Abstract
During development and in cancer, cells often move together in small to large collectives. To move as a unit, cells within collectives need to stay coupled together and coordinate their motility. How cell collectives remain interconnected and migratory, especially when moving through in vivo environments, is not well understood. The genetically tractable border cell group undergoes a highly polarized and cohesive cluster-type migration in the Drosophila ovary. Here we report that the small GTPase Rap1, through activation by PDZ-GEF, regulates border cell collective migration. We find that Rap1 maintains cell contacts within the cluster, at least in part by promoting the organized distribution of E-cadherin at specific cell–cell junctions. Rap1 also restricts migratory protrusions to the front of the border cell cluster and promotes the extension of protrusions with normal dynamics. Further, Rap1 is required in the outer migratory border cells but not in the central nonmigratory polar cells. Such cell specificity correlates well with the spatial distribution of the inhibitory Rapgap1 protein, which is higher in polar cells than in border cells. We propose that precisely regulated Rap1 activity reinforces connections between cells and polarizes the cluster, thus facilitating the coordinated collective migration of border cells.
INTRODUCTION
Many cells that migrate to form and remodel tissues and organs during development move in small to large groups, known as collectives (Scarpa and Mayor, 2016). Collective cell movement also occurs in cancer and may contribute to invasion and metastasis (Yamamoto et al., 1983; Friedl et al., 1995, 2012; Cheung et al., 2013; Cheung and Ewald, 2016; Khalil et al., 2017). Both single cells and cells in collectives undergo a motility cycle that consists of several stereotypical steps (reviewed in Ridley et al., 2003; Friedl and Gilmour, 2009). First, cells polarize to produce a major F-actin–enriched protrusion from the plasma membrane at the front, which helps pull the cell forward. Second, cells adhere to a migratory substrate, made up of either other cells or extracellular matrix (ECM). Finally, cells break rearward adhesions to retract the cell rear, allowing the cell body to move. In single-cell movement, individual epithelial cells need to lose cell–cell adhesions with the adjacent epithelium to become motile (reviewed in Thiery et al., 2009). In contrast, cells that migrate in collectives retain connections with neighboring cells to facilitate their movement as coordinated multicellular units (reviewed in Etienne-Manneville, 2014; Mayor and Etienne-Manneville, 2016; De Pascalis and Etienne-Manneville, 2017; Friedl and Mayor, 2017). Cell–cell contacts, typically through adherens junction (AJ) proteins such as E-cadherin, further facilitate transmission of information among the connected cells (Bazellières et al., 2015; Collins and Nelson, 2015; Friedl and Mayor, 2017). Mechanical coupling of cell adhesions to the cytoskeleton in turn helps coordinate the entire cell group so that one (or more) cell at the front becomes the protrusive leader cell, while the cells at the back become nonprotrusive followers (Etienne-Manneville, 2014; Llense and Etienne-Manneville, 2015; Mayor and Etienne-Manneville, 2016; Friedl and Mayor, 2017). The mechanisms that promote precise cell–cell communication within collectives to establish and maintain this front–back polarity remain poorly understood, especially for those cells that migrate inside tissues.
The relatively simple Drosophila border cells provide a genetically accessible model to investigate how cell collectives form and move in vivo (reviewed in Montell et al., 2012; Saadin and Starz-Gaiano, 2016). Border cells migrate as a group in the developing egg chamber, which is the functional unit of the ovary (Montell et al., 1992; Spradling, 1993). During late oogenesis, four to eight epithelial follicle cells at the anterior are specified to become motile border cells through activation of janus kinase/signal transducer and activator of transcription (JAK-STAT) signaling (Silver and Montell, 2001; Beccari et al., 2002; Ghiglione et al., 2002; Xi et al., 2003). The border cells coalesce around the central pair of polar cells to form a migratory cluster. Subsequently, the border cell cluster detaches (delaminates) from the epithelium (Figure 1A). The timing of migration is regulated by a pulse of the ecdysone steroid hormone (Bai et al., 2000; Jang et al., 2009). Border cells then move between the large germline-derived nurse cells (Figure 1A). A combination of apical cell polarity proteins and adhesion proteins, including Par-3 (Bazooka; Baz), aPKC, and E-cadherin, keep the border cells attached to the central polar cells and organized tightly into a cohesive cluster (Pinheiro and Montell, 2004; Llense and Martín-Blanco, 2008; Cai et al., 2014; Wang et al., 2018). During migration, the border cell cluster is clearly polarized, producing a forward-directed protrusion that keeps the collective motile (Fulga and Rørth, 2002; Prasad and Montell, 2007; Poukkula et al., 2011). Eventually the border cells reach the oocyte at the posterior (Figure 1A). Once there, border cells contribute to formation of the micropyle, the sperm-entry pore used for fertilization of the oocyte (Montell et al., 1992; Spradling, 1993).
FIGURE 1:
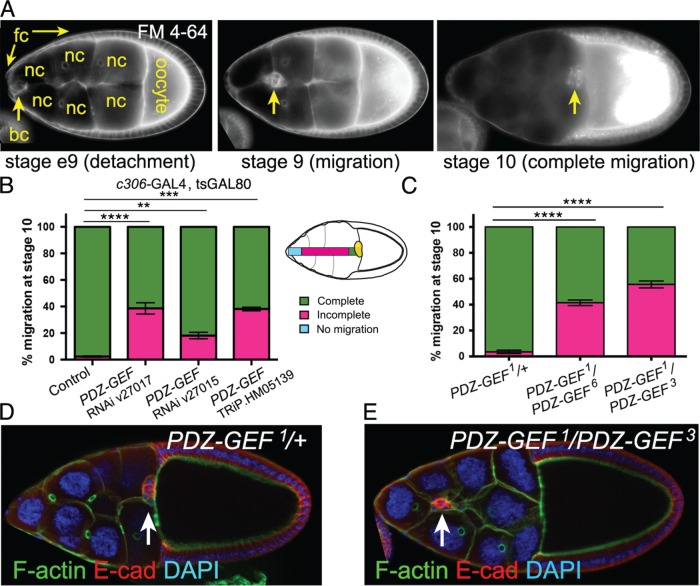
PDZ-GEF is required for border cell migration. (A) Wild-type border cell migration at the indicated ovarian developmental stages. Frames from live time-lapse videos of wild-type egg chambers stained for the lipophilic dye FM 4-64. Border cells (bc; arrows) detach from the follicle cell (fc) epithelium at early stage 9 (e9), migrate between the 15 nurse cells (nc) at stage 9, and reach the large oocyte at the posterior by stage 10. (B) PDZ-GEF RNAi knockdown prevents border cell migration. Quantification of border cell migration at stage 10, shown as the percentage of complete (green), incomplete (pink), or no (blue) border cell migration in control (c306-GAL4, tsGAL80/+) and PDZ-GEF RNAi (c306-GAL4, tsGAL80/+; +/PDZ-GEF RNAi) egg chambers, using three PDZ-GEF RNAi lines: 27017, 27105, and TRiP.HM05139. The egg chamber schematic illustrates the migration distance categories (no migration, incomplete, complete) of border cells (yellow). Values consist of four trials, with each trial assaying n ≥ 75 egg chambers (total n ≥ 310 egg chambers per genotype); **p < 0.01; ***p < 0.001; ****p < 0.0001; unpaired two-tailed t test comparing “complete” migration. (C) Loss of PDZ-GEF, using strong transallelic combinations of mutant alleles (Lee et al., 2002; Singh et al., 2006; Wang et al., 2006), disrupts border cell migration. Quantification of migration at stage 10, as shown in B. Genotypes: PDZ-GEF1/+ (control), PDZ-GEF1/PDZ-GEF6, and PDZ-GEF1/PDZ-GEF3. Values consist of five trials, with each trial assaying n ≥ 50 egg chambers (total n ≥ 255 egg chambers per genotype); ****p < 0.0001; unpaired two-tailed t test comparing “complete” migration. Error bars in B and C: ± SEM. (D, E) Loss of PDZ-GEF impairs border cell migration. E-cadherin (E-cad; red) labels cell membranes of border cells (arrows) and follicle cells, phalloidin (green) labels F-actin and DAPI (blue) labels nuclear DNA in stage 10 PDZ-GEF1/+ (control, D) and PDZ-GEF1/PDZ-GEF3 mutant (E) egg chambers. Anterior is to the left in this and all following figures.
Recent work in border cells has produced critical insights into the cellular and molecular mechanisms that establish and reinforce the formation of leader and follower cells in collectives (reviewed in Montell et al., 2012; Mayor and Etienne-Manneville, 2016; Saadin and Starz-Gaiano, 2016). Signaling through two receptor tyrosine kinases (RTKs), epidermal growth factor receptor (EGFR) and platelet-derived growth factor/vascular-endothelial growth factor (PDGF/VEGF) receptor related (PVR), polarizes the border cell cluster in response to guidance ligands secreted by the oocyte (Duchek and Rørth, 2001; Duchek et al., 2001; McDonald et al., 2006; Prasad and Montell, 2007; Wang et al., 2010). These RTKs activate the small GTPase Rac at the cluster front, thus promoting an enrichment of F-actin in the front (“leader”) border cell, which then induces formation of a stable protrusion. Rab11, through the actin regulator Moesin, helps restrict Rac activity to the front, and communicates this information to the other cells so that nonleader (“follower”) cells cannot form stable protrusions (Ramel et al., 2013). The cell–cell adhesion protein E-cadherin, through its function in AJs and coupling to F-actin (Baum and Georgiou, 2011), further mechanically links border cells, stabilizing the lead protrusion and suppressing protrusions from the other cells, thus reinforcing their status as follower cells (Cai et al., 2014). Jun amino-terminal kinase (JNK) signaling also promotes normal cell–cell contacts between border cells for cluster cohesion, as well as communication between cells (Llense and Martín-Blanco, 2008; Wang et al., 2010). Protrusions thus restricted to the lead border cell help the cluster navigate its way to the oocyte. Currently, it is unclear whether additional molecular mechanisms work together with this RTK-mediated pathway, or in parallel, to polarize the cluster and help establish leader versus follower cells. In a screen to uncover new regulators of cell polarity and migration of border cells (Aranjuez et al., 2012), we recently identified PDZ-GEF, a canonical guanine nucleotide exchange factor (GEF) for Rap1 (Raaijmakers and Bos, 2009).
Rap1, a highly conserved member of the Ras family of small GTPases, regulates many morphogenetic events during development through control of cell–cell and cell–extracellular matrix (ECM) adhesion, cell polarity, and/or the actin cytoskeleton (reviewed in Kooistra et al., 2007; Boettner and Van Aelst, 2009; Frische and Zwartkruis, 2010; Gloerich and Bos, 2011). Like all GTPases, Rap1 undergoes an activity cycle, consisting of activation by GEFs and inactivation by GTPase-activating proteins (GAPs). Highly specific functions of Rap1 occur through its downstream effectors, such as Canoe/Afadin, Riam, Rasip1, and others (Boettner et al., 2003; Lafuente et al., 2004; Kooistra et al., 2007; Raaijmakers and Bos, 2009; Post et al., 2013). In the early Drosophila embryo, Rap1 promotes establishment of epithelial polarity through positioning of AJs via Canoe (Choi et al., 2013; Bonello et al., 2018). Later in embryogenesis, differing levels of activated Rap1, through the spatially expressed GAP Rapgap1, positions where epithelia will fold and create invaginations (Wang et al., 2013). Here Rap1 regulates adhesion strength and location by coupling AJs to F-actin. Similarly, in the developing wing, Rap1 stabilizes E-cadherin-containing AJs (Knox and Brown, 2002). Additional functions for Rap1 in fly development include invagination of the mesoderm, dorsal closure, anchoring of testis stem cells to their niche, neuroblast polarity, and eye morphogenesis (Asha et al., 1999; Wang et al., 2006; Sawyer et al., 2009; Carmena et al., 2011; Spahn et al., 2012; Walther et al., 2018). Developmental roles for Rap1 are conserved in vertebrates, where Rap1 participates in neural tube closure, convergent extension during gastrulation, as well as neuronal differentiation, polarity, and axon pathfinding (Haigo et al., 2003; Tsai et al., 2007; Shah and Püschel, 2016; Shah et al., 2016, 2017). There is emerging evidence that Rap1-mediated signaling also regulates migration of single cells (Huelsmann et al., 2006; Jossin and Cooper, 2011; Lee and Jeon, 2012; Magliozzi et al., 2013; Zhang et al., 2017). The function for Rap1 in collective cell migration, however, is relatively unexplored. Here we show that Rap1, activated by PDZ-GEF and inactivated by Rapgap1, is a major regulator of border cell collective movement. Specifically, Rap1 promotes the organization of cell–cell contacts within the border cell cluster and facilitates the polarized extension of lead cell protrusions.
RESULTS
PDZ-GEF is required for border cell migration
To identify new regulators of cell–cell junctions, cell polarity and other critical parameters of border cell collective migration, we previously performed an RNA interference (RNAi) screen that targeted the majority of Drosophila PDZ (Psd95/Dlg/ZO-1) domain-containing proteins (Aranjuez et al., 2012). One of the strongest candidates from the original screen was PDZ-GEF (also known as dizzy or GEF26). PDZ-GEF encodes a Rapgef1/2 homologue with single cyclic nucleotide monophosphate-binding (cNMP-binding), Ras-like guanine nucleotide exchange factor N-terminal (also called Ras exchanger motif or REM), PDZ, Ras-association (RA), and catalytic GEF domains (Lee et al., 2002; Boettner and Van Aelst, 2007). To confirm a requirement in border cell migration, we first obtained additional independent upstream activating sequence (UAS)-RNAi lines that targeted PDZ-GEF. We drove UAS-RNAi knockdown in the entire border cell cluster using c306-GAL4, which drives expression early in anterior and posterior follicle cells and maintains expression in both border cells and the central polar cells throughout their migration (Manseau et al., 1997; Silver and Montell, 2001). Control border cells normally finish migrating to the oocyte by stage 10 (Figure 1, A and B). In contrast, each of the three PDZ-GEF RNAi lines consistently disrupted border cell migration when driven by c306-GAL4 (Figure 1B). Specifically, ∼20–40% of border cells deficient for PDZ-GEF stopped along the migration pathway (Figure 1B). We also validated the ability of these RNAi lines to knock down PDZ-GEF. Each of the three PDZ-GEF RNAi lines reduced the levels of PDZ-GEF RNA when driven ubiquitously in vivo (Supplemental Figure 1A). We further verified the requirement for PDZ-GEF using two strong but viable transallelic combinations of PDZ-GEF mutant alleles, PDZ-GEF1/PDZ-GEF3 and PDZ-GEF1/PDZ-GEF6 (Figure 1, C–E) (see Materials and Methods; Singh et al., 2006; Wang et al., 2006). While control border cells (PDZ-GEF1/+ heterozygotes) migrated to the oocyte, ∼40–50% of border cells in PDZ-GEF mutant egg chambers failed to complete their migration (Figure 1, C and E). Similarly to what we observed for PDZ-GEF RNAi, border cells mutant for PDZ-GEF initiated migration but stopped partway along the migration pathway (Figure 1, B, C, and E).
We next confirmed that PDZ-GEF was expressed during the stages of border cell migration. A lacZ enhancer trap in the PDZ-GEF gene (PDZ-GEF-lacZ; genotype: PDZ-GEF1/+) was ubiquitously expressed in all border cells during their entire migration, as well as in follicle cells and nurse cells (Supplemental Figure 1B). PDZ-GEF transcript was similarly detected in a ubiquitous pattern at these stages of ovarian development (Supplemental Figure 1C; Jambor et al., 2015). Finally, PDZ-GEF protein, as visualized using a functional green fluorescent protein (GFP)-tagged transgene driven by the endogenous PDZ-GEF promoter (Boettner and Van Aelst, 2007; Spahn et al., 2012), was present in all cells of the ovary, including border cells (Supplemental Figure 1, D and D′). Together these data show that PDZ-GEF is expressed in and required for border cell migration.
Rap1 is regulated by PDZ-GEF and is required for border cell migration
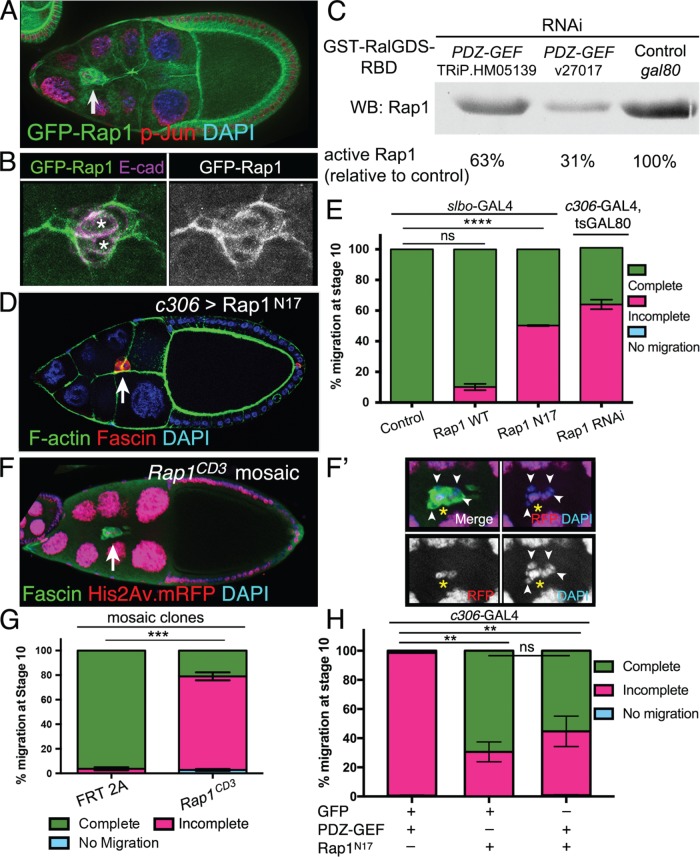
PDZ-GEF typically functions as a GEF for the small GTPase Rap1 (de Rooij et al., 1999; Liao et al., 1999). Therefore, we next asked whether Rap1 was expressed in the ovary during the stages when border cells migrate (stages 9–10). We made use of a functional GFP-Rap1 transgene driven by the endogenous Rap1 regulatory sequences (Knox and Brown, 2002). Rap1 was detected in all follicle cells and nurse cells in the ovary (Figure 2A). Moreover, Rap1 was expressed in border cells during initiation of cluster delamination/detachment (Supplemental Figure 2, A–A″), during migration (Figure 2, A and B), and at the end of migration. Specifically, Rap1 was enriched at the cell cortex of border cells and polar cells (Figure 2B), consistent with membrane-recruited active Rap1 (Bivona et al., 2004; Gloerich and Bos, 2011). Previous studies provided genetic evidence that Drosophila PDZ-GEF and Rap1 act in the same pathway and demonstrated that the two proteins could bind in a yeast two-hybrid assay (Lee et al., 2002; Huelsmann et al., 2006; Singh et al., 2006; Wang et al., 2006; Boettner and Van Aelst, 2007). We wanted to more directly test the extent to which PDZ-GEF regulates Rap1 activity in Drosophila. Therefore, we performed a GTPase activity assay in cultured S2 cells, designed to specifically pull down activated Rap1 (see Materials and Methods). When PDZ-GEF was knocked down by RNAi, using either of two double-stranded RNAs (dsRNAs; see Materials and Methods), the amount of activated Rap1 pulled down was markedly reduced compared with control dsRNA-treated cells (Figure 2C and Supplemental Figure 2B). Specifically, Rap1 activity was 63 and 31% of control levels (Figure 2C), closely matching the efficiency of the respective PDZ-GEF RNAi lines in vivo (Figure 1B and Supplemental Figure 1A). We independently repeated the experiment and observed a similar reduction in Rap1 activity due to PDZ-GEF dsRNA-mediated knockdown (Supplemental Figure 2B).
FIGURE 2:
Rap1 is regulated by PDZ-GEF and is required for border cell migration. (A, B) GFP-Rap1 is expressed in border cells. Representative examples of stage 9 egg chambers expressing GFP-Rap1 (green) and costained for DAPI (blue in A) to label nuclear DNA, phospho-Jun (red in A) to label nuclei, or E-cadherin (E-cad; magenta in B) to label cell membranes. (A) Arrow points to border cells. GFP-Rap1 is also expressed in follicle cells and nurse cells. (B) Close-up view of a border cell cluster showing that GFP-Rap1 is membrane-enriched and partly colocalizes with E-cadherin in border cells and polar cells (left panel: asterisks; colocalization in white). (C) Activity pull-down assay demonstrates that PDZ-GEF regulates Rap1 activity in Drosophila S2 cells. GST-RalGDS-RBD beads were used to pull down GTP-bound active Rap1 from S2 cells in the presence of wild-type levels of PDZ-GEF (control gal80 RNAi) or when PDZ-GEF was knocked down (v27107 and TRiP.HM05139 RNAi; see Materials and Methods). The relative amount of active Rap1 pulled down was identified by Western blot using a Rap1 antibody. Relative band intensities were measured as a percentage of the control, which represents the amount of maximally active Rap1 in this assay. (D, E) Inhibition of Rap1 activity by dominant-negative Rap1 (Rap1N17) or knockdown by RNAi disrupts border cell migration. (D) Expression of Rap1N17 disrupts border cell migration. Example of a stage 10 c306>Rap1N17 egg chamber (c306-GAL4 tsGAL80/+; UAS-Rap1N17/+) stained for Fascin (red) to label border cells (arrow), phalloidin to label F-actin (green) and DAPI (blue) to label nuclear DNA. (E) Quantification of complete (green), incomplete (pink), and no (blue) migration in stage 10 control (slbo-Gal4/+), Rap1WT (slbo-Gal4/+; +/UAS-Rap1WT), Rap1N17 (slbo-Gal4/UAS-Rap1N17), and Rap1 RNAi (c306-Gal4 tsGAL80/+; +/UAS-Rap1 RNAi v33437) egg chambers. Migration distance as in Figure 1B. Values consist of five trials, with each trial assaying n ≥ 50 egg chambers (total n ≥ 250 egg chambers per genotype); ns, not significant, p ≥ 0.05; ****p < 0.0001; unpaired two-tailed t test comparing “complete” migration. (F, F′) Rap1 mosaic mutant border cells do not complete their migration to the oocyte. Stage 10 Rap1CD3 mosaic mutant egg chamber stained for Fascin (green) to label the border cells (arrow) and DAPI (blue) to visualize nuclear DNA. His2Av.mRFP (red fluorescent protein, RFP; red) marks wild-type cells; colocalization with DAPI appears as magenta. Loss of RFP marks the homozygous mutant cells, including border cells (arrowheads in F′). (F′) Magnified view of the Rap1CD3 mosaic mutant border cell cluster from (F). Three cells, likely the pair of polar cells (yellow asterisk) and one border cell, are wild-type (red), while the remaining border cells are mutant (loss of red fluorescence). (G) Extent of migration when border cells are mosaic mutant for a loss-of-function allele of Rap1. Quantification of complete (green), incomplete (pink), and no (blue) migration in stage 10 control (FRT 2A) and Rap1CD3 FRT 2A mosaic mutant egg chambers. Migration distance as in Figure 1B. Values consist of four trials, with each trial assaying n ≥ 75 egg chambers (total n ≥ 300 egg chambers per genotype); ***p = 0.0001; unpaired two-tailed t test comparing “complete” migration. (H) Expression of Rap1N17 partially suppresses the migration defects caused by PDZ-GEF overexpression. Quantification of complete (green), incomplete (pink), and no (blue) migration in stage 10 egg chambers expressing PDZ-GEF and GFP (c306-GAL4/+; UAS-PLC∆PH-GFP/UAS-PDZ-GEF), Rap1N17 and GFP (c306-GAL4/+; UAS-Rap1N17/+; +/UAS-PLC∆PH-GFP), or Rap1N17 and PDZ-GEF (c306-GAL4/+; UAS-Rap1N17/+; +/UAS-PDZ-GEF). Values consist of three trials, with each trial assaying n ≥ 42 egg chambers per genotype (total n ≥ 176 egg chambers per genotype); ns, p ≥ 0.05; **p < 0.01; one-way analysis of variation (ANOVA) comparing “incomplete” migration. Error bars in E, G, and H: ± SEM.
We next investigated Rap1 function in border cells using three different approaches: expression of dominant-negative- (DN-) Rap1N17, Rap1 RNAi, and a Rap1 loss-of-function mutant allele. Expression of Rap1N17, using either of two different border cell GAL4 drivers, the earlier-expressing c306-GAL4 (Figure 2, D and H) and the later- (but generally higher) expressing slbo-GAL4 (Figure 2E), prevented 30–50% of border cells from reaching the oocyte by the correct stage. Rap1 RNAi driven by c306-GAL4 similarly disrupted border cell migration (Figure 2E). DN constructs and RNAi do not always represent true loss-of-function situations. Therefore, we next analyzed border cells mutant for a null allele of Rap1 (Rap1CD3, a deletion of the entire Rap1 gene) (Asha et al., 1999). Because complete loss of Rap1 is lethal, we used the mosaic FLP recombinase-FLP recombination target (FLP-FRT) system (Xu and Rubin, 1993) to generate clones consisting of homozygous Rap1 mutant cells in an otherwise heterozygous animal. Here, wild-type cells were marked by the presence of nuclear red fluorescent protein (RFP), while mutant cells were marked by absence of nuclear RFP. As with the other genetic manipulations, border cell clusters containing Rap1 mosaic mutant cells fully delaminated (detached) from the epithelium but stopped along the migration pathway (Figure 2, F and G). Most phenotypic border cell clusters were composed of both wild-type and mutant cells (Figure 2, F and F′). Nonetheless, 80% of clusters that contained a mix of Rap1 mutant and wild-type border cells failed to reach the oocyte (Figure 2G). Taken together, these data demonstrate that the small GTPase Rap1 is required for border cell migration.
The above-described migration defects caused by loss of PDZ-GEF and loss of Rap1 were similar, and PDZ GEF is a known GEF for Rap1 (Boettner and Van Aelst, 2007; Raaijmakers and Bos, 2009; Spahn et al., 2012). To test more directly whether PDZ-GEF was a major regulator of Rap1 in border cells, we next examined whether there was a genetic interaction between Rap1 and PDZ-GEF. The Rap1N17 mutation titrates away specific GEF activity for Rap1 in cells (Feig, 1999; Boettner et al., 2003; Huelsmann et al., 2006). In Drosophila embryonic hemocytes, coexpression of PDZ-GEF rescued the migration defects caused by Rap1N17, likely due to overcoming the loss of GEF activity induced by DN-Rap1 (Huelsmann et al., 2006). We drove expression of PDZ-GEF in border cells using c306-GAL4, either with a control UAS line (UAS-PLC∆PH-GFP, a neutral membrane GFP; Verstreken et al., 2009) or with UAS-Rap1N17 (Figure 2H). Expression of PDZ-GEF strongly inhibited border cell migration (Figure 2H, and below [see Figure 3, A and D]). Coexpression of PDZ-GEF with Rap1N17, however, more closely resembled that found when Rap1N17 was coexpressed with the control UAS-GFP transgene. Thus, expression of Rap1N17 suppressed the migration defects caused by high levels of PDZ-GEF, likely by titrating away the exogenously expressed PDZ-GEF.
FIGURE 3:
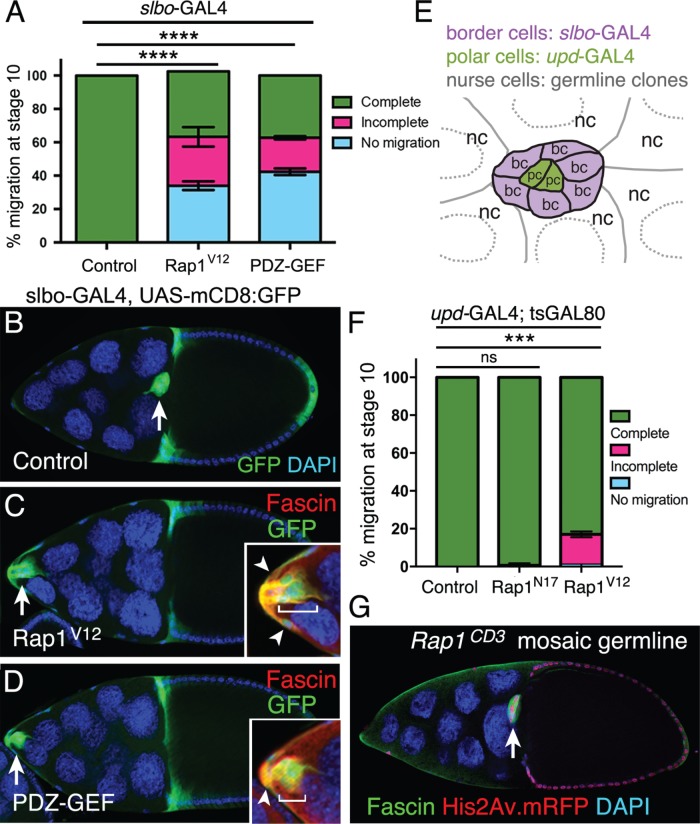
Defined levels of activated Rap1 are required in specific cells for border cell migration. (A–D) Expression of constitutively activated Rap1, or elevated activation of Rap1 through PDZ-GEF, in border cells impairs border cell migration. (A) Quantification of complete (green), incomplete (pink), and no (blue) migration in stage 10 control, Rap1V12, and UAS-PDZ-GEF overexpression egg chambers. Genotypes: control (slbo-GAL4, UAS-mCD8:GFP/+), Rap1V12 (slbo-GAL4 UAS-mCD8:GFP/+; +/UAS-Rap1V12), PDZ-GEF (slbo-GAL4 UAS-mCD8:GFP/+; +/UAS-PDZ-GEF). Migration distance as in Figure 1B. Values consist of three trials, with each trial assaying n ≥ 100 egg chambers (total n ≥ 310 egg chambers per genotype); ****p < 0.0001; unpaired two-tailed t test, comparing “no migration.” Error bars: ± SEM. (B–D) Stage 10 control (B), Rap1V12 (C), and PDZ-GEF (D) overexpression egg chambers. slbo-GAL4 drives expression of UAS-Rap1V12 and UAS-PDZ-GEF, along with UAS-mCD8:GFP (green), in border cells (arrow), adjacent follicle cells, and centripetal cells (cells at the anterior side of the oocyte). DAPI (blue) labels nuclear DNA. Genotypes as in A. Insets, magnified view of the same border cell cluster costained with Fascin (red) to further label border cells (brackets) and adjacent follicle cells (arrowheads). (E) Schematic drawing of the border cell cluster, with the central polar cells, and surrounding nurse cells. Different GAL4 drivers can be used to test gene function in border cells (bc) and central polar cells (pc). Germline mosaic mutant clones can be used to test function in nurse cells (nc). (F) Rap1 function in polar cells. Quantification of migration at stage 10 when the polar cells express a control GFP (upd-GAL4/+; tsGAL80/UAS-PLC∆PH-GFP), Rap1N17 (upd-GAL4/+; +/UAS-Rap1N17; tsGAL80/+) or Rap1V12 (upd-GAL4/+; tsGAL80/UAS-Rap1V12), shown as complete (green), incomplete (pink), and no (blue) border cell migration. Migration distance as in Figure 1B. Values consist of three trials, with each trial assaying n ≥ 27 egg chambers per genotype (total n ≥ 134 per genotype); ns, p ≥ 0.05; ***p < 0.001; unpaired two-tailed t test comparing “complete” migration. Error bars in A and F: ± SEM. (G) Border cells complete their migration to the oocyte when nurse cells are mutant for a loss-of-function allele of Rap1. Representative example of a stage 10 Rap1CD3 mosaic mutant egg chamber stained for Fascin (green) to label the border cells (arrow) and DAPI (blue) to visualize nuclear DNA. His2Av.mRFP (red) marks the wild-type cells; loss of RFP marks homozygous mutant cells. In this egg chamber, all nurse cells are mutant (loss of red signal); the border cells and most follicle cells are wild type (colocalization of DAPI in blue and RFP in red appears as magenta).
Although the data so far suggest that PDZ-GEF is a GEF for Rap1 in border cells, additional GEFs are known to regulate Rap1 activity (Raaijmakers and Bos, 2009; Gloerich and Bos, 2011). Two other Rap1 GEFs have been reported in Drosophila, Exchange protein directly activated by cAMP (Epac) and C3G guanyl-nucleotide exchange factor (C3G) (Dupuy et al., 2005; Shirinian et al., 2010). We obtained several RNAi lines for each gene and drove expression in border cells using c306-GAL4 (Supplemental Figure 2C). Decreased expression of Epac and C3G only mildly disrupted border cell migration, with ∼5–15% of border cells failing to reach the oocyte. The strong phenotypes observed with both PDZ-GEF and Rap1 mutants suggest that PDZ-GEF is the major GEF for Rap1 in border cells, but we do not rule out minor and/or redundant roles for Epac and C3G.
Defined levels of Rap1 activity promote border cell migration
Like other small GTPases, Rap1 activity levels are tightly regulated to produce distinct cellular outcomes (Gloerich and Bos, 2011). Therefore, we next tested the impact of elevated Rap1 activity on border cell migration. We increased the levels of activated Rap1 either by expressing a constitutively active (CA) mutant Rap1 (Rap1V12) or by overexpressing the activator PDZ-GEF in border cells and adjacent follicle cells using the slbo-GAL4 driver. Normally, at early stage 9, border cells detach from the anterior follicle cell epithelium before migrating between nurse cells and eventually reaching the oocyte (Figures 1A and 3, A and B). In contrast, ∼60% of border cells with higher Rap1 activity failed to complete their migration to the oocyte, with 30-40% of border cells remaining at the anterior tip of the egg chamber (Figure 3, A, C, and D). Border cells retained at the anterior end appeared to be tightly connected to the neighboring epithelial follicle cells (Figure 3, C and D, insets), suggesting a failure to detach from the epithelium. These data support the idea that precise levels of Rap1 activity are needed so that border cells can detach from the epithelium, initiate migration, and move between the nurse cells.
Cell-specific requirement for Rap1 in border cell migration
We next asked in which cells Rap1 activity was required for border cell migration. The border cell cluster consists of two cell types, the central polar cells and the outer migratory border cells (Figure 3E). In addition, border cells migrate on and between nurse cells (Figures 1A and 3E). Several key regulatory genes and pathways function in more than one of these cell types to control distinct aspects of border cell movement. For example, E-cadherin has multiple cell-specific roles: 1) in polar cells, E-cadherin maintains adhesion of border cells to the cluster; 2) in border cells, E-cadherin transmits mechanical tension so that only the lead border cell forms a protrusion; and 3) in nurse cells, E-cadherin provides traction for border cell movement (Niewiadomska et al., 1999; Fulga and Rørth, 2002; Cai et al., 2014). Moreover, activation of JAK/STAT in border cells relies on secretion of the cytokine Unpaired from the polar cells, which then specifies and recruits epithelial follicle cells to become motile border cells (Silver and Montell, 2001; Beccari et al., 2002; Ghiglione et al., 2002; Xi et al., 2003). Subsequently, JAK/STAT signaling is required in border cells for sustained migration (Silver et al., 2005). Because Rap1 protein is uniformly expressed in border cells, polar cells, and nurse cells (Figure 2, A and B, and Supplemental Figure 2A), this raised the possibility that Rap1 similarly functions in more than one cell type for successful migration.
To determine whether Rap1 was required in specific cells, we took advantage of two GAL4 drivers that have distinct expression patterns within the border cell cluster (Figure 3E); upd-GAL4 is expressed only in polar cells (Bai and Montell, 2002), whereas slbo-GAL4 is restricted to border cells (Rørth et al., 1998). We drove expression of UAS-Rap1N17 to inhibit Rap1 activity and UAS-Rap1V12 to activate Rap1. As described above, loss or gain of Rap1 activity in border cells using slbo-GAL4 strongly disrupted migration (Figures 2E and 3A). We next tested the requirement for Rap1 activity in polar cells using upd-GAL4. We included tsGAL80 to bypass potential lethality that could result from driving high expression of Rap1 mutants with upd-GAL4 during earlier stages of development (McGuire et al., 2003, 2004; Xiang et al., 2016; see Materials and Methods). We confirmed that under these experimental conditions the upd-GAL4 driver was functional. Expression of the transcriptional coactivator and protein tyrosine phosphatase Eyes Absent (Eya) caused frequent loss of anterior polar cells (unpublished data), in agreement with the role for Eya in suppressing polar cell specification (Bai and Montell, 2002). Loss of Rap1 activity in polar cells, however, did not affect cluster migration or the ability of border cells to attach to polar cells (Figure 3F). In contrast, increased Rap1 activity in polar cells by Rap1V12 mildly disrupted migration. Specifically, activated Rap1V12 expression in polar cells blocked border cell migration in ∼15% of egg chambers (Figure 3F).
Finally, we tested whether Rap1 was required in nurse cells for border cell migration. We produced germline clones with the FLP-FRT method (Xu and Rubin, 1993) using a null allele of Rap1, Rap1CD3 (see Materials and Methods). As expected, control clones in nurse cells (FRT alone) had no effect on border cell migration (n = 19 egg chambers). Similarly, loss of Rap1 in all nurse cells did not impair border cell movement (n = 31 egg chambers; Figure 3G). Rap1 function is thus required in border cells but does not appear to be necessary in polar cells or nurse cells for migration. However, having too much Rap1 activity, either in border cells alone or in polar cells alone, prevents border cell migration.
Rapgap1 modulates Rap1 activity during border cell collective migration
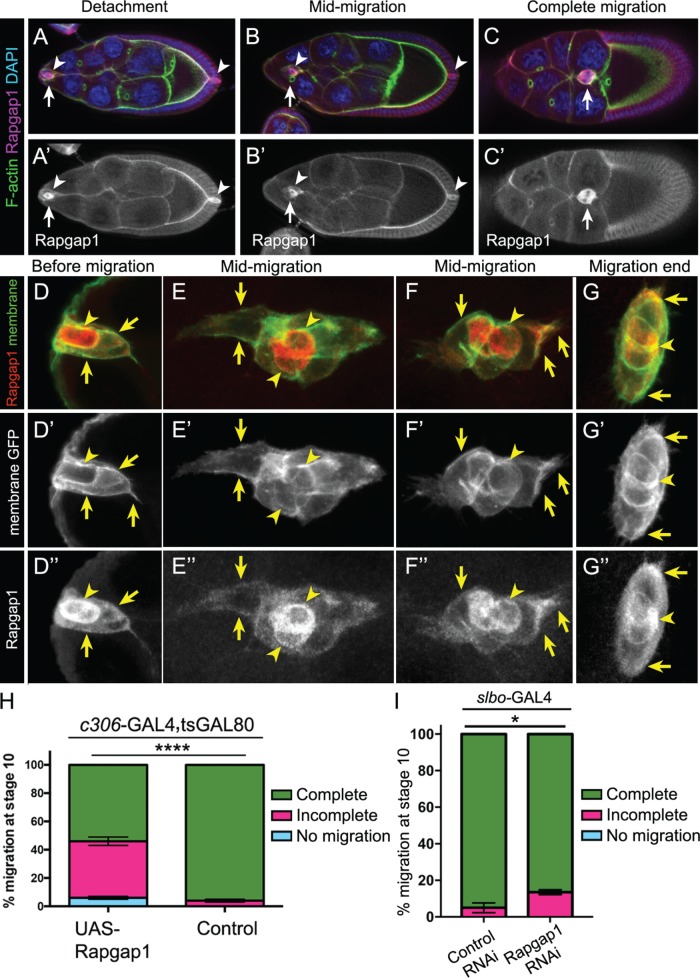
The results described above indicate that active Rap1 primarily functions in border cells, but not in polar cells, whereas ectopic activation of Rap1 in polar cells can inhibit migration. These data thus raised the possibility that Rap1 activity was restricted in some way within the border cell cluster. Both Rap1 and its activator PDZ-GEF are broadly expressed in migrating border cells (Figure 2, A and B, and Supplemental Figures 1, B–D′, and 2A). Thus, another mechanism likely restricts Rap1 activity to border cells. We focused our attention on Rapgap1, which is a major GTPase-activating protein (GAP) for Rap1 in Drosophila (Chen et al., 1997). GAPs hydrolyze guanosine triphosphate (GTP) to guanosine diphosphate (GDP), thus switching small GTPases from an active to an inactive state (Bos et al., 2007). Notably, Rapgap1 protein was expressed in the developing ovary during the stages that border cells migrate (Figure 4, A–C′). Throughout oogenesis, Rapgap1 levels appeared to be high in both the anterior and posterior pairs of polar cells and was expressed in border cells (Figure 4, A–C′, and unpublished data). To confirm the polar cell staining of Rapgap1, we costained egg chambers with a specific marker of polar cells, Fasciclin III (FasIII; Ruohola et al., 1991). Coexpression with FasIII confirmed that Rapgap1 was expressed in polar cells during the entire migration of border cells (Supplemental Figure 3, A–C′). Notably, prior to migration, we observed relatively lower levels of Rapgap1 protein in border cells compared with polar cells (Figure 4, A, A′, D, and D″). Once border cells moved into the egg chamber, Rapgap1 levels remained lower in border cells than in polar cells (Figure 4, B, B′, E, E″, F, and F″). However, Rapgap1 reached maximal levels in border cells by late phases of migration, particularly after the border cell cluster had reached the oocyte (Figure 4, C, C′, G, and G″).
FIGURE 4:
Rapgap1 is important for border cell migration. (A–G″) Rapgap1 protein expression and subcellular localization during the stages of oogenesis when border cells migrate. Wild-type egg chambers stained for Rapgap1 (magenta, A–C; red, D–G; white, A′–C′, D″–G″), DAPI to visualize nuclear DNA (blue, A–C), and costained for phalloidin to label F-actin (green, A–C) or expressing UAS-PLC∆PH:GFP to visualize cell membranes (green in D–G and white in D′–G′). (A–C′) Rapgap1 is expressed in anterior and posterior polar cells (arrowheads), as well as border cells (arrows), throughout the stages when border cells migrate, from detachment to complete migration. (D–G″) Close-up view of border cells coexpressing membrane-GFP and Rapgap1. Rapgap1 levels are highest in polar cells (arrowheads) at the start of migration (D, D‴) and at mid-migration stages (E, E‴, F, and F‴). By the end of migration, polar cells and border cells exhibit more equivalent levels of Rapgap1 (G, G‴). Rapgap1 exhibits diffuse subcellular localization in the cytoplasm and at cell membranes of border cells (arrows, D–G‴) and polar cells (arrowheads, D–G‴). The colocalization of Rapgap1 with membrane-GFP was confirmed by measuring normalized pixel intensities across border cell (arrows) and polar cell (arrowheads) membranes using the “plot profile” feature of FIJI (see Materials and Methods and Supplemental Figure 3, F and G). Maximum intensity projections of four to six merged z-stack sections are shown here but were not used for quantification. The direction of migration is to the right. (H, I) Raising or lowering the levels of Rapgap1 in border cells impairs migration. Quantification of complete (green), incomplete (pink), and no (blue) migration in stage 10 egg chambers. (H) Overexpression of Rapgap1 in border cells and polar cells driven by c306-GAL4. Genotypes are as follows: control (c306-GAL4, tsGAL80/+) and UAS-Rapgap1 (c306-GAL4, tsGAL80/+; +/UAS-Rapgap1). Migration distance as in Figure 1B. Values consist of four trials, with each trial assaying n ≥ 50 egg chambers (total n ≥ 230 egg chambers per genotype); ****p < 0.0001; unpaired two-tailed t test comparing “complete” migration. (I) Rapgap1 RNAi in border cells driven by slbo-GAL4. Genotypes are control (slbo-GAL4/UAS-mCherry RNAi) and Rapgap1 RNAi (slbo-GAL4/UAS-Rapgap1 RNAi v102659). Values consist of three trials, with each trial assaying n ≥ 44 egg chambers (total n ≥ 186 egg chambers per genotype); *p = 0.016; unpaired two-tailed t test comparing “incomplete” migration. Error bars in H and I: ± SEM.
In other cells, Rapgap1 homologues are associated with a variety of subcellular compartments (Su et al., 2003; Gloerich and Bos, 2011). Therefore, we examined the subcellular localization of Rapgap1 in border cells. We expressed a GFP-tagged membrane marker (UAS-PLC∆PH-GFP) in border cells and polar cells using c306-GAL4 (Figure 4, D–G″). Rapgap1 protein was detected in the cytoplasm of border cells and polar cells (e.g., Figure 4, D and D″). However, a fraction of Rapgap1 was also associated with the cell membrane (Figure 4, D–G″; Supplemental Figure 3, F and G). Additionally, Rapgap1 protein was detected at the cell cortex of nurse cells and the oocyte (Figure 4, A–C′). The membrane-associated Rapgap1, as well as differential polar cell versus border cell enrichment, suggests that Rapgap1 normally limits Rap1 activity in border cells and polar cells, although likely to different extents.
To test this idea further, we next raised and lowered the levels of Rapgap1 in border cells using the GAL4/UAS system. Overexpression of Rapgap1 prevented ∼45% of border cell clusters from completing their migration (Figure 4H), consistent with a loss of Rap1 activity. Next, we knocked down Rapgap1 by RNAi only in border cells using slbo-GAL4 (Figure 4I). Rapgap1 RNAi significantly reduced the levels of Rapgap1 protein in border cells (Supplemental Figure 3, D–E′). Knockdown of Rapgap1 in border cells produced a mild but significant impairment of border cell migration (∼15%; Figure 4I). Together these results support the idea that Rapgap1 modulates Rap1 activity in border cells. Moreover, the polar cell enrichment of Rapgap1 protein, along with the mild but reproducible disruption of border cell migration on expression of activated Rap1 in polar cells (Figure 3F), supports a model in which Rapgap1 prevents Rap1 from being fully active in polar cells.
Rap1 promotes the distribution of junctional E-cadherin within the border cell collective
Rap1 promotes the formation and maturation of cell–cell junctions and can regulate cell-extracellular matrix (ECM) adhesion (Bos, 2005; reviewed in Kooistra et al., 2007; Boettner and Van Aelst, 2009; Pannekoek et al., 2009). Maintaining integrity of junctional contacts between border cells is critical to coordinate their collective migration (Pinheiro and Montell, 2004; Llense and Martín-Blanco, 2008; Melani et al., 2008; Cai et al., 2014; Felix et al., 2015; Wang et al., 2018). E-cadherin-containing AJs are established and/or stabilized by Rap1 in many cell types, including the Drosophila wing, embryo, and eye (Knox and Brown, 2002; O’Keefe et al., 2009; Spahn et al., 2012; Choi et al., 2013; Bonello et al., 2018; Walther et al., 2018). E-cadherin itself is a critical regulator of border cell movement, cell–cell communication, and stabilization of front-directed protrusions (Niewiadomska et al., 1999; Fulga and Rørth, 2002; Cai et al., 2014). Given the importance of having organized cell–cell contacts, which occurs at least partly through the proper distribution of E-cadherin during border cell collective migration, we next determined whether Rap1 activity regulates junctional E-cadherin.
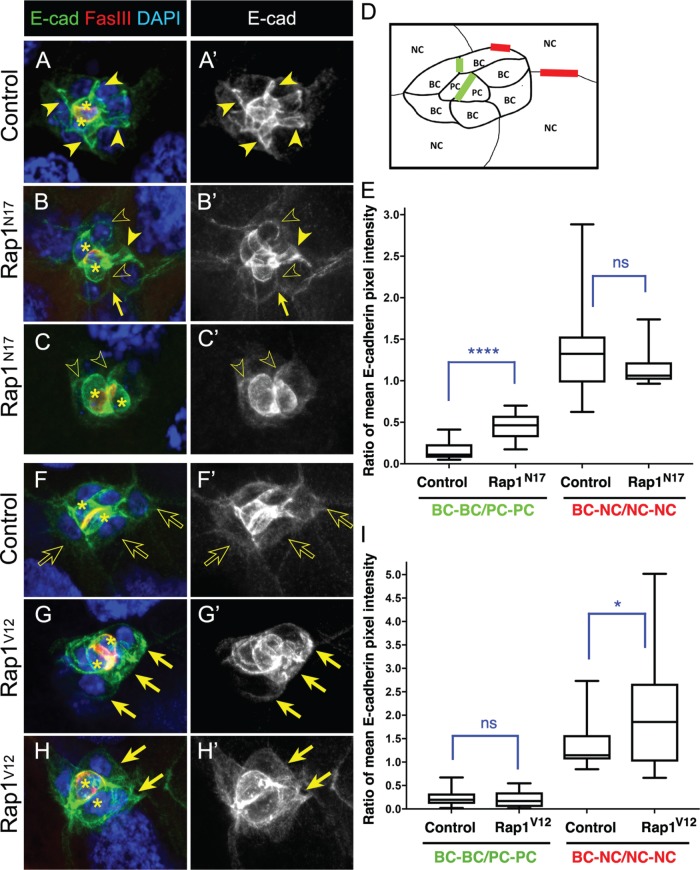
High levels of E-cadherin are normally found at cell–cell contacts within the migrating border cell cluster (Niewiadomska et al., 1999; Pinheiro and Montell, 2004; Cai et al., 2014), particularly at border cell–border cell (BC-BC) junctions, at junctions between the central polar cells and surrounding border cells (PC-BC), and between polar cells (PC-PC; Figure 5, A, A′, D, F, and F′). Low levels of E-cadherin are found at junctions between border cells and nurse cells (BC-NC; Figure 5, A, A′, D, F, and F′), where it provides traction for border cell movement on nurse cells (Niewiadomska et al., 1999; Fulga and Rørth, 2002; Cai et al., 2014). Because of the similar migration defects observed on using different genetic manipulations (Figure 2, D–G), we analyzed Rap1 function using the Rap1N17 construct. To quantify changes to the levels and/or distribution of E-cadherin, we measured the fluorescence intensity at specific junctions in control and Rap1N17 border cell clusters. Specifically, within the same cluster we quantified the E-cadherin fluorescence intensity ratio at BC-BC junctions compared with PC-PC junctions and the ratio of BC-NC junctions compared with NC-NC junctions (Figure 5, A, A′, and D). Rap1N17 border cell clusters on average accumulated higher E-cadherin levels at BC-BC junctions (Figure 5, B, B′, and E), compared with control (Figure 5, A, A′, and E). However, E-cadherin levels at BC-BC junctions were also quite variable and sometimes were visibly lower than normal (Figure 5, B–C′ and E). In contrast, E-cadherin was not generally altered at BC-NC contacts, although the overall levels of E-cadherin at BC-NC junctions were variable in both control and Rap1N17 border cell clusters (Figure 5E). Border cells were often rounder than normal (Figure 5, B–C′). Additionally, some border cells were less tightly adhered to each other and appeared to partially separate (Figure 5, B and B′).
FIGURE 5:
Rap1 regulates E-cadherin levels at specific cell–cell junctions. (A–C′, F–H′) Representative stage 9 control (A, A′, F, F′), stage 10 Rap1N17 (B–C’), and stage 10 Rap1V12 (G–H′) border cell clusters stained for E-cadherin (green in A–C and F–H; white in A′–C′ and F′–H′), Fas III to label the central polar cells (red in A–C and F–H; colocalization with E-cadherin appears yellow), and DAPI to visualize nuclear DNA (blue in A–C and F–H). Polar cells are marked with asterisks in A–C and F–H. The direction of migration is to the right in all panels. Maximum intensity projections of five merged z-stack sections are shown. (A–C′) Solid arrowheads mark high E-cadherin expression at border cell–border cell (BC-BC) junctions; open arrowheads indicate lower E-cadherin at BC-BC junctions. Arrows in B and B′ indicate altered contacts between border cells. (F–H′) Solid arrows mark high E-cadherin expression at BC-nurse cell (NC) junctions; open arrows indicate low E-cadherin at BC-NC junctions. (D) Schematic of a migrating border cell cluster showing cell–cell contacts measured for E-cadherin mean pixel fluorescence intensity in E and I; BC-BC and PC-PC (green) and BC-NC and NC-NC (red). (E, I) Quantification, represented as box-and-whisker plots, of the mean pixel intensity of E-cadherin as a ratio of BC-BC/PC-PC (green) and BC-NC/NC-NC (red) in matched control and Rap1N17 egg chambers (E) and matched control and Rap1V12 egg chambers (I). The whiskers represent the minimum and maximum pixel intensity; the box extends from the 25th to the 75th percentiles; the line indicates the median. ns, not significant (p ≥ 0.05); *p < 0.05; ****p < 0.0001; two-tailed unpaired t test. Genotypes are as follows: matched controls (c306-GAL4 tsGal80/+; UAS-PLC-∆PH-GFP/+ in A and A′; slbo-GAL4, UAS-mCD8-GFP/+ in D and D′); Rap1N17 (c306-GAL4 tsGal80/+; +/UAS-Rap1N17); Rap1V12 (slbo-GAL4/+; UAS-Rap1V12/+). (E) For control, 31 BC-BC contacts and 22 BC-NC contacts were measured from 15 border cell clusters. For Rap1N17, 22 BC-BC contacts were measured from 12 border cell clusters and 24 BC-NC contacts were measured from 11 border cell clusters. (I) For control, 40 BC-BC contacts and 22 BC-NC contacts were measured from 20 border cell clusters. For Rap1V12, 46 BC-BC contacts and 30 BC-NC contacts were measured from 25 border cell clusters.
We next examined the impact on E-cadherin and cell–cell junctions when Rap1 activity was elevated. E-cadherin levels at BC-BC junctions in Rap1V12 border cell clusters were overall unchanged, compared with control (Figure 5, F–I). However, there was a significant, though variable, elevation of E-cadherin at Rap1V12 BC-NC junctions (Figure 5I), ranging from quite high (Figure 5, G and G′) to a more moderate increase (Figure 5, H and H′). Thus, increased Rap1 activity produced higher E-cadherin at the cluster periphery, where border cells contact the nurse cells. Taken together, these data support the idea that Rap1 controls the proper distribution and levels of E-cadherin at specific border cell junctions, as well as cell shape and cluster organization.
Rap1 coordinates protrusions in migrating border cells
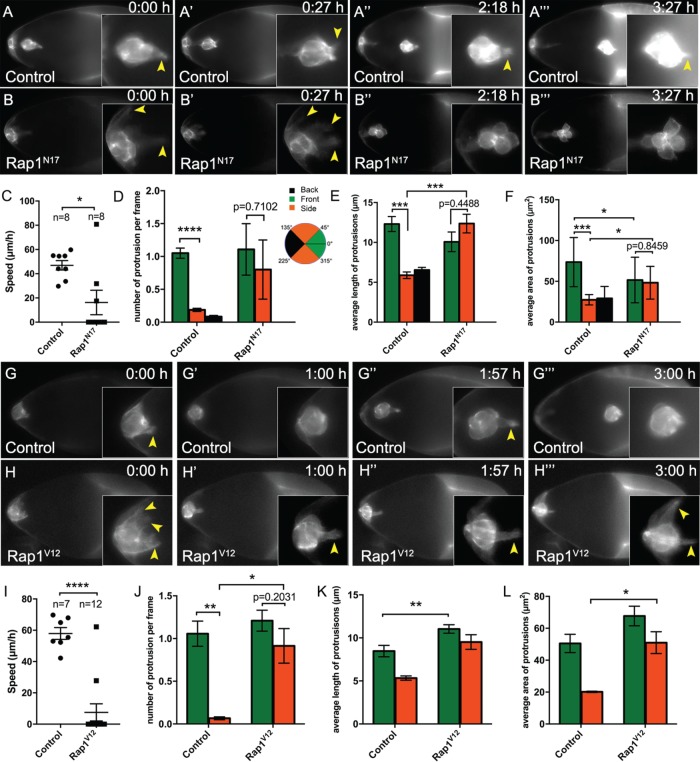
The results described above indicate that having the correct levels of active Rap1 is critical for migration and the normal junctional E-cadherin distribution in border cells. To investigate why border cells with altered Rap1 activity often could not complete their migration, we next performed live time-lapse imaging (Figure 6; Supplemental Videos 1–7). We visualized border cells with slbo-LifeAct-GFP, which specifically labels F-actin in border cells and a few adjacent follicle cells (Cai et al., 2014). This marker allowed us to examine border cell membranes and protrusions in more detail when Rap1 activity was inhibited (Figure 6, A–B‴; Supplemental Videos 1–3). At the start of migration, control border cells formed an organized cluster, with one or two prominent forward-directed protrusions that extended and retracted (Figure 6A; Supplemental Video 1; n = 8 videos) (Bianco et al., 2007; Prasad and Montell, 2007). During the remainder of migration, control border cell clusters stayed in a fairly tight group, with a leader cell continuing to extend and retract protrusions at the front (Figure 6, A–A‴; Supplemental Video 1). Rap1N17 border cells, in contrast, had trouble moving forward, which was reflected in decreased migration compared with control border cells (Figure 6, B and C; Supplemental Videos 2 and 3; n = 8 videos). Many Rap1N17 border cell clusters extended protrusions, consistent with the ability of some mutant clusters to leave the follicular epithelium and begin migrating (Figure 6, B–D, Supplemental Videos 2 and 3; n = 8 videos). When Rap1N17 border cells were able to move into the egg chamber, they generally had a decreased migration speed compared with control (Figure 6C). Rap1N17 border cell clusters often extended multiple protrusions (Supplemental Videos 2 and 3; Figure 6D), rather than the typical single protrusion found in control border cell clusters (Supplemental Video 1). Additionally, control border cells had significantly more front-directed protrusions than those at the “side” of the cluster (Figure 6D). In contrast, Rap1N17 border cells had almost as many front and side protrusions (Figure 6D). Moreover, some Rap1N17 border cell clusters stopped extending protrusions after migrating a short distance (Figure 6, B″ and B‴; Supplemental Video 2). Further, Rap1N17 border cells were rounder overall than normal (Figure 6, B″ and B‴; Supplemental Videos 2 and 3). Consistent with a role in promoting cell–cell contacts (Figure 5, B and B′), Rap1N17 border cells often appeared to partially separate from each other and were less tightly connected within the cluster compared with control (Figure 6B‴; Supplemental Video 2; unpublished data).
FIGURE 6:
Rap1 promotes collective motility and the proper formation of a single lead protrusion. (A–B‴, G–H‴) Frames from matched control (A–A‴; Supplemental Video 1) and Rap1N17 (B–B‴; Supplemental Video 2), and matched control (G–G‴; Supplemental Video 4) and Rap1V12 (H–H‴; Supplemental Video 5) live time-lapse videos showing migrating border cells (slbo-LifeAct-GFP) at the indicated times (h:min). Insets, close-up views of the same border cell clusters from the indicated video frame. Arrowheads indicate protrusions. (A–A‴, G–G‴) Representative control border cell clusters with major front protrusions. Cells within the cluster stay tightly cohesive throughout migration. (B–B‴) Representative Rap1N17 border cell cluster with multiple protrusions during early migration (B, B′). Later, the border cells become round (B″ and B‴). (H-H‴) Representative Rap1V12 border cell cluster that failed to initiate migration. Border cells remain at the anterior of the egg chamber. Multiple “side” protrusions extend (H, H‴), in addition to prominent “front” protrusions (H′, H″). (C, I) Measurement of migration speed in individual videos. (C) Matched control (n = 8) and Rap1N17 (n = 8). (I) Matched control (n = 7) and Rap1V12 (n = 12). (D–F, J–L) Measurements of protrusions within the first hour of matched control and Rap1N17 (D–F) and matched control and Rap1V12 (J–L) videos. (D, J) Number of protrusions from migrating clusters, per frame of the video, at the front, back, or side of the cluster. (E, F, K, L) Quantification of the average length (E, K) and average area (F, L) of protrusions from time-lapse videos of the indicated genotypes. See Supplemental Figure 4A for a schematic showing how protrusion length and area were measured. (D–F) N = 8 videos for control: 22 front protrusions, 7 side protrusions, and 3 back protrusions were analyzed; n = 7 videos for Rap1N17: 16 front protrusions and 3 side protrusions were analyzed; no back protrusions were observed. (J–L) N = 7 videos for control: 19 front protrusions and 3 side protrusions were analyzed; n = 12 videos for Rap1V12: 26 front protrusions and 12 side protrusions. Error bars: SEM; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; all other values were not significant (p ≥ 0.05), with the exception of D, E, F, and J, where the p values are shown to compare front and side protrusions within the same genotype; unpaired two-tailed t test. (A–F) Genotypes: control (c306-GAL4, tsGal80/+; +/slbo-LifeAct-GFP) and Rap1N17 (c306-GAL4, tsGal80/+; UAS-Rap1N17/slbo-LifeAct-GFP). (G–L) Genotypes: control (slbo-GAL4/slbo-LifeAct-GFP) and Rap1V12 (slbo-GAL4/slbo-LifeAct-GFP; +/UAS-Rap1V12).
Video S1.
Migrating control border cells marked with LifeAct-GFP. Representative example of a live stage 9 control (c306-GAL4, tsGAL80/+; +/slbo-LifeAct-GFP) egg chamber. The Video shows that the control border cell cluster stays organized and that one or two border cells extend protrusions as they migrate to the oocyte (time in h:min). Frames were acquired every 3 min with a 20x objective. Anterior is to the left.
Video S2.
Rap1N17 border cells are less organized and fail to complete their migration. Example of a live stage 9 Rap1N17 (c306-GAL4, tsGAL80/+; UAS-Rap1N17/slbo-LifeAct-GFP) egg chamber in which the rear border cell remains attached to an anterior follicle cell. Border cells extend longer side protrusions during the first half of the Video (until ~1:18; time in h:min), after which border cells become round. By the end of the Video, border cells in the cluster are less organized than in control. Border cell migration is slower than in the control and the cluster does not reach the oocyte by the correct stage. Frames were acquired every 3 min with a 20x objective. Anterior is to the left.
Video S3.
Rap1N17 border cells are initially highly protrusive but migrate slower than control. Example of a live stage 9 Rap1N17 (c306-GAL4/+; UAS-Rap1N17/slbo-LifeAct-GFP) egg chamber in which the cluster completely detaches from the anterior follicle cells and migrates partway to the oocyte. Similar to Video 2, multiple cells extend protrusions during the first half of the Video, especially "side" cells (until ~1:30; time in h:min). Border cells eventually become rounder than normal. Frames were acquired every 2 min with a 20x objective. Anterior is to the left.
Video S4.
Migrating control border cells marked with LifeAct-GFP. Representative example of a live stage 9 control (slbo-GAL4/slbo-LifeAct-GFP) egg chamber. Control border cells detach from the anterior follicle cells, extend forward-directed protrusions and migrate to the oocyte (time in h:min). Frames were acquired every 3 min with a 20x objective. Anterior is to the left.
Video S5.
Rap1V12 border cells do not migrate but extend multiple front and side protrusions. Representative example of a live stage 9 Rap1V12 (slbo-GAL4/slbo-LifeAct-GFP; UAS-Rap1V12/+) egg chamber. Rap1V12 border cells stay at the anterior end of the egg chamber and do not move forward, but extend both front protrusions and prominent side protrusions (time in h:min). Frames were acquired every 3 min with a 20x objective. Anterior to the left.
Video S6.
Migrating control border cells marked with mCD8::GFP. Representative example of a live stage 9 control (slbo-GAL4, UAS-mCD8::GFP/+) egg chamber. The mCD8::GFP marker appears primarily cytoplasmic in migrating border cells but not in central polar cells because slbo-GAL4 does not drive expression in polar cells (Geisbrecht and Montell, 2002). Control border cells can extend several protrusions, but the forward protrusion is usually more stable (time in h:min). Frames were acquired every 3 min with a 20x objective. Anterior is to the left.
Video S7.
PDZ-GEF-overexpressing (OE) border cells do not detach from the follicle cell epithelium. Representative example of live stage 9 egg chamber overexpressing PDZ-GEF (slbo-GAL4, UAS-mCD8::GFP/+; +/UAS-PDZ-GEF). PDZ-GEF-overexpressing border cells stay attached to the adjacent anterior follicle cells, which also express slbo-GAL4 UAS-mCD8::GFP, but extend front-directed protrusions (time in h:min). Frames were acquired every 3 min with a 20x objective. Anterior is to the left.
The formation of dynamic protrusions is critical for cells to successfully migrate (Ridley, 2011). Given that Rap1N17 border cells did not complete their migration, yet many clusters extended extra protrusions, we next analyzed how protrusions were affected by loss of Rap1 activity. We measured the length and area of protrusions produced by Rap1N17 border cell clusters compared with control (Figure 6, E and F; Supplemental Figure 4, A and B). Interestingly, we observed an increase in the average length of non-forward-directed protrusions (“side”) but an overall decrease in the maximum length of forward protrusions (Figure 6E; Supplemental Figure 4A). The size of protrusions was also altered, with forward protrusions decreasing and side protrusions increasing in area compared with control (Figure 6F; Supplemental Figure 4B). We further noticed that Rap1N17 side protrusions often resembled front protrusions (Figure 6B-B‴; Supplemental Video 2). Indeed, the length and area of Rap1N17 side protrusions were predominantly similar to the ones at the front, whereas in control the front and side protrusions were significantly different (Figure 6, E and F; Supplemental Figure 4B). These phenotypes together suggest that decreased Rap1 activity impairs the ability of border cells to produce productive lead protrusions and to suppress side protrusions.
Next, we analyzed the impact of elevated Rap1 activity on live border cells. We imaged matched control (Figure 6, G–G‴; Supplemental Video 4; n = 13) and Rap1V12-expressing border cell clusters (Figure 6, H–H‴; Supplemental Video 5; n = 20) labeled with either mCD8::GFP (unpublished data) or slbo-LifeAct-GFP. In fixed egg chambers, ∼35–40% of border cells with increased Rap1 activity (Rap1V12 or PDZ-GEF overexpression) did not migrate away from the anterior tip of the egg chamber (Figure 3A). Most live Rap1V12-expressing border cells also did not move forward during imaging, in contrast to control border cells that always completed their migration (Figure 6, G–I; Supplemental Videos 4 and 5). Border cells overexpressing PDZ-GEF also did not migrate and strongly resembled Rap1V12 border cells (Supplemental Video 7, n = 8; see Supplemental Video 6 for matched control, n = 6). This lack of forward movement was despite the ability of Rap1V12 (Figure 6J; Supplemental Video 5) and PDZ-GEF-overexpressing (Supplemental Video 7) border cells to extend front-directed protrusions.
Inclusion of slbo-LifeAct-GFP allowed us to further analyze protrusions in Rap1V12 border cells compared with control (Figure 6, J–L; Supplemental Figure 4, C and D; n = 7 videos for control; n = 12 videos for Rap1V12). Notably, Rap1V12 border cells extended more side-directed protrusions than control, with almost equal numbers of total protrusions produced at the side as at the front (Figure 6J). Rap1V12 front- and side-directed protrusions were longer (Figure 6K; Supplemental Figure 4C), and had an increased area (Figure 6L; Supplemental Figure 4D), compared with control protrusions. These data together indicate that having the proper levels of activated Rap1 are necessary for producing front-directed protrusions with the correct length and size. The abnormal protrusions produced by both loss and gain of Rap1 activity, and the failure to restrict protrusions to the front, could account for the inability of these border cell clusters to complete their migration. Together, our data suggest that having an optimal level of Rap1 controls the shape of border cells, maintains cell–cell contacts within the cluster, and promotes the formation of polarized protrusions with normal dynamics.
DISCUSSION
Rap1 is required for organ and tissue morphogenesis in developing organisms, often through its roles in modulating the cytoskeleton, cell polarity, and/or cell–cell or cell–matrix adhesions (Boettner and Van Aelst, 2009; Frische and Zwartkruis, 2010). While Rap1 has been implicated in the motility of some single cells (Huelsmann et al., 2006; Jossin and Cooper, 2011; Lee and Jeon, 2012; Magliozzi et al., 2013; Zhang et al., 2017), whether or how Rap1 regulates migration of cells that move as collectives is poorly understood. Here we report Rap1 as a new coordinator of Drosophila border cell collective migration. Specifically, Rap1 promotes connections between cells in the cluster, which occurs at least partly through regulation of the proper distribution of E-cadherin and potentially through maintenance of normal border cell shape. Rap1 further controls the extension of polarized, front-directed protrusions. Optimal levels of Rap1 activity restricts the number of protrusions produced by the cluster and ensures that these protrusions have the proper length and size to sustain movement. Thus, we propose a model in which precise levels of activated Rap1, controlled by PDZ-GEF and Rapgap1, promotes the organization, shape, and polarity of the entire border cell cluster; this in turn drives the coordinated migration of the collective.
Our study indicates that Rap1 promotes the maintenance of cell–cell contacts within the border cell cluster during migration. Border cells, like other cell collectives, require tight cellular connections so that cells stay interconnected and move together in vivo. E-cadherin-based AJs are used by many epithelial-derived collectives to keep cells together during migration (reviewed in Collins and Nelson, 2015; Mayor and Etienne-Manneville, 2016; Friedl and Mayor, 2017). In border cells this is achieved through the proper localization and levels of multiple junctional proteins, including E-cadherin but also the apical polarity proteins aPKC, Par-3/Baz, and Par-6 (Niewiadomska et al., 1999; Pinheiro and Montell, 2004; Llense and Martín-Blanco, 2008; Melani et al., 2008; Cai et al., 2014; Wang et al., 2018). Intriguingly, we found that either loss or gain of Rap1 activity was sufficient to disrupt the levels and distribution of E-cadherin at specific cell–cell junctions. Inhibition of Rap1 altered E-cadherin at BC-BC junctions, resulting in higher E-cadherin at some junctions and lower levels at others; in some cases, there was an apparent rounding and partial separation of the cells within the cluster. Border cells with activated Rap1, in contrast, had elevated E-cadherin at BC-NC contacts, and many failed to detach from the epithelium. Low levels of E-cadherin at the BC-NC interface provides traction for border cells to migrate on nurse cells (Niewiadomska et al., 1999; Cai et al., 2014). Elevation of E-cadherin at BC-NC contacts when Rap1 is constitutively activated could prevent forward movement of border cells as seen in other mutants that disrupt distribution of E-cadherin within the cluster (Pinheiro and Montell, 2004; Schober et al., 2005; Anllo and Schupbach, 2016). Previous studies showed that abnormal elevation of apical polarity proteins and a failure to downregulate E-cadherin at junctions between border cells and follicle cells in turn prevents complete border cell detachment from the epithelium (Schober et al., 2005; McDonald et al., 2008; Anllo and Schupbach, 2016). Although not directly tested here, our results suggest that in order for border cells to detach from the epithelium, Rap1 activity must transiently be low so that junctions between border cells and follicle cells can be remodeled or broken.
Altogether our results suggest that having the correct levels of active Rap1 in border cells may impact E-cadherin junctional positioning, distribution and/or stability, similar to what has been seen in the wing and other epithelial tissues (Knox and Brown, 2002; Spahn et al., 2012; Choi et al., 2013; Wang et al., 2013). Notably, loss of E-cadherin does not disrupt cell–cell contacts or the shape of cells within the border cell cluster (Niewiadomska et al., 1999; Fulga and Rørth, 2002; Cai et al., 2014). Instead, E-cadherin-deficient border cells fail to extend major protrusions but can migrate “off-track” for short distances, indicating that directional guidance to the oocyte is lost (Fulga and Rørth, 2002; Cai et al., 2014). Thus, it is reasonable to predict that Rap1 promotes cell–cell contacts within the migrating border cell cluster through additional cell junction or cell polarity proteins. For example, Rap1 could more directly regulate connection of AJs to the F-actin cytoskeleton, possibly through junctional components such as alpha-Catenin, Vinculin, and/or Canoe/Afadin (Mandai et al., 2013; Lecuit and Yap, 2015). Further work is needed to test these different possibilities.
We have shown that Rap1 promotes the formation of polarized protrusions within the border cell cluster. Increasing or decreasing Rap1 activity caused an overall increase in the number of protrusions, especially those produced by nonleading border cells. These results suggest that altering Rap1 activity disrupts polarization of the migrating cluster. Cells that migrate in collectives need to establish one or more cells that will become protrusive leaders, and then reinforce this information among the group so that follower cells do not extend extra protrusions (Mayor and Etienne-Manneville, 2016). Such leader–follower orientation facilitates efficient directional movement. Border cells establish cluster polarity through a signaling cascade that begins with long-range secretion of chemoattractant guidance ligands from the oocyte (Duchek et al., 2001; McDonald et al., 2003, 2006). The border cell in front presumably receives the highest levels of ligands, triggering RTK-mediated activation of the Rac small GTPase and enrichment of F-actin, thus forming a stable lead protrusion (Duchek et al., 2001; Wang et al., 2010). This information is then communicated to follower cells through a combination of Rab11, Moesin, and JNK signaling, which prevent follower cells from extending protrusions (Llense and Martín-Blanco, 2008; Wang et al., 2010; Ramel et al., 2013). Loss of any one of these components results in all border cells, both leader and follower cells, extending protrusions (Prasad and Montell, 2007; Wang et al., 2010; Ramel et al., 2013), similar to what we observed when Rap1 activity was impaired.
Intracollective adhesions also couple cells together to communicate and stabilize the front-rear polarity of the migrating collective (Bazellières et al., 2015; Collins and Nelson, 2015; Mayor and Etienne-Manneville, 2016). In border cells, E-cadherin-based AJs facilitate this communication of leader-follower protrusion position in response to Rac GTPase signaling through mechanical linkage of cells in the cluster (Cai et al., 2014). We propose that Rap1 participates in this collective-wide communication of leader-follower protrusion formation, although the mechanism is currently unknown. Loss of Rap1 disrupted contacts between border cells and the normal distribution of E-cadherin. Thus, it is possible that Rap1 mediates reinforcement of protrusion extension from the front border cell through stabilization of cell–cell junctions. A recent study also found that Rap1 is required for the formation of a single leading border cell protrusion (Chang et al., 2018), in agreement with our study. Supporting a role for Rap1 in border cell cluster polarization, Chang and colleagues (2018) found that disruption of Rap1 resulted in spatially uniform Rac activation. This depolarized Rac activity is consistent with a failure to restrict Rac-induced protrusions to the cluster front (Wang et al., 2010). Whether Rap1 functions more directly as part of this canonical RTK-Rac-E-cadherin polarization pathway, however, remains to be determined.
Our data also demonstrate a role for Rap1 in protrusion formation. While many border cells deficient for Rap1 activity initially produced a burst of additional protrusions, eventually these protrusions retracted and did not reform. Notably, both loss and gain of Rap1 activity disrupted protrusion length and shape. Thus, having optimal levels of Rap1 activity is required for the proper morphology and dynamics of protrusions, which in turn is required for normal migration. Chang et al. (2018) further investigated the role for Rap1 in border cell protrusions. Similarly to the findings reported here, Chang et al. (2018) found that Rap1 influenced protrusion formation and number. Moreover, Rap1 promoted the proper distribution of F-actin and myosin within the cluster. In this context, Rap1 inhibits the Hippo/Warts pathway (Chang et al., 2018). Hippo suppresses F-actin enrichment in border cells through inhibition of Enabled (Ena), a regulator of F-actin polymerization (Lucas et al., 2013). Rap1 binds to Hippo and suppresses Hippo activation, relieving inhibition of Ena (Chang et al., 2018), thus potentially accounting for the effects on protrusion dynamics (Gates et al., 2009). The Hippo/Warts-Ena pathway also polarizes F-actin within the cluster (Lucas et al., 2013). However, there are distinct differences in the effects on protrusions, and cluster polarity, caused by loss of Hippo/Warts versus gain of Rap1 activity (Lucas et al., 2013; Chang et al., 2018; this study). Therefore, it is likely that Rap1 functions with additional downstream molecular targets in border cell migration.
Because Rap1 has multiple functions in border cells, an open question is where and when Rap1 is active. Both Rap1 and the major GEF, PDZ-GEF, are uniformly expressed during migration. Rap1 is required in border cells for migration, but its activity needs to be low or off in the central polar cells. These Rap1 functions correlate well with the expression pattern of Rapgap1, a GAP for Rap1 (Chen et al., 1997). Rapgap1 protein is high in polar cells, but is expressed at lower levels in migrating border cells. However, Rapgap1 cannot simply turn off Rap1 activity in border cells as it likely does in polar cells, because this would be expected to block migration. Instead, Rapgap1 may induce rapid cycling of Rap1 in border cells, leading to dynamic or differential activation of this pathway. In Drosophila gastrulation, spatially different levels of Rapgap1 produces two distinct outcomes: 1) low Rapgap1 results in high Rap1 activation, which tightly links cell–cell junctions to F-actin, thus resulting in shallow invagination of the epithelium, and 2) high Rapgap1 results in rapid cycling of Rap1, which decouples cell–cell junctions from F-actin, thus allowing deeper invagination of the epithelium and further folding of the tissue (Wang et al., 2013). More work, however, is needed to determine whether Rapgap1 influences Rap1 activity in a similar manner in border cells versus polar cells. Further, it is unknown why Rapgap1 levels dramatically increase in border cells as they finish their migration and what consequence this has, if any, for border cells once they arrive at the oocyte. In the future, development of a more direct readout of Rap1 activity in vivo will help to clarify the spatial and temporal functions of Rap1. Likewise, identification of specific downstream effectors of Rap1 in border cells will be needed to further reveal the precise mechanisms by which Rap1 controls cluster organization, cell–cell contacts, and polarized protrusion extension. Given the molecular and cellular similarities found in diverse cells that migrate as collectives (Friedl and Gilmour, 2009; Mayor and Etienne-Manneville, 2016; Scarpa and Mayor, 2016), along with high conservation of the Rap1 protein (Frische and Zwartkruis, 2010), our study in border cells suggests that Rap1 may be a conserved regulator of collective cell movements in vivo.
MATERIALS AND METHODS
Drosophila genetics and strains
Crosses were generally performed at 25°C. Crosses with temperature-sensitive GAL80 (“tsGAL80”) were placed at 18°C to suppress GAL4/UAS during earlier developmental stages (McGuire et al., 2003, 2004). For slbo-GAL4 or c306-GAL4 crosses, flies were incubated at 29°C for ≥14 h prior to dissection to produce optimal GAL4/UAS transgene expression and to inactivate tsGAL80. For upd-GAL4, flies were incubated at 29°C for 3 d prior to dissection (Lin et al., 2014). c306-GAL4 is expressed early in border cells, polar cells, and anterior follicle cells (Manseau et al., 1997; Silver and Montell, 2001) and was used to drive UAS-RNAi and other UAS constructs earlier in oogenesis before border cells are specified. c306-GAL4 is generally more efficient at driving RNAi-dependent knockdown, likely because of earlier expression than other drivers (Aranjuez et al., 2012). slbo-GAL4 drives later, high expression in border cells, but not polar cells, after border cell cluster formation; it is also expressed in a few anterior and posterior follicle cells (Rørth et al., 1998). upd-GAL4 drives expression solely in polar cells throughout oogenesis (Bai and Montell, 2002; Pinheiro and Montell, 2004). GAL4 lines were generally outcrossed to w1118 to serve as controls.
Mosaic mutant clones of Rap1CD3 FRT 2A, and FRT 2A (control), were generated using the FLP-FRT system (Xu and Rubin, 1993). To produce somatic clones in border cells, flies were crossed to hs-FLP; FRT 2A His2Av-mRFP. Adult progeny of the correct genotype was heat shocked for 1 h at 37°C, two to three times a day for 3 d, followed by 6 d at 25°C prior to dissection. Mosaic mutant clones were marked by loss of the His2Av-mRFP (nuclear RFP) signal. For production of germline clones, Rap1CD3 FRT 2A, and FRT 2A (control), flies were mated to hs-FLP; FRT 2A, His2Av-mRFP flies and allowed to lay eggs for 2 d. The progeny was then heat shocked on days 3 and 4 (approximately second and third instar larval stages), followed by incubation at 25°C. Adult flies were dissected 5–7 d after eclosion. Dissected ovaries were analyzed for loss of nuclear RFP in the germline of individual egg chambers, indicating that clones had been made and nurse cells were mutant for Rap1.
The following Drosophila strains in this study were obtained from the Bloomington Drosophila Stock Center [BDSC], unless otherwise indicated: tub-GAL80ts (“tsGAL80”), hsp70-GAL4 (“hs-GAL4”), c306-GAL4, c306-GAL4 tsGAL80 (Aranjuez et al., 2016), slbo-GAL4, slbo-GAL4 UAS-mCD8::GFP (from D. Montell, University of California, Santa Barbara, Santa Barbara, CA), upd-GAL4; tsGal80 (from D. Montell), UAS-GFP RNAi (dsRNA GFP; line 9331), UAS-mCherry RNAi (dsRNA mCherry; line 35785), UAS-PDZ-GEF RNAi (line 27017; Vienna Drosophila Resource Center [VDRC]), UAS-PDZ-GEF RNAi (line 27015; VDRC), UAS-PDZ-GEF RNAi (line TRiP.HM05139), UAS-Epac RNAi (line 50372; VDRC), UAS-Epac RNAi (line 50373; VDRC), UAS-Epac RNAi (line 110077; VDRC), UAS-C3G RNAi (line 21306; VDRC), UAS-C3G RNAi (line 21307; VDRC), UAS-C3G RNAi (line 105664; VDRC), UAS-Rap1 RNAi (line 33437; VDRC), UAS-Rapgap1 RNAi (line 102659; VDRC), PDZ-GEF1 (PDZ-GEFk13720; P-element enhancer trap insertion line, from Kyoto Stock Center), PDZ-GEF3 (P-element insertion line, from S. Hou, National Cancer Institute, National Institutes of Health, Frederick, MD), PDZ-GEF6 (P-element excision line, from S. Hou), FRT 2A, FRT 2A Rap1CD3 (deletes the entire Rap1 gene; from J. Curtiss, New Mexico State University, Las Cruces, NM), UAS-PDZ-GEF on third (from B. Boettner, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY), UAS-Rap1N17 (DN-Rap1 mutation; from B. Boettner), UAS-Rap1WT (wild-type Rap1; from B. Boettner), UAS-Rap1V12 (constitutively active- [CA-] Rap1; from B. Boettner), UASp-Rapgap1 (from Y.-C. Wang, RIKEN Center for Developmental Biology, Kobe, Japan), UAS-Eya.II (Bai and Montell, 2002), UAS-PLC∆PH-GFP (“membrane GFP”; Verstreken et al., 2009), slbo-LifeAct-GFP on second (from D. Montell), PDZ-GEF-GFP-PDZ-GEF reporter (“GFP-PDZ-GEF”; from R. Reuter, University of Tübingen, Tübingen, Germany; Boettner and Van Aelst, 2007), and Rap1-GFP-Rap1 reporter (“GFP-Rap1”; from D. Siekhaus, Institute of Science and Technology Austria, Klosterneuburg, Austria; Knox and Brown, 2002). Detailed information on Drosophila strains can be found at FlyBase (http://flybase.org/). The PDZ-GEF RNAi line TRiP.HM05139 targets independent PDZ-GEF sequences from PDZ-GEF RNAi lines 27015 and 27017 (construct GD14231). UAS-Rap1N17 and UAS-Rap1V12 mutations within the respective transgenic fly lines were PCR amplified and sequence verified using a UAS primer and a Rap1 gene-specific primer.
Immunostaining
Ovaries from 3- to 5-d-old females were dissected in Schneider’s media (Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (FBS) and either kept whole or further dissected into individual ovarioles as described (McDonald and Montell, 2005). This was followed by fixation for 10 min with 4% methanol-free formaldehyde (Polysciences) in 0.1 M potassium phosphate buffer, pH 7.4, and washes with NP40 block (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.5% NP40, 5 mg/ml bovine serum albumin). All primary and secondary antibody incubations were performed in NP40 block. The following primary antibodies from the Developmental Studies Hybridoma Bank (DSHB) were used at the indicated concentrations: rat anti-E-cadherin 1:10 (DCAD2), mouse anti-GFP 1:10 (12A6), mouse anti-Fasciclin 3 1:10 (FasIII; 7G10), and mouse anti-Fascin 1:25 (Sn7C). Additional antibodies used were rat anti-Rapgap1 1:1000 (a gift of Y.-C. Wang and E. Wieschaus, Princeton University, Princeton, NJ) (Wang et al., 2013); rabbit anti-phosphorylated c-Jun (p-Jun) 1:200 (KM-1; Santa Cruz); rabbit anti-β-galactosidase 1:1000 (Cappel, MP Biomedicals); rabbit anti-GFP Tag polyclonal 1:1000–1:2000 (A-11122; Thermo Fisher Scientific). Alexa Fluor 488, 568, or 647 secondary antibodies (Thermo Fisher Scientific) were used at 1:400 dilution. Alexa Fluor 568 phalloidin and Alexa Fluor 488 phalloidin were used at 1:400 dilution. 4’,6-Diamidino-2-phenylindole (DAPI) was used at 0.05 µg/ml. To amplify GFP signal in some experiments, GFP booster (ChromoTek) was used according to manufacturer’s protocol. Dissected egg chambers were mounted on slides in Aqua-Poly/Mount (Polysciences) or FluorSave Reagent (Millipore Sigma) prior to imaging.
Live time-lapse imaging
For live imaging of border cells inhibited for Rap1, c306-GAL4 tsGAL80 (control) and c306-GAL4 tsGAL80; UAS-Rap1N17 stocks were each crossed to slbo-LifeAct-GFP. To obtain optimal GAL4/UAS expression, flies of the correct genotypes were incubated at 29°C for ≥14 h prior to dissection. To image the overall effects of activated Rap1 on live border cell migration, w1118 (control), UAS-Rap1V12 (CA-Rap1), and UAS-PDZ-GEF were each crossed to slbo-Gal4 UAS-mCD8:GFP. To image protrusion dynamics, slbo-GAL4; UAS-Rap1V12 and slbo-GAL4 (control) were each crossed to slbo-LifeAct-GFP. Flies were incubated at 28°–29°C for ≥14 h prior to dissection. Live imaging was performed essentially as described (Prasad and Montell, 2007; Prasad et al., 2007; Manning and Starz-Gaiano, 2015; Dai and Montell, 2016). Briefly, ovarioles were dissected in live imaging media (Schneider’s media, pH 6.95, supplemented with 15–20% FBS and 0.2 µg/ml bovine insulin) and mounted on a lumox Dish 50 (Sarstedt, cat. no. 94.6077.410), a gas-permeable culture dish. Fresh live imaging media was added to the sample just prior to imaging. For imaging cell membranes (Figure 1A), the lipophilic dye FM 4-64 (Thermo Fisher Scientific) was added at 9 µM concentration to dissected egg chambers in live imaging media as described (Bianco et al., 2007; Prasad et al., 2007). In all cases, time-lapse images were acquired at intervals of 2–3 min for up to 4 h using a 20× Plan-Apochromat 0.75 NA objective, a Zeiss Colibri LED light source and either a Zeiss MRm or Axiocam 503 mono camera. Light intensity of the LED was adjusted to minimize phototoxicity of the sample. In some cases, multiple z-stacks were acquired and merged in Zeiss AxioVision, Zeiss ZEN, or FIJI to produce a single in-focus time-lapse video of border cell migration.
Microscopy and analyses
Images of fixed egg chambers were mainly acquired with an upright Zeiss AxioImager Z1 microscope using Apotome.2, an Axiocam 503 mono camera, and either a 20× Plan-Apochromat 0.75 numerical aperture (NA) or a 40× Plan-Neofluar 1.3 NA oil-immersion objective. Some images were acquired on a Zeiss LSM 880 confocal microscope using the 405-, 488-, 561-, and/or 633-nm laser lines and either a 20 × 0.8 NA or 40 × 1.3 NA oil-immersion objective (KSU-CVM Confocal Core). Image brightness/contrast and measurements were performed using Zeiss AxioVision 4.8, Zeiss ZEN or ImageJ (FIJI) (Schneider et al., 2012). FIJI was used to merge z-stacks; maximum intensity projections are shown in the figures but only single sections were used to measure fluorescence intensity.
Analyses of live border cell migration time-lapse videos was performed using Zeiss ZEN software. The migration speed was calculated from the duration of its movement. For protrusion quantification, a circle was drawn around the cell cluster, and extensions greater than 4 µm were defined as protrusions (see Supplemental Figure 4A). Protrusions were classified into front (0° to 45° and 0° to 315°), side (45° to 135°and 225° to 315°), and back (135° to 225°) based on position within the cluster. The first 1 h of each video was used for quantification.
Measurements of E-cadherin intensity at cell–cell junctions were performed on egg chambers stained using identical conditions for E-cadherin (Alexa-568), FasIII (Alexa-647), and DAPI. Identical confocal laser settings were used for each channel, based on a control border cell cluster, and imaged with a 40 × 1.3 NA objective. Acquired images were then processed in FIJI. First, the center of each z-stack was manually identified using the polar cells as a landmark. Two z-stacks above and below this central section (five total) were then used to create a sum intensity projection. Cell–cell contacts (BC-BC, PC-PC, BC-NC, and NC-NC; see Figure 5D) were manually identified, a line drawn, and mean fluorescence intensity across the line was obtained using the “measure” tool. The PC-PC contact was defined by the FasIII staining, which is highest between polar cells. A ratio of BC-BC intensity versus PC-PC intensity was calculated to normalize “within-cluster” E-cadherin levels; a ratio of BC-NC intensity versus NC-NC (always a NC-NC contact in front of the cluster) was calculated to normalize the E-cadherin levels at the outer edge of the cluster.
To measure Rapgap1 colocalization with membrane GFP, at least one line was drawn using FIJI across each border cell cluster at clearly visible cell and polar cell membranes. The “plot profile” function was used to obtain graph curves for Rapgap1 pixel intensity and membrane-GFP pixel intensity across the line. Only a single z-section was measured for each border cell cluster (n = 7 border cell clusters). The values for each channel were normalized to the highest pixel value, and a scatter plot showing both channels was generated in Microsoft Excel (Supplemental Figure 3G). In most cases, the curves for Rapgap1 and membrane-GFP overlapped across all measured cell membranes within the border cell cluster.
RT-PCR
To measure RNAi efficiency of PDZ-GEF in flies, the three UAS-PDZ-GEF RNAi lines (v27017, v27015, and TRiP.HM05139) were each crossed to hs-GAL4. Adult progeny from this cross were heat shocked to achieve ubiquitous knockdown, followed by RNA extraction and cDNA synthesis, as described (Aranjuez et al., 2012). Briefly, RNA from whole flies was extracted using Trizol (Thermo Fisher Scientific) and used for cDNA synthesis. Reverse transcription-PCR (RT-PCR) was performed with the Superscript III One-Step RT-PCR system (Thermo Fisher Scientific). PCR amplification was as follows: 50°C for 30 min during the cDNA synthesis step, 55°C for 30 s during the annealing step, and 72°C for 1 min during the extension step. PCR was performed for 29 cycles to avoid the plateau phase of amplification. Band intensities of the RT-PCR products were measured using ImageJ (Schneider et al., 2012) and compared with GAPDH, which was used as the reference gene. The gene-specific primers were as follows: PDZ-GEF fwd, AGGAACGCGTCTCACTCAAG; PDZ-GEF rev, AGGAACGCGTCTCACTCAAG; GAPDH fwd, ACTCATCAACCCTCCCCCG; GAPDH rev, GCGGACGGTAAGATCCACAA.
Production of dsRNA for RNAi treatment of Drosophila S2 cells and Rap1 activity pull down
To make double-stranded RNA (dsRNA) for RNAi treatment of S2 cells, PCR primers were designed with the T7 RNA polymerase sequence at the 5′ end (underlined, below) as described (Rogers and Rogers 2008). Templates for dsRNA were amplified by PCR using genomic DNA from w1118 flies. RNA was produced using the T7 MEGAscript kit (Thermo Fisher Scientific), followed by annealing and purification as described (Rogers and Rogers, 2008). PDZ-GEF primers were designed to match the sequences of the in vivo RNAi lines, v27017 (GD 14231) and TRiP.HM05139. The following primers were used: gal80 fwd, TAATACGACTCACTATAGGGAGAATTAAGCGGCCGCAACATGGAC; gal80 rev TAATACGACTCACTATAGGGAGAGCGTGTCTAGATTATAAACTAT; PDZ-GEF v27107 fwd, TAATACGACTCACTATAGGCGCGAATTCGTCGCCATCCAACGCTCTCTTCTC; PDZ-GEF v27107 rev, TAATACGACTCACTATAGGGCGTCTAGAGCTGCCTCCACCACCGCTTC; PDZ-GEF TRiP.HM05139 fwd, TAATACGACTCACTATAGGAAGAATTCACCGCGGATAACTACGTGAC; PDZ-GEF TRiP.HM05139 rev, TAATACGACTCACTATAGGAAGAATTCACCGCGGATAACTACGTGAC
Drosophila S2 cells were treated with dsRNA designed to the two independent PDZ-GEF RNAi sequences (see above). Cells were then treated with dsRNA for 4 d as described (Rogers and Rogers, 2008). Cells were harvested and lysates prepared according to manufacturer’s instructions using the Active Rap1 Pull-Down and Detection Kit (Thermo Fisher Scientific, cat. no. 16120). Halt protease inhibitor cocktail (Thermo Fisher Scientific) was added to the lysis buffer. Lysates were incubated with GST-RalGDS-RBD purified protein and glutathione resin for 1 h followed by affinity precipitation. Western blot analysis was used to determine the amount of Rap1 present in the pull down, bound to GST-RalGDS-RBD according to manufacturer’s instructions. Briefly, samples were prepared for Western blot analysis by addition of 2X SDS sample buffer, heated for 5 min at 100°C, separated by SDS–PAGE on a 12% polyacrylamide gel, transferred to nitrocellulose membrane, and followed by incubation with anti-Rap1 antibody at 1:1000 dilution. A single band corresponding to ∼24 kDa was recognized by anti-human Rap1A antibody, consistent with the predicted size of Drosophila Rap1 (22 kDa). Drosophila Rap1 and human Rap1A are 90% identical and 96% similar. Rap1 band intensities were measured using ImageJ.
Figures, graphs, and statistics
Figures were assembled in Adobe Photoshop CS4 and CC 2018. Illustrations were created in Adobe Illustrator CC 2018. Videos were assembled in Zeiss AxioVision 4.8, Zeiss ZEN, or FIJI (Schindelin et al., 2012, 2015). Graphs and statistical tests were performed with GraphPad Prism 7. Statistical tests and p values are listed in the figure legends.
Supplementary Material
ACKNOWLEDGMENTS
We thank B. Boettner, L. Van Aelst, R. Reuter, S. Hou, Y.-C. Wang, E. F. Wieschaus, J. Curtiss, R. Lehmann, D. Siekhaus, D. Montell, the Transgenic RNAi Project (TRiP; Harvard Medical School [funded by National Institutes of Health/National Institute of General Medical Sciences R01 GM084947]), the Kyoto Stock Center, the Bloomington Drosophila Stock Center (BDSC), the Vienna Drosophila Resource Center (VDRC), and the Developmental Studies Hybridoma Bank (University of Iowa) for fly stocks and antibodies. We also thank C. Luke Messer, A. Burtscher, M. Lawas, and O. Athukorala Arachchige for technical contributions to this study and J. Campanale for advice on imaging analyses. Special thanks to A. C.-C. Jang for discussion and sharing results prior to publication. We thank the Confocal Core, funded by the Kansas State University College of Veterinary Medicine, for use of the Zeiss LSM 880 confocal microscope. This work was supported by the National Institutes of Health (R21 CA198254 to J.A.M., Kansas INBRE P20GM103418 Postdoctoral Award to Y.C.), the National Science Foundation (1456053 to J.A.M.), and institutional funds from Kansas State University (to J.A.M.). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations used:
- AJ
adherens junction
- BC
border cell
- CA
constitutively active
- DN
dominant-negative
- E-cad
E-cadherin
- ECM
extracellular matrix
- FC
follicle cell
- GAP
GTPase-activating protein
- GEF
guanine nucleotide exchange factor
- GFP
green fluorescent protein
- NC
nurse cell
- PC
polar cell
- RFP
red fluorescent protein
- RNAi
RNA interference
- RTK
receptor tyrosine kinase
- UAS
upstream activating sequence.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-12-0752) on August 29, 2018.
REFERENCES
- Anllo L, Schupbach T. (2016). Signaling through the G-protein-coupled receptor Rickets is important for polarity, detachment, and migration of the border cells in Drosophila. Dev Biol , 193–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranjuez G, Burtscher A, Sawant K, Majumder P, McDonald JA. (2016). Dynamic myosin activation promotes collective morphology and migration by locally balancing oppositional forces from surrounding tissue. Mol Biol Cell , 1898–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranjuez G, Kudlaty E, Longworth MS, McDonald JA. (2012). On the role of PDZ domain-encoding genes in Drosophila border cell migration. G3 (Bethesda) , 1379–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asha H, de Ruiter ND, Wang MG, Hariharan IK. (1999). The Rap1 GTPase functions as a regulator of morphogenesis in vivo. EMBO J , 605–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai J, Montell D. (2002). Eyes absent, a key repressor of polar cell fate during Drosophila oogenesis. Development , 5377–5388. [DOI] [PubMed] [Google Scholar]
- Bai J, Uehara Y, Montell DJ. (2000). Regulation of invasive cell behavior by Taiman, a Drosophila protein related to AIB1, a steroid receptor coactivator amplified in breast cancer. Cell , 1047–1058. [DOI] [PubMed] [Google Scholar]
- Baum B, Georgiou M. (2011). Dynamics of adherens junctions in epithelial establishment, maintenance, and remodeling. J Cell Biol , 907–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazellières E, Conte V, Elosegui-Artola A, Serra-Picamal X, Bintanel-Morcillo M, Roca-Cusachs P, Muñoz JJ, Sales-Pardo M, Guimerà R, Trepat X. (2015). Control of cell–cell forces and collective cell dynamics by the intercellular adhesome. Nat Cell Biol , 409–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beccari S, Teixeira L, Rørth P. (2002). The JAK/STAT pathway is required for border cell migration during Drosophila oogenesis. Mech Dev , 115–123. [DOI] [PubMed] [Google Scholar]
- Bianco A, Poukkula M, Cliffe A, Mathieu J, Luque CM, Fulga TA, Rørth P. (2007). Two distinct modes of guidance signalling during collective migration of border cells. Nature , 362–365. [DOI] [PubMed] [Google Scholar]
- Bivona TG, Wiener HH, Ahearn IM, Silletti J, Chiu VK, Philips MR. (2004). Rap1 up-regulation and activation on plasma membrane regulates T cell adhesion. J Cell Biol , 461–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettner B, Harjes P, Ishimaru S, Heke M, Fan HQ, Qin Y, Van Aelst L, Gaul U. (2003). The AF-6 homolog canoe acts as a Rap1 effector during dorsal closure of the Drosophila embryo. Genetics , 159–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettner B, Van Aelst L. (2007). The Rap GTPase activator Drosophila PDZ-GEF regulates cell shape in epithelial migration and morphogenesis. Mol Cell Biol , 7966–7980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettner B, Van Aelst L. (2009). Control of cell adhesion dynamics by Rap1 signaling. Curr Opin Cell Biol , 684–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonello TT, Perez-Vale KZ, Sumigray KD, Peifer M. (2018). Rap1 acts via multiple mechanisms to position Canoe and adherens junctions and mediate apical-basal polarity establishment. Development , dev157941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos JL. (2005). Linking Rap to cell adhesion. Curr Opin Cell Biol , 123–128. [DOI] [PubMed] [Google Scholar]
- Bos JL, Rehmann H, Wittinghofer A. (2007). GEFs and GAPs: critical elements in the control of small G proteins. Cell , 865–877. [DOI] [PubMed] [Google Scholar]
- Cai D, Chen S-C, Prasad M, He L, Wang X, Choesmel-Cadamuro V, Sawyer JK, Danuser G, Montell DJ. (2014). Mechanical feedback through E-cadherin promotes direction sensing during collective cell migration. Cell , 1146–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmena A, Makarova A, Speicher S. (2011). The Rap1-Rgl-Ral signaling network regulates neuroblast cortical polarity and spindle orientation. J Cell Biol , 553–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y-C, Wu J-W, Hsieh Y-C, Huang T-H, Liao Z-M, Huang Y-S, Mondo JA, Montell D, Jang AC-C. (2018). Rap1 negatively regulates the hippo pathway to polarize directional protrusions in collective cell migration. Cell Rep , 2160–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen FF, Barkett MM, Ram KTK, Quintanilla AA, Hariharan IKI. (1997). Biological characterization of Drosophila Rapgap1, a GTPase activating protein for Rap1. Proc Natl Acad Sci USA , 12485–12490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung KJ, Ewald AJ. (2016). A collective route to metastasis: seeding by tumor cell clusters. Science , 167–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung KJ, Gabrielson E, Werb Z, Ewald AJ. (2013). Collective invasion in breast cancer requires a conserved basal epithelial program. Cell , 1639–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi W, Harris NJ, Sumigray KD, Peifer M. (2013). Rap1 and Canoe/afadin are essential for establishment of apical-basal polarity in the Drosophila embryo. Mol Biol Cell , 945–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins C, Nelson WJ. (2015). Running with neighbors: coordinating cell migration and cell–cell adhesion. Curr Opin Cell Biol , 62–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai W, Montell DJ. (2016). Live imaging of border cell migration in Drosophila. Methods Mol Biol , 153–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Pascalis C, Etienne-Manneville S. (2017). Single and collective cell migration: the mechanics of adhesions. Mol Biol Cell , 1833–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rooij J, Boenink NM, van Triest M, Cool RH, Wittinghofer A, Bos JL. (1999). PDZ-GEF1, a guanine nucleotide exchange factor specific for Rap1 and Rap2. J Biol Chem , 38125–38130. [DOI] [PubMed] [Google Scholar]
- Duchek P, Rørth P. (2001). Guidance of cell migration by EGF receptor signaling during Drosophila oogenesis. Science , 131–133. [DOI] [PubMed] [Google Scholar]
- Duchek P, Somogyi K, Jékely G, Beccari S, Rørth P. (2001). Guidance of cell migration by the Drosophila PDGF/VEGF receptor. Cell , 17–26. [DOI] [PubMed] [Google Scholar]
- Dupuy AG, L’Hoste S, Cherfils J, Camonis J, Gaudriault G, de Gunzburg J. (2005). Novel Rap1 dominant-negative mutants interfere selectively with C3G and Epac. Oncogene , 4509–4520. [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S. (2014). Neighborly relations during collective migration. Curr Opin Cell Biol , 51–59. [DOI] [PubMed] [Google Scholar]
- Feig LA. (1999). Tools of the trade: use of dominant-inhibitory mutants of Ras-family GTPases. Nat Cell Biol , E25–E27. [DOI] [PubMed] [Google Scholar]
- Felix M, Chayengia M, Ghosh R, Sharma A, Prasad M. (2015). Pak3 regulates apical-basal polarity in migrating border cells during Drosophila oogenesis. Development , 3692–3703. [DOI] [PubMed] [Google Scholar]
- Friedl P, Gilmour D. (2009). Collective cell migration in morphogenesis, regeneration and cancer. Nat Rev Mol Cell Biol , 445–457. [DOI] [PubMed] [Google Scholar]
- Friedl P, Locker J, Sahai E, Segall JE. (2012). Classifying collective cancer cell invasion. Nat Cell Biol , 777–783. [DOI] [PubMed] [Google Scholar]
- Friedl P, Mayor R. (2017). Tuning collective cell migration by cell-cell junction regulation. Cold Spring Harb Perspect Biol , a029199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl P, Noble PB, Walton PA, Laird DW, Chauvin PJ, Tabah RJ, Black M, Zänker KS. (1995). Migration of coordinated cell clusters in mesenchymal and epithelial cancer explants in vitro. Cancer Res , 4557–4560. [PubMed] [Google Scholar]
- Frische EW, Zwartkruis FJT. (2010). Rap1, a mercenary among the Ras-like GTPases. Dev Biol , 1–9. [DOI] [PubMed] [Google Scholar]
- Fulga TA, Rørth P. (2002). Invasive cell migration is initiated by guided growth of long cellular extensions. Nat Cell Biol , 715–719. [DOI] [PubMed] [Google Scholar]
- Gates J, Nowotarski SH, Yin H, Mahaffey JP, Bridges T, Herrera C, Homem CCF, Janody F, Montell DJ, Peifer M. (2009). Enabled and Capping protein play important roles in shaping cell behavior during Drosophila oogenesis. Dev Biol , 90–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiglione C, Devergne O, Georgenthum E, Carballès F, Médioni C, Cérézo D, Noselli S. (2002). The Drosophila cytokine receptor Domeless controls border cell migration and epithelial polarization during oogenesis. Development , 5437–5447. [DOI] [PubMed] [Google Scholar]
- Gloerich M, Bos JL. (2011). Regulating Rap small G-proteins in time and space. Trends Cell Biol , 615–623. [DOI] [PubMed] [Google Scholar]
- Haigo SL, Hildebrand JD, Harland RM, Wallingford JB. (2003). Shroom induces apical constriction and is required for hingepoint formation during neural tube closure. Curr Biol , 2125–2137. [DOI] [PubMed] [Google Scholar]
- Huelsmann S, Hepper C, Marchese D, Knöll C, Reuter R. (2006). The PDZ-GEF dizzy regulates cell shape of migrating macrophages via Rap1 and integrins in the Drosophila embryo. Development , 2915–2924. [DOI] [PubMed] [Google Scholar]
- Jambor H, Surendranath V, Kalinka AT, Mejstrik P, Saalfeld S, Tomancak P. (2015). Systematic imaging reveals features and changing localization of mRNAs in Drosophila development. Elife , R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang AC-C, Chang Y-C, Bai J, Montell D. (2009). Border-cell migration requires integration of spatial and temporal signals by the BTB protein Abrupt. Nat Cell Biol , 569–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jossin Y, Cooper JA. (2011). Reelin, Rap1 and N-cadherin orient the migration of multipolar neurons in the developing neocortex. Nat Neurosci , 697–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AA, Ilina O, Gritsenko PG, Bult P, Span PN, Friedl P. (2017). Collective invasion in ductal and lobular breast cancer associates with distant metastasis. Clin Exp Metastasis , 421–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox AL, Brown NH. (2002). Rap1 GTPase regulation of adherens junction positioning and cell adhesion. Science , 1285–1288. [DOI] [PubMed] [Google Scholar]
- Kooistra MRH, Dubé N, Bos JL. (2007). Rap1: a key regulator in cell-cell junction formation. J Cell Sci , 17–22. [DOI] [PubMed] [Google Scholar]
- Lafuente EM, van Puijenbroek AAFL, Krause M, Carman CV, Freeman GJ, Berezovskaya A, Constantine E, Springer TA, Gertler FB, Boussiotis VA. (2004). RIAM, an Ena/VASP and profilin ligand, interacts with Rap1-GTP and mediates Rap1-induced adhesion. Dev Cell , 585–595. [DOI] [PubMed] [Google Scholar]
- Lecuit T, Yap AS. (2015). E-cadherin junctions as active mechanical integrators in tissue dynamics. Nat Cell Biol , 533–539. [DOI] [PubMed] [Google Scholar]
- Lee JH, Cho KS, Lee J, Kim D, Lee SB, Yoo J, Cha GH, Chung J. (2002). Drosophila PDZ-GEF, a guanine nucleotide exchange factor for Rap1 GTPase, reveals a novel upstream regulatory mechanism in the mitogen-activated protein kinase signaling pathway. Mol Cell Biol , 7658–7666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M-R, Jeon TJ. (2012). Cell migration: regulation of cytoskeleton by Rap1 in Dictyostelium discoideum. J Microbiol , 555–561. [DOI] [PubMed] [Google Scholar]
- Liao Y, Kariya K-I, Hu C-D, Shibatohge M, Goshima M, Okada T, Watari Y, Gao X, Jin T-G, Yamawaki-Kataoka Y, et al. (1999). RA-GEF, a novel Rap1A guanine nucleotide exchange factor containing a Ras/Rap1A-associating domain, is conserved between nematode and humans. J Biol Chem , 37815–37820. [DOI] [PubMed] [Google Scholar]
- Lin T-H, Yeh T-H, Wang T-W, Yu J-Y. (2014). The hippo pathway controls border cell migration through distinct mechanisms in outer border cells and polar cells of the Drosophila ovary. Genetics , 1087–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llense F, Etienne-Manneville S. (2015). Front-to-rear polarity in migrating cells. In: Cell Polarity 1, ed. Ebnet K, Cham, Switzerland: Springer. [Google Scholar]
- Llense F, Martín-Blanco E. (2008). JNK signaling controls border cell cluster integrity and collective cell migration. Curr Biol , 538–544. [DOI] [PubMed] [Google Scholar]
- Lucas EP, Khanal I, Gaspar P, Fletcher GC, Polesello C, Tapon N, Thompson BJ. (2013). The Hippo pathway polarizes the actin cytoskeleton during collective migration of Drosophila border cells. J Cell Biol , 875–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magliozzi R, Low TY, Weijts BGMW, Cheng T, Spanjaard E, Mohammed S, van Veen A, Ovaa H, de Rooij J, Zwartkruis FJT, et al. (2013). Control of epithelial cell migration and invasion by the IKKβ- and CK1α-mediated degradation of RAPGEF2. Dev Cell , 574–585. [DOI] [PubMed] [Google Scholar]
- Mandai K, Rikitake Y, Shimono Y, Takai Y. (2013). Afadin/AF-6 and canoe: roles in cell adhesion and beyond. Prog Mol Biol Transl Sci , 433–454. [DOI] [PubMed] [Google Scholar]
- Manning L, Starz-Gaiano M. (2015). Culturing Drosophila egg chambers and investigating developmental processes through live imaging. Methods Mol Biol , 73–88. [DOI] [PubMed] [Google Scholar]
- Manseau L, Baradaran A, Brower D, Budhu A, Elefant F, Phan H, Philp AV, Yang M, Glover D, Kaiser K, et al. (1997). GAL4 enhancer traps expressed in the embryo, larval brain, imaginal discs, and ovary of Drosophila. Dev Dyn , 310–322. [DOI] [PubMed] [Google Scholar]
- Mayor R, Etienne-Manneville S. (2016). The front and rear of collective cell migration. Nat Rev Mol Cell Biol , 97–109. [DOI] [PubMed] [Google Scholar]
- McDonald JA, Khodyakova A, Aranjuez G, Dudley C, Montell DJ. (2008). PAR-1 kinase regulates epithelial detachment and directional protrusion of migrating border cells. Curr Biol , 1659–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JA, Montell DJ. (2005). Analysis of cell migration using Drosophila as a model system. Methods Mol Biol , 175–202. [DOI] [PubMed] [Google Scholar]
- McDonald JA, Pinheiro EM, Kadlec L, Schupbach T, Montell DJ. (2006). Multiple EGFR ligands participate in guiding migrating border cells. Dev Biol , 94–103. [DOI] [PubMed] [Google Scholar]
- McDonald JA, Pinheiro EM, Montell DJ. (2003). PVF1, a PDGF/VEGF homolog, is sufficient to guide border cells and interacts genetically with Taiman. Development , 3469–3478. [DOI] [PubMed] [Google Scholar]
- McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL. (2003). Spatiotemporal rescue of memory dysfunction in Drosophila. Science , 1765–1768. [DOI] [PubMed] [Google Scholar]
- McGuire SE, Mao Z, Davis RL. (2004). Spatiotemporal gene expression targeting with the TARGET and gene-switch systems in Drosophila. Sci Signal , l6. [DOI] [PubMed] [Google Scholar]
- Melani M, Simpson KJ, Brugge JS, Montell D. (2008). Regulation of cell adhesion and collective cell migration by hindsight and its human homolog RREB1. Curr Biol , 532–537. [DOI] [PubMed] [Google Scholar]
- Montell DJ, Rørth P, Spradling AC. (1992). slow border cells, a locus required for a developmentally regulated cell migration during oogenesis, encodes Drosophila CEBP. Cell , 51–62. [DOI] [PubMed] [Google Scholar]
- Montell DJ, Yoon WH, Starz-Gaiano M. (2012). Group choreography: mechanisms orchestrating the collective movement of border cells. Nat Rev Mol Cell Biol , 631–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewiadomska P, Godt D, Tepass U. (1999). DE-Cadherin is required for intercellular motility during Drosophila oogenesis. J Cell Biol , 533–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Keefe DD, Gonzalez-Niño E, Burnett M, Dylla L, Lambeth SM, Licon E, Amesoli C, Edgar BA, Curtiss J. (2009). Rap1 maintains adhesion between cells to affect Egfr signaling and planar cell polarity in Drosophila. Dev Biol , 143–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannekoek W-J., Kooistra MRH, Zwartkruis FJT, Bos JL. (2009). Cell-cell junction formation: the role of Rap1 and Rap1 guanine nucleotide exchange factors. Biochim Biophys Acta , 790–796. [DOI] [PubMed] [Google Scholar]
- Pinheiro EM, Montell DJ. (2004). Requirement for Par-6 and Bazooka in Drosophila border cell migration. Development , 5243–5251. [DOI] [PubMed] [Google Scholar]
- Post A, Pannekoek W-J, Ross SH, Verlaan I, Brouwer PM, Bos JL. (2013). Rasip1 mediates Rap1 regulation of Rho in endothelial barrier function through ArhGAP29. Proc Natl Acad Sci USA , 11427–11432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poukkula M, Cliffe A, Changede R, Rørth P. (2011). Cell behaviors regulated by guidance cues in collective migration of border cells. J Cell Biol , 513–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad M, Jang AC-C, Starz-Gaiano M, Melani M, Montell DJ. (2007). A protocol for culturing Drosophila melanogaster stage 9 egg chambers for live imaging. Nat Protoc , 2467–2473. [DOI] [PubMed] [Google Scholar]
- Prasad M, Montell DJ. (2007). Cellular and molecular mechanisms of border cell migration analyzed using time-lapse live-cell imaging. Dev Cell , 997–1005. [DOI] [PubMed] [Google Scholar]
- Raaijmakers JH, Bos JL. (2009). Specificity in Ras and Rap signaling. J Biol Chem , 10995–10999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramel D, Wang X, Laflamme C, Montell DJ, Emery G. (2013). Rab11 regulates cell-cell communication during collective cell movements. Nat Cell Biol , 317–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley AJ. (2011). Life at the leading edge. Cell , 1012–1022. [DOI] [PubMed] [Google Scholar]
- Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. (2003). Cell migration: integrating signals from front to back. Science , 1704–1709. [DOI] [PubMed] [Google Scholar]
- Rogers SL, Rogers GC. (2008). Culture of Drosophila S2 cells and their use for RNAi-mediated loss-of-function studies and immunofluorescence microscopy. Nat Protoc , 606–611. [DOI] [PubMed] [Google Scholar]
- Rørth P, Szabo K, Bailey A, Laverty T, Rehm J, Rubin GM, Weigmann K, Milán M, Benes V, Ansorge W, et al. (1998). Systematic gain-of-function genetics in Drosophila. Development , 1049–1057. [DOI] [PubMed] [Google Scholar]
- Ruohola H, Bremer KA, Baker D, Swedlow JR, Jan LY, Jan YN. (1991). Role of neurogenic genes in establishment of follicle cell fate and oocyte polarity during oogenesis in Drosophila. Cell , 433–449. [DOI] [PubMed] [Google Scholar]
- Saadin A, Starz-Gaiano M. (2016). Circuitous genetic regulation governs a straightforward cell migration. Trends Genet , 660–673. [DOI] [PubMed] [Google Scholar]
- Sawyer JK, Harris NJ, Slep KC, Gaul U, Peifer M. (2009). The Drosophila afadin homologue Canoe regulates linkage of the actin cytoskeleton to adherens junctions during apical constriction. J Cell Biol , 57–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpa E, Mayor R. (2016). Collective cell migration in development. J Cell Biol , 143–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods , 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Rueden CT, Hiner MC, Eliceiri KW. (2015). The ImageJ ecosystem: an open platform for biomedical image analysis. Mol Reprod Dev , 518–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat Methods , 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schober M, Rebay I, Perrimon N. (2005). Function of the ETS transcription factor Yan in border cell migration. Development , 3493–3504. [DOI] [PubMed] [Google Scholar]
- Shah B, Lutter D, Bochenek ML, Kato K, Tsytsyura Y, Glyvuk N, Sakakibara A, Klingauf J, Adams RH, Püschel AW. (2016). C3G/Rapgef1 is required in multipolar neurons for the transition to a bipolar morphology during cortical development. PLoS One , e0154174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah B, Lutter D, Tsytsyura Y, Glyvuk N, Sakakibara A, Klingauf J, Püschel AW. (2017). Rap1 GTPases are master regulators of neural cell polarity in the developing neocortex. Cereb Cortex , 1253–1269. [DOI] [PubMed] [Google Scholar]
- Shah B, Püschel AW. (2016). Regulation of Rap GTPases in mammalian neurons. Biol Chem , 1055–1069. [DOI] [PubMed] [Google Scholar]
- Shirinian M, Grabbe C, Popovic M, Varshney G, Hugosson F, Bos H, Rehmann H, Palmer RH. (2010). The Rap1 guanine nucleotide exchange factor C3G is required for preservation of larval muscle Integrity in Drosophila melanogaster. PLoS One , e9403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver DL, Geisbrecht ER, Montell DJ. (2005). Requirement for JAK/STAT signaling throughout border cell migration in Drosophila. Development , 3483–3492. [DOI] [PubMed] [Google Scholar]
- Silver DL, Montell DJ. (2001). Paracrine signaling through the JAK/STAT pathway activates invasive behavior of ovarian epithelial cells in Drosophila. Cell , 831–841. [DOI] [PubMed] [Google Scholar]
- Singh SR, Oh S-W, Liu W, Chen X, Zheng Z, Hou SX. (2006). Rap-GEF/Rap signaling restricts the formation of supernumerary spermathecae in Drosophila melanogaster. Dev Growth Differ , 169–175. [DOI] [PubMed] [Google Scholar]
- Spahn P, Ott A, Reuter R. (2012). The PDZ-GEF protein Dizzy regulates the establishment of adherens junctions required for ventral furrow formation in Drosophila. J Cell Sci , 3801–3812. [DOI] [PubMed] [Google Scholar]
- Spradling AC. (1993). Developmental genetics of oogenesis. In: The Development of Drosophila Melanogaster, ed. Bate M, Martinez-Arias A, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 1–70. [Google Scholar]
- Su L, Hattori M, Moriyama M, Murata N, Harazaki M, Kaibuchi K, Minato N. (2003). AF-6 controls integrin-mediated cell adhesion by regulating Rap1 activation through the specific recruitment of Rap1GTP and SPA-1. J Biol Chem , 15232–15238. [DOI] [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RYJ, Nieto MA. (2009). Epithelial-mesenchymal transitions in development and disease. Cell , 871–890. [DOI] [PubMed] [Google Scholar]
- Tsai I-C, Amack JD, Gao Z-H, Band V, Yost HJ, Virshup DM. (2007). A Wnt-CKIvarepsilon-Rap1 pathway regulates gastrulation by modulating SIPA1L1, a Rap GTPase activating protein. Dev Cell , 335–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstreken P, Ohyama T, Haueter C, Habets RLP, Lin YQ, Swan LE, Ly CV, Venken KJT, De Camilli P, Bellen HJ. (2009). Tweek, an evolutionarily conserved protein, is required for synaptic vesicle recycling. Neuron , 203–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther RF, Burki M, Pinal N, Rogerson C, Pichaud F. (2018). Rap1, Canoe and Mbt cooperate with Bazooka to promote zonula adherens assembly in the fly photoreceptor. J Cell Sci , jcs207779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Qiu Z, Xu Z, Chen SJ, Luo J, Wang X, Chen J. (2018). aPKC is a key polarity determinant in coordinating the function of three distinct cell polarities during collective migration. Development , dev158444. [DOI] [PubMed] [Google Scholar]
- Wang H, Singh SR, Zheng Z, Oh S-W., Chen X, Edwards K, Hou SX. (2006). Rap-GEF signaling controls stem cell anchoring to their niche through regulating DE-cadherin-mediated cell adhesion in the Drosophila testis. Dev Cell , 117–126. [DOI] [PubMed] [Google Scholar]
- Wang X, He L, Wu YI, Hahn KM, Montell DJ. (2010). Light-mediated activation reveals a key role for Rac in collective guidance of cell movement in vivo. Nat Cell Biol , 591–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y-C, Khan Z, Wieschaus EF. (2013). Distinct Rap1 activity states control the extent of epithelial invagination via α-catenin. Dev Cell , 299–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi R, McGregor JR, Harrison DA. (2003). A gradient of JAK pathway activity patterns the anterior-posterior axis of the follicular epithelium. Dev Cell , 167–177. [DOI] [PubMed] [Google Scholar]
- Xiang W, Zhang D, Montell DJ. (2016). Tousled-like kinase regulates cytokine-mediated communication between cooperating cell types during collective border cell migration. Mol Biol Cell , 12–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Rubin GM. (1993). Analysis of genetic mosaics in developing and adult Drosophila tissues. Development , 1223–1237. [DOI] [PubMed] [Google Scholar]
- Yamamoto E, Kohama G-I, Sunakawa H, Iwai M, Hiratsuka H. (1983). Mode of invasion, bleomycin sensitivity, and clinical course in squamous cell carcinoma of the oral cavity. Cancer , 2175–2180. [DOI] [PubMed] [Google Scholar]
- Zhang Y-L, Wang R-C, Cheng K, Ring BZ, Su L. (2017). Roles of Rap1 signaling in tumor cell migration and invasion. Cancer Biol Med , 90–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.