We identified a protein phosphatase 1 (PP1)–targeting compound, C31, that inhibited Ebola virus transcription and replication. C31 bound to the PP1 C-terminal groove and increased VP30 phosphorylation. C31 has better pharmacological properties than 1E7-03 and is a good candidate for future antifiloviral therapy.
Keywords: Ebola virus, protein phosphatase 1, transcription inhibitor
Abstract
Background
Ebola virus (EBOV) infection causes severe hemorrhagic fever. EBOV transcription is controlled by host protein phosphatase 1 (PP1), which dephosphorylates VP30 protein. We previously developed 1E7-03, a compound targeting a noncatalytic site of PP1 that induced VP30 phosphorylation and inhibited EBOV transcription. Here, we attempted to further improve 1E7-03, which was not stable in murine serum.
Results
High-throughput screening with EBOV–green fluorescent protein was conducted on 72 1E7-03 analogs and identified 6 best inhibitory and the least toxic compounds. A parallel in silico screening of compounds from the ZINC database by docking to PP1 identified the best-binding compound C31, which was also present among the top 6 compounds found in the viral screen. C31 showed the best EBOV inhibitory activity among the top 6 compounds and also inhibited EBOV minigenome. C31 bound to the PP1 C-terminal groove in vitro and increased VP30 phosphorylation in cultured cells. C31 demonstrated improved stability in mouse plasma and cell permeability, compared with 1E7-03. It was also detected for 24 hours after injection in mice.
Conclusion
C31 represents a novel PP1-targeting EBOV inhibitor with improved pharmacological properties that can be further evaluated for future antifiloviral therapy.
Ebola virus (EBOV) disease is an emerging infection for which there is no approved vaccine or an established postexposure treatment [1]. Thus, novel anti filoviral drugs are urgently needed to curb filovirus infections. We recently showed that 1E7-03, a small molecule targeted to host protein phosphatase 1 (PP1), efficiently inhibited EBOV infection in cultured cells [2]. We also showed that PP1 is involved in EBOV transcription by controlling VP30 dephosphorylation [2]. Recently, NP was shown to recruit host PP2A-B56 protein phosphatase to facilitate dephosphorylation of the VP30 N-terminus and upregulate EBOV transcription [3]. Thus, both PP1 and PP2A control VP30 phosphorylation. PP1 is a major serine/threonine phosphatase that dephosphorylates many cellular proteins and is essential for cell division, glycogen metabolism, muscle contractility and protein synthesis [4]. One of the 3 isoforms of PP1 catalytic subunit binds to a host of regulatory subunits through a combination of short binding motifs, including the ubiquitous RVxF motif present on most PP1 regulatory subunits [5].
Our initial studies, which involved human immunodeficiency virus type 1 (HIV-1) transcription, identified a compound, 1H4, that disrupted the interaction of PP1 with HIV-1 Tat protein and inhibited HIV-1 transcription [2]. In a follow-up study, we identified the compound 1E7-03, a cyclopentane quinoline derivative that inhibited HIV-1 more efficiently than 1H4 [6]. As PP1 was proposed to control EBOV VP30 phosphorylation [7], we tested the effect of 1E7-03 on EBOV and showed that it induced overall VP30 protein phosphorylation, blocked transcription of EBOV genome, and inhibited EBOV replication [2]. We recently tested 1E7-03 in vivo in HIV-1–infected humanized mice and found that the compound was unstable in mouse serum and quickly degraded when injected in mice [8]. In mouse serum, 1E7-03 was converted into degradation product 1 [8], which was found to have no anti-EBOV activity (Supplementary Figure 1).
In the present study, we aimed to develop more-stable and more-efficient EBOV-eGFP inhibitory compounds by screening a library of 1E7-03 analogues in a high-throughput assay with EBOV expressing enhanced green fluorescent protein (EBOV-eGFP). We also conducted an in silico screen of a ZINC database (available at: http://zinc.docking.org) for 1E7-03 homologues, using Internal Coordinate Mechanics (ICM) software, which demonstrated a superior PP1 binding score for several compounds as compared to that for 1E7-03. We identified a new compound, C31, which bound stronger to PP1 in vitro, compared with 1E7-03. We tested its EBOV inhibition properties and its effect on VP30 phosphorylation in cultured cells and examined its toxicity, stability, and cellular permeability.
METHODS
Virtual Docking of Small Molecules to the Catalytic Subunit of PP1α
Ligand ZINC Database
ZINC is a noncommercial free database for virtual screening that contains over 21 million compounds in ready-to-dock 3D formats. ZINC provides several search criteria, such as molecular property constraints, ZINC codes, vendor-based searches, and molecular substructure searches [9]. Molecular modeling and ligand docking were performed using ICM-Pro software package, version 3.8 (Molsoft) [10]. 3D atomic structures of PP1α were taken from the Protein Data Bank database (identifiers 1FJM, 3E7A, 3E7B, 3EGG, 3EGH, 3HVQ, and 3N5U). Since analysis showed very high similarity of these structures (the mean pair-wise root mean square deviation of Cα atoms of the main protein chain was <0.5 Å), 3E7A structure with the best resolution of 1.6 Å was selected to build the molecular model of PP1α [11]. The model was regularized using standard ICM-Pro protocol, and amino acid residues were renumbered in accordance to the sequence PP1A_HUMAN (UniProt identifier P62136). ICM docking was performed for the PP1 C-terminal docking site, using the ICM-Pro flexible ligand docking protocol. The thoroughness parameter was set to 10. The ICM score was calculated for the best position of the ligand in the protein-binding site.
EBOV Minigenome System
The minigenome was assembled as described elsewhere [2] by using plasmids pCEZ-NP, pCEZ-VP35, pCEZ-VP30, pCEZ-L, and pC-T7, which were kindly provided by Dr Yoshihiro Kawaoka [12]. Briefly, plasmids pCEZ-NP (0.25 μg), pCEZ-VP35 (0.25 μg), pCEZ-VP30 or its mutants (0.15 μg), pCEZ-L (2.0 μg), pCT7 (0.5 μg), and monocistronic minigenome (0.5 μg) were cotransfected in 293T cells, using Mirus transfection reagent (Mirus Bio). At 48 hours after transfection, transcription was measured by the luciferase assay (Promega).
EBOV-eGFP Titration
The method was previously described [2]. Briefly, experiments with live EBOV-eGFP were performed in biosafety level 4 facilities of the Galveston National Laboratory and Robert E. Shope Laboratory, University of Texas Medical Branch. To measure titers of EBOV-eGFP in supernatants of infected Vero E6 cells, aliquots were collected every 24 hours, frozen, and titrated in Vero E6 cell monolayers under a 0.9% methylcellulose/minimal essential medium (MEM) overlay. After 3 days at 37ºC, fluorescent viral plaques were counted under a UV microscope.
C31 Synthesis
Synthesis of compound C31 is described in the Supplementary Materials, and a schematic of the synthesis is presented in Supplementary Figure 2.
Analysis of C31 Stability in Mouse Serum
Stability of C31 in mouse serum and its pharmacokinetics were analyzed using liquid chromatography/Fourier transform mass spectrometry (LC/FT-MS) as described previously [8] (see the detailed procedure in the Supplementary Methods).
Surface Plasmon Resonance (SPR)
The SPR measurements were conducted on a Biacore T200 instrument (GE Healthcare, Piscataway, NJ) as previously described [2, 8] (see the detailed procedure in the Supplementary Methods).
VP30 Phosphorylation in Cultured Cells
293T cells were grown to 40% confluence and transfected with Flag-VP30 expression vector, using Lipofectamine 3000/Plus in OPTI-MEM as directed by the manufacturer. At 24 hours after transfection, the cells were treated overnight with 10 µM C31 and, as a control, with a corresponding concentration of dimethyl sulfoxide (DMSO). Also, in an additional control, VP30-transfected cells were treated with 100 nM okadaic acid for 2 hours. VP30 was resolved by 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), with the gel digested and processed for mass spectrometry as previously described [2]. Samples were loaded onto an in-house–prepared and eluted for 80 minutes with a 2%–80% gradient of acetonitrile and a flow rate of 600 nL/minute, using Shimadzu Prominence Nano high-performance liquid chromatography. The 1 FT-MS scan and 3 data-dependent FT-MS/MS scans were performed on a Thermo LTQ Orbitrap XL mass spectrometer on major multicharged MS peaks with a resolution of 60 000 in each event set. Samples were run in triplicate. The resulting set of MS/MS spectra were analyzed by Proteome Discoverer 1.4 with the SEQUEST (Thermo) search engine (precursor tolerance, 30 ppm; and fragments tolerance, 0.1 Da), using FASTA database containing EBOV proteins. The FASTA database was concatenated with proteins of African green monkey and common contaminants. For quantification of VP30 phosphorylation, SIEVE 2.1 software (Thermo Fisher, Waltham, MA) was used in combination with Proteome Discoverer 1.4 for label-free quantitative analysis of the high-resolution MS spectra produced by Orbitrap MS scans.
PP1 C-terminal Groove Variability Analysis
Alignment of PP1 sequences from human, mouse, rat, bovine, and primate species was conducted using Seqlogos software (available at: http://imed.med.ucm.es/Tools/seqlogo.html). Single-nucleotide polymorphism (SNP) analysis for PP1 was conducted using the National Center for Biotechnology Information SNP server and PPP1CA as input.
RESULTS AND DISCUSSION
C31 Is a New Compound Derived From a Library of 1E7-03 Analogues
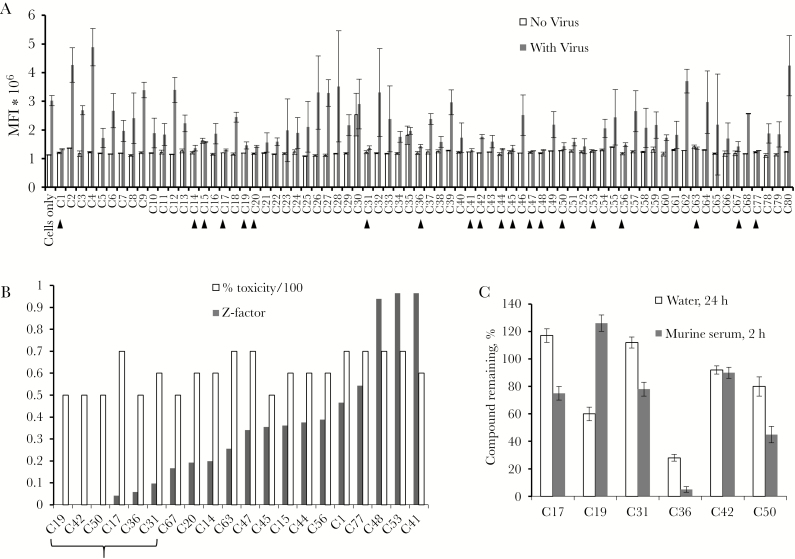
In a previous study, we demonstrated that 1E7-03 binds to PP1α in vitro, using a SPR assay [6, 8]. A library of 1E7-03 analogues was assembled from compounds tested in our previous studies [6, 13] and included 72 unique compounds (Supplementary Table 1). We screened this library in EBOV-infected Vero E6 cells, using a high-throughput assay. All experiments using infectious EBOV were performed at the Galveston National Laboratory biosafety level 4 facility. The anti-EBOV activity of the compounds was initially screened with a fluorescence-based assay in Vero E6 cells infected with live EBOV-eGFP as described elsewhere [14]. Compounds (10 µM) were added 1 hour prior to the infection with EBOV-eGFP (multiplicity of infection, 0.3 plaque-forming units [PFU]/cell). At day 3 after infection, the mean fluorescence intensity (MFI) was measured using a Wallac-EnVision plate reader (Figure 1A). Z-factor was calculated to determine the separation band between a mock-treated control sample and test samples [15]. Twenty compounds that inhibited EBOV with a Z-factor of ≥0.5, indicating the largest separation band, were selected and further tested for toxicity, using a Viral ToxGlo assay. The top 6 compounds that showed <20% toxicity were selected from the top 20 inhibitory compounds and included C17, C19, C31, C36, C42, and C50 (Figure 1B and Supplementary Figure 3). We analyzed the toxicity of C31 by use of the MTT assay in 293T cells and peripheral blood mononuclear cells (PBMCs) and found <30% toxicity in 293T cells at a 100 M concentration and no toxicity in PBMCs (Supplementary Figure 4). Overall, C31 demonstrated low cytotoxicity in cultured cells, although it precipitated at a high concentration and formed crystals (Supplementary Figure 5).
Figure 1.
Screening of 1E7-03 analogues. A, High-throughput assays with Ebola virus expressing enhanced green fluorescent protein (EBOV-eGFP). Mean fluorescence intensity (MFI) of Vero E6 cell monolayer treated with 10 µM compound alone (white bars) and 10 µM compound followed by EBOV-eGFP infection (multiplicity of infection, 0.03 plaque-forming units/cell; dark gray bars). Compounds that showed EBOV inhibition are indicated with arrows. B, Antiviral effect and percentage toxicity of the top 20 inhibitory compounds with Z-scores of ≥0.5. Shown are Z-factors (white bars) and percentage toxicity (scale 1/100; dark gray bars). Vero E6 cells were treated with 10 µM concentrations of compound and screened for toxic effect, using the ViralToxGlo (Promega) kit. Z-factors were calculated from MFI data from panel A. Marked are 6 compounds with a percentage toxicity of <10% that were chosen for further analysis. C, Stability of the 6 top EBOV inhibitory compound in water solution (24 hours; white bars) and murine serum (2 hours; dark gray bars).
The top 6 compounds were further tested for stability in water or mouse serum after incubation for 2 hours. Concentrations of the compounds were determined by LC/FT-MS as described in Methods. We chose C17, C31, and C42 for further testing, based on their overall stability (Figure 1C).
C31 Inhibits EBOV in Cultured Cells
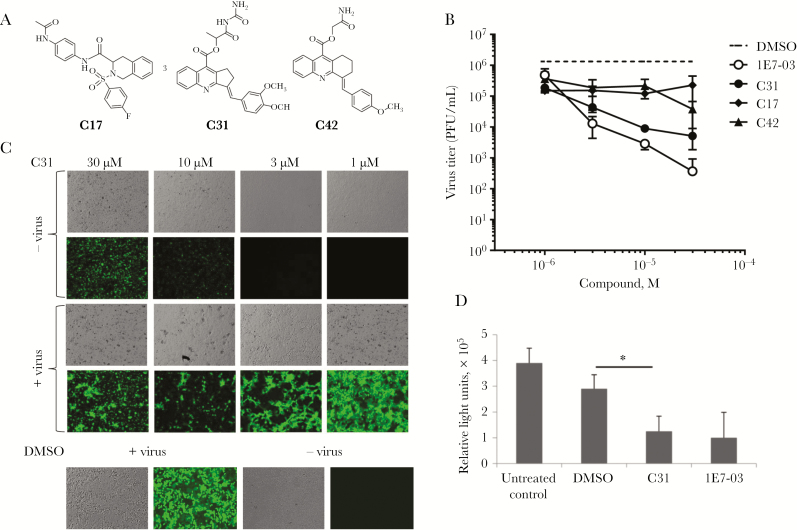
Next, we tested the selected 3 compounds for EBOV inhibition at different concentrations in Vero E6 cells plated in a 24-well antiviral confirmation assay. These 3 compounds had very distinct structures, and only C31 resembled 1E7-03 (Figure 2A). Vero E6 cell monolayers were treated with 4 different concentrations of each compound for 1 hour. Plates were transported to biosafety level 4 facility and infected with EBOV-eGPF (multiplicity of infection, 2 PFU/cell) for 3 hours. Monolayers were washed and supplemented with fresh medium containing the compounds. At 48 hours after infection, supernatant samples were collected and titrated as described elsewhere [16] and also in Methods. Only C31 showed a strong antiviral effect, inhibiting EBOV replication by 2–3 logs, comparable to data for 1E7-03 (Figure 2B). Photographs were taken at 24 hours after treatment with compounds, to evaluate monolayers (data for C31 are in Figure 2C; data for other compounds are not shown). We observed fluorescent crystals of C31, especially at higher (10 µM and 30 µM) concentrations (Figure 2C), which were also seen in the absence of viral infection (Figure 2C and Supplementary Figure 5). To determine viral titers, we collected aliquots of supernatant and infected fresh monolayers of cells. This procedure eliminated any compound fluorescence interference with the GFP signal (Figure 2B). C31 effectively inhibited EBOV replication at 3 µM and higher C31 concentrations (Figure 2C). Thus, compound C31 inhibits EBOV replication at a level comparable to that for 1E7-03 and displayed similarly low toxicity.
Figure 2.
Inhibition of Ebola virus (EBOV) replication by protein phosphatase 1 (PP1)–targeting compounds. A, Chemical structures of 3 top inhibitory compounds: C17, C31, and C42. B, EBOV inhibition. Vero E6 cell monolayers were pretreated where indicated with 1, 3, 10, and 30 μM concentrations of C17, C31, and C42 for 1 hour before infection with EBOV expressing enhanced green fluorescent protein (EBOV-eGFP; multiplicity of infection, 2 plaque-forming units [PFU]/cell). Following incubation for 1 hour, medium was removed, and fresh medium containing the original concentrations of the compounds was added. For the samples in which no virus was detected, values 2-fold below the limit of detection were assigned. Shown are mean EBOV-eGFP titers (±SD) in the medium, based on triplicate monolayers. C, Vero E6 cell GFP fluorescence and monolayers were photographed 48 hours after infection. Upper panels show noninfected cells treated with C31. D, C31 inhibits EBOV minigenome replication. 293T cells were transfected with monocistronic minigenome, nucleoprotein, VP35, L, VP30, and T7 plasmids. They also were treated with 10 µM C31, 1E7-03, or treated with dimethyl sulfoxide (DMSO) as a vehicle control. At 48 hours after transfection, the minigenome transcriptional activity was analyzed by a luciferase assay. The experiment was repeated 3 times, and a P value of <.05 was considered statistically significant.
C31 Inhibits EBOV Minigenome Replication
We further tested the effect of C31 with the monocistronic EBOV minigenome system in 293T cells. Cells were transfected with monocistronic minigenome, NP, VP35, L, VP30, and T7 plasmids. The cells were treated with 10 µM C31 or 1E7-03 or with DMSO as a vehicle control. At 48 hours after transfection, the minigenome transcriptional activity was analyzed by a luciferase assay. In this assay, C31 inhibited EBOV minigenome transcription at a level similar to that for 1E7-03 (Figure 2D), with levels of inhibition about 3 times greater than that for DMSO-treated cells. Thus, compounds 1E7-03 and C31 have comparable inhibitory effects on EBOV transcription and replication.
Analysis of C31 Binding to PP1
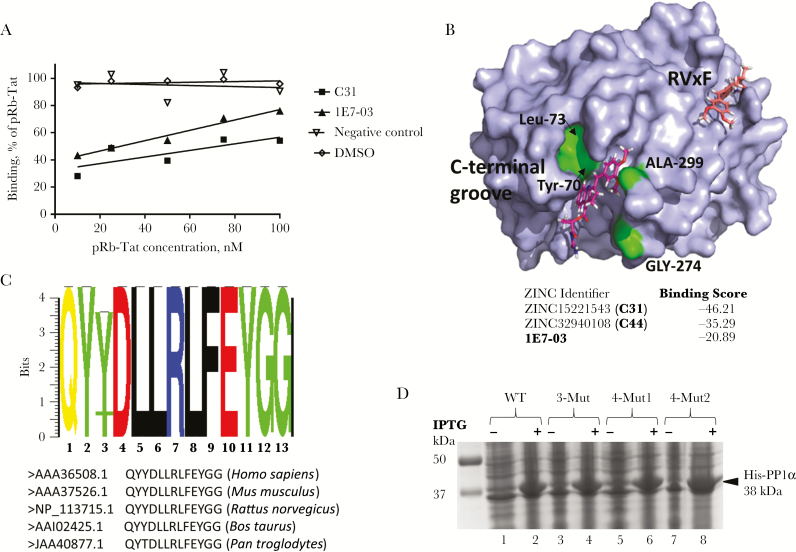
To test the binding of C31 to PP1, recombinant PP1 protein was expressed and purified as described previously [6] and immobilized on a SPR CM5 sensor chip. Binding of C31 was analyzed on a Biacore T-200 instrument as described in Methods. Different concentrations of C31 and, as a control, 1E7-03 were injected over the surface of the chip. A non-PP1 binding small molecule (negative control; compound G3 [13]) was also injected. Direct binding of the compounds to PP1 was measured in real time, and binding affinities were calculated on the basis of a 1:1 binding model. C31 bound to PP1 3-fold stronger than 1E7-03 (Table 1 and Supplementary Figure 6), with equilibrium dissociation constant (KD) values in the same micromolar range, suggesting that C31 also targets PP1. Previously, we showed that pRb-Tat peptide, which has a retinoblastoma protein–derived phosphopeptide sequence fused to an HIV-1 Tat–derived peptide sequence containing QVCF (an RVxF-binding motif), bound PP1 with a KD of 0.15 × 10–6. To determine whether C31 binding to PP1 occurs through an RVxF-binding motif, a competition assay was performed with pRB-Tat peptide. In the competition assay, increasing concentrations of pRb-Tat peptide (10 nM, 25 nM, 50 nM, 75 nM, and 100 nM) were added to the compounds that were kept a constant concentration (1 µM). Both C31 and 1E7-03 competed with pRb-Tat peptide for the binding to PP1, whereas the nonrelated compound, G3 (a negative control), did not compete for binding (Figure 3A). These data suggest that C31 binds PP1 at the RVxF binding site.
Table 1.
Binding of 1E7-03 Analogs to Protein Phosphatase 1 (PP1), Using Surface Plasmon Resonance
| Compound Identifier | PP1 Wild Type | PP1 Triple Mutanta | PP1 Quadruple Mutantb |
|---|---|---|---|
| KD (M) | KD (M) | KD (M) | |
| 1E7-03 | 6.16 × 10 -6 | 9.757 × 10 -6 | 9.042 × 10 -6 |
| C17 | 9.6 × 10-7 | No data | No data |
| C25 | 6.702 × 10-6 | 5.667 × 10-6 | 9.975 × 10-6 |
| C31 | 1.88 × 10 -6 | No data | 8.01 × 10 -6 |
| C33 | 1.412 × 10-5 | 1.300 × 10-5 | 1.006× 10-5 |
| C34 | 2.55 × 10-5 | 2.533 × 10-5 | 6.671 × 10-5 |
| C42 | 18.8 × 10-6 | No data | No data |
| C44 | 5.26 × 10-6 | No data | 15.12 × 10-6 |
| C46 | 3.245 × 10-4 | 2.756 × 10-4 | 3.676 × 10-4 |
The equilibrium dissociation constant (KD) was calculated on the basis of a 1:1 binding model.
aContains the mutations Y70W, L73Y, and A299E.
bContains the mutations Y70W, L73Y, G274E, and A299E.
Figure 3.
C31 binds to protein phosphatase 1 (PP1) in vitro. A, Binding of C31 to recombinant PP1 was measured by surface plasmon resonance (SPR) in a binding competition assay. Increasing concentrations of chimeric pRb-Tat peptide (10 nM, 25 nM, 50 nM, 75 nM, and 100 nM), which consists of a short phosphorylated sequence from retinoblastoma protein (PP1 substrate) and the Tat protein sequence that resembles the RVxF regulatory motif for PP1, were added to the compounds, which were kept at a constant concentration (1 µM). Binding was measured in HBS-P buffer (GE Life Sciences) containing 100 μM MnCl2. The negative control was a compound with molecular weight around 450 Da that did not bind PP1. B, Virtual docking of C31 onto the crystal structure of PP1α. C31 docked to the C-terminal groove of PP1 is shown in purple. Amino acids that formed the C-terminal binding groove are indicated (Tyr-70, Lys-73, Glu-274, and Arg-99) and marked in green. C31 docked to the RVxF-binding cavity is shown in orange. Molecules from the ZINC database with the top scores for binding into the C-terminal groove of PP1 are shown and include 1E7-03 and C31. C, PP1 C-terminal groove alignment. Virtual docking of C31 onto the crystal structure of PP1α. C31 docked to the C-terminal groove of PP1 is shown in purple. Amino acids that formed the C-terminal binding groove are indicated (Tyr-70, Lys-73, Glu-274, and Arg-99) and marked in green. C31 docked to the RVxF-binding cavity is shown in orange. Molecules from the ZINC database with the top scores for binding into the C-terminal groove of PP1 are shown and include 1E7-03 and C31. D, Expression of PP1 for SPR analysis. Lanes 1 and 2, WT PP1; lanes 3 and 4, the triple PP1 mutants Y70W, L73Y, and G274E; lanes 5–8, the PP1 quadruple mutants Y70W, L73Y, G274E, and A299E. Where indicated, IPTG was added to induce expression of His-tagged PP1 (amino acids 7–300), which was further purified on a Ni-NTA column as described in Methods. Abbreviations: DMSO, dimethyl sulfoxide; WT, wild type.
Virtual Docking of ZINC Database Compounds to the Catalytic Subunit of PP1α
Virtual docking of 1E7-03 to the PP1α crystal structure showed several possible binding sites, including an RVxF-binding site. However, we found that the C-terminal groove, described elsewhere [5, 17, 18], binds the compound much stronger than any other possible binding site. For the C-terminal binding site, 1E7-03 showed a binding score of −20.89 (Figure 3B). Several compounds from the ZINC database showed a binding score considerably better than that of 1E7-03, with scores ranging from −35 to −45. One of the best binding compounds was identified as C31, which showed a score of −46.21 (Figure 3B). To analyze variability of the PP1 C-terminal groove, we analyzed PP1α residues 68–80 and also residues G274 and A299, using PP1α sequences from human, mouse, rat, bovine, and primate species (Figure 3C). Among these species, residues 68–80 and also 274 and 299 were conserved, except for changes in Pan troglodytes (Y70T and A299S) (Figure 3C). Analysis of SNPs in humans showed several SNPs, including G274L, A299T, and A299I (data not shown). Of these SNPs, only G274T had a notable heterozygote frequency of 0.003 and a minor allele frequency (MAF) of 0.0004. Thus, the PP1 C-terminal groove sequence is highly conserved among mammals, as well as among humans. These observations further strengthen the utility of the C-terminal groove of PP1 for targeting with small molecules.
Binding of C31 to the C-terminal Groove of PP1 In Vitro
We next analyzed the ability of C31 to bind PP1 in vitro. To determine whether C31 binds to the C-terminal groove, we introduced 4 mutations in PP1 that were identified in in silico analysis: Y70W, L73Y, G274E, and A299E (Figure 3B and 3D). We expressed and purified 2 PP1 mutants, a triple mutant containing Y70W, L73Y, and A299E mutations and a quadruple mutant containing Y70W, L73Y, G274E, and A299E mutations (Figure 3D). Both PP1 mutants maintained enzymatic activity similar to wild-type PP1, indicating that mutations within the C-terminal groove of PP1 had no effect on PP1 enzymatic activity (Supplementary Figure 7). This observation also suggested that C-terminal groove mutations do not change the structure of PP1. We analyzed C31 binding to wild-type and mutant PP1 proteins, using SPR technology as described above. Direct binding to PP1 was measured in real time, and the binding constants were calculated using a 1:1 binding model. Along with C31, we analyzed several additional compounds from the 1E7-03 analogues library (Table 1 and Supplementary Table 2). C31 was the strongest PP1 binding compound, with a 3-fold lower KD than 1E7-03 (Table 1). Importantly, both 1E7-03 and C31 showed reduced binding to the mutant PP1, suggesting that the C-terminal groove represents a major PP1 binding site for the compounds. Remarkably, mutation of the PP1 C-terminal binding groove reduced C31 binding by about 4-fold, whereas 1E7-03 binding was only reduced by 1.5-fold (Table 1), suggesting that the C-terminal binding groove is the major PP1 binding site for C31. Thus, unlike 1E7-03 which is likely to bind to PP1 through the C-terminal groove and the RVxF binding site, C31 binds primarily to the C-terminal groove of PP1.
Effect of C31 on VP30 Phosphorylation
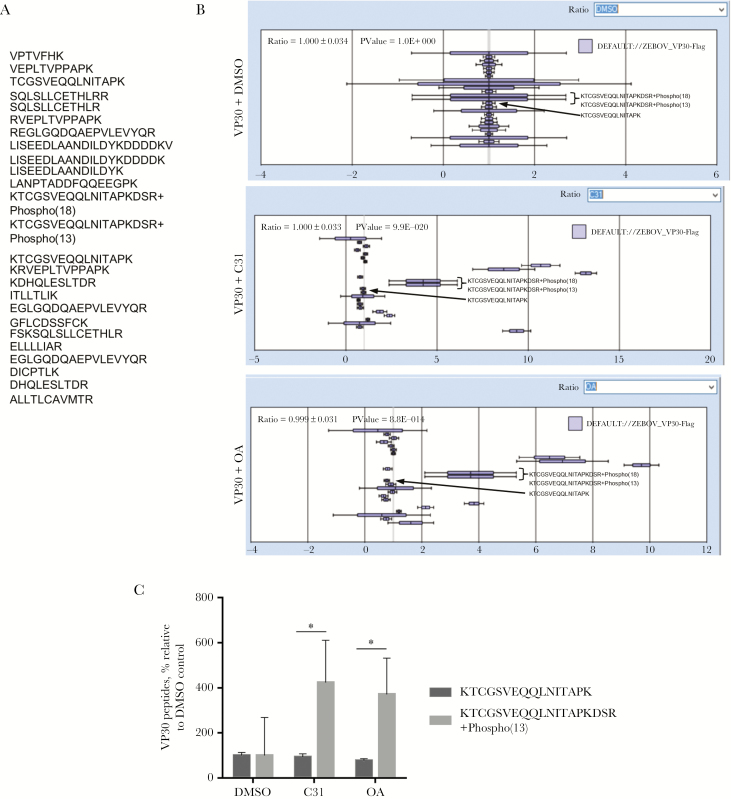
As we previously showed that VP30 phosphorylation was increased in the cells treated with 1E7-03 or okadaic acid [2], we tested the effect of C31 and, as control, okadaic acid on VP30 phosphorylation in cultured cells. Flag-VP30 was expressed in 293T cells, and the cells were treated overnight with 10 µM C31 or DMSO as a vehicle control or for 2 hours with 0.1 M okadaic acid. VP30 was immunoprecipitated from cellular lysates with anti-Flag antibodies, resolved by SDS-PAGE, and analyzed by LC-MS/MS. Mass spectra were analyzed with Proteome Discoverer 1.4 and quantified using a label-free approach with SIEVE 2.1 software. We detected 25 Flag-VP30 peptides (Figure 4A). VP30 expression was normalized using global normalization to equalize ratios for each experimental group. After normalization, relative ratios of VP30 expression in C31 and okadaic acid treatment groups were 1 (Figure 4B and 4C). We did not detect N-terminal peptides of VP30 and, instead, focused on the KTCGSVEQQLNITpAPKDSR phosphopeptide, which was identified with high confidence. Nonphosphorylated KTCGSVEQQLNITAPK peptide was present at similar ratios in the C31 and okadaic acid treatment groups (Figure 4B and 4C). In contrast, levels of KTCGSVEQQLNITpAPKDSR phosphopeptide were increased by about 3-fold in the samples treated with C31 or okadaic acid, suggesting increased VP30 phosphorylation (Figure 4B and 4C). Thus, C31 induced VP30 phosphorylation in vivo.
Figure. 4.
Effect of C31 on VP30 phosphorylation. A, VP30-derived peptides identified by mass spectrometry. B, Label-free quantitative analysis of the high-resolution mass spectrometry spectra produced by Orbitrap MS scans for VP30 by SIEVE 2.1 software. Mean intensities (±SD) of the indicated VP30 peptides are shown. C, Quantification of nonphosphorylated KTCGSVEQQLNITAPK and phosphorylated KTCGSVEQQLNITAPKDSR peptides derived from the data in panel B. *P < .05.
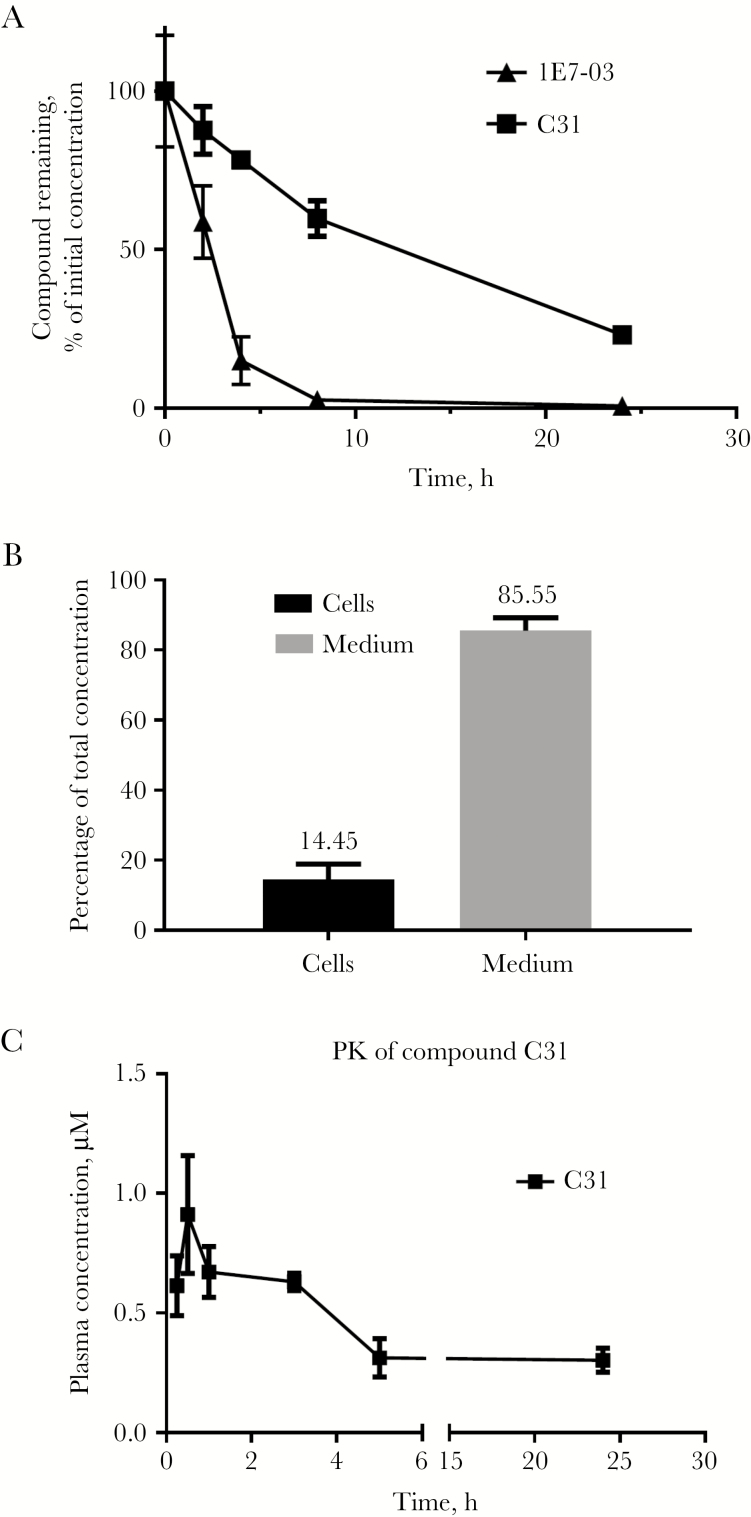
C31 Stability and Pharmacokinetics Analysis
First, we tested the stability of C31 (10 µM) in mouse serum. The samples were incubated at 37 and aliquots were collected at 0, 2, 6, 8, and 24 hours. Total protein was precipitated with acetone, and the compounds were recovered from the supernatant and quantified by LC/FT-MS. The half-life of C31 in murine serum was determined to be 11 hours, compared with only 2 hours for 1E7-03 (Figure 5A). Thus, C31 exhibited much higher stability than previously reported for 1E7-03 [8]. Intracellular accumulation of compound C31 was determined in 293T cells treated overnight with 10 µM C31 (Figure 5B). We observed 14.5% accumulation of C31 (Figure 5B), which was significantly higher than the approximately 3% accumulation of 1E7-03 reported previously [8]. Next, we analyzed the pharmacokinetics of C31 in mice. The compound was injected intraperitoneally, and the time-dependent plasma concentrations of C31 were determined. C31 was present in plasma 24 hours after injection (Figure 5C). However, we could only reach a submicromolar plasma concentration, which may limit the efficacy of C31 in vivo.
Figure 5.
Analysis of the pharmacological properties of C31. A, Stability of C31 in murine serum. C31 and 1E7-03 were dissolved in dimethyl sulfoxide (DMSO; 10 mM), and then mixed with murine serum at 100 μM concentrations. Aliquotes were collected after incubation for 0, 2, 6, 8, and 24 hours. Each sample (70 μL) was mixed with 280 μL of cold acetone, vortexed for 2 minutes, incubated at −20°C for 30 minutes, and centrifuged at 13000 × g for 5 minutes to remove proteins. The supernatant was transferred to a clean test tube and evaporated to dryness, using a SpeedVac concentrator. The residue was reconstituted in 70 μL of acetonitrile, and a 10 μL aliquot was injected for liquid chromatography/Fourier transform-mass spectrometry (LC/FT-MS) analysis. B, Intracellular accumulation of C31 and the compound left in the cell culture medium after overnight treatment of CEM T cells. Compound was extracted from cell lysates as described in panel A and quantified by LC/FT-MS. C, Pharmacokinetics of C31 in mice. Mice were injected intraperitoneally with 100 mg/kg C31. The concentrations of C31 were quantified by LC/FT-MS. Three mice were used for each time point. Mean values (±SD) are shown.
In summary, we screened for EBOV inhibitory compounds by using an EBOV-GFP high-throughput assay and in silico docking. We found that C31 had the best EBOV inhibitory activity and low toxicity. C31 bound to PP1 C-terminal groove in vitro and increased VP30 phosphorylation in cultured cells. C31 was more stable and bioavailable than previously described for 1E7-03. Taken together, C31 represents a novel PP1-targeting EBOV inhibitor with improved pharmacological properties that can be further evaluated for future antifiloviral therapeutics.
Supplementary Data
Supplementary Methods and Materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We thank Dr Elke Muhlberger (Boston University), for providing the EBOV minigenome plasmid; Dr Yoshihiro Kawaoka (University of Wisconsin), for providing plasmids expressing EBOV NP, VP35, VP30, and L and the plasmid expressing T7 polymerase; Dr Ryan Leib (Stanford University), for generating and providing the FASTA database containing EBOV proteins concatenated with proteins of African green monkey and common contaminants; the Biacore Molecular Interaction Shared Resource at the Lombardi Comprehensive Cancer Center (Georgetown University); and Anton Nekhai, for language corrections.
T. A., C. A. P., Y. S., A. I., and N. K. performed experiments. A. I. and M. P. conducted in silico screening. M. J. and X. L. conducted stability and pharmacokinetics analysis. A. K. synthesized C31. A. Ü., A. B., and S. N. supervised the study and wrote and revised the manuscript.
Disclaimer. The content is solely the responsibility of the authors and does not necessarily represent the official view of NHLBI, NIAID, NIMHD, NCI or NIH.
Financial support. This work was supported by the National Institutes of Health (grants 1P50HL118006, 1R01HL125005, 5G12MD007597, and P30AI117970 to S. N.; grant U19AI109664 to A. B. and S. N.; and grant P30CA51008 to A. U.).
Potential conflicts of interest. All authors: No reported conflicts.All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Jerebtsova M, Nekhai S. Therapeutics for postexposure treatment of Ebola virus infection. Future Virol 2015; 10:221–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ilinykh PA, Tigabu B, Ivanov A, et al. . Role of protein phosphatase 1 in dephosphorylation of Ebola virus VP30 protein and its targeting for the inhibition of viral transcription. J Biol Chem 2014; 289:22723–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kruse T, Biedenkopf N, Hertz EPT, et al. . The Ebola virus nucleoprotein recruits the host PP2A-B56 phosphatase to activate transcriptional support activity of VP30. Mol Cell 2018; 69:136–45 e6. [DOI] [PubMed] [Google Scholar]
- 4. Bollen M, Peti W, Ragusa MJ, Beullens M. The extended PP1 toolkit: designed to create specificity. Trends Biochem Sci 2010; 35:450–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Heroes E, Lesage B, Görnemann J, Beullens M, Van Meervelt L, Bollen M. The PP1 binding code: a molecular-lego strategy that governs specificity. FEBS J 2013; 280:584–95. [DOI] [PubMed] [Google Scholar]
- 6. Ammosova T, Platonov M, Ivanov A, et al. . 1E7-03, a low MW compound targeting host protein phosphatase-1, inhibits HIV-1 transcription. Br J Pharmacol 2014; 171:5059–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Modrof J, Mühlberger E, Klenk HD, Becker S. Phosphorylation of VP30 impairs ebola virus transcription. J Biol Chem 2002; 277:33099–104. [DOI] [PubMed] [Google Scholar]
- 8. Lin X, Kumari N, DeMarino C, et al. . Inhibition of HIV-1 infection in humanized mice and metabolic stability of protein phosphatase-1-targeting small molecule 1E7-03. Oncotarget 2017; 8:76749–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Irwin JJ, Shoichet BK. ZINC–a free database of commercially available compounds for virtual screening. J Chem Inf Model 2005; 45:177–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Totrov M, Abagyan R. Flexible protein-ligand docking by global energy optimization in internal coordinates. Proteins 1997; Suppl 1:215–20. [DOI] [PubMed] [Google Scholar]
- 11. Kelker MS, Page R, Peti W. Crystal structures of protein phosphatase-1 bound to nodularin-R and tautomycin: a novel scaffold for structure-based drug design of serine/threonine phosphatase inhibitors. J Mol Biol 2009; 385:11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Watanabe S, Watanabe T, Noda T, et al. . Production of novel ebola virus-like particles from cDNAs: an alternative to ebola virus generation by reverse genetics. J Virol 2004; 78:999–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ammosova T, Platonov M, Yedavalli VR, et al. . Small molecules targeted to a non-catalytic “RVxF” binding site of protein phosphatase-1 inhibit HIV-1. PLoS One 2012; 7:e39481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Edwards MR, Pietzsch C, Vausselin T, Shaw ML, Bukreyev A, Basler CF. High-throughput minigenome system for identifying small-molecule inhibitors of Ebola virus replication. ACS Infect Dis 2015; 1:380–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang JH, Chung TD, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen 1999; 4:67–73. [DOI] [PubMed] [Google Scholar]
- 16. Towner JS, Paragas J, Dover JE, et al. . Generation of eGFP expressing recombinant Zaire Ebolavirus for analysis of early pathogenesis events and high-throughput antiviral drug screening. Virol 2005; 332:20–7. [DOI] [PubMed] [Google Scholar]
- 17. Zhang L, Qi Z, Gao Y, Lee EYC. Identification of the interaction sites of Inhibitor-3 for protein phosphatase-1. Biochem Biophys Res Commun 2008; 377:710–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Peti W, Nairn AC, Page R. Structural basis for protein phosphatase 1 regulation and specificity. FEBS J 2013; 280:596–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.