Abstract
Although aneuploidy usually results in severe abnormalities in multicellular eukaryotes, recent data suggest that it could be beneficial for unicellular eukaryotes, such as yeast and trypanosomatid parasites, providing increased survival under stressful conditions. Among characterized trypanosomatids, Trypanosoma cruzi, Trypanosoma brucei and species from the genus Leishmania stand out due to their importance in public health, infecting around 20 million people worldwide. The presence of aneuploidies in T. cruzi and Leishmania was recently confirmed by analysis based on next generation sequencing (NGS) and fluorescence in situ hybridization, where they have been associated with adaptation during transmission between their insect vectors and mammalian hosts and in promoting drug resistance. Although chromosomal copy number variations (CCNVs) are present in the aforementioned species, PFGE and fluorescence cytophotometry analyses suggest that aneuploidies are absent from T. brucei. A re-evaluation of CCNV in T. b gambiense based on NGS reads confirmed the absence of aneuploidies in this subspecies. However, the presence of aneuploidies in the other two T. brucei subspecies, T. b. brucei and T. b. rhodesiense, has not been evaluated using NGS approaches. In the present work, we tested for aneuploidies in 26 T. brucei isolates, including samples from the three T. brucei subspecies, by both allele frequency and read depth coverage analyses. These analyses showed that none of the T. brucei subspecies presents aneuploidies, which could be related to differences in the mechanisms of DNA replication and recombination in these parasites when compared with Leishmania.
Keywords: Trypanosoma brucei, aneuploidy, genome, chromosomal copy number variation, genome replication
Data Summary
All read libraries used in this work are listed and characterized in Table S1. All were downloaded from NCBI Sequence Read Archive.
Impact Statement.
Aneuploidy, the gain or loss of copies of chromosomes, is usually detrimental. For instance, in humans, an extra copy of chromosome 21 results in Down syndrome, a number of conditions arise from imbalanced numbers of sexual chromosomes, and aneuploidies are associated with many types of tumours. On the other hand, some single-celled microbes, such as yeast and trypanosomatid parasites, the latter an important group of pathogens that infect around 20 million people worldwide, appear to rely on aneuploidy as a mechanism to allow adaptation to changing environments, such as different hosts and drug exposure. Surprisingly, among characterized trypanosomatid parasites, some species present aneuploidies, while others do not. Therefore, comparing the presence of aneuploidy in members of the trypanosomatid family with processes associated with genomic stability could reveal cellular mechanisms that lead to aneuploidy and allow it to be tolerated. Understanding the variations in ploidy in these organisms could provide insights into important processes that affect the infectivity of parasites (and other pathogens) and contribute to better knowledge of the cellular processes that dictate the stability and propagation of genomes in all cells.
Introduction
Trypanosoma brucei, a protozoan parasite from the family Trypanosomatidae, is the causative agent of sleeping sickness, an endemic disease in 36 sub-Saharan African countries [1]. Other members of this family include parasites of medical relevance such as Trypanosoma cruzi and Leishmania, the aetiological agents of Chagas disease and leishmaniasis, respectively. T. brucei is subdivided into three subspecies, T. brucei gambiense, T. brucei rhodesiense and T. brucei brucei. T. b. gambiense is responsible for ~90 % of reported human cases of infection and is mainly found in central and western Africa, whereas T. b. rhodesiense is primarily found in eastern and southern Africa [1, 2]. T. b. brucei is usually restricted to non-human animal infections, although human cases have been reported [3].
As cytogenetic analyses are hampered in trypanosomatids by the lack of chromosome condensation during mitosis, karyotype studies in these parasites were initially evaluated by PFGE and fluorescence cytophotometry [4–8]. The development of next generation sequencing (NGS) methodologies and the availability of chromosomal-level assemblies of protozoans from the family Trypanosomatidae [9–12] have enabled a re-evaluation of chromosomal copy number variation (CCNV) occurrence based on read depth coverage (RDC) and allele frequency [13–16]. This new methodology was able to confirm the presence of mosaic aneuploidy in Leishmania, explaining the non-stoichiometric staining intensities of different chromosomal bands observed in PFGE analysis [4]. CCNV was observed in several species of the genus Leishmania, where the pattern of aneuploidies varies among species and even within a population [13, 14, 17–19]. Aneuploidies were shown to greatly impact the phenotype of the parasite, altering gene expression levels, promoting drug resistance and influencing host interchange adaptations [20–22]. Therefore, while aneuploidy is usually lethal or results in severe abnormalities in multicellular eukaryotes [23–25], it could provide rapid adaptation to stressful conditions in unicellular eukaryotes. Chromosomal gain/loss events were also observed among and within T. cruzi discrete typing units (DTUs) [15, 26], suggesting that aneuploidies are a common event in trypanosomatids.
In contrast to what has been observed for the genus Leishmania and T. cruzi DTUs, PFGE and fluorescence cytophotometry analyses suggest that T. brucei is mainly diploid [5, 6, 8, 27–29]. Although triploid T. brucei parasites have been observed following experimental crossing [30, 31], no triploid T. brucei isolates have been previously reported from the field. From the three subspecies, only T. b. gambiense ploidy has been evaluated based on whole genome sequencing and RDC, and no aneuploidies were detected [32]. However, the occurrence of aneuploidies in T. b. brucei and T. b. rhodesiense has not been evaluated using NGS approaches. Therefore, the objective of this work was to estimate CCNV in all three T. brucei subspecies using RDC analysis, compare this with what has already been reported for other trypanosomatids and correlate the pattern of aneuploidy with cellular processes associated with genomic stability.
Methods
Read libraries and processing
A total of 27 T. brucei whole genome read libraries were downloaded from the NCBI Sequence Read Archive (SRA). These included four samples from T. b. brucei, two from T. b. gambiense and 21 from T. b. rhodesiense (Table S1, available in the online version of this article). The reads from each library were submitted to a quality check using the FASTQc tool (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Read libraries were filtered using Trimmomatic [33], with a minimum threshold of phred quality 20 and a minimal length of 40 nucleotides.
Read mapping
The 27 quality trimmed read libraries were mapped on the 11 chromosomal-size T. b. brucei 927 version 28 scaffolds (TritrypDB, http://tritrypdb.org/tritrypdb/), using the BWA-mem software [34, 35]. The mapped reads were trimmed based on a mapping quality of 30 using SAMtools v1.1 [36]. The percentage of mapped reads was estimated with SAMtools flagstat, where only libraries in which more than 70 % of the reads mapped were used (Table S1).
Chromosomal copy number variation estimations
Two methodologies were used to evaluate the occurrence of CCNV in T. brucei: allele frequency of heterozygous positions and RDC analysis.
To evaluate CCNV based on allele frequency, we performed SNP calling using GATK v3.3 [37, 38] in each read library mapped to the T. b. brucei 927 reference genome. Initially, duplicated reads were marked using Picard v1.119 (https://github.com/broadinstitute/picard). Illumina reads were then re-aligned with GATK RealignerTargetCreator and SNP records were obtained with GATK HaplotypeCaller, with minimal confidence threshold for calling of 30 (Phred scale) and minimum threshold confidence for emission of 10 (Phred scale). Next, GATK SelectVariants was used to report only SNPs, excluding insertion/deletions, and VCFfilter was used to select SNP positions with read depth of at least 10 and quality higher than 10. Next, the heterozygous SNP positions in all genes, excluding variant surface glycoproteins (VSGs) and expression site associated genes (ESAGs), were retrieved using in-house Perl scripts, where only SNPs with a support of at least five reads in each variant allele were reported. For each chromosome, the proportion of read depth in alleles in each variant of a predicted heterozygous site was obtained and rounded to the second decimal place, ranging from 0.01 to 1.00, and an approximate distribution of base frequencies for each chromosome was obtained based on Perl scripts and plotted in R (www.r-project.org, R Development 2010). To estimate the overall ploidy of each genome, the same methodology was applied, but the heterozygous positions of all coding sequences (CDS) from all chromosomes were computed simultaneously. Disomic chromosomes are expected to have a peak of 0.50, while trisomic chromosomes are expected to have peaks of 0.33 and 0.66, and tetrasomic chromosomes a combination of 0.25, 0.50 and 0.75 peaks.
The estimation of ploidy variation based on RDC assumes that if the median RDC of a chromosome is higher or lower than the median RDC of the whole genome, this could represent chromosomal gains or losses, respectively. To that end, the chromosomal copy numbers were estimated by the median of dc/dg in a chromosome using in-house Perl scripts and BEDtools, where dc represents the median RDC of all genes in a given chromosome, excluding the multigene VSG and ESAG families, and dg corresponds to the median genome coverage. Initially, the median RDC of all genes, excluding VSGs and ESAGs, for each chromosome was obtained. Next, genes with a coverage lower than 50 % of the gene length were excluded. Then, genes with outlier coverage for each chromosome were excluded, based on an iterative Grubb’s test, with P<0.05. Finally, the median and quantile coverages of each chromosome were plotted in boxplots using R. Median values close to 1 confirm that the chromosomal somy is close to the genome ploidy. This methodology is similar to the one used by Downing et al. [14] for Leishmania, Reis-Cunha et al. [15] for T. cruzi and Tihon et al. for Trypanosoma congolense [39]. Genome coverage was estimated based on the median RDC of all genes in the genome, excluding VSGs and ESAGs, using Perl scripts.
Principal component analysis (PCA) and maximum-likelihood phylogeny of the T. brucei SNPs
To estimate the distance among the 26 T. brucei samples based on whole genome differential SNPs, a consensus nuclear genomic sequence was generated for each sample, using GATK FastaAlternateReferenceMaker (https://software.broadinstitute.org/gatk/documentation/tooldocs/current/org_broadinstitute_gatk_tools_walkers_fasta_FastaAlternateReferenceMaker.php). This genomic sequence was used as input to generate PCA plots and maximum-likelihood phylogeny estimations.
To generate the PCA plot, a distance matrix based on differential SNPs was generated and loaded in the R caret package (http://topepo.github.io/caret/index.html). To evaluate the maximum-likelihood phylogeny, the best-fitting nucleotide substitution model for the phylogenetic analysis was determined using Jmodeltest [40]. The maximum-likelihood phylogenetic tree was built using PhyML [41], with the Generalized Time Reversible model with 1000 bootstrap replicates, proportion of invariable sites of 0.96 and gamma distribution of 0.56. The final phylogenetic tree images were built using FigTree v.1.4.2 software (http://tree.bio.ed.ac.uk/software/figtree/).
Results and Discussion
Chromosomal copy number evaluations
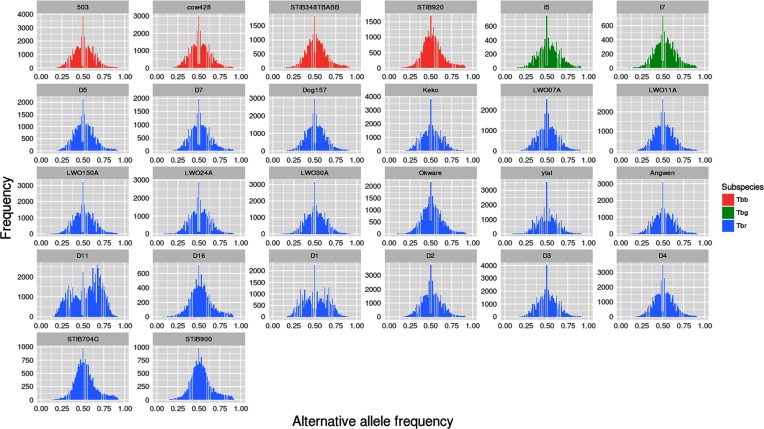
From the 27 T. brucei read libraries evaluated in this work, 26 presented with more than 70 % of the trimmed reads mapping to the 11 T. b. brucei 927 chromosomal-size scaffolds and were used in the parasite ploidy estimations (Table S1). Initially, the overall genome ploidy of each T. brucei strain/isolate was estimated based on the allele frequency of heterozygous positions in all non-VSG and non-ESAG genes, comprising 9295 genes (Fig. 1, Table S2). Based on this analysis, 25 samples presented a single mode of 0.5, suggesting that they are mainly diploid, while T. b. rhodesiense isolate D11 had modes of ~0.33, ~0.5 and ~0.66, suggesting mixed disomic/trisomic chromosomes, or that isolate D11 corresponds to a mixed infection, with more than one parasite population in the same isolate (Fig. 1). This isolate clustered together with other T. b. rhodesiense isolates in a PCA based on SNP data (Fig. S1). Although mainly diploid, T. brucei isolate D1 also presented discrete peaks of ~0.33 and ~0.66, suggesting that a part of its population could also be triploid. The absence of any strong evidence for triploidy in D1 could be due to the lower genome coverage when compared to D11 (Table S1 and Fig. S2).
Fig. 1.
T. brucei subspecies whole genome allele frequency ratio. In each image, the x-axis denotes the allele frequency ratio of heterozygous positions from 0 (0 %) to 1 (100 %), while the y-axis denotes the occurrence of that allele frequency in the genome. An allele frequency ratio peak of 0.5 denotes that the majority of heterozygous positions in the genome had a similar depth of coverage for both alleles, suggesting diploidy. Peaks of 0.33 and/or 0.66 suggest triploidy, and peaks of 0.25, 0.5 and 0.75 suggest tetraploidy. T. b. brucei samples are in red, T. b. gambiense in green and T. b. rhodesiense in blue.
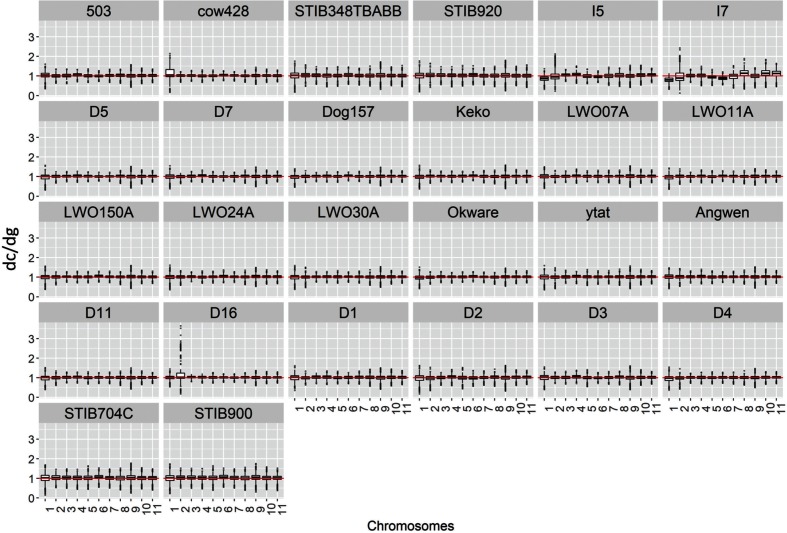
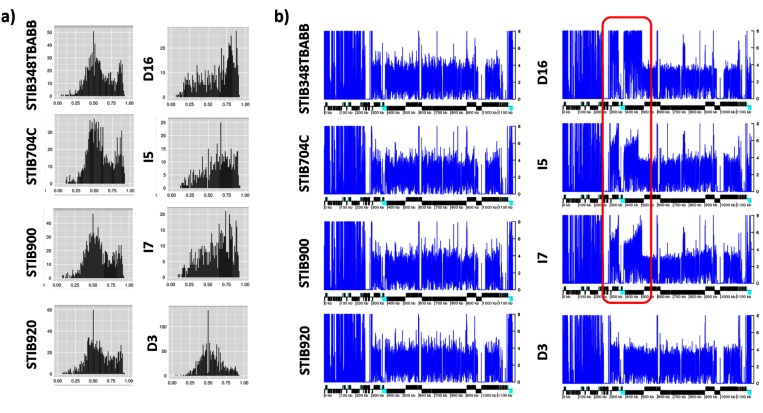
An evaluation of the somy of each chromosome from each isolate by RDC revealed that the chromosomal somy is in accordance with the whole genome ploidy, where the median RDC of each chromosome was similar to the median genome coverage, even for isolate D11 (Fig. 2, and Table S3). Similar results were obtained based on chromosomal somy estimations by allele frequency (Fig. S3). The only exceptions were for chromosome 2 from isolates D16, I5 and I7, which showed a major skewed allele frequency ratio, and isolates STIB348TBABB, STIB704C, STIB900 and STIB920, which presented a minor skewed allele frequency ratio (Fig. 3a). The evaluation of the RDC variations alongside each chromosome (Fig. S4) revealed a large segmental duplication in chromosome 2 from isolates D16, I5 and I7, which was not observed in the other T. brucei isolates (Fig. 3b). Interestingly, I5 and I7 are T. b. gambiense isolates, while D16 was previously classified as a T. b. rhodesiense isolate [42]. In our PCA evaluations (Fig. S1) as well as maximum-likelihood phylogenetic analysis (Fig. S5), isolate D16 clustered together with I5 and I7, suggesting that it could actually be a T. b. gambiense isolate. Alternatively, D16 could actually be a T. b. rhodesiense isolate and the three T. brucei subspecies may not be monophyletic as previously suggested [42].
Fig. 2.
T. brucei subspecies chromosome somy estimated based on dc/dg. In each image, the y-axis corresponds to a boxplot of the median coverage of all genes in a chromosome normalized by genome coverage, where the median value corresponds to the chromosome-predicted somy. Each bar on the x-axis represents a T. brucei chromosome, numbered from 1 to 11. A median dc/dg value of ~1 means that the chromosomally estimated copy number was similar to the genome ploidy.
Fig. 3.
T. brucei chromosome 2 allele frequency ratio and segmental duplication. (a) Allele frequency ratio distribution for chromosome 2. The x-axis denotes the allele frequency ratio of heterozygous positions from 0 (0 %) to 1 (100 %), while the y-axis denotes the occurrence of that allele frequency in chromosome 2. (b) In this image, the blue line corresponds to the normalized RDC of each position in a chromosome, estimated by the ratio between the RDC and the genome coverage. Below, the protein-coding genes are depicted as rectangles drawn proportional to their length, and their coding strand is indicated by their position above (top strand) or below (bottom strand) the central line. Cyan boxes represent VSGs and ESAGs. Black boxes represent all other genes. Segmental duplications not located in sub-telomeric regions are highlighted by a red box. Isolates D16, I5 and I7 show a major skewed allele frequency distribution, while STIB348TBABB, STIB704C, STIB900 and STIB920 show a minor skewed allele frequency distribution. Isolate D3 was added as an example of a disomic chromosome.
These combined results confirm an overall absence of aneuploidies in all three T. brucei subspecies, as previously observed by RDC analysis in T. b. gambiense [32] and by PFGE and fluorescence cytophotometry analyses for the three subspecies [6, 27]. The only isolate with a non-diploid pattern in our analysis, T. b. rhodesiense D11, presents all its chromosomes in the same polysomic state (Fig. S3). Triploid T. brucei parasites have already been obtained in experimental mating in the tsetse fly, suggesting that T. brucei can sustain whole genome aneuploidies in laboratory conditions [30, 31]. However, polyploidy has not yet been documented in T. brucei field isolates [43]. Recent ploidy estimations in 56 Trypanosoma congolense field isolates led to the identification of one triploid lineage, BANANCL2 [39]. This triploid isolate was viable and stable during all the life-cycle stages of the parasite and was efficiently transmitted to mice, resulting in systemic infections [43]. These results suggest that parasites from the Salivarian evolutionary branch, T. brucei and T. congolense, can sustain polysomy, but do not appear to be aneuploid as observed in T. cruzi [15, 26] and in Leishmania [13], where different chromosomes present different somies. CCNV evaluations in other parasites from the Salivaria clade, such as Trypanosoma vivax and Trypanosoma evansi, as well as from protozoans closely related to T. cruzi (e.g. Trypanosoma rangeli, Trypanosoma grayi) and Leishmania (e.g. Crithidia fasciculata, Leptomonas pyrrhicoris) would be valuable to understand this potential dichotomy in genome structure in kinetoplastids.
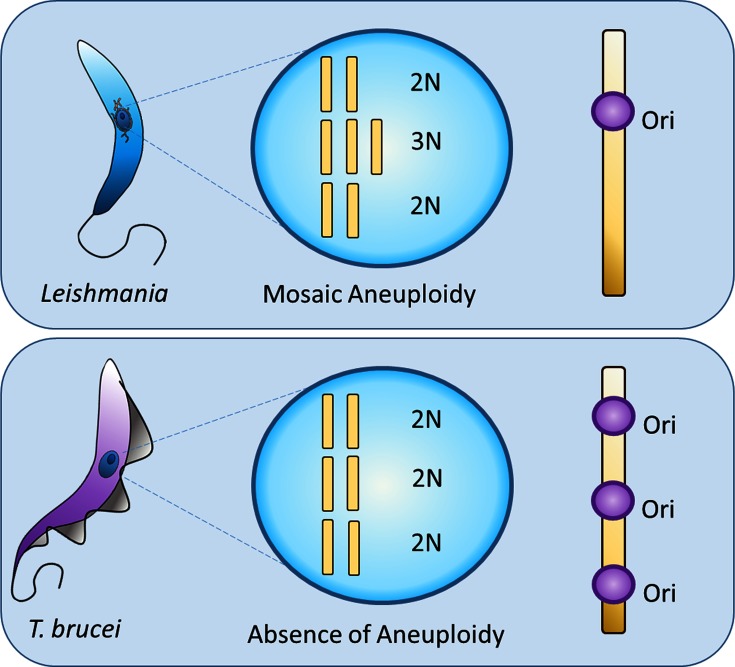
Transcription in trypanosomatids is polycistronic, sharing a profound overlap with DNA replication, where both processes frequently start at strand-switch regions [44, 45]. However, whereas each of the 11 megabase-sized T. brucei chromosomes have several origins of DNA replication [44], the same mapping strategy in Leishmania revealed either a single or a highly preferred origin of replication per chromosome [46]. It is possible that this differing pattern of DNA replication initiation accounts for the distinct ploidies of the two related parasites. For instance, a clash between the transcription and replication machineries at the preferential or singular origin in Leishmania may result in a duplication failure of a given chromosome, which could compromise survival of the parasite. For this reason, the retention of extra chromosomal copies in Leishmania could mitigate against eventual chromosomal loss (Fig. 4). In fact, it has already been suggested that CCNV in Leishmania is generated by asymmetric chromosomal replications, yielding chromosome gains and losses after a number of mitotic generations [47, 48]. The presence of several origins of replication in each T. brucei chromosome would shield this parasite from chromosomal losses due to clashes between the replication and transcription machineries, as DNA replication that emanates from one origin but suffers a blockade could still be completed using replication from another origin [44]. Alternatively, aneuploidies could be generated by recombination events, as observed in the Candida albicans parasexual cycle [49]. In this model, the fusion of parental cells is followed by karyogamy, resulting in a polyploid progeny that undergoes reductional mitotic divisions and genome erosion [49]. Environmental pressures could then select parasite cells that present extra copies of chromosomes whose amplification could be advantageous for parasite survival due to increased copy number of their genes. This mechanism is supported by the subtetraploidy found in T. cruzi experimental hybrids, which present 70 % higher DNA content compared to parental strains [50, 51], and by chromosomal amplification variations as Leishmania migrates from the insect vector to the mammalian host [20].
Fig. 4.
Schematic representation of aneuploidy in trypanosomatids. Parasites from the genus Leishmania that present mosaic aneuploidy have a preferential origin of DNA replication (Ori) in each chromosome, while all three T. brucei subspecies lack aneuploidies and present several origins of replication in each chromosome.
Conclusion
The putative presence of aneuploidies in Leishmania and T. cruzi but not in T. brucei or T. congolense suggests that although evolutionarily related, these parasites present different tolerance to CCNVs. Whether Salivarian parasites lost an ancestral ability to sustain aneuploidy, or whether Leishmania and T. cruzi evolved this mechanism independently is still unknown. The evaluation of CCNV in a larger number of trypanosomatids and kinetoplastids, and correlating this property with DNA replication initiation and recombination, will shed light on the biological mechanisms behind aneuploidy generation in this evolutionary grouping, which will have relevance for all eukaryotes.
Supplementary Data
Funding information
Work in RM’s lab is supported by the BBSRC [BB/K006495/1, BB/M028909/1, BB/N016165/1], the European Commission [RECREPEMLE] and the Wellcome Trust [206815/Z/17/Z]. The Wellcome Centre for Molecular Parasitology, Glasgow, is supported by core funding from the Wellcome Trust [104111]. DCB’s group is supported by Fundação de Amparo a Pesquisa do Estado de Minas Gerais (FAPEMIG), Instituto Nacional de Ciência e Tecnologia de Vacinas (INCTV) – Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES). DCB is a CNPq research fellow. JLRC and LVA received scholarships from CNPq, and GFRL and ACS received scholarships from CAPES.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: CCNV, chromosomal copy number variation; CDS, coding sequence; dc, median read depth coverage of all genes in a chromosome; dg, median genome coverage; ESAG, expression site associated gene; FISH, fluorescence in situ hybridization; NCBI, National Center for Biotechnology Information; NGS, next generation sequencing; PCA, principal component analysis; RDC, read depth coverage; SRA, Sequence Read Archive; VCF, Variant Call Format; VSG, variant surface glycoprotein.
Three supplementary tables and four supplementary figures are available with the online version of this article.
References
- 1.Brun R, Blum J, Chappuis F, Burri C. Human African trypanosomiasis. Lancet. 2010;375:148–159. doi: 10.1016/S0140-6736(09)60829-1. [DOI] [PubMed] [Google Scholar]
- 2.Malvy D, Chappuis F. Sleeping sickness. Clin Microbiol Infect. 2011;17:986–995. doi: 10.1111/j.1469-0691.2011.03536.x. [DOI] [PubMed] [Google Scholar]
- 3.Deborggraeve S, Koffi M, Jamonneau V, Bonsu FA, Queyson R, et al. Molecular analysis of archived blood slides reveals an atypical human Trypanosoma infection. Diagn Microbiol Infect Dis. 2008;61:428–433. doi: 10.1016/j.diagmicrobio.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 4.Bastien P, Blaineau C, Pages M. Leishmania: sex, lies and karyotype. Parasitol Today. 1992;8:174–177. doi: 10.1016/0169-4758(92)90016-U. [DOI] [PubMed] [Google Scholar]
- 5.Gibson WC, Osinga KA, Michels PA, Borst P. Trypanosomes of subgenus Trypanozoon are diploid for housekeeping genes. Mol Biochem Parasitol. 1985;16:231–242. doi: 10.1016/0166-6851(85)90066-0. [DOI] [PubMed] [Google Scholar]
- 6.Hope M, Macleod A, Leech V, Melville S, Sasse J, et al. Analysis of ploidy (in megabase chromosomes) in Trypanosoma brucei after genetic exchange. Mol Biochem Parasitol. 1999;104:1–9. doi: 10.1016/S0166-6851(99)00103-6. [DOI] [PubMed] [Google Scholar]
- 7.Tait A. Evidence for diploidy and mating in trypanosomes. Nature. 1980;287:536–538. doi: 10.1038/287536a0. [DOI] [PubMed] [Google Scholar]
- 8.Borst P, Fase-Fowler F, Frasch ACC, Hoeijmakers JHJ, Weijers PJ. Characterization of DNA from Trypanosoma brucei and related trypanosomes by restriction endonuclease digestion. Mol Biochem Parasitol. 1980;1:221–246. doi: 10.1016/0166-6851(80)90030-4. [DOI] [Google Scholar]
- 9.El-Sayed NM, Myler PJ, Blandin G, Berriman M, Crabtree J, et al. Comparative genomics of trypanosomatid parasitic protozoa. Science. 2005;309:404–409. doi: 10.1126/science.1112181. [DOI] [PubMed] [Google Scholar]
- 10.El-Sayed NM, Myler PJ, Bartholomeu DC, Nilsson D, Aggarwal G, et al. The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science. 2005;309:409–415. doi: 10.1126/science.1112631. [DOI] [PubMed] [Google Scholar]
- 11.Berriman M, Ghedin E, Hertz-Fowler C, Blandin G, Renauld H, et al. The genome of the African trypanosome Trypanosoma brucei. Science. 2005;309:416–422. doi: 10.1126/science.1112642. [DOI] [PubMed] [Google Scholar]
- 12.Ivens AC, Peacock CS, Worthey EA, Murphy L, Aggarwal G, et al. The genome of the kinetoplastid parasite, Leishmania major. Science. 2005;309:436–442. doi: 10.1126/science.1112680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rogers MB, Hilley JD, Dickens NJ, Wilkes J, Bates PA, et al. Chromosome and gene copy number variation allow major structural change between species and strains of Leishmania. Genome Res. 2011;21:2129–2142. doi: 10.1101/gr.122945.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Downing T, Imamura H, Decuypere S, Clark TG, Coombs GH, et al. Whole genome sequencing of multiple Leishmania donovani clinical isolates provides insights into population structure and mechanisms of drug resistance. Genome Res. 2011;21:2143–2156. doi: 10.1101/gr.123430.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reis-Cunha JL, Rodrigues-Luiz GF, Valdivia HO, Baptista RP, Mendes TA, et al. Chromosomal copy number variation reveals differential levels of genomic plasticity in distinct Trypanosoma cruzi strains. BMC Genomics. 2015;16:499. doi: 10.1186/s12864-015-1680-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dujardin JC, Mannaert A, Durrant C, Cotton JA. Mosaic aneuploidy in Leishmania: the perspective of whole genome sequencing. Trends Parasitol. 2014;30:554–555. doi: 10.1016/j.pt.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 17.Mannaert A, Downing T, Imamura H, Dujardin JC. Adaptive mechanisms in pathogens: universal aneuploidy in Leishmania. Trends Parasitol. 2012;28:370–376. doi: 10.1016/j.pt.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 18.Valdivia HO, Reis-Cunha JL, Rodrigues-Luiz GF, Baptista RP, Baldeviano GC, et al. Comparative genomic analysis of Leishmania (Viannia) peruviana and Leishmania (Viannia) braziliensis. BMC Genomics. 2015;16:715. doi: 10.1186/s12864-015-1928-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lachaud L, Bourgeois N, Kuk N, Morelle C, Crobu L, et al. Constitutive mosaic aneuploidy is a unique genetic feature widespread in the Leishmania genus. Microbes Infect. 2014;16:61–66. doi: 10.1016/j.micinf.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 20.Dumetz F, Imamura H, Sanders M, Seblova V, Myskova J, et al. Modulation of aneuploidy in Leishmania donovani during adaptation to different in vitro and in vivo environments and its impact on gene expression. MBio. 2017;8:1–14. doi: 10.1128/mBio.00599-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prieto Barja P, Pescher P, Bussotti G, Dumetz F, Imamura H, et al. Haplotype selection as an adaptive mechanism in the protozoan pathogen Leishmania donovani. Nat Ecol Evol. 2017;1:1961–1969. doi: 10.1038/s41559-017-0361-x. [DOI] [PubMed] [Google Scholar]
- 22.Iantorno SA, Durrant C, Khan A, Sanders MJ, Beverley SM, et al. Gene Expression in Leishmania Is Regulated Predominantly by Gene Dosage. MBio. 2017;8:e01393-17. doi: 10.1128/mBio.01393-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Torres EM, Williams BR, Amon A. Aneuploidy: cells losing their balance. Genetics. 2008;179:737–746. doi: 10.1534/genetics.108.090878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hassold T, Hunt P. To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet. 2001;2:280–291. doi: 10.1038/35066065. [DOI] [PubMed] [Google Scholar]
- 25.Stankiewicz P, Lupski JR. Structural variation in the human genome and its role in disease. Annu Rev Med. 2010;61:437–455. doi: 10.1146/annurev-med-100708-204735. [DOI] [PubMed] [Google Scholar]
- 26.Minning TA, Weatherly DB, Flibotte S, Tarleton RL. Widespread, focal copy number variations (CNV) and whole chromosome aneuploidies in Trypanosoma cruzi strains revealed by array comparative genomic hybridization. BMC Genomics. 2011;12:139. doi: 10.1186/1471-2164-12-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Borst P, van der Ploeg M, van Hoek JF, Tas J, James J. On the DNA content and ploidy of trypanosomes. Mol Biochem Parasitol. 1982;6:13–23. doi: 10.1016/0166-6851(82)90049-4. [DOI] [PubMed] [Google Scholar]
- 28.Tait A. Evidence for diploidy and mating in trypanosomes. Nature. 1980;287:536–538. doi: 10.1038/287536a0. [DOI] [PubMed] [Google Scholar]
- 29.Tait A, Turner CM, Le Page RW, Wells JM. Genetic evidence that metacyclic forms of Trypanosoma brucei are diploid. Mol Biochem Parasitol. 1989;37:247–255. doi: 10.1016/0166-6851(89)90156-4. [DOI] [PubMed] [Google Scholar]
- 30.Gibson W, Garside L, Bailey M. Trisomy and chromosome size changes in hybrid trypanosomes from a genetic cross between Trypanosoma brucei rhodesiense and T. b. brucei. Mol Biochem Parasitol. 1992;51:189–199. doi: 10.1016/0166-6851(92)90069-V. [DOI] [PubMed] [Google Scholar]
- 31.Gibson W, Peacock L, Ferris V, Williams K, Bailey M. The use of yellow fluorescent hybrids to indicate mating in Trypanosoma brucei. Parasit Vectors. 2008;1:4–15. doi: 10.1186/1756-3305-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weir W, Capewell P, Foth B, Clucas C, Pountain A, et al. Population genomics reveals the origin and asexual evolution of human infective trypanosomes. Elife. 2016;5:1–14. doi: 10.7554/eLife.11473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv Prepr arXiv. 2013 [Google Scholar]
- 36.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmidt S. The genome analysis toolkit: a mapreduce framework for analyzing next-generation DNA sequencing data. Proc Int Conf Intellect Capital, Knowl Manag Organ Learn. 2009;20:254–260. [Google Scholar]
- 38.Mckenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tihon E, Imamura H, Dujardin JC, van den Abbeele J, van den Broeck F. Discovery and genomic analyses of hybridization between divergent lineages of Trypanosoma congolense, causative agent of Animal African Trypanosomiasis. Mol Ecol. 2017;26:6524–6538. doi: 10.1111/mec.14271. [DOI] [PubMed] [Google Scholar]
- 40.Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 41.Guindon S, Delsuc F, Dufayard JF, Gascuel O. Estimating maximum likelihood phylogenies with PhyML. Methods Mol Biol. 2009;537:113–137. doi: 10.1007/978-1-59745-251-9_6. [DOI] [PubMed] [Google Scholar]
- 42.Richardson JB, Lee KY, Mireji P, Enyaru J, Sistrom M, et al. Genomic analyses of African Trypanozoon strains to assess evolutionary relationships and identify markers for strain identification. PLoS Negl Trop Dis. 2017;11:e0005949. doi: 10.1371/journal.pntd.0005949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tihon E, Imamura H, Dujardin JC, van den Abbeele J. Evidence for viable and stable triploid Trypanosoma congolense parasites. Parasit Vectors. 2017;10:1–8. doi: 10.1186/s13071-017-2406-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tiengwe C, Marcello L, Farr H, Dickens N, Kelly S, et al. Genome-wide analysis reveals extensive functional interaction between DNA replication initiation and transcription in the genome of Trypanosoma brucei. Cell Rep. 2012;2:185–197. doi: 10.1016/j.celrep.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marques CA, Mcculloch R. Conservation and variation in strategies for DNA replication of kinetoplastid nuclear genomes. Curr Genomics. 2018;19:98–109. doi: 10.2174/1389202918666170815144627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marques CA, Dickens NJ, Paape D, Campbell SJ, Mcculloch R, et al. Genome-wide mapping reveals single-origin chromosome replication in Leishmania, a eukaryotic microbe. Genome Biol. 2015;16:230. doi: 10.1186/s13059-015-0788-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sterkers Y, Lachaud L, Crobu L, Bastien P, Pagès M. FISH analysis reveals aneuploidy and continual generation of chromosomal mosaicism in Leishmania major. Cell Microbiol. 2011;13:274–283. doi: 10.1111/j.1462-5822.2010.01534.x. [DOI] [PubMed] [Google Scholar]
- 48.Sterkers Y, Crobu L, Lachaud L, Pagès M, Bastien P. Parasexuality and mosaic aneuploidy in Leishmania: alternative genetics. Trends Parasitol. 2014;30:429–435. doi: 10.1016/j.pt.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 49.Bennett RJ. The parasexual lifestyle of Candida albicans. Curr Opin Microbiol. 2015;28:10–17. doi: 10.1016/j.mib.2015.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Messenger LA, Miles MA. Evidence and importance of genetic exchange among field populations of Trypanosoma cruzi. Acta Trop. 2015;151:150–155. doi: 10.1016/j.actatropica.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lewis MD, Llewellyn MS, Gaunt MW, Yeo M, Carrasco HJ, et al. Flow cytometric analysis and microsatellite genotyping reveal extensive DNA content variation in Trypanosoma cruzi populations and expose contrasts between natural and experimental hybrids. Int J Parasitol. 2009;39:1305–1317. doi: 10.1016/j.ijpara.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.