes were found in all Butyrivibrio species except B. crossotus. Interestingly, exoglucanase genes were absent, not only from INBov1, but from all Butyrivibrio species. As expected, no lignin degradation genes, PMO or LPMO genes were found in any of the Butyrivibrio species.
Chromid replicon
An important feature of the INBov1 genome was the presence of a single large unplaced contig that we identified initially as a mega-replicon (pINBov266). As a result of a detailed analysis of this contig and its gene content, we have reclassified this replicon as the first chromid to be identified in B. fibrisolvens. Chromids are defined as replicons with a G+C content that is similar to the main chromosome. However, they have plasmid-type maintenance and replication systems and are significantly smaller than the main chromosome [14].
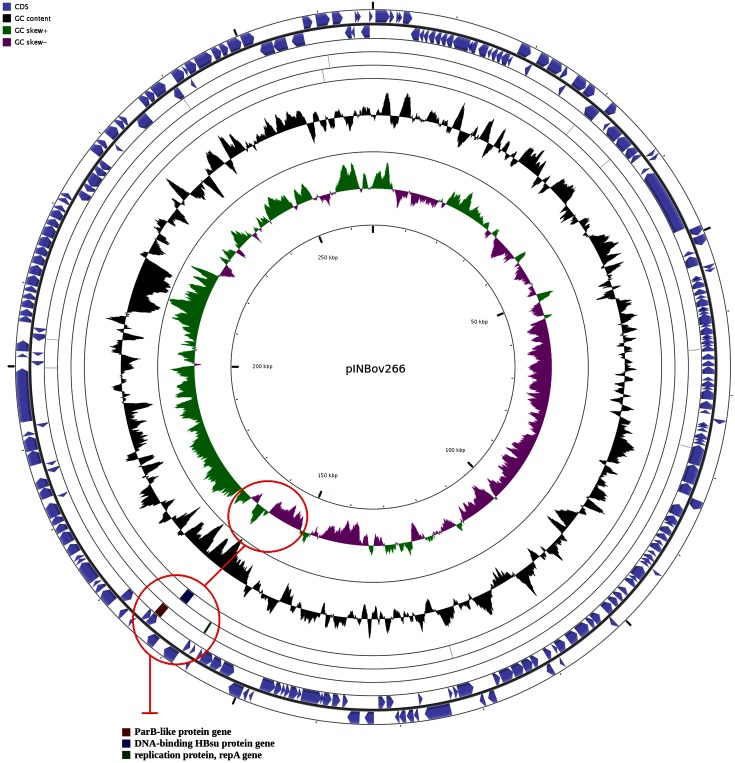
The G+C content of the chromid is 38.9 % and it contains 238 coding sequences, including the genes related to plasmid replication systems (e.g. repA, parB and hbs). RepA is a motor protein that acts as an initiator factor for plasmid replication [13, 14]. The parB gene encodes a centromere-binding protein (CBP), an element characteristic of type 1 partition systems [27]. The hbs protein was shown to participate in controlling DNA gyrase activity [28, 29], playing a role in the initiation of oriC-dependent DNA replication [30, 31]. The repA, parB and hbs genes are co-localized in a region where the GC skew switches the nucleotide frequency polarity (Fig. 3), suggesting that the origin of replication might be located in that area. No conjugation-related genes (e.g. tra and trb genes) were found in the chromid, main chromosome or any unplaced contig sequences. Moreover, use of the oriTfinder tool [32] failed to find evidence of an origin of transfer (oriT) in the pINBov266 sequence. As a result, we conclude there is no conjugative system in pINBov266, suggesting that it is likely to be non-mobile.
Fig. 3.
Circular visualization of the chromid sequence (pINBov266) in Cgview. The location of parB, hbs and repA genes are shown by red circles. The upper circle shows that these genes are located in a region where the GC skew switches the nucleotide frequency polarity. The lower circle corresponds to the blast hits of the genes parB (red), hbs (blue) and repA (green).
pINBov266 encodes several genes involved in antibiotic resistance, and the production of bacteriocins and toxins. We found genes encoding multi-antimicrobial extrusion protein genes from the MATE family of MDR efflux pumps, known to be crucial for resistance to antimicrobial compounds [33]. The chromid also encodes genes for β-lactamase, VanZ and ABC-transporters, which are also involved in antibiotic resistance, and for MerR and SpaF/MutF genes, which are involved in cobalt–zinc–cadmium and lantibiotic bacteriocin resistance, respectively [34, 35].
Chromid pINBov266 encodes 35 putative carbohydrate degradation enzymes, including 17 different types of hydrolases (including a serine hydrolase, α/β hydrolase and glycoside hydrolase). Genes encoding four key enzymes in the glycolysis/gluconeogenesis pathways were also identified: phosphotransferase (EC 2.7.1.90), phosphohexokinase (EC 2.7.1.11), l-lactate dehydrogenase (EC 1.1.1.27), aldehyde dehydrogenase [NAD(P)+] (EC 1.2.1.5), and the only aldehyde dehydrogenase (NAD) (EC 1.2.1.3) gene present in the entire genome. An interesting finding was the presence of genes on the INBov1 genome encoding two PLs of which the PL9 and PL11 genes are encoded only in the chromid. SignalP [36] analysis indicates that the products of these PL genes appear to be secreted.
Genes encoding proteins of the NiFe hydrogenase maturation system (HypD, HypE, HypF and Ferredoxin subunit A) are also only present in pINBov266. Four of the five total genes, which play a role in the hydrogenase maturation system, are found only on the chromid. This system is known to provide a mechanism to store and utilize energy by reversibly converting molecular hydrogen, one of the key products of rumen fermentation [37, 38]. Furthermore, pINBov266 encodes proteins from the TldE/TldD proteolytic complex, which have been reported to play a key role in the maturation and exportation of antibiotics and other proteins [39]. Other genes that were unique to pINBov266 included genes encoding the nitric oxide reductase activation proteins NorD and NorQ, which play a role in denitrification, and carbon starvation protein A (CspA) involved in the carbon starvation stress response.
In view of the above findings, we propose this sequence (CM009897.1) as a chromid, consistent with a previous study [7]. Recently, the presence of chromids has also been reported in other species of the genus Butyrivibrio, namely in B. hungatei MB2003 [40] and B. proteoclasticus B316T [41].
Conclusion
At present, there are nine genome assembly projects of B. fibrisolvens deposited in the NCBI database. Each genome assembled is in more than 60 unordered sequences. The exception is strain 16/4, which is available as a single chromosome, although it appears to be incorrectly classified as a B. fibrisolvens strain. An analysis of its 16S rRNA gene sequence and significant differences of its genome metrics when compared with the other B. fibrisolvens genomes deposited in the NCBI database support this conclusion.
We consider that INBov1 may serve as a reference genome for B. fibrisolvens and propose pINBov266 as a chromid as well. We assembled 96 % of the B. fibrisolvens genome in one sequence of 4 398 850 bp and a total of 163 074 unidentified bases, providing the first nearly complete genome sequence and complete genomic structure of B. fibrisolvens INBov1. Additionally, we identified and assembled the chromid sequence of 266 542 bp (pINBov266) for this organism by finding the presence of elements – repA, parB and hbs genes – characteristic of a plasmidic replication system. These genes are also found co-localized in a region predicted by the GC skew as the probable origin of replication. The designation of pINBov266 as a chromid is also supported by the presence of multiple genes involved in antibiotic resistance and bacteriocin and toxin production. Moreover, the pINBov266 restriction map reveals the absence of any possible alignment between the chromid and chromosome restriction maps. These and other data confirm for the first time the presence of a non-mobilizable chromid in this species. That several genes and functional subsystems are only present in pINBov266 [e.g. aldehyde dehydrogenase (NAD), lyases PL9 and PL11, NiFe hydrogenase maturation, TldE–TldD proteolytic complex, carbon starvation and denitrification genes] suggests that this chromid plays an important and potentially essential role in B. fibrisolvens. However, further assays are required to understand the importance of this chromid in the ecology of this bacterium.
As expected, we found that the INBov1 genome encodes a large set of genes involved in the cellulolytic process but does not encode an exoglucanase gene. This is indicative of B. fibrisolvens playing an important role in the ruminal fermentation of cellulose as part of the gut microbiome community rather than it being an autonomous cellulolytic microbe. With respect to the hydrogenation of unsaturated fatty acids, no linoleate isomerase gene was found. Nonetheless, the presence of oleate hydratase and enolase genes in the INBov1 genome is consistent with previous studies, indicating that this strain participates in the biohydrogenation of unsaturated fatty acids in the rumen [3]. The oleate hydratase encoded by the INBov1 genome could be part of a resistance mechanism against the bactericidal effect of unsaturated acids, as has been proposed previously [3, 42].
The work described here provides new insight into the genome of B. fibrisolvens, contributing to our understanding of a species with high potential in the development of biotechnological applications.
Data bibliography
Rodríguez Hernáez et al. Experiment sequencing data. NCBI BioProject PRJNA412083. https://www.ncbi.nlm.nih.gov/bioproject/412083 Sequence Read Archive (SRA) accession SRP128053 (2017).
Rodríguez Hernáez, et al. Final genome sequences. GenBank Accession Number GCA_003175155.1 (2017).
Supplementary Data
Funding information
Funding for the project was by the AECID D/024562/09; D/031348/10 Projects, National Institute for Agronomic Sciences through PNBIO1131043, and by MinCyT through the PPL 2011 004 ‘Consorcio Argentino de Tecnología Genomica’.
Acknowledgements
Sequencing services using MiSeq (Illumina) were performed at INTA, Consorcio Argentino de Tecnología Genómica (CATG) funded by grant MinCyT PPL 2011 004 Genómica, AECID PCI_ARG109 A1/041041/11 and INTA. This work used computational resources from BioCAD – Instituto de Biotecnología CICVyA, INTA, Consorcio Argentino de Tecnología Genómica, MinCyT PPL 2011 004; AECID PCI_ARG109, D/024562/09.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: LPMO, lytic polysaccharide monooxygenase; MP, mate pair; PE, paired-end; PL, polysaccharide lyase; PMO, polysaccharide monooxygenase; SE, single-end.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Supplementary material is available with the online version of this article.
References
- 1.Bryant MP, Small N. The anaerobic monotrichous butyric acid-producing curved rod-shaped bacteria of the rumen. J Bacteriol. 1956;72:16–21. doi: 10.1128/jb.72.1.16-21.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown DW, Moore WEC. Distribution of Butyrivibrio fibrisolvens in nature. J Dairy Sci. 1960;43:1570–1574. doi: 10.3168/jds.S0022-0302(60)90377-5. [DOI] [Google Scholar]
- 3.Maia MR, Chaudhary LC, Bestwick CS, Richardson AJ, McKain N, et al. Toxicity of unsaturated fatty acids to the biohydrogenating ruminal bacterium, Butyrivibrio fibrisolvens. BMC Microbiol. 2010;10:52. doi: 10.1186/1471-2180-10-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ohkawara S, Furuya H, Nagashima K, Asanuma N, Hino T. Effect of oral administration of Butyrivibrio fibrisolvens MDT-1 on experimental enterocolitis in mice. Clin Vaccine Immunol. 2006;13:1231–1236. doi: 10.1128/CVI.00267-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ohkawara S, Furuya H, Nagashima K, Asanuma N, Hino T. Oral administration of Butyrivibrio fibrisolvens, a butyrate-producing bacterium, decreases the formation of aberrant crypt foci in the colon and rectum of mice. J Nutr. 2005;135:2878–2883. doi: 10.1093/jn/135.12.2878. [DOI] [PubMed] [Google Scholar]
- 6.Creevey CJ, Kelly WJ, Henderson G, Leahy SC. Determining the culturability of the rumen bacterial microbiome. Microb Biotechnol. 2014;7:467–479. doi: 10.1111/1751-7915.12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Teather RM. Isolation of plasmid DNA from Butyrivibrio fibrisolvens. Appl Environ Microbiol. 1982;43:298–302. doi: 10.1128/aem.43.2.298-302.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andrews S. FastQC: a quality control tool for high throughput sequence data. 2010 www.bioinformatics.babraham.ac.uk/projects/fastqc Available online at. [accessed 10 July 2017]
- 9.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Magoč T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vincze T, Posfai J, Roberts RJ. NEBcutter: a program to cleave DNA with restriction enzymes. Nucleic Acids Res. 2003;31:3688–3691. doi: 10.1093/nar/gkg526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo R, Liu B, Xie Y, Li Z, Huang W, et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 2012;1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chattoraj DK, Snyder KM, Abeles AL. P1 plasmid replication: multiple functions of RepA protein at the origin. Proc Natl Acad Sci USA. 1985;82:2588–2592. doi: 10.1073/pnas.82.9.2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harrison PW, Lower RP, Kim NK, Young JP. Introducing the bacterial 'chromid': not a chromosome, not a plasmid. Trends Microbiol. 2010;18:141–148. doi: 10.1016/j.tim.2009.12.010. [DOI] [PubMed] [Google Scholar]
- 15.Aziz RK, Bartels D, Best AA, Dejongh M, Disz T, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu S, Zhu Z, Fu L, Niu B, Li W. WebMGA: a customizable web server for fast metagenomic sequence analysis. BMC Genomics. 2011;12:444. doi: 10.1186/1471-2164-12-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, et al. Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol. 2017;67:1613–1617. doi: 10.1099/ijsem.0.001755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stackebrand E, Ebers J. Taxonomic parameters revisited: tarnished gold standards. Microbiol Today. 2006;8:6–9. [Google Scholar]
- 19.Yarza P, Yilmaz P, Pruesse E, Glöckner FO, Ludwig W, et al. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat Rev Microbiol. 2014;12:635–645. doi: 10.1038/nrmicro3330. [DOI] [PubMed] [Google Scholar]
- 20.Grant JR, Stothard P. The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acids Res. 2008;36:W181–W184. doi: 10.1093/nar/gkn179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lobry JR. A simple vectorial representation of DNA sequences for the detection of replication origins in bacteria. Biochimie. 1996;78:323–326. doi: 10.1016/0300-9084(96)84764-X. [DOI] [PubMed] [Google Scholar]
- 22.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, et al. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 2009;37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yin Y, Mao X, Yang J, Chen X, Mao F, et al. dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2012;40:W445–W451. doi: 10.1093/nar/gks479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ortega-Anaya J, Hernández-Santoyo A. Production of bioactive conjugated linoleic acid by the multifunctional enolase from Lactobacillus plantarum. Int J Biol Macromol. 2016;91:524–535. doi: 10.1016/j.ijbiomac.2016.05.105. [DOI] [PubMed] [Google Scholar]
- 25.Seshadri R, Leahy SC, Attwood GT, Teh KH, Lambie SC, et al. Cultivation and sequencing of rumen microbiome members from the Hungate1000 collection. Nat Biotechnol. 2018;36:359–367. doi: 10.1038/nbt.4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dimarogona M, Topakas E, Christakopoulos P. Cellulose degradation by oxidative enzymes. Comput Struct Biotechnol J. 2012;2:e201209015. doi: 10.5936/csbj.201209015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vecchiarelli AG, Neuman KC, Mizuuchi K. A propagating ATPase gradient drives transport of surface-confined cellular cargo. Proc Natl Acad Sci USA. 2014;111:4880–4885. doi: 10.1073/pnas.1401025111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dri AM, Moreau PL, Rouvière-Yaniv J. Role of the histone-like proteins OsmZ and HU in homologous recombination. Gene. 1992;120:11–16. doi: 10.1016/0378-1119(92)90003-8. [DOI] [PubMed] [Google Scholar]
- 29.Kamashev D, Balandina A, Mazur AK, Arimondo PB, Rouviere-Yaniv J. HU binds and folds single-stranded DNA. Nucleic Acids Res. 2008;36:1026–1036. doi: 10.1093/nar/gkm667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kamashev D, Rouviere-Yaniv J. The histone-like protein HU binds specifically to DNA recombination and repair intermediates. Embo J. 2000;19:6527–6535. doi: 10.1093/emboj/19.23.6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ryan VT, Grimwade JE, Nievera CJ, Leonard AC. IHF and HU stimulate assembly of pre-replication complexes at Escherichia coli oriC by two different mechanisms. Mol Microbiol. 2002;46:113–124. doi: 10.1046/j.1365-2958.2002.03129.x. [DOI] [PubMed] [Google Scholar]
- 32.Li X, Xie Y, Liu M, Tai C, Sun J, et al. oriTfinder: a web-based tool for the identification of origin of transfers in DNA sequences of bacterial mobile genetic elements. Nucleic Acids Res. 2018;46:W229–W234. doi: 10.1093/nar/gky352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rahman T, Yarnall B, Doyle DA. Efflux drug transporters at the forefront of antimicrobial resistance. Eur Biophys J. 2017;46:647–653. doi: 10.1007/s00249-017-1238-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee SW, Glickmann E, Cooksey DA. Chromosomal locus for cadmium resistance in Pseudomonas putida consisting of a cadmium-transporting ATPase and a MerR family response regulator. Appl Environ Microbiol. 2001;67:1437–1444. doi: 10.1128/AEM.67.4.1437-1444.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yonezawa H, Kuramitsu HK. Genetic analysis of a unique bacteriocin, Smb, produced by Streptococcus mutans GS5. Antimicrob Agents Chemother. 2005;49:541–548. doi: 10.1128/AAC.49.2.541-548.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nielsen H. Predicting secretory proteins with SignalP. In: Kihara D, editor. Protein Function Prediction (Methods in Molecular Biology) vol. 1611. New York, NY: Humana Press; 2017. pp. 59–73. (editor) [DOI] [PubMed] [Google Scholar]
- 37.Lacasse M, Zamble D. [NiFe]-hydrogenase maturation biochemistry. 2016;55:1689–1701. doi: 10.1021/acs.biochem.5b01328. [DOI] [PubMed] [Google Scholar]
- 38.van Lingen HJ, Plugge CM, Fadel JG, Kebreab E, Bannink A, et al. Thermodynamic driving force of hydrogen on rumen microbial metabolism: a theoretical investigation. PLoS One. 2016;11:e0161362. doi: 10.1371/journal.pone.0161362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Allali N, Afif H, Couturier M, van Melderen L. The highly conserved TldD and TldE proteins of Escherichia coli are involved in microcin B17 processing and in CcdA degradation. J Bacteriol. 2002;184:3224–3231. doi: 10.1128/JB.184.12.3224-3231.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palevich N, Kelly WJ, Leahy SC, Altermann E, Rakonjac J, et al. The complete genome sequence of the rumen bacterium Butyrivibrio hungatei MB2003. Stand Genomic Sci. 2017;12:72. doi: 10.1186/s40793-017-0285-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kelly WJ, Leahy SC, Altermann E, Yeoman CJ, Dunne JC, et al. The glycobiome of the rumen bacterium Butyrivibrio proteoclasticus B316T highlights adaptation to a polysaccharide-rich environment. PLoS One. 2010;5:e11942. doi: 10.1371/journal.pone.0011942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O'Connell KJ, Motherway MO, Hennessey AA, Brodhun F, Ross RP, et al. Identification and characterization of an oleate hydratase-encoding gene from Bifidobacterium breve. Bioengineered. 2013;4:313–321. doi: 10.4161/bioe.24159. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.