Abstract
Short interfering RNAs (siRNAs) and antisense oligonucleotides (ASOs) are the most clinically advanced oligonucleotide-based platforms. A number of N-acetylgalactosamine (GalNAc)-conjugated siRNAs (GalNAc-siRNAs), also referred to as RNA interference (RNAi) therapeutics, are currently in various stages of development, though none is yet approved. While the safety of ASOs has been the subject of extensive review, the nonclinical safety profiles of GalNAc-siRNAs have not been reported. With the exception of sequence differences that confer target RNA specificity, GalNAc-siRNAs are largely chemically uniform, containing limited number of phosphorothioate linkages, and 2’-O-methyl and 2’-deoxy-2’-fluoro ribose modifications. Here, we present the outcomes of short-term (3–5 week) rat and monkey weekly repeat-dose toxicology studies of six Enhanced Stabilization Chemistry GalNAc-siRNAs currently in clinical development. In nonclinical studies at supratherapeutic doses, these molecules share similar safety signals, with histologic findings in the organ of pharmacodynamic effect (liver), the organ of elimination (kidney), and the reticuloendothelial system (lymph nodes). The majority of these changes are nonadverse, partially to completely reversible, correlate well with pharmacokinetic parameters and tissue distribution, and often reflect drug accumulation. Furthermore, all GalNAc-siRNAs tested to date have been negative in genotoxicity and safety pharmacology studies.
Keywords: cell(ular) pathology, drug development, liver, monkey pathology, preclinical research and development, preclinical safety assessment/risk management, rat pathology
Drugs based on nucleic acids are emerging as an important class of human therapeutics due to their ability to harness natural cellular mechanisms that can potentially regulate the expression of any RNA transcript. In contrast to antibody-based agents that target synthesized proteins that are secreted or expressed on the cell surface, oligonucleotide-based drugs have the ability to silence production of disease-associated proteins by intervening at the transcript level. Of the various classes of oligonucleotide-based therapies, short interfering RNAs (siRNAs) and antisense oligonucleotides (ASOs) have demonstrated the most clinical potential to date (Chery 2016; Watts and Corey 2012). Four ASO drugs have been approved, and four RNA interference (RNAi) therapeutics are currently in late-stage clinical trials, with multiple N-acetylgalactosamine (GalNAc)-conjugated siRNAs (GalNAc-siRNA) achieving proof of concept status in the clinic (Fitzgerald, Kallend, and Simon 2017; Pasi et al. 2017; Zimmermann et al. 2017). Recent review articles have discussed the nonclinical safety profiles of ASOs (Engelhardt et al. 2015; Frazier 2015; Zanardi et al. 2018), but differences in structure, chemical modifications, and modes of action result in distinct safety profiles for siRNAs. Here, we review the platform-wide nonclinical safety profiles of Enhanced Stabilization Chemistry (ESC) RNAi therapeutics conjugated to GalNAc (Nair et al 2014; Foster et al 2018), which are currently in clinical development (givosiran, fitusiran, inclisiran, lumasiran, cemdisiran, and ALN-TTRSC02). Overall, successful efficacy and safety screening strategies, advances in backbone chemistries, and targeted delivery of GalNAc-siRNAs to hepatocytes have resulted in few toxicities observed in rats and cynomolgus monkeys in short-term (3–5 week) repeat-dose toxicology studies at doses 30- to 300-fold greater than the efficacious pharmacologic dose.
Overview of GalNAc-siRNAs
siRNAs are synthetic double-stranded RNA molecules, typically 19 to 23 nucleotides in length, that utilize the endogenous RNAi pathway to mediate target RNA knockdown (Martinez, Jimenez, and Paneda 2015; Watts and Corey 2012). They are highly polar molecules that do not effectively cross cellular membranes by passive diffusion, and therefore, delivery systems such as lipid nanoparticles have been used to facilitate intracellular delivery (Jayaraman et al. 2012; Maier et al. 2013). More recently, targeted delivery of siRNAs to hepatocytes has been achieved by direct conjugation to a triantennary GalNAc sugar, for receptor-mediated endocytosis via the asialoglycoprotein receptor (ASGPR) that is primarily expressed on the surface of hepatocytes (Nair et al. 2014; Zatsepin, Kotelevtsev, and Koteliansky 2016). This method of siRNA delivery has been used successfully in nonclinical species and in humans to suppress the expression of many hepatocellular target RNAs (Chan et al. 2015; Liebow et al. 2017; Nair et al. 2014; Sehgal et al. 2015; Pasi et al. 2017; Ray et al. 2017; Adityanjee 1987; Fitzgerald, Kallend, and Simon 2017; Zimmermann et al. 2017).
The ASGPR is a C-type lectin receptor that facilitates the clearance of desialylated glycoproteins from the blood (Cummings and McEver 2009; Geffen and Spiess 1992). It is expressed at high copy number (0.5–1 million per cell) on the surface of hepatocytes (Baenziger and Fiete 1980; Schwartz, Rup, and Lodish 1980), and each hepatocyte may endocytose up to 5 million copies of ASGPR per hour, providing significant excess receptor availability for drug binding and uptake (Dancygier et al. 2010). Its expression is largely restricted to hepatocytes (Wu, Nantz, and Zern 2002; Baenziger and Fiete 1980), although individual ASGPR subunits have been inconsistently reported in other tissues including human thyroid, large intestine, renal epithelium, testis, and human blood monocytes (Harris, van den Berg, and Bowen 2012; Seow, Tan, and Woo 2002). However, we found that ASGPR1 messenger RNA is uniquely expressed in the liver across species and that ASGPR labeling by immunohistochemistry (IHC) is likely due to antibody cross-reactivity with other closely related receptors (manuscript in preparation). Consistent with these data, tissue distribution of GalNAc-siRNAs is typically less than 5% beyond the target organ (liver) and the main organ of elimination (kidney). Furthermore, nonclinical screening can identify GalNAc-siRNAs with minimal toxicities at exaggerated doses in animal models, indicating high receptor capacity and therefore little to no interference with endogenous substrates in the liver and lack of effects in other tissues.
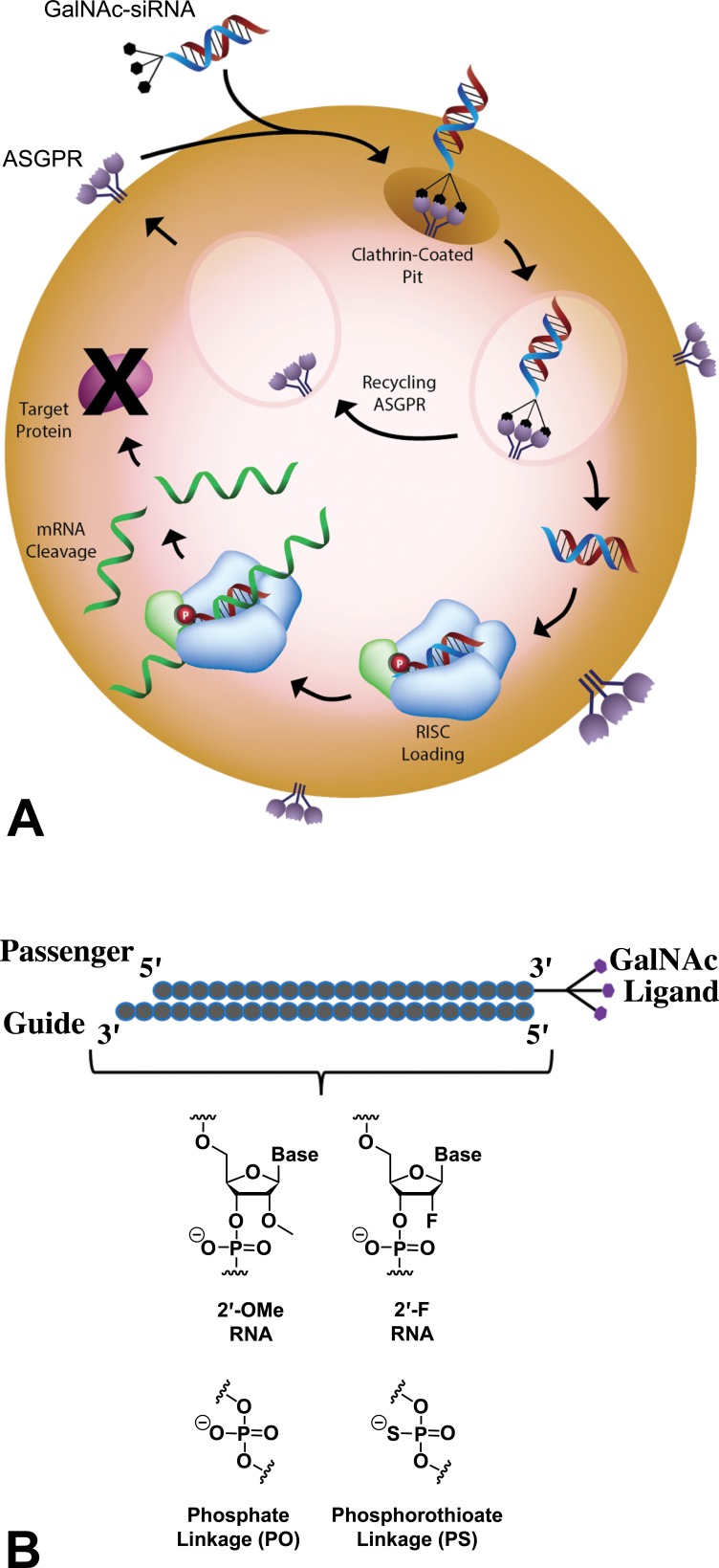
Upon uptake, a fraction of GalNAc-siRNA molecules reaches the cytoplasm and loads into the Argonaute protein in the RNA-induced silencing complex (RISC; Figure 1A). Subsequently, the sense (or “passenger”) strand of the siRNA is released, and the antisense (or “guide”) strand facilitates sequence-specific enzymatic cleavage by guiding RISC to the complementary RNA, thereby decreasing protein expression in a highly targeted manner (Elbashir et al. 2001).
Figure 1.
GalNAc-siRNA targeted delivery mechanism. (A) Following GalNAc-mediated delivery to hepatocytes via ASGPR and endolysosomal escape, siRNAs are loaded into a multi-subunit RNAi-induced silencing complex (RISC). After siRNA duplex unwinding, the antisense strand (red) remains bound to RISC and directs site-specific cleavage of the complementary target RNA sequence (green), resulting in RNA degradation and reduced expression of the target protein. (B) siRNA chemical modifications enhance stability and conjugation with a trivalent GalNAc ligand at the 3’ end of the sense strand allows for targeting to the ASGPR on hepatocytes. Alnylam’s Enhanced Stabilization Chemistry (ESC) GalNAc-siRNAs are fully modified at the 2’ position of the ribose with 2′-O-methyl (2′-OMe) and 2′-deoxy-2′-fluoro (2′-F) and contain limited number of strategically placed phosphorothioate (PS) linkages (Nair et al. 2014; Foster et al. 2018). ASGPR = asialoglycoprotein receptor; GalNAc = N-acetylgalactosamine; siRNA = short interfering RNA.
Unmodified siRNAs are rapidly eliminated by the kidney and degraded by exo- and endonucleases and therefore cannot achieve significant tissue distribution upon systemic administration (Soutschek et al. 2004). To confer optimal drug-like properties, siRNAs use common chemical modifications of the phosphodiester backbone and of the 2’-position of the ribose moiety. Although the siRNA backbone predominantly consists of natural phosphodiester linkages, a small number (up to six) of terminal phosphorothioate (PS) linkages, in which a nonbridging phosphate oxygen is replaced by sulfur, have been shown to enhance metabolic stability of the double-stranded structure (Berman et al. 2016; Eckstein 2000; Nair et al. 2014; Nair et al. 2017; Foster et al. 2018). In addition to strategically placed PS linkages, Alnylam’s ESC siRNAs are fully modified at the 2’ position of the ribose sugar with 2’-O-methyl (2’-OMe) and 2’-deoxy-2’-fluoro (2’-F) modifications (Figure 1B), which improve nuclease stability, reduce immunostimulatory properties, and increase binding affinity to the complementary RNA (Berman et al. 2016; Prakash and Bhat 2007). ESC GalNAc-siRNAs are generally chemically uniform with the exception of nucleobases that confer specificity for the target RNA. Consequently, in the absence of pharmacology-related adverse effects, GalNAc-siRNAs selected for clinical development have shown relatively consistent nonclinical safety profiles due to class effects across the platform.
siRNAs and ASOs have different structures, mechanisms of action, and chemical modifications and are thus expected to have different safety profiles. In contrast to double-stranded siRNAs, ASOs are single-stranded, typically 14 to 20 nucleotides in length, and their hybridization to complementary RNA leads to target downregulation via RNase H-mediated cleavage, translational repression by steric hindrance, or other mechanisms (Dias and Stein 2002; Watts and Corey 2012). While siRNAs have low PS content, most ASO backbones are fully PS-modified to reduce enzymatic cleavage by nucleases ubiquitous in both plasma and tissue (Deleavey and Damha 2012; Dias and Stein 2002; Eckstein 2000). ESC siRNAs employ 2’-OMe and 2’-F sugar modifications, while the 2’-O-methoxyethyl modification is most commonly used in ASOs (Berman et al. 2016; Martin 1995). In addition, conformationally restricted locked nucleic acid (LNA) or constrained ethyl modifications can be incorporated into ASOs to increase the binding affinity but are not used in GalNAc-siRNAs (Braasch and Corey 2001; Manoharan 2014).
Potential Mechanisms of GalNAc-siRNA Toxicities
On-target Effects
Toxicities of GalNAc-siRNAs may be mediated by the knockdown of the target RNA and should be considered within the context of their application. For example, exaggerated pharmacological effects were observed in nonclinical studies with fitusiran (ALN-AT3), a GalNAc-siRNA targeting antithrombin for the treatment of hemophilia (Sehgal et al. 2015). In wild-type mice, administration of fitusiran resulted in adverse events such as thrombosis, disseminated intravascular coagulation, and premature death. However, in mouse models of hemophilia, administration of fitusiran ameliorated the phenotypes of hemophilia and promoted hemostasis, resulting in prolonged survival of hemophiliac mice administered fitusiran compared to vehicle-treated hemophiliac mice. Thus, the toxicity associated with exaggerated pharmacology in nondiseased animals was consistent with the therapeutic hypothesis for fitusiran. Most on-target toxicities are predictable based on the expected functions of target genes and mouse and human genetic data.
Hybridization-dependent Off-target Effects
Similar to most oligonucleotide-based therapies, GalNAc-siRNAs can mediate hybridization-dependent off-target effects via the silencing of unintended RNAs with partial complementarity to the antisense strand (Birmingham et al. 2006; Jackson et al. 2003, 2006; Burel et al. 2016; Janas et al. 2018). A common toxicity observed at pharmacologically exaggerated doses in the early stages of drug development candidate screening in 3-week exploratory toxicology studies in rats consists of centrilobular hepatocellular degeneration and/or coagulative necrosis, associated with liver enzyme elevations that can exceed twofold the upper limit of normal. When this degree of toxicity is observed, compounds are eliminated as potential development candidates. These effects are most likely RNAi-mediated and are largely driven by the seed region of the antisense strand, analogous to endogenous microRNA activity (Janas et al. 2018). For instance, blocking siRNA RISC loading, blocking off-target binding of siRNA–RISC complexes with REVERSIR compounds (Zlatev et al. 2018), and swapping seed regions of known toxic and nontoxic siRNAs without changing the 2’-F, 2’-OMe, and PS content or liver exposure mitigated rat hepatotoxicity at pharmacologically exaggerated doses (Janas et al. 2018). In contrast, altering siRNA chemistry by partially substituting 2’-F with 2’-OMe modifications had no effect on hepatotoxicity (Janas et al. 2018). Importantly, thermally destabilizing chemical modifications in the seed region can reduce the risk of these sequence-dependent off-target effects (Bramsen et al. 2010; Mook et al. 2010; Vaish et al. 2011; Lee et al. 2015; Seok, Jang, and Chi 2016; Janas et al. 2018) and may minimize the occurrence of hepatotoxic siRNAs across species.
Chemical Modifications that Influence Sequence-independent Effects
Chemical modifications can substantially affect the toxicity profiles of oligonucleotides (Frazier 2015; Stanton et al. 2012). High PS content in single-stranded oligonucleotides has been linked to high protein binding that may result in hepatotoxicity or nephrotoxicity (Brown et al. 1994; Guvakova et al. 1995; Rockwell et al. 1997; Sewing et al. 2017). In addition, immunostimulatory effects may be associated with high PS content and can include complement activation, vasculitis, and glomerulopathy, as well as coagulopathies such as target independent severe thrombocytopenias (Engelhardt et al. 2015; Ferdinandi et al. 2011; Frazier 2015; Frazier et al. 2014; Hartmann et al. 1996; Henry et al. 1997, 2017; Senn, Burel, and Henry 2005; Crooke et al. 2017). Because only short consecutive stretches of PS are needed for optimal stability of double-stranded GalNAc-siRNAs, these toxicities are largely avoided (Perry et al. 2016). To date, extensive platform-wide comparisons and mechanistic toxicity studies of GalNAc-siRNAs have not found any associations between number or pattern of 2’-OMe or 2’-F modifications and in vivo hepatotoxicity in rodents (Janas et al. 2018) or in vitro cytotoxicity, including DNA double-strand breaks (Janas et al. 2017).
Oligonucleotides that employ LNA modifications, such as microRNA inhibitors, have also been associated with toxicities, most notably degeneration or necrosis in the liver (Burdick et al. 2014; Hagedorn et al. 2013; Kakiuchi-Kiyota et al. 2014). In mice, administration of LNA-modified oligonucleotides has been associated with significantly elevated liver transaminases and histopathologic evidence of hepatotoxicity (Swayze et al. 2007). The pro-inflammatory effects and protein binding properties of molecules containing high LNA content may contribute to the observed hepatic injury (Burdick et al. 2014; Hagedorn et al. 2013; Kakiuchi-Kiyota et al. 2014). Although LNA backbones were incorporated into some early variants of siRNA molecules (Elmen et al. 2005), current GalNAc-siRNAs do not contain LNA modifications and therefore avoid these associated risks.
In addition to backbone modifications, specific sequence motifs may contribute to hybridization-independent toxicity, likely via protein binding (Frazier 2015; Burdick et al. 2014). Using machine-learning techniques combined with validation experiments, studies have reported sequence motifs in LNA-modified ASOs associated with hepatotoxicity, suggesting that these predictable and potentially toxic motifs could be removed at the design stage (Burdick et al. 2014; Hagedorn et al. 2013). Although preliminary and ongoing, our lead-finding and bioinformatics activities to date have not identified any specific sequence motifs that were associated with liver safety signals of GalNAc-siRNAs.
Safety Profiles of GalNAc-siRNAs in Short-term Repeat-dose Studies in Rats and Monkeys
In vivo pharmacology and toxicology testing of GalNAc-siRNAs is typically conducted in two species, Sprague-Dawley rats and cynomolgus macaques. Across the platform, rats have generally proven to be toxicologically the most sensitive species to potential hepatotoxic effects of GalNAc-siRNAs. However, as ASGPR-mediated uptake of GalNAc-siRNAs in hepatocytes can approach saturation at high doses, and excess circulating GalNAc-siRNA is eliminated by the kidneys (without demonstrable impact on renal function), maximum tolerated doses often cannot be achieved in either species. Also of note for toxicity screening, GalNAc-siRNAs designed for human RNA sequences may not be active for on-target pharmacology in rats due to lack of target sequence homology between humans and rodents. In such cases, surrogate siRNA sequences have been used in proof of concept studies to derisk the potential for pharmacology-related on-target toxicities. All tested GalNAc-siRNAs based on human sequences have been pharmacologically active in monkeys to date, and thus, the development of cynomolgus monkey surrogates has not been required.
The toxicity findings described herein have been observed in weekly repeat-dose rat studies (three weeks in duration) and monkey studies (five weeks in duration) with ESC GalNAc-siRNAs that are currently in clinical development (givosiran, fitusiran, inclisiran, lumasiran, cemdisiran, and ALN-TTRSC02). All test articles are formulated in saline and administered subcutaneously. In typical short-term toxicology studies, doses 30- to 300-fold greater (30–300 mg/kg in both species) than the efficacious pharmacologic dose (typically 1–3 mg/kg) are administered weekly despite the fact that the expected clinical dosing regimen may be more infrequent. Data are currently being generated for several programs in subchronic, chronic, and carcinogenicity studies and will be reported separately.
Hepatocellular Vacuolation
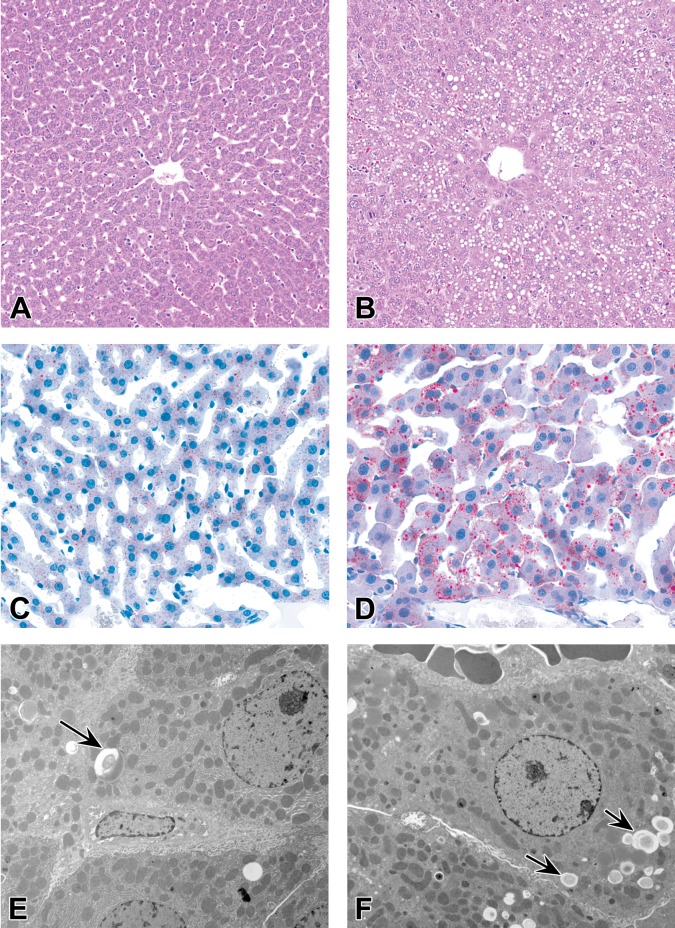
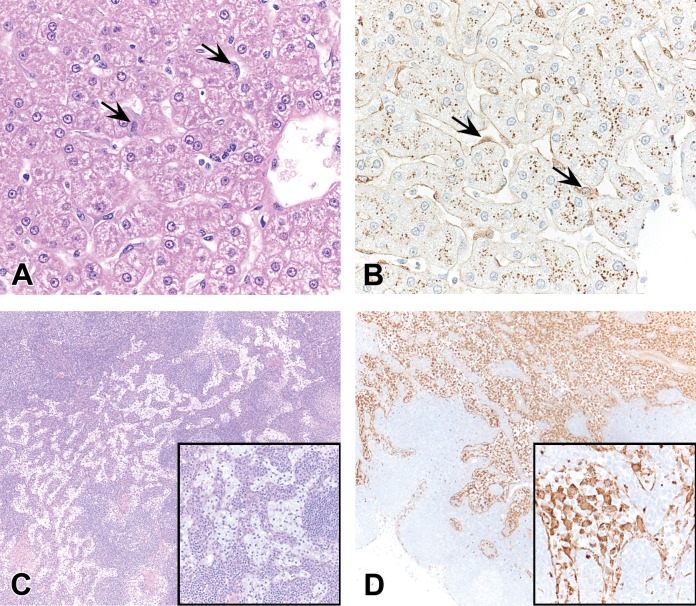
The most consistent finding in rats subcutaneously administered GalNAc-siRNAs is a dose-dependent increase in the number and size of hepatocellular vacuoles compared with control rats administered saline (Figure 2A and B). The six ESC GalNAc-siRNAs described here have the same linker, ligand, and chemical modifications (Figure 1B), but not all are associated with this finding at toxicologic doses of ≥30 mg/kg (four of six). Thus, it may be related to sequence-based hybridization effects or sequence-dependent effects on the natural RNAi pathways (Hsu et al. 2012; Wen and Friedman 2012). Oil Red O staining indicates that the majority of vacuoles contain neutral lipids (Figure 2C and D); in addition, transmission electron microscopy confirms the vacuoles to be lipid droplets, which are also observed to a lesser extent in vehicle control rats (Figure 2E and F). Hepatocellular vacuolation has occasionally correlated with minimal increases in liver weights but has not been associated with any significant clinical pathology changes (Figure 3). As such, it is considered nonadverse when observed as an isolated histologic finding and has not contributed to the determination of the no-observed-adverse-effect-level (NOAEL) in rats. Hepatocellular vacuolation has not been observed to date in monkey studies with GalNAc-siRNAs.
Figure 2.
Hepatocellular vacuolation in rats administered GalNAc-siRNAs once weekly in 3-week repeat-dose toxicity studies at doses up to 300 mg/kg. (A, B) Hematoxylin and eosin stained liver sections in 0.9% sodium chloride–treated control rats (A) and GalNAc-siRNA-treated rats (B). (C, D) Oil Red O staining of the liver shows neutral lipids in control rats (C) and increased amounts in GalNAc-siRNA-treated rats (D). (E, F) Transmission electron micrographs of the liver of control rats (E) and GalNAc-siRNA-treated rats (F). Arrows indicate vacuoles containing lipid droplets. GalNAc = N-acetylgalactosamine; siRNA = short interfering RNA.
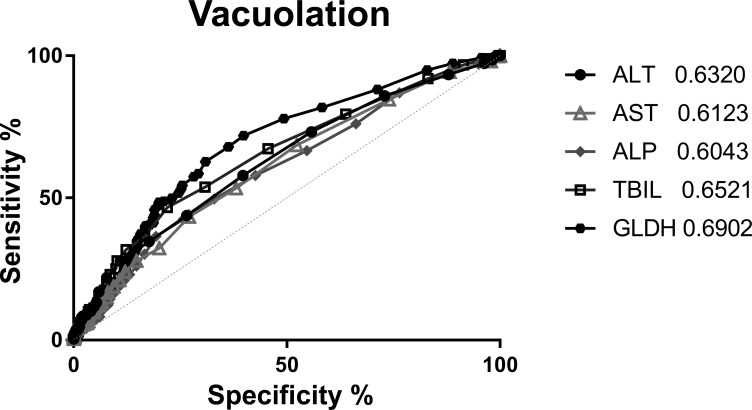
Figure 3.
Receiver operating characteristic (ROC) curve for hepatocellular vacuolation. Diagnostic performance of clinical chemistry parameters was evaluated for hepatocellular vacuolation by ROC analysis (performed using GraphPad Prism version 6.07 for Windows, GraphPad Software, La Jolla, CA, www.graphpad.com). The method of Hanley (Hanley and McNeil 1982) was used to calculate the area under the curve (AUC) values with a 95% confidence interval. A diagnostic test with an AUC of 0.8–1.0 is considered to have good predictive ability, whereas a test with an AUC of 0.5–0.7 indicates a random or nondiagnostic test result with poor predictive ability (Ennulat et al. 2010). Clinical and anatomical pathology data from 933 animals over 28 studies were compiled. Chemistry parameters were normalized to the respective control mean and expressed as the fold change of each individual compared to the control mean. ROC analysis was performed on five liver parameters: Alanine Aminotransferase (ALT), Aspartate Aminotransferase (AST), Alkaline Phosphatase (ALP), Total Bilirubin (TBIL), and Glutamate Dehydrogenase (GLDH). The observation of hepatocellular vacuolation was separated into two bins, no observation and grades 1 to 4. AUC for all five parameters were below 0.7, indicating they were not predictive as biomarkers for hepatocellular vacuolation.
Hepatocellular Single-cell Necrosis (SCN)
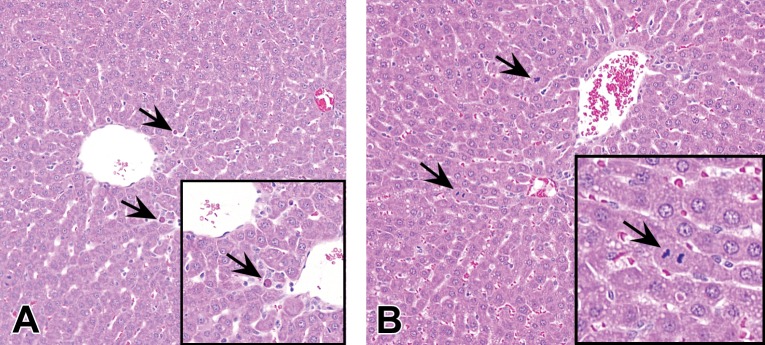
In rats, minimal to mild increases in hepatocellular SCN may be observed after the administration of GalNAc-siRNAs (Figure 4A). SCN is histologically characterized by rounded smaller hepatocytes with hypereosinophilic cytoplasm and a small pyknotic nucleus, suggestive of apoptotic cell death. Hepatic SCN and vacuolation are generally independent but not exclusive events. SCN is commonly detected in conjunction with signs of regeneration, including minimally to mildly increased mitotic figures (Figure 4B) and minimal karyomegaly. These findings occurred in subacute rat studies with two of the six ESC GalNAc-siRNAs described here, at supratherapeutic doses of ≥30 mg/kg in a dose-dependent manner, and generally only after repeat doses. The precise pathophysiology of hepatocellular SCN is unknown. Given that ESC GalNAc-siRNAs have the same linker and ligand and contain the same chemical modifications (Figure 1B), these findings may be related to sequence. Necrotic cells do not coalesce to form foci of overt hepatocellular coagulative necrosis and are not associated with any apparent local or systemic inflammatory changes. As an isolated event, minimal hepatocellular SCN has been considered nonadverse and does not affect the NOAEL in rat toxicology studies. However, when the severity grade increases, it often correlates with clinical pathology changes such as >2-fold elevations in ALT or AST and then is considered adverse. SCN as a result of GalNAc-siRNA administration appears to be largely specific to rats, as minimal SCN is only rarely observed in monkeys.
Figure 4.
Increased hepatocellular SCN and mitosis in rats administered GalNAc-siRNAs once weekly in 3-week repeat-dose toxicity studies at doses up to 300 mg/kg. (A) Instances of hepatocellular SCN are indicated by arrows. (B) Mitotic events (arrows) are often observed with SCN, consistent with regenerative activity. Hematoxylin and eosin stained liver sections are shown. GalNAc = N-acetylgalactosamine; SCN = single-cell necrosis; siRNA = short interfering RNA.
Drug Accumulation
Although some species-specific differences exist, the distribution of GalNAc-siRNAs is largely restricted to hepatocytes, with little to no distribution to nonparenchymal cells of the liver and limited delivery to other tissues. Functional RISC loading of GalNAc-siRNAs and RNA knockdown in other tissues have not been observed. Tissue pharmacokinetics are driven primarily by the GalNAc ligand and the metabolic stability conferred by the ESC chemistry (Nair et al. 2014; Foster et al. 2018) and are therefore similar across compounds.
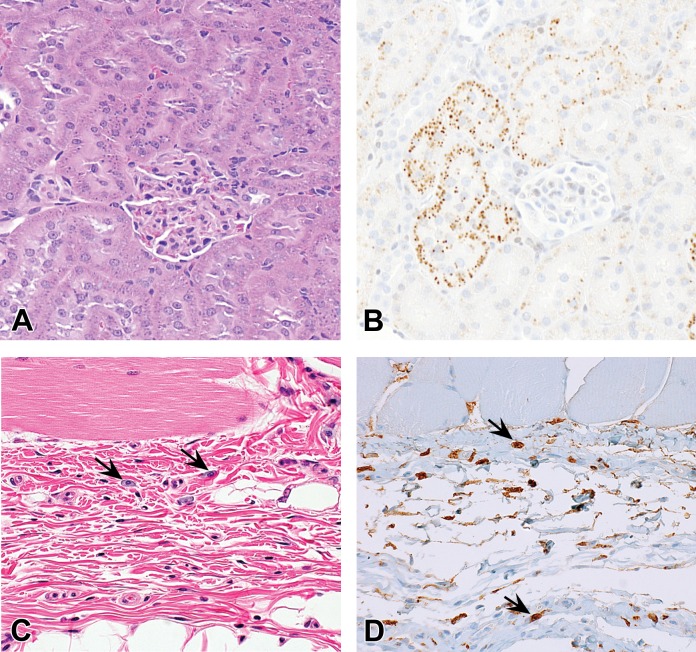
In rats, at repeat doses of ≥30 mg/kg, GalNAc-siRNA accumulation is commonly observed in the proximal renal tubular cells on hematoxylin and eosin sections as punctate cytoplasmic basophilic granules (Figure 5A). IHC confirms the identity of these granules as drug and/or metabolites (Figure 5B). Uptake into kidney proximal tubules is unlikely mediated by the GalNAc ligand, as this accumulation at toxicologic doses is observed both with conjugated and unconjugated siRNAs. Administration of GalNAc-siRNAs does not result in any microscopic evidence of degeneration or any clinical pathology changes that would suggest nephrotoxicity, even at doses up to 300 mg/kg administered weekly in both rat and monkey.
Figure 5.
Drug accumulation in rats administered GalNAc-siRNAs once weekly in 3-week repeat-dose toxicity studies at doses up to 300 mg/kg. (A, B) Proximal renal tubular cells of rats administered GalNAc-siRNAs. H&E staining (A) shows cytoplasmic basophilic granules and IHC (B) using an antibody recognizing 2’F-containing oligonucleotides confirms these granules to be GalNAc-siRNA drug. (C, D) The subcutaneous injection site of rats administered GalNAc-siRNAs. H&E staining (C) shows vacuolated mononuclear cells (arrows) in the superficial dermis, and IHC (D) confirms the presence of GalNAc-siRNA drug in these cells. GalNAc = N-acetylgalactosamine; H&E = hematoxylin and eosin; IHC = immunohistochemistry; siRNA = short interfering RNA.
In monkeys, following GalNAc-siRNA administration at toxicologic doses ≥100-fold higher than pharmacologic doses, drug accumulation most commonly manifests as basophilic granules in the cytoplasm of hepatic Kupffer cells and less commonly in hepatocytes (Figure 6A and B). Basophilic granules are rarely observed in proximal renal tubular cells in monkeys, which contrasts to their high occurrence in rats. Conversely, basophilic granules in Kupffer cells and hepatocytes are only rarely observed in rats.
Figure 6.
Drug accumulation in monkeys administered GalNAc-siRNAs once weekly in 5-week repeat-dose toxicity studies at doses up to 300 mg/kg. (A, B) Hepatic Kupffer cells of monkeys administered GalNAc-siRNAs. H&E staining (A) reveals basophilic granules (arrows), and IHC (B) using an antibody recognizing 2’F-containing oligonucleotides confirms these granules to be GalNAc-siRNA drug (arrows). Positive IHC labeling is also observable in the cytoplasm of hepatocytes. (C, D) Macrophages in the medulla of systemic lymph nodes of monkeys administered GalNAc-siRNAs. H&E staining (C) demonstrates vacuolated macrophages in the lymph node medulla, and IHC (D) confirms the presence of GalNAc-siRNA drug in these cells. The mesenteric lymph node often contains the most prominent granules versus the draining lymph nodes from the subcutaneous injection site or other lymph nodes. GalNAc = N-acetylgalactosamine; H&E = hematoxylin and eosin; IHC = immunohistochemistry; siRNA = short interfering RNA.
In reticuloendothelial cells such as macrophages in the medulla of lymph node and mononuclear cells at the injection site, drug accumulation manifests more prominently as cytoplasmic vacuolation rather than distinct basophilic granules. In monkeys, macrophage vacuolation is commonly observed in one or more lymph nodes (axillary, mesenteric, inguinal, and/or cervical; Figure 6C) and is accompanied by subtle basophilic stippling confirmed to be drug by IHC (Figure 6D). This finding is less frequently observed in rats but appears histologically similar. In both species, cytoplasmic vacuolation is also found at subcutaneous injection sites following repeat-dose GalNAc-siRNA administration, specifically in cells of presumptive macrophage and/or dendritic cell lineage (Figure 5C and D).
In contrast to the reversibility of hepatocellular vacuolation and hepatic SCN, manifestations of GalNAc-siRNA accumulation (cytoplasmic basophilic granules in the kidneys, hepatocytes, and Kupffer cells; cytoplasmic vacuolation of lymph node macrophages) generally persist throughout the recovery period. However, a decrease in incidence and/or severity often occurs, indicating a partial recovery. The persistence of most findings associated with GalNAc-siRNA accumulation reflects the long tissue half-life (typically weeks) of these compounds and also correlates to the long duration of pharmacologic activity observed in the liver. The plasma half-life, in contrast, is short (hours) and relatively noninformative for predicting toxicity.
Safety Profiles of GalNAc-siRNAs in Specialty Toxicology Evaluations
Genotoxicity Studies
Similar to other oligonucleotides (Berman et al. 2016), siRNAs consistently yield negative results in both in vitro and in vivo genotoxicity studies up to dose limits set by the International Conference on Harmonisation guidance S2(R1). Intracellular drug exposure has been demonstrated in the Ames assay, in the in vitro chromosome aberration assay in Chinese hamster ovary cells and in CD3+ human peripheral blood lymphocytes, and in the in vivo micronucleus assay in rat bone marrow, confirming that the negative genotoxicity results were valid and not the result of limited intracellular exposure (Janas et al. 2016).
Safety Pharmacology Studies
Cardiovascular, respiratory, and central nervous system safety pharmacology studies in monkeys have been performed for several GalNAc-siRNA clinical candidates to date. No drug-related effects were identified in these studies at both pharmacological and toxicological doses, consistent with extremely low drug concentrations and lack of detectable siRNA activity in these tissues.
Conclusions
ESC GalNAc-siRNAs containing limited PS linkages and fully modified with 2’-OMe and 2’-F are a promising class of oligonucleotide-based investigational therapeutics with the potential to impact rare genetic diseases, cardiometabolic diseases, and hepatic infectious diseases by targeted knockdown of disease-causing liver-expressed RNAs. In short-term (3–5 week) nonclinical weekly repeat-dose toxicology studies over a wide range of doses 30- to 300-fold greater than the predicted efficacious pharmacologic dose in both rat and monkey, GalNAc-siRNAs that are currently in clinical development (givosiran, fitusiran, inclisiran, lumasiran, cemdisiran, and ALN-TTRSC02) induced few consistent platform-wide effects independent of molecular target. The most common effects are histologic manifestations of drug accumulation in the liver, kidney, and lymph nodes, which are generally considered nonadverse up to the highest doses tested. In these studies, no kidney toxicities, immunostimulatory effects, thrombocytopenia, or injection site reactions were observed. Advances that minimize hybridization-dependent off-target effects (Janas et al. 2018) and a deeper understanding of hepatotoxic pathways will further enhance the safety and performance of GalNAc-siRNAs and other oligonucleotide-based therapeutics.
GalNAc-siRNAs that advance into the clinic are well-tolerated in nonclinical toxicology studies. Evaluations of the safety of GalNAc-conjugated RNAi therapeutics in humans will rely primarily on close monitoring during clinical trials, including any clinical symptoms or alterations in laboratory tests. There are currently six GalNAc-siRNA molecules undergoing testing in human clinical trials, with additional programs in development. Owing to their high predicted therapeutic indices, efficacy, and duration of action, GalNAc-conjugated RNAi therapeutics have the potential to have a major impact in a variety of high unmet need patient populations.
Acknowledgments
The authors thank Wendell Davis, Joseph Dybowski, Kevin Fitzgerald, Vasant Jadhav, Martin Maier, Peter Smith, and Tracy Zimmermann for critical reading of this article.
Authors’ Note: Professional medical writing and editorial assistance were provided by Laura Yee, PhD, of Caudex.
Author Contributions: Authors contributed to conception or design (MJ, CH, JS, AV, NK, GW); data acquisition, analysis, or interpretation (CH, VP, BC, NK); drafting the manuscript (MJ, CH, NK); and critically revising the manuscript (VP, BC, JS, AV, GW). Authors contributed equally to this work (MJ, CH). All authors gave final approval and agreed to be accountable for all aspects of work in ensuring that questions relating to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Declaration of Conflicting Interests: The author(s) declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article. Maja M. Janas, Victoria K. Perry, Brenda Carito, Jessica E. Sutherland, and Akshay K. Vaishnaw are employees of Alnylam Pharmaceuticals. Carole E. Harbison, Natalie D. Keirstead, and Garvin Warner are former employees of Alnylam Pharmaceuticals.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This article is funded by Alnylam Pharmaceuticals.
References
- Adityanjee. (1987). Role of electroconvulsive therapy in neuroleptic malignant syndrome (NMS). Acta Psychiatr Scand 76, 603–4. [DOI] [PubMed] [Google Scholar]
- Baenziger J. U., Fiete D. (1980). Galactose and N-acetylgalactosamine-specific endocytosis of glycopeptides by isolated rat hepatocytes. Cell 22, 611–20. [DOI] [PubMed] [Google Scholar]
- Berman C. L., Barros S. A., Galloway S. M., Kasper P., Oleson F. B., Priestley C. C., Sweder K. S., Schlosser M. J., Sobol Z. (2016). OSWG recommendations for genotoxicity testing of novel oligonucleotide-based therapeutics. Nucleic Acid Ther 26, 73–85. [DOI] [PubMed] [Google Scholar]
- Birmingham A., Anderson E. M., Reynolds A., Ilsley-Tyree D., Leake D., Fedorov Y., Baskerville S., et al. (2006). 3’ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat Methods 3, 199–204. [DOI] [PubMed] [Google Scholar]
- Braasch D. A., Corey D. R. (2001). Locked nucleic acid (LNA): Fine-tuning the recognition of DNA and RNA. Chem Biol 8, 1–7. [DOI] [PubMed] [Google Scholar]
- Bramsen J. B., Pakula M. M., Hansen T. B., Bus C., Langkjaer N., Odadzic D., Smicius R., et al. (2010). A screen of chemical modifications identifies position-specific modification by UNA to most potently reduce siRNA off-target effects. Nucleic Acids Res 38, 5761–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D. A., Kang S. H., Gryaznov S. M., DeDionisio L., Heidenreich O., Sullivan S., Xu X., Nerenberg M. I. (1994). Effect of phosphorothioate modification of oligodeoxynucleotides on specific protein binding. J Biol Chem 269, 26801–5. [PubMed] [Google Scholar]
- Burdick A. D., Sciabola S., Mantena S. R., Hollingshead B. D., Stanton R., Warneke J. A., Zeng M., et al. (2014). Sequence motifs associated with hepatotoxicity of locked nucleic acid–modified antisense oligonucleotides. Nucleic Acids Res 42, 4882–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burel S. A., Hart C. E., Cauntay P., Hsiao J., Machemer T., Katz M., Watt A., et al. (2016). Hepatotoxicity of high affinity gapmer antisense oligonucleotides is mediated by RNase H1 dependent promiscuous reduction of very long pre-mRNA transcripts. Nucleic Acids Res 44, 2093–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan A., Liebow A., Yasuda M., Gan L., Racie T., Maier M., Kuchimanchi S., et al. (2015). Preclinical development of a subcutaneous ALAS1 RNAi therapeutic for treatment of hepatic porphyrias using circulating RNA quantification. Mol Ther Nucleic Acids 4, e263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chery J. (2016). RNA therapeutics: RNAi and antisense mechanisms and clinical applications. Postdoc J 4, 35–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooke S. T., Baker B. F., Witztum J. L., Kwoh T. J., Pham N. C., Salgado N., McEvoy B. W., et al. (2017). The effects of 2’-O-methoxyethyl containing antisense oligonucleotides on platelets in human clinical trials. Nucleic Acid Ther 27, 121–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings R. D., McEver R. P. (2009). C-type Lectins. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [PubMed] [Google Scholar]
- Dancygier H., Merle U., Stremmel W., Niederau C. (2010). Hepatic metabolism In Clinical Hepatology: Principles and Practice of Hepatobiliary Diseases (Dancygier H., ed.), pp. 75–102. Springer, Berlin Heidelberg. [Google Scholar]
- Deleavey G. F., Damha M. J. (2012). Designing chemically modified oligonucleotides for targeted gene silencing. Chem Biol 19, 937–54. [DOI] [PubMed] [Google Scholar]
- Dias N., Stein C. A. (2002). Antisense oligonucleotides: Basic concepts and mechanisms. Mol Cancer Ther 1, 347–55. [PubMed] [Google Scholar]
- Eckstein F. (2000). Phosphorothioate oligodeoxynucleotides: What is their origin and what is unique about them? Antisense Nucleic Acid Drug Dev 10, 117–21. [DOI] [PubMed] [Google Scholar]
- Elbashir S. M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. (2001). Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411, 494–98. [DOI] [PubMed] [Google Scholar]
- Elmen J., Thonberg H., Ljungberg K., Frieden M., Westergaard M., Xu Y., Wahren B., et al. (2005). Locked nucleic acid (LNA) mediated improvements in siRNA stability and functionality. Nucleic Acids Res 33, 439–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhardt J. A., Fant P., Guionaud S., Henry S. P., Leach M. W., Louden C., Scicchitano M. S., et al. (2015). Scientific and regulatory policy committee points-to-consider paper*: Drug-induced vascular injury associated with nonsmall molecule therapeutics in preclinical development: Part 2. antisense oligonucleotides. Toxicol Pathol 43, 935–44. [DOI] [PubMed] [Google Scholar]
- Ennulat D., Magid-Slav M., Rehm S., Tatsuoka K. S. (2010). Diagnostic performance of traditional hepatobiliary biomarkers of drug-induced liver injury in the rat. Toxicol Sci 116, 397–412. [DOI] [PubMed] [Google Scholar]
- Ferdinandi E. S., Vassilakos A., Lee Y., Lightfoot J., Fitsialos D., Wright J. A., Young A. H. (2011). Preclinical toxicity and toxicokinetics of GTI-2040, a phosphorothioate oligonucleotide targeting ribonucleotide reductase R2. Cancer Chemother Pharmacol 68, 193–205. [DOI] [PubMed] [Google Scholar]
- Fitzgerald K., Kallend D., Simon A. (2017). A highly durable RNAi therapeutic inhibitor of PCSK9. N Engl J Med 376, e38. [DOI] [PubMed] [Google Scholar]
- Foster D. J., Brown C. R., Shaikh S., Trapp C., Schlegel M. K., Qian K., Sehgal A., et al. (2018). Advanced siRNA designs further improve in vivo performance of GalNAc-siRNA conjugates. Mol Ther 26, 708–717. https://www.ncbi.nlm.nih.gov/pubmed/29456020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazier K. S. (2015). Antisense oligonucleotide therapies: The promise and the challenges from a toxicologic pathologist’s perspective. Toxicol Pathol 43, 78–89. [DOI] [PubMed] [Google Scholar]
- Frazier K. S., Sobry C., Derr V., Adams M. J., Besten C. D., De Kimpe S., Francis I., et al. (2014). Species-specific inflammatory responses as a primary component for the development of glomerular lesions in mice and monkeys following chronic administration of a second-generation antisense oligonucleotide. Toxicol Pathol 42, 923–35. [DOI] [PubMed] [Google Scholar]
- Geffen I., Spiess M. (1992). Asialoglycoprotein receptor. Int Rev Cytol 137B, 181–219. [DOI] [PubMed] [Google Scholar]
- Guvakova M. A., Yakubov L. A., Vlodavsky I., Tonkinson J. L., Stein C. A. (1995). Phosphorothioate oligodeoxynucleotides bind to basic fibroblast growth factor, inhibit its binding to cell surface receptors, and remove it from low affinity binding sites on extracellular matrix. J Biol Chem 270, 2620–27. [DOI] [PubMed] [Google Scholar]
- Hagedorn P. H., Yakimov V., Ottosen S., Kammler S., Nielsen N. F., Hog A. M., Hedtjarn M., et al. (2013). Hepatotoxic potential of therapeutic oligonucleotides can be predicted from their sequence and modification pattern. Nucleic Acid Ther 23, 302–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanley J. A., McNeil B. J. (1982). The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 143, 29–36. [DOI] [PubMed] [Google Scholar]
- Harris R. L., van den Berg C. W., Bowen D. J. (2012). ASGR1 and ASGR2, the genes that encode the asialoglycoprotein receptor (ashwell receptor), are expressed in peripheral blood monocytes and show interindividual differences in transcript profile. Mol Biol Int 2012, 283974 doi: 10.1155/2012/283974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann G., Krug A., Waller-Fontaine K., Endres S. (1996). Oligodeoxynucleotides enhance lipopolysaccharide-stimulated synthesis of tumor necrosis factor: Dependence on phosphorothioate modification and reversal by heparin. Mol Med 2, 429–38. [PMC free article] [PubMed] [Google Scholar]
- Henry S. P., Narayanan P., Shen L., Bhanot S., Younis H. S., Burel S. A. (2017). Assessment of the effects of 2’-methoxyethyl antisense oligonucleotides on platelet count in cynomolgus nonhuman primates. Nucleic Acid Ther 27, 197–208. [DOI] [PubMed] [Google Scholar]
- Henry S. P., Novotny W., Leeds J., Auletta C., Kornbrust D. J. (1997). Inhibition of coagulation by a phosphorothioate oligonucleotide. Antisense Nucleic Acid Drug Dev 7, 503–10. [DOI] [PubMed] [Google Scholar]
- Hsu S. H., Wang B., Kota J., Yu J., Costinean S., Kutay H., Yu L., et al. (2012). Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J Clin Invest 122, 2871–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson A. L., Bartz S. R., Schelter J., Kobayashi S. V., Burchard J., Mao M., Li B., Cavet G., Linsley P. S. (2003). Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol 21, 635–37. [DOI] [PubMed] [Google Scholar]
- Jackson A. L., Burchard J., Schelter J., Chau B. N., Cleary M., Lim L., Linsley P. S. (2006). Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA 12, 1179–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janas M. M., Jiang Y., Duncan R. G., Hayes A. N., Liu J., Kasperkovitz P. V., Placke M. E., Barros S. A. (2016). Exposure to siRNA-GalNAc conjugates in systems of the standard test battery for genotoxicity. Nucleic Acid Ther 26, 363–71. [DOI] [PubMed] [Google Scholar]
- Janas M. M., Jiang Y., Schlegel M. K., Waldron S., Kuchimanchi S., Barros S. A. (2017). Impact of oligonucleotide structure, chemistry, and delivery method on in vitro cytotoxicity. Nucleic Acid Ther 27, 11–22. [DOI] [PubMed] [Google Scholar]
- Janas M. M., Schlegel M. K., Harbison C. E., Yilmaz V. O., Jiang Y., Parmar R., Zlatev I., et al. (2018). Selection of GalNAc-conjugated siRNAs with limited off-target-driven rat hepatotoxicity. Nat Commun 9, 723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraman M., Ansell S. M., Mui B. L., Tam Y. K., Chen J., Du X., Butler D., et al. (2012). Maximizing the potency of siRNA lipid nanoparticles for hepatic gene silencing in vivo . Angew Chem Int Ed Engl 51, 8529–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakiuchi-Kiyota S., Koza-Taylor P. H., Mantena S. R., Nelms L. F., Enayetallah A. E., Hollingshead B. D., Burdick A. D., et al. (2014). Comparison of hepatic transcription profiles of locked ribonucleic acid antisense oligonucleotides: Evidence of distinct pathways contributing to non-target mediated toxicity in mice. Toxicol Sci 138, 234–48. [DOI] [PubMed] [Google Scholar]
- Lee H. S., Seok H., Lee D. H., Ham J., Lee W., Youm E. M., Yoo J. S., et al. (2015). Abasic pivot substitution harnesses target specificity of RNA interference. Nat Commun 6, 10154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebow A., Li X., Racie T., Hettinger J., Bettencourt B. R., Najafian N., Haslett P., et al. (2017). An investigational RNAi therapeutic targeting glycolate oxidase reduces oxalate production in models of primary hyperoxaluria. J Am Soc Nephrol 28, 494–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier M. A., Jayaraman M., Matsuda S., Liu J., Barros S., Querbes W., Tam Y. K., et al. (2013). Biodegradable lipids enabling rapidly eliminated lipid nanoparticles for systemic delivery of RNAi therapeutics. Mol Ther 21, 1570–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin P. (1995). Ein neuer Zugang zu 2’-O-Alkylribonucleosiden und Eigenschaften deren Oligonucleotide. Helvetica Chimica Acta 78, 486–504. [Google Scholar]
- Martinez T., Jimenez A. I., Paneda C. (2015). Short-interference RNAs: Becoming medicines. EXCLI J 14, 714–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mook O., Vreijling J., Wengel S. L., Wengel J., Zhou C., Chattopadhyaya J., Baas F., Fluiter K. (2010). In vivo efficacy and off-target effects of locked nucleic acid (LNA) and unlocked nucleic acid (UNA) modified siRNA and small internally segmented interfering RNA (sisiRNA) in mice bearing human tumor xenografts. Artif DNA PNA XNA 1, 36–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair J. K., Attarwala H., Sehgal A., Wang Q., Aluri K., Zhang X., Gao M., et al. (2017). Impact of enhanced metabolic stability on pharmacokinetics and pharmacodynamics of GalNAc-siRNA conjugates. Nucleic Acids Res 45, 10969–10977. doi: 10.1093/nar/gkx818 https://www.ncbi.nlm.nih.gov/pubmed/28981809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair J. K., Willoughby J. L., Chan A., Charisse K., Alam M. R., Wang Q., Hoekstra M., et al. (2014). Multivalent N-acetylgalactosamine-conjugated siRNA localizes in hepatocytes and elicits robust RNAi-mediated gene silencing. J Am Chem Soc 136, 16958–61. [DOI] [PubMed] [Google Scholar]
- Pasi K. J., Rangarajan S., Georgiev P., Mant T., Creagh M. D., Lissitchkov T., Bevan D., et al. (2017). Targeting of antithrombin in hemophilia A or B with RNAi therapy. N Engl J Med 377, 819–28. [DOI] [PubMed] [Google Scholar]
- Perry K., Hettinger J., Carito B., Keirstead N., Placke M. (2016). Review of GalNAc-siRNA conjugates across multiple programs demonstrates no evidence of thrombocytopenia or pro-inflammatory effects. Presented at 12th Annual Meeting of the Oligonucleotide Therapeutics Society (OTS), September 25–28, Montreal, Canada. [Google Scholar]
- Prakash T. P., Bhat B. (2007). 2’-Modified oligonucleotides for antisense therapeutics. Curr Top Med Chem 7, 641–49. [DOI] [PubMed] [Google Scholar]
- Ray K. K., Landmesser U., Leiter L. A., Kallend D., Dufour R., Karakas M., Hall T., et al. (2017). Inclisiran in patients at high cardiovascular risk with elevated LDL cholesterol. N Engl J Med 376, 1430–40. [DOI] [PubMed] [Google Scholar]
- Rockwell P., O’Connor W. J., King K., Goldstein N. I., Zhang L. M., Stein C. A. (1997). Cell-surface perturbations of the epidermal growth factor and vascular endothelial growth factor receptors by phosphorothioate oligodeoxynucleotides. Proc Natl Acad Sci U S A 94, 6523–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz A. L., Rup D., Lodish H. F. (1980). Difficulties in the quantification of asialoglycoprotein receptors on the rat hepatocyte. J Biol Chem 255, 9033–36. [PubMed] [Google Scholar]
- Sehgal A., Barros S., Ivanciu L., Cooley B., Qin J., Racie T., Hettinger J., et al. (2015). An RNAi therapeutic targeting antithrombin to rebalance the coagulation system and promote hemostasis in hemophilia. Nat Med 21, 492–97. [DOI] [PubMed] [Google Scholar]
- Senn J. J., Burel S., Henry S. P. (2005). Non-CpG-containing antisense 2’-methoxyethyl oligonucleotides activate a proinflammatory response independent of Toll-like receptor 9 or myeloid differentiation factor 88. J Pharmacol Exp Ther 314, 972–79. [DOI] [PubMed] [Google Scholar]
- Seok H., Jang E. S., Chi S. W. (2016). Rationally designed siRNAs without miRNA-like off-target repression. BMB Rep 49, 135–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seow Y. Y., Tan M. G., Woo K. T. (2002). Expression of a functional asialoglycoprotein receptor in human renal proximal tubular epithelial cells. Nephron 91, 431–38. [DOI] [PubMed] [Google Scholar]
- Sewing S., Roth A. B., Winter M., Dieckmann A., Bertinetti-Lapatki C., Tessier Y., McGinnis C., et al. (2017). Assessing single-stranded oligonucleotide drug-induced effects in vitro reveals key risk factors for thrombocytopenia. PLoS One 12, e0187574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutschek J., Akinc A., Bramlage B., Charisse K., Constien R., Donoghue M., Elbashir S., et al. (2004). Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature 432, 173–78. [DOI] [PubMed] [Google Scholar]
- Stanton R., Sciabola S., Salatto C., Weng Y., Moshinsky D., Little J., Walters E., et al. (2012). Chemical modification study of antisense gapmers. Nucleic Acid Ther 22, 344–59. [DOI] [PubMed] [Google Scholar]
- Swayze E. E., Siwkowski A. M., Wancewicz E. V., Migawa M. T., Wyrzykiewicz T. K., Hung G., Monia B. P., Bennett C. F. (2007). Antisense oligonucleotides containing locked nucleic acid improve potency but cause significant hepatotoxicity in animals. Nucleic Acids Res 35, 687–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaish N., Chen F., Seth S., Fosnaugh K., Liu Y., Adami R., Brown T., et al. (2011). Improved specificity of gene silencing by siRNAs containing unlocked nucleobase analogs. Nucleic Acids Res 39, 1823–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts J. K., Corey D. R. (2012). Silencing disease genes in the laboratory and the clinic. J Pathol 226, 365–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J., Friedman J. R. (2012). miR-122 regulates hepatic lipid metabolism and tumor suppression. J Clin Invest 122, 2773–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Nantz M. H., Zern M. A. (2002). Targeting hepatocytes for drug and gene delivery: Emerging novel approaches and applications. Front Biosci 7, d717–25. [DOI] [PubMed] [Google Scholar]
- Zanardi T. A., Kim T. W., Shen L., Serota D., Papagiannis C., Park S. Y., Kim Y., Henry S. P. (2018). Chronic toxicity assessment of 2’-O-methoxyethyl antisense oligonucleotides in mice. Nucleic Acid Ther 28, 233–241. doi: 10.1089/nat.2017.0706 https://www.ncbi.nlm.nih.gov/pubmed/29708844. [DOI] [PubMed] [Google Scholar]
- Zatsepin T. S., Kotelevtsev Y. V., Koteliansky V. (2016). Lipid nanoparticles for targeted siRNA delivery—going from bench to bedside. Int J Nanomedicine 11, 3077–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann T. S., Karsten V., Chan A., Chiesa J., Boyce M., Bettencourt B. R., Hutabarat R., et al. (2017). Clinical proof of concept for a novel hepatocyte-targeting GalNAc-siRNA conjugate. Mol Ther 25, 71–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlatev I., Castoreno A., Brown C. R., Qin J., Waldron S., Schlegel M. K., Degaonkar R., et al. (2018). Reversal of siRNA-mediated gene silencing in vivo. Nat Biotechnol 36, 509–11. [DOI] [PubMed] [Google Scholar]