Abstract
Determining sex is a binary developmental decision that most metazoans must make. Like many organisms, Caenorhabditis elegans specifies sex (XO male or XX hermaphrodite) by tallying X-chromosome number. We dissected this precise counting mechanism to determine how tiny differences in concentrations of signals are translated into dramatically different developmental fates. Determining sex by counting chromosomes solved one problem but created another—an imbalance in X gene products. We found that nematodes compensate for the difference in X-chromosome dose between sexes by reducing transcription from both hermaphrodite X chromosomes. In a surprising feat of evolution, X-chromosome regulation is functionally related to a structural problem of all mitotic and meiotic chromosomes: achieving ordered compaction of chromosomes before segregation. We showed the dosage compensation complex is a condensin complex that imposes a specific three-dimensional architecture onto hermaphrodite X chromosomes. It also triggers enrichment of histone modification H4K20me1. We discovered the machinery and mechanism underlying H4K20me1 enrichment and demonstrated its pivotal role in regulating higher-order X-chromosome structure and gene expression.
INTRODUCTION
I feel honored to receive the unexpected tribute of an E. B. Wilson Medal. I am extremely lucky to be a scientist during an explosive period of discovery in cell biology and to have carried out research with wonderfully talented and dedicated students, postdoctoral fellows, staff scientists, and collaborators at both the Massachusetts Institute of Technology and the University of California, Berkeley.
The work being honored began during my postdoctoral years when I joined the small cadre of scientists led by Sydney Brenner who saw the potential of the nematode Caenorhabditis elegans for understanding eukaryotic developmental mechanisms. I was drawn to the question of how the nematode specifies its sex. Determining sex is a fundamental, binary developmental decision that most metazoans must make (Cline and Meyer, 1996).
I was stimulated to tackle this problem by my graduate work with Mark Ptashne, which demonstrated the power of a simple virus and a binary developmental decision to yield important lessons in gene regulation and development. My work was instrumental in elucidating the binary genetic switch by which bacteriophage lambda triggers one of two developmental fates: a quiescent lysogenic state or a replicating state (Meyer et al., 1975, 1980; Meyer and Ptashne, 1980; Johnson et al., 1978; Maurer et al., 1980). The work revealed fundamentally new principles in gene regulation, including cooperativity in repressor binding to turn off gene expression (Johnson et al., 1979).
Before I joined the nematode field, Victor Nigon and Robert Herman had shown that C. elegans determines sex with high fidelity by tallying X-chromosome number relative to the ploidy, the sets of autosomes (X:A signal; Figure 1A; Nigon, 1951; Madl and Herman, 1979). The process is finely tuned and executed with remarkable accuracy, allowing embryos with ratios of 1X:2A (0.5) and 2X:3A (0.67) to develop into fertile males and embryos with ratios of 3X:4A (0.75) and 2X:2A (1.0) to develop into self-fertile hermaphrodites (Figure 1A). I wanted to determine the molecular mechanisms by which C. elegans counts its X chromosomes to determine how small changes in the concentrations of molecular signals can be translated into dramatically different developmental fates. Jonathan Hodgkin had found that mutations in individual genes caused diploid XX animals to develop into males or XO animals into hermaphrodites, showing the problem might be genetically tractable (Hodgkin and Brenner, 1977; Hodgkin, 1980).
FIGURE 1:
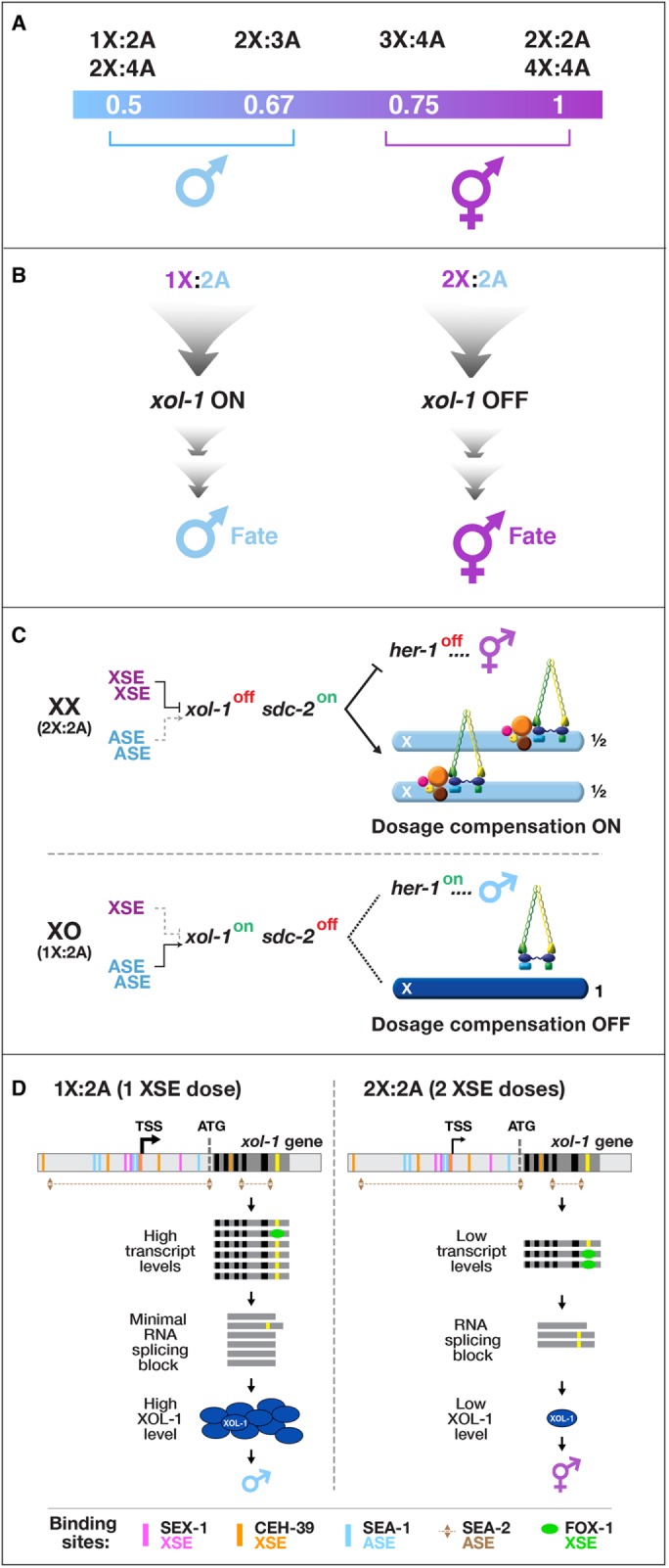
Chromosome-based sex determination. (A) The ratio of X chromosomes to sets of autosomes (X:A signal) determines nematode sex. (B) xol-1 is the direct target of the X:A signal and the master sex-determination switch gene. It activates male fate when turned on and permits hermaphrodite fate when turned off. (C) Overview of the genetic hierarchy controlling both sex determination and dosage compensation in C. elegans. In diploid XX hermaphrodites, the dosage compensation complex (DCC) binds to both X chromosomes to reduce transcription by half, thereby equalizing it with that of the single male X. The hermaphrodite-specific gene sdc-2 triggers assembly of the DCC onto X and activates the hermaphrodite sexual fate by repressing the male-determining gene her-1, which is the first gene in the sex-determination branch of the genetic hierarchy. In diploid XO males, xol-1 represses sdc-2, thereby preventing the DCC from binding to the male X and preventing sdc-2 from repressing her-1. (D) Model for X:A signal assessment. XSEs and ASEs bind directly to numerous sites in xol-1 regulatory regions to antagonize each other and thereby control xol-1 transcription. Molecular rivalry at the xol-1 promoter between XSE transcriptional repressors (nuclear receptor SEX-1, magenta binding sites; homeodomain protein CEH-39, orange binding sites) and ASE transcriptional activators (T-box transcription factor SEA-1, blue binding sites; zinc-finger protein SEA-2, numerous brown binding sites) causes high xol-1 transcript levels in 1X:2A animals with one dose of XSEs and low levels in 2X:2A embryos, with two doses of XSEs. The RNA binding protein FOX-1, an XSE, then enhances the fidelity of X counting by binding to an alternatively spliced xol-1 intron (yellow), thereby blocking proper splicing and causing mRNA splice variants with in-frame stop codons. High XOL-1 protein levels induce male fate and low XOL-1 levels permit hermaphrodite fate. Black rectangles, xol-1 exons; dark-gray rectangles, xol-1 introns; light-gray rectangles, 5′ and 3′xol-1 regulatory regions.
I was also mindful of a possible complication. Many organisms that use sex chromosomes to determine sexual fate evolved the essential, chromosome-wide regulatory process called dosage compensation to balance X-chromosome gene expression between the sexes (Muller, 1932; Lyon, 1962). The failure to do so is lethal (Cline, 1978; Cline and Meyer, 1996). We had no knowledge of whether the worm had a dosage compensation mechanism and whether its regulation might be genetically linked to the sex determination decision. If so, the phenotype caused by disrupting the X:A signal might be sex-specific lethality due to improper X gene expression, thus masking any reversal of sexual fate. While I considered how to approach this problem, Thomas Cline showed that a single developmental switch gene coordinately controls both sex determination and dosage compensation in Drosophila melanogaster, which also uses an X:A signal to determine sex (Bridges, 1921; Cline, 1978, 1979). Null mutations in the switch gene caused the masculinization and death of 2X:2A animals, but 1X:2A mutants were wild type. The path to take became clear.
Early work as a postdoc and professor demonstrated that nematodes compensate for the difference in X-chromosome dosage between sexes by repressing X transcript levels in hermaphrodites and revealed, via extensive genetic screens, the identity of numerous genes that implement dosage compensation (Meyer and Casson, 1986; Miller et al., 1988; Plenefisch et al., 1989). We found that dosage compensation and sex determination were linked by genes that coordinately controlled both processes, a finding that allowed us to discover the master sex-determination switch gene xol-1 (XO lethal) and the molecular basis of the X:A signal, which controls xol-1 directly (Figure 1B; Villeneuve and Meyer, 1987; Miller et al., 1988; Nusbaum and Meyer, 1989; DeLong et al., 1993; Klein and Meyer, 1993; Rhind et al., 1995; Davis and Meyer, 1997; Carmi et al., 1998; Dawes et al., 1999).
COUNTING CHROMOSOMES TO DETERMINE SEX
We discovered that a set of X-linked genes called X-signal elements (XSEs) communicates X-chromosome dose by repressing xol-1 in a cumulative, dose-dependent manner (Figure 1C; Akerib and Meyer, 1994; Nicoll et al., 1997; Carmi et al., 1998; Gladden et al., 2007; Gladden and Meyer, 2007; Farboud et al., 2013). In addition, a set of genes encoded on autosomes called autosomal signal elements (ASEs) communicates the ploidy by stimulating xol-1 activity in a cumulative, dose-dependent manner to counter XSEs (Figure 1C; Powell et al., 2005; Farboud et al., 2013). XOL-1, a GHMP kinase, is activated in 1X:2A embryos to set the male fate but repressed in 2X:2A embryos to promote the hermaphrodite fate, including the activation of dosage compensation (Rhind et al., 1995; Luz et al., 2003). Both XSEs (nuclear receptors and homeodomain proteins) and ASEs (T-box and zinc-finger proteins) bind directly to multiple, distinct sites in xol-1 regulatory DNA in vitro to counteract each other’s activities and regulate xol-1 transcription (Figure 1D; Farboud et al., 2013). Disrupting ASE and XSE binding sites in vivo recapitulated the misregulation of xol-1 transcription caused by disrupting the cognate signal element genes. The X:A signal is thus transmitted in part through multiple antagonistic molecular interactions carried out on a single promoter (Farboud et al., 2013). Fidelity of X:A counting is then enhanced by a second tier of repression, via an XSE that binds a xol-1 intron to prevent proper RNA splicing (Figure 1D; Hodgkin et al., 1994; Nicoll et al., 1997; Skipper et al., 1999). The concept of a sex signal using competing XSEs and ASEs arose as a theory for D. melanogaster one century ago (Bridges, 1921). Recent work showed the fruit fly signal does not fit this simple paradigm, but our work showed the worm signal does (Erickson and Quintero, 2007; Farboud et al., 2013).
X-CHROMOSOME DOSAGE COMPENSATION: REPRESSING X CHROMOSOMES VIA MOLECULAR MACHINES
Strategies for dosage compensation differ from worms to mammals, but invariably a regulatory complex is targeted to X chromosomes of one sex to modulate transcription along the entire chromosome (Meyer, 2010; Kuroda et al., 2016; da Rocha and Heard, 2017; Samata and Akhtar, 2018). This heritable regulation of X-chromosome expression during development is exemplary for dissecting the coordinate regulation of gene expression over large chromosome territories and the role of chromosome structure and chromatin modification in regulating gene expression.
We discovered the C. elegans dosage compensation complex (DCC) and showed its homology to condensin, a complex that mediates the compaction, resolution, and segregation of mitotic and meiotic chromosomes from yeast to humans (Figure 2A; Chuang et al., 1994, 1996; Hsu et al., 1995; Lieb et al., 1996, 1998; Davis and Meyer, 1997; Dawes et al., 1999; Tsai et al., 2008; Csankovszki et al., 2009; Mets and Meyer, 2009; Pferdehirt et al., 2011; Hirano, 2016). We found the DCC binds to both hermaphrodite X chromosomes to reduce transcription by half, thus equalizing X expression between the sexes (Figure 1C; Chuang et al., 1996; Jans et al., 2009; Kruesi et al., 2013). Failure to do so causes elevated X expression and hermaphrodite-specific lethality. Ectopic binding of the DCC to the single male X chromosome in xol-1 mutants causes male-specific lethality.
FIGURE 2:
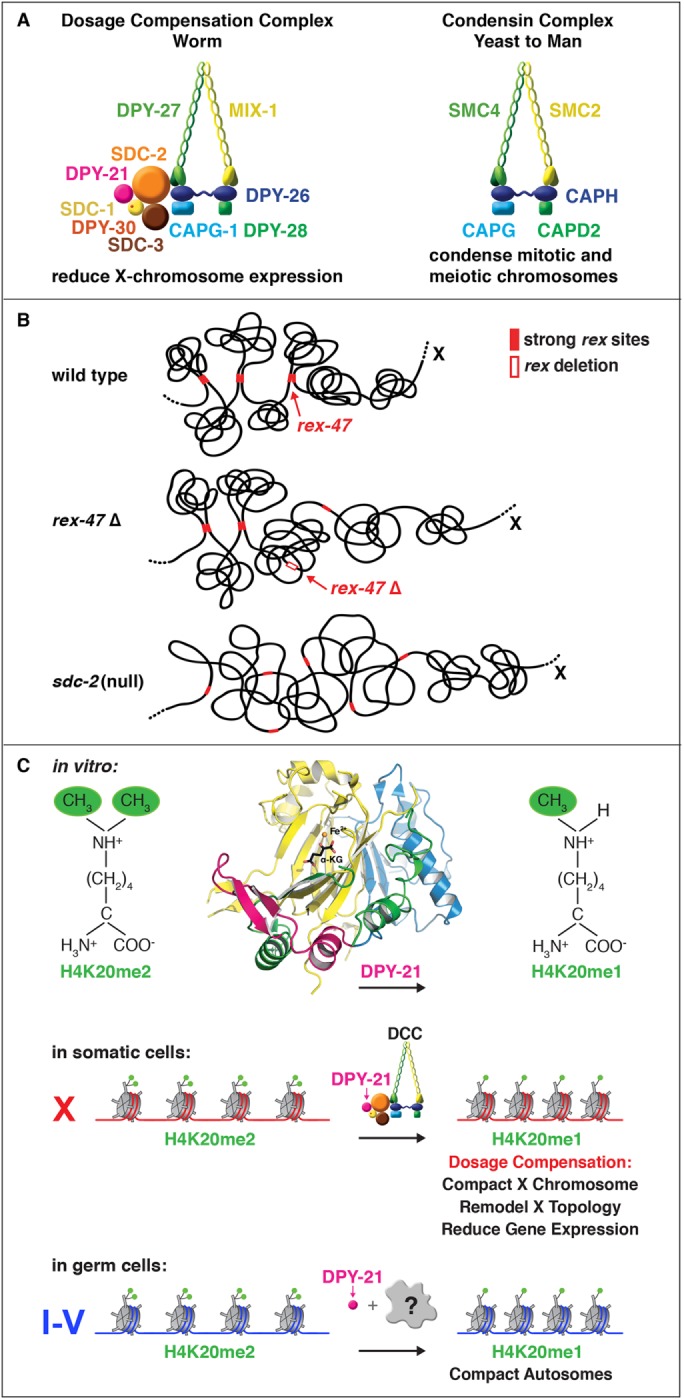
Overview of dosage compensation in C. elegans. (A) The DCC is compared with condensin I of other eukaryotes. The DCC condensin subunits (MIX-1, DPY-27, DPY-26, DPY-28, and CAPG-1) are color matched to their condensin I homologues. All DCC condensin subunits except DPY-27 also act in other distinct condensins that function in C. elegans mitosis and meiosis. The DPY-27 paralogue SMC-4 (Hagstrom et al., 2002) replaces DPY-27 in mitotic and meiotic condensins. The DCC likely arose by duplicating and modifying the gene encoding SMC-4 to create DPY-27 for a specific role in gene expression. The DCC also includes the XX-specific protein SDC-2 that triggers DCC assembly onto X. Two DCC subunits aid SDC-2 in recruiting the complex to X: SDC-3 (a zinc-finger protein) and DPY-30 (a subunit of the MLL/COMPASS H3K4me3 methyltransferase complex). Two subunits, SDC-1 (a zinc-finger protein) and DPY-21 (Jumonji C H4K20me2 demethylase), are required for DCC activity but not DCC assembly. (B) Cartoon model of TAD formation on a segment of X. Top, the DCC remodels the topology of X into a hermaphrodite-specific conformation by forming topologically associating domains (TADs). DCC-dependent looping interactions are found between high-affinity rex sites (filled red rectangles) located at TAD boundaries. Middle, deletion of the high-affinity DCC binding site rex-47 (open red rectangle) located at a DCC-dependent TAD boundary eliminates boundary formation. Bottom, severe disruption of DCC binding by an sdc-2 mutation eliminates formation of all DCC-dependent TADs on X. (C) The DPY-21 H4K20me2 histone demethylase regulates three-dimensional chromosome structure and gene expression by modulating enrichment of H4K20me1. The 1.8 Å crystal structure and biochemical activity of DPY-21 revealed a novel, highly conserved H4K20me2 JmjC demethylase subfamily that converts H4K20me2 to H4K20me1 in vitro in an Fe2±-dependent and α-ketoglutarate–dependent manner. In somatic cells, DPY-21 binds to X chromosomes via the DCC and enriches H4K20me1 to repress gene expression. The H4K20me1 enrichment controls the higher-order structure of X chromosomes by facilitating compaction and TAD formation. In germ cells, DPY-21 enriches HK20me1 on autosomes in a DCC-independent manner to promote chromosome compaction.
Most DCC condensin subunits also control the structure and function of mitotic and meiotic chromosomes by participating in two other distinct condensin complexes, whose roles in recombination and chromosome segregation were dissected (Hagstrom et al., 2002; Chan et al., 2004; Tsai et al., 2008; Csankovszki et al., 2009; Mets and Meyer, 2009; Meyer, 2010). The promiscuous DCC condensin subunits are initially recruited to hermaphrodite X chromosomes by a large (350 kDa) hermaphrodite-specific DCC subunit called SDC-2 (sex determination and dosage compensation) that triggers binding to cis-acting elements on X called rex (recruitment elements on X) sites (Dawes et al., 1999; Csankovszki et al., 2004; McDonel et al., 2006). These rex sites recruit the DCC in an autonomous, sequence-dependent manner using DNA motifs enriched on X (Jans et al., 2009; Pferdehirt et al., 2011).
CONDENSIN-DRIVEN REMODELING OF X-CHROMOSOME TOPOLOGY DURING DOSAGE COMPENSATION
Involvement of condensin subunits in dosage compensation suggested the DCC might alter the topology of X to regulate gene expression. Furthermore, other observations promoted the view that the DCC acts globally through a chromosome-wide mechanism to repress transcription rather than locally on a gene-by-gene basis. For example, a nearby DCC binding site is neither necessary nor sufficient to compensate an endogenous X-linked gene, and all transgenes integrated randomly on X, whether near or far from a DCC binding site, become dosage compensated (Jans et al., 2009; Wheeler et al., 2016). We found the DCC remodels hermaphrodite X chromosomes into a unique, sex-specific spatial conformation, distinct from autosomes, using its highest-affinity rex sites (Figure 2B; Crane et al. 2015). Dosage-compensated X chromosomes have self-interacting domains (∼1 Mb) resembling mammalian topoplogically associating domans (TADs; Crane et al., 2015; Galupa and Heard, 2017). TADs on X have stronger boundaries and more regular spacing than those on autosomes. Most TAD boundaries on X coincide with the highest-affinity rex sites. Those boundaries are lost in DCC mutants, causing X structure to resemble that of autosomes (Figure 2B; Crane et al., 2015). Deleting a single endogenous rex site at a DCC-dependent boundary disrupted the boundary, indicating the DCC forms TADs using its strongest binding sites (Figure 2B; Crane et al., 2015). Our study was the first to identify the machinery and DNA sites that create chromosome-wide TAD structure and to use specific mutations to disrupt it. The structure of dosage-compensated X’s offers fertile ground to decipher the mechanistic relationship between higher-order chromosome structure and gene expression.
DYNAMIC CONTROL OF X-CHROMOSOME CONFORMATION AND REPRESSION BY A HISTONE H4K20 DEMETHYLASE
The role of chromatin modification in establishing higher-order chromosome structure during gene regulation has been elusive. Dosage-compensated X chromosomes of C. elegans are enriched in H4K20me1 (Liu et al., 2011; Vielle et al., 2012; Wells et al., 2012; Kramer et al., 2015). We dissected the mechanism underlying H4K20me1 enrichment on X during dosage compensation and discovered a key role for H4K20me1 in regulating X-chromosome topology and chromosome-wide gene expression (Bian et al., 2017; Brejc et al., 2017). Structural and functional analysis of DCC subunit DPY-21 revealed a novel Jumonji C demethylase subfamily that converts H4K20me2 to H4K20me1 in worms and mammals via an Fe2+-dependent and α-ketoglutarate–dependent mechanism (Figure 2C; Brejc et al., 2017). H4K20me1 is also enriched on the inactive X chromosome of female mice, relating our studies to mammalian development (Kohlmaier et al., 2004). Inactivation of demethylase activity in C. elegans eliminated H4K20me1 enrichment on X in nematode somatic cells, increased X-linked gene expression, reduced X compaction, and disrupted X conformation by diminishing TAD formation (Figure 2C; Bian et al., 2017; Brejc et al., 2017). DPY-21 demethylase binding to X is both DCC-dependent and cell-cycle–dependent, enriching H4K20me1 on X during interphase, but not mitosis. Thus, the DCC recruits an “eraser” of a histone modification, enriching H4K20me1 by activating an H4K20me2 demethylase.
DPY-21 also binds to autosomes (but not X) of germ cells in a DCC-independent manner to enrich H4K20me1 and compact chromosomes (Figure 2C; Brejc et al., 2017). Thus, DPY-21 is an adaptable chromatin regulator. Its demethylase activity can be harnessed during development for distinct biological functions by targeting it to diverse genomic locations. In both somatic and germ cells, H4K20me1 modulates three-dimensional topology, showing a direct link between chromatin modification and higher-order chromosome structure.
THE NEXT STEPS
Deeper mechanistic insights into sex determination and dosage compensation await future experiments with single-cell and single-molecule resolution as well as continued structural analysis of the DCC.
ACKNOWLEDGMENTS
I am indebted to my courageous students, postdoctoral fellows, research staff, administrative assistants, and collaborators whose enormous dedication and talent enabled us to overcome numerous technical and intellectual obstacles in tackling our ambitious research directions. I am also grateful for the unwavering support and inspiration from my husband Thomas Cline. I thank the Howard Hughes Medical Institute, the National Institute of General Medical Sciences, the American Cancer Society, and the Miller Institute at Berkeley for generously supporting our research.
Footnotes
REFERENCES
- Akerib CC, Meyer BJ. (1994). Identification of X chromosome regions in Caenorhabditis elegans that contain sex-determination signal elements. Genetics , 1105–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian Q, Anderson EC, Brejc K, Meyer BJ. (2017). Dynamic control of chromosome topology and gene expression by a chromatin modification. Cold Spring Harb Symp Quant Biol , 279–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brejc K, Bian Q, Uzawa S, Wheeler BS, Anderson EC, King DS, Kranzusch PJ, Preston CG, Meyer BJ. (2017). Dynamic control of X chromosome conformation and repression by a histone H4K20 demethylase. Cell , 85–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges CB. (1921). Triploid intersexes in Drosophila melanogaster. Science , 252–254. [DOI] [PubMed] [Google Scholar]
- Carmi I, Kopczynski JB, Meyer BJ. (1998). The nuclear hormone receptor SEX-1 is an X-chromosome signal that determines nematode sex. Nature , 168–173. [DOI] [PubMed] [Google Scholar]
- Chan RC, Severson AF, Meyer BJ. (2004). Condensin restructures chromosomes in preparation for meiotic divisions. J Cell Biol , 613–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang PT, Albertson DG, Meyer BJ. (1994). DPY-27: a chromosome condensation protein homolog that regulates C. elegans dosage compensation through association with the X chromosome. Cell , 459–474. [DOI] [PubMed] [Google Scholar]
- Chuang PT, Lieb JD, Meyer BJ. (1996). Sex-specific assembly of a dosage compensation complex on the nematode X chromosome. Science , 1736–1739. [DOI] [PubMed] [Google Scholar]
- Cline TW. (1978). Two closely linked mutations in Drosophila melanogaster that are lethal to opposite sexes and interact with daughterless. Genetics , 683–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline TW. (1979). A male-specific lethal mutation in Drosophila melanogaster that transforms sex. Dev Biol , 266–275. [DOI] [PubMed] [Google Scholar]
- Cline TW, Meyer BJ. (1996). Vive la différence: males vs females in flies vs worms. Annu Rev Genet , 637–702. [DOI] [PubMed] [Google Scholar]
- Crane E, Bian Q, McCord RP, Lajoie BR, Wheeler BS, Ralston EJ, Uzawa S, Dekker J, Meyer BJ. (2015). Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature , 240–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csankovszki G, Collette K, Spahl K, Carey J, Snyder M, Petty E, Patel U, Tabuchi T, Liu H, McLeod I, et al (2009). Three distinct condensin complexes control C. elegans chromosome dynamics. Curr Biol , 9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csankovszki G, McDonel P, Meyer BJ. (2004). Recruitment and spreading of the C. elegans dosage compensation complex along X chromosomes. Science , 1182–1185. [DOI] [PubMed] [Google Scholar]
- da Rocha ST, Heard E. (2017). Novel players in X inactivation: insights into Xist-mediated gene silencing and chromosome conformation. Nat Struct Mol Biol , 197–204. [DOI] [PubMed] [Google Scholar]
- Davis TL, Meyer BJ. (1997). SDC-3 coordinates the assembly of a dosage compensation complex on the nematode X chromosome. Development , 1019–1031. [DOI] [PubMed] [Google Scholar]
- Dawes HE, Berlin DS, Lapidus DM, Nusbaum C, Davis TL, Meyer BJ. (1999). Dosage compensation proteins targeted to X chromosomes by a determinant of hermaphrodite fate. Science , 1800–1804. [DOI] [PubMed] [Google Scholar]
- DeLong L, Plenefisch JD, Klein RD, Meyer BJ. (1993). Feedback control of sex determination by dosage compensation revealed through Caenorhabditis elegans sdc-3 mutations. Genetics , 875–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson JW, Quintero JJ. (2007). Indirect effects of ploidy suggest X chromosome dose, not the X:A ratio, signals sex in Drosophila. PLoS Biol , e332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farboud B, Nix P, Jow MM, Gladden JM, Meyer BJ. (2013). Molecular antagonism between X-chromosome and autosome signals determines nematode sex. Genes Dev , 1159–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galupa R, Heard E. (2017). Topologically associating domains in chromosome architecture and gene regulatory landscapes during development, disease, and evolution. Cold Spring Harb Symp Quant Biol , 267–278. [DOI] [PubMed] [Google Scholar]
- Gladden JM, Farboud B, Meyer BJ. (2007). Revisiting the X:A signal that specifies Caenorhabditis elegans sexual fate. Genetics , 1639–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladden JM, Meyer BJ. (2007). A ONECUT homeodomain protein communicates X chromosome dose to specify Caenorhabditis elegans sexual fate by repressing a sex switch gene. Genetics , 1621–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagstrom K, Holmes V, Cozzarelli N, Meyer BJ. (2002). C. elegans condensin promotes mitotic chromosome architecture, centromere organization, and sister chromatid segregation during mitosis and meiosis. Genes Dev , 729–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T. (2016). Condensin-based chromosome organization from bacteria to vertebrates. Cell , 847–857. [DOI] [PubMed] [Google Scholar]
- Hodgkin J. 1980. More sex-determination mutants of Caenorhabditis elegans. Genetics , 649–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin JA, Brenner S. 1977. Mutations causing transformation of sexual phenotype in the nematode Caenorhabditis elegans. Genetics , 275–287. [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J, Zellan JD, Albertson DG. (1994). Identification of a candidate primary sex determination locus, fox-1, on the X chromosome of Caenorhabditis elegans. Development , 3681–3689. [DOI] [PubMed] [Google Scholar]
- Hsu DR, Chuang PT, Meyer BJ. (1995). DPY-30, a nuclear protein essential early in embryogenesis for Caenorhabditis elegans dosage compensation. Development , 3323–3334. [DOI] [PubMed] [Google Scholar]
- Jans J, Gladden JM, Ralston EJ, Pickle CS, Michel AH, Pferdehirt RR, Eisen MB, Meyer BJ. (2009). A condensin-like dosage compensation complex acts at a distance to control expression throughout the genome. Genes Dev , 602–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A, Meyer BJ, Ptashne M. (1978). Mechanism of action of the cro protein of bacteriophage lambda. Proc Natl Acad Sci USA , 1783–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AD, Meyer BJ, Ptashne M. (1979). Interactions between DNA-bound repressors govern regulation by the λ phage repressor. Proc Natl Acad Sci USA , 5061–5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein RD, Meyer BJ. (1993). Independent domains of the SDC-3 protein control sex determination and dosage compensation in C. elegans. Cell , 349–364. [DOI] [PubMed] [Google Scholar]
- Kohlmaier A, Savarese F, Lachner M, Martens J, Jenuwein T, Wutz A. (2004). A chromosomal memory triggered by Xist regulates histone methylation in X inactivation. PLoS Biol , E171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer M, Kranz AL, Su A, Winterkorn LH, Albritton SE, Ercan S. (2015). Developmental dynamics of X-chromosome dosage compensation by the DCC and H4K20me1 in C. elegans. PLoS Genet , e1005698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruesi WS, Core LJ, Waters CT, Lis JT, Meyer BJ. (2013). Condensin controls recruitment of RNA polymerase II to achieve nematode X-chromosome dosage compensation. eLife , e00808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda MI, Hilfiker A, Lucchesi JC. (2016). Dosage compensation in Drosophila—a model for the coordinate regulation of transcription. Genetics , 435–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieb JD, Albrecht MR, Chuang PT, Meyer BJ. (1998). MIX-1: an essential component of the C. elegans mitotic machinery executes X chromosome dosage compensation. Cell , 265–277. [DOI] [PubMed] [Google Scholar]
- Lieb JD, Capowski EE, Meneely P, Meyer BJ. (1996). DPY-26, a link between dosage compensation and meiotic chromosome segregation in the nematode. Science , 1732–1736. [DOI] [PubMed] [Google Scholar]
- Liu T, Rechtsteiner A, Egelhofer TA, Vielle A, Latorre I, Cheung MS, Ercan S, Ikegami K, Jensen M, Kolasinska-Zwierz P, et al (2011). Broad chromosomal domains of histone modification patterns in C. elegans. Genome Res , 227–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luz JG, Hassig CA, Pickle C, Godzik A, Meyer BJ, Wilson IA. (2003). XOL-1, primary determinant of sexual fate in C. elegans, is a GHMP kinase family member and a structural prototype for a class of developmental regulators. Genes Dev , 977–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon MF. (1962). Sex chromatin and gene action in the mammalian X-chromosome. Am J Hum Genet , 135–148. [PMC free article] [PubMed] [Google Scholar]
- Madl JE, Herman R. (1979). Polyploids and sex determination in Caenorhabditis elegans. Genetics , 393–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer R, Meyer BJ, Ptashne M. (1980). Gene regulation at the right operator (OR) of bacteriophage lambda. I. OR3 and autogenous negative control by repressor. J Mol Biol , 147–161. [DOI] [PubMed] [Google Scholar]
- McDonel P, Jans J, Peterson BK, Meyer BJ. (2006). Clustered DNA motifs mark X chromosomes for repression by a dosage compensation complex. Nature , 614–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mets DG, Meyer BJ. (2009). Condensins regulate meiotic DNA break distribution, thus crossover frequency, by controlling chromosome structure. Cell , 73–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer BJ. (2010). Targeting X chromosomes for repression. Curr Opin Genet Dev , 179–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer BJ, Casson LP. (1986). Caenorhabditis elegans compensates for the difference in X chromosome dosage between the sexes by regulating transcript levels. Cell , 871–881. [DOI] [PubMed] [Google Scholar]
- Meyer BJ, Kleid DG, Ptashne M. (1975). Lambda repressor turns off transcription of its own gene. Proc Natl Acad Sci USA , 4785–4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer BJ, Maurer R, Ptashne M. (1980). Gene regulation at the right operator (OR) of bacteriophage lambda. II. OR1, OR2, and OR3: their roles in mediating the effects of repressor and cro. J Biol Mol , 163–194. [DOI] [PubMed] [Google Scholar]
- Meyer BJ, Ptashne M. (1980). Gene regulation at the right operator (OR) of bacteriophage lambda. III. Lambda repressor directly activates gene transcription. J Mol Biol , 195–205. [DOI] [PubMed] [Google Scholar]
- Miller LM, Plenefisch JD, Casson LP, Meyer BJ. (1988). xol-1: a gene that controls the male modes of both sex determination and X chromosome dosage compensation in C. elegans. Cell , 167–183. [DOI] [PubMed] [Google Scholar]
- Muller HJ. (1932), Further studies on the nature and causes of gene mutations. Int Congr Genet , 213–255. [Google Scholar]
- Nicoll M, Akerib CC, Meyer BJ. (1997). X-chromosome-counting mechanisms that determine nematode sex. Nature , 200–204. [DOI] [PubMed] [Google Scholar]
- Nigon V. (1951). Polyploidie experimentale chez un nematode libre, Rabditis elegans Maupas. Bull Sci Fr Belg , 187–225. [Google Scholar]
- Nusbaum C, Meyer BJ. (1989). The Caenorhabditis elegans gene sdc-2 controls sex determination and dosage compensation in XX animals. Genetics , 579–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pferdehirt RR, Kruesi WS, Meyer BJ. (2011). An MLL/COMPASS subunit functions in the C. elegans dosage compensation complex to target X chromosomes for transcriptional regulation of gene expression. Genes Dev , 499–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plenefisch JD, DeLong L, Meyer BJ. (1989). Genes that implement the hermaphrodite mode of dosage compensation in Caenorhabditis elegans. Genetics , 57–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell JR, Jow MM, Meyer BJ. (2005). The T-box transcription factor SEA-1 is an autosomal element of the X:A signal that determines C. elegans sex. Dev Cell , 339–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhind NR, Miller LM, Kopczynski JB, Meyer BJ. (1995). xol-1 acts as an early switch in the C. elegans male/hermaphrodite decision. Cell , 71–82. [DOI] [PubMed] [Google Scholar]
- Samata M, Akhtar A. (2018). Dosage compensation of the X chromosome: a complex epigenetic assignment involving chromatin regulators and long noncoding RNAs. Annu Rev Biochem , 323–350. [DOI] [PubMed] [Google Scholar]
- Skipper M, Milne CA, Hodgkin J. (1999). Genetic and molecular analysis of fox-1, a numerator element involved in Caenorhabditis elegans primary sex determination. Genetics , 617–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CJ, Mets DG, Albrecht MR, Nix P, Chan A, Meyer BJ. (2008). Meiotic crossover number and distribution are regulated by a dosage compensation protein that resembles a condensin subunit. Genes Dev , 194–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vielle A, Lang J, Dong Y, Ercan S, Kotwaliwale C, Rechtsteiner A, Appert A, Chen QB, Dose A, Egelhofer T, et al (2012). H4K20me1 contributes to downregulation of X-linked genes for C. elegans dosage compensation. PLoS Genet , e1002933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve AM, Meyer BJ. (1987). sdc-1: a link between sex determination and dosage compensation in C. elegans. Cell , 25–37. [DOI] [PubMed] [Google Scholar]
- Wells MB, Snyder MJ, Custer LM, Csankovszki G. (2012). Caenorhabditis elegans dosage compensation regulates histone H4 chromatin state on X chromosomes. Mol Cell Biol , 1710–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler BS, Anderson E, Frokjaer-Jensen C, Bian Q, Jorgensen E, Meyer BJ. (2016). Chromosome-wide mechanisms to decouple gene expression from gene dose during sex-chromosome evolution. eLife , e17365. [DOI] [PMC free article] [PubMed] [Google Scholar]


