Abstract
Background
Ebola virus (EBOV) mainly targets myeloid cells; however, extensive death of T cells is often observed in lethal infections. We have previously shown that EBOV VP40 in exosomes causes recipient immune cell death.
Methods
Using VP40-producing clones, we analyzed donor cell cycle, extracellular vesicle (EV) biogenesis, and recipient immune cell death. Transcription of cyclin D1 and nuclear localization of VP40 were examined via kinase and chromatin immunoprecipitation assays. Extracellular vesicle contents were characterized by mass spectrometry, cytokine array, and western blot. Biosafety level-4 facilities were used for wild-type Ebola virus infection studies.
Results
VP40 EVs induced apoptosis in recipient T cells and monocytes. VP40 clones were accelerated in growth due to cyclin D1 upregulation, and nuclear VP40 was found bound to the cyclin D1 promoter. Accelerated cell cycling was related to EV biogenesis, resulting in fewer but larger EVs. VP40 EV contents were enriched in ribonucleic acid-binding proteins and cytokines (interleukin-15, transforming growth factor-β1, and interferon-γ). Finally, EBOV-infected cell and animal EVs contained VP40, nucleoprotein, and glycoprotein.
Conclusions
Nuclear VP40 upregulates cyclin D1 levels, resulting in dysregulated cell cycle and EV biogenesis. Packaging of cytokines and EBOV proteins into EVs from infected cells may be responsible for the decimation of immune cells during EBOV pathogenesis.
Keywords: apoptosis, cell cycle, Ebola virus, extracellular vesicles, VP40
Ebola virus (EBOV) is a negative sense, single-stranded ribonucleic acid (RNA) virus capable of causing severe hemorrhagic fever in humans and nonhuman primates (NHPs). Until 2013, EBOV was responsible for approximately 1600 deaths; however, the most recent outbreak in Western Africa (Sierra Leone, Guinea, and Liberia), leading to eventual spread to Europe and the United States, resulted in over 28600 cases and 11300 deaths as of April 2016 [1, 2]. The progression of EBOV disease (EVD) within infected individuals involves nonspecific influenza-like symptoms, cytokine storm, and potential hemorrhagic manifestations including coagulopathy, leaky blood vessels, and multiorgan failure. Ebola virus mainly targets host cells of the myeloid lineage (monocytes, macrophages, and dendritic cells), but it does not productively infect T cells. Despite this targeting, lethal infections are accompanied by extensive cell death of CD4 and CD8 T-lymphocytes [3–10]. There are several proposed mechanisms for the induction of apoptosis in bystander lymphocytes during EBOV pathogenesis, including the following: Fas/Fas ligand and tumor necrosis factor (TNF)-TNF-related apoptosis-inducing ligand (TRAIL) interactions, impaired dendritic cell/T-cell contacts, and nitric oxide or viral glycoprotein (GP)-induced apoptosis [5, 9–11]. Likewise, we have recently proposed an additional potential mechanism for the induction of bystander T-cell death in the form of the EBOV matrix protein transferred to recipient cells via exosomes [12, 13]. However, the extent to which each of these mechanisms may contribute to bystander lymphocyte apoptosis and EBOV pathogenesis is not well characterized. Nevertheless, the loss of circulating T-cell populations in infected individuals may contribute to unchecked replication of the virus, potentially correlating with poor patient prognosis [13, 14].
In recent years, it has become clear that exosomes—small (~50–120 nm in diameter), membrane-bound extracellular vesicles (EVs) produced from the late endosomal pathway—are important for intercellular communication, particularly during disease [15–18]. Various specific tetraspanin proteins such as CD63, CD81, and CD9 mark the surface of exosomes and can be used to distinguish them from other EVs [19]. The content of exosomes can vary based upon cell type and environment; however, the selective packaging of cargo into exosomes is largely regulated by the Endosomal Sorting Complexes Required for Transport (ESCRT) proteins. The ESCRT proteins can be classified as ESCRT-I, -II, -III complex components and VPS4, which sequentially act to recognize cargo, package it into nascent vesicles, and pinch off the membrane while recycling the machinery back for future cargo packaging [20]. During infection, various viral proteins and nucleic acids can become integrated into exosomes, often by utilizing the ESCRT pathway [21–25]. Exosomes originating from infected cells can then induce proviral effects in recipient cells [12, 13, 16, 17, 21–28]. In addition to exosomes, other EVs such as exosome-like vesicles, microvesicles, and apoptotic bodies are produced from host cells by various mechanisms. Microvesicles, sometimes called ectosomes, are typically larger (100–1000 nm) in diameter and differ from exosomes mainly in that they arise from the direct outward budding and fission of the plasma membrane, or ectocytosis [29–31]. Microvesicles do not contain tetraspanins CD9 or CD81, and they have a similar lipid composition as the plasma membrane from which they budd [28, 29, 32]. Apoptotic bodies, unlike the other secreted vesicles, are formed only during programmed cell death. They are generally larger (500–4000 nm in diameter) than microvesicles and may contain deoxyribonucleic acid (DNA) and histones [28, 33, 34].
The EBOV matrix protein VP40 is one of the most abundant viral proteins, and, therefore, it is commonly used as a diagnostic marker for patients [35]. Alone or in conjunction with the GP and nucleoprotein (NP) of EBOV, VP40 can independently bud from cells to form virion-like particles (VLPs) that are morphologically similar to infectious virions [36–41]. Recently, we found the first evidence that VP40 is also able to exit cells by becoming packaged into exosomes [12, 13]. This finding is consistent with previous studies that have shown filoviral VP40 concentrating in late endosomal compartments and multivesicular exosomal bodies [42, 43]. Another study by Steele et al [44] showed that in EBOV-infected NHP and guinea pig models, VP40 antigen alone is concentrated in renal tubule epithelial cells, indicating that VP40 can exist outside of the virion (ie, potentially in exosomes) [45].
In the current study, we focused on (1) the mechanisms involved in the packaging of EBOV VP40 into exosomes and other EVs and, (2) the characterization of these vesicles. We have found that VP40 was able to affect cell cycle through cyclin D1 (which binds to cdk4 or cdk6) modulation to regulate the synthesis of EVs in donor cells. This regulation was done by the translocation of VP40 into the nucleus to stimulate overtranscription of cyclin D1 at its promoter, thus producing an activated phenotype in VP40-producing cells. We tested the efficacy of cdk4/6 inhibitors for their ability to hinder donor cell EV production and found that Fascaplysin and Ribociclib (Ribociclib is US Food and Drug Administration [FDA]-approved) may impact the biogenesis of EVs. In previous studies, we showed that VP40 was secreted in exosomes [12, 13], and here we additionally found that VP40 and other viral proteins (ie, NP and GP) were associated with EVs from wild-type EBOV-infected cells and macaques. Finally, mass spectrometry (MS) and cytokine analysis showed that exosomes originating from VP40-producing cells were enriched in several RNA-binding proteins and cytokines, which may play a pivotal role in recipient cell outcome.
MATERIALS AND METHODS
Cell Culture and Reagents
U937, CEM, and Jurkat cells were obtained from ATCC (Manassas, VA) and maintained in Roswell Park Memorial Institute (RPMI) 1640 media containing 10% heat-inactivated fetal bovine serum (FBS), 1% l-glutamine, and 1% streptomycin/penicillin (Quality Biological, Gaithersburg, MD). 293T cells, VP40 clones, and HeLa cells were sustained in Dulbecco's modified Eagle medium (DMEM) containing 10% heat-inactivated fetal bovine serum (FBS), 1% l-glutamine, and 1% streptomycin/penicillin (Quality Biological). Peripheral blood mononuclear cells (PBMCs) were maintained in RPMI 1640 media containing 20% FBS, 1% l-glutamine, and 1% streptomycin/penicillin. The PBMCs were also maintained with 50 IU/mL human recombinant interleukin (IL)-2 (Sigma) before transfections, but they were incubated in the absence of exogenous growth factors once transfected. All cells were incubated at 37°C with 5% CO2. For antibiotic selection of transfected 293T cells, hygromycin B (Invitrogen) was added to the culture the next day. Cdk4/6 inhibitors Fascaplysin (0.1–1 µM; Abcam) and Ribociclib (LEEO11; 0.1–10.0 µM; MedChemExpress) were used for the treatment of 293T and VP40 clone cell cultures for the analysis of cyclin D1 inhibition, cell viability, and EV production.
Plasmids, Transfections, and Generation of Resistant Clones
EBOV Mayinga VP40 isolate was expressed from a plasmid (Invitrogen) with cytomegalovirus (CMV) promoter and specific antibiotic selection marker (pcDNA3.1/Hygro). Twenty micrograms of Escherichia coli-purified DNA was transfected into 293T cells using electroporation as previously described [46]. Transfected cells were treated the next day with hygromycin B (200–400 µg/mL) for plasmid selection. To generate resistant clones, transfected cells were cultured for >3 weeks, followed by isolation of surviving colonies and multiple passages under specific antibiotic selection. A total of 21 new resistant clones were generated, which produced varying amounts of VP40 (select clones are shown in Supplementary Figure 1). Two clones, an intermediate and a high VP40-producing clone, were selected to complement an existing low VP40-producing clone that we previously published (EVTR2C) [12]. For clarity, we renamed this clone V2CL (VP40 293T Clone Low), whereas we designated the medium and high clones V2CI (VP40 293T Clone Intermediate) and V2CH (VP40 293T Clone High), respectively. All resistant clones were kept under constant hygromycin B (200 µg/mL for V2CL; 400 µg/mL for V2CI and V2CH) antibiotic selection.
Transfections of U937 cells, HeLa cells, and PBMCs were performed with log-phase cells and attractene reagent (QIAGEN) according to the manufacturer’s instructions. In brief, 1.65 × 105 log-phase cells were incubated with transfection complexes formed from 1.5 µg of VP40-producing plasmid DNA and 1.5 µL attractene reagent in serum-free media. Cells and complexes were incubated for 3 days in 100 µL, followed by analysis of cell viability by CellTiter-Glo assay.
Capture of Extracellular Vesicles, Virus-Like Particles, and Proteins With Nanotrap Particles
The Nanotrap (NT) particles we used here were provided by Ceres Nanosciences, Inc. and have been described in detail previously [47]. In brief, NT particles are highly porous, multifunctional hydrogel particles, approximately 700–800 nm in diameter and are composed of high-affinity aromatic dye compounds covalently linked to an interior hydrogel particle structure. The 2 particles used frequently by our laboratory include NT80 particles (Ceres no. CN1030), which contain a Reactive Red 120 core bait, and NT82 particles (Ceres no. CN2010), which have Cibacron Blue F3GA. Both particle types have a NIPAm-Bis-AA matrix with a shell lacking vinyl sulfonic acid [47]. For the capture and isolation of EVs from cell culture supernatants, a 30-µL slurry (30%) of 1:1 NT80 and NT82 particles (Ceres Nanosciences) was incubated with 1 mL cell-free, filtered (0.22 μm) supernatant. Samples were bound at 4°C for 24–72 hours. The NT pellets were isolated and washed with 500 µL of sterile 1× phosphate-buffered saline (PBS) without calcium and magnesium, followed by preparation for downstream assays. For western blot analysis, washed NT pellets were resuspended in 10 µL of Laemmli buffer and then loaded onto a 4–20% Tris-glycine sodium dodecyl sulfate (SDS) gel.
For the analysis and characterization of VLP, VLPs containing EBOV VP40, NP, and GP produced in Sf9 insect cells through recombinant baculovirus infection were used (obtained from IBT Bioservices). For analysis of VLP passage through 0.22-µm filters, 5 µg of VLP was spiked into 1 mL of sterile 1× PBS, followed by filtration (through 0.22 µm) alongside a PBS control. The resulting filtrate was then incubated with 30 µL of NT80/82 particles (30% slurry) overnight at 4°C. NT pellets were used the next day for SDS-polyacrylamide gel electrophoresis (PAGE) and western blot analysis for presence of EBOV proteins. For the analysis of VLP in comparison to EV distribution upon passage through size exclusion fractionation before and after filtration, 10 µg of VLP was spiked into 10 mL of 5-day V2CH supernatants produced in exosome-free media. Supernatants were either filtered (through 0.22 µm filter) or directly incubated with ExoMAX Opti Enhancer (1:1 reagent/filtered supernatant; Systems Biosciences) overnight at 4ºC to concentrate EVs. Extracellular vesicles were pelleted, resuspended in 0.5 mL of sterile 1× PBS, and loaded onto qEVoriginal size exclusion columns (IZON) according to the manufacturer’s instructions. Fraction numbers 2–11 (0.5 mL each) were collected and incubated with 30 µL of NT80/82 30% slurry overnight at 4°C. The resulting NT pellets were then resuspended in Laemmli buffer for SDS-PAGE and western blot analysis of EBOV protein and exosomal marker levels.
Extracellular vesicles were obtained from EBOV-infected cell cultures under biosafety level (BSL)-4 conditions. Human umbilical vein endothelial cells (HUVECs) were infected with EBOV H.sapiens-tc/COD/1995/Kikwit (multiplicity of infection [MOI] of 1) and incubated for 3 days. Two milliliters of supernatants were harvested, passed through a 0.22-μm filter, and incubated with ExoMAX (1:1 reagent/filtered supernatant) reagent overnight at 4°C to concentrate EVs. Extracellular vesicles were pelleted, resuspended in 0.5 mL of sterile 1× PBS, and loaded onto qEVoriginal size exclusion columns according to the manufacturer’s instructions. Purified vesicles were collected as 0.5-mL fraction numbers 7–10. Each fraction was incubated with 30 μL of NT80/82 (30% slurry) at room temperature for 1 hour. Extracellular vesicle-bound NTs were then washed once with 1× PBS and resuspended in 10 μL of 2× NuPAGE LDS sample buffer (Thermo Fisher) for SDS-PAGE and western blot analysis of EBOV protein levels. Samples were heated at 95°C for 10 minutes twice before electrophoresis.
For the capture and isolation of EVs from inactivated (gamma irradiation) rhesus monkey (Macaca mulatta) serum samples (obtained from US Army Medical Research Institute of Infectious Diseases), 100 µL of serum from each animal (n = 3; 2 EBOV+, 1 EBOV−) were diluted in 400 µL of sterile 1× PBS without calcium or magnesium and passed through a 0.22-μm filter. Samples were incubated with 25 µL of NT80/82 30% slurry for 48 hours at 4°C. The NT pellets were washed with sterile 1× PBS without calcium and magnesium and resuspended in 10 µL Laemmli buffer for western blot analysis of exosomal markers and VP40 protein. Nonhuman primate details are as follows: NHP 1 (EBOV−) = day 0 prebleed serum; NHP 2 (EBOV+) = pooled serum from days 4 to 5 postinfection (pi), animal died on day 7; NHP 3 (EBOV+) = pooled serum from days 8 to 11 pi, animal died on day 12. All experiments involving NHPs and inactivated EBOV samples were carried out under the Institutional Biosafety Committee-approved institutional biosafety guidelines and were performed at BSL-2/2+ level.
Cell Cycle Analysis
The 293T, V2CL, or V2CH cells were either plated in 0.1% FBS DMEM, blocked with hydroxyurea for 2–5 days, or preblocked with hydroxyurea for 18 hours (20 mM final concentration; Sigma). Cells were placed in fresh DMEM for 1 hour, followed by addition of nocodazole (50 ng/mL; Sigma) for 2–5 days. After the block, the cells were either (1) imaged with EVOS XL Core Imaging System (Life Technologies) and subsequently assayed for cell viability or (2) harvested, lysed, and used for downstream western blot analysis. Extracellular vesicles from blocked cells were obtained after 5 days of incubation in exosome-free media and isolated with NT80/82 particles. Extracellular vesicle-bound particles were then used for either western blot, acetylcholinesterase (AChE) assay, or ZetaView analysis.
Cell Treatment and Viability Assay
Cells were seeded into 96-well plates at 50000 cells per well in fresh media followed by treatment. All wells were plated and treated in triplicate. For treatment of CEM, Jurkat, U937, and 293T cells with cell culture supernatants, control cells or resistant clones were cultured for 5 days in exosome-free media to maximize EV concentration within the media, as previously described [26]. Media were then harvested from the cells, centrifuged for 10 minutes at 20800 ×g to remove cells and cellular debris, and filtered (0.22 μm). Seeded cells in fresh media (50 µL total) were then treated with 50 µL of supernatant. All treatments of CEM, Jurkat, U937, and 293T cells were incubated for 5 days, followed by measurement of cell viability using CellTiter-Glo Cell Luminescence Viability kit (Promega, Madison, WI) as per manufacturer’s instructions. In brief, 100 µL of CellTiter-Glo reagent were added to the wells (1:1 reagent/cell suspension). The plate was shaken for 2 minutes, incubated at room temperature for 10 minutes, and followed by detection of luminescence using the GloMax Explorer multidetection system (Promega). For the viability analysis of cell cycle blocks, 293T, V2CL, and V2CH cells were seeded as described above. Starvation wells were plated in fresh 0.1% FBS DMEM, whereas all others were in 10% FBS DMEM. Cells were then treated with either hydroxyurea (20 mM) or nocodazole (50 ng/mL). Cells receiving nocodazole treatment were pretreated for 18 hours with hydroxyurea (20 mM). It should be noted that hydroxyurea is normally used at 1–2 mM to block cells, but here we have tested higher concentrations to block cells without cell death. Cells were incubated for 5 days followed by measurement of cell viability using CellTiter-Glo assay as described above.
Preparation of Whole-Cell Extracts and Western Blot Analysis
Cell pellets were harvested, washed twice with 1× PBS without calcium and magnesium, and resuspended in lysis buffer (50 mM Tris-HCl [pH 7.5], 120 mM NaCl, 5 mM EDTA, 0.5% Nonidet P-40, 50 mM NaF, 0.2 mM Na3VO4, 1 mM dithiothreitol [DTT], and 1 complete protease inhibitor mixture table/50 mL [Roche Applied Science]). The suspension was incubated on ice for 20 minutes with gentle vortexing every 5 minutes, followed by centrifugation at 10000 ×g at 4°C for 10 minutes. Protein concentration from the lysate supernatant was quantified using Bradford protein assay according to the manufacturer’s instructions (Bio-Rad).
Preparation of EBOV-infected HUVEC lysates was performed as previously described [48]. Infected HUVECs were fixed in 10% neutral-buffer formalin at 4°C in 6-well plates. Fixed HUVEC monolayers were washed in PBS, a solution of 300 mM of Tris pH 8.0 containing 2% SDS was added, and cells were scraped off and transferred to polypropylene tubes. Samples were heated at 100°C for 30 minutes, then 60°C for 2 hours. Excess SDS was dialyzed using spin filtration, and protein content was determined by BCA assay.
For western blot analysis, whole-cell extracts (10–30 µg) were resuspended in 10 µL of Laemmli buffer, heated at 95°C for 3 minutes, and loaded onto a 4–20% Tris-glycine SDS gel. NT particle pellets were resuspended in 10 µL of Laemmli buffer, heated at 95°C for 3 minutes, and vortexed 3 times until fully resuspended. The eluted material was then loaded onto a 4–20% Tris-glycine gel. Gels were run at a maximum 150 V and wet-transferred overnight at 50 mA onto polyvinylidene difluoride (PVDF) membranes. Membranes were blocked in 5% milk in 1× PBS containing 0.1% Tween 20 for 1 hour at 4°C, then incubated overnight at 4°C with appropriate primary antibody: α-Caspase 3, α-poly(ADP-ribose)polymerase-1 (PARP-1), α-Alix, α-VPS4, α-CHMP6, α-TSG101, α-EAP20, α-EAP45, α-cyclin D1, α-cyclin E, α-cyclin A, α-cdk2, α-cdk4, α-cdk6, α-cdc2 p34 (α-cdk1), α- histone deacetylase 1 ([HDAC1] Santa Cruz Biotechnology); α-β-actin (Abcam); α-CD63, α-CD81, α-CD9 (Systems Biosciences); α-cyclin B1 (Cell Signaling); and α-GP, α-NP, and α-VP40 (IBT Bioservices). Extracellular vesicles from EBOV-infected HUVECs and lysates from EBOV-infected cells were dry-transferred to PVDF membranes with the iBlot system (Thermo Fisher). VP40 and NP were detected as described above, whereas GP was detected with murine monoclonal EBOV GP-specific antibody 6D8 [49]. Membranes were then incubated with the appropriate horseradish peroxidase (HRP)-conjugated secondary antibody for 2 hours at 4°C and developed using Clarity or Clarity Max Western ECL Substrate (Bio-Rad). Luminescence was visualized on a ChemiDoc Touch Imaging System (Bio-Rad).
Isolation of Exosomes for Characterization and Functional Assays
Our laboratory has substantial experience in the isolation and preparation of exosomes and EVs for characterization and functional analysis, particularly in the context of viral infection [12, 21, 22, 26, 27, 50–52]. For the experiments used here, 293T cells and VP40 clones were grown in exosome-free DMEM containing 10% heat-inactivated FBS (ultracentrifuged at 100000 ×g for 90 minutes to remove vesicles), 1% l-glutamine, and 1% streptomycin/penicillin (Quality Biological). Exosome preparations were made from exosome-free cell culture supernatant (filtered; 0.22 μm) produced from fully confluent cells grown for 5 days. For isolation of exosomes for western blot analysis and AChE assay, NT80/82 particles were used as described above. For the characterization of exosomes by gradient separation and western blot, 10 mL of unfiltered cell culture supernatant was incubated with 10 mL of ExoMAX Opti Enhancer at 4°C overnight. The vesicle pellet was spun down at 1500 ×g for 30 minutes and resuspended in 300 μL sterile 1× PBS without calcium or magnesium. Resuspended vesicles were then placed on top of iodixanol (OptiPrep; Sigma) gradients prepared in 1× PBS in 1.2% increments ranging from 6% to 18%. The gradient was ultracentrifuged for 90 minutes at 100000 ×g in a SW41 Ti rotor (Beckman). Gradient fractions were collected from the top of the gradient in 1-mL increments and transferred to sterile 1.5-mL microcentrifuge tubes. Separated iodixanol fractions were incubated with 30 μL of NT80/82 30% slurry at 4°C overnight, followed by processing for western blot or MS analysis. Alternatively, select iodixanol fractions were pooled and resuspended in sterile 1× PBS, followed by ultracentrifugation for 90 minutes at 100000 ×g in a Ti-70 rotor (Beckman) to pellet EVs for the removal of iodixanol for downstream functional assays. Direct isolation of EVs from small volumes of cell-free supernatants from 293T and V2CH cells (grown in exosome-free media) was completed by ultracentrifugation for 90 minutes at 100000 ×g in a TL-100 rotor (Beckman). Resulting EV pellets were resuspended in 40 μL of sterile 1× PBS and used for ZetaView analysis and subsequent functional assays.
Acetylcholinesterase Assay
ACHE protein levels of the exosomes were determined with the FLUOROCET Ultrasensitive Exosome Quantitation Assay Kit (Systems Biosciences) following the manufacturer’s instructions. In brief, a negative control containing buffer solution (exosome negative) and standard curve were prepared. Nanoparticle pellet samples (from filtered cell culture media) were plated and treated. Fluorescence corresponding to AChE activity was measured with a GloMax Explorer multidetection system (Promega). The number of exosomes was calculated using the standard curve.
ZetaView Nanoparticle Tracking Analysis
Nanoparticle tracking analysis (NTA) was performed using the ZetaView Z-NTA (Particle Metrix) and its corresponding software (ZetaView 8.04.02). One hundred nanometer polystyrene nanostandard particles (Applied Microspheres) were used to calibrate the instrument before sample readings at a sensitivity of 65 and a minimum brightness of 20. Automated quality control measurements including, but not limited to, cell quality check and instrument alignment and focus were also performed before the use of the ZetaView for sample measurements. For each measurement, the instrument preacquisition parameters were set to a temperature of 23°C, a sensitivity of 85, a frame rate of 30 frames per second, and a shutter speed of 250. For each sample, 1 mL of the sample, diluted in deionized water, was loaded into the cell, and the instrument measured each sample at 11 different positions throughout the cell, with 3 cycles of readings at each position. After automated analysis and removal of any outliers from the 11 positions, the mean, median, and mode (indicated as diameter) sizes and the concentration of the sample were calculated by the machine software. Measurement data from the ZetaView were analyzed using the corresponding software, ZetaView 8.04.02, and Microsoft Excel 2016. We selected the mode, defined as the size of the most abundant particles, as the measurement for size in our analysis.
Cytoplasmic Versus Nuclear Isolation
Cytoplasmic and nuclear compartments were isolated and extracted using the NE-PER Nuclear and Cytoplasmic Extraction Reagent kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. In brief, log-phase 293T and V2CH cells (1 × 106) were harvested, washed once in 1× PBS, and incubated with ice cold CER I (100 μL). Samples were vortexed for 15 seconds and incubated on ice for 10 minutes. Next, ice cold CER II was added (5.5 μL) to each sample, vortexed for 5 seconds, and incubated on ice for 1 minute. Tubes were then vortexed, spun for 5 minutes at 16000 ×g, followed by transfer of cytoplasmic fractions to clean prechilled tubes. Insoluble nuclei pellets were suspended in ice cold NER (50 μL), vortexed for 15 seconds, and incubated on ice for 40 minutes with additional vortexing every 10 minutes. Samples were centrifuged for 10 minutes at 16000 ×g, and resulting nuclear fractions were transferred to clean prechilled tubes. Resulting fractions were analyzed for protein concentration by Bradford assay (Bio-Rad) and used for SDS-PAGE and subsequent western blot analysis.
Kinase Assay
Peptide kinase assays were performed as described previously [12]. In brief, immunoprecipitation (IP) was performed by incubation of 500 µg of 293T or V2CH whole-cell extracts with 10 µg of appropriate primary antibody (α-cyclin D1, α-normal rabbit immnunoglobulin [Ig]G; Santa Cruz Biotechnology) and 1 mL of TNE50 + 0.1% NP-40 for 24 hours at 4°C. The next day, complexes were precipitated with 50 µL of a 30% slurry of A/G beads (Calbiochem) for 2 hours at 4°C and washed twice with TNE50 + 0.1% NP-40 and once with kinase buffer. Phosphorylation reactions were performed with IPed material (15 µL), 50 µg of appropriate peptide (780S [775RPPTLSPIPHIPR787], 780A [775RPPTLAPIPHIPR787] [ABclonal Science]), and [γ-32P] ATP (2 µL) as substrates in TTK kinase buffer containing 50 mM HEPES (pH 7.9), 10 mM MgCl2, 6 mM EGTA, and 2.5 mM DTT. Some samples were also incubated with cdk4/6 inhibitor Fascaplysin (1 µM; Abcam). The reaction mixtures contained the following final concentrations: 40 mM β-glycerophosphate (pH 7.4), 7.5 mM MgCl2, 7.5mM EGTA, 5% glycerol, [γ-32P] adenosine triphosphate (ATP) (0.4 mM, 1 µCi), 50 mM NaF, 1 mM orthovanadate, and 0.1% (v/v) β-mercaptoethanol. Reactions were incubated at 37°C for 1 hour, and samples (10 µL) were dotted onto Whatman glass microfiber filters, dried for 30 minutes, and subsequently submerged in 1× TE buffer with gentle agitation for 48 hours. Samples were quantified using a scintillation counter (QuantaSmart CPM assay count).
Chromatin Immunoprecipitation and Quantitative Polymearse Chain Reaction for Cyclin D1 Promoter DNA
Log-phase 293T and V2CH cells (3 × 106 cells) were harvested and processed for chromatin IP (ChIP) and cyclin D1 promoter PCR using the Imprint Chromatin Immunoprecipitation Kit (Sigma) according to the manufacturer’s instructions. In brief, samples were cross-linked using 1% formaldehyde and incubated for 1 hour, followed by quenching with 1.25 M glycine (9:1 cell suspension/glycine). Samples were sonicated and used for IP with 1 µg of α-RNA Pol II, α-p300 (Santa Cruz), or α-VP40 (IBT Bioservices) at 4°C overnight. The next day, a 30% slurry of A/G beads (Calbiochem) was added and incubated for 2 hours at 4ºC. The samples were washed once with each TNE300 + 0.1% NP40, TNE150 + 0.1% NP40, TNE50 + 0.1% NP40, and IP Wash Buffer (Sigma) before the addition of proteinase K (800 units/mL). After a 15-minute incubation at 65°C, Reversing Solution (Sigma) was added, and samples were incubated at 65°C for an additional 90 minutes. The DNA was purified, and quantitative PCR analysis was performed with 2 μL of undiluted aliquots of DNA using SYBR Green (Bio-Rad) with the following pair of primers that amplify the cyclin D1 promoter region containing the RNA Pol II start site (+1): cyclin D1-Reverse (−180), (5’-CAC TTC GCA GCA CAG GAG-3’, Tm = 56.3°C); and cyclin D1-Forward (+238), (5’-CGG ACT ACA GGG GCA ACT-3’, Tm = 57.0°C). Serial dilutions of DNA from 293T cells were used as quantitative standards. The PCR conditions were as follows: 1 cycle at 50°C for 2 minutes, followed by 1 cycle at 95°C for 2 minutes, and 42 cycles at 95°C for 15 seconds and 48.7°C for 40 seconds. The absolute quantification of the samples was determined based on the cycle threshold value relative to the standard curve. Quantitative PCR reactions were carried out in triplicate using the CFX96 Real Time System (Bio-Rad).
Mass Spectrometry
The EVs from NT80/82 NT particle-processed 10.8 and 12.0 iodixanol fractions (as described above) were treated with 8 M of urea with 0.00005% DTT to lyse the vesicles. The samples were reduced with 10 mM of DTT and alkylated with 50 mM of iodoacetamide. The samples were next diluted by a solution of equal parts water and 500 mM of NH4HCO3, followed by trypsin (Promega) digestion for 4 hours at 37°C. Samples were then centrifuged at 12000 ×g at room temperature for 10 minutes, and supernatants were collected into a microcentrifuge tube. ZipTip was then used to collect the peptide samples, which were dried and resuspended in 10 μL of 0.1% trifluoroacetic acid solution before loading into an Orbitrap Fusion mass spectrometer. Bioinformatic searches from Swiss-Prot were used to identify peptides, and a label-free precursor ion detection method (Proteome Discoverer, version 1.3; Thermo Fisher Scientific) was used for accurate mass measurements on proteins and peptides with specific retention times on precursors and fragments.
Cytokine Analysis and Densitometry
293T and V2CH cells were cultured in exosome-free DMEM for 5 days, followed by harvesting and filtering (0.22 μm) of supernatants. Ten milliliters of supernatant samples were incubated 1:1 with ExoMAX at 4°C overnight, spun down, resuspended, and separated on an OptiPrep iodixanol gradient, as described above. The 10.8 and 12.0 iodixanol fractions for each cell type were combined and used side-by-side with unprocessed cell supernatants (filtered; 0.22 μm). For analysis of cytokine profiles in whole supernatant versus in association with exosomes, human cytokine array membranes (Abcam) were used according to the manufacturer’s instructions. In brief, membranes were blocked at room temperature for 30 minutes, followed by incubation with sample overnight at 4°C with gentle rocking. The next day, samples were aspirated, and membranes were washed and then subsequently incubated overnight at 4°C with biotin-conjugated α-cytokines. Membranes were then washed, incubated with HRP-conjugated streptavidin for 2 hours at room temperature, and washed again. Detection of chemiluminescence was accomplished via ChemiDoc Touch Imaging System (Bio-Rad). Raw densitometry counts were obtained using ImageJ software. Exposures were matched between cytokine membranes according to positive control signals. Densitometry data were normalized by subtracting the background represented by the negative control on each membrane.
Statistical Analysis
Standard deviation was calculated in all quantitative experiments done in triplicate. All P values were calculated using 2-tailed Student’s t tests (Microsoft Excel) and were considered statistically significant when P < .05.
RESULTS
Cells Producing VP40 Are Dysregulated in Their Cell Cycle
In our previous work, we used a stably transfected VP40-producing clone cell line that we named V2CL (EVTR2C [12]; see Methods) to show that VP40 can be packaged into exosomes and thereby impact recipient cells. To further verify our findings, we transfected a new series of 293T cells with EBOV VP40 plasmid (pcDNA3.1/Hygro) and treated them with hygromycin B for plasmid selection. After >3 weeks culture under constant antibiotic selection, surviving colonies (total of 21 clones) were isolated and cultured for 5 days. EVs by NT80/82 particle incubation and western blot analysis for VP40 levels and exosomal markers. Results in Supplementary Figure 1 show a representative panel of 6 of the resulting clones: V2C1–4, V2CI, and V2CH. Most of the V2C lines (19 of 21) produced similar levels of VP40 in EV western blot analysis. However, 2 lines, V2CI and V2CH, produced lower and higher levels of VP40, respectively, in comparison to the rest of the clones, while still exhibiting higher levels of exosomal marker Alix and Actin. This finding indicates that these 2 clones may produce higher levels of EVs with differential amounts of VP40 packaged. Compared side-by-side to the pre-existing V2CL line, which produced very low levels of extracellular VP40, V2CI produced intermediate levels, whereas V2CH released high levels of VP40 in EVs (data not shown). For this reason, we chose these 2 lines to utilize in future experiments to determine dose dependency of VP40 on donor and recipient cell responses.
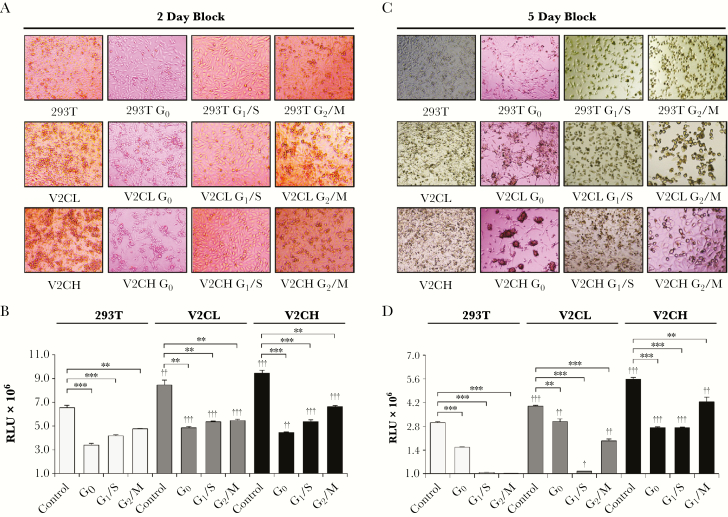
In the process of generating these clones, we observed a notably accelerated growth in cells producing VP40 (data not shown). It is well documented for many viruses, including human immunodeficiency virus type 1 (HIV-1), human cytomegalovirus, human T-cell leukemia virus type 1 (HTLV-1), Epstein-Barr virus (EBV), human papillomaviruses (HPVs), Kaposi sarcoma herpesvirus, hepatitis B virus, hepatitis C virus, adenoviruses, influenza viruses, and infectious bronchitis virus, that infected cells are altered in their cell cycle to benefit viral replication [53–56]. Therefore, we asked whether a similar phenotype occurred within VP40 clones. To address this question, we blocked control 293T, V2CL, and V2CH cells at the G0, G1/S, and G2/M stages of the cell cycle for either 2 or 5 days to observe changes in cell morphology and viability (Figure 1). Data in Figure 1A show that after 2 days of cell cycle blocks, all cell types had the expected morphology with minimal cell death. More specifically, G0 cells appeared adherent and stretched, G1/S cells were adherent and enlarged, whereas G2/M cells were smaller and rounded. Results from the corresponding 2-day cell viability assay (Figure 1B) indicated that all blocked cells had reduced viability, as would also be expected with functioning cell cycle blocks. After 5 days, a different pattern between cell types was observed. Control 293T cells retained similar phenotypes at 5 days compared with 2 days. However, it should be noted that significant cell death was observed for 293T cells at both G1/S and G2/M after 5 days (Figure 1C and D). In contrast, VP40 donor cells expressed greater cell viability at G1/S and G2/M, in line with increasing amounts of VP40 produced (Figure 1D). Strikingly, with increased levels of VP40 produced, V2C cells adhered less to the expected phenotype exhibited by 293T cells. At G0 after 5 days, V2C cells were more viable compared with 293T cells and had clumped together to form structures that closely resembled viral synapses (Figure 1C), similar to those seen with viruses such as HIV-1, Japanese encephalitis virus, influenza A virus, and HTLV-1 [57–61]. It is possible that these clumped structures may represent a potential synaptic mechanism used by EBOV in epithelial cell infection when nutrients are depleted and cell cycling is hindered. In addition, V2C cells at G1/S and G2/M continued to “slip through” cell cycle blocks, especially in V2CH cells, as demonstrated by many rounded cells (G2/M) present in G1/S-blocked cultures and many adherent cells (G1/S) in G2/M-blocked cultures (Figure 1C). This pattern was further demonstrated by an increase in cell viability of V2C cells compared with 293T at G1/S and G2/M (Figure 1D). Finally, regardless of the length of incubation, there was a VP40 production level-dependent increase in the number and viability of V2C control cells compared with 293T, supporting our previous observation of an increased growth rate in VP40-producing cells. Taken together, these results indicate that cells producing VP40 may have accelerated rates of growth directly proportional to the amount of VP40 being produced. Furthermore, VP40-producing cells may potentially be able to push past cell cycle blocks imposed by external or internal influences, indicating a dysregulation of cell cycle checkpoints and resulting in increased survival under various environmental stresses.
Figure 1.
Cell cycle analysis of VP40 clones. 293T, V2CL, and V2CH cells were seeded in a 96-well plate at 5 × 105 cells in 100 μL of fresh media. Cells were blocked at G0 (starvation; 0.1% fetal bovine serum DMEM), G1/S (20 mM of hydroxyurea), and G2/M (18-hour 20 mM of hydroxyurea pretreatment, followed by release for 1 hour, and subsequent treatment with 50 ng/mL of nocodazole) for 2 days. Blocked cells were then imaged (A) and assayed for cell viability with CellTiter-Glo (B). The same experiment was repeated; however, cells were allowed to incubate for 5 days after blocking. Cells were subsequently imaged (C) and assayed for cell viability via CellTiter-Glo (D). Statistical analysis by Student’s 2-tailed t test compares V2CL and V2CH cell cycle-blocked groups with corresponding 293T groups (†, P < .05; ††, P < .01; †††, P < .001). Additional Student’s 2-tailed t test compares cell cycle-blocked groups with controls of their own cell type (**, P < .01; ***, P < .001).
VP40 Increases Functional Cyclin D1 Levels in Donor Cells
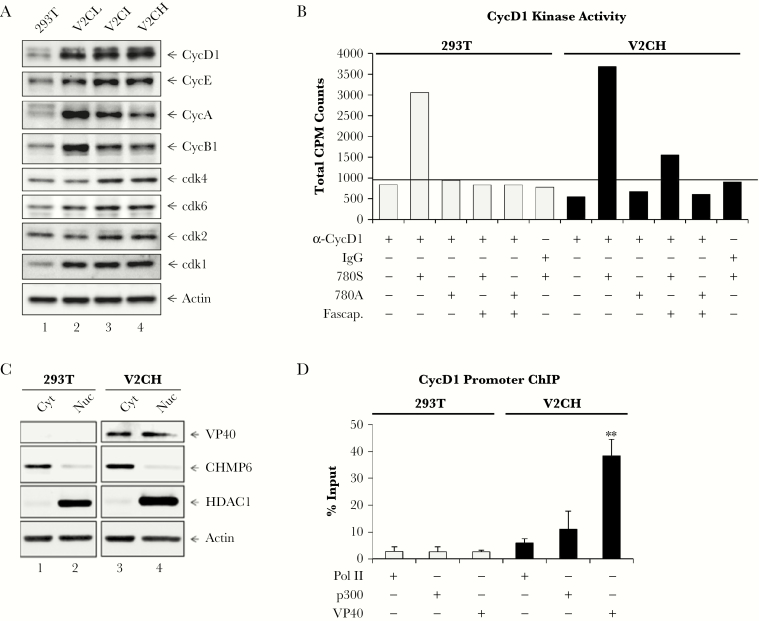
As V2C cells had been observed to have accelerated growth, we next explored the effect of VP40 production on levels of cyclin and cdk proteins. Western blot analysis of cyclin and cdk levels in 293T, V2CL, V2CI, and V2CH cells showed that levels of cdk1 increased in all V2C cells, whereas cdk2, -4, -6, and cyclin E showed only a small upregulation with increasing VP40 produced (Figure 2A). Cyclins A and B1 were initially increased with low levels of VP40 but then decreased in a dose-dependent manner with increasing levels of VP40 produced. On the other hand, cyclin D1 increased in a VP40 dose-dependent manner. Increased levels of cyclin D1 have previously been shown to result in accelerated cell cycling and many types of oncogenic phenotypes [62–64]. In addition, several viruses, including HTLV-1, HPV, EBV, human neurotropic JC virus, and human respiratory syncytial virus, have previously been shown to influence the levels of cyclin D in infected cells to aid in their viral life cycle [65–70].
Figure 2.
Effect of nuclear VP40 on cyclin and cdk regulation and activity. (A) Log phase 293T, V2CL, V2CI, and V2CH cells were harvested, lysed, and subjected to SDS/PAGE for western blot analysis of cyclin D1 (CycD1), cyclin E (CycE), cyclin A (CycA), cyclin B1 (CycB1), cdk4, cdk6, cdk2, cdk1, and actin levels. (B) Five hundred micrograms of 293T or V2CH whole-cell extracts were used for IP with 10 µg of either normal rabbit immunoglobulin G (IgG) or α-CycD1. IPed material was incubated with 50 µL 30% of Protein A/G for 2 hours, followed by two 1× PBS washes and 1 kinase buffer wash. Pellets were resuspended in kinase buffer, and 15 µL samples were incubated with 50 µg of either no peptide, 780S, or 780A peptide, along with 2 µL of [γ-32P] ATP. Some samples were also treated with Fascaplysin ([Fascap.] 1 µM). All samples incubated for 1 hour, followed by dotting onto Whatman glass microfiber filters and drying for 30 minutes. Filters were incubated in 1× TE buffer with gentle agitation for 2 days, dried, and then quantified with a scintillation counter. Background levels of CycD1 kinase activity (α-CycD1 IP with 780A peptide substrate) are indicated by black bar. (C) Log phase 293T and V2CH cells were harvested for separation of cytoplasmic (Cyt) and nuclear (Nuc) compartments with the NE-PER Nuclear and Cytoplasmic Extraction Reagent Kit (Thermo Fisher Scientific). Western blot analysis for levels of VP40, CHMP6, HDAC1, and Actin was performed. (D) Log phase 293T and V2CH cells (3 × 106) were harvested and cross-linked with 1% formaldehyde for 1 hour followed by quenching with 1.25 glycine (9:1 cell suspension/glycine). Samples were sonicated, and 100 µL of each sample (corresponding to approximately 5 × 105 cells) was used for IP with 1 µg of anti-Pol II, α-p300, or α-VP40 at 4°C overnight. The next day, Protein A/G (30% slurry) was added and incubated for 2 hours at 4°C. Complexes were washed once with each TNE300 + 0.1% NP40, TNE150 + 0.1% NP40, TNE50 + 0.1% NP40, and IP wash buffer before the addition of proteinase K (800 units/mL). Samples were incubated for 15 minutes at 65°C, reversing solution was added, and samples were incubated for an additional 90 minutes at 65°C. DNA was purified, and qPCR was performed with 2 μL of undiluted DNA with primers for the CycD1 promoter region (spanning −180 to +238 from the messenger RNA start site at +1). The absolute quantification of the samples was determined based on the cycle threshold value relative to the standard curve generated from serial dilutions of DNA from 293T cells. Data are presented as percentage (%) of the input (DNA purified from sonicated samples before IP with specific antibodies). Student’s 2-tailed t test compares V2CH ChIPed DNA with corresponding 293T ChIP samples (**, P < .01).
Cyclin D1’s primary role during cell cycle is to bind to its cdk partner(s), cdk4 or cdk6, to activate its kinase activity, and thereby phosphorylate its substrate Rb to induce cell cycle progression from G1 to S phase [62]. Therefore, we investigated the ability of cyclin D1 from V2C cells to initiate phosphorylation of its substrate Rb at the Serine-780 residue. Two 13-mer peptides were generated: one 780S peptide mapping to a cyclin D1/cdk4 phosphorylation site of Rb, and one 780A peptide (S780A point mutation) [71]. Control 293T and V2CH whole-cell extracts were immunoprecipitated with α-cyclin D1 or IgG. Precipitated complexes were incubated with either 780S or 780A peptide, and some were incubated with Fascaplysin (cdk4/6 inhibitor). In vitro kinase assays were then performed using [γ-32P] ATP. Results in Figure 2B show that in 293T cells, active cyclin D1 complexes phosphorylated the 780S peptide as expected, whereas the 780A peptide and the 780S peptide incubated with Fascaplysin had only minimal background levels of phosphorylation. It is interesting to note that V2CH cells had an increased level of phosphorylation of 780S by IPed cyclin D1 compared with 293T cells. Moreover, incubation with Fascaplysin did not completely ameliorate the cyclin D1 activity on the 780S peptide. Taken together, these results indicate that cyclin D1 has a higher level of activity in V2CH cells compared with 293T cells, and that inhibition of active cyclin D1/cdk4/6 complexes in V2CH cells by Fascaplysin is less effective. This finding could perhaps be due to an overall greater amount of cyclin D1 present in V2CH cells, and therefore a higher concentration of Fascaplysin may be needed to prevent kinase activity.
VP40 Is Present in the Nucleus and Binds the Cyclin D1 Promoter
Based upon our observations of accelerated growth and increased levels of cyclin D1 in cells producing VP40, we next wished to investigate potential factors contributing to cyclin D1 transcription. Previous studies have shown the intriguing presence of EBOV VP40 within the nucleus, especially at early time points in infection [72–74]. To confirm VP40 presence in the nuclear compartment of our VP40 clones, cytoplasm and nuclear fractions of 293T and V2CH cells were isolated by NE-PER Nuclear and Cytoplasmic Extraction kit. CHMP6 (cytoplasmic protein; ESCRT-III component) and HDAC1 (nuclear protein involved in chromatin remodeling) were used as controls for cytoplasmic and nuclear compartments, respectively. Actin was also used to measure even loading of samples, because Actin has been shown to be abundantly present in both the cytoplasm and the nucleus [75, 76]. Results in Figure 2C show that VP40 protein was present in both the cytoplasm and the nucleus of V2CH cells. We were surprised to find that the levels of VP40 in the nuclear fraction were nearly equivalent with those seen in the cytoplasmic compartment, representing a much higher proportion of nuclear VP40 than we had anticipated. Therefore, we hypothesized that VP40 may potentially act as a transcription factor for the cyclin D1 promoter to elicit increased levels of cyclin D1, thereby accelerating the growth of host cells. RNA Polymerase II (Pol II) is found on the promoters of actively transcribed genes, and p300 has previously been shown to be involved in the activation of transcription of cyclin D1 and other mitogen-induced genes involved in cell growth and differentiation [77, 78]. Along these lines, to ascertain whether Pol II, p300, or VP40 were potentially involved in the transcriptional upregulation of cyclin D1, we performed ChIP analysis of the cyclin D1 promoter in both 293T and V2CH cells. Data in Figure 2D show that in 293T cells, background levels of Pol II, p300, and VP40 were <3% of the DNA input (2.8%, 2.6%, and 2.7%, respectively). The background levels of these, particularly VP40, may be partially attributed to nonspecific binding of proteins during the IP procedure during ChIP. Interestingly, Pol II on the cyclin D1 promoter more than doubled to 5.9%, whereas p300 increased more than 4-fold to 11.11% of DNA input in V2CH cells. Even more surprising, VP40 was present on the cyclin D1 promoter in V2CH cells at an astounding 38.4% of DNA input. Collectively, these data suggest that VP40, Pol II, and p300 are present at elevated levels on the cyclin D1 promoter in cells producing VP40, which may result in the upregulated transcription of cyclin D1. This in turn may be responsible for the accelerated cell cycling of VP40-producing cells. Additional consequences of this abundance of nuclear VP40 are likely, which should be the focus of future studies.
Altered Growth Dynamics Occur in Other Cell Types Producing VP40
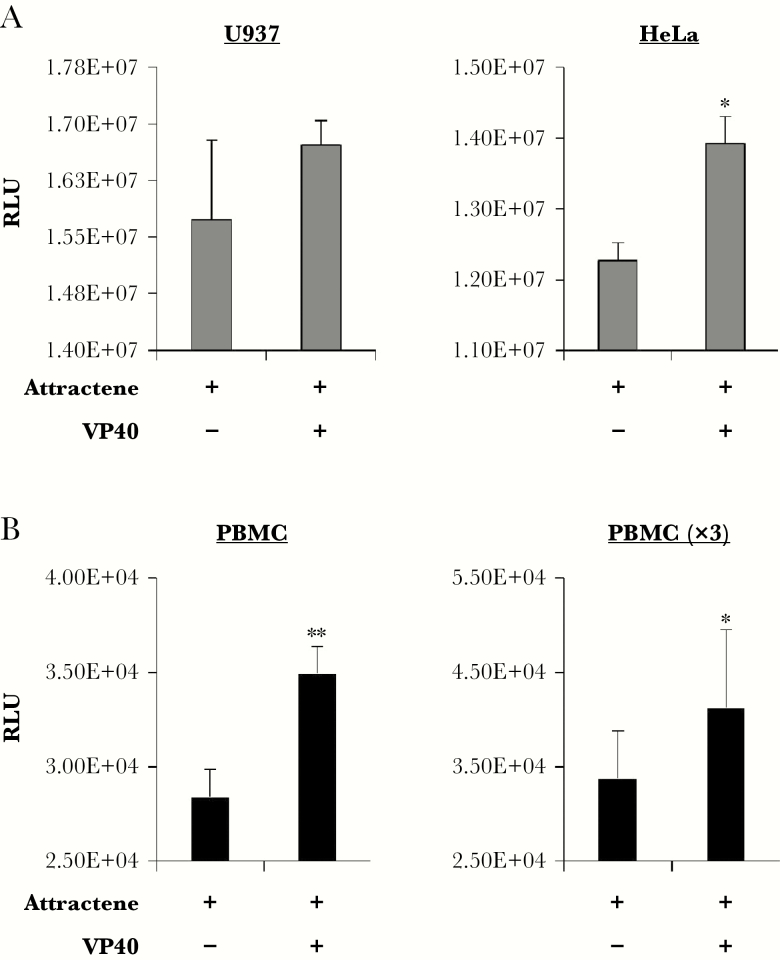
To determine whether production of VP40 resulted in upregulated growth in other cell types, we transfected U937 monocytes and HeLa cells with VP40-producing plasmid using attractene reagent alongside attractene-treated controls (no plasmid) for 3 days. Cells were then analyzed for differences in viability. Data in Figure 3A show that transfection with VP40 had little effect on U937 cells, whereas HeLa cells were significantly increased in viability. Next, to explore the effect of VP40 production in primary cells, PBMCs were transfected with CMV-VP40 for 3 days with attractene reagent, followed by analysis of cell viability. Results in Figure 3B show one representative PBMC sample, as well as the pooled data for PBMCs tested in biological triplicate (PBMC ×3). For all PBMCs tested, an increase in cellular viability was observed upon transfection with VP40 in comparison with control-treated cells. Collectively, these data point to an increased rate of growth in some cell types other than 293T, including primary cells, and thereby potentially indicating a cell type-dependent effect of VP40 on growth regulation.
Figure 3.
Alteration of cell viability by VP40 in multiple cell types. Cells including (A) U937, HeLa, and (B) 3 peripheral blood mononuclear cells (PBMCs) log-phase cultures (~1.65 × 105 cells) were transfected with attractene and 1.5 µg of cytomegalovirus-VP40 plasmid. The PBMCs received a 1-time treatment of 50 IU/mL of interleukin-2 the day before transfection. Control cells received attractene treatment alone. Cell viability was assayed 3 days post-transfection. Statistical analysis by Student’s 2-tailed t test compares control cells with transfected cells (*, P < .05; **, P < .01).
VP40 Alters Exosome Biogenesis in a Cell Cycle-Dependent Manner
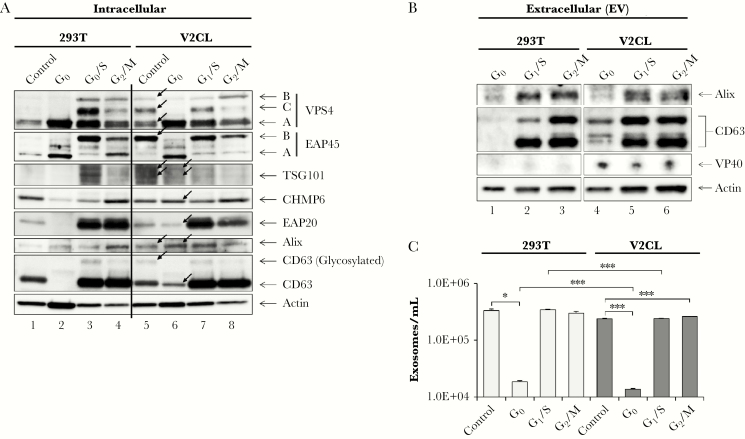
We previously showed that exosomes from V2CL cells are enriched in specific markers CD63 and Alix, whereas donor cells contain higher levels of ESCRT proteins [12, 13]. Therefore, we asked whether the altered cell cycle dynamics in VP40 donor cells could impact EV production. Control 293T and V2CL cells were blocked at G0, G1/S, and G2/M stages of the cell cycle and analyzed for levels of ESCRT proteins and exosomal markers (Figure 4A). Side-by-side analyses of EV contents and AChE activity of the exosomes produced from cell cycle blocked cells were likewise performed (Figure 4B and C). Data in Figure 4A show that, interestingly, unsynchronized control V2CL cells had drastic differences in ESCRT protein levels compared with control 293T cells (Figure 4A, compare lane 5 vs 1). Consistent with our previous findings [12], levels of TSG101 and Alix were also increased in unsynchronized V2CL cells. In addition, V2CL cells had a greater abundance of a modified form of EAP45 (EAP45 B) and glycosylated CD63 when unsynchronized. Moreover, control V2CL cells contained a distinct pattern of VPS4 bands between 50 and 70 kDa (VPS4A-C) that resembled 293T cells at the G1/S or G2/M phases. The functional significance of these larger forms of EAP45 and VPS4 is currently uncertain. Changes at G0 were also present, with increased levels of TSG101, CHMP6, EAP20, Alix, and CD63 in V2CL compared with 293T cells (Figure 4A, compare lanes 6 vs 2). Changes between 293T and V2CL cells at G1/S or G2/M phases were less drastic with only minor differences. Collectively, these data indicate that V2CL cells may have a different cell cycle dynamic compared with 293T control cells, particularly at G0, resulting in altered patterns of ESCRT proteins and exosomal markers.
Figure 4.
Differential biogenesis of exosomes at different phases of the cell cycle. 293T and V2CL cells were blocked at G0 (starvation; 0.1% fetal bovine serum DMEM), G1/S (20 mM of hydroxyurea), and G2/M (18-hour 20 mM of hydroxyurea pretreatment, followed by release for 1 hour and subsequent treatment with 50 ng/mL of nocodazole) for 5 days. Control (unsynchronized) cells were also incubated for 5 days. Black arrows point to bands of noticeable difference between V2CL and 293T cells. (A) Blocked cells and controls were harvested, washed twice in 1× PBS, and lysed. Samples were run on a 4–20% Tris-glycine gel and analyzed by western blot for the presence of ESCRT pathway proteins (VPS4, EAP45, TSG101, CHMP6, and EAP20), exosomal markers (Alix and CD63), and Actin. (B) Cell-free supernatants from blocked cells were harvested and passed through a 0.22-µm filter. One milliliter of filtered supernatant was incubated with 30 μL of NT80/82 particles overnight at 4°C. The next day, the NT pellet was washed once in 1× PBS and resuspended in 10 μL Laemmli buffer, followed by SDS/PAGE and western blot analysis for VP40 protein, exosomal markers CD63 and Alix, and Actin. (C) One milliliter of filtered supernatant from blocked and control cells was incubated with 30 μL of NT80/82 particles overnight at 4°C. The next day, the NT pellets were isolated, washed, and subjected to AChE assay for quantification of exosomes. Statistical analysis was completed by Student’s 2-tailed t test (*, P < .05; ***, P < .001).
Analysis of exosomal markers from cell cycle-blocked V2CL cell EVs revealed an increase in all forms of CD63 at G0 compared with 293T cells (Figure 4B). When testing for AChE activity (indicative of exosomal abundance), the total number of exosomes from unsynchronized (control) 293T vs V2CL cells was not significantly different (Figure 4C). However, it was interesting to note that at G0, the number of exosomes was reduced by >1 log compared with unsynchronized control exosomes for both cell types. This finding was corroborated by the distinct reduction of most ESCRT (Figure 4A) and exosomal proteins at G0 compared with levels at G1/S and G2/M phases (Figure 4A and B). Both cell types had similar levels of exosomes produced when blocked at G1/S compared with unsynchronized (control) cells; however, V2CL cells produced significantly more exosomes at G2/M compared with control, whereas 293T cells did not. In addition, V2CL cells produced significantly fewer exosomes at G0 and at G1/S phases compared with 293T cells at the same phases (Figure 4C). Together, these results suggest that exosomes and EVs are produced in a cell cycle-dependent manner, with cells at G0 phase producing significantly fewer vesicles. Moreover, cells producing VP40 may have an altered pattern of cell cycle-driven exosomal production, as demonstrated by differential patterns of ESCRT and exosomal marker protein expression.
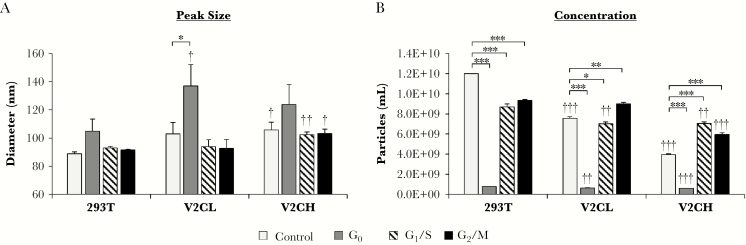
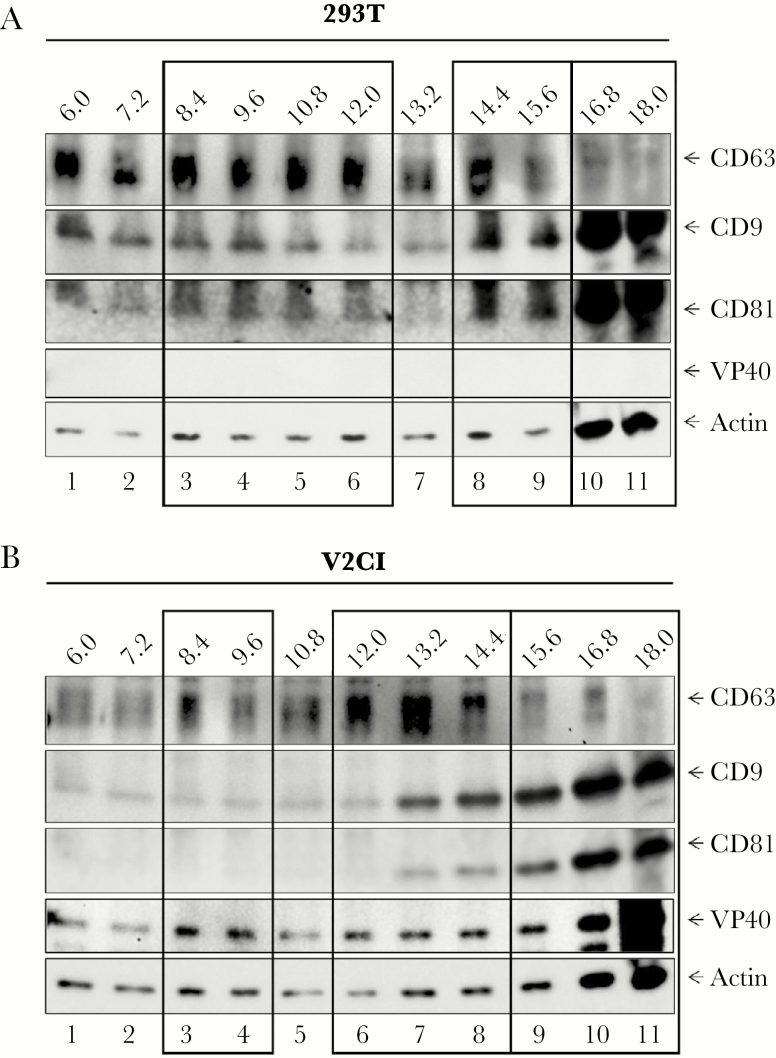
Next, we wished to more closely examine the effects of cell cycle blocks on EV production in cells producing VP40 at low and high levels. 293T, V2CL, and V2CH cells were blocked at G0, G1/S, and G2/M phases for 5 days, followed by harvesting of the cell culture supernatants, filtration (through 0.22 μm), and analysis of the resulting EVs by ZetaView. Data in Figure 5A display the peak diameter size (mode) of vesicles released. V2CL cells had significantly larger vesicles produced at G0 compared with 293T cells, whereas V2CH cells had larger EVs when not only blocked at G1/S and G2/M phases, but also in untreated cultures when compared with 293T cells. In addition, larger vesicles were produced from all 3 cell types at G0 in comparison with untreated controls, although this difference was not consistently significant. The overall trend regarding EV concentration was that vesicles from V2C cells decreased in number in a VP40 level-dependent manner when compared against 293T cell groups (Figure 5B). Furthermore, amongst all cell types, the number of EVs present at G0 phase were significantly reduced from unsynchronized (control) cultures. This finding supports our earlier conclusions that exosomes may be produced at lower levels when cells are resting at G0 (Figure 4B and C). It should be noted that ZetaView analysis detects the total number of vesicles or large particles present, with exosomes comprising only a fraction of these. Therefore, to determine the distribution of true exosomes containing VP40, we used our previously published strategy of exosome characterization by density gradient ultracentrifugation [50]. Through this methodology, we have shown that it is possible to clearly separate out exosomes from potentially contaminating viral particles, thus making more detailed characterizations of specific EV populations possible. Here, we grew control 293T and V2CI cells for 5 days, harvested the supernatants, and incubated the unfiltered supernatants in ExoMAX, followed by separation of resulting extracellular material through iodixanol density gradient fractionation. Fractions were subjected to western blot analysis for exosomal marker proteins and VP40. Results in Figure 6A show that in 293T cells, CD63+CD9+CD81+ (exosome) fractions fell into 3 distinct categories. First, fractions 8.4–12.0 contained high levels of CD63 and moderate levels of CD9 and CD81. Previous publications have classified this group as true exosomes, distinct from viral particles and apoptotic bodies [16, 50, 79]. Second, in fractions 14.4–15.6, CD63 was present in moderate amounts, whereas levels of CD9 and CD81 increased. This population was also consistent with an exosomal category containing viral particles demonstrated in previous publications [16, 50, 79], although, in our case, this group is likely to be exosomes that are larger in size or more dense than the smaller ones seen in fractions 8.4–12.0. Finally, the 16.8–18.0 fractions showed a distinct reduction of CD63 but a drastic increase in CD9, CD81, and Actin. It is likely that these larger or denser vesicles are not true exosomes but either microvesicles or apoptotic bodies. In contrast, V2CI cells had a noticeably different distribution of exosomal markers, although 3 distinct populations were still present (Figure 6B). Fractions 8.4–9.6 were almost absent in CD9 and CD81, although CD63 was still present in higher amounts compared with background. In addition, a shift to the right of the major CD63+CD9+CD81+ population from fractions 10.8–12.0 in 293T EVs to fractions 12.0–14.4 was observed. Finally, a population with high levels of CD9 and CD81 and low levels of CD63 was seen from fractions 15.6–18.0, similar to 293T cells. VP40 protein, although present in all fractions to various extents, was seen to be enriched in each of the 3 vesicle populations, especially in fraction 18.0. This abundance of VP40 in fraction 18.0 is likely to be due to the presence of VLPs, beecause there was no filtration through 0.22 µm of the supernatants before vesicle enrichment and fractionation in this experiment. Altogether, these results suggest that cells producing VP40 release fewer EVs that are slightly larger in size, and that these cells in general release significantly fewer (although larger) exosomes at G0. Furthermore, outside of VLPs and virions, VP40 is released in EVs (primarily exosomes) consistent with our previous observations [12, 13], but it may also be released in other larger vesicles and potentially as free protein or within protein complexes.
Figure 5.
Extracellular vesicles released by VP40-producing cells at different phases of the cell cycle. 293T, V2CL, and V2CH cells were blocked at G0 (starvation; 0.1% fetal bovine serum DMEM), G1/S (20 mM of hydroxyurea), and G2/M (18-hour 20 mM of hydroxyurea pretreatment, followed by release for 1 hour and subsequent treatment with 50 ng/mL of nocodazole) for 5 days. Supernatants were harvested, filtered (0.22 μm), and analyzed by ZetaView for size (peak [mode] diameter) (A) and concentration of particles (B). Statistical analysis by Student’s 2-tailed t test compares V2CL and V2CH cell cycle-blocked groups with corresponding 293T groups (†, P < .05; ††, P < .01; †††, P < .001). Additional Student’s 2-tailed t test compares cell cycle-blocked groups with controls of their own cell type (*, P < .05; **, P < .01; ***, P < .001).
Figure 6.
Iodixanol gradient separation of extracellular vesicles (EVs) from 293T and VP40-producing cells. 293T and V2CI cells were grown in exosome-free media for 5 days, followed by harvesting of the supernatant and incubation with ExoMAX (1:1 reagent/filtered supernatant) reagent overnight at 4°C. The EVs were pelleted, resuspended in 300 µL of sterile 1× PBS, and loaded onto a 6–18% iodixanol density gradient (1.2% increments). Samples were ultracentrifuged for 90 minutes at 100000 ×g, followed by harvesting and isolation of each fraction, and incubation with 30 µL of NT80/82 particles overnight at 4°C. The NT pellets were washed in 1× PBS, resuspended in 12 µL of Laemmli buffer, and loaded onto a 4–20% Tris-glycine gel. Western blot of 293T (A) and V2CI (B) fractions were analyzed for levels of VP40, CD63, CD81, CD9, and Actin. Major groups of EVs or exosome type are indicated by black boxes.
Filtration Effectively Removes Virion Particles From Samples, Leaving Viral Protein-Containing Extracellular Vesicles
Our rationale for utilizing 0.22-μm filters in our experiments was based upon the knowledge that Ebola virions and VLPs are commonly 1–2 μm in size or larger, are rarely straight, and therefore should not be able to effectively pass through a 0.22-μm filter [36, 37, 39, 42, 80]. In fact, Beniac et al [80] previously showed that approximately 53% of infectious virions were 982 ± 79 nm in length, with all other virions occurring in multiples of that size. No smaller virions containing complete genomes were observed in their investigation of over 2000 virions by electron microscopy. On the other hand, EVs and exosomes below 220 nm in diameter will be able to pass through 0.22-μm filtration for downstream analysis. To test this concept, we used VLPs containing VP40, NP, and GP EBOV proteins. A simple filtration (0.22 μm) of VLPs spiked into 1 mL PBS is shown in Supplementary Figure 2A, alongside filtered PBS alone (mock) and unfiltered VLP controls. Filtered samples were incubated with NT80/82 particles and processed for western blot analysis of EBOV proteins. The results indicate that filtration of VLPs in small volumes drastically reduced the levels of EBOV proteins present to undetectable levels.
Next, to further characterize the distribution of VLPs in comparison to EVs by size, we spiked 10 μg of VLPs into 10 mL of V2CH supernatants. VLP-spiked supernatants were then either filtered (0.22 μm) or left unfiltered and incubated with a vesicle-enrichment agent (ExoMAX). The resulting material was then loaded on top of qEV (IZON) size exclusion columns, and fractions were collected corresponding to the column void volume (fraction nos. 2–6) and those known to contain vesicles larger than 70 nm in diameter (fraction nos. 7–11). Fractions were next incubated with NT80/82 particles to concentrate material, followed by western blot analysis for viral proteins, exosomal markers, and Actin. Results in Supplementary Figure 2B show that, unsurprisingly, proteins originating from unfiltered VLPs mostly appeared in the first fraction after the void volume of the column (fraction no. 7), with some overlap into the next 1–2 fractions. This was shown by the strong presence of NP and GP, particularly the larger glycosylated form of GP, and the low levels of Actin, which we have not observed to be present in our control VLP but is present in EVs (Supplementary Figure 2B, compare lane 1 to lanes 9–12). However, upon filtration through 0.22 μm, approximately all EBOV proteins vanished from fraction number 7, and there was a sizeable reduction in the levels of GP and NP in fraction numbers 8 and 9. Notably, not all NPs and GPs1,2 (originating from VLPs) were eliminated from the fractions after filtration.
Another interesting observation centered around the visible forms of GP. Specifically, in the unfiltered fractions, with the higher fraction numbers (corresponding to smaller vesicles), the larger glycosylated form of GP decreased while the levels of lower molecular weight forms of GP increased. In the filtered fractions, the glycosylated larger form of GP was present only at very low levels, whereas lower molecular weight GP levels were relatively high. In comparison to the VLP control (lane 1), the predominant form of GP present was the glycosylated form. Together, these results suggest that glycosylated GPs may be the principal form within VLPs, whereas the lower molecular weight forms of GP may be more likely to be associated with EVs. Collectively, these data may indicate that VLPs appear mainly in the larger fraction number 7 and to a lesser extent in fraction numbers 8 and 9, whereas EVs and true exosomes will be present in fraction numbers 8–11.
VP40 Is Present at Varying Levels in Extracellular Vesicles In Vitro and In Vivo
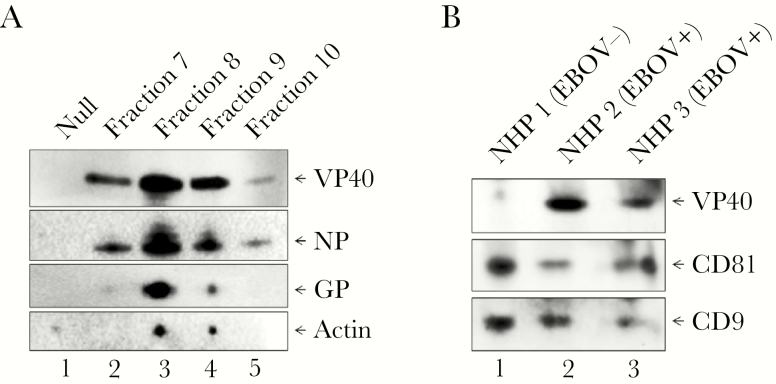
We have previously shown that exosomes from VP40-producing transfected cells contain VP40 in vitro [12, 13], but, to our knowledge, VP40 has never been shown to be present in EVs in the context of full viral infection. To address this, we infected HUVECs with EBOV/Kik (MOI of 1) under BSL-4 containment for 3 days, followed by harvesting of the supernatants, filtration through 0.22 μm, and incubation with ExoMAX. Enriched vesicles were then loaded onto qEV size exclusion columns. Fraction numbers 7–10 were then collected, incubated with NT80/82 particles, and analyzed by western blot for EBOV proteins VP40, NP, and GP, and Actin. Results in Figure 7A demonstrate that VP40, NP, and GP were all present in the EVs concentrated from EBOV-infected cells. In addition, all 3 EBOV proteins were observed to be most highly concentrated in fraction number 8, followed closely by fraction number 9, which we had shown to be the fractions where most EVs and exosomes appear (Supplementary Figure 2B). VP40 and NP were also present in fraction number 7, whereas GP was present at very low levels in this fraction. It is possible that this fraction may represent slightly larger vesicles that do not incorporate GP. All 3 proteins were much less abundant in fraction number 10, which contained smaller vesicles. Actin followed a similar trend, with the most Actin present in fraction numbers 8–9, and little to no detectable protein present in fraction numbers 7 or 10. Collectively, these results indicate that not only VP40 but GP and NP are also packaged into vesicles in cells actively infected with EBOV.
Figure 7.
The presence of VP40 in exosomes in in vitro and in vivo EBOV-infected cells. (A) HUVECs were cultured and infected with EBOV (MOI of 1) and incubated for 3 days under BSL-4 containment. Two milliliters supernatant were harvested, passed through a 0.22-µm filter, and incubated with ExoMAX (1:1 reagent/filtered supernatant) reagent overnight at 4°C. EVs were pelleted, resuspended in 0.5 mL 1× PBS, and loaded on qEV columns. Fraction numbers 7–10 (0.5 mL each) were collected and separately incubated with 30 μL NT80/82 at room temperature for 1 hour. The EV-bound NTs were washed with 1× PBS, followed by resuspension in 10 μL 2× NuPAGE LDS sample buffer, heating at 95°C for 10 minutes, and loading onto a 4–12% Tris-glycine gel for subsequent western blot analysis for VP40, GP, NP, and Actin levels. Negative control (Null) samples consisted of purified exosomes from uninfected HUVECs. (B) Gamma-irradiated and inactivated NHP (rhesus monkey) serum samples were obtained. NHP 1: day 0 prebleed sample. NHP 2: pool of day 4 and day 5 postinfection (pi); NHP 2 died on day 7 post-EBOV infection. NHP 3: pool of day 8–11 pi; NHP 3 died on day 12 post-EBOV infection. One hundred microliters of serum were diluted with 400 μL sterile 1× PBS and filtered (0.22 µm). Twenty-five microliters NT80/82 particles were incubated with the filtered samples at 4°C overnight. The next day, NT pellets were washed once in 1× PBS, resuspended in 12 μL Laemmli buffer, run on 4–20% SDS/PAGE, and analyzed by western blot for VP40 protein and exosomal markers CD81 and CD9.
To take this one step further, we next used inactivated NHP serum samples from 2 infected macaques and 1 uninfected prebleed animal. Inactivated samples were filtered (0.22 μm), EVs were concentrated with NT80/82 particles, and samples were analyzed by western blot for levels of VP40 and exosomal proteins. Data in Figure 7B show that exosomes from all 3 NHP serum samples were concentrated, as demonstrated by the presence of CD81 and CD9 tetraspanins. In addition, VP40 appeared at fairly high levels in the 2 serum samples from infected animals. Quantitative densitometry was used to calculate the approximate amounts of VP40 present in our V2CH cell EVs and how much was released from infected NHP samples. V2CH cells released approximately 13.7 ng of EV-associated VP40/mL, whereas NHP 2 and NHP 3 released ~319 and ~206.5 ng of EV-associated VP40/mL serum, respectively. In addition, we determined that VP40 composed on average approximately 55.8% of purified VLPs (containing GP, NP, and VP40; data not shown). It is interesting to note that more VP40 was present from NHP 2 serum, which succumbed to infection sooner (day 7 vs day 12) and whose serum was collected at earlier time points in infection in comparison with NHP 3 (days 4–5 vs days 8–11). Therefore, these results indicate that VP40 is present in EVs from in vivo EBOV infection, with higher levels potentially being prevalent during earlier time points or with more robust infection. Although western blot confirmed the presence of captured exosomes, we cannot be certain that we did not capture other EVs or free proteins in our analysis of NHP samples. Therefore, it cannot be ruled out that VP40 may be present in not only exosomes, but also a combination of EVs and/or as a free protein in vivo. Taken together, these results indicate that relatively high levels of VP40, NP, and GP1,2 are present in EVs from cells infected with EBOV both in vitro and in vivo.
VP40 in Extracellular Vesicles Induces Apoptosis in Recipient T Cells and Monocytes
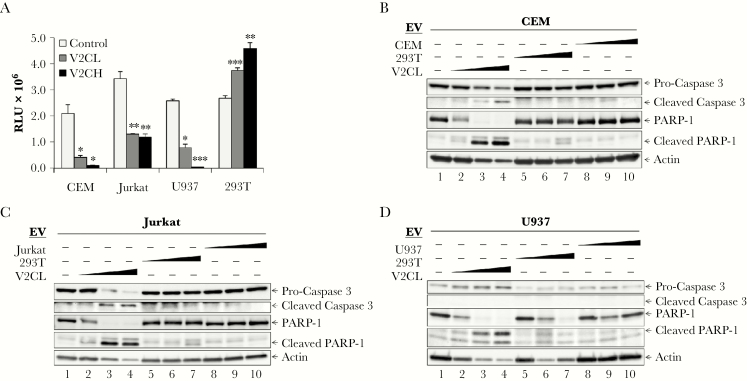
During EVD, bystander T cells mainly die by apoptosis, whereas monocytes/macrophages appear to die by necrosis [5, 7, 9, 10, 81–83]. We have previously shown that EVs from V2CL cells can induce death in recipient immune cells [12, 13]. Therefore, to test the ability of recipient cells to withstand low versus high levels of EV-associated VP40, we incubated CEM, Jurkat, U937, and 293T cells with 5-day filtered supernatants from 293T, V2CL, and V2CH cells, followed by measurement of cell viability 5 days later. Results in Figure 8A show that CEM and Jurkat T cells had a similar reduction in cellular viability from treatment with supernatants from both V2CL and V2CH cells. However, treatment of U937 monocytes with supernatants from these 2 lines showed a dose-dependent reduction in cellular viability. These data suggest that the T cells were equally sensitive to both low and high levels of extracellular VP40, whereas monocytes were more sensitive to higher doses of EV-associated VP40. It is interesting to note that when 293T cells were incubated with supernatants from VP40-expressing cells, 293T cells showed a significant increase in viability, potentially indicating a cell-type dependent mechanism of cell death induction by EV-associated VP40.
Figure 8.
Recipient monocyte and T-cell apoptosis. (A) Supernatants from 293T (Control), V2CL, and V2CH cells (5 days, 0.22 µm filtered, in exosome-free media) were used to treat CEM, Jurkat, U937, and 293T recipient cells. Recipient cells were seeded in a 96-well plate at 5 × 105 cells in 50 μL of fresh media, treated with 50 μL of filtered supernatants, and incubated 5 days, followed by analysis by CellTiter-Glo for cell viability. Statistical analysis by Student’s 2-tailed t test compares supernatant-treated groups with control cells of the same type (*, P < .05; **, P < .01; ***, P < .001). 293T, V2CL, CEM, Jurkat, and U937 cells were grown in exosome-free media for 5 days, followed by harvesting and filtering (0.22 µm) of the supernatant. Log-phase CEM, Jurkat, and U937 cells were plated at a density of 1 × 106 cells/mL and treated with increasing concentrations (100, 250, and 500 μL) of supernatant from either 293T, V2CL, or their own cell type. Cells were incubated for 5 days, followed by harvesting of the cells and lysis. Lysates of CEM (B), Jurkat (C), and U937 (D) cells were then run on a 4–20% Tris-glycine gel and subjected to western blot analysis for apoptotic markers procaspase 3 and PARP-1 and their cleaved forms, and Actin.
The mechanism of cell death under these conditions has not been previously explored. Along these lines, we incubated recipient CEM and Jurkat T cells and U937 monocytes with increasing concentrations of EVs from V2CL cells, EVs from control 293T cells, and EVs from their own cell type that were produced in the absence of VP40. Western blots were performed for cell death markers including Caspase 3 and PARP-1. Data in Figure 8B–D show that treatment with increasing concentrations of EVs from V2CL cells induced cell death by apoptosis, as shown by cleavage of Caspase 3 and PARP-1, in recipient T cells and monocytes. This phenotype was not observed with 293T cell EV treatment or EVs from the same cell type. It should be noted that very high levels of EVs from 293T cells stimulated some low levels of PARP-1 cleavage in all cell types; however, there was no cleavage of Caspase 3 detected. Together, these data indicate that EVs from VP40-producing cells cause death by apoptosis in recipient T cells and monocytes.
Because our V2C cells were kept under constant antibiotic selection, we wished to verify that induction of cell death in recipient cells was not due to residual hygromycin B within the 5-day supernatants. Therefore, 293T and V2CH cells were grown for 5 days in exosome-free media, followed by harvesting of the supernatants, filtration (0.22 μm), and ultracentrifugation. The resulting EV pellets were then resuspended in sterile PBS and used to treat recipient CEM cells in increasing concentrations. Cells were treated for 3 days followed by analysis of cell viability. Data in Figure 9A show that in comparison with CEM cells treated with the highest concentration of 293T EVs (75000 particles/cell), there was a significant dose-dependent decrease in recipient cell viability when treated with purified EVs from V2CH cells. This finding further verifies that EVs from V2C cells negatively impact recipient immune cells.
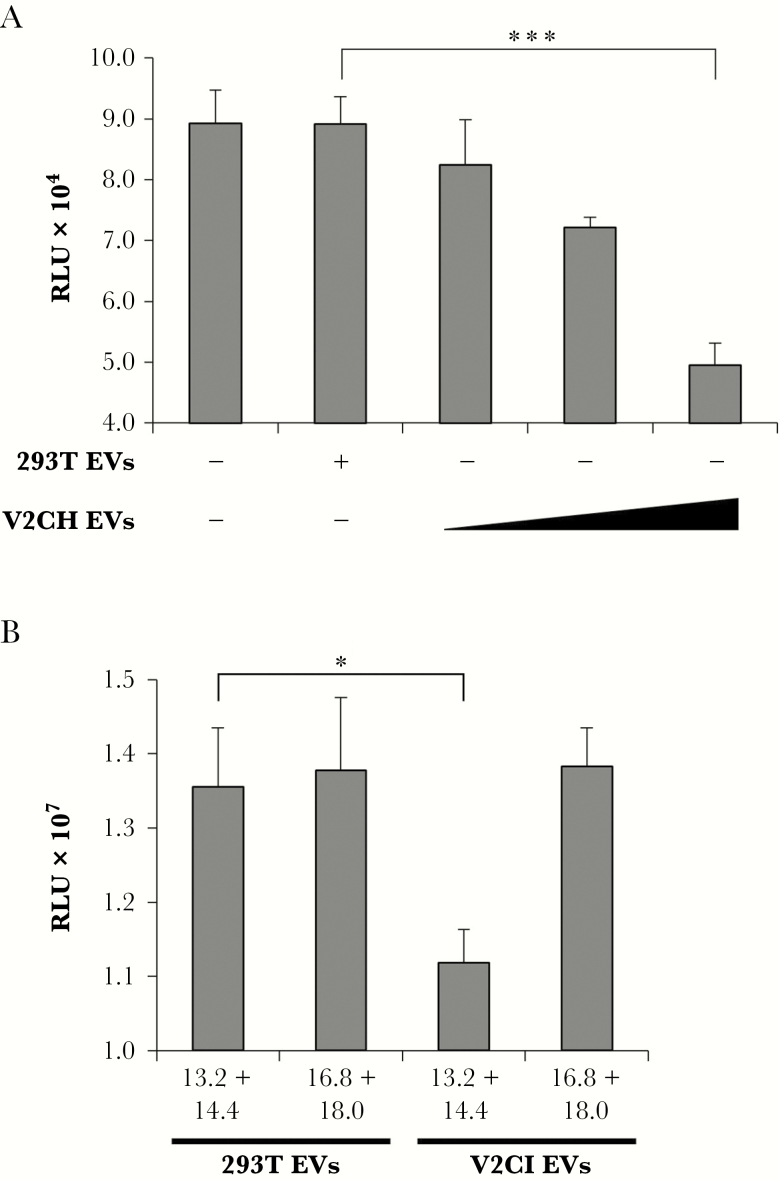
Figure 9.
Induction of recipient T-cell death by purified VP40 EVs. (A) 293T and V2CH cells were grown in exosome-free media for 5 days, followed by harvesting of cell-free supernatants and filtration through 0.22 μm. Supernatants were then spun at 100000 ×g for 90 minutes to pellet EVs, followed by resuspension in sterile 1× PBS. Concentrations of resulting ultracentrifuged EVs were determined with ZetaView analysis, followed by treatment of CEM cells with increasing concentrations of EVs (10000, 25000, or 75000 particles/cell) from V2CH cell type. Controls included CEM cells that were left untreated and CEM cells that received a treatment of the highest concentration of 293T cells (75000 particles/cell). Cells were incubated for 3 days followed by analysis of cell viability by CellTiter-Glo. (B) 293T and V2CI cells were grown in exosome-free media for 5 days. Cell-free supernatants were harvested and incubated with equal volumes of ExoMAX overnight at 4°C. The EVs were pelleted, resuspended in 400 µL sterile 1× PBS, and loaded onto a 6–18% iodixanol density gradient (1.2% increments). Samples were ultracentrifuged for 90 minutes at 100000 ×g, followed by harvesting and isolation of each fraction. Select fractions (13.2 + 14.4 and 16.8 + 18.0) were pooled and subjected to a second ultracentrifuge spin for 90 minutes at 100000 ×g diluted in 1× PBS to pellet the EVs away from residual iodixanol. Resulting EV pellets were resuspended in 100 µL sterile 1× PBS and used to treat recipient CEM cells at a concentration of 10000 particles per cell (concentrations determined by ZetaView analysis). Cells were incubated for 5 days followed by analysis of viability by CellTiter-Glo assay. Statistical analysis by Student’s 2-tailed t test compares groups treated with EVs from V2C cells to those treated with EVs from 293T cells (*, P < .05; ***, P < .001).
Given our previous characterization of EVs from V2CI cells in comparison with 293T cells (Figure 6), we next wished to determine which EV fractions were responsible for the induction of cell death in recipient cells. Supernatants from 293T and V2CI cells were harvested and separated through an iodixanol gradient, followed by pooling of select fractions from each cell type. Pooling of fractions 13.2 + 14.4 and 16.8 + 18.0 were chosen because, in V2CI cells, these were the 2 groups with the highest levels of VP40 and exosomal markers. Pooled fractions were then diluted in sterile PBS and subjected to a second ultracentrifuge spin to isolate the EVs away from residual iodixanol. The resulting EV pellets were then resuspended in PBS and used to treat recipient CEM cells at a concentration of 10000 particles/cell for 5 days, followed by analysis of cell viability. Treatment with EVs from the 13.2 + 14.4 fractions from V2CI cells resulted in a significant decrease in viability in CEM cells in comparison with those treated with 293T EVs from the same fractions (Figure 9B). On the other hand, T cells treated with the 16.8 + 18.0 fractions from either cell type did not differ in their viability. Collectively, these data may signify that the EVs within the 13.2 and 14.4 fractions from V2C cells can induce cell death in recipient immune cells.
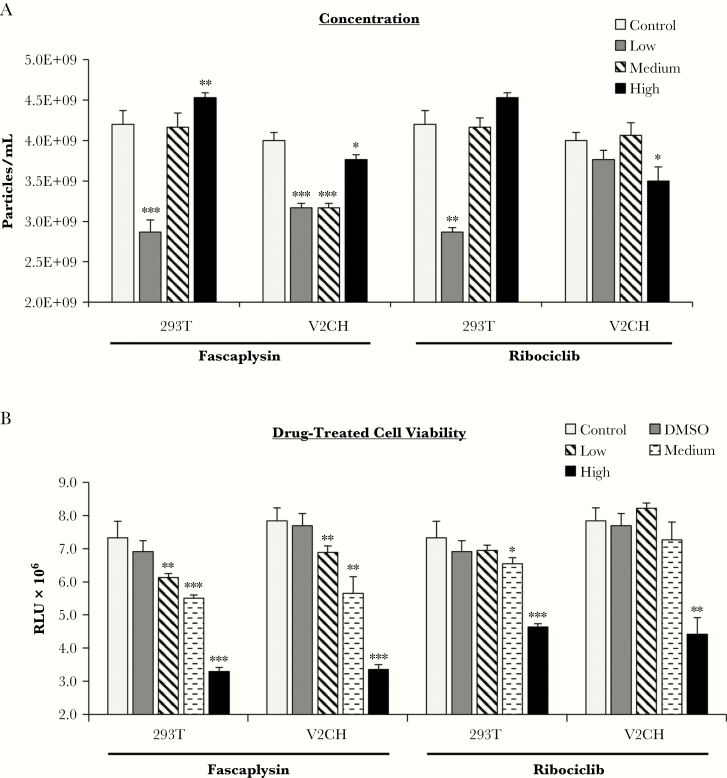
Cdk4/6 Inhibitor Treatment of VP40-Producing Cells Affects Extracellular Vesicle Release
Our data showed that EVs from VP40-producing cells were capable of causing apoptosis in recipient immune cells (Figure 8) and that the biogenesis of these EVs was potentially tied to regulation of the cell cycle (Figures 4 and 5). Furthermore, we demonstrated that accelerated cell cycle was potentially driven by the upregulation of cyclin D1 by nuclear VP40 (Figure 2). Therefore, we next investigated the utility of cdk4/6 inhibitors Fascaplysin and Ribociclib (LEE011; FDA-approved) in reducing EV production from 293T and V2CH cells. Cells were treated for 5 days with increasing concentrations of Fascaplysin (0.1, 0.5, and 1.0 µM) and Ribociclib (0.1, 1.0, and 10.0 µM), followed by analysis of cell viability and EV concentration. When 293T cells were treated with low levels of either inhibitor, the number of EVs released significantly decreased. However, with increasing concentrations of drugs, the production of vesicles increased in a dose-dependent manner (Figure 10A). These results may be due to the toxicity of the drugs at high levels, as the viability of both 293T and V2CH cells were significantly decreased with increased doses of either drug (Figure 10B). More specifically, low levels of cdk4/6 inhibitors in 293T cells may result in the blocking of cell cycle at G1, and thereby result in lower levels of EVs released, whereas higher levels of the drugs may become toxic, and result in the increased release of vesicles. In contrast, V2CH cells showed significantly decreased levels of EV production with all doses of Fascaplysin, although with the highest dose EV production decreased less significantly in comparison to the lowest dose tested (Figure 10A). These EV reductions occurred despite significant decreases in cell viability in V2CH cells (Figure 10B). Ribociclib treatment of V2CH cells showed less significant results compared with Fascaplysin treatment, with only the highest dose resulting in any significant reduction of EV numbers or cellular viability (Figure 10A and B). Together, these results suggest that Fascaplysin treatment results in fewer EVs from V2CH cells compared with 293T cells with higher doses, although high doses also result in significant decreases in viability in both cell types. In addition, Ribociclib treatment may impact the cell viability and EV biogenesis of VP40-producing cells less than 293T cells. The indication that 293T cells react to both cdk4/6 inhibitors in a similar way, but V2CH cells do not, hints that these drugs may impact cell cycle regulation and EV biogenesis pathways in distinctly different ways. Although the mechanism of action for Fascaplysin is currently uncharacterized, Ribociclib is known to interact directly with cdk4 and cdk6 molecules at their ATP pocket [84, 85]. Future study into the divergent mechanisms used by these drugs and how they may interact with pathways modulated by VP40 may potentially be advantageous to determine the potential therapeutic utility of cdk4/6 inhibitors in infection.
Figure 10.
Effect of cdk4/6 inhibitors on VP40-producing cell viability and extracellular vesicle biogenesis. 293T and V2CH cells were treated with low, medium, and high concentrations of Fascaplysin (0.1, 0.5, 1.0 µM) or Ribociclib (0.1, 1.0, 10.0 µM) for 5 days. DMSO was also used at a final concentration of 1%. (A) Supernatants were harvested, filtered (0.22 µm), and analyzed by ZetaView for concentration of particles. (B) Cell viability was also measured by CellTiter-Glo. Statistical analysis by Student’s 2-tailed t test compares drug-treated groups to untreated controls of their own cell type (*, P < .05; **, P < .01; ***, P < .001).
Extracellular Vesicles From VP40 Donor Cells Contain Cytokines Associated With Ebola Virus Disease Pathogenesis
We previously showed that exosomes from VP40-producing cells are capable of inducing cell death in recipient T cells and monocytes [12, 13]. We also confirmed here that the mechanism of cell death is by apoptosis (Figure 8). Along these lines, we attempted to gain insight into proteins contained in VP40 exosomes that may potentially point to a mechanism for the induction of cell death in recipient cells. 293T, V2CL, and V2CH cells were cultured for 5 days in exosome-free media. Supernatants were harvested, filtered (0.22 µm), and incubated with ExoMAX to enrich for EVs. Samples were then loaded onto a 6%–18% OptiPrep iodixanol gradient, and 10.8 and 12.0 fractions were harvested for use. We chose the 10.8 and 12.0 fractions because previous publications have shown that these fractions are specifically enriched for exosomes, with fewer contaminating vesicles or viral particles [16, 79]. Fractions were then incubated with NT80/82 particles and processed for analysis of tandem mass spectra. The data presented in Supplementary Table 1 show the proteins significantly upregulated in V2C EVs, with protein scores from 10.8 and 12.0 fractions for each cell type summated. Overall, there was an interesting trend gleaned from the number of proteins identified from each sample. The 10.8 fractions from 293T, V2CL, and V2CH cells had 826, 537, and 261 detected proteins, respectively, potentially showing an overall decrease in the number of proteins present with increase in VP40 production levels (data not shown). However, for the 12.0 fraction, the opposite trend was seen: 293T, V2CL, and V2CH EVs had 387, 1393, and 1680 proteins identified, respectively (data not shown). These results could potentially indicate that the total number of EVs had shifted from the 10.8 fraction more towards the 12.0 fraction for V2C cells, whereas 293T cells possibly possessed more EVs in the 10.8 fraction in comparison to the 12.0.
These data support our previous conclusions that the EVs from V2C cells are shifted more towards the right (ie, are more dense; Figure 6), and therefore they may contain a greater and more diverse quantity of cargo. The presence of heat shock protein (HSP)90, HSP70, and heat shock cognate 71 seen in abundance across all 3 cell types was indicative of all samples containing exosomes. A large increase in the number of proteins related to RNA binding was seen with increasing levels of VP40. Specifically, numerous RNA binding proteins, including heterogeneous nuclear ribonucleoproteins, were found in EVs from V2C cells, whereas only 1 was detected in 293T cell EVs. In addition, several DEAD-box protein (DDX)-type helicases were shown to be upregulated in EVs from VP40-producing cells. Other categories of interest that were upregulated with the presence of VP40 included additional vesicle-associated, translation-associated, nuclear or transcription-associated, proteasome or cell death-related, protein transport, and RNA splicing proteins. Of particular interest, 3 cytokines (vascular endothelial growth factor [VEGF], transforming growth factor beta-1 [TGF-β1] precursor, and insulin-like growth factor II [IGF-II]) and 2 corresponding binding proteins (latent-transforming growth factor beta-binding protein 1 precursor and insulin-like growth factor-binding protein 4 precursor) were found to be present in the EVs.
It has been well documented that an eruption of cytokines, or a “cytokine storm,” often accompanies fatal cases of EBOV infection [6, 8, 86, 87]. Specifically, cytokines including granulocyte-macrophage colony-stimulating factor (GM-CSF), growth-regulated oncogene (GRO)-α, IL-1β, IL-2, IL-6, IL-8, IL-10, IL-15, IL-18, interferon (IFN)-α, -β, and -γ, IFN-inducible protein-10 (IP10), Eotaxin, monocyte chemoattractant protein (MCP)-1, macrophage-inflammatory protein (MIP)-1α and -1β, macrophage colony-stimulating factor (MCSF), RANTES, TGF-β1, TNF-α, and VEGF have all been implicated as differentially up- or downregulated during EBOV pathogenesis [8, 86–89]. This led us to question whether additional cytokines could potentially be present or upregulated in VP40 exosomes or EVs. To this end, filtered 5-day supernatants and pooled 10.8–12.0 exosome fractions from 293T and V2CH cells were run side-by-side on human cytokine arrays. Filtered exosome-free DMEM media was also used as control. Densitometry was then performed, and cytokine counts above the DMEM background levels were graphed in Supplementary Figure 3. In the supernatants, some cytokines (ENA-78, IL-12, IL-15, RANTES, TNF-α, TNF-β, epidermal growth factor [EGF], IGF-1, and Oncostatin M) were present at higher levels in V2CH cells compared with 293T cells (Supplementary Figure 3A). In contrast, 293T cell supernatants were slightly more enriched in several different cytokines (GRO, GRO-α, I-309, IL-1α, IL-1β, and IFN-γ). However, when cytokines from the enriched exosomal fractions were analyzed, an increase in almost all cytokine levels was observed compared with those seen in whole supernatants (Supplementary Figure 3B vs A). In particular, V2CH exosomes possessed higher levels of ENA-78, GRO, IL-12, IL-15, IFN-γ, MCP-1, MCSF, MIG, TARC, TGF-β1, TNF-β, EGF, and Oncostatin M and lower amounts of MIP-1 δ and leptin compared with 293T exosomes. Additional cytokines that were undetectable in whole supernatants that were found in exosomal samples included IL-2, IL-3, IL-4, IL-5, MCP-1, -2, and -3, MCSF, macrophage-derived chemokine (MDC), stem cell factor (SCF), angiogenin, platelet-derived growth factor (PDGF) BB, and leptin. This finding could potentially be due to our selective enrichment workflow, such that concentrations of cytokine-associated exosomes were much higher than those in whole supernatant samples and therefore more easily detected by the assay. Cytokines such as granulocyte colony-stimulating factor (GCSF), GM-CSF, GRO-α, I-309, IL-1α, IL-1β, IL-2, IL-3, IL-4, IL-5, IL-8, IL-10, IL-13, MCP-2, MCP-3, MDC, RANTES, SCF, SDF-1, TNF-α, IGF-1, angiogenin, thrombopoietin, VEGF, and PDGF BB were present in both 293T and V2CH exosomes at similar levels. It is interesting to note that exosomes from both cell types were shown to possess VEGF, TGF-β1, and IGF-1, further supporting our MS results (Supplementary Table 1). Collectively, these data indicate that EVs, specifically exosomes, from VP40-producing cells may contain a diverse set of unique or upregulated proteins. In addition, a plethora of cytokines are associated with vesicles released from virally affected and healthy host cells. Moreover, specific cytokines may be concentrated within EVs from cells producing VP40, which may potentially play a role in the cytokine storm associated with the pathogenesis of EBOV, induction of cell death in recipient immune cells, and poor outcome of infected individuals.
DISCUSSION
We previously showed that EBOV VP40 can become incorporated into exosomes and thereby negatively impact recipient T cells and monocytes [12, 13]. In line with this, we have additionally shown here that VP40-containing vesicles can induce cell death by apoptosis in these recipient cells (Figure 8). This cell death was more exacerbated in monocytes treated with EVs from cells that produced higher levels of VP40. On the other hand, EVs from cells that produced both low and high levels of VP40 were equally destructive to T cells. Bystander T-cell death by apoptosis is one of the hallmarks of EVD, especially fatal cases [5, 7, 9, 10, 81–83]. Furthermore, we determined that VP40 EVs that sediment within the 13.2 and 14.4 fractions of iodixanol density gradients are able to significantly reduce recipient T-cell death, but EVs that fall in the 16.8 and 18.0 fractions do not (Figure 9B). This finding may be noteworthy, because we believe the 16.8 + 18.0 fractions mainly contain apoptotic bodies, large microvesicles, and VLPs in the case of V2C cells, thus potentially indicating that these other extracellular particles may not be a significant contributor to the overall decreased health of recipient immune cells. Therefore, VP40 EVs of intermediate density may comprise a potential mechanism of bystander immune cell apoptosis in EVD. Interestingly, in contrast to immune cells, 293T (human embryonic kidney epithelial) cells that were treated with EVs from both low and high V2C cells showed an increased cell viability. This could potentially be due to a cell type-dependent mechanism of action on recipient cells by VP40-associated vesicles. In addition, 293T cells may be altogether resistant to VP40 EV components responsible for cell death in immune cells and may conversely be positively stimulated by the vesicles. Further study into the specific components responsible for these phenotypes in recipient cells is needed, but the resistance of certain cell types could be quite significant in the context of infection. Monocytes or macrophages and dendritic cells are the first sustained target of EBOV, and T cells die relatively early in infection; however, epithelial and endothelial cells are also productively infected by the virus, albeit at later time points in infection [3–10]. If these uninfected nonimmune cells (endothelial and epithelial) are bombarded with vesicles containing viral components such as VP40 during early infection, it is possible that the recipient cells may be stimulated and primed for future infection. This type of mechanism has been seen before in the case of HIV-1 TAR, in which exosomes containing this viral RNA predisposed recipient cells to be more receptive to infection [26]. Should this type of pathogenic process take place, inhibition of vesicle biogenesis from infected cells should be further explored to determine potential benefits in recipient cells.
To determine pathways contributing to EV biogenesis in donor cells, we investigated the cell cycle of VP40-producing cells. Our rationale for doing these experiments was that when cells contained VP40, they replicated faster than control cells, leading us to ask whether cell cycle patterns were affected. While exploring differences in cell cycle dynamics with the VP40-producing clones, it is important to note that although our vectors have SV40 origin of replication and cells contain T-antigen, it is possible that over time few copies of the plasmid can be integrated into the host genome because these cells are Rec+ and allow recombination. However, we have not observed any differences between these clones that cannot be attributed to a dose response of VP40 production and cell cycle effects. Here, V2C cells were observed to be altered in a cell cycle-dependent manner in not only their exosomal biogenesis pathway components (ESCRT proteins), but also in their morphology, growth patterns, and EV content and abundance (Figures 1, 4, 5, and 6). Moreover, V2C cells exhibited increased levels of cyclin D1 protein and activity (Figure 2). Cyclin D1 is a well described cell cycle regulator and a potential oncogene known to be upregulated in several cancers [62–64]. In addition, cyclin D1 was previously shown to be produced at elevated levels as a result of many viral infections, especially those known to cause cancers [65–70]. Although EBOV has not been shown to result in higher incidences of cancer in survivors, it is possible that this phenotype may be a transient one and only in effect for a brief time before other virulence factors and the replication of the virus results in the infected cells’ death. Because EBOV has been shown to best thrive in actively growing cells [90], upregulation of cyclin D1 may simply be a strategy used by the virus to aid its own replication. However, larger proportions of recurrent, persistent, and asymptomatic EBOV infections have come to light with study of the increasing numbers of EBOV survivors, which is a significant topic of concern, as this can potentially lead to transmission of virus and future outbreaks [91–100]. How the virus manages to remain long term within human and NHP hosts has not yet been conclusively determined; however, viral persistence and replication within CD68+ monocyte or macrophage cell types has been detected within the brain, eyes, and testes of NHP survivors of infection [101]. If these central nervous system-resident myeloid cells are also dysregulated in cell cycle, this could potentially represent a method used by EBOV to counteract its own cytotoxicity to support prolonged viral replication or latency.
Previous studies have shown VP40 to be transiently present in the nucleus of infected cells at early time points [72–74]. Here, we have shown that VP40 is present at relatively high levels within the nucleus of stably transfected cells, and that VP40 is found on a large proportion of cyclin D1 promoters (Figure 2C and D). It may be possible that early in infection, VP40 translocation into the nucleus may serve a role in transcriptional regulation of host genes to best benefit the virus. In all likelihood, nuclear VP40 action as a transcriptional regulator is not limited to the cyclin D1 gene. Investigation into the direct interactions and far-reaching effects of VP40 in the nucleus could be vitally important for elucidating the molecular basis of EBOV pathogenesis in host cells.
Cell cycle dysregulation of VP40-producing cells also resulted in an altered pattern of EV biogenesis (Figures 4 and 5). It is interesting that all cells tested at G0 produced significantly fewer EVs that were also larger than unsynchronized controls. Because many eukaryotic cells are quiescent in normal, healthy, in vivo environments, this could represent an important trend that inherently differentiates EVs from cells that are not frequently dividing versus actively replicating or cancer cells. Our results also showed that EVs from VP40-producing cells were generally fewer in number but denser and larger in size (Figures 5 and 6). Larger vesicles of higher density would imply an enrichment of cargo within VP40 EVs, which was supported by a greater abundance of proteins detected by MS analysis (Supplementary Table 1). The increased numbers and diversity of proteins found in EVs from VP40-producing cells could influence the outcomes of recipient cells, particularly if those proteins were associated with nucleic acids. A collection of RNA-binding proteins was found from MS analysis of EVs from V2C cells, and although we do not yet know the RNAs associated with these proteins, they may be important for activities in recipient cells. An assortment of cytokines was also found, with an upregulation of IFN-γ, IL-15, MCP-1, and TGF-β1 seen in VP40 EVs (Supplementary Figure 3). That we were able to detect these cytokines at elevated levels in exosomal preparations from VP40-producing cells may indicate that VP40 stimulates the production of some of these, potentially as other direct or indirect targets of nuclear VP40 transcriptional regulation. Moreover, the fact that we were able to detect cytokines in exosomal preps in all cell types is a significant finding in and of itself. In our study, the majority of cytokines tested were enriched in exosomes compared with whole supernatants. If this is the case in vivo, vesicles heavily laden with cytokines originating from infected cells could act as a “cytokine bomb” on recipient immune cells. In this way, a few EVs from a single diseased cell would be able to enact devastating effects on a recipient cell. This may also be of substantial importance for many other viral or inflammatory diseases in which cytokines play an inflammatory or damaging role.
Due to the observed upregulation of cyclin D1 protein and activity, we also explored the ability of Ribociclib and Fascaplysin to inhibit cdk4/6 activity and thereby slow the release of vesicles in a cell cycle-driven manner from VP40 cells (Figure 10). Results showed that Fascaplysin was able to significantly decrease the number of vesicles produced from both 293T and V2CH cells with low, nontoxic doses. In contrast, Ribociclib was only able to decrease levels of EVs produced from 293T cells at doses that did not affect cell viability. The differences between 293T and V2CH cell responses to the inhibitors suggest that VP40-producing cells may be less susceptible to the effects of Ribociclib. Future mechanistic studies into the molecular action of these drugs in connection with VP40 may elucidate potential therapeutic utility or other more directed drug targets.
We previously showed that EBOV VP40 is packaged into exosomes in VP40-transfected cells [12, 13]. Here, we show that VP40 is found within EVs isolated from both in vitro and in vivo wild-type EBOV infection (Figure 7A and B). Furthermore, EVs from EBOV-infected cells contained both NP and GP proteins of the virus. NP is responsible for encapsidating the RNA genome of the virus as soon as it is synthesized and for formation of the nucleocapsid (in conjunction with other viral proteins) [102]. Because NP is the main viral protein responsible for binding of the RNA genome (each NP is bound to 6 bases of RNA [103]), the presence of NP within EVs could also potentially hint at the presence of bound viral RNA within EVs as well. On the other hand, GP is the sole surface protein of EBOV and is important for both viral entry and egress [104]. GP has also been implicated in pathogenesis, because GP itself has shown cytotoxic effects in infected cells. Furthermore, soluble GP is found in large quantities in the blood of infected patients, and it has been proposed to potentially play a role in immune evasion by absorption of neutralizing antibodies [105, 106]. Our experiments involving filtration of VLPs followed by size exclusion separation of extracellular materials showed that VLPs were able to be removed from samples by filtration, whereas smaller vesicles in the VLP preparations that contained EBOV proteins were able to pass through 0.22-µm filtration (Supplementary Figure 2B). These results also interestingly hinted towards a potential role for GP in EVs, as unfiltered VLPs showed enrichment of primarily glycosylated GP in the fractions corresponding to larger vesicles (fraction number 7), whereas filtered VLPs showed mainly lower molecular weight forms of GP, which colocalized with exosomal markers. Therefore, if EVs are studded with GP proteins, this could form an additional previously unknown source of immune subversion and viral masking used by EBOV. Presence of other viral proteins or RNAs in EVs were not explored here and will be analyzed in future studies.
CONCLUSIONS
In conclusion, we show that VP40 dysregulated the cell cycle in donor cells through translocation to the nucleus and upregulation of functional cyclin D1 protein. In turn, cell cycle was shown to be a regulator of EV biogenesis. Extracellular vesicles from VP40 cells were also altered in their content, which included RNA-binding proteins and cytokines important for EBOV pathogenesis. In addition, NP and GP proteins were detected within EVs from EBOV-infected cells. Collectively, the cell cycle-related effects of VP40 within donor cells may have repercussions on the virulence and replication efficiency of EBOV, the upregulation and packaging of proteins into EVs, and the overall outcome of EV-recipient cells in infected individuals.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Presented in part: American Society for Exosomes and Microvesicles, Asilomar Conference Center, 2017, Pacific Beach, CA; 9th International Symposium on Filoviruses, Welcome Hotel Marburg, 2017, Marburg, Germany; Cell Symposia on Emerging and Re-Emerging Viruses, Sheraton Pentagon City, 2017, Arlington, VA; Bio-Trac Exosomes: Principles, Methods and Applications Workshop, 2018, Montgomery College Germantown Campus, Germantown, MD; 2nd annual Extracellular Vesicles and Infections Conference, 2018, Potomac, MD.
Notes
Acknowledgments. We thank all members of the Kashanchi laboratory, especially Gwen Cox, Angela Schwab, and Yao Akpamagbo. We also thank Laura Bollinger for thorough grammatical editing of the manuscript.
Disclaimer. The views and conclusions contained in this document are those of the authors and should not be interpreted as necessarily representing the official policies, either expressed or implied, of the US Department of the Army, the US Department of Defense, the US Department of Health and Human Services, or of the institutions and companies affiliated with the authors.
Financial support. This work was funded by National Institutes of Health Grants AI078859, AI074410, AI127351-01, AI043894, and NS099029 (to F. K.) and R33 CA206937 and R01AR068436 (to L. A. L.). This work was supported in part through Battelle Memorial Institute’s prime contract with the US National Institute of Allergy and Infectious Diseases under Contract No. HHSN272200700016I (to J. L., J. H. K.). Support was also kindly provided by the Defense Threat Reduction Agency (Grant CB4088; to J. M. D.).
Potential conflicts of interest. B. L. is employed by Ceres Nanosciences, Inc. M. J. A. is the President and Chief Scientific Officer of Integrated BioTherapeutics, Inc. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Centers of Disease Control. Outbreaks chronology: Ebola virus disease | Ebola hemorrhagic fever. Available at: https://www.cdc.gov/vhf/ebola/outbreaks/history/chronology.html#modalIdString_outbreaks. Accessed 9 March 2017. [Google Scholar]
- 2. World Health Organization. Ebola virus disease. Available at: http://www.who.int/mediacentre/factsheets/fs103/en/. Accessed 9 March 2017. [Google Scholar]
- 3. Feldmann H, Geisbert TW. Ebola haemorrhagic fever. Lancet 2011; 377:849–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Martines RB, Ng DL, Greer PW, Rollin PE, Zaki SR. Tissue and cellular tropism, pathology and pathogenesis of Ebola and Marburg viruses. J Pathol 2015; 235:153–74. [DOI] [PubMed] [Google Scholar]
- 5. Wauquier N, Becquart P, Padilla C, Baize S, Leroy EM. Human fatal Zaire Ebola virus infection is associated with an aberrant innate immunity and with massive lymphocyte apoptosis. PLoS Negl Trop Dis 2010; 4:pii: e837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Messaoudi I, Basler CF. Immunological features underlying viral hemorrhagic fevers. Curr Opin Immunol 2015; 36:38–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bradfute SB, Braun DR, Shamblin JD, et al. Lymphocyte death in a mouse model of Ebola virus infection. J Infect Dis 2007; 196(Suppl 2):S296–304. [DOI] [PubMed] [Google Scholar]
- 8. Rougeron V, Feldmann H, Grard G, Becker S, Leroy EM. Ebola and Marburg haemorrhagic fever. J Clin Virol 2015; 64:111–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Falasca L, Agrati C, Petrosillo N, et al. Molecular mechanisms of Ebola virus pathogenesis: focus on cell death. Cell Death Differ 2015; 22:1250–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gupta M, Spiropoulou C, Rollin PE. Ebola virus infection of human PBMCs causes massive death of macrophages, CD4 and CD8 T cell sub-populations in vitro. Virology 2007; 364:45–54. [DOI] [PubMed] [Google Scholar]
- 11. Yaddanapudi K, Palacios G, Towner JS, et al. Implication of a retrovirus-like glycoprotein peptide in the immunopathogenesis of Ebola and Marburg viruses. FASEB J 2006; 20:2519–30. [DOI] [PubMed] [Google Scholar]
- 12. Pleet ML, Mathiesen A, DeMarino C, et al. Ebola VP40 in exosomes can cause immune cell dysfunction. Front Microbiol 2016; 7:1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pleet ML, DeMarino C, Lepene B, Aman MJ, Kashanchi F. The role of exosomal VP40 in Ebola virus disease. DNA Cell Biol 2017; 36:243–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bradfute SB, Warfield KL, Bavari S. Functional CD8+ T cell responses in lethal Ebola virus infection. J Immunol 2008; 180:4058–66. [DOI] [PubMed] [Google Scholar]
- 15. Fleming A, Sampey G, Chung MC, et al. The carrying pigeons of the cell: exosomes and their role in infectious diseases caused by human pathogens. Pathog Dis 2014; 71:109–20. [DOI] [PubMed] [Google Scholar]
- 16. Schwab A, Meyering SS, Lepene B, et al. Extracellular vesicles from infected cells: potential for direct pathogenesis. Front Microbiol 2015; 6:1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Anderson MR, Kashanchi F, Jacobson S. Exosomes in viral disease. Neurotherapeutics 2016; 13:535–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Keller S, Sanderson MP, Stoeck A, Altevogt P. Exosomes: from biogenesis and secretion to biological function. Immunol Lett 2006; 107:102–8. [DOI] [PubMed] [Google Scholar]
- 19. Andreu Z, Yáñez-Mó M. Tetraspanins in extracellular vesicle formation and function. Front Immunol 2014; 5:442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Henne WM, Buchkovich NJ, Emr SD. The ESCRT pathway. Dev Cell 2011; 21:77–91. [DOI] [PubMed] [Google Scholar]
- 21. Ahsan NA, Sampey GC, Lepene B, et al. Presence of viral RNA and proteins in exosomes from cellular clones resistant to rift valley fever virus infection. Front Microbiol 2016; 7:139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jaworski E, Narayanan A, Van Duyne R, et al. Human T-lymphotropic virus type 1-infected cells secrete exosomes that contain Tax protein. J Biol Chem 2014; 289:22284–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pegtel DM, van de Garde MD, Middeldorp JM. Viral miRNAs exploiting the endosomal-exosomal pathway for intercellular cross-talk and immune evasion. Biochim Biophys Acta 2011; 1809:715–21. [DOI] [PubMed] [Google Scholar]
- 24. Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, et al. Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci U S A 2010; 107:6328–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Votteler J, Sundquist WI. Virus budding and the ESCRT pathway. Cell Host Microbe 2013; 14:232–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Narayanan A, Iordanskiy S, Das R, et al. Exosomes derived from HIV-1-infected cells contain trans-activation response element RNA. J Biol Chem 2013; 288:20014–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sampey GC, Saifuddin M, Schwab A, et al. Exosomes from HIV-1-infected cells stimulate production of pro-inflammatory cytokines through trans-activating response (TAR) RNA. J Biol Chem 2016; 291:1251–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sampey GC, Meyering SS, Zadeh MA, Saifuddin M, Hakami RM, Kashanchi F. Exosomes and their role in CNS viral infections. J Neurovirol 2014; 20:199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol 2013; 200:373–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Colombo M, Raposo G, Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol 2014; 30:255–89. [DOI] [PubMed] [Google Scholar]
- 31. van der Pol E, Coumans FA, Grootemaat AE, et al. Particle size distribution of exosomes and microvesicles determined by transmission electron microscopy, flow cytometry, nanoparticle tracking analysis, and resistive pulse sensing. J Thromb Haemost 2014; 12:1182–92. [DOI] [PubMed] [Google Scholar]
- 32. Kastelowitz N, Yin H. Exosomes and microvesicles: identification and targeting by particle size and lipid chemical probes. Chembiochem 2014; 15:923–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vader P, Breakefield XO, Wood MJ. Extracellular vesicles: emerging targets for cancer therapy. Trends Mol Med 2014; 20:385–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Akers JC, Gonda D, Kim R, Carter BS, Chen CC. Biogenesis of extracellular vesicles (EV): exosomes, microvesicles, retrovirus-like vesicles, and apoptotic bodies. J Neurooncol 2013; 113:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Broadhurst MJ, Brooks TJ, Pollock NR. Diagnosis of Ebola virus disease: past, present, and future. Clin Microbiol Rev 2016; 29:773–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Timmins J, Scianimanico S, Schoehn G, Weissenhorn W. Vesicular release of Ebola virus matrix protein VP40. Virology 2001; 283:1–6. [DOI] [PubMed] [Google Scholar]
- 37. Kallstrom G, Warfield KL, Swenson DL, et al. Analysis of Ebola virus and VLP release using an immunocapture assay. J Virol Methods 2005; 127:1–9. [DOI] [PubMed] [Google Scholar]
- 38. Licata JM, Johnson RF, Han Z, Harty RN. Contribution of Ebola virus glycoprotein, nucleoprotein, and VP24 to budding of VP40 virus-like particles. J Virol 2004; 78:7344–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bavari S, Bosio CM, Wiegand E, et al. Lipid raft microdomains. J Exp Med 2002; 195:593–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jasenosky LD, Neumann G, Lukashevich I, Kawaoka Y. Ebola virus VP40-induced particle formation and association with the lipid bilayer. J Virol 2001; 75:5205–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Noda T, Sagara H, Suzuki E, Takada A, Kida H, Kawaoka Y. Ebola virus VP40 drives the formation of virus-like filamentous particles along with GP. J Virol 2002; 76:4855–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kolesnikova L, Bamberg S, Berghöfer B, Becker S. The matrix protein of Marburg virus is transported to the plasma membrane along cellular membranes: exploiting the retrograde late endosomal pathway. J Virol 2004; 78:2382–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kolesnikova L, Bugany H, Klenk HD, Becker S. VP40, the matrix protein of Marburg virus, is associated with membranes of the late endosomal compartment. J Virol 2002; 76:1825–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Steele K, Crise B, Kuehne A, Kell W. Ebola virus glycoprotein demonstrates differential cellular localization in infected cell types of nonhuman primates and Guinea pigs. Arch Pathol Lab Med 2001; 125:625–30. [DOI] [PubMed] [Google Scholar]
- 45. Reynard O, Reid SP, Page A, et al. Unconventional secretion of Ebola virus matrix protein VP40. J Infect Dis 2011; 204(Suppl 3):S833–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kashanchi F, Duvall JF, Brady JN. Electroporation of viral transactivator proteins into lymphocyte suspension cells. Nucleic Acids Res 1992; 20:4673–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jaworski E, Saifuddin M, Sampey G, et al. The use of Nanotrap particles technology in capturing HIV-1 virions and viral proteins from infected cells. PLoS One 2014; 9:e96778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sadick JS, Boutin ME, Hoffman-Kim D, Darling EM. Protein characterization of intracellular target-sorted, formalin-fixed cell subpopulations. Sci Rep 2016; 6:33999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wilson JA, Hevey M, Bakken R, et al. Epitopes involved in antibody-mediated protection from Ebola virus. Science 2000; 287:1664–6. [DOI] [PubMed] [Google Scholar]
- 50. DeMarino C, Pleet ML, Cowen M, et al. Antiretroviral drugs alter the content of extracellular vesicles from HIV-1-infected cells. Sci Rep 2018; 8:7653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Barclay RA, Pleet ML, Akpamagbo Y, Noor K, Mathiesen A, Kashanchi F. Isolation of exosomes from HTLV-infected cells. Methods Mol Biol 2017; 1582:57–75. [DOI] [PubMed] [Google Scholar]
- 52. Barclay RA, Schwab A, DeMarino C, et al. Exosomes from uninfected cells activate transcription of latent HIV-1. J Biol Chem 2017; 292:11682–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nascimento R, Costa H, Parkhouse RM. Virus manipulation of cell cycle. Protoplasma 2012; 249:519–28. [DOI] [PubMed] [Google Scholar]
- 54. Fehr AR, Yu D. Control the host cell cycle: viral regulation of the anaphase-promoting complex. J Virol 2013; 87:8818–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dove B, Brooks G, Bicknell K, Wurm T, Hiscox JA. Cell cycle perturbations induced by infection with the coronavirus infectious bronchitis virus and their effect on virus replication. J Virol 2006; 80:4147–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chaurushiya MS, Weitzman MD. Viral manipulation of DNA repair and cell cycle checkpoints. DNA Repair (Amst) 2009; 8:1166–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bracq L, Xie M, Benichou S, Bouchet J. Mechanisms for cell-to-cell transmission of HIV-1. Front Immunol 2018; 9:260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sylwester A, Wessels D, Anderson SA, et al. HIV-induced syncytia of a T cell line form single giant pseudopods and are motile. J Cell Sci 1993; 106(Pt 3):941–53. [DOI] [PubMed] [Google Scholar]
- 59. Wang P, Li M, Lu W, Zhang D, Hu Q, Liu Y. DC-SIGN promotes Japanese encephalitis virus transmission from dendritic cells to T cells via virological synapses. Virol Sin 2017; 32:495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ceccaldi PE, Delebecque F, Prevost MC, et al. DC-SIGN facilitates fusion of dendritic cells with human T-cell leukemia virus type 1-infected cells. J Virol 2006; 80:4771–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kononova AA, Cheresiz SV, Chechushkov AV, Razumova YV, Pokrovskii AG. Comparative study of fusogenic activity of H1 and H5 subtypes influenza virus hemagglutinins. Bull Exp Biol Med 2017; 164:85–9. [DOI] [PubMed] [Google Scholar]
- 62. Casimiro MC, Velasco-Velázquez M, Aguirre-Alvarado C, Pestell RG. Overview of cyclins D1 function in cancer and the CDK inhibitor landscape: past and present. Expert Opin Investig Drugs 2014; 23:295–304. [DOI] [PubMed] [Google Scholar]
- 63. Pestell RG. New roles of cyclin D1. Am J Pathol 2013; 183:3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Klein EA, Assoian RK. Transcriptional regulation of the cyclin D1 gene at a glance. J Cell Sci 2008; 121:3853–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Santiago F, Clark E, Chong S, et al. Transcriptional up-regulation of the cyclin D2 gene and acquisition of new cyclin-dependent kinase partners in human T-cell leukemia virus type 1-infected cells. J Virol 1999; 73:9917–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kehn K, Deng L, de la Fuente C, et al. The role of cyclin D2 and p21/waf1 in human T-cell leukemia virus type 1 infected cells. Retrovirology 2004; 1:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zhou L, Kang D, Xu C, Zhao W, Tian B, Chen L. Expression of cyclin D1 and cyclin E significantly associates with human papillomavirus subtypes in Bowenoid papulosis. Acta Histochem 2013; 115:339–43. [DOI] [PubMed] [Google Scholar]
- 68. Ripple MJ, Parker Struckhoff A, Trillo-Tinoco J, et al. Activation of c-Myc and cyclin D1 by JCV T-antigen and β-catenin in colon cancer. PLoS One 2014; 9:e106257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Xiang Z, Liang Z, Yanfeng H, Leitao K. Persistence of RSV promotes proliferation and epithelial-mesenchymal transition of bronchial epithelial cells through Nodal signaling. J Med Microbiol 2017; 66:1499–505. [DOI] [PubMed] [Google Scholar]
- 70. Xu Y, Shi Y, Yuan Q, et al. Epstein-Barr virus encoded LMP1 regulates cyclin D1 promoter activity by nuclear EGFR and STAT3 in CNE1 cells. J Exp Clin Cancer Res 2013; 32:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kitagawa M, Higashi H, Jung HK, et al. The consensus motif for phosphorylation by cyclin D1-Cdk4 is different from that for phosphorylation by cyclin A/E-Cdk2. EMBO J 1996; 15:7060–9. [PMC free article] [PubMed] [Google Scholar]
- 72. Björndal AS, Szekely L, Elgh F. Ebola virus infection inversely correlates with the overall expression levels of promyelocytic leukaemia (PML) protein in cultured cells. BMC Microbiol 2003; 3:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Nanbo A, Watanabe S, Halfmann P, Kawaoka Y. The spatio-temporal distribution dynamics of Ebola virus proteins and RNA in infected cells. Sci Rep 2013; 3: doi: 10.1038/srep01206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Del Vecchio K, Frick CT, Gc JB, et al. A cationic, C-terminal patch and structural rearrangements in Ebola virus matrix VP40 protein control its interactions with phosphatidylserine. J Biol Chem 2018; 293:3335–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Moore HM, Vartiainen MK. F-actin organizes the nucleus. Nat Cell Biol 2017; 19:1386–8. [DOI] [PubMed] [Google Scholar]
- 76. Falahzadeh K, Banaei-Esfahani A, Shahhoseini M. The potential roles of actin in the nucleus. Cell J 2015; 17:7–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Albanese C, D’Amico M, Reutens AT, et al. Activation of the cyclin D1 gene by the E1A-associated protein p300 through AP-1 inhibits cellular apoptosis. J Biol Chem 1999; 274:34186–95. [DOI] [PubMed] [Google Scholar]
- 78. Byun JS, Wong MM, Cui W, et al. Dynamic bookmarking of primary response genes by p300 and RNA polymerase II complexes. Proc Natl Acad Sci U S A 2009; 106:19286–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Cantin R, Diou J, Bélanger D, Tremblay AM, Gilbert C. Discrimination between exosomes and HIV-1: purification of both vesicles from cell-free supernatants. J Immunol Methods 2008; 338:21–30. [DOI] [PubMed] [Google Scholar]
- 80. Beniac DR, Melito PL, Devarennes SL, et al. The organisation of Ebola virus reveals a capacity for extensive, modular polyploidy. PLoS One 2012; 7:e29608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Olejnik J, Alonso J, Schmidt KM, et al. Ebola virus does not block apoptotic signaling pathways. J Virol 2013; 87:5384–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Geisbert TW, Hensley LE, Larsen T, et al. Pathogenesis of Ebola hemorrhagic fever in cynomolgus macaques: evidence that dendritic cells are early and sustained targets of infection. Am J Pathol 2003; 163:2347–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Bradfute SB, Swanson PE, Smith MA, et al. Mechanisms and consequences of ebolavirus-induced lymphocyte apoptosis. J Immunol 2010; 184:327–35. [DOI] [PubMed] [Google Scholar]
- 84. Chen P, Lee NV, Hu W, et al. Spectrum and degree of CDK drug interactions predicts clinical performance. Mol Cancer Ther 2016; 15:2273–81. [DOI] [PubMed] [Google Scholar]
- 85. Tripathy D, Bardia A, Sellers WR. Ribociclib (LEE011): mechanism of action and clinical impact of this selective cyclin-dependent kinase 4/6 inhibitor in various solid tumors. Clin Cancer Res 2017; 23:3251–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Messaoudi I, Amarasinghe GK, Basler CF. Filovirus pathogenesis and immune evasion: insights from Ebola virus and Marburg virus. Nat Rev Microbiol 2015; 13:663–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Villinger F, Rollin PE, Brar SS, et al. Markedly elevated levels of interferon (IFN)-gamma, IFN-alpha, interleukin (IL)-2, IL-10, and tumor necrosis factor-alpha associated with fatal Ebola virus infection. J Infect Dis 1999; 179(Suppl 1):S188–91. [DOI] [PubMed] [Google Scholar]
- 88. Bixler SL, Goff AJ. The role of cytokines and chemokines in Filovirus infection. Viruses 2015; 7:5489–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Baize S, Leroy EM, Georges AJ, et al. Inflammatory responses in Ebola virus-infected patients. Clin Exp Immunol 2002; 128:163–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Kota KP, Benko JG, Mudhasani R, et al. High content image based analysis identifies cell cycle inhibitors as regulators of Ebola virus infection. Viruses 2012; 4:1865–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Jacobs M, Rodger A, Bell DJ, et al. Late Ebola virus relapse causing meningoencephalitis: a case report. Lancet 2016; 388:498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Varkey JB, Shantha JG, Crozier I, et al. Persistence of Ebola virus in ocular fluid during convalescence. N Engl J Med 2015; 372:2423–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Deen GF, Broutet N, Xu W, et al. Ebola RNA persistence in semen of Ebola virus disease survivors—final report. N Engl J Med 2017; 377:1428–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Uyeki TM, Erickson BR, Brown S, et al. Ebola virus persistence in semen of male survivors. Clin Infect Dis 2016; 62:1552–5. [DOI] [PubMed] [Google Scholar]
- 95. Diallo B, Sissoko D, Loman NJ, et al. Resurgence of Ebola virus disease in guinea linked to a survivor with virus persistence in seminal fluid for more than 500 days. Clin Infect Dis 2016; 63:1353–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Christie A, Davies-Wayne GJ, Cordier-Lassalle T, et al. Possible sexual transmission of Ebola virus—Liberia, 2015. MMWR Morb Mortal Wkly Rep 2015; 64:479–81. [PMC free article] [PubMed] [Google Scholar]
- 97. Mate SE, Kugelman JR, Nyenswah TG, et al. Molecular evidence of sexual transmission of Ebola virus. N Engl J Med 2015; 373:2448–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Blackley DJ, Wiley MR, Ladner JT, et al. Reduced evolutionary rate in reemerged Ebola virus transmission chains. Sci Adv 2016; 2:e1600378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Arias A, Watson SJ, Asogun D, et al. Rapid outbreak sequencing of Ebola virus in Sierra Leone identifies transmission chains linked to sporadic cases. Virus Evol 2016; 2:vew016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Rodriguez LL, Roo AD, Guimard Y, et al. Persistence and genetic stability of Ebola virus during the outbreak in Kikwit, Democratic Republic of the Congo, 1995. J Infect Dis 1999; 179(Supp 1):S170–6. [DOI] [PubMed] [Google Scholar]
- 101. Zeng X, Blancett CD, Koistinen KA, et al. Identification and pathological characterization of persistent asymptomatic Ebola virus infection in rhesus monkeys. Nat Microbiol 2017; 2:17113. [DOI] [PubMed] [Google Scholar]
- 102. Martin B, Canard B, Decroly E. Filovirus proteins for antiviral drug discovery: Structure/function bases of the replication cycle. Antiviral Res 2017; 141:48–61. [DOI] [PubMed] [Google Scholar]
- 103. Bharat TA, Noda T, Riches JD, et al. Structural dissection of Ebola virus and its assembly determinants using cryo-electron tomography. Proc Natl Acad Sci U S A 2012; 109:4275–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Martin B, Reynard O, Volchkov V, Decroly E. Filovirus proteins for antiviral drug discovery: structure/function of proteins involved in assembly and budding. Antiviral Res 2018; 150:183–92. [DOI] [PubMed] [Google Scholar]
- 105. Lee JE, Saphire EO. Ebolavirus glycoprotein structure and mechanism of entry. Future Virol 2009; 4:621–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Martin B, Hoenen T, Canard B, Decroly E. Filovirus proteins for antiviral drug discovery: a structure/function analysis of surface glycoproteins and virus entry. Antiviral Res 2016; 135:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.