Abstract
Dysregulation in the miRNA-21 expression has been previously observed in a number of malignancies and not only in the tumor cell itself but also in the body fluids of the cancer patients. The present study aimed to find out the clinical significance of cell-free circulating miRNA-21 as an efficient non-invasive biomarker for the screening of lung cancer patients. The present case-control study included plasma samples from 80 lung cancer patients and 80 healthy controls. Magnetic bead technology was used for efficient miRNA isolation and advanced TaqMan miRNA assays were used for the quantification of miRNA-21 level in the plasma of the lung cancer patients and healthy individuals. The overall mean relative expression level of plasma miRNA-21 among lung cancer patients (2.32±1.7) was higher when compared to healthy individuals (0.715 ± 0.48) and it showed a significant difference of p<0.0001. The area under ROC curve was 0.8913 [95% confidence interval (CI): 0.8394 to 0.9431, p< 0.0001] and the sensitivity and specificity were both 80.0% when the cut-off value was 1.207. In conclusion, plasma miRNA-21 can be efficiently extracted by the magnetic bead technology and quantified by the advanced TaqMan miRNA assay. Plasma miRNA-21 showed a high ability to distinguish between lung cancer patients and healthy individuals, therefore can be used as an efficient non-invasive biomarker for the screening of Lung cancer patients.
Keywords: Lung cancer, biomarker, plasma miRNA-21
Introduction
Lung cancer is one the most common malignancies and the leading cause of cancer-related deaths worldwide. Despite the advanced imaging and cytology-based screening strategies employed for early detection of tumors, the majority of the lung cancer cases are diagnosed in late stages which are a determining factor for the high mortality rate for the disease. Therefore, there is an urgent need for the development of an effective screening test for the diagnosis of lung cancer. Many promising non-invasive biomarkers have been observed; however, their clinical use is still limited due to the paucity of validating studies currently available.
One of the prime focuses of the present cancer research is to find out the cell-free circulatory miRNAs as non-invasive biomarkers for the diagnosis and prognosis of solid tumors. MicroRNAs are small noncoding RNA molecules (~22 nucleotides) encoded by specific microRNA genes, which function in negative regulation of gene expression at the post-transcriptional level by binding to the respective mRNA target transcripts at 3′ untranslated region (Castro et al., 2017; Yong et al., 2016). Dysregulation in the expression of various miRNAs has been observed in a number of malignancies including lung cancer (Palanichamy et al., 2014). The dysregulation of the miRNAs has been observed not only in the tumor cell itself but also in the body fluids of the cancer patients, such as plasma, serum, pleural effusion and urine etc. (Zhao et al., 2015;Cappellesso et al., 2016; Kao et al., 2016).
A well know miRNA, the miRNA-21 have been observed to play a pivotal role in the tumor development by regulating various molecular mechanisms such as cell proliferation, metastasis, angiogenesis, and anti-apoptosis etc. (Tao et al., 2015; Yin Hsun et al., 2016). Various studies have observed the dysregulation of miRNA-21 in the circulation of lung cancer patients (Yang et al., 2015; Wei et al., 2015), however, in reality, the clinical significance of circulatory miRNA-21 as a biomarker in Lung cancer is still at its preliminary level. The goal of the current study was to find out the clinical significance of cell-free circulating miRNA-21 as an efficient non-invasive biomarker for the screening of Lung cancer patients.
Materials and Methods
Study Population
The present study was approved by the Research Ethics committee at the University of Tabuk, Ministry of Health, Kingdom of Saudi Arabia and the Institutional Review Board (IRB) of King Hussein Cancer Center, Jordan. The study included two study groups:1) Histopathologically confirmed Lung cancer patients (n=80) and healthy controls (n=80). Study subjects were collected from King Fahd specialist hospital, Tabuk, Saudi Arabia, and King Hussein Cancer Center, Jordan. Blood samples were withdrawn from each participant after obtaining the written informed consent.
Plasma microRNA isolation
Blood samples up to 5 ml were collected in EDTA vials and plasma was isolated by centrifugation at 2,500 rpm for 15 minutes and stored at –80°C until use. MicroRNAs from plasma were extracted by MagMAX™ magnetic-bead technology using MagMAX™ mirVana™ RNA Isolation Kit (Applied Biosystems, USA). Protocol provided by the manufacturer was followed for the miRNA isolation and all the solutions provided in the kit were prepared as per the given instructions. In a processing plate provided in the kit, 100 μL of plasma sample was mixed with 50μL of Protienase K digestion mix and incubated at 65°C for 30 minutes. Followed by addition of 100 μL of lysis binding mix and 20 μL of RNA binding beads to the sample. After proper mixing 270 μL of isopropanol was added and the processing plate was placed on the magnetic stand-96 for 5 minutes or until clear solution was observed. The supernatant was carefully aspirated and discard without disturbing the RNA binding beads. Processing plate was removed from the magnetic stand-96 and 150 μL of wash solution-1 provided in the kit was added to the sample. Processing plate was again placed on the magnetic stand-96 for 1 minute or until the solution is clear and the supernatant was carefully aspirated and discarded without disturbing the RNA binding beads. The above wash step was repeated using 150 μL of wash solution-2 provided in the kit. After drying the RNA binding beads, 50 μL of TURBO DNase™ solution was added to the sample. Sample was properly mixed and 50 μL of rebinding buffer and 100μL of isopropanol was added. The processing plate was placed on the magnetic stand-96 for 5 minutes or until clear solution was observed. The supernatant was carefully aspirated and discarded without disturbing the RNA binding beads. Processing plate was removed from the magnetic stand-96 and 150 μL of wash solution-2 was added to the sample. Processing plate was again placed on the magnetic stand-96 for 1 minute or until the solution is clear and the supernatant was carefully aspirated and discarded without disturbing the RNA binding beads. After drying the RNA binding beads, 50 μL of pre-heated elution buffer was added to the sample and incubated at 65°C for 5 minutes. The processing plate was placed on the magnetic stand-96 for 3 minutes or until the solution is clear. The supernatant containing the miRNA was carefully transfered to the elution plate and stored at –20°C until further process.
Synthetic cel-miR-39-3p miRNA (Spike-In Control; QIAGEN, USA) of 1.6 x 108 copies/μL concentration was added to each plasma sample during the extraction process as an exogenous control to normalize and monitor the extraction efficiency.
Plasma miRNA quantification
TaqMan MicroRNA reverse transcription Kit (Applied Biosystems, USA) was used to convert the plasma miRNA to cDNA by RT-PCR. The cDNA synthesis process included four important reaction steps; poly (A) tailing reaction, adapter ligation reaction, reverse transcription (RT) reaction and finally the miR-Amp reaction. All of the above steps were performed according to the protocol provided by the reverse transcription Kit manufacturer (Applied Biosystems, USA). The expression level of the miRNA-21 in plasma was quantified using quantitative real-time PCR analysis in an ABI 7500 real-time PCR system. TaqMan-advanced miRNA assay (Applied Biosystems, USA) specific for miRNA-21 and TaqMan® Fast Advanced Master Mix was used in the real-time PCR. Real-time PCR reactions were performed in a 20 μL reaction mixture containing 10μL TaqMan® Fast Advanced Master Mix (2X), 1.0 μL Taqman® Advanced miRNA-21 Assay (20x), 5.0 μL diluted (1:10) RT cDNA template and remaining 4μL of RNase-free water. The cycling conditions were as follows: 95°C for 20s, followed by 40 cycles of 95°C for 3s and 60°C for 30s. For each sample, the quantitative real-time PCR analysis was performed in triplicates. We observed miRNA-16 as the most stable endogenous reference in the plasma of our study population of lung cancer patients and healthy controls. The expression data were then analyzed by relative quantification (2–ΔΔCT) method using miRNA-16 as an endogenous control and synthetic cel-miR-39-3p as an endogenous control for normalization. Plasma miRNA-21 concentration was quantified as described by Ioanna S. et al., 2013; first by normalizing the expression levels of both target miRNA-21 and the endogenous control miRNA-16 with respect to the exogenous control cel-miR-39, and then the relative quantification (RQ) was estimated.
Statistical Analysis
Statistical analysis was performed by using the SPSS statistical software version -21 (SPSS Inc., Chicago, IL) and GraphPad Prism version 5.0 (GraphPad Software, San Diego, CA, USA). The difference in plasma miRNA-21 levels between groups was estimated by using Mann-Whitney U test. ROC curve was used to analyze the diagnosis value, including AUC, cutoff point, sensitivity, and specificity. A p-value of < 0.05 was considered statistically significant.
Results
Demographic characteristics of the study population
A summary of the baseline characteristics of the lung cancer patients and healthy controls are presented in Table 1. Present study included 80 Lung cancer patients and 80 healthy controls. Among the patient group, 60 were adenocarcinoma and 20 were squamous cell carcinoma. Lung cancer patients were diagnosed in different stages (Stage I = 2.5%, Stage II= 8.75%, Stage III=32.5% and Stage IV= 57.5%) with varying histological grades (Grade 1=2.5%, Grade 2=41.25% and Grade 3=28.75%). Majority of the lung cancer patients were males (72.5%) and the overall mean age for the patient group was 57.6 years in the range of 28 to 87 years.
Table 1.
Distribution of Selected Characteristics among Lung Cancer Patients and Healthy Controls
| Variable | Lung Cancer patients (%) | Healthy Controls (%) |
|---|---|---|
| Study population | ||
| Total No. | 80 | 80 |
| Tabuk | 20 | 20 |
| Jordan | 60 | 60 |
| Gender | ||
| Males | 58 (72.5) | 53 (66.25) |
| Females | 22 (27.5) | 27 (33.75) |
| Smoking status | ||
| Non-Smokers | 26 (32.5) | 80 (100.0 %) |
| Smokers | 54 (67.5) | 0 (0.0 %) |
| Age at diagnosis (Years) | ||
| Mean +SD age (Y) | 57.6 + 11.49 | 26.8+ 4.01 |
| (range, 28-87 Years) | (range, 21- 40 Years) | |
| Histological type | ||
| ADC* | 60 (75.0) | |
| SCC** | 20 (25.0) | |
| TNM Stage | ||
| I | 2 (2.5) | |
| II | 7 (8.75) | |
| III | 26 (32.5) | |
| IV | 46 (57.5) | |
| Distant metastases | ||
| No | 35 (43.75) | |
| Yes | 45 (56.25) | |
| Histological grade | ||
| Grade 1 | 2 (2.5) | |
| Grade 2 | 33 (41.25) | |
| Grade 3 | 23 (28.75) | |
ADC- adenocarcinoma,
SCC-squamous cell carcinoma
Plasma miRNA-21 level among Lung cancer patients and healthy controls
Real-time relative quantification analysis showed that the overall mean relative expression level of plasma miRNA-21 among lung cancer patients (2.32 ± 1.7) was significantly higher compared to that of the healthy individuals (0.715 ± 0.48) (Figure 1).
Figure 1.
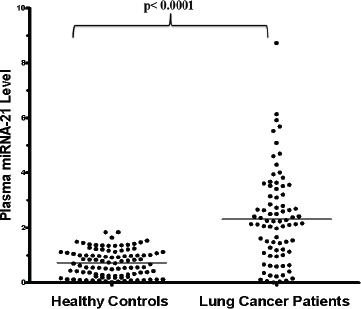
The Relative Expression Level of Plasma miRNA-21 in Lung Cancer Patients and Healthy Controls.
Plasma miRNA-21 level with respect to the clinical parameters of Lung cancer patients
In comparison to the healthy controls, we observed that there was a higher detectable level of plasma miRNA-21 even among the patients with early stage (I and II) of lung cancer disease. A higher level of plasma miRNA-21 was also observed among the patients with distant organ metastasis in comparison to the lung cancer patients with local tumor invasion, however, the difference was not observed to be statistically significant. Squamous cell carcinoma patients showed the slightly higher level of plasma miRNA-21 than the patients with adenocarcinoma. In addition, while comparing the plasma miRNA-21 in different histological grades, moderately and poorly differentiated (Grade 2 and 3) cell type reflected higher plasma miRNA-21 level than grade 1 differentiation; but the difference doesn’t reach to the value of statistical significance. Smoking is one of the major risk factors for lung cancer, however in the present study we didn’t observed any significant difference (p=0.147) in the level of miRNA-21 among smoker (2.71 ±1.92) and non-smoker (2.13 ±1.49) lung cancer patients (Table 2).
Table 2.
Relative Quantification of Plasma miRNA-21 level (Mean ±SD) among the Study Population
| Variables | Plasma miRNA-21 level (Mean ±SD) | 95% Confidence interval | p value |
|---|---|---|---|
| Study Population | |||
| Lung cancer patients | 2.32 ± 1.7 | 1.95 to 2.69 | <0.0001 |
| Healthy controls | 0.715 ± 0.48 | 0.60 to 0.82 | |
| Lung cancer patients (n=80) | |||
| Gender | |||
| Male | 2.15 ±1.49 | 1.75 to 2.55 | 0.153 |
| Female | 2.75 ±1.98 | 1.86 to 3.63 | |
| TNM Stage | |||
| I | 3.14 ±0.71 | -3.21 to 9.49 | 0.829 |
| II | 2.32 ±1.55 | 0.89 to 3.76 | |
| III | 2.13 ±1.77 | 0.35 to 1.41 | |
| IV | 2.40 ±1.66 | 1.89 to 2.89 | |
| Distant Metastases | |||
| M0 | 2.21 ±1.68 | 1.65 to 2.80 | 0.667 |
| M1 | 2.4 ±1.66 | 1.89 to 2.89 | |
| Histological type | |||
| ADC | 2.16 ±1.5 | 1.77 to 2.56 | 0.159 |
| SCC | 2.77 ±1.98 | 1.84 to 3.69 | |
| Histological Grades | |||
| Grade 1 | 1.15 ±1.26 | -10.2 to 12.5 | 0.333 |
| Grade 2 | 2.53 ±1.72 | 1.92 to 3.14 | |
| Grade 3 | 2.09 ±1.55 | 1.54 to 2.64 | |
| Smoking status | |||
| Smoker | 2.71 ±1.92 | 1.93 to 3.48 | 0.147 |
| Non-Smoker | 2.13 ±1.49 | 1.72 to 2.54 |
Diagnostic value of plasma miRNA-21 in lung cancer patient screening
In order to determine the diagnostic value of plasma miRNA-21 for the screening of lung cancer patients, receiver operating characteristic (ROC) curve was created by GraphPad Prism (version 5.0) software. We observed that the area under curve (AUC) was 0.8913 [95% confidence interval (CI): 0.8394 to 0.9431, p< 0.0001] and the sensitivity and specificity were both 80.0%, when the cut-off value was 1.207, as shown in Figure 2.
Figure 2.
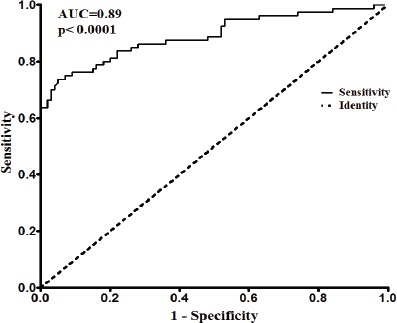
ROC Curve for Diagnostic Value of Plasma miRNA-21 in Lung Cancer Patient Screening
Discussion
Currently available advanced imaging and cytology-based screening strategies are very helpful for the detection of lung tumors; however, the majority of the lung cancer cases are still diagnosed at late stages. The recent findings of the potential role of miRNAs in various biological pathways and their presence in blood circulation have given a positive hope towards the identification of some efficient circulatory biomarkers for the diagnosis of lung cancer. miRNA-21 is one of the most extensively studied microRNAs, encoded by MIR21 gene in humans, located on chromosome 17q23.2. miRNA-21 has been observed to regulate many cell functions by targeting multiple genes associated with cell proliferation, metastasis, angiogenesis, and anti-apoptosis etc. (Tao et al., 2015; Yin Hsun et al., 2015). miRNA-21 has been observed to be the most commonly upregulated miRNA in solid and hematological malignancies (Volinia et al., 2006). miRNA-21 targets Spry2, BTG2 and PDCD4 thereby enhances oncogenic K-ras activation and modulates tumorigenesis in lung cancer (Hatley et al., 2010). miR-21 also targets PTEN, RECK, and Bcl-2 in lung squamous cell carcinoma that regulates cell proliferation, apoptosis, and tumor invasiveness (XuLF et al., 2014) EGFR mutation-positive lung adenocarcinoma have been found to be associated with significantly increased miRNA-21 expression and the blockade of miRNA-21 reverses the EMT phenotype associated with EGFR-TKI resistance (Seike et al., 2009; Byers et al., 2013). miRNA-21 also induces EGFR-TKI resistance either by activating the PI3K/Akt signal pathway or by inhibiting the PTEN and PDCD4 expression (Li et al., 2014). MiRNA-21 was observed to down-regulate the expression of SOCS1, SOCS6, and PTEN thereby promoting the development of Lung cancer (Xinying et al., 2016). In NSCLC cell line (A549), inhibition of miRNA-21 expression upregulates the expression of PDCD4 and ultimately reduced the proliferation, migration, and invasion (Yang et al., 2015).
In the present study, we used magnetic bead technology for efficient miRNA isolation and highly sensitive and specific advanced TaqMan miRNA assay for the quantification of plasma miRNA-21 expression in lung cancer patients and healthy individuals. In order to prevent amplification bias, the ligation-based cDNA synthesis kit included miR-Amp primers which efficiently recognize the universal sequences added to every miRNA on the 5’ and 3’ end. We observed a significantly high level of miRNA-21 in the plasma of lung cancer patients than in healthy controls, reflecting its possible involvement in the development of lung cancer. A significant value of AUC (0.891) with 81.25% sensitivity and 80% specificity of plasma miRNA-21 for the screening of lung cancer patients were observed, which determines its potential utility as a circulatory marker for lung cancer diagnosis. Our results are consistent with the previous findings of Wei Zhao et al observed significantly higher serum miRNA-21 level in non small cell lung cancer patients than healthy people with 73.8% sensitivity and 71.7% specificity (Wei Zhao et al., 2015). Hankui Chen et al observed up-regulation of plasma miRNA-21 in patients with benign lung tumors as compared to normal controls (Chen et al., 2016). Over expression of miRNA-21 level in serum of non small cell lung cancer patients was observed especially in an advanced stage of lung cancer (Yang et al., 2015a; Yang et al., 2015b).
In conclusion, we observed that plasma miRNA-21 can be efficiently extracted by the magnetic bead technology and quantified by the advanced TaqMan miRNA assay. Plasma miRNA-21 showed a high ability to distinguish between lung cancer patients and healthy individuals, therefore can be used as an efficient non-invasive biomarker for the screening of Lung cancer patients. However, further studies are required in order to establish the circulatory miRNA-21 as a potential biomarker for the lung cancer screening in clinical practice.
Conflict of interest
The authors declare that there is no conflict of interest in relation to this article.
Acknowledgments
The authors acknowledge Deanship of Scientific Research, University of Tabuk, Saudi Arabia for the financial support and also acknowledge the study subjects especially lung cancer patients for their participation in this study.
References
- Byers LA, Diao L, Wang J, et al. An epithelial–mesenchymal transition gene signature predicts resistance to EGFR and PI3K inhibitors and identifies axl as a therapeutic target for overcoming EGFR inhibitor resistance. Clin Cancer Res. 2013;19:279–90. doi: 10.1158/1078-0432.CCR-12-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappellesso R, Nicole L, Caroccia B, et al. Young investigator challenge:MicroRNA-21/ MicroRNA-126 profiling as a novel tool for the diagnosis of malignant mesothelioma in pleural effusion cytology. Cancer Cytopathol. 2016;124:28–37. doi: 10.1002/cncy.21646. [DOI] [PubMed] [Google Scholar]
- Castro D, Moreira M, Gouveia AM, et al. MicroRNAs in lung cancer. Oncotarget. 2017;8:81679–85. doi: 10.18632/oncotarget.20955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Liu H, Zou H, et al. Evaluation of plasma miR-21 and miR-152 as diagnostic biomarkers for common types of human cancers. J Cancer. 2016;7:490–9. doi: 10.7150/jca.12351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatley ME, Patrick DM, Garcia MR, et al. Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell. 2010;18:282–93. doi: 10.1016/j.ccr.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao HW, Pan CY, Lai CH, et al. Urine miR-21-5p as a potential non-invasive biomarker for gastric cancer. Oncotarget. 2017;8:56389–97. doi: 10.18632/oncotarget.16916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Ren S, Li X, et al. miR-21 over expression is associated with acquired resistance of EGFR-TKI in non-small cell lung cancer. Lung Cancer. 2014;83:146–53. doi: 10.1016/j.lungcan.2013.11.003. [DOI] [PubMed] [Google Scholar]
- Palanichamy JK, Rao DS. miRNA dysregulation in cancer:towards a mechanistic understanding. Front Genet. 2014;18:54. doi: 10.3389/fgene.2014.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seike M, Goto A, Okano T, et al. miR-21 is an EGFR regulated anti-apoptotic factor in lung cancer in never smokers. Proc Natl Acad Sci U S A. 2009;106:12085–90. doi: 10.1073/pnas.0905234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourvinou IS, Markou A, Lianidou ES. Quantification of circulating miRNAs in plasma:effect of preanalytical and analytical parameters on their isolation and stability. J Mol Diagn. 2013;15:827–34. doi: 10.1016/j.jmoldx.2013.07.005. [DOI] [PubMed] [Google Scholar]
- Tao Y.J, Li Y.J, Zheng W, et al. Antisense oligonucleotides against microRNA-21 reduced the proliferation and migration of human colon carcinoma cells. Cancer Cell Int. 2015;15:77. doi: 10.1186/s12935-015-0228-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Zhao, Jun-Jie Zhao, Long Zhang, et al. Serum miR-21 level:a potential diagnostic and prognostic biomarker for non-small cell lung cancer. Int J Clin Exp Med. 2015;8:14759–63. [PMC free article] [PubMed] [Google Scholar]
- Xinying X, Yuxia L, Yong W, et al. MiR-21 and MiR-155 promote non-small cell lung cancer progression by down regulating SOCS1, SOCS6, and PTEN. Oncotarget. 2016;7:84508–19. doi: 10.18632/oncotarget.13022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu LF, Wu ZP, Chen Y, et al. MicroRNA21(miR21) regulates cellular proliferation, invasion, migration, and apoptosis by targeting PTEN, RECK and Bcl2 in lung squamous carcinoma, Gejiu City, China. PLoS One. 2014;9:e103698. doi: 10.1371/journal.pone.0103698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JS, Li BJ, Lu HW, et al. Serum miR-152, miR-148a, miR-148b, and miR-21 as novel biomarkers in non-small cell lung cancer screening. Tumour Biol. 2015a;36:3035–42. doi: 10.1007/s13277-014-2938-1. [DOI] [PubMed] [Google Scholar]
- Yang Y, Meng H, Peng Q, et al. Downregulation of microRNA-21 expression restrains non-small cell lung cancer cell proliferation and migration through upregulation of programmed cell death 4. Cancer Gene Ther. 2015b;22:23–9. doi: 10.1038/cgt.2014.66. [DOI] [PubMed] [Google Scholar]
- Yin-Hsun F, Chao-Jung T. Emerging role of microRNA-21 in cancer. Biomed Rep. 2016;5:395–402. doi: 10.3892/br.2016.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong P, Carlo MC. The role of MicroRNAs in human cancer. Signal Transduct Target Ther. 2016;1:15004. doi: 10.1038/sigtrans.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Zhao JJ, Zhang L, et al. Serum miR-21 level:a potential diagnostic and prognostic biomarker for non-small cell lung cancer. Int J Clin Exp Med. 2015;8:14759–63. [PMC free article] [PubMed] [Google Scholar]


