Abstract
Post-translational histone modifications such as acetylation and methylation can affect gene expression. Histone acetylation is commonly associated with activation of gene expression whereas histone methylation is linked to either activation or repression of gene expression. Depending on the site of histone modification, several histone marks can be present throughout the genome. A combination of these histone marks can shape global chromatin architecture, and changes in patterns of marks can affect the transcriptomic landscape. Alterations in several histone marks are associated with different types of cancers, and these alterations are distinct from marks found in original normal tissues. Therefore, it is hypothesized that patterns of histone marks can change during the process of tumorigenesis.
This review focuses on histone methylation changes (both removal and addition of methyl groups) in malignant melanoma, a deadly skin cancer, and the implications of specific inhibitors of these modifications as a combinatorial therapeutic approach.
Keywords: Histone methylation, Melanoma, Epigenetics, Histone demethylase, Histone methyltransferase, Small molecule inhibitor
Background
Histone modifications
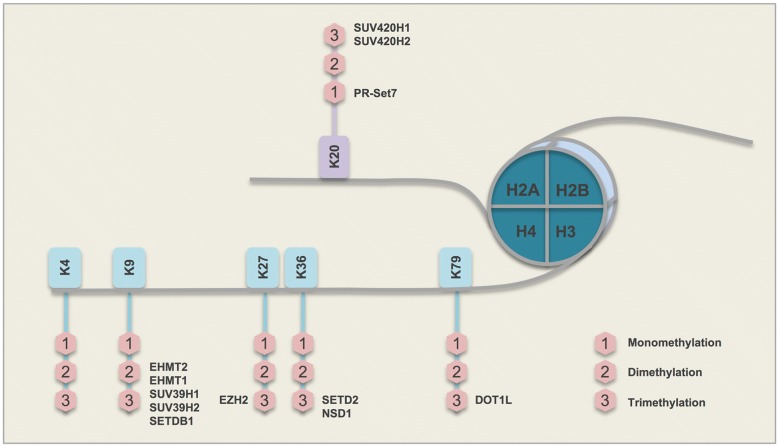
The nucleosome, the fundamental structural unit of chromosomes, is composed of two copies each of four core histones (i.e., H2A, H2B, H3, and H4), around which 147 bp of DNA is coiled [1, 2]. N-terminal tails of histone polypeptides can be altered by a variety of post-translational modifications, including methylation, acetylation, phosphorylation, ubiquitylation, glycosylation, ADP-ribosylation, carbonylation, and SUMOylation (collectively known as histone modifications) [3–6]. The acetylation of histones is controlled by the balanced action of histone acetyltransferases (HATs) and histone deacetylases (HDACs). Acetylated histones have been associated with actively expressed genes. However, histone methylation may have both repressive (H3K9, H3K27) or enhancing (H3K4) effects on transcription, depending on the residue that is modified [7]. Histone methylation can occur on lysine and/or arginine residues. Lysine residues at various positions along the histone N-terminal tail are common sites for methylation (Fig. 1). Lysine (K) methylation occurs by stepwise addition of one to three methyl groups, which can lead to unique functions of a genomic region. This stepwise conversion, from an unmethylated to a trimethylated lysine residue, is facilitated by histone methyltransferases (writers), and the backward demethylation process is catalyzed by histone demethylases (erasers). Further, a group of proteins (readers) recognize methyl-lysines throughout the genome [8]. Epigenetic changes along with genetic alterations can determine cell fate, either to maintain cell homeostasis or to promote tumorigenesis. Because epigenetic changes are reversible, understanding such changes is of crucial significance for drug development and exploring therapeutic strategies [9].
Fig. 1.
Major histone H3 and H4 lysine methyltransferase enzymes (H3K4, H3K9, H3K27, H3K36, H3K79, and H4K20) and their common sites of action
Several histone methylation alterations are known to play a role in the transition of melanocytes to melanoma cells. These alterations are the consequence of deregulation of their corresponding enzymes. In this review, we focus on major erasers and writers of histone lysine methylation as regulated via two categories of demethylase and methyltransferase enzymes and discuss compounds available to target these enzymes and reverse their impact.
Main body
Histone demethylases
JARID1B (H3K4-demethylase)
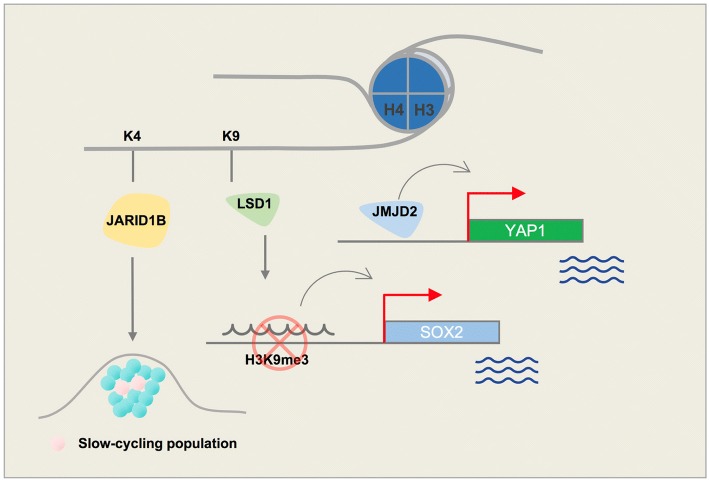
Several enzymes demethylate histone 3 at the position 4 lysine residue. One such enzyme, H3K4 demethylase JARID1B (PLU-1, KDM5B), plays a role in the development of melanoma, and regulates gene transcription and cell differentiation [10]. It has three plant homeodomain (PHD) finger domains in its structure (Fig. 2). PHD fingers are approximately 50–80 aa long and are usually involved in chromatin-mediated gene regulation. Two of these PHD fingers in JARID1B can bind to histones. JARID1B binds to its substrate through distinct PHD fingers. Finger PHD1 is highly specific for unmethylated H3K4 (H3K4me0). PHD3 is more specific to H3K4me3 [11]. The H3K4 demethylase JARID1B is highly expressed in nevi but not in melanoma; however, it marks a small subpopulation of melanoma cells that can cycle very slowly throughout the tumor mass and is essential for continuous tumor growth [12, 13] (Fig. 3). Interestingly, different treatments, including BRAF inhibitors (e.g., vemurafenib) as well as cytotoxic agents (e.g., cisplatin), lead to enrichment of slow-cycling, long-lasting melanoma tumor cells that express H3K4-demethylase JARID1B [14]. Eliminating this subpopulation of tumor cells might help overcome resistance to conventional treatments.
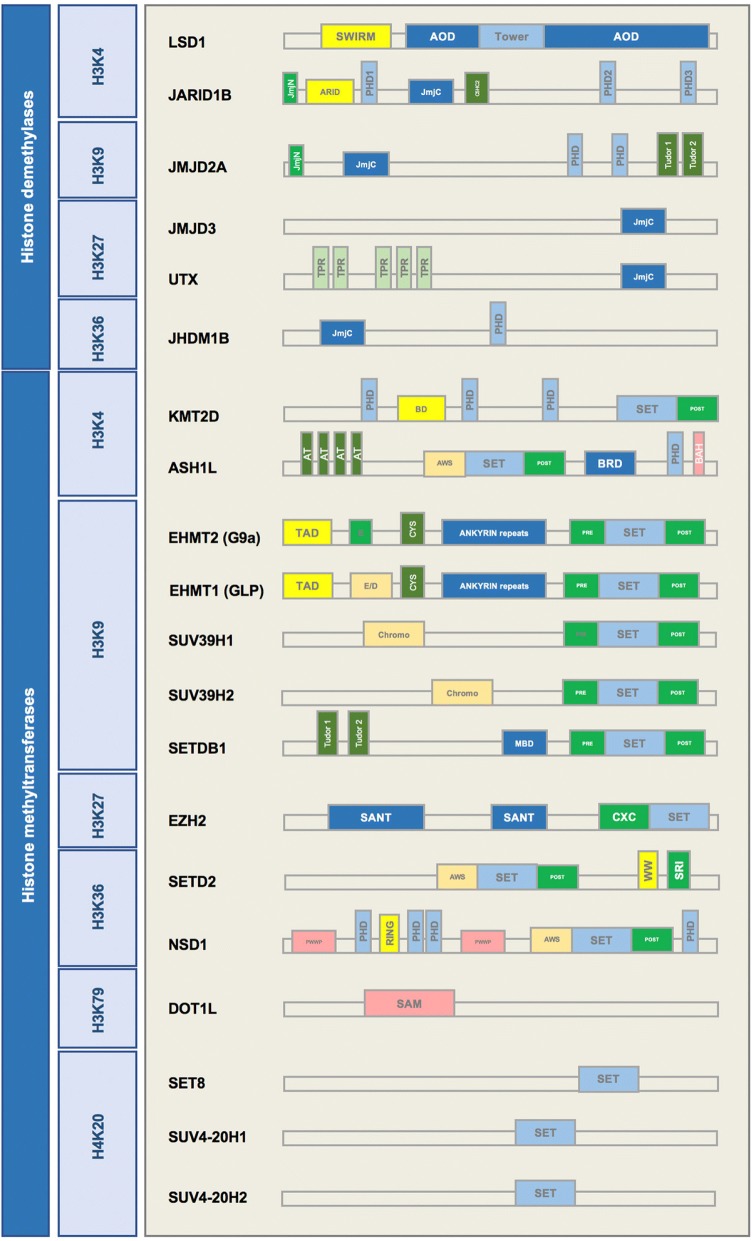
Fig. 2.
Schematic structures of histone lysine demethylases (H3K4, H3K9, H3K27, H3K36), histone lysine methyltransferases (H3K4, H3K9, H3K27, H3K36, H4K20) with SET domain, and histone methyltransferases without the SET domain (H3K79)
Fig. 3.
Schematic figure demonstrating key roles of histone lysine demethylases (H3K4 and H3K9) in melanoma
Treatment of melanoma tumors with MAPK inhibitors increases the H3K4 demethylase JARID1B-positive subpopulation of melanoma cells [15]. These cells are slow-cycling and treatment-resistant. Thus, in addition to curbing tumor growth with MAPK inhibitor, a supplementary agent is needed to effectively treat this subpopulation of cells. Several compounds inhibit JARID1B activity. KDM5-C49 is a potent inhibitor of the JARID1 enzyme family; however, its cellular permeability is limited. Therefore, KDM5-C70 was developed to be used in cellular assays and in vivo [16]. KDM5-C70 inhibits myeloma cells, increasing levels of H3K4me3 at the whole genome level [16]. 2,4-PDCA (2,4-pyridinedicarboxylic acid) acts in vitro to inhibit JARID1B and KDM4 demethylases. A high throughput screen of > 15,000 compounds conducted to identify inhibitors of JARID1B confirmed the activity of 2,4-PDCA and also revealed a novel compound, 2-4(4-methylphenyl)-1,2-benzisothiazol-3(2H)-one (PBIT) that can effectively inhibit JARID1B in vitro with no activity toward H3K27 demethylases, UTX or JMJD3 [17]. KDOAM-25 was recently introduced as a potent and selective JARID1 inhibitor with the ability to impact multiple myeloma cells [18].
LSD1 (H3K4-demethylase | H3K9-demethylase)
H3K4 methyl groups can also be removed by members of the LSD (lysine-specific histone demethylase) family [19, 20]. LSD1, also known as KDM1A, demethylates histone 3 on lysine residues at positions 4 and 9 (H3K4 and H3K9). LSD1 specifically removes 1 or 2 methyl groups from H3K4me2, converting it to H3K4me1/0. Monomethylation of H3K4 is a histone modification that marks enhancers throughout the genome. The histone demethylase role of LSD1 could change the gene transcription status by generating a H3K4me1 mark and activating enhancers, or by removing another methyl group and generating unmethylated H3K4. LSD1 has a SWIRM domain and 2 amino oxidase domains (AODs) in its structure (Fig. 2). A SWIRM domain is usually present in proteins with a role in chromatin remodeling and in expression of genes such as SMARC2 and SWI3.
LSD1 can bind to both gene promoters and distal elements. This feature of LSD1 is in line with its demethylation role on H3K4 (promoter) as well as H3K9 (distal) [21]. Trimethylated H3K9 is a well-known heterochromatin mark, which often highlights closed chromatin regions and is present on distal regions of genes. H3K9me3-active demethylase LSD1 is reported to disable senescence in melanocytes. Senescence halts the proliferation of melanocytes and further progression to melanoma cells; this role of LSD1 may cause Ras/Braf-induced transformation. Yu Y. et al. showed that enforced LSD1 expression in vivo can promote BRAFV600E-driven melanomagenesis [22]. Differentiated melanoma cells have an elevated level of H3K9 methylation. The presence of this repressive histone mark at the promoter region of pluripotency marks (e.g., SOX2) can inhibit expression that is essential to maintaining self-renewal and tumorigenicity. The H3K9 demethylase activity of LSD1 can epigenetically control SOX2 transcription and thus maintain cancer stem cell pluripotency [23, 24] (Fig. 3).
Depletion of histone demethylase LSD1 in cancer cells increases repetitive element expression that can stimulate anti-tumor T cell immunity and inhibit tumor growth [25]. LSD1 inhibition also suppresses colony formation and the growth of melanoma xenografts; LSD1 has not yet been pharmacologically targeted for treatment of melanoma [26].
Several known inhibitors of LSD1 have been identified, one of which is GSK2879552. This small molecule is an irreversible, selective, orally bioavailable inhibitor of LSD1. Anti-proliferative effects of this inhibitor have been reported in small cell lung carcinoma (SCLC) [27]. LSD1 interacts with pluripotency factors in human embryonic stem cells (hESCs) and is also critical for hematopoietic differentiation. Several studies have reported on effects of LSD1 inhibitors on hematological malignancies. In acute myeloblastic leukemia (AML) cell lines, GSK2879552 treatment inhibited proliferation [28, 29]. Treatment of neuroblastoma or breast cancer cell lines with tranylcypromine (TCP), another LSD1 inhibitor, or its analogs also resulted in the inhibition of cell proliferation [30]. Although LSD1 inhibition does not affect global levels of H3K4me1/2, its impact is observed near the TSS of LSD1 target genes. In GBM cell lines, the combination of TCP and an HDAC inhibitor led to a synergistic increase of apoptosis [30]. Recently, compound S2101, a new LSD1 inhibitor, is shown to have a greater effect than TCPs. Several other LSD1 inhibitors including T-3775440, NCD38, and RN1 are in pre-clinical stages of investigation [31–34].
JMJD2A (H3K9-demethylase | H3K36-demethylase)
H3K9/36me3 lysine demethylase JMJD2A (KDM4A) overexpression leads to copy gain of certain loci in chromosomes without global chromosome instability. JMJD2A-amplified tumors demonstrate copy gains for these regions [35]. Histone demethylase JMJD2A is involved in regulation of cell proliferation, interacts with RNA polymerase I, and is associated with active ribosomal RNA genes. Cellular localization of JMJD2A is controlled through the PI3K signaling pathway [36]. JMJD2A upregulation is observed in various cancers including the breast, colon, lung, and prostate [37–40]. Its overexpression in prostate cancer is positively correlated with tumor metastasis. ETV1 recruits JMJD2A to the YAP1 (a Hippo pathway component) promoter, leading to changes in histone lysine methylation in prostate cancer cells [40]. Yap1 also promotes melanoma metastasis through its TEAD-interaction domain [41]. A potential mechanism for this activity could be recruitment of JMJD2A to the Yap1 promoter, thus modulating its methylation (Fig. 3). Interestingly, another member of the JMJD2 family, histone demethylase JMJD2C, is increased in a subset of melanoma patients and promotes growth of aggressive tumors with metastasis [22].
JMJD3 (H3K27-demethylase)
H3K27 demethylase JMJD3 (KDM6B) plays a key role in transcriptional elongation and cell differentiation. Complexes of JMJD3 and proteins involved in transcriptional elongation, demethylate H3K27me3 on their target genes. This demethylation can abolish transcriptional repression of genes initially maintained by a H3K27me3 mark. JMJD3 modulates melanoma tumor microenvironment and promotes tumor progression and metastasis. JMJD3 does not alter proliferation in melanoma cells but does enhance other tumorigenic features of melanoma cells such as clonogenicity, self-renewal, and trans-endothelial migration. JMJD3 also promotes angiogenesis and macrophage recruitment [42]. Finally, JMJD3 can neutralize polycomb-mediated silencing at the INK4b-ARF-INK4a locus in melanoma. JMJD3 expression is upregulated in melanocytic nevi in response to oncogenic RAS signaling that leads to oncogenic-induced senescence [43, 44].
UTX (H3K27-demethylase)
Another mechanism of tumorigenesis is the activity of polycomb repressive complex 2 (PRC2). Any disruption in this complex can inhibit this process. Mutations in histone methyltransferase MLL3 (a subunit of the COMPASS complex with H3K4me1 methyltransferase activity) or BAP1 (a tumor suppressor) in cancer cells can inhibit H3K27 demethylase UTX and MLL3 recruitment to gene enhancers. Thus, inhibition of H3K27 methyltransferase PRC2 in these cells can restore normal transcription patterns [45]. A recent study proposed a mechanism for the role of UTX in suppressing myeloid leukemogenesis through deregulation of ETS and GATA programs [46]. H3K27-demethylase UTX activates gene transcription in melanoma at sites with poised promoters that are marked with trimethylated H3K27. Recruitment of UTX and another activating histone modifier, P300, is promoted through MEK1-mediated phosphorylation of RNF2 [47], indicating that UTX-mediated histone demethylation is a histone modification that may activate melanomagenesis.
GSK-J1 is a selective inhibitor of H3K27 demethylases JMJD3 and UTX and is inactive against a panel of other demethylases in JMJ family [48]. GSK-J4 is an ethyl ester pro-drug produced by masking the polarity of acidic groups of GSK-J1. GSK-J4 administration increases total nuclear H3K27me3 levels in cells [48]. High throughput screening of epigenetic compounds revealed sensitivity of neuroblastoma tumor cells to GSK-J4, a dual inhibitor of H3K27 demethylases UTX and JMJD3 [49].
JHDM1B (H3K36-demethylase | H3K79-demethylase)
JHDM1B (KDM2B), which is a known H3K36 demethylase member of the Jumonji C family of proteins, also plays a role as an H3K79 demethylase and a transcriptional repressor via SIRT1-mediated chromatin silencing [50]. JHDM1B specifically recognizes non-methylated DNA in CpG islands and recruits the polycomb repressive complex 1 (PRC1) that is required for sustaining synovial sarcoma cell transformation [51, 52]. JHDM1B is found to be highly expressed in glioblastoma compared to normal brain tissue and regulates the apoptotic response of GBM cells to TNF-related apoptosis-inducing ligand [53]. JHDM1B also plays a major role early in the reprogramming process for generation of induced pluripotent stem cells [54]. JHDM1B contributes to embryonic neural development by regulating cell proliferation and cell death [55] and plays a key role in cellular senescence and tumorigenesis [56].
A concise schematic picture of the role of major histone lysine demethylases that are involved in melanoma is provided in Fig. 3, and a list of these enzymes along with their known inhibitors are shown in Table 1.
Table 1.
Histone lysine demethylases (KDMs) and their selective inhibitors
| Enzyme | Alias | Structure | Inhibitor | Reference | ||
|---|---|---|---|---|---|---|
| H3K4 | LSD1 | KDM1A | AOF2 | SWIRM/AOD | GSK2879552 Tranylcypromine (TCP) Compound S2101 T-3775440 NCD38 OG-L002 RN1 |
[24–27] |
| JARID1A | KDM5A | RBP2 | PHD/ARID/ZF | |||
| JARID1B | KDM5B | PLU-1 | PHD/ARID/ZF | KDM5-C49 KDM5-C70 2,4-PDCA PBIT KDOAM-25 |
[13–15] | |
| JARID1C | KDM5C | SMCX | PHD/ARID/ZF | |||
| JARID1D | KDM5D | SMCY | PHD/ARID/ZF | |||
| H3K9 | LSD1 | KDM1A | AOF2 | SWIRM/AOD | ||
| JMJD1A | KDM3A | JHDM2A | JmjC | |||
| JMJD1B | KDM3B | JHDM2B | JmjC | |||
| JMJD2A | KDM4A | JHDM3A | PHD/Tudor | 2,4-PDCA | [14] | |
| JMJD2B | KDM4B | JHDM3B | PHD/Tudor | |||
| JMJD2C | KDM4C | JHDM3C | PHD/Tudor | |||
| H3K27 | UTX | KDM6A | TPR/JmjC | GSK-J1 GSK-J4 |
[41, 42] | |
| JMJD3 | KDM6B | JmjC | ||||
| H3K36 | FBXL11 | KDM2A | JHDM1A | PHD/ZF/LRR | ||
| FBXL10 | KDM2B | JHDM1B | PHD/ZF/LRR | |||
| JMJD2A | KDM4A | JHDM3A | PHD/Tudor | |||
| JMJD2B | KDM4B | JHDM3B | PHD/Tudor | |||
| JMJD2C | KDM4C | JHDM3C | PHD/Tudor | |||
| H4K20 | LSD1n | neuroLSD1 |
Histone methyltransferases
H3K4 methyltransferases
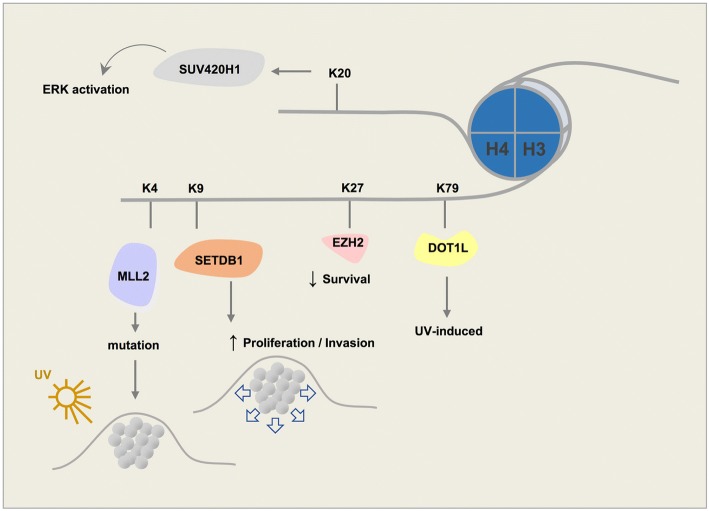
KMT2D (MLL2). Monomethylation of H3K4 is a widely known feature of enhancers and gene promoters [57]. In mammals, KMT2D is part of a huge complex that induces monomethylation of promoter/enhancer regions [58, 59]. KMT2D is a member of the histone lysine methyltransferase (HKMTase) family that can induce genome accessibility and transcription and promotes an open chromatin state. The Cancer Genome Atlas (TCGA) data reveal that KMT2D is mutated in 15% of melanoma patients. Despite no clear association in TCGA cases between KMT2D and conventional subtypes of melanoma such as NRAS or BRAF, a subsequent study showed that KMT2D deregulates specific enhancers and genes in NRAS-mutant melanoma [60]. KMT2D is encoded by genes with the highest number of UVB signature mutations [61] (Fig. 4).
Fig. 4.
Schematic figure demonstrating key roles of histone lysine methyltransferases involved in the tumorigenesis and survival of melanoma
H3K4me2 is also an active histone mark enriched in sites toward the 5′-end of transcribing genes and present in several melanoma cell lines. An increase in the level of H3K4me2 has been observed in melanoma samples compared to normal skin of the same patients [62]. Interestingly, H3K4me2 levels were found to be lower in metastatic melanoma compared to primary tumors [63].
H3K4me3 is generally associated with promoters of actively transcribing genes [64, 65] or with genes that are poised for activation. H3K4me3 is linked to promoter activity. Global changes in H3K4me3 were observed in zebrafish when comparing melanoma and normal skin [66]. Comparison of metastatic to primary melanoma revealed that decreased levels of H3K4me3 in metastases are significantly associated with repressed expression of genes in embryonic stem cells (ESCs), including targets of PRC. Further, increased levels of H3K4me3 are associated with interferon and inflammatory response genes [59]. Finally, HDAC inhibitors that promote an open chromatin state also increase H3K4 trimethylation in vitro [67].
ASH1L (ASH1, KMT2H). ASH1L is an H3K4/H3K36 methyltransferase. Nucleotide excision repair (NER) is a known mechanism to prevent formation of UV-induced melanoma tumors. The NER mechanism is in place upon recruitment of H3K4 histone methyltransferase ASH1L [68]. Loss of Ash1l leads to increased proliferation of keratinocytes [69]. ASH1L can act as an H3K36 dimethyltransferase to recruit MLL to chromatin and maintain hematopoiesis. This action supports a role of ASH1L in MLL-mediated leukemia oncogenesis and indicates that it could be a target for leukemia therapy [70].
WDR5 is an essential part of the KMT2 (MLL) complex that trimethylates H3K4. An interaction exists between this protein and the catalytic domains of KMT2. Despite the lack of chemical compounds that directly target the SET domain in KMT2 complexes, several compounds can specifically disrupt the interaction between WDR5 and MLL [71]. MM-102 specifically targets MLL-WDR5 interaction, inhibits cell growth, and induces apoptosis in leukemia cells [72]. MM-401 also specifically inhibits MLL activity by blocking MLL-WDR5 interaction [73]. A third compound, WDR5-0103, antagonizes the interaction of WDR5 with MLL by competing with MLL for their mutual binding site on WDR5 [74].
H3K9 methyltransferases
Monomethylated H3K9 is shown to be enriched in the TSS of active genes [75]. Unlike H3K9me1, both di- and trimethylated H3K9 are often found in silenced genes; however, monomethylated H3K9 may also mark distinct regions of silent chromatin in the presence of a monomethylated H4K20 mark [76].
EHMT. Dimethylated H3K9 has a higher global level in melanoma samples compared to the normal skin tissue [62]. EHMT2 (G9A) mono- and dimethylates H3K9. Methylated H3K9 recruits HP1 proteins that cause transcriptional repression [77]. HP1 proteins harbor a methyl lysine binding chromodomain that binds to methylated H3K9 [78]. HP1 proteins in human cells include HP1α, β, and γ that play key roles in the formation of transcriptionally inactive heterochromatin. Interestingly, in several cancer types (not including melanoma), inhibition of EHMT2 resulted in the arrest of cancer cell proliferation and a decrease in cell survival [79]. Another study proposed that G9A contributes to ovarian cancer metastasis [80]. EHMT1 (GLP) is also a histone methyltransferase that specifically mono- and dimethylates H3K9 in euchromatin.
SUV39. Three known members in this family have been identified. SUV39H1 is a histone methyltransferase that specifically trimethylates H3K9me1 and plays a crucial role in establishment of constitutive heterochromatin at pericentromeric and telomeric regions. Formation of various cancer types including rhabdomyosarcoma and melanoma are sensitive to the loss of SUV39H1 [81]. In differentiated melanoma cells, silencing SUV39H1 or the other H3K9 methyltransferase, G9A, can elevate their self-renewal capabilities [24]. SUV39H2, a second member of the family, has a similar structure to SUV39H1 (Fig. 2). SUV39H2 may play a role in higher order chromatin structure during spermatogenesis [82, 83]. SUV39H2 trimethylates histone lysine demethylase LSD1 and prevents degradation of this protein due to polyubiquitination, thus stabilizing its structure in the cell [84]. SETDB1 (ESET) is the third member of the SUV39 family of proteins. It is overexpressed in several cancers including human melanomas [85]. SUV39 proteins, unlike the HP1 chromodomain of the EHMT complex that shows high affinity for dimethyl, prefer the trimethyl state [86]. Hence, histone lysine methyltransferase SETDB1 also primarily trimethylates H3K9. SETDB1 is known to accelerate melanoma progression [85] (Fig. 4). Previously, SETDB1 was reported to be highly amplified in melanoma and to be associated with a more aggressive phenotype through regulation of thrombospondin 1 [87].
Chaetocin is a competitive inhibitor of S-adenosyl-L-methionine (SAM) and was the first histone lysine methyltransferase inhibitor to be identified. It specifically inhibits lysine histone methyltransferases of the SUV39 family, making the compound useful in the study of heterochromatin-mediated gene repression [88]. BIX01294 is an inhibitor of GLP and G9A methyltransferase [89]. This compound does not compete with cofactor SAM but acts as a competitive inhibitor of N-terminal peptides of H3 [90]. It selectively impairs the generation of H3K9me2 in vitro. UNC0321 is a newer modification of BIX12094 with a greater potency against G9A and GLP [91].
H3K27 methyltransferases
EZH2. Unlike monomethylated H3K27, which is associated with gene activation, di-/trimethylated H3K27 act as repressive marks [75, 92]. Histone lysine methyltransferase EZH2 is mostly responsible for trimethylation of lysine 27 on histone 3 (H3K27me3). EZH2 is the catalytic subunit of the polycomb repressive complex 2 (PRC2). PRC2 has 3 other subunits EED, SUZ12, and RBBP4. PRC2 mediates gene repression through chromatin reorganization by methylating H3K27. Therefore, H3K27me3 is associated with repressed chromatin. Mutations in EZH2 could result from the replacement of a tyrosine in its SET domain (Tyr641). This change can reduce enzymatic activity of EZH2 resulting in a higher affinity to mono- or dimethylated H3K27 [93]. In addition, in various melanoma cell lines, Tyr641 mutations of EZH2 have shown to alter the substrate specificity of EZH2 to H3K27me2, causing an increase in H3K27me3 and depletion of H3K27me2 [94]. H3K27me1/me2 produced by wild-type EZH2 could be methylated by Tyr641-mutant EZH2 to generate elevated levels of H3K27me3 that can promote tumorigenesis [95]. High levels of EZH2 activation are observed in neuroblastoma, hepatocellular carcinoma, small cell lung cancer, and melanoma [96]. Increased expression of EZH2 is associated with melanoma progression and decreased overall patient survival [97]. Protein levels of EZH2 increase incrementally from benign nevi to cutaneous malignant melanoma, which also suggests that EZH2 may play a role in the pathogenesis and tumorigenesis of melanoma [98–100] (Fig. 4). EZH2 suppresses senescence in melanoma by repressing CDKN1A expression independent of p16INK4a expression or p53 function [98]. In addition, aggressive melanoma cells have excessive levels of H3K27me3 accompanied by EZH2 overexpression [101], in which EZH2 controls melanoma progression and metastasis by silencing tumor suppressors [102].
Several available EZH2 inhibitors have been used in numerous studies. EPZ-6438 (Tazemetostat) is an EZH2 inhibitor in phase 1 and 2 clinical trials. This drug has been investigated for multiple cancer types, including B cell non-Hodgkin lymphoma and advanced solid tumors [103]. EPZ011989 is a potent and orally bioavailable EZH2 inhibitor [104]. GSK126 is a potent and also highly selective inhibitor of EZH2 that decreases global H3K27me3 levels and reactivates silenced PRC2 target genes. GSK126 inhibits proliferation of EZH2-mutant diffuse large B cell lymphoma (DLBCL) cell lines and mouse xenografts [105]. EZH2 inhibition using this small molecule is proposed to increase NK cell death suggesting an immunosuppressive effect of EZH2 [106]. Treatment with another EZH2 inhibitor, GSK503, in a melanoma mouse model, blocked tumor growth and metastasis formation. In human melanoma cells, GSK503 impaired cell proliferation and invasiveness, along with re-expression of tumor suppressors associated with increased patients’ survival [102]. In osteosarcoma, GSK343, another inhibitor of EZH2, restricts cell viability and promotes apoptosis. GSK343 inhibits EZH2 expression and its substrate, H3K27me3. It also inhibits fuse binding protein 1 (FBP1) expression, a c-Myc regulator [107]. In cancers with genetic alteration in EZH2, such as various forms of lymphoma, EPZ005687 reduces H3K27 methylation. Mutations in EZH2 result in a dependency on its enzymatic activity for proliferation that could make EPZ005687 a treatment for cancers in which EZH2 is genetically altered [108].
Another approach to inhibit the PRC2 complex and subsequently H3K27 methylation is to block another component of this complex, such as EED. In a recent study, a compound, EED226, was developed to block PRC2. EED226 is a potent and selective PRC2 inhibitor that directly binds to the H3K27me3 binding pocket of EED. It induces regression of human lymphoma xenograft tumors [109, 110].
H3K36 methyltransferases
Di-/trimethylation of H3K36 is usually associated with transcriptional activation and double-strand break repair. H3K36me2 can be deposited near the breaks and can recruit early repair factors [111]. H3K36me3 is considered as a hallmark of active gene transcription. It is commonly enriched at the body of active genes and particularly at their 3′-terminal regions. Its activity produces a methylation pattern distinct from that of other histone methylations [112]. H3K36me3 is involved in defining exons which are enriched in nucleosomes. Nucleosomes are more frequent in areas with this particular histone mark [113]. H3K36 trimethylation is also shown to play a role in homologous recombination (HR) repair in human cells [114]. Another study proposes a less known role of H3K36me3 in combination with other histone marks in contributing to the composition of heterochromatin [115]. Also, H3K36 methylation antagonizes PRC2-mediated H3K27 methylation [116].
SETD2. SETD2 is the only known gene in human cells responsible for trimethylation of lysine 36 of Histone H3 (H3K36) [117]. This enzyme is mutated in 11.5% of renal clear cell carcinoma tumors [118]. The H3K36me3 mark is associated with DNA methylation in actively transcribed gene regions [119]. A TCGA study also suggests crosstalk between H3K36 methylation and DNA methylation. Renal clear cell carcinoma tumors with SETD2 mutations had DNA hypomethylation at non-promoter regions that are marked by H3K36me3 [118]. A similar pattern is found in sarcoma where H3K36 mutation prevents methylation as well as mesenchymal progenitor cell differentiation and induces sarcoma [120]. SETD2 counteracts Wnt signaling and signaling loss promotes intestinal tumorigenesis in vivo. Mechanistically, SETD2 downregulation affects alternative gene splicing leading to tumor inhibition [121].
NSD1. NSD1 binds near various promoter elements and interacts with H3K36 and RNA Pol II, to regulate transcription [122]. H3K36me2 methyltransferase NSD1 is a modulator of PRC2 function and demarcates H3K27me2 and H3K27me3 domains in ESCs [123]. NSD1 inactivating mutations define a hypomethylated subtype in different cancer types such as head and neck squamous cell carcinoma (HNSC) and lung squamous cell carcinoma (LUSC) [124–126]. This subtype is also characterized by low T cell infiltration into the tumor microenvironment, suggesting an identification of this subtype prior to immunotherapy [125]. NSD1 inactivation along with SETD2 mutation that causes a distorted H3K36 is a key feature of clear cell renal cell carcinoma [127]. NSD1 mutations may have a genome-wide impact on DNA methylation [128]. NSD1 expression in patient-derived metastatic cell lines is significantly higher compared to normal melanocytes; however, NSD1 upregulation does not apply to primary tumors developing to metastatic lesions [129].
N-propyl sinefungin (Pr-SNF) is shown to interact preferentially with SETD2 and act as an inhibitor of this protein [130]. However, no compounds that can target NSD proteins selectively have been identified. Due to the high similarity of the SET domain of NSDs to that of GLP and G9A, BIX01294 would be capable of inhibiting H3K36 methyltransferase NSD1/2/3 [131].
H3K79 methyltransferases
DOT1L. H3K79 is a histone mark associated with active chromatin and transcriptional elongation [132]. DOT1L is the only enzyme identified that methylates H3K79. Interestingly, DOT1L has no SET domain in its protein structure (Fig. 2). It is linked to both active and repressed genes. However, H3K79 methylation is mainly a mark of active transcription. DOT1L methylates at 3 levels, mono-di-, and trimethylation. H3K79 methylation has a crucial role in heterochromatin formation and chromosome integrity [133]. An oncogenic role of DOT1L histone H3 lysine 79 (H3K79) methyltransferase in MLL-rearranged leukemogenesis has been established. Unlike leukemia, DOT1L plays a repressive role in UV-induced melanoma development. DOT1L is frequently mutated in human melanoma, leading to a reduced level of H3K79 methylation. DOT1L depletion will cause UV-induced DNA damage not to be efficiently repaired, thus encouraging progression of melanoma [134] (Fig. 4).
Pinometostat (EPZ5676) is a selective, small molecule inhibitor of DOT1L. MLL-rearranged cells and xenograft models treated with this inhibitor showed reduced levels of H3K79me2 [135, 136]. This drug is currently in phase 1 clinical trials and has modest clinical activity [137, 138]. EPZ004777 is a specific, SAM-competitive inhibitor of DOT1L. It selectively kills leukemia cells with MLL rearrangements. A chemical analog of this drug, SGC 0946, with improved solubility and potency has also been developed [139].
H4K20 methyltransferases
PR-Set7 (SET8). H4K20me1 is linked to transcriptional activation, a modification that is present in highly transcribed genes. Histone H4 lysine methyltransferase PR-Set7 (SET8) mono-methylates H4K20 [140, 141]. This enzyme plays a role in several processes including DNA damage response, chromatin compaction, DNA replication, transcriptional regulation, and tumorigenesis [140, 142]. Loss of SET8 could cause cell cycle defects and promote DNA damage [143]. Depletion of SET8 in melanoma cells under treatment with the NEDD inhibitor, pevonedistat, indicated that preventing degradation of SET8 is essential for effective impact of this drug in melanoma suppression because of its role in DNA re-replication and senescence [144].
SUV420H1/H2. Dimethylation of H4K20 is the most abundant methylation state and is present in about 80% of H4 histones [145]. H4K20me2 plays a role in cell cycle control and DNA damage response whereas H4K20me3 takes part in transcriptional repression and is a hallmark of silenced heterochromatin regions. Loss of trimethylated H4K20 is considered a common mark of cancer. Both these methyltransferases have a Zn-binding post-SET domain [146]. SUV420H1 and SUV420H2 play roles in NHEJ-directed DNA repair by di- and trimethylation of H4K20 [146]. Further, overexpression of SUV420H1 may lead to activation of ERK through enhancement of ERK phosphorylation and transcription [147] (Fig. 4).
A-196 is a selective inhibitor of SUV420H1 and SUV420H2. This drug induces a genome-wide decrease in H4K20me2/me3 and increases H4K20me1 [148].
A concise schematic picture of the role of major histone lysine methyltransferase involved in melanoma is provided in Fig. 4, and a list of these enzymes along with their known inhibitors is shown in Table 2.
Table 2.
Histone lysine methyltransferases (KMTs) and their selective inhibitors
| Enzyme | Alias | Structure | Inhibitor | Reference | ||
|---|---|---|---|---|---|---|
| H3K4 | MLL1 | KMT2A | TRX1 | SET/PHD/FYRC/BD | MM102* MM-401* WDR5-0103* |
[66–68] |
| MLL2 | KMT2B | MLL4 | SET/PHD/FYRC/BD | |||
| MLL3 | KMT2C | HALR | SET/PHD/FYRC | |||
| MLL4 | KMT2D | MLL2 | SET/PHD/FYRC | |||
| MLL5 | KMT2E | SET/PHD | ||||
| SET1A | KMT2F | SETD1A | SET/RRM | |||
| SET1B | KMT2G | SETD1B | SET/RRM | |||
| ASH1L | KMT2H | ASH1 | SET/PHD | |||
| SET7/SET9 | KMT7 | SETD7 | SET | |||
| H3K9 | G9A | KMT1C | EHMT2 | SET/TAD/ANK | BIX01294 UNC0321 |
[83, 85] |
| GLP | KMT1D | EHMT1 | SET/TAD/ANK | |||
| SUV39H1 | KMT1A | H3-K9-HMTase 1 | SET/CD | Chaetocin | [82] | |
| SUV39H2 | KMT1B | H3-K9-HMTase 2 | SET/CD | |||
| SETDB1 | KMT1E | H3-K9-HMTase 4 | SET/Tudor/MBD | |||
| RIZ1 | KMT8 | PRDM2 | PR/ZF | |||
| H3K27 | EZH2 | KMT6 | SET/SANT | EPZ-6438 (Tazemetostat) EPZ011989 GSK126 GSK503 GSK343 EPZ005687 CPI-1205 CPI-169 EI1 EED226** |
[96–99, 101–104] | |
| H3K36 | SET2 | KMT3A | SETD2 | SET/SRI | N-propyl sinefungin (Pr-SNF) |
[124] |
| NSD1 | KMT3B | STO | SET/PHD | BIX01294 | [125] | |
| SMYD2 | KMT3C | ZMYND14 | SET/MYND | |||
| ASH1L | KMT2H | ASH1 | SET/PHD/BD | |||
| H3K79 | DOT1L | KMT4 | LRR | EPZ5676 (Pinometostat) EPZ004777 SGC 0946 |
[129–133] | |
| H4K20 | PR-Set7 | KMT5A | SETD8 | SET | ||
| SUV420H1 | KMT5B | CGI85 | SET | A-196 | [142] | |
| SUV420H2 | KMT5C | SET | ||||
| NSD1 | KMT3B | STO | SET/PHD |
*These compounds target MLL-WDR5 interaction
**This compound blocks EED, another component of PRC2 to inhibit H3K27 methylation
Discussion
Future direction
Despite the emergence of several new targeted therapies and immunotherapy drugs, tumor resistance to these novel therapies is a major hurdle that reflects a need for more efficient therapeutic strategies. Numerous studies have been published in recent years to clarify the role of histone methylation and associated enzymes in tumorigenesis. Because of the reversible nature of histone methylation, demethylase and methyltransferase enzymes as well as chemical compounds that inhibit the activities of the enzymes and can therefore change the level of methylation at specific histone sites are under comprehensive investigation as targets for cancer therapy. Current understanding of the role of these enzymes suggests that they regulate transcription activation and repression based on the histone involved and the specific amino acid residue being modified.
Various compounds that target the epigenome, such as HDAC inhibitors (e.g., the entinostat, NCT00185302) or DNMT inhibitors (e.g., the decitabine, NCT00030615), have been used in clinical trials to treat melanoma patients, but the use of histone methylating/demethylating agents requires substantial additional investigation. As discussed in this review, different histone demethylases play a role in cell transformation, although H3K4 and H3K9 demethylases are most consistently reported [12–14, 22–24]. This result could be due to an insufficient body of evidence for lesser known erasers, but investigation of JARID1 or LSD1 inhibitors in extensive in vivo experiments, and eventually, in clinical trials, could be of clinical significance.
A different approach to clinical application of histone demethylase function might be used in these enzymes in removing methylation in regions of the genome that are unmethylated in normal cells. An enzyme recently used in this manner is LSD1. This enzyme was used in fusion with dCas9 to modulate histone methylation. It has been applied on certain gene enhancers in the genome to suppress specific gene transcription without disrupting local genomic architecture [149]. Lack of impact to the genomic structure is key because disruption of the chromatin structure can change DNA-protein interactions at the global level with unpredictable consequences. The use of such enzymes in combination with gene editing technology could allow development of novel treatment modalities involving removal of active histone marks on promoter regions of oncogenes or repressive histone marks on distal elements of tumor suppressor genes, both of which are common features seen particularly in cutaneous melanoma.
As we have summarized here, past literature leads us to several epigenetic patterns that seem to be frequently repeated in distinct types of cancer, including the role of histone methylation modifiers on the promoters of major known drivers of a particular type of cancer. These drivers (i.e., MITF, c-Myc in melanoma) have been recently associated with clusters of non-coding regions on the genome called super enhancers that are usually marked with enhancer marks such as H3K4me1 as well as H3K27ac [150]. Inactivation of these frequently methylated histone tails could help to restore cell homeostasis.
Lastly, epigenome-wide studies have partly elucidated the function of combinations of histone methylation marks in melanoma, although understanding of the melanoma epigenome is still unclear. These studies reveal that in a given region of DNA, more than one histone methylation mark could be active in regulating gene expression. Further, the 3D structure of the genome can juxtapose certain regions of the DNA harboring specific histone marks and other regions, thereby activating or repressing genes that may be mega-bases down or upstream.
As per the available information, a comprehensive therapeutic approach, involving minimal disruption of the genome architecture, seems to be essential. Such a therapeutic strategy could help reverse or halt the function of a combination of histone modifications that lead to cell transformation.
Acknowledgements
Not applicable.
Funding
This work has been supported by the RTG2099 Hallmarks of Skin Cancer.
Availability of data and materials
Not applicable.
Abbreviations
- AOD
Amino oxidase domain
- DLBCL
Diffuse large B cell lymphoma
- ESC
Embryonic stem cell
- ESET
ERG-associated protein with SET domain
- EZH2
Enhancer of zeste homolog 2
- HAT
Histone acetyltransferase
- HDAC
Histone deacetylase
- HKMTase
Histone lysine methyltransferase
- HNSC
Head and neck squamous cell carcinoma
- LUSC
Lung squamous cell carcinoma
- MAPK
Mitogen-activated protein kinase
- NER
Nucleotide excision repair
- NHEJ
Non-homologous end joining
- PHD
Plant homeodomain
- PRC
Polycomb repressive complex
- SAM
S-adenosyl-l-methionine
- SCLC
Small cell lung carcinoma
- TCGA
The Cancer Genome Atlas
- TCP
Tranylcypromine
- TSS
Transcription start site
Authors’ contributions
EO and JU wrote the manuscript, prepared the figures, and revised the manuscript to its final form. Both authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Portela A, Esteller M. Epigenetic modifications and human disease. Nat Biotechnol. 2010;28(10):1057–1068. doi: 10.1038/nbt.1685. [DOI] [PubMed] [Google Scholar]
- 2.Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011;11(10):726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4(2):143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 4.Gronbaek K, Hother C, Jones PA. Epigenetic changes in cancer. APMIS. 2007;115(10):1039–1059. doi: 10.1111/j.1600-0463.2007.apm_636.xml.x. [DOI] [PubMed] [Google Scholar]
- 5.Gal-Yam EN, Saito Y, Egger G, Jones PA. Cancer epigenetics: modifications, screening, and therapy. Annu Rev Med. 2008;59:267–280. doi: 10.1146/annurev.med.59.061606.095816. [DOI] [PubMed] [Google Scholar]
- 6.Rodriguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nat Med. 2011;17(3):330–339. doi: 10.1038/nm.2305. [DOI] [PubMed] [Google Scholar]
- 7.Sigalotti L, Covre A, Fratta E, Parisi G, Colizzi F, Rizzo A, et al. Epigenetics of human cutaneous melanoma: setting the stage for new therapeutic strategies. J Transl Med. 2010;8:56. doi: 10.1186/1479-5876-8-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol. 2005;6(11):838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- 9.Yoo CB, Jones PA. Epigenetic therapy of cancer: past, present and future. Nat Rev Drug Discov. 2006;5(1):37–50. doi: 10.1038/nrd1930. [DOI] [PubMed] [Google Scholar]
- 10.Schmitz SU, Albert M, Malatesta M, Morey L, Johansen JV, Bak M, et al. Jarid1b targets genes regulating development and is involved in neural differentiation. EMBO J. 2011;30(22):4586–4600. doi: 10.1038/emboj.2011.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klein BJ, Piao L, Xi Y, Rincon-Arano H, Rothbart SB, Peng D, et al. The histone-H3K4-specific demethylase KDM5B binds to its substrate and product through distinct PHD fingers. Cell Rep. 2014;6(2):325–335. doi: 10.1016/j.celrep.2013.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roesch A, Mueller AM, Stempfl T, Moehle C, Landthaler M, Vogt T. RBP2-H1/JARID1B is a transcriptional regulator with a tumor suppressive potential in melanoma cells. Int J Cancer. 2008;122(5):1047–1057. doi: 10.1002/ijc.23211. [DOI] [PubMed] [Google Scholar]
- 13.Roesch A, Fukunaga-Kalabis M, Schmidt EC, Zabierowski SE, Brafford PA, Vultur A, et al. A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell. 2010;141(4):583–594. doi: 10.1016/j.cell.2010.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roesch A, Vultur A, Bogeski I, Wang H, Zimmermann KM, Speicher D, et al. Overcoming intrinsic multidrug resistance in melanoma by blocking the mitochondrial respiratory chain of slow-cycling JARID1B (high) cells. Cancer Cell. 2013;23(6):811–825. doi: 10.1016/j.ccr.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trousil S, Chen S, Mu C, Shaw FM, Yao Z, Ran Y, et al. Phenformin enhances the efficacy of ERK inhibition in NF1-mutant melanoma. J Invest Dermatol. 2017;137(5):1135–1143. doi: 10.1016/j.jid.2017.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johansson C, Velupillai S, Tumber A, Szykowska A, Hookway ES, Nowak RP, et al. Structural analysis of human KDM5B guides histone demethylase inhibitor development. Nat Chem Biol. 2016;12(7):539–545. doi: 10.1038/nchembio.2087. [DOI] [PubMed] [Google Scholar]
- 17.Sayegh J, Cao J, Zou MR, Morales A, Blair LP, Norcia M, et al. Identification of small molecule inhibitors of Jumonji AT-rich interactive domain 1B (JARID1B) histone demethylase by a sensitive high throughput screen. J Biol Chem. 2013;288(13):9408–9417. doi: 10.1074/jbc.M112.419861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tumber A, Nuzzi A, Hookway ES, Hatch SB, Velupillai S, Johansson C, et al. Potent and selective KDM5 inhibitor stops cellular demethylation of H3K4me3 at transcription start sites and proliferation of MM1S myeloma cells. Cell Chem Biol. 2017;24(3):371–380. doi: 10.1016/j.chembiol.2017.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cloos PA, Christensen J, Agger K, Helin K. Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes Dev. 2008;22(9):1115–1140. doi: 10.1101/gad.1652908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Secombe J, Eisenman RN. The function and regulation of the JARID1 family of histone H3 lysine 4 demethylases: the Myc connection. Cell Cycle. 2007;6(11):1324–1328. doi: 10.4161/cc.6.11.4269. [DOI] [PubMed] [Google Scholar]
- 21.Whyte WA, Bilodeau S, Orlando DA, Hoke HA, Frampton GM, Foster CT, et al. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature. 2012;482(7384):221–225. doi: 10.1038/nature10805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu Y, Schleich K, Yue B, Ji S, Lohneis P, Kemper K, et al. Targeting the senescence-overriding cooperative activity of structurally unrelated H3K9 demethylases in melanoma. Cancer Cell. 2018;33(2):322–336. doi: 10.1016/j.ccell.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang X, Lu F, Wang J, Yin F, Xu Z, Qi D, et al. Pluripotent stem cell protein Sox2 confers sensitivity to LSD1 inhibition in cancer cells. Cell Rep. 2013;5(2):445–457. doi: 10.1016/j.celrep.2013.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan Y, Tajik A, Chen J, Jia Q, Chowdhury F, Wang L, et al. Matrix softness regulates plasticity of tumour-repopulating cells via H3K9 demethylation and Sox2 expression. Nat Commun. 2014;5:4619. doi: 10.1038/ncomms5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sheng W, LaFleur MW, Nguyen TH, Chen S, Chakravarthy A, Conway JR, et al. LSD1 ablation stimulates anti-tumor immunity and enables checkpoint blockade. Cell. 2018;18 PubMed PMID: 29937226. Epub 2018/06/26. eng. [DOI] [PMC free article] [PubMed]
- 26.Lian CG, Fang R, Zhan Q, Ma J, Lee C-W, Frank MH, et al. Inhibition of lysine-specific histone demethylase LSD1 suppresses melanoma growth. FASEB J. 2013;27(1_supplement):1088. [Google Scholar]
- 27.Mohammad HP, Smitheman KN, Kamat CD, Soong D, Federowicz KE, Van Aller GS, et al. A DNA Hypomethylation signature predicts antitumor activity of LSD1 inhibitors in SCLC. Cancer Cell. 2015;28(1):57–69. doi: 10.1016/j.ccell.2015.06.002. [DOI] [PubMed] [Google Scholar]
- 28.Cusan M, Cai SF, Mohammad HP, Krivtsov A, Chramiec A, Loizou E, et al. LSD1 inhibition exerts its antileukemic effect by recommissioning PU.1- and C/EBPalpha-dependent enhancers in AML. Blood. 2018;131(15):1730–1742. doi: 10.1182/blood-2017-09-807024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mould DP, McGonagle AE, Wiseman DH, Williams EL, Jordan AM. Reversible inhibitors of LSD1 as therapeutic agents in acute myeloid leukemia: clinical significance and progress to date. Med Res Rev. 2015;35(3):586–618. doi: 10.1002/med.21334. [DOI] [PubMed] [Google Scholar]
- 30.Lynch JT, Harris WJ, Somervaille TC. LSD1 inhibition: a therapeutic strategy in cancer? Expert Opin Ther Targets. 2012;16(12):1239–1249. doi: 10.1517/14728222.2012.722206. [DOI] [PubMed] [Google Scholar]
- 31.Cui S, Lim KC, Shi L, Lee M, Jearawiriyapaisarn N, Myers G, et al. The LSD1 inhibitor RN-1 induces fetal hemoglobin synthesis and reduces disease pathology in sickle cell mice. Blood. 2015;126(3):386–396. doi: 10.1182/blood-2015-02-626259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ishikawa Y, Gamo K, Yabuki M, Takagi S, Toyoshima K, Nakayama K, et al. A novel LSD1 inhibitor T-3775440 disrupts GFI1B-containing complex leading to transdifferentiation and impaired growth of AML cells. Mol Cancer Ther. 2017;16(2):273–284. doi: 10.1158/1535-7163.MCT-16-0471. [DOI] [PubMed] [Google Scholar]
- 33.Sugino N, Kawahara M, Tatsumi G, Kanai A, Matsui H, Yamamoto R, et al. A novel LSD1 inhibitor NCD38 ameliorates MDS-related leukemia with complex karyotype by attenuating leukemia programs via activating super-enhancers. Leukemia. 2017;31(11):2303–2314. doi: 10.1038/leu.2017.59. [DOI] [PubMed] [Google Scholar]
- 34.Yamamoto R, Kawahara M, Ito S, Satoh J, Tatsumi G, Hishizawa M, et al. Selective dissociation between LSD1 and GFI1B by a LSD1 inhibitor NCD38 induces the activation of ERG super-enhancer in erythroleukemia cells. Oncotarget. 2018;9(30):21007–21021. doi: 10.18632/oncotarget.24774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Black JC, Manning AL, Van Rechem C, Kim J, Ladd B, Cho J, et al. KDM4A lysine demethylase induces site-specific copy gain and rereplication of regions amplified in tumors. Cell. 2013;154(3):541–555. doi: 10.1016/j.cell.2013.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salifou K, Ray S, Verrier L, Aguirrebengoa M, Trouche D, Panov KI, et al. The histone demethylase JMJD2A/KDM4A links ribosomal RNA transcription to nutrients and growth factors availability. Nat Commun. 2016;7:10174. doi: 10.1038/ncomms10174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li LL, Xue AM, Li BX, Shen YW, Li YH, Luo CL, et al. JMJD2A contributes to breast cancer progression through transcriptional repression of the tumor suppressor ARHI. Breast Cancer Res. 2014;16(3):R56. doi: 10.1186/bcr3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim TD, Fuchs JR, Schwartz E, Abdelhamid D, Etter J, Berry WL, et al. Pro-growth role of the JMJD2C histone demethylase in HCT-116 colon cancer cells and identification of curcuminoids as JMJD2 inhibitors. Am J Transl Res. 2014;6(3):236–247. [PMC free article] [PubMed] [Google Scholar]
- 39.Xu W, Jiang K, Shen M, Qian Y, Peng Y. SIRT2 suppresses non-small cell lung cancer growth by targeting JMJD2A. Biol Chem. 2015;396(8):929–936. doi: 10.1515/hsz-2014-0284. [DOI] [PubMed] [Google Scholar]
- 40.Kim TD, Jin F, Shin S, Oh S, Lightfoot SA, Grande JP, et al. Histone demethylase JMJD2A drives prostate tumorigenesis through transcription factor ETV1. J Clin Invest. 2016;126(2):706–720. doi: 10.1172/JCI78132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lamar JM, Stern P, Liu H, Schindler JW, Jiang ZG, Hynes RO. The hippo pathway target, YAP, promotes metastasis through its TEAD-interaction domain. Proc Natl Acad Sci U S A. 2012;109(37):E2441–E2450. doi: 10.1073/pnas.1212021109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park WY, Hong BJ, Lee J, Choi C, Kim MY. H3K27 demethylase JMJD3 employs the NF-kappaB and BMP signaling pathways to modulate the tumor microenvironment and promote melanoma progression and metastasis. Cancer Res. 2016;76(1):161–170. doi: 10.1158/0008-5472.CAN-15-0536. [DOI] [PubMed] [Google Scholar]
- 43.Barradas M, Anderton E, Acosta JC, Li S, Banito A, Rodriguez-Niedenfuhr M, et al. Histone demethylase JMJD3 contributes to epigenetic control of INK4a/ARF by oncogenic RAS. Genes Dev. 2009;23(10):1177–1182. doi: 10.1101/gad.511109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Agger K, Cloos PA, Rudkjaer L, Williams K, Andersen G, Christensen J, et al. The H3K27me3 demethylase JMJD3 contributes to the activation of the INK4A-ARF locus in response to oncogene- and stress-induced senescence. Genes Dev. 2009;23(10):1171–1176. doi: 10.1101/gad.510809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang L, Zhao Z, Ozark PA, Fantini D, Marshall SA, Rendleman EJ, et al. Resetting the epigenetic balance of Polycomb and COMPASS function at enhancers for cancer therapy. Nat Med. 2018;24(6):758–769. doi: 10.1038/s41591-018-0034-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gozdecka M, Meduri E, Mazan M, Tzelepis K, Dudek M, Knights AJ, et al. UTX-mediated enhancer and chromatin remodeling suppresses myeloid leukemogenesis through noncatalytic inverse regulation of ETS and GATA programs. Nat Genet. 2018;50(6):883–894. doi: 10.1038/s41588-018-0114-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rai K, Akdemir KC, Kwong LN, Fiziev P, Wu CJ, Keung EZ, et al. Dual roles of RNF2 in melanoma progression. Cancer Discov. 2015;5(12):1314–1327. doi: 10.1158/2159-8290.CD-15-0493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kruidenier L, Chung CW, Cheng Z, Liddle J, Che K, Joberty G, et al. A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature. 2012;488(7411):404–408. doi: 10.1038/nature11262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lochmann TL, Powell KM, Ham J, Floros KV, Heisey DAR, Kurupi RIJ, et al. Targeted inhibition of histone H3K27 demethylation is effective in high-risk neuroblastoma. Sci Transl Med. 2018;16:10(441). doi: 10.1126/scitranslmed.aao4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kang JY, Kim JY, Kim KB, Park JW, Cho H, Hahm JY, et al. KDM2B is a histone H3K79 demethylase and induces transcriptional repression via sirtuin-1-mediated chromatin silencing. FASEB J. 2018; fj201800242R. PubMed PMID: 29763382. Epub 2018/05/16. eng. [DOI] [PubMed]
- 51.Farcas AM, Blackledge NP, Sudbery I, Long HK, McGouran JF, Rose NR, et al. KDM2B links the polycomb repressive complex 1 (PRC1) to recognition of CpG islands. elife. 2012;1:e00205. doi: 10.7554/eLife.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Banito A, Li X, Laporte AN, Roe JS, Sanchez-Vega F, Huang CH, et al. The SS18-SSX oncoprotein hijacks KDM2B-PRC1.1 to drive synovial sarcoma. Cancer Cell. 2018;33(3):527–541. doi: 10.1016/j.ccell.2018.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Staberg M, Rasmussen RD, Michaelsen SR, Pedersen H, Jensen KE, Villingshoj M, et al. Targeting glioma stem-like cell survival and chemoresistance through inhibition of lysine-specific histone demethylase KDM2B. Mol Oncol. 2018;12(3):406–420. doi: 10.1002/1878-0261.12174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liang G, He J, Zhang Y. Kdm2b promotes induced pluripotent stem cell generation by facilitating gene activation early in reprogramming. Nat Cell Biol. 2012;14(5):457–466. doi: 10.1038/ncb2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fukuda T, Tokunaga A, Sakamoto R, Yoshida N. Fbxl10/Kdm2b deficiency accelerates neural progenitor cell death and leads to exencephaly. Mol Cell Neurosci. 2011;46(3):614–624. doi: 10.1016/j.mcn.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 56.He J, Kallin EM, Tsukada Y, Zhang Y. The H3K36 demethylase Jhdm1b/Kdm2b regulates cell proliferation and senescence through p15(Ink4b) Nat Struct Mol Biol. 2008;15(11):1169–1175. doi: 10.1038/nsmb.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39(3):311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 58.Herz HM, Mohan M, Garruss AS, Liang K, Takahashi YH, Mickey K, et al. Enhancer-associated H3K4 monomethylation by Trithorax-related, the Drosophila homolog of mammalian Mll3/Mll4. Genes Dev. 2012;26(23):2604–2620. doi: 10.1101/gad.201327.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cheng CS, Rai K, Garber M, Hollinger A, Robbins D, Anderson S, et al. Semiconductor-based DNA sequencing of histone modification states. Nat Commun. 2013;4:2672. doi: 10.1038/ncomms3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bossi D, Cicalese A, Dellino GI, Luzi L, Riva L, D'Alesio C, et al. In vivo genetic screens of patient-derived tumors revealed unexpected frailty of the transformed phenotype. Cancer Discov. 2016;6(6):650–663. doi: 10.1158/2159-8290.CD-15-1200. [DOI] [PubMed] [Google Scholar]
- 61.Lee JJ, Sholl LM, Lindeman NI, Granter SR, Laga AC, Shivdasani P, et al. Targeted next-generation sequencing reveals high frequency of mutations in epigenetic regulators across treatment-naive patient melanomas. Clin Epigenetics. 2015;7:59. doi: 10.1186/s13148-015-0091-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Uzdensky A, Demyanenko S, Bibov M, Sharifulina S, Kit O, Przhedetski Y, et al. Expression of proteins involved in epigenetic regulation in human cutaneous melanoma and peritumoral skin. Tumour Biol. 2014;35(8):8225–8233. doi: 10.1007/s13277-014-2098-3. [DOI] [PubMed] [Google Scholar]
- 63.Kampilafkos P, Melachrinou M, Kefalopoulou Z, Lakoumentas J, Sotiropoulou-Bonikou G. Epigenetic modifications in cutaneous malignant melanoma: EZH2, H3K4me2, and H3K27me3 immunohistochemical expression is enhanced at the invasion front of the tumor. Am J Dermatopathol. 2015;37(2):138–144. doi: 10.1097/DAD.0b013e31828a2d54. [DOI] [PubMed] [Google Scholar]
- 64.Bernstein BE, Humphrey EL, Erlich RL, Schneider R, Bouman P, Liu JS, et al. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc Natl Acad Sci U S A. 2002;99(13):8695–8700. doi: 10.1073/pnas.082249499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, et al. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419(6905):407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 66.Anelli V, Santoriello C, Distel M, Koster RW, Ciccarelli FD, Mione M. Global repression of cancer gene expression in a zebrafish model of melanoma is linked to epigenetic regulation. Zebrafish. 2009;6(4):417–424. doi: 10.1089/zeb.2009.0612. [DOI] [PubMed] [Google Scholar]
- 67.Bjornsson HT, Benjamin JS, Zhang L, Weissman J, Gerber EE, Chen YC, et al. Histone deacetylase inhibition rescues structural and functional brain deficits in a mouse model of Kabuki syndrome. Sci Transl Med. 2014;6(256):256ra135. doi: 10.1126/scitranslmed.3009278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Balbo Pogliano C, Gatti M, Ruthemann P, Garajova Z, Penengo L, Naegeli H. ASH1L histone methyltransferase regulates the handoff between damage recognition factors in global-genome nucleotide excision repair. Nat Commun. 2017;8(1):1333. doi: 10.1038/s41467-017-01080-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li G, Ye Z, Shi C, Sun L, Han M, Zhuang Y, et al. The histone methyltransferase Ash1l is required for epidermal homeostasis in mice. Sci Rep. 2017;7:45401. doi: 10.1038/srep45401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhu L, Li Q, Wong SH, Huang M, Klein BJ, Shen J, et al. ASH1L links histone H3 lysine 36 dimethylation to MLL leukemia. Cancer Discov. 2016;6(7):770–783. doi: 10.1158/2159-8290.CD-16-0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hyun K, Jeon J, Park K, Kim J. Writing, erasing and reading histone lysine methylations. Exp Mol Med. 2017;49(4):e324. doi: 10.1038/emm.2017.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Karatas H, Townsend EC, Cao F, Chen Y, Bernard D, Liu L, et al. High-affinity, small-molecule peptidomimetic inhibitors of MLL1/WDR5 protein-protein interaction. J Am Chem Soc. 2013;135(2):669–682. doi: 10.1021/ja306028q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cao F, Townsend EC, Karatas H, Xu J, Li L, Lee S, et al. Targeting MLL1 H3K4 methyltransferase activity in mixed-lineage leukemia. Mol Cell. 2014;53(2):247–261. doi: 10.1016/j.molcel.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Senisterra G, Wu H, Allali-Hassani A, Wasney GA, Barsyte-Lovejoy D, Dombrovski L, et al. Small-molecule inhibition of MLL activity by disruption of its interaction with WDR5. Biochem J. 2013;449(1):151–159. doi: 10.1042/BJ20121280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129(4):823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 76.Sims JK, Houston SI, Magazinnik T, Rice JC. A trans-tail histone code defined by monomethylated H4 Lys-20 and H3 Lys-9 demarcates distinct regions of silent chromatin. J Biol Chem. 2006;281(18):12760–12766. doi: 10.1074/jbc.M513462200. [DOI] [PubMed] [Google Scholar]
- 77.El Gazzar M, Yoza BK, Chen X, Hu J, Hawkins GA, McCall CE. G9a and HP1 couple histone and DNA methylation to TNFalpha transcription silencing during endotoxin tolerance. J Biol Chem. 2008;283(47):32198–32208. doi: 10.1074/jbc.M803446200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jacobs SA, Taverna SD, Zhang Y, Briggs SD, Li J, Eissenberg JC, et al. Specificity of the HP1 chromo domain for the methylated N-terminus of histone H3. EMBO J. 2001;20(18):5232–5241. doi: 10.1093/emboj/20.18.5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ding J, Li T, Wang X, Zhao E, Choi JH, Yang L, et al. The histone H3 methyltransferase G9A epigenetically activates the serine-glycine synthesis pathway to sustain cancer cell survival and proliferation. Cell Metab. 2013;18(6):896–907. doi: 10.1016/j.cmet.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hua KT, Wang MY, Chen MW, Wei LH, Chen CK, Ko CH, et al. The H3K9 methyltransferase G9a is a marker of aggressive ovarian cancer that promotes peritoneal metastasis. Mol Cancer. 2014;13:189. doi: 10.1186/1476-4598-13-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Albacker CE, Storer NY, Langdon EM, Dibiase A, Zhou Y, Langenau DM, et al. The histone methyltransferase SUV39H1 suppresses embryonal rhabdomyosarcoma formation in zebrafish. PLoS One. 2013;8(5):e64969. doi: 10.1371/journal.pone.0064969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ait-Si-Ali S, Guasconi V, Fritsch L, Yahi H, Sekhri R, Naguibneva I, et al. A Suv39h-dependent mechanism for silencing S-phase genes in differentiating but not in cycling cells. EMBO J. 2004;23(3):605–615. doi: 10.1038/sj.emboj.7600074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.O'Carroll D, Scherthan H, Peters AH, Opravil S, Haynes AR, Laible G, et al. Isolation and characterization of Suv39h2, a second histone H3 methyltransferase gene that displays testis-specific expression. Mol Cell Biol. 2000;20(24):9423–9433. doi: 10.1128/MCB.20.24.9423-9433.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Piao L, Suzuki T, Dohmae N, Nakamura Y, Hamamoto R. SUV39H2 methylates and stabilizes LSD1 by inhibiting polyubiquitination in human cancer cells. Oncotarget. 2015;6(19):16939–16950. doi: 10.18632/oncotarget.4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ceol CJ, Houvras Y, Jane-Valbuena J, Bilodeau S, Orlando DA, Battisti V, et al. The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature. 2011;471(7339):513–517. doi: 10.1038/nature09806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Al-Sady B, Madhani HD, Narlikar GJ. Division of labor between the chromodomains of HP1 and Suv39 methylase enables coordination of heterochromatin spread. Mol Cell. 2013;51(1):80–91. doi: 10.1016/j.molcel.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Orouji E, Federico A, Larribere L, Novak D, Utikal J. Abstract A180: the histone methyltransferase SETDB1 contributes to melanoma tumorigenesis. Mol Cancer Ther. 2018;17(1 Supplement):A180.
- 88.Greiner D, Bonaldi T, Eskeland R, Roemer E, Imhof A. Identification of a specific inhibitor of the histone methyltransferase SU(VAR)3-9. Nat Chem Biol. 2005;1(3):143–145. doi: 10.1038/nchembio721. [DOI] [PubMed] [Google Scholar]
- 89.Kubicek S, O'Sullivan RJ, August EM, Hickey ER, Zhang Q, Teodoro ML, et al. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Molecular Cell. 2007;25(3):473–481. doi: 10.1016/j.molcel.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 90.Chang Y, Ganesh T, Horton JR, Spannhoff A, Liu J, Sun A, et al. Adding a lysine mimic in the design of potent inhibitors of histone lysine methyltransferases. J Mol Biol. 2010;400(1):1–7. doi: 10.1016/j.jmb.2010.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liu F, Chen X, Allali-Hassani A, Quinn AM, Wigle TJ, Wasney GA, et al. Protein lysine methyltransferase G9a inhibitors: design, synthesis, and structure activity relationships of 2,4-diamino-7-aminoalkoxy-quinazolines. J Med Chem. 2010;53(15):5844–5857. doi: 10.1021/jm100478y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tiffen J, Gallagher SJ, Hersey P. EZH2: an emerging role in melanoma biology and strategies for targeted therapy. Pigment Cell Melanoma Res. 2015;28(1):21–30. doi: 10.1111/pcmr.12280. [DOI] [PubMed] [Google Scholar]
- 93.Morin RD, Johnson NA, Severson TM, Mungall AJ, An J, Goya R, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet. 2010;42(2):181–185. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Barsotti AM, Ryskin M, Zhong W, Zhang WG, Giannakou A, Loreth C, et al. Epigenetic reprogramming by tumor-derived EZH2 gain-of-function mutations promotes aggressive 3D cell morphologies and enhances melanoma tumor growth. Oncotarget. 2015;6(5):2928–2938. doi: 10.18632/oncotarget.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sneeringer CJ, Scott MP, Kuntz KW, Knutson SK, Pollock RM, Richon VM, et al. Coordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas. Proc Natl Acad Sci U S A. 2010;107(49):20980–20985. doi: 10.1073/pnas.1012525107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cohen AL, Piccolo SR, Cheng L, Soldi R, Han B, Johnson WE, et al. Genomic pathway analysis reveals that EZH2 and HDAC4 represent mutually exclusive epigenetic pathways across human cancers. BMC Med Genet. 2013;6:35. doi: 10.1186/1755-8794-6-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Asangani IA, Harms PW, Dodson L, Pandhi M, Kunju LP, Maher CA, et al. Genetic and epigenetic loss of microRNA-31 leads to feed-forward expression of EZH2 in melanoma. Oncotarget. 2012;3(9):1011–1025. doi: 10.18632/oncotarget.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fan T, Jiang S, Chung N, Alikhan A, Ni C, Lee CC, et al. EZH2-dependent suppression of a cellular senescence phenotype in melanoma cells by inhibition of p21/CDKN1A expression. Mol Cancer Res. 2011;9(4):418–429. doi: 10.1158/1541-7786.MCR-10-0511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.McHugh JB, Fullen DR, Ma L, Kleer CG, Su LD. Expression of polycomb group protein EZH2 in nevi and melanoma. J Cutan Pathol. 2007;34(8):597–600. doi: 10.1111/j.1600-0560.2006.00678.x. [DOI] [PubMed] [Google Scholar]
- 100.Bachmann IM, Halvorsen OJ, Collett K, Stefansson IM, Straume O, Haukaas SA, et al. EZH2 expression is associated with high proliferation rate and aggressive tumor subgroups in cutaneous melanoma and cancers of the endometrium, prostate, and breast. J Clin Oncol. 2006;24(2):268–273. doi: 10.1200/JCO.2005.01.5180. [DOI] [PubMed] [Google Scholar]
- 101.Sengupta D, Byrum SD, Avaritt NL, Davis L, Shields B, Mahmoud F, et al. Quantitative histone mass spectrometry identifies elevated histone H3 lysine 27 (Lys27) trimethylation in melanoma. Mol Cell Proteomics. 2016;15(3):765–775. doi: 10.1074/mcp.M115.053363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zingg D, Debbache J, Schaefer SM, Tuncer E, Frommel SC, Cheng P, et al. The epigenetic modifier EZH2 controls melanoma growth and metastasis through silencing of distinct tumour suppressors. Nat Commun. 2015;6:6051. doi: 10.1038/ncomms7051. [DOI] [PubMed] [Google Scholar]
- 103.Italiano A, Soria JC, Toulmonde M, Michot JM, Lucchesi C, Varga A, et al. Tazemetostat, an EZH2 inhibitor, in relapsed or refractory B-cell non-Hodgkin lymphoma and advanced solid tumours: a first-in-human, open-label, phase 1 study. Lancet Oncol. 2018;19(5):649–659. doi: 10.1016/S1470-2045(18)30145-1. [DOI] [PubMed] [Google Scholar]
- 104.Campbell JE, Kuntz KW, Knutson SK, Warholic NM, Keilhack H, Wigle TJ, et al. EPZ011989, a potent, orally-available EZH2 inhibitor with robust in vivo activity. ACS Med Chem Lett. 2015;6(5):491–495. doi: 10.1021/acsmedchemlett.5b00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492(7427):108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 106.Hersey P, Gallagher S, Tiffen J, Shklovskaya E, Wilson S, Filipp F. Abstract 1676: EZH2 inhibitors in immunotherapy of melanoma. Cancer Res. 2017;77(13 Supplement):1676. doi: 10.1158/1538-7445.AM2017-1676. [DOI] [Google Scholar]
- 107.Xiong X, Zhang J, Liang W, Cao W, Qin S, Dai L, et al. Fuse-binding protein 1 is a target of the EZH2 inhibitor GSK343, in osteosarcoma cells. Int J Oncol. 2016;49(2):623–628. doi: 10.3892/ijo.2016.3541. [DOI] [PubMed] [Google Scholar]
- 108.Knutson SK, Wigle TJ, Warholic NM, Sneeringer CJ, Allain CJ, Klaus CR, et al. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nat Chem Biol. 2012;8(11):890–896. doi: 10.1038/nchembio.1084. [DOI] [PubMed] [Google Scholar]
- 109.Qi W, Zhao K, Gu J, Huang Y, Wang Y, Zhang H, et al. An allosteric PRC2 inhibitor targeting the H3K27me3 binding pocket of EED. Nat Chem Biol. 2017;13(4):381–388. doi: 10.1038/nchembio.2304. [DOI] [PubMed] [Google Scholar]
- 110.Huang Y, Zhang J, Yu Z, Zhang H, Wang Y, Lingel A, et al. Discovery of first-in-class, potent, and orally bioavailable embryonic ectoderm development (EED) inhibitor with robust anticancer efficacy. J Med Chem. 2017;60(6):2215–2226. doi: 10.1021/acs.jmedchem.6b01576. [DOI] [PubMed] [Google Scholar]
- 111.Fnu S, Williamson EA, De Haro LP, Brenneman M, Wray J, Shaheen M, et al. Methylation of histone H3 lysine 36 enhances DNA repair by nonhomologous end-joining. Proc Natl Acad Sci U S A. 2011;108(2):540–545. doi: 10.1073/pnas.1013571108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448(7153):553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Schwartz S, Meshorer E, Ast G. Chromatin organization marks exon-intron structure. Nat Struct Mol Biol. 2009;16(9):990–995. doi: 10.1038/nsmb.1659. [DOI] [PubMed] [Google Scholar]
- 114.Pfister SX, Ahrabi S, Zalmas LP, Sarkar S, Aymard F, Bachrati CZ, et al. SETD2-dependent histone H3K36 trimethylation is required for homologous recombination repair and genome stability. Cell Rep. 2014;7(6):2006–2018. doi: 10.1016/j.celrep.2014.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chantalat S, Depaux A, Hery P, Barral S, Thuret JY, Dimitrov S, et al. Histone H3 trimethylation at lysine 36 is associated with constitutive and facultative heterochromatin. Genome Res. 2011;21(9):1426–1437. doi: 10.1101/gr.118091.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yuan W, Xu M, Huang C, Liu N, Chen S, Zhu B. H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J Biol Chem. 2011;286(10):7983–7989. doi: 10.1074/jbc.M110.194027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Li J, Duns G, Westers H, Sijmons R, van den Berg A, Kok K. SETD2: an epigenetic modifier with tumor suppressor functionality. Oncotarget. 2016;7(31):50719–50734. doi: 10.18632/oncotarget.9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.TCGA Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499(7456):43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bannister AJ, Schneider R, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Spatial distribution of di- and tri-methyl lysine 36 of histone H3 at active genes. J Biol Chem. 2005;280(18):17732–17736. doi: 10.1074/jbc.M500796200. [DOI] [PubMed] [Google Scholar]
- 120.Lu C, Jain SU, Hoelper D, Bechet D, Molden RC, Ran L, et al. Histone H3K36 mutations promote sarcomagenesis through altered histone methylation landscape. Science. 2016;352(6287):844–849. doi: 10.1126/science.aac7272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yuan H, Li N, Fu D, Ren J, Hui J, Peng J, et al. Histone methyltransferase SETD2 modulates alternative splicing to inhibit intestinal tumorigenesis. J Clin Invest. 2017;127(9):3375–3391. doi: 10.1172/JCI94292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lucio-Eterovic AK, Singh MM, Gardner JE, Veerappan CS, Rice JC, Carpenter PB. Role for the nuclear receptor-binding SET domain protein 1 (NSD1) methyltransferase in coordinating lysine 36 methylation at histone 3 with RNA polymerase II function. Proc Natl Acad Sci U S A. 2010;107(39):16952–16957. doi: 10.1073/pnas.1002653107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Streubel G, Watson A, Jammula SG, Scelfo A, Fitzpatrick DJ, Oliviero G, et al. The H3K36me2 methyltransferase Nsd1 demarcates PRC2-mediated H3K27me2 and H3K27me3 domains in embryonic stem cells. Mol Cell. 2018;70(2):371–379. doi: 10.1016/j.molcel.2018.02.027. [DOI] [PubMed] [Google Scholar]
- 124.Papillon-Cavanagh S, Lu C, Gayden T, Mikael LG, Bechet D, Karamboulas C, et al. Impaired H3K36 methylation defines a subset of head and neck squamous cell carcinomas. Nat Genet. 2017;49(2):180–185. doi: 10.1038/ng.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Brennan K, Shin JH, Tay JK, Prunello M, Gentles AJ, Sunwoo JB, et al. NSD1 inactivation defines an immune cold, DNA hypomethylated subtype in squamous cell carcinoma. Sci Rep. 2017;7(1):17064. doi: 10.1038/s41598-017-17298-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Peri S, Izumchenko E, Schubert AD, Slifker MJ, Ruth K, Serebriiskii IG, et al. NSD1- and NSD2-damaging mutations define a subset of laryngeal tumors with favorable prognosis. Nat Commun. 2017;8(1):1772. doi: 10.1038/s41467-017-01877-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Su X, Zhang J, Mouawad R, Comperat E, Roupret M, Allanic F, et al. NSD1 inactivation and SETD2 mutation drive a convergence toward loss of function of H3K36 writers in clear cell renal cell carcinomas. Cancer Res. 2017;77(18):4835–4845. doi: 10.1158/0008-5472.CAN-17-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Choufani S, Cytrynbaum C, Chung BH, Turinsky AL, Grafodatskaya D, Chen YA, et al. NSD1 mutations generate a genome-wide DNA methylation signature. Nat Commun. 2015;6:10207. doi: 10.1038/ncomms10207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.de Souza CF, Xander P, Monteiro AC, Silva AG, da Silva DC, Mai S, et al. Mining gene expression signature for the detection of pre-malignant melanocytes and early melanomas with risk for metastasis. PLoS One. 2012;7(9):e44800. doi: 10.1371/journal.pone.0044800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zheng W, Ibanez G, Wu H, Blum G, Zeng H, Dong A, et al. Sinefungin derivatives as inhibitors and structure probes of protein lysine methyltransferase SETD2. J Am Chem Soc. 2012;134(43):18004–18014. doi: 10.1021/ja307060p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Morishita M, Mevius DEHF, Shen Y, Zhao S, di Luccio E. BIX-01294 inhibits oncoproteins NSD1, NSD2 and NSD3. Med Chem Res. 2017;26(9):2038–2047. doi: 10.1007/s00044-017-1909-7. [DOI] [Google Scholar]
- 132.Steger DJ, Lefterova MI, Ying L, Stonestrom AJ, Schupp M, Zhuo D, et al. DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Mol Cell Biol. 2008;28(8):2825–2839. doi: 10.1128/MCB.02076-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Nguyen AT, Zhang Y. The diverse functions of Dot1 and H3K79 methylation. Genes Dev. 2011;25(13):1345–1358. doi: 10.1101/gad.2057811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhu B, Chen S, Wang H, Yin C, Han C, Peng C, et al. The protective role of DOT1L in UV-induced melanomagenesis. Nat Commun. 2018;9(1):259. doi: 10.1038/s41467-017-02687-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Campbell CT, Haladyna JN, Drubin DA, Thomson TM, Maria MJ, Yamauchi T, et al. Mechanisms of pinometostat (EPZ-5676) treatment-emergent resistance in MLL-rearranged leukemia. Mol Cancer Ther. 2017;16(8):1669–1679. doi: 10.1158/1535-7163.MCT-16-0693. [DOI] [PubMed] [Google Scholar]
- 136.Daigle SR, Olhava EJ, Therkelsen CA, Basavapathruni A, Jin L, Boriack-Sjodin PA, et al. Potent inhibition of DOT1L as treatment of MLL-fusion leukemia. Blood. 2013;122(6):1017–1025. doi: 10.1182/blood-2013-04-497644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Stein EM, Garcia-Manero G, Rizzieri DA, Tibes R, Berdeja JG, Savona MR, et al. The DOT1L inhibitor pinometostat reduces H3K79 methylation and has modest clinical activity in adult acute leukemia. Blood. 2018;131(24):2661–2669. doi: 10.1182/blood-2017-12-818948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Shukla N, Wetmore C, O'Brien MM, Silverman LB, Brown P, Cooper TM, et al. Final report of phase 1 study of the DOT1L inhibitor, pinometostat (EPZ-5676), in children with relapsed or refractory MLL-r acute leukemia. Blood. 2016;128(22):2780. [Google Scholar]
- 139.Yu W, Chory EJ, Wernimont AK, Tempel W, Scopton A, Federation A, et al. Catalytic site remodelling of the DOT1L methyltransferase by selective inhibitors. Nat Commun. 2012;3:1288. doi: 10.1038/ncomms2304. [DOI] [PubMed] [Google Scholar]
- 140.Beck DB, Oda H, Shen SS, Reinberg D. PR-Set7 and H4K20me1: at the crossroads of genome integrity, cell cycle, chromosome condensation, and transcription. Genes Dev. 2012;26(4):325–337. doi: 10.1101/gad.177444.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Couture JF, Collazo E, Brunzelle JS, Trievel RC. Structural and functional analysis of SET8, a histone H4 Lys-20 methyltransferase. Genes Dev. 2005;19(12):1455–1465. doi: 10.1101/gad.1318405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wang Z, Dai X, Zhong J, Inuzuka H, Wan L, Li X, et al. SCF(beta-TRCP) promotes cell growth by targeting PR-Set7/Set8 for degradation. Nat Commun. 2015;6:10185. doi: 10.1038/ncomms10185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Oda H, Hubner MR, Beck DB, Vermeulen M, Hurwitz J, Spector DL, et al. Regulation of the histone H4 monomethylase PR-Set7 by CRL4(Cdt2)-mediated PCNA-dependent degradation during DNA damage. Mol Cell. 2010;40(3):364–376. doi: 10.1016/j.molcel.2010.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Benamar M, Guessous F, Du K, Corbett P, Obeid J, Gioeli D, et al. Inactivation of the CRL4-CDT2-SET8/p21 ubiquitylation and degradation axis underlies the therapeutic efficacy of pevonedistat in melanoma. EBioMed. 2016;10:85–100. doi: 10.1016/j.ebiom.2016.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Schotta G, Sengupta R, Kubicek S, Malin S, Kauer M, Callen E, et al. A chromatin-wide transition to H4K20 monomethylation impairs genome integrity and programmed DNA rearrangements in the mouse. Genes Dev. 2008;22(15):2048–2061. doi: 10.1101/gad.476008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wu H, Siarheyeva A, Zeng H, Lam R, Dong A, Wu XH, et al. Crystal structures of the human histone H4K20 methyltransferases SUV420H1 and SUV420H2. FEBS Lett. 2013;587(23):3859–3868. doi: 10.1016/j.febslet.2013.10.020. [DOI] [PubMed] [Google Scholar]
- 147.Vougiouklakis T, Sone K, Saloura V, Cho HS, Suzuki T, Dohmae N, et al. SUV420H1 enhances the phosphorylation and transcription of ERK1 in cancer cells. Oncotarget. 2015;6(41):43162–43171. doi: 10.18632/oncotarget.6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Bromberg KD, Mitchell TR, Upadhyay AK, Jakob CG, Jhala MA, Comess KM, et al. The SUV4-20 inhibitor A-196 verifies a role for epigenetics in genomic integrity. Nat Chem Biol. 2017;13(3):317–324. doi: 10.1038/nchembio.2282. [DOI] [PubMed] [Google Scholar]
- 149.Kearns NA, Pham H, Tabak B, Genga RM, Silverstein NJ, Garber M, et al. Functional annotation of native enhancers with a Cas9-histone demethylase fusion. Nat Methods. 2015;12(5):401–403. doi: 10.1038/nmeth.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Eliades P, Abraham BJ, Ji Z, Miller DM, Christensen CL, Kwiatkowski N, et al. High MITF expression is associated with super-enhancers and suppressed by CDK7 inhibition in melanoma. J Invest Dermatol. 2018;138(7):1582–1590. doi: 10.1016/j.jid.2017.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.