Abstract
Objective:
The aim of this study was to investigate the BRCA1 promoter methylation and clinicopathological characteristics in sporadic breast cancer patients in Indonesia.
Methods:
In this cohort study, we selected 90 patients with stage I-III who had definitive surgery at our institution in 2011-2014. Demographic and clinical data regarding pathological stage, breast cancer treatment, outcome etc. were collected from the medical records. Twelve patients had incomplete information on follow up and 18 samples had insufficient tissues for the experiment. Sixty patients with adequate cancer tissues and complete follow up record were analyzed, only 56 patients were analyzed because 4 samples mRNA expression could not be detected. The Mann–Whitney U tests for non-normally distributed groups were used to compare the levels expression of BRCA1 mRNA between methylated and non-methylated samples. Chi-square tests were used to compare methylation status, BRCA1 mRNA expression and clinicopathological characteristics. P value < 0.05 was considered as statistically significant correlation. Data analysis was held by using the GraphPad PRISM 7 (GraphPad Software Inc., USA).
Results:
DNA and RNA were isolated from primary tumor tissues of 56 breast cancer patients. BRCA1 promoter methylation was detected in 48 of 56 patients (85%). Level of BRCA1 mRNA expression was associated with decreased methylation level in the BRCA1 promoter regions suggesting the role of epigenetic silencing. However, there was no statistically significant association among methylation levels, BRCA1 mRNA transcript level with clinicopathological factors.
Conclusion:
To our knowledge, this is the first study investigating methylation status and level of BRCA1 mRNA transcripts among breast cancer patients in Indonesia. We found that the prevalence of BRCA1 promoter methylation is higher than other studies from different populations. However, further investigation involving larger number of patients is required.
Keywords: Breast cancer, BRCA1, DNA methylation, BRCA1 promoter methylation, gene expression
Introduction
Breast and cervical cancers have the highest cancer prevalence rates that cause the most common cancer-related mortality among women in Indonesia. The incidence of breast cancer tends to increase and it is suggested that it largely contributes to the rise of newly diagnosed cancers among young patients (Youlden et al., 2014; Nindrea et al., 2017). Several studies have shown that the peak age of onset in Indonesia is 45–50 years of age, whereas it is 55–60 years in the West countries. The characteristic of breast cancer among Indonesian patients are more aggressive with lymph node involvement, larger tumor size, higher histological grade, higher mitotic index, higher c-erbB2 and p53 expression and higher MIB-1 proliferation index than West patients (Aryandono et al., 2006; Ng et al., 2010; Prajoko and Aryandono, 2014). Molecular and cellular heterogenesity combinated with sedentary lifestyle, high fat diet and exposure to environmental chemicals have been predicted as the risk factors of breast cancer (Leong et al., 2010; Bhoo-Pathy et al., 2013). Understanding tumor characteristics of breast cancer patients is important to select effective therapies and reduce treatment resistance (Alizadeh et al., 2015; Nindrea et al., 2018).
BRCA1 is a human tumor suppressor gene with play crucial role in repairing DNA damage and has been intensively be a subject of investigation since discovered 1990 (Wooster et al., 1994). BRCA1deficiency caused either by germ-line mutations or by down-regulation of gene expression, leads to tumor formation inappropriate target tissue. Decreased expression of the BRCA1 gene has been contributes in both inherited and sporadic breast cancer, and the magnitude of the decreased is correlated with tumor progression. The lowest level of BRCA1 protein commonly found in high-grade tumor and exhibit higher proliferation rates. There are several possible molecular mechanisms that could lead to permanent decreased of BRCA1 levels in tumor cell including allelic loss of heterozygosity (LOH) and hypermethylation BRCA1 gene promoter region (Mueller and Roskelley, 2002; Venkitaraman, 2002).
It was reported that DNA methylation was the major cause of transcriptional silence of BRCA1, ranging from 13–40% in sporadic breast cancer (Butcher and Rodenhiser, 2007). Several studies has been reported that BRCA1 methylation was correlated with the prognosis of breast cancer in Caucasian patients (Baldwin et al., 2000; Cho et al., 2012). However, the role of BRCA1 methylation is not well characterized in the prognosis of sporadic breast cancers among other populations. In the present study, we determined the methylation status of BRCA1 in 90 Indonesian patients with sporadic breast cancer and investigated whether the BRCA1 methylation was associated with clinical outcomes.
Materials and Methods
Study design and research sample
In this cohort study, the data were collected from Dr. M Djamil General Hospital Padang. We selected 90 patients with stage I-III who had definitive surgery in 2011-2014. Demographic and clinical data regarding pathological stages, breast cancer treatment, outcome etc. were collected from the medical records. Twelve patients had incomplete follow up data. Frozen tumor tissue samples were retrieved from the Biobank Faculty of Medicine Universitas Andalas. Eighteen samples had insufficient tissue materials for this experiment. Sixty patients with adequate cancer tissues and complete follow up data were further analyzed. Unfortunately, BRCA1 mRNA expression from 4 samples cannot be detected. Therefore, the final analysis included 56 patients (Figure 1).
Figure 1.
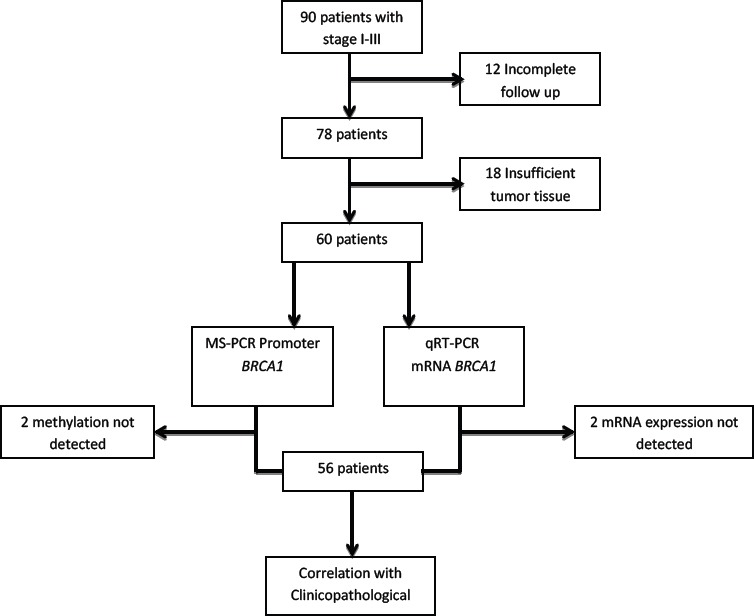
Number of Selected Specimens for Analysis
Ethics statement
The study was approved by the ethical committee board of Faculty of Medicine Universitas Andalas, Padang City, Indonesia. Written informed consent was obtained from all patients.
DNA extraction and bisulfite modification
Genomic DNA was extracted from frozen breast cancer tissues using PureLink® Genomic DNA Mini Kit (Thermo Fisher Scientific, catalog number K182001) according to the manufacturer’s instructions. Genomic DNA concentration and purity was quantified in duplicate using NanoDrop spectrophotometer. DNA samples were stored at -20°C.
Genomic DNA (200-500 ng) from each patient was converted using MethylCodeTM Bisulfite Conversion Kit (Invitrogen, catalog number MECOV-50) according to the manufacturer’s manual instruction. The bisulfite-converted DNA was then stored at -20°C until further analysis.
Methylation-specific PCR
Methylation-Specific PCR was performed using primers specific to methylated DNA sequence in the promoter region of BRCA1 (MF: 5’-TCGTGGTAACGGAAAAGCGC-3’ and MR: 5’-AAATCTCAACGAACTCACGCCG-3’, PCR product size: 75 bp). The unmethylated DNA sequence was amplified using primer specific to unmethylated-bisulfite-converted DNA sequence, which the C’s (cytosines) in the template should be treated as T’s (UF: 5’-TTGGTTTTTGTGGTAATGGAAAAGTGT-3’ and UR: 5’-AAAAAATCTCAACAAACTCACACCA-3’, PCR product size: 86 bp). The methylation and unmethylation-specific primers were adopted from Esteller et al., (2000) and Butcher and Rodenhiser, (2007). These primers annealed in the promoter region and flanked the transcription site of BRCA1 (Figure 2).
Figure 2.
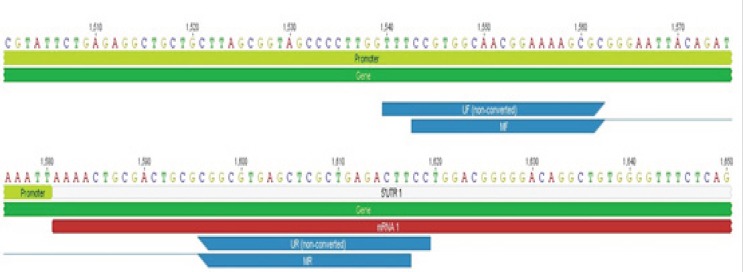
Location of Methylation-Specific Primers (MF-MR) and Unmethylation-Specific Primers (UF-UR) in BRCA1 Sequence (NCBI Accession Number: NG_005905.2)
MS-PCR reactions were performed in 25 μl total volume which contained 400 nM MF primer and 400 nM MR primer (synthesized by Integrated DNA Technologies, Singapore), 1x GoTaq Green Master mix (Promega, catalog number M7122), and 2 μl template of bisulfite-converted DNA. The cycling conditions were initial denaturation at 96°C for 4 minutes followed by 35 cycles of denaturation at 95°C for 50 seconds, annealing at 62°C for 30 seconds and extension at 72°C for 30 seconds, and followed by final extension at 72°C for 5 minutes, using T100 thermocycler (Bio-Rad, USA). The reaction of unmethylated-specific PCR was also performed in different PCR tube with the same reaction except the primer. The PCR products were separated in 2% agarose gel and visualized after staining with GelRed (Biotium).
RNA extraction and cDNA synthesis
Total RNA was extracted from frozen breast cancer tissues, using TRIzol Reagent with the PureLink® RNA Mini Kit (Ambion, Life Technologies, catalog number 15596-026 and 12183018A, respectively) according to the manufacturer’s instructions. Total RNA concentration and purity were quantified in duplicate using NanoDrop spectrophotometer. RNA samples were stored at -80°C.
In total 1 μg RNA was used for cDNA conversion using iScript cDNA synthesis kit (Bio-Rad, catalog number 1708890). The total volume reaction 20 μl was incubated according to the manufacturer’s instruction. The converted cDNA was stored at −20°C.
BRCA1 expression analysis by quantitative real-time reversetranscription-PCR
Quantitative reverse-transcription PCR primers were designed to span an exon-exon junction to reduce the risk of false positives from amplification of any contaminating genomic DNA (Figure 3). BRCA1-Forward (5’-CAGGAGTGGAAAGGTCATC-3’) annealed in exon 13-exon 14 junctions and BRCA1-Reverse (5’-TCCCTCTAGATCTTGCCTT-3’) annealed in exon 14-exon 15 junctions. GAPDH mRNA expression level also quantified as a housekeeping gene for normalization using GAPDH-Forward (5’-CATTGACCTCAACTACATGGTTT -3’) and GAPDH-Reverse (5’-GAAGATGGTGATGGGATTTCC-3’). Both primers were designed using bioinformatics software Geneious 7.0.6.
Figure 3.
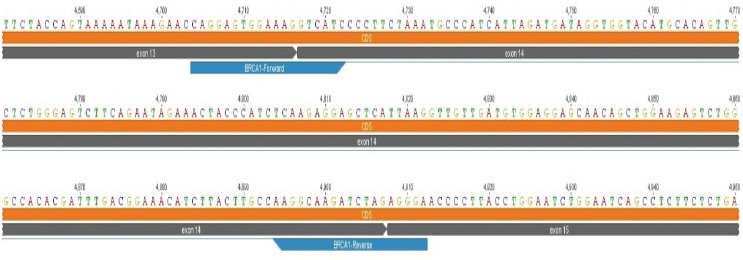
Location of BRCA1 Forward and Reverse Primers in BRCA1 mRNA Sequence (NCBI Accession Number: NM_007294.3)
Real-time PCR was performed in optical strip PCR tubes in duplicate which contained 500 nM BRCA1-Forward and 500 nM BRCA1-Reverse (synthesized by Integrated DNA Technologies, Singapore), 1x Sso Evagreen Supermix (Bio-Rad, catalog number 172-5200), and 2 μl template (cDNA or BRCA1 plasmid standard) in a final volume 20 μl. The cycling conditions were enzyme activation at 95°C for 30 seconds followed by 40 cycles of denaturation at 95°C for 5 seconds, annealing/extension at 52.5°C for 5 seconds, and followed by melt curve from 65°C to 95°C, using CFX96 Touch Real-Time PCR Detection System (Bio-Rad, USA). The BRCA1 standard curve in linear regression was automatically generated by software CFX96 touch system by plotting the quantification cycle (Cq) against the log of copy number, so that the BRCA1 copy number from samples will be known. Quantification cycle (Cq) or threshold cycle (CT) is the number of PCR cycles for which enough fluorescence was detected above background. Real-time PCR for GAPDH also conducted in a different plate with the same reactions above except the primer and the annealing/extension temperature (57.1°C).
Statistical analysis
The Mann–Whitney U test for non-normally distributed groups was used to compare the levels expression of BRCA1 mRNA between methylated and unmethylated samples. The association among methylation status, BRCA1 mRNA expression and clinicopathological characteristics was examined by chi-square test. P value < 0.05 was considered statistically significant correlation. Data analysis was carried out with the GraphPad PRISM 7 (GraphPad Software Inc., USA).
Results
The Methylation-Specific PCR was used to investigate methylation status of BRCA1 promoter. It was successfully performed in 97% (58/60) samples to detect the methylation status of BRCA1 promoter region. The typical results of methylated and unmethylated status were shown in Figure 4. BRCA1 promoter methylation was detected in 48 out of 60 (80%) sporadic breast tumors. The methylation status was not statistically significant associated with any clinicopathological parameters (p > 0.05) (Table 1).
Figure 4.
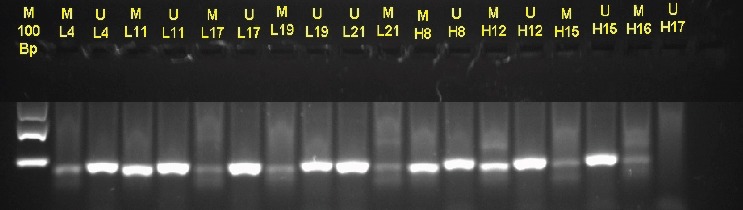
Electrophoresis of Amplification Products Specific Promoter Region BRCA1 from Bisulfite-Treated DNA in Human Tumor Tissue. Each line consists amplification products from methylated (U), 75bp or unmethylated (M), 86bp primer
Table 1.
Association between Methylation Status of BRCA1 and Clinicopathological Characteristics Breast Cancer Patients
| Characteristics | f | (%) | |
|---|---|---|---|
| Age (years) | 47,5 years | 56 | 100 |
| Tumor Size | 8.3 cm | 56 | 100 |
| Clinical stage | Early BC (I-II) | 22 | 39.3 |
| Advance (III-IV) | 34 | 60.7 | |
| Histological grade | I | 1 | 1.8 |
| II | 51 | 91 | |
| III | 4 | 7.2 | |
| Lymph node status | N0 | 19 | 34 |
| N1 | 28 | 50 | |
| N2 | 9 | 16 | |
| ER status | Negative | 33 | 59 |
| Positive | 23 | 41 | |
| PR status | Negative | 39 | 69.6 |
| Positive | 17 | 30.4 | |
| KI67 | < 14% | 28 | 50 |
| > 14% | 28 | 50 | |
| HER2 | HER2 (-) | 35 | 62.5 |
| HER2 (+++) | 21 | 37.5 | |
| TNBC | TNBC | 15 | 26.8 |
| Relapse status | No relapse | 43 | 76.7 |
| Relapse | 13 | 23.3 | |
| Current status | Die | 49 | 87.5 |
| Survive | 7 | 12.5 |
Level of BRCA1 mRNA expression in tissue tumor specimen was successfully detected by using qRT-PCR. The median mRNA expression level of BRCA1 relative to reference gene of GADPH were 2.27 (range 0.014-13.335). There was statistically significant association between methylation status promoter region and mRNA expression level of BRCA1 (p-value 0.042), suggesting epigenetic silencing activities in the BRCA1 promoter region. However there was no statistically significant correlation between levels of mRNA BRCA1 and clinicopathological parameters including estrogen receptor (p=0.839), progesterone receptor (p=0.382), Ki67 (p=0.644), histological grade (p=0.329), stage (p=0.764), tumor size (p=0.148), and lymph node status (p=0.730) (Table 2).
Table 2.
The Relationship between BRCA1 mRNA Expression and Clinicopathological Factors
| Clinicopathology | n | Mean ± SD mRNA BRCA1 |
p-value |
|---|---|---|---|
| Estrogen receptor | |||
| Negative | 33 | 3.43 ± 3.18 | 0.839 |
| Positive | 23 | 3.26 ± 3.11 | |
| Progesterone receptor | |||
| Negative | 39 | 3.61 ± 3.17 | 0.382 |
| Positive | 17 | 2.81 ± 3.03 | |
| KI67 | |||
| < 14% | 28 | 3.56 ± 3.17 | 0.644 |
| > 14% | 28 | 3.17± 3.11 | |
| HER2 | |||
| HER2 (-) | 27 | 3.06 ± 2.92 | 0.840 |
| HER2 (+) | 6 | 3.05 ± 3.10 | |
| HER2 (++) | 2 | 4.54 ± 4.45 | |
| HER2 (+++) | 21 | 3.72 ± 3.44 | |
| TNBC | |||
| Luminal A | 23 | 3.26 ± 3.10 | 0.977 |
| HER2 | 18 | 3.47 ± 3.25 | |
| TNBC | 15 | 3.39 ± 3.20 | |
| Histological Grade | |||
| I | 1 | 7.9 | 0.329 |
| II | 51 | 3.31 ± 3.18 | |
| III | 4 | 3.36 ± 3.12 | |
| Stage | |||
| Stage I | 3 | 2.30 ± 3.12 | 0.764 |
| Stage II | 19 | 3.67 ± 3.36 | |
| Stage III | 34 | 3.28 ± 3.05 | |
| Tumor size | |||
| T1 | 3 | 2.30 ± 3.12 | 0.198 |
| T2 | 11 | 4.79 ± 3.78 | |
| T3 | 10 | 1.95 ± 1.88 | |
| T4 | 32 | 3.42 ± 3.09 | |
| Lymph node status | |||
| N0 | 19 | 3.31 ± 3.28 | 0.730 |
| N1 | 28 | 3.16 ± 2.95 | |
| N2 | 9 | 4.12 ± 3.55 |
There was an inverse relationship between BRCA1 promoter methylation and mRNA transcript levels (p-value< 0,05) (Figure 5).
Figure 5.
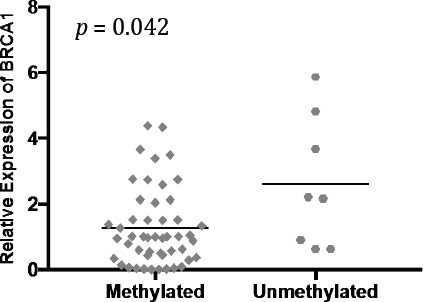
The Relationship between BRCA1 Promoter Methylation and mRNA Transcript Levels. Lines note median expression value, p-value represents Mann-Whitney test.
Discussion
Cancer is resulted from the accumulation of altered genetic regulations which cause abnormal cell growth and expansion. In addition to genetic mutations, epigenetic alteration also play an important role in breast carcingenesis involving changes in DNA methylation (global hypomethylation and locus-specific hypermethylation), altered histone tail modifications patterns and nucleosomal remodeling. Methylation of BRCA1 promoter tumor suppressor gene has been known as one of the gene expression loss mechanisms, and has been identified in 10–30% of sporadic early-onset breast cancer with aggressive pathological features (Esteller et al., 2000; Wei et al., 2005; Matros et al., 2005; Turner et al., 2007).
We report for the first time from Indonesian population, promoter methylation of BRCA1 gene is present in 48 of 60 (80%) patients with early-stage sporadic breast carcinomas. The promoter methylation status is significantly higher than previously reported from other population such as Taiwanese present in 56%, Thailand 24.6%, India 45% but present similar highest status with Vietnamese, 82,1% and France, 89,1% (Bosviel et al., 2012; Hsu et al., 2013; Hasan et al., 2013; Saelee et al., 2014; Truong et al., 2014). Our study supports previous premise that the promoter methylation status is depending on race/ethnicity and population, which also relates to tumor characterization (Wiencke, 2004). The promoter methylation of BRCA1 gene relates to tumor grade III and is frequently found in stage IIB. The data of 536 sporadic breast cancer studied at Chinese population found 26% hypermethylation on BRCA1 promoters has a poor prognosis effect (Chen et al., 2009).
In our study, expression of mRNA in tissue tumor specimen was successfully detected by using qRT-PCR. There was statistically significant association between methylation status promoter region and mRNA expression level of BRCA1 (p-value 0.042), suggesting epigenetic silencing activities in the BRCA1 promoter region. However there was no statistically significant correlation between levels of mRNA BRCA1 and clinicopathological parameters including estrogen receptor, progesterone receptor and expression of KI 67.
Hypermethylation of the BRCA1 gene promoter is present in 56% (78 of 139) of Taiwanese women with early-stage sporadic breast carcinomas, which is significantly higher than previously reported frequencies for this alteration in unselected sporadic breast tumors (Hsu et al., 2013). The result of meta-analysis study showed BRCA1 methylation prevalence in Asians population (OR= 4.03, 95%CI 1.07–15.18, P= 0.04) was higher than in Caucasians populations (OR= 3.16, 95%CI 1.78–5.62, P< 0.001) and in Australians populations (OR= 3.27, 95% CI 1.37–7.84, P= 0.008) in breast cancers compared with non-cancer controls (Zhang and Long, 2015). The prevalence of BRCA1 mutations in a cohort of Nigerian (predominantly Yoruba) breast cancer patients unselected for age of onset or family history. Surprisingly, 31/434 (7.1%) patients carried BRCA1 mutations. This result show consistently lower frequencies of BRCA1 mutations in non-founder populations, and especially low frequencies in African Americans (Fackenthal et al., 2011). Another study examined 1,628 population-based breast cancer cases and showed that mutations occurred in 2.9% of White cases and only 1.4% of Black cases (Malone et al., 2006). Our study supports the role of BRCA1 methylation in the aggressiveness of breast cancer.
In our study, there was no any significant correlation between the hypermethylated BRCA1 promoter and patient’s age, tumor grades, and clinical stages which was similar with a study from Vietnamese breast cancer cohort (Truong et al., 2007). Decreased estrogen receptor expression is found in BRCA1 promoter hypermethylated patients. More than 25% (15/56) patients with BRCA1 promoter methylation were triple-negative breast cancers, even though a significant number of triple-negative breast cancer patients do not carry BRCA1 mutations. Our study finding supported that hypermethylation at BRCA1 promoter region playing a role on etiological of triple-negative phenotype breast cancer (Miyoshi, 2008). Some study indicated that hypermethylation of promoter BRCA1 gen was associated with the pathogenesis of breast-cancer subtype. Breast cancer with have hypermethylation on BRCA1 promoter region are more likely to be of high grade or estrogen-receptor negative, and p53 positive (Johannsson et al., 1997; Hsu et al., 2013). It has been postulated that breast cancer with has hypermethylation on BRCA1 promoter region is more aggressive. The benefits of epigenetic research, especially on gene promoter methylation are linked to the prevention and treatment of breast cancer.
The use of antioxidants as an antimethylation agent can reduce the occurrence of methylation in certain genes especially the proliferative gen and then can prevent the occurrence of breast cancer in the community. Likewise in the treatment of breast cancer, the use of demethylation drugs will be able to reduce the recurrence and mortality in cancer patients (Fleischauer et al., 2003).
This study is the first study involving Indonesian breast cancer. But there are are some limitations of this study including relatively small number of patient cohort, no survival or progression-free survival data comparing hypermethylated and non- methylated, and as focused in sporadic cancer. This study is lack of environmental-associated data including diet, smoking habit, Body Mass Index (BMI), obesity, menarche, hormonal contraception use, and physical activity levels. The genetic and environmental factors are postulated to be playing a role for breast cancer risk. Environmental factors may be associated with the methylation of the promoter regions within tumor suppressor and DNA repair genes BRCA1.
In conclusion, our study found that methylation rates in Indonesian breast cancer women is higher than in the literature, possibly due to advanced cancer stages. The presence of hypermethylation in the BRCA1 promoter does not affect the type of breast cancer. This finding indicates that BRCA1 methylation is involved in the late-stage progression of breast-cancer and can be used as one of the prognostic marker in breast cancer.
Conflict of interest
The authors declare no conflict of interest.
Acknowledgments
The authors would thanks to Syifa’u Warahmah for collecting data and Dessy Arisanti for laboratory analysis.
References
- Alizadeh AA, Aranda V, Bardelli A, et al. Toward understanding and exploiting tumor heterogeneity. Nat Med. 2015;21:846–53. doi: 10.1038/nm.3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aryandono T, Harijadi S. Breast cancer in young women:prognostic factors and clinicopathological features. Asian Pac J Cancer Prev. 2006;7:451–4. [PubMed] [Google Scholar]
- Baldwin RL, Nemeth E, Tran H, et al. BRCA1 promoter region hypermethylation in ovarian carcinoma:a population-based study. Cancer Res. 2000;60:5329–33. [PubMed] [Google Scholar]
- Bhoo-Pathy N, Yip CH, Hartman M, et al. Breast cancer research in Asia:adopt or adapt Western knowledge? Eur J Cancer. 2013;49:703–9. doi: 10.1016/j.ejca.2012.09.014. [DOI] [PubMed] [Google Scholar]
- Bosviel R, Garcia S, Lavediaux G, et al. BRCA1 promoter methylation in peripheral blood DNA was identified in sporadic breast cancer and controls. Cancer Epidemiol. 2012;36:e177–82. doi: 10.1016/j.canep.2012.02.001. [DOI] [PubMed] [Google Scholar]
- Butcher DT, Rodenhiser DI. Epigenetic inactivation of BRCA1 is associated with aberrant expression of CTCF and DNA methyltransferase (DNMT3B) in some sporadic breast tumours. Eur J Cancer. 2007;43:210–9. doi: 10.1016/j.ejca.2006.09.002. [DOI] [PubMed] [Google Scholar]
- Chen Y, Zhou J, Xu Y, et al. BRCA1 promoter methylation associated with poor survival in Chinese patients with sporadic breast cancer. Cancer Sci. 2009;100:1663–7. doi: 10.1111/j.1349-7006.2009.01225.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YH, Shen J, Gammon MD, et al. Prognostic significance of gene-specific promoter hypermethylation in breast cancer patients. Breast Cancer Res Treat. 2012;131:197–205. doi: 10.1007/s10549-011-1712-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M, Silva JM, Dominguez G, et al. Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors. J Natl Cancer Inst. 2000;92:564–9. doi: 10.1093/jnci/92.7.564. [DOI] [PubMed] [Google Scholar]
- Fackenthal JD, Zhang J, Zhang B, et al. High prevalence of BRCA1 and BRCA2 mutations in unselected Nigerian breast cancer patients. Int J Cancer. 2012;131:114–23. doi: 10.1002/ijc.27326. [DOI] [PubMed] [Google Scholar]
- Fleischauer AT, Simonsen N, Arab L. Antioxidant supplements and risk of breast cancer recurrence and breast cancer-related mortality among postmenopausal women. Nutr Cancer. 2003;46:15–22. doi: 10.1207/S15327914NC4601_02. [DOI] [PubMed] [Google Scholar]
- Hasan TN, Leena Grace B, Shafi G, Syed R. Association of BRCA1 promoter methylation with rs11655505 (c.2265C>T) variants and decreased gene expression in sporadic breast cancer. Clin Transl Oncol. 2013;15:555–62. doi: 10.1007/s12094-012-0968-y. [DOI] [PubMed] [Google Scholar]
- Hsu NC, Huang YF, Yokoyama KK, et al. Methylation of BRCA1 promoter region is associated with unfavorable prognosis in women with early-stage breast cancer. PLoS One. 2013;8:e56256. doi: 10.1371/journal.pone.0056256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannson OT, Idvall I, Anderson C, et al. Tumour biological features of BRCA1-induced breast and ovarian cancer. Eur J Cancer. 1997;33:362–71. doi: 10.1016/s0959-8049(97)89007-7. [DOI] [PubMed] [Google Scholar]
- Leong SP, Shen ZZ, Liu TJ, et al. Is breast cancer the same disease in Asian and Western countries? World J Surg. 2010;34:2308–24. doi: 10.1007/s00268-010-0683-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone KE, Daling JR, Doody DR, et al. Prevalence and predictors of BRCA1 and BRCA2 mutations in a population-based study of breast cancer in white and black American women ages 35 to 64 years. Cancer Res. 2006;66:8297–308. doi: 10.1158/0008-5472.CAN-06-0503. [DOI] [PubMed] [Google Scholar]
- Matros E, Wang ZC, Lodeiro G, et al. BRCA1 promoter methylation in sporadic breast tumors:relationship to gene expression profiles. Breast Cancer Res Treat. 2005;91:179–86. doi: 10.1007/s10549-004-7603-8. [DOI] [PubMed] [Google Scholar]
- Miyoshi Y, Murase K, Oh K. Basal-like subtype and BRCA1 dysfunction in breast cancers. Int J Clin Oncol. 2008;13:395–400. doi: 10.1007/s10147-008-0831-x. [DOI] [PubMed] [Google Scholar]
- Mueller CR, Roskelley CD. Regulation of BRCA1 expression and its relationship to sporadic breast cancer. Breast Cancer Res. 2003;5:45–52. doi: 10.1186/bcr557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng CH, Pathy NB, Taib NA, et al. Comparison of breast cancer in Indonesia and Malaysia--a clinico-pathological study between Dharmais Cancer Centre Jakarta and University Malaya Medical Centre, Kuala Lumpur. Asian Pac J Cancer Prev. 2011;12:2943–6. [PubMed] [Google Scholar]
- Nindrea RD, Aryandono T, Lazuardi L. Breast cancer risk from modifiable and non-modifiable risk factors among women in Southeast Asia:a meta-analysis. Asian Pac J Cancer Prev. 2017;18:3201–6. doi: 10.22034/APJCP.2017.18.12.3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nindrea RD, Harahap WA, Aryandono T, Lazuardi L. Association of BRCA1 Promoter methylation with breast cancer in Asia:a meta- analysis. Asian Pac J Cancer Prev. 2018;19:885–9. doi: 10.22034/APJCP.2018.19.4.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prajoko YW, Aryandono T. Expression of nuclear factor kappa B (NF-κB) as a predictor of poor pathologic response to chemotherapy in patients with locally advanced breast cancer. Asian Pac J Cancer Prev. 2014;15:595–8. doi: 10.7314/apjcp.2014.15.2.595. [DOI] [PubMed] [Google Scholar]
- Saelee P, Chaiwerawattana A, Ogawa K, et al. Clinicopathological significance of BRCA1 promoter hypermethylation in Thai breast cancer patients. Asian Pac J Cancer Prev. 2014;15:10585–9. doi: 10.7314/apjcp.2014.15.24.10585. [DOI] [PubMed] [Google Scholar]
- Truong PK, Lao TD, Doan TP, Le TA. BRCA1 promoter hypermethylation signature for early detection of breast cancer in the Vietnamese population. Asian Pac J Cancer Prev. 2014;15:9607–10. doi: 10.7314/apjcp.2014.15.22.9607. [DOI] [PubMed] [Google Scholar]
- Turner NC, Reis-Filho JS, Russell AM, et al. BRCA1 dysfunction in sporadic basal-like breast cancer. Oncogene. 2007;26:2126–32. doi: 10.1038/sj.onc.1210014. [DOI] [PubMed] [Google Scholar]
- Venkitaraman AR. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell. 2002;108:171–82. doi: 10.1016/s0092-8674(02)00615-3. [DOI] [PubMed] [Google Scholar]
- Wei M, Grushko TA, Dignam J, et al. BRCA1 promoter methylation in sporadic breast cancer is associated with reduced BRCA1 copy number and chromosome 17 aneusomy. Cancer Res. 2005;65:10692–9. doi: 10.1158/0008-5472.CAN-05-1277. [DOI] [PubMed] [Google Scholar]
- Wiencke JK. Impact of race/ethnicity on molecular pathways in human cancer. Nat Rev Cancer. 2004;4:79–84. doi: 10.1038/nrc1257. [DOI] [PubMed] [Google Scholar]
- Wooster R, Neuhausen SL, Mangion L, et al. Localization of a breast cancer susceptibility gene, BRCA2, to chromosome 13q12-13. Science. 1994;265:2088–90. doi: 10.1126/science.8091231. [DOI] [PubMed] [Google Scholar]
- Youlden DR, Cramb SM, Yip CH, Baade PD. Incidence and mortality of female breast cancer in the Asia-Pacific region. Cancer Biol Med. 2014;11:101–15. doi: 10.7497/j.issn.2095-3941.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Long X. Association of BRCA1 promoter methylation with sporadic breast cancers:Evidence from 40 studies. Sci Rep. 2015;5:17869. doi: 10.1038/srep17869. [DOI] [PMC free article] [PubMed] [Google Scholar]


