Abstract
In yeast, protein kinase A (PKA) adjusts transcriptional profiles, metabolic rates, and cell growth in accord with carbon source availability. PKA affects gene expression mostly via the transcription factors Msn2 and Msn4, two key regulators of the environmental stress response. Here we analyze the role of the PKA-Msn2 signaling module using an Msn2 allele that harbors serine-to-alanine substitutions at six functionally important PKA motifs (Msn2A6). Expression of Msn2A6 mimics low PKA activity, entails a transcription profile similar to that of respiring cells, and prevents formation of colonies on glucose-containing medium. Furthermore, Msn2A6 leads to high oxygen consumption and hence high respiratory activity. Substantially increased intracellular concentrations of several carbon metabolites, such as trehalose, point to a metabolic adjustment similar to diauxic shift. This partial metabolic switch is the likely cause for the slow-growth phenotype in the presence of glucose. Consistently, Msn2A6 expression does not interfere with growth on ethanol and tolerated is to a limited degree in deletion mutant strains with a gene expression signature corresponding to nonfermentative growth. We propose that the lethality observed in mutants with hampered PKA activity resides in metabolic reprogramming that is initiated by Msn2 hyperactivity.
INTRODUCTION
Cells constantly monitor their environments and translate information based on a variety of extracellular parameters into adjusted rates of gene expression and metabolic fluxes. Growth rates of the yeast Saccharomyces cerevisiae are mainly determined by complex regulatory networks centered on the preferential utilization of glucose as a carbon source (Schüller, 2003; Zaman et al., 2008; Conrad et al., 2014). Fluctuations in glucose availability result in characteristic gene expression patterns affecting the transcription of thousands of genes, many involved in ribosome biogenesis and carbohydrate metabolism (DeRisi et al., 1997; Gasch et al., 2000; Causton et al., 2001; Brauer et al., 2005; Zid and O’Shea, 2014). The glucose-sensing network uses several signaling pathways to achieve optimal use of the preferred carbon source and regulated switching to other carbon sources (Galdieri et al., 2010). The main protein kinases involved are the protein kinase A (PKA), the AMP-activated kinase (AMPK) Snf1, the target of rapamycin kinase complex 1 (TORC1), and the Greatwall-family protein kinase Rim15. PKA plays a prominent role in regulation of transcription of almost all genes that change expression concordant with glucose availability (Wang et al., 2004; Slattery et al., 2008; Zaman et al., 2009), whereas AMPK Snf1 regulates most of the PKA-independent genes (Zaman et al., 2009). Two major types of gene expression pattern are present in mutants lacking genes for various components of the carbon-sensing network (Apweiler et al., 2012). Mutations leading to lower PKA activity, such as deletions of genes regulating intracellular cAMP levels (e.g., the GTP-binding protein Ras2, the guanine nucleotide exchange factor Cdc25, or adenylate cyclase Cyr1), exhibit a gene expression profile similar to that of wild-type cells experiencing low glucose availability. Mutants with high PKA activity, such as strains lacking BCY1 (encoding the regulatory subunit of PKA), express a reciprocal pattern resembling growth in glucose-rich medium.
PKA affects the global gene expression pattern mostly via the transcription factor Msn2 and its paralog Msn4, the main regulators of stress response and diauxic shift in budding yeast (Martinez-Pastor et al., 1996; Boy-Marcotte et al., 1998; Gasch et al., 2000; Elfving et al., 2014). Depending on the gene context, the function of Msn4 as a transcription factor is similar to that of Msn2 (AkhavanAghdam et al., 2016). Both proteins cycle constantly between cytoplasm and nucleus, reflecting an equilibrium achieved by regulated import and export processes (Görner et al., 2002; Jacquet et al., 2003; Durchschlag et al., 2004; Garmendia-Torres et al., 2007; Logg et al., 2009; Kuang et al., 2017; Chatterjee and Acar, 2018). Nuclear export of Msn2 requires the interaction of a nuclear export sequence (NES) with the exportin Msn5. This NES is activated by phosphorylation of key residues and inactivated by stress conditions (Durchschlag et al., 2004). Nuclear import of Msn2 is regulated by two nuclear localization sequences (NLS); one is PKA-regulated and requires interaction with the karyopherin Kap123, whereas the other is constitutive, importin α-dependent (Srp1), and closely associated to the protein’s zinc-finger DNA-binding domain (Görner et al., 2002; Garmendia-Torres et al., 2007). PKA-dependent phosphorylation of multiple residues of Msn2 restricts nuclear import and activation (Görner et al., 2002; Reiter et al., 2013). Six of these sites are embedded in previously described functionally important regions of Msn2 (Görner et al., 1998, 2002; Hao et al., 2013; Reiter et al., 2013), namely S288 of the NES (Görner et al., 1998; Hao et al., 2013), S582, S620, S625, and S633 of the NLS (Görner et al., 1998, 2002; Hao et al., 2013), and S686 of the zinc-finger DNA-binding domain (Reiter et al., 2013). Owing to its easily observable activation properties, Msn2 has become a powerful tool for studying mechanisms of transcriptional regulation in a variety of single cell studies (Garmendia-Torres et al., 2007; Cai et al., 2008; Capaldi et al., 2008; Logg et al., 2009; Hao and O’Shea, 2012; Hao et al., 2013; Hansen and O’Shea, 2013, 2015, 2016; Hansen et al., 2015; Elfving et al., 2014; Lin et al., 2015; Chatterjee and Acar, 2018).
Msn2 and Msn4 exhibit a dual role in activating carbohydrate metabolism genes and stress response genes (Kuang et al., 2017). Msn2/4 activation by most physicochemical stress types is not completely understood. The Msn2 phosphorylation pattern changes transiently in stressed cells. Only conditions that negatively affect PKA activity, such as acute glucose starvation and exposure to H2O2, cause dramatic dephosphorylation of PKA sites (Reiter et al., 2013; Bodvard et al., 2017). Moreover, constitutive nuclear localization of the transcription factor (e.g., by elimination of its nuclear export) does not lead to a corresponding transcriptional output (Durchschlag et al., 2004), even though induced nuclear accumulation of Msn2 is usually correlated with stress-induced transcription. Thus, additional mechanisms are needed to activate Msn2. Posttranslational activation might be caused by modifications apart from phosphorylation (Gallmetzer et al., 2015) or interaction with additional stress-dependent transcription factors.
Low PKA activity delays the cell cycle (Amigoni et al., 2015). Triple deletion of TPK1, TPK2, and TPK3, encoding the catalytic subunits of PKA, or absence of cAMP causes cell cycle arrest in G1 (Toda et al., 1987; Baroni et al., 1994; Tokiwa et al., 1994). Growth defects caused by PKA depletion can be suppressed by eliminating MSN2 and MSN4 (Smith et al., 1998), suggesting that cell cycle progression might be negatively affected by the Msn2/4-mediated transcriptional response (e.g., by induction of a cell cycle regulator). A factor suggested to be involved is the serine–threonine protein kinase Yak1, since activation of Msn2/4 induces YAK1 expression and a yak1∆ deletion rescues a tpk1∆tpk2∆tpk3∆ mutant lacking all catalytic PKA subunits (Garrett and Broach, 1989; Lee et al., 2008). Yak1 activity has been connected to G1 cyclin regulation. However, the network downstream of PKA is complex as a result of the combined effects of PKA activity on transcription, translation, and posttranslational modification patterns of kinase substrates.
Here we sought to examine the consequences of the activation of the PKA-Msn2 signaling module. Expression of a constitutively active Msn2 allele harboring alanine substitutions at six PKA consensus sites (Msn2A6) was shown to cause growth inhibition. Using conditional expression of the Msn2A6 mutant, we determined gene expression characteristics, as well as the metabolic state in the respective mutant. Our results suggest that prevention of phosphorylation and thus constitutive activation of Msn2 is sufficient to induce a partial shift of metabolism toward glucose exhaustion (diauxic shift), a condition in which yeast cells are adjusted to respiratory metabolism and to utilizing ethanol as their primary carbon source. We propose that the growth arrest caused by PKA depletion results from a maladjusted metabolic state. Our data support the role of the transcription factor Msn2 as a key determinant of metabolic adaptation to carbon sources.
RESULTS
Msn2-related proteins share conserved functional regions
In budding yeast, PKA deficiency is lethal and can be rescued by inactivation of both MSN2 and MSN4 (Smith et al., 1998). PKA-driven phosphorylation of Msn2 at several key regulatory sites represses its activity, suggesting that Msn2/4-induced transcription is a major signaling output downstream of PKA (Zaman et al., 2009). If this is the case, an Msn2 allele mimicking the dephosphorylated state and thus absence of PKA-dependent phosphorylation should result in a growth phenotype similar to the absence of PKA. To be able to test this, we first wanted to determine all direct and indirect PKA-responsive domains of the transcription factor required for constructing a constitutively active, kinase-unresponsive allele.
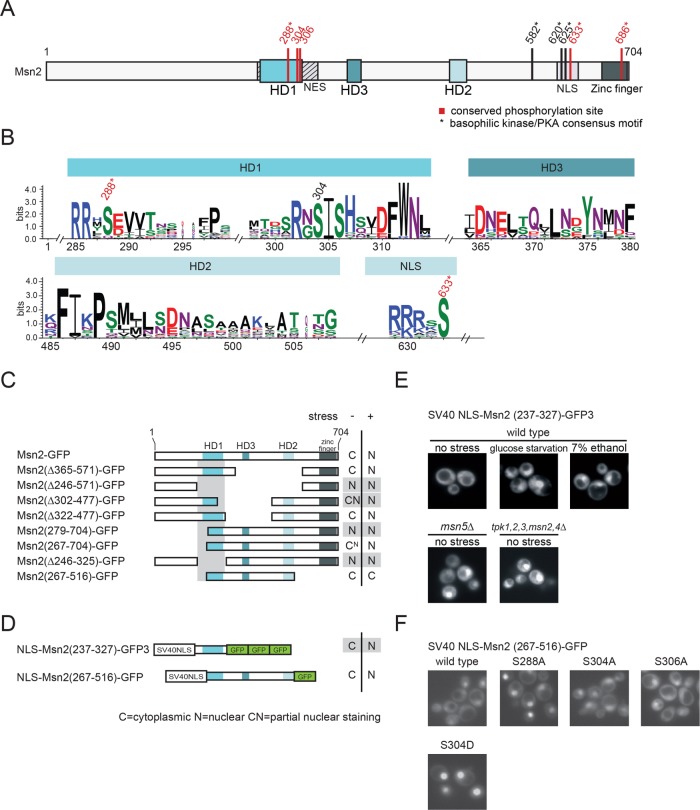
To identify functional and regulatory determinants within Msn2, we performed a similarity footprinting analysis to define conserved domains of the protein. Numerous recently published genome sequences allowed us to screen fungal species for Msn2-related transcription factors. In total, we identified 22 Msn2 and 6 Msn4 orthologues sharing three short conserved sequences, designated homology domains HD1–3 (Figure 1, A and B; Supplemental Figure S1). HD1 and HD2 of S. cerevisiae Msn2, encompassing aa 259–314 and aa 485–508, respectively, have been described previously (Görner et al., 1998). In addition, we identified a third conserved region (HD3) encompassing aa 364–380. Notably, of the three homology domains, only HD1 overlaps with a previously established functionally important region of Msn2, as it is located within the NES of the protein (Görner et al., 1998). Within HD1, we observed a high degree of conservation of an acidic stretch followed by a PKA consensus motif (RRxS288) and a distinctive motif (S304IS306HxxDFW).
Figure 1:
Functional analysis of conserved regions of Msn2-related proteins. (A) Scheme of S. cerevisiae Msn2. Locations of the homology domains HD1 and HD2 and the newly identified HD3, as well as the NES, the NLS, and the zinc-finger DNA-binding domain are indicated. Conserved phosphorylation sites highlighted in red. * indicates a canonical PKA consensus motif (R/K-R/K-X-S/T). (B) Similarity plot of 19 Msn2 and 5 Msn4 homologues. Only results obtained for HD1, HD2, HD3 and a highly conserved phosphoaccepting residue (S633) of the NLS are shown. Conserved phosphorylation sites are highlighted. (C–F) The Msn2 region, comprising residues 237–327, constitutes a stress– and PKA signaling–regulated minimal NES. (C, D) Localization analysis of a series of Msn2-GFP truncations and derivatives. W303 msn2∆ msn4∆ cells carrying plasmids expressing Msn2-GFP derivatives were grown to logarithmic phase (–) and subsequently stressed with 7.5% ethanol (+). N: a nuclear GFP signal. C: cytoplasmic. CN and CN: partial nuclear staining. (E) Msn2 residues 237–327 were fused to the SV40 NLS and three tandem GFP units. A variety of physicochemical stresses (here 7% ethanol or glucose starvation) promote accumulation of SV40 NLS-Msn2(237-327)-GFP in the nucleus. Nucleocytoplasmic transport of the fusion protein also requires Msn5 and PKA signaling. (F) Effects of serine–to–alanine or –aspartic acid substitutions at conserved residues of the Msn2 NES.
We also observed conservation of a single PKA consensus motif within a region corresponding to the S. cerevisiae Msn2-NLS upstream of the DNA binding domain (Figure 1, A and B). It is noteworthy that in S. cerevisiae Msn2 this site seems to be expanded to three PKA consensus motifs, all of which have been connected to regulation of nuclear import (Görner et al., 2002). Also, the closely located PKA/AMPK motif (corresponding to S582 in S. cerevisiae Msn2) (De Wever et al., 2005) and the PKA consensus motif within the zinc-finger domain (S686) are well conserved in all Msn2/4-related sequences (Supplemental Figures S1 and S2).
In summary, several PKA consensus sites close to or embedded within functional regions are highly conserved between the Msn2 orthologues of different fungal species.
The Msn2NES is sufficient for Msn2 nuclear export
We next tested whether the three HD domains are of functional relevance. PKA-mediated activation of the NES might be a major determinant for intracellular distribution of the protein during exponential growth (Görner et al., 1998). To functionally identify the region required for Msn2 export regulation, we performed a deletion analysis using truncated variants or internal deletion mutants of the transcription factor fused to one or multiple copies of GFP (Figure 1, C and D). Msn2 derivatives truncated for the first 279 amino acids showed nuclear staining (truncation of the first 267 aa led to a faint nuclear staining) in unstressed cells, suggesting a partly defective NES (assuming continuous nuclear import). A region encompassing aa 267–322 is apparently essential for nuclear export regulation. Msn2 constructs with internal deletions eliminating the HD2 (Msn2-(Δ365-571)-GFP) and HD3 (Msn2-(Δ322-477)-GFP) showed no apparent localization defects.
We set out to define a minimal sequence sufficient for Msn2 export regulation. We created protein fusions of internal Msn2 sequences with the unregulated heterologous SV40 NLS. In addition, we used three GFP units to increase the size of the fusion proteins to prevent passive diffusion through the nuclear pore. In the context of the constitutive SV40 NLS, we defined a region of Msn2 (aa 237–327) showing sufficient nuclear export activity to counteract the SV40 NLS (Figure 1, D and E). A protein fusion of the SV40 NLS with the Msn2 sequence from 267–516, encompassing all three of HD1–3, showed localization behavior more similar to that of wild-type Msn2 (Figure 1, D and F). In response to stress or glucose starvation, the SV40 NLS-Msn2-237-327-3GFP protein accumulated rapidly in the nucleus (Figure 1E). In addition, cytoplasmic distribution of this fusion protein was dependent on PKA activity and on the exportin Msn5 (Figure 1E). Taking these results together, we defined a minimal sequence that mediates PKA- and stress-regulated Msn2 NES activity. Other conserved elements (including HD2 and HD3) might possibly be required to fine tune nuclear export control; however, our deletion analysis did not support a decisive function of these elements.
Phosphomimetic mutants suggest a PKA- and stress-regulated NES
All conserved PKA consensus sites of S. cerevisiae Msn2 have previously been shown to be phosphorylated in vivo and have also been functionally characterized (Görner et al., 1998, 2002; Durchschlag et al., 2004; De Wever et al., 2005; Hao et al., 2013; Reiter et al., 2013; Bodvard et al., 2017). Within HD1, S304 and S308 (and probably also S306) have been found as glucose starvation responsive phosphorylation sites, even though they do not fit the canonical R/K-R/K-X-S/T PKA consensus motif (Hao et al., 2013; Reiter et al., 2013).
In the context of NLS-Msn2-267-516-GFP, serine-to-alanine substitutions at either position 288 or 304 conferred predominant nuclear localization. However, we found that the phosphorylation-mimetic mutation S304D (Figure 1F) also abrogates NES function. Thus, S304, similarly to S288 (Görner et al., 1998), is required for the integrity of the NES; however, phosphorylation has not been demonstrated to have a regulatory role. Furthermore, in the context of full-length Msn2, a combinatorial S288A/S304A mutation showed constitutive nuclear accumulation, indicating severe down-regulation of NES activity when both residues are not phosphorylated (Hao et al., 2013). Hence, phosphorylation of the conserved sites of HD1 is a prerequisite for Msn2 NES activity, which is in accordance with previous reports (Görner et al., 1998, 2002; Hao et al., 2013; Bodvard et al., 2017).
To uncouple Msn2 from PKA activity, we initially combined point mutations at five PKA sites, namely S288, S582, S620, S625, and S633. We generated substitutions with aspartic acid (Msn2D5) or alanine (Msn2A5). Msn2D5-GFP localized to the cytoplasm under unstressed conditions and rapidly accumulated in the nucleus upon exposure to various stress types such, as heat shock, hyperosmotic shock, and ethanol stress (Supplemental Figure S3A). Moreover, we observed nuclear accumulation of Msn2D5-GFP in response to low PKA activity (Supplemental Figure S3B), suggesting that PKA either targets atypical motifs (such as S304) or regulates Msn2 localization via an additional indirect mechanism. In an msn5∆ mutant strain, Msn2D5-GFP showed a constitutively nuclear distribution, confirming that the phosphomimetic mutations did not interfere with the exportin–cargo interaction. The observation that stress conditions seem to interfere with the Msn2-NES function despite the phosphorylation-mimetic mutations shows that stress and PKA partially regulate Msn2 nuclear export in parallel. Consistently, the dephosphorylation-mimetic mutant Msn2A5-GFP showed constitutive nuclear localization, indicating constitutive inactivation of the NES (Figure S3A and Durchschlag et al., 2004). A similar constitutive nuclear localization of Msn2A6-GFP has been noted by us (unpublished data) and observed by life microscopy (Elfving et al. 2014; Bodvard et al. 2017). Therefore, absence of PKA activity overrides the Msn2-NES function, suggesting that direct modification via phosphorylation sites is a main way of action for PKA and that these sites are sufficient for creation of a dominant active MSN2 allele.
Mutation of six Msn2 PKA-targeted serines to alanine is detrimental for growth
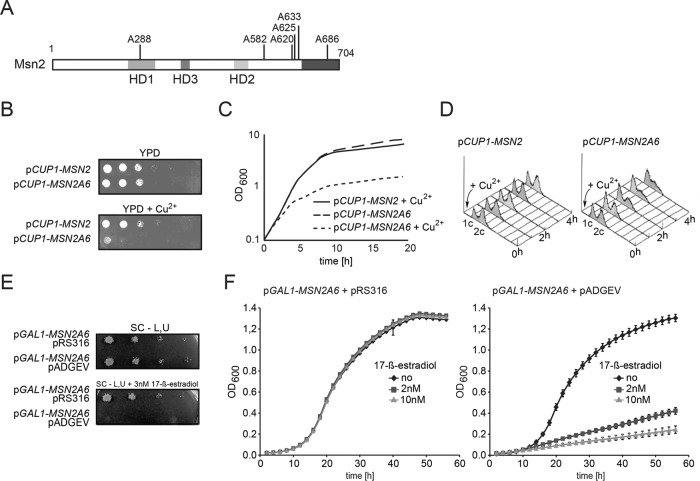
Next, we created an Msn2 mutant (designated as Msn2A6) harboring serine-to-alanine substitutions at the six functionally important PKA motifs, namely S288 of the NES, S582, S620, S625, S633 of the NLS and S686 of the DNA binding domain (Figure 2A). Most key PKA target residues of this allele can no longer be directly modified. Hence, cells expressing Msn2A6 should develop a phenotype similar to a PKA-deficient strain. Indeed, we could not obtain viable transformants expressing Msn2A6 driven by the ADH1 promoter. We constructed a conditional MSN2A6 allele under the control of the inducible CUP1 promoter. Expression of MSN2A6 was stimulated with low concentrations of copper sulfate. Growth of cells expressing CUP1-regulated MSN2 or MSN2A6 was monitored by optical density (OD600nm) measurements, drop tests, and fluorescence-activated cell sorting (FACS) analysis (Figure 2, B–D). Msn2A6-expressing cells showed a severe impairment of growth compared with cells expressing the wild-type form of the protein (Figure 2, B and C), suggesting that the six serine-to-alanine substitutions sufficiently prevent integration of PKA signaling. To address the type of arrest caused by Msn2A6, expression of MSN2 and MSN2A6 was induced in liquid cultures and analyzed for DNA content by FACS. We observed an increasing number of cells with 1c DNA content, indicating a prolonged G1/S phase of the cell cycle (Figure 2D).
Figure 2:
Mutation of 6 Msn2 PKA–targeted serines to alanine is detrimental for growth. (A) Scheme of Msn2A6. Induced expression of Msn2A6 by a copper-regulated promoter (CUP1) represses growth on (B) solid medium and in (C) liquid culture (50 µM CuSO4). (D) FACS analysis of the terminal phenotype of arrested Msn2A6 cells. Expression of CUP1-MSN2 and CUP1-MSN2A6 was induced in W303 msn2∆msn4∆ cells by addition of CuSO4 (6.25 µM) to liquid cultures at an OD600nm of 0.7. Cells were grown further to an OD600nm of 1 (first time point T = 0) and kept in logarithmic growth by continuously diluting the culture with prewarmed media containing copper sulfate. At four additional time points (T = 1–4 h), samples were taken. An increasing number of cells expressing Msn2A6 are in the G1/S phase of the cell cycle. Induced expression of Msn2A6 in W303 msn2∆msn4∆ cells by a 17-ß-estradiol–induced system represses growth on (E) solid medium and in (F) liquid culture, showing a transition from exponential to linear growth.
We additionally constructed a GAL1-10 promoter–driven version of MSN2A6 that can be induced by 17-ß-estradiol in strains carrying the ADGEV construct (Louvion et al., 1993). ADGEV is an ADH1 promoter–driven human estradiol receptor fused to the Gal4 DNA–binding domain and to the VP16 activation domain. A similar 17-ß-estradiol–triggered conditional expression system of Msn2A6 based on Z4EV was reported earlier (Elfving et al., 2014). In contrast to the ADGEV GAL1-10 promoter–based expression, Z4EV has no target in the yeast genome and thus expression of the GAL genes is prevented. However, it has previously been established that ADGEV activation is biologically inert at the used concentration of 17-ß-estradiol and does not lead a growth effect, as observed with Msn2A6–expressing cells (O’Duibhir et al., 2014). Drop tests on plates containing 3 nM 17-ß-estradiol showed severe reduction of growth of the Msn2A6–expressing cells as opposed to control cells, without the ADGEV construct where the Msn2A6 allele is not estradiol–inducible (Figure 2E). Growth curves in the presence of 2 or 10 nM 17-ß-estradiol measured over 56 h showed that expression of Msn2A6 leads to a rapid and dose-dependent shift from exponential to very slow linear growth (Figure 2F). This indicates that Msn2A6-expressing cells do not become arrested for growth, but rather that their cell cycle is dramatically slowed.
The Msn2A6 expression profile deviates from glucose-grown cells
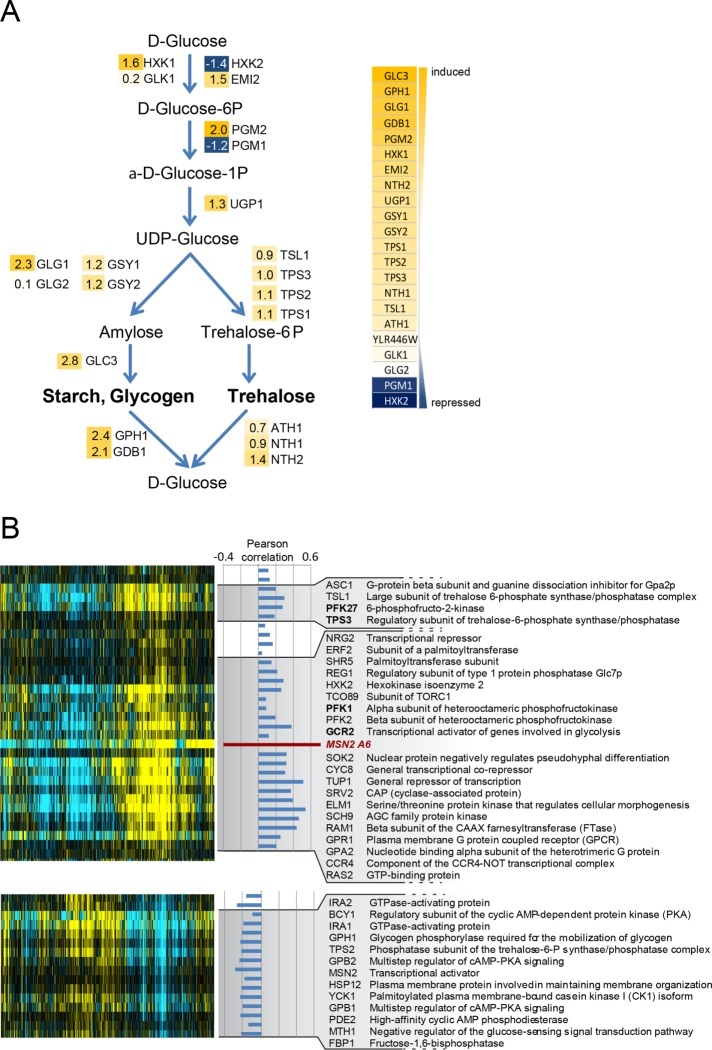
To address the basis of the growth-inhibitory effect observed in Msn2A6-expressing cells, we determined the global transcription pattern caused by induction of the allele. To minimize the impact of indirect effects, a short but effective treatment had to be applied. Therefore, we initially analyzed the Msn2A6-induced up-regulation of Catalase T (CTT1) (Wieser et al., 1991; Marchler et al., 1993) transcripts by Northern analysis and found that treatment with 17-ß-estradiol for 90 min resulted in substantial induction. Using microarrays, we identified 194 induced and 338 repressed genes following MSN2A6 expression for 90 min (Figure 3A and Supplemental Figure S4; Supplemental Data File S1). The functional analysis highlighted the induction of hallmark genes of the Msn2/4 regulon, such as HXK1, ALD3, DDR2, and CTT1. Genes involved in glucose metabolism showed a mixed pattern. Some hexose transporter genes were repressed (HXT1, HXT2), while others (HXT5, HXT6, and HXT7) were induced. Expression of genes encoding enzymes involved in early and rate-limiting steps of glycolysis, namely glucose-6-phosphate isomerase 1 (PGI1) and phosphofructo-kinase 1 and 2 (PFK1 and PFK2), was repressed. Also, HXK2, encoding the predominant hexokinase during growth on glucose, and PGM1, encoding phosphoglucomutase, were repressed, indicating a change in glycolytic flux in Msn2A6-expressing cells. In addition, we observed that Msn2A6 expression induced several genes involved in glycogen metabolism. For example, GPH1, GDB1, and GIP2, the gene products of which catalyze glycogen degradation, but also several genes required for glycogen synthesis, such as GLC3 (glycogen branching enzyme), GLG1 (glycogenin glucosyltransferase; Cheng et al., 1995), and GAC1 (required for tethering Glc7 to glycogen synthase Gsy2), as well as two genes required for glycerol utilization (DAK1 and GUT2), were found to be up-regulated. Gene ontology (GO) analysis pointed to significant expression changes of genes involved in hexose metabolism. Several of these genes were clustered in the glycogen and trehalose metabolism (Figure 3A). Expression levels of genes involved in glycolysis, as well as citric acid cycle and oxidative phosphorylation components, were not generally affected with the exception of the succinate dehydrogenase subunits (SDH1, SDH2, SDH3, SDH4, SHH3, SHH4, YJL045W). Expression of genes of the gluconeogenesis pathway and the pentose phosphate cycle enzymes was induced (Supplemental Figure S4). Key steps of the pentose phosphate cycle, transaldolase (NQM1), phosphogluconate dehydrogenase (GND2), transketolase (TKL2), and 6-phosphogluconolactonase (SOL4), showed increased expression levels.
Figure 3:
The Msn2A6 expression profile resembles that of glucose-starved cells. (A) Expression changes of glycogen and trehalose synthesis genes (log2 values indicated). (B) Microarray analysis of W303 msn2∆msn4∆ cells expressing Msn2A6 under control of the GAL1-10 promoter driven by the 17-ß-estradiol–inducible ADGEV transcription factor. The normalized Msn2A6 expression profile was aligned by hierarchical clustering to a compendium of microarrays derived from mutants involved in glucose metabolism and signaling. Pearson´s correlation coefficient (PC) values are indicated between the Msn2A6 expression profile and the respective genes; the most significant correlated and anti-correlated genes are selected. The full description and data represented in the figure are provided as supplemental files (Supplemental Figure S4; Supplemental Data File S2).
Taken together, the observed transcriptional pattern fits to cells with a decreased rate of glucose uptake, utilization, or sensing. In addition, we observed strong repression of genes involved in amino acid and protein biosynthesis, which might be the cause or a consequence of reduced growth rates. We concluded that the observed expression pattern probably represents a combination of direct induction of transcripts caused by Msn2A6 and indirect regulation as a consequence of the slow-growth effect on global transcription.
We further analyzed the Msn2A6 expression pattern in the context of previously published profiles (Apweiler et al., 2012; O’Duibhir et al., 2014). The microarray data were downloaded from the Holstege Lab website (http://deleteome.holstegelab.nl/) and used for hierarchical clustering with the normalized Msn2A6 expression data (Figure 3B; Supplemental Figure S5; Supplemental Data File S2). Interestingly, the Msn2A6-induced expression pattern was clustering with profiles obtained from deletion mutants of genes influencing the activity of the PKA pathway (Figure 3B; Supplemental Figure S5), such as Srv2, which binds the adenylate cyclase Cyr1 and facilitates its activation by Ras1/2. Shr5 and Erf2 (palmitoyltransferase subunits) and also the beta subunit of the CAAX farnesyltransferase Ram1 had similar expression patterns and are required for activity of Ras1/2. Furthermore, the expression patterns of mutants lacking GPR1 and GPA2, encoding nutrient response factors (Zaman et al., 2008), showed a positive correlation to the Msn2A6-regulated set (Figure 3B; Supplemental Figure S5). Gpa2 and Ras2 work in parallel (via activation of PKA) in mediating a transcriptional response to glucose (Xue et al., 1998; Wang et al., 2004). Also present in this cluster was Sch9, the orthologue of mammalian S6 kinase, which is part of a nutrient-sensing pathway in parallel to the PKA signaling pathway (Fabrizio et al., 2001; Wang et al., 2004), Ccr4, a component of the Ccr4-Not complex that has previously been connected to a broad variety of cellular processes and has been suggested to activate Msn2-dependent genes even under high-PKA conditions (Lenssen et al., 2002, 2005; Wang et al., 2004) and the genes encoding the general repressor complex Tup1-Cyc8, necessary for repression of glucose-repressed genes by Mig1 (Treitel and Carlson, 1995). Mutants lacking PFK1 and PFK2 as well as PFK27 (phosphofructo-2 kinase) also expressed a profile corresponding to low glucose availability. Other mutants lacking BCY1, IRA1/2, MSN2, and PDE2 had a profile reflecting high glucose growth and inverse to that of Msn2A6-expressing cells. In summary, the expression profile evoked by Msn2A6 largely resembled that of glucose-starved cells.
Msn2A6 is partially tolerated in certain metabolic settings
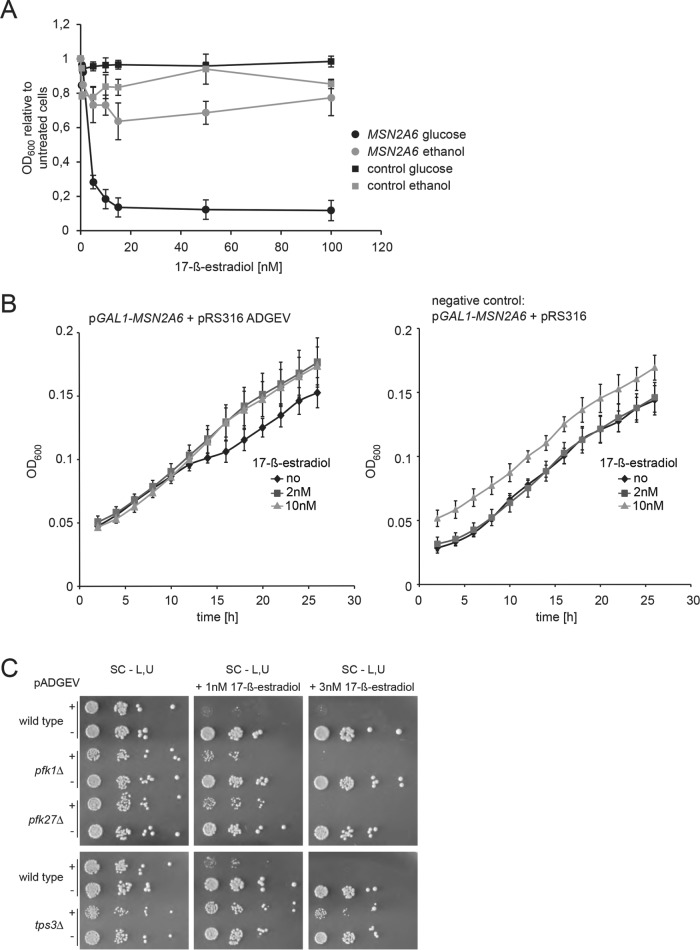
Consequently, we investigated whether growth is similarly affected by Msn2A6 in cells grown on nonfermentative carbon sources and thereby resembles the induced metabolic state. We performed growth assays with liquid selective medium containing 2% ethanol and 1% glycerol as carbon sources. ADGEV-driven MSN2A6 expression was induced by increasing concentrations of 17-ß-estradiol, and OD600 values were determined after 48 h of incubation at 30°C. Indeed, cells growing on nonfermentative carbon sources were significantly less sensitive to Msn2A6 expression than those growing in glucose-containing medium (Figure 4A). Interestingly, we observed no significant change of growth rates of Msn2A6-expressing cells and control cells (no pADGEV plasmid) within the first 24 h after induction when cells were grown on nonfermentative carbon sources (Figure 4B). Hence, Msn2A6 expression does not interfere with utilization of a nonfermentative carbon source.
Figure 4:
Cells grown on a nonfermentative carbon source tolerate Msn2A6 expression. (A) W303 wild-type cells carrying plasmids pGAL1-10-MSN2A6 and pADGEV were grown to logarithmic phase in liquid synthetic media containing either glucose or ethanol/glycerol as a carbon source and subsequently exposed to a range of 17-ß-estradiol concentrations. Growth (OD600nm) was recorded for 48 h. Ratios of treated vs. untreated growth are shown. Cells transformed with plasmids pGAL1-10-MSN2A6 and plasmids pRS316 (instead of pADGEV) served as negative controls. (B) W303 wild-type cells carrying plasmid pGAL1-10-MSN2A6 and pADGEV (control: pRS316) were grown in liquid synthetic media in the presence of ethanol/glycerol as sole carbon source. Msn2A6 expression was induced by 2 or 10 nM 17-ß-estradiol. Growth was recorded for 26 h and compared with that of negative controls (pRS316 instead of pADGEV). (C) Mutants with a predisposition for utilization of nonfermentative carbon sources (pfk1∆, pfk27∆, tps3∆) have reduced growth inhibition upon induction of Msn2A6 with 17-ß-estradiol.
These results suggested that the Msn2A6-induced growth phenotype might originate in the inability to use glucose as a carbon source as if a nonfermentable carbon source is expected. We reasoned that deletion mutant strains expressing a transcriptional pattern similar to that of Msn2A6 might exert an adapted metabolic state and could therefore be less sensitive to the expression of the allele. To test this idea, we selected mutants with deletions in genes involved in glucose metabolism–encoding enzymes not related to signaling, or to regulation of transcription upstream of Msn2: phosphofructokinase pfk1∆ and pfk27∆ and tps3∆, a subunit of the trehalose-6-phosphate-synthase. Expression of Msn2A6 was induced in these mutants using the ADGEV system and drop tests were performed to determine its effect on growth of the mutant cells. Indeed, all mutants showed a growth defect compared with cells lacking pADGEV; however, the defect was strikingly less severe, as in the wild-type control (Figure 4C).
Taken together, our data suggest that the transcriptional pattern induced by Msn2A6 forces cells to—at least partially—transit into respiratory growth and prevents the utilization of glucose as a carbon source.
Oxygen consumption per generation increases as a consequence of Msn2A6 induction
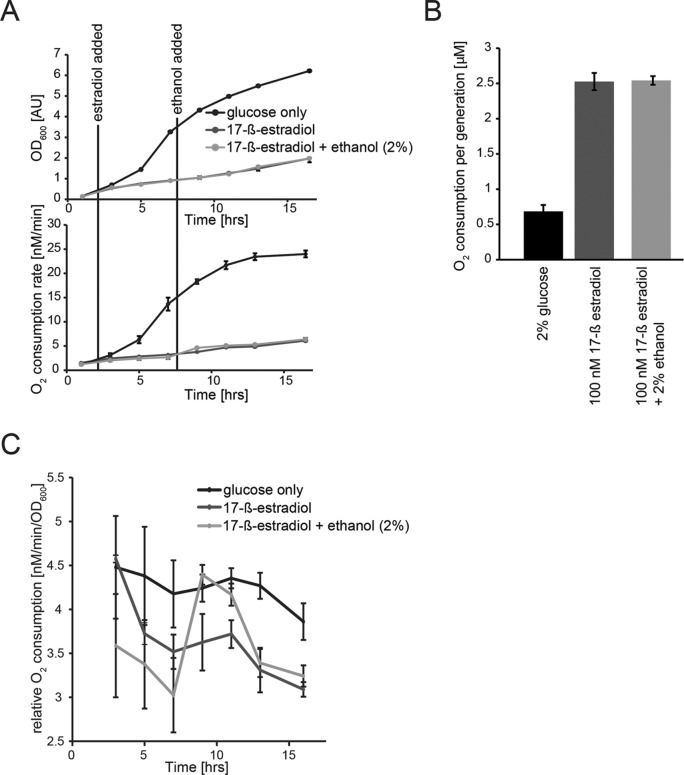
Msn2A6 expression did not influence growth of cells on ethanol as sole carbon source. We speculated that these cells would be in a similar metabolic state, however, in the presence of glucose. A hallmark of growth on nonfermentable carbon sources is a higher respiration rate than for fermenting cells and thus also a higher rate of oxygen consumption. Therefore, we measured oxygen consumption of cells expressing Msn2A6 in the presence of glucose polarographically, using a Clark-type oxygen electrode. Activation of Msn2A6 expression immediately reduced growth as well as oxygen consumption in relation to the that in the uninduced control (Figure 5, A–C). However, oxygen consumption per cell division of the Msn2A6-expressing cells (12 h) was constant and at an elevated rate compared with that of exponential phase cells (Figure 5, A and B). The oxygen consumption per generation of the uninduced cells reached comparable levels after glucose exhaustion (unpublished data). Thus, even in the presence of glucose, Msn2A6 expressing cells consume oxygen at rates like those for cells on a nonfermentable carbon source.
Figure 5:
Oxygen consumption rates increase in Msn2A6-expressing cells. (A) W303 wild-type cells carrying plasmids pGAL1-10-MSN2A6 and pADGEV were grown in liquid synthetic media containing glucose. Black lines indicate addition of 17-ß-estradiol (100 nM) and 2% ethanol to the cultures. Growth (OD600) was recorded every 2 h. (B) Molecular oxygen consumption [µM s–1] of all cultures was determined from the linear decrease in oxygen concentration followed polarographically for 1 min using a Clark-type oxygen electrode at 30°C. (B) Oxygen consumption normalized to growth rate. (C) Oxygen consumption rate relative to cell density.
This raised the question of whether a nonfermentable carbon source becomes a limiting factor for Msn2A6-expressing cells in the glucose-rich medium. We addressed this idea by adding ethanol (2% final concentration) to the induced cells. As a consequence, we observed a transient increase in oxygen consumption, followed by a return to basal levels, similar to those in cells growing in the absence of ethanol after 6 h without significant change of growth (Figure 5C). In addition, the addition of ethanol did not affect the oxygen consumption rate, indicating that the carbon source is not limiting growth (Figure 5C). These results underpin the notion that this transcription factor can radically change the metabolism of yeast cells.
Metabolite patterns are shifted due to Msn2A6 expression
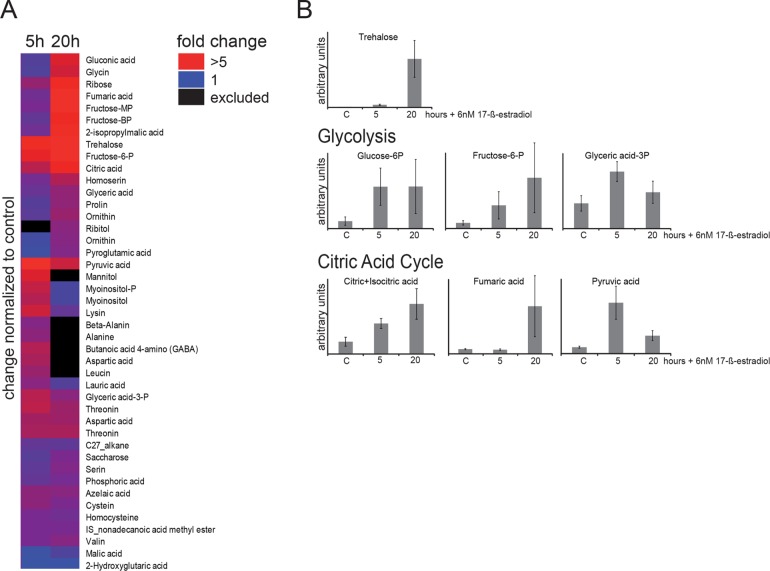
To test whether the global transcriptional pattern induced by Msn2A6 expression also influences the metabolic states of cells, we monitored intracellular levels of a range of metabolites. To do so, cells carrying the inducible MSN2A6 gene were treated with 17-ß-estradiol and subsequently grown for 5 and 20 h in the presence of glucose. Intracellular levels of metabolites were determined via gas chromatography–mass spectrometry (GC-MS) measurements (Figure 6, A and B; Supplemental Data File S3). More than 40 metabolites were affected by Msn2A6 expression (Figure 6A). We observed a more than 70-fold increase in trehalose levels after 20 h (fivefold increase after 5 h) compared with the uninduced control sample (Figure 6B). Trehalose (as well as glycogen) is known to accumulate in the event of nutritional depletion and also during growth on nonfermentable carbon sources (Lillie and Pringle, 1980; Sillje et al., 1999). In addition, we observed a fivefold increase in glucose-6-phosphate levels, suggesting a restriction of glycolytic flux. Also, the decrease of mRNAs of glycolysis-committing enzymes (phosphofructokinase and hexokinase) might contribute to the repression of glycolysis, depending on the stability of the enzymes. Finally, increased concentrations of isocitric/citric acid and fumaric acid suggest an increase of flux through the TCA cycle. Msn2A6 might induce a metabolic state similar to that of cells undergoing or having passed the diauxic shift, which is in partial accordance with a previous analysis of the metabolic consequences of the transition from glycolytic to gluconeogenic growth (Zampar et al., 2013).
Figure 6:
Metabolite concentrations are affected by Msn2A6 expression. W303 wild-type cells carrying plasmids pGAL1-10-MSN2A6 and pADGEV were grown in liquid synthetic media. Msn2A6 expression was induced by treatment with 6 nM 17-ß-estradiol for 5 and 20 h and levels of hallmark metabolites of glycolysis and the citric acid cycle were captured by LC-MS. Control cells were grown without 17-ß-estradiol and harvested at the same optical density. (A) Metabolite changes due to expression of Msn2A6. Data are expressed as fold change to untreated controls and sorted by hierarchical clustering. (B) Peak intensity is expressed as arbitrary units.
We conclude that Msn2A6 expression provokes a deregulation of metabolic activity toward respiratory growth despite the presence of glucose, most likely resulting in a fatal starvation condition. Therefore, the growth arrest caused by PKA depletion, which leads to Msn2 dephosphorylation and hyperactivation, is possibly provoked by a maladjusted metabolic state, rather than by a direct signaling process.
DISCUSSION
In S. cerevisiae, the glucose-responsive PKA pathway is a key regulator of cellular growth. PKA directly regulates enzyme activities but also has a large impact on the transcription pattern. Here we analyzed effects caused by PKA-dependent transcription mediated by the PKA-Msn2 module, using a PKA-independent Msn2 allele. We find that the dephosphorylated mimetic variant of Msn2 partly mimics the global phenotype of low kinase activity. Expression of Msn2A6 not only is changing the gene expression pattern but also is sufficient to induce a physiological state similar to cells grown on nonfermentative carbon sources.
Is the Msn2A6 allele a model for dephosphorylated Msn2?
Our comparison of Msn2 homologues revealed a striking similarity of functional domains (NES, NLS, and DNA-binding) and closely associated PKA consensus motifs. Among these domains, the NES was found to be unusually large compared with the prototypic PKA-inhibitor (PKI) NES (Wen et al., 1995). The smallest Msn2 fragment mediating export comprised 90 amino acid residues. The phosphoacceptor property of a single serine residue (S288) within the NES was required for regulation. Additional conserved phosphorylation sites are embedded within the NLS and the Zn-finger DNA-binding domain. In summary, a small set of PKA consensus sites are conserved, which likely establish the regulatory core of the Msn2 inhibition by PKA. Also, previous studies have shown that phosphorylation at most of these sites is a prerequisite for Msn2 regulation (De Wever et al., 2005; Hao et al., 2013; Reiter et al., 2013).
Our suggestion that Msn2A6 expression mimics PKA depletion is based on the following observations: Msn2A6 expression induces a growth defect and a transcriptional profile similar to cells with hampered PKA activity (such as ras2∆) and leads to an increase in levels of metabolites indicative for respiratory growth. We show that cells that express Msn2A6, similarly to cells that are deficient in PKA activity, accumulate in G1/S and show physiological changes normally associated with nutrient deprivation, including the accumulation of trehalose. In contrast, cells carrying mutations that yield elevated PKA activity fail to arrest in G1 and are defective for trehalose and glycogen accumulation (Tatchell, 1986; Cameron et al., 1988; Thevelein, 1994). Previous reports have shown that control of intracellular trehalose levels via posttranslational (Ewald et al., 2016; Zhao et al., 2016) and Msn2/4-dependent transcriptional (Zahringer et al., 2000) regulation of neutral trehalase, Nth1, seems to be one of the key events in the interplay between the cell cycle and metabolism. A comparison of our results on the Msn2A6 transcriptome with those of a previous study (Elfving et al., 2014), analyzing the transcriptional response mediated by a Z4EV promoter–driven version of the hyperactive MSN2 allele (McIsaac et al., 2013) revealed a high level of correlation (r = 0.61). However, we focused on the question of how the transcriptional pattern induced by the PKA-Msn2 module can affect physiology.
Does Msn2A6 inhibit the cell cycle?
Firm genetic evidence places Msn2 and Msn4 downstream of PKA. Deletion of both genes (Smith et al., 1998) rescues the effects of PKA depletion (Toda et al., 1987; Cameron et al., 1988; Garrett and Broach, 1989). Thus, one or more Msn2/4 targets might be detrimental for growth. Several factors, such as the protein kinases Yak1 (Garreau et al., 2000; Chi et al., 2001) and Rim15 (Reinders et al., 1998; Pedruzzi et al., 2003), have been described previously in this context. YAK1 expression is controlled by Msn2/4 (Smith et al., 1998) and activity of the kinase is directly inhibited by phosphorylation by PKA (Lee et al., 2008). Furthermore, Yak1 phosphorylates and activates Msn2/4 in vitro (Lee et al., 2008) and also phosphorylates the PKA regulatory subunit Bcy1, shifting its localization to the cytoplasm (Griffioen et al., 2001). Bcy1 localization is likely to interfere with Msn2 phosphorylation. Under high glucose growth conditions, Bcy1 is located in the nucleus and PKA activity is as a consequence tightly regulated within the nuclear compartment. Decreased levels of Bcy1 in the nucleus promote free and active PKA catalytic subunits in this compartment and therefore increased phosphorylation of Msn2. In addition to Yak1, kinase Rim15 has been shown to promote the entry of cells into a quiescent state in G1 in response to nutrient starvation (Reinders et al., 1998; Pedruzzi et al., 2003). Deletion of either of the kinases rescues a tpk1∆, tpk2∆, tpk3∆ triple deletion. Additional potentially interesting factors in the context of stress- or starvation-induced cell cycle arrest include Xbp1, a transcriptional repressor important for maintaining stress- or starvation-induced G1 arrests (Mai and Breeden, 1997; Miles et al., 2013), and Whi2, a complex partner of plasma membrane phosphatase Psr1, which regulates the phosphorylation status and activity of Msn2 (Kaida et al., 2002).
A simple model for how the PKA-Msn2 module affects cellular growth would therefore place these factors in the position of cell cycle regulators that become up-regulated upon low PKA activity and directly antagonize growth. However, xbp1∆, yak1∆, or rim15∆ mutations could not rescue cellular growth from the effect mediated by Msn2A6. We therefore speculated that the growth effect might not be mediated via the up-/down-regulation of a single specific regulator and pursued a global transcriptomic approach. The Msn2A6 expression pattern showed similarities to previously published transcription profiles of cells that are impaired in the sensing or utilization of glucose (Apweiler et al., 2012). This is in line with a transcriptional switch of carbon source utilization leading to a cascade of consequences, perhaps via adjustment of metabolism, as has been proposed earlier (Apweiler et al., 2012).
Is the observed transcript profile a consequence of retarded growth?
Recently, a systematic microarray analysis of a compendium of single–gene deletion mutants revealed that many of the mutant strains showed one of two stereotypical gene expression profiles during unperturbed growth in rich medium, resembling either respiratory or fermentative growth. Mutants with a respiratory signature commonly have a partially delayed cell cycle (O’Duibhir et al., 2014). Various types of acute stress generally cause a transient cell cycle arrest and have a common transcriptional signature similar to the pattern present in slowly growing cells. Gene expression profiles in response to physicochemical stress consist of a stress type–specific (e.g., temperature shock, osmotic shock) as well as the generic growth-dependent pattern (O’Duibhir et al., 2014). The overall high transcript levels of many genes induced by Msn2A6 expression suggest that retarded growth contributes to the pattern only to a minor degree. On the other hand, it has been noted before that such a construct causes induction of many genes apparently lacking Msn2 binding sites (Elfving et al., 2014). The observed pattern might reflect a perceived carbon starvation, combined with Msn2/4- and growth-specific components.
The trehalose content of cells growing on a nonfermentable carbon source is low compared with that of cells approaching the stationary phase (∼20% of dry weight). The amount measured in our long term–treated samples (20 h) resembles the stationary content. We did not measure the amount of the alternative storage carbohydrate glycogen in our conditional mutants, but suspect an increase also. The expression patterns of the relevant enzymes of the primary carbon metabolism and the oxygen consumption of the MsnA6-expressing cells point to an adaptation toward fermentation. Nevertheless, Msn2 (and also Msn4) have an influence on the carbon metabolism equilibrium and a role in the reentry into growth from quiescence. Msn2 and Msn4 mutants display aberrant metabolic cycling in nutrient source–limited chemostat cultures and have reduced transcripts of glycolytic genes and reduced Acteyl-CoA levels (Kuang et al., 2017). The seeming contradiction to our results where a constitutive active version of Msn2 represses glycolysis and the results of Kuang et al. (2017), where Msn2 acts as an activator of glycolytic genes, may possibly be attributed to the different growth conditions. Together, the results imply an important contribution of this transcription factor to fermentative and respiratory growth, depending on its phosphorylation status.
Yeast cells are equipped with a range of transcription factors steering carbon utilization. These are, for example, Gcr1, which promotes glycolysis genes (Holland et al., 1987; Barbara et al., 2007), and Mig1, a Snf1-regulated repressor of gluconeogenic genes (Nehlin et al., 1991), and the Hap complex (Hap2-5), which activates genes required for respiration and Rtg1/3-mediated retrograde signaling of the mitochondrial status to the nucleus. Furthermore, the zinc cluster transcription factors Cat8, Sip4, Ert1, Rds2, and Adr1 have been shown to mediate reprogramming of gene expression toward respiration patterns (Schüller, 2003). Snf1/4 is repressed by glucose, which in turn represses Sip4 and Cat8 and their gluconeogenic target genes. Oaf1 and Pip2 are zinc cluster transcription factors regulating peroxisomal ß-oxidation (Rottensteiner et al., 1996). Of these transcription factors, only Adr1 overexpression is sufficient for derepression of its target genes (Simon et al., 1991). Overlapping and redundant signaling systems, such as the combination of nuclear accumulation and posttranslational modification, ensure tight control in these systems. It is therefore not surprising that the metabolite profile observed by us does not exactly match postdiauxic shift or respiratory state (Zampar et al., 2013). Because the conditional expression of Msn2A6 occurred in the presence of 2% glucose, glucose-triggered degradation of FBP1 and PCK1 mRNAs (both increase only slightly) might interfere with correct adaptation to gluconeogenesis (Yin et al., 2000).
Taking these results together, we suggest that Msn2 hyperactivity triggers a metabolic maladjustment rather than a dysregulation of the cell cycle. Two observations reported here support our conclusion. First, deletion mutants impaired in glucose sensing are tolerant for Msn2A6 expression, and second, ethanol-growing cells are not affected by Msn2A6 expression. Somewhat surprising, but nevertheless fitting, is our observation that the oxygen consumption of the Mns2A6-expressing cells resembles that of ethanol-growing cells. We propose that the growth arrest caused by PKA depletion results from a maladjusted metabolic state triggered by Msn2-dependent change of gene expression.
MATERIALS AND METHODS
Strains and growth conditions
Cells were grown in complete medium (YEP) or buffered synthetic complete (SC) medium (pH 5.8) containing 2% glucose. Standard growth conditions were applied (30°C, shaking at 180 rpm). Yeast strains have been described earlier. W303 msn2∆msn4∆ and W303 msn2∆msn4∆cdc35∆pde2∆ are derivatives of W303-1A (Görner et al., 1998, 2002; Durchschlag et al., 2004). BY4741 and related gene deletion strains were obtained from Euroscarf (http://web.uni-frankfurt.de/fb15/mikro/euroscarf/). Plasmid-carrying strains were grown in minimal SC medium, selecting for the respective auxotrophic markers, and shifted to prewarmed YPD before the experimental condition was applied. To assay stress responses, cells were grown to exponential growth phase (OD600nm 0.8–1) followed by exposure to 7% ethanol or 0.5 M NaCl (final concentration). Heat shock was induced by resuspending cells in prewarmed (42°C) media. Glucose starvation was achieved by resuspension of the cells in prewarmed media lacking glucose. cAMP deprivation and add-back experiments were described in (Görner et al., 1998, 2002; Durchschlag et al., 2004). Conditional promoter–regulated alleles were induced by adding 6.25–50 µM (liquid culture) or 50 µM (plates) of CuSO4 or 2–100 nM 17-ß-estradiol for times indicated.
Plasmids
Plasmids expressing internal deletions and point mutations of the MSN2 gene are derivatives of pAMG (ADH1-MSN2-GFP; Görner et al., 1998). Point mutations were introduced either by using a QuickChange Multi Site-directed Mutagenesis kit (Stratagene) or by amplifying the corresponding DNA fragment by PCR with mutagenic primers. Primer sequences are available on request. Plasmids encoding truncated versions and internal deletions of Msn2 were created by inserting SalI/NcoI-cut PCR fragments into SalI/NcoI-cut pAMG (Görner et al., 1998). pADH1-MSN2(279-704)-GFP was generated using primers O6 and O18, pADH1-MSN2(267-704)-GFP with primers O6 and O19, and pADH1-MSN2(267-516)-GFP with primers O19 and O4. Further deletion plasmids are derivatives of plasmid pADH1-MSN2(∆521-571)-GFP, which was obtained by a triple ligation of a SalI/BamHI-cut PCR amplification using primers O12 and O1, a BamH1/NcoI-cut PCR fragment obtained with primers O13 and O6, and a SalI/NcoI-cut plasmid pAMG. The following constructs were generated by inserting SalI/BamHI-cut PCR fragments obtained with the following primer pairs into SalI-BamHI-cut plasmid pADH1-MSN2(∆521-571)-GFP: pADH1-MSN2(∆365-571)-GFP using primers O1 and O15 and pADH1-MSN2(∆246-571)-GFP using primers O1 and O16. To create pADH1-MSN2(∆246-325)-GFP, a BamHI/NcoI-cut PCR fragment obtained with primers O20(dB325) and O6 was ligated into BamHI/NcoI-cut pADH1-MSN2(∆246-571)-GFP. pADH1-MSN2(∆302-477)-GFP was generated by excision of the EcoRI fragment of pAMG. Plasmid pADH1-MSN2(∆322-477)-GFP was created by ligating a SalI-EcoRI-cut PCR fragment obtained with the oligonucleotides O17 and O1 into SalI-EcoRI-cut pAMG. The SV40-NLS-Msn2-GFP plasmids were constructed as described in (Görner et al., 1998).
To obtain pCUP1-MSN2A6, an NdeI/HindIII fragment from pAMG-S686A (Reiter et al., 2013) was inserted into NdeI/HindIII-cut pAMGX12A5 (Durchschlag et al., 2004), resulting in pAMGX12A6. pAMGA6 (ADH1-MSN2A6-GFP) was created by introducing fragment MSN2× 12-(aa7-aa264)-SalI (Görner et al., 1998), into SalI-cut pAMGX12A6. This construct was further cut with XhoI, and the obtained fragment was introduced into pCUP1-MSN2 (Durchschlag et al., 2004) to generate pCUP1-MSN2A6. Plasmid pAMGD5 was obtained by fusing XhoI-cut plasmid pAMGX5S288D with an XhoI-HindIII-cut PCR fragment, received by amplifying a S582, 620, 625, 633D-containing sequence (Görner et al., 1998). An NdeI-HindIII-cut fragment of pAMG-S686D (Reiter et al., 2013) was inserted into NdeI-HindIII-cut pAMGD5, resulting in formation of pAMGD6. The plasmid pRS316-ADGEV was provided by the Picard lab (University of Geneva) and has been described earlier (Louvion et al., 1993).
FACS analysis
A cell culture at OD600nm = 1 (1 ml) was harvested, resuspended with 1 ml 70% EtOH for fixation, and shaken at room temperature overnight. Fixed cells were suspended in 200 mM Tris, pH 7.8, with 0.4 mg/ml RNase and incubated for 4 h at 37°C, washed with FACS buffer (200 mM Tris, pH 7.8, 211 mM NaCl, 78 mM MgCl2), and subsequently resuspended in FACS buffer containing 0.05 mg/ml propidium iodide. Samples were sonicated and FACS analysis was performed with 1:20–1:10 dilutions of cell suspension in FACS buffer using a Becton Dickinson FACScan machine according to the manufacturers’ protocol. Fluorescence microscopy experiments were performed as described previously (Görner et al., 1998, 2002; Durchschlag et al., 2004).
Growth analysis
Cells were grown either in 10-ml SC media lacking leucine and uracil (SC-LU) containing 2% glucose (glucose growth experiments) or in SC-LU containing 2% ethanol, 1% glycerol, and 0.3% glucose (nonfermentative carbon source) overnight at 30°C. Cells were then diluted to a final density of OD600 = 0.1 in SC-LU and the appropriate carbon source in a 96-well format. The cell suspension was mixed with 17-ß-estradiol stock solution (final concentration 2 nM, 10 nM) and subsequently incubated with shaking for 26 or 56 h at 30°C. OD600 was measured every 2 h using the SynergyH1 multimode plate reader (BioTeck Industries). For all samples, two clones were measured in triplicate. Background values were subtracted from all measurements and the average and SD were calculated.
17-ß-Estradiol sensitivity assay
Cells were grown overnight in 10 ml SC containing 2% ethanol, 1% glycerol, and 0.3% glucose at 30°C, harvested by centrifugation (2 min, 2000 rpm), resuspended to OD600 = 1 in sterile water and 10 µl of distributed in a 96-well plate format containing 180 µl SC plus 2% ethanol and 1% glycerol or SC with 2% glucose. To the cell suspension, 17-ß-estradiol was added to a final concentration of 0.5, 1, 5, 10, 15, 50, or 100 nM or sterile water (untreated control) and subsequently incubated for 36 h at 30°C. OD600 values were determined using an Enspire multimode plate reader (PerkinElmer). For all experimental conditions, three independent clones were measured in duplicate. For data analysis, background signals were subtracted from all measurements, samples were normalized relative to the mean OD600 of the untreated samples, and the mean growth and SD were calculated.
Microarrays
For microarray gene expression profiling, strains were grown for four generations in 50-ml cultures in YPD at 30°C to OD600 of ∼1 before 17-ß-estradiol was added to a final concentration of 6 nM. After 90 min, cells were harvested and immediately frozen. Total RNA (1 µg) was used for labeling the reaction (Agilent Quick Amp labeling kit, two-color, Cat.Nr. 5190-0444). The standard Agilent protocol for one-color labeling was used (G4140-90041). Cy3-labeled cRNA (325 ng) was hybridized to standard S. cerevisiae GE 8 × 15 K arrays. Samples were hybridized for 17 h at 65°C, 10 rpm. An Agilent G2505C microarray scanner system was used to scan the arrays.
The Agilent Feature Extraction program (Version FE 10.5.1.1) was used to analyze the array images. The raw intensities were imported into Bioconductor and processed and normalized with the limma R package. Genes with all values higher than 50 (untreated) were retained and fold changes were centered according to median and then normalized, analyzed with cluster analysis (Eisen et al., 1998; Nadon and Shoemaker, 2002) using cluster3, and visualized with JavaTreeView (Saldanha, 2004) (http://jtreeview.sourceforge.net). Significant associations to either GO-terms or transcription factors were obtained by GO-Term finder at SGD. TreeView files corresponding to the figures and the raw data are supplied (Supplemental Data Files S1 and S2). Data have been deposited in the GEO database (Accession number GSE81890).
Metabolite analysis
Cells were grown under shaking (180 rpm) in 10-ml SC-LU overnight at 30°C, diluted to OD600 = 1 in prewarmed (30°C) 100-ml SC-LU, and incubated while being shaken (180 rpm) until the mid–logarithmic phase (OD600 = 1). Induction of ADGEV-driven genes was achieved by addition of 6 nM (final concentration) 17-ß-estradiol. Samples were harvested 5 and 20 h postinduction (OD600 = 2). Control samples were harvested at OD600 = 2. For sample preparation/extraction, cells were harvested by centrifugation (5 min, 4000 rpm, 4°C), immediately quenched with 5 ml of ice-cold 30% methanol, and centrifuged again. The cell pellet was resuspended in 700 µl of ice-cold 75% methanol and 0.1% formic acid. Cell breakage was achieved by bead beating (100-µl glass beads) on an IKA-VIBRAX-VXR shaker at 4°C. Removal of lipid components was achieved by chloroform extraction. The polar phase (300 µl) was dried overnight using a vacuum concentrator at 15°C before analysis. For automated derivatization on a PAL-CTC-autosampler, the dried samples were resuspended in 40 µl of MOX reagent (20 mg/ml methoxyamine hydrochloride in pyridine, prepared fresh daily) and incubated while being shaken for 90 min in an agitator at 60°C. After addition of 50 µl of MS-TFA and 10 µl of internal standard (370 mg/l nonadecanoic acid methyl ester), the samples were again incubated while being shaken at 60°C for 60 min and subsequently incubated at 4°C for 10 min before injection into the GC-MS system. For GC-MS measurement, analytes were separated and detected using an Agilent 7890A gas chromatograph coupled to a 5975C inert XL MSD detector (Agilent, Waldbronn, Germany). An HP5-MS column (30 m × 0.25 mm × 0.25 μm; Agilent Technologies, Waldbronn, Germany) and helium as carrier gas were used (constant flow of 1 ml/min). The derivatized sample (1 µl) was injected at 250°C into the split/splitless injector (split of 1:25). The oven temperature was set to 50°C for 2 min and then increased by 10°C/min to 310°C, which was maintained for15 min. Measurement was carried out in scan mode at m/z 50–800. The EI source of the MS was set to 230°C, the quadrupole to 150°C. The samples were measured in randomized order. Raw data were processed with the Metabolite Detector software (version 2.5.20130704-Linux; Hiller et al., 2009). GC-EI-MS spectra and retention indices of recognized features were compared with an in-house established library. Results are provided as Supplemental Data File S3.
Oxygen consumption
Oxygen consumption was measured polarographically using a Clark-type oxygen electrode (Oxygraph plus; Hansatech Instruments, Norfolk, UK) at 30°C. The electrode was equilibrated to 100% oxygen saturation with air and 0% oxygen with nitrogen. Molecular oxygen consumption [µM s–1] was determined from the linear decrease in oxygen concentration followed for 1 min. All samples were measured in biological triplicate. Cultures were grown in YPD (2% glucose, 180 rpm, 30°C, 20 ml) inoculated to OD600 = 0.1 from overnight precultures. After 1.5 h, 17-ß estradiol was added to a final concentration of 100 nM to all samples except control. After 4.5 h (7 h after inoculation), ethanol was added to the estradiol/ethanol samples to a final concentration of 2%. Every 2 h, 600 µl samples were taken and analyzed for OD600 and oxygen consumption.
Msn2 sequence analysis
Sequences homologous to S. cerevisiae Msn2 (excluding the zinc-finger domains) were identified using Uniprot Blast (effective June 2018). Thirty-four unique Msn2 and Msn4 sequences were identified and 28 of these aligned using MEGA-X (Muscle algorithm, cluster method: neighbor joining). For better visualization, the alignment was visualized using ClustalX2 without modifications. Accession numbers are listed in Supplemental Table S1. The alignment is provided as Supplemental Figures S1 and S2 and Supplemental Data Files S4 and S5. The alignment file was used as input to calculate the similarity plot using plotcon (http://emboss.bioinformatics.nl/cgi-bin/emboss/plotcon).
Supplementary Material
Acknowledgments
Jillian Augustine, Marion Janschitz, and David Hollenstein are acknowledged for critically reading the manuscript. We thank Gustav Ammerer for support throughout this study. Design of the study, data collection, and interpretation of data and writing the manuscript were supported by Austrian Science Fund (FWF) Grant SFB Fusarium F3715 (to R.S.) and Grant P23355-B12 (to C.S.), by the Herzfelder Foundation (to C.S.), and by the Austrian Science Fund, FWF (Doctoral Program BioToP-Molecular Technology of Proteins [W1224]).
Abbreviations used:
- AMPK
AMP-activated kinase
- HD
homology domain
- NES
nuclear export sequence
- NLS
nuclear localization sequence
- PKA
protein kinase A
- TORC1
rapamycin kinase complex 1
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-06-0389) on September 26, 2018.
REFERENCES
- AkhavanAghdam Z, Sinha J, Tabbaa OP, Hao N. (2016). Dynamic control of gene regulatory logic by seemingly redundant transcription factors. eLife , e18458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amigoni L, Colombo S, Belotti F, Alberghina L, Martegani E. (2015). The transcription factor Swi4 is target for PKA regulation of cell size at the G1 to S transition in Saccharomyces cerevisiae . Cell Cycle , 2429–2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apweiler E, Sameith K, Margaritis T, Brabers N, van de Pasch L, Bakker LV, van Leenen D, Holstege FC, Kemmeren P. (2012). Yeast glucose pathways converge on the transcriptional regulation of trehalose biosynthesis. BMC Genomics , 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbara KE, Haley TM, Willis KA, Santangelo GM. (2007). The transcription factor Gcr1 stimulates cell growth by participating in nutrient-responsive gene expression on a global level. Mol Genet Genomics , 171–188. [DOI] [PubMed] [Google Scholar]
- Baroni MD, Monti P, Alberghina L. (1994). Repression of growth-regulated G1 cyclin expression by cyclic AMP in budding yeast. Nature , 339–342. [DOI] [PubMed] [Google Scholar]
- Bodvard K, Peeters K, Roger F, Romanov N, Igbaria A, Welkenhuysen N, Palais G, Reiter W, Toledano MB, Kall M, Molin M. (2017). Light-sensing via hydrogen peroxide and a peroxiredoxin. Nat Commun , 14791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boy-Marcotte E, Perrot M, Bussereau F, Boucherie H, Jacquet M. (1998). Msn2p and Msn4p control a large number of genes induced at the diauxic transition which are repressed by cyclic AMP in Saccharomyces cerevisiae . J Bacteriol , 1044–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauer MJ, Saldanha AJ, Dolinski K, Botstein D. (2005). Homeostatic adjustment and metabolic remodeling in glucose-limited yeast cultures. Mol Biol Cell , 2503–2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L, Dalal CK, Elowitz MB. (2008). Frequency-modulated nuclear localization bursts coordinate gene regulation. Nature , 485–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron S, Levin L, Zoller M, Wigler M. (1988). cAMP-independent control of sporulation, glycogen metabolism, and heat shock resistance in S. cerevisiae . Cell , 555–566. [DOI] [PubMed] [Google Scholar]
- Capaldi AP, Kaplan T, Liu Y, Habib N, Regev A, Friedman N, O’Shea EK. (2008). Structure and function of a transcriptional network activated by the MAPK Hog1. Nat Genet , 1300–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causton HC, Ren B, Koh SS, Harbison CT, Kanin E, Jennings EG, Lee TI, True HL, Lander ES, Young RA. (2001). Remodeling of yeast genome expression in response to environmental changes. Mol Biol Cell , 323–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee M, Acar M. (2018). Heritable stress response dynamics revealed by single-cell genealogy. Sci Adv , e1701775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C, Mu J, Farkas I, Huang D, Goebl MG, Roach PJ. (1995). Requirement of the self-glucosylating initiator proteins Glg1p and Glg2p for glycogen accumulation in Saccharomyces cerevisiae . Mol Cell Biol , 6632–6640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi Y, Huddleston MJ, Zhang X, Young RA, Annan RS, Carr SA, Deshaies RJ. (2001). Negative regulation of Gcn4 and Msn2 transcription factors by Srb10 cyclin-dependent kinase. Genes Dev , 1078–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad M, Schothorst J, Kankipati HN, Van Zeebroeck G, Rubio-Texeira M, Thevelein JM. (2014). Nutrient sensing and signaling in the yeast Saccharomyces cerevisiae . FEMS Microbiol Rev , 254–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wever V, Reiter W, Ballarini A, Ammerer G, Brocard C. (2005). A dual role for PP1 in shaping the Msn2-dependent transcriptional response to glucose starvation. EMBO J , 4115–4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRisi JL, Iyer VR, Brown PO. (1997). Exploring the metabolic and genetic control of gene expression on a genomic scale. Science , 680–686. [DOI] [PubMed] [Google Scholar]
- Durchschlag E, Reiter W, Ammerer G, Schüller C. (2004). Nuclear localization destabilizes the stress-regulated transcription factor Msn2. J Biol Chem , 55425–55432. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. (1998). Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA , 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfving N, Chereji RV, Bharatula V, Bjorklund S, Morozov AV, Broach JR. (2014). A dynamic interplay of nucleosome and Msn2 binding regulates kinetics of gene activation and repression following stress. Nucleic Acids Res , 5468–5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald JC, Kuehne A, Zamboni N, Skotheim JM. (2016). The yeast cyclin-dependent kinase routes carbon fluxes to fuel cell cycle progression. Mol Cell , 532–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P, Pozza F, Pletcher SD, Gendron CM, Longo VD. (2001). Regulation of longevity and stress resistance by Sch9 in yeast. Science , 288–290. [DOI] [PubMed] [Google Scholar]
- Galdieri L, Mehrotra S, Yu S, Vancura A. (2010). Transcriptional regulation in yeast during diauxic shift and stationary phase. OMICS , 629–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallmetzer A, Silvestrini L, Schinko T, Gesslbauer B, Hortschansky P, Dattenbock C, Muro-Pastor MI, Kungl A, Brakhage AA, Scazzocchio C, Strauss J. (2015). Reversible oxidation of a conserved methionine in the nuclear export sequence determines subcellular distribution and activity of the fungal nitrate regulator NirA. PLoS Genet , e1005297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garmendia-Torres C, Goldbeter A, Jacquet M. (2007). Nucleocytoplasmic oscillations of the yeast transcription factor Msn2: evidence for periodic PKA activation. Curr Biol , 1044–1049. [DOI] [PubMed] [Google Scholar]
- Garreau H, Hasan RN, Renault G, Estruch F, Boy-Marcotte E, Jacquet M. (2000). Hyperphosphorylation of Msn2p and Msn4p in response to heat shock and the diauxic shift is inhibited by cAMP in Saccharomyces cerevisiae. Microbiology (Pt 9), 2113–2120. [DOI] [PubMed] [Google Scholar]
- Garrett S, Broach J. (1989). Loss of Ras activity in Saccharomyces cerevisiae is suppressed by disruptions of a new kinase gene, YAKI, whose product may act downstream of the cAMP-dependent protein kinase. Genes Dev , 1336–1348. [DOI] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. (2000). Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell , 4241–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görner W, Durchschlag E, Martinez-Pastor MT, Estruch F, Ammerer G, Hamilton B, Ruis H, Schüller C. (1998). Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev , 586–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görner W, Durchschlag E, Wolf J, Brown EL, Ammerer G, Ruis H, Schüller C. (2002). Acute glucose starvation activates the nuclear localization signal of a stress-specific yeast transcription factor. EMBO J , 135–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffioen G, Branduardi P, Ballarini A, Anghileri P, Norbeck J, Baroni MD, Ruis H. (2001). Nucleocytoplasmic distribution of budding yeast protein kinase A regulatory subunit Bcy1 requires Zds1 and is regulated by Yak1-dependent phosphorylation of its targeting domain. Mol Cell Biol , 511–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen AS, Hao N, O’Shea EK. (2015). High-throughput microfluidics to control and measure signaling dynamics in single yeast cells. Nat Protoc , 1181–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen AS, O’Shea EK. (2013). Promoter decoding of transcription factor dynamics involves a trade-off between noise and control of gene expression. Mol Syst Biol , 704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen AS, O’Shea EK. (2015). cis determinants of promoter threshold and activation timescale. Cell Rep , 1226–1233. [DOI] [PubMed] [Google Scholar]
- Hansen AS, O’Shea EK. (2016). Encoding four gene expression programs in the activation dynamics of a single transcription factor. Curr Biol , R269–R271. [DOI] [PubMed] [Google Scholar]
- Hao N, Budnik BA, Gunawardena J, O’Shea EK. (2013). Tunable signal processing through modular control of transcription factor translocation. Science , 460–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao N, O’Shea EK. (2012). Signal-dependent dynamics of transcription factor translocation controls gene expression. Nat Struct Mol Biol , 31–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller K, Hangebrauk J, Jäger C, Spura J, Schreiber K, Schomburg D. (2009). MetaboliteDetector: comprehensive analysis tool for targeted and nontargeted GC/MS based metabolome analysis . Anal Chem , 3429–3439. [DOI] [PubMed] [Google Scholar]
- Holland MJ, Yokoi T, Holland JP, Myambo K, Innis MA. (1987). The GCR1 gene encodes a positive transcriptional regulator of the enolase and glyceraldehyde-3-phosphate dehydrogenase gene families in Saccharomyces cerevisiae . Mol Cell Biol , 813–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquet M, Renault G, Lallet S, De Mey J, Goldbeter A. (2003). Oscillatory nucleocytoplasmic shuttling of the general stress response transcriptional activators Msn2 and Msn4 in Saccharomyces cerevisiae . J Cell Biol , 497–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaida D, Yashiroda H, Toh-e A, Kikuchi Y. (2002). Yeast Whi2 and Psr1-phosphatase form a complex and regulate STRE-mediated gene expression. Genes Cells , 543–552. [DOI] [PubMed] [Google Scholar]
- Kuang Z, Pinglay S, Ji H, Boeke JD. (2017). Msn2/4 regulate expression of glycolytic enzymes and control transition from quiescence to growth. eLife 6, e29938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P, Cho BR, Joo HS, Hahn JS. (2008). Yeast Yak1 kinase, a bridge between PKA and stress-responsive transcription factors, Hsf1 and Msn2/Msn4. Mol Microbiol , 882–895. [DOI] [PubMed] [Google Scholar]
- Lenssen E, James N, Pedruzzi I, Dubouloz F, Cameroni E, Bisig R, Maillet L, Werner M, Roosen J, Petrovic K, et al. (2005). The Ccr4-Not complex independently controls both Msn2-dependent transcriptional activation—via a newly identified Glc7/Bud14 type I protein phosphatase module—and TFIID promoter distribution. Mol Cell Biol , 488–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenssen E, Oberholzer U, Labarre J, De Virgilio C, Collart MA. (2002). Saccharomyces cerevisiae Ccr4-not complex contributes to the control of Msn2p-dependent transcription by the Ras/cAMP pathway. Mol Microbiol , 1023–1037. [DOI] [PubMed] [Google Scholar]
- Lillie SH, Pringle JR. (1980). Reserve carbohydrate metabolism in Saccharomyces cerevisiae: responses to nutrient limitation. J Bacteriol , 1384–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Sohn CH, Dalal CK, Cai L, Elowitz MB. (2015). Combinatorial gene regulation by modulation of relative pulse timing. Nature , 54–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logg K, Bodvard K, Blomberg A, Kall M. (2009). Investigations on light-induced stress in fluorescence microscopy using nuclear localization of the transcription factor Msn2p as a reporter. FEMS Yeast Res , 875–884. [DOI] [PubMed] [Google Scholar]
- Louvion JF, Havaux-Copf B, Picard D. (1993). Fusion of GAL4-VP16 to a steroid-binding domain provides a tool for gratuitous induction of galactose-responsive genes in yeast. Gene , 129–134. [DOI] [PubMed] [Google Scholar]
- Mai B, Breeden L. (1997). Xbp1, a stress-induced transcriptional repressor of the Saccharomyces cerevisiae Swi4/Mbp1 family. Mol Cell Biol , 6491–6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler G, Schuller C, Adam G, Ruis H. (1993). A Saccharomyces cerevisiae UAS element controlled by protein kinase A activates transcription in response to a variety of stress conditions. EMBO J , 1997–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Pastor MT, Marchler G, Schüller C, Marchler-Bauer A, Ruis H, Estruch F. (1996). The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE). EMBO J , 2227–2235. [PMC free article] [PubMed] [Google Scholar]
- McIsaac RS, Oakes BL, Wang X, Dummit KA, Botstein D, Noyes MB. (2013). Synthetic gene expression perturbation systems with rapid, tunable, single-gene specificity in yeast. Nucleic Acids Res , e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles S, Li L, Davison J, Breeden LL. (2013). Xbp1 directs global repression of budding yeast transcription during the transition to quiescence and is important for the longevity and reversibility of the quiescent state. PLoS Genet , e1003854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadon R, Shoemaker J. (2002). Statistical issues with microarrays: processing and analysis. Trends Genet , 265–271. [DOI] [PubMed] [Google Scholar]
- Nehlin JO, Carlberg M, Ronne H. (1991). Control of yeast GAL genes by MIG1 repressor: a transcriptional cascade in the glucose response. EMBO J , 3373–3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Duibhir E, Lijnzaad P, Benschop JJ, Lenstra TL, van Leenen D, Groot Koerkamp MJ, Margaritis T, Brok MO, Kemmeren P, Holstege FC. (2014). Cell cycle population effects in perturbation studies. Mol Syst Biol , 732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedruzzi I, Dubouloz F, Cameroni E, Wanke V, Roosen J, Winderickx J, De Virgilio C. (2003). TOR and PKA signaling pathways converge on the protein kinase Rim15 to control entry into G0. Mol Cell , 1607–1613. [DOI] [PubMed] [Google Scholar]
- Reinders A, Burckert N, Boller T, Wiemken A, De Virgilio C. (1998). Saccharomyces cerevisiae cAMP-dependent protein kinase controls entry into stationary phase through the Rim15p protein kinase. Genes Dev , 2943–2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter W, Klopf E, De Wever V, Anrather D, Petryshyn A, Roetzer A, Niederacher G, Roitinger E, Dohnal I, Gorner W, et al. (2013). Yeast protein phosphatase 2A-Cdc55 regulates the transcriptional response to hyperosmolarity stress by regulating Msn2 and Msn4 chromatin recruitment. Mol Cell Biol , 1057–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottensteiner H, Kal AJ, Filipits M, Binder M, Hamilton B, Tabak HF, Ruis H. (1996). Pip2p: a transcriptional regulator of peroxisome proliferation in the yeast Saccharomyces cerevisiae . EMBO J , 2924–2934. [PMC free article] [PubMed] [Google Scholar]
- Saldanha AJ. (2004). Java Treeview—extensible visualization of microarray data. Bioinformatics , 3246–3248. [DOI] [PubMed] [Google Scholar]
- Schüller HJ. (2003). Transcriptional control of nonfermentative metabolism in the yeast Saccharomyces cerevisiae . Curr Genet , 139–160. [DOI] [PubMed] [Google Scholar]
- Sillje HH, Paalman JW, ter Schure EG, Olsthoorn SQ, Verkleij AJ, Boonstra J, Verrips CT. (1999). Function of trehalose and glycogen in cell cycle progression and cell viability in Saccharomyces cerevisiae . J Bacteriol , 396–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M, Adam G, Rapatz W, Spevak W, Ruis H. (1991). The Saccharomyces cerevisiae ADR1 gene is a positive regulator of transcription of genes encoding peroxisomal proteins. Mol Cell Biol , 699–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slattery MG, Liko D, Heideman W. (2008). Protein kinase A, TOR, and glucose transport control the response to nutrient repletion in Saccharomyces cerevisiae . Eukaryot Cell , 358–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A, Ward MP, Garrett S. (1998). Yeast PKA represses Msn2p/Msn4p-dependent gene expression to regulate growth, stress response and glycogen accumulation. EMBO J , 3556–3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatchell K. (1986). RAS genes and growth control in Saccharomyces cerevisiae . J Bacteriol , 364–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevelein JM. (1994). Signal transduction in yeast. Yeast , 1753–1790. [DOI] [PubMed] [Google Scholar]
- Toda T, Cameron S, Sass P, Zoller M, Wigler M. (1987). Three different genes in S. cerevisiae encode the catalytic subunits of the cAMP-dependent protein kinase. Cell , 277–287. [DOI] [PubMed] [Google Scholar]
- Tokiwa G, Tyers M, Volpe T, Futcher B. (1994). Inhibition of G1 cyclin activity by the Ras/cAMP pathway in yeast. Nature , 342–345. [DOI] [PubMed] [Google Scholar]
- Treitel MA, Carlson M. (1995). Repression by SSN6-TUP1 is directed by MIG1, a repressor/activator protein. Proc Natl Acad Sci USA , 3132–3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Pierce M, Schneper L, Guldal CG, Zhang X, Tavazoie S, Broach JR. (2004). Ras and Gpa2 mediate one branch of a redundant glucose signaling pathway in yeast. PLoS Biol , E128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen W, Taylor SS, Meinkoth JL. (1995). The expression and intracellular distribution of the heat-stable protein kinase inhibitor is cell cycle regulated. J Biol Chem , 2041–2046. [DOI] [PubMed] [Google Scholar]
- Wieser R, Adam G, Wagner A, Schüller C, Marchler G, Ruis H, Krawiec Z, Bilinski T. (1991). Heat shock factor-independent heat control of transcription of the CTT1 gene encoding the cytosolic catalase T of Saccharomyces cerevisiae . J Biol Chem , 12406–12411. [PubMed] [Google Scholar]
- Xue Y, Batlle M, Hirsch JP. (1998). GPR1 encodes a putative G protein-coupled receptor that associates with the Gpa2p Galpha subunit and functions in a Ras-independent pathway. EMBO J , 1996–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z, Hatton L, Brown AJ. (2000). Differential post-transcriptional regulation of yeast mRNAs in response to high and low glucose concentrations. Mol Microbiol , 553–565. [DOI] [PubMed] [Google Scholar]
- Zahringer H, Thevelein JM, Nwaka S. (2000). Induction of neutral trehalase Nth1 by heat and osmotic stress is controlled by STRE elements and Msn2/Msn4 transcription factors: variations of PKA effect during stress and growth. Mol Microbiol , 397–406. [DOI] [PubMed] [Google Scholar]
- Zaman S, Lippman SI, Schneper L, Slonim N, Broach JR. (2009). Glucose regulates transcription in yeast through a network of signaling pathways. Mol Syst Biol , 245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaman S, Lippman SI, Zhao X, Broach JR. (2008). How Saccharomyces responds to nutrients. Annu Rev Genet , 27–81. [DOI] [PubMed] [Google Scholar]
- Zampar GG, Kummel A, Ewald J, Jol S, Niebel B, Picotti P, Aebersold R, Sauer U, Zamboni N, Heinemann M. (2013). Temporal system-level organization of the switch from glycolytic to gluconeogenic operation in yeast. Mol Syst Biol , 651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G, Chen Y, Carey L, Futcher B. (2016). Cyclin-dependent kinase co-ordinates carbohydrate metabolism and cell cycle in S. cerevisiae . Mol Cell , 546–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zid BM, O’Shea EK. (2014). Promoter sequences direct cytoplasmic localization and translation of mRNAs during starvation in yeast. Nature , 117–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.