Abstract
The huntingtin protein participates in several cellular processes that are disrupted when the polyglutamine tract is expanded beyond a threshold of 37 CAG DNA repeats in Huntington’s disease (HD). Cellular biology approaches to understand these functional disruptions in HD have primarily focused on cell lines with synthetically long CAG length alleles that clinically represent outliers in this disease and a more severe form of HD that lacks age onset. Patient-derived fibroblasts are limited to a finite number of passages before succumbing to cellular senescence. We used human telomerase reverse transcriptase (hTERT) to immortalize fibroblasts taken from individuals of varying age, sex, disease onset, and CAG repeat length, which we have termed TruHD cells. TruHD cells display classic HD phenotypes of altered morphology, size and growth rate, increased sensitivity to oxidative stress, aberrant adenosine diphosphate/adenosine triphosphate (ADP/ATP) ratios, and hypophosphorylated huntingtin protein. We additionally observed dysregulated reactive oxygen species (ROS)-dependent huntingtin localization to nuclear speckles in HD cells. We report the generation and characterization of a human, clinically relevant cellular model for investigating disease mechanisms in HD at the single-cell level, which, unlike transformed cell lines, maintains functions critical for huntingtin transcriptional regulation and genomic integrity.
INTRODUCTION
Huntington’s disease (HD) is a late-onset, autosomal-dominant neurodegenerative disorder characterized by a triad of motor, cognitive, and psychiatric symptoms. The disease is caused by a CAG trinucleotide expansion of >37 repeats in the huntingtin gene, manifesting as polyglutamine-expanded huntingtin protein (Huntington’s Disease Collaborative Research Group, 1993). The functional implications of this expanded, mutant huntingtin are not fully understood. Much of the existing research on HD cell biology in relevant neuronal cell types has been limited to primary postmitotic neurons from murine brain tissue or transformed cell lines, which have several limitations, including the use of synthetically long CAG lengths to model human disease in mice (Mangiarini et al., 1996; Hodgson et al., 1999; Schilling et al., 1999; Shelbourne et al., 1999; Trettel et al., 2000; Lin et al., 2001; Wyttenbach et al., 2001; Menalled et al., 2003, Slow et al., 2003; Gray et al., 2008; Menalled et al., 2012; Ferris et al., 2014; Southwell et al., 2016; Singer et al., 2017). These alleles actually genetically model juvenile or Westphal variant HD, which are not age-onset diseases. The mean clinical CAG allele length is 43 repeats, with even >50 repeats representing statistical outliers (Myers, 2004). Disease models in Neuro-2A cells, HEK293, or HeLa cells rely on cell transformation to maintain line longevity, but transformation typically involves initiating genomic instability and “shattering” of genomes (Landry et al., 2013; Mittelman and Wilson, 2013), affecting intra- and interlaboratory reproducibility.
The pursuit of investigating human cells has driven researchers to culture patient fibroblast cells that can be extracted from a skin biopsy (Barkley et al., 1977; Goetz et al., 1981; del Hoyo et al., 2006). Primary fibroblasts from HD patients possess clinically relevant polyglutamine expansion lengths, making them an attractive model for studying HD human cell biology. Further, they can be reprogrammed into induced pluripotent stem cells (iPSCs) (HD iPSC Consortium, 2012), which can be differentiated into various neuronal cell lineages (Juopperi et al., 2012; Martinez et al., 2012; Zhang et al., 2013) or directly reprogrammed to medium spiny neurons (Yoo et al., 2011; Xue et al., 2013; Victor et al., 2014, 2018). However, primary fibroblasts are subject to telomere-controlled senescence, which limits the number of passages as telomeres shorten with each cell division (Hayflick and Moorhead, 1961; Hayflick, 1965). Senescent cells display altered gene expression, decreased proliferation, and resistance to apoptotic mechanisms (Dimri et al., 1995; Marcotte et al., 2004), hindering long-term use and consistency between trials.
We sought to overcome these limitations by immortalizing patient fibroblasts with human telomerase reverse transcriptase (hTERT). hTERT has been extensively used to immortalize human cell types to study cell biology in a number of diseases (Jiang et al., 1999; Morales et al., 1999; Ouellette et al., 2000; Wood et al., 2001; Lee et al., 2004; Young et al., 2004). Like primary cells, hTERT-immortalized cells mimic in vivo tissue phenotypes, with the added benefits of proliferative capacity, karyotypic stability, and interexperimental reproducibility (hTERT Immortalized Cell Lines, n.d.).
We sought to generate a panel of human cell lines with polyglutamine expansions in the 40- to 50-CAG range, reflective of those seen in the clinic. We immortalized fibroblasts from three individuals and termed the cell lines TruHD cells: control female (TruHD-Q21Q18F), heterozygous male (TruHD-Q43Q17M), and homozygous female (TruHD-Q50Q40F) (Table 1). To validate these cells as HD models, we examined known model and patient phenotypes. Consistent with previous reports, we found that HD cells could be distinguished from wild type based on morphology (Trettel et al., 2000; Singer et al., 2017), size (Singer et al., 2017), growth rate (Menkes and Stein, 1973; Kirk et al., 1977; Goetz et al., 1981), sensitivity to stress (Slow et al., 2003; Atwal et al., 2007; Reis et al., 2011; Maiuri et al., 2017), aberrant adenosine diphosphate/adenosine triphosphate (ADP/ATP) ratios (Trettel et al., 2000; Milakovic and Johnson, 2005; Acuña et al., 2013), hypophosphorylated huntingtin protein (Gu et al., 2009; Atwal et al., 2011; Di Pardo et al., 2012; Bowie et al., 2018), and altered reactive oxygen species (ROS)-dependent localization to nuclear speckles (DiGiovanni et al., 2016). We have therefore generated and characterized a panel of clinically relevant cellular models for investigating disease mechanisms in HD.
TABLE 1:
TruHD patient fibroblast information.
| Primary cell name | Cell line name | Allele 1 | Allele 2 | Sex | Age at sampling | Disease onset age (yr) |
|---|---|---|---|---|---|---|
| ND30013 | TruHD-Q43Q17M | 43 CAG | 17 CAG | Male | 54 | 50 |
| ND30014 | TruHD-Q21Q18F | 21 CAG | 18 CAG | Female | 52 | — |
| GM04857 | TruHD-Q50Q40F | 50 CAG | 40 CAG | Female | 23 | 28 |
Genome-wide association studies (GWAS) for HD modifiers of age of onset revealed DNA damage and oxidative stress mechanisms as critical modifiers of the disease onset in patients (Genetic Modifiers of Huntington’s Disease [GeM-HD] Consortium, 2015). One of the most important regulators of DNA repair, cell stress, and cell death responses is the TP53 protein (Fritsche et al., 1993; Smith et al., 1995; Liu and Kulesz-Martin, 2001), which also directly regulates huntingtin transcription via a response element in the HTT promoter region (Bae et al., 2005; Feng et al., 2006). To date, commonly used HD cell lines are SV40 large T-antigen-transformed mouse striatal cell lines (STHdh) (Trettel et al., 2000), which have become an invaluable tool for studying HD cell biology. However, conditional cell immortalization by transformation requires inhibited TP53 function. Investigation of the role of huntingtin in DNA damage and cell stress pathways may therefore be confounded by TP53 inactivation. In contrast, hTERT immortalization does not alter TP53 function (Bodnar et al., 1998; Vaziri and Benchimol, 1998; Ouellette et al., 2000), making TruHD cells an attractive model for these disease mechanisms in particular. Immortalized fibroblasts provide the added benefits of interexperimental and interlaboratory reproducibility and long-term applications such as generation of stable cell lines and direct conversion to patient-specific human neurons. These cell models are readily available to the HD research community and will be distributed.
RESULTS
Immortalization of primary fibroblasts with hTERT
Primary fibroblasts from various patients were obtained from the Coriell Institute for Medical Research and transduced with TERT Human Lentifect Purified Lentiviral Particles. Three immortalized cell lines were generated from patients with various CAG repeat lengths and disease onset ages (Table 1) as representatives of control (TruHD-Q21Q18F), heterozygous HD (TruHD-Q43Q17M), and homozygous HD (TruHD-Q50Q40F). We standardized the line naming to a format compatible with digital file annotation, defining the CAG length of each HTT allele and the sex of the donor.
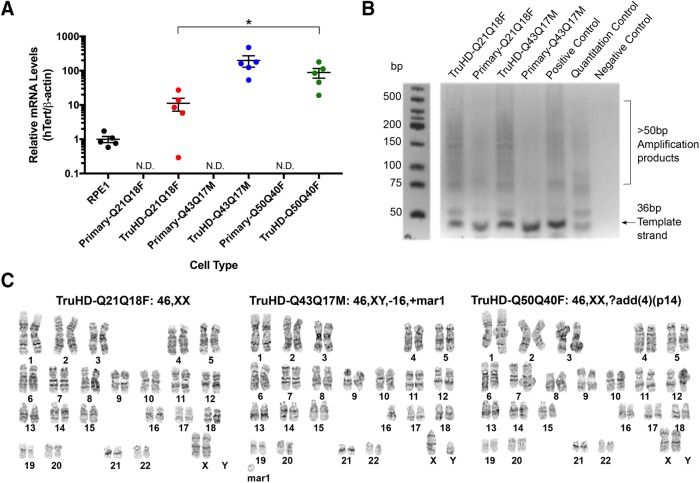
To verify that cells were successfully overexpressing hTERT, RNA levels in primary cells and TruHD cells were compared by quantitative PCR (qPCR), showing detectable hTERT mRNA levels in TruHD cells compared with primary cells normalized to commercially available hTERT-immortalized retinal pigment epithelial (RPE1) cells (Figure 1A). To ensure that the increased hTERT expression was associated with increased hTERT catalytic activity, telomerase activity was tested in TruHD-Q21Q18F and TruHD-Q43Q17M cells using a telomeric repeat amplification protocol (TRAP) assay. As shown in Figure 1B, multiple amplification products resulting from active hTERT were observed in TruHD cells but not primary cells, indicating that the transduced hTERT is catalytically active in TruHD cells.
FIGURE 1:
Generation of TruHD-immortalized cell lines. (A) hTERT mRNA levels normalized to β-actin mRNA levels in RPE1 cells (positive control), primary cells, and TruHD cells. hTERT levels in primary cells were not detectable (ND). n = 5. Error bars represent SEM. *p = 0.0369 comparing TruHD-Q21Q18F, TruHD-Q43Q17M, and TruHD-Q50Q40F by one-way analysis of variance (ANOVA). (B) Telomeric repeat amplification product (TRAP) assay. Amplification products run on 10% TBE gel after telomere extension reaction, showing telomeric repeats >50 base pairs in increments of 6 base pairs. Template strand is 36 base pairs. Negative control contains no Taq polymerase or template strand. (C) Representative karyotypes of TruHD-Q21Q18F, TruHD-Q43Q17M, and TruHD-Q50Q40F cells. “mar” denotes marker chromosomes, “+” are additional chromosomes and “?add(4)(p14)” denotes additional patterns observed on chromosome 4 at band p14. Results from full karyotype shown in Table 2.
Unlike immortalization by transformation, hTERT immortalization maintains karyotypic stability in normal, human diploid cells (Bodnar et al., 1998; Jiang et al., 1999; Ouellette et al., 2000). Chromosomal instability leading to polyploidy and aneuploidy can affect gene expression and cell viability, which is a hallmark of transformed cancer cells (Potapova et al., 2013; Dürrbaum and Storchová, 2016). To confirm karyotypic stability in TruHD cells after >25 passages, we compared the karyotypes of STHdh cells and TruHD cells. Large chromosomal abnormalities were detected in transformed HD mouse striatal derived cells (STHdhQ111/Q111) cells (Supplemental Figure 1A and Table 2), consistent with a recently published study (Singer et al., 2017). No chromosomal changes were recorded for control TruHD-Q21Q18F, and minor chromosomal changes were recorded in TruHD-Q43Q17M and TruHD-Q50Q40F cell lines (Figure 1C and Table 2). In TruHD-Q43Q17M cells, the majority of the analyzed cells were missing one chromosome 16, and in TruHD-Q50Q40F cells, most of the cells had an abnormal banding pattern on chromosome 4. These changes should be considered when interpreting results of phenotypic analysis.
TABLE 2:
G-band karyotyping.
| Cell line | Karyotype |
|---|---|
| TruHD-Q21Q18F | 46,XX[13], 92,XXXX[2] |
| TruHD-Q43Q17M | 46,XY,-16,+mar[9], 47,XY,+7[3], 46,XY[3] |
| TruHD-Q50Q40F | 46,XX,?add(4)(p14)[9] |
| STHdhQ111/Q111 | 70-82<4n>, XX or XXX or XXXX, -X[12], -X[4],add(X)(F1)[2],-1[9],add(1)(F)[3],del(1)(?G)[3],+2[2],-2[7],-4[7],?add(4)(C4)[3],dic(4;?)(?D;?)[2],+5[3],-6[8],-7[19],-7[5],+8[4],-8[4],rob(8;8)(A1;A1)[2],-9[12]+11[4],-11[3],-12[19],-12[6],-13[6],-14[15],-14[6],+15[18],+15[15],+15[4],-17[11],-17[3],-18[15],-18[3],+19[17],+1-15mar[20][cp20] |
[] = number of analyzed cells with specified karyotype. Bold indicates representative result.
Validation of CAG repeats in TruHD cell lines
To verify that the CAG repeats of the fibroblasts matched the clinical information reported after successful immortalization, the CAG repeats were sized using a standardized HTT CAG repeat sizing assay (Warner et al., 1993; Keum et al., 2016). The length of each CAG tract was as expected (Table 3). The human HTT gene typically bears an additional CAACAG sequence beyond the pure CAG DNA tract sequence (Huntington’s Disease Collaborative Research Group, 1993). These two additional codons encoding glutamine residues were not considered in the annotation by the Coriell Institute. Therefore, TruHD-Q21Q18F, for example, refers only to the polyglutamine tract that corresponds to the pure CAG tract, but the full polyglutamine tract lengths are actually Q23Q20. The true polyglutamine lengths corresponding to each TruHD cell line are listed in Table 3.
TABLE 3:
Sizing of CAG repeats in TruHD fibroblasts.
| Cell line name | CAGn repeats | Q lengths (= CAG +2, based on genetic structure of CAGnCAACAG in humans) |
|---|---|---|
| TruHD-Q21Q18F | 21 CAG/18 CAG | 23Q/20Q |
| TruHD-Q43Q17M | 43 CAG/17 CAG | 45Q/19Q |
| TruHD-Q50Q40F | 50 CAG/40 CAG | 52Q/42Q |
Defining senescence in TruHD cell lines
Primary fibroblasts are typically cultured for ∼15 passages from the initial skin biopsy before reaching senescence, while the successfully immortalized cells reported here can be passaged beyond 80 passages without reaching senescence. Senescent cells show changes in cell growth, morphology, and gene expression, which can be detected by an associated β-galactosidase activity (Walen, 2006; Debacq-Chainiaux et al., 2009). Senescence was not detectable in TruHD-immortalized cell lines (Supplemental Figure 1B) under normal culture conditions. We did note, however, that control TruHD-Q21Q18F fibroblasts seeded too sparsely exhibited senescence-associated β-galactosidase activity (Supplemental Figure 1C) and stopped dividing. Normal, adherent cells in culture undergo contact inhibition or postconfluence inhibition of cell mitosis (Walen, 2006; Küppers et al., 2010; Lemons et al., 2010). Essentially, once the cells become too confluent and make contact with nearby cells, they stop dividing and do not grow because of contact inhibition, unlike transformed cell lines. Cells that are left in this state for too long can become senescent and do not recover in culture (Goldstein and Singal, 1974). Specific protocols for freeze/thaw were therefore considered and explained in detail under Materials and Methods. After 7 d of confluence without media changes, control cells were more susceptible to senescence compared with the HD cell line (Supplemental Figure 1D). On karyotypic analysis of TruHD-Q21Q18F cells cultured under these conditions, a small percentage of control cells displayed tetraploidy (Supplemental Figure 1E), a phenomenon that has been reported to be a result of cellular senescence (Schwarzacher and Schnedl, 1965; Walen, 2006). In contrast, tetraploidy did not occur for either of the HD cell lines. Overall, we observed that cells did not senesce after extended passaging (over 80 passages), but control cells did senesce if cultured too sparsely or at high confluence, unlike HD cells, which have a more senescence-resistant phenotype. This aspect of these cell lines could provide utility to study huntingtin biology in senescent cells, or cells in transition from mitotic to senescent. As with any cell lines though, it is important to note that prolonged culturing of cells (beyond 100 passages), even if immortalized, can result in abnormal behavior (Noble et al., 2004; van Waarde-Verhagen et al., 2006), especially if cell confluence and senescence are not monitored regularly.
Cell morphology, size, growth, and viability
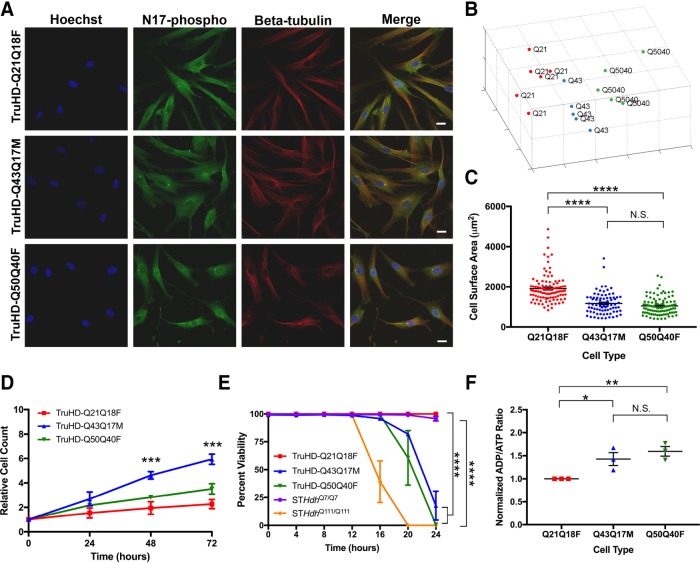
Mutant STHdh cells exhibit altered morphology (Trettel et al., 2000; Reis et al., 2011; Singer et al., 2017). To probe whether TruHD cells could be distinguished by their morphology, nuclei were DNA stained with Hoechst dye, and immunofluorescence was performed with antibodies against phosphorylated huntingtin at serines 13 and 16 (N17-phospho) and β-tubulin (Figure 2A). Images were analyzed with Phenoripper open software (www.phenoripper.org), which defines textures of the images in a nonsupervised manner and plots vectors of the three most variant textures in unitless three-dimensional space using principal component analysis (PCA). This allows for identification of similarity between the images based on those defined features. Merged images of TruHD-Q21Q18F, TruHD-Q43Q17M, and TruHD-Q50Q40F, considering the three parameters Hoechst, N17-phospho, and β-tubulin, clustered apart from each other on a PCA plot (Figure 2B). The clustering pattern from each individual channel is shown in Supplemental Figure 2A, where β-tubulin alone showed the greatest separation in the clusters compared with Hoechst and N17-phospho. This unbiased detection of a morphology phenotype between control and HD cells may be attributed to differences in cell size. We therefore compared cell size in control and mutant TruHD cells. Quantification of cell surface area shows that HD cells are smaller than control cells (Figure 2C). Therefore, consistent with numerous independent previous reports, huntingtin may be involved in cytoskeletal regulation (Hoffner et al., 2002; Muchowski et al., 2002; Munsie et al., 2012; Singer et al., 2017).
FIGURE 2:
TruHD cell properties. (A) Immunofluorescence images of TruHD-Q21Q18F, TruHD-Q43Q17M, and TruHD-Q50Q40F. Scale bar = 10 μm. (B) PCA plot of images sorted with Phenoripper. (C) Cell surface area comparison in TruHD cells. n = 3, N > 200. Error bars represent SEM; *p < 0.0001. (D) Relative cell count measured every 24 h. n = 3, N > 200. Error bars represent SEM. ***p = 0.0003 at 48 h; ***p = 0.0001 at 72 h by one-way ANOVA. (E) Percentage cell viability of TruHD cells compared with STHdh cells. n = 3, N > 200. Error bars represent SEM. At 24 h, ****p < 0.0001 for STHdhQ7/Q7 vs. STHdhQ111/Q111 by two-ANOVA and ****p < 0.0001 for TruHD-Q21Q18F vs. TruHD-Q43Q17M and TruHD-Q50Q40F by two-way ANOVA. (F) Normalized ADP/ATP ratio in TruHD cells at ∼75% confluency 24 h after seeding. n = 3, N > 200. Error bars represent SEM. *p = 0.0371 and **p = 0.0048.
Reentry of postmitotic neuronal cells into the cell cycle is a potential mechanism in the neurodegenerative process (see review by Folch et al. [2012]). A consistent observation when culturing TruHD fibroblasts is that the mutant fibroblasts divide more rapidly, as seen with primary HD fibroblasts (Goetz et al., 1981). Monitoring proliferation of TruHD cells over 72 h showed that TruHD-Q43Q17M cells double after ∼20 h, TruHD-Q50Q40F cells double after ∼24 h, while TruHD-Q21Q18F double after ∼72 h (Figure 2D). These observations are consistent with reports implicating huntingtin in cell-cycle regulation and DNA damage repair mechanisms (Hoffner et al., 2002; Lu et al., 2015; Maiuri et al., 2017) and that these functions are aberrant in HD cells.
Susceptibility to various types of cell stress is a well-documented phenomenon in HD cellular models (Atwal et al., 2007, 2011; Munsie et al., 2012; Nath et al., 2015). We have previously shown that huntingtin responds to oxidizing agents by becoming phosphorylated, translocating from the endoplasmic reticulum (ER) to the nucleus, and interacting with chromatin (DiGiovanni et al., 2016; Maiuri et al., 2017). We therefore tested TruHD cell viability on oxidative stress with potassium bromate (KBrO3) treatment over 24 h. As shown in Figure 2E, both HD cell lines were most susceptible to cell death compared with the control line. Dose-dependent response of treatments in each individual TruHD cell line can be found in Supplemental Figure 2B.
Besides response to cell stress, another well-documented HD phenotype is an energy deficit as measured by ADP/ATP ratio (Trettel et al., 2000; Milakovic and Johnson, 2005; Reis et al., 2011). The detection of ADP/ATP ratio in TruHD cells demonstrated an energy deficit in HD cell lines compared with the wild-type cell line (Figure 2F) similarly to the trend in STHdh cells (Supplemental Figure 2C), which is consistent with previously reported studies in HD models and synthetic allele lengths (Trettel et al., 2000; Acuña et al., 2013).
These phenotypes described in TruHD cells such as cell morphology, size, growth rate, susceptibility to oxidative stress, and energy levels, demonstrate their utility as a cellular model with clinically relevant CAG allele lengths as phenotypes in mutant TruHD cells were all consistent with previous HD cellular models.
Total huntingtin and phosphorylated huntingtin protein levels
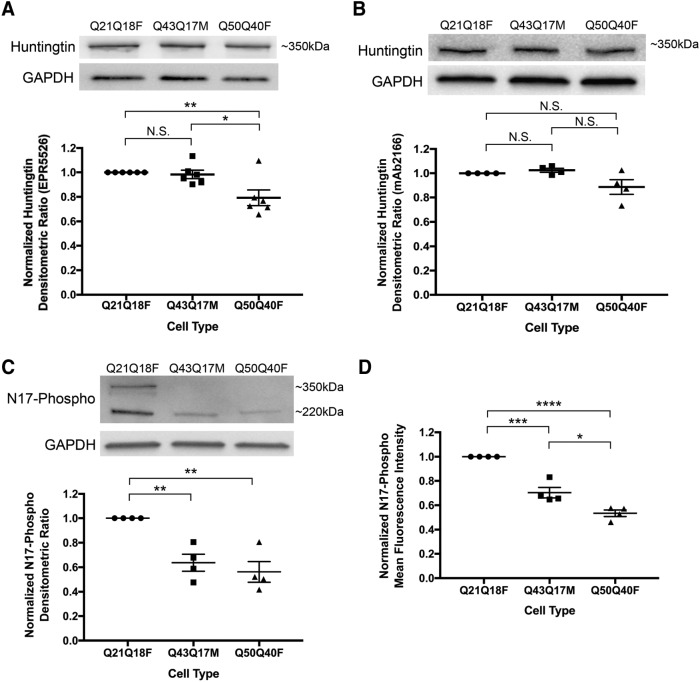
Since the cells were taken from various patients and are not isogenic, the amount of total huntingtin was quantified. Validated antibodies to different epitopes of full-length huntingtin were used (EPR5526 and mAb2166), showing a minor decrease in total huntingtin levels in TruHD-Q50Q40F compared with TruHD-Q21Q18F and TruHD-Q43Q17M (Figure 3, A and B; full immunoblots Supplemental Figure 4, A and B).
FIGURE 3:
Huntingtin protein levels in TruHD cells. (A) Densitometric analysis of total huntingtin levels using Western blot with EPR5526 antibody. Immunoblots were cut horizontally at the 75-kDa marker so that GAPDH loading control was probed for separately. Normalized to control TruHD-Q21Q18F cells. n = 6. Error bars represent SEM. **p = 0.0087 and *p = 0.0254 by unpaired Student’s t test. (B) Densitometric analysis of total huntingtin levels using Western blot with mAb2166 antibody. Immunoblots were cut horizontally at the 75-kDa marker so that GAPDH loading control was probed for separately. Normalized to control TruHD-Q21Q18F cells. n = 4. Error bars represent SEM. (C) Densitometric analysis of N17-phospho levels using Western blot. Immunoblots were cut horizontally at the 75-kDa marker so that GAPDH loading control was probed for separately. Normalized to control TruHD-Q21Q18F cells. n = 4. Error bars represent SEM. **p = 0.0020 by unpaired t test. (D) Mean fluorescence intensity analysis of N17-phospho using flow cytometry. Normalized to control TruHD-Q21Q18F. n = 4. Error bars represent SEM. ***p = 0.0005, *p = 0.0159 and ****p < 0.0001 by unpaired t test.
Phosphorylation is a protective posttranslational modification in HD cells (Gu et al., 2009; Atwal et al., 2011; Di Pardo et al., 2012). Restoration of N17 phosphorylation is a therapeutic target in HD that has been explored because mutant, polyglutamine-expanded huntingtin is hypophosphorylated at serines 13 and 16 (S13p and S16p, respectively) (Gu et al., 2009; Atwal et al., 2011; Di Pardo et al., 2012). Immunoblotting performed with a validated antibody against both serines (α-N17-phospho) (Supplemental Figure 3) showed decreased levels of N17 phosphorylation in mutant TruHD fibroblast cell lines compared with wild type (Figure 3C; full immunoblot in Supplemental Figure 4C). Two distinct molecular-weight bands are seen with the antibody, at ∼350 and ∼220 kDa. Degradation products are often reported with other huntingtin antibodies as well, due to the large size of the protein and the rigorous processing steps of immunoblotting (Landles et al., 2012; Sapp et al., 2012). Both bands were considered in quantifications, and this confirms hypophosphorylation of mutant huntingtin in a human HD. These results were verified by measuring whole-cell mean fluorescence intensity by flow cytometry (Figure 3D). Therefore, N17-phospho levels vary, but total huntingtin levels are invariant. This phenotype is consistent with previous reports (Gu et al., 2009; Atwal et al., 2011; Di Pardo et al., 2012) and further validates N17-phosphorylation restoration as a target for HD therapeutic development.
Huntingtin stress response in human patient fibroblasts
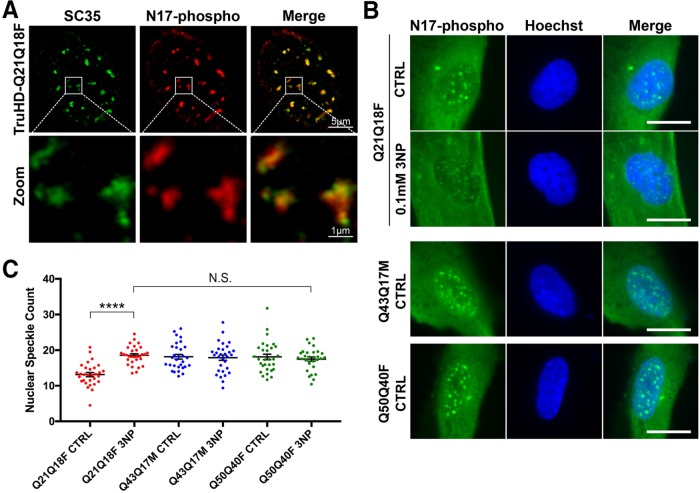
Huntingtin is a stress response protein and is involved in DNA damage repair, oxidative stress, and ER stress pathways (Atwal et al., 2007; Atwal and Truant, 2008; Vidal et al., 2012; DiGiovanni et al., 2016; Maiuri et al., 2017). Previous studies from our lab show that huntingtin is bound to the ER membrane in steady-state conditions and is released under conditions of stress, particularly ROS stress (Atwal et al., 2007, 2011). Once soluble, huntingtin is phosphorylated at serines 13 and 16 (S13,S16), translocates to the nucleus, and localizes to nuclear puncta (Atwal et al., 2011; Maiuri et al., 2013; DiGiovanni et al., 2016). Using superresolution structured-illumination microscopy, we have now identified that these previously reported nuclear puncta are SC35 positive nuclear speckles: dynamic RNA/protein structures that are rich in mRNA splice factors that are important in cell stress responses (Hall et al., 2006; Campalans et al., 2007; Lin et al., 2008) (Figure 4A).
FIGURE 4:
Huntingtin localizes to nuclear speckles in response to stress. (A) N17-phospho localizes to SC35+ nuclear speckles. (B) Nuclear speckles are increased in TruHD-Q21Q18F control cells on treatment with 0.1 mM 3NP but not for TruHD-Q43Q17M or TruHD-Q50Q40F. Scale bar = 10 μm. (C) Quantification of nuclear speckles. n = 3, N = 180. Error bars represent SEM. ****p < 0.0001 by unpaired t test. Comparison of TruHD-Q21Q18F 3NP, TruHD-Q43Q17M CTRL, TruHD-Q43Q17M 3NP, TruHD-Q50Q40F CTRL, and TruHD-Q50Q40F 3NP by one-way ANOVA shows no significant difference (p = 0.8475).
We previously reported that stress-dependent phosphorylation of S13 and S16 promotes huntingtin localization to nuclear speckles in RPE1 (DiGiovanni et al., 2016). We therefore tested this phenomenon in TruHD cells. Cells were treated with 0.1 mM 3-nitroproprionic acid (3NP), a mitochondrial complex II inhibitor, for 1 h to induce oxidative stress. We observed a significant increase in the number of nuclear speckles in TruHD-Q21Q18F cells (Figure 4, B and C). However, in both the mutant TruHD-Q43Q17M and TruHD-Q50Q40F lines, there was no significant difference in the number of nuclear speckles between treated and untreated conditions. The number of nuclear speckles in both heterozygote and homozygote HD cells were similar to that of TruHD-Q21Q18F in the presence of 3NP (N.S., p = 0.8475), suggesting that in HD cells are under a chronic stress load.
Characterization of several HD phenotypes, combined with the establishment of methods to easily detect these disease-relevant phenotypes, in TruHD cells demonstrate their utility as a cellular model and will hopefully facilitate further investigation into pathological mechanisms.
DISCUSSION
Patient fibroblasts have been used previously by us and others (Menkes and Stein, 1973; Barkley et al., 1977; Kirk et al., 1977; Maiuri et al., 2017) to study HD cell biology, but currently there are no defined cell lines that are used consistently between projects. After generation of hTERT-immortalized TruHD cells, we focused on defining characteristics of these cells to facilitate their use. Our wild-type line (TruHD-Q21Q18F), heterozygous HD line (TruHD-Q43Q17M), and homozygous HD line (TruHD-Q50Q40F) were chosen for this study as their CAG repeat lengths were most representative of annotated lengths but also because they cultured well and the observed phenotypes were consistent throughout the study. A homozygote TruHD-Q50Q40F was chosen, because although a rare clinical example, this line could have utility as both alleles of huntingtin are mutant expanded and thus can help resolve data in heterozygote lines, where mutant huntingtin phenotypes could be confounded by the presence of the normal allele.
We observed HD phenotypes in both heterozygous TruHD-Q43Q17M and homozygous TruHD-Q50Q40F cells at clinically relevant CAG repeat lengths. Typical HD cell phenotypes include reduced cell size (Singer et al., 2017), decreased cell viability on cellular stress (Atwal et al., 2007, 2011; Munsie et al., 2011), altered cell proliferation (Singer et al., 2017), decreased ADP/ATP ratio (Trettel et al., 2000), and hypophosphorylation of huntingtin N17 at serines 13 and 16 (Gu et al., 2009; Atwal et al., 2011; Di Pardo et al., 2012). Here, we have also demonstrated that HD cells showed altered susceptibility to cellular senescence and deficient response to oxidative stress as seen by SC35 nuclear speckle counts. Additionally, TruHD cells can be distinguished in an unbiased manner using nonsupervised image texture analysis and principal component analysis via software such as Phenoripper. Phenoripper can be used as a readout for future high-content drug screening assays.
Improvement in interexperimental and interlaboratory reproducibility have also been observed with these immortalized lines and may be beneficial for long-term applications such as generation of stable cell lines and cellular reprogramming to generate patient-specific neurons that maintain age-associated signatures (Yoo et al., 2011; Victor et al., 2014, 2018). However, as with all cell lines, strict culturing practices are needed to maintain proper cellular function and genomic stability, as critical cellular pathways involving TP53 should be monitored as they can change over a prolonged period of time, even in immortalized cells, especially over 100 passages (Noble et al., 2004; van Waarde-Verhagen et al., 2006).
Recent developments in understanding cell biology throughout the course of HD progression highlight the need for improved methods for disease modeling. DNA damage repair pathways have been implicated as the predominant modifiers of HD pathogenesis (Genetic Modifiers of Huntington’s Disease (GeM-HD) Consortium, 2015). Previous observations in our lab show that huntingtin can sense oxidative stress and that huntingtin is involved in the DNA damage response (Maiuri et al., 2017; Bowie et al., 2018). These processes require TP53, which an important transcription factor that integrates various cellular stress signals and is widely considered the master regulator of genomic integrity due to its roles in DNA damage sensing, cell-cycle checkpoint control, and apoptotic regulation (see reviews by Shaw [1996], Wang and El-Deiry [2007], and Reinhardt and Schumacher [2012]).
The choice of model system for studying certain aspects of cell biology is therefore critical. Historically, synthetically long CAG repeat alleles of HTT have been used in cell models because of the apparent lack of obvious phenotypes of clinical HD alleles in animal models. Cell biology research in HD has been primarily focused on neurons from HD mouse models and easily accessible cell lines, each with their own restrictions and limitations. The main limitation of cells taken from mouse models are the synthetically long polyglutamine tracts used to mimic a late-onset human disease within the lifespan of a mouse. In these models, it can be overlooked that the majority of patients have CAG repeats between 40 and 50 and that these patients have varying age onset that is not attributed to just the number of repeats. Additionally, some models are transgenic and thus do not have an accurate gene dosage, while others express huntingtin at superphysiological levels, confounding data with incorrect protein stoichiometry. This is a significant concern for the huntingtin scaffolding protein, which been shown to cause gene dosage phenotypes even with a wild-type allele (Van Raamsdonk et al., 2006). Additionally, the role of brain somatic CAG allele expansion is not fully understood in HD or other CAG expansion diseases (Schmidt and Pearson, 2016), but synthetic hyperallele models circumvent this phenomenon seen longitudinally in human brains (Telenius et al., 1994) and thus model out what could be an essential mechanistic step in humans to initiate disease.
The fact that these lines are derived from the HD periphery could present great utility to define precise molecular biomarkers of HD, as these cells are adherent and ideal for microscopy and high content analysis and represent a cell population that can be resampled in clinic.
This is the first characterized human HD-immortalized cell line model and can therefore be used to test therapeutic reagents that are designed specifically for human cells and will be a tool for the HD research community.
MATERIALS AND METHODS
Cell culture and generation of hTERT-immortalized fibroblasts
Patient fibroblasts were purchased from the Coriell Institute from the NINDS repository. HD patient fibroblasts (ND30013, GM04857) and control patient fibroblasts (ND30014) were obtained. Cells were cultured in MEM (Life Technologies #10370) with 15% fetal bovine serum (FBS; Life Technologies) and 1X GlutaMAX (Life Technologies #35050). Cells were infected with 1 × 106 TERT Human Lentifect Purified Lentiviral Particles (GeneCopoeia, LPP-Q0450-Lv05-200-S). To aid in infection, 10 μg/ml polybrene was added. After 8 h, cells were infected again and left for 24 h. Media was changed, and cells were left for an additional 48 h. Successfully transduced cells were selected in media with 1 μg/ml puromycin. Cells were grown at 37°C with 5% CO2.
STHdh cells (a kind gift from Marcy E. Macdonald, Center for Genomic Medicine at the Massachusetts General Hospital) were cultured in DMEM (Life Technologies #11995) with 10% FBS. Cells were grown at 33°C with 5% CO2. RPE1 cells (American Type Culture Collection) were cultured in 1:1 DMEM/Nutrient Mixture F-12 (DMEM/F12; Life Technologies #11330) with 10% FBS and 0.01% hygromycin. Cells were grown at 37°C with 5% CO2.
Quantitative PCR to measure hTERT mRNA levels
Primary, TruHD and RPE1 cells were grown to ∼85% confluence. Total RNA was obtained from frozen cell pellets lysed in 1 ml of Trizol (Thermo Fisher Scientific) per ∼1 × 106 cells, followed by phenol-chloroform extraction. RNA was treated with DNase I (Thermo Fisher Scientific), and cDNA was prepared using 1000 ng total RNA and SuperScript III Reverse Transcriptase (Thermo Fisher Scientific). Transcript expression was measured using TaqMan Assays with AmpliTaq Gold DNA polymerase (Thermo Fisher Scientific), and hTERT (Hs00972650_m1) was compared with ACTB (Hs01060665_g1) housekeeping gene using the ΔΔCT method.
Telomeric repeat amplification protocol
Primary fibroblasts and corresponding TruHD cells were grown to ∼85% confluence. A two-step PCR method (TRAPeze Telomerase Detection Kit S7700; Millipore) was used to evaluate hTERT catalytic activity. Briefly, in the first step, telomerases from lysed cells add telomeric repeats (AG followed by repetitive GGTTAG sequences) on the 3′ end of a substrate oligonucleotide (TS). In the second step, the extended products are amplified by PCR using a primer specific for the beginning of TS and a reverse primer specific for the end of the repeats. The lowest amplification product should be 50 base pairs, and increases by 6-base-pair-long repeat increments are visualized on a 10% Tris-borate ethylene diamine tetra-acetic acid (TBE) polyacrylamide gel.
Cryogenic storage of TruHD cells
For freezing one vial of TruHD cells, a plate was grown to 90% confluence (∼1 × 105/ml) on a 10-cm dish and was split in half. The next day, two ∼60% confluent plates, that were still in growth phase, were trypsinized and combined. Cells were centrifuged at 1500 rpm, and pellets were resuspended in culture media with 1 ml of 5% dimethyl sulfoxide (DMSO). Vials were put into a slow-freeze unit in the –80°C to ensure optimal cell preservation. After 24–48 h, vials were moved to –150°C for long-term storage.
Vials were thawed slowly at 37°C for around 2–5 min. Using a 10 ml pipette, 1 ml of cells was moved directly into a 10-cm plate already preincubated with media. After 24 h, media was changed to remove residual DMSO.
Sizing of CAG repeat
TruHD cells were grown to ∼90% confluence in a 10-cm plate. Cells were scraped and centrifuged at 4°C at 1500 rpm. Genomic DNA was extracted using the PureLink Genomic DNA kit (ThermoFisher). A fluorescence-based assay was used to size the CAG repeats, based on the originally described assay by Warner et al. (1993) and further described in Keum et al. (2016)
Karyotyping
Karyotyping was performed by The Centre for Applied Genomics (The Hospital for Sick Children, Toronto, Canada). Karyotype analysis via G-banding was performed on cells from two T25 flasks per cell line. When cells reached 80–90% confluence, Karyomax Colcemid was added to each flask to a final concentration of 0.1 μg/ml (Life Technologies #15212-012) and incubated in a 37°C CO2 incubator for 1.5–2 h (for TruHD-Q43Q17M and TruHD-Q50Q40F) and 3–4 h for TruHD-Q21Q18F. Cells were then collected and suspended in 6 ml of 0.075 M KCl and incubated at 37°C for 20 min. Eight drops of Carnoy’s Fixative (methanol/acetic acid, 3:1) were added and mixed together. The cells were centrifuged at 1000 rpm for 10 min at room temperature, and cell pellets were collected. After three rounds of fixations (add 8 ml fixative and centrifuge at 1000 rpm for 10 min), cells were resuspended in 0.5–1 ml of fixative, and cells from each suspension were dispensed onto glass slides and baked at 90°C for 1.5 h. Routine G-banding analysis was then carried out. Approximately 15–20 metaphases per cell line were examined.
Senescence-associated β-galactosidase activity assay
TruHD and primary fibroblasts were seeded into a six-well plate. Primary fibroblasts reached ∼20 passages before analysis, and TruHD cells reached ∼50 passages. When cells reached ∼90% confluency, media was aspirated and cells were washed 1X with phosphate-buffered saline (PBS), and experiments were carried out using Senescence Detection Kit (Abcam; ab65351) per manufacturer instructions.
Phenoripper and cell surface area measurement
Immunofluorescence was performed with Hoechst 33342 (ThermoFisher), beta-tubulin antibody (E7, DSHB, 1:250 dilution in 2% FBS in PBS with 0.02% Tween), and N17-phospho antibody (NEP, validated in Supplemental Figure 3). Images were analyzed using Phenoripper. Five images per trial of TruHD fibroblasts acquired on a Nikon TiEclipse inverted epifluorescence widefield using a 20× objective (NA = 0.75) and Spectra X LED lamp (Lumencor) capture using an Orca-Flash 4.0 complementary metal oxide semiconductor (CMOS) camera (Hamamatsu). Cell surface area was calculated with ImageJ. Cells were thresholded to remove background to identify the whole-cell region of interest, and the area of each cell was measured and plotted.
Cell counting
Cells were seeded into a 24-well plate (105/ml). After 24 h, nuclei were stained for 15 min with NucBlue Live ReadyProbes Reagent (1 drop/ml media) (ThermoFisher). Cells were imaged using a Nikon TiEclipse inverted widefield epifluorescence microscope, and, using an automated round object detector in NIS Elements Advanced Research 4.30.02v software (Nikon), cell nuclei were counted. This was repeated; repeat plates were left for 48 and 72 h for all cell lines.
ADP/ATP ratio assay
The ADP/ATP Ratio Assay Kit (Sigma MAK135) was used according to the protocol, except the first step to seed TruHD cells directly into a 96-well plate on which the rest of the assay is performed. If seeded directly into a 96-well plate, then there is not enough room to grow the suggested 104 fibroblast cells because of their large size. Therefore, TruHD cells were seeded into a 24-well plate, for no more than 48 h, to ∼80% confluency. After reaching confluency, the cells are lysed with the working reagent (provided in the kit). After being lysed, cells from the 24-well plate were moved directly into a 96-well plate to continue the rest of the assay according to the protocol. For STHdh cells, assay was performed according to the protocol with no changes.
Cell viability assay
Cells were seeded into a 96-well plate. After 24 h, cells were stained for 15 min with NucBlue Live ReadyProbes Reagent (1 drop/ml media) (ThermoFisher) in Hank’s balanced salt solution (HBSS) (Life Technologies). Cells were washed once with HBSS and treated with 100 μl of potassium bromate (KBrO3) (Millipore) at concentrations of 0, 1, 10, 100, and 200 mM in HBSS with NucGreen Dead 488 ReadyProbes Reagent (1 drop/ml media) (ThermoFisher).
The 96-well plate was imaged immediately over the course of 24 h every 20 min at 37°C using a Nikon TiEclipse inverted A1 confocal microscope equipped with a 20× objective (NA = 0.75) and driven by NIS Elements AR 4.30.02v 64-bit acquisition software (Nikon). Cells were imaged simultaneously in the fluorescein isothiocyanate (FITC) (NucGreen) and 4′6-diamidino-2-phenylindole (DAPI) (NucBlue) channels. A cell was defined as undergoing cell death when 50% or more of the nucleus, as defined by NucBlue-positive pixels, was overlapped by NucGreen-positive pixels. Cell death was multiplied by 100% and subtracted from 100 to calculate % Viability. Images were analyzed using Python.
Immunoblotting
TruHD cells were grown to ∼90% confluence. Cells were scraped and centrifuged at 4°C at 1500 rpm. Cell pellets were lysed in radioimmunoprecipitation assay buffer with 10% phosphatase (Roche) and 10% protease inhibitors (Roche) for 12 min on ice and centrifuged at 10,000 × g at 4°C for 12 min. Supernatant was collected, and 40 µg of protein was loaded into a precast 4–20% polyacrylamide gradient gel (Biorad). Proteins were separated by SDS–PAGE and electroblotted onto 0.45-µm polyvinyl difluoride (PVDF) membrane (EMD Millipore).
Blots were blocked with 5% nonfat dry milk in TBS-T (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1% Tween-20) for 1 h at room temperature. Blots were cut horizontally at a 75-kDa marker to probe for huntingtin (∼350 kDa) or GAPDH (loading control, ∼37 kDa) separately. Blots were then incubated with primary N17-phospho antibody (1:1250), EPR5526 (1:2500; Abcam ab106115), mAb2166 (1:2500; Millipore), or GAPDH (1:10,000; Abcam ab8425) overnight at 4°C. Blots were washed three times for 10 min with TBS-T and then incubated with anti-rabbit or anti-mouse horseradish peroxidase (HRP) secondary (1:50,000; Abcam) for 45 min at room temperature. Finally, blots were washed three times for 10 min with TBS-T and visualized with enhanced chemiluminescent HRP substrate (EMD Millipore) on a MicroChemi system (DNR Bio-imaging Systems). Huntingtin bands were quantified using National Institutes of Health ImageJ and normalized to the GAPDH loading control.
Flow cytometry
TruHD cells were grown to ∼90% confluence. Cells were scraped and centrifuged at 4°C at 1500 rpm. Cells (∼106) were resuspended and fixed in ice-cold methanol for 12 min, inverting every 4 min. Cells were centrifuged at 4°C at 10,000 × g for 5 min followed by two washes in flow buffer (PBS with 2.5 mM EDTA and 0.5% bovine serum albumin [BSA]) and blocked in flow buffer with 2% FBS for 1 h at room temperature. Cells were incubated overnight in AlexaFluor488-conjugated N17-phospho antibody, diluted 1:15 in flow buffer with 0.02% Tween-20, rotating at 4°C. Cells were washed twice and resuspended in flow buffer.
3NP treatment and nuclear speckle count
TruHD cells were treated with 0.1 mM 3NP for 1 h at 37°C and then fixed and permeabilized with ice-cold methanol for 12 min. Cells were washed in PBS and blocked in antibody solution (2% FBS, 0.1% [vol/vol] Triton X-100 in 1X TBS) at room temperature for 10 min. AlexaFluor488-conjugated N17-phospho antibody was diluted 1:15 in antibody solution and incubated overnight at 4°C. Cells were washed in PBS and stained with Hoechst 33342 dye for 12 min at room temperature and left in PBS prior to imaging.
Cells were imaged using Nikon TiEclipse inverted widefield epifluorescence microscope using a Plan Apo 60X (NA = 1.4) oil objective and Spectra X LED lamp (Lumencor) captured on an Orca-Flash 4.0 CMOS camera (Hamamatsu). A z-stack was obtained for each image and displayed as a maximum projection prior to image analysis. Image acquisition was completed using the NIS-Elements Advanced Research 4.30.02v 64-bit acquisition software (Nikon). Nuclear speckles were quantified in over 200 cells over three trials using an open-source speckle counting pipeline in CellProfiler (www.cellprofiler.org).
Dot blot assay for antibody validation
Various concentrations (from 25 to 1000 ng) of different synthetic N17 peptides (N17, N17S13p, N17S16p, and N17S13pS16p) were spotted onto a nitrocellulose membrane (Pall Life Sciences) and allowed to dry at room temperature for 45 min. Immunoblotting was carried out as described earlier.
Immunofluorescence peptide competition assay for antibody validation
The N17-phospho antibody (1:250) was incubated, with rotation, with 1000 ng of synthetic N17 peptides (N17, N17S13p, N17S16p, N17S13pS16p, and a control peptide, p53 [371–393]; New England Peptides) at room temperature for 1 h prior to overnight incubation with RPE1 cells fixed with methanol. Cells were washed three times with 2% FBS in PBS and then incubated in anti-rabbit AlexaFluor488 secondary antibody (1:500, Molecular Probes) for 45 min at room temperature and then washed and left in PBS before imaging using a Nikon TiEclipse inverted epifluorescence microscope.
Huntingtin siRNA knockdown for antibody validation
Endogenous huntingtin knockdown was established with huntingtin small interfering RNA (siRNA) (Santa Cruz; sc35617) in RPE1 cells. siRNA was transfected using Lipofectamine RNAiMax (Invitrogen) according to manufacturer instructions. Control dishes were transfected with scrambled siRNA. Protein was extracted using as described earlier, and 60 µg protein was loaded. Immunoblotting was carried out as described earlier.
Statistics
Data with a normal distribution were analyzed by unpaired Student’s t test unless otherwise stated. Error bars represent SEM.
Data availability
The raw data sets generated and analyzed for this study are available from the corresponding author on reasonable request.
Supplementary Material
Acknowledgments
We thank Raymond Wong of The Centre for Applied Genomics, The Hospital for Sick Children, Toronto, Canada, for assistance with G-banding karyotype analysis and Alina Lelic of the Human Immune Testing Suite, McMaster Immunology Research Centre, Hamilton, Canada, for assistance with flow cytometry experiments. This work was supported by the Canadian Institutes of Health Research, grant MOP-119391, the Krembil Foundation, and the Huntington Society of Canada.
Abbreviations used:
- 3NP
3-nitroproprionic acid
- ADP
adenosine diphosphate
- ANOVA
analysis of variance
- ATP
adenosine triphosphate
- BSA
bovine serum albumin
- CAA
cytosine-adenine-adenine
- CAG
cytosine-adenine-guanine
- CMOS
complementary metal oxide semiconductor
- DAPI
4′,6-diamidino-2-phenylindole
- DMSO
dimethyl sulfoxide
- ER
endoplasmic reticulum
- FBS
fetal bovine serum
- FITC
fluorescein isothiocyanate
- GWAS
genome-wide association studies
- HBSS
Hank’s balanced salt solution
- HD
Huntington’s disease
- HRP
horseradish peroxidase
- hTERT
human telomerase reverse transcriptase
- HTT
huntingtin gene
- iPSC
induced pluripotent stem cell
- N17
first 17 amino acids of huntingtin
- N17-phospho
N17 phosphorylated at serines 13 and 16
- PCA
principal component analysis
- PVDF
polyvinyl difluoride
- qPCR
quantitative PCR
- ROS
reactive oxygen species
- RPE1
retinal pigment epithelial cells
- S13 and S16
phosphorylated serines 13 and 16
- ST Hdh
mouse striatal-derived cell lines
- TBE
Tris-borate EDTA
- TRAP
telomeric repeat amplification protocol
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-09-0590) on September 26, 2018.
REFERENCES
- Acuña AI, Esparza M, Kramm C, Beltrán FA, Parra AV, Cepeda C, Toro CA, Vidal RL, Hetz C, Concha II, et al (2013). A failure in energy metabolism and antioxidant uptake precede symptoms of Huntington’s disease in mice. Nat Commun , 2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwal RS, Desmond CR, Caron N, Maiuri T, Xia J, Sipione S, Truant R. (2011). Kinase inhibitors modulate huntingtin cell localization and toxicity. Nat Chem Biol , 453–460. [DOI] [PubMed] [Google Scholar]
- Atwal RS, Truant R. (2008). A stress sensitive ER membrane-association domain in Huntingtin protein defines a potential role for Huntingtin in the regulation of autophagy. Autophagy , 91–93. [DOI] [PubMed] [Google Scholar]
- Atwal RS, Xia J, Pinchev D, Taylor J, Epand RM, Truant R. (2007). Huntingtin has a membrane association signal that can modulate huntingtin aggregation, nuclear entry and toxicity. Hum Mol Genet , 2600–2615. [DOI] [PubMed] [Google Scholar]
- Bae BI, Xu H, Igarashi S, Fujimuro M, Agrawal N, Taya Y, Hayward SD, Moran TH, Montell C, Ross CA, et al (2005). p53 mediates cellular dysfunction and behavioral abnormalities in Huntington’s disease. Neuron , 29–41. [DOI] [PubMed] [Google Scholar]
- Barkley DS, Hardiwidjaja S, Menkes JH. (1977). Abnormalities in growth of skin fibroblasts of patients with Huntington’s disease. Ann Neurol , 426–430. [DOI] [PubMed] [Google Scholar]
- Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu CP, Morin GB, Harley CB, Shay JW, Lichtsteiner S, Wright WE. (1998). Extension of life-span by introduction of telomerase into normal human cells. Science , 349–352. [DOI] [PubMed] [Google Scholar]
- Bowie LE, Maiuri T, Alpaugh M, Gabriel M, Arbez N, Galleguillos D, Hung CLK, Patel S, Xia J, Hertz NT, et al (2018). N6-Furfuryladenine is protective in Huntington’s disease models by signaling huntingtin phosphorylation. Proc Natl Acad Sci USA , E7081–E7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campalans A, Amouroux R, Bravard A, Epe B, Radicella JP. (2007). UVA irradiation induces relocalisation of the DNA repair protein hOGG1 to nuclear speckles. J Cell Sci , 23–32. [DOI] [PubMed] [Google Scholar]
- Debacq-Chainiaux F, Erusalimsky JD, Campisi J, Toussaint O. (2009). Protocols to detect senescence-associated beta-galactosidase (SA-βgal) activity, a biomarker of senescent cells in culture and in vivo. Nat Protoc , 1798. [DOI] [PubMed] [Google Scholar]
- del Hoyo P, García-Redondo A, de Bustos F, Molina JA, Sayed Y, Alonso-Navarro H, Caballero L, Arenas J, Jiménez-Jiménez FJ. (2006). Oxidative stress in skin fibroblasts cultures of patients with Huntington’s disease. Neurochem Res , 1103–1109. [DOI] [PubMed] [Google Scholar]
- DiGiovanni LF, Mocle AJ, Xia J, Truant R. (2016). Huntingtin N17 domain is a reactive oxygen species sensor regulating huntingtin phosphorylation and localization. Hum Mol Genet , 3937–3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, Medrano EE, Linskens M, Rubelj I, Pereira-Smith O. (1995). A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA , 9363–9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pardo A, Maglione V, Alpaugh M, Horkey M, Atwal RS, Sassone J, Ciammola A, Steffan JS, Fouad K, Truant R, et al (2012). Ganglioside GM1 induces phosphorylation of mutant huntingtin and restores normal motor behavior in Huntington disease mice. Proc Natl Acad Sci USA , 3528–3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dürrbaum M, Storchová Z. (2016). Effects of aneuploidy on gene expression: implications for cancer. FEBS J , 791–802. [DOI] [PubMed] [Google Scholar]
- Feng Z, Jin S, Zupnick A, Hoh J, de Stanchina E, Lowe S, Prives C, Levine AJ. (2006). p53 tumor suppressor protein regulates the levels of huntingtin gene expression. Oncogene , 1–7. [DOI] [PubMed] [Google Scholar]
- Ferris CF, Kulkarni P, Toddes S, Yee J, Kenkel W, Nedelman M. (2014). Studies on the Q175 knock-in model of Huntington’s disease using functional imaging in awake mice: evidence of olfactory dysfunction. Front Neurol , 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folch J, Junyent F, Verdaguer E, Auladell C, Pizarro JG, Beas-Zarate C, Pallàs M, Camins A. (2012). Role of cell cycle re-entry in neurons: a common apoptotic mechanism of neuronal cell death. Neurotox Res , 195–207. [DOI] [PubMed] [Google Scholar]
- Fritsche M, Haessler C, Brandner G. (1993). Induction of nuclear accumulation of the tumor-suppressor protein p53 by DNA-damaging agents. Oncogene , 307–318. [PubMed] [Google Scholar]
- Genetic Modifiers of Huntington’s Disease (GeM-HD) Consortium (2015). Identification of genetic factors that modify clinical onset of Huntington’s disease. Cell , 516–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz IE, Roberts E, Warren J. (1981). Skin fibroblasts in Huntington disease. Am J Hum Genet , 187–196. [PMC free article] [PubMed] [Google Scholar]
- Goldstein S, Singal DP. (1974). Senescence of cultured human fibroblasts: mitotic versus metabolic time. Exp Cell Res , 359–364. [DOI] [PubMed] [Google Scholar]
- Gray M, Shirasaki DI, Cepeda C, André VM, Wilburn B, Lu XH, Tao J, Yamazaki I, Li SH, Sun YE, et al (2008). Full-length human mutant huntingtin with a stable polyglutamine repeat can elicit progressive and selective neuropathogenesis in BACHD mice. J Neurosci , 6182–6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Greiner ER, Mishra R, Kodali R, Osmand A, Finkbeiner S, Steffan JS, Thompson LM, Wetzel R, Yang XW. (2009). Serines 13 and 16 are critical determinants of full-length human mutant huntingtin induced disease pathogenesis in HD mice. Neuron , 828–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall LL, Smith KP, Byron M, Lawrence JB. (2006). Molecular anatomy of a speckle. Anat Rec A Discov Mol Cell Evol Biol , 664–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayflick L. (1965). The limited in vitro lifetime of human diploid cell strains. Exp Cell Res , 614–636. [DOI] [PubMed] [Google Scholar]
- Hayflick L, Moorhead PS. (1961). The serial cultivation of human diploid cell strains. Exp Cell Res , 585–621. [DOI] [PubMed] [Google Scholar]
- HD iPSC Consortium (2012). Induced pluripotent stem cells from patients with Huntington’s disease show CAG-repeat-expansion-associated phenotypes. Cell Stem Cell , 264–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson JG, Agopyan N, Gutekunst CA, Leavitt BR, LePiane F, Singaraja R, Smith DJ, Bissada N, McCutcheon K, Nasir J, et al. (1999). A YAC mouse model for Huntington’s disease with full-length mutant huntingtin, cytoplasmic toxicity, and selective striatal neurodegeneration. Neuron , 181–192. [DOI] [PubMed] [Google Scholar]
- Hoffner G, Kahlem P, Djian P. (2002). Perinuclear localization of huntingtin as a consequence of its binding to microtubules through an interaction with beta-tubulin: relevance to Huntington’s disease. J Cell Sci , 941–948. [DOI] [PubMed] [Google Scholar]
- hTERT Immortalized Cell Lines (n.d.). Available at www.atcc.org/Products/Cells_and_Microorganisms/hTERT_Immortalized_Cell_Lines.aspx (accessed January 28, 2018).
- Huntington’s Disease Collaborative Research Group (1993). A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell , 971–983. [DOI] [PubMed] [Google Scholar]
- Jiang XR, Jimenez G, Chang E, Frolkis M, Kusler B, Sage M, Beeche M, Bodnar AG, Wahl GM, Tlsty TD, et al (1999). Telomerase expression in human somatic cells does not induce changes associated with a transformed phenotype. Nat Genet , 111–114. [DOI] [PubMed] [Google Scholar]
- Juopperi TA, Kim WR, Chiang C-H, Yu H, Margolis RL, Ross CA, Ming G-L, Song H. (2012). Astrocytes generated from patient induced pluripotent stem cells recapitulate features of Huntington’s disease patient cells. Mol Brain , 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keum JW, Shin A, Gillis T, Mysore JS, Abu Elneel K, Lucente D, Hadzi T, Holmans P, Jones L, Orth M, et al (2016). The HTT CAG-expansion mutation determines age at death but not disease duration in Huntington disease. Am J Hum Genet , 287–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk D, Parrington JM, Corney G, Bolt JM. (1977). Anomalous cellular proliferation in vitro associated with Huntington’s disease. Hum Genet , 143–154. [DOI] [PubMed] [Google Scholar]
- Küppers M, Ittrich C, Faust D, Dietrich C. (2010). The transcriptional programme of contact-inhibition. J Cell Biochem , 1234–1243. [DOI] [PubMed] [Google Scholar]
- Landles C, Weiss A, Franklin S, Howland D, Bates G. (2012). Caspase-6 does not contribute to the proteolysis of mutant huntingtin in the HdhQ150 knock-in mouse model of Huntington’s disease. PLoS Curr , e4fd085bfc9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry JJ, Pyl PT, Rausch T, Zichner T, Tekkedil MM, Stütz AM, Jauch A, Aiyar RS, Pau G, Delhomme N, et al (2013). The genomic and transcriptomic landscape of a HeLa cell line. G3 , 1213–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KM, Choi KH, Ouellette MM. (2004). Use of exogenous hTERT to immortalize primary human cells. Cytotechnology , 33–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemons JMS, Feng X-J, Bennett BD, Legesse-Miller A, Johnson EL, Raitman I, Pollina EA, Rabitz HA, Rabinowitz JD, Coller HA. (2010). Quiescent fibroblasts exhibit high metabolic activity. PLoS Biol , e1000514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CH, Tallaksen-Greene S, Chien WM, Cearley JA, Jackson WS, Crouse AB, Ren S, Li XJ, Albin RL, Detloff PJ. (2001). Neurological abnormalities in a knock-in mouse model of Huntington’s disease. Hum Mol Genet , 137–144. [DOI] [PubMed] [Google Scholar]
- Lin S, Coutinho-Mansfield G, Wang D, Pandit S, Fu X-D. (2008). The splicing factor SC35 has an active role in transcriptional elongation. Nat Struct Mol Biol , 819–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Kulesz-Martin M. (2001). p53 protein at the hub of cellular DNA damage response pathways through sequence-specific and non-sequence-specific DNA binding. Carcinogenesis , 851–860. [DOI] [PubMed] [Google Scholar]
- Lu M, Boschetti C, Tunnacliffe A. (2015). Long term aggresome accumulation leads to DNA damage, p53-dependent cell cycle arrest, and steric interference in mitosis. J Biol Chem , 27986–28000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiuri T, Mocle AJ, Hung CL, Xia J, van Roon-Mom WMC, Truant R. (2017). Huntingtin is a scaffolding protein in the ATM oxidative DNA damage response complex. Hum Mol Genet , 395–406. [DOI] [PubMed] [Google Scholar]
- Maiuri T, Woloshansky T, Xia J, Truant R. (2013). The huntingtin N17 domain is a multifunctional CRM1 and Ran-dependent nuclear and cilial export signal. Hum Mol Genet , 1383–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangiarini L, Sathasivam K, Seller M, Cozens B, Harper A, Hetherington C, Lawton M, Trottier Y, Lehrach H, Davies SW, et al (1996). Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell , 493–506. [DOI] [PubMed] [Google Scholar]
- Marcotte R, Lacelle C, Wang E. (2004). Senescent fibroblasts resist apoptosis by downregulating caspase-3. Mech Ageing Dev , 777–783. [DOI] [PubMed] [Google Scholar]
- Martinez Y, Dubois-Dauphin M, Krause K-H. (2012). Generation and applications of human pluripotent stem cells induced into neural lineages and neural tissues. Front Physiol , 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menalled LB, Kudwa AE, Miller S, Fitzpatrick J, Watson-Johnson J, Keating N, Ruiz M, Mushlin R, Alosio W, McConnell K, et al (2012). Comprehensive behavioral and molecular characterization of a new knock-in mouse model of Huntington’s disease: zQ175. PLoS One , e49838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menalled LB, Sison JD, Dragatsis I, Zeitlin S, Chesselet M-F. (2003). Time course of early motor and neuropathological anomalies in a knock-in mouse model of Huntington’s disease with 140 CAG repeats. J Comp Neurol , 11–26. [DOI] [PubMed] [Google Scholar]
- Menkes JH, Stein N. (1973). Fibroblast cultures in Huntington’s disease. N Engl J Med , 856–857. [DOI] [PubMed] [Google Scholar]
- Milakovic T, Johnson GVW. (2005). Mitochondrial respiration and ATP production are significantly impaired in striatal cells expressing mutant huntingtin. J Biol Chem , 30773–30782. [DOI] [PubMed] [Google Scholar]
- Mittelman D, Wilson JH. (2013). The fractured genome of HeLa cells. Genome Biol , 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales CP, Holt SE, Ouellette M, Kaur KJ, Yan Y, Wilson KS, White MA, Wright WE, Shay JW. (1999). Absence of cancer–associated changes in human fibroblasts immortalized with telomerase. Nat Genet , 115. [DOI] [PubMed] [Google Scholar]
- Muchowski PJ, Ning K, D’Souza-Schorey C, Fields S. (2002). Requirement of an intact microtubule cytoskeleton for aggregation and inclusion body formation by a mutant huntingtin fragment. Proc Natl Acad Sci USA , 727–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munsie L, Caron N, Atwal RS, Marsden I, Wild EJ, Bamburg JR, Tabrizi SJ, Truant R. (2011). Mutant huntingtin causes defective actin remodeling during stress: defining a new role for transglutaminase 2 in neurodegenerative disease. Hum Mol Genet , 1937–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munsie LN, Desmond CR, Truant R. (2012). Cofilin nuclear–cytoplasmic shuttling affects cofilin–actin rod formation during stress. J Cell Sci , 3977–3988. [DOI] [PubMed] [Google Scholar]
- Myers RH. (2004). Huntington’s disease genetics. NeuroRx , 255–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath S, Munsie LN, Truant R. (2015). A huntingtin-mediated fast stress response halting endosomal trafficking is defective in Huntington’s disease. Hum Mol Genet , 450–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble JR, Zhong Z-H, Neumann AA, Melki JR, Clark SJ, Reddel RR. (2004). Alterations in the p16(INK4a) and p53 tumor suppressor genes of hTERT-immortalized human fibroblasts. Oncogene , 3116–3121. [DOI] [PubMed] [Google Scholar]
- Ouellette MM, McDaniel LD, Wright WE, Shay JW, Schultz RA. (2000). The establishment of telomerase-immortalized cell lines representing human chromosome instability syndromes. Hum Mol Genet , 403–411. [DOI] [PubMed] [Google Scholar]
- Potapova TA, Zhu J, Li R. (2013). Aneuploidy and chromosomal instability: a vicious cycle driving cellular evolution and cancer genome chaos. Cancer Metastasis Rev , 377–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt HC, Schumacher B. (2012). The p53 network: cellular and systemic DNA damage responses in aging and cancer. Trends Genet , 128–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis SA, Thompson MN, Lee JM, Fossale E, Kim HH, Liao JK, Moskowitz MA, Shaw SY, Dong L, Haggarty SJ, et al (2011). Striatal neurons expressing full-length mutant huntingtin exhibit decreased N-cadherin and altered neuritogenesis. Hum Mol Genet , 2344–2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapp E, Valencia A, Li X, Aronin N, Kegel KB, Vonsattel J-P, Young AB, Wexler N, DiFiglia M. (2012). Native mutant huntingtin in human brain: evidence for prevalence of full-length monomer. J Biol Chem , 13487–13499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling G, Becher MW, Sharp AH, Jinnah HA, Duan K, Kotzuk JA, Slunt HH, Ratovitski T, Cooper JK, Jenkins NA, et al (1999). Intranuclear inclusions and neuritic aggregates in transgenic mice expressing a mutant N-terminal fragment of huntingtin. Hum Mol Genet , 397–407. [DOI] [PubMed] [Google Scholar]
- Schmidt MHM, Pearson CE. (2016). Disease-associated repeat instability and mismatch repair. DNA Repair , 117–126. [DOI] [PubMed] [Google Scholar]
- Schwarzacher HG, Schnedl W. (1965). Endoreduplication in human fibroblast cultures. Cytogenetics , 1–18. [DOI] [PubMed] [Google Scholar]
- Shaw PH. (1996). The role of p53 in cell cycle regulation. Pathol Res Pract , 669–675. [DOI] [PubMed] [Google Scholar]
- Shelbourne PF, Killeen N, Hevner RF, Johnston HM, Tecott L, Lewandoski M, Ennis M, Ramirez L, Li Z, Iannicola C, et al (1999). A Huntington’s disease CAG expansion at the murine Hdh locus is unstable and associated with behavioural abnormalities in mice. Hum Mol Genet , 763–774. [DOI] [PubMed] [Google Scholar]
- Singer E, Walter C, Weber JJ, Krahl A-C, Mau-Holzmann UA, Rischert N, Riess O, Clemensson LE, Nguyen HP. (2017). Reduced cell size, chromosomal aberration and altered proliferation rates are characteristics and confounding factors in the STHdh cell model of Huntington disease. Sci Rep , 16880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slow EJ, van Raamsdonk J, Rogers D, Coleman SH, Graham RK, Deng Y, Oh R, Bissada N, Hossain SM, Yang YZ, et al (2003). Selective striatal neuronal loss in a YAC128 mouse model of Huntington disease. Hum Mol Genet , 1555–1567. [DOI] [PubMed] [Google Scholar]
- Smith ML, Chen IT, Zhan Q, O’Connor PM, Fornace AJ., Jr (1995). Involvement of the p53 tumor suppressor in repair of u.v.-type DNA damage. Oncogene , 1053–1059. [PubMed] [Google Scholar]
- Southwell AL, Smith-Dijak A, Kay C, Sepers M, Villanueva EB, Parsons MP, Xie Y, Anderson L, Felczak B, Waltl S, et al (2016). An enhanced Q175 knock-in mouse model of Huntington disease with higher mutant huntingtin levels and accelerated disease phenotypes. Hum Mol Genet , 3654–3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telenius H, Kremer B, Goldberg YP, Theilmann J, Andrew SE, Zeisler J, Adam S, Greenberg C, Ives EJ, Clarke LA. (1994). Somatic and gonadal mosaicism of the Huntington disease gene CAG repeat in brain and sperm. Nat Genet , 409–414. [DOI] [PubMed] [Google Scholar]
- Trettel F, Rigamonti D, Hilditch-Maguire P, Wheeler VC, Sharp AH, Persichetti F, Cattaneo E, MacDonald ME. (2000). Dominant phenotypes produced by the HD mutation in STHdh (Q111) striatal cells. Hum Mol Genet , 2799–2809. [DOI] [PubMed] [Google Scholar]
- Van Raamsdonk JM, Gibson WT, Pearson J, Murphy Z, Lu G, Leavitt BR, Hayden MR. (2006). Body weight is modulated by levels of full-length huntingtin. Hum Mol Genet , 1513–1523. [DOI] [PubMed] [Google Scholar]
- Vaziri H, Benchimol S. (1998). Reconstitution of telomerase activity in normal human cells leads to elongation of telomeres and extended replicative life span. Curr Biol , 279–282. [DOI] [PubMed] [Google Scholar]
- Victor MB, Richner M, Hermanstyne TO, Ransdell JL, Sobieski C, Deng P-Y, Klyachko VA, Nerbonne JM, Yoo AS. (2014). Generation of human striatal neurons by microRNA-dependent direct conversion of fibroblasts. Neuron , 311–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victor MB, Richner M, Olsen HE, Lee SW, Monteys AM, Ma C, Huh CJ, Zhang B, Davidson BL, Yang XW, et al (2018). Striatal neurons directly converted from Huntington’s disease patient fibroblasts recapitulate age-associated disease phenotypes. Nat Neurosci , 341–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal RL, Figueroa A, Court FA, Thielen P, Molina C, Wirth C, Caballero B, Kiffin R, Segura-Aguilar J, Cuervo AM, et al (2012). Targeting the UPR transcription factor XBP1 protects against Huntington’s disease through the regulation of FoxO1 and autophagy. Hum Mol Genet , 2245–2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Waarde-Verhagen MAWH, Kampinga HH, Linskens MHK. (2006). Continuous growth of telomerase-immortalised fibroblasts: how long do cells remain normal? Mech Ageing Dev , 85–87. [DOI] [PubMed] [Google Scholar]
- Walen KH. (2006). Human diploid fibroblast cells in senescence; cycling through polyploidy to mitotic cells. In Vitro Cell Dev Biol Anim , 216–224. [DOI] [PubMed] [Google Scholar]
- Wang S, El-Deiry WS. (2007). p53, cell cycle arrest and apoptosis. In: 25 Years of p53 Research, Dordrecht, Netherlands: Springer, 141–163. [Google Scholar]
- Warner JP, Barron LH, Brock DJ. (1993). A new polymerase chain reaction (PCR) assay for the trinucleotide repeat that is unstable and expanded on Huntington’s disease chromosomes. Mol Cell Probes , 235–239. [DOI] [PubMed] [Google Scholar]
- Wood LD, Halvorsen TL, Dhar S, Baur JA, Pandita RK, Wright WE, Hande MP, Calaf G, Hei TK, Levine F, et al (2001). Characterization of ataxia telangiectasia fibroblasts with extended life-span through telomerase expression. Oncogene , 278–288. [DOI] [PubMed] [Google Scholar]
- Wyttenbach A, Swartz J, Kita H, Thykjaer T, Carmichael J, Bradley J, Brown R, Maxwell M, Schapira A, Orntoft TF, et al (2001). Polyglutamine expansions cause decreased CRE-mediated transcription and early gene expression changes prior to cell death in an inducible cell model of Huntington’s disease. Hum Mol Genet , 1829–1845. [DOI] [PubMed] [Google Scholar]
- Xue Y, Ouyang K, Huang J, Zhou Y, Ouyang H, Li H, Wang G, Wu Q, Wei C, Bi Y, et al (2013). Direct conversion of fibroblasts to neurons by reprogramming PTB-regulated microRNA circuits. Cell , 82–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo AS, Sun AX, Li L, Shcheglovitov A, Portmann T, Li Y, Lee-Messer C, Dolmetsch RE, Tsien RW, Crabtree GR. (2011). MicroRNA-mediated conversion of human fibroblasts to neurons. Nature , 228–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young ATL, Lakey JRT, Moore RB. (2004). Transient telomerase expression in normal somatic cells leads to telomere extension and increased proliferation in the absence of malignant transformation. Transplantation , 108. [Google Scholar]
- Zhang Y, Pak C, Han Y, Ahlenius H, Zhang Z, Chanda S, Marro S, Patzke C, Acuna C, Covy J, et al (2013). Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron , 785–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data sets generated and analyzed for this study are available from the corresponding author on reasonable request.