Abstract
The gut has a central role in digestion and nutrient absorption, but it also serves in defending against pathogens, engages in mutually beneficial interactions with commensals, and is a major source of endocrine signals. Gut homeostasis is necessary for organismal health and changes to the gut are associated with conditions like obesity and diabetes and inflammatory illnesses like Crohn’s disease. We report that peroxisomes, organelles involved in lipid metabolism and redox balance, are required to maintain gut epithelium homeostasis and renewal in Drosophila and for survival and development of the organism. Dysfunctional peroxisomes in gut epithelial cells activate Tor kinase-dependent autophagy that increases cell death and epithelial instability, which ultimately alter the composition of the intestinal microbiota, compromise immune pathways in the gut in response to infection, and affect organismal survival. Peroxisomes in the gut effectively function as hubs that coordinate responses from stress, metabolic, and immune signaling pathways to maintain enteric health and the functionality of the gut–microbe interface.
INTRODUCTION
The intestinal epithelium absorbs nutrients, maintains energy homeostasis, and manages interactions with microorganisms to provide resistance to pathogens and to promote beneficial contacts with commensals (Clemente et al., 2012; Hooper et al., 2012; Nicholson et al., 2012; Lemaitre and Miguel-Aliaga, 2013; Guo et al., 2014). Understanding how enteric health is maintained therefore has important implications for overall human health and requires knowledge of the signaling pathways that control gut metabolism, stress response, and interactions with microbes.
Peroxisomes are ubiquitous organelles conserved across the breadth of eukaryotes. Peroxin (Pex) genes encode proteins called peroxins that are required for peroxisome formation and maintenance of peroxisome populations (Smith and Aitchison, 2013). Peroxisomes perform many important metabolic functions and maintain cellular redox homeostasis (Wanders and Waterham, 2006; Nguyen et al., 2008). Peroxisome deficiency or functional impairments directly due to defective peroxisome formation result in devastating genetic conditions known as the peroxisome biogenesis disorders (PBDs) (Waterham et al., 2016); contribute to the pathology of Alzheimer’s and Parkinson’s diseases, aging, cancer, diabetes, and heart failure; and affect immunity (Dixit et al., 2010; Beach et al., 2012; Fransen et al., 2012; Colasante et al., 2015; Di Cara et al., 2017). Emerging evidence suggests that peroxisomes are pivotal to the development and survival of different tissues; for example, peroxisomes are recognized mediators of brain development (Berger et al., 2016) and of skeletal muscle formation and integrity (Braverman et al., 2013). Because peroxisomes are notably abundant in gut epithelial cells (Novikoff and Novikoff, 1972; Beard and Holtzman, 1987; Faust et al., 2014; Morvay et al., 2017) and because gastrointestinal bleeding has been reported in PBD patients (Lodhi and Semenkovich, 2014), we investigated a potential requirement for peroxisomes in maintaining the structure and function of the gut epithelium using Drosophila as a model. Studies of the Drosophila gut have been at the forefront of recent research on host–commensal and host–pathogen interactions, innate immune signaling, and the regenerative capacity of the intestinal epithelia (Buchon et al., 2013; Lemaitre and Miguel-Aliaga, 2013). Differentiated cells in the Drosophila gut epithelium undergo normal turnover, but turnover is more rapid in damaged tissue (Amcheslavsky et al., 2009; Buchon et al., 2009). Therefore, stress response programs and increased epithelial renewal need to be activated to repair the intestinal epithelium when damaged and thereby maintain the integrity of the gut barrier.
Here we report that peroxisomes in the Drosophila gut modulate target of rapamycin (Tor) kinase-dependent autophagy, stress signaling and tissue regeneration to maintain gut epithelium homeostasis, promote gut epithelium renewal, and ultimately influence host–commensal and host–pathogen interactions needed for the survival and development of Drosophila.
RESULTS
Dysfunctional peroxisomes lead to increased lysosomal and autophagic activity in the gut epithelium
Given that peroxisomes are notably abundant in gut epithelial cells and that peroxisomes have been shown to be important for the development and maintenance of other body tissues, we investigated a potential role for peroxisomes in maintenance of the gut epithelium in health, in gut repair, and in the interaction between gut and microbiota. We used the Gal4/UAS system (Brand and Perrimon, 1993) to compromise peroxisome function in Drosophila midgut epithelial cells via RNA interference (RNAi) by expressing a double-stranded RNA targeting the mRNA for Pex5. Pex5 is the conserved receptor that recognizes peroxisomal proteins made in the cytosol and targets them to the peroxisomal matrix (Klein et al., 2001; Baron et al., 2016; Di Cara et al., 2017). We targeted gut cells specifically by using a line expressing Gal4 under control of the Mex promoter (Phillips and Thomas, 2006). The efficiency of RNAi for Pex5 (Mex >Pex5-i) was confirmed by quantitative real-time PCR (qRT-PCR) (Supplemental Figure S1A) and Western blotting (Supplemental Figure S1D) using an antibody to human Pex5 that recognizes Drosophila Pex5 as demonstrated by its ability to recognize a fusion between EGFP and Drosophila Pex5 by Western blotting (Supplemental Figure S1C). Immunofluorescence microscopy also showed reduced import of peroxisome targeting signal 1 (PTS1)-containing proteins into peroxisomes in Mex >Pex5-i enterocytes compared with control cells (Supplemental Figure S1B). Mex >Pex5-i animals exhibited lethality during development with only 60% reaching adulthood compared with control (Mex >w1118) flies (Figure 1A). Thus, flies with dysfunctional peroxisomes in the midgut epithelium had reduced viability.
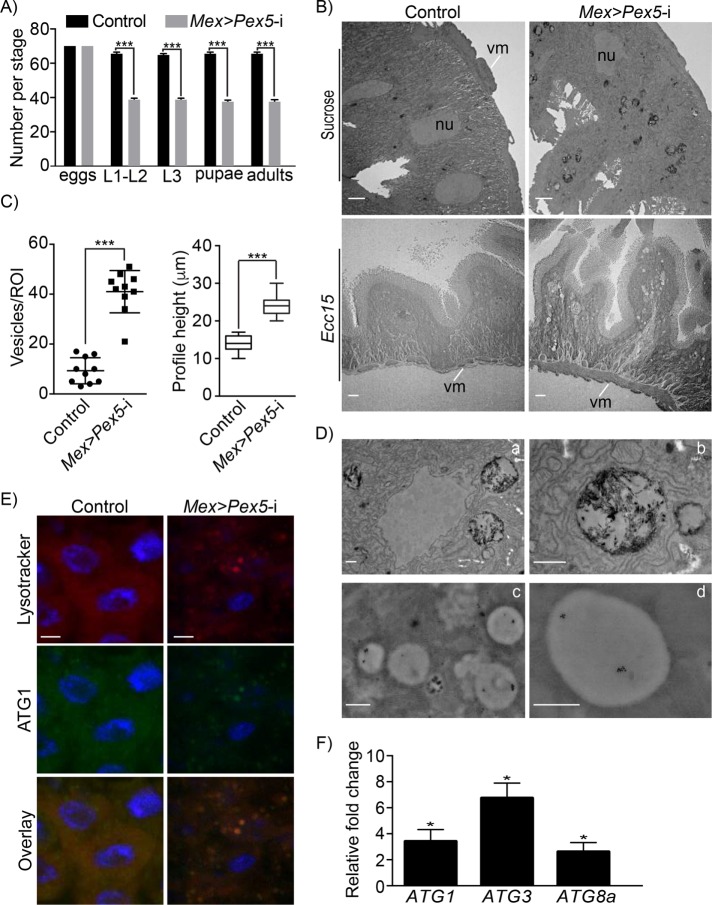
FIGURE 1:
Dysfunctional peroxisomes lead to structural disorganization and increased lysosomal/autophagic activity in the fly gut. (A) Pex5 depletion in the midgut causes increased lethality during fly development. Embryos were followed through development, and survival to larval, pupal, and adult stages were scored for Mex >Pex5-i flies and control flies; n = 70 eggs for each genotype in a single experiment. Values reported represent the averages of three independent experiments ± SD. Statistical significance was determined using Student’s t test; ***p < 0.001. (B) Representative electron microscopy images of midguts from control flies and Mex >Pex5-i flies uninfected (top panels) or infected with Ecc15 (bottom panels). nu, nucleus; vm, visceral muscle. Scale bar, 2 µm. (C) Number of vesicles containing electron dense material per region of interest (ROI) observed in midguts from control flies and Mex >Pex5-i flies (left graphic); profile height of midgut longitudinal sections (µm ± SD) from control flies and Mex >Pex5-i flies (right graphic). Flies were subjected to chronic oral infection with Ecc15. Twenty-five cells of each genotype were analyzed. Statistical significance was determined using Student’s t test; ***p < 0.001. (D) Immunogold labeling of epithelial cells with anti-Lamp1 antibodies. Panels a and b show higher magnifications of the vesicular structures seen in epithelial cells of infected Mex >Pex5-i flies presented in B. Panels c and d show immunogold labeling of round, electron-dense vesicles with anti-Lamp1 antibodies, confirming that these structures are autophagosomes or lysosomes. Scale bar, 500 nm. (E) Fluorescence detection of the autophagosome markers, Lysotracker (red) and ATG1 (green), in midguts from infected control flies and Mex >Pex5-i flies. Scale bar, 10 µm. (F) Quantification of ATG1, ATG3, and ATG8a mRNA transcript levels in midguts from Mex >Pex5-i flies relative to midguts from control flies. The values reported represent the average of four independent experiments. Statistical significance was determined using Student’s t test; *p < 0.05.
We compared the ultrastructure of midguts of control and Mex >Pex5-i flies. Cells of Mex >Pex5-i midguts showed numerous membranous vesicles of different sizes (Figure 1, B and C) containing electron-dense material, presumably products of degradation (Figure 1D, panels a and b). These vesicles were rarely observed in cells of midguts from control flies (Figure 1, B and C). Midguts from Mex >Pex5-i flies challenged by infection with the Gram-negative bacterium Erwinia carotovora (Ecc15) showed accumulation of vacuole-like structures in cells, epithelial cell hypertrophy, and an overall disorganization of the epithelial structure as compared with control midguts (Figure 1, B and C).
Immunogold detection of lysosomal-associated membrane protein 1 (LAMP1) (Eskelinen, 2006) demonstrated that the vesicles in cells from Mex >Pex5-i flies were lysosomes (Figure 1D, panels c and d). Lysosomes are responsible for the degradation of macromolecules derived from the extracellular space by endocytosis or phagocytosis, as well as macromolecules from the cytoplasm produced by autophagy (Eskelinen, 2006; Luzio et al., 2007). We stained midguts from infected control flies and Mex >Pex5-i flies with the lysosome marker Lysotracker (Juhasz and Neufeld, 2008; Kim et al., 2013). Lysotracker-positive vesicles accumulated in Mex >Pex5-i midgut cells but not in control midgut cells (Supplemental Figure S1E). Similarly, immunofluorescence detection of the autophagy-specific protein ATG1 showed increased ATG1 levels in Mex >Pex5-i midgut cells compared with control midgut cells (Supplemental Figure S1F). Lysotracker-positive and ATG1-positive puncta largely overlapped in Mex >Pex5-i midgut cells (Figure 1E).
Tor kinase signaling is reduced in gut epithelial cells with dysfunctional peroxisomes
There is growing evidence of lipotoxicity and oxidative stress acting as important intracellular signal transducers that sustain autophagy (Wu et al., 2009; Martino et al., 2012; Filomeni et al., 2015). Midguts from Mex >Pex5-i flies with dysfunctional peroxisomes showed greater expression of the autophagy genes ATG1, ATG3 and ATG8a compared with control midguts (Figure 1F). Induction of ATG genes in response to chemically induced oxidative stress has been reported to be dependent on the c-Jun N-terminal kinase (JNK) pathway in Drosophila gut (Wu et al., 2009). Activation of the JNK pathway can be measured by antibody detection of phosphorylated JNK (P-JNK). The amounts of P-JNK were comparable in midguts from control flies and Mex >Pex5-i flies (Supplemental Figure S1G), suggesting that the increased transcript levels of ATG genes observed in midguts from Mex >Pex5-i flies were not due to activation of the JNK pathway.
Tor kinase is a conserved regulator of translation and autophagy that responds to growth and stress stimuli (Reiling and Sabatini, 2006; Cebollero and Reggiori, 2009; Gonzalez and Rallis, 2017). Tor kinase activity has been shown to be modulated by peroxisomes in mammalian cell cultures (Zhang et al., 2013). Inhibition of Tor kinase suppresses general translation and activates autophagy. To determine whether Tor kinase was inhibited in Drosophila guts with dysfunctional peroxisomes, we compared the global translation rate in control midguts and Mex >Pex5-i midguts by quantifying the in situ incorporation of L-azidohomoalanine (AHA), a methionine analogue. We observed that global translation was reduced ∼70% in Mex >Pex5-i midguts compared with control midguts, similarly to the reduction observed in midguts from control flies or Mex >Pex5-i flies infected with Pseudomonas entomophila (Figure 2A), a condition that has been reported to dampen global translation in the gut (Chakrabarti et al., 2012). Our results are consistent with inactivation of Tor kinase in midguts of flies with dysfunctional peroxisomes. Inactivation of Tor kinase also causes dephosphorylation of two of Tor kinase’s targets, S6K and 4E-BP (Dennis et al., 1999; Hay and Sonenberg, 2004). These proteins regulate ribosome biogenesis and cap-dependent mRNA translation. We therefore examined the requirement for functional peroxisomes to modulate the activity of Tor kinase in the midgut by comparing the amounts of phosphorylated S6K (P-S6K) and phosphorylated 4E-BP1 (P-4E-BP) in Mex >Pex5-i guts compared with control midguts. Western blotting showed greatly reduced amounts of P-4E-BP and P-S6K in Mex >Pex5-i midguts compared with control midguts (Figure 2, B and C), confirming that Tor kinase activity requires functional peroxisomes.
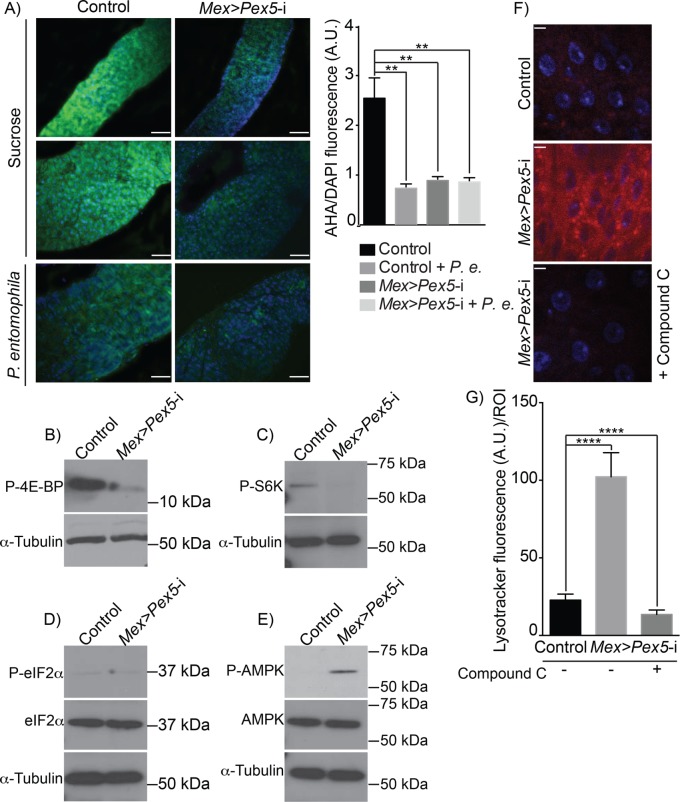
FIGURE 2:
Loss of functional peroxisomes leads to AMPK-dependent Tor kinase inhibition. (A) Microscopic evaluation of global protein synthesis by incorporation of the fluorescent initiator methionyl-tRNA analogue, L-azidohomoalanine (AHA, green), by midguts from control flies and Mex >Pex5-i flies. Infection with P. entomophila has been reported to dampen global translation in the gut and is used here as a positive control for the assay. DNA was stained by DAPI (blue). Scale bar, 50 µm. Quantification of global protein synthesis was done on representative fluorescence microscopy images of midguts from control flies and Mex >Pex5-i flies. The ratio of AHA signal (green) to DAPI signal (blue) was determined to normalize for cell number. Values represent the median of 20 midgut regions ± SD for each genotype under each condition. Statistical significance was determined using one-way ANOVA; **p < 0.01. P.e., P. entomophila. (B) Representative Western blot showing that P-4E-BP amounts are reduced in Mex >Pex5-i midguts compared with control midguts. (C) Representative Western blot showing that P-S6K amounts are reduced in Mex >Pex5-i midguts compared with control midguts. (D) Representative Western blot showing that total eIF2α amounts and P-eIF2α amounts are unchanged between control midguts and Mex >Pex5-i midguts. (E) Representative Western blot showing that total AMPK amounts are unchanged between control midguts and Mex >Pex5-i midguts, but P-AMPK amounts are greater in Mex >Pex5-i midguts. (B–E) α-Tubulin served as a control for protein loading. (F) Lysotracker staining (red) of midguts from flies of the designated genotypes. DNA was stained by DAPI (blue). Scale bar, 10 µm. (G) Quantification of Lysotracker fluorescence per ROI of midguts of flies of the designated genotypes. Values reported represent the averages of 20 independent ROIs ± SD for each genotype. Statistical significance was determined using two-way ANOVA; ****p < 0.0001. Compound C functions as an AMPK inhibitor (F, G).
Another pathway that can arrest cap-dependent mRNA translation in response to stress depends on phosphorylation of eukaryotic initiation factor 2α (eIF2α) (Holcik and Sonenberg, 2005). Under resting conditions, eIF2α is not phosphorylated and is part of a complex that recruits the initiator methionyl-tRNA to the start codon. However, phosphorylated eIF2α (P-eIF2α) acts as an inhibitor of general translation (Holcik and Sonenberg, 2005). Western blot analysis showed no change in the levels of P-eIF2α between control midguts and Mex >Pex5-i midguts (Figure 2D), consistent with the scenario in which inhibition of translation in Mex >Pex5-i midguts is due to inhibition of Tor kinase.
Nutrient and redox stress are major factors that inhibit Tor kinase, which in turn inhibits global translation and activates autophagy (Reiling and Sabatini, 2006; Filomeni et al., 2015). Inhibition of the Tor kinase pathway by nutrients or redox stress can be caused by activation of antimicrobial peptide (AMP) kinase (AMPK) (Reiling and Sabatini, 2006; Chakrabarti et al., 2012; Inokuchi-Shimizu et al., 2014; Li et al., 2014), which in turn activates Tuberous sclerosis 2 protein (Tsc2) (Chakrabarti et al., 2012; Hwang et al., 2014; Kim and Lee, 2015). The tuberous sclerosis complex, composed of Tsc1 and Tsc2 subunits, is a negative regulator of Tor kinase activity (Hay and Sonenberg, 2004), and activation of Tsc2 results in silencing of Tor kinase activity. We investigated whether the absence of functional peroxisomes in the midgut activates AMPK by analyzing the extent of AMPK phosphorylation in midguts from control flies and Mex >Pex5-i flies. Western blot analysis showed that AMPK is phosphorylated in Mex >Pex5-i midguts but not control midguts (Figure 2E; Supplemental Figure S2, A and B), suggesting that the presence of dysfunctional peroxisomes induces AMPK-dependent inactivation of Tor kinase in the fly gut. We also inhibited AMPK phosphorylation by maintaining control flies and Mex >Pex5-i flies on medium supplemented with the AMPK inhibitor, compound C, and found that inhibition of AMPK phosphorylation by compound C (Supplemental Figure S2, A and B) reduced autophagy in Mex >Pex5-i midguts as measured by reduced Lysotracker staining (Figure 2, F and G).
Increased Tor kinase activity leads to reduced autophagy in midgut epithelial cells of Mex >Pex5-i flies
We evaluated the effect of Tor kinase on autophagy in control midguts and Mex >Pex5-i midguts by manipulating the levels of Tor kinase and assessing the extent of autophagy by Lysotracker staining. Depletion of Tor gene transcript in midguts was achieved by expression of a double-stranded RNA (dsRNA) transgene to Tor via the Mex promoter (Mex >Tor-i). Lysotracker staining was increased when Tor expression was reduced in midguts with dysfunctional peroxisomes (Mex >Pex5-i; Tor-i) and was greater in amount than the lysosomal staining observed in Mex >Pex5-i midguts and control midguts (Figure 3, A and B). When we overexpressed Tor in Mex >Pex5-i midguts (Mex >Pex5-i; UAS-Tor) or down-regulated the gene TSC2 for the Tor kinase inhibitor in Mex >Pex5-i midguts (Mex >Pex5-i; TSC2-i), we observed a large reduction in Lysotracker staining in the midgut (Figure 3, A and B), comparable in its intensity to Lysotracker staining in control midguts or to midguts in which we overexpressed Tor (Mex >UAS-Tor) or reduced TSC2 expression (Mex >TSC2-i) to enhance Tor kinase activity (Figure 3, A and B).
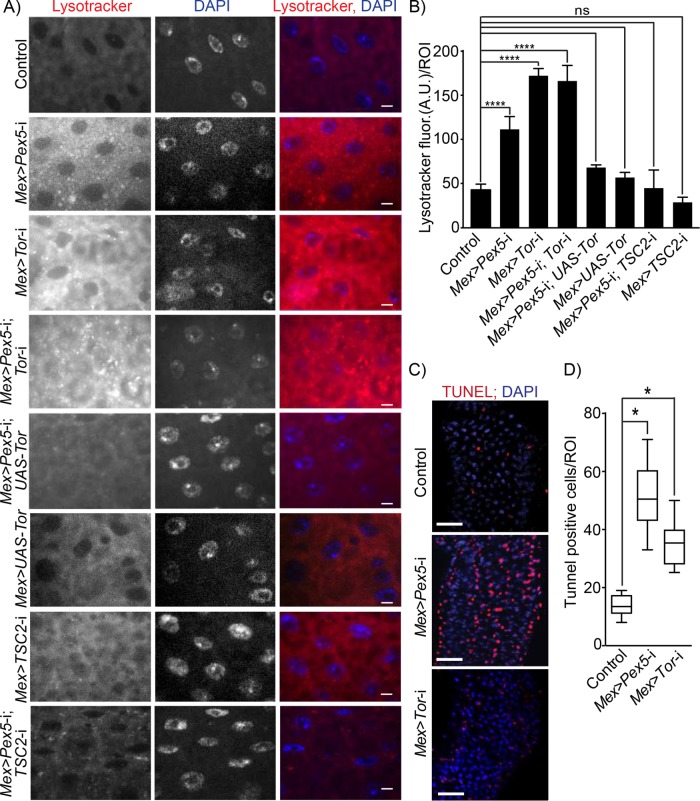
FIGURE 3:
Dysfunctional peroxisomes in the gut lead to increased Tor kinase-dependent autophagy and increased epithelial cell death. (A) Lysotracker staining (red) of midguts from flies of the designated genotypes. DNA was stained by DAPI (blue). Scale bar, 10 µm. (B) Quantification of Lysotracker fluorescence per ROI of midguts of flies of the designated genotypes. Values reported represent the averages of 20 independent ROIs ± SD for each genotype. Statistical significance was determined using two-way ANOVA; ****p < 0.0001; ns = not significant. (C) Midguts from Mex >Pex5-i and Mex >Tor-i flies exhibit increased numbers of apoptotic cells relative to midguts from control flies. Apoptotic cells were detected by TUNEL staining (red). Nuclei were stained by DAPI (blue). Scale bar, 25 μm. (D) Values reported represent the average number of TUNEL-positive cells per ROI per genotype for 25 midguts from each genotype. Statistical significance was determined using one-way ANOVA; *p < 0.05.
Autophagy is intimately associated with eukaryotic cell death and apoptosis (Yonekawa and Thorburn, 2013). We therefore measured the amount of cell death in midguts isolated from control, Mex >Pex5-i, and Mex >Tor-i flies by staining with the apoptotic marker terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) (Figure 3, C and D) and detected increased TUNEL staining in Mex >Pex5-i and Mex >Tor-i midguts compared with midguts from control flies (Figure 3, C and D), confirming that the absence of functional peroxisomes leads to increased cell death in the gut.
Peroxisome dysfunction induces a compensatory proliferation of intestinal stem cells for gut repair
Gut homeostasis is maintained through regulation of stem cell activity (Amcheslavsky et al., 2009; Buchon et al., 2009; Jiang et al., 2009). The adult Drosophila gut contains multipotent intestinal stem cells (ISCs) scattered along its basement membrane (Ohlstein and Spradling, 2007; Li and Jasper, 2016). Increased apoptosis in the gut epithelium causes damage that in turn stimulates the underlying ISCs to proliferate for tissue repair (Amcheslavsky et al., 2009; Buchon et al., 2009; Jiang et al., 2009).
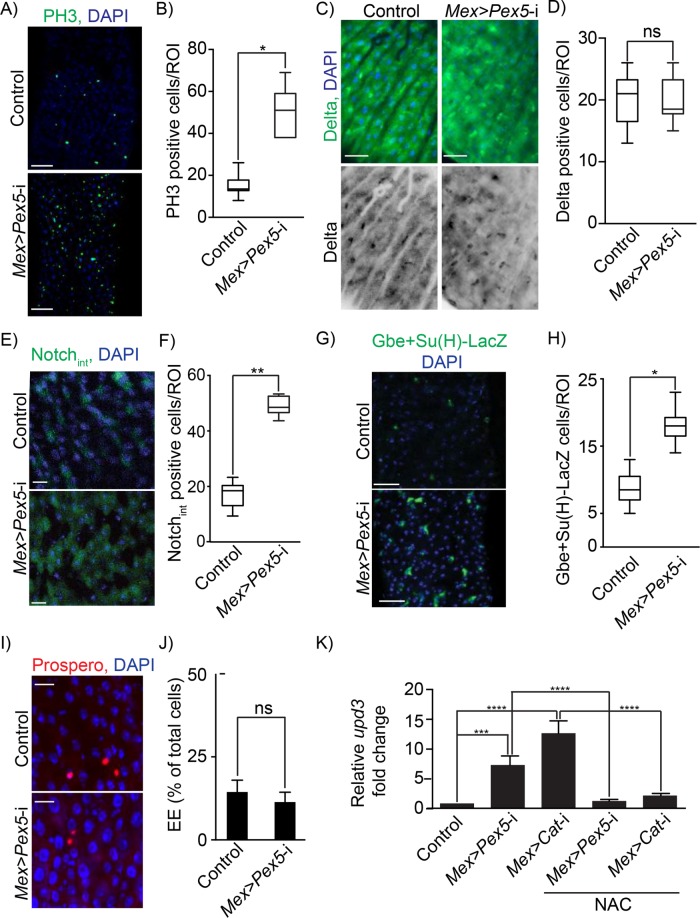
Since peroxisome dysfunction in the midgut of Mex >Pex5-i flies led to increased apoptosis in the midgut epithelium (cf. Figure 3, C and D), we checked whether this increased epithelial damage activated a compensatory proliferation of ISCs. Phospho-Ser10-histone 3 (PH3) is a specific marker for condensed chromosomes and can be used to detect mitotic cells (Wei et al., 1999). The number of PH3-positive cells in midguts dissected from Mex >Pex5-i flies was ∼ -times greater than in control samples (Figure 4, A and B). On cell division, each ISC produces a daughter cell that retains the ISC fate and a postmitotic enteroblast (EB) that differentiates into either an absorptive enterocyte (EC) or a secretory enteroendocrine cell (EE) (Micchelli and Perrimon, 2006; Ohlstein and Spradling, 2007). Drosophila ISCs can be identified by expression of the protein Delta (Ohlstein and Spradling, 2007). The number of Delta-positive cells detected by indirect immunofluorescence was the same in control and Mex >Pex5-i midguts (Figure 4, C and D). EBs and differentiating ECs can be identified by indirect immunofluorescence using antibodies to the intracellular domain of the protein Notch (Notchint) (Micchelli and Perrimon, 2006; Ohlstein and Spradling, 2007). Notchint-positive cells were increased in number in Mex >Pex5-i midguts compared with control midguts (Figure 4, E and F). Transgenic control and Mex >Pex5-i flies containing the Notch-reporter element Gbe+Su(H)-LacZ were used to mark exclusively the EB cell lineage (Ohlstein and Spradling, 2007), and the number of EBs was found to be greater in Mex >Pex5-i midguts than control midguts (Figure 4, G and H). Prospero is a homeodomain protein expressed specifically in gut EEs (Micchelli and Perrimon, 2006). We observed no increase in the number of Prospero-expressing EEs in Mex >Pex5-i midguts compared with control midguts (Figure 4, I and J). Our results suggest that the high rate of proliferation of ISCs in the Mex >Pex5-i midgut gives rise to an increase in the EB-EC lineage and support a scenario in which dysfunctional peroxisomes in mature ECs induce cellular stress and epithelial damage (Figure 3, C and D) that in turn can stimulate ISCs to proliferate to promote tissue repair.
FIGURE 4:
Functional peroxisomes are required for gut epithelium homeostasis. (A) Midguts from Mex >Pex5-i flies exhibit increased numbers of dividing ISCs compared with midguts from control flies. Dividing ISCs were detected by indirect immunofluorescence of PH3 (green). DNA was stained by DAPI (blue). Scale bar, 30 μm. (B) Number of PH3-positive cells per ROI of midguts from control flies and Mex >Pex5-i flies. Values represent the median of 25 midgut ROIs ± SD for each genotype and each condition. Statistical significance was determined using two-way ANOVA; *p < 0.05. (C) Indirect immunofluorescence of Delta (green) in midguts from control flies and Mex >Pex5-i flies. DNA was stained by DAPI (blue). Scale bar, 20 μm. (D) Number of Delta-positive cells per ROI in midguts from control flies and Mex >Pex5-i flies. Values represent the median of 25 midgut ROIs ± SD. Statistical significance was determined using one-way ANOVA; ns = not significant. (E) Indirect immunofluorescence of Notchint (green) in midguts from control flies and Mex >Pex5-i flies. DNA was stained by DAPI (blue). Scale bar, 20 μm. (F) Number of Notchint-positive cells per ROI of midguts from control flies and Mex >Pex5-i flies. Values represent the median of 20 midgut ROIs ± SD. Statistical significance was determined using one-way ANOVA; **p < 0.01. (G) Indirect immunofluorescence of β-galactosidase (green) in midguts from control (Gbe+Su(H)-LacZ) flies and Mex >Pex5-i; Gbe+Su(H)-LacZ flies. DNA was stained by DAPI (blue). Scale bar, 20 μm. (H) Number of β-galactosidase positive cells per ROI of midguts from control flies and Mex >Pex5-i flies. Values represent the median of 20 midgut ROIs ± SD. Statistical significance was determined using one-way ANOVA; *p < 0.05. (I) Indirect immunofluorescence of Prospero (red) to detect EEs in midguts from control flies and Mex >Pex5-i flies. DNA was stained by DAPI (blue). Scale bar, 20 μm. (J) EEs as a percentage of all cells in midguts from control flies and Mex >Pex5-i flies. Values represent the median of 20 midgut ROIs ± SD. Statistical significance was determined using one-way ANOVA; ns = not significant. (K) Quantification of upd3 transcript in midguts from control, Mex >Pex5-i, and Mex >Cat-i flies fed cornmeal or cornmeal with 1 mM NAC, as designated. Values reported are the averages of three independent experiments ± SD. Statistical significance was determined using one-way ANOVA; ****p < 0.0001; ***p < 0.001.
ECs that are stressed and are going to undergo apoptosis produce cytokines that activate Jak/Stat signaling in ISCs to promote their compensatory division (Buchon et al., 2009; Jiang et al., 2009). Activation of the JAK-STAT pathway can be monitored by increased expression of the upd3 gene encoding the cytokine, Unpaired 3. Expression of upd3 increased greatly in midguts from Mex >Pex5-i (Figure 4K). These results confirm that the epithelial damage in the midgut caused by dysfunctional peroxisomes induces a compensatory proliferation of ISCs for damage repair.
We next investigated the effects of having dysfunctional peroxisomes in ISCs on tissue homeostasis. We depleted Pex5 in ISCs and EBs by expressing dsRNA targeting Pex5 from the escargot-promoter-driven Gal4 (esg-Gal4) to give esg>Pex5-i flies. We found that there were approximately three times more PH3-positive mitotic cells in dissected midguts from esg>Pex5-i flies than in midguts from control flies (Supplemental Figure S3, A and B). esg-Gal4 flies have also a UAS-GFP construct in their genomic background that marks both ISCs and EBs in the midgut by GFP fluorescence. GFP fluorescence showed greater numbers of ISCs and EBs in midguts from esg>Pex5-i flies than in midguts from control flies (Supplemental Figure S3, C and D), as well as increased numbers of cell clusters marked by GFP fluorescence in midguts from esg>Pex5-i flies than from control flies (Supplemental Figure S3C, arrowheads). In contrast, there was no difference in the number of EEs stained by antibodies to Prospero in midguts of esg>Pex5-i flies and control flies (Supplemental Figure S3, E and F), and cell death as measured by TUNEL staining was not significantly different in the midguts of control flies and esg>Pex5-i flies (Supplemental Figure S3, G and H).
Gut epithelial cells with dysfunctional peroxisomes accumulate fatty acids and are subject to redox stress
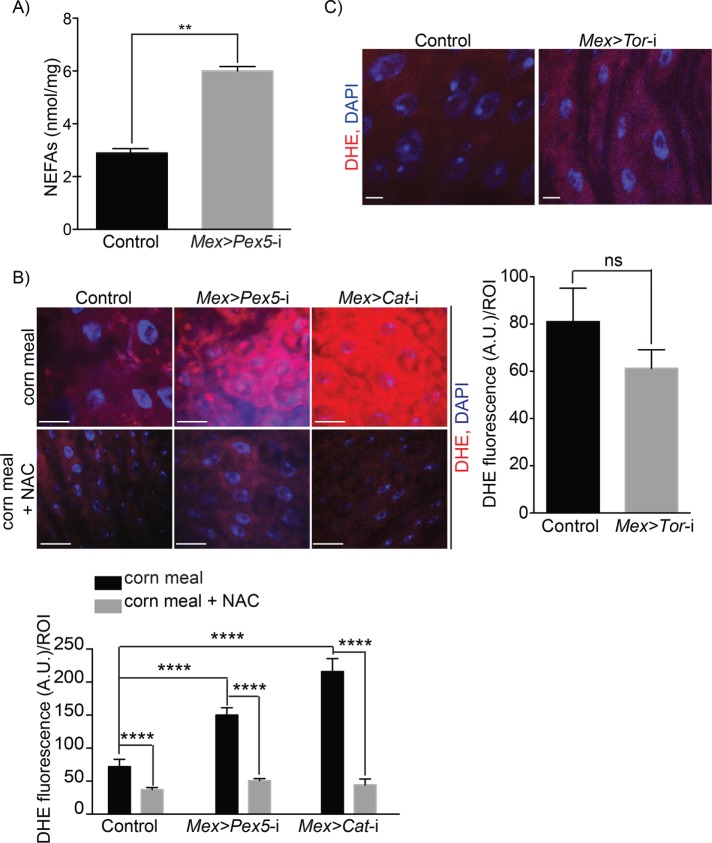
Peroxisomes regulate the synthesis and turnover of complex lipids and reactive species (Wanders and Waterham, 2006). Cells lacking peroxisomes accumulate nonesterified fatty acids (NEFAs), which results in lipotoxicity and contributes to oxidative stress (Titorenko and Terlecky, 2011; Song et al., 2014; Bulow et al., 2018), two factors that can lead to AMP-dependent induction of autophagy (Martino et al., 2012; Hasnain et al., 2016). As expected, we found greater amounts of NEFAs in midguts from Mex >Pex5-i flies than in midguts from control flies (Figure 5A). Dysfunctional peroxisomes also generate cellular redox stress, to which the accumulation of NEFAs contributes. We assessed the redox status of midguts by staining with dihydroethidium (DHE). DHE is used primarily in assessing superoxide production (Owusu-Ansah and Banerjee, 2009), but it can also be used to monitor the redox status of cells or tissues (Zielonka and Kalyanaraman, 2010). We observed that DHE signal was greater in midguts from Mex >Pex5-i flies than in midguts from control flies (Figure 5B). The increase in redox observed in midguts from Mex >Pex5-i flies was comparable to the increase in redox observed in midguts with reduced peroxisomal catalase (Mex >Cat-i) (Figure 5B and Supplemental S1A), the enzyme that converts H2O2 to water and molecular oxygen (Wanders and Waterham, 2006). Defects in peroxisomal catalase can also cause mitochondrial damage in mammals, including damage to the respiratory chain with the consequent accumulation of redox species including superoxide (Koepke et al., 2008; Ivashchenko et al., 2011; Hwang et al., 2012; Walton and Pizzitelli, 2012). However, DHE signals dropped to control amounts in midguts from Mex >Pex5-i flies and Mex >Cat-i flies fed for 48 h on cornmeal medium supplemented with the antioxidant, N-acetylcysteine (NAC) (Figure 5B). Mex >Cat-i flies also showed increased apoptosis in the gut (Supplemental Figure S2, C and D) like Mex >Pex5-i flies and Mex >Tor-i flies and increased expression of upd3 (Figure 4K). The expression of upd3 in midguts of Mex >Pex5-i flies and Mex >Cat-i flies fed cornmeal supplemented with 1 mM NAC was similar to upd3 expression in control midguts (Figure 4K). These results confirm that redox stress induced by dysfunctional peroxisomes produces gut epithelial damage.
FIGURE 5:
Gut epithelium with dysfunctional peroxisomes accumulates NEFAs and exhibits increased redox stress. (A) Amounts of NEFAs in midguts from control flies and Mex >Pex5-i flies. Values reported represent the averages of three independent experiments ± SD. Statistical significance was determined using Student’s t test; **p < 0.01. (B) DHE staining (red) showing increased amounts of ROS in midguts from Mex >Pex5-i flies and Mex >Cat-i flies compared with ROS amounts in midguts from control flies (top panels). Feeding the antioxidant NAC to flies reduces ROS amounts in midguts of Mex >Pex5-i flies and Mex >Cat-i flies to amounts observed in control fly midguts (bottom panels). DNA was stained by DAPI (blue). Scale bar, 20 μm. Quantification of DHE fluorescence per ROI in midguts from flies of the reported genotypes and under the given conditions. n = 20 midguts for each genotype under each condition. Values reported represent the averages of three independent experiments ± SD. Statistical significance was determined using two-way ANOVA; ****p < 0.0001. (C) Fluorescence microscopy images of DHE staining (red) showing equal amounts of ROS in midguts from control flies (left image) and Mex >Tor-i flies (right image). DNA was stained by DAPI (blue). Scale bar, 10 μm. Quantification of DHE fluorescence per ROI of midguts of control flies and Mex >Tor-i flies. Values reported represent the average ± SD of 20 midgut ROIs for each genotype. Statistical significance was determined using one-way ANOVA; ns = not significant.
To evaluate whether peroxisomal redox stress acts upstream or downstream of Tor kinase-induced autophagy and epithelial damage, we monitored redox levels in Mex >Tor-i midgut cells by staining with DHE. We observed no significant change in the amount of DHE staining in Mex >Tor-i midgut cells compared with control midgut cells (Figure 5C). Overall, these results suggest that Tor kinase-induced autophagy leads to increased cell death and epithelial instability in the gut in response to possible lipotoxicity and greater redox stress in flies whose gut epithelial cells contain dysfunctional peroxisomes.
Redox stress in gut epithelium cells with dysfunctional peroxisomes causes AMPK-dependent Tor kinase inhibition and increased autophagy
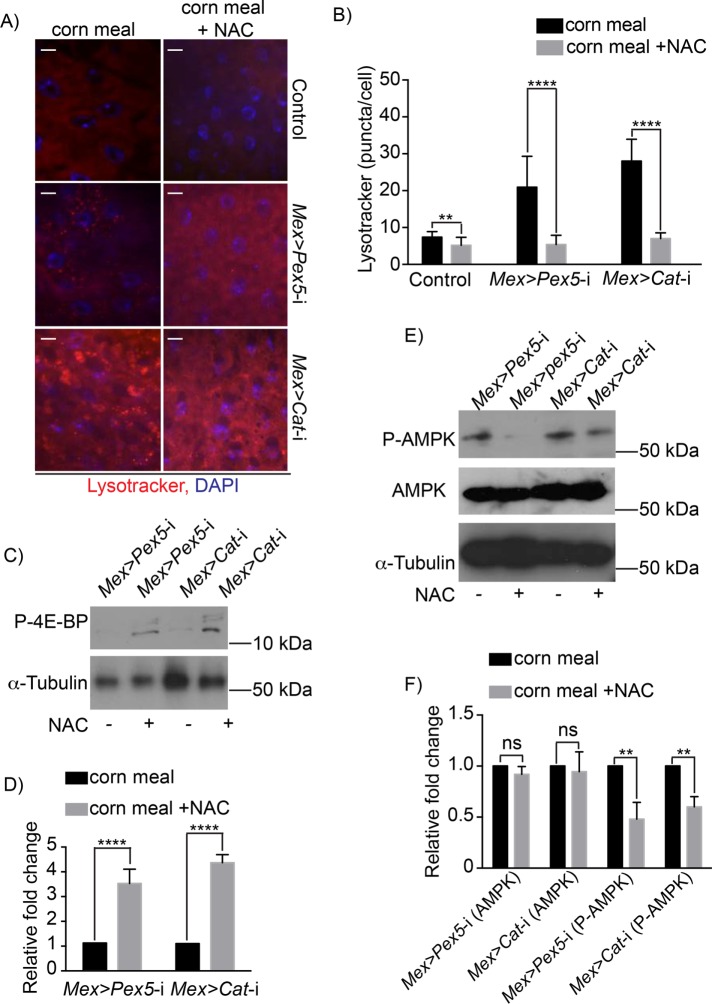
To determine whether the increased redox stress observed in Mex >Pex5-i midguts resulted in inhibition of Tor kinase and increased autophagy, we fed Mex >Pex5-i flies and Mex >Cat-i flies with the antioxidant NAC. Ingestion of NAC reduced the number of Lysotracker-positive vesicles in midguts from Mex >Pex5-i flies and Mex >Cat-i flies fed cornmeal alone (Figure 6, A and B). NAC ingestion also rescued the phosphorylation of 4E-BP, the target of Tor kinase, in midguts from Mex >Pex5-i flies and Mex >Cat-i flies (Figure 6, C and D), indicating that reducing the oxidative stress releases the inhibition on Tor kinase activity in midguts of Mex >Pex5-i flies and Mex >Cat-i flies.
FIGURE 6:
Increased redox stress in fly guts with dysfunctional peroxisomes enhances AMPK-Tor-mediated autophagy. (A) Lysotracker staining (red) of midguts from flies of the designated genotypes. Note increased Lysotracker staining in midguts of Mex >Pex5-i flies and Mex >Cat-i flies compared with control flies. The increase in Lysotracker staining in midguts of Mex >Pex5-i flies and Mex >Cat-i flies is reversed on oral administration of NAC. DNA was stained by DAPI (blue). Scale bar, 10 µm. (B) Quantification of Lysotracker fluorescence. The values reported represent the average of 20 independent images ± SD for each midgut under each condition. Statistical significance was determined using two-way ANOVA; ****p < 0.0001; **p < 0.01. (C) Representative Western blot showing that P-4E-BP amounts are increased in midguts of MexPex5-i flies and Mex >Cat-i flies given food supplemented with NAC. α-Tubulin served as a control for protein loading. (D) Quantification of P-4E-BP amounts normalized to the amounts of α-tubulin. Values represent the median of four independent experiments ± SD. Statistical significance was determined using Student’s t test; ****p < 0.0001. (E) Representative Western blots showing that while total AMPK amounts do not change in midguts of Mex >Pex5-i flies and Mex >Cat-i flies fed food with or without NAC, the amounts of P-AMPK are reduced in the midguts of both Mex >Pex5-i flies and Mex >Cat-i flies fed food supplemented with NAC. α-Tubulin served as a control for protein loading. (F) Quantification of P-AMPK amounts normalized to amounts of total AMPK. Values represent the median of four independent experiments ± SD. Statistical significance was determined using Student’s t test; **p < 0.01; ns = not significant.
We next investigated whether redox-dependent inhibition of Tor kinase in the midgut is mediated by AMPK activation in response to oxidative stress by analyzing the effects of administering NAC to Mex >Pex5-i flies and Mex >Cat-i flies on AMPK phosphorylation in the midgut. Western blot analysis showed that P-AMPK levels are high in Mex >Pex5-i and Mex >Cat-i midguts from flies fed cornmeal alone but significantly reduced in midguts from flies fed cornmeal containing NAC (Figure 6, E and F).
Peroxisomes modulate host-microbe interaction in the gut
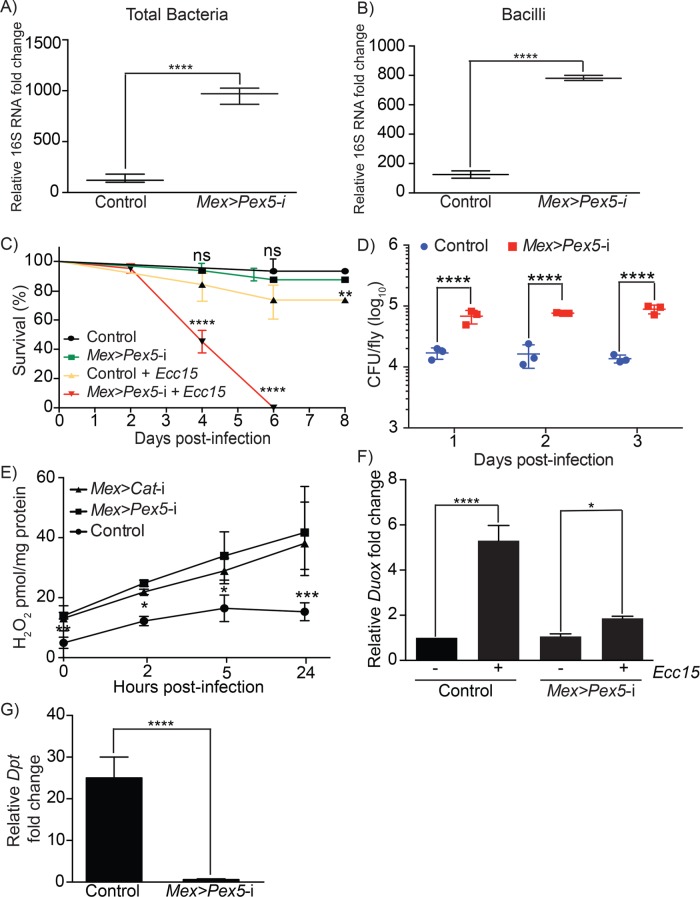
The composition of the intestinal microbiota can be influenced by genetics, metabolic status, redox status, and diet (Nicholson et al., 2012). We characterized the overall bacterial load and the bacillus-specific bacterial load in midguts from control flies and Mex >Pex5-i flies by quantitative PCR using universal primers or bacillus-specific primers to bacterial 16S rRNA. Bacilli constitute one of the largest bacterial classes in the Drosophila gut (Broderick and Lemaitre, 2012). Significant increases in overall bacterial load and in bacillus-specific load were observed in midguts from Mex >Pex5-i flies compared with midguts from control flies (Figure 7, A and B). Our data show that altered peroxisomal function in the gut epithelium results in dysbiosis of the gut microbiota.
FIGURE 7:
Dysfunctional peroxisomes in gut epithelial cells lead to dysbiosis of the gut microbiota and compromised host-pathogen interaction. Quantification of total bacteria (A) and bacillus-specific bacteria (B) assayed by qPCR of the 16S rRNA gene in dissected intestines from control and Mex >Pex5-i flies. Values reported represent the average of four independent experiments. Statistical significance was determined using Student’s t test; ****p < 0.0001. (C) Survival of adult control and Mex >Pex5-i flies fed 5% sucrose or under chronic oral infection with Ecc15. Values reported represent the averages of three independent experiments ± SD. Statistical significance was determined using the log-rank test (Mantel-Cox); ****p < 0.0001; **p < 0.01; ns = not significant. (D) Adult flies of the indicated genotypes were infected with Ecc15. Bacteria were isolated from flies and plated onto solid nutrient medium, and the numbers of CFUs were counted 1, 2, and 3 d after infection. Note the significant increases in bacterial load in infected Mex >Pex5-i flies. Data report the mean ± SD. Statistical significance was determined using two-way ANOVA; ****p < 0.0001. (E) Relative amounts of endogenous H2O2 in midguts of adult control flies, Mex >Pex5-i flies and Mex >Cat-i flies before infection (0 h) and at 2, 5, and 24 h postinfection with Ecc15. n = 15 for each genotype in a single experiment. Values reported represent the average of three independent experiments ± SD for each genotype at each time point. Statistical significance was determined using two-way ANOVA; ***p < 0.001; **p < 0.01; *p < 0.05. (F) Quantification of the fold change in Duox expression in adult guts from control and Mex >Pex5-i flies after infection with Ecc15 relative to guts from uninfected flies of the corresponding genotype. Values reported are the averages of three independent experiments ± SD. Statistical significance was determined using one-way ANOVA; ****p < 0.0001; *p < 0.05. (G) Quantification of the fold change in Dpt expression in adult midguts from control and Mex >Pex5-i flies after infection with Ecc15 relative to midguts from uninfected flies of the corresponding genotype. Values reported are the averages of three independent experiments ± SD. Statistical significance was determined using one-way ANOVA; ****p < 0.0001.
We next probed whether the survival of Mex >Pex5-i flies was affected by immune challenge. Mex >Pex5-i flies that eclosed as adults showed a survival rate similar to that of control Mex >w1118flies; however, when challenged by chronic oral infection with the nonpathogenic Gram-negative bacterium, E. carotovora (Ecc15), Mex >Pex5-i flies died at a higher rate than control flies until 6 d postinfection when no Mex >Pex5-i flies survived (Figure 7C). Similarly, Mex >Pex5-i flies under chronic oral infection with the pathogenic Gram-negative bacterium, P. entomophila, were more sensitive to infection than control flies (Supplemental Figure S4A). Quantitative assessment showed that the pathogen load was greater for Mex >Pex5-i flies than for control flies when infected with Ecc15 (Figure 7D) or P. entomophila (Supplemental Figure S4B). We also tested the ability of esg>Pex5-i flies to survive chronic infection by P. entomophila. Uninfected and infected adult esg>Pex5-i flies showed rates of survival similar to those of corresponding uninfected and infected control flies (Supplemental Figure S4C).
We also determined whether manipulation of Tor kinase impacts fly survival in response to chronic oral infection with Ecc15. Uninfected flies of every analyzed genotype did not show reduced viability compared with uninfected control flies (Supplemental Figure S4D). When infected, half of Mex >Pex5-i and Mex >Tor-i flies died after 3.5 d (Supplemental Figure S4E). After 6 d, all Mex >Pex5-i flies and Mex >Pex5-i; Tor-i flies died, while all Mex >Tor-i flies died at day 8 postinfection. However, Mex >UAS-Tor flies and Mex >Pex5-i; UAS-Tor flies did not show reduced viability compared with infected control flies. Together our data support a scenario in which gut cells with dysfunctional peroxisomes exhibit inhibition of Tor kinase, leading to increased autophagy in cells and increased sensitivity of flies to chronic bacterial infection.
Peroxisomes affect barrier epithelium immunity in the gut
Two conserved innate immune systems act in the Drosophila gut. One produces microbicidal reactive oxygen species (ROS) that act as a first line of defense to combat opportunistic pathogens (Ha et al., 2005a, 2009; Bae et al., 2010); the other, which is termed the immune deficiency (IMD) pathway and is the Drosophila homologue of the mammalian TNF pathway, acts as a second line of defense that is responsible for the activation and nuclear translocation of Relish (Rel), the Drosophila NF-κB, to produce a battery of antimicrobial peptides (AMPs) (Lemaitre and Hoffmann, 2007; Kleino and Silverman, 2014).
One immediate epithelial innate immune response strategy in both Drosophila and mammals involves the generation of sufficient ROS to combat the pathogen while concurrently eliminating residual ROS to protect the host from oxidative damage (Kinnula et al., 1992; Geiszt et al., 2003; Ha et al., 2005b). Failure to balance the synthesis and elimination of ROS can lead to chronic epithelial inflammatory diseases (Hoidal, 2001). We assessed the amounts of H2O2 in midguts dissected from 4- or 5-d-old control flies or Mex >Pex5-i flies before infection with the bacterium Ecc15 and at 2, 5, and 24 h after infection (Figure 7E). The amounts of endogenous ROS were significantly elevated in the midgut of Mex >Pex5-i flies compared with the midgut of control flies at every time analyzed. The elevated ROS amounts observed in midguts from Mex >Pex5-i flies were comparable to those observed in midguts with reduced peroxisomal catalase (Mex >Cat-i) (Figure 7E).
In the gut, microbicidal ROS are produced as a consequence of the increased expression of Dual Oxidase (Duox) encoding nicotinamide adenine dinucleotide phosphate (NADPH) oxidase (Ha et al., 2005b). We measured the amounts of Duox mRNA in midguts from adult flies under chronic infection by Ecc15. Midguts from control flies exhibited strong induction of Duox expression after infection, while midguts from Mex >Pex5-i flies showed a much-reduced induction of Duox expression after infection (Figure 7F), suggesting that DUOX is not the source of the increase in ROS in midguts of Mex >Pex5-i flies.
An elevated redox state and activation of the stress response pathway have been proposed to reduce an organism’s fitness during infection (Ha et al., 2005b; Cuenda and Rousseau, 2007; Chakrabarti et al., 2014; Cheesman et al., 2016). We therefore tested whether oral administration of the antioxidant NAC could improve the survival of Mex >Pex5-i flies and Mex >Cat-i flies when challenged by chronic infection with Ecc15. Feeding NAC did not change the survival of uninfected control flies, Mex >Pex5-i flies and Mex >Cat-i flies, which all survived like uninfected control flies not treated with NAC (Supplemental Figure S4F). When infected with Ecc15, 60% of Mex >Pex5-i flies died after 4 d and 50% of Mex >Cat-i flies died after 6 d (Supplemental Figure S4G). Infected Mex >Pex5-i flies and Mex >Cat-i flies died at a faster rate than control flies until day 6 and day 8 postinfection when all Mex >Pex5-i flies and all Mex >Cat-i flies, respectively, were dead (Supplemental Figure S4G). However, infected Mex >Pex5-i flies and Mex >Cat-i flies fed NAC did not show reduced viability compared with infected control flies not exposed to NAC (Supplemental Figure S4G). Conversely, NAC ingestion made control flies extremely sensitive to bacterial infection, as 100% of these flies were dead 2 d postinfection (Supplemental Figure S4G). This is not unexpected, since a rise in ROS in the gut is necessary as a first line of defense during infection, and the inability to increase ROS might cause flies to be highly vulnerable to microbial challenge.
To evaluate whether peroxisomes have a role in modulating IMD-dependent immune signaling in the fly gut, as was previously demonstrated for other tissues (Di Cara et al., 2017), we measured expression of the Drosophila AMP gene, Dpt, in midguts dissected from adult flies under chronic infection by Ecc15. Midguts from control flies exhibited strong induction of Dpt expression after infection, while midguts from Mex >Pex5-i flies showed essentially no induction of Dpt expression after infection (Figure 7G). Dpt expression is regulated by Rel, which translocates to the nucleus of cells on their infection (Lemaitre and Hoffmann, 2007). Indirect immunofluorescence showed nuclear accumulation of Rel in midgut cells of adult flies 4 h after oral infection with Ecc15, while no nuclear localization of Rel was detected in midgut cells of Mex >Pex5-i adult flies (Supplemental Figure S5A), confirming down-regulation of immune signaling in these cells. Forkhead box, subgroup O (FOXO) signaling has been reported also to drive the production of AMPs in response to infection and metabolic stress (Becker et al., 2010; Fink et al., 2016). Indirect immunofluorescence showed nuclear accumulation of FOXO in both control and Mex >Pex5-i midgut cells 1 h after metabolic stress (feeding 5% sucrose) (Supplemental Figure S5B). In contrast, nuclear localization of FOXO was observed in unstressed (20% sucrose) control midgut cells but not in Mex >Pex5-i midgut cells 1 h after oral infection with Ecc15 (Supplemental Figure S5B), demonstrating that FOXO activity is affected in the immune response but not the metabolic stress response of midgut cells with dysfunctional peroxisomes.
We next tested whether the elevated redox state in midguts of Mex >Pex5-i flies and Mex >Cat-i flies affected their enteric immune response by determining whether NAC feeding was able to rescue the increase in Dpt expression in the midgut observed on infection of control flies with Ecc15. Midguts from control flies exhibited a large increase in Dpt transcript amounts 4 h after oral infection with Ecc15 (Supplemental Figure S5C). In contrast, midguts from infected Mex >Pex5-i flies and Mex >Cat-i flies did not exhibit the same increase in Dpt transcript amounts. Oral administration of NAC rescued Dpt transcript amounts in midguts of Mex >Pex5-i flies and Mex >Cat-i flies (Supplemental Figure S5C), demonstrating that redox imbalance in midgut cells from Mex >Pex5-i flies and Mex >Cat-i flies results in defects in the NF-κB-mediated immune response.
The mitogen-activated protein kinase p38 phosphorylates various substrates in response to a wide range of physical, chemical, and biological stresses to regulate cellular adaptation to stress (Qi and Elion, 2005) and host defense (Chen et al., 2010; Chakrabarti et al., 2014). In Drosophila, activation of p38 regulates the transcription of Duox (Ha et al., 2009). Activation of the conserved p38 MAPK pathway can be measured by antibody detection of phosphorylated p38 (P-p38). Indirect immunofluorescence analysis showed that P-p38 is not detectable in midgut cells from control uninfected flies but is increased in amount and detectable in the nuclei of midgut cells from uninfected Mex >Pex5-i adult flies (Supplemental Figure S5E). P-p38 was also readily detectable in the nuclei of midgut cells from control flies and Mex >Pex5-i flies chronically infected with Ecc15 (Supplemental Figure S5D) (Ha et al., 2009). The redox state of midgut cells has an apparent role in p38 activation, because reduction in the redox state of midgut cells isolated from uninfected Mex >Pex5-i flies fed NAC reduced P-p38 to amounts comparable to those observed in midgut cells from uninfected control flies (Supplemental Figure S5E).
DISCUSSION
The gastrointestinal tract functions in digestion and absorption of nutrients and provides the first line of defense against ingested pathogens (Lemaitre and Miguel-Aliaga, 2013). The intestinal epithelium also takes part in mutually beneficial interactions with commensals that help shape the host immune system (Clemente et al., 2012; Hooper et al., 2012; Nicholson et al., 2012; Guo et al., 2014). Changes in gut epithelium physiology are associated with a variety of disorders, including inflammatory bowel disease, autoimmune and allergenic diseases, obesity, and diabetes (Clemente et al., 2012), and can potentially influence cancer (Kaser et al., 2010) where changes in gut epithelium correlate with patients’ capacity to respond to cancer immunotherapy (Gopalakrishnan et al., 2018). Understanding how enteric health is maintained requires understanding the signaling that controls metabolism and stress response in the gut, as well as the interaction between gut and microbes. The Drosophila intestine is an excellent model with which to characterize enteric metabolic signaling, host-pathogen/commensal interactions, innate immune signaling, and tissue regeneration (Buchon et al., 2013; Lemaitre and Miguel-Aliaga, 2013). Here we report that increased cell death and Tor kinase-dependent autophagy present in gut epithelial cells with dysfunctional peroxisomes compromise enteric structure and function, and reduce organismal health.
There is mounting evidence that the different functions of peroxisomes are critical for the maintenance of human health and that peroxisomes are pivotal in the development, activity, and survival of different tissues. For example, peroxisomes are increasingly recognized as mediators of neural trophic survival and function in the brain, in the production of specialized lipid species like plasmalogens, and for muscle function (Braverman et al., 2013). The notable abundance of peroxisomes in gut epithelial cells (Novikoff and Novikoff, 1972; Beard and Holtzman, 1987; Faust et al., 2014; Morvay et al., 2017) prompted us to investigate a potential role for peroxisomes in the maintenance and regulation of gut epithelial physiology.
Pex5 functions as a shuttling receptor for peroxisomal protein import, delivering proteins from the cytosol to the peroxisome matrix. Reduction in Pex5 levels affects peroxisome biogenesis and peroxisome function across organisms, including Drosophila (Baron et al., 2016). We used RNAi-mediated Pex5 gene silencing to produce flies (Mex >Pex5-i) lacking functional peroxisomes in the midgut epithelium. Mex >Pex5-i flies are affected in their development, as approximately half of Mex >Pex5-i flies die by the third larval stage. Analysis of the ultrastructure of control and Mex >Pex5-i adult fly midguts showed that cells of Mex >Pex5-i midguts accumulate lysosomes. Lysosomes are responsible for the degradation of macromolecules derived from the extracellular space by endocytosis or phagocytosis, as well as from the cytoplasm by autophagy (Eskelinen, 2006; Luzio et al., 2007). Immunofluorescence detection of the autophagy-specific protein ATG1 confirmed that the vesicles in Mex >Pex5-i midgut cells participate in autophagy. Autophagy is a self-degradative cellular process that is important for balancing sources of energy during development and in response to different types of stress (Glick et al., 2010). Primary metabolic activities of peroxisomes are to regulate the synthesis and turnover of complex lipids and reactive species (Wanders and Waterham, 2006). Cells lacking peroxisomes accumulate NEFAs that lead to lipotoxicity and contribute to oxidative stress (Titorenko and Terlecky, 2011; Song et al., 2014; Bulow et al., 2018). Adult midguts dissected from Mex >Pex5-i flies showed abnormal peroxisomal metabolism and exhibited elevated amounts of NEFAs and high redox status. Interestingly, liver-specific knockout of peroxisomes in mouse did not result in increased oxidative stress (Dirkx et al., 2005).
Although NEFAs are a key energy source in the body, increased NEFA load can lead to chronic inflammation and increased oxidative stress that ultimately affect tissue stability and animal health (Wood et al., 2009; Soardo et al., 2011; Phielix et al., 2014; Song et al., 2014; Rodriguez-Carrio et al., 2016; Bulow et al., 2018). The increased autophagy in the gut epithelium of Mex >Pex5-i flies argues for a rapid cellular response to NEFA lipotoxicity and augmented redox production that is mediated by stress-sensitive proteins, among which AMPK could be a candidate. Experiments in mammals have demonstrated that AMPK controls autophagy through Tor kinase (Filomeni et al., 2015), and this signaling pathway has recently been shown to be active in the gut of Drosophila (Chakrabarti et al., 2012). Moreover, NEFAs can activate AMPK-Tor-dependent autophagy and promote cell death in mammals (Martino et al., 2012; Hasnain et al., 2016). Indeed, we observed activation of AMPK and inhibition of Tor kinase activity in midguts from Mex >Pex5-i flies that exhibited increased autophagy and apoptosis in epithelial cells. Interestingly, peroxisome-dependent modulation of Tor kinase has been proposed to promote homeostasis in mammalian cells (Zhang et al., 2013; Tripathi and Walker, 2016).
Treatment with antioxidants can partially or completely reverse the autophagic response (Filomeni et al., 2015). Increased AMPK activation, Tor kinase inhibition, and autophagy activation with its consequent elevated epithelial cell death can be reversed in Mex >Pex5-i flies and Mex >Cat-i flies by their ingestion of the antioxidant NAC, consistent with dysfunctional peroxisomes causing redox stress in the gut. One mechanism by which AMPK can inhibit Tor kinase in fly and mammalian cells involves phosphorylation of Tsc2, which subsequently suppresses the activity of Tor kinase through deactivation of Rheb-GTPase (Hay and Sonenberg, 2004). Intriguingly, reduction in TSC2 expression in gut epithelial cells with dysfunctional peroxisomes reversed the increased autophagy in these cells.
Intestinal epithelia renew as a normal course, as well as in response to tissue damage. Impaired intestinal renewal can lead to inflammatory bowel disease and cancer in humans (Amcheslavsky et al., 2009). The division of Drosophila ISCs can be regulated by tissue damage, as ingestion of damaging substances, metabolic stress, or pathogenic bacteria trigger epithelial injury that in turn increases the division of ISCs to compensate for the damage-induced loss of cells (Amcheslavsky et al., 2009; Jiang et al., 2009; Opota et al., 2011). Dysfunctional peroxisomes in Drosophila gut epithelium enhanced apoptosis, which in its turn activated ISCs to proliferate in an attempt to replace damaged ECs. However, while the proliferation of ISCs in the absence of infection is sufficient to maintain intestinal homeostasis and animal health under normal conditions, flies with dysfunctional peroxisomes in their gut do not survive microbial stress, probably because of increased epithelial damage and dysplasia caused by microbially induced damage.
Mex >Pex5-i flies that complete development and reach adulthood had a survival rate similar to that of control flies; however, when challenged by chronic oral infection with a nonpathogenic (Ecc15) or a pathogenic (P. entomophila) bacterium, Mex >Pex5-i flies rapidly succumbed to the bacteria. We also observed that DUOX and NF-κB-mediated pathways of defense against pathogenic infection in the gut (Lemaitre and Hoffmann, 2007; Kleino and Silverman, 2014) were reduced in guts with dysfunctional peroxisomes. Administration of the anti-oxidant NAC to Mex >Pex5-i flies reactivated immune signaling in their gut, reversed their sensitivity to bacterial infection, and restored their ability to survive chronic bacterial infection. However, inhibition of immune signaling in guts of Mex >Pex5-i flies appears not to be the only reason for their sensitivity to microbial challenge. Mex >Tor-i flies were more sensitive to chronic oral infection with the bacterium Ecc15 than control flies, while overexpression of Tor kinase in Mex >Pex5-i flies rescued their ability to survive infection. Therefore, the increased sensitivity of Mex >Pex5-i flies to infection depends strongly on the inactivation of Tor kinase, which in its turn leads to hyperactivation of autophagy and subsequent accelerated cell death in the intestine. In addition, we found that compromised peroxisome function in gut epithelial cells resulted in Tor kinase-dependent inhibition of global translation, a process that has been reported to reduce the immune response (Jaramillo et al., 2011; Chakrabarti et al., 2012; Dunbar et al., 2012). Altogether, impaired metabolic signaling, increased autophagy-induced epithelial cell death, and a reduced immune response make Mex >Pex5-i flies less able to respond to immune challenge and to die earlier.
The intestinal epithelium also takes part in mutually beneficial interactions with commensal bacteria (Clemente et al., 2012; Hooper et al., 2012; Nicholson et al., 2012; Guo et al., 2014). The composition of the intestinal microbiota can be influenced by genetics, metabolic status, redox status, and diet (Nicholson et al., 2012). We found that the overall bacterial load and the bacillus-specific bacterial load were significantly increased in midguts of Mex >Pex5-i flies. Bacilli constitute one of the largest bacterial classes in the Drosophila gut, and their load was reported to increase during aging and in response to gut epithelial damage (Broderick and Lemaitre, 2012). Taken together, our data show that altered peroxisomal function in the gut epithelium affects host–commensal interactions and results in dysbiosis of the gut microbiota that could aggravate the effects of epithelial damage and cellular stress.
In conclusion, our study demonstrates that peroxisomes, ubiquitous organelles involved in lipid metabolism and redox balance, are required in the gut to maintain metabolic homeostasis needed for the survival and development of Drosophila and for gut epithelium homeostasis and renewal in adult flies. Dysfunctional peroxisomes in intestinal epithelial cells activate Tor kinase-dependent autophagy that leads to increased cell death and epithelial instability, which ultimately alter the composition of the intestinal microbiota, compromise immune pathways in the gut in response to infection, and affect the survival of the organism. Peroxisomes in the gut therefore effectively function as hubs that coordinate the responses from stress, metabolic, and immune pathways to maintain enteric health and to regulate the gut’s response to a constantly changing microbiological environment. Given that peroxisomes, peroxisomal functions, and AMPK-Tor autophagic signaling are conserved across the breadth of eukaryotes, our findings on the roles of peroxisomes in tissue homeostasis, metabolic balance, and immune response in the Drosophila gut are germane to an increased understanding of how peroxisomes contribute to the maintenance and modulation of gut homeostasis and mucosal epithelial immunity in mammals, including humans, under conditions of health or disease.
MATERIALS AND METHODS
Fly stocks, husbandry, and infection
Stocks.
Pex5 dsRNA expressing lines y1 v1; PattP40 (Tripathi and Walker, 2016) and w1118; P{GD14972}v42332 were from the Bloomington Drosophila Stock Center (BDSC) and the Vienna Drosophila Resource Center (VDRC), respectively. The Catalase expressing line y1 sc* v1; P{y[+t7.7] v[+t1.8] = TRiP.HMS00990}attP2, the Tor dsRNA expressing line y1 sc* v1; P{y[+t7.7] v[+t1.8] = TRiP.HMS00904}attP2, and the Tor cDNA expressing line y1 w1; P{ry[+t7.2] = hsFLP}12; P{w[+mC] = UAS-Tor. WT were from the BDSC. The TSC2 dsRNA expressing line w1118; P{KK100646}VIE-260Bv 103417 was from the VDRC. The midgut-specific driver, Mex-GAL4, line w1118; P{mex1-GAL4.2.1} (Phillips and Thomas, 2006) was a gift from Kirst King-Jones, University of Alberta. The esg-GAL4, UAS-GFP; Gal80ts was a gift from Edan Foley, University of Alberta. The GBE+Su(H)-LacZ line was a gift from Benjamin Ohlstein, Columbia University.
Husbandry.
Drosophila lines were maintained at 25°C on standard BDSC cornmeal medium; 3- to 5-d-old adult female flies were used.
Infection.
For oral infection, flies were cultured on filter paper soaked in 5% sucrose or 5% sucrose containing Ecc15 (OD600 = 200) (Basset et al., 2000) or P. entomophila (OD600 = 35) (Vodovar et al., 2005). Flies were transferred to fresh vials every 2 d, and the number of dead flies was determined daily. Survival experiments were done with 20–30 flies tested for each group. For studies of FOXO localization, flies were cultured on filter paper soaked in 5% sucrose (metabolically stressed) or 20% sucrose (unstressed). For treatments with NAC or compound C, flies were fed a mixture of Ecc15 (OD600 = 200) and 5% sucrose to which NAC (Sigma) or compound C (Merck) was added to 20 mM and 100 μM final concentrations, respectively.
Ecc15 was a kind gift from Edan Foley, University of Alberta. P. entomophila was a kind gift of Nicolas Buchon, Cornell University. Bacteria were grown in Luria–Bertani medium at 29°C for 24 h before flies were infected.
Drosophila S2 cell lineage.
Drosophila S2 cells were obtained from the Drosophila Genomics Resource Center and grown in SFX-Insect Cell Culture Medium supplemented with 50 U penicillin/ml and 50 μg streptomycin sulfate/ml. The sex of animals from which S2 cells were derived is unknown.
Reagents
Rabbit antibodies to Lamp1 (ab30687), and Alexa Fluor 488 and Alexa Fluor 568 donkey anti-mouse (ab150101 and ab175699, respectively) or donkey anti-rabbit (ab150069 and ab175693, respectively) secondary antibodies, were from Abcam. Antibodies to P-JNK (#4668), P-p38 (#9211), P-4E-BP (#2855), P-p70S6 kinase (#9209), P-eIF2α (#3597), and P-AMPK (#2535) were from Cell Signaling. Antibodies to eIF2α (AV41041) and to α-tubulin (T5168) were from Sigma-Aldrich. Antibodies to AMPK (GTX42788) were from GeneTex. Antibodies to Notch intracellular domain (Notchint) (C.17.9C6) and to Delta extracellular domain (C.594.9B) were raised by Spyros Artavanis-Tsakonas, Harvard Medical School, and obtained from the Developmental Studies Hybridoma Bank. Antibodies to Prospero (MR1A) were raised by Chris Doe, University of Oregon, and obtained from the Developmental Studies Hybridoma Bank. Antibodies to Relish (C 21F3) were raised by Svenja Stoven at Umeå University and obtained from the Developmental Studies Hybridoma Bank. Antibodies to ATG1 were a kind gift of Jun Hee Lee, University of Michigan. Antibodies to phosphohistone H3 (06-570) were from Upstate Biotechnology. Antibodies to FOXO were raised by Robert Tjian, University of California, Berkeley. Antibodies to β-galactosidase (40-1a) were raised by Joshua Sanes, Harvard Medical School, and obtained from the Developmental Studies Hybridoma Bank. Antibodies to amino acids 416–430 of human Pex5 (SAB1100459) were from Sigma-Aldrich. Antibodies to the PTS1 Ser-Lys-Leu (SKL) were raised by Richard Rachubinski.
DHE staining
Flies were dissected and incubated with 30 μM DHE and Hoechst 33342 for 5 min and fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) for 4 min. Guts were then washed three times in PBS and imaged.
Lysotracker staining
All incubations and washes were conducted in the dark and at room temperature. Flies were dissected and incubated with 0.8 µM Lysotracker for 3 min, washed three times for 5 min each with PBS, incubated in 4% paraformaldehyde in PBS for 20 min room temperature, washed three times in 0.1% Triton X-100 in PBS for 5 min each, and imaged immediately by confocal microscopy.
Measurement of H2O2
For H2O2 measurement, guts from uninfected or infected flies were homogenized in PBS and clarified by centrifugation. H2O2 amounts in the resultant supernatants were measured using the Amplex Red Hydrogen Peroxide/Peroxidase Assay Kit (Thermo Fisher) and normalized to protein amounts. Protein amounts were measured using a Qubit II fluorimeter (Thermo Fisher). Experiments were done in triplicate.
Measurement of nascent protein synthesis
The Click-iT AHA for Nascent Protein Synthesis kit (Invitrogen) was used to measure the global level of translation in fly midgets. Reactions were performed according to the manufacturer’s instructions.
Measurement of NEFAs
NEFAs from isolated guts, whole flies, or larvae were measured using the copper-triethanolamine method, as modified (Tinnikov and Boonstra, 1999). Tissue was homogenized in 20 µl chloroform + 1% Triton X-100 per mg of tissue and subjected to centrifugation at 13,000 × g for 10 min. The supernatant was removed and evaporated at 60°C. Lipids were taken up in the same volume of phosphate buffer, and 25 µl of sample was transferred to a glass vial with 500 µl of chloroform/heptane (4:3). Vials were shaken for 2 min and subjected to centrifugation for 5 min at 2000 × g. The amount of 300 µl of the organic phase was transferred to a glass vial containing 250 µl of copper-triethanolamine, shaken for 2 min, and subjected to centrifugation for 5 min at 2000 × g. The amount of 150 µl of the organic phase was removed and evaporated at 60°C. Lipids were taken up in 150 µl of ethanol, and vials were shaken for 15 min at 37°C. Copper was detected by complexation with a mixture of dicarbazone-dicarbazide, and color intensity was measured in a 96-well plate at 550 nm in a microplate reader (TECAN).
CFU counting
To measure the bacterial burden of the fly gut during infection, an individual gut was isolated 48 h after infection, dipped into 70% ethanol, and air dried. An individual gut was mashed in 100 μl of sterile microorganism culture medium, and the mash was subjected to centrifugation to remove debris. The resultant supernatant was serially diluted, plated onto solid nutrient medium, and incubated at 29°C until there were visible colonies, which were counted.
Genomic DNA isolation
Genomic DNA was extracted using the Blood and Tissue DNA isolation kit (Qiagen). Flies were sterilized on their surfaces by addition of 50% sodium hypochlorite and dissected over ice in sterile PBS. Dissected intestines included all but the anterior foregut, from the point at which the crop diverges, and including the crop, to the rectal papilla. The gut was kept intact to prevent loss of lumenal content. Dissected intestines were stored in sterile tubes at –80°C prior to DNA extraction.
RNA extraction and qRT-PCR
Guts from adult flies were dissected in PBS, transferred to TRIzol reagent (Thermo Fisher), and snap-frozen in liquid nitrogen. Total RNA was extracted using the RNeasy-Micro Kit (Qiagen) following the supplied protocol. RNA was reverse transcribed using the iScript cDNA Synthesis kit (Bio-Rad), and the synthesized cDNA was used for qPCR using the SYBR-Green PCR master mix (Kapa Biosystems) using a Realplex qPCR machine (Eppendorf). Samples were normalized to Rpl23 gene expression using the 2-ΔΔCT method (Livak and Schmittgen, 2001). Forward and reverse primer sequences used in qRT-PCR are, respectively,
Rpl23, 5′-GACAACACCGGAGCCAAGAACC, 5′-GTTTGCGCTGCCGAATAACCAC
Dpt, 5′-ACCGCAGTACCCACTCAATC, 5′-ACTTTCCAGCTCGGTTCTGA
Duox, 5′-TAGCAAGCCGGTGTCGCAATCAAT, 5′-ACGGCCAGAGCACTTGCACATAG
Pex5, 5′-AAATGCGAAGACATGGAACC, 5′-TGTAACGCACACGGATGAAG
Catalase, 5′-GATGCGGCTTCCAATCAGTTG, 5′-GCAGCAGGATAGGTCCTCG
upd3, 5′-GAGCACCAAGACTCTGGACA, 5′-CCAGTGCAACTTGATGTTGC
ATG1, 5′-GAGTATTGCAATGGCGGCGACT, 5′-CAGGAATCGCGCAAACCCAA
ATG3, 5′-TCTTCCAGGTCCCAATATGGCC, 5′-TGAAAAGCATGGCGGGTCTT
ATG8a, 5′-GCAAATATCCAGACCGTGTGCC, 5′-AGCCCATGGTAGCCGATGTT
Bacilli, 5′-CGACCTGAGAGGGTAATCGGC, 5′-GTAGTTAGCCGTGGCTTTCTGG
V1/V2, 5′-AGAGTTTGATCCTGGCTCAG, 5′-CTGCTGCCTYCCGTA
Expression cloning of EGFP-Pex5 in S2 cells
Drosophila Pex5 cDNA was amplified by PCR using the upstream oligonucleotide 5′-CCAAGCTTATGGTGCAGTCGGGGATTTG and the downstream oligonucleotide 5′-CCAAGCTTTTAATCCTTAAAGGCCTCAT, which contain sites for cleavage by HindIII (underlined). The amplified fragment was cloned into the HindIII site of the expression vector, Act-STABLE2-neo (Gonzalez et al., 2011), downstream and in-frame of sequence encoding EGFP. One million S2 cells were transfected with vector using CellFectin II (Thermofisher) according to the manufacturer’s protocol. Cells were checked for EGFP-Pex5 expression 3 d after transfection by fluorescence microscopy, and lysates were prepared from cells for Western blot analysis with anti-human Pex5 antibodies.
Preparation of protein extracts for SDS–PAGE
Cold lysis buffer (70 µl) (Ephrussi-Beadle Ringer’s solution containing 10 mM EDTA, 10 mM dithiothreitol (DTT), and Roche complete protease inhibitor) was added to an isolated fly gut or to a pellet containing 200,000 S2 cells, which was then homogenized. Thirty microliters of hot (70°C) 3 × SDS–PAGE sample buffer containing 10 mM DTT was added to the homogenate, followed by boiling for 10 min. Particulate matter was pelleted by centrifugation at 16,000 × g for 1 min, and the supernatant was transferred to a fresh tube for analysis by SDS–PAGE.
Western blotting
Protein was resolved by SDS–PAGE and transferred to nitrocellulose membranes. Membranes were blocked with 5% bovine serum albumin (BSA) for 1 h at room temperature, incubated for 1 h with primary antibody (1:1000 final dilution in TBST [150 mM NaCl, 20 mM Tris-HCl, pH 7.5, 0.05% Tween 20]) and washed three times for 5 min each with TBST. The washed membranes were incubated with an appropriate secondary antibody for 1 h at room temperature. Membranes were then washed three times for 5 min each with TBS containing 0.2% Triton X-100, and immunocomplexes were detected by enhanced chemiluminescence (ECL; Amersham Biosciences) using an appropriate horseradish peroxidase–linked secondary antibody (Amersham Biosciences).
Immunofluorescence microscopy
Guts were fixed in 4% paraformaldehyde in PBS for 30 min, washed three times in PBST (PBS containing 0.3% Triton X-100), and incubated for 1 h at room temperature in 5% goat serum in PBST and for 16 h at 4°C with primary antibody generally at 1:100 dilution in 5% normal goat serum, although anti-Prospero antibodies were used at 1:50 dilution, anti-FOXO antibodies at 1:1000 dilution, and anti-Notchint antibodies at 1:10 dilution. Tissues were then washed five times in PBST and incubated in the appropriate Alexa Fluor secondary antibody at 1:1000 dilution in 5% normal goat serum. After four washes in PBSTw (PBS containing 0.2% Tween 20), guts were mounted in 4′,6-diamidino-2-phenylindole (DAPI) Prolong-Gold Antifade Reagent (Thermo Fisher) and imaged using a 20× (NA = 0.5) or a 63× (NA = 1.4) oil immersion objective mounted onto an AxioObserver M1 microscope (Zeiss) coupled to an Ultraview ERS spinning disk confocal imager controlled by Volocity imaging software, v6.0 (PerkinElmer). Images were captured using a C9100 electron-multiplying charge-coupled device camera (Hamamatsu) at 130-μm vertical (z) spacing. Values of signal intensity are the average of green or red signal measured on representative fields of 20–25 guts and quantified with Fiji imaging software by recording the median intensity values. Cell numbers expressing different markers on representative fields of 20–25 guts were counted using the “Analyze Particles” module of Fiji. The software automatically counted the nuclei and then automatically counted the respective marker positive spots (Prospero or Delta or Notch) to identify the percentage of Hoechst-positive cells that were also Prospero-, Delta-, or Notch-positive.
Electron microscopy
Guts were fixed in 0.16 M sodium cacodylate buffer, pH 7.4, containing 4% glutaraldehyde, 2% paraformaldehyde, 0.2 M sucrose, and 4 mM CaCl2 at 37°C for 60 min. Cells were then washed with 0.05 M sodium cacodylate buffer, pH 7.4, stained with ice-cold 1% OsO4 in 0.05 M sodium cacodylate buffer, pH 7.4, and washed in 0.05 M sodium cacodylate buffer, pH 7.4. To increase contrast, guts were stained with 1% uranyl acetate in 0.1 M sodium acetate buffer, pH 5.2, for 15 min and washed first in 0.1 M sodium acetate buffer, pH 5.2, and then in distilled, deionized water. Guts were next dehydrated by solutions of increasing ethanol concentration (30, 50, 70, 80, 90, 95, and 100% ethanol), followed by washing in propylene oxide. Guts were infiltrated with a mixture of EMbed 812 and Araldite 502 resins and embedded in gelatin capsules. Resin polymerization was at 60°C for 48 h. Sections (60-nm thickness) were cut on a Leica UC7 ultramicrotome and contrasted by staining in 2% uranyl acetate and Reynold’s lead citrate. Sections were imaged using a Hitachi H-7650 transmission electron microscope at 80 kV and a 16 megapixel XR111 camera (Advanced Microscopy Techniques).
Immunoelectron microscopy
Guts were fixed in in 0.1 M sodium cacodylate buffer, pH 7.4, containing 0.5% glutaraldehyde and 2% paraformaldehyde for 1 h at 4°C, rinsed in 0.075 M sodium cacodylate buffer, pH 7.4, dehydrated with a graded ethanol series (30, 50, 70, and 80% ethanol), and infiltrated with LR White resin (London Resin). Infiltrated samples were embedded in gelatin capsules and polymerized under UV light for 24 h at 4°C. Following polymerization, ultrathin sections (60-nm thickness) were cut and loaded onto a 300-mesh nickel grid without coating.
Before incubation with antibodies, dried sections were blocked overnight at 4°C with 8% BSA in TBS, pH 7.4. Sections were incubated with rabbit anti-Lamp1 (1:10 dilution) for 16 h at 4°C and then with 12-nm colloidal gold-conjugated donkey anti-rabbit immunoglobulin G antibody (1:20 dilution) for 60 min at room temperature. All antibodies were diluted with TBS, pH 7.4, containing 1% BSA. After incubation with antibodies, sections were contrasted by staining with 2% aqueous uranyl acetate for 15 min. Sections were observed on a Hitachi H-7650 transmission electron microscope at 80 kV and equipped with a 16-megapixel XR111 camera (Advanced Microscopy Techniques).
Quantification and statistical analysis
Statistical analysis.
All analyses were done in Prism (Graph-Pad). Statistical significance was determined using two-tailed Student’s t test, two-way analysis of variance (ANOVA), or the log-rank test (Mantel-Cox).
Quantification of Lysotracker-stained puncta.
Average number of puncta per cell were calculated using ImageJ software, applying the following steps to each image:
-
Open image.
File → Open…
-
Filter to remove noise.
Process → Filters → Gaussian Blur…
-
Subtract background.
Process → Subtract Background…
The box marked “Light Background” was unticked.
-
Set measurements to use later for filtering the puncta.
Analyze → Set Measurements …
Tick the boxes marked “Area” and “Shape Descriptors.” Then click on the “OK” button
In this step, manually set in FIJI measurements to use as “filter” later in the workflow [which features to include or exclude in the counting (area, shape descriptors, centroid, perimeter)].
-
Threshold image.
Image → Adjust → Threshold…
Box labeled “Dark Background” is ticked. Adjust the sliders so that features are red colored, but the rest of the image is not. Click “Apply” button. This will replace grayscale image with an “8-bit binary image.” All “red” pixels are converted to a value of “255,″ while all nonred pixels will be given a value of “0.”
-
Fill in any holes in the nuclei.
Process → Binary → Fill Holes
-
Separate “Touching” puncta.
Process → Binary → Watershed
-
Perform the analysis.
Analyze → Analyze Particles…
In this dialogue box the algorithm starts to include or exclude puncta based on their attributes. “Size” smaller than 1 square pixel. “Circularity” set range: 200-1.
Quantification of Western blots
Quantification was performed on scanned images of each film using Fiji as reported below:
Setting the Measurement Criteria: Under the “Analyze” menu select “Set Measurements.” Check ONLY the “Grey Mean Value.”
Go to the “File” menu and open the JPEG file format for the film. Maximize the window.
Select the “rectangle” tool from ImageJ and draw a frame around the largest band of that row. Drag it around and resize the frame. Adjust it so that it covers the minimum area to contain the whole of the largest band of the row.
Once the frame is sized properly, click the “File” menu, “Save as,” and “Selection,” and save this frame with the protein name.
Do the same for the loading control bands.
Place the frame on the first band. The frame should fit all the bands since previously sized according to the largest one. Centre the band inside the frame and record a measurement by clicking “Measure” under the “Analyze” menu.
Move the frame to the next lane and make measurement for that protein for all the samples (across the row).
Using the same frame, take a background measurement.
Repeat the previous step for the respective loading control picture.
Export the data into a spreadsheet.
Invert the pixel density for all data (bands/controls + their backgrounds) in new columns in the spreadsheet. The inverted value is expressed as 255 – X, where X is the value recorded by Fiji.
For the protein bands and loading controls, express the net value by deducting the inverted background from the inverted band value.
When the net bands and loading controls are calculated, take a ratio of a net band value over the net relative loading control.
The final relative quantification values are the ratio of net band to net loading control.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by a Collaborative Research Innovation Opportunities grant from Alberta Innovates–Health Solutions to R.A.R. and A.J.S., a Canadian Institutes of Health Research Foundation Grant to R.A.R., and charitable support from The Edgar Foundation and the Ladies Auxiliary of the Fraternal Order of Eagles #3395 to R.A.R. We thank the Simmonds laboratory for insightful comments; Bruce Edgar, Nicolas Buchon, and Bruno Lemaitre for advice and reagents; and Woo Jung Cho for help with microscopy.
Abbreviations used:
- AMP
antimicrobial peptide
- DHE
dihydroethidium
- EB
enteroblast
- EC
enterocyte
- EE
enteroendocrine cell
- IMD
immune deficiency
- ISC
intestinal stem cell
- NAC
N-acetylcysteine
- NEFA
nonesterified fatty acid
- Pex
gene encoding a peroxin
- RNAi
RNA interference
- ROS
reactive oxygen species
- Tor
target of rapamycin.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-07-0434) on September 6, 2018.
REFERENCES
- Amcheslavsky A, Jiang J, Ip YT. (2009). Tissue damage-induced intestinal stem cell division in Drosophila . Cell Stem Cell , 49–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae YS, Choi MK, Lee WJ. (2010). Dual oxidase in mucosal immunity and host-microbe homeostasis. Trends Immunol , 278–287. [DOI] [PubMed] [Google Scholar]
- Baron MN, Klinger CM, Rachubinski RA, Simmonds AJ. (2016). A systematic cell-based analysis of localization of predicted Drosophila peroxisomal proteins. Traffic , 536–553. [DOI] [PubMed] [Google Scholar]
- Basset A, Khush RS, Braun A, Gardan L, Boccard F, Hoffmann JA, Lemaitre B. (2000). The phytopathogenic bacteria Erwinia carotovora infects Drosophila and activates an immune response. Proc Natl Acad Sci USA , 3376–3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach A, Burstein MT, Richard VR, Leonov A, Levy S, Titorenko VI. (2012). Integration of peroxisomes into an endomembrane system that governs cellular aging. Front Physiol , 283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beard ME, Holtzman E. (1987). Peroxisomes in wild-type and rosy mutant Drosophila melanogaster . Proc Natl Acad Sci USA , 7433–7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker T, Loch G, Beyer M, Zinke I, Aschenbrenner AC, Carrera P, Inhester T, Schultze JL, Hoch M. (2010). FOXO-dependent regulation of innate immune homeostasis. Nature , 369–373. [DOI] [PubMed] [Google Scholar]
- Berger J, Dorninger F, Forss-Petter S, Kunze M. (2016). Peroxisomes in brain development and function. Biochim Biophys Acta , 934–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development , 401–415. [DOI] [PubMed] [Google Scholar]
- Braverman NE, D’Agostino MD, Maclean GE. (2013). Peroxisome biogenesis disorders: biological, clinical and pathophysiological perspectives. Dev Disabil Res Rev , 187–196. [DOI] [PubMed] [Google Scholar]
- Broderick NA, Lemaitre B. (2012). Gut-associated microbes of Drosophila melanogaster . Gut Microbes , 307–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchon N, Broderick NA, Lemaitre B. (2013). Gut homeostasis in a microbial world: insights from Drosophila melanogaster . Nat Rev Microbiol , 615–626. [DOI] [PubMed] [Google Scholar]
- Buchon N, Broderick NA, Poidevin M, Pradervand S, Lemaitre B. (2009). Drosophila intestinal response to bacterial infection: activation of host defense and stem cell proliferation. Cell Host Microbe , 200–211. [DOI] [PubMed] [Google Scholar]
- Bulow MH, Wingen C, Senyilmaz D, Gosejacob D, Sociale M, Bauer R, Schulze H, Sandhoff K, Teleman AA, Hoch M, et al (2018). Unbalanced lipolysis results in lipotoxicity and mitochondrial damage in peroxisome-deficient Pex19 mutants. Mol Biol Cell , 396–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebollero E, Reggiori F. (2009). Regulation of autophagy in yeast Saccharomyces cerevisiae . Biochim Biophys Acta , 1413–1421. [DOI] [PubMed] [Google Scholar]
- Chakrabarti S, Liehl P, Buchon N, Lemaitre B. (2012). Infection-induced host translational blockage inhibits immune responses and epithelial renewal in the Drosophila gut. Cell Host Microbe , 60–70. [DOI] [PubMed] [Google Scholar]
- Chakrabarti S, Poidevin M, Lemaitre B. (2014). The Drosophila MAPK p38c regulates oxidative stress and lipid homeostasis in the intestine. PLoS Genet , e1004659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheesman HK, Feinbaum RL, Thekkiniath J, Dowen RH, Conery AL, Pukkila-Worley R. (2016). Aberrant activation of p38 MAP kinase-dependent innate immune responses is toxic to Caenorhabditis elegans . G3 (Bethesda) , 541–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Xie C, Tian L, Hong L, Wu X, Han J. (2010). Participation of the p38 pathway in Drosophila host defense against pathogenic bacteria and fungi. Proc Natl Acad Sci USA , 20774–20779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemente JC, Ursell LK, Parfrey LW, Knight R. (2012). The impact of the gut microbiota on human health: an integrative view. Cell , 1258–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colasante C, Chen J, Ahlemeyer B, Baumgart-Vogt E. (2015). Peroxisomes in cardiomyocytes and the peroxisome/peroxisome proliferator-activated receptor-loop. Thromb Haemost , 452–463. [DOI] [PubMed] [Google Scholar]
- Cuenda A, Rousseau S. (2007). p38 MAP-kinases pathway regulation, function and role in human diseases. Biochim Biophys Acta , 1358–1375. [DOI] [PubMed] [Google Scholar]
- Dennis PB, Fumagalli S, Thomas G. (1999). Target of rapamycin (TOR): balancing the opposing forces of protein synthesis and degradation. Curr Opin Genet Dev , 49–54. [DOI] [PubMed] [Google Scholar]
- Di Cara F, Sheshachalam A, Braverman NE, Rachubinski RA, Simmonds AJ. (2017). Peroxisome-mediated metabolism is required for immune response to microbial infection. Immunity , 93–106. [DOI] [PubMed] [Google Scholar]
- Dirkx R, Vanhorebeek I, Martens K, Schad A, Grabenbauer M, Fahimi D, Declercq P, Van Veldhoven PP, Baes M. (2005). Absence of peroxisomes in mouse hepatocytes causes mitochondrial and ER abnormalities. Hepatology , 868–878. [DOI] [PubMed] [Google Scholar]
- Dixit E, Boulant S, Zhang Y, Lee AS, Odendall C, Shum B, Hacohen N, Chen ZJ, Whelan SP, Fransen M, et al (2010). Peroxisomes are signaling platforms for antiviral innate immunity. Cell , 668–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunbar TL, Yan Z, Balla KM, Smelkinson MG, Troemel ER. (2012). C. elegans detects pathogen-induced translational inhibition to activate immune signaling. Cell Host Microbe , 375–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskelinen EL. (2006). Roles of LAMP-1 and LAMP-2 in lysosome biogenesis and autophagy. Mol Aspects Med , 495–502. [DOI] [PubMed] [Google Scholar]
- Faust JE, Manisundaram A, Ivanova PT, Milne SB, Summerville JB, Brown HA, Wangler M, Stern M, McNew JA. (2014). Peroxisomes are required for lipid metabolism and muscle function in Drosophila melanogaster . PLoS One , e100213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filomeni G, De Zio D, Cecconi F. (2015). Oxidative stress and autophagy: the clash between damage and metabolic needs. Cell Death Differ , 377–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink C, Hoffmann J, Knop M, Li Y, Isermann K, Roeder T. (2016). Intestinal FoxO signaling is required to survive oral infection in Drosophila . Mucosal Immunol , 927–936. [DOI] [PubMed] [Google Scholar]
- Fransen M, Nordgren M, Wang B, Apanasets O. (2012). Role of peroxisomes in ROS/RNS-metabolism: implications for human disease. Biochim Biophys Acta , 1363–1373. [DOI] [PubMed] [Google Scholar]
- Geiszt M, Witta J, Baffi J, Lekstrom K, Leto TL. (2003). Dual oxidases represent novel hydrogen peroxide sources supporting mucosal surface host defense. FASEB J , 1502–1504. [DOI] [PubMed] [Google Scholar]
- Glick D, Barth S, Macleod KF. (2010). Autophagy: cellular and molecular mechanisms. J Pathol , 3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González M, Martin-Ruíz I, Jiménez S, Pirone L, Barrio R, Sutherland JD. (2011). Generation of stable Drosophila cell lines using multicistronic vectors. Sci Rep , 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez S, Rallis C. (2017). The TOR signaling pathway in spatial and temporal control of cell size and growth. Front Cell Dev Biol , 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, Prieto PA, Vicente D, Hoffman K, Wei SC, et al (2018). Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science , 97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Karpac J, Tran SL, Jasper H. (2014). PGRP-SC2 promotes gut immune homeostasis to limit commensal dysbiosis and extend lifespan. Cell , 109–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha EM, Lee KA, Seo YY, Kim SH, Lim JH, Oh BH, Kim J, Lee WJ. (2009). Coordination of multiple dual oxidase-regulatory pathways in responses to commensal and infectious microbes in Drosophila gut. Nat Immunol , 949–957. [DOI] [PubMed] [Google Scholar]
- Ha EM, Oh CT, Bae YS, Lee WJ. (2005a). A direct role for dual oxidase in Drosophila gut immunity. Science , 847–850. [DOI] [PubMed] [Google Scholar]
- Ha EM, Oh CT, Ryu JH, Bae YS, Kang SW, Jang IH, Brey PT, Lee WJ. (2005b). An antioxidant system required for host protection against gut infection in Drosophila . Dev Cell , 125–132. [DOI] [PubMed] [Google Scholar]
- Hasnain SZ, Prins JB, McGuckin MA. (2016). Oxidative and endoplasmic reticulum stress in β-cell dysfunction in diabetes. J Mol Endocrinol , R33–R54. [DOI] [PubMed] [Google Scholar]
- Hay N, Sonenberg N. (2004). Upstream and downstream of mTOR. Genes Dev , 1926–1945. [DOI] [PubMed] [Google Scholar]
- Hoidal JR. (2001). Reactive oxygen species and cell signaling. Am J Respir Cell Mol Biol , 661–663. [DOI] [PubMed] [Google Scholar]
- Holcik M, Sonenberg N. (2005). Translational control in stress and apoptosis. Nat Rev Mol Cell Biol , 318–327. [DOI] [PubMed] [Google Scholar]
- Hooper LV, Littman DR, Macpherson AJ. (2012). Interactions between the microbiota and the immune system. Science , 1268–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang AB, Ryu EA, Artan M, Chang HW, Kabir MH, Nam HJ, Lee D, Yang JS, Kim S, Mair WB, et al (2014). Feedback regulation via AMPK and HIF-1 mediates ROS-dependent longevity in Caenorhabditis elegans . Proc Natl Acad Sci USA , E4458–E4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Lee J, Huh JY, Park J, Lee HB, Ho YS, Ha H. (2012). Catalase deficiency accelerates diabetic renal injury through peroxisomal dysfunction. Diabetes , 728–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inokuchi-Shimizu S, Park EJ, Roh YS, Yang L, Zhang B, Song J, Liang S, Pimienta M, Taniguchi K, Wu X, et al (2014). TAK1-mediated autophagy and fatty acid oxidation prevent hepatosteatosis and tumorigenesis. J Clin Invest , 3566–3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivashchenko O, Van Veldhoven PP, Brees C, Ho YS, Terlecky SR, Fransen M. (2011). Intraperoxisomal redox balance in mammalian cells: Oxidative stress and interorganellar cross-talk. Mol Biol Cell , 1440–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaramillo M, Gomez MA, Larsson O, Shio MT, Topisirovic I, Contreras I, Luxenburg R, Rosenfeld A, Colina R, McMaster RW, et al (2011). Leishmania repression of host translation through mTOR cleavage is required for parasite survival and infection. Cell Host Microbe , 331–341. [DOI] [PubMed] [Google Scholar]
- Jiang H, Patel PH, Kohlmaier A, Grenley MO, McEwen DG, Edgar BA. (2009). Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell , 1343–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhasz G, Neufeld TP. (2008). Experimental control and characterization of autophagy in Drosophila . Methods Mol Biol , 125–133. [DOI] [PubMed] [Google Scholar]
- Kaser A, Zeissig S, Blumberg RS. (2010). Inflammatory bowel disease. Annu Rev Immunol , 573–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Lee JH. (2015). Identification of an AMPK phosphorylation site in Drosophila TSC2 (gigas) that regulate cell growth. Int J Mol Sci , 7015–7026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Park HL, Park HW, Ro SH, Nam SG, Reed JM, Guan JL, Lee JH. (2013). Drosophila Fip200 is an essential regulator of autophagy that attenuates both growth and aging. Autophagy , 1201–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnula VL, Adler KB, Ackley NJ, Crapo JD. (1992). Release of reactive oxygen species by guinea pig tracheal epithelial cells in vitro. Am J Physiol , L708–L712. [DOI] [PubMed] [Google Scholar]
- Klein AT, Barnett P, Bottger G, Konings D, Tabak HF, Distel B. (2001). Recognition of peroxisomal targeting signal type 1 by the import receptor Pex5p. J Biol Chem , 15034–15041. [DOI] [PubMed] [Google Scholar]
- Kleino A, Silverman N. (2014). The Drosophila IMD pathway in the activation of the humoral immune response. Dev Comp Immunol , 25–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koepke JI, Wood CS, Terlecky LJ, Walton PA, Terlecky SR. (2008). Progeric effects of catalase inactivation in human cells. Toxicol Appl Pharmacol , 99–108. [DOI] [PubMed] [Google Scholar]
- Lemaitre B, Hoffmann J. (2007). The host defense of Drosophila melanogaster . Annu Rev Immunol , 697–743. [DOI] [PubMed] [Google Scholar]
- Lemaitre B, Miguel-Aliaga I. (2013). The digestive tract of Drosophila melanogaster . Annu Rev Genet , 377–404. [DOI] [PubMed] [Google Scholar]
- Li H, Jasper H. (2016). Gastrointestinal stem cells in health and disease: from flies to humans. Dis Models Mech , 487–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Min Q, Ouyang C, Lee J, He C, Zou MH, Xie Z. (2014). AMPK activation prevents excess nutrient-induced hepatic lipid accumulation by inhibiting mTORC1 signaling and endoplasmic reticulum stress response. Biochim Biophys Acta , 1844–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods , 402–408. [DOI] [PubMed] [Google Scholar]
- Lodhi IJ, Semenkovich CF. (2014). Peroxisomes: a nexus for lipid metabolism and cellular signaling. Cell Metab , 380–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzio JP, Pryor PR, Bright NA. (2007). Lysosomes: fusion and function. Nat Rev Mol Cell Biol , 622–632. [DOI] [PubMed] [Google Scholar]
- Martino L, Masini M, Novelli M, Beffy P, Bugliani M, Marselli L, Masiello P, Marchetti P, De Tata V. (2012). Palmitate activates autophagy in INS-1E β-cells and in isolated rat and human pancreatic islets. PLoS One , e36188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micchelli CA, Perrimon N. (2006). Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature , 475–479. [DOI] [PubMed] [Google Scholar]
- Morvay PL, Baes M, Van Veldhoven PP. (2017). Differential activities of peroxisomes along the mouse intestinal epithelium. Cell Biochem Funct , 144–155. [DOI] [PubMed] [Google Scholar]
- Nguyen SD, Baes M, Van Veldhoven PP. (2008). Degradation of very long chain dicarboxylic polyunsaturated fatty acids in mouse hepatocytes, a peroxisomal process. Biochim Biophys Acta , 400–405. [DOI] [PubMed] [Google Scholar]
- Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S. (2012). Host-gut microbiota metabolic interactions. Science , 1262–1267. [DOI] [PubMed] [Google Scholar]
- Novikoff PM, Novikoff AB. (1972). Peroxisomes in absorptive cells of mammalian small intestine. J Cell Biol , 532–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlstein B, Spradling A. (2007). Multipotent Drosophila intestinal stem cells specify daughter cell fates by differential notch signaling. Science , 988–992. [DOI] [PubMed] [Google Scholar]
- Opota O, Vallet-Gély I, Vincentelli R, Kellenberger C, Iacovache I, Gonzalez MR, Roussel A, van der Goot FG, Lemaitre B. (2011). Monalysin, a novel β-pore-forming toxin from the Drosophila pathogen Pseudomonas entomophila, contributes to host intestinal damage and lethality. PLoS Pathog , e1002259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owusu-Ansah E, Banerjee U. (2009). Reactive oxygen species prime Drosophila haematopoietic progenitors for differentiation. Nature , 537–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phielix E, Jelenik T, Nowotny P, Szendroedi J, Roden M. (2014). Reduction of non-esterified fatty acids improves insulin sensitivity and lowers oxidative stress, but fails to restore oxidative capacity in type 2 diabetes: a randomised clinical trial. Diabetologia , 572–581. [DOI] [PubMed] [Google Scholar]
- Phillips MD., Thomas GH. (2006). Brush border spectrin is required for early endosome recycling in Drosophila . J Cell Sci , 1361–1370. [DOI] [PubMed] [Google Scholar]
- Qi M, Elion EA. (2005). MAP kinase pathways. J Cell Sci , 3569–3572. [DOI] [PubMed] [Google Scholar]
- Reiling JH, Sabatini DM. (2006). Stress and mTORture signaling. Oncogene , 6373–6383. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Carrio J, Alperi-Lopez M, Lopez P, Ballina-Garcia FJ, Suarez A. (2016). Non-esterified fatty acids profiling in rheumatoid arthritis: associations with clinical features and Th1 response. PLoS One , e0159573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JJ, Aitchison JD. (2013). Peroxisomes take shape. Nat Rev Mol Cell Biol , 803–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soardo G, Donnini D, Domenis L, Catena C, De Silvestri D, Cappello D, Dibenedetto A, Carnelutti A, Bonasia V, Pagano C, et al (2011). Oxidative stress is activated by free fatty acids in cultured human hepatocytes. Metab Syndr Relat Disord , 397–401. [DOI] [PubMed] [Google Scholar]
- Song Y, Li X, Li Y, Li N, Shi X, Ding H, Zhang Y, Li X, Liu G, Wang Z. (2014). Non-esterified fatty acids activate the ROS-p38-p53/Nrf2 signaling pathway to induce bovine hepatocyte apoptosis in vitro. Apoptosis , 984–997. [DOI] [PubMed] [Google Scholar]
- Tinnikov AA, Boonstra R. (1999). Colorimetric micro-determination of free fatty acids in plasma using microplate readers. Clin Chim Acta , 159–162. [DOI] [PubMed] [Google Scholar]
- Titorenko VI, Terlecky SR. (2011). Peroxisome metabolism and cellular aging. Traffic , 252–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi DN, Walker CL. (2016). The peroxisome as a cell signaling organelle. Curr Opin Cell Biol , 109–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vodovar N, Vinals M, Liehl P, Basset A, Degrouard J, Spellman P, Boccard F, Lemaitre B. (2005). Drosophila host defense after oral infection by an entomopathogenic Pseudomonas species. Proc Natl Acad Sci USA , 11414–11419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton PA, Pizzitelli M. (2012). Effects of peroxisomal catalase inhibition on mitochondrial function. Front Physiol , 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanders RJ, Waterham HR. (2006). Biochemistry of mammalian peroxisomes revisited. Annu Rev Biochem , 295–332. [DOI] [PubMed] [Google Scholar]
- Waterham HR, Ferdinandusse S, Wanders RJ. (2016). Human disorders of peroxisome metabolism and biogenesis. Biochim Biophys Acta , 922–933. [DOI] [PubMed] [Google Scholar]
- Wei Y, Yu L, Bowen J, Gorovsky MA, Allis CD. (1999). Phosphorylation of histone H3 is required for proper chromosome condensation and segregation. Cell , 99–109. [DOI] [PubMed] [Google Scholar]
- Wood LG, Scott HA, Garg ML, Gibson PG. (2009). Innate immune mechanisms linking non-esterified fatty acids and respiratory disease. Prog Lipid Res , 27–43. [DOI] [PubMed] [Google Scholar]
- Wu H, Wang MC, Bohmann D. (2009). JNK protects Drosophila from oxidative stress by trancriptionally activating autophagy. Mech Dev , 624–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekawa T, Thorburn A. (2013). Autophagy and cell death. Essays Biochem , 105–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Kim J, Alexander A, Cai S, Tripathi DN, Dere R, Tee AR, Tait-Mulder J, Di Nardo A, Han JM, et al (2013). A tuberous sclerosis complex signalling node at the peroxisome regulates mTORC1 and autophagy in response to ROS. Nat Cell Biol , 1186–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielonka J, Kalyanaraman B. (2010). Hydroethidine- and MitoSOX-derived red fluorescence is not a reliable indicator of intracellular superoxide formation: another inconvenient truth. Free Radic Biol Med , 983–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.