Abstract
Cargo-selective and nonselective autophagy pathways employ a common core autophagy machinery that directs biogenesis of an autophagosome that eventually fuses with the lysosome to mediate turnover of macromolecules. In yeast (Saccharomyces cerevisiae) cells, several selective autophagy pathways fail in cells lacking the dimeric Snx4/Atg24 and Atg20/Snx42 sorting nexins containing a BAR domain (SNX-BARs), which function as coat proteins of endosome-derived retrograde transport carriers. It is unclear whether endosomal sorting by Snx4 proteins contributes to autophagy. Cells lacking Snx4 display a deficiency in starvation induced, nonselective autophagy that is severely exacerbated by ablation of mitochondrial phosphatidylethanolamine synthesis. Under these conditions, phosphatidylserine accumulates in the membranes of the endosome and vacuole, autophagy intermediates accumulate within the cytoplasm, and homotypic vacuole fusion is impaired. The Snx4-Atg20 dimer displays preference for binding and remodeling of phosphatidylserine-containing membrane in vitro, suggesting that Snx4-Atg20-coated carriers export phosphatidylserine-rich membrane from the endosome. Autophagy and vacuole fusion are restored by increasing phosphatidylethanolamine biosynthesis via alternative pathways, indicating that retrograde sorting by the Snx4 family sorting nexins maintains glycerophospholipid homeostasis required for autophagy and fusion competence of the vacuole membrane.
INTRODUCTION
Cellular health and homeostasis requires autophagic processes that break down macromolecules to provide nutrients, defend the cell against pathogens, and promote clearing of toxic cellular debris (Levine and Kroemer, 2008; Kaur and Debnath, 2015; Farré and Subramani, 2016). In starvation-induced autophagy, cytoplasmic constituents are nonselectively captured and delivered to the lysosome-like vacuole where they are degraded to provide nutrients for the cell that promote survival. The autophagy machinery is also utilized by the cell during nutrient-replete conditions to specifically degrade certain organelles, protein aggregates, and other constituents in a process termed “selective” autophagy. In addition to the core autophagy machinery, selective autophagy also requires additional selectivity factors, which are specific to different autophagy cargos (Farré and Subramani, 2016).
In budding yeast (Saccharomyces cerevisiae) more than 30 autophagy-related proteins have been identified and the core steps of the autophagy pathway have been established. Initiation of autophagy occurs at the preautophagosomal structure (PAS), followed by expansion and sealing of the phagophore membrane, and finally fusion of the autophagosome with the lysosome (Lynch-Day and Klionsky, 2010). These key steps and the autophagy-related proteins that mediate and regulate them are evolutionarily conserved and shared across all autophagy pathways, including starvation-induced bulk autophagy and cargo-selective autophagy pathways (Kaur and Debnath, 2015).
Two related yeast proteins, Snx4/Atg24 and Atg20/Snx42, have been shown to play crucial roles in several selective autophagy pathways, including the cytoplasm-to-vacuole targeting (CVT) pathway, pexophagy, mitophagy, and starvation-induced selective autophagy of fatty acid synthase and of the proteasome (Nice et al., 2002; Kanki et al., 2009; Okamoto et al., 2009; Shpilka et al., 2015; Nemec et al., 2017). Interestingly, Atg20 has been shown to interact with Atg11, a scaffold protein required for the CVT pathway and for selective autophagy of organelles, including mitochondria, peroxisomes, nuclear envelope, and endoplasmic reticulum (Yorimitsu and Klionsky, 2005; Farré and Subramani, 2016). Snx4 and Atg20 are peripheral membrane proteins that dimerize via their Bin-Amphiphysin-Rvs (BAR) domains to form a composite membrane binding surface (van Weering et al., 2012; Ma et al., 2017; Popelka et al., 2017). A third Snx4-related protein, Snx41, also dimerizes with Snx4, but it has not been implicated in autophagy, although it is required for proper trafficking of Atg27, a fungal integral membrane protein of unknown function that promotes general and selective autophagy in yeast (Yen et al., 2007; Bean et al., 2017; Ma et al., 2017). All three Snx4-related proteins also possess a phosphatidylinositol 3-phosphate–binding PHOX homology (PX) domain that is required for organelle targeting in vivo (Nice et al., 2002; Ma et al., 2017). Snx4-Atg20 localizes to the endosome and the PAS (Nice et al., 2002; Reggiori and Klionsky, 2013; Ma et al., 2017) and has been shown to associate with the autophagy-initiation complex via binding to Atg17 (Ito et al., 2001; Nice et al., 2002; Vollert and Uetz, 2004), suggesting that they play roles in early events in autophagosome biogenesis. Interestingly, however, Atg17 is not required for the CVT pathway (Nice et al., 2002) but rather acts as a scaffold protein for initiation of general autophagy (Kabeya et al., 2005; Suzuki et al., 2007), raising the possibility that Snx4-Atg20 may have further uncharacterized roles in bulk autophagy that are distinct from its role(s) in selective autophagy.
The best characterized functions of Snx4-Atg20 are as sorting factors that mediate export of particular integral membrane proteins and lipids from the endosome and vacuole via retrograde trafficking pathways (Hettema et al., 2003; Bean et al., 2017; Ma et al., 2017; Suzuki and Emr, 2018). One Snx4-Atg20 pathway cargo is Snc1, a v-SNARE that is trafficked from the endosome to the Golgi, but Snc1 has no known role in autophagy. Accordingly, it has been proposed that Snx4’s role in autophagy is to mediate trafficking of the integral membrane protein Atg9 from the endosome to the Golgi, from where it is packaged into a vesicle that may associate with the PAS (Mari et al., 2010; Ohashi and Munro, 2010; Yamamoto et al., 2012). However, a role for Snx4-Atg20 in retrograde trafficking of Atg9 is revealed only when the retromer pathway is eliminated (Shirahama-Noda et al., 2013), indicating that this cannot fully account for the roles of Snx4-Atg20 in autophagy. Thus, the role(s) of the Snx4-Atg20 complex in autophagy, and whether or how sorting at the endosome by Snx4-Atg20 contributes to autophagy, are unknown.
In this study, we establish that snx4Δ cells have a previously unappreciated deficiency in starvation-induced bulk autophagy that is greatly exacerbated by reducing phosphatidylethanolamine (PE) within the cell. Under these conditions, autophagy intermediates accumulate within the cytoplasm as a result of deficient autophagosome maturation and fusion with the vacuole. In addition, phosphatidylserine accumulates on organelles of the endovacuolar system, and both deficiencies are mitigated by increasing phosphatidylethanolamine synthesis via alternative PE biosynthetic pathways. These results bring to light the role of the Snx4 family of sorting nexins in controlling local pools of glycerophospholipids needed to maintain fusion competence of endovacuolar organelles.
RESULTS AND DISCUSSION
Snx4-Atg20 contributes to nonselective autophagy
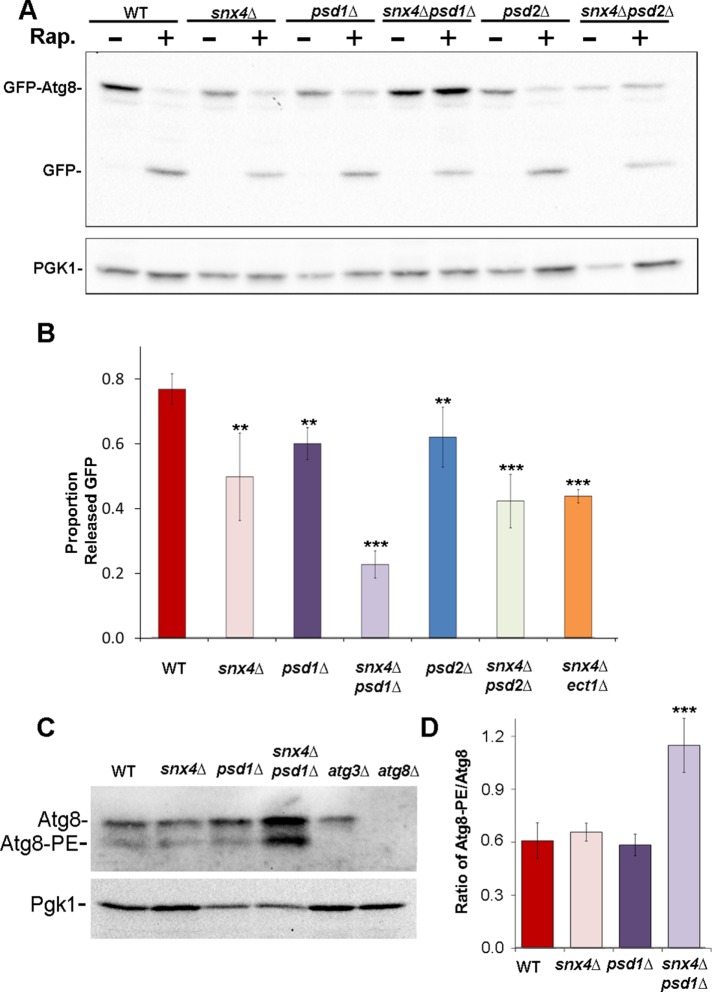
Previously, it was shown that vacuole-mediated processing of Ape1, a cargo of the CVT selective autophagy pathway, but not a marker of bulk autophagy (Pho8Δ60), is ablated in snx4Δ or atg20Δ cells, prompting the conclusion that Snx4-Atg20 functions solely in the CVT pathway (Nice et al., 2002). In the course of our analysis of autophagy in snx4Δ cells, we observed using a green fluorescent protein (GFP)-Atg8 processing assay (Klionsky et al., 2016) that proteolytic processing of GFP-Atg8 in the vacuole of snx4Δ cells is reduced by ∼30% compared with wild-type cells, 4 h after inducing autophagy by addition of rapamycin to the culture medium (Figure 1, A and B), a result that is corroborated by a recent study (Popelka et al., 2017). The results indicate that Snx4 plays a role in the starvation-induced autophagy pathway that was not previously appreciated.
FIGURE 1:
snx4Δpsd1Δ cells display a synthetic autophagy defect. (A) Representative immunoblot analysis of GFP-Atg8 processing in cells incubated with rapamycin (RAP) for 4 h to induce autophagy. Anti-GFP was used to detect GFP-Atg8 and the released GFP proteolytic fragment. Note the large increase in the proportion of full-length GFP-Atg8 to free GFP in snx4Δpsd1Δ cells. Loading control is anti-PGK immunoblot. (B) Quantitation of GFP-Atg8 processing. “Proportion released GFP” is measured as the ratio of free GFP/(free GFP + GFP-Atg8 signal within the same lane). The results from three experiments were averaged and standard error of the mean indicated. The proportion of processed GFP-Atg8 is reduced in snx4Δpsd1Δ cells compared with wild-type or to single mutations. **p < 0.01; ***p < 0.001. (C) Immunoblot analysis of native Atg8. The positions of nonlipidated (Atg8) and lipidated Atg8 (Atg8-PE) are indicated. (D) Quantification of three independent experiments with standard error of the mean is shown in the graph. Anti-PGK immunoblot was used to normalize loads. ***p < 0.001.
Autophagy has a specific requirement for phosphatidylethanolamine (PE), which is reversibly and covalently conjugated to Atg8, a protein required for multiple aspects of autophagy (Ichimura et al., 2000; Reggiori and Klionsky, 2013). Systematic genetic studies of autophagy reported genetic interactions between snx4Δ or atg20Δ deletion alleles with deletion alleles of ATG8 (atg8Δ) (Kramer et al., 2017) and phosphatidylserine decarboxylase 1 (psd1Δ), a mitochondrial enzyme that produces most of the PE in cells grown in standard complete growth medium (Hoppins et al., 2011). We therefore considered the possibility that Snx4-Atg20 may influence the biogenesis and/or trafficking of Atg8. Immunoblotting for Atg8 in wild-type and snx4Δ cell lysates shows no difference in the proportions or amounts of precursor or lipidated Atg8 in snx4Δ cells (Figure 1, C and D), indicating that PE for Atg8 lipidation is not limited by loss of Snx4-Atg20. Because PE is an abundant lipid (∼15% of total lipid of a yeast cell) that is broadly distributed throughout the cell (Zinser et al., 1991; Ejsing et al., 2009), a role for Snx4-Atg20 trafficking of PE to sustain autophagy may be masked by trafficking via other pathways. We therefore assayed GFP-Atg8 processing in snx4Δ cells with reduced amounts of PE. Cells grown in standard complete medium produce nearly all PE by decarboxylation of phosphatidylserine by two enzymes, Psd1 and Psd2, which localize to the inner membrane of the mitochondrion and to Golgi/endosome organelles, respectively (Schuiki et al., 2010). Accordingly, we examined GFP-Atg8 processing in double mutant cells, where snx4Δ was combined with psd1Δ or psd2Δ mutations (Figure 1A). In cells with a single deletion of PSD1, PSD2, or SNX4, there is a modest (∼15–30%) reduction of GFP-Atg8 processing induced by rapamycin; in snx4Δpsd1Δ and snx4Δpsd2Δ double mutant cells, additive decreases in rapamycin-induced processing of GFP-Atg8 are observed (Figure 1, A and B). However, there is a striking accumulation of full-length GFP-Atg8 fusion protein in snx4Δpsd1Δ, but not snx4Δpsd2Δ, cells (Figure 1, A and B). Immunoblotting of endogenous Atg8 in lysates of wild-type and snx4Δpsd1Δ cells confirmed that lipidation of endogenous (i.e., untagged) Atg8 is unaffected by loss of SNX4 and PSD1, and in fact, there is an increase in the amount of lipidated Atg8 in snx4Δpsd1Δ cells (Figure 1, C and D). We conclude that Snx4-Atg20, Psd1, and Psd2 make distinct contributions to autophagy and that their functions do not converge on Atg8 lipidation. Rather, it appears that a step of the autophagy pathway lying downstream of Atg8 lipidation is deficient in snx4Δpsd1Δ cells.
Examination of GFP-Atg8 in snx4Δpsd1Δ cells by fluorescence microscopy reveals a possible basis for Atg8-PE accumulation; there is a sevenfold increase in the number of GFP-Atg8 decorated compartments, presumably autophagy intermediates, in the cytoplasm of snx4Δpsd1Δ cells, compared with wild-type cells. The GFP-Atg8 decorated compartments vary in size and shape, from that of a diffraction limited spot to compartments of up to 0.5 μm in diameter (Figure 2). Many of the larger compartments appear to fully enclose a lumen (Figure 2, inset) while others we cannot discriminate. The accumulation of unprocessed GFP-Atg8 in snx4Δpsd1Δ cells is not affected by addition of rapamycin to the culture medium (Figures 1 and 2), consistent with a role for SNX4 and PSD1 in starvation-induced autophagy.
FIGURE 2:
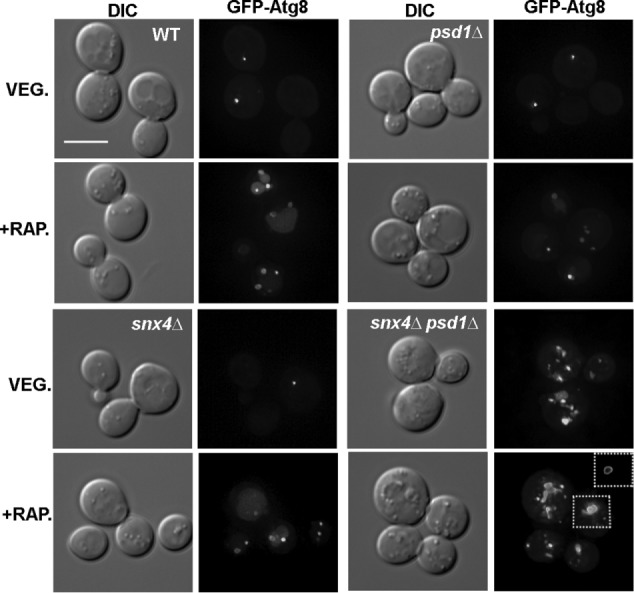
snx4Δpsd1Δ cells accumulate autophagosomes independent of autophagy induction. Maximum projection micrographs showing cells expressing GFP-Atg8 after (4 h) induction of autophagy with rapamycin (+RAP) are shown. The inset of snx4Δpsd1Δ cells show a medial Z slice to visualize the lumen of autophagosomes. The sale bar indicates 5 μm.
The results indicate Snx4 is required to prevent accumulation of autophagy intermediates in the cytoplasm of psd1Δ cells. Snx4 forms functionally distinct dimers with Atg20 and Snx41 (Nice et al., 2002; Ma et al., 2017; Popelka et al., 2017), so we examined GFP-Atg8 in psd1Δ cells also deleted for ATG20, SNX41, or both genes (Figure 3). Surprisingly, GFP-Atg8 appears as a single punctum in atg20Δpsd1Δ and snx41Δpsd1Δ cells, just as in wild-type cells. However, GFP-Atg8 decorated compartments accumulate in atg20Δsnx41Δpsd1Δ triple mutant cells, indicating that the Snx4-Atg20 and Snx4-Snx41 dimers are redundant with respect to this phenotype. Importantly, genetic interactions between psd1Δ and null alleles of SNX4 family genes reflects a specific requirement for Snx4 family proteins, as deletion of VPS5 (vps5Δpsd1Δ), encoding a retromer SNX-BAR, does not lead to the accumulation of autophagy intermediates (Figure 3).
FIGURE 3:
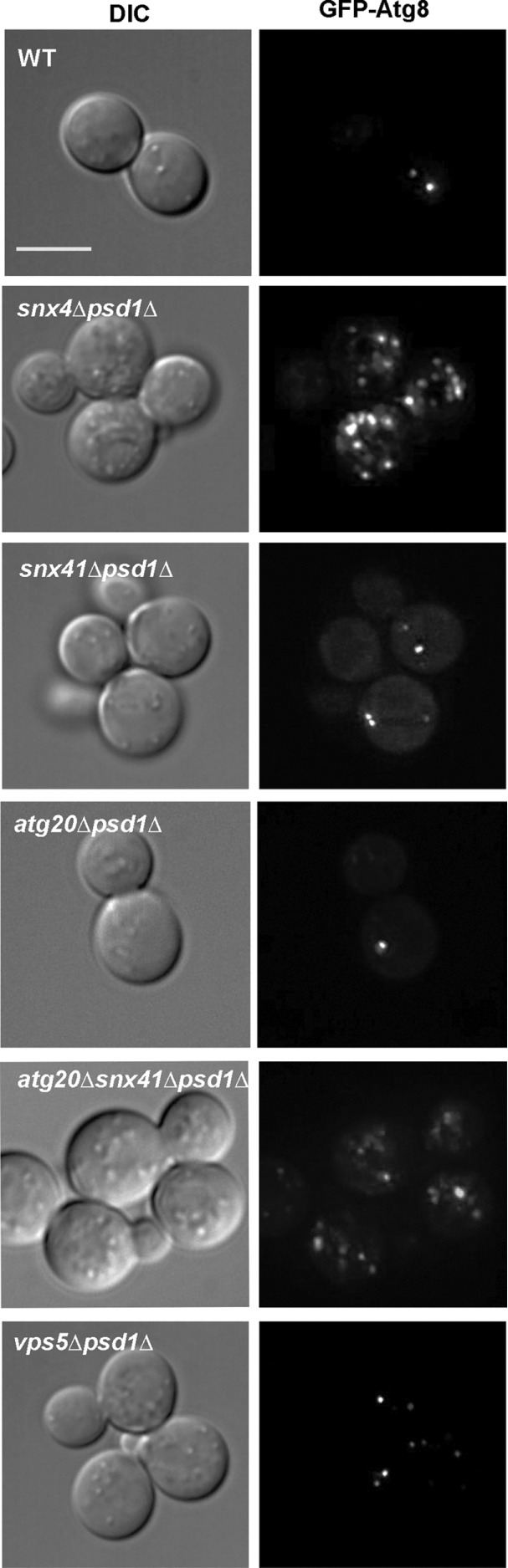
Autophagosome accumulation defect is specific to the Snx4 family SNX-BAR proteins. Maximum projection micrographs showing wild-type, snx4Δpsd1Δ, snx41Δpsd1Δ, atg20Δpsd1Δ, atg20Δsnx41Δpsd1Δ, and vps5Δpsd1Δ cells expressing GFP-Atg8 grown in standard growth medium. The sale bar indicates 5 μm.
Perturbed aminophospholipid distribution in snx4Δpsd1Δ cells results in autophagosome accumulation
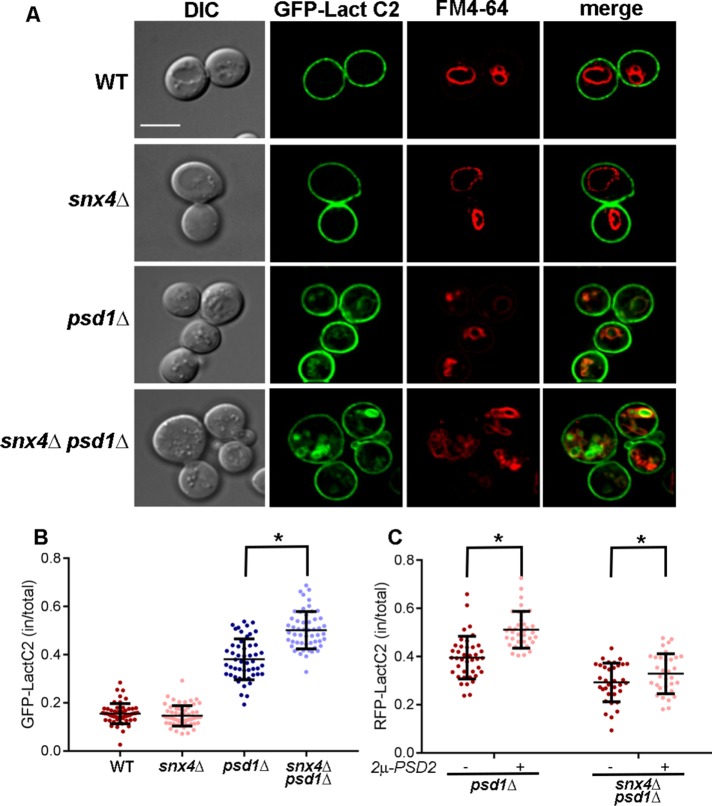
The results reveal an unexpected link between phospholipid metabolism, autophagy, and interorganelle trafficking mediated by Snx4 family proteins. Loss of PSD1 results in an ∼50% decrease in the amount of PE, and an ∼20% increase in the amount of PS, in the cell (Trotter et al., 1993; Birner et al., 2001; Gulshan et al., 2010). In light of the accumulation of autophagy intermediates and fragmented vacuoles in snx4Δpsd1Δ cells, we speculate that changes in the lipid composition of the vacuole membrane and/or the autophagosome outer membrane reduces the fusion competence of these membranes, resulting in either deficient sealing of the autophagosome membrane and/or fusion of the autophagosome with the vacuole. To examine the distribution of lipids affected by deletion of PSD1 and SNX4, we first examined the intracellular distribution of PS, the substrate of phosphatidylserine decarboxylase, using the GFP-tagged C2 domain of lactadherin (GFP-LactC2), which specifically recognizes PS (Yeung et al., 2008; Leventis and Grinstein, 2010). Whereas GFP-LactC2 is largely restricted to the cytoplasmic leaflet of the plasma membrane in wild-type (Yeung et al., 2008; Fairn et al., 2011a) and snx4Δ cells, internal cytoplasmic organelles are decorated by GFP-LactC2 in psd1Δ and snx4Δpsd1Δ cells (Figure 4). Colabeling with GFP-LactC2 and FM4-64, a lipophilic dye that accumulates on the vacuole membrane (Vida and Emr, 1995), reveals that GFP-LactC2 decorates the vacuole membrane, as well as other organelle membranes in these cells (Figure 4A). This is particularly interesting because the presence of an electrostatic gradient along the membranes of the endocytic pathway, with the plasma membrane being the most electrostatic, is a fundamental feature of all cells and the maintenance of this gradient plays a crucial role in promoting proper protein targeting (Bigay and Antonny, 2012; Platre et al., 2018). To quantify changes in GFP-LactC2 distribution, we calculated the mean proportion of GFP-LactC2 fluorescence within the interior volume of the cell (i.e., excluding the cell cortex) versus the total GFP-LactC2 fluorescence (Figure 4B). We did not observe GFP-LactC2 to colocalize with cytoplasmic GFP-Atg8 or GFP-Ape1 puncta in snx4Δpsd1Δ cells, suggesting that PS does not accumulate on membrane of the PAS or autophagosomes (not shown). In wild-type and snx4Δ cells, ∼15% of GFP-LactC2 localizes to internal organelles, and in psd1Δ cells this is increased to 38 ± 8% (Figure 4B). These data suggest that loss of PSD1 results in an increase of PS in the membranes of organelles of the endomembrane system, including the vacuole. Importantly, the intracellular pool of PS is further increased (to ∼50%) in snx4Δpsd1Δ cells, revealing a contribution of Snx4 family proteins in restricting the accumulation of PS on internal organelles, possibly via a role in recycling of plasma membrane.
FIGURE 4:
Snx4 contributes to phosphatidylserine distribution within the endovacuolar system. (A) Micrographs (medial Z slice) of wild-type and mutant cells expressing the phosphatidylserine sensor, GFP-LactC2, are shown. Vacuoles are labeled with FM4-64. The sale bar indicates 5 μm. (B) Quantitation of GFP-LactC2 distribution. The proportion of GFP-LactC2 within the cell vs. at the cell cortex were measured. Each point is a single cell and the mean and SD are indicated. (C) Quantitation of RFP-LactC2 distribution in psd1Δ and snx4Δpsd1Δ cells with or without overexpression of PSD2. Same analysis was conducted as in B. *p < 0.0001. Standard deviations are indicated in B and C.
Snx4-Atg20 binds preferentially to PS-containing membrane
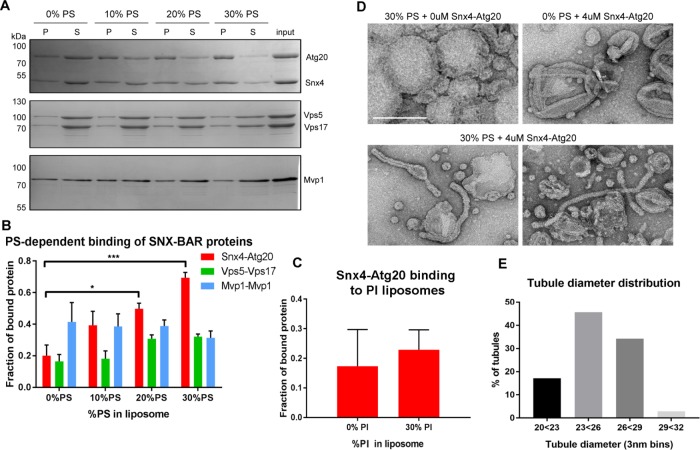
We speculate that retrograde trafficking of PS-enriched carriers coated with Snx4 family proteins supports plasma membrane restriction of PS by promoting export of PS from the endosome via a plasma membrane recycling pathway. To test whether Snx4 proteins have lipid binding specificity, we examined the binding of Snx4-Atg20 and other SNX-BAR proteins to liposomes containing different amounts of PS (Figure 5A). Purified SNX-BAR dimers, Snx4-Atg20, Vps5-Vps17, and Mvp1-Mvp1 were presented with liposomes containing 0, 10, 20, or 30 mol percent PS. Liposomes were recovered by centrifugation and the bound and unbound fractions of SNX-BARs were determined. This analysis shows that the amount of Snx4-Atg20 bound to the liposomes tracks in direct proportion to PS content, but this is not the case for the Vps5-Vps17 retromer SNX-BAR, or for Mvp1, a homodimeric SNX-BAR that functions on the retromer pathway (Figure 5B). The other abundant anionic lipid on the yeast endosome is phosphatidylinositol (PI) (van Meer et al., 2008). Accordingly, we also tested binding of Snx4-Atg20 to liposomes containing 0 or 30 mol percent PI and observed no difference in the amount of Snx4-Atg20 bound (Figure 5C). Similar experiments using liposomes with varying amounts of PE showed no effect on Snx4-Atg20 binding (not shown). Therefore, we conclude that PS is the preferred lipid for Snx4-Atg20 binding.
FIGURE 5:
The Snx4-Atg20 heterodimer exhibits membrane remodeling activity. (A) Representative Coomassie stained gels of liposome sedimentation reactions containing SNX-BAR complexes and liposomes with indicated amounts of PS. The position of mass standards (kDa) are indicated. (B) Quantification of Snx4-Atg20 binding to liposomes. Fraction of bound protein is calculated as a fraction of SNX-BAR proteins in the pellet fraction (P) over the sum of pellet and supernatant (S) fraction and is averaged over at least three independent experiments. *p ≤ 0.05; ***p ≤ 0.001. (C) Quantification of Snx4-Atg20 binding to liposomes containing 0% or 30% PI. Fraction of bound protein is calculated as a fraction of SNX-BAR proteins in the pellet fraction (P) over the sum of pellet and supernatant (S) fraction and is averaged over at least three independent experiments. The difference is not statistically significant. (D) A gallery of micrographs of negative stained liposomes with (30%) or without PS (0%) incubated with (4 μM) or without (0 μM) Snx4-Atg20 heterodimer is shown. The scale bar represents 200 nm. (E) Histogram of the diameters of tubules generated by 4 μM Snx4-Atg20 heterodimer (n = 35) in 3 nm bins is shown. Snx4-Atg20 generated average tubule diameters of 25.2 ± 2.1 nm.
Many SNX-BAR proteins have the ability to topologically remodel membrane into coated tubules resembling endosome-derived transport carriers. To test Snx4-Atg20 for this activity, and to determine whether PS content of the membrane influences remodeling activity, we examined the various Snx4-Atg20 liposome preparations by negative stain electron microscopy (Figure 5D). We observed membrane tubules in Snx4-Atg20 samples incubated with liposome preparations containing 30 mol percent PS but not with liposomes that lack PS. The tubular structures that formed were uniform in diameter, which we measured to be 25.2 ± 2.1 nm, further demonstrating that this is a property of the Snx4-Atg20 complex that is dependent on the presence of PS in the membrane (Figure 5E). Thus, our observations suggest that Snx4-Atg20 preferentially coats and exports PS-enriched membrane from the endosome. Interestingly, a recent study of plant endomembranes demonstrated that an electrostatic gradient maintained in part by PS is important for targeting of proteins to organelles of the endovacuolar system (Platre et al., 2018).
Increased PE synthesis restores clearance of autophagy intermediates in snx4Δpsd1Δ cells
The data indicate that PS is more widely distributed within the endovacuolar system in snx4Δpsd1Δ cells compared with wild-type cells, and this is correlated with fragmented vacuoles and accumulated cytoplasmic autophagy intermediates. Comparisons of differential interference contrast and GFP-Atg8 fluorescence micrographs of snx4Δpsd1Δ cells suggests that, in addition to an accumulation of GFP-Atg8-decorated autophagy intermediates (Figure 2), organelles that are not decorated by GFP-Atg8 are also impacted (Figure 6). Staining of snx4Δpsd1Δ cells with 7-aminochloromethylcoumarin (CMAC), a dye that accumulates in the acidic lumen of the vacuole, revealed that these cells possess numerous (>2) small vacuoles or vacuoles that do not stain well with CMAC in contrast to wild-type cells, which typically possess one to two uniformly stained vacuoles per cell (Figure 6). Numerous small vacuoles accumulate in cells when the vacuole fusion machinery is disabled, leading us to speculate that the fusion competence of the vacuole membrane is deficient in snx4Δpsd1Δ cells (Figure 6D).
FIGURE 6:
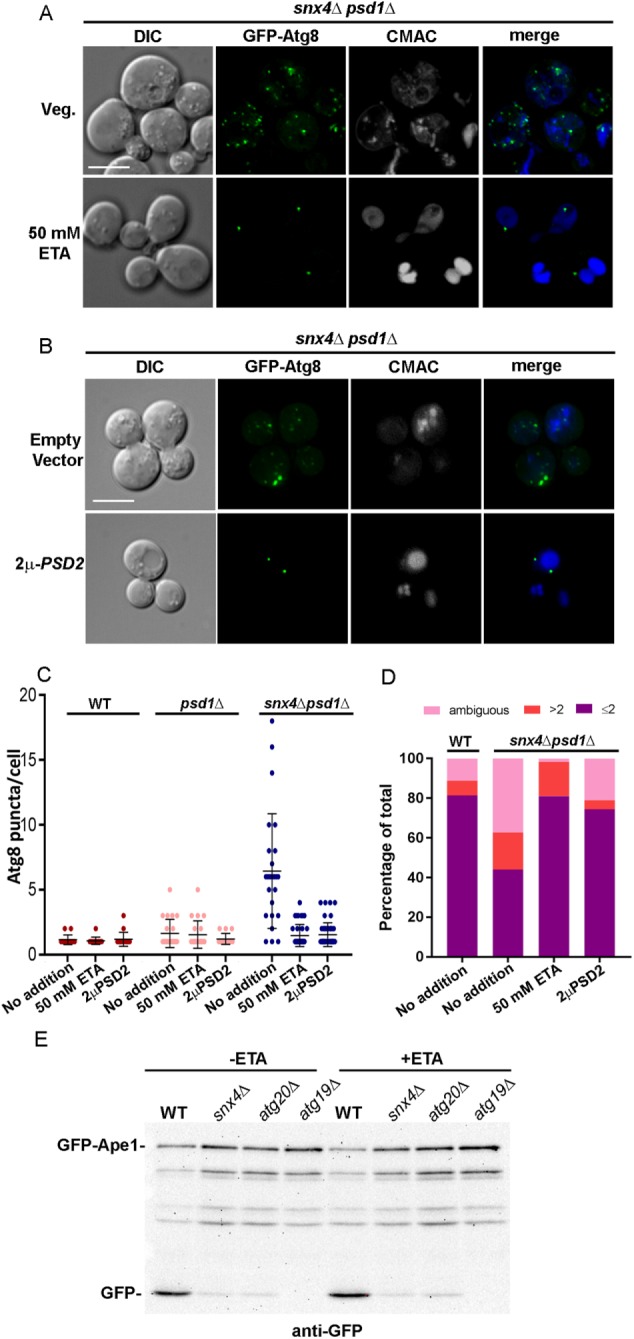
snx4Δpsd1Δ cells are deficient in PE trafficking to the autophagosome resulting in the loss of fusion to the vacuole. (A) Maximum projection micrographs showing snx4Δpsd1Δ cells expressing GFP-Atg8 grown in standard synthetic medium or medium supplemented with 50 mM ethanolamine. Vacuoles were visualized using CMAC. The sale bar indicates 5 μm. (B) Maximum projection micrographs showing snx4Δpsd1Δ cells expressing GFP-Atg8 and overexpressing PSD2 (2μ-PSD2) are shown. The sale bar indicates 5 μm. (C) The number of Atg8 puncta per cell was quantified in wild-type and mutant cells in the absence and presence of 50 mM ethanolamine (ETA), or overexpression of PSD2. Each point is a single cell and the mean and SD are indicated. (D) The number of vacuoles per cell was quantified in wild-type and snx4Δpsd1Δ cells in the absence of 50 mM ETA or 2μ-PSD2 and snx4Δpsd1Δ cells either grown in 50 mM ETA or expressing 2μ-PSD2. The cells were binned into three categories, one or two CMAC-stained vacuoles (a single vacuole may contain multiple lobes that are clearly connected), more than two CMAC-stained vacuoles, or “ambiguous,” which are cells that do not stain well with CMAC, and presented as percentages of the total number of cells quantified. (E) Representative immunoblot analysis of GFP-Ape1 processing in cells grown to log phase in standard or 50 mM ETA-supplemented medium in wild-type or indicated mutant cells. Anti-GFP was used to detect full length GFP-Ape1 and free GFP proteolytic fragment. Note the reduction in GFP-Ape1 processing in snx4Δ and atg20Δ cells compared with wild-type cells, which is still maintained when cells are grown in ETA. The atg19Δ mutant lacks the Ape1 sorting receptor and serves as a control.
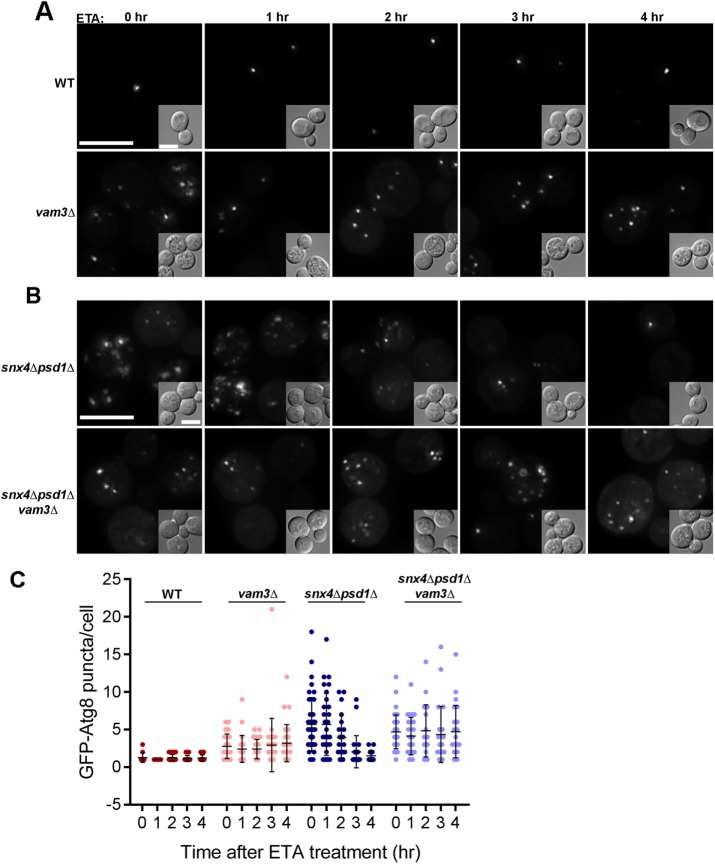
Genetic studies of Neo1, an aminophospholipid flippase that maintains lipid bilayer asymmetry of membranes of organelles of the endovacuolar system, show that the vacuoles in mutant neo1-2 cells, like the vacuoles in snx4Δpsd1Δ cells, are fragmented (Wu et al., 2016). For neo1-2 cells, the vacuole fusion defect can be rescued by metabolically forcing cells to produce more PE (Wu et al., 2016). Thus, perturbations to aminophospholipid trafficking and trans-bilayer distribution have a profound consequence on pathways that rely on vacuole fusion. Given the similar vacuole morphologies of snx4Δpsd1Δ and neo1-2 cells, we cultured snx4Δpsd1Δ cells in medium supplemented with ethanolamine (50 mM) to drive PE synthesis via the Kennedy salvage pathway and assayed accumulation of GFP-Atg8 decorated compartments. In snx4Δpsd1Δ cells cultured overnight in this medium, the appearance of GFP-Atg8 is indistinguishable from that of wild-type cells (one to two puncta per cell), and vacuole morphology is rescued (Figure 6, A, C, and D). However, the block in GFP-Ape1 processing in snx4Δpsd1Δ cells is not rescued by ethanolamine supplementation, indicating that this does not bypass the requirement for Snx4-Atg20 in the CVT pathway (Figure 6E). A time-course experiment showed that the reduction in the number of GFP-Atg8 decorated organelles decreased gradually after ethanolamine addition and was complete after 4 h (Figure 7B). Importantly, ethanolamine supplementation also rescued vacuole fragmentation in snx4Δpsd1Δ cells (Figure 6, A and D), indicating that increased PE promotes fusion competence of the vacuole membrane. In addition to ethanolamine supplementation, overexpression of PSD2, encoding a cytoplasmic PS decarboxylase, also rescues the accumulation of GFP-Atg8 decorated organelles and vacuole fragmentation in snx4Δpsd1Δ cells (Figure 6, B–D), just as it rescues the vacuole morphology of neo1-2 cells (Wu et al., 2016). Rescue is due to conversion of PS to PE in the cytoplasmic leaflet of internal organelles, as overexpression of PSD2 reduces the proportion of red fluorescent protein (RFP)-LactC2 on these organelles (Figure 4C). This is consistent with the finding that Psd2 activity modulates the amounts of PS and PE in the vacuole membrane (Gulshan et al., 2010).
FIGURE 7:
Vam3-dependent vacuole fusion is a requirement of ETA mediated autophagy rescue in snx4Δpsd1Δ cells. (A) Maximum projection micrographs of WT and vam3Δ cells expressing GFP-Atg8 before and after addition of ethanolamine (50 mM) are shown. The scale bars indicate 5 μm. (B) Maximum projection micrographs of snx4Δpsd1Δ and snx4Δpsd1Δvam3Δ cells expressing GFP-Atg8 treated similarly are shown. (C) The number of GFP-Atg8 puncta per cell was quantified at the indicated time points after ETA addition in each of the four strains. A minimum of 19 cells were analyzed in each condition and plotted as individual dots as indicated and the mean and SD are indicated.
SNARE-dependent fusion is required for clearance of autophagy intermediates from the cytoplasm
Our findings suggest that the balance of PE and PS in the vacuole membrane is critical to sustain homotypic vacuole–vacuole fusion and autophagosome biogenesis and/or fusion with the vacuole. When assayed by in vitro reconstitution, membrane fusion catalyzed by the yeast vacuolar SNAREs requires that the membranes contain physiological levels of neutral, small headgroup lipids, such as PE (Zick et al., 2014). With this in mind, the results suggest that changes in the lipid composition of the vacuole membrane resulting from loss of SNX4 and PSD1 results in a deficit of PE that decreases fusion competence of the vacuole, resulting in the accumulation of autophagy intermediates in the cytoplasm and fragmented vacuoles. Fusion of a mature autophagosome with the vacuole membrane requires the Rab-regulated tethering complex and SNARE proteins that mediates homotypic vacuole fusion (Darsow et al., 1997; Hyttinen et al., 2013). To directly assay for a requirement for vacuolar fusion in clearance of autophagy intermediates in snx4Δpsd1Δ cells, we deleted VAM3, encoding a SNARE fusion protein that is required for fusion of autophagosomes with the vacuole (Darsow et al., 1997), in the snx4Δpsd1Δ background (snx4Δpsd1Δvam3Δ) and then monitored the number of GFP-Atg8 decorated compartments over time after addition of ethanolamine (Figure 7). In these cells, the number of GFP-Atg8 decorated organelles remained constant for the duration of the experiment, indicating that SNARE-mediated fusion with the vacuole is required for their clearance.
Lipid trafficking by Snx4 proteins promotes autophagy
Previously, Snx4-Atg20 was reported to be strictly required for several selective autophagy pathways (CVT pathway, mitophagy, pexophagy) but not for starvation-induced nonselective autophagy (Nice et al., 2002; Kanki et al., 2009; Okamoto et al., 2009; Shpilka et al., 2015). Contrary to these prior reports, and in agreement with Popelka et al. (2017), we find that Snx4-Atg20 does in fact play a role in starvation-induced nonselective autophagy, albeit a nonessential role. At present, it is unclear whether Snx4-Atg20 fulfills the same function(s) in selective and nonselective autophagy because deletion of SNX4 or ATG20 completely blocks the CVT selective autophagy pathway but has only a modest effect on nonselective autophagy. Snx4-Atg20 has been shown to associate with proteins of the core autophagy initiation complex, leading to the suggestion that it plays a role in autophagy initiation and/or phagophore expansion (Nice et al., 2002; Yorimitsu and Klionsky, 2005; Popelka et al., 2017).
In this study, we discovered that Snx4-Atg20 promotes nonselective autophagy indirectly through its role in exporting lipids from the endosome and/or vacuole via retrograde trafficking, which maintains the fusion competence of the vacuole membrane. A role for lysosome and autophagosome membrane lipid composition in maintaining fusion competence was previously suggested by in vitro studies of autophagosome and lysosome fusion (Koga et al., 2010). In this study, the authors demonstrated that in cellular fractions enriched in autophagosome and lysosome, reduced levels of cholesterol in autophagosome and lysosome membranes resulted in reduced fusion (Koga et al., 2010), corroborating that lipid composition of organelle membranes must be tightly regulated to maintain fusion competence. Our study extends these observations regarding the roles of lipids in promoting fusion of the autophagosome with the vacuole/lysosome in yeast cells.
Increased PE synthesis has been previously noted to suppress deficiencies in vacuolar processing of autophagy cargo resulting from mutations in genes encoding multivesicular body (MVB) pathway components, a phenomenon that was proposed to be due to an increase in the amount of lipidated Atg8 (Nebauer et al., 2007). Considered in light of the data presented herein, we suggest that deficiencies in the endosome maturation pathway (e.g., loss of recycling, MVB vesicle formation) impact the fusion competence of the vacuole membrane through changes in its lipid composition. This is apparent in the accumulation of fragmented vacuoles in snx4Δpsd1Δ cells, indicating defects in homotypic vacuole fusion. Furthermore, this vacuole phenotype can be rescued by increasing PE production through either ethanolamine hydrochloride (ETA) supplementation, which drives PE synthesis via the Kennedy pathway, or overexpression of PSD2. However, while overexpression of PSD2 decreases PS levels (Figure 4C), ETA supplementation has been demonstrated to slightly increase PS levels in the cells deleted for PSD1 (Storey et al., 2001), suggesting that it is the balance of PE/PS on organelle membranes, rather than absolute amounts, that is important in maintaining membrane fusion competence. A second, related defect that we have also characterized as also a result of lipid imbalance is the accumulation of GFP-Atg8 decorated autophagy intermediates in the cytosol of snx4Δpsd1Δ cells. The accumulated compartments vary in size and shape, some appearing to enclose a lumen, suggesting that autophagosome-vacuole fusion is defective in snx4Δpsd1Δ cells. The accumulation of these GFP-Atg8 organelles are also rescued by increasing PE synthesis via either ETA supplementation or PSD2 overexpression. Because homotypic vacuole fusion defects in snx4Δpsd1Δ cells are also rescued on a timescale similar to clearance of GFP-Atg8-decorated compartments (Figure 6), autophagosome–vacuole fusion is likely deficient in snx4Δpsd1Δ cells. However, the variability in size and shape of the accumulated GFP-Atg8 compartments also suggest defective autophagosome biogenesis, which may reflect deficient phagophore expansion and/or sealing of the autophagosome. Therefore, the clearance of these autophagy intermediates indicates that a PE/PS balance maintained by Psd1 and Snx4-mediated interorganelle trafficking is also required for promoting fusion competence of the autophagosome membrane, required for autophagosome growth and closure. The results suggest that Snx4-Atg20 mediated export of PS from the endosome ensures that PS does not accumulate in the membranes of organelles of the endolysosomal/vacuolar system required to sustain fusion of the autophagosome with the vacuole and for homotypic vacuole fusion (Figure 8). Our results also highlight the importance of maintaining an electrostatic PS gradient along the endomembrane system, which has been previously demonstrated for endomembrane systems of mammalian and plant cells (Fairn et al., 2011a,b; Platre et al., 2018). When this gradient is disrupted in snx4Δpsd1Δ cells, severe and pleiotropic defects in cell physiology homeostasis emerge, ranging from defects in autophagy to defects in vacuole dynamics. Furthermore, we demonstrate that the presence of the anionic gradient, contributed in large part by PS, is maintained by both retrograde trafficking pathways that remove PS from late endocytic membranes and by enzymes, such as Psd2, that contribute locally to metabolism of PS on endomembranes. Together, these findings highlight a lipid-based perspective on SNX-BAR function, which has typically been considered a protein-centric view of sorting nexin function.
FIGURE 8:
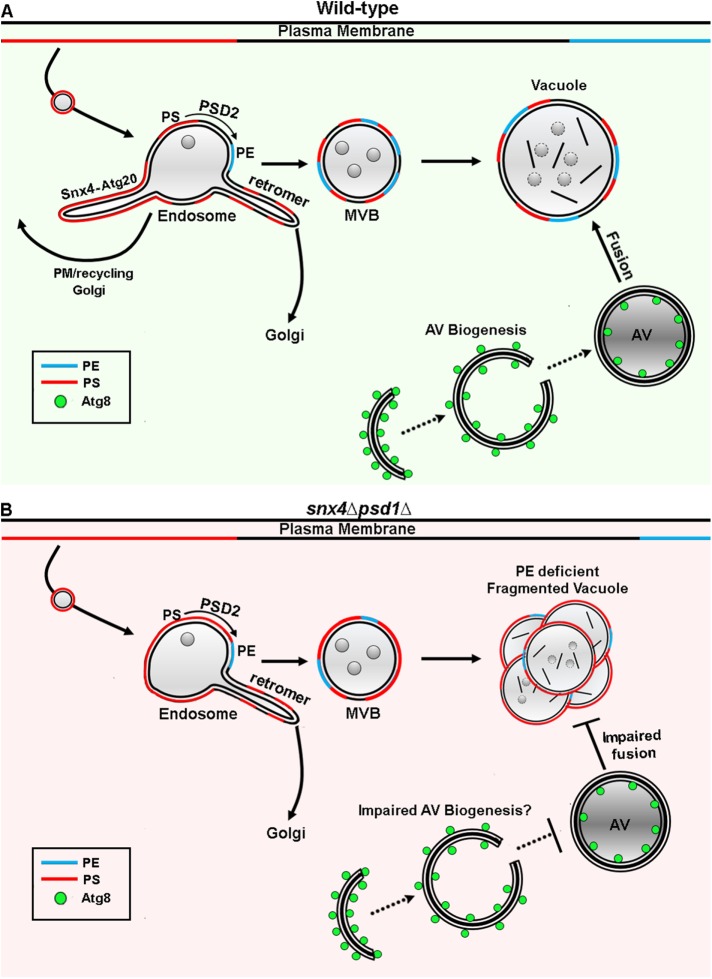
Retrograde trafficking by Snx4 family proteins maintains lipid homeostasis of the vacuole membrane. The endosomal maturation pathway and autophagosome–vacuole fusion are depicted. The relative abundances and topology of PS and PE in organelle membranes are depicted. (A) We propose that during endosome maturation, the ratio of PE to PS on the membrane increases as a result of Snx4-mediated export of membrane enriched in PS (compared with retromer) and the activity of Psd2, which converts PS to PE on the endosome (Gulshan et al., 2010). This results in a PE/PS ratio on the vacuole membrane that is permissive for SNARE-mediated fusion of autophagosomes (AV) with the vacuole. Also depicted are the Snx4 family- and retromer-dependent retrograde pathways that originate from a common endosome (Ma et al., 2017). (B) In snx4Δpsd1Δ cells, the amount of PE in the cell is substantially (∼60%) reduced (Trotter et al., 1993) and PS is not adequately removed from the endosomal system, resulting in a decreased PE/PS ratio on the vacuole membrane. This manifests as a decrease in fusion competence of the vacuole, resulting in impaired fusion of autophagosomes with the vacuole and also homotypic vacuole fusion. Furthermore, it is likely that the membranes that supply biogenesis of autophagosomes also have a fusion-deficient ratio of PE/PS, resulting in an accumulation of autophagy intermediates.
MATERIALS AND METHODS
Yeast strains and culture conditions
Yeast strains were constructed in BY4742 (MATαhis3-1, leu2-0, met15-0, and ura3-0) by homologous recombination of gene-targeted, PCR-generated DNAs using the method of Longtine et al. (1998). Mutant strains were either derived from the EUROSCARF KANMX deletion collection (Open Biosystems/Thermo Scientific, Waltham, MA) or produced by replacement of the complete reading frame with the HIS3MX6 or URA3 cassette. Gene deletions were confirmed by PCR amplification of the deleted locus. Cells were grown in standard synthetic complete medium lacking nutrients required to maintain selection for auxotrophic markers and/or plasmids (Sherman et al., 1979), unless indicated otherwise. To induce nonselective autophagy, cells were grown to early log phase, concentrated, and resuspended in standard synthetic complete medium containing 0.2 μg/ml rapamycin (R-5000; LC laboratories) for 2–4 h at 30°C. For experiments rescuing autophagosomes-vacuole fusion defects, cells were grown overnight to mid–log phase in synthetic complete medium or in synthetic medium containing 50 mM ETA (Sigma-Aldrich; E6133). For time-course experiments, mid–log phase cells grown in synthetic medium were directly added ETA to a final concentration of 50 mM and then imaged using 60-min time intervals.
Light microscopy and image analysis
Yeast cells from cultures grown to OD600 ≈ 0.5 were mounted in growth medium, and three-dimensional image stacks were collected at 0.3-µm z increments on a DeltaVision workstation (Applied Precision) based on an inverted microscope (IX-70; Olympus) using a 100 × 1.4NA oil immersion lens. Images were captured at 24°C with a front illuminated scientific complementary metal–oxide–semiconductor, 2560 × 2160 pixels camera and deconvolved using the iterative-constrained algorithm and the measured point spread function. To visualize vacuole morphology, yeast cells were labeled with CMAC (Life Technologies) at a concentration of 100 μM for 30 min in synthetic medium at room temperature. To visualize the vacuole membrane, FM4-64 (32 nM) was added to cell cultures for 20 min at 30°C. Cells were then washed, resuspended in fresh medium, and then incubated for 60 min to allow FM4-64 to accumulate in the vacuole membrane (adapted from Vida and Emr, 1995). Image analysis and preparation was done using Softworx 6.1 (Applied Precision Instruments) and ImageJ v1.50d (Rasband). To quantify GFP-LactC2 fluorescence signal, two to three 0.3-µm medial Z planes from the approximate center of a cell were used to calculate the integrated density of the selected region of the cell subtracted by the corrected background of the same region. Analysis was performed with ImageJ v1.50d (Rasband).
Atg8 processing and immunoblotting
For quantitative Western blot analysis of GFP-Atg8 or GFP-Ape1 processing, cells were grown under standard vegetative (or supplemented with 50 mM ETA) or autophagy inducing conditions to OD600 ≈ 0.5, as described above. Typically, 3.0 × 107 cells were harvested by centrifugation and lysed by glass bead agitation in SDS–PAGE sample buffer. Polyacrylamide (10%) gels were loaded with 0.5 × 107 cell equivalents and transferred onto standard 0.45-μm nitrocellulose. Anti-GFP primary mouse monoclonal antibody (1814460; Roche) was diluted 1:2500, and Santa Cruz (sc-2055) goat anti-mouse HRP-conjugated antibody was used at 1:5000. Anti-Pgk1 at 1:5000 (Life Technologies) was used as loading controls. For native Atg8 Western blots, 13.5% polyacrylamide gels containing 6 M urea were prepared as indicated (Nakatogawa and Ohsumi, 2012) and loaded with 1.5 × 107 cell equivalents. Anti-Atg8 primary rabbit antibody was generously provided by Claudine Kraft (University of Freiburg) and used at 1:1000, and goat anti-rabbit HRP-conjugated antibody was used at 1:5000. All enhanced chemiluminescence (ECL) blots were development on a Chemidoc-XRS+ (Bio-Rad), and band intensities were quantified using Quantity One 1D analysis software (Bio-Rad).
Plasmids
Centromeric GFP-Atg8 and GFP-Ape1 (Shintani et al., 2002) plasmids were used in processing assays. Centromeric plasmids encoding GFP-LactC2 and RFP-LactC2 are described in Yeung et al. (2008).
A pRS426-based high copy PSD2 expression vector was made by homologous recombination of PCR-amplified wild-type genomic PSD2 locus with the following primers: CCCCCCCTCGAGGTCGACGGTATCGATAAGCTTGATATCGTAGCTGCTCCATTTGTGCTC and GCTCCACCGCGGTGGCGGCCGCTCTAGAACTAGTGGATCCGGGAGAGGGAATTAGATCAAC.
SNX-BAR dimers purification and liposome binding/tubulation assay
Snx4-Atg20 and Snx4-Snx41 complexes were purified from yeast TVY614 strain background. Endogenous promoters of Snx4 and Atg20 or Snx4 and Snx41 were replaced with galactose-inducible promoters using the method described in Longtine et al. (1998). Tandem affinity purification tag was fused to the C-terminus of Atg20 or Snx41 using the method described in Puig et al. (2001). Protein expression was induced using galactose, where 50-ml precultures were grown overnight in yeast peptone (YP) medium containing 2% raffinose and 0.1% glucose, 1-l cultures were inoculated the next morning and grown in YP medium containing 2% raffinose and 0.1% glucose, and then 2% galactose was added after 4–5 h of growth. Snx4-Atg20 and Snx4-Snx41 complexes were immunopurified using IgG Sepharose (GE Healthcare) in 50 mM Tris, pH 7.4, buffer containing 300 mM NaCl, 2 mM MgCl2, 1 mM dithiothreitol, and cOmplete Protease Inhibitor Cocktail (Roche). The complexes were cleaved using His-tagged recombinant tobacco etch virus (TEV) protease overnight at 4°C. The His-TEV was removed using Ni-NTA agarose. Vps5-Vps17 complex was purified using the same strategy.
Mvp1 was purified using His-Mvp1 construct from Escherichia coli BL21 cells as previously described (Chi et al., 2014).
Purified protein concentration was quantified by BCA assay (Pierce)
Liposome binding/tubulation assays were carried out as previously described (Chi et al., 2014). Briefly, liposomes (2.5 mM lipid) (0%, 10%, 20%, or 30% PS; 1% PtdIns(3)P; 20% ergosterol; PC) were incubated with 4 μM Snx4-Atg20, Vps5-Vps17, or Mvp1-Mvp1 heterodimer for 30 min at 30°C and sedimented at 100,000 × g for 20 min. Pellet (P) and supernatant (S) fractions were loaded onto 10% polyacrylamide gels and visualized using Coomassie Brilliant Blue stain. To test Snx4-Atg20 binding to PI containing liposomes, liposome compositions were 0% or 30% soy PI; 1% PtdIns(3)P; 10% ergosterol; PC. Band intensities were quantified using Quantity One 1D analysis software (Bio-Rad) and proportion of SNX-BAR proteins in pellet fraction was quantified. Two-way analysis of variance was used to determine statistical significance (GraphPad Prism 7.01). Samples for EM analysis were spotted onto a carbon-coated copper mesh grid. Liposomes were negative stained by 1% uranylacetate and analyzed on a FEI Tecnai F20 transmission electron microscope (200 kV). Tubules were quantified in ImageJ analysis software as an average of three measurements across the tubule width.
Acknowledgments
We are grateful to colleagues, especially Tom Melia, for discussions and critical reading of the manuscript, Scott Emr for sharing unpublished data, and Claudine Kraft for anti-Atg8 antiserum. We also thank Sandy Lemmon for sharing reagents. Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number GM060221 and in part by the National Institute of General Medical Sciences of the National Institutes of Health under award number T32GM007223. R.C. was supported in part by the UNC-Charlotte Faculty Research Grants Program. C.U. was supported by the Deutsche Forschungsgemeinschaft (SFB 944, project P11).
Abbreviations used:
- BAR
Bin-Amphiphysin-Rvs161
- PGK
phosphoglycerate kinase
- PE
phosphatidylethanolamine
- PS
phosphatidylserine
- PX
Phox homology
- SNX-BAR
sorting nexin containing a BAR domain
- WT
wild type
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-12-0743) on June 27, 2018.
REFERENCES
- Bean BD, Davey M, Conibear E. (2017). Cargo selectivity of yeast sorting nexins. Traffic , 110–122. [DOI] [PubMed] [Google Scholar]
- Bigay J, Antonny B. (2012). Curvature, lipid packing, and electrostatics of membrane organelles: defining cellular territories in determining specificity. Dev Cell , 886–895. [DOI] [PubMed] [Google Scholar]
- Birner R, Burgermeister M, Schneiter R, Daum G. (2001). Roles of phosphatidylethanolamine and of its several biosynthetic pathways in Saccharomyces cerevisiae. Mol Biol Cell , 997–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi RJ, Liu J, West M, Wang J, Odorizzi G, Burd CG. (2014). Fission of SNX-BAR-coated endosomal retrograde transport carriers is promoted by the dynamin-related protein Vps1. J Cell Biol , 793–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darsow T, Rieder SE, Emr SD. (1997). A multispecificity syntaxin homologue, Vam3p, essential for autophagic and biosynthetic protein transport to the vacuole. J Cell Biol , 517–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejsing CS, Sampaio JL, Surendranath V, Duchoslav E, Ekroos K, Klemm RW, Simons K, Shevchenko A. (2009). Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc Natl Acad Sci USA , 2136–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairn GD, Hermansson M, Somerharju P, Grinstein S. (2011a). Phosphatidylserine is polarized and required for proper Cdc42 localization and for development of cell polarity. Nat Cell Biol , 1424–1430. [DOI] [PubMed] [Google Scholar]
- Fairn GD, Schieber NL, Ariotti N, Murphy S, Kuerschner L, Webb RI, Grinstein S, Parton RG. (2011b). High-resolution mapping reveals topologically distinct cellular pools of phosphatidylserine. J Cell Biol , 257–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farré J-C, Subramani S. (2016). Mechanistic insights into selective autophagy pathways: lessons from yeast. Nat Rev Mol Biol Cell , 537–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulshan K, Shahi P, Moye-Rowley WS. (2010). Compartment-specific synthesis of phosphatidylethanolamine is required for normal heavy metal resistance. Mol Biol Cell , 443–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettema EH, Lewis MJ, Black MW, Pelham HRB. (2003). Retromer and the sorting nexins Snx4/41/42 mediate distinct retrieval pathways from yeast endosomes. EMBO J , 548–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppins S, Collins SR, Cassidy-Stone A, Hummel E, DeVay RM, Lackner LL, Westermann B, Schuldiner M, Weissman JS, Nunnari J. (2011). A mitochondrial-focused genetic interaction map reveals a scaffold-like complex required for inner membrane organization in mitochondria. J Cell Biol , 323–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyttinen JMT, Niittykoski M, Salminen A, Kaarniranta K. (2013). Maturation of autophagosomes and endosomes: a key role for Rab7. Biochim Biophys Acta , 503–510. [DOI] [PubMed] [Google Scholar]
- Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M. et al (2000). A ubiquitin-like system mediates protein lipidation. Nature , 488. [DOI] [PubMed] [Google Scholar]
- Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. (2001). A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci USA , 4569–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Kamada Y, Baba M, Takikawa H, Sasaki M, Ohsumi Y. (2005). Atg17 Functions in Cooperation with Atg1 and Atg13 in Yeast Autophagy. Mol Biol Cell , 2544–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T, Wang K, Baba M, Bartholomew CR, Lynch-Day MA, Du Z, Geng J, Mao K, Yang Z, Yen WL, Klionsky DJ. (2009). A genomic screen for yeast mutants defective in selective mitochondria autophagy. Mol Biol Cell , 4730–4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur J, Debnath J. (2015). Autophagy at the crossroads of catabolism and anabolism. , 461. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, Adachi H, Adams CM, Adams PD, Adeli K. et al (2016). Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy , 1–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga H, Kaushik S, Cuervo AM. (2010). Altered lipid content inhibits autophagic vesicular fusion. FASEB J , 3052–3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer MH, Farre JC, Mitra K, Yu MK, Ono K, Demchak B, Licon K, Flagg M, Balakrishnan R, Cherry JM. et al (2017). Active interaction mapping reveals the hierarchical organization of autophagy. Mol Cell , 761–774.e765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leventis PA, Grinstein S. (2010). The distribution and function of phosphatidylserine in cellular membranes. Annu Rev Biophys , 407–427. [DOI] [PubMed] [Google Scholar]
- Levine B, Kroemer G. (2008). Autophagy in the pathogenesis of disease. Cell , 27–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast , 953–961. [DOI] [PubMed] [Google Scholar]
- Lynch-Day MA, Klionsky DJ. (2010). The Cvt pathway as a model for selective autophagy. FEBS Lett , 1359–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma M, Burd CG, Chi RJ. (2017). Distinct complexes of yeast Snx4 family SNX-BARs mediate retrograde trafficking of Snc1 and Atg27. Traffic , 134–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mari M, Griffith J, Rieter E, Krishnappa L, Klionsky DJ, Reggiori F. (2010). An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J Cell Biol , 1005–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatogawa H, Ohsumi Y. (2012). SDS-PAGE techniques to study ubiquitin-like conjugation systems in yeast autophagy. In Ubiquitin Family Modifiers and the Proteasome: Reviews and Protocols, ed Dohmen RJ, Scheffner M, Totowa, NJ: Humana Press, 519–529. [DOI] [PubMed] [Google Scholar]
- Nebauer R, Schuiki I, Kulterer B, Trajanoski Z, Daum G. (2007). The phosphatidylethanolamine level of yeast mitochondria is affected by the mitochondrial components Oxa1p and Yme1p. FEBS J. , 6180–6190. [DOI] [PubMed] [Google Scholar]
- Nemec AA, Howell LA, Peterson AK, Murray MA, Tomko RJ., Jr (2017). Autophagic clearance of proteasomes in yeast requires the conserved sorting nexin Snx4. J Biol Chem , 21466–21480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nice DC, Sato TK, Stromhaug PE, Emr SD, Klionsky DJ. (2002). Cooperative binding of the cytoplasm to vacuole targeting pathway proteins, Cvt13 and Cvt20, to phosphatidylinositol 3-phosphate at the pre-autophagosomal structure is required for selective autophagy. J Biol Chem , 30198–30207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi Y, Munro S. (2010). Membrane delivery to the yeast autophagosome from the Golgi-endosomal system. Mol Biol Cell , 3998–4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K, Kondo-Okamoto N, Ohsumi Y. (2009). Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev Cell , 87–97. [DOI] [PubMed] [Google Scholar]
- Platre MP, Noack LC, Doumane M, Bayle V, Simon MLA, Maneta-Peyret L, Fouillen L, Stanislas T, Armengot L, Pejchar P. et al (2018). A combinatorial lipid code shapes the electrostatic landscape of plant endomembranes. Dev Cell , 465–480.e411. [DOI] [PubMed] [Google Scholar]
- Popelka H, Damasio A, Hinshaw JE, Klionsky DJ, Ragusa MJ. (2017). Structure and function of yeast Atg20, a sorting nexin that facilitates autophagy induction. Proc Natl Acad Sci USA , E10112–E10121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Seraphin B. (2001). The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods , 218–229. [DOI] [PubMed] [Google Scholar]
- Reggiori F, Klionsky DJ. (2013). Autophagic processes in yeast: mechanism, machinery and regulation. Genetics. , 341–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuiki I, Schnabl M, Czabany T, Hrastnik C, Daum G. (2010). Phosphatidylethanolamine synthesized by four different pathways is supplied to the plasma membrane of the yeast Saccharomyces cerevisiae. Biochim Biophys Acta , 480–486. [DOI] [PubMed] [Google Scholar]
- Sherman F, Fink GR, Lawrence LW. (1979). Methods in Yeast Genetics: A Laboratory Manual, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Shintani T, Huang WP, Stromhaug PE, Klionsky DJ. (2002). Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev Cell , 825–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirahama-Noda K, Kira S, Yoshimori T, Noda T. (2013). TRAPPIII is responsible for vesicular transport from early endosomes to Golgi, facilitating Atg9 cycling in autophagy. J Cell Sci , 4963–4973. [DOI] [PubMed] [Google Scholar]
- Shpilka T, Welter E, Borovsky N, Amar N, Shimron F, Peleg Y, Elazar Z. (2015). Fatty acid synthase is preferentially degraded by autophagy upon nitrogen starvation in yeast. Proc Natl Acad Sci USA , 1434–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey MK, Clay KL, Kutateladze T, Murphy RC, Overduin M, Voelker DR. (2001). Phosphatidylethanolamine has an essential role in Saccharomyces cerevisiae that is independent of its ability to form hexagonal phase structures. J Biol Chem , 48539–48548. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Kubota Y, Sekito T, Ohsumi Y. (2007). Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells , 209–218. [DOI] [PubMed] [Google Scholar]
- Suzuki SW, Emr SD. (2018). Membrane protein recycling from the vacuole/lysosome membrane. J Cell Biol , 1623–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotter PJ, Pedretti J, Voelker DR. (1993). Phosphatidylserine decarboxylase from Saccharomyces cerevisiae. Isolation of mutants, cloning of the gene, and creation of a null allele. J Biol Chem , 21416–21424. [PubMed] [Google Scholar]
- van Meer G, Voelker DR, Feigenson GW. (2008). Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol , 112–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Weering JR, Sessions RB, Traer CJ, Kloer DP, Bhatia VK, Stamou D, Carlsson SR, Hurley JH, Cullen PJ. (2012). Molecular basis for SNX-BAR-mediated assembly of distinct endosomal sorting tubules. EMBO J , 4466–4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vida TA, Emr SD. (1995). A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol , 779–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollert CS, Uetz P. (2004). The phox homology (PX) domain protein interaction network in yeast. Mol Cell Proteomics , 1053–1064. [DOI] [PubMed] [Google Scholar]
- Wu Y, Takar M, Cuentas-Condori AA, Graham TR. (2016). Neo1 and phosphatidylethanolamine contribute to vacuole membrane fusion in Saccharomyces cerevisiae. Cell Logist , e1228791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Kakuta S, Watanabe TM, Kitamura A, Sekito T, Kondo-Kakuta C, Ichikawa R, Kinjo M, Ohsumi Y. (2012). Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J Cell Biol , 219–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen W-L, Legakis JE, Nair U, Klionsky DJ. (2007). Atg27 is required for autophagy-dependent cycling of Atg9. Mol Biol Cell , 581–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung T, Gilbert GE, Shi J, Silvius J, Kapus A, Grinstein S. (2008). Membrane phosphatidylserine regulates surface charge and protein localization. Science , 210–213. [DOI] [PubMed] [Google Scholar]
- Yorimitsu T, Klionsky DJ. (2005). Atg11 links cargo to the vesicle-forming machinery in the cytoplasm to vacuole targeting pathway. Mol Biol Cell , 1593–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zick M, Stroupe C, Orr A, Douville D, Wickner WT. (2014). Membranes linked by trans-SNARE complexes require lipids prone to non-bilayer structure for progression to fusion. eLife , e01879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinser E, Sperka-Gottlieb C.D., Fasch EV, Kohlwein SD, Paltauf F, Daum G. (1991). Phospholipid synthesis and lipid composition of subcellular membranes in the unicellular eukaryote Saccharomyces cerevisiae. J Bacteriol , 2026–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]