Abstract
Cytokinesis is a complicated yet conserved step of the cell-division cycle that requires the coordination of multiple proteins and cellular processes. Here we describe a previously uncharacterized protein, Ync13, and its roles during fission yeast cytokinesis. Ync13 is a member of the UNC-13/Munc13 protein family, whose animal homologues are essential priming factors for soluble N-ethylmaleimide-sensitive factor attachment protein receptor complex assembly during exocytosis in various cell types, but no roles in cytokinesis have been reported. We find that Ync13 binds to lipids in vitro and dynamically localizes to the plasma membrane at cell tips during interphase and at the division site during cytokinesis. Deletion of Ync13 leads to defective septation and exocytosis, uneven distribution of cell-wall enzymes and components of cell-wall integrity pathway along the division site and massive cell lysis during cell separation. Interestingly, loss of Ync13 compromises endocytic site selection at the division plane. Collectively, we find that Ync13 has a novel function as an UNC-13/Munc13 protein in coordinating exocytosis, endocytosis, and cell-wall integrity during fission yeast cytokinesis.
INTRODUCTION
Cytokinesis partitions a mother cell into two daughter cells. From yeast to humans, cytokinesis starts with the assembly of an actomyosin contractile ring. The ring then constricts and guides plasma membrane invagination and extracellular matrix formation/remodeling (Pollard and Wu, 2010; Lee et al., 2012; D’Avino et al., 2015; Meitinger and Palani, 2016; Rincon and Paoletti, 2016). To ensure proper cell separation and integrity, cytokinesis requires the coordination of multiple pathways, including actomyosin ring constriction, the Rho GTPase-dependent cell integrity pathway (CIP), exocytosis, and endocytosis (Arellano et al., 1999; Albertson et al., 2005; Martín-Cuadrado et al., 2005; Wu et al., 2010; Sanchez-Mir et al., 2014). However, the mechanisms and coordination of these events remain poorly understood.
Membrane trafficking during cytokinesis relies on the balance between exocytosis and endocytosis to expand the plasma membrane and remodel the extracellular matrix at the division site. Exocytosis delivers and fuses secretory vesicles to the plasma membrane (Albertson et al., 2005; Echard, 2008). It starts with the budding of secretory vesicles from late Golgi or endosomes. Once vesicles are delivered to the target site, they are kept in close proximity to the plasma membrane by tethering complexes (Guo et al., 1999b; Donovan and Bretscher, 2012; James and Martin, 2013). It was thought that most vesicles are tethered at or within a narrow zone adjacent to the leading edge of the cleavage furrow via the exocyst, an octomeric tethering complex, during cytokinesis (TerBush et al., 1996; Guo et al., 1999b; Shuster and Burgess, 2002; Danilchik et al., 2003; VerPlank and Li, 2005; He and Guo, 2009; Neto et al., 2013). Surprisingly, we recently found that the exocytic vesicles are delivered all over the division plane in the fission yeast S. pombe, and the Transport Particle Protein II (TRAPP-II) complex and the exocyst work together to tether the vesicles (Wang et al., 2016). In synaptic vesicle exocytosis, syntaxin-1, the soluble N-ethylmaleimide–sensitive factor (NSF) attachment protein receptor (SNARE) protein from the plasma membrane that binds tightly to SM protein Munc18-1, is converted to an open conformation during the “priming” stage by UNC-13/Munc13 proteins (Hata et al., 1993; Garcia et al., 1994; Pevsner et al., 1994; Ma, Li, et al., 2011; Yang, Wang, Sheng, et al., 2015). Then the tight SNARE complex assembles to provide energy and to bring the membrane layers close enough to trigger fusion of tethered vesicles to the plasma membrane (James and Martin, 2013; Feyder et al., 2015; Martin, 2015). Blockage of vesicle secretion impairs cytokinesis from yeast to human cells (Skop et al., 2001; Liu et al., 2002; Gromley et al., 2005; Giansanti et al., 2015). However, roles of UNC-13/Munc13 proteins in cytokinesis have not been reported.
Endocytosis is also essential for cytokinesis (McCollum et al., 1996; Wienke et al., 1999; Gerald et al., 2001; Feng et al., 2002; Schweitzer et al., 2005; Baluska et al., 2006; Wu et al., 2006; Boucrot and Kirchhausen, 2007; Montagnac et al., 2008; de Leon et al., 2013). Clathrin-mediated endocytosis (CME) is the best characterized and main endocytic pathway, which occurs in a stepwise manner (Kaksonen et al., 2005; Berro et al., 2010; Sirotkin et al., 2010; Feyder et al., 2015). The early (Ede1, F-BAR protein Syp1, AP-2, and clathrin), middle (Sla2, Epsins, and YAP1801), and late (Sla1, Pan1, End3, and WASP) coat complexes are recruited to the endocytic site sequentially. They trigger branched actin filament assembly, membrane invagination, and vesicle scission (Berro et al., 2010; Sirotkin et al., 2010; Weinberg and Drubin, 2012; Idrissi and Geli, 2014; Goode et al., 2015). The internalized vesicles are released from the clathrin coats and are sorted to vacuoles/lysosomes for degradation or recycled back to the plasma membrane with the help of Rab11 GTPase (Grant and Donaldson, 2009; Neto et al., 2013; Feyder et al., 2015). In addition to CME, clathrin independent endocytosis (CIE) mediated by Rho GTPases has also been found in yeast and animal cells (Lamaze et al., 2001; Prosser et al., 2011; Prosser and Wendland, 2012). The exact role of endocytosis during cytokinesis remains poorly understood. Nevertheless, the temporal and spatial coordination between endocytosis and exocytosis is critical for proper cellular functions (Wu et al., 2014). In budding yeast, Sec4 Rab GTPase couples exocytosis with endocytosis to maintain the polarized Cdc42 GTPase for bud growth (Layton et al., 2011; McCusker et al., 2012; Johansen et al., 2016). During mating, focused pheromone secretion and shmoo formation can be explained in a mathematical model with exocytosis corralled by endocytosis (Chou, Moore, et al., 2012). In neuronal cells, inhibition of exocytosis abolishes endocytosis (Xie et al., 2017). In reverse, the compensatory endocytosis after synaptic vesicle exocytosis is essential for maintaining synaptic structures and restoring membrane tension (Lou, 2018; Maritzen and Haucke, 2018). In fission yeast, the balanced membrane trafficking is vital for regulating the antibiotic tolerance and the number of antibiotic-sensitive sterol-rich domains on the plasma membrane (Nishimura et al., 2014). We recently found that endocytosis happens preferentially at the rim of the division plane while exocytic vesicles are deposited evenly along the cleavage furrow during fission yeast cytokinesis, raising the question of how the temporal and spatial regulation of membrane trafficking affects cytokinesis (Wang et al., 2016).
In this study, we explored the roles of Ync13, an uncharacterized fission yeast protein from the UNC-13/Munc13 protein family, in cytokinesis. The UNC-13/Munc13 proteins often have a characteristic minimal Munc13 region (MUN domain) and various C1 and C2 domains (Pei et al., 2009; Li, Ma, et al., 2011). The MUN domain contains four subdomains (A, B, C, and D) and forms an elongated α-helix structure. The C-D region resembles some vesicle tethering factors such as the exocyst subunit Sec6 both in structure and sequence (Pei et al., 2009; Li, Ma, et al., 2011). The full-length (FL) MUN domain opens the closed conformation of the syntaxin-Munc18 complex and promotes SNARE complex formation in a Ca2+-dependent manner during the priming stage of fast neurotransmitter release (Guan et al., 2008; Ma, Li, et al., 2011; Yang, Wang, Sheng, et al., 2015; Wang, Choi, Gong, et al., 2017). C1 and C2 domains interact with lipids, Ca2+, or other binding partners to facilitate MUN domain functions by tethering vesicles to the plasma membrane (Shen et al., 2005; Lu et al., 2006; Dimova et al., 2009; Shin, Lu, Rhee, et al., 2010; Herbst et al., 2014; Liu, Seven, et al., 2016a; Xu, Camacho, et al., 2017). Additionally, Munc13 helps the proper syntaxin/synaptosomal associated protein (SNAP)-25 subconfiguration during SNARE assembly (Lai, Choi, et al., 2017). The Caenorhabditis elegans UNC-13, the founding member of the family, is recruited by RHO-1 to the presynaptic active zone and is crucial for the priming of both fast and slow transmitter release in neuronal cells (Maruyama et al., 2001; Madison et al., 2005; McMullan et al., 2006; Hu et al., 2013; Zhou et al., 2013). Besides their vital roles in neuronal cells, the mammalian Munc13s also contribute to exocytosis in other cell types (Brose et al., 1995; Betz et al., 1996; Ma, Li, et al., 2011; Yang, Wang, Sheng, et al., 2015). Munc13-1 mediates the priming for secretory amyloid precursor protein processing in brain and insulin secretion in pancreatic cells (Sheu, Pasyk, Ji, Haung, et al., 2003; Rossner et al., 2004; Kwan, Xie, et al., 2007; Hartlage-Rubsamen et al., 2013). Munc13-3 is expressed in the visual cortex for neuronal plasticity besides its function in the cerebellum (Yang et al., 2002; Yang, Kiser, et al., 2007; Netrakanti et al., 2015). Munc13-4, an effector of Rab27 GTPase, promotes the exocytosis of lytic granules in cytotoxic T-cells and neutrophils, secretory lysosomes in hematopoietic cells, and dense granules in platelets and mast cells (Brzezinska et al., 2008; Chang, Jaffray, et al., 2011; Elstak et al., 2011; Johnson et al., 2011; Dudenhoffer-Pfeifer et al., 2013; Chicka et al., 2016; Hiejima, Shibata, et al., 2018; Rodarte et al., 2018). Recently, Munc13-4 has been found to affect late endosome maturation and interact with Rab11 for recycling endosome delivery (He et al., 2016; Johnson et al., 2016). However, UNC-13/Munc13 proteins have no reported functions in cytokinesis. The fungal members of the protein family are rarely studied. Intriguingly, the MUN domains in fungi are separated into two Munc13 homology domains (MHD1 and MHD2) by a C2 domain (Pei et al., 2009). Schizosaccharomyces pombe has two UNC-13/Munc13 homologues, Git1 and Ync13. Git1 is a critical component in the cAMP signaling pathway for glucose sensing (Kao et al., 2006). However, Ync13 does not share the function of Git1 in this signaling pathway (Kao et al., 2006). Thus, despite the homology, Ync13 may play a different role from Git1 in fission yeast.
Here we find that Ync13 is essential for late stages of cytokinesis in fission yeast. Ync13 localizes to the expanding plasma membrane including the division site. Deletion of Ync13 causes massive cell lysis during cell division due to defective septation. Ync13 plays overlapping roles with the vesicle tethers exocyst and the TRAPP-II complex during cytokinesis. Unexpectedly, Ync13 is important for endocytic site selection to ensure the normal localization and dynamics of cell-wall enzymes. Collectively, our study reveals an essential and novel function of an UNC-13/Munc13 protein in cytokinesis.
RESULTS
Ync13 localizes to the division site during cytokinesis
We identified Ync13 (yeast UNC-13/Munc13; SPAC11E3.02c) as a novel candidate for cytokinesis from previous genomewide localization and deletion-phenotype studies (Matsuyama, Arai, Yashiroda, et al., 2006; Hayles, Wood, Jeffery, Hoe, Kim, Park, et al., 2013). Most members of the UNC-13/Munc13 family in animals contain several C2 domains and a characteristic MUN domain (Pei et al., 2009) (Supplemental Figure S1A). As a fungal member, Ync13 has only one C2 domain, which separates the MUN domain into MHD1 and MHD2 (Figure 1A).
FIGURE 1:
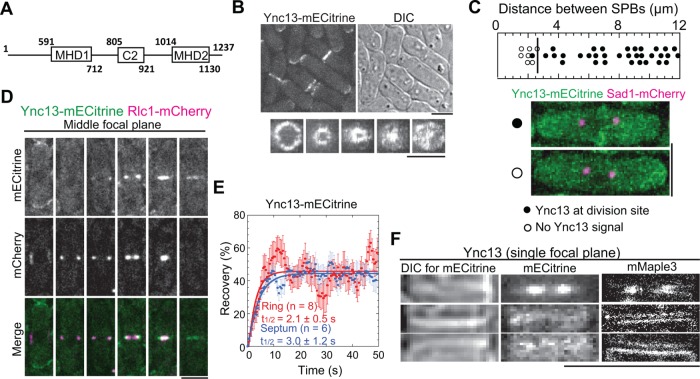
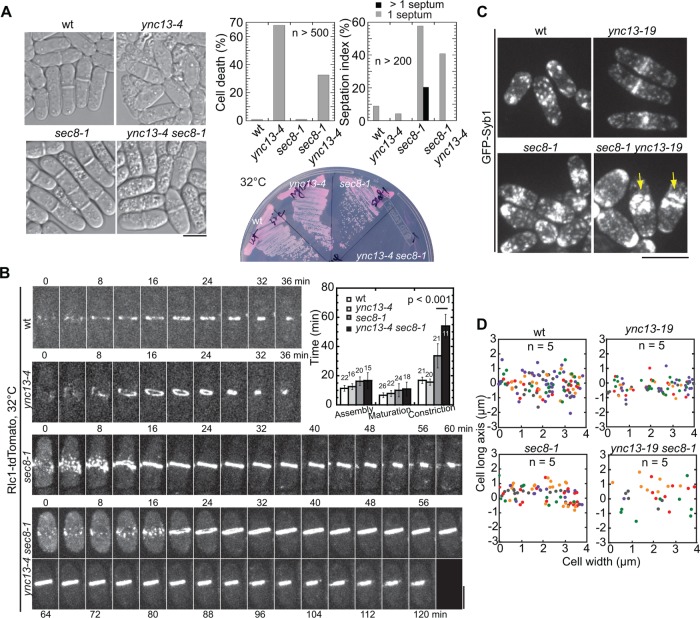
Ync13 localizes to the division site. (A) The schematics of Ync13. (B) Localization of Ync13. Top, mECitrine and DIC images of cells expressing Ync13-mECitrine. Bottom, vertical views of division-site localization of Ync13 at different stages of cytokinesis. (C) Timing of Ync13 appearance at the division site (marked by the vertical line) relative to the distance between two spindle pole bodies (SPBs). Representative cells with (filled circles) or without (empty circles) Ync13 at division site are shown with Sad1-mCherry as the SPB marker. (D) Micrographs (middle focal plane) of Ync13 (green) localization relative to the contractile ring marked with Rlc1 (magenta) during cytokinesis. (E) Dynamics of Ync13 at the contractile ring (red) and septum (blue) in FRAP assays. (F) Comparison of Ync13 localization in confocal microscopy (middle) and superresolution PALM (right) at the division site during ring constriction (top) and septum maturation (between the end of ring constriction and initiation of cell separation, during which the primary septum is closed and the secondary septa are still being built) stages (middle and bottom panels). DIC (left) is used to judge the stages of septum. Bars, 5 µm.
We first examined the cellular localization of Ync13 expressed at the endogenous level. Ync13-mECitrine localized to the cell tips during interphase and to the cell-division site at early anaphase B, when the spindle pole bodies were ∼2.6 µm apart (Figure 1, B and C). Ync13 partially colocalized with the myosin-II light chain Rlc1 labeled contractile ring and followed ring constriction before distributing along the division plane (Figure 1, B and D). It was highly dynamic at the division site as it recovered with halftimes of 2.1 ± 0.5 s and 3.0 ± 1.2 s at the contractile ring and septation site, respectively, in fluorescence recovery after photobleaching (FRAP) assays (Figure 1E).
To dissect Ync13 localization with higher spatial resolution, we tagged it with mMaple3 for photoactivated localization microscopy (PALM). Ync13-mMaple3 localized all over the cleavage furrow with higher concentration at the leading edge during ring constriction and displayed double layer structures during septum maturation (from the end of ring constriction to the initiation of daughter-cell separation; Figure 1F). These data suggested that Ync13 localizes to the plasma membrane and concentrates at the leading edge of the cleavage furrow during cytokinesis.
Ync13 depends on the cues from the contractile ring for localization to the division site and interacts with membrane lipids
To figure out how Ync13 localizes to the division site, we examined its localization in various mutants or under drug treatments. Ync13 did not require actin filaments, microtubules, or membrane trafficking for localization or maintenance of the localization (Supplemental Figure S1, B–E). We next tested whether the contractile ring provides the cues for Ync13 localization by examining Ync13 localization in mutants of key ring components Cdc15, Myo2, and Cdc12. F-BAR protein Cdc15 binds to the plasma membrane using its F-BAR domain and is one of the key components that bridge the contractile ring and the plasma membrane (Fankhauser et al., 1995; Takeda et al., 2004; McDonald et al., 2015; Ren et al., 2015). In cdc15-140 mutant, cells can still assemble contractile rings at the elevated temperatures. However, the rings are unstable and cause cytokinesis failure (Wachtler et al., 2006; Arasada and Pollard, 2014). At 36°C, Ync13-mECitrine appeared in additional cytoplasmic puncta (maybe some of the fluorescent proteins aggregated) besides normal localization to cell tips and the division site as in wt cells at 25°C (Supplemental Figure S1F). Because no morphological defects were found in Ync13-mECitrine–expressing cells, it is suggested that Ync13-mECitrine was still functional at 36°C. However, Ync13 failed to localize to the division site in cdc15-140 cells during anaphase B (inferred from the positions of nucleus; Supplemental Figure S1F, arrows). In contrast, Ync13 still localized to the division site as aberrant strings in myosin-II mutant myo2-E1 or foci in formin mutants cdc12-112 and cdc12-299 at the restrictive temperatures (unpublished data). Thus, Cdc15 is important for Ync13 localization. The fission yeast Its3 encodes a PI(4)P-5-kinase, which produces PI(4,5)P2, an essential lipid component at the division site with important biological functions (Zhang et al., 2000; Deng et al., 2005). We found that Ync13 localization to the plasma membrane was significantly reduced in its3-1 mutant, suggesting that Ync13 may require lipid binding for its division site localization (Supplemental Figure S1G, arrows).
We used truncation analysis to dissect Ync13 domains required for localization (Supplemental Figure S2). Unexpectedly, cells with any of the four truncations we made were inviable on rich medium YE5S but viable on YE5S + sorbitol or minimal medium EMM5S (Supplemental Figure S2A). Interestingly, Ync13 N terminal (Ync13[591-1237]) or C terminal (Ync13[1-1013], [1-804], and [1-590]) truncations still localized to the division site, although with varied intensity (Supplemental Figure S2, B–E). Moreover, after ring constriction, the truncated Ync13 was more concentrated at the center of the division plane compared with the FL Ync13 (Supplemental Figure S2B). The MHD2 domain (and aa 1131–1237) was critical for Ync13 stability or expression level as all C terminal truncations led to significant reduction in Ync13 global level (Supplemental Figure S2, C and D). The C2 domain was important for Ync13 localization as we found only ∼50 Ync13(1-804) molecules at the division site compared with ∼140 Ync13(1-1013) although their global levels were similar (Supplemental Figure S2, C and E). While truncating the N terminus of Ync13 had no strong effect on its global level (Supplemental Figure S2, C and D), only ∼50 Ync13(591-1237) molecules were at division site (Supplemental Figure S2E). Thus, these data suggest that the MHD2 domain contributes to Ync13 stability or expression level, while the C2 domain and the N terminal 1–590 aa are important for Ync13 localization to the division site.
The C2 domains in UNC-13/Munc13 proteins interact with lipids or protein partners (Lu et al., 2006; Shin, Lu, Rhee, et al., 2010). Consistently, purified Ync13 C2 and MHD1C2MHD2 (Supplemental Figure S3A) interacted with lipids. In lipid cofloatation assays, both fragments cofloated with liposomes that mimicked plasma membrane composition after density gradient centrifugations (Supplemental Figure S3B). Specifically, Ync13 interacted with PS and PIP2 (Kd of ∼82 µM and ∼3 µM, respectively) but not with PC and PE in liposome copelleting and protein–lipid overlay assays (Supplemental Figure S3, C and D). Thus, the C2 domain may contribute to Ync13 localization by its interaction with membrane lipids.
The major function of the UNC-13/Munc13 proteins is to help SNARE complex assembly and promote vesicle fusion (Rizo and Xu, 2015; Yang, Wang, Sheng, et al., 2015; Liu, Seven, et al., 2016a; Xu, Camacho, et al., 2017). We thus tested whether Ync13 promotes lipid and content mixing in reconstitution assays using the proteins involved in synaptic vesicle exocytosis, including the Sec1/Munc18 (SM) protein Munc18-1, the v-SNARE synaptobrevin, the t-SNAREs syntaxin-1 and SNAP-25, and the SNARE complex disassembly factors NSF and α-SNAP (Liu et al., 2017). As a positive control, a fragment spanning the C1, C2B, MUN, and C2C domains of Munc13-1 (C1C2BMUNC2C, Supplemental Figure S1A; simplified as Munc13-1 in Supplemental Figure S3, E and F) promoted efficient lipid and content mixing between T- and V-proteoliposomes on calcium stimulation. In contrast, neither C2 nor MHD1C2MHD2 (simplified as MHDC2 in Supplemental Figure S3, E–G) of Ync13 supported lipid or content mixing, even without NSF, α-SNAP, and Munc18-1. We next tested whether Ync13 can tether the liposomes. Although the Ync13 C2 and MHDC2 fragments interacted with T-liposomes (Supplemental Figure S3B), neither of them promoted liposome clustering to increase the particle size in dynamic light scattering (DLS) assays (Supplemental Figure S3G). These results contrast with those obtained for Munc13-1, which efficiently clusters T-liposomes (Liu, Seven, et al., 2016a). This activity is critical for Munc13-1 to stimulate liposome fusion (Yang, Wang, Sheng, et al., 2015). Although our results need to be interpreted with caution because the fusion assays were performed with neuronal proteins and Ync13 fragments, the finding that Ync13 C2 and MHDC2 cannot cluster liposomes suggests that Ync13 may not be involved in promoting membrane fusion, at least through a membrane bridging activity similar to that of Munc13-1.
Ync13 is essential for cell integrity
To explore Ync13 function in vivo, we examined the ync13∆ phenotype. ync13 is an essential gene (Hayles, Wood, Jeffery, Hoe, Kim, Park, et al., 2013), so we deleted one copy of ync13 gene from a diploid strain. Germinating spores on YE5S medium confirmed that ync13 is indeed essential for cell survival (Figure 2A, left). ync13∆ cells could grow and divide for ∼7 cell cycles (n = 37) before most, if not all, cells lysed (Figure 2A). Interestingly, sorbitol rescued ync13∆ cells for growth and colony formation with drastically reduced cell lysis (Figure 2B). Moreover, ync13∆ cells were also viable on EMM5S with ∼33% cell lysis (n > 500 cells; Supplemental Figure S2A). Despite cell lysis, the morphology (including cell shape, length, and width) of ync13∆ cells was similar to wt cells in both rich and minimal medium. Thus, we cultured ync13∆ cells using YE5S medium with sorbitol or EMM5S for the rest of the experiments.
FIGURE 2:
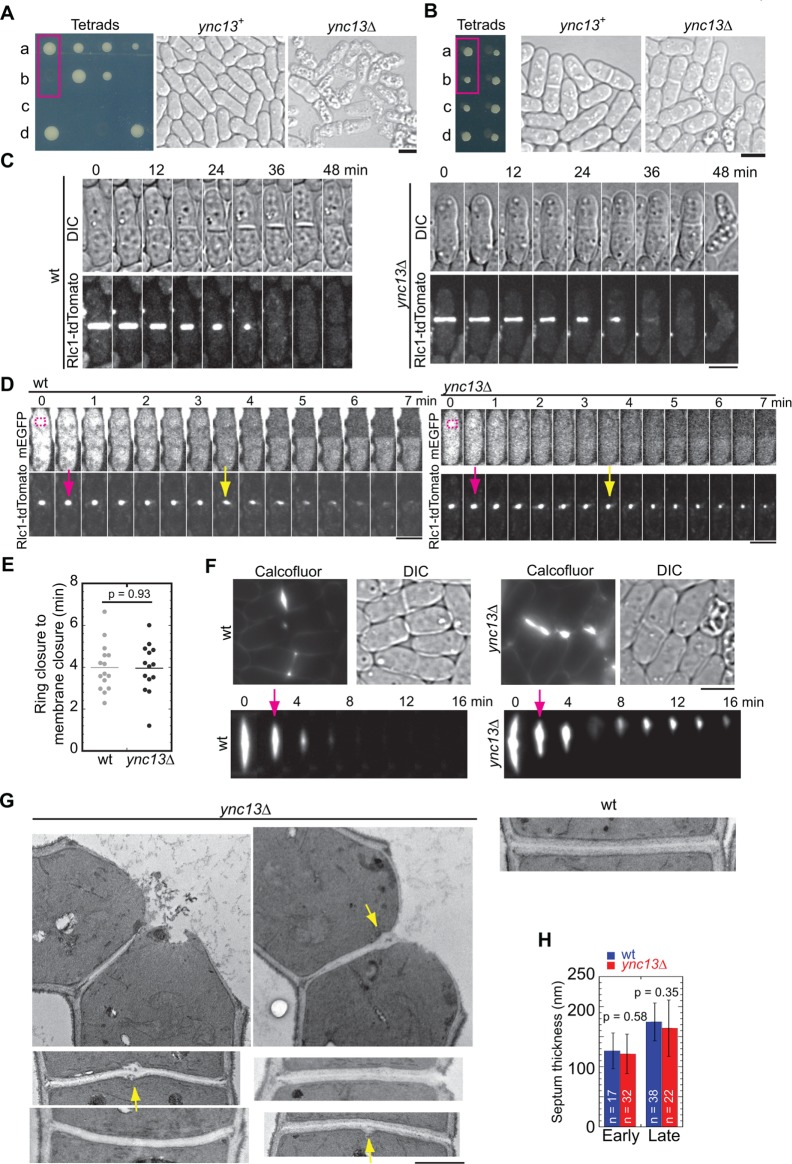
ync13∆ is lethal due to cell lysis. (A, B) Tetrad analyses of ync13∆/ync13+ diploid cells that were sporulated and dissected to positions a, b, c, and d on YE5S (A) or YE5S + 1.2 M sorbitol medium (B). DIC images of wt and ync13∆ cells from the boxed positions are shown on the right. (C) ync13∆ cells lyse during cell separation. Time courses of wt (left) and ync13∆ (right) cells labeled with Rlc1-tdTomato during cytokinesis. Cells were grown at log phase in YE5S + 1.2 M sorbitol and washed into YE5S 2 h before imaging, as described under Materials and Methods. Cells were imaged every minute and time 0 is the start of ring constriction. (D, E) Plasma membrane closure is normal in ync13∆ cells during cytokinesis in FLIP assays. The magenta boxes show bleached ROI (D). Time from the end of ring constriction (magenta arrows) to membrane closure (yellow arrows) is shown in E. (F) Micrographs (top) and time courses (bottom) showing Calcofluor staining of ync13∆ cells. Arrows mark the initiation of cell separation. (G) ync13∆ cells form bulges at septal center. EM images of the division site of wt and ync13∆ cells are shown. The arrows point out the bulges on the septa. (H) Quantification of septum thickness for cells with closed septa. Septa are grouped into early (no clear three layers) and late (clear three layers) stages. Bars, 5 µm for A–F, 0.5 µm for G.
To elucidate how ync13∆ causes cell death, we examined the contractile ring and cell separation in wt and ync13∆ cells. It took a similar amount of time for the contractile ring to assemble, mature, constrict, disappear, and for the daughter cells to initiate cell separation (Figure 2C and Supplemental Figure S4A). We also found no or weak synthetic interactions between mutations in ync13 and contractile ring components (Table 1), suggesting that the contractile ring machinery is not affected. However, ∼50% of ync13∆ cells lysed during cell separation under these growth conditions (Figure 2C and Supplemental Movie 1), which could result from defects in membrane closure or septation.
TABLE 1:
Genetic interactions between mutations in ync13 and others affecting cytokinesis and membrane trafficking.a
| Mutations | 25°C | 30°C | 32°C | 36°C |
|---|---|---|---|---|
| ync13-4 | +++b | +++ | ++c | +d |
| Contractile ring | ||||
| myo2-E1 | +++ | ++ | + | + |
| myo2-E1 ync13-4 | ++ | + | –e | – |
| cdc15-140 | +++ | + | – | – |
| cdc15-140 ync13-4 | +++ | + | – | – |
| imp2∆ | + | /f | / | / |
| imp2∆ ync13-4 | – | / | / | / |
| fic1∆ | +++ | +++ | ++ | ++ |
| fic1∆ ync13-4 | +++ | +++ | ++ | + |
| pxl1∆ | +++ | +++ | +++ | ++ |
| pxl1∆ ync13-4 | +++ | ++ | + | + |
| Glucan synthases and glucanases | ||||
| bgs1-191 | +++ | +++ | ++ | – |
| bgs1-191 ync13-4 | ++ | ++ | – | – |
| cwg1-2 | +++ | +++ | ++ | + |
| cwg1-2 ync13-4 | +++ | ++ | + | – |
| mok1-664 | +++ | +++ | + | – |
| mok1-664 ync13-4 | +++ | +++ | – | – |
| eng1∆ | +++ | +++ | +++ | +++ |
| eng1∆ ync13-4 | +++ | +++ | +++ | ++ |
| agn1∆ | +++ | +++ | +++ | +++ |
| agn1∆ ync13-4 | +++ | +++ | ++ | + |
| Rho GTPase-dependent CIP | ||||
| rho1-596 | +++ | / | / | / |
| rho1-596 ync13-4 | – | / | / | / |
| rho2∆ | +++ | +++ | +++ | +++ |
| rho2∆ ync13-4 | +++ | +++ | ++ | + |
| art1∆ | ++ | / | / | / |
| art1∆ ync13-4 | – | / | / | / |
| rga7∆ | ++ | / | / | / |
| rga7∆ ync13-4 | – | / | / | / |
| pck1∆ | +++ | +++ | +++ | ++ |
| pck1∆ ync13-4 | + | – | – | – |
| pck2∆ | +++ | +++ | ++ | ++ |
| pck2∆ ync13-4 | +++ | +++ | + | + |
| Exocytosis | ||||
| sec8-1 | +++ | ++ | ++ | – |
| ync13-19 | +++ | + | – | – |
| sec8-1 ync13-4 | ++ | + | – | – |
| sec8-1 ync13-19 | ++ | + | – | – |
| exo70∆ | +++ | +++ | +++ | ++ |
| exo70∆ ync13-4 | +++ | +++ | ++ | – |
| sec3-913 | +++ | +++ | ++ | + |
| sec3-913 ync13-4 | +++ | +++ | + | – |
| sec3-916 | +++ | +++ | ++ | – |
| sec3-916 ync13-4 | + | – | – | – |
| myo52∆ | +++ | ++ | ++ | + |
| myo52∆ ync13-4 | +++ | ++ | + | + |
| cdc42-1625 | +++ | / | / | / |
| cdc42-1625 ync13-4 | – | / | / | / |
| rho3∆ | +++ | +++ | +++ | +++ |
| rho3∆ ync13-4 | +++ | +++ | + | + |
| for3∆ | ++ | ++ | ++ | ++ |
| for3∆ ync13-19 | ++ | + | – | – |
| trs120-ts1 | +++ | +++ | +++ | – |
| trs120-ts1 ync13-4 | +++ | +++ | ++ | – |
| Endocytosis | ||||
| end4∆ | + | – | – | – |
| end4∆ ync13-4 | + | – | – | – |
| pan1∆ACV | +++ | +++ | +++ | +++ |
| pan1∆ACV ync13-4 | +++ | +++ | ++ | + |
| fim1∆ | +++ | +++ | +++ | ++ |
| fim1∆ ync13-4 | +++ | +++ | ++ | + |
| arp2-1 | +++ | +++ | ++ | + |
| arp2-1 ync13-4 | +++ | +++ | ++ | + |
| acp2∆ | +++ | +++ | +++ | +++ |
| acp2∆ ync13-4 | +++ | +++ | ++ | + |
| wsp1∆ | +++ | +++ | +++ | +++ |
| wsp1∆ ync13-4 | +++ | +++ | ++ | ++ |
| Other mutants | ||||
| its3-1 | +++ | ++ | + | – |
| its3-1 ync13-4 | +++ | ++ | + | – |
| rho4∆ | +++ | +++ | +++ | +++ |
| rho4∆ ync13-4 | +++ | +++ | +++ | ++ |
| Grown on EMM5S | ||||
| ync13∆ | ++ | ++ | ++ | ++ |
| lad1-1(rgf3) | +++ | +++ | ++ | ++ |
| lad1-1(rgf3) ync13∆ | ++ | + | – | – |
| Grown on YE5S | ||||
| ync13∆ | – | / | / | / |
| eng1∆ | +++ | / | / | / |
| eng1∆ ync13∆ | ++ | / | / | / |
| agn1∆ | +++ | / | / | / |
| agn1∆ ync13∆ | – | / | / | / |
aGrowth and color of colonies on YE5S + phloxin B plates except where noted at various temperatures.
b+++, similar to wt.
c++, some cell lysis or cytokinesis defects.
d+, massive cell lysis, severe cytokinesis defects with reduced growth.
e–, inviable.
f/, not tested.
Movie S1.
ync13Δ cells lyse during or after cell separation. Time lapse fluorescence microscopy (UltraVIEW Vox CSUX1 system; PerkinElmer) of wt (top) and ync13Δ (bottom) cells expressing Rlc1-tdTomato as the contractile ring marker. Images of Rlc1 (left) and DIC (right) are shown. Interval, 1 min. Photobleaching during image acquisition was not corrected. This video corresponds to Figure 2C. Display rate is 8 fps. Bar, 5 μm.
We first investigated whether ync13∆ cells are defective in membrane closure. Fluorescence loss in photobleaching (FLIP) assays were performed in wt and ync13∆ cells expressing free monomeric enhanced green fluorescent protein (mEGFP) driven by the strong promoter P3nmt1 (Maundrell, 1990; Moreno et al., 2000). We chose cells close to the end of ring constriction as is indicated by Rlc1 marker for photobleaching. As we bleached the mEGFP signal (every 30 s) in one daughter cell (Figure 2D, dashed boxes), we monitored the decrease of mEGFP intensity in the other daughter cell due to diffusion of mEGFP between them. It took both wt and ync13∆ cells ∼4 min to close the plasma membrane (yellow arrows) and stop mEGFP exchange after the end of contractile ring constriction (Figure 2, D and E). Thus, membrane closure appears normal, which is confirmed using t-SNARE Psy1 to label the plasma membrane in time-lapse microscopy (Supplemental Figure S4B and Supplemental Movie 2).
Movie S2.
Plasma membrane closure in ync13Δ cells. Wt (left) and ync13Δ (right) cells expressing GFP-Psy1 (green) and Rlc1-tdTomato (magenta) are shown. Interval, 1 min. This video corresponds to Figure S4B. Display rate is 8 fps. Bar, 5 μm.
We next compared the primary septum in wt and ync13∆ cells by Calcofluor staining (Figure 2F and Supplemental Movie 3). Interestingly, while cell-wall materials from the primary septum disappeared quickly after cell separation in wt (Figure 2F, left), Calcofluor signal accumulated at the division site in ync13∆ cells (Figure 2F, montage). It is possible that the two daughter cells remained connected to each other by undigested primary septum after cell separation (Supplemental Movie 3). Under electron microscopy (EM), although we found no significant differences in septum thickness during early and late septation stages in ync13∆ cells compared with wt cells (Figure 2H), cell-wall breakage at or near the center of the septum was often observed (Figure 2G and Supplemental Figure S5, A and C). Intriguingly, we saw a bulge at the center of the septum in EM thin sections in some images (Figure 2G, arrows), which could correspond to the strongly Calcofluor stained materials (Figure 2F). To further examine the central septum structure, we attempted serial EM thin sections (100-nm spacing) to capture the “middle plane” of septum. Of 11 ync13∆ cells with closed septa, we observed similar bulge structures in five cells (Supplemental Figure S5A). This could still be an underestimation as we are not sure whether the sepal center was captured in the serial thin sections in the other six cells. Thus, ync13∆ causes uneven distribution of cell-wall materials, which may lead to cell lysis during cell separation. These data supported that Ync13 is essential for cell integrity.
Movie S3.
Calcofluor staining of ync13Δ cells during cytokinesis. Calcofluor stained wt (left) and ync13Δ (right) cells were imaged in time lapse microscopy with 1 min interval. The video corresponds to Figure 2F. Display rate is 8 fps.
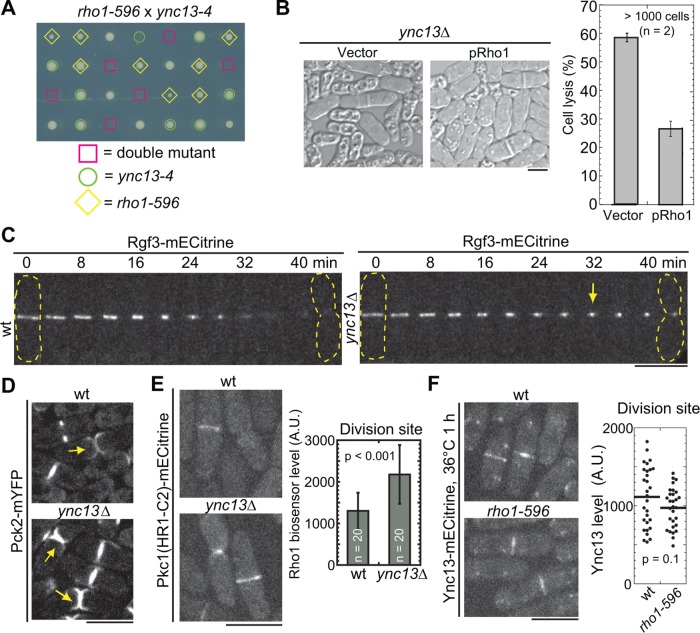
The CIP in yeasts regulates cell-wall formation and integrity (Levin et al., 1994; Garcia et al., 2006; Levin, 2011; Sanchez-Mir et al., 2014). Rho1 GTPase and its downstream effector, protein kinase C (PKC), activate cell-wall synthases for septum synthesis (Levin et al., 1994; Nonaka et al., 1995; Tajadura et al., 2004; Sanchez-Mir et al., 2014). To explore the relationship between Ync13 and Rho1-dependent CIP, we generated ync13 temperature-sensitive mutants, ync13-4 and ync13-19, by marker reconstitution mutagenesis (Tang et al., 2011). ync13-4 caused ∼2% at 25°C but ∼55% cell lysis at 36°C for 6 h (Supplemental Figure S4C), while the ync13-19 could not survive above 32°C (Table 1). We found that ync13-4 was synthetic lethal with the Rho1 temperature-sensitive mutant, rho1-596 (Viana, Pinar, et al., 2013) at 25°C (Figure 3A). In addition, ync13-4 had strong or lethal interactions with mutations in other putative components of the CIP including arrestin Art1 (Davidson et al., 2015), PKC Pck1 (Arellano et al., 1999), and GTPase activating protein Rga7 (Martin-Garcia et al., 2014) (Table 1). In reverse, Rho1 overexpression dramatically reduced cell lysis in ync13∆ cells (Figure 3B).
FIGURE 3:
Ync13 coordinates with Rho1 GTPase-dependent CIP during cytokinesis. (A) Synthetic lethality between ync13-4 and rho1-596. The four spores from each tetrad were dissected to the same column on YE5S plate and grown at 25°C. The genotype of each colony except wt cells is indicated. (B) Rho1 overexpression rescues cell lysis in ync13∆ cells. ync13∆ cells with pUR19-Rho1 or pUR19 empty vector were grown in EMM5S-uracil and then washed into YE5S for 4 h before imaging. (C, D) Rgf3 and Pck2 localization in ync13∆ cells. The dashed lines mark cell boundary and arrows mark the accumulated Rgf3 or Pck2. (C) Montages of wt and ync13∆ cells with time 0 as the start of ring constriction. (E) Localization and level (at the division site during septum maturation) of the Rho1 biosensor in ync13∆ cells. Cells were grown in YE5S + 1.2 M sorbitol and washed into YE5S for 2 h before imaging. (F) Ync13 localization in rho1-596 after shifting to 36°C for 1 h. Right, quantifications of Ync13 levels at the division site during septum maturation. Bars, 5 µm.
To test how Ync13 affects the CIP, we examined the localizations of the pathway components in ync13∆ cells. Interestingly, both the Rho1 activator Rho GEF Rgf3 and the effector PKC Pck2 were more concentrated at the center of the division site in ync13∆ cells (Figure 3, C and D). In addition, the active Rho1 level in ync13∆ cells as visualized by the Rho1 biosensor (budding yeast Pkc1[HR1-C2] domain; Denis and Cyert, 2005; Davidson et al., 2015) was elevated (Figure 3E). In contrast, Ync13 localization and level at the division site was not obviously affected in rho1 mutant (Figure 3F). Thus, loss of Ync13 likely activates CIP. Together, our data suggest that Ync13 coordinates with CIP for cell integrity during cytokinesis.
Roles of Ync13 in localization of the cell-wall enzymes glucan synthases and glucanase
Because ync13∆ cells have septum defects and Ync13 is important for cell integrity, we studied how Ync13 affected septum formation and degradation. The septum is a three-layer structure composed of mainly β- and α-glucan synthesized by the glucan synthases Bgs1(Cps1), Bgs4(Cwg1), and Ags1(Mok1) during cytokinesis (Ribas et al., 1991; Ishiguro et al., 1997; Katayama et al., 1999; Liu et al., 1999; Cortés et al., 2005, 2016; Perez et al., 2016). We first tested the genetic interactions between ync13 and cell-wall synthase mutants (Table 1). ync13-4 showed moderate synthetic interactions with β-glucan synthase mutants bgs1-191 and cwg1(bgs4)-2 but strong interactions with α-glucan synthase ags1 mutant mok1-664 (Supplemental Figure S4D). These data suggest that Ync13 works together with glucan synthases for cell-wall synthesis.
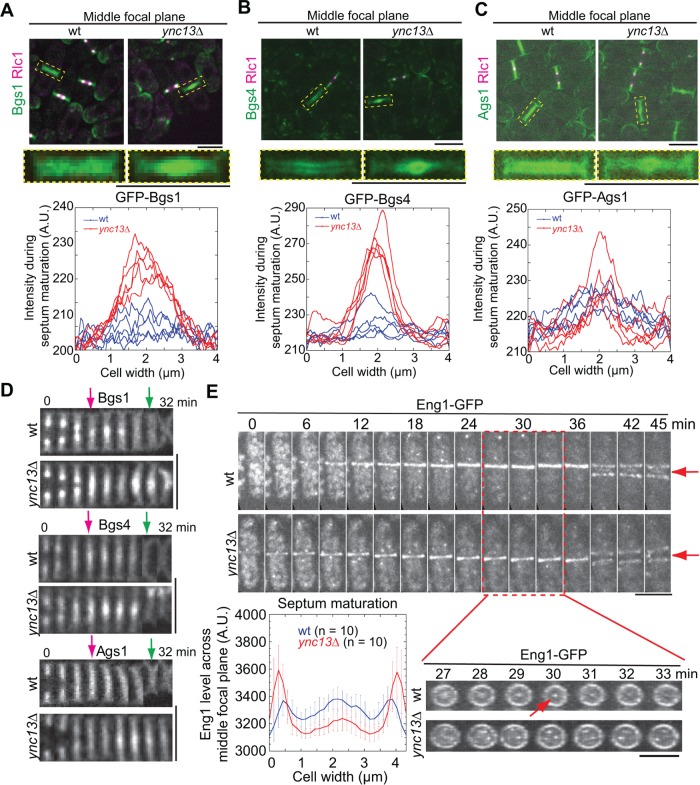
We next examined the expression and localization of the glucan synthases in ync13∆ cells. Bgs4 and Ags1 had higher global protein levels in ync13∆ cells while the Bgs1 level was similar to wt (Supplemental Figure S4E). In wt cells, all three enzymes distributed almost evenly along the division plane during (except Bgs1) and after ring constriction (Figure 4, A–D and Supplemental Movie 4). In ync13∆ cells, however, they were more concentrated at the leading edge of the invaginating septa during ring constriction and continued to accumulate at the center of the division site during septum maturation, forming a flattened bubblelike structure in cells labeled with Bgs4 and Ags1 (Figure 4, A–D, and Supplemental Movie 4). These abnormal accumulations persisted even after cell separation (Figure 4D, green arrows). Together, these data suggest that ync13∆ leads to mislocalization of glucan synthases at the division plane.
FIGURE 4:
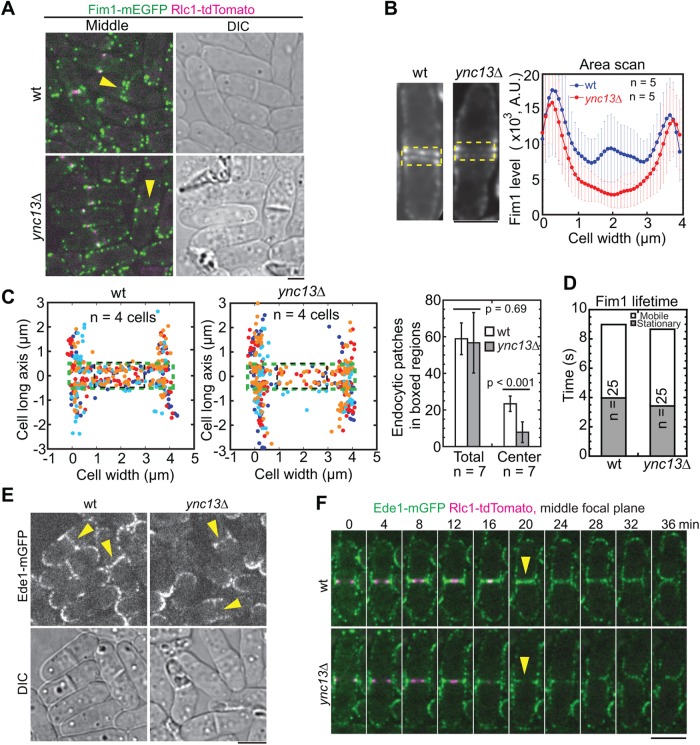
Ync13 regulates the distribution of cell-wall enzymes on the division plane. (A–C) Glucan synthases Bgs1 (A), Bgs4 (B), and Ags1 (C) are mislocalized in ync13∆ cells. Top, the middle focal plane of merged images; middle, enlarged images of localization of glucan synthases during septum maturation from representative cells in top panels (dashed boxes); bottom, area scans of Bgs1, Bgs4, and Ags1 intensity along the septum in five wt or ync13∆ cells during septum maturation. (D) Montages of glucan synthase distribution along the division plane (vertical) during late cytokinesis. Magenta arrows, the end of ring constriction; Green arrows, the initiation of cell separation. (E) Localization and distribution of glucanase Eng1 in ync13∆ cells. Top, time courses with time 0 as Eng1 ring appearance. Bottom right, vertical views of the cell from late stage of septation to just before cell separation (red box). Arrows mark the signal from the central spot in wt cells, which is missing in ync13∆ cells. Bottom left, quantification of Eng1 level across the division plane during septum maturation. Bars, 5 µm.
Movie S4.
Localization of glucan synthases Bgs1, Bgs4, and Ags1 in ync13Δ cells during cytokinesis. Wt (left) and ync13Δ (right) cells expressing both GFP tagged glucan synthases (green) and Rlc1-tdTomato (magenta) are shown. Interval, 1 min. This video corresponds to Figure 4, AC. Display rate is 8 fps. Bar, 5 μm.
The glucanases Eng1 and Agn1 digest primary septum and some adjacent cell wall on cell sides to trigger cell separation (Martin-Cuadrado et al., 2003; Dekker et al., 2004; Cortes et al., 2016; Perez et al., 2016). Deletion of β-glucanase eng1 rescued ync13∆ cells (Table 1). The ync13∆ eng1∆ cells survived in YE5S medium with only 6% cell lysis compared with 54% in ync13∆ cells (Supplemental Figure S4F). By contrast, deletion of α-glucanase agn1 failed to rescue ync13∆ cells (Table 1). We previously found that Eng1 localizes to a nonconstricting ring at the rim and a dot at the center of the division plane (Wang et al., 2015, 2016). In ync13∆ cells, Eng1 was abolished from the central dot and formed dispersed spots along the division plane, while the rim localization appeared normal but with higher intensity during septum maturation (Figure 4E). Together, these results indicate that Ync13 plays a role in the normal distribution of cell-wall enzymes glucan synthases and glucanase along the division plane.
Ync13 collaborates with the exocyst complex to mediate exocytosis at the division site
Exocytosis delivers cell-wall enzymes to the division site for septum formation and cell separation (Albertson et al., 2005; Wang et al., 2016). The exocyst is an octameric complex that tethers vesicles to the plasma membrane at the division site during cytokinesis (He and Guo, 2009; Giansanti et al., 2015). Failure in late stages of exocytosis or a defective exocyst causes significant vesicle accumulation and delayed cytokinesis (Wang, Tang, et al., 2002; Wang et al., 2016). We asked whether Ync13 regulates septum and cell-wall integrity through exocyst-mediated exocytosis. Unlike exocyst mutants and Ync13 animal homologues (Richmond et al., 1999), ync13∆ did not cause vesicle accumulation or delay in cytokinesis (Supplemental Figures S4A and S5, B and C), which is consistent with our in vitro reconstitution experiments that the Ync13 fragments could not support vesicle fusion and clustering (Supplemental Figure S3, E–G). However, ync13-4 had strong synthetic interactions with mutations in exocyst subunits Sec8, Sec3, and Exo70 (Figure 5A, Supplemental Figure S6A, and Table 1). At 36°C, sec8-1 cells were elongated and multiseptated while ync13-4 cells lysed. In contrast, ync13-4 sec8-1 cells seemed to halt the cell division or growth and appeared vacuolated, as 41% of cells had one septa and <1% cells were multiseptated (Figure 5A). Consistently, the contractile ring constricted significantly slower in ync13-4 sec8-1 cells while the ring assembly and maturation were similar to sec8-1 cells (Figure 5B).
FIGURE 5:
Cooperation between Ync13 and the exocyst during cytokinesis. (A) Synthetic interaction between mutations in ync13 and the exocyst subunit sec8. DIC images (left), and cell viability and septation index (top right) of cells grown for 4 h at 36°C; bottom right, growth test on YE5S + phloxin B at 32°C for 2 d. (B) Contractile ring constriction is delayed in ync13-4 sec8-1 cells. Time courses and quantification of cells imaged at 32°C after shifting to 32°C for 2 h. (C, D) Vesicle deposition rates to the division site is reduced in ync13-19 and sec8-1 ync13-19 cells during septum maturation. (C) Micrographs and (D) vesicle deposition sites along the septum (x-axis) in color-coded cells. the division plane for this and other vesicle tracking graphs is y = 0. Bars, 5 µm.
Normal ring constriction and plasma membrane invagination depend on exocytosis (Wang et al., 2016). We investigated whether the defects in ync13 sec8-1 cells are due to impaired exocytosis. We tracked vesicle delivery using the v-SNARE synaptobrevin Syb1 as marker in temperature-sensitive mutant ync13-19, which had similar synthetic interactions with sec8-1 but had fewer mutations than ync13-4 (Table 1). ync13-19 sec8-1 cells resembled ync13-4 sec8-1 in morphology (Figure 5, A and C). Syb1 accumulated close to the division site and cell tips in both sec8-1 and ync13-19 sec8-1 cells (Figure 5C, arrows). In addition, trackable exocytic vesicles delivered to the division site decreased in ync13-19 and ync13-19 sec8-1 cells compared with wt (Figure 5D and Supplemental Movie 5). Thus, exocytosis was impaired during cytokinesis in ync13-19 and ync13-19 sec8-1 cells, indicating a role of Ync13 in vesicle delivery to the division site. However, Ync13 did not affect the exocyst localization (Supplemental Figure S6B), suggesting that Ync13 and the exocyst complex function in parallel pathways to mediate exocytosis during cytokinesis.
Movie S5.
Vesicle tracking in ync13-19 sec8-1 cells during septum maturation. GFP-Syb1 was used as a vesicle marker for tracking vesicle movement in representative cells in continuous 2 min time-lapse movies. Tracked vesicle movements are shown in colored lines. We recorded the location of the vesicles in each frame and connected these locations to visualize the tracks of vesicles. The vesicles are present at or near the leading end of the tracks in each frame until they were delivered to the division plane. Time in min : sec. This video corresponds to Figure 5D. Display rate is 8 fps. Bar, 5 μm.
Does Ync13 regulate exocytosis mediated by the TRAPP-II complex?
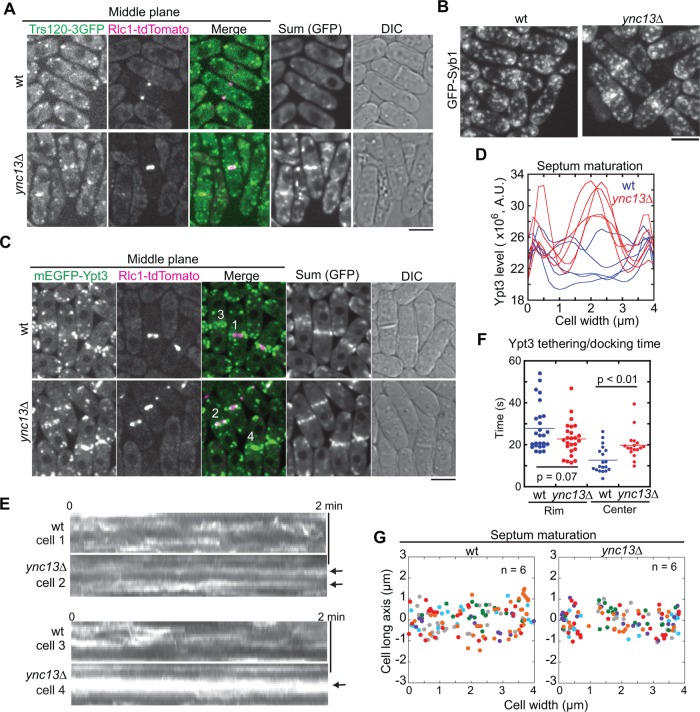
We recently reported that the TRAPP-II complex and the exocyst tether vesicles to different locations at the division site during fission yeast cytokinesis. The exocyst predominantly tethers vesicles at the rim and the TRAPP-II at the interior of the division site (Wang et al., 2016). We thus investigated how Ync13 affects TRAPP-II–mediated exocytosis. As reported (Wang et al., 2016), the TRAPP-II subunit Trs120 dynamically localizes to the division site during and after ring constriction (Figure 6A). In ync13∆ cells, however, Trs120 concentrated at the center of the division plane after ring constriction (Figure 6A). Syb1 and Rab11 GTPase Ypt3, two proteins that work with the TRAPP-II complex (Wang et al., 2016), had similar accumulation in ync13∆ and ync13-19 cells during septum maturation (Figures 5C and 6, B–D).
FIGURE 6:
Deletion of ync13 affects localization and dynamics of TRAPP-II complex and Rab11 GTPase Ypt3 at the division site during cytokinesis. (A) Trs120 accumulates at the center of division site in ync13∆ cells. The middle focal planes are shown with the sum projection (GFP channel) of a 2-min continuous movie and DIC. (B) Syb1 accumulates at the center of division plane in ync13∆ cells. (C–F) ync13∆ affects Ypt3 distribution (C, D), dynamics (E), and tethering/docking time (F). (C) The sum projection in GFP channel is from a 2-min continuous movie. (D) Area scans of Ypt3 distribution in the sum projection during septum maturation. (E) Kymographs of the division site (vertical) of the numbered cells in C during ring constriction (cells 1 and 2) or septum maturation (cells 3 and 4). The arrows mark stable Ypt3 at the leading edge (cell 2) or the center of septum (cell 4). (F) Ypt3 tethering/docking time at the center of division plane is lengthened in ync13∆ cells. The division plane was divided to the center half and rim half. (G) The final deposition site of Ypt3 along the division site during septum maturation. Bars, 5 µm.
We thus used Ypt3 as a marker to track the delivery and docking of vesicles to the division plane (Supplemental Movie 6). In wt cells, Ypt3 was highly dynamic both during and after ring constriction at the division site (Figure 6E, cells 1 and 3). In ync13∆ cells, however, Ypt3 signal was stable at the leading edge of the cleavage furrow (Figure 6E, cell 2) or at the center of division site after ring constriction (Figure 6E, cell 4). Consistently, the docking/tethering time of Ypt3 vesicles was significantly longer at the center, but not at the rim, of the division plane in ync13∆ than in wt cells (Figure 6F). The uneven distribution of Trs120 and Ypt3 at the division site suggested that ync13∆ affected TRAPP-II–mediated exocytosis at the interior of the division site. However, the delivery or docking sites of Ypt3 vesicles were not dramatically affected in ync13∆ cells during ring constriction and septum maturation and likely slightly biased toward the center and rim of the division site during septum maturation (Figure 6G and Supplemental Figure S6C). Together, these results suggest the Ync13 coordinates with the exocyst and TRAPP-II complexes in regulating exocytosis in S. pombe, although the mechanism is unknown.
Movie S6.
Vesicle tracking in ync13Δ cells during septum maturation. mEGFP-Ypt3 was used as a vesicle marker for tracking vesicle movement in representative wt (left) and ync13Δ (right) cells in continuous 2 min time-lapse movies. Tracked vesicle movements are shown in colored lines as in Movie 5. Time in min : sec. This video corresponds to Figure 6G. Display rate is 8 fps.
Ync13 is important for sites of endocytosis during cell division
The balance between exocytosis and endocytosis is critical for plasma membrane dynamics (Johansen et al., 2016; Xie et al., 2017). The CME is the major pathway for retrieving lipids and proteins from the plasma membrane (Schweitzer et al., 2005; Weinberg and Drubin, 2012; Goode et al., 2015). We previously reported that endocytosis occurs along the division site albeit with a preference at the rim (Wang et al., 2016). Our data presented so far indicate that Ync13 plays a role in protein distribution at the division site, but the defects in exocytosis cannot provide an obvious mechanism. We speculated that Ync13 may also affect protein distribution at the division site by regulating endocytosis. Thus, we asked whether endocytic sites are altered in ync13∆ cells.
We tracked the initiation sites of endocytic patches using the actin cross-linker fimbrin Fim1 (Figure 7, A–D, and Supplemental Movie 7). As reported (Wang et al., 2016), endocytic patches formed predominantly on the plasma membrane at the rim of the division site and adjacent cortex, with some patches along the division plane in wt cells. In contrast, patches at the center of the division site reduced significantly in ync13∆ cells although the number of patches across the entire division site and patch lifetimes were similar (Figure 7, A–D). Then we asked whether the locations of proteins involved in endocytic site selection are affected. The Eps15 protein Ede1 is one of the first coat proteins localized to the endocytic sites in the CME pathway (Kaksonen et al., 2005; Lu and Drubin, 2017). As expected, Ede1 localized to active growth sites such as the cell tips, division site, and its adjacent cortex in wt cells (Figure 7E). During cytokinesis, Ede1 followed the constricting ring and distributed evenly across the division plane during septum maturation (Figure 7F). However, Ede1 barely localized to the center of the division site after ring constriction in ync13∆ cells (Figure 7, E and F, arrowheads), suggesting that Ync13 plays a role in endocytic site selection. Consistently, the middle coat protein End4 and the late coat protein Pan1 displayed the same defects in ync13∆ cells (Supplemental Figure S6D). Thus, Ync13 plays a role in selection of endocytic sites and therefore regulates protein distribution at the division site during cytokinesis.
FIGURE 7:
Sites of endocytosis at the division plane are altered in ync13∆ cells. (A–D) Localization (A), intensity (B), tracking of initiation sites (C), and lifetimes (D) of fimbrin Fim1 at the division site in wt and ync13∆ cells during septum maturation. (A) The arrowheads indicate the loss of Fim1 at the center of division site in the middle focal plane in ync13∆ cells. (B) Sum projections of middle-focal images from 2-min continuous movies of representative cells. Fim1 intensity in the boxed regions was measured by area scans. (C) Initiation sites of endocytic patches along the division plane (left and middle) and quantification of the numbers of Fim1 patches in the boxed (green, total; black, center) regions (right). (D) Lifetime of Fim1 patches. (E, F) Micrographs (E) and time courses (F) of Ede1 localization in ync13∆ cells. The arrowheads indicate the loss of Ede1 at the center of division site in ync13∆ cells. Bars, 5 µm.
Movie S7.
Visualization of endocytic patches in ync13Δ cells. Wt (left) and ync13Δ (right) cells expressing Fim1-mEGFP Rlc1-tdTomato were used in the 2 min continuous time-lapse movie in green channel. Representative cells during septum maturation are shown. Time in min : sec. This video corresponds to Figure 7, B and C. Display rate is 8 fps.
DISCUSSION
In this study, we identified the UNC-13/Munc13 protein Ync13 as a novel regulator of membrane trafficking during cytokinesis (Figure 8). Ync13 coordinates with exocytosis and guides distribution of endocytic events at the division site, which regulates the localization and dynamics of cell-wall enzymes and CIP to ensure normal cell division.
FIGURE 8:
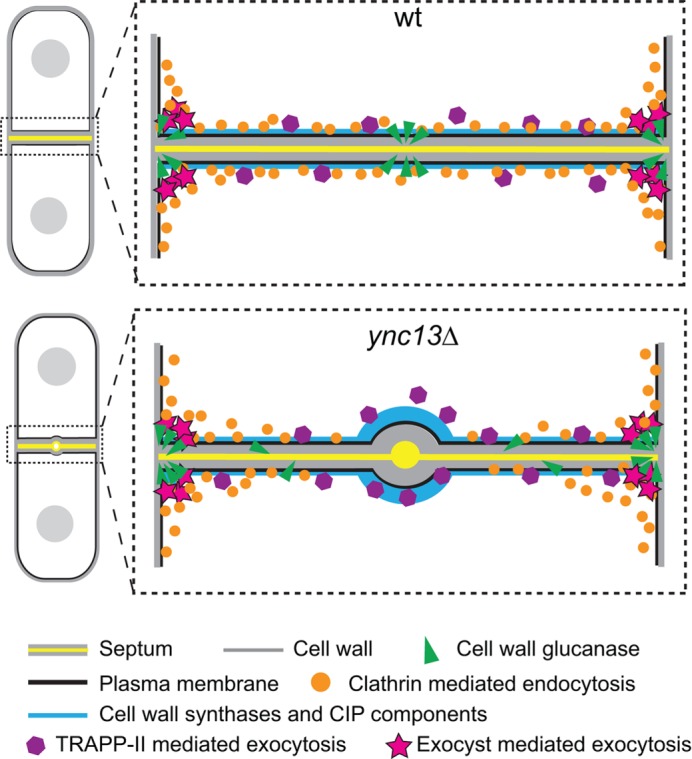
Models for roles of Ync13 in cytokinesis. Ync13 ensures proper distribution of cell-wall enzymes at the division site. In ync13∆ cells, the TRAPP-II–mediated exocytosis is defective. More importantly, the endocytosis at the center of the division site is drastically reduced without Ync13. As a result, the vesicle cargoes, glucan synthases and glucanase, are mislocalized. These lead to impaired septum synthesis, cell separation, and cell lysis.
A novel role of UNC-13/Munc13 protein family in cytokinesis
A main function of UNC-13/Munc13 proteins is to facilitate SNARE complex assembly by opening the closed syntaxin-1 conformation through their MUN domains during vesicle priming (Betz et al., 1996; Aravamudan et al., 1999; Yang, Wang, Sheng, et al., 2015). However, recent in vitro reconstitution and crystal structure have suggested that Munc13-1 can also bridge T and V liposomes (Liu, Seven, et al., 2016a; Xu, Camacho, et al., 2017). Such tethering brings vesicles and the target membrane in close proximity and helps MUN domain function. Despite the well-studied function of UNC-13/Munc13 proteins in vesicle fusion, no roles of this protein family in cytokinesis have been reported.
The fungi subfamily is distinguished from the rest of UNC-13/Munc13 proteins by its unique domain organization where the MUN domain is separated into MHD1 and MHD2 domains by a C2 domain (Pei et al., 2009). The MHD1C2MHD2 of Ync13 cannot promote membrane fusion in assays performed with neuronal SNAREs or liposome bridging in vitro (Supplemental Figure S3). However, whether the FL Ync13 assists SNARE complex assembly with the cognate yeast proteins is unknown. Nevertheless, ync13∆ cells do not accumulate vesicles at the division site during cytokinesis (Supplemental Figure S5, B and C), a phenotype commonly seen in exocyst or TRAPP-II mutants in yeasts or Munc13 deficient neurons (TerBush et al., 1996; Aravamudan et al., 1999; Guo et al., 1999a; Zhou et al., 2013; Wang et al., 2016). What is the exact role of Ync13 in membrane trafficking in fission yeast? In mammalian cells, SNARE proteins are usually restricted to specific cellular locations to fulfill their functions, as predicted by the SNARE hypothesis (Chen and Scheller, 2001). In S. pombe, SNARE proteins are only known to be involved in forespore formation during meiosis (Nakamura et al., 2005; Maeda et al., 2009; Yamaoka, Imada, et al., 2013). The plasma membrane targeting syntaxin, Psy1, localizes all over the cell cortex during vegetative growth (Maeda et al., 2009; Kashiwazaki et al., 2011), and the other t-SNARE Sec9 is not studied in detail. To direct vesicle delivery to active growth sites like the cell tips and division site, additional proteins like Ync13 may serve as receptors or landmarks. Future work is needed to explore the relationship between Ync13 and the SNARE complex in fission yeast. In addition, we found that Ync13 affects the localization and dynamics of Rab11 Ypt3 (Figure 6). The animal Munc13-4 binds to Rab11 and Rab27 GTPases to regulate vesicle trafficking and docking in neutrophil cells and lysosome secretion in hematopoietic cells (Neeft, Wieffer, de Jong, et al., 2005; Johnson et al., 2016). It is possible that Ync13 serves as an effector of Rab GTPases to spatially regulate membrane trafficking during cytokinesis. Alternatively, the main role of Ync13 may be in endocytosis (see below). Nevertheless, Ync13 is essential for fission yeast cytokinesis by regulating the localization and dynamics of key proteins during septum formation and daughter-cell separation.
Ync13 domains for its localization and functions
We found that Ync13 depends on F-BAR protein Cdc15 but not actin filaments in the contractile ring to localize to and maintain its localization at the division site (Supplemental Figure S1). Cdc15 is an important scaffold for late stage of cytokinesis and serves as a linker between the contractile ring and plasma membrane (Takeda et al., 2004; Arasada and Pollard, 2014; McDonald et al., 2015; Ren et al., 2015). Whether Ync13 localization depends on Cdc15 or any downstream targets of Cdc15 remains to be tested. The PI(4)P-5-kinase Its3 produces PI(4,5)P2, a plasma membrane component that played essential biological roles (Zhang et al., 2000; Echard, 2012). The Its3 temperature-sensitive mutant its3-1 cannot localize to the division site, and causes reduction in PI(4,5)P2 level and increase in septation index and defects in actin patch polarization (Zhang et al., 2000). We showed that Ync13 interacts strongly with PI(4,5)P2 in vitro, and its3-1 mutant greatly reduces Ync13 division site localization (Supplemental Figures S1G and S3). Since Ync13 does not depend on actin in the contractile ring for localization (Supplemental Figure S1, B and C), we conclude that Cdc15 and PI(4,5)P2 are important for Ync13 localization.
The MHD2 domain and the rest of C terminal sequence of Ync13 are vital for its functions (Supplemental Figure S2). This domain corresponds to MUN-CD region in Munc13-1, which contains multiple helix structures that share structural homology with the exocyst complex subunit Sec6 (Pei et al., 2009; Li, Ma, et al., 2011). MUN-CD, together with C1, C2B, and MUN-AB, form an elongated rod domain composed of mostly α-helical bundles, which interacts with both SNARE complex and lipids (Xu, Camacho, et al., 2017). Solving the structures of MHD domains will be helpful in understanding Ync13 functions.
We showed that the Ync13 C2 domain interacts with lipids and is important for Ync13 localization (Supplemental Figures S2 and S3). The N-terminal residues 1–590 of Ync13 are also critical for Ync13 localization and function. The N termini of UNC-13/Munc13 family proteins interact with various binding partners for localization (Lu et al., 2006; Kawabe et al., 2017). Munc13-1 interacts with itself through its C2A domain at its N terminus in an autoinhibitory status (Lu et al., 2006). Despite no predicted domain in Ync13(1-590), it still weakly localizes to the division site (Supplemental Figure S2, B and E). It is of great interest to identify the binding partners of Ync13 besides lipids, especially through its N terminus.
Roles of Ync13 in spatial regulation of cell-wall enzymes, CIP, and membrane trafficking during cytokinesis
Exocytosis and endocytosis are essential for cytokinesis as disruption of membrane trafficking by mutants or inhibitory drugs such as brefeldin A (BFA) causes slowed furrow ingression, furrow regression, and cytokinesis failure (Wu et al., 2006; Neto et al., 2013; Giansanti et al., 2015; Wang et al., 2016). By studying Ync13, we show, from another aspect, that how the spatial regulation of membrane trafficking affects cytokinesis.
In ync13 mutant cells, we observed abnormal accumulation of CIP components and cell-wall synthases Bgs1, Bgs4, and Ags1, which leads to defective septum formation and frequent bulge appearance at the septal center (Figures 2–4). However, the glucanase Eng1 becomes dispersed and absent from the center of division site while retaining a slightly higher concentration at the rim of division plane during septum maturation (Figure 4E). It is possible that the cell lysis in ync13 mutant cells results from a combination of malformed septum and the aberrant digestion of septum by Eng1. Because the septum thickness and the initiation timing of daughter-cell separation are normal, it is less likely that cell lysis in ync13 mutant cells is caused by premature cell separation.
The strong genetic interactions between ync13 and CIP mutants suggest that they both contribute to cell-wall integrity. Yet the relationship between the two is not fully established. Although Ync13 affects the localization of CIP and Rho1 activity at the division site (Figure 3), we failed to detect any interaction between Ync13 and CIP components. Thus, Ync13 could indirectly regulate CIP, possibly through its effect on membrane trafficking. Alternatively, CIP is up-regulated at the division site due to the defects in Ync13 cells. There are several possible reasons to explain the rescue of ync13∆ by Rho1 overexpression. The Rho GTPase-dependent CIP activates cell-wall synthases, and the extra activity may compensate for the loss of cell-wall synthases in the Rho1 overexpression cells (Figure 3B) (Arellano et al., 1996, 1999). In addition, Rho1/RhoA also coordinates actin filament nucleation, vesicle trafficking by exocyst complex, and CIE in budding yeast and animals (Imamura et al., 1997; Guo et al., 2001; Prosser et al., 2011; Prosser and Wendland, 2012; Jordan and Canman, 2012). All these processes can regulate membrane dynamics and make up for the loss of membrane dynamics at the division site, although these Rho1 functions have not yet been validated in fission yeast.
We previously proposed that the TRAPP-II complex and exocyst complex mediate vesicle delivery in independent pathways (Wang et al., 2016). Our study on Ync13 supported this idea. The deletion of Ync13 affects the localization of TRAPP-II components but not exocyst (Figure 6A and Supplemental Figure S6B). The TRAPP-II complex and Rab GTPase Ypt3 stably accumulate at the leading edge of the septum during the ring constriction and subsequently at the center of the division site during septum maturation in ync13∆ cells (Figure 6). We suggest that the absence of endocytosis at the leading edge/center of the septum disrupts, or at least slows down, the recycle of TRAPP-II complex and its regulators and cargoes. To test this idea, future experiments could be done by ectopically targeting endocytosis coat protein such as Ede1 to the center of division site in ync13∆ cells and examining whether the mislocalization and lysis phenotype are rescued.
The reduced endocytosis at the division site in ync13 mutant cells is intriguing. CME and CIE in fission yeast are not as systematically studied as their counterparts in budding yeast (Goode et al., 2015). Moreover, endocytosis during cytokinesis is rarely studied in either yeasts. It was reported that the deletion of clathrin light chain Clc1 blocks Bgs1 delivery to the cortex in S. pombe, which provides a direct link between endocytosis and cell-wall synthesis (de Leon et al., 2013). Why are endocytic sites mislocalized in ync13∆? Ync13 may not directly recruit CME as we did not detect interactions between Ync13 and the early coat protein Ede1 in immunoprecipitation (IP) assays or ectopic targeting assays using GFP binding protein (GBP). Exocytosis and endocytosis are often tightly linked. In neuronal cells, the coupled exo/endocytosis can be visualized by pH-sensitive GFP-tagged vesicular proteins (Hua et al., 2011). Inhibition of exocytosis by neurotoxin botulinum also blocks endocytosis (Yamashita et al., 2005). In budding yeast, the polarized exocytosis and endocytosis are coupled by Rab GTPase Sec4 (Johansen et al., 2016). Thus, the defects in endocytosis could instead be a response to the reduced exocytosis in ync13∆ cells. However, it is hard to explain why endocytic defects and excess accumulation of glucan synthases occur at the center of division site. Thus, we favor a model that Ync13 plays an active role in endocytosis. Further studies, especially identification of Ync13 binding partners, are needed to elucidate the exact roles of Ync13 in endocytosis.
In conclusion, we identified the fission yeast UNC-13/Munc13 protein Ync13 as an essential regulator of cytokinesis. Ync13 regulates exocytosis and endocytosis to ensure proper distribution of cell-wall enzymes and other regulators at the division site for successful cytokinesis. It will be interesting to explore whether other UNC-13/Munc13 proteins are also involved in endocytosis and cytokinesis.
MATERIALS AND METHODS
Strains and genetic, molecular, and cellular methods
Strains used in this study are listed in Supplemental Table S1. We constructed strains using PCR-based gene targeting and standard yeast genetic methods (Moreno et al., 1991; Bähler, Wu, et al., 1998). All tagged or truncated Ync13 are expressed from native chromosomal loci and regulated under endogenous promoters. The strains with Ync13 truncations were grown and selected on medium with 1.2 M sorbitol. Ync13 C-terminal truncations were constructed as previously described (Bähler, Wu, et al., 1998). For N-terminal truncations, we cloned -305 to +6 base pairs of ync13 into pFA6a-kanMX6-P3nmt1-mECitrine at BglII and PacI sites to replace the 3nmt1 promoter. The resulting plasmid (JQW745) was used as the template for PCR amplification and gene targeting. The C terminal tagged FL Ync13 was fully functional, revealed by growth tests on YE5S + phloxin B plates at various temperatures.
To delete ync13, a diploid strain rlc1-tdTomato-natMX6/rlc1-tdTomato-natMX6 ade6-210/ade6-216 leu1-32/leu1-32 ura4-18/ura4-18 was made by ade6 intragenic complementation (Kohli et al., 1977; Moreno et al., 1991). One copy of ync13 was then replaced by kanMX6 using primers immediately upstream and downstream of ync13 ORF (Bähler, Wu, et al., 1998). The resulting diploid strain YZ3-2 ync13∆::kanMX6/ync13+ rlc1-tdTomato-natMX6/rlc1-tdTomato-natMX6 ade6-210/ade6-216 leu1-32/leu1-32 ura4-18/ura4-18 was sporulated on a SPA5S plate, and tetrads were dissected onto YE5S rich medium or YE5S + 1.2 M sorbitol for spore germination or microscopy. The colonies grown on sorbitol medium were replica plated to selection medium YE5S + G418 + 1.2 M sorbitol to verify the ync13∆ genotype. For crosses involving ync13∆ strains, the parent strains were mixed on SPA5S + 1.2 M sorbitol, and the formed tetrads were dissected and germinated on YE5S + 1.2 M sorbitol.
To construct ync13 temperature-sensitive alleles, we used marker reconstitution mutagenesis method (Tang et al., 2011; Lee et al., 2014). A his5∆C-kanMX6 construct was inserted after the 3′UTR of ync13 to obtain strain JW5750 (h– ync13-his5ΔC-kanMX6 his5Δ ade6-M210 leu1-32 ura4). The ync13 FL gene with its 3′UTR was cloned onto pHis5C, which was then served as the template for error-prone PCR using Taq polymerase (NEB), 0.2 mM dNTPs, and mutagenesis cocktail (8 mM dTTP, 8 mM dCTP, 48 mM MgCl2, and 5 mM MnCl2). The PCR products were purified and transformed into JW5750. The transformants were selected on an EMM5S–histidine plate and then examined for temperature-sensitive growth (on YE5S + phloxin B) and phenotype at 36°C. The selected mutants were sequenced. The mutations in ync13-4 are E365G, I373T, K581E, M593L, L744S, I1013T, I1031V, and V1081E, and an “A” deletion 99 base pairs downstream of the stop codon; and the mutations in ync13-19 are L916H and W1048C.
For Calcofluor staining, 1 ml cell culture was washed with 1 ml EMM5S and 1 ml EMM5S + 5 µM n-propyl gallate and concentrated to 100 µl. One microliter of 1 mg/ml Calcofluor stock solution was added, and the samples were incubated in the dark for 1 min before imaging. For methyl benzimidazole-2-yl carbamate (MBC) treatment, 1 ml cells were incubated with 5 µl 5 mg/ml MBC or DMSO for 15 min before mounting on the gelatin pad with the same concentration of MBC or DMSO for imaging (Wu et al., 2011; Wang et al., 2016). For other drug treatments, cells were spotted on bare slide for imaging. The treatment conditions are as follows: latrunculin A, 100 µM for 10 min (Wu et al., 2001; Coffman et al., 2013); BFA, 50 µg/ml for 5 min (Wang et al., 2016); CK666, 100 µM for 5 min (Nolen, Tomasevic, et al., 2009; Laporte et al., 2012). The same volumes of corresponding solvents DMSO or ethanol were used as controls.
Confocal microscopy and image analyses
We grew cells from –80°C stocks on YE5S plates at 25°C for about 2 d and then grew them in YE5S liquid medium at log phase for ∼48 h at 25°C before imaging except where noted. For strains with ync13∆ and ync13 truncations, 1.2 M sorbitol was included in the growth medium and was washed out with fresh medium 2 h before imaging except where noted. Alternatively, some ync13∆ strains were grown in EMM5S without sorbitol for 48 h before imaging.
Microscopy samples were prepared largely as described (Zhu et al., 2013; Davidson et al., 2016). Briefly, cells (1 ml) were centrifuged at 3000–5000 rpm for 30 s, and washed once with 1 ml EMM5S and once with 1 ml EMM5S + 5 µM n-propyl gallate before being mounted on an EMM5S + 20% gelatin pad with 5 µM n-propyl gallate. For long movies, cells were washed and resuspended in 50 µl YE5S + 5 µM n-propyl gallate before spotting onto a coverglass-bottom dish (Delta TPG Dish; Biotechs Inc., Butler, PA). Then an agar pad cut from a YE5S plate was placed onto cells to immobilize them and provide nutrients. For Tetrad Fluorescence Microscopy (Coffman et al., 2013; Lee et al., 2014; Davidson et al., 2016), spores from tetrads were dissected onto a YE5S plate at 2.5 mm apart and were incubated at 25°C for 12–18 h. The YE5S agar containing cells from geminated spores was then incised from the plate and placed upside down onto a coverglass-bottom dish or a 24 × 60 mm coverglass. The top of agar piece was then covered by a piece of coverslip to slow drying. Air bubbles were removed by gently pressing on the coverslip before microscopy.
Cells were imaged at ∼23°C or at restrictive temperatures in a climate chamber (Stage Top Incubator INUB-PPZI2-F1 equipped with UNIV2-D35 dish holder; Tokai Hit, Shizuoka-ken, Japan) with 100×/1.4 numerical aperture (NA) Plan-Apo objectives. For observing cell morphology only, samples were imaged on a Nikon Eclipse Ti inverted microscope (Nikon, Melville, NY) equipped with a Nikon cooled digital camera DS-Ql1. For fluorescence imaging, cells were observed on a spinning disk confocal microscope (UltraVIEW ERS; Perkin Elmer Life and Analytical Sciences, Waltham, MA) with 440- and 568-nm solid -state lasers and 488- and 514-nm argon ion lasers and an ORCA-AG camera (Hamamatsu, Bridgewater, NJ) with 2 × 2 binning, or on a spinning disk confocal microscope (UltraVIEW Vox CSUX1 system; Perkin Elmer Life and Analytical Sciences, Waltham, MA) with 440-, 488-, 515-, and 561-nm solid-state lasers and a back-thinned electron-multiplying charge-coupled device (EMCCD) camera (Hamamatsu C9100-13, Bridgewater, NJ) without binning.
Images and data were collected and analyzed using Volocity (Perkin Elmer), UltraVIEW, and ImageJ software (Schneider et al., 2012). For measuring the levels of proteins across the division plane, the cells during septum maturation (judged by Rlc1 signal and/or differential interference contrast [DIC]; the stage between the end of ring constriction when Rlc1 signal reaches highest pixel intensity and the initiation of daughter-cell separation) were chosen and rotated so that the septa were horizontal. An 85 × 20 pixel region of interest (ROI) was drawn to cover the whole septum area, and the plot profile of the box was recorded. For Figure 7B, a 2× ROI of the previous ROI (elongated along the cell’s long axis) was used for calculating and subtracting background (Wu and Pollard, 2005; Zhu et al., 2013; Coffman and Wu, 2014). We tracked vesicles similarly as before (Wang et al., 2016). The movements of Syb1 or Ypt3 marked exocytic vesicles and the initiation sites of Fim1 labeled endocytic patches were tracked manually using ImageJ plug-in mTrackJ (Meijering et al., 2012). To track Syb1 vesicles, the GFP-Syb1 signal at the division site was bleached before taking a 2-min continuous movie. The locations of endocytic patch initiation sites were recorded after Fim1 reached maximum intensity and just before its internalization (Berro et al., 2010; Sirotkin et al., 2010). The coordinates of data were then transformed by Matlab software so that the septa were horizontal. The cell width was normalized to 4 µm before plotting.
To count protein molecules of Ync13 and its truncations, strains tagged with mECitrine were mixed with wt cells without fluorescence before imaging (Davidson et al., 2016). Cells were imaged at 0.5-µm spacing for 13 slices. The global mean intensity from the sum projection was measured and subtracted by that of wt cells as background. For local intensity, an ROI was drawn to measure the mean intensity at the division site. A 2× ROI was used to calculate and subtract background (Wu and Pollard, 2005; Zhu et al., 2013; Coffman and Wu, 2014). To obtain molecule numbers, the mean intensities of Ync13 and truncations were plotted on the standard curve generated with proteins (Gef2, Nod1, Rng8, and Rng9) with known molecule numbers (Zhu et al., 2013; Wang et al., 2014).
Micrographs shown in the figures are maximum intensity projections except where noted. The error bars in figures are ±1 SD. The statistical analyses were done using a two-tailed Student’s t test.
FRAP, photobleaching for vesicle tracking, and FLIP assays
We performed FRAP assays using the Photokinesis unit on UltraVIEW Vox CSUX1 confocal system as described (Coffman et al., 2009; Laporte et al., 2011; Zhu et al., 2013). Briefly, a single focal plane was chosen to perform FRAP. Selected ROIs were bleached to <50% of the original fluorescence intensity after four to five prebleach images were collected; 100 postbleach images with 1 s delay were collected. After correcting images for background and photobleaching during image acquisition at nonbleached sites, we normalized the prebleach intensity of the ROI to 100% and the intensity just after bleaching to 0%. Rolling average of every three consecutive postbleaching time points was used to plot and fit using an exponential equation y = m1 + m2 exp (–m3 * x), where m3 is the off-rate (KaleidaGraph; Synergy Software, PA). The half-time of recovery was calculated as t1/2 = (ln2)/m3.
To monitor the vesicle delivery, we bleached Syb1 signals at the division site before taking a 2-min continuous movie with a speed of 5 frame per second (fps) on a single focal plane as described previously (Wang et al., 2016).
The FLIP experiments were also performed on the Photokinesis unit (Liu et al., 2016b). Briefly, we selected cells expressing cytoplasmic mEGFP and Rlc1-tdTomato at the final stage of ring constriction for FLIP. A 2 × 2 pixel ROI in one of the daughter cells was photobleached using the 488-nm laser every 30 s. Recovery images were taken immediately before the next round of photobleaching. Ring closure was defined as when Rlc1 reached highest pixel intensity as a dot at the middle of the cleavage furrow; and plasma membrane closure was defined as when the cytoplasmic mEGFP stopped exchange between the two daughter cells. The time between ring closure and membrane closure was quantified.
Superresolution microscopy
Superresolution microscopy PALM was performed on an inverted microscopy (IX71; Olympus, Tokyo, Japan) with a 100 ×/1.49 NA oil immersion total internal reflection fluorescence objective and an EMCCD camera (iXon Ultra 897; AndorTechnologies, Belfast, United Kingdom) with EM gain of 255 and exposure time of 0.02 s (Liu et al., 2016b). We used 405- and 488-nm diode lasers (Vortran Laser Technology, Sacramento, CA) and a 561-nm diode-pumped solid-state laser (CrystaLaser, Reno, NV) for photoactivation and imaging. Sample preparation was the same as previously described (Liu et al., 2016b). After we selected an ROI by GFP channel, Ync13-mMaple3 was photoactivated by the 405-nm laser with increasing power for optimization of fluorophore activation efficiency, and images were taken using the 561-nm laser with a 593-/40-nm emission filter. DIC images were taken to monitor focus drift. Usually we took 4500–8000 images for each field. Superresolution images were reconstructed using Memp-STORM (Huang, Sun, et al., 2015).
Electron microscopy
EM was performed at the Campus Microscopy and Imaging facility at The Ohio State University (OSU) or the Boulder Electron Microscopy Services at the University of Colorado (Davidson et al., 2015; Liu et al., 2016b; Wang et al., 2016). For Figure 2G, we grew wt and ync13∆ cells in YE5S with 1.2 M sorbitol at log phase for 48 h. Cells were washed into YE5S for 2 h before fixation with 2.5% glutaraldehyde and 0.1 M sucrose in 0.1 M sodium phosphate, pH 7.4. The samples were then submitted to the imaging facility at OSU for further processing and imaging. For EM in Supplemental Figure S5 performed at Boulder, wt (strain JW81), sec8-1, and ync13∆ cells were grown in EMM5S at log phase for 48 h. Cells were then prepared and imaged as described previously (Giddings et al., 2001; Lee et al., 2014).
To quantify the septum thickness in EM images, cells with closed septa were chosen. For each cell, the thickness of the septum was averaged from three measurements at both the edge and the center of the septum. For ync13∆ cells, the bulged area was avoided. The data in Figure 2G were further categorized based on the visibility of the three-layer primary and secondary septum structure (early, no three-layer structure; late, with three-layer structure). To quantify the number of vesicles, we defined secretory vesicles as membrane enclosed circular structures with a diameter of 60–90 nm. Only vesicles within 500 nm from the septum were counted for each section.
Plasmid construction and protein purification
Ync13 FL cDNA amplified from a cDNA library or corresponding fragments were cloned into pQE80L vector at the BamHI and SalI restriction sites. The plasmids were then transformed into BL21 (DE3) pLysS cells. Protein expression was induced with 1 mM IPTG at 25°C for 6 h.
To purify FL Ync13, frozen cells from 1 l cell culture were homogenized in 50 ml Tris extraction buffer (20 mM Tris, 150 mM NaCl, 10% glycerol, 0.5% NP-40, 10 mM imidazole, 1 mM phenylmethane sulfonyl fluoride [PMSF], 10 mM β-ME, and protease inhibitor tablet [Roche], pH 9) and sonicated for 4× at output 9, 50% duty cycle, and 20 pulses. The cell lysate was then centrifuged at 25,000 rpm for 15 min and 38,000 rpm for 30 min, before incubaion with Talon Metal Affinity Resin (635501; Clontech, Mountain View, CA) at 4°C for 1 h. The mixture was then packed onto a column and washed with 50 ml washing buffer (20 mM Tris, 150 mM NaCl, 10% glycerol, 0.5% NP-40, 20 mM imidazole, 1 mM PMSF, and 10 mM β-ME, pH 9) per liter cell culture, and eluted with elution buffer (20 mM Tris, 150 mM NaCl, 10% glycerol, 0.1% NP-40, 200 mM imidazole, 1 mM PMSF, and 10 mM β-ME, pH 9). The eluted protein was dialyzed against HEPES (25 mM HEPES, 150 mM NaCl or KCl, 10% glycerol, 1 mM dithiothreitol [DTT], and 0.01% NaN3, pH 7.4) or TBS buffer (20 mM Tris-HCl, 150 mM NaCl, pH 7.5) for imidazole removal and future experiments.
Purification of Ync13 MHD1C2MHD2 (aa591–1130) and C2 (aa805–921) fragments were similar to those described before (Zhu et al., 2013). The phosphate buffer (50 mM sodium phosphate, 300 mM NaCl, 1 mM PMSF, 10 mM β-ME, and protease inhibitor tablet [Roche], pH 8) with various concentrations of imidazole (10 mM for extraction, 20 mM for washing, and 200 mM for elution) was used for purification. After purification, the C2 domain fragment was dialyzed into low salt buffer (50 mM sodium phosphate, 25 mM NaCl, 1 mM DTT, and 0.01% NaN3, pH 6.4) and further purified through MonoS 5/50 GL cation exchange column on AKTA Explorer 10 system (GE Healthcare). MHD1C2MHD2 domain was dialyzed into low salt buffer (50 mM sodium phosphate, 100 mM NaCl, 1 mM DTT, and 0.01% NaN3, pH 7.5) and loaded onto a MonoQ 5/50 GL anion exchange column on the same AKTA system. Both Ync13 fragments were then dialyzed against the TBS or HEPES buffer for experiments.
Protein–lipid overlay assays
Protein–lipid overlay assays were performed as described previously (Lee and Wu, 2012). Briefly, the PIP membrane strips (Invitrogen, Cat. no. P23751) were first blocked using TBS (20 mM Tris-HCl, 150 mM NaCl, pH 7.5) with 3% fatty-acid-free BSA (Sigma, cat. no. A7030) at 23°C for 1 h with shaking. Purified proteins were incubated with the membrane at a final concentration of 50 nM for 1 h at 23°C or overnight at 4°C. The membrane was then washed with TBST (20 mM Tris-HCl, 150 mM NaCl, 0.1% Tween 20, pH 7.5) to remove the unbound protein. Lipid bindings were examined by immunoblot with anti-His antibody (Clontech, cat. no. 631212; 1:20,000 dilution) as primary antibody and anti-mouse IgG as secondary antibody (1:5000 dilution) in TBS + 3% fatty-acid-free BSA.
Liposome cofloatation, clustering, and copelleting assays
Lipids from Avanti Lipids (Alabaster, AL) were used to make liposomes of single lipids, or to make T liposomes by mixing POPC 38%, DOPS 18%, POPE 20%, PIP2 2%, DAG 2%, and cholesterol 20% (Liu, Seven, et al., 2016a). We used rhodamine-PE to visualize the lipids. We dried the lipids in glass tubes by nitrogen gas and vacuum and resuspended lipids in HEPES buffer (25 mM HEPES, 150 mM NaCl or KCl, 10% glycerol, pH 7.4) to make a final lipid concentration of 5 mM. The suspension was then frozen/thawed in liquid nitrogen and a 42°C water bath 5× and extruded through a 0.1 µm filter 19×. All protein samples were dialyzed into the HEPES buffer before experiments.
For cofloatation assays, 1 mM T liposomes, and 1 µM final concentration of Munc13-1 C1C2BMUNC2C, Ync13 C2, or Ync13 MHD1C2MHD2 were mixed in HEPES buffer to a total volume of 165 µl and incubated at 23°C for 1 h. The samples were then mixed with an equal volume of 80% histodenz and added to the ultracentrifuge tube. A layer of 150 µl 35% and 150 µl 30% histodenz was then carefully loaded to the top of the samples to from a histodenz gradient (30%:35%:40%). HEPES buffer (50 µl) was then added to the top of the ultracentrifuge tubes and centrifuged at 48,000 rpm for 4 h at 4°C. The floating liposome samples were taken from the top of the gradient after ultracentrifuge and used to run SDS–PAGE gel with 1 µg of protein samples as reference.
For clustering assays, 1 µl of T liposomes or T liposomes mixed with 0.5 µM of Ync13 C2 or MHD1C2MHD2 fragments were loaded to a DynaPro instrument to measure dynamic light scattering (DLS) at 30°C (Liu, Seven, et al., 2016a).
For copelleting assays, 5 µM of Ync13 C2 fragment was mixed and incubated with a range of concentrations of PS, PIP2, or PC 80%/PE 20% liposomes in a total volume of 100 µl at 23°C for 1 h before centrifugation at 90,000 rpm for 30 min or 1 h at 4°C (Sun, Guan, et al., 2015). For PS and PIP2, supernatants (S) were carefully removed, and the pellets (P) were resuspended with the HEPES buffer with the same reaction volume. For PC/PE, the sample below the floating lipid layer was taken as soluble fraction (S), and the lipid (L) layer was resuspended. Samples from lipid and non-lipid fractions were run on SDS–PAGE gel and compared with the total input protein. The intensities of the C2 protein in supernatant (S) samples were used to calculate bound C2 and generate the curves for determining the Kd using the Michaelis–Menton equation y = m1 * x/(m2 + x), where Kd is m2.
Simultaneous lipid mixing and content mixing assays
The assays were performed similarly as previously described (Liu, Seven, et al., 2016a; Liu et al., 2017). Briefly, lipid mixtures for T liposomes (38% PC/18% PS/20% PE/20% Cholesterol/2% PIP2/2% DAG) and V liposomes (39% PC/19% PS/19% PE/20% Cholesterol/1.5% NBD-PE/1.5% MB-DHPE) were resuspended in the HEPES buffer by vortex and sonication. Purified syntaxin and SNAP25 proteins were mixed with the T liposome lipids, and synaptobrevin with the V liposome lipids in the presence of 1% octyl-β-glucoside (β-OG) to make T and V proteoliposomes. The detergent was then removed by dialysis in HEPES buffer, and the proteoliposomes were purified by lipid floatation using a histodenz gradient (0%:25%:35%).
For lipid mixing and content mixing assays, 100 µl T proteolipsomes (0.25 mM total lipid concentration) that had been incubated with Munc18-1, α-SNAP, and NSF (unless otherwise indicated) were mixed with 100 µl V proteoliposomes (0.125 mM total lipid concentration). Munc13-1 C1C2BMUNC2C, Ync13 C2, or Ync13 MHD1C2MHD2 was added to examine their ability to promote lipid mixing and content mixing. The experiments were performed at 30°C, and 0.5 mM final concentration of CaCl2 was added 5 min after the experiment started. We measured lipid mixing by the dequenching of Marina Blue labeled V liposomes (excitation 370 nm and emission 465 nm), and content mixing by the FRET between the PhycoE-Biotin wrapped in T liposomes and Cy5-Streptavidin wrapped in V liposomes (excitation 565 nm and emission 670 nm). Octyl-β-glucoside (1% wt/vol) was added to solubilize all liposomes and obtain the maximum lipid mixing signal as reference.
Supplementary Material
Acknowledgments
We thank Mohan Balasubramanian, Juan Carlos, Kathy Gould, Sophie Martin, Dan Mulvihill, Beatriz Santos, Shelley Sazer, Vladimir Sirotkin, and Takashi Toda for yeast strains; Jianjie Ma and Mingzhai Sun for PALM; the Campus Microscopy and Imaging facility at OSU and the Boulder Electron Microscopy Services at the University of Colorado for help with EM; the Anita Hopper, Joseph Krzycki, Dmitri Kudryashov, and Stephen Osmani laboratories for equipment; Victoria Esser in the Rizo lab for help with liposome cofloatation assays; and the current and former members of the Wu lab and rotation student Vedud Purde for helpful suggestions and technical supports. This study was supported by a Pelotonia Graduate Fellowship to Y.-H. Z., a Pelotonia Undergraduate Fellowship to J. H., and by National Institutes of Health grants R35 NS097333 (to J.R.) and R01 GM118746 (to J.-Q.W). This work is dedicated to James E. Hopper, a caring, generous, thoughtful, outstanding mentor and colleague.
Abbreviations used:
- BFA
brefeldin A
- CIP
cell integrity pathway
- CME
clathrin-mediated endocytosis
- DIC
differential interference contrast
- EMM5S
Edinburgh minimal medium plus five supplements
- FL
full length
- FLIP
fluorescence loss in photobleaching
- FRAP
fluorescence recovery after photobleaching
- MBC
methyl benzimidazole-2-yl carbamate
- mEGFP
monomeric enhanced green fluorescent protein
- MHD
Munc13 homology domain
- PALM
photoactivated localization microscopy
- ROI
region of interest
- SNARE
soluble NSF ( N-ethylmaleimide–sensitive factor) attachment protein receptor
- TRAPP-II
transport protein particle II
- wt
wild type
- YE5S
yeast extract plus five supplements.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-04-0225) on July 25, 2018.
REFERENCES
- Albertson R, Riggs B, Sullivan W. (2005). Membrane traffic: a driving force in cytokinesis. Trends Cell Biol , 92–101. [DOI] [PubMed] [Google Scholar]
- Arasada R, Pollard TD. (2014). Contractile ring stability in S. pombe depends on F-BAR protein Cdc15p and Bgs1p transport from the Golgi complex. Cell Rep , 1533–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravamudan B, Fergestad T, Davis WS, Rodesch CK, Broadie K. (1999). Drosophila Unc-13 is essential for synaptic transmission. Nat Neurosci , 965–971. [DOI] [PubMed] [Google Scholar]
- Arellano M, Durán A, Pérez P. (1996). Rho 1 GTPase activates the (1–3)β-D-glucan synthase and is involved in Schizosaccharomyces pombe morphogenesis. EMBO J , 4584–4591. [PMC free article] [PubMed] [Google Scholar]
- Arellano M, Valdivieso MH, Calonge TM, Coll PM, Durán A, Pérez P. (1999). Schizosaccharomyces pombe protein kinase C homologues, pck1p and pck2p, are targets of rho1p and rho2p and differentially regulate cell integrity. J Cell Sci , 3569–3578. [DOI] [PubMed] [Google Scholar]
- Bähler J, Wu J-Q, Longtine MS, Shah NG, McKenzie A, III, Steever AB, Wach A, Philippsen P, Pringle JR. (1998). Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe . Yeast , 943–951. [DOI] [PubMed] [Google Scholar]
- Baluska F, Menzel D, Barlow PW. (2006). Cytokinesis in plant and animal cells: endosomes “shut the door.” Dev Biol , 1–10. [DOI] [PubMed] [Google Scholar]
- Berro J, Sirotkin V, Pollard TD. (2010). Mathematical modeling of endocytic actin patch kinetics in fission yeast: disassembly requires release of actin filament fragments. Mol Biol Cell , 2905–2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betz A, Telemenakis I, Hofmann K, Brose N. (1996). Mammalian Unc-13 homologues as possible regulators of neurotransmitter release. Biochem Soc Trans , 661–666. [DOI] [PubMed] [Google Scholar]
- Boucrot E, Kirchhausen T. (2007). Endosomal recycling controls plasma membrane area during mitosis. Proc Natl Acad Sci USA , 7939–7944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brose N, Hofmann K, Hata Y, Sudhof TC. (1995). Mammalian homologues of Caenorhabditis elegans unc-13 gene define novel family of C2-domain proteins. J Biol Chem , 25273–25280. [DOI] [PubMed] [Google Scholar]
- Brzezinska AA, Johnson JL, Munafo DB, Crozat K, Beutler B, Kiosses WB, Ellis BA, Catz SD. (2008). The Rab27a effectors JFC1/Slp1 and Munc13–4 regulate exocytosis of neutrophil granules. Traffic , 2151–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TY, Jaffray J, Woda B, Newburger PE, Usmani GN. (2011). Hemophagocytic lymphohistiocytosis with MUNC13–4 gene mutation or reduced natural killer cell function prior to onset of childhood leukemia. Pediatr Blood Cancer , 856–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YA, Scheller RH. (2001). SNARE-mediated membrane fusion. Nat Rev Mol Cell Biol , 98–106. [DOI] [PubMed] [Google Scholar]
- Chicka MC, Ren Q, Richards D, Hellman LM, Zhang J, Fried MG, Whiteheart SW. (2016). Role of Munc13–4 as a Ca2+-dependent tether during platelet secretion. Biochem J , 627–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou CS, Moore TI, Chang SD, Nie Q, Yi TM. (2012). Signaling regulated endocytosis and exocytosis lead to mating pheromone concentration dependent morphologies in yeast. FEBS Lett , 4208–4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffman VC, Nile AH, Lee I-J, Liu HY, Wu J-Q. (2009). Roles of formin nodes and myosin motor activity in Mid1p-dependent contractile-ring assembly during fission yeast cytokinesis. Mol Biol Cell , 5195–5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffman VC, Sees JA, Kovar DR, Wu J-Q. (2013). The formins Cdc12 and For3 cooperate during contractile ring assembly in cytokinesis. J Cell Biol , 101–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffman VC, Wu J-Q. (2014). Every laboratory with a fluorescence microscope should consider counting molecules. Mol Biol Cell , 1545–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortés JC, Ramos M, Osumi M, Pérez P, Ribas JC. (2016). Fission yeast septation. Commun Integr Biol , e1189045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortés JCG, Carnero E, Ishiguro J, Sanchéz Y, Durán A, Ribas JC. (2005). The novel fission yeast (1,3)β-D-glucan synthase catalytic subunit Bgs4p is essential during both cytokinesis and polarized growth. J Cell Sci , 157–174. [DOI] [PubMed] [Google Scholar]
- Danilchik MV, Bedrick SD, Brown EE, Ray K. (2003). Furrow microtubules and localized exocytosis in cleaving Xenopus laevis embryos. J Cell Sci , 273–283. [DOI] [PubMed] [Google Scholar]
- Davidson R, Laporte D, Wu J-Q. (2015). Regulation of Rho-GEF Rgf3 by the arrestin Art1 in fission yeast cytokinesis. Mol Biol Cell , 453–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson R, Liu Y, Gerien KS, Wu J-Q. (2016). Real-time visualization and quantification of contractile ring proteins in single living cells. Methods Mol Biol , 9–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Avino PP, Giansanti MG, Petronczki M. (2015). Cytokinesis in animal cells. Cold Spring Harb Perspect Biol , a015834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leon N, Sharifmoghadam MR, Hoya M, Curto MA, Doncel C, Valdivieso MH. (2013). Regulation of cell wall synthesis by the clathrin light chain is essential for viability in Schizosaccharomyces pombe . PLoS One , e71510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker N, SpeI-Jer D, Grun CH, van den Berg M, de Haan A, Hochstenbach F. (2004). Role of the α-glucanase Agn1p in fission-yeast cell separation. Mol Biol Cell , 3903–3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Sugiura R, Ohta K, Tada K, Suzuki M, Hirata M, Nakamura S, Shuntoh H, Kuno T. (2005). Phosphatidylinositol-4-phosphate 5-kinase regulates fission yeast cell integrity through a phospholipase C-mediated protein kinase C-independent pathway. J Biol Chem , 27561–27568. [DOI] [PubMed] [Google Scholar]
- Denis V, Cyert MS. (2005). Molecular analysis reveals localization of Saccharomyces cerevisiae protein kinase C to sites of polarized growth and Pkc1p targeting to the nucleus and mitotic spindle. Eukaryot Cell , 36–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimova K, Kalkhof S, Pottratz I, Ihling C, Rodriguez-Castaneda F, Liepold T, Griesinger C, Brose N, Sinz A, Jahn O. (2009). Structural insights into the calmodulin-Munc13 interaction obtained by cross-linking and mass spectrometry. Biochemistry , 5908–5921. [DOI] [PubMed] [Google Scholar]
- Donovan KW, Bretscher A. (2012). Myosin-V is activated by binding secretory cargo and released in coordination with Rab/Exocyst function. Dev Cell , 769–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudenhoffer-Pfeifer M, Schirra C, Pattu V, Halimani M, Maier-Peuschel M, Marshall MR, Matti U, Becherer U, Dirks J, Jung M, et al. (2013). Different Munc13 isoforms function as priming factors in lytic granule release from murine cytotoxic T lymphocytes. Traffic , 798–809. [DOI] [PubMed] [Google Scholar]
- Echard A. (2008). Membrane traffic and polarization of lipid domains during cytokinesis. Biochem Soc Trans , 395–399. [DOI] [PubMed] [Google Scholar]
- Echard A. (2012). Phosphoinositides and cytokinesis: the “PIP” of the iceberg. Cytoskeleton , 893–912. [DOI] [PubMed] [Google Scholar]
- Elstak ED, Neeft M, Nehme NT, Voortman J, Cheung M, Goodarzifard M, Gerritsen HC, Henegouwen PMPVE, Callebaut I, de Saint Basile G, van der SluI-Js P. (2011). The munc13–4-rab27 complex is specifically required for tethering secretory lysosomes at the plasma membrane. Blood , 1570–1578. [DOI] [PubMed] [Google Scholar]
- Fankhauser C, Reymond A, Cerutti L, Utzig S, Hofmann K, Simanis V. (1995). The S. pombe cdc15 gene is a key element in the reorganization of F-actin at mitosis. Cell , 435–444. [DOI] [PubMed] [Google Scholar]
- Feng B, Schwarz H, Jesuthasan S. (2002). Furrow-specific endocytosis during cytokinesis of zebrafish blastomeres. Exp Cell Res , 14–20. [DOI] [PubMed] [Google Scholar]
- Feyder S, De Craene JO, Bar S, Bertazzi DL, Friant S. (2015). Membrane trafficking in the yeast Saccharomyces cerevisiae model. Int J Mol Sci , 1509–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García EP, Gatti E, Butler M, Burton J, De Camilli P. (1994). A rat brain Sec1 homologue related to Rop and UNC18 interacts with syntaxin. Proc Natl Acad Sci USA , 2003–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García P, Tajadura V, García I, Sanchéz Y. (2006). Role of Rho GTPases and Rho-GEFs in the regulation of cell shape and integrity in fission yeast. Yeast , 1031–1043. [DOI] [PubMed] [Google Scholar]
- Gerald NJ, Damer CK, O’Halloran TJ, De Lozanne A. (2001). Cytokinesis failure in clathrin-minus cells is caused by cleavage furrow instability. Cell Motil Cytoskeleton , 213–223. [DOI] [PubMed] [Google Scholar]
- Giansanti MG, Vanderleest TE, Jewett CE, Sechi S, Frappaolo A, Fabian L, Robinett CC, Brill JA, Loerke D, Fuller MT, Blankenship JT. (2015). Exocyst-dependent membrane addition is required for anaphase cell elongation and cytokinesis in Drosophila . PLoS Genet , e1005632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giddings TH, Jr, O’Toole ET, Morphew M, Mastronarde DN, McIntosh JR, Winey M. (2001). Using rapid freeze and freeze-substitution for the preparation of yeast cells for electron microscopy and three-dimensional analysis. Methods Cell Biol , 27–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode BL, Eskin JA, Wendland B. (2015). Actin and endocytosis in budding yeast. Genetics , 315–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant BD, Donaldson JG. (2009). Pathways and mechanisms of endocytic recycling. Nat Rev Mol Cell Biol , 597–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromley A, Yeaman C, Rosa J, Redick S, Chen CT, Mirabelle S, Guha M, Sillibourne J, Doxsey SJ. (2005). Centriolin anchoring of exocyst and SNARE complexes at the midbody is required for secretory-vesicle-mediated abscission. Cell , 75–87. [DOI] [PubMed] [Google Scholar]
- Guan R, Dai H, Rizo J. (2008). Binding of the Munc13-1 MUN domain to membrane-anchored SNARE complexes. Biochemistry , 1474–1481. [DOI] [PubMed] [Google Scholar]
- Guo W, Grant A, Novick P. (1999a). Exo84p is an exocyst protein essential for secretion. J Biol Chem , 23558–23564. [DOI] [PubMed] [Google Scholar]
- Guo W, Roth D, Walch-Solimena C, Novick P. (1999b). The exocyst is an effector for Sec4p, targeting secretory vesicles to sites of exocytosis. EMBO J , 1071–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Tamanoi F, Novick P. (2001). Spatial regulation of the exocyst complex by Rho1 GTPase. Nat Cell Biol , 353–360. [DOI] [PubMed] [Google Scholar]
- Hartlage-Rubsamen M, Waniek A, Rossner S. (2013). Munc13 genotype regulates secretory amyloid precursor protein processing via postsynaptic glutamate receptors. Int J Dev Neurosci , 36–45. [DOI] [PubMed] [Google Scholar]
- Hata Y, Slaughter CA, Südhof TC. (1993). Synaptic vesicle fusion complex contains unc-18 homologue bound to syntaxin. Nature , 347–351. [DOI] [PubMed] [Google Scholar]
- Hayles J, Wood V, Jeffery L, Hoe KL, Kim DU, Park HO, Salas-Pino S, Heichinger C, Nurse P. (2013). A genome-wide resource of cell cycle and cell shape genes of fission yeast. Open Biol , 130053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B, Guo W. (2009). The exocyst complex in polarized exocytosis. Curr Opin Cell Biol , 537–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Johnson JL, Monfregola J, Ramadass M, Pestonjamasp K, Napolitano G, Zhang J, Catz SD. (2016). Munc13-4 interacts with syntaxin 7 and regulates late endosomal maturation, endosomal signaling, and TLR9-initiated cellular responses. Mol Biol Cell , 572–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbst S, Lipstein N, Jahn O, Sinz A. (2014). Structural insights into calmodulin/Munc13 interaction. Biol Chem , 763–768. [DOI] [PubMed] [Google Scholar]
- Hiejima E, Shibata H, Yasumi T, Shimodera S, Hori M, Izawa K, Kawai T, Matsuoka M, Kojima Y, Ohara A, et al. (2018). Characterization of a large UNC13D gene duplication in a patient with familial hemophagocytic lymphohistiocytosis type 3. Clin Immunol , 63–66. [DOI] [PubMed] [Google Scholar]
- Hu Z, Tong XJ, Kaplan JM. (2013). UNC-13L, UNC-13S, and Tomosyn form a protein code for fast and slow neurotransmitter release in Caenorhabditis elegans . eLife , e00967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua , Leal-Ortiz S, Foss SM, Waites CL, Garner CC, Woglmaier SM, Edwards RH. (2011). v-SNARE composition distinguishes synaptic vesicle pools. Neuron , 474–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang , Sun MZ, Gumpper K, Chi YJ, Ma JJ. (2015). 3D multifocus astigmatism and compressed sensing (3D MACS) based superresolution reconstruction. Biomed Opt Express , 902–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idrissi FZ, Geli MI. (2014). Zooming in on the molecular mechanisms of endocytic budding by time-resolved electron microscopy. Cell Mol Life Sci , 641–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura H, Tanaka K, Hihara T, Umikawa M, Kamei T, Takahashi K, Sasaki T, Takai Y. (1997). Bni1p and Bnr1p: downstream targets of the Rho family small G-proteins which interact with profilin and regulate actin cytoskeleton in Saccharomyces cerevisiae . EMBO J , 2745–2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro J, Saitou A, Durán A, Ribas JC. (1997). cps1+, a Schizosaccharomyces pombe gene homolog of Saccharomyces cerevisiae FKS genes whose mutation confers hypersensitivity to cyclosporin A and papulacandin B. J Bacteriol , 7653–7662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James DJ, Martin TF. (2013). CAPS and Munc13: CATCHRs that SNARE vesicles. Front Endocrinol (Lausanne) , 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen J, Alfaro G, Beh CT. (2016). Polarized exocytosis induces compensatory endocytosis by Sec4p-regulated cortical actin polymerization. PLoS Biol , e1002534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JL, He J, Ramadass M, Pestonjamasp K, Kiosses WB, Zhang J, Catz SD. (2016). Munc13-4 is a Rab11-binding protein that regulates Rab11-positive vesicle trafficking and docking at the plasma membrane. J Biol Chem , 3423–3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JL, Hong H, Monfregola J, Kiosses WB, Catz SD. (2011). Munc13-4 restricts motility of Rab27a-expressing vesicles to facilitate lipopolysaccharide-induced priming of exocytosis in neutrophils. J Biol Chem , 5647–5656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan SN, Canman JC. (2012). Rho GTPases in animal cell cytokinesis: an occupation by the one percent. Cytoskeleton (Hoboken) , 919–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaksonen M, Toret CP, Drubin DG. (2005). A modular design for the clathrin- and actin-mediated endocytosis machinery. Cell , 305–320. [DOI] [PubMed] [Google Scholar]
- Kao RS, Morreale E, Wang L, Ivey FD, Hoffman CS. (2006). Schizosaccharomyces pombe Git1 is a C2-domain protein required for glucose activation of adenylate cyclase. Genetics , 49–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwazaki J, Yamasaki Y, Itadani A, Teraguchi E, Maeda Y, Shimoda C, Nakamura T. (2011). Endocytosis is essential for dynamic translocation of a syntaxin 1 orthologue during fission yeast meiosis. Mol Biol Cell , 3658–3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama S, Hirata D, Arellano M, Pérez P, Toda T. (1999). Fission yeast α-glucan synthase Mok1 requires the actin cytoskeleton to localize the sites of growth and plays an essential role in cell morphogenesis downstream of protein kinase C function. J Cell Biol , 1173–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabe H, Mitkovski M, Kaeser PS, Hirrlinger J, Opazo F, Nestvogel D, Kalla S, Fejtova A, Verrier SE, Bungers SR, et al. (2017). ELKS1 localizes the synaptic vesicle priming protein bMunc13-2 to a specific subset of active zones. J Cell Biol , 1143–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli J, Hottinger H, Munz P, Strauss A, Thuriaux P. (1977). Genetic-mapping in Schizosaccharomyces pombe by mitotic and meiotic analysis and induced haploidization. Genetics , 471–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan EP, Xie L, Sheu L, Ohtsuka T, Gaisano HY. (2007). Interaction between munc13-1 and RIM is critical for glucagon-like peptide-1-mediated rescue of exocytotic defects in munc13-1-deficient pancreatic beta-cells. Diabetes , 2579–2588. [DOI] [PubMed] [Google Scholar]
- Lai Y, Choi UB, Leitz J, Rhee HJ, Lee C, Altas B, Zhao M, Pfuetzner RA, Wang AL, Brose N, et al. (2017). Molecular mechanisms of synaptic vesicle priming by Munc13 and Munc18. Neuron , 591–607 e510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamaze C, Dujeancourt A, Baba T, Lo CG, Benmerah A, Dautry-Varsat A. (2001). Interleukin 2 receptors and detergent-resistant membrane domains define a clarthin-independent endocytic pathway. Mol Cell , 661–671. [DOI] [PubMed] [Google Scholar]
- Laporte D, Coffman VC, Lee I-J, Wu J-Q. (2011). Assembly and architecture of precursor nodes during fission yeast cytokinesis. J Cell Biol , 1005–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte D, Ojkic N, Vavylonis D, Wu J-Q. (2012). α-Actinin and fimbrin cooperate with myosin II to organize actomyosin bundles during contractile-ring assembly. Mol Biol Cell , 3094–3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layton AT, Savage NS, Howell AS, Carroll SY, Drubin DG, Lew DJ. (2011). Modeling vesicle traffic reveals unexpected consequences for Cdc42p-mediated polarity establishment. Curr Biol , 184–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I-J, Coffman VC, Wu J-Q. (2012). Contractile-ring assembly in fission yeast cytokinesis: recent advances and new perspectives. Cytoskeleton (Hoboken) , 751–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I-J, Wang N, Hu W, Schott K, Bähler J, Giddings TH, Jr, Pringle JR, Du L-L, Wu J-Q. (2014). Regulation of spindle pole body assembly and cytokinesis by the centrin-binding protein Sfi1 in fission yeast. Mol Biol Cell , 2735–2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I-J, Wu J-Q. (2012). Characterization of Mid1 domains for targeting and scaffolding in fission yeast cytokinesis. J Cell Sci , 2973–2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin DE. (2011). Regulation of cell wall biogenesis in Saccharomyces cerevisiae: the cell wall integrity signaling pathway. Genetics , 1145–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin DE, Bowers B, Chen CY, Kamada Y, Watanabe M. (1994). Dissecting the protein kinase C/MAP kinase signalling pathway of Saccharomyces cerevisiae . Cell Mol Biol Res , 229–239. [PubMed] [Google Scholar]
- Li W, Ma C, Guan R, Xu YB, Tomchick DR, Rizo J. (2011). The crystal structure of a Munc13 C-terminal module exhibits a remarkable similarity to vesicle tethering factors. Structure , 1443–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JH, Tang X, Wang HY, Oliferenko S, Balasubramanian MK. (2002). The localization of the integral membrane protein Cps1p to the cell division site is dependent on the actomyosin ring and the septation-inducing network in Schizosaccharomyces pombe . Mol Biol Cell , 989–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JH, Wang HY, McCollum D, Balasubramanian MK. (1999). Drc1p/Cps1p, a 1,3-β-glucan synthase subunit, is essential for division septum assembly in Schizosaccharomyces pombe . Genetics , 1193–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Seven AB, Xu J, Esser V, Su L, Ma C, Rizo J. (2017). Simultaneous lipid and content mixing assays for in vitro reconstitution studies of synaptic vesicle fusion. Nat Protoc , 2014–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XX, Seven AB, Camacho M, Esser V, Xu JJ, Trimbuch T, Quade B, Su LJ, Ma C, Rosenmund C, Rizo J. (2016a). Functional synergy between the Munc13 C-terminal C-1 and C-2 domains. eLife , e13696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Lee I-J, Sun M, Lower CA, Runge KW, Ma J, Wu J-Q. (2016b). Roles of the novel coiled-coil protein Rng10 in septum formation during fission yeast cytokinesis. Mol Biol Cell , 2528–2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou XL. (2018). Sensing exocytosis and triggering endocytosis at synapses: synaptic vesicle exocytosis-endocytosis coupling. Front Cell Neurosci , 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Machius M, Dulubova I, Dai H, Sudhof TC, Tomchick DR, Rizo J. (2006). Structural basis for a Munc13-1 homodimer to Munc13-1/RIM heterodimer switch. PLoS Biol , 1159–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Drubin DG. (2017). Selection and stabilization of endocytic sites by Ede1, a yeast functional homologue of human Eps15. Mol Biol Cell , 567–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C, Li W, Xu Y, Rizo J. (2011). Munc13 mediates the transition from the closed syntaxin-Munc18 complex to the SNARE complex. Nat Struct Mol Biol , 542–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison JM, Nurrish S, Kaplan JM. (2005). UNC-13 interaction with syntaxin is required for synaptic transmission. Curr Biol , 2236–2242. [DOI] [PubMed] [Google Scholar]
- Maeda Y, Kashiwazaki J, Shimoda C, Nakamura T. (2009). The Schizosaccharomyces pombe syntaxin 1 homolog, Psy1, is essential in the development of the forespore membrane. Biosci Biotechnol Biochem , 339–345. [DOI] [PubMed] [Google Scholar]
- Maritzen T, Haucke V. (2018). Coupling of exocytosis and endocytosis at the presynaptic active zone. Neurosci Res , 45–52. [DOI] [PubMed] [Google Scholar]
- Martin TF. (2015). PI(4,5)P2-binding effector proteins for vesicle exocytosis. Biochim Biophys Acta , 785–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Cuadrado AB, Duenas E, Sipiczki M, de Aldana CRV, del Rey F. (2003). The endo-β-1,3-glucanase eng1p is required for dissolution of the primary septum during cell separation in Schizosaccharomyces pombe . J Cell Sci , 1689–1698. [DOI] [PubMed] [Google Scholar]
- Martín-Cuadrado AB, Morrell JL, Konomi M, An H, Petit C, Osumi M, Balasubramanian M, Gould KL, Del Rey F, de Aldana CR. (2005). Role of septins and the exocyst complex in the function of hydrolytic enzymes responsible for fission yeast cell separation. Mol Biol Cell , 4867–4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-García R, Coll PM, Pérez P. (2014). F-BAR domain protein Rga7 collaborates with Cdc15 and Imp2 to ensure proper cytokinesis in fission yeast. J Cell Sci , 4146–4158. [DOI] [PubMed] [Google Scholar]
- Maruyama H, Rakow TL, Maruyama IN. (2001). Synaptic exocytosis and nervous system development impaired in Caenorhabditis elegans unc-13 mutants. Neuroscience , 287–297. [DOI] [PubMed] [Google Scholar]
- Matsuyama A, Arai R, Yashiroda Y, Shirai A, Kamata A, Sekido S, Kobayashi Y, Hashimoto A, Hamamoto M, Hiraoka Y, et al. (2006). ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe . Nat Biotechnol , 841–847. [DOI] [PubMed] [Google Scholar]
- Maundrell K. (1990). nmt1 of fission yeast. A highly transcribed gene completely repressed by thiamine. J Biol Chem , 10857–10864. [PubMed] [Google Scholar]
- McCollum D, Feoktistova A, Morphew M, Balasubramanian M, Gould KL. (1996). The Schizosaccharomyces pombe actin-related protein, Arp3, is a component of the cortical actin cytoskeleton and interacts with profilin. EMBO J , 6438–6446. [PMC free article] [PubMed] [Google Scholar]
- McCusker D, Royou A, Velours C, Kellogg D. (2012). Cdk1-dependent control of membrane-trafficking dynamics. Mol Biol Cell , 3336–3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald NA, Vander Kooi CW, Ohi MD, Gould KL. (2015). Oligomerization but not membrane bending underlies the function of certain F-BAR proteins in cell motility and cytokinesis. Dev Cell , 725–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullan R, Hiley E, Morrison P, Nurrish SJ. (2006). Rho is a presynaptic activator of neurotransmitter release at pre-existing synapses in C. elegans . Genes Dev , 65–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijering E, Dzyubachyk O, Smal I. (2012). Methods for cell and particle tracking. Methods Enzymol , 183–200. [DOI] [PubMed] [Google Scholar]
- Meitinger F, Palani S. (2016). Actomyosin ring driven cytokinesis in budding yeast. Semin Cell Dev Biol ,19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagnac G, Echard A, Chavrier P. (2008). Endocytic traffic in animal cell cytokinesis. Curr Opin Cell Biol , 454–461. [DOI] [PubMed] [Google Scholar]
- Moreno MB, Duran A, Ribas JC. (2000). A family of multifunctional thiamine-repressible expression vectors for fission yeast. Yeast , 861–872. [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. (1991). Molecular genetic analysis of fission yeast Schizosaccharomyces pombe . Methods Enzymol , 795–823. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Kashiwazaki J, Shimoda C. (2005). A fission yeast SNAP-25 homologue, SpSec9, is essential for cytokinesis and sporulation. Cell Struct Funct , 15–24. [DOI] [PubMed] [Google Scholar]
- Neeft M, Wieffer M, de Jong AS, Negroiu G, Metz CHG, van Loon A, Griffith J, KrI-Jgsveld J, Wulffraat N, Koch H, et al. (2005). Munc13–4 is an effector of Rab27a and controls secretion of lysosomes in hematopoietic cells. Mol Biol Cell , 731–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neto H, Balmer G, Gould G. (2013). Exocyst proteins in cytokinesis: regulation by Rab11. Commun Integr Biol , e27635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netrakanti PR, Cooper BH, Dere E, Poggi G, Winkler D, Brose N, Ehrenreich H. (2015). Fast cerebellar reflex circuitry requires synaptic vesicle priming by Munc13-3. Cerebellum , 264–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura S, Tokukura M, Ochi J, Yoshida M, Kakeya H. (2014). Balance between exocytosis and endocytosis determines the efficacy of sterol-targeting antibiotics. Chem Biol , 1690–1699. [DOI] [PubMed] [Google Scholar]
- Nolen BJ, Tomasevic N, Russell A, Pierce DW, Jia Z, McCormick CD, Hartman J, Sakowicz R, Pollard TD. 2009. Characterization of two classes of small molecule inhibitors of Arp2/3 complex. Nature , 1031–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka H, Tanaka K, Hirano H, Fujiwara T, Kohno H, Umikawa M, Mino A, Takai Y. (1995). A downstream target of Rho1 small GTP-binding protein is Pkc1, a homolog of protein-kinase-C, which leads to activation of the MAP kinase cascade in Saccharomyces cerevisiae . EMBO J , 5931–5938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei JM, Ma C, Rizo J, Grishin NV. (2009). Remote homology between Munc13 MUN domain and vesicle tethering complexes. J Mol Biol , 509–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez P, Cortés JC, Martin-García R, Ribas JC. (2016). Overview of fission yeast septation. Cell Microbiol , 1201–1207. [DOI] [PubMed] [Google Scholar]
- Pevsner J, Hsu SC, Braun JE, Calakos N, Ting AE, Bennett MK, Scheller RH. (1994). Specificity and regulation of a synaptic vesicle docking complex. Neuron , 353–361. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Wu J-Q. (2010). Understanding cytokinesis: lessons from fission yeast. Nat Rev Mol Cell Biol , 149–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser DC, Drivas TG, Maldonado-Baez L, Wendland B. (2011). Existence of a novel clathrin-independent endocytic pathway in yeast that depends on Rho1 and formin. J Cell Biol , 657–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser DC, Wendland B. (2012). Conserved roles for yeast Rho1 and mammalian RhoA GTPases in clathrin-independent endocytosis. Small GTPases , 229–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren L, Willet AH, Roberts-Galbraith RH, McDonald NA, Feoktistova A, Chen JS, Huang H, Guillen R, Boone C, Sidhu SS, et al. (2015). The Cdc15 and Imp2 SH3 domains cooperatively scaffold a network of proteins that redundantly ensure efficient cell division in fission yeast. Mol Biol Cell , 256–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribas JC, Diaz M, Durán A, Pérez P. (1991). Isolation and characterization of Schizosaccharomyces pombe mutants defective in cell wall (1-3)β-D-glucan. J Bacteriol , 3456–3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond JE, Davis WS, Jorgensen EM. (1999). UNC-13 is required for synaptic vesicle fusion in C. elegans . Nat Neurosci , 959–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rincón SA, Paoletti A. (2016). Molecular control of fission yeast cytokinesis. Semin Cell Dev Biol , 28–38. [DOI] [PubMed] [Google Scholar]
- Rizo J, Xu J. (2015). The synaptic vesicle release machinery. Annu Rev Biophys , 339–367. [DOI] [PubMed] [Google Scholar]
- Rodarte EM, Ramos MA, Davalos AJ, Moreira DC, Moreno DS, Cardenas EI, Rodarte AI, Petrova Y, Molina S, Rendon LE, et al. (2018). Munc13 proteins control regulated exocytosis in mast cells. J Biol Chem , 345–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossner S, Fuchsbrunner K, Lange-Dohna C, Hartlage-Rubsamen M, Bigl V, Betz A, Reim K, Brose N. (2004). Munc13-1-mediated vesicle priming contributes to secretory amyloid precursor protein processing. J Biol Chem , 27841–27844. [DOI] [PubMed] [Google Scholar]
- Sanchéz-Mir L, Soto T, Franco A, Madrid M, Viana RA, Vicente J, Gacto M, Pérez P, Cansado J. (2014). Rho1 GTPase and PKC ortholog Pck1 are upstream activators of the cell integrity MAPK pathway in fission yeast. PLoS One , e88020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat Methods , 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweitzer JK, Burke EE, Goodson HV, D’Souza-Schorey C. (2005). Endocytosis resumes during late mitosis and is required for cytokinesis. J Biol Chem , 41628–41635. [DOI] [PubMed] [Google Scholar]
- Shen N, Guryev O, Rizo J. (2005). Intramolecular occlusion of the diacylglycerol-binding site in the C1 domain of Munc13-1. Biochemistry , 1089–1096. [DOI] [PubMed] [Google Scholar]
- Sheu L, Pasyk EA, Ji JZ, Haung XH, Gao XD, Varoqueaux F, Brose N, Gaisano HY. (2003). Regulation of insulin exocytosis by Munc13-1. J Biol Chem , 27556–27563. [DOI] [PubMed] [Google Scholar]
- Shin OH, Lu J, Rhee JS, Tomchick DR, Pang ZP, Wojcik SM, Camacho-Pérez M, Brose N, Machius M, Rizo J, et al. (2010). Munc13 C2B domain is an activity-dependent Ca2+ regulator of synaptic exocytosis. Nat Struct Mol Biol , 280–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuster CB, Burgess DR. (2002). Targeted new membrane addition in the cleavage furrow is a late, separate event in cytokinesis. Proc Natl Acad Sci USA , 3633–3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirotkin V, Berro J, Macmillan K, Zhao L, Pollard TD. (2010). Quantitative analysis of the mechanism of endocytic actin patch assembly and disassembly in fission yeast. Mol Biol Cell , 2894–2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skop AR, Bergmann D, Mohler WA, White JG. (2001). Completion of cytokinesis in C. elegans requires a brefeldin A-sensitive membrane accumulation at the cleavage furrow apex. Curr Biol , 735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Guan R, Lee I-J, Liu Y, Chen M, Wang J, Wu J-Q, Chen Z. (2015). Mechanistic insights into the anchorage of the contractile ring by anillin and Mid1. Dev Cell , 413–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajadura V, García B, García I, García P, Sanchéz Y. (2004). Schizosaccharomyces pombe Rgf3p is a specific Rho1 GEF that regulates cell wall β-glucan biosynthesis through the GTPase Rho1p. J Cell Sci , 6163–6174. [DOI] [PubMed] [Google Scholar]
- Takeda T, Kawate T, Chang F. (2004). Organization of a sterol-rich membrane domain by cdc15p during cytokinesis in fission yeast. Nat Cell Biol , 1142–1144. [DOI] [PubMed] [Google Scholar]
- Tang X, Huang J, Padmanabhan A, Bakka K, Bao Y, Tan BY, Cande WZ, Balasubramanian MK. (2011). Marker reconstitution mutagenesis: a simple and efficient reverse genetic approach. Yeast , 205–212. [DOI] [PubMed] [Google Scholar]
- TerBush DR, Maurice T, Roth D, Novick P. (1996). The exocyst is a multiprotein complex required for exocytosis in Saccharomyces cerevisiae . EMBO J , 6483–6494. [PMC free article] [PubMed] [Google Scholar]
- VerPlank L, Li R. (2005). Cell cycle-regulated trafficking of Chs2 controls actomyosin ring stability during cytokinesis. Mol Biol Cell , 2529–2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viana RA, Pinar M, Soto T, Coll PM, Cansado J, Perez P. (2013). Negative functional interaction between cell integrity MAPK pathway and Rho1 GTPase in fission yeast. Genetics , 421–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachtler V, Huang Y, Karagiannis J, Balasubramanian MK. (2006). Cell cycle-dependent roles for the FCH-domain protein Cdc15p in formation of the actomyosin ring in Schizosaccharomyces pombe . Mol Biol Cell , 3254–3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HY, Tang X, Liu JH, Trautmann S, Balasundaram D, McCollum D, Balasubramanian MK. (2002). The multiprotein exocyst complex is essential for cell separation in Schizosaccharomyces pombe . Mol Biol Cell , 515–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Lee I-J, Rask G, Wu J-Q. (2016). Roles of the TRAPP-II complex and the exocyst in membrane deposition during fission yeast cytokinesis. PLoS Biol , e1002437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Lo Presti L, Zhu Y-H, Kang M, Wu Z, Martin SG, Wu J-Q. (2014). The novel proteins Rng8 and Rng9 regulate the myosin-V Myo51 during fission yeast cytokinesis. J Cell Biol , 357–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Wang M, Zhu Y-H, Grosel TW, Sun D, Kudryashov DS, Wu J-Q. (2015). The Rho-GEF Gef3 interacts with the septin complex and activates the GTPase Rho4 during fission yeast cytokinesis. Mol Biol Cell , 238–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Choi UB, Gong J, Yang X, Li Y, Wang AL, Yang X, Brunger AT, Ma C. (2017). Conformational change of syntaxin linker region induced by Munc13s initiates SNARE complex formation in synaptic exocytosis. EMBO J , 816–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg J, Drubin DG. (2012). Clathrin-mediated endocytosis in budding yeast. Trends Cell Biol , 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienke DC, Knetsch MLW, Neuhaus EM, Reedy MC, Manstein DJ. (1999). Disruption of a dynamin homologue affects endocytosis, organelle morphology, and cytokinesis in Dictyostelium discoideum . Mol Biol Cell , 225–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J-Q, Bähler J, Pringle JR. (2001). Roles of a fimbrin and an α-actinin-like protein in fission yeast cell polarization and cytokinesis. Mol Biol Cell , 1061–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J-Q, Pollard TD. (2005). Counting cytokinesis proteins globally and locally in fission yeast. Science , 310–314. [DOI] [PubMed] [Google Scholar]
- Wu J-Q, Sirotkin V, Kovar DR, Lord M, Beltzner CC, Kuhn JR, Pollard TD. (2006). Assembly of the cytokinetic contractile ring from a broad band of nodes in fission yeast. J Cell Biol , 391–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J-Q, Ye Y, Wang N, Pollard TD, Pringle JR. (2010). Cooperation between the septins and the actomyosin ring and role of a cell-integrity pathway during cell division in fission yeast. Genetics , 897–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu LG, Hamid E, Shin W, Chiang HC. (2014). Exocytosis and endocytosis: modes, functions, and coupling mechanisms. Annu Rev Physiol , 301–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P, Zhao R, Ye Y, Wu J-Q. (2011). Roles of the DYRK kinase Pom2 in cytokinesis, mitochondrial morphology, and sporulation in fission yeast. PLoS One , e28000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Long J, Liu J, Chai Z, Kang X, Wang C. (2017). Molecular mechanisms for the coupling of endocytosis to exocytosis in neurons. Front Mol Neurosci , 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Camacho M, Xu Y, Esser V, Liu X, Trimbuch T, Pan YZ, Ma C, Tomchick DR, Rosenmund C, Rizo J. (2017). Mechanistic insights into neurotransmitter release and presynaptic plasticity from the crystal structure of Munc13–1 C1C2BMUN. eLife , e22567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaoka T, Imada K, Fukunishi K, Yamasaki Y, Shimoda C, Nakamura T. (2013). The fission yeast synaptobrevin ortholog Syb1 plays an important role in forespore membrane formation and spore maturation. Eukaryot Cell , 1162–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita T, Hige T, Takahashi T. (2005). Vesicle endocytosis requires dynamin-dependent GTO hydrolysis at a fast CNS synapse. Science , 124–127. [DOI] [PubMed] [Google Scholar]
- Yang CB, Kiser PJ, Zheng YT, Varoqueaux F, Mower GD. (2007). Bidirectional regulation of Munc13-3 protein expression by age and dark rearing during the critical period in mouse visual cortex. Neuroscience , 603–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CB, Zheng YT, Li GY, Mower GD. (2002). Identification of Munc13-3 as a candidate gene for critical-period neuroplasticity in visual cortex. J Neurosci , 8614–8618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Wang S, Sheng Y, Zhang M, Zou W, Wu L, Kang L, Rizo J, Zhang R, Xu T, Ma C. (2015). Syntaxin opening by the MUN domain underlies the function of Munc13 in synaptic-vesicle priming. Nat Struct Mol Biol , 547–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Sugiura R, Lu Y, Asami M, Maeda T, Itoh T, Takenawa T, Shuntoh H, Kuno T. (2000). Phosphatidylinositol 4-phosphate 5-kinase Its3 and calcineurin Ppb1 coordinately regulate cytokinesis in fission yeast. J Biol Chem , 35600–35606. [DOI] [PubMed] [Google Scholar]
- Zhou K, Stawicki TM, Goncharov A, Jin Y. (2013). Position of UNC-13 in the active zone regulates synaptic vesicle release probability and release kinetics. eLife , e01180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y-H, Ye Y, Wu Z, Wu J-Q. (2013). Cooperation between Rho-GEF Gef2 and its binding partner Nod1 in the regulation of fission yeast cytokinesis. Mol Biol Cell , 3187–3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.