Abstract
Prior to the sequencing of the human genome, it was presumed that most of the DNA coded for proteins. However, with the advent of next-generation sequencing, it has now been recognized that most complex eukaryotic genomes are in fact transcribed into noncoding RNAs (ncRNAs), including a family of transcripts referred to as long noncoding RNAs (lncRNAs). LncRNAs have been implicated in many biological processes ranging from housekeeping functions such as transcription to more specialized functions such as dosage compensation or genomic imprinting, among others. Interestingly, lncRNAs are not limited to a defined set of functions but can regulate varied activities such as messenger RNA degradation, translation, and protein kinetics or function as RNA decoys or scaffolds. Although still in its infancy, research into the biology of lncRNAs has demonstrated the importance of lncRNAs in development and disease. However, the specific mechanisms through which these lncRNAs act remain poorly defined. Focused research into a small number of these lncRNAs has provided important clues into the heterogeneous nature of this family of ncRNAs. Due to the complex diversity of lncRNA function, in this review, we provide an update on the platforms available for investigators to aid in the identification of lncRNA function.
Keywords: long noncoding RNA, lncRNA interactome, tools for lncRNA functional annotation
Introduction
Commencing with the initial discovery that coding exons of genes account for only 1.5% to 2% of the entire genome, a veritable revolution has been undertaken to understand the functional relevance of the nonprotein coding part of the genome.1,2 Undoubtedly, these studies have been greatly simplified by the advent of novel DNA sequencing technologies, which permit accurate whole-genome transcriptome analysis.3 This perpetual effort has resulted in the identification and cataloging of thousands of noncoding RNAs (ncRNAs).4
In general, ncRNAs can be divided into two broad classes based on their functions, as either housekeeping ncRNAs or regulatory ncRNAs. Housekeeping ncRNAs primarily regulate generic cellular functions such as messenger RNA (mRNA) translation (rRNA/tRNA), splicing (snRNAs), or rRNA modification (snoRNA). Regulatory ncRNAs can be further classified based on transcript length, with short noncoding transcripts comprising fewer than 200 nucleotides and long noncoding RNAs comprising transcripts greater than 200 nucleotides. A number of short ncRNAs subclasses exist, including microRNA (miRNA), small interfering RNA (siRNA), piwi interacting RNA (piRNA), transcription initiation RNA, and small cajal body-specific RNA (scaRNA).5–7 Long noncoding RNAs (lncRNAs) represent the largest class of ncRNAs. However, in contrast to short ncRNAS, which are mostly attributed to gene regulation, the mechanistic function of lncRNAs is highly diverse, adding to the increased complexity of this family of genes.8 In addition, the lack of insight into the function of lncRNAs may also be attributed to the low expression levels and tissue specificity of lncRNAs, resulting in an incomplete understanding of lncRNA regulation. Nevertheless, through genomic initiatives like ENCODE, FANTOM, GTEx, and GENCODE, over 60,000 lncRNAs have been predicted, a number of which have been demonstrated to be altered in certain diseases, underscoring the importance of these set of transcripts.9–12 However, to date, only a small percentage of these lncRNAs has been described in the literature, with an even smaller number being attributed to a specific mechanistic function. Furthermore, like proteins, many lncRNAs can employ more than one mechanism of action.
Increasing evidence points toward an important role played by lncRNAs in regulating multiple processes of gene expression, with instances of their transcription leading to gene silencing or gene activation. Studies have also found that regulation of lncRNAs can affect mRNA transcription, splicing, translation, export, import, and stability.13 LncRNAs have been demonstrated to function as transcription factor recruiters through their interaction with transcriptional start sites, act as transcriptional coactivators, or function as scaffold for proteins in general. Equivalently, lncRNAs may function as molecular decoys trapping transcription factors and thus limiting the ability of transcription factors to associate with DNA binding sites.14,15 Furthermore, lncRNAs have been shown to be involved in chromatin looping, nuclear body formation and function, or transcriptional read-through.15 Apart from transcriptional regulation, lncRNAs also play a role in mRNA processing, maturation, and stability through the regulation of mRNA splicing, inhibiting translation, operating as miRNAs sponges, or competing for miRNA binding sites on mRNA.15 Some lncRNAs can also code for small peptides.16,17 This fact does not necessarily disqualify a subset of lncRNAs as nonprotein coding transcripts but rather indicates that these lncRNAs can act as a bifunctional transcripts serving as either an lncRNA or a protein template. Last, lncRNAs can regulate protein and transcript trafficking and shuttling. The overall outcome of this is the association of lncRNAs with a myriad of biological processes, including imprinting, cell cycle regulation, pluripotency, dosage compensation, retro-transposon silencing, and telomere lengthening. In this review, we outline the platforms required to help define and catalogue newly discovered lncRNAs and discuss relevant techniques required to ascertain mechanistic functions of lncRNAs.
Platforms for Annotating lncRNA Functions
LncRNAs in recent times have debunked the ancient theory of junk DNA owing mainly to the recent developments in sequencing technologies and have been attributed to be essential for a number of physiological processes.18–20 In addition, lncRNAs interface with DNA, RNA, and proteins to exert their functions, raising their complexity to a higher level. The conventional methods used to study mRNA functions are inefficient for lncRNAs studies. LncRNAs have unique features, have developmental and tissue-specific peaks in expression, and are also available in very low copy numbers. These unique properties of lncRNAs make their detection extremely difficult as well as more amenable to investigate certain lncRNAs. Furthermore, certain lncRNAs provide for allele-specific epigenetic modification of gene expression in cis, which is possible through the limited spatial exposure at the site of transcription. While their tissue-specific occurrence can aid toward the development of biomarkers, their occurrence inside the nucleus in certain cases in a similar fashion can pose a problem when employing RNA interference for loss of function studies. Therefore, methods used to investigate lncRNAs should be highly efficient with enhanced targeting and higher resolution and increased maneuverability at the molecular level. We address here some of the strategies used to study the functions of lncRNAs.
RNA Sequencing
RNA sequencing (RNA-Seq) has seen a rapid growth over the past decades, allowing for the generation of quality in-depth sequencing and providing extensive information in a short time. Presently, a single run on any of the mainstream RNA-Seq platforms can yield up to a billion reads, with an individual read length of about 10 to 300 bp to longer read lengths of 10 to 20 kb. RNA-Seq provides a quantitative as well as descriptive scenario of the complex cellular content. It is important to note here that the data obtained herein are of high resolution compared to microarray technologies.21 In addition to obtaining high-quality RNA transcriptome profiles, RNA sequencing also aids in providing a top-up information on the 3′ end processing, alternative splicing regions, and RNA editing sites. RNA-Seq technologies have likewise helped to annotate lncRNAs, revealing global properties and even specific subclasses of lncRNAs. RNA sequencing has been advancing at a very rapid pace, with the rapid development of newer sequencing technologies serving different applications. RNA sequencing and next-generation sequencing technologies are a whole realm in themselves and have not been included in this review as a result of space constraints (refer to reviews on next-generation sequencing22–25). A short overview of the databases and tools used to study long noncoding RNA is presented in Table 1 .
Table 1.
Databases and Tools Used to Study Long Noncoding RNA.
| Database/Tools | Application | Reference |
|---|---|---|
| LncRNAdb v2.0 (lncRNA Database) | Reference database for functional long noncoding RNAs and provides comprehensive annotations of eukaryotic lncRNAs. | 106,107 |
| FANTOM (Functional Annotation of the mammalian Genome) | Database as a resource for experimentally supported lncRNA-disease association data. Database also has platform with integrated tools for predicting novel lncRNA-disease associations. | 108 |
| ENCODE (Encyclopaedia of DNA Elements) | The ENCODE database is a comprehensive collection of functional elements in the human genome, including elements that act at the protein and RNA levels and regulatory elements that control cells and circumstances in which a gene is active. | 109 |
| The GENCODE Project | The repository contains comprehensive gene annotations on reference chromosomes, scaffolds, assembly patches, and alternate loci. There is also comprehensive gene annotation of lncRNA genes. | 10 |
| lncRNAMap | A repository to investigate the putative regulatory functions of human lncRNAs and expression profiles for lncRNAs and their homologous protein coding genes. In addition, information regarding miRNA regulators of lncRNA is also available. | 110 |
| LNCipedia 3.0 | A repository for annotated human lncRNA sequences. | 111,112 |
| The LncRNA and Disease Database | A repository for curated and experimentally supported lncRNA-disease association data. The database also hosts integrated tools for predicting novel disease associations. Interactions at various levels such as protein, RNA, miRNA, and DNA are also available. | 113,114 |
| lnCeDB | A database of human lncRNAs that can act as ceRNAs. Database also provides information on lncRNA-mRNA pairs having common targeting miRNAs. The expression of lncRNA can be compared across 22 human tissues to estimate the chances of the pair for actually being ceRNAs. | 115 |
| starBASE v2.0 | Database designed for decoding pan-cancer and interaction networks of lncRNAs, miRNAs, ceRNAs, RNA binding proteins, and mRNAs from large-scale CLIP-Seq (HITS-CLIP, PAR-CLIP, iCLIP, CLASH) data and tumor samples comprising 14 cancer types spanning more than 6000 samples. Starbase also provides miR and ceRNA function web tools to predict the function of ncRNAs and protein coding genes from the miRNA-mediated (ceRNA) regulatory networks. | 116 |
| DIANA TOOLS LncBase v.2 | Tool for determining experimentally verified and computationally predicted miRNA targets on long noncoding RNAs. The experimental module engages miRNA and lncRNA interactions pertaining to the experimental validation and outcomes. The prediction module contains information for more than 10 million interactions and provides information of interaction sites, graphical representation of their binding, and the predicted score. | 117 |
| GeneCards | A human gene database that provides comprehensive information on all annotated and predicted human genes, including lncRNAs. An overall integrated data comprising linked genomic, transcriptomic proteomic, genetic, clinical, and functional information. | 118 |
| LincSNP2.0 | Database that stores and annotates disease-associated single-nucleotide polymorphisms in human long noncoding RNA and their transcription factor binding sites. | 119 |
| LncRNA2Target | A repository for differentially expressed genes after lncRNA knockdown or overexpression. | 120 |
| ChIP Base v2.0 | Open database for studying transcription factor binding sites and motifs and decoding the transcriptional regulatory networks of lncRNAs, miRNAs, other noncoding RNAs, and protein coding genes. | 121 |
| NRED | Database for lncRNA expression from microarray and in situ hybridization data. In addition, provides information on the evolutionary conservation, secondary structure, genomic context links, and antisense relationships. | 122 |
| NONCODE | An integrated database dedicated to noncoding RNA and in particular long noncoding RNA with more accurate annotations. The recent update provides additional features such as conservation annotation, lncRNA-disease relationships, and an interface to choose high-quality data sets through predicted scores, literature support, and long-read sequencing method support. | 123 |
| HGNC (HUGO Gene Nomenclature Committee) | Database aimed at approving unique names and symbols for human loci, including protein coding genes, noncoding genes, and pseudogenes, to allow unambiguous scientific communication. | 124 |
| PhyloCSF (Phylogenetic Codon Substitution Frequency) | Tool used to distinguish between protein coding and noncoding regions based on a formal statistical comparison of phylogenetic codon models. | 125 |
ceRNA, competing endogenous RNA; CLASH, cross-linking, ligation, and sequencing hybrids; CLIP-Seq, crosslinked immunoprecipitation sequencing; HITS-CLIP, high-throughput sequencing of RNA isolated by crosslinking immunoprecipitation; iCLIP, individual nucleotide resolution crosslinked immunoprecipitation; lncRNA, long noncoding RNA; miRNA, microRNA; mRNA, messenger RNA; ncRNA, noncoding RNA; PAR-CLIP, photoactivable ribonucleoside-enhanced crosslinked immunoprecipitation.
RNA Interference (RNAi)
Conventional gene modifications involving RNA interference (RNAi) techniques such as small interfering RNA (siRNA) have been used extensively to study functions of lncRNAs and function by suppressing the RNA expression by cleaving the RNA molecules.26 These strategies are transient and can be used to study quick effects of the lncRNAs upon depletion. LncRNAs are found to localize in various cellular components such as the nucleus, cytoplasm, or both, and targeting lncRNAs in the nucleus using these strategies has been less effective and also highly debated.27,28 Recently, knocking down of lncRNAs with higher efficiencies and reduced off-target effects have been achieved through the preparation of endoribonuclease-prepared siRNA (esiRNA) transcripts.29–31 Alternate ways of silencing include chemical synthesis of siRNAs but have been demonstrated to exhibit increased off-target effects despite comparable suppression levels.29
Short hairpin RNA (shRNA) is another class of molecules used in RNAi. shRNAs overcame the limitation of siRNAs in their transfection ability. The introduction of shRNA through viral vectors allows for its stable integration and long-term knockdown of the target gene. Another added advantage of using shRNAs is that they can be inducible and used for functional studies demanding a tight regulation of the gene. Evidence of successful RNAi knockdown of lncRNA has been documented in many reports such as siRNA-mediated knockdown of second chromosome locus associated with prostrate 1, UCA1, and hnRNP1.32–34
CRISPR/Cas9
The introduction of the CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats) nucleases has drastically propelled scientific research. CRISPR tools can be employed to ablate lncRNA expression or function by direct mutagenesis of the DNA sequence. However, it must be noted that unlike protein coding mRNA, changes made to the sequences coding ncRNA tend to be ineffective as mutations in the 5′ end of the ncRNA may not affect lncRNA activity, while in protein coding genes, this may result in loss of expression. Furthermore, as in many cases, lncRNAs overlap protein coding genes, and therefore the use of CRISPR may affect the function of both the lncRNA and the protein coding gene. As such, stringent designing needs to be considered in particular to alter the lncRNA structure or the specific lncRNA binding sites. Nevertheless, CRISPR technology has been used to efficiently delete large fragments of lncRNA as well.35 CRISPR can also be used as a targeting module to recruit activators and inhibitors to affect the transcription of lncRNAs. Furthermore, it also aids in the modulation of chromatin structure at specific regions neighboring lncRNAs, ultimately perturbing its expression. CRISPR can also be used to target the lncRNA under investigation to particular gene loci or cellular compartments to study their spatial influence.36
CRISPR Interference (CRISPRi) and CRISPR Activation (CRISPRa)
The explosion of CRISPR technology has seen the transition of CRISPR used in genome editing to genome regulation. CRISPR technologies modulate the expression of genes from their endogenous promoter and have been extensively demonstrated to affect activating or silencing lncRNAs.37–39 A modified CRISPR system, referred to as CRISPR interference/activation (CRISPR i/a), can effectively downregulate or upregulate gene expression by blocking or activating transcription, respectively.40–42 CRISPRi comprises a catalytically dead Cas9 protein (dCas9) and a guide RNA (gRNA) targeting the target gene to be knocked down.40 It is important to note here that gRNAs targeting the nontemplated DNA strand in the promoter or −35 regions exhibited greater downregulation than guides targeting 100 bp upstream of the promoter in the targeting template strand. Further modifications of CRISPRi include fusions of the dCas9 proteins with KRAB (Kruppel-associated box), which promotes heterochromatin and is also used to also bring about epigenetic silencing.40 This strategy leads to the error-prone nonhomologous end-joining pathway in incorporating mutations in the form of frame shifts of INDELS (insertions/deletions) and disrupting the gene function. CRISPRi technology has been used successfully to repress the expression of green fluorescent protein (GFP) in HEK293 cells and even endogenous genes like CXCR4 and CD71.41 Similarly, CRISPRa covers gain-of-function strategies by the overexpression of open reading frames (ORFs) and functions by deploying transcriptional activators through single guide RNAs (sgRNAs) and dCas9 to the transcriptional start sites (TSSs). This involves the fusion of dCas9 to transcriptional activator domains such as VP64, p65, and RTA (VPR-tripartite fusion of VP64 and the activation domains of the p65 subunit of NFκB and Epstein-Barr virus R transactivator, Rta).43–45 A second form of transcriptional activator module is a protein tagging system for signal amplification and fluorescence imaging, consisting of an array of repeating peptides and an antibody fusion protein, and is referred to as the SunTag.42,46 This system comprises VP64 fused to superfolder GFP (sfGFP) and an antibody single-chain variable fragment (scFv) that targets GCN4 epitope and is recruited in a tandem array of 10 copies of GCN4 epitope. A third variant of the activation systems includes an RNA scaffold and is referred to as the synergistic activation mediator (SAM) approach. The components of this system include p65 and HSF1 transcriptional activation domains fused to the MS2 coat protein. Dimers of these MS2 coat proteins are recruited to MS2 RNA hairpins.37 A recent genome-wide approach using a dual protein coding and noncoding integrated CRISPRa screen (DICaS) to identify functional coding and long noncoding RNA using the CaLR library (CRISPR activation of long noncoding RNA) revealed putative resistance genes toward cytarabine (Ara-C), a chemotherapeutic used in the treatment of patients with acute myeloid leukemia (AML).47
Antisense Oligonucleotides (ASO)
Antisense oligonucleotides (ASOs), as its name suggests, are antisense oligonucleotides that are highly effective in depletion of lncRNAs present in the nucleus.48,49 ASOs comprise modified or unmodified single-stranded deoxyribonucleotides that can hybridize to their respective complementary transcript targets followed by the ability of RNaseH to degrade the RNA component of the RNA-DNA duplex.50 LncRNAs have a wide range of cellular localization, with a predominant portion of them residing in the nucleus (such as MALAT1 and NEAT1), some in the cytoplasm (such as DANCR), and some in both (such as HOTAIR and TUG1). ASOs rank above siRNA and shRNAs in their ability to access the lncRNAs inside the nucleus.27 However, ASOs are also subjected to lower stability inside the cells due to their single-stranded nature and the action of nucleases. To overcome this, an advanced version of the ASOs, consisting of 15 to 20 nucleotides with a phosphorothioate modification in the backbone, can limit degradation by cellular nucleases. Yet another modification of the ASOs at the 20 position with an O-methoxy-ethyl group providing drug-like properties to ASOs showed improved binding affinity and sustained pharmacokinetics.51,52 While these second-generation ASOs confer resistance to nucleases, their modifications lead to inefficient binding of targets, and hence using higher concentrations leads to off-target effects.27 ASOs have been successfully used in loss-of-function studies. MALAT1 is one such lncRNA that was systematically knocked down using two different gapmers targeting two different parts of MALAT1, suggesting a potential therapy for inhibiting breast cancer progression.53 A few ASOs, such as nusinersen and mipomersen, also have been clinically approved.54,55 While the former is used to correct a splicing switch in SMN2 (survival of motor neuron 2) in patients with spinal muscular atrophy, the latter is used to knock down APOB100 mRNA to treat patients with familial hypercholesterolemia.54,55
Studying the functionality of lncRNAs is complicated owing to their complex genomic architecture and the added complexity of structure and shape they might harbor. Despite available techniques to uncover the functionality of lncRNAs, major limitations and challenges still remain such as the specificity of Cas9-sgRNAs and their ability to affect the neighboring or overlapping genes in the targeted loci.34 Strikingly, discrepancies have been observed between RNAi-based techniques and CRISPR-based techniques, underlining the necessity to pay attention to genes sharing promoters or overlapping transcripts to obtain biologically significant and relevant results for the targeted genes.42,56 Furthermore, the functionality of lncRNAs can be assigned with higher confidence when RNAi techniques such as siRNAs, siPOOLs (short interfering RNA pools, comprising a pool of 30 siRNAs), shRNAs, ASOs, and GapmeRs are complemented with CRISPR-based experiments.
Investigating lncRNA-Protein Interaction
RNA binding proteins (RBPs), ribonucleoproteins (RNPs), and a number of RNA species, including lncRNAs, are involved in this complex regulatory network.57,58 The spatiotemporal arrangement of the mRNA transcripts and the structural dynamicity of the RNPs are precisely correlated inside the cell.59 To identify the diverse regulatory interactions between RNA and proteins or other genetic elements, a combination of genetic, biochemical, and computation techniques can be applied to identify the complex RNA interactome.
RNA Immunoprecipitation (RIP)
The association of proteins with specific RNA species in vivo can be studied with the help of approaches such as RNA immunoprecipitation (RIP). Through an extension of protein-protein immunoprecipitation and techniques using an antibody of choice, proteins complexed with RNA can be pulled down. The association of the RNA with proteins or other associated RNA species can be quantified with real-time PCR or extensively with RNA sequencing.60 However, depending on the mode of protein interaction with RNA, RNA pulldown can be achieved through native RNA precipitation methods or by RNA crosslinking methods, with each technique having its own advantages. While the native RNA immunoprecipitation is indicative of a strong and direct RNA-protein binding, the crosslinking approach may be used to investigate indirect or weak binding of proteins to RNA. Importantly, one must not neglect that a binding event between two components could take place even after the lysis of the cells. The native RIP helps pull down kinetically stable interactions, but it is not conclusive if the interaction is still direct or indirect through a complex binding with the RNA.61 Furthermore, the signature binding motifs of the proteins cannot be determined owing to the long stretches of RNA in the antibody targeting the protein pulldown.62 In addition, native RIPs require multiple biological replicates because of their reduced reproducibility and the myriad of complex reactions taking place.63
Crosslinked Immunoprecipitation (CLIP)
Crosslinked immunoprecipitation (CLIP), on the other hand, engages both the RNA and protein via crosslinking. Crosslinking is mainly achieved with the help of ultraviolet light (UV), forming strong and specific crosslinks, followed by RNase treatment to shorten the RNA fragments. It is worthwhile to mention here that along with increased specificity, one can perform stringent washes on the same to reduce any background signals.64 Even though the UV-crosslinking in CLIP increases the specificity of protein and RNA interaction, false positives are frequent, and determining the exact binding consensus remains unanswered.65 Modification of the CLIP protocol, such as the iCLIP (individual nucleotide resolution CLIP) and PAR-CLIP (photoactivable ribonucleoside-enhanced CLIP), helps evade such drawbacks.66–70 iCLIP helps identify RNA binding motifs by identifying the exact crosslinking site and allows mapping the RNA and protein contacts at a nucleotide resolution. This is mainly achieved by the introduction of an adapter at the 5′ end by the primer used for reverse transcription, wherein the complementary DNA (cDNA) is circularized and subsequently linearized in the following steps capturing both truncated cDNA and read-through cDNAs.69 PAR-CLIP, on the other hand, uses 4-SU (4-thiouridine) or 6-SG (6-thiguanosine) infused in the culture media, wherein these moieties are incorporated in the RNA. The advantage of this approach is the elimination of the nonspecific targets and boosting the identification of exact binding sites at a single-nucleotide resolution.71 Nevertheless, the use of the photoreactive ribonucleoside analogues 4-SU and 6-SG might prove toxic to the cells and needs to be optimized at the right concentrations for the cell line used. Recently, a new technique, digestion-optimized RIP sequencing (DO-RIP Seq), has the added advantage of quantifying the binding at both the whole transcript level and the binding site level. DO-RIP Seq employs micrococcal nuclease and helps generate global protein RNA interactions with added information on their binding strength in cells or tissues.72
Interestingly, many groups have used RNA immunoprecipitation to understand the role between lncRNA and protein binding partners.73–75 Zhao and colleagues75 showed that the polycomb repressive complex binds to the RepA of Xist along with other lncRNAs involved in X chromosome inactivation, including Tsix (a lncRNA lying antisense of Xist). Another lncRNA, Fendrr, was also identified in a similar fashion to be associated with the PRC2 complex and WDR5 protein.74 With advances in technology and with native RIPs coupled to RNA sequencing, it has become possible to uncover many PRC2-interacting RNAs, including some already reported RNAs and some unannotated RNAs in embryonic stem cells.75 CLIP also has been used extensively to study lncRNA and protein associations. One such example is the association of the lncRNA air to methyl transferase G9a.76 In combination with high-throughput sequencing, a number of RNA binding proteins like Nova, TDP-43, Ago2, and Piwi proteins were identified.77–79 iCLIP coupled with deep sequencing revealed the global regulatory roles of hnRPN L protein.80 Furthermore, the PAR-CLIP approach has been successfully applied to RNA binding proteins like HuR, Ataxin2, AUF1, and FMRP.81–84
RAP, ChIRP, and CHART
Other robust methods of studying the lncRNA and protein interaction are through approaches that target the RNA directly. These include the RNA pulldown approaches like RNA antisense purification (RAP), chromatin isolation by RNA purification (ChIRP), and capture hybridization analysis of RNA targets (CHART). These methods can provide us information depending on the experiment used, and investigators can look at the DNA, RNA, or the proteins it is interacting with coupled with an added extension of a Western blot, quantitative real-time PCRs, mass spectrometry, and high-throughput sequencing. These RNA pulldown approaches use probes targeting the lncRNA under investigation, coupled with an affinity tag, such as biotin, subsequently pulled down using streptavidin-coated agarose or magnetic beads. One such lncRNA that was discovered using the RNA pulldown method was HOTAIR, which has been attributed for its role in cancers and importantly shown to interact with the PRC2 complex.85,86
ChIRP aids in understanding the interactions between proteins and chromatin by using biotinylated oligo probes as a bait, designed antisense to the lncRNA under investigation. It should be noted here that the cell is crosslinked, resulting in a snapshot of the interactions at that precise moment. Following up with the same experiment performed over a course of time on the cells or drug effects on the cells would help us to get a concise understanding of how the proteins interact on the given stretch of nucleic acids. Pulling down the whole chromatin using this approach provides immense information on the state of chromatin and the regions it interacts with when subsequently followed by quantitative PCRs, sequencing or mass spectrometry, or even simple Western blots when suspecting particular proteins involved in the process. A slight yet significant variation of the ChIRP is the domain-specific chromatin isolation by RNA purification (dChIRP), which was developed with the idea of investigating specific domains of the target RNA. This approach uses specific biotinylated probe pools targeting the specific domain, which in turn reduces the signal-to-noise ratio and enhances the localization and specificity, subsequently helping in the characterization of the lncRNA architecture and function.15,87,88 The same authors showed the direct binding of the MSL protein to the 3D structure of the roX1 lncRNA.87 Importantly, ChIRP coupled with mass spectrometry has been instrumental in the discovery of the proteome embracing the lncRNA.89 This approach helped pave the way to understand the spread of the lncRNA Xist and the silencing flow-through mechanism by the identification of 81 endogenous proteins. Proteins such as hnRNPK were shown to participate in chromatin modification and Xist-mediated gene silencing, but it did not play a role in the localization of Xist or its biogenesis.89
Another approach that enables the localization of the lncRNA in the chromatin and its association with proteins, based on the hybridization purification strategy, is capture hybridization analysis of RNA targets (CHART). Although highly similar to ChIRP, it differs from the former approach in its design criteria for the probes. While the probes for the ChIRP span across the complete lncRNA, CHART requires specially designed capture oligonucleotides, capable of hybridizing to accessible regions of the lncRNA. This is mainly achieved when formaldehyde-crosslinked nuclei lysates are subjected to RNAse H treatment, exposing potential hybridization regions. A follow-up mass spectrometry would help identify proteins associated with and similar to a ChIP, and CHART helps identify regions in the genome where the RNA is bound.90,91 CHART coupled with RNA sequencing was used to identify binding sites for MALAT1 and NEAT1, and CHART coupled with mass spectrometry helped unveil a repertoire of proteins that were associated with nuclear speckle and paraspeckle components and proteins associated with active chromatin.92
RAP is another approach of capturing the lncRNAs under investigation. RAP requires a crosslinking step but is not restricted to any particular kind of crosslinking. Any of the following crosslinking agents psoralens, formaldehyde, and UV crosslinking can be used. While psoralens are more suitable for RNA-RNA interactions, formaldehyde and UV cross-linking is preferred to study protein and nucleic acid (both RNA and DNA) interactions. RAP differs from the other RNA-centric methods in its use of long capture biotinylated probes usually greater than 60 nucleotides and the formation of very stable RNA-DNA hybrids.93 lncRNAs such as FIRRE subsequently showed their association with a nuclear matrix factor hnRNPU.94 The authors were also able to show that upon the genetic deletion of FIRRE and hnRNPU, the localization of this lncRNA was lost at the transchromosomal interacting loci, also highlighting the fact that RAP can be used to study the nuclear architecture even across chromosomes.94 Like the previous approaches, RAP can be coupled with mass spectrometry (MS) to study the proteins interacting and with RNA/DNA sequencing to identify potential interacting regions or binding regions. Recently, RAP-MS helped identify about 10 proteins associated with Xist, particularly elucidating its direct interaction with SHARP to alleviate silencing transcription through the HDAC3 complex and subsequently mediating the recruitment of PRC2 in a SHARP- and HDAC3-dependent manner.95
Identification of lncRNA Structural-Functional Relationships
Biochemical strategies provide information on the structural-functional relationship of lncRNAs through the study of their structures with techniques such as dimethyl sulfate sequencing (DMS-Seq), selective 2′-hydroxyl acylation analyzed by primer extension sequencing (SHAPE-Seq), fragmentation sequencing (FRAG-Seq), and parallel analysis of RNA structure (PARS). SHAPE is one such technique that is widely used and is based on the 2′-hydroxyl moiety. When this moiety is acetylated, 1-methyl-7-nitroisatoic anhydride (1M7) and N-methylisatoic anhydride (NMIA), the main reagents used in SHAPE, block the reverse transcription forming a 2′-O-adduct. This RNA is then subjected to cDNA synthesis. SHAPE in itself can, however, be used for a limited number of RNA or single-strand region analyses but has become powerful with the addition of next-generation sequencing to the SHAPE technique, enabling genome-wide structure probing.96,97 Another recent advance in the RNA SHAPE technique is the SHAPE MAP (Mutational Profiling), which is less cumbersome and does not involve any RNA ligation steps or library preparation steps.98 RNA interaction groups by mutational profiling (RING-MaP), another technique used for RNA structure-function studies, also aid in understanding the 3D RNA structure. DMS, a reagent used in the RING-MaP technique, has a limitation in that it can only modify the cytosine and adenosine nucleotides and could lead to bias in the interpretation of the results. PARS is another technique that is employed to study genome-wide analysis of RNA structures. This method employs the use of RNAse V1 and S1 followed subsequently by RNA sequencing. In addition, this method also suggested a stark difference between the coding regions and the untranslated regions. The coding regions pertain to fewer conformational changes owing to their structured regions, whereas the unstructured UTR regions expose their functional elements and their structural coding regions.99–101 Frag Seq is another technique that employs the P1 endonuclease to digest single-stranded RNA followed by high-throughput sequencing and bioinformatics analysis of the generated fragments.102 Table 2 summarizes some of the techniques used in understanding the functions of lncRNAs, their respective probes, and the advantages of using the techniques. Figure 1 provides a snapshot of the diverse lncRNA interactome and the protein-, RNA-, and DNA-centric approaches that can be used to further investigate the long noncoding RNA.
Table 2.
Techniques Used to Investigate lncRNAs.
| Technique | Bait | Crosslinking | Interaction | Technical Concept | Scope | Reference |
|---|---|---|---|---|---|---|
| nRIP | Protein | No | Direct/indirect | Captures transcriptome and its targets. RNA and protein components associated with the protein of interest. | Genome-wide | 75 |
| CLIP-Seq | Protein | UV 254 nm | Direct | Captures protein-RNA interactions in vivo. RNA components associated with protein of interest. | Genome-wide | 64 |
| CLIP–mass spectrometry | Protein | UV 254 nm | Direct/indirect | Captures protein-RNA interactions in vivo. Proteins complexes associated with protein of interest and the RNA targets it interacts with. | Genome-wide | 126,127 |
| PAR-CLIP | Protein | UV 365 nm | Direct T/C or G/A | Captures protein-RNA covalent binding enabled by efficient crosslinking from 4-SU or 6-SG. | Genome-wide | 71,127 |
| iCLIP | Protein | UV 254 nm | Direct; bound to a barcode sequence | Circularization of reverse transcribed products after the ligation of cleavable adaptors. | Genome-wide | 68,72 |
| RNA pulldown | lncRNA | Optional | Direct | Special aptamers such as biotin or MS2 fused to the lncRNA pulls down interactome of lncRNA. This includes the targets of lncRNA and complexes interacting. Proteins can be studied by immunoblotting or mass spectrometry. | lncRNA-specific interactions | 85,128,129 |
| RAP50,83 | Antisense-RNA | Disuccinimidyl glutarate-formaldehyde-aminomethyl-trioxsalen | Direct/indirect | 120-nt long nucleotide probes antisense to the target RNA and tiled across the entire RNA target. The probes are biotinylated and captures the lncRNA enrichment amidst protein-RNA interactions, RNA degradations and RNA secondary structures. | Genome-wide | 93,130 |
| ChIRP | DNA | Glutaraldehyde | Direct | Antisense DNA probes that hybridize to target RNA. Pulls down endogenous RNA and associated genomic DNA. | Genome-wide | 131 |
| ChIRP-domain | DNA | Glutaraldehyde-formaldehyde | Direct | Enables the pulldown of endogenous RNA-chromatin interactions in living cells. Similar to ChIRP, also provides functional information on the architecture and domains of the RNA under investigation. | Genome-wide | 88 |
| SHAPE-Seq | RNA | 1-Methyl-7 (1M7)–nitroisatoic anhydride (NMIA) | Architecture/structure | The method uses selective 2′-hydroxyl acylation analyzed by primer extension sequencing (SHAPE-Seq), which measures nucleotide resolution flexibility information for RNAs in vitro and in vivo. | Structural information | 97 |
| SHAPE-MaP | RNA | 1-methyl-7 (1M7)–nitroisatoic anhydride (NMIA)–1M6 | Architecture/structure | Similar to SHAPE, but SHAPE-MaP provides additional information on the mutations and yields accurate and high-resolution secondary-structure models and disentangles sequence polymorphisms. | Structural and mutation information | 98 |
| DMS-Seq | RNA | Dimethyl sulfate | Architecture/structure | Can be performed in vivo and in vitro. Interacts with unpaired adenine and cytosine residues followed by deep sequencing to identify modifications. | Structural modifications | 132 |
| FRAG-Seq | RNA | RNaseP1 | Architecture/structure | High-throughput RNA structure probing method that uses high-throughput RNA sequencing of fragments generated by digestion with nuclease P1, which specifically cleaves single-stranded nucleic acids. | Genome-wide | 133 |
| PARS, PARTE | RNA | RNase V1, RNase S1 | Architecture/structure | High-throughput deep sequencing of RNA fragments that are treated with structure-specific enzymes providing in vitro profiling of secondary structures at single-nucleotide resolution. | Genome-wide | 99,134 |
| icSHAPE | RNA | 2-methylnicotinic acid imidazolide N3 | Architecture/structure | Living cells are treated with the icSHAPE chemical NAI-N3 followed by selective chemical enrichment of NAI-N3–modified RNA, which provides an improved signal-to-noise ratio compared with similar methods leveraging deep sequencing. Purified RNA is then reverse-transcribed to produce cDNA, with SHAPE-modified bases leading to truncated cDNA. | Genome-wide | 135,136 |
4-SU, 4-thiouridine; 6-SG, 6-thiguanosine; cDNA, complementary DNA; ChIRP, chromatin isolation by RNA purification; CLIP, crosslinked immunoprecipitation; CLIP-Seq, crosslinked immunoprecipitation sequencing; iCLIP, individual nucleotide resolution crosslinked immunoprecipitation; DMS-Seq, dimethyl sulfate sequencing; FRAG-Seq, fragmentation sequencing; icSHAPE, in vivo click selective 2′-hydroxyl acylation analyzed by primer extension; lncRNA, long noncoding RNA; nRIP, native RNA immunoprecipitation; PAR-CLIP, photoactivable ribonucleoside-enhanced crosslinked immunoprecipitation; PARS, parallel analysis of RNA structure; PARTE, parallel analysis of RNA structure with temperature elevation; RAP, RNA antisense purification; SHAPE-MaP, selective 2′-hydroxyl acylation analyzed by primer extension mutational profiling; SHAPE-Seq, selective 2′-hydroxyl acylation analyzed by primer extension sequencing; UV, ultraviolet.
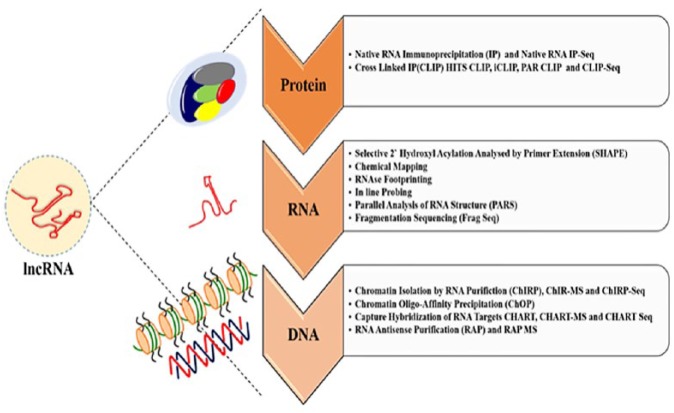
Figure 1.
Long noncoding RNA (lncRNAs) interactome and strategies. The lncRNA interactome is complex and involves DNA, RNA, and/or proteins. It is important to understand the mechanisms and functions of lncRNAs and the role they play in normal and diseased states. The methods or strategies employed for studying lncRNAs can be achieved by either of the following techniques. Protein-lncRNA interactions broadly represent the protein partners of lncRNAs and suggest their functional mechanisms and pathways. RNA immunoprecipitation (RIP) and crosslinked immunoprecipitation (CLIP) techniques provide clues of the associated RNAs when ribonucleoprotein complexes are pulled down based on the antibody of interest. The coupling of these techniques with high-throughput RNA sequencing and mass spectrometry could help identify the protein interactions to lncRNAs genome-wide or simply other proteins associated in the RNA binding protein complex or the protein of interest, respectively. Techniques that shed information based on the structural features like the secondary and tertiary structure of the lncRNAs eventually aid toward understanding the lncRNA function. The structural features can be harnessed through techniques and chemical reagents that cleave RNA at specific nucleotides or attack the regions that are exposed to the solvent, avoiding the RNA regions that are buried inside or are covered by proteins. Crosslinking also could reveal the intramolecular interactions that could be extended over a long range. Ribonucleases with different cleavage specificities can be used to obtain a RNAse footprint of potential regions covered by the proteins. Methods such as selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) and in-line probing aim at providing information on the local nucleotide flexibility. Coupling of SHAPE with sequencing could provide details of binding regions. Fragment sequencing (FragSeq) and parallel analysis of RNA structure (PARS), on the other hand, also employ RNase digestion to provide information on the RNA structure. Several techniques have been developed to identify the genomic DNA targets of lncRNAs. Based on the workflow backbone of chromatin immunoprecipitation (ChIP), chromatin isolation by RNA purification (ChIRP) helps identify lncRNAs associated with unique chromatin marks, whereas techniques such as chromatin oligo-affinity purification (ChOP) and capture hybridization of RNA targets (CHART) are basically used to identify the complementary DNA regions that interact with the RNA of interest. In addition, coupling with RNA sequencing, quantitative PCRs and mass spectrometry could yield important information regarding the RNA and protein interactome, respectively.
Future Directions
Since the majority of lncRNAs are present in cells in very low levels, studying their interactions with proteins or with nucleic acids poses a major challenge for techniques employed to study the lncRNA interactome such as RIP, RAP, CLIP, and other related techniques. Initially, the low abundance of lncRNAs was considered a limitation, but with the advance in sequencing technologies and molecular and biochemical techniques, it is possible to investigate lncRNAs that are sparsely populated in cells. Harnessing the properties of CRISPRa could help in overcoming this challenge by the endogenous overexpression of the lncRNA. While recent studies focus primarily on lncRNA expression in the total cell populations, lncRNAs expressed in different subcellular components of the cell can also be investigated considering the necessity to scale up the experiment. With single-cell transcriptomics on the rise, expression of the lncRNA at the single cell level could provide substantial information on its function.103 While substantial information regarding the genetic annotation of lncRNAs is obtained from advanced sequencing technologies, their subcellular localization still remains evasive. High-resolution quantification and spatial position of lncRNAs are possible with single-molecule RNA fluorescence in situ hybridization and could aid in functionally classifying them based on subcellular localization.104 In addition, these noncoding RNAs are present at low levels, possibly due to their unstable nature and quick degradation posttranscription.105 The functionality of lncRNAs in cells needs to be thoroughly analyzed through gene perturbation experiments such as overexpression and downregulation, followed by real-time quantitative PCRs or deep sequencing, to observe any differential gene expressions. Importantly, not all lncRNAs demonstrate functions in cells and laboratory model animals as most of the preliminary screenings are done in cell lines and not in physiological conditions of living organisms. Further challenges could also arise from the lack of measurability by the technique used or simply failure to knock down or overexpress the lncRNA due to its genomic architecture. A combination of computational and functional analysis such as those mentioned earlier is required for the identification of novel lncRNAs involved in both normal physiological conditions and diseased states.
Upon the identification of regulatory ncRNAs, it has now become readily apparent that regulation of proteins has been completely underestimated. We are now only beginning to understand that signal transduction is dependent upon, as it appears to us today, an almost incalculable level of regulation within a signaling cascade if not at the individual protein level. This continuous dynamic regulation is necessary for cells to finetune external cellular signaling cues into appropriate transcriptional responses. The identification that loss-of-function mutations within ncRNAs contribute to the genesis and progression of human disorders further highlights their importance. Nevertheless, defining the specific modes of action of lncRNAs will be a daunting task, possibly even greater than defining the functional relevance of approximately 20,000 predicted proteins. Based upon next-generation sequencing technology, large genome efforts such as ENCODE have begun to uncover an outline of the genome and equally important corresponding transcriptional profiles. This has permitted the identification of not only lncRNAs but also other overlapping sense transcripts. Although in these cases, sequence overlap may indicate the function of these lncRNAs as transcriptional activators or repressors, sequence homology is also essential to identify miRNA binding partners or 3′-UTR overlaps. Arguably, the identification of these lncRNAs functions is relatively simple compared to elaborate mechanisms involving protein complexes or subcellular localization. Understanding the mechanism of action of these lncRNAs will remain the key challenge to the identification of therapeutics targeting these molecules. Presently, the best method to target lncRNAs is through oligonucleotide-based therapies. While major progress has been made in terms of oligonucleotide design through either in silico–based crystallography or the generation of chemically modified analogues, effective targeting of these molecules remains inefficient. As mechanisms of action, oligonucleotide design and targeting methods continue to be researched, and it is expected that a number of a compounds targeting lncRNAs will be included in a physician’s vademecum for the treatment of a wide variety of diseases.
Footnotes
Declaration of Conflicting Interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was in part supported by the National Research Foundation Singapore and the Singapore Ministry of Education under its Research Centers of Excellence initiative, Koninklijke Philips N.V., and the Ministry of Education Academic Research Fund Tier 1 grants (T1-2013 Sep-10 and T1-2014 Oct-08).
ORCID iDs: John Lalith Charles Richard  https://orcid.org/0000-0002-1186-6390
https://orcid.org/0000-0002-1186-6390
Pieter Johan Adam Eichhorn  https://orcid.org/0000-0001-5840-943X
https://orcid.org/0000-0001-5840-943X
References
- 1. Kapranov P., Cheng J., Dike S., et al. RNA Maps Reveal New RNA Classes and a Possible Function for Pervasive Transcription. Science 2007, 316, 1484–1488. [DOI] [PubMed] [Google Scholar]
- 2. Lander E. S., Linton L. M., Birren B., et al. Initial Sequencing and Analysis of the Human Genome. Nature 2001, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 3. Bertone P., Stolc V., Royce T. E., et al. Global Identification of Human Transcribed Sequences with Genome Tiling Arrays. Science 2004, 306, 2242–2246. [DOI] [PubMed] [Google Scholar]
- 4. Mattick J. S., Rinn J. L. Discovery and Annotation of Long Noncoding RNAs. Nat. Struct. Mol. Biol. 2015, 22, 5–7. [DOI] [PubMed] [Google Scholar]
- 5. Mattick J. S. Non-Coding RNA. Hum. Mol. Genet. 2006, 15, R17–R29. [DOI] [PubMed] [Google Scholar]
- 6. Li L., Liu Y. Diverse Small Non-Coding RNAs in RNA Interference Pathways. Methods Mol. Biol. 2011, 764, 169–182. [DOI] [PubMed] [Google Scholar]
- 7. Kutter C., Svoboda P. MiRNA, SiRNA, PiRNA: Knowns of the Unknown. RNA Biol. 2008, 5, 181–188. [DOI] [PubMed] [Google Scholar]
- 8. Esteller M. Non-Coding RNAs in Human Disease. Nat. Rev. Genet. 2011, 12, 861–874. [DOI] [PubMed] [Google Scholar]
- 9. ENCODE Project Consortium. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science 2004, 306, 636–640. [DOI] [PubMed] [Google Scholar]
- 10. Harrow J., Frankish A., Gonzalez J. M., et al. GENCODE: The Reference Human Genome Annotation for the ENCODE Project. Genome Res. 2012, 22, 1760–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Forrest A. R. R., Kawaji H., Rehli M., et al. A Promoter-Level Mammalian Expression Atlas. Nature 2014, 507, 462–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. GTEx Consortium. The Genotype-Tissue Expression (GTEx) Project. Nat. Genet. 2013, 45, 580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kornienko A. E., Guenzl P. M., Barlow D. P., et al. Gene Regulation by the Act of Long Non-Coding RNA Transcription. BMC Biol. 2013, 11, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang K. C., Chang H. Y. Molecular Mechanisms of Long Noncoding RNAs. Mol. Cell 2011, 43, 904–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lau E. Non-Coding RNA: Zooming in on LncRNA Functions. Nat. Rev. Genet. 2014, 15, 3795. [DOI] [PubMed] [Google Scholar]
- 16. Ruiz-Orera J., Messeguer X., Subirana J. A., et al. Long Non-Coding RNAs as a Source of New Peptides. Elife 2014, 3, e03523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Richard J. L. C., Eichhorn P. J. A. Deciphering the Roles of LncRNAs in Breast Development and Disease. Oncotarget 2018, 9, 20179–20212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Geisler S., Coller J. RNA in Unexpected Places: Long Non-Coding RNA Functions in Diverse Cellular Contexts. Nat. Rev. Mol. Cell Biol. 2013, 14, 699–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dey B. K., Mueller A. C., Dutta A. Long Non-Coding RNAs as Emerging Regulators of Differentiation, Development, and Disease. Transcription 2014, 5, e914044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fatica A., Bozzoni I. Long Non-Coding RNAs: New Players in Cell Differentiation and Development. Nat. Rev. Genet. 2014, 15, 7–21. [DOI] [PubMed] [Google Scholar]
- 21. Wang Z., Gerstein M., Snyder M. RNA-Seq: A Revolutionary Tool for Transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mardis E. R. Next-Generation Sequencing Platforms. Annu. Rev. Anal. Chem. 2013, 6, 287–303. [DOI] [PubMed] [Google Scholar]
- 23. Metzker M. L. Sequencing Technologies—The Next Generation. Nat. Rev. Genet. 2010, 11, 31–46. [DOI] [PubMed] [Google Scholar]
- 24. Goodwin S., McPherson J. D., McCombie W. R. Coming of Age: Ten Years of Next-Generation Sequencing Technologies. Nat. Rev. Genet. 2016, 17, 333–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Buermans H. P. J., den Dunnen J. T. Next Generation Sequencing Technology: Advances and Applications. Biochim. Biophys. Acta 2014, 1842, 1932–1941. [DOI] [PubMed] [Google Scholar]
- 26. Castel S. E., Martienssen R. A. RNA Interference in the Nucleus: Roles for Small RNAs in Transcription, Epigenetics and Beyond. Nat. Rev. Genet. 2013, 14, 100–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lennox K. A., Behlke M. A. Cellular Localization of Long Non-Coding RNAs Affects Silencing by RNAi More Than by Antisense Oligonucleotides. Nucleic Acids Res. 2016, 44, 863–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gagnon K. T., Li L., Chu Y., et al. RNAi Factors Are Present and Active in Human Cell Nuclei. Cell Rep. 2014, 6, 211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Buchholz F., Kittler R., Slabicki M., et al. Enzymatically Prepared RNAi Libraries. Nat. Methods 2006, 3, 696–700. [DOI] [PubMed] [Google Scholar]
- 30. Chakraborty D., Kappei D., Theis M., et al. Combined RNAi and Localization for Functionally Dissecting Long Noncoding RNAs. Nat. Methods 2012, 9, 360–362. [DOI] [PubMed] [Google Scholar]
- 31. Kittler R., Surendranath V., Heninger A. K., et al. Genome-Wide Resources of Endoribonuclease-Prepared Short Interfering RNAs for Specific Loss-of-Function Studies. Nat. Methods 2007, 4, 337–344. [DOI] [PubMed] [Google Scholar]
- 32. Prensner J. R., Iyer M. K., Sahu A., et al. The Long Noncoding RNA SChLAP1 Promotes Aggressive Prostate Cancer and Antagonizes the SWI/SNF Complex. Nat. Genet. 2013, 45, 1392–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Huang J., Zhou N., Watabe K., et al. Long Non-Coding RNA UCA1 Promotes Breast Tumor Growth by Suppression of P27 (Kip1). Cell Death Dis. 2014, 5, e1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Goyal A., Myacheva K., Groß M., et al. Challenges of CRISPR/Cas9 Applications for Long Non-Coding RNA Genes. Nucleic Acids Res. 2017, 45, e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Han J., Zhang J., Chen L., et al. Efficient in Vivo Deletion of a Large Imprinted LncRNA by CRISPR/Cas9. RNA Biol. 2014, 11, 829–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wright A. V., Nuñez J. K., Doudna J. A. Biology and Applications of CRISPR Systems: Harnessing Nature’s Toolbox for Genome Engineering. Cell 2016, 164, 29–44. [DOI] [PubMed] [Google Scholar]
- 37. Konermann S., Brigham M. D., Trevino A. E., et al. Genome-Scale Transcriptional Activation by an Engineered CRISPR-Cas9 Complex. Nature 2015, 517, 583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Joung J., Engreitz J. M., Konermann S., et al. Genome-Scale Activation Screen Identifies a LncRNA Locus Regulating a Gene Neighbourhood. Nature 2017, 548, 343–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liu S. J., Horlbeck M. A., Cho S. W., et al. CRISPRi-Based Genome-Scale Identification of Functional Long Noncoding RNA Loci in Human Cells. Science 2017, 355, aah7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Qi L. S., Larson M. H., Gilbert L. A., et al. Repurposing CRISPR as an RNA-Guided Platform for Sequence-Specific Control of Gene Expression. Cell 2013, 152, 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gilbert L., Larson M. H., Morsut L., et al. CRISPR-Mediated Modular RNA-Guided Regulation of Transcription in Eukaryotes. Cell 2013, 154, 442–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gilbert L. A., Horlbeck M. A., Adamson B., et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell 2014, 159, 647–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Maeder M. L., Linder S. J., Cascio V. M., et al. CRISPR RNA-Guided Activation of Endogenous Human Genes. Nat. Methods 2013, 10, 977–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Perez-Pinera P., Kocak D. D., Vockley C. M., et al. RNA-Guided Gene Activation by CRISPR-Cas9-Based Transcription Factors. Nat. Methods 2013, 10, 973–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chavez A., Scheiman J., Vora S., et al. Highly Efficient Cas9-Mediated Transcriptional Programming. Nat. Methods 2015, 12, 326–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tanenbaum M. E., Gilbert L. A., Qi L. S., et al. A Protein Tagging System for Signal Amplification in Gene Expression and Fluorescence Imaging: Suntag. Cell 2014, 159, 635–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bester A. C., Lee J. D., Chavez A., et al. An Integrated Genome-Wide CRISPRa Approach to Functionalize LncRNAs in Drug Resistance. Cell 2018, 173, 649–664.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Dias N., Stein C. A. Antisense Oligonucleotides: Basic Concepts and Mechanisms. Mol. Cancer Ther. 2002, 1, 347–355. [PubMed] [Google Scholar]
- 49. Kole R., Krainer A. R., Altman S. RNA Therapeutics: Beyond RNA Interference and Antisense Oligonucleotides. Nat. Rev. Drug Discov. 2012, 11, 125–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bennett C. F., Baker B. F., Pham N., et al. Pharmacology of Antisense Drugs. Annu. Rev. Pharmacol. Toxicol. 2017, 57, 81–105. [DOI] [PubMed] [Google Scholar]
- 51. Yoo B. H. 2′-O-Methyl-Modified Phosphorothioate Antisense Oligonucleotides Have Reduced Non-Specific Effects in Vitro. Nucleic Acids Res. 2004, 32, 2008–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Crooke S. T. Molecular Mechanisms of Antisense Oligonucleotides. Nucleic Acid Ther. 2017, 27, 70–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Arun G., Diermeier S., Akerman M., et al. Differentiation of Mammary Tumors and Reduction in Metastasis upon Malat1 LncRNA Loss. Genes Dev. 2016, 30, 34–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Corey D. R. Nusinersen, an Antisense Oligonucleotide Drug for Spinal Muscular Atrophy. Nat. Neurosci. 2017, 20, 497–499. [DOI] [PubMed] [Google Scholar]
- 55. Geary R. S., Baker B. F., Crooke S. T. Clinical and Preclinical Pharmacokinetics and Pharmacodynamics of Mipomersen (Kynamro®): A Second-Generation Antisense Oligonucleotide Inhibitor of Apolipoprotein B. Clin. Pharmacokinet. 2015, 54, 133–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bassik M. C., Kampmann M., Lebbink R. J., et al. A Systematic Mammalian Genetic Interaction Map Reveals Pathways Underlying Ricin Susceptibility. Cell 2013, 152, 909–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yoon J.-H., Abdelmohsen K., Gorospe M. Posttranscriptional Gene Regulation by Long Noncoding RNA. J. Mol. Biol. 2013, 425, 3723–3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Quinodoz S., Guttman M. Long Noncoding RNAs: An Emerging Link between Gene Regulation and Nuclear Organization. Trends Cell Biol. 2014, 24, 651–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Keene J. RNA Regulons: Coordination of Post-Transcriptional Events. Nat. Rev. Genet. 2007, 8, 533–543. [DOI] [PubMed] [Google Scholar]
- 60. Selth L. A., Close P., Svejstrup J. Q. Studying RNA-Protein Interactions In Vivo by RNA Immunoprecipitation. Methods Mol. Biol. 2011, 791, 253–264. [DOI] [PubMed] [Google Scholar]
- 61. Gong C., Popp M. W.-L., Maquat L. E. Biochemical Analysis of Long Non-Coding RNA-Containing Ribonucleoprotein Complexes. Methods 2012, 58, 88–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Niranjanakumari S., Lasda E., Brazas R., et al. Reversible Cross-Linking Combined with Immunoprecipitation to Study RNA-Protein Interactions in Vivo. Methods 2002, 26, 182–190. [DOI] [PubMed] [Google Scholar]
- 63. Gagliardi M., Matarazzo M. R. RIP: RNA Immunoprecipitation. Methods Mol. Biol. 2016, 1480, 73–86. [DOI] [PubMed] [Google Scholar]
- 64. Darnell R. CLIP (Cross-Linking and Immunoprecipitation) Identification of RNAs Bound by a Specific Protein. Cold Spring Harb. Protoc. 2012, 7, 1146–1160. [DOI] [PubMed] [Google Scholar]
- 65. Li Q., Uemura Y., Kawahara Y. Cross-Linking and Immunoprecipitation of Nuclear RNA-Binding Proteins. Methods Mol. Biol. 2015, 1262, 247–263. [DOI] [PubMed] [Google Scholar]
- 66. Hafner M., Lianoglou S., Tuschl T., et al. Genome-Wide Identification of MiRNA Targets by PAR-CLIP. Methods 2012, 58, 94–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hafner M., Landthaler M., Burger L., et al. Transcriptome-Wide Identification of RNA-Binding Protein and MicroRNA Target Sites by PAR-CLIP. Cell 2010, 141, 129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. König J., Zarnack K., Rot G., et al. ICLIP Reveals the Function of HnRNP Particles in Splicing at Individual Nucleotide Resolution. Nat. Struct. 2010, 17, 909–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Huppertz I., Attig J., D’Ambrogio A., et al. ICLIP: Protein-RNA Interactions at Nucleotide Resolution. Methods 2014, 65, 274–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Hauer C., Curk T., Anders S., et al. Improved Binding Site Assignment by High-Resolution Mapping of RNA-Protein Interactions Using ICLIP. Nat. Commun. 2015, 6, 7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hafner M., Landthaler M., Burger L., et al. PAR-CLIP—a Method to Identify Transcriptome-Wide the Binding Sites of RNA Binding Proteins. J. Vis. Exp. 2010, 41, 2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Nicholson C. O., Friedersdorf M. B., Keene J. D. Quantifying RNA Binding Sites Transcriptome-Wide Using DO-RIP-Seq. RNA 2016, 23, 32–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zhao J., Sun B. K., Erwin J. A., et al. Polycomb Proteins Targeted by a Short Repeat RNA to the Mouse X Chromosome. Science 2008, 322, 750–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Grote P., Wittler L., Hendrix D., et al. The Tissue-Specific LncRNA Fendrr Is an Essential Regulator of Heart and Body Wall Development in the Mouse. Dev. Cell 2013, 24, 206–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zhao J., Ohsumi T. K., Kung J. T., et al. Genome-Wide Identification of Polycomb-Associated RNAs by RIP-Seq. Mol. Cell 2010, 40, 939–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Nagano T., Mitchell J. A., Sanz L. A., et al. The Air Noncoding RNA Epigenetically Silences Transcription by Targeting G9a to Chromatin. Science 2008, 322, 1717–1720. [DOI] [PubMed] [Google Scholar]
- 77. Licatalosi D. D., Mele A., Fak J. J., et al. HITS-CLIP Yields Genome-Wide Insights into Brain Alternative RNA Processing. Nature 2008, 456, 464–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Chi S. W., Zang J. B., Mele A., et al. Argonaute HITS-CLIP Decodes MicroRNA-MRNA Interaction Maps. Nature 2009, 460, 479–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Vourekas A., Mourelatos Z. HITS-CLIP (CLIP-Seq) for Mouse Piwi Proteins. Methods Mol. Biol. 2014, 1093, 73–95. [DOI] [PubMed] [Google Scholar]
- 80. Rossbach O., Hung L.-H., Khrameeva E., et al. Crosslinking-Immunoprecipitation (ICLIP) Analysis Reveals Global Regulatory Roles of HnRNP L. RNA Biol. 2014, 11, 146–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Lebedeva S., Jens M., Theil K., et al. Transcriptome-Wide Analysis of Regulatory Interactions of the RNA-Binding Protein HuR. Supplemental Information. Mol. Cell 2011, 43, 1–34. [DOI] [PubMed] [Google Scholar]
- 82. Yokoshi M., Li Q., Yamamoto M., et al. Direct Binding of Ataxin-2 to Distinct Elements in 3′ UTRs Promotes MRNA Stability and Protein Expression. Mol. Cell 2014, 55, 186–198. [DOI] [PubMed] [Google Scholar]
- 83. Yoon J., De S., Srikantan S., et al. PAR-CLIP Analysis Uncovers AUF1 Impact on Target RNA Fate and Genome Integrity. Nat. Commun. 2014, 5, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Ascano M., Jr., Mukherjee N., Bandaru P., et al. FMRP Targets Distinct MRNA Sequence Elements to Regulate Protein Expression. Nature 2012, 492, 382–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Rinn J. L., Kertesz M., Wang J. K., et al. Functional Demarcation of Active and Silent Chromatin Domains in Human HOX Loci by Noncoding RNAs. Cell 2007, 129, 1311–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Wu Y., Zhang L., Wang Y., et al. Long Noncoding RNA HOTAIR Involvement in Cancer. Tumor Biol. 2014, 35, 9531–9538. [DOI] [PubMed] [Google Scholar]
- 87. Quinn J. J., Ilik I. A., Qu K., et al. Revealing Long Noncoding RNA Architecture and Functions Using Domain-Specific Chromatin Isolation by RNA Purification. Nat. Biotechnol. 2014, 32, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Quinn J. J., Chang H. Y. In Situ Dissection of RNA Functional Subunits by Domain-Specific Chromatin Isolation by RNA Purification (DChIRP). Methods Mol. Biol. 2015, 1262, 199–213. [DOI] [PubMed] [Google Scholar]
- 89. Chu C., Zhang Q. C., Da Rocha S. T., et al. Systematic Discovery of Xist RNA Binding Proteins. Cell 2015, 161, 404–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Simon M. D., Wang C. I., Kharchenko P. V, et al. The Genomic Binding Sites of a Noncoding RNA. Proc. Natl. Acad. Sci. U. S. A. 2011, 108, 20497–20502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Simon M. D. Insight into LncRNA Biology Using Hybridization Capture Analyses. Biochim. Biophys. Acta 2016, 1859, 121–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. West J. A., Davis C. P., Sunwoo H., et al. The Long Noncoding RNAs NEAT1 and MALAT1 Bind Active Chromatin Sites. Mol. Cell 2014, 55, 791–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Engreitz J., Lander E. S., Guttman M. RNA Antisense Purification (RAP) for Mapping RNA Interactions with Chromatin. Methods Mol. Biol. 2015, 1262, 183–197. [DOI] [PubMed] [Google Scholar]
- 94. Hacisuleyman E., Goff L. A., Trapnell C., et al. Topological Organization of Multichromosomal Regions by the Long Intergenic Noncoding RNA Firre. Nat Struct Mol Biol 2014, 21, 198–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. McHugh C. A., Chen C.-K., Chow A., et al. The Xist LncRNA Interacts Directly with SHARP to Silence Transcription through HDAC3. Nature 2015, 521, 232–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Spitale R. C., Crisalli P., Flynn R. A., et al. RNA SHAPE Analysis in Living Cells. Nat. Chem. Biol. 2013, 9, 18–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Lucks J. B., Mortimer S. A., Trapnell C., et al. Multiplexed RNA Structure Characterization with Selective 2′-Hydroxyl Acylation Analyzed by Primer Extension Sequencing (SHAPE-Seq). Proc. Natl. Acad. Sci. U. S. A. 2011, 108, 11063–11068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Siegfried N., Busan S., Rice G. M., et al. RNA Motif Discovery by SHAPE and Mutational Profiling (SHAPE-MaP). Nat. Methods 2014, 11, 959–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Kertesz M., Wan Y., Mazor E., et al. Genome-Wide Measurement of RNA Secondary Structure in Yeast. Nature 2010, 467, 103–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Wan Y., Qu K., Ouyang Z. Q., et al. Genome-Wide Mapping of RNA Structure Using Nuclease Digestion and High-Throughput Sequencing. Nat. Protoc. 2013, 8, 849–869. [DOI] [PubMed] [Google Scholar]
- 101. Wan Y., Qu K., Zhang Q. C., et al. Landscape and Variation of RNA Secondary Structure across the Human Transcriptome. Nature 2014, 505, 706–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Underwood J. G., Uzilov A. V, Katzman S., et al. FragSeq: Transcriptome-Wide RNA Structure Probing Using High-Throughput Sequencing. Nat. Methods 2010, 7, 995–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Kim D. H., Marinov G. K., Pepke S., et al. Single-Cell Transcriptome Analysis Reveals Dynamic Changes in LncRNA Expression during Reprogramming. Cell Stem Cell 2015, 16, 88–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Cabili M. N., Dunagin M. C., McClanahan P. D., et al. Localization and Abundance Analysis of Human LncRNAs at Single-Cell and Single-Molecule Resolution. Genome Biol. 2015, 16, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Palazzo A. F., Gregory T. R. The Case for Junk DNA. PLoS Genet. 2014, 10, e1004351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Quek X. C., Thomson D. W., Maag J. L. V., et al. LncRNAdb v2.0: Expanding the Reference Database for Functional Long Noncoding RNAs. Nucleic Acids Res. 2015, 43, D168–D173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Amaral P. P., Clark M. B., Gascoigne D. K., et al. LncRNAdb: A Reference Database for Long Noncoding RNAs. Nucleic Acids Res. 2011, 39, D146–D151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Lizio M., Harshbarger J., Abugessaisa I., et al. Update of the FANTOM Web Resource: High Resolution Transcriptome of Diverse Cell Types in Mammals. Nucleic Acids Res. 2017, 45, D737–D743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Bernstein B. E., Birney E., Dunham I., et al. An Integrated Encyclopedia of DNA Elements in the Human Genome. Nature 2012, 489, 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Chan W. L., Huang H. D., Chang J. G. LncRNAMap: A Map of Putative Regulatory Functions in the Long Non-Coding Transcriptome. Comput. Biol. Chem. 2014, 50, 41–49. [DOI] [PubMed] [Google Scholar]
- 111. Volders P. J., Helsens K., Wang X., et al. LNCipedia: A Database for Annotated Human IncRNA Transcript Sequences and Structures. Nucleic Acids Res. 2013, 41, D246–D251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Volders P. J., Verheggen K., Menschaert G., et al. An Update on LNCipedia: A Database for Annotated Human LncRNA Sequences. Nucleic Acids Res. 2015, 43, D174–D180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Chen G., Wang Z., Wang D., et al. LncRNADisease: A Database for Long-Non-Coding RNA-Associated Diseases. Nucleic Acids Res. 2013, 41, D983–D986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Chen X., Yan G. Y. Novel Human LncRNA-Disease Association Inference Based on LncRNA Expression Profiles. Bioinformatics 2013, 29, 2617–2624. [DOI] [PubMed] [Google Scholar]
- 115. Das S., Ghosal S., Sen R., et al. LnCeDB: Database of Human Long Noncoding RNA Acting as Competing Endogenous RNA. PLoS One 2014, 9, e98965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Li J. H., Liu S., Zhou H., et al. StarBase v2.0: Decoding MiRNA-CeRNA, MiRNA-NcRNA and Protein-RNA Interaction Networks from Large-Scale CLIP-Seq Data. Nucleic Acids Res. 2014, 42, D92–D97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Paraskevopoulou M. D., Georgakilas G., Kostoulas N., et al. DIANA-LncBase: Experimentally Verified and Computationally Predicted MicroRNA Targets on Long Non-Coding RNAs. Nucleic Acids Res. 2013, 41, D239–D245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Safran M., Dalah I., Alexander J., et al. GeneCards Version 3: The Human Gene Integrator. Database 2010, 2010, baq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Ning S., Yue M., Wang P., et al. LincSNP 2.0: An Updated Database for Linking Disease-Associated SNPs to Human Long Non-Coding RNAs and Their TFBSs. Nucleic Acids Res. 2017, 45, D74–D78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Jiang Q., Wang J., Wu X., et al. LncRNA2Target: A Database for Differentially Expressed Genes after LncRNA Knockdown or Overexpression. Nucleic Acids Res. 2015, 43, D193–D196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Zhou K. R., Liu S., Sun W. J., et al. ChIPBase v2.0: Decoding Transcriptional Regulatory Networks of Non-Coding RNAs and Protein-Coding Genes from ChIP-Seq Data. Nucleic Acids Res. 2017, 45, D43–D50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Dinger M. E., Pang K. C., Mercer T. R., et al. NRED: A Database of Long Noncoding RNA Expression. Nucleic Acids Res. 2009, 37, D122–D126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Zhao Y., Li H., Fang S., et al. NONCODE 2016: An Informative and Valuable Data Source of Long Non-Coding RNAs. Nucleic Acids Res. 2016, 44, D203–D208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Eyre T. A. The HUGO Gene Nomenclature Database, 2006 Updates. Nucleic Acids Res. 2006, 34, D319–D321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Lin M. F., Jungreis I., Kellis M. PhyloCSF: A Comparative Genomics Method to Distinguish Protein Coding and Non-Coding Regions. Bioinformatics 2011, 27, i272–i282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Ule J., Jensen K., Mele A., et al. CLIP: A Method for Identifying Protein-RNA Interaction Sites in Living Cells. Methods 2005, 37, 376–386. [DOI] [PubMed] [Google Scholar]
- 127. Scheibe M., Butter F., Hafner M., et al. Quantitative Mass Spectrometry and PAR-CLIP to Identify RNA-Protein Interactions. Nucleic Acids Res. 2012, 40, 9897–9902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Tsai M.-C., Manor O., Wan Y., et al. Long Noncoding RNA as Modular Scaffold of Histone Modification Complexes. Science 2010, 329, 689–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Marín-Béjar O., Huarte M. RNA Pulldown Protocol for In Vitro Detection and Identification of RNA-Associated Proteins. In Regulatory Non-Coding RNAs: Methods and Protocols; 2014; pp. 87–95. Humana Press: New York, NY: 10.1007/978-1-4939-1369-5. [DOI] [PubMed] [Google Scholar]
- 130. Engreitz J. M., Sirokman K., McDonel P., et al. RNA-RNA Interactions Enable Specific Targeting of Noncoding RNAs to Nascent Pre-MRNAs and Chromatin Sites. Cell 2014, 159, 188–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Chu C., Quinn J., Chang H. Y. Chromatin Isolation by RNA Purification (ChIRP). J. Vis. Exp. 2012, 61, 4–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Rouskin S., Zubradt M., Washietl S., et al. Genome-Wide Probing of RNA Structure Reveals Active Unfolding of MRNA Structures In Vivo. Nature 2013, 505, 701–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Underwood J. G., Uzilov A. V, Katzman S., et al. FragSeq: Transcriptome-Wide RNA Structure Probing Using High-Throughput Sequencing. Nat. Methods 2010, 7, 995–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Wan Y., Qu K., Ouyang Z., et al. Genome-Wide Measurement of RNA Folding Energies. Mol. Cell 2012, 48, 169–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Flynn R. A., Zhang Q. C., Spitale R. C., et al. Transcriptome-Wide Interrogation of RNA Secondary Structure in Living Cells with IcSHAPE. Nat. Protoc. 2016, 11, 273–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Chan D., Feng C., Spitale R. C. Measuring RNA Structure Transcriptome-Wide with IcSHAPE. Methods 2017, 120, 85–90. [DOI] [PMC free article] [PubMed] [Google Scholar]