Abstract
Background
Several different IgA-proteases exist in Haemophilus influenzae. The variants have been suggested to play differential roles in pathogenesis, but there is limited information on their distribution in clinical isolates. The objective of this study was to investigate the distribution of IgA-protease genotypes in H. influenzae and assess the association between IgA-protease genotype and type of clinical infection.
Methods
We performed PCR-screening of the IgA-protease gene variants in two cohorts of clinical H. influenzae. The first cohort consisted of 177 isolates from individuals with respiratory tract infection in January 2010, 2011 and 2012. Information on age, gender and clinical infection was available in this cohort. The second cohort comprised 53 isolates, including NTHi from bloodstream, cerebrospinal fluid (CSF) and urogenital origin as well as encapsulated isolates respresenting all capsule types. We assessed associations between IgA protease genotype and clinical predictors using basic statistical tests of association as well as regression analysis.
Results
The igaB gene was found in 46% of isolates in the respiratory tract cohort, and no evident trend could be seen during the study years. However, the igaB gene was significantly less common among invasive isolates (19%), p = 0.003 (Fischer’s exact test), even when encapsulated isolates were excluded (21%), p = 0.012. A significantly negative association between bacteraemia and igaB genotype remained after adjusting for covariates. We did not identify a significant association between IgA-protease gene variants and type of respiratory tract infection, but isolates with an igaA2 genotype were overrepresented in pre-school children.
Conclusions
The distribution of IgA-protease gene variants in Swedish H. influenzae highlighted the widespread abundance of the igaB in isolates from cases of respiratory tract infection, but the igaB gene variant was significantly less common in invasive (bloodstream and CSF) isolates of H. influenzae compared with respiratory tract isolates.
Electronic supplementary material
The online version of this article (10.1186/s12879-018-3464-3) contains supplementary material, which is available to authorized users.
Keywords: Genotype, Human infection, Haemophilus influenzae, IgA protease
Background
A virulence factor common to most successful bacterial respiratory tract pathogens is the ability to degrade human IgA1, the dominant IgA subclass in the nasopharyngeal cavity [1]. Cleavage of IgA1 at different sites of the heavily glycosylated hinge region is performed by secreted endopeptidases that are structurally heterogenous in different pathogens. IgA-proteases are produced by Streptococcus pneumoniae [2], and presence of IgA proteases distinguish pathogenic from non-pathogenic species of Neisseria [3] and Haemophili [4]. The exact biological significance of microbial IgA-proteases is not understood, as animal models are of little use due to the specificity for human IgA1, but IgA-proteases are widely believed to facilitate host mucosal colonization and tissue invasion [5].
In H. influenzae, at least three patterns of IgA1 cleavage exist, defined by what bond they cleave in the hinge region; IgA can be cleaved at either of two distinct sites or at both sites simultaneously [6, 7]. Encapsulated H. influenzae are generally clonal and have specific cleavage patterns, even though data are conflicting regarding the cleavage pattern of type f isolates, the only encapsulated type associated with IgA-protease antigenic diversity [6–8]. In contrast, non-typeable H. influenzae isolates are heterogenous, may have any of the three cleavage patterns and have a high degree of IgA-protease antigenic diversity [7]. The cleavage patterns correspond to different gene variants of the common IgA-protease gene, igaA, and of a second IgA-protease gene, igaB, with a high degree of homology with the IgA1 protease of Neisseria spp. [9]. The two genes both code for autotransporters with autoproteolytic cleavage of the passenger domain responsible for the protease activity [10].
Virtually all H. influenzae isolates have the iga gene, while only a subgroup carries both igaA and igaB. The presence of igaB is associated with higher levels of protease activity and is considered to be a virulence factor on its own [11]. Altogether, four distinct gene variants of IgA-proteases have been discovered based on two variants of the two genes [12], and their respective expression results in at least three different cleavage patterns. In the only major study that has been performed regarding the IgA-protease gene distribution in a substantial collection of isolates, 30–40% of NTHi causing disease carried the igaB gene [11], with a distinct variation between disease entities.
The purposes of this study were: 1) to investigate the distribution of IgA-protease genotypes in a collection of consecutive clinical respiratory tract H. influenzae isolates from patients with respiratory tract infections as well as in a more diverse collection of encapsulated and invasive isolates, and 2) to assess the association between IgA-protease subtype and type of clinical infection.
Results
A total of 230 isolates were included in the screening analysis. Of these, 177 isolates were from the collection of respiratory tract isolates; 43 isolates from January 2010, 52 isolates from January 2011 and 82 isolates from January 2012. In the 53 isolates comprising cohort 2, 42 isolates were from cases of invasive disease (26 from the bloodstream and 16 from cerebrospinal fluid (CSF)), 3 were from urogenital infections and the remaining 8 were encapsulated isolates from different origins. Of the invasive isolates, 33 were non-typeable (18 CSF and 15 bloodstream isolates). For a more detailed description of cohort 2, see Additional file 1: Table S1.
Clinical descriptive features of patients in the respiratory tract cohort
The 177 isolates were collected from 177 unique patients. The descriptive features of patients from cohort 1 are presented in Table 1. The gender distribution was almost balanced, with a slight overrepresentation of women. The median age in the cohort was 32 years of age, 66 samples (37.7%) were from pre-school children (< 6 years of age). Approximately two thirds of isolates were collected in primary care facilities and one third in hospital care. Nineteen % of isolates were from cases of acute otitis media and 15% were from cases with lower respiratory tract infection. In 22% of patients, there was insufficient information in the referral text to identify the type of clinical infection.
Table 1.
Cohort 1 comprising all respiratory tract isolates (n = 177). The distribution of clinical and laboratory characteristics per study year
| Culture year | 2010 | 2011 | 2012 | Total |
|---|---|---|---|---|
| Number of isolates | 43 | 52 | 82 | 177 |
| igaA1, n (%) | 31 (72%) | 39 (75%) | 51 (62%) | 121 (68%) |
| igaA1 alone, n (%) | 14 (33%) | 29 (56%) | 20 (24%) | 63 (36%) |
| igaA2a, n (%) | 12 (28%) | 13 (25%) | 30 (37%) | 55 (31%) |
| igaB1a, n (%) | 23 (53%) | 15 (29%) | 43 (52%) | 81 (46%) |
| igaB2, n (%) | 0 | 0 | 1 (1.2%) | 1 (0.6%) |
| Age (years), mean (median) | 24 (6) | 32 (36) | 29 (34) | 29 (32) |
| Gender, % female | 53% | 67% | 55% | 58% |
| Primary care referrals, n (%) | 31 (72%) | 33 (63%) | 55 (67%) | 119 (67%) |
| Acute otitis media, n (%) | 10 (24%) | 8 (15%) | 15 (18%) | 33 (19%) |
| Sinusitis, n (%) | 5 (12%) | 5 (10%) | 11 (13%) | 21 (12%) |
| Upper respiratory tract infection, n (%) | 17 (41%) | 21 (40%) | 18 (22%) | 56 (32%) |
| Lower respiratory tract infection, n (%) | 7 (17%) | 8 (15%) | 12 (15%) | 27 (15%) |
| Unclear diagnosis from referral, n (%) | 2 (5%) | 10 (19%) | 26 (32%) | 38 (22%) |
aSome isolates have both the igaA and igaB gene, resulting in a total percentage of > 100%
The iga gene distribution in consecutive isolates from the respiratory tract
The PCR screening results performed for all 177 isolates of cohort 1 is presented in Table 1. The igaA gene PCR was positive in all but one isolate (176/177; 99%). The igaA1 gene variant was found in 121/177 (68%) whereas the igaA2 gene variant was found in 55/177 (31%) of isolates. Isolates also carrying the igaB gene were common throughout the study period, as 81/177 (46%) of isolates tested positive in the PCR screening. Only one of the isolates was igaB2 positive. There was a year-to-year variation of the distribution, but no clear trend over time, with a lower proportion of igaB-positive isolates in 2011 compared with both 2010 and 2012, likely reflecting temporal variations in circulating strains.
The iga gene distribution in cohort 2
The PCR screening results for all 53 isolates of cohort 2 is presented in Table 2. The igaA gene PCR was positive in all isolates. The capsule type-associated IgA-cleavage pattern from prior studies was mainly confirmed in this genotype analysis, as all included type b and d isolates had an igaA1 genotype, and type c and e isolates had an igaA2 genotype. Data on type f isolates have been conflicting as judged by prior studies, but in our study all four type f isolates included carried an igaA2 genotype. In our two capsule type a isolates tested, the PCR for igaB screening was positive. Among the tested isolates in cohort 2, igaB was found in 11 isolates (21%), including 3 igaB2. Among all invasive isolates and invasive NTHi isolates igaB was found in 8/42 (19%) and 7/33 (21%) isolates, respectively. Two of the three included urogenital isolates had an igaB genotype. Information regarding isolation site and capsule type for cohort 2 is outlined in Additional file 1: Table S1. Information on the clinical descriptive features was, however, only available in a subset of the cohort.
Table 2.
The distribution of laboratory characteristics per isolation site of the 53 isolates comprising cohort 2
| Isolation site | Bloodstream | CSF | Otherb | Total |
|---|---|---|---|---|
| Number of isolates | 26 | 16 | 11 | 53 |
| Encapsulated | 8 | 1 | 6 | 15 |
| Nonencapsulated | 18 | 15 | 5 | 38 |
| igaA1, n (%) | 19 (73%) | 11 (69%) | 6 (55%) | 36 (68%) |
| igaA1 alone, n (%) | 16 (62%) | 8 (50%) | 5 (45%) | 26 (49%) |
| igaA2a, n (%) | 7 (27%) | 5 (31%) | 5 (45%) | 17 (32%) |
| igaB1a, n (%) | 1 (4%) | 4 (24%) | 3 (27%) | 8 (15%) |
| igaB2, n (%) | 2 (8%) | 1 (6%) | 0 | 3 (6%) |
aSome isolates have both the igaA and igaB gene, resulting in a total percentage of > 100%
bAvailable details on all isolates are given in Additional file 1: Table S1
IgA1 cleavage pattern as compared to the diagnosed genotype
It is well established that a positive PCR screening does not necessarily correlate with protease activity. Therefore, we determined the IgA1 cleavage patterns by Western blot in a subset of 54 isolates. The isolates were chosen to represent all different genotypes, all years in cohort 1, in addition to all capsule types and isolation sites. Forty isolates were from cohort 1 (14 from 2010, 9 from 2011 and 17 from 2012), and 14 isolates from cohort 2 (representing all capsule types as well as isolation sites). Cleavage pattern A1 correlates with IgA-A1, cleavage pattern A2/B can be either IgA-A2 or IgA-B, and cleavage pattern A1 + A2/B indicates expression of two proteases simultaneously. Among 22 isolates tested with an igaA1 genotype without igaB, 20 had indeed an A1 cleavage pattern. Among the 14 isolates analysed with an igaA2 genotype without igaB, 13 had an A2/B cleavage pattern. In 11 out of 14 isolates tested with an igaA1 + igaB genotype, the igaB gene was expressed; 9 had an A2/B cleavage pattern, while two had an A1 + A2/B cleavage pattern. Two isolates with an igaB2 genotype were examined and only one of them had an A2/B cleavage pattern. Both analysed isolates with a combined igaA2 and igaB genotype had an A2/B cleavage pattern. All tested encapsulated isolates had cleavage patterns corresponding to the genotype except for capsule type a, which had a A1 cleavage pattern, suggesting that the igaB gene is not expressed in H. influenzae type a. Taken together, in the majority of isolates the cleavage pattern phenotype corresponded to the genotype.
Associations between the iga genotype and clinical predictors
Data of association and results from the regression analysis are presented in Tables 3 and 4, respectively. The igaB genotype was less common in invasive (bloodstream and CSF) isolates compared with isolates from other isolation sites. The difference was significant using Fischer’s test, 8/42 (19%) of invasive isolates compared with 84/188 (45%) of non-invasive isolates (p = 0.003), as well as for invasive non-typeable H. influenzae isolates alone, 7/33 (21%) compared with 83/182 (46%) of non-invasive isolates (p = 0.012). The negative association between igaB gene presence and bloodstream infection remained independently significant even when adjusting for covariates in a multinomial multivariate regression (Table 4). Younger age was associated with acute otitis media, whereas increasing age was associated with bloodstream infection in the same model. There was no significant association between iga genotype and type of respiratory tract infection (Table 3). A significant association between igaA2 genotype and young age was also observed (Table 3).
Table 3.
The iga protease gene variants among respiratory tract isolates with different properties
| iga genotype | igaA1 alone | igaA2 | igaB gene present | p-value |
|---|---|---|---|---|
| Age (continuous, year), mean (median) | 29.4 (21) | 18.1 (4) | 33.5 (40) | < 0.001 |
| Age category (n = 175, 2 missing) | 0.042 | |||
| > 70 years, n (% within genotype) | 5 (8%) | – | 5 (6%) | |
| 50–70 years, n (% within genotype) | 14 (22%) | 7 (13%) | 20 (25%) | |
| 20–50 years, n (% within genotype) | 15 (24%) | 14 (25%) | 27 (34%) | |
| 6–20 years, n (% within genotype) | 5 (8%) | 3 (5%) | 6 (8%) | |
| < 6 years, n (% within genotype) | 24 (38%) | 31 (56%) | 21 (27%) | |
| Gender, % female | 56% | 53% | 62% | 0.684 |
| Primary care referrals, % | 71% | 65% | 64% | 0.966 |
| Clinical diagnosis (n = 175, 2 missing) | 0.172 | |||
| AOM, n (% within genotype) | 11 (17%) | 13 (24%) | 13 (16%) | |
| Sinusitis, n (% within genotype) | 6 (10%) | 6 (11%) | 10 (13%) | |
| Upper respiratory tract infection, n (% within genotype) | 26 (41%) | 16 (29%) | 24 (30%) | |
| Lower respiratory tract infection, n (% within genotype) | 11 (17%) | 3 (5%) | 16 (20%) | |
| Unclear from referral, n (% within genotype) | 9 (14%) | 17 (31%) | 16 (20%) |
The Kruskal-Wallis test for continuous variables and the chi-2 test for categorical variables were used. Significant p-values are indicated in bold
Table 4.
Multinomial univariate and multivariate regressions with clinical diagnosis as the outcome (URTI was used as base category)
| Univariate analysis | Multivariate analysis | |
|---|---|---|
| Covariate | Beta-coefficient (95% CI), p-value | Beta-coefficient (95% CI), p-value |
| Age | AOM vs URTI −0.087 (−0.13 - (−0.044)), < 0.001 Sepsis vs URTI 0.038 (0.016–0.060), 0.001 |
AOM vs URTI - 0.091 (− 0.14 – (− 0.047), < 0.001 Sepsis vs URTI 0.041 (0.018–0.063), < 0.001 |
| igaB vs all | Sepsis vs URTI − 1.69 (−3.00 – (− 0.38)), 0.012 | Sepsis vs URTI −1.86 (−3.22- (− 0.49)), 0.008 |
| igaA2 vs all | no significant category |
The analysis was performed for isolates with igaB genotype vs all other isolates as well as those with igaA2 genotype vs all other isolates. Only statistically significant associations are presented
Discussion
In Sweden, the distribution of iga genotypes in H. influenzae was surprisingly consistent with the distribution described in the prior study performed on selected cohorts [11], as more than 40% of isolates from consecutive respiratory tract isolates were positive in the igaB gene screening. There was no significant association between the presence of the igaB gene and type of respiratory tract infection. We found a significant negative association between the presence of the igaB gene and invasive disease. This corresponds to the results in the study by Fernaays et al., where less than 20% invasive isolates carried the igaB gene compared with 30% of middle ear isolates and more than 40% of sputum isolates [11]. In the present material, 30% of CSF isolates possess the igaB allele, which is similar to middle ear isolates.
It has been suggested that IgA-A and IgA-B proteases have different roles in H. influenzae pathogenesis, and may be variably expressed during different phases of host colonization and invasion [13]. While IgA-A is implied in tissue colonization and invasion, IgA-B cleaves Lysosomal-associated membrane protein 1 (LAMP-1), which has been suggested to be important for bacterial persistence [14]. Bacterial persistence and colonization is likely less important than tissue invasion in cases of invasive disease. In addition, the expression of an additional immunogenic protein may hypothetically even be a negative factor in bloodstream survival of H. influenzae considering the limited serum survival capacity of the bacterium and potent humoral response elicited in most individuals [15]. One study has related the capacity to cause disease to differences in actual protease activity levels, where increased protease activity was suggested to be associated with increasing disease severity [16].
In prior studies, no association with specific pathogenesis or targeted population has been correlated with the igaA2 variant. In this study, we found that igaA2 was associated with young age, and was comparatively uncommon among individuals above 6 years of age. Furthermore, most H. influenzae type f isolates seem to have igaA2. Not much is known regarding the relative immunogenicity of the specific IgA protease variants. In children, a larger variety of strains have been suggested as necessary since antigenic type variation is important for colonization in children, in contrast to adults with chronic obstructive pulmonary disease (COPD) [17]. Thus, it could be speculated that IgA-A2 may have a smaller range of antigenic variation, a range potentially exhausted in early life for most patients. It has been suggested that strains with cleavage pattern A2/B has a smaller range of antigenic variation [8].
The distribution of iga genotypes in this study is similar to the distribution from a prior study from North America [9]. Non-typeable H. influenzae is often described as quite genetically diverse due to its natural competence, and subsequent horizontal gene transfer. This variation seems unlimited based on descriptions in the literature, but the core genome in H. influenzae has been successfully divided into a limited number of clades using Multi Locus Sequence Typing (MLST) or whole genome sequencing, with no clear geographical difference between clade distribution [18, 19]. Encapsulated isolates sort into specific clades while NTHi are scattered throughout the phylogenic tree, which still contains a finite number of clades. In the work by De Chiara and colleagues, all NTHi isolates with igaB were sorted into two distinct clades [19]. Our results, in comparison with results from the North American study, may suggest that the distribution of genetic clades of respiratory tract NTHi might be relatively consistent across geographical regions.
The main strengths of the study include the tested population-based isolate collection, including non-selected consecutive respiratory tract isolates from three different years with associated clinical information as well as a collection of encapsulated and non-encapsulated isolates of varying origin. Another strength is the statistical comparisons included. Limitations of the study include its modest size and the differences in origin and isolation years between cohort 1 and cohort 2. This is also the reason why the cohorts have been treated separately.
Conclusions
To summarize, our data demonstrate that the igaB genotype is frequently found in respiratory tract isolates from patients in the County of Skåne, Sweden with respiratory tract infections, a finding consistent over a 3-year period of observations. We also demonstrate that the igaB genotype was less frequently seen in invasive isolates compared to isolates from the respiratory tract. Finally, the igaA2 genotype seems to be particularly relevant for respiratory tract infections in children (< 6 years).
Methods
Study setting and isolate selection
This study was performed in Malmö, Sweden, at a laboratory of clinical microbiology serving an area with a population of approximately 500,000 patients. It was the only microbiology laboratory in the region. Two cohorts were investigated. In the first cohort, all H. influenzae isolated from respiratory tract samples in January 2010, 2011 and 2012 (all tested as non-typeable) were selected for further testing. This strategy was chosen to allow for temporal variations in the distribution. The second cohort comprised of a collection of well-characterized invasive and encapsulated isolates from cases with different clinical infections. The cohort includes Swedish isolates from 1997 to 2010 and international isolates representing all capsule types. Details on isolation sites and origins of isolates in cohort 2 are given in Additional file 1: Table S1.
Culture conditions and DNA preparation
All isolates were grown on chocolate agar plates overnight (18 h) in a humid atmosphere at 37 °C and 5% CO2 before any experiments were conducted. All isolates had been confirmed as H. influenzae using standard microbiology techniques or MALDI-TOF (Matrix Assisted Laser Desorption and Ionization - Time-of-flight mass spectrometry). DNA was prepared by adding a few colonies of bacteria to distilled water. After heating at 98 °C for 10 min, each sample was centrifuged (16,000 x g) followed by collection of the supernatant.
PCR screening
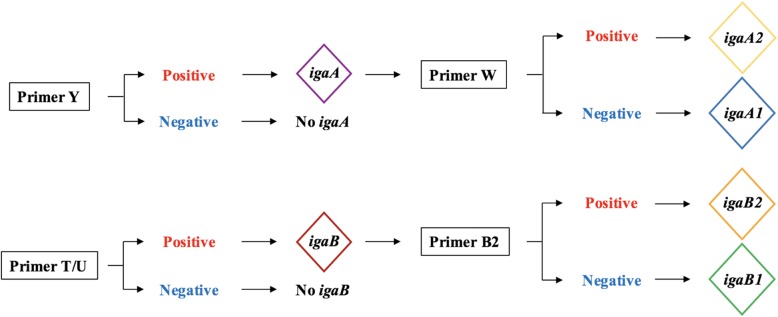
All isolates were screened using four different PCR primer pairs as described in prior studies [12], Y (identifying the igaA gene), W (identifying the igaA2 variant of the igaA gene) as well as T and U (identifying the igaB gene). Isolates positive in the igaB screening were subjected to a fifth PCR, identifying the igaB2 variant. The genotyping algorithm is visualized in Fig 1. Conditions and primer sequences are outlined in Additional file 2: Table S2. DNA was separated using a 1xTAE 1% agarose gel with 0.625 mg/mL ethidium bromide. Bands were visualized with UV light using a BioRad molecular Imager (Hercules, CA).
Fig. 1.
Sequential genotyping used to identify the different iga alleles
IgA1 cleavage analysis by Western blot
A subset of 54 isolates, representing all IgA protease gene variants, was chosen for confirmation of IgA1 cleavage by Western blot. The strains were grown in brain-heart-infusion (BHI) broth supplemented with 10 μg/ml NAD and hemin at 37 °C 5% CO2 overnight. The bacteria were spun down and 20 μl of supernatant was incubated with 3 μl of 1 mg/ml human IgA1 (Calbiochem, Solna, Sweden) at 37 °C overnight. The proteins were separated by SDS-PAGE and transferred to nitrocellulose membranes. The membranes were incubated with polyclonal rabbit anti-human IgA antibodies at dilution 1:1000 overnight at 4 °C (A0262, Dako; Glostrup Denmark) followed by polyclonal HRP (horseradish peroxidase)-conjugated swine anti-rabbit IgG (dilution 1:1000) (P0217, Dako; Glostrup, Denmark) 2 h at room temperature and developed.
Information on clinical infection
In the cohort of respiratory tract isolates, the information regarding type of clinical infection was gathered from the referral text. The disease types were lower respiratory tract infection, acute otitis media, sinusitis or unspecfic upper respiratory tract infection. We also collected data on patient age, gender and whether the culture had been taken in primary care or at a hospital facility. In the second cohort, information on capsule type and isolation site (blood, CSF or other) was available, and in a subset of cases also age and gender.
Statistical analysis
Data analysis was performed using STATA, version14 (College Station, TX). The presence or absence of igaB between invasive and non-invasive isolates was assessed using a two-sided Fischer’s exact test. Comparison of clinical traits among isolates with different genotypes was performed using the Kruskal-Wallis test for continuous variables and the chi-2 test for categorical variables. To assess the relevance of confounders in the associations between IgA1 protease genotypes and clinical diagnoses, multinomial multivariate logistic regression models using categorical clinical diagnosis as outcome were fitted using the purposeful selection algorithm [20]. Briefly, the main predictor (IgA protease genotype, igaA2 vs all and igaB vs all, respectively) and all covariates that in a univariate analysis had a p-value of < 0.2 were included in a crude model. The least significant covariate was then stepwise removed until only significant covariates or covariates that substantially affected the odds ratio of a remaining covariate were retained in the final model.
Additional files
Table S1. A more detailed description of isolates comprising cohort 2. (DOCX 20 kb)
Table S2. Primers and PCR conditions. (DOCX 18 kb)
Acknowledgements
We would like to thank Mrs. Kerstin Norrman for technical assistance, and the Clinical Microbiology laboratory at Labmedicin Skåne (Lund, Sweden) for providing clinical isolates.
Funding
This study was supported by Anna and Edwin Berger foundation (KR), the Swedish Medical Research Council (KR: grant number K2015-57X-03163-43-4, www.vr.se), the Cancer Foundation at the University Hospital in Malmö (KR), the Royal Physiographical Society (GM), Skåne County Council’s research and development foundation (KR), and the Heart Lung Foundation (KR: grant number 20150697, www.hjart-lungfonden.se). The various funding bodies did not interfere with the research project, and did not influence the interpretation of results obtained during the study.
Availability of data and materials
The dataset used during the current study is available from the corresponding author on reasonable request.
Abbreviations
- BHI
Brain-heart-infusion
- COPD
Chronic obstructive pulmonary disease
- CSF
Cerebrospinal fluid
- DNA
Deoxyribonucleic acid
- HRP
Horseradish peroxidase
- IgA
Immunoglobulin A
- IgG
Immunoglobulin G
- LAMP-1
Lysosomal-associated membrane protein 1
- MALDI-TOF
Matrix assisted laser desorption and ionization - Time-of-flight mass spectrometry
- MLST
Multi locus sequence typing
- NTHi
Non-typeable Haemophilus influenzae
- PCR
Polymerase chain reaction
Authors’ contributions
This study was conceived by FR, KR and TM. The laboratory work was performed by VL and GM. All authors contributed to the writing of the manuscript.
Ethics approval and consent to participate
The study was approved by the regional ethical review board (EPN) in Malmö/ Lund, Sweden (2018/244). According to national regulations and guidelines, all samples analysed were anonymous and any connection with a particular patient could not be traced.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Fredrik Resman, Email: fredrik.resman@med.lu.se.
Guillaume Manat, Email: guillaume.manat@med.lu.se.
Victor Lindh, Email: vctrlndh@gmail.com.
Timothy F. Murphy, Email: murphyt@buffalo.edu
Kristian Riesbeck, Phone: +467030377433, Email: kristian.riesbeck@med.lu.se.
References
- 1.Kett K, Brandtzaeg P, Radl J, Haaijman JJ. Different subclass distribution of IgA-producing cells in human lymphoid organs and various secretory tissues. J Immunol. 1986;136(10):3631–3635. [PubMed] [Google Scholar]
- 2.Male CJ. Immunoglobulin A1 protease production by Haemophilus influenzae and Streptococcus pneumoniae. Infect Immun. 1979;26(1):254–261. doi: 10.1128/iai.26.1.254-261.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mulks MH, Plaut AG. IgA protease production as a characteristic distinguishing pathogenic from harmless neisseriaceae. N Engl J Med. 1978;299(18):973–976. doi: 10.1056/NEJM197811022991802. [DOI] [PubMed] [Google Scholar]
- 4.Kilian M, Mestecky J, Schrohenloher RE. Pathogenic species of the genus Haemophilus and Streptococcus pneumoniae produce immunoglobulin A1 protease. Infect Immun. 1979;26(1):143–149. doi: 10.1128/iai.26.1.143-149.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kilian M, Reinholdt J, Lomholt H, Poulsen K, Frandsen EV. Biological significance of IgA1 proteases in bacterial colonization and pathogenesis: critical evaluation of experimental evidence. APMIS. 1996;104(5):321–338. doi: 10.1111/j.1699-0463.1996.tb00724.x. [DOI] [PubMed] [Google Scholar]
- 6.Kilian M, Thomsen B, Petersen TE, Bleeg H. Molecular biology of Haemophilus influenzae IgA1 proteases. Mol Immunol. 1983;20(9):1051–1058. doi: 10.1016/0161-5890(83)90046-9. [DOI] [PubMed] [Google Scholar]
- 7.Mulks MH, Kornfeld SJ, Frangione B, Plaut AG. Relationship between the specificity of IgA proteases and serotypes in Haemophilus influenzae. J Infect Dis. 1982;146(2):266–274. doi: 10.1093/infdis/146.2.266. [DOI] [PubMed] [Google Scholar]
- 8.Kilian M, Thomsen B. Antigenic heterogeneity of immunoglobulin A1 proteases from encapsulated and non-encapsulated Haemophilus influenzae. Infect Immun. 1983;42(1):126–132. doi: 10.1128/iai.42.1.126-132.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fernaays MM, Lesse AJ, Sethi S, Cai X, Murphy TF. Differential genome contents of nontypeable Haemophilus influenzae strains from adults with chronic obstructive pulmonary disease. Infect Immun. 2006;74(6):3366–3374. doi: 10.1128/IAI.01904-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spahich NA, St Geme JW., 3rd Structure and function of the Haemophilus influenzae autotransporters. Front Cell Infect Microbiol. 2011;1:5. doi: 10.3389/fcimb.2011.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fernaays MM, Lesse AJ, Cai X, Murphy TF. Characterization of igaB, a second immunoglobulin A1 protease gene in nontypeable Haemophilus influenzae. Infect Immun. 2006;74(10):5860–5870. doi: 10.1128/IAI.00796-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murphy TF, Kirkham C, Jones MM, Sethi S, Kong Y, Pettigrew MM. Expression of IgA proteases by Haemophilus influenzae in the respiratory tract of adults with chronic obstructive pulmonary disease. J Infect Dis. 2015;212(11):1798–1805. doi: 10.1093/infdis/jiv299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murphy Timothy F, Kirkham Charmaine, Gallo Mary C, Yang Yang, Wilding Gregory E, Pettigrew Melinda M. Immunoglobulin A Protease Variants Facilitate Intracellular Survival in Epithelial Cells By Nontypeable Haemophilus influenzae That Persist in the Human Respiratory Tract in Chronic Obstructive Pulmonary Disease. The Journal of Infectious Diseases. 2017;216(10):1295–1302. doi: 10.1093/infdis/jix471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clementi CF, Hakansson AP, Murphy TF. Internalization and trafficking of nontypeable Haemophilus influenzae in human respiratory epithelial cells and roles of IgA1 proteases for optimal invasion and persistence. Infect Immun. 2014;82(1):433–444. doi: 10.1128/IAI.00864-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.King PT, Ngui J, Gunawardena D, Holmes PW, Farmer MW, Holdsworth SR. Systemic humoral immunity to non-typeable Haemophilus influenzae. Clin Exp Immunol. 2008;153(3):376–384. doi: 10.1111/j.1365-2249.2008.03697.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vitovski S, Dunkin KT, Howard AJ, Sayers JR. Nontypeable Haemophilus influenzae in carriage and disease: a difference in IgA1 protease activity levels. JAMA. 2002;287(13):1699–1705. doi: 10.1001/jama.287.13.1699. [DOI] [PubMed] [Google Scholar]
- 17.Lomholt H, van Alphen L, Kilian M. Antigenic variation of immunoglobulin A1 proteases among sequential isolates of Haemophilus influenzae from healthy children and patients with chronic obstructive pulmonary disease. Infect Immun. 1993;61(11):4575–4581. doi: 10.1128/iai.61.11.4575-4581.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Erwin AL, Sandstedt SA, Bonthuis PJ, Geelhood JL, Nelson KL, Unrath WC, Diggle MA, Theodore MJ, Pleatman CR, Mothershed EA, Sacchi CT, Mayer LW, Gilsdorf JR, Smith AL. Analysis of genetic relatedness of Haemophilus influenzae isolates by multilocus sequence typing. J Bacteriol. 2008;190(4):1473–1483. doi: 10.1128/JB.01207-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Chiara M, Hood D, Muzzi A, Pickard DJ, Perkins T, Pizza M, Dougan G, Rappuoli R, Moxon ER, Soriani M, Donati C. Genome sequencing of disease and carriage isolates of nontypeable Haemophilus influenzae identifies discrete population structure. Proc Natl Acad Sci U S A. 2014;111(14):5439–5444. doi: 10.1073/pnas.1403353111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bursac Z, Gauss CH, Williams DK, Hosmer DW. Purposeful selection of variables in logistic regression. Source code for biology and medicine. 2008;3:17. doi: 10.1186/1751-0473-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. A more detailed description of isolates comprising cohort 2. (DOCX 20 kb)
Table S2. Primers and PCR conditions. (DOCX 18 kb)
Data Availability Statement
The dataset used during the current study is available from the corresponding author on reasonable request.