Abstract
Background
Chicken erythrocytes are involved in immunity through binding of toll-like receptors (TLRs) with their ligands to activate downstream signaling and lead to cytokine production in erythrocytes. Some avian β-defensins (AvBDs) are constitutively expressed in tissues and some others can be induced by various bacteria and viruses. However, the expression of AvBDs in erythrocytes has not yet been studied extensively.
Results
The transcripts of eight AvBDs (AvBD1 to AvBD7, and AvBD9) and liver-expressed antimicrobial peptide-2 (LEAP-2) were found in normal chicken erythrocytes. The expression levels of AvBD2, 4 and 7 were significantly increased (P < 0.01), whereas the levels of AvBD1, 6 and 9 were significantly decreased (P < 0.01) after Marek’s disease virus (MDV) infection. The mRNA expression level of LEAP-2 was not significantly changed after MDV infection. Highest viral nucleic acid (VNA) of MDV in the feather tips among the tested time points was found at 14 days post-infection (d.p.i.). In addition, 35 MD5-related gene segments were detected in the erythrocytes at 14 d.p.i. by transcriptome sequencing.
Conclusions
These results suggest that the AvBDs in chicken erythrocytes may participate in MDV-induced host immune responses.
Keywords: Chicken, Erythrocytes, AvBDs, TLRs, MDV
Background
Erythrocytes are the most abundant cells in circulation and the most important cells in the transportation and exchange of gases. In addition, several studies have reported that chicken erythrocytes play an important role in immunology [1]. For example, erythrocytes of vertebrates are nucleated and contain organelles in their cytoplasm, except in mammals [2]. It has been reported that the expression of cytokine transcripts in salmon erythrocytes were induced by infection with infectious salmon anaemia virus (ISAV). Similarly, cytokines, produced by trout erythrocytes when exposed to Candida albicans, enhanced the phagocytic capabilities [3, 4]. Moreover, the transcripts for many toll-like receptors (TLRs) were constitutively expressed in erythrocytes [1]. Chicken erythrocytes can induce the expression of transcripts for some cytokines and secrete thermo-labile molecules, which can enhance the antiviral capabilities of macrophages in response to TLR ligand treatments, such as type I interferons (IFNs) and interleukin (IL)-8 [1, 5].
Marek’s disease (MD) is a contagious lymphoproliferative disease caused by Marek’s disease virus (MDV), and can lead to tumors, lymphocyte proliferation, and ultimately death in chickens [6]. MDV has been evolving worldwide over the past few decades. The emerging MDV strains have changed from mild MDV (mMDV) to very virulent MDV (vvMDV) and very virulent plus MDV (vv + MDV) [7]. Host defense peptides (HDPs), also known as antimicrobial peptides, are a group of cationic amphipathic peptides that are produced by the host itself; they play a vital role in innate immunity and could kill various pathogenic microorganisms such as bacteria, fungi and even enveloped viruses [8–11]. Avian β-defensins (AvBDs) is the only type of defensin found in chickens [12–14]. Researchers are more interested in HDPs or their derivatives, because they believe HDPs could fight and protect against infectious diseases [15].
Recent studies have determined that TLRs, AvBDs and cytokines are involved in the response of the immune system in various tissues [16]. But the changes of expression level of AvBDs in chicken erythrocytes after MDV infection has not yet been investigated. The aim of the present study was to explore the transcriptional levels of AvBDs and liver-expressed antimicrobial peptide-2 (LEAP-2) in erythrocytes of chickens infected by MDV.
Results
AvBDs and LEAP-2 genes expression detected in chicken erythrocytes
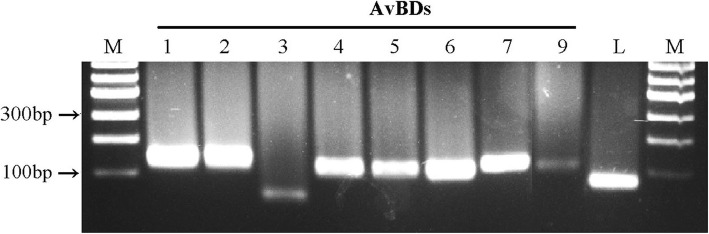
The results of the reverse transcription-polymerase chain reaction (RT-PCR) showed that the genes expression of AvBD1 to AvBD7, AvBD9, and LEAP-2 was detected in the normal commercial chicken erythrocytes, but not for AvBD8, and AvBD10 to AvBD14 (Fig. 1), and no bands were found in the negative controls. AvBD1 to AvBD7, AvBD9 and LEAP-2 genes expression was also detected in specific pathogen-free (SPF) chicken erythrocytes (data not shown). These detected genes expression was further confirmed by Sanger sequencing and Real-time PCR.
Fig. 1.
Expression pattern of AvBDs and LEAP-2 genes in the erythrocytes of 3-week-old roosters. All of the AvBDs and LEAP-2 were detected by RT-PCR. M = size marker, L = LEAP-2
Virus load in the feather tips of MDV-infected chickens
No severe clinical symptom except late phase depression was observed in 14 d experimental period. The copy numbers of MDV were tested at 4, 7, 9 and 14 days post-infection (d.p.i.) by duplex real-time PCR (Fig. 2). The results showed that the MDV was detected after 7 d.p.i., and the MDV copy number at 14 d.p.i. was significantly higher than that of all other time points. MDV nucleic acid was not detected in the control group.
Fig. 2.
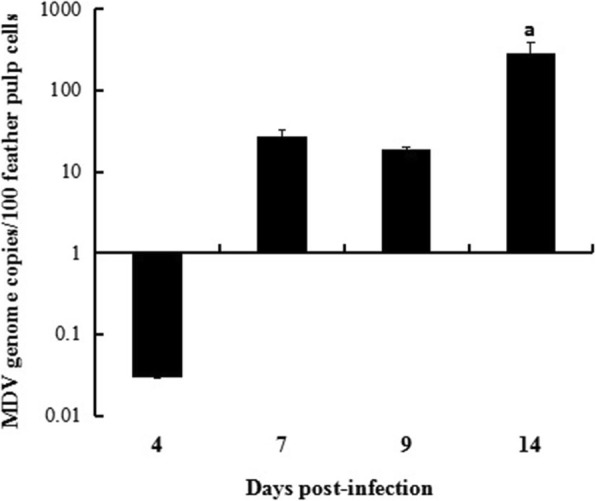
MDV load in feather tips on 4, 7, 9 and 14 d.p.i. (n = 3). Chickens were infected with MDV. Feather tips were obtained on 4, 7, 9 and 14 d.p.i., and the virus loads were measured by duplex real-time PCR. The error bars represent standard error of the mean value. The data of virus load was subjected to one-way ANOVA and then analyzed by the Tukey’s pairwise comparison to identify treatment differences, a = significant when compared to MDV-infected chickens sampled on 4, 7 and 9 d.p.i. (P < 0.05)
Detection of MDV-related genes in erythrocytes
To determine whether MDV invaded chicken erythrocytes which could activate AvBDs expression against the infection, we performed a search for virus genomes in the transcriptome of erythrocytes on 14 d.p.i. (SRA accession No: SRP136403) when the MDV load was significantly higher than that of all other time points. The results showed that the transcripts of 35 MD5-related genes were detected (Table 1). These genes were mainly phosphoprotein pp24, major capsid protein, assembly protein, major DNA-binding protein, deneddylase, DNA polymerase processivity factor, tegument protein VP16, and deoxyuridine triphosphatase, in which the copy number of phosphoprotein pp24 that act as an integral component of membrane was the most abundant. These results indicated that erythrocytes had been invaded by MDV.
Table 1.
The identities of 35 MD5-related gene transcripts detected in chicken erythrocytes after MDV infection
| ID | Length(nt) | Count (Reads numbers) | Swissprot_annotation (strain Chicken/Md5/ATCC VR-987) | nr_annotation [Gallid herpesvirus 2] |
|---|---|---|---|---|
| MDV008 | 2608 | 17 | Phosphoprotein pp24 | protein pp24 |
| MDV018 | 2169 | 1 | Portal protein UL6 | capsid portal protein |
| MDV021 | 2526 | 1 | Replication origin-binding protein | DNA replication origin-binding helicase |
| MDV023 | 3786 | 5 | Tegument protein UL11 | myristylated tegument protein |
| MDV030 | 960 | 1 | Triplex capsid protein VP23 | capsid triplex subunit 2 |
| MDV031 | 4182 | 8 | Major capsid protein | HSV-1 UL19-like protein |
| MDV034 | 2442 | 1 | Envelope glycoprotein H | envelope glycoprotein H |
| MDV038 | 1992 | 6 | Assembly protein | capsid maturation protease |
| MDV040 | 2598 | 1 | Envelope glycoprotein B | envelope glycoprotein B |
| MDV042 | 3576 | 6 | Major DNA-binding protein | single-stranded DNA-binding protein |
| MDV043 | 3663 | 3 | DNA polymerase catalytic subunit | DNA polymerase catalytic subunit |
| MDV044 | 903 | 1 | Virion egress protein UL31 | nuclear egress lamina protein |
| MDV046 | 1926 | 1 | Packaging protein UL32 | DNA packaging protein UL32 |
| MDV047 | 834 | 1 | Virion egress protein UL34 | nuclear egress membrane protein |
| MDV049 | 10,029 | 7 | Deneddylase | large tegument protein |
| MDV050 | 3141 | 1 | Capsid assembly protein UL37 | tegument protein UL37 |
| MDV051 | 1413 | 1 | Triplex capsid protein | capsid triplex subunit 1 |
| MDV052 | 2469 | 2 | Ribonucleoside-diphosphate reductase large subunit | ribonucleotide reductase subunit 1 |
| MDV053 | 1032 | 1 | Ribonucleoside-diphosphate reductase small chain | ribonucleotide reductase subunit 2 |
| MDV055 | 1506 | 6 | DNA polymerase processivity factor | DNA polymerase processivity subunit |
| MDV059 | 1707 | 4 | Tegument protein UL46 | tegument protein VP11/12 |
| MDV060 | 2427 | 3 | Tegument protein UL47 | tegument protein VP13/14 |
| MDV061 | 1284 | 6 | Tegument protein VP16 | transactivating tegument protein VP16 |
| MDV062 | 750 | 1 | Tegument protein VP22 | tegument protein VP22 |
| MDV063 | 1311 | 9 | Deoxyuridine 5' −triphosphate nucleotidohydrolase | deoxyuridine triphosphatase |
| MDV065 | 750 | 1 | Tegument protein UL51 | tegument protein UL51 |
| MDV068 | 1422 | 3 | mRNA export factor ICP27 | multifunctional expression regulator |
| MDV069 | 810 | 1 | Uncharacterized gene 69 protein | protein LORF4 |
| MDV070 | 501 | 1 | Tegument protein UL55 | nuclear protein UL55 |
| MDV071 | 585 | 1 | Uncharacterized gene 71 protein | myristylated tegument protein CIRC |
| MDV072 | 2712 | 5 | Uncharacterized gene 72 protein | protein LORF5 |
| MDV072.5 | 873 | 3 | – | membrane protein UL56 |
| MDV088 | 540 | 3 | Transcriptional regulator ICP22 | regulatory protein ICP22 |
| MDV089 | 642 | 2 | Virion protein US10 | virion protein US10 |
| MDV094 | 4716 | 3 | Envelope glycoprotein D | Envelope glycoprotein D |
Quantitative PCR analysis for AvBDs and LEAP-2 expression in the chicken erythrocytes in response to MDV stimulation
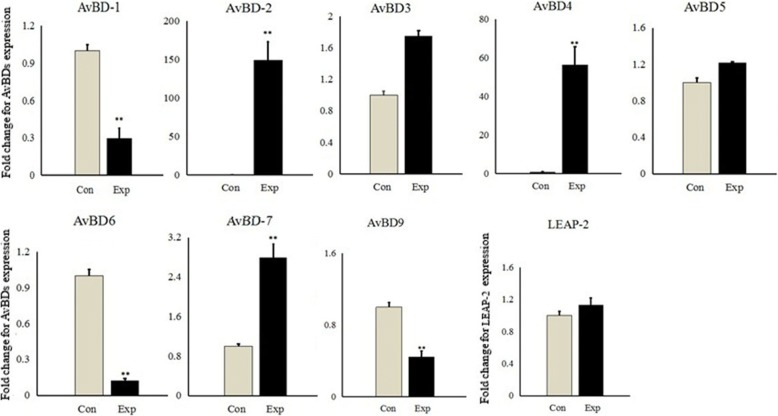
To further investigate the functions of these genes during MDV infections, their expression levels in the erythrocytes were analyzed. We utilized real-time RT-PCR to quantify the repertoire of AvBDs expressed in chicken erythrocytes in response to MDV on 14 d.p.i.(Fig. 3). The expression levels of AvBD2, 4 and 7 were significantly upregulated compared to the control group (P < 0.01). However, the gene transcript levels of AvBD1, 6 and 9 in the experimental group were significantly lower than those in the control group (P < 0.01). There was no significant difference for AvBD3, AvBD5 and LEAP-2 between the two groups.
Fig. 3.
The expression pattern of mRNAs of AvBDs in chicken erythrocytes on 14 d.p.i. The experimental groups of this study were as follows: the control group (Con, PBS-injection, n = 3) and the infection group (Exp, MDV-injection, n = 3). Expression levels of AvBDs were calculated relative to that of the housekeeping gene 18S rRNA by using quantitative real-time PCR. The data represent mean fold changes from PBS-treated controls ± standard error. Statistical significance between the infection group and the control group was analyzed using t-test. *P < 0.05, **P < 0.01
Discussion
Recent studies have reported that erythrocytes play an important role in immune responses. It has been found that trout and chicken erythrocytes can detect and specifically respond to different pathogen-associated molecular patterns (PAMPs) [5]. In the present study, it was observed that AvBDs and LEAP-2 transcripts are expressed not only in tissues but also in erythrocytes. This result provides a new perspective for poultry health, as the expression of these transcripts may be linked to the protection of chickens from pathogens. Primary literatures showed that AvBDs are not only largely associated to reproductive organs but also to bone marrow [17–19], and more generally to mucosal tissues. The transcripts for 13 types of AvBDs were expressed in chicken reproductive organs [20]. It has been reported that AvBD1, 2 and AvBD4 to AvBD7 were highly expressed in bone marrow of chicken [18, 19, 21]. Strong expression of AvBDs transcripts was found throughout the digestive tract such as in the esophagus, craw, proventriculus, and intestinal tissues, except for AvBD11 [16, 22, 23]. This is similar to our results that AvBD1 to AvBD7 and AvBD9 transcripts were detected in chicken erythrocytes. Moreover, many AvBDs also were found and regulated by both pathogenic and attenuated infectious bronchitis virus (IBV) strains in many highly vascularized tissues, such as liver, spleen and kidney [24, 25]. The expression of AvBDs was also observed in other tissues such as the respiratory tract and skin [12, 17, 21]. As the most abundant circulating cells, erythrocytes are distributed in all of these tissues. All of the above support the hypothesis that erythrocytes can participate in the immune response in various tissues, especially highly vascularized tissues, by releasing AvBDs to fight pathogens.
The results from current study indicated that MDV genome load in the feather tips was significantly increased at 14 d.p.i.. Similar results were reported by Abdul-Careem et al [26]. This could be because some latent MDV was reactivated and started the second cytolytic phase by 14 d.p.i. [6]. The transcripts of 35 MD5-related genes in erythrocytes indicated that MDV invaded the membrane or intracellular of erythrocytes. The explanation for the results could be 1) the viral PAMPs were recognized by the pattern recognition receptors (PRRs) on the surface of erythrocytes; 2) MDV invaded and replicated in the erythrocytes; 3) there was an interaction between MDV and erythrocytes. However, the molecular mechanism needs to be further elucidated.
TLRs are associated with immune cells and other tissue cells [17]. It was recently found that chicken erythrocytes constitutively express transcripts for many TLRs as well as for some cytokines and other immune-related genes such as IFN-α, IFN-β, and IL-8 [1]. AvBDs can be induced by the interaction of TLRs with their ligands and proinflammatory cytokines [27, 28]. The results of the present study show that it is possible that erythrocytes can participate in some of these aspects. Furthermore, many MDV-related genes and differentially expressed AvBD2 were detected in the transcriptome of erythrocytes, suggesting the expression of AvBDs in erythrocytes may participate in the antiviral response against MDV infection. The significant upregulation of AvBD2, 4 and 7 was found after MDV infection, which may result from the activation of broader innate immune signaling pathways after PRRs-viral PAMPs interaction, and this still needs further researches to confirm. We found that the upregulated expression of AvBD2 is the most significant result obtained after MDV infection. Similarly, some studies found that AvBD2 was also upregulated by the infections of pathogenic IBV and pigeon paramyxovirus type 1 (PPMV-1) in most tissues [24]. Furthermore, it has been reported that AvBD2 is the major defensin in heterophiles [27, 29]. We speculate that AvBD2 may be expressed as functional proteins. In duck tissues, significant antiviral activities of three AvBDs (Apl_AvBD4, 7 and 12) were shown against duck hepatitis virus (DHV) [30]. In another study, the recombinant AvBD2, 6 and 12 displayed the obvious anti-viral activity against IBV in vitro [24]. These suggested that AvBD2, 4, 6 and 7 may have strong antiviral activities in chicken erythrocytes. Moreover, we also found that the transcripts of AvBD1, 6 and 9 were significantly downregulated after MDV infection; this may be associated to the viral immunomodulators that fight against these AvBDs after MDV infection. The LEAP-2 expression in erythrocytes during MDV infection has also been found; thus, we speculate that LEAP-2 might also be an effector of innate immune signaling as a response to some pathogens [31].
The modulation of AvBDs in response to the bacterial components or ligands for TLRs in chickens have already been studied [1, 20, 23], but the reaction of AvBDs in response to MDV in chicken erythrocytes has not yet been investigated. Based on the expressed repertoire of AvBDs, it is possible that erythrocytes can respond both virus and bacteria, which is supported by the fact that AvBD2 which is highly overexpressed in response to MDV possess a strong antibacterial activity [19]. This finding is also supported by another fact that the transcript of type I IFNs are also expressed in chicken erythrocytes and are upregulated after the treatment with poly I:C or CpG ODN [1], such that a second wave of signaling pathways would be activated to synthesize IFN-regulated proteins contributing to the antiviral response and inhibiting virus replication [32]. Another study showed the expression of several AvBDs was closely correlated to the expression of TLR3, TLR7 and TLR15 in tissues [33]. All the above clues supported the possible defensive mechanism of chicken erythrocytes that the PAMPs could be recognized by various PRRs, such as TLRs and scavenger receptor (SR). Cytokines, AvBDs and other molecules will be activated and modulate host immune response when pathogens invade into the blood.
Conclusions
In conclusion, the transcripts of eight AvBDs and LEAP-2 were identified in chicken erythrocytes. The transcripts of 35 MD5-related genes were found in erythrocytes after MDV infection. MDV induced an immune response wherein AvBD2, 4 and 7 were significantly upregulated and AvBD1, 6 and 9 were significantly downregulated. Further studies are needed on the mechanism of AvBDs in the response of chicken erythrocytes to MDV infections, which could help to prevent and control MDV in the future.
Methods
Experiment 1. Detection of AvBDs and LEAP-2 genes expression in chicken erythrocytes
Experimental animals
Three 3-week-old roosters (Hy-Line Brown) were purchased from a commercial supplier in Taigu County. The roosters were kept in cages, under an artificial light regimen of 14 h day-time/10 h night-time. A diet appropriate for their age was provided ad libitum. Before exsanguination and necropsy, injection of pentobarbital was used along with the standard protocols of euthanasia to minimize animals suffering.
Erythrocyte collection
Venous blood was drawn from these chickens (2 mL per bird) and mixed with the same volume of Alsever’s solution (Solarbio, Beijing, China). The diluted blood was added carefully into a 4 mL Histopaque-1119 solution (Sigma–Aldrich, Oakville, ON) and centrifuged at 500 g for 20 min; subsequently, the supernatant containing platelets and leukocytes-was removed. Subsequent procedures were performed as described earlier [34]. Isolated erythrocytes were determined to be > 99.9% pure by Wright–Giemsa staining.
Extraction of total RNA from chicken erythrocytes, and cDNA synthesis
Total RNA was purified from erythrocytes isolated from blood samples using RNAiso Plus (Takara Bio Inc., Dalian, China) according to the manufacturer instructions, and dissolved in 20 μL RNase-free water. Purified RNAs were examined by 1.5% agarose gel electrophoresis and a NanoDrop Bioanalyzer ND1000 (Labtech, Uckfield, UK). The criteria for assessing the quality of RNA were that 28S and 18S rRNA could be seen in the gel and the A260/A280 ratio was between 1.9–2.1. cDNA was synthesized from 500 ng total RNAs using the PrimeScript RT reagent Kit with gDNA Eraser (Takara Bio Inc., Dalian, China) according to manufacturer instructions. The cDNA samples were diluted at 1:10 in sterile water and stored at − 20 °C.
Primer design and PCR
Primers were designed by Primer Express 3.0 (Applied Biosystems, Foster City, CA, USA) and synthesized by Shanghai Generay Biotech Co., Ltd. (Shanghai, China). GenBank accession numbers of the reference sequences used for primer design are presented in Table 2. The PCR was conducted in a volume of 20 μL of reaction mixture containing 2 μL of cDNA, 0.4 μL at 10 pmol/μL of each of forward and reverse primers, 10 μL 2 × MasterMix (Real-Times Biotechnology Co., Ltd., Beijing, China) and 7.2 μL double-distilled H2O. The cDNA was replaced by water as negative control for each gene. Amplification for AvBDs or LEAP-2 was performed in a Gradient Thermocycler (Eppendorf AG, Germany). The optimum thermal cycling procedure was as follows: 95 °C for 3 min; 35 cycles of 95 °C for 30 s, 55 to 57 °C (the optimal temperature of AvBDs, listed in Table 2) for 30 s, and 72 °C for 1 min, followed by 72 °C for 5 min. The PCR results were detected by electrophoresis in 1.5% TAE agarose gels, and imaged by GelDoc-It Imaging Systems (Ultra-Violet Products, USA).
Table 2.
Primer sequences and reference genes used for PCR
| Target Genes | Primer sequence(5′ → 3′) | Annealing temperature | Sizes(bp) | GenBank Accession number of the reference gene |
|---|---|---|---|---|
| AvBD1 | F: GGCTCTAGGAAGGAAGTCA R: GCAGCAGAGGTAAAATCTTG |
57 °C | 112 | NM_204993.1 |
| AvBD2 | F: GCACTCCAGGTTTCTCCA R: GGCGTCCGACTTTGATTA |
57 °C | 110 | DQ677633.1 |
| AvBD3 | F: AGAGGAGGATTCTGTCGTGTT R: CAACCTCATATGCTCTTCCA |
55 °C | 100 | NM_204650.2 |
| AvBD4 | F: TCATGGAGCTGTGGGCTTTT R: AGCATTCCCATAAGGGCATT |
55 °C | 100 | NM_001001610.2 |
| AvBD5 | F: ATTACCCCAGGACTGTGA R: GCAGCAGAAGTCTTCCTT |
55 °C | 100 | NM_001001608.2 |
| AvBD6 | F: CTGCTGCTGTCTGTCCTCTT R: TGCAGACACCCCTTTGATAT |
55 °C | 100 | NM_001001193.1 |
| AvBD7 | F: CTATTGATACTTGTTGGCTTCG R: AACTCCTCCATCCCCTTG |
57 °C | 122 | AY621322.1 |
| AvBD9 | F: CCAGCTTACAGCCAAGAAGA R: TCCCAATGTCAACTGAAGGA |
55 °C | 100 | NM_001001611.2 |
| LEAP-2 | F: ACTCTGGAATTCTGCCTGATGACA R: CATCTGCATCCGTGCCTGA |
57 °C | 66 | NM_001001606.1 |
| 18S rRNA | F: TTCCGATAACGAACGACAC R: GACATCTAAGGGCATCACAG |
55 °C | 139 | FM165414 |
Experiment 2. AvBDs and LEAP-2 gene expression and virus load in erythrocytes and feather tips of MDV-infected chickens
Virus and animals
Twenty 1-day-old SPF chickens (B15 haplotype), very virulent MDV strain MD5, and recombinant plasmid pMD-O containing the ovotransferrin gene and recombinant plasmid pMD-M containing the MDV Eco Q (meq) gene were provided by Dr. Lingxia Han of Harbin Veterinary Research Institute, Chinese Academy of Agriculture Sciences (Harbin, China). The strain of MDV (MD5) was preserved in liquid nitrogen until use.
Animal infection
The chickens were equally divided into two groups: an experimental group and a control group. The animal experiment was carried out in an Animal Biosafety Level 2 Laboratory. The MDV was diluted properly with sterile phosphate buffered saline (PBS) (Solarbio, Beijing, China). Each chicken in the experimental group was intra-abdominally infected with MDV at a dose of 500 plaque-forming units (PFU)/500 μL. The control group was injected with the same volume of PBS. On 4, 7, 9 and 14 d.p.i., 4–5 feather tips were collected from each bird in the experimental and control groups and preserved in liquid nitrogen immediately. Anticoagulated blood samples were collected at 14 d.p.i. for both groups and used for total RNA extraction as described in Experiment 1. Before exsanguination and necropsy, injection of pentobarbital was used along with the standard protocols of euthanasia to minimize animals suffering. All these tests were performed by Lingxia Han at Harbin Veterinary Research Institute, Chinese Academy of Agriculture Sciences (Harbin, China).
Library preparation and sequencing
RNA was extracted from the erythrocytes. A total of 1 μg purified RNA per sample was used for next-generation sequencing (NGS). Sequencing libraries were generated using NEBNext®Ultra™ RNA Library Prep Kit for Illumina® (NEB, USA) following the manufacturer’s instruction, and index codes were added to attribute sequences to each sample as described previously [35]. The libraries were sequenced by Illumina HiSeq 2500.
Transcriptome analysis
To identify the viral sequence, the clean reads were mapped to the reference genomic sequence by Tophat2 tools software [36], and each unigene was then transformed into FPKM (Fragments Per Kilo base per Million mapped reads) by using RSEM software [37]. Annotations of the unigenes were searched using BLAST against the Nr (non-redundant) and the Swiss-Prot databases (E-value ≤10− 5).
Detection of AvBDs and LEAP-2 mRNA expression in erythrocytes
RNA was extracted from the erythrocytes on 14 d.p.i.. The quantitative expression analysis of AvBDs and LEAP-2 genes were performed with real-time RT-PCR using a TaKaRa SYBR Premix Ex Taq™II (Takara Bio Inc.). The total volume of real-time PCR mixture contained 6 μL 1× SYBR Premix Ex TaqII, 0.1 μL ROX dye II, 1.5 pmol each of forward and reverse primers, 1 μL of diluted cDNA, and 2.6 μL of double distilled H2O. The reaction mixture was placed into MicroAmp® Optical 8-Cap Strip (Applied Biosystems), and real-time PCR was performed following user instructions of the QuantStudio™ 6 (Applied Biosystems). Briefly, each reaction involved a pre-incubation at 95 °C for 3 min, followed by 42 cycles of 95 °C for 30 s, 55 °C - 57 °C (TA as per primer) for 30 s, and extension at 72 °C for 10 s. The expression level of AvBDs and LEAP-2 were calculated relative to that of the housekeeping gene 18S rRNA using the QuantStudio™ 6 Flex Real-Time PCR System Software (Applied Biosystems, USA).
DNA extraction from feather tips
DNA was extracted from feather tips collected at 4, 7, 9 and 14 d.p.i. using Trizol reagent (Invitrogen, Carlsbad, California, USA) as described previously [38]. The purity and concentration of purified DNAs were detected by a NanoDrop Bioanalyzer ND1000 (Labtech). The feather tips DNA was then diluted to 100 ng/μL for future use.
Primers and probes for meq and ovotransferrin genes
Primers and probes of the duplex real-time PCR (Table 3) for MDV meq gene and ovotransferrin gene followed that by Islam et al. [39] and Baigent et al. [40], respectively. Chicken ovotransferrin genomes gene was used as DNA extraction reference.
Table 3.
Sequences of the primers, probes and reference genes used for the duplex real-time PCR assay
| Primer/probe name | Primer/probe sequence(5′ → 3′) | Target gene | GenBank Accession number |
|---|---|---|---|
| meq-F | GGAGCCGGAGAGGCTTTATG | ||
| meq-R | ATCTGGCCCGAATACAAGGAA | MDV Eco Q | M89471 |
| meq-P | (FAM)CGTCTTACCGAGGATCCCGAACAGG(TAMRA) | ||
| ovo-F | CACTGCCACTGGGCTCTGT | ||
| ovo-R | GCAATGGCAATAAACCTCCAA | ovotransfenrin | Y00407 |
| ovo-P | (ROX)AGTCTGGAGAAGTCTGTGCAGCCTCCA(BHQ2) |
FAM: Carboxy fluorescein, TAMRA: Carboxy tetramethyl rhodamine, ROX: Carboxy-X-rhodamine, BHQ2: Black Hole Quencher 2
Quantification of virus loads by real-time PCR
Serial 10-fold dilutions (104–1010 copies/μL) of the recombinant plasmids of pMD-O and pMD-M were used to generate a standard curve for ovotransferrin and meq gene. The duplex real-time PCR was performed in a 25-μL volume. The reaction mixture included 2.5 μL 10 × Ex Taq buffer; 1 μL dNTPs; 0.2 μL HS Ex Taq; 8 pM meq-P and ovo-P probes; 10 pM meq-F, meq-R, ovo-F and ovo-R primers; 1 μL pMD-O and pMD-M. Sterilized H2O was added to bring the final volume to 25 μL. The protocol for the reaction was as follows: 95 °C for 5 min; 40 cycles of 94 °C for 15 s, 59.5 °C for 50 s. The fluorescence intensity of the reporter dye (FAM and ROX) was measured during the 59.5 °C annealing/extension step. The data analysis was performed using an iCycler Thermal Cycler (BioRad, Hercules, CA, USA) generating the standard curve. For the detection of the copy numbers of MDV in feather tips on 4, 7, 9 and 14 d.p.i., the DNAs in feather tips were used as templates for the duplex real-time PCR assay to detect the meq and ovotransferrin. The copy numbers of MDV were calculated using the standard curve.
Statistical analysis
The 2−ΔΔCt method was used to calculate the real-time PCR data. Data represent mean fold change from PBS-treated controls ± standard error. Statistical analyses were performed using SPSS statistical software 17.0 (IBM Company, New York, NY, USA). The data of virus load was subjected to one-way ANOVA and then analyzed by the Tukey’s pairwise comparison to identify treatment differences which were considered significant when P < 0.05. Statistical significance of each gene expression between the experimental group and the control group was analyzed using t-test for independent samples. Differences were considered significant when the P-value was *P < 0.05 or **P < 0.01.
Acknowledgements
The authors are very thankful to Gary Wong (Institute of Microbiology, Chinese Academy of Sciences, Beijing, China) and Tuoying Ao (Alltech, USA) for help with English language quality of the manuscript.
Funding
This work was supported by the National Key R & D Program (2016YFD0500800), Research Project Supported by Shanxi Scholarship Council of China (2017–073), Key scientific and technological innovation platform of Jinzhong (P171002–3), the National Natural Science Foundation of China (31072179), Key Technologies R and D Program of Shanxi Province (20130311027–3) and “131” Leading Talent Project for Colleges and Universities of Shanxi Province.
Availability of data and materials
Most data supporting findings are provided within the text. Original data are available from the corresponding author on reasonable request. All transcriptome sequences have been submitted in Sequence Read Archive under the SRA accession number: SRP136403.
Abbreviations
- AvBDs
Avian β-defensins
- HDPs
Host defense peptides
- IBV
Infectious bronchitis virus
- IFNs
Type I interferons
- IL
Interleukin
- ISAV
Infectious salmon anaemia virus
- LEAP-2
Liver-expressed antimicrobial peptide-2
- MD
Marek’s disease
- MDV
Marek’s disease virus
- meq
MDV Eco Q
- mMDV
Mild MDV
- NGS
Next-generation sequencing
- PAMPs
Pathogen associated molecular patterns
- PBS
Phosphate buffered saline
- PFU
Plaque-forming units
- PPMV-1
Pigeon paramyxovirus type 1
- PRRs
Pattern recognition receptors
- RT-PCR
Reverse transcription polymerase chain reaction
- SPF
Specific pathogen-free
- SR
Scavenger receptor
- TLRs
Toll-like receptors
- VNA
Viral nucleic acid
- vvMDV
Very virulent MDV
Authors’ contributions
SN drafted the basic manuscript and analyzed the data. WXT, LXH, GFG and YHB contributed to conception and design of the research and reviewed the manuscript. ARJ, FJJ, XL, GBN, DZ, HLM and WFH participated in sample collection and laboratory testing. WWG, YJZ, SMG, GLL, JHL, FY and RKG participated in the data analysis and revising the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The protocols for animal studies was approved by the ethical committee of College of Animal Science and Veterinary Medicine of Shanxi Agricultural University, China (Number 88, 2010) and approved by the Committee on the Ethics of Animal Experiments of the Harbin Veterinary Research Institute of the Chinese Academy of Agricultural Sciences (approval no. SYXK (Hei) 2011–022).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Sheng Niu, Email: niusheng93@126.com.
Ali Raza Jahejo, Email: arjahejo@gmail.com.
Fa-jie Jia, Email: 2251651836@qq.com.
Xin Li, Email: 397044807@qq.com.
Guan-bao Ning, Email: ningguanbao034@163.com.
Ding Zhang, Email: zdlucky2014@sina.com.
Hai-li Ma, Email: mahaili1718@126.com.
Wei-fang Hao, Email: 316072115@qq.com.
Wen-wei Gao, Email: sxndgaowenwei@163.com.
Yu-jun Zhao, Email: yujun_zhao@126.com.
Shi-min Gao, Email: smgao@126.com.
Gui-lan Li, Email: lgl.8000@163.com.
Jian-hui Li, Email: jianhui19840717@163.com.
Fang Yan, Email: yanfang6615@163.com.
Rong-kun Gao, Email: gaorongkun@sohu.com.
Yu-hai Bi, Email: yuhai_bi@qq.com.
Ling-xia Han, Email: 11703120@qq.com.
George F. Gao, Email: gaof@im.ac.cn
Wen-xia Tian, Email: wenxiatian@126.com.
References
- 1.Paolucci S, Barjesteh N, Wood RD, Sharif S. Chicken erythrocytes respond to toll-like receptor ligands by up-regulating cytokine transcripts. Res Vet Sci. 2013;95(1):87–91. doi: 10.1016/j.rvsc.2013.01.024. [DOI] [PubMed] [Google Scholar]
- 2.Claver JA, Quaglia AI. Comparative morphology, development, and function of blood cells in nonmammalian vertebrates. J Exot Pet Med. 2009;18(2):87–97. doi: 10.1053/j.jepm.2009.04.006. [DOI] [Google Scholar]
- 3.Passantino L, Massaro MA, Jirillo F, Di Modugno D, Ribaud MR, Di Modugno G, Passantino GF, Jirillo E. Antigenically activated avian erythrocytes release cytokine-like factors: a conserved phylogenetic function discovered in fish. Immunopharmacol Immunotoxicol. 2007;29(1):141–152. doi: 10.1080/08923970701284664. [DOI] [PubMed] [Google Scholar]
- 4.Workenhe ST, Kibenge MJ, Wright GM, Wadowska DW, Groman DB, Kibenge FS. Infectious salmon anaemia virus replication and induction of alpha interferon in Atlantic salmon erythrocytes. Virol J. 2008;5(1):36. doi: 10.1186/1743-422X-5-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morera D, Roher N, Ribas L, Balasch JC, Doñate C, Callol A, Boltaña S, Roberts S, Goetz G, Goetz FW. RNA-Seq reveals an integrated immune response in nucleated erythrocytes. PLoS One. 2011;6(10):e26998. doi: 10.1371/journal.pone.0026998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calnek B. W. Current Topics in Microbiology and Immunology. Berlin, Heidelberg: Springer Berlin Heidelberg; 2001. Pathogenesis of Marek’s Disease Virus Infection; pp. 25–55. [DOI] [PubMed] [Google Scholar]
- 7.Witter R. L. Characteristics of Marek's Disease Viruses Isolated from Vaccinated Commercial Chicken Flocks: Association of Viral Pathotype with Lymphoma Frequency. Avian Diseases. 1983;27(1):113. doi: 10.2307/1590377. [DOI] [PubMed] [Google Scholar]
- 8.Eade CR, Wood MP, Cole AM. Mechanisms and modifications of naturally occurring host defense peptides for anti-HIV microbicide development. Curr HIV Res. 2012;10(1):61–72. doi: 10.2174/157016212799304580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hancock RE, Diamond G. The role of cationic antimicrobial peptides in innate host defences. Trends Microbiol. 2000;8(9):402–410. doi: 10.1016/S0966-842X(00)01823-0. [DOI] [PubMed] [Google Scholar]
- 10.Brogden KA. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol. 2005;3(3):238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 11.Yacoub HA, Elazzazy AM, Abuzinadah OA, Al-Hejin AM, Mahmoud MM, Harakeh SM. Antimicrobial activities of chicken β-defensin (4 and 10) peptides against pathogenic bacteria and fungi. Front Cell Infect Microbiol. 2015;5:36. doi: 10.3389/fcimb.2015.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiao Y, Hughes AL, Ando J, Matsuda Y, Cheng J-F, Skinner-Noble D, Zhang G. A genome-wide screen identifies a single β-defensin gene cluster in the chicken: implications for the origin and evolution of mammalian defensins. BMC Genomics. 2004;5(1):56. doi: 10.1186/1471-2164-5-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Higgs R, Lynn DJ, Gaines S, McMahon J, Tierney J, James T, Lloyd AT, Mulcahy G, O’Farrelly C. The synthetic form of a novel chicken β-defensin identified in silico is predominantly active against intestinal pathogens. Immunogenetics. 2005;57(1–2):90–98. doi: 10.1007/s00251-005-0777-3. [DOI] [PubMed] [Google Scholar]
- 14.Lynn DJ, Higgs R, Lloyd AT, O’Farrelly C, Hervé-Grépinet V, Nys Y, Brinkman FS, Yu PL, Soulier A, Kaiser P, Zhang G. Avian beta-defensin nomenclature: a community proposed update. Immunol Lett. 2007;110(1):86–89. doi: 10.1016/j.imlet.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 15.Cuperus T, Coorens M, van Dijk A, Haagsman HP. Avian host defense peptides. Dev Comp Immunol. 2013;41(3):352–369. doi: 10.1016/j.dci.2013.04.019. [DOI] [PubMed] [Google Scholar]
- 16.van Dijk A, Veldhuizen EJ, Haagsman HP. Avian defensins. Vet Immunol Immunopathol. 2008;124:1):1–1)18. doi: 10.1016/j.vetimm.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoshimura Y. Avian β-defensins expression for the innate immune system in hen reproductive organs. Poult Sci. 2015;94(4):804–809. doi: 10.3382/ps/peu021. [DOI] [PubMed] [Google Scholar]
- 18.Lynn DJ, Higgs R, Gaines S, Tierney J, James T, Lloyd AT, Fares MA, Mulcahy G, O’Farrelly C. Bioinformatic discovery and initial characterisation of nine novel antimicrobial peptide genes in the chicken. Immunogenetics. 2004;56(3):170–177. doi: 10.1007/s00251-004-0675-0. [DOI] [PubMed] [Google Scholar]
- 19.Derache C, Labas V, Aucagne V, Meudal H, Landon C, Delmas AF, Magallon T, Lalmanach AC. Primary structure and antibacterial activity of chicken bone marrow-derived β-defensins. Antimicrob Agents Chemother. 2009;53(11):4647–4655. doi: 10.1128/AAC.00301-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Subedi Kalpana, Isobe Naoki, Nishibori Masahide, Yoshimura Yukinori. Changes in the expression of gallinacins, antimicrobial peptides, in ovarian follicles during follicular growth and in response to lipopolysaccharide in laying hens (Gallus domesticus) Reproduction. 2007;133(1):127–133. doi: 10.1530/REP-06-0083. [DOI] [PubMed] [Google Scholar]
- 21.Zhao C, Nguyen T, Liu L, Sacco RE, Brogden KA, Lehrer RI. Gallinacin-3, an inducible epithelial β-defensin in the chicken. Infect Immun. 2001;69(4):2684–2691. doi: 10.1128/IAI.69.4.2684-2691.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Derache C, Esnault E, Bonsergent C, Le Vern Y, Quéré P, Lalmanach A-C. Differential modulation of β-defensin gene expression by Salmonella Enteritidis in intestinal epithelial cells from resistant and susceptible chicken inbred lines. Dev Comp Immunol. 2009;33(9):959–966. doi: 10.1016/j.dci.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 23.Lu L, Li S, Zhang L, Liu X, Li D, Zhao X, Liu Y. Expression of β-defensins in intestines of chickens injected with vitamin D3 and lipopolysaccharide. Genet Mol Res. 2015;14(2):3330–3337. doi: 10.4238/2015.April.13.12. [DOI] [PubMed] [Google Scholar]
- 24.Xu Y, Zhang T, Xu Q, Han Z, Liang S, Shao Y, Ma D, Liu S. Differential modulation of avian β-defensin and toll-like receptor expression in chickens infected with infectious bronchitis virus. Appl Microbiol Biotechnol. 2015;99(21):9011–9024. doi: 10.1007/s00253-015-6786-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li S, Ouyang L, Zhou D. Effects of vitamin D3 on expression of defensins, toll-like receptors, and vitamin D receptor in liver, kidney, and spleen of silky fowl. Czech J Anim Sci. 2013;58:1):1–1):7. [Google Scholar]
- 26.Abdul-Careem MF, Hunter BD, Lee LF, Fairbrother JH, Haghighi HR, Read L, Parvizi P, Heidari M, Sharif S. Host responses in the bursa of Fabricius of chickens infected with virulent Marek's disease virus. Virology. 2008;379(2):256–265. doi: 10.1016/j.virol.2008.06.027. [DOI] [PubMed] [Google Scholar]
- 27.Kannan L, Liyanage R, Lay JO, Rath NC. Evaluation of beta defensin 2 production by chicken heterophils using direct MALDI mass spectrometry. Mol Immunol. 2009;46(15):3151–3156. doi: 10.1016/j.molimm.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 28.Abdel-Mageed AM, Isobe N, Yoshimura Y. Effects of different TLR ligands on the expression of proinflammatory cytokines and avian β-defensins in the uterine and vaginal tissues of laying hens. Vet Immunol Immunopathol. 2014;162(3):132–141. doi: 10.1016/j.vetimm.2014.10.013. [DOI] [PubMed] [Google Scholar]
- 29.Kannan L, Rath N, Liyanage R, Lay J. Direct screening identifies mature β-defensin 2 in avian heterophils. Poult Sci. 2009;88(2):372–379. doi: 10.3382/ps.2008-00366. [DOI] [PubMed] [Google Scholar]
- 30.Ma D, Lin L, Zhang K, Han Z, Shao Y, Liu X, Liu S. Three novel Anas platyrhynchos avian β-defensins, upregulated by duck hepatitis virus, with antibacterial and antiviral activities. Mol Immunol. 2011;49(1):84–96. doi: 10.1016/j.molimm.2011.07.019. [DOI] [PubMed] [Google Scholar]
- 31.Michailidis G. Expression of chicken LEAP-2 in the reproductive organs and embryos and in response to Salmonella enterica infection. Vet Res Commun. 2010;34(5):459–471. doi: 10.1007/s11259-010-9420-3. [DOI] [PubMed] [Google Scholar]
- 32.Samuel CE. Antiviral actions of interferons. Clin Microbiol Rev. 2001;14(4):778–809. doi: 10.1128/CMR.14.4.778-809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y, Xu Q, Zhang T, Gao M, Wang Q, Han Z, Shao Y, Ma D, Liu S. Host avian Beta-Defensin and toll-like receptor responses of pigeons following infection with pigeon paramyxovirus type 1. Appl Environ Microbiol. 2015;81(18):6415–6424. doi: 10.1128/AEM.01413-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kabanova S, Kleinbongard P, Volkmer J, Andrée B, Kelm M, Jax TW. Gene expression analysis of human red blood cells. Int J Med Sci. 2009;6(4):156. doi: 10.7150/ijms.6.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shao ES, Lin GF, Liu S, Ma XL, Chen MF, Lin L, Wu SQ, Sha L, Liu ZX, Hu XH, Guan X. Identification of transcripts involved in digestion, detoxification and immune response from transcriptome of Empoasca vitis (Hemiptera: Cicadellidae) nymphs. Genomics. 2017;109(1):58–66. doi: 10.1016/j.ygeno.2016.11.006. [DOI] [PubMed] [Google Scholar]
- 36.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC bioinformatics. 2011;12(1):323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abdul-Careem MF, Hunter BD, Sarson AJ, Mayameei A, Zhou H, Sharif S. Marek’s disease virus–induced transient paralysis is associated with cytokine gene expression in the nervous system. Viral Immunol. 2006;19(2):167–176. doi: 10.1089/vim.2006.19.167. [DOI] [PubMed] [Google Scholar]
- 39.Islam A, Harrison B, Cheetham BF, Mahony TJ, Young PL, Walkden-Brown SW. Differential amplification and quantitation of Marek’s disease viruses using real-time polymerase chain reaction. J Virol Methods. 2004;119(2):103–113. doi: 10.1016/j.jviromet.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 40.Baigent SJ, Petherbridge LJ, Howes K, Smith LP, Currie RJ, Nair VK. Absolute quantitation of Marek's disease virus genome copy number in chicken feather and lymphocyte samples using real-time PCR. J Virol Methods. 2005;123(1):53–64. doi: 10.1016/j.jviromet.2004.08.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Most data supporting findings are provided within the text. Original data are available from the corresponding author on reasonable request. All transcriptome sequences have been submitted in Sequence Read Archive under the SRA accession number: SRP136403.