Abstract
We classified the genes encoding carbohydrate-active enzymes (CAZymes) in 17 sequenced genomes representing 16 evolutionarily diverse Aspergillus species. We performed a phylogenetic analysis of the encoding enzymes, along with experimentally characterized CAZymes, to assign molecular function to the Aspergilli CAZyme families and subfamilies. Genome content analysis revealed that the numbers of CAZy genes per CAZy family related to plant biomass degradation follow closely the taxonomic distance between the species. On the other hand, growth analysis showed almost no correlation between the number of CAZyme genes and the efficiency in polysaccharide utilization. The exception is A. clavatus where a reduced number of pectinolytic enzymes can be correlated with poor growth on pectin. To gain detailed information on the enzymes used by Aspergilli to breakdown complex biomass, we conducted exoproteome analysis by mass spectrometry. These results showed that Aspergilli produce many different enzymes mixtures in the presence of sugar beet pulp and wheat bran. Despite the diverse enzyme mixtures produced, species of section Nigri, A. aculeatus, A. nidulans and A. terreus, produce mixtures of enzymes with activities that are capable of digesting all the major polysaccharides in the available substrates, suggesting that they are capable of degrading all the polysaccharides present simultaneously. For the other Aspergilli, typically the enzymes produced are targeted to a subset of polysaccharides present, suggesting that they can digest only a subset of polysaccharides at a given time.
Key words: Aspergillus, Cellulose, Pectin, Plant biomass degradation, Sugar beet pulp, Wheat bran, Xylan, Xyloglucan
Introduction
Plant biomass is an abundant renewable source of sugar and aromatic compounds, which makes it an attractive feedstock to replace fossil resources in the manufacture of bio-based products in several industries, including fuel, pulp and paper, food and feed, and chemical industries (Mäkelä et al. 2016). Polysaccharides, i.e. cellulose, hemicelluloses and pectin, and aromatic lignin are the main polymers of plant biomass forming a highly complex matrix in plant cell walls (Harris & Stone 2008). In addition, plant biomass contains proteins and storage polysaccharides, such as starch and inulin.
Fungi produce extracellular enzymes for degradation of plant biomass polymers and use the resulting mono- and oligomers for their growth and reproduction. Therefore, these organisms have an intrinsic role in the global carbon cycle and are rich sources of biotechnologically relevant enzymes. Based on their amino acid sequence and structural similarity, the plant biomass modifying enzymes are catalogued into Carbohydrate-Active enZymes (CAZymes) database (www.cazy.org), in which they are organized into families of glycoside hydrolases (GH), carbohydrate esterases (CE), polysaccharide lyases (PL), glycosyltransferases (GT) and auxiliary activities (AA) (Lombard et al. 2014). The production of these enzymes is under control of a complex regulatory system (Kowalczyk et al., 2014, Benocci et al., 2017). While most of these regulators are present in all analyzed Aspergilli, there are exceptions to this (Benocci et al. 2017). The galactose-responsive regulator GalR is only present in section Nidulantes (Christensen et al. 2011), while GalX is present in most Aspergilli (Gruben et al. 2012). The amylolytic regulator MalR is only present in A. oryzae (Hasegawa et al. 2010) and some other species. However, conserved presence of a transcriptional activator does not guarantee similar regulation. It has been demonstrated that the (hemi-)cellulolytic regulator XlnR regulates different gene sets in different species (Klaubauf et al. 2014), while a previous study showed significant differences in enzyme production during growth on plant biomass of several Aspergilli (Benoit et al. 2015). To provide a general impression of the regulatory system in Aspergilli, Table 2 provides an overview of the identified transcriptional activators involved in plant biomass utilization, indicating which polysaccharides and metabolic pathways they are related to.
Table 2.
Comparison of numbers of genes per CAZy family related to cellulose degradation. If an activity is only present in a single family then the values are in boldface. If the activity is present in multiple families, these are summed up in the ‘Total’ column and in boldface. LPMO = lytic polysaccharide mono-oxygenase, EGL = endoglucanase, XEG = xyloglucan-active endoglucanase, BGL = β-glucosidase, BXL = β-xylosidase, CBH = cellobiohydrolase.
| Species/strain | LPMO |
EGL EGL/XEG |
BGL BGL/BXL |
CBH |
Total cellulose | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA9 | GH5 | GH7 | GH9 | GH45 | GH131 | GH12 | Total | GH1 | GH3 | GH3 | Total | GH6 | GH7 | Total | ||
| A. luchuensis | 6 | 3 | 0 | 0 | 0 | 1 | 2 | 6 | 2 | 11 | 2 | 15 | 2 | 2 | 4 | 31 |
| A. tubingensis | 6 | 3 | 0 | 0 | 0 | 1 | 2 | 6 | 2 | 10 | 2 | 14 | 2 | 2 | 4 | 30 |
| A. niger ATCC1015 | 7 | 3 | 0 | 0 | 0 | 1 | 2 | 6 | 3 | 12 | 3 | 18 | 2 | 2 | 4 | 35 |
| A. niger CBS 513.88 | 7 | 3 | 0 | 0 | 0 | 1 | 2 | 6 | 3 | 10 | 3 | 16 | 2 | 2 | 4 | 33 |
| A. brasiliensis | 7 | 3 | 0 | 0 | 0 | 1 | 2 | 6 | 3 | 12 | 3 | 18 | 2 | 2 | 4 | 35 |
| A. carbonarius | 3 | 3 | 0 | 0 | 0 | 0 | 1 | 4 | 3 | 8 | 2 | 13 | 2 | 2 | 4 | 24 |
| A. aculeatus | 7 | 3 | 0 | 0 | 0 | 1 | 2 | 6 | 3 | 12 | 2 | 17 | 1 | 2 | 3 | 33 |
| A. nidulans | 10 | 3 | 1 | 0 | 1 | 1 | 0 | 6 | 3 | 13 | 2 | 18 | 2 | 2 | 4 | 38 |
| A. sydowii | 10 | 2 | 1 | 0 | 1 | 1 | 1 | 6 | 3 | 18 | 5 | 26 | 1 | 2 | 3 | 45 |
| A. versicolor | 10 | 2 | 1 | 0 | 1 | 1 | 2 | 7 | 5 | 19 | 4 | 28 | 1 | 2 | 3 | 48 |
| A. flavus | 7 | 3 | 1 | 0 | 1 | 3 | 3 | 11 | 3 | 15 | 4 | 22 | 1 | 2 | 3 | 43 |
| A. oryzae | 8 | 3 | 1 | 0 | 0 | 1 | 3 | 8 | 3 | 15 | 4 | 22 | 1 | 2 | 3 | 41 |
| A. terreus | 12 | 4 | 2 | 0 | 0 | 1 | 5 | 12 | 3 | 14 | 3 | 20 | 2 | 2 | 4 | 48 |
| A. fumigatus | 7 | 4 | 2 | 0 | 1 | 2 | 3 | 12 | 5 | 11 | 1 | 17 | 1 | 2 | 3 | 39 |
| A. fischeri | 8 | 5 | 2 | 0 | 1 | 2 | 4 | 14 | 5 | 12 | 3 | 20 | 2 | 3 | 5 | 47 |
| A. clavatus | 7 | 3 | 2 | 0 | 0 | 1 | 2 | 8 | 3 | 6 | 1 | 8 | 2 | 2 | 4 | 27 |
| A. wentii | 8 | 2 | 1 | 0 | 0 | 1 | 2 | 6 | 3 | 9 | 3 | 15 | 1 | 2 | 3 | 32 |
Plant biomass degradation by filamentous ascomycete fungi has been intensively studied for several decades. The genus Aspergillus is one of the most extensively studied fungal genera, not only due to its relevance to human health and economic importance, but also in terms of plant biomass degradation (de Vries et al. 2017). The Aspergilli are cosmopolitan fungi that inhabit diverse environments. Their genomes possess a wide repertoire of plant polysaccharides targeted CAZymes, which makes them interesting species to study plant biomass degradation (Benoit et al. 2015). While several Aspergillus species are already used for the production of commercial enzyme formulations, identification of complementary enzyme sets produced by different species provides valuable information in the development of efficient enzyme formulations to expand the use of plant-based feedstocks in industrial applications.
Studies in which different species of a fungal genus are compared at the genome and post-genomic level are relatively rare and usually only cover a small number of species. We previously compared eight Aspergillus species for their plant biomass degradation ability (Benoit et al. 2015). A broad comparative genomic study of three Trichoderma species has also been reported, which included comparison of their CAZyme genome content (Schmoll et al. 2016), as well as a study in which genomes of five plant pathogenic species of the genus Mycosphaerella were compared (Zeng et al. 2017). In this study, we compared 17 genome-sequenced Aspergilli representing 16 evolutionary diverse species at both the genome and exoproteome level with respect to their approaches to degrade plant biomass. While our previous study showed that eight closely related Aspergillus species possess diverse enzymatic approaches for plant biomass degradation (Benoit et al. 2015) the current work significantly expands our understanding of the capabilities within this fungal genus, both in the diversity of fungi examined and in the details of enzymes identified in the exoproteomes. Of the studied species, six are from the industrially relevant section Nigri of the black Aspergilli including A. luchuensis (formerly A. foetidus) (de Vries et al. 2017), A. tubingensis (de Vries et al. 2017), A. niger (Pel et al., 2007, Andersen et al., 2011), A. brasiliensis (de Vries et al. 2017), A. carbonarius and A. aculeatus (de Vries et al. 2017). From the section Flavi, another widely used industrial workhorse A. oryzae (Machida et al., 2005, Arnaud et al., 2012) was included together with plant pathogenic, aflatoxin producing A. flavus (Payne et al., 2006, Arnaud et al., 2012). Species from the section Nidulantes are A. nidulans that has a highly divergent genome sequence compared to those of other Aspergilli (Galagan et al., 2005, Arnaud et al., 2012, Wortman et al., 2009, de Vries et al., 2017), a marine fungus A. sydowii that is a pathogen of Gorgonian corals (Alker et al., 2001, de Vries et al., 2017) and A. versicolor that is a well-known producer of industrially relevant metabolites (Davies et al., 1956, Fremlin et al., 2009, de Vries et al., 2017). The two species from the section Fumigati are the opportunistic human pathogen A. fumigatus (Nierman et al. 2005) and A. fischeri (Arnaud et al., 2012, Lonial et al., 1997) that is only rarely reported as a human pathogen, while A. clavatus (section Clavati) (Arnaud et al. 2012) is a source of allergen and produces several mycotoxins (Fedorova et al. 2008). The industrially relevant A. terreus (section Terrei) (Arnaud et al. 2012) is a producer of itaconic acid and enzymes (Okabe et al. 2009), whereas A. wentii (section Cremei) has been used for industrial enzyme production, is most distantly related to the other genome-sequenced Aspergilli (de Vries et al. 2017). We decided to include both A. niger strains due to the significant amount of work done on them and their relevance for biotechnology. Aspergillus niger ATCC1015 is a citric acid-producing wild-type strain (Andersen et al. 2011), while A. niger CBS 513.88 is an industrial enzyme-producing strain (Pel et al. 2007). The strains have a very high number of single nucleotide polymorphisms per kilobase (SNPs/kb), with an average of 7.8 and a maximum of 160 and also have highly variable exo-metabolite profiles (Andersen et al. 2011). Although CBS 513.88 is unable to grow well on minimal medium with pure sugars, likely due to an auxotrophic mutation, it grows well on crude substrates (de Vries et al. 2017) and therefore was not reduced in growth in this study.
We assigned functions to the catalogued CAZymes of the Aspergilli by phylogenetic analysis, which enabled us to study in detail how these species use their genomic potential to degrade complex lignocellulosic biomass. For this, we performed an exoproteomic analysis on wheat bran (WB) and sugar beet pulp (SBP) that differ significantly in their chemical composition, and compared the enzymatic approaches of the diverse Aspergillus species for decomposition of plant biomass.
Materials and methods
Fungal strains and cultures
The fungal strains used in this study are shown in Suppl. Table 2. The strains were grown on Aspergillus minimal medium (MM) (de Vries et al. 2004) agar plates with 25 mM mono- or oligosaccharides, 1 % pure plant polysaccharides or 3 % crude plant biomass substrates as carbon sources. Casein and lignin were used as control substrates. The pH of the medium was adjusted to 6.0. Agar plates were inoculated by pipetting 2 μL of a spore suspension (500 spores/μL) to the center of the plates and incubated for 5 d at 30 °C. In addition to the sequenced strain, a second isolate of each species was cultivated to confirm that the detected differences were species specific. Due to different growth rates of the tested strains, growth on 25 mM glucose that among the monosaccharides supported the fastest growth for all species was used as a reference. Growth on the other substrates relative to growth on glucose was then compared among the species.
Liquid cultures in Aspergillus MM supplemented with 1 % wheat bran or 1 % sugar beet pulp were inoculated with 106 spores/mL (final concentration) and incubated at 30 °C, 250 rpm for 3 d. All cultures were grown in duplicate.
Glucose, α-cellulose, inulin, beech wood xylan, guar gum, apple pectin and polygalacturonic acid (PGA) were from Sigma–Aldrich. Soluble starch was from Difco. Sugar beet pulp was obtained from De Nederlandse Suikerunie and wheat bran from windmill ‘de Vlijt’ (Wageningen, the Netherlands).
Identification of CAZymes
All protein sequences of six Aspergillus species (A. fischeri, A. clavatus, A. flavus, A. fumigatus, A. oryzae and A. terreus) were downloaded from Broad Institute’s database (https://www.broadinstitute.org/). For the remaining 11 genomes included in the analysis (A. luchuensis, A. aculeatus, A. brasiliensis, A. carbonarius, A. niger ATCC 1015, A. niger CBS 513.88, A. nidulans, A. sydowii, A. tubingensis, A. versicolor and A. wentii), the amino acid sequences of their proteomes were obtained from the Fungal Genomics Resource of the Joint Genome Institute (JGI, https://jgi.doe.gov/). The carbohydrate-active enzyme (CAZyme) domains were detected by running hmmscan from the HMMER v3.1b1 package (http://hmmer.org/) with parameters –E 1E-05 –domE 1E-05. Most of the hidden Markov models (HMMs) of the domains were obtained from the dbCAN v6.0 database (Yin et al. 2012). For carbohydrate-binding module 10 (CBM10), the HMM was acquired from the PFAM database (http://pfam.xfam.org/). For glycoside hydrolase family 74 (GH74), an in-house HMM was constructed using experimentally characterized GH74 proteins from the CAZy database (Lombard et al. 2014). Predicted domains covering less than 30 % of the HMMs were discarded. For overlapping domains, the one with the lowest e-value was retained. The results obtained from this analysis were compared to the CAZyme annotations for these 17 genomes reported previously (de Vries et al. 2017). Discrepancies between the two analyses were further examined using Blastp against annotated databases including UniProtKB and mycoCLAP (Murphy et al., 2011, Strasser et al., 2015).
Correlation analysis
The heatmap was made by the "gplots" package of R software, with the parameters "Complete-linkage clustering method and Pearson correlation distance".
Functional assignment of CAZyme orthologs
The full-length amino acid sequences of identified CAZymes in the same CAZy family were extracted to build the multiple sequence alignment (MSA) profiles using MUSCLE (Edgar 2004). Problematic gene models were revealed from the MSA profiles as those displaying sequence gaps and/or insertions. These problematic models were manually corrected where possible. The MSA profiles with the corrected models were then used to construct the maximum likelihood phylogenetic trees by executing the FastTree program with default parameters (Price et al. 2010). In addition to the phylogenetic tree, a matrix showing pairwise percent of identity of all sequence pairs was generated by using in-house perl scripts for each CAZy family of interest in the analysis. The tree and the matrix were used in combine with initial ortholog groups predicted by the OrthoFinder program (Emms & Kelly 2015) to determine ortholog groups in each family. To assign function to the ortholog groups, protein sequences of biochemically characterized CAZymes curated in mycoCLAP and the CAZy database were included in building the phylogenetic trees. Proteins that fall within clearly defined clades that contain characterized proteins were assigned function based on the biochemical activity of the characterized proteins. In some cases, two activities were assigned (e.g., BGL/BXL) because the clades contain characterized proteins with two biochemical activities.
Proteomics analysis
Extracellular proteins from 3 mL of culture filtrate, prepared by centrifugation to remove residual mycelia and insoluble substrate, were precipitated with cold TCA/acetone. The amount of protein recovered was determined using the RCDC kit assay (BioRad, Mississauga, ON, Canada). Five micrograms of protein were digested with trypsin for proteomic analysis as previously described (Budak et al. 2014). Dried peptide digest samples were solubilized in a solution of 5 % acetonitrile, 0.1 % formic acid and 4 fmol/μL of predigested Bovine Serum Albumin (BSA; Michrom, Auburn, CA) used as internal standard. The peptides were analyzed by LC-MS/MS using an Easy-LC II Nano-HPLC system connected in-line with a Velos LTQ-Orbitrap mass spectrometer (Thermo-Fisher, San Jose, CA). LC-MS/MS data were matched to protein sequence databases from separate Aspergillus strains. The protein databases used contain only all annotated CAZymes excluding the glycosyltransferases as well as the common Repository of Adventitious Proteins (cRAP) sequences from the proteome machine organization (ftp://ftp.thegpm.org/fasta/cRAP). In cases where the gene models are problematic, manually corrected gene models were used to deduce protein sequences for the databases. Protein identification and quantification were done using the Proteome Discoverer Quant v1.4 (Thermo-Fisher) precursor ion quantitation workflow. Normalized Individual protein area values were expressed as a fold value of the protein area value determined for the BSA internal standard. Relative CAZyme enzyme production values were calculated as a percent of the summed total normalized protein area values for each sample. The mass spectrometry data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE (Vizcaíno et al. 2016) partner repository with the dataset identifiers PXD000982 and PXD005563.
Results and discussion
Genomic diversity of plant biomass degradation potential among the Aspergilli correlates with taxonomy
In this study, we catalogued the genes encoding carbohydrate-active enzymes (CAZymes) in 17 sequenced genomes representing 16 evolutionarily diverse Aspergillus species. Based on the phylogenetic analysis of these enzymes, along with experimentally characterized CAZymes, we assigned molecular functions to the Aspergilli CAZyme families and subfamilies (summarized in Table 2, details in Suppl. Table 3). The results of these analyses provided us with the background knowledge to examine in detail the potential of various Aspergillus species in the degradation of polysaccharides and the enzymatic approaches that they use in the decomposition of complex lignocellulosic biomass.
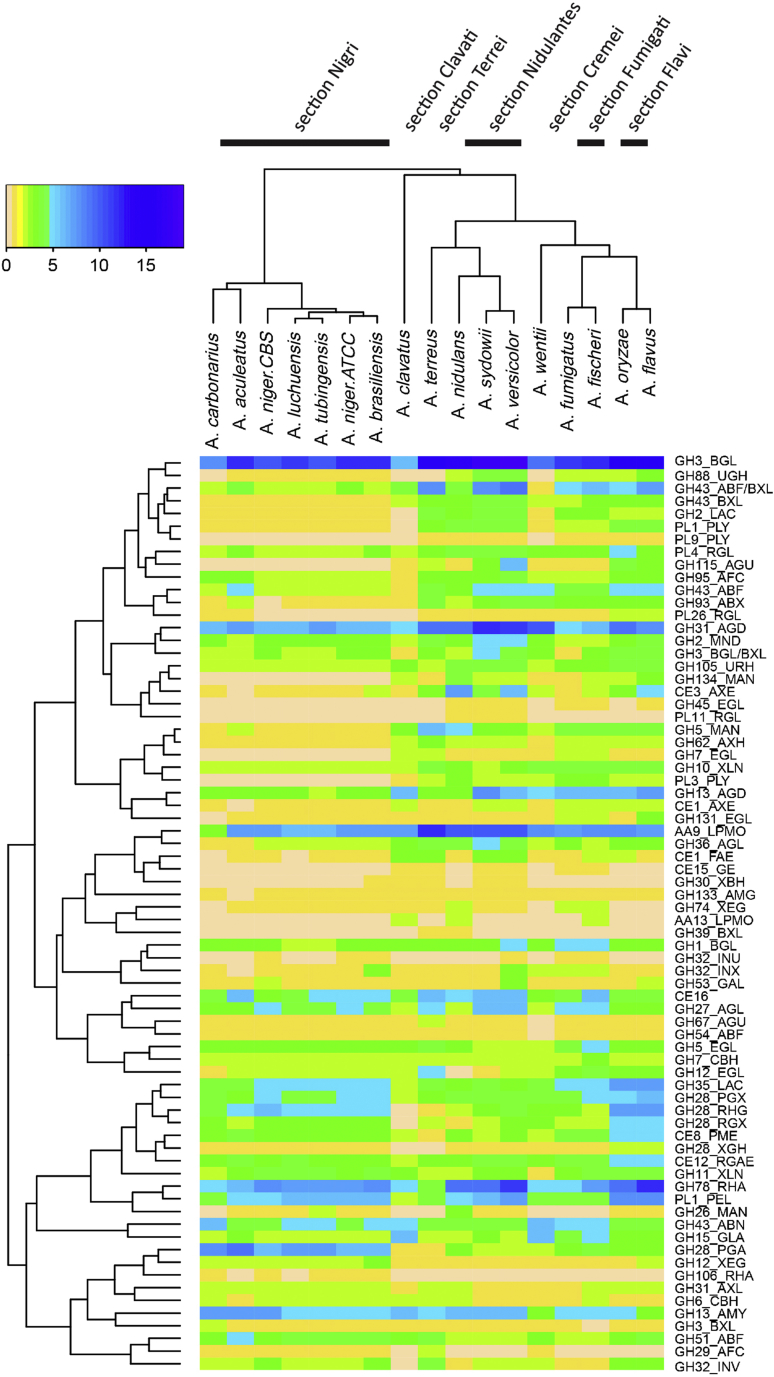
Correlation analysis revealed that the numbers of CAZy genes per CAZy family related to plant biomass degradation follow closely the taxonomic distance between the species (Fig. 1). The black Aspergilli (section Nigri) cluster together with A. aculeatus and A. carbonarius being slightly more distant from the other species of this section. Similarly, A. flavus and A. oryzae cluster together, as well as A. fumigatus and A. fischeri, and A. nidulans, A. sydowii and A. versicolor.
Fig. 1.
Correlation analysis of the number of genes per activity per CAZy family for the studied species. The number of genes is indicated by the color graph.
A closer look at the ability of these Aspergilli to degrade the different plant polysaccharides provides more insight into the evolution of their approaches and confirms in general the high similarity between related species, even at the individual enzyme (sub-)family level.
The number of genes related to cellulose degradation varies from 27 to 48, with the largest numbers in the species of section Flavi (Table 1). These are still lower numbers compared to the very good cellulose degrading ascomycetes, such as Podospora anserina, which has a significantly higher number of genes related to cellulose degradation and also grows much better on cellulose (Espagne et al. 2008) than any of the Aspergilli evaluated here (Fig. 2). In contrast, the Aspergilli have relatively little variation in the gene numbers related to xyloglucan degradation (Table 3). Most noteworthy is the absence of GH74 xyloglucan-active endoglucanases (XEG) and GH29 α-fucosidases (AFC) in several species, indicating a different enzymatic toolbox for the degradation of this polysaccharide.
Table 1.
Transcriptional activators involved in plant biomass degradation in Aspergilli. Details on the regulators and their roles can be found in previous publications (Kowalczyk et al., 2014, Benocci et al., 2017).
| Regulator | Degradation of polysaccharides | Metabolic pathways | Original reference in Aspergillus |
|---|---|---|---|
| AmyR | Starch | Petersen et al. 1999 | |
| MalR | Starch | Hasegawa et al. 2010 | |
| XlnR | Xylan, xyloglucan, galactomannan, cellulose | Pentose catabolic pathway, pentose phosphate pathway | van Peij et al. 1998a |
| AraR | Xylan, pectin | Pentose catabolic pathway, pentose phosphate pathway, D-galacturonic acid pathway | Battaglia et al. 2011 |
| ClrA | Cellulose | Raulo et al. 2016 | |
| ClrB/ManR | Cellulose, galactomannan | Ogawa et al., 2012, Ogawa et al., 2013 | |
| ClbR | Cellulose, xylan | Kunitake et al. 2013 | |
| GalR | D-galactose oxido-reductive pathway | Christensen et al. 2011 | |
| GalX | Galactomannan | Leloir pathway | Gruben et al. 2012 |
| GaaR | Pectin | D-galacturonic acid pathway | Alazi et al. 2016 |
| RhaR | Pectin | L-rhamnose pathway | Gruben et al. 2014 |
| InuR | Inulin | Yuan et al. 2008 |
Fig. 2.
Comparative growth profiles of the studies species on plant-based polysaccharides and the two crude substrates used for exoproteomic analysis. As the growth rate of the species varies, glucose is included as an internal control.
Table 3.
Comparison of numbers of genes per CAZy family related to xyloglucan degradation. If an activity is only present in a single family then the values are in boldface. If the activity is present in multiple families, these are summed up in the ‘Total’ column and in boldface. EGL = endoglucanase, XEG = xyloglucan-active endoglucanase, AFC = α-fucosidase, AXL = α-xylosidase.
| Species/strain | EGL/XEG XEG |
AFC |
AXL |
Total xyloglucan | |||||
|---|---|---|---|---|---|---|---|---|---|
| GH12 | GH12 | GH74 | Total | GH29 | GH95 | Total | GH31 | ||
| A. luchuensis | 2 | 2 | 1 | 5 | 1 | 2 | 3 | 2 | 10 |
| A. tubingensis | 2 | 2 | 1 | 5 | 1 | 2 | 3 | 2 | 10 |
| A. niger ATCC1015 | 2 | 2 | 1 | 5 | 1 | 2 | 3 | 2 | 10 |
| A. niger CBS 513.88 | 2 | 2 | 1 | 5 | 1 | 2 | 3 | 2 | 10 |
| A. brasiliensis | 2 | 3 | 1 | 6 | 1 | 2 | 3 | 2 | 11 |
| A. carbonarius | 1 | 2 | 0 | 3 | 1 | 3 | 4 | 2 | 9 |
| A. aculeatus | 2 | 2 | 1 | 5 | 1 | 3 | 4 | 2 | 11 |
| A. nidulans | 0 | 1 | 2 | 3 | 0 | 3 | 3 | 2 | 8 |
| A. sydowii | 1 | 1 | 1 | 3 | 0 | 3 | 3 | 1 | 7 |
| A. versicolor | 2 | 1 | 1 | 4 | 0 | 4 | 4 | 1 | 9 |
| A. flavus | 3 | 2 | 0 | 5 | 0 | 3 | 3 | 2 | 10 |
| A. oryzae | 3 | 1 | 0 | 4 | 0 | 3 | 3 | 2 | 9 |
| A. terreus | 5 | 1 | 1 | 7 | 2 | 3 | 5 | 2 | 14 |
| A. fumigatus | 3 | 1 | 2 | 6 | 0 | 2 | 2 | 1 | 9 |
| A. fischeri | 4 | 1 | 2 | 7 | 0 | 2 | 2 | 2 | 11 |
| A. clavatus | 2 | 1 | 1 | 4 | 0 | 1 | 1 | 1 | 6 |
| A. wentii | 2 | 1 | 0 | 3 | 1 | 2 | 3 | 1 | 7 |
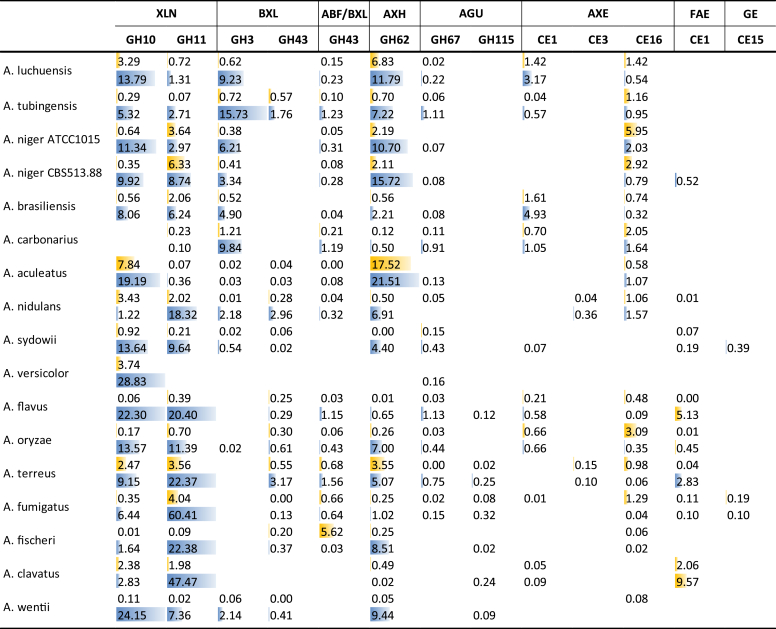
A larger variation in gene numbers is observed for genes related to xylan degradation (Table 4), with only 19 present in A. carbonarius while A. versicolor has 52. This variation does not correlate with the ability of the species to use xylan as a carbon source compared to glucose (Fig. 2). This could be due to the fact that commercial beechwood xylan does not contain all the side chains that can be found in nature, such as acetyl- or arabinofuranosyl-residues, which may affect the result of the growth profile. The diversity in gene numbers is visible for nearly all xylan-related enzyme families and activities, with e.g. endoxylanases (XLN) numbers varying from four to eight, β-xylosidases (BXL) from six to 18, and α-glucuronidases (AGU) from one to seven. One of the AGU CAZy families (GH115) is absent in section Nigri, but present in all other species, suggesting gene loss in the black Aspergilli. The presence of GH39 BXLs in only two distantly related species (A. nidulans and A. terreus) and GH30 XBHs in six mostly distantly related species (e.g. only A. brasiliensis from section Nigri), would suggest that these may have originated from horizontal gene transfer to the Aspergilli. However, the GH39 genes are syntenic, suggesting that instead these enzymes may have little impact on the overall plant biomass degrading ability and have been lost in most species. The situation is less clear for GH30 as the A. terreus and A. clavatus genes are syntenic, whereas the A. versicolor and A. sydowii genes are in a separate syntenic group. Of note is that neither GH30 XBHs nor GH39 BXLs were detected in the exoproteomes of fungi cultured on complex biomass (see below). Interestingly, from the Aspergilli most commonly used for industrial xylanase production (Polizeli et al., 2005, Ahmed et al., 2009), A. niger and A. tubingensis are among the species with relatively low xylan-related gene numbers, while A. oryzae has almost twice as many (Table 4).
Table 4.
Comparison of numbers of genes per CAZy family related to xylan degradation. If an activity is only present in a single family then the values are in boldface. If the activity is present in multiple families, these are summed up in the ‘Total’ column and in boldface. XLN = endoxylanase, BGL = β-glucosidase, BXL = β-xylosidase, ABF = α-arabinofuranosidase, XBH = xylobiohydrolase, AXE = acetyl xylan esterase, AXH = arabinoxylan arabinofuranohydrolase, FAE = feruloyl esterase, GE = glucuronoyl esterase, AGU = α-glucuronidase.
| Species/strain | XLN |
BGL/BXL |
BXL |
BXL/ABF |
XBH |
AXE |
AXH |
FAE |
GE |
AGU |
Total xylan | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GH10 | GH11 | Total | GH3 | GH3 | GH39 | GH43 | GH43 | Total | GH30 | CE1 | CE3 | CE16 | Total | GH62 | CE1 | CE15 | GH67 | GH115 | Total | ||
| A. luchuensis | 2 | 4 | 6 | 2 | 1 | 0 | 1 | 2 | 6 | 0 | 1 | 1 | 4 | 6 | 1 | 0 | 0 | 1 | 0 | 1 | 20 |
| A. tubingensis | 2 | 4 | 6 | 2 | 1 | 0 | 1 | 2 | 6 | 0 | 1 | 1 | 5 | 7 | 1 | 1 | 0 | 1 | 0 | 1 | 22 |
| A. niger ATCC1015 | 2 | 3 | 5 | 3 | 1 | 0 | 1 | 3 | 8 | 0 | 1 | 1 | 5 | 7 | 1 | 1 | 0 | 1 | 0 | 1 | 23 |
| A. niger CBS 513.88 | 2 | 4 | 6 | 3 | 1 | 0 | 1 | 2 | 7 | 0 | 1 | 1 | 3 | 5 | 1 | 1 | 0 | 1 | 0 | 1 | 21 |
| A. brasiliensis | 2 | 4 | 6 | 3 | 1 | 0 | 1 | 2 | 7 | 1 | 1 | 2 | 5 | 8 | 1 | 1 | 0 | 1 | 0 | 1 | 25 |
| A. carbonarius | 2 | 2 | 4 | 2 | 2 | 0 | 1 | 2 | 7 | 0 | 1 | 1 | 4 | 6 | 1 | 0 | 0 | 1 | 0 | 1 | 19 |
| A. aculeatus | 2 | 3 | 5 | 2 | 1 | 0 | 1 | 4 | 8 | 0 | 0 | 0 | 6 | 6 | 1 | 1 | 0 | 1 | 0 | 1 | 22 |
| A. nidulans | 3 | 2 | 5 | 2 | 1 | 1 | 3 | 4 | 11 | 0 | 1 | 7 | 5 | 13 | 2 | 1 | 0 | 1 | 1 | 2 | 34 |
| A. sydowii | 2 | 3 | 5 | 5 | 1 | 0 | 4 | 8 | 18 | 1 | 2 | 4 | 6 | 12 | 2 | 3 | 1 | 1 | 3 | 4 | 45 |
| A. versicolor | 3 | 3 | 6 | 4 | 1 | 0 | 4 | 9 | 18 | 1 | 2 | 6 | 6 | 14 | 2 | 3 | 1 | 1 | 6 | 7 | 52 |
| A. flavus | 4 | 4 | 8 | 4 | 1 | 0 | 3 | 7 | 15 | 0 | 2 | 5 | 4 | 11 | 2 | 1 | 0 | 1 | 3 | 4 | 41 |
| A. oryzae | 4 | 4 | 8 | 4 | 1 | 0 | 3 | 5 | 13 | 0 | 2 | 3 | 4 | 9 | 2 | 2 | 0 | 1 | 4 | 5 | 39 |
| A. terreus | 4 | 2 | 6 | 3 | 1 | 1 | 3 | 8 | 16 | 1 | 1 | 3 | 6 | 10 | 3 | 3 | 1 | 2 | 2 | 4 | 44 |
| A. fumigatus | 4 | 3 | 7 | 1 | 1 | 0 | 2 | 5 | 9 | 0 | 2 | 1 | 4 | 7 | 2 | 1 | 1 | 1 | 1 | 2 | 28 |
| A. fischeri | 4 | 4 | 8 | 3 | 0 | 0 | 3 | 6 | 12 | 0 | 2 | 2 | 6 | 10 | 2 | 2 | 1 | 1 | 1 | 2 | 36 |
| A. clavatus | 2 | 3 | 5 | 1 | 1 | 0 | 2 | 4 | 8 | 1 | 2 | 1 | 4 | 7 | 2 | 3 | 1 | 1 | 1 | 2 | 28 |
| A. wentii | 4 | 1 | 5 | 3 | 1 | 0 | 2 | 1 | 7 | 0 | 1 | 2 | 3 | 6 | 1 | 1 | 0 | 0 | 1 | 1 | 21 |
High variation in gene numbers was also detected for genes related to galacto(gluco)mannan degradation, with only eight present in A. carbonarius while A. sydowii has 22 (Table 5). As in the case for xylan, no direct correlation was observed between the number of genes and the ability of the species to grow on guar gum (a galactomannan). However, it should be noted that this substrate is similar, but not identical to plant cell wall galactoglucomannans, which may affect the results. Similar to the GH115 (AGU), section Nigri lacks members of GH134 endomannanases (MAN), which are present in all other Aspergilli, suggesting that gene loss may also have occurred in the black Aspergilli for these enzymes.
Table 5.
Comparison of numbers of genes per CAZy family related to galacto(gluco)mannan degradation. If an activity is only present in a single family then the values are in boldface. If the activity is present in multiple families, these are summed up in the ‘Total’ column and in boldface. MAN = endomannanase, AGL = α-galactosidase, MND = β-mannosidase.
| Species/strain | MAN |
AGL |
MND |
Total galactomannan | |||||
|---|---|---|---|---|---|---|---|---|---|
| GH5 | GH26 | GH134 | Total | GH27 | GH36 | Total | GH2 | ||
| A. luchuensis | 1 | 1 | 0 | 2 | 4 | 2 | 6 | 3 | 11 |
| A. tubingensis | 1 | 2 | 0 | 3 | 4 | 2 | 6 | 3 | 12 |
| A. niger ATCC1015 | 1 | 1 | 0 | 2 | 5 | 2 | 7 | 3 | 12 |
| A. niger CBS 513.88 | 1 | 1 | 0 | 2 | 5 | 2 | 7 | 3 | 12 |
| A. brasiliensis | 1 | 1 | 0 | 2 | 4 | 2 | 6 | 3 | 11 |
| A. carbonarius | 1 | 0 | 0 | 1 | 3 | 1 | 4 | 3 | 8 |
| A. aculeatus | 2 | 1 | 0 | 3 | 3 | 1 | 4 | 2 | 9 |
| A. nidulans | 5 | 3 | 4 | 12 | 2 | 4 | 6 | 3 | 21 |
| A. sydowii | 3 | 1 | 2 | 6 | 6 | 5 | 11 | 5 | 22 |
| A. versicolor | 3 | 1 | 2 | 6 | 6 | 4 | 10 | 5 | 21 |
| A. flavus | 4 | 1 | 3 | 8 | 3 | 3 | 6 | 4 | 18 |
| A. oryzae | 4 | 1 | 2 | 7 | 3 | 3 | 6 | 3 | 16 |
| A. terreus | 6 | 0 | 1 | 7 | 5 | 4 | 9 | 4 | 20 |
| A. fumigatus | 4 | 0 | 1 | 5 | 5 | 3 | 8 | 2 | 15 |
| A. fischeri | 4 | 0 | 2 | 6 | 5 | 2 | 7 | 2 | 15 |
| A. clavatus | 3 | 0 | 2 | 5 | 2 | 3 | 5 | 2 | 12 |
| A. wentii | 2 | 0 | 1 | 3 | 2 | 2 | 4 | 3 | 10 |
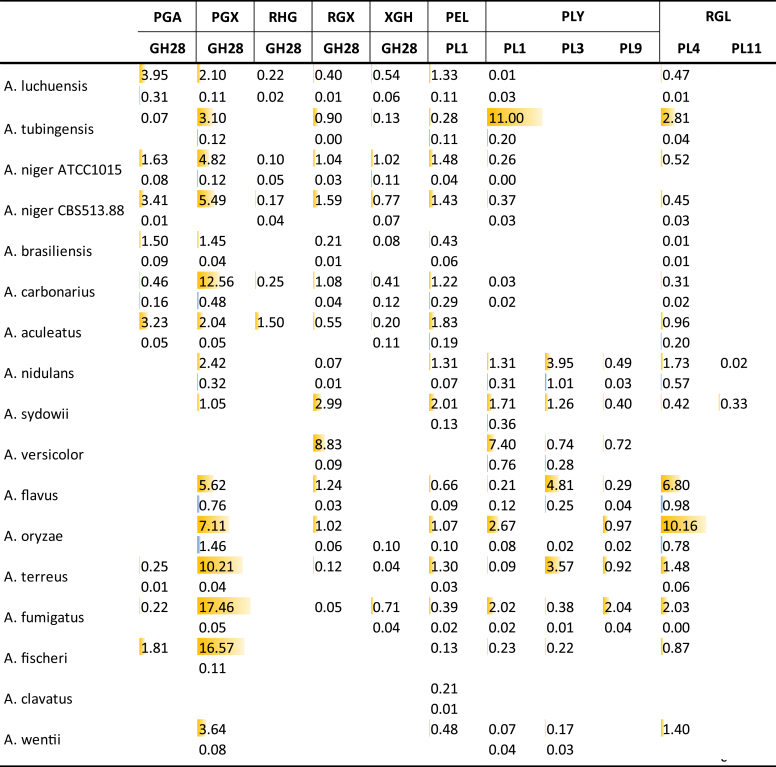
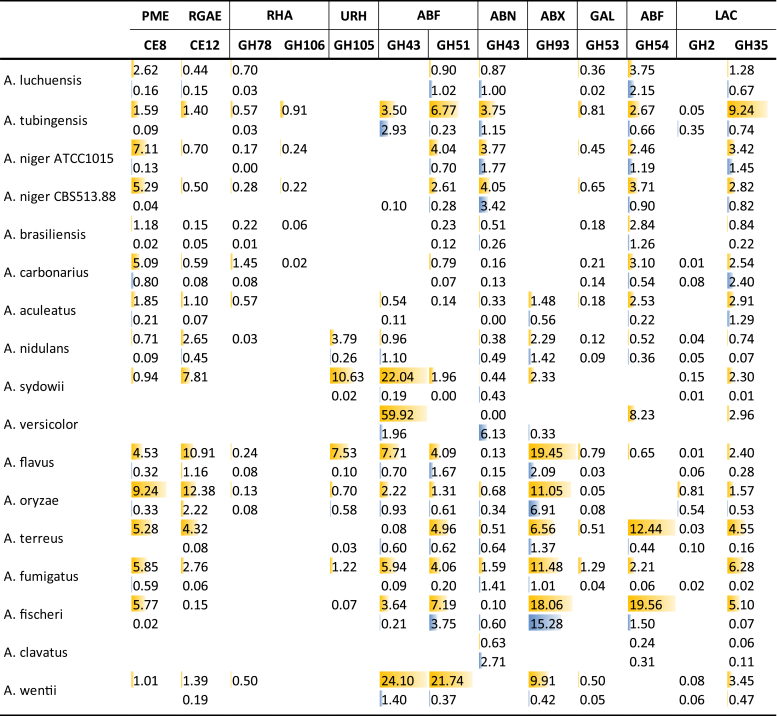
Pectin is a highly complex polysaccharide with defined substructures (Voragen et al. 2009), resulting a wide range of enzymes that are involved in pectin degradation. The number of genes related to pectin degradation also vary significantly between the Aspergilli, with genes encoding main chain acting enzymes varying from 16 for A. clavatus to 60 for A. flavus, while the gene numbers encoding side chain active enzymes are between 18 (A. clavatus) and 38 (A. flavus) (Table 6, Table 7). Aspergillus clavatus has significantly fewer genes encoding pectinolytic enzymes, which is correlated with very poor growth on pectin. This reduction extends beyond the enzyme-encoding genes as it also lacks ortholog for the galacturonic acid transporter GatA (Sloothaak et al. 2014). There is no clear correlation between growth and gene numbers for the other species in the utilization of pectin. The black Aspergilli have a clearly reduced number of pectin-active lyases, but an expanded number of endopolygalacturonases, compared to the other Aspergilli. This is likely due to the acidification commonly observed in section Nigri, in particular for A. niger, which would reduce the pH of the local environment. Since pectin-related hydrolases are more active at low pH, while pectin-related lyases prefer neutral pH, this genomic evolution correlates directly to the overall physiology of the species. PL26 rhamnogalacturonan lyase (RGL) is absent in A. clavatus and part of section Nigri, suggesting another example of gene loss in this group of Aspergilli. In contrast, PL11 RGLs are only present in section Nidulantes, suggesting that the ancestor of these Aspergillus species may have obtained this gene through horizontal gene transfer. Similarly, GH106 α-rhamnosidases (RHA) are only present in section Nigri, with the exception of A. luchuensis, also arguing for horizontal gene transfer of this gene to the ancestor of the black Aspergilli or alternatively gene loss in all other sections of the Aspergilli.
Table 6.
Comparison of numbers of genes per CAZy family related to pectin main chain degradation. If an activity is only present in a single family then the values are in boldface. If the activity is present in multiple families, these are summed up in the ‘Total’ column and in boldface. PGA = endopolygalacturonase, PGX = exopolygalacturonases, RHG = endorhamnogalacturonase, RGX = exorhamnogalacturonase, XGH = xylogalacturonase, PEL = pectin lyase, PLY = pectate lyase, RGL = rhamnogalacturonan lyase, RHA = α-rhamnosidase, UGH = unsaturated galacturonan hydrolase, URH = unsaturated rhamnogalacturonan hydrolase.
| Species/strain | PGA |
PGX |
RHG |
RGX |
XGH |
PEL |
PLY |
RGL |
RHA |
UGH |
URH |
Total pectin main chain | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GH28 | GH28 | GH28 | GH28 | GH28 | PL1 | PL1 | PL3 | PL9 | Total | PL4 | PL11 | PL26 | Total | GH78 | GH106 | Total | GH88 | GH105 | ||
| A. luchuensis | 7 | 3 | 5 | 3 | 1 | 6 | 1 | 0 | 0 | 1 | 2 | 0 | 0 | 2 | 7 | 0 | 7 | 1 | 2 | 38 |
| A. tubingensis | 7 | 3 | 5 | 3 | 1 | 6 | 1 | 0 | 0 | 1 | 2 | 0 | 0 | 2 | 7 | 1 | 8 | 1 | 2 | 39 |
| A. niger ATCC1015 | 6 | 4 | 5 | 3 | 1 | 6 | 1 | 0 | 0 | 1 | 2 | 0 | 0 | 2 | 7 | 1 | 8 | 1 | 2 | 39 |
| A. niger CBS 513.88 | 6 | 4 | 6 | 3 | 1 | 5 | 1 | 0 | 0 | 1 | 2 | 0 | 0 | 2 | 8 | 1 | 9 | 1 | 2 | 40 |
| A. brasiliensis | 6 | 4 | 5 | 3 | 1 | 6 | 1 | 0 | 0 | 1 | 3 | 0 | 0 | 3 | 8 | 1 | 9 | 1 | 2 | 42 |
| A. carbonarius | 8 | 3 | 4 | 3 | 1 | 4 | 1 | 0 | 0 | 1 | 2 | 0 | 1 | 3 | 5 | 1 | 6 | 0 | 2 | 35 |
| A. aculeatus | 9 | 2 | 5 | 2 | 1 | 5 | 1 | 0 | 0 | 1 | 3 | 0 | 1 | 4 | 6 | 0 | 6 | 1 | 2 | 38 |
| A. nidulans | 3 | 3 | 2 | 1 | 1 | 5 | 4 | 3 | 1 | 8 | 4 | 1 | 1 | 6 | 9 | 0 | 9 | 2 | 4 | 44 |
| A. sydowii | 2 | 3 | 3 | 2 | 1 | 6 | 4 | 2 | 1 | 7 | 4 | 1 | 1 | 6 | 9 | 0 | 9 | 4 | 4 | 47 |
| A. versicolor | 2 | 3 | 4 | 3 | 1 | 7 | 4 | 2 | 1 | 7 | 4 | 1 | 1 | 6 | 11 | 0 | 11 | 4 | 4 | 52 |
| A. flavus | 3 | 4 | 7 | 5 | 2 | 8 | 4 | 2 | 1 | 7 | 3 | 0 | 2 | 5 | 12 | 0 | 12 | 3 | 4 | 60 |
| A. oryzae | 3 | 3 | 7 | 5 | 2 | 8 | 4 | 2 | 1 | 7 | 5 | 0 | 2 | 7 | 9 | 0 | 9 | 2 | 4 | 57 |
| A. terreus | 1 | 3 | 1 | 2 | 0 | 4 | 3 | 2 | 1 | 6 | 3 | 0 | 1 | 4 | 4 | 0 | 4 | 0 | 2 | 27 |
| A. fumigatus | 3 | 3 | 3 | 2 | 1 | 4 | 2 | 3 | 1 | 6 | 3 | 0 | 1 | 4 | 5 | 0 | 5 | 2 | 3 | 36 |
| A. fischeri | 4 | 4 | 2 | 2 | 1 | 4 | 2 | 3 | 1 | 6 | 3 | 0 | 1 | 4 | 7 | 0 | 7 | 2 | 3 | 39 |
| A. clavatus | 1 | 2 | 0 | 0 | 0 | 2 | 0 | 1 | 0 | 1 | 2 | 0 | 0 | 2 | 5 | 0 | 5 | 0 | 3 | 16 |
| A. wentii | 2 | 3 | 3 | 3 | 1 | 4 | 1 | 2 | 0 | 3 | 3 | 0 | 1 | 4 | 5 | 0 | 5 | 0 | 2 | 30 |
Table 7.
Comparison of numbers of genes per CAZy family related to pectin side chain degradation. If an activity is only present in a single family then the values are in boldface. If the activity is present in multiple families, these are summed up in the ‘Total’ column and in boldface. PME = pectin methyl esterase, RGAE = rhamnogalacturonan acetyl esterase, ABN = endoarabinanase, ABX = exoarabinanase, ABF = α-arabinofuranosidase, LAC = β-galactosidase, GAL = endogalactanase.
| Species/strain | PME |
RGAE |
ABN |
ABX |
ABF |
LAC |
GAL |
Total pectin side chain | Total pectin main chain | Total pectin | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CE8 | CE12 | GH43 | GH93 | GH43 | GH51 | GH54 | Total | GH2 | GH35 | Total | GH53 | ||||
| A. luchuensis | 3 | 4 | 5 | 1 | 2 | 4 | 1 | 7 | 1 | 5 | 6 | 1 | 27 | 38 | 65 |
| A. tubingensis | 3 | 3 | 5 | 1 | 2 | 4 | 1 | 7 | 1 | 5 | 6 | 1 | 26 | 39 | 65 |
| A. niger ATCC1015 | 3 | 3 | 4 | 1 | 2 | 4 | 1 | 7 | 1 | 5 | 6 | 1 | 25 | 39 | 64 |
| A. niger CBS 513.88 | 3 | 3 | 4 | 0 | 2 | 3 | 1 | 6 | 1 | 5 | 6 | 2 | 24 | 40 | 64 |
| A. brasiliensis | 3 | 3 | 5 | 1 | 2 | 4 | 1 | 7 | 1 | 5 | 6 | 1 | 26 | 42 | 68 |
| A. carbonarius | 3 | 3 | 6 | 1 | 2 | 3 | 1 | 6 | 1 | 4 | 5 | 1 | 25 | 35 | 60 |
| A. aculeatus | 4 | 4 | 4 | 2 | 5 | 5 | 1 | 11 | 1 | 4 | 5 | 1 | 31 | 38 | 69 |
| A. nidulans | 3 | 3 | 4 | 2 | 3 | 2 | 1 | 6 | 3 | 3 | 6 | 1 | 23 | 44 | 67 |
| A. sydowii | 2 | 4 | 4 | 3 | 5 | 2 | 1 | 8 | 4 | 4 | 8 | 1 | 30 | 47 | 77 |
| A. versicolor | 3 | 4 | 4 | 4 | 5 | 2 | 1 | 8 | 4 | 3 | 7 | 3 | 33 | 52 | 85 |
| A. flavus | 5 | 5 | 4 | 3 | 5 | 4 | 1 | 10 | 2 | 7 | 9 | 2 | 38 | 60 | 98 |
| A. oryzae | 5 | 5 | 4 | 3 | 5 | 3 | 1 | 9 | 2 | 7 | 9 | 1 | 36 | 57 | 93 |
| A. terreus | 1 | 3 | 4 | 4 | 4 | 4 | 1 | 9 | 4 | 4 | 8 | 1 | 30 | 27 | 57 |
| A. fumigatus | 4 | 4 | 5 | 3 | 4 | 2 | 1 | 7 | 3 | 5 | 8 | 1 | 32 | 36 | 68 |
| A. fischeri | 4 | 4 | 5 | 3 | 4 | 2 | 1 | 7 | 3 | 5 | 8 | 1 | 32 | 39 | 71 |
| A. clavatus | 2 | 2 | 5 | 1 | 1 | 3 | 1 | 5 | 0 | 2 | 2 | 1 | 18 | 16 | 34 |
| A. wentii | 2 | 3 | 6 | 4 | 5 | 3 | 0 | 8 | 1 | 4 | 5 | 1 | 29 | 30 | 59 |
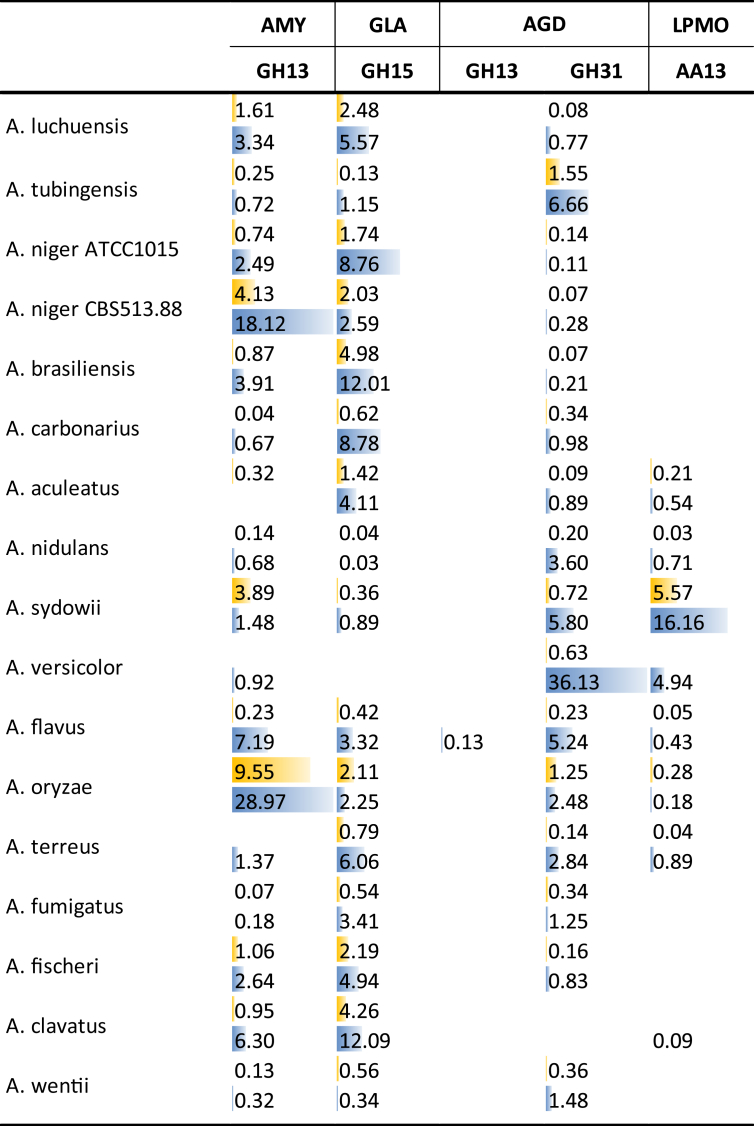
Gene numbers related to starch degradation are less varied between the Aspergilli (Table 8). The exception is a lower number of GH13 α-glucosidases (AGD) in particular in section Nigri, despite the well-established role of A. niger as a producer of industrial starch degrading enzyme cocktails (Tsukagoshi et al. 2001). As glucoamylase (GLA) has sufficient exo-acting activity, a lower AGD activity may be favorable for industrial enzyme cocktails, as this also implies a lower transglycosylation activity (Armbruster 1960). The low variation in gene numbers correlates with the ability of nearly all strains to grow well on starch (Fig. 2). The absence of growth for A. carbonarius is surprising, as it contains the same number of genes as most other species from section Nigri. A possible explanation could be that the starch-related transcriptional activator AmyR (Petersen et al. 1999) is no longer functional or expressed in this species. This inactivation would likely be a recent event, as in the absence of a functional regulatory system for starch utilization, there would be no selection pressure for maintenance of starch utilization enzymes and they would quickly be lost. However, the inactivation must have already happened before the strain was deposited, as also other samples of this strain in the collection show this phenotype (data not shown).
Table 8.
Comparison of numbers of genes per CAZy family related to starch and inulin degradation. If an activity is only present in a single family then the values are in boldface. If the activity is present in multiple families, these are summed up in the ‘Total’ column and in boldface. AMY = α-amylase, GLA = glucoamylase, AGD = α-glucosidase, AMG = amylo-α-1,6-glucosidase, LPMO = lytic polysaccharide mono-oxygenase, INU = endoinulinase, INX = exoinulinase, INV = invertase/β-fructofuranosidase.
| Species/strain | AMY |
GLA |
AGD |
AMG |
LPMO |
Total starch | INU |
INX |
INV |
Total inulin | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GH13 | GH15 | GH13 | GH31 | Total | GH133 | AA13 | GH32 | GH32 | GH32 | |||
| A. luchuensis | 5 | 2 | 3 | 6 | 9 | 1 | 0 | 17 | 0 | 1 | 2 | 3 |
| A. tubingensis | 5 | 2 | 2 | 7 | 9 | 1 | 0 | 17 | 0 | 1 | 2 | 3 |
| A. niger ATCC1015 | 5 | 2 | 3 | 6 | 9 | 1 | 0 | 17 | 1 | 1 | 2 | 4 |
| A. niger CBS 513.88 | 7 | 2 | 3 | 6 | 9 | 1 | 0 | 19 | 1 | 1 | 3 | 5 |
| A. brasiliensis | 5 | 2 | 3 | 6 | 9 | 1 | 0 | 17 | 1 | 3 | 3 | 7 |
| A. carbonarius | 7 | 2 | 3 | 6 | 9 | 1 | 0 | 19 | 0 | 1 | 2 | 3 |
| A. aculeatus | 7 | 3 | 3 | 7 | 10 | 0 | 0 | 20 | 0 | 0 | 2 | 2 |
| A. nidulans | 6 | 2 | 3 | 9 | 12 | 1 | 2 | 23 | 0 | 1 | 1 | 2 |
| A. sydowii | 6 | 3 | 8 | 12 | 20 | 1 | 0 | 30 | 0 | 1 | 2 | 3 |
| A. versicolor | 6 | 2 | 6 | 11 | 17 | 1 | 0 | 26 | 1 | 3 | 2 | 6 |
| A. flavus | 3 | 3 | 7 | 8 | 15 | 1 | 0 | 22 | 0 | 1 | 3 | 4 |
| A. oryzae | 5 | 3 | 6 | 9 | 15 | 1 | 0 | 24 | 0 | 1 | 3 | 4 |
| A. terreus | 5 | 2 | 3 | 9 | 12 | 1 | 0 | 20 | 0 | 1 | 4 | 5 |
| A. fumigatus | 5 | 4 | 6 | 5 | 11 | 1 | 0 | 21 | 1 | 2 | 1 | 4 |
| A. fischeri | 5 | 5 | 6 | 6 | 12 | 1 | 2 | 25 | 1 | 2 | 1 | 4 |
| A. clavatus | 6 | 6 | 6 | 5 | 11 | 1 | 2 | 24 | 0 | 1 | 0 | 1 |
| A. wentii | 3 | 6 | 5 | 10 | 15 | 1 | 0 | 25 | 0 | 2 | 2 | 4 |
In contrast, high variation is observed in numbers for inulin degradation related genes. While nearly all species contain at least two GH32 invertases (INV), the number of exoinulinases (INX) varies more strongly even between related species. Only six species (three from section Nigri, two from section Fumigati and A. versicolor) contain a gene encoding an endoinulinase (INU). However, this cannot explain all the diversity in the ability to grow on inulin (Fig. 2). The poor growth of A. aculeatus is likely due to absence of both INU and INX, while for A. clavatus this could be explained by absence of INU and INV. All other species contain at least two of the three GH32 activities, and all grow better on inulin than the aforementioned species. However, the number of genes per activity and presence or absence of INU do not correlate to the ability to grow on inulin for all these species.
Overall, the CAZy genome content related to plant biomass degradation shows high correlation with the taxonomic relationship of the species. The presence of additional genes in specific species could be either due to recent gene duplications, gene loss in the other species or horizontal gene transfer, but additional analyses are needed to confirm this. The absence of a gene in a specific species could be due to a recent gene deletion. However, it cannot be fully excluded that the gene may have been missed in the genome annotation of this species or that the gene is present in a gap in the assembly of the genome sequence. The most remarkable difference observed is the reduction in pectin degradation related genes in A. clavatus that has also been reported previously (Benoit et al., 2015, de Vries et al., 2017). In addition, several gene deletions in section Nigri were observed, suggesting that this group of Aspergilli may have undergone specialization after they split from the other Aspergillus sections.
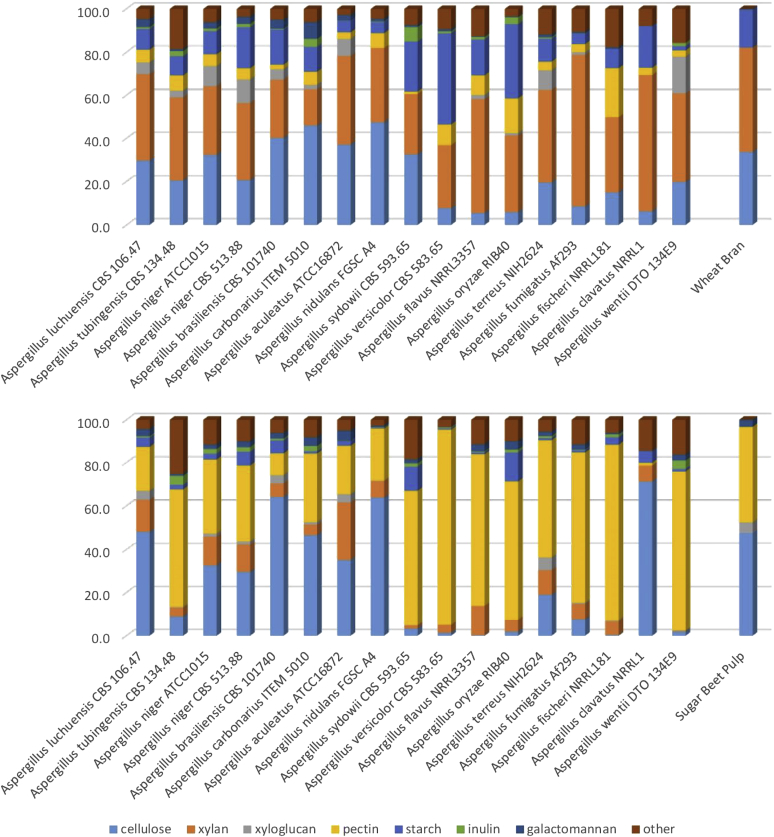
Exoproteomic analysis reveals high diversity in the plant biomass degradation approach among the Aspergilli that does not follow taxonomic relationships between the species
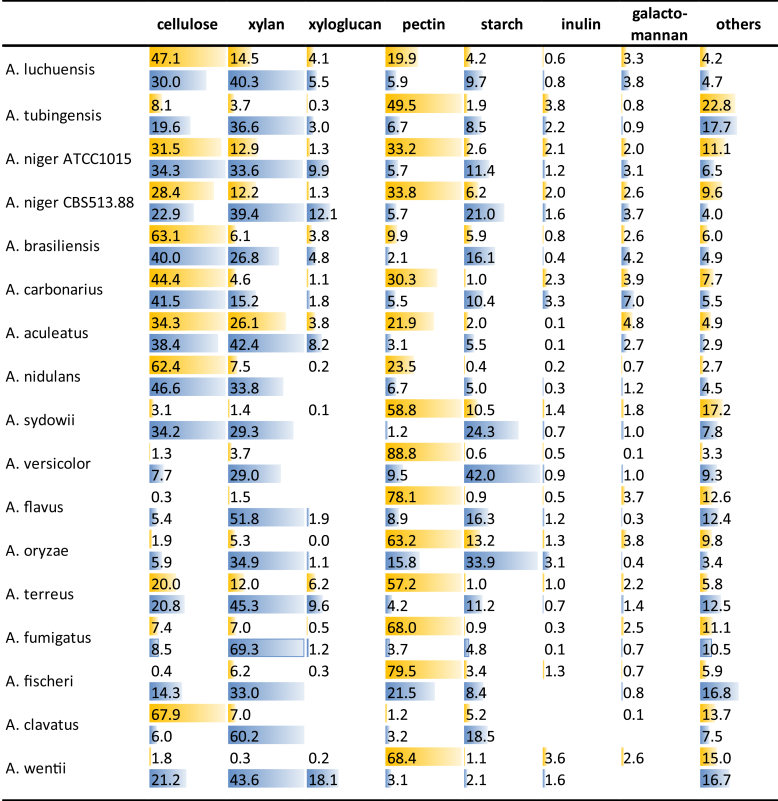
As shown above, the differences in genome content cannot explain the relative growth differences of the Aspergilli on plant biomass related polysaccharides. We postulate that regulation of the expression of these genes may be differently organized. To test this hypothesis, we grew the species on two crude plant biomass substrates, wheat bran (WB) and sugar beet pulp (SBP). These two substrates differ significantly in their composition (Suppl. Table 4), with WB consisting mainly of arabinoxylan and cellulose with some starch while SBP consisting mainly of pectin and cellulose with some xyloglucan. All species of this study were able to grow very well on these two substrates (Fig. 2), with only somewhat reduced growth for A. clavatus on SBP, likely due to the absence of many pectinolytic genes in its genome. The enzymes produced by the Aspergilli reflect very well the overall compositions of the substrates. In the presence of SBP, cellulose-, xyloglucan- and pectin-degrading enzymes represent 58–90 % of the CAZymes in the exoproteomes. Similarly, in the presence of WB, cellulose-, xylan- and starch-degrading enzymes represent 56–88 % of the CAZymes in the exoproteomes (Table 9). However, large differences between the species were detected in the division of the secreted enzymes over the polysaccharides they act on (Fig. 3, Table 9), which confirms a previous study on a smaller set of species (Benoit et al. 2015). The exoproteome analysis reveals that in response to WB and SBP, the species produce significantly different sets of enzymes that may indicate the polysaccharide(s) they most aim for. Most species of section Nigri as well as A. nidulans, A. sydowii (only WB) and A. clavatus (only SBP) produce high levels of cellulolytic enzymes, while this is much less pronounced for the other species. For A. clavatus this can be explained by the reduced pectinolytic system (see above), which makes cellulose the primary available substrate. However, for the other species, the disparate enzymes produced suggests regulatory adaptations. Nearly all species produce significantly more xylanolytic and amylolytic enzymes on WB than on SBP, and more pectinolytic enzymes on SBP than on WB, which matches the composition of the substrates. The exception to this is A. aculeatus that produces highly similar levels of xylanolytic enzymes on both substrates. In fact, this species has the lowest difference in enzyme production between the two substrates. Interestingly, xyloglucan related enzymes are in general produced more highly on WB than on SBP, despite xyloglucan being mainly a SBP component. It has been reported that regulation of xyloglucanolytic genes is under control of XlnR in A. niger and A. oryzae (Hasper et al., 2002, Noguchi et al., 2009). If this is a common phenomenon in Aspergilli that would explain the similar production profile to xylanolytic enzymes. Only very low amounts of CAZymes related to galactomannan or inulin degradation were detected (Fig. 3). This is likely due to the absence of these two polysaccharides in both WB and SBP and therefore they will not be discussed in detail.
Table 9.
Comparison of the percentage of enzymes produced on sugar beet pulp and wheat bran in relation to the composition of these substrates (Suppl. Table 3).
| Species/strains | Sugar beet pulp |
Wheat bran |
||||||
|---|---|---|---|---|---|---|---|---|
| Cellulases | Xyloglucanases | Pectinases | Total | Cellulases | Xylanases | Amylases | Total | |
| A. luchuensis | 47.1 | 4.1 | 19.9 | 71.2 | 30.0 | 40.3 | 9.7 | 80.0 |
| A. tubingensis | 8.1 | 0.3 | 49.5 | 58.0 | 19.6 | 36.6 | 8.5 | 64.7 |
| A. niger ATCC1015 | 31.5 | 1.3 | 33.2 | 66.0 | 34.3 | 33.6 | 11.4 | 79.3 |
| A. niger CBS 513.88 | 28.4 | 1.3 | 33.8 | 63.5 | 22.9 | 39.4 | 21.0 | 83.3 |
| A. brasiliensis | 63.1 | 3.8 | 9.9 | 76.8 | 40.0 | 26.8 | 16.1 | 82.9 |
| A. carbonarius | 44.4 | 1.1 | 30.3 | 75.8 | 41.5 | 15.2 | 10.4 | 67.1 |
| A. aculeatus | 34.3 | 3.8 | 21.9 | 60.1 | 38.4 | 42.4 | 5.5 | 86.4 |
| A. nidulans | 62.4 | 0.2 | 23.5 | 86.1 | 46.6 | 33.8 | 5.0 | 85.4 |
| A. sydowii | 3.1 | 0.1 | 58.8 | 62.0 | 34.2 | 29.3 | 24.3 | 87.9 |
| A. versicolor | 1.3 | 88.8 | 90.1 | 7.7 | 29.0 | 42.0 | 78.7 | |
| A. flavus | 0.3 | 78.1 | 78.4 | 5.4 | 51.8 | 16.3 | 73.6 | |
| A. oryzae | 1.9 | 63.2 | 65.0 | 5.9 | 34.9 | 33.9 | 74.7 | |
| A. terreus | 20.0 | 6.2 | 57.2 | 83.4 | 20.8 | 45.3 | 11.2 | 77.2 |
| A. fumigatus | 7.4 | 0.5 | 68.0 | 75.9 | 8.5 | 69.3 | 4.8 | 82.7 |
| A. fischeri | 0.4 | 0.3 | 79.5 | 80.2 | 14.3 | 33.0 | 8.4 | 55.7 |
| A. clavatus | 67.9 | 1.2 | 69.1 | 6.0 | 60.2 | 18.5 | 84.7 | |
| A. wentii | 1.8 | 0.2 | 68.4 | 70.5 | 21.2 | 43.6 | 2.1 | 66.9 |
Fig. 3.
Summary of the secretion of CAZymes related to degradation of different plant polysaccharides during growth on wheat bran (blue) or sugar beet pulp (yellow). Peptide counts were summed up for all enzymes related to a specific polysaccharide.
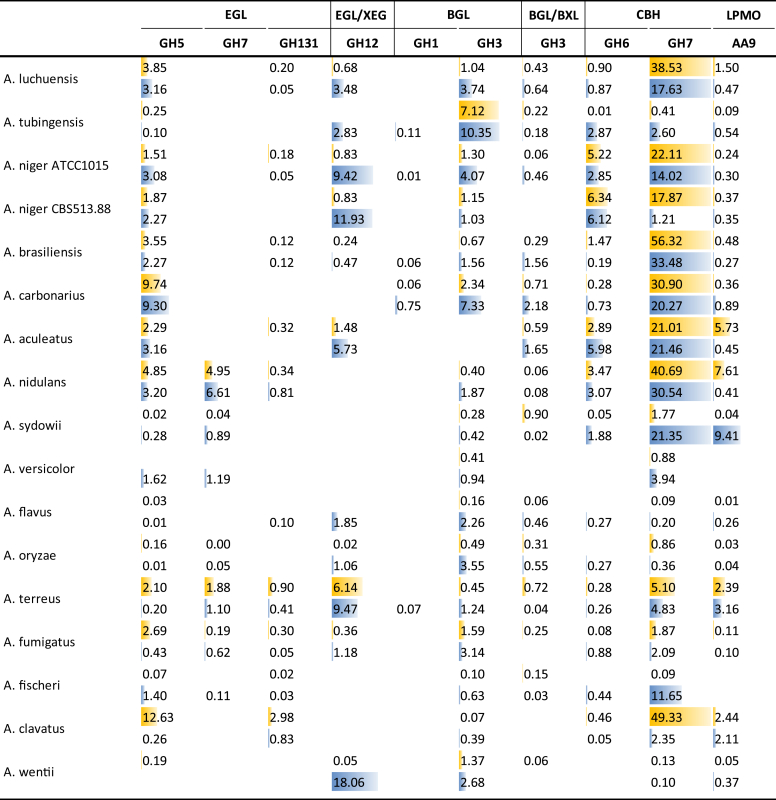
Significant differences were observed in the activities related to cellulose degradation between the species (Fig. 4). GH7 cellobiohydrolases (CBH) are the most prominent enzymes produced on both WB and SBP for nearly all species from section Nigri and Nidulantes, with the exception of A. tubingensis (WB and SBP), A. niger CBS 513.88 (WB) and A. sydowii (SBP). The similar protein levels confirm a previous study in which other strains from A. niger, A. luchuensis and A. aculeatus were compared for several plant biomass degrading enzyme activities (Li et al. 2017). For A. tubingensis the predominant activity is GH3 β-glucosidase (BGL), which is a minor activity in most other species. Aspergillus niger CBS 513.88 as well as A. niger ATCC1015 has high production of GH12 (xyloglucan-active) endoglucanases (EGL/XEG) on WB, which is also prominently produced by A. terreus on both substrates and in A. wentii on WB. No high levels of any cellulolytic enzyme were detected for A. sydowii on SBP. GH7 CBHs are also highly produced by A. terreus and A. clavatus on SBP and by A. fischeri on WB. Overall, production of other cellulolytic enzymes is low in the Aspergilli, which confirms an earlier report of the relatively low cellulolytic activity of Aspergilli compared to Trichoderma reesei (Jiang et al 2016). GH7 CBH-encoding genes have been shown to be controlled by the (hemi-)cellulolytic regulator XlnR in A. niger and A. oryzae (van Peij et al., 1998a, van Peij et al., 1998b, Hasper et al., 2002). XlnR responds to the presence of xylose (van Peij et al., 1998a, van Peij et al., 1998b), which could explain CBH production on wheat bran in these and related species. However, the xylose amount in SBP is much lower and mainly linked to xyloglucan, suggesting that a different regulator drives the production of these CBHs on SBP. It has been shown that many cellulases are under the control of one of the major cellulolytic activators, ClrB, in A. nidulans (Coradetti et al. 2013), while A. niger cbhA (GH7) was shown to be under control of both ClrA and ClrB (Raulo et al. 2016). Since cellulose is present in both substrates, this could imply that under our conditions, the production of GH7 CBHs depends more on ClrA and ClrB, than on XlnR. The higher abundance for exo-acting than for endo-acting cellulolytic enzymes suggests that while the Aspergilli do obtain carbon from cellulose (cellobiose, glucose), they do not appear to focus on full degradation of this polymer.
Fig. 4.
Secretion of CAZymes related to cellulose degradation during growth on wheat bran (blue) or sugar beet pulp (yellow). Peptide counts were summed up for all iso-enzymes with the same activity of the same CAZy family.
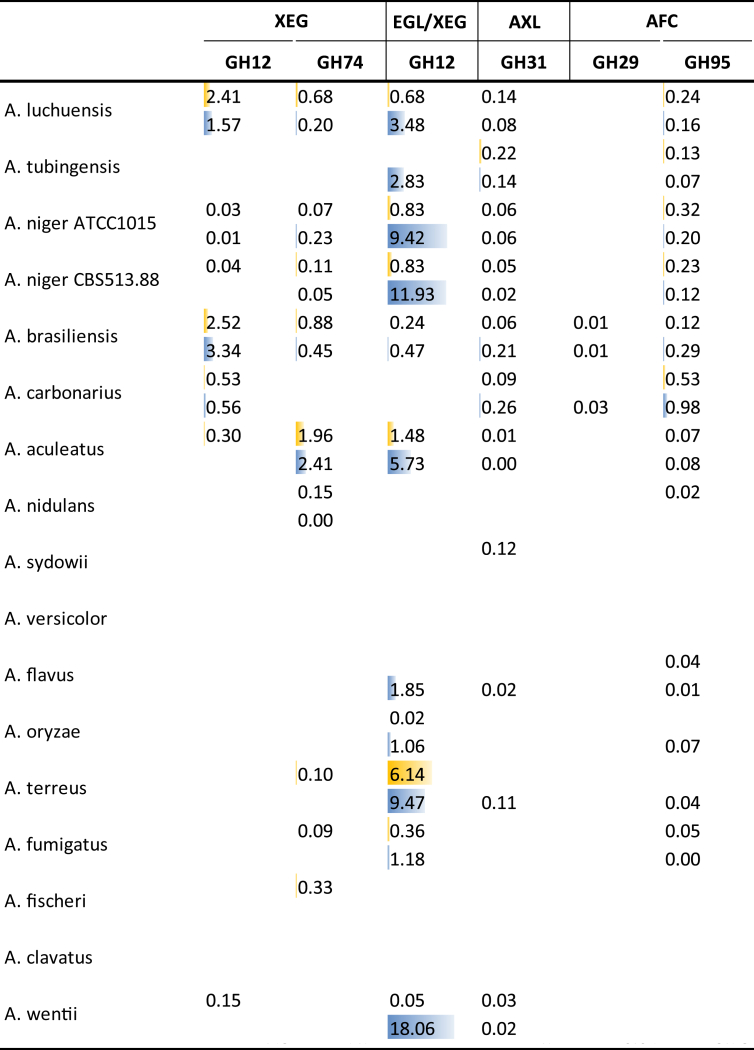
With the exception of the GH12 EGL/XEG enzymes mentioned above, only low levels of other xyloglucan-related enzymes were detected in our study (Fig. 5), suggesting that xyloglucan degradation is not a major part of the enzymatic response during growth on these substrates.
Fig. 5.
Secretion of CAZymes related to xyloglucan degradation during growth on wheat bran (blue) or sugar beet pulp (yellow). Peptide counts were summed up for all iso-enzymes with the same activity of the same CAZy family.
The production of enzymes related to xylan degradation was higher on WB than on SBP for all species, except A. aculeatus (Fig. 6). The most abundant enzymes are GH10 and GH11 endoxylanases (XLN), GH3 β-xylosidases (BXL) and GH62 arabinoxylan arabinofuranohydrolases (AXH). The balance between GH10 and GH11 XLN differs strongly among the species, and they fall into three groups: 1) A. luchuensis, A. niger ATCC1015 and A. aculeatus producing mainly GH10, 2) A. nidulans, A. fischeri, A. fumigatus and A. clavatus producing mainly GH11, and 3) A. niger CBS 513.88, A. sydowii, A. flavus and A. oryzae producing GH10 and GH11 endoxylanases at similar levels. This has a significant effect on the enzymatic mechanism of xylan degradation as these two XLN classes of have different substrate specificities. While GH11 XLNs mainly cleave non-substituted regions of xylan, GH10 XLNs can also cleave substituted regions and are also active on smaller oligosaccharides (Biely et al., 1997, Vardakou et al., 2003, Pollet et al., 2010). The growth substrate may affect the balance in GH10 and GH11 XLN production as a proteomic study of A. nidulans on sorghum stover revealed higher levels of GH10 than GH11 XLNs (Saykhedkar et al. 2012), which is opposite to our results on WB.
Fig. 6.
Secretion of CAZymes related to xylan degradation during growth of wheat bran (blue) or sugar beet pulp (yellow). Peptide counts were summed up for all iso-enzymes with the same activity of the same CAZy family.
Production of GH3 BXL is particularly high for A. tubingensis, A. luchuensis, A. niger ATCC1015 and A. carbonarius, all members of section Nigri. They are also detected in two other members of this section, A. brasiliensis and A. niger CBS 513.88, but is barely present in A. aculeatus. The other Aspergilli seem to in part compensate for the lower production of GH3 BXLs by producing BXLs of GH43. AXH was most abundant in A. luchuensis, A. tubingensis, both A. niger strains and A. aculeatus, but was also detected in several other Aspergilli. Low abundance of xylanolytic enzymes was detected in SBP, with the exception of AXH and GH10 XLN in A. aculeatus, demonstrating a strong link between composition of the substrate and production of the enzymes, most likely due to the role of XlnR as mentioned above. While the general function of XlnR is conserved among ascomycete fungi, it has been shown that the specific gene sets under its control can differ strongly (Klaubauf et al. 2014). This may explain the different xylanolytic enzyme sets observed on WB.
Pectinolytic enzymes acting on the pectin main chain are mainly detected on SBP, with much lower abundance in WB (Fig. 7). Similar to what was observed for cellulolytic enzymes, the highest abundance is for exo-acting enzymes, with only low abundance for major endo-acting enzymes such as endopolygalacturonase (PGA). Whether this indicates a much higher need for exo-acting enzymes to release the monomeric compounds or that the specific activity of the endo-acting enzymes is much higher than that of the exo-acting enzymes will require further study. However, the same effect was not observed for xylanolytic, xyloglucanolytic and amylolytic enzymes (Fig. 5, Fig. 6, Fig. 9). The difference in abundance between endo- and exopolygalacturonases also occurred in two previous studies. In a secretome study of A. niger on galacturonic acid, only exopolygalacturonases were detected, and no endopolygalacturonases (Braaksma et al. 2010). Similarly, in a transcriptome study on sugar beet pectin, expression levels of exopolygalacturonase-encoding genes were significantly higher than the levels of endopolygalacturonase-encoding genes (Kowalczyk et al. 2017).
Fig. 7.
Secretion of CAZymes related to degradation of the pectin main chain during growth on wheat bran (blue) or sugar beet pulp (yellow). Peptide counts were summed up for all iso-enzymes with the same activity of the same CAZy family.
Fig. 9.
Secretion of CAZymes related to degradation of starch during growth on wheat bran (blue) or sugar beet pulp (yellow). Peptide counts were summed up for all iso-enzymes with the same activity of the same CAZy family.
The difference between SBP and WB is not as pronounced for all pectinolytic side-chain active enzymes (Fig. 8). This is likely due to a more diverse role for some of these enzymes, acting also on other polysaccharides (e.g. xylan, xyloglucan). Interestingly, high abundance of exopolygalacturonases (PGX) and some other main chain and side chain active enzymes was observed in species with low abundance of cellulolytic enzymes, such as A. oryzae, A. fischeri and A. fumigatus (Fig. 4). This is also obvious for GH93 exoarabinanases (ABX), which are present at high abundance in these species, but absent in the samples of the species from section Nigri. GH93 ABX has been suggested to be essential for degradation of rhamnogalacturonan I (Benoit et al. 2012), a pectin substructure abundantly present in SBP. The production of GH93 ABX on WB in several species may indicate that it could also act on L-arabinose side chains (with the degree of polymerization of 2 or larger) of xylan. However, so far only activity on arabinan has been reported for this enzyme (Sakamoto and Thibault, 2001, Sakamoto et al., 2004, Kühnel et al., 2010, Mardones et al., 2015). These differences may indicate that A. oryzae, A. fischeri and A. fumigatus focus in particular on the pectin component of SBP, while some others (e.g. some species from section Nigri) degrade cellulose and pectin simultaneously.
Fig. 8.
Secretion of CAZymes related to degradation of pectin side chains during growth on wheat bran (blue) or sugar beet pulp (yellow). Peptide counts were summed up for all iso-enzymes with the same activity of the same CAZy family.
While abundance of pectin-related lyases is typically low in species from section Nigri, this is not the case for A. tubingensis. Our previous experiments indicated that this species only weakly acidifies the medium (data not shown), which would favor the action of lyases over hydrolases.
In general, amylolytic enzymes are mainly produced on WB, correlating with the composition of this substrate (Fig. 9). Highest abundance of α-amylases (AMY) was observed for A. niger CBS 513.88 and A. oryzae, both strains that have been selected and developed for industrial enzyme production, in particular amylolytic enzymes. Interestingly, both strains also produce AMY on SBP, and have similar GLA abundance on both substrates, but low levels of AGD. As mentioned above, AGD can perform undesired transglycosylation reactions and is therefore typically preferred to be low in amylolytic enzyme formulations, where the exo-activity is provided by GLA. In contrast, A. tubingensis, A. versicolor and A. sydowii have high AGD abundance on WB, but low AMY abundance. High abundance of AA13 (Lo Leggio et al. 2015) lytic polysaccharide mono-oxygenases (LPMO) is only observed for A. sydowii. Together with the low abundance of AMY in this species, this could indicate that A. sydowii has switched to a more oxidative approach to starch degradation.
The diverse Aspergilli genomes examined in this study all possess genes predicted to encode a broad spectrum of CAZymes that are capable of breaking down all major polysaccharides found in plant-derived biomass. In general, there is no correlation between the number of CAZyme genes and the efficiency in polysaccharide utilization. The notable exception is A. clavatus where the substantially reduced number of pectinolytic enzymes (16 enzymes as compared to 27–60 enzymes for the other Aspergilli targeting the pectin main chain) is correlated with poor growth on pectin. As well, A. clavatus produces negligible amount of pectinases in the presence of SBP, a pectin-rich feedstock. The fewer genes for pectinolytic enzymes have previously been proposed to be underlying cause for the poor growth of A. clavatus on pectin (Benoit et al. 2015). However there maybe another factor affecting the nonresponsive pectinolytic enzyme genes in A. clavatus. The orthologs of the transcription factor GaaR (Alazi et al. 2016) and repressor GaaX (Niu et al. 2017) regulating the expression of pectinolytic enzyme genes are present in the A. clavatus genome (JGI protein ID 4581 for GaaR ortholog and ID 4582 for GaaX ortholog), suggesting that the gene regulatory system for pectinases is functional. The inducer for most main chain pectinolytic enzyme encoding genes is D-galacturonic acid and the ortholog of its transporter GatA (Martens-Uzunova & Schaap 2008) is missing in the A. clavatus genome (de Vries et al. 2017). It is therefore possible that the missing transporter for the inducer of pectinolytic enzyme genes also contributes, together with the reduced number of pectinolytic enzyme genes, to poor growth of A. clavatus on pectin.
For the Aspergilli examined, species of section Nigri, together with A. aculeatus, A. nidulans and A. terreus, produce mixtures of enzymes with activities that are capable of digesting all the major polysaccharide in the available substrates (Fig. 10); for example, cellulolytic, xylanolytic and amylolytic enzymes are produced on WB whereas cellulolytic, xylanolytic and pectinolytic enzymes are produced on SBP. This finding suggests that these species are able to degrade simultaneously all the polysaccharides present in the substrate. For the other Aspergilli, typically the enzymes produced are capable of digesting a subset of polysaccharides present; for example, primarily pectinolytic enzymes on SBP. The exception is A. clavatus where mainly cellulolytic enzymes are produced on SBP, likely due to the significant reduction in its pectinolytic machinery. These results suggest that this group of Aspergilli digests only a subset of polysaccharides at a given time. Confirmation of these conclusions however would require further time-course experiments on transcriptomes and exoproteomes. However, it is clear that the match of the enzyme mixture produced by these species to the composition of the substrate they are growing on differs significantly (Fig. 10).
Fig. 10.
Overview of the enzymatic response of the selected Aspergilli to the presence of Wheat Bran (WB) and Sugar Beet Pulp (SBP). Peptide counts have been added up per polysaccharide they act on and expressed as percentage of total peptide counts. On the right the polysaccharide composition of WB and SBP is plotted (in %) to allow comparison of the enzymatic response to the composition of the substrates.
Acknowledgements
Part of this work was supported by Genome Canada and Génome Québec. Funding from the Academy of Finland (grant no. 308284) to MRM is acknowledged. The work conducted by the U.S. Department of Energy Joint Genome Institute (JGI), a DOE Office of Science User Facility, was supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231.
Footnotes
Peer review under responsibility of Westerdijk Fungal Biodiversity Institute.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.simyco.2018.09.001.
Contributor Information
A. Tsang, Email: adrian.tsang@concordia.ca.
R.P. de Vries, Email: r.devries@westerdijkinstitute.nl.
Appendix A. Supplementary data
The following are the supplementary data to this article:
References
- Ahmed S., Riaz S., Jamil A. Molecular cloning of fungal xylanases: an overview. Applied Microbiology and Biotechnology. 2009;84:19–35. doi: 10.1007/s00253-009-2079-4. [DOI] [PubMed] [Google Scholar]
- Alazi E., Niu J., Kowalczyk J.E., Peng M. The transcriptional activator GaaR of Aspergillus niger is required for release and utilization of D-galacturonic acid from pectin. FEBS Letters. 2016;590:1804–1815. doi: 10.1002/1873-3468.12211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alker A., Smith G., Kim K. Characterization of Aspergillus sydowii (Thom et Church), a fungal pathogen of Caribbean sea fan corals. Hydrobiology. 2001;460:105–111. [Google Scholar]
- Andersen M.R., Salazar M.P., Schaap P.J. Comparative genomics of citric-acid-producing Aspergillus niger ATCC 1015 versus enzyme-producing CBS 513.88. Genome Research. 2011;21:885–897. doi: 10.1101/gr.112169.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbruster FC (1960). Enzyme preparation. US Patent Application US3012944A.
- Arnaud M.B., Cerqueira G.C., Inglis D.O. The Aspergillus Genome Database (AspGD): recent developments in comprehensive multispecies curation, comparative genomics and community resources. Nucleic Acids Research. 2012;40:D653–D659. doi: 10.1093/nar/gkr875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battaglia E., Visser L., Nijssen A. Analysis of regulation of pentose utilization in Aspergillus niger reveals evolutionary adaptations in the Eurotiales. Studies in Mycology. 2011;69:31–38. doi: 10.3114/sim.2011.69.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benocci T., Aguilar-Pontes M.V., Zhou M. Regulators of plant biomass degradation in ascomycetous fungi. Biotechnology for Biofuels. 2017;10:152. doi: 10.1186/s13068-017-0841-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit I., Coutinho P.M., Schols H.A. Degradation of different pectins by fungi: correlations and contrasts between the pectinolytic enzyme sets identified in genomes and the growth on pectins of different origin. BMC Genomics. 2012;13:321. doi: 10.1186/1471-2164-13-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit I., Culleton H., Zhou M. Closely related fungi employ diverse enzymatic strategies to degrade plant biomass. Biotechnology for Biofuels. 2015;8:107. doi: 10.1186/s13068-015-0285-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biely P., Vrsanska M., Tenkanen M. Endo-β-1,4-xylanase families: differences in catalytic properties. Journal of Biotechnology. 1997;57:151–166. doi: 10.1016/s0168-1656(97)00096-5. [DOI] [PubMed] [Google Scholar]
- Braaksma M., Martens-Uzunova E.S., Punt P.J. An inventory of the Aspergillus niger secretome by combining in silico predictions with shotgun proteomics data. BMC Genomics. 2010;11:584. doi: 10.1186/1471-2164-11-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budak S.O., Zhou M., Brouwer C. A genomic survey of proteases in Aspergilli. BMC Genomics. 2014;523:1–15. doi: 10.1186/1471-2164-15-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen U., Gruben B.S., Madrid S. Unique regulatory mechanism for D-galactose utilization in Aspergillus nidulans. Applied and Environmental Microbiology. 2011;77:7084–7087. doi: 10.1128/AEM.05290-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coradetti S.T., Xiong Y., Glass N.L. Analysis of a conserved cellulase transcriptional regulator reveals inducer-independent production of cellulolytic enzymes in Neurospora crassa. MicrobiologyOpen. 2013;2:595–609. doi: 10.1002/mbo3.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J., Roberts J., Wallwork S. Sterigmatocystin, a metabolic product of Aspergillus versicolor. Chemistry & Industry. 1956;178 [Google Scholar]
- de Vries R.P., Burgers K., van de Vondervoort P. A new black Aspergillus species, A. vadensis, is a promising host for homologous and heterologous protein production. Applied and Environmental Microbiology. 2004;70:3954–3959. doi: 10.1128/AEM.70.7.3954-3959.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries R.P., Riley R., Wiebenga A. Comparative genomics reveals high biological diversity and specific adaptations in the industrially and medically important fungal genus Aspergillus. Genome Biology. 2017;18:28. doi: 10.1186/s13059-017-1151-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research. 2004;32:1792–1997. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms D.M., Kelly S. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biology. 2015;16:157. doi: 10.1186/s13059-015-0721-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espagne E., Lespinet O., Malagnac F. The genome sequence of the model ascomycete fungus Podospora anserina. Genome Biology. 2008;9:R77. doi: 10.1186/gb-2008-9-5-r77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorova N.D., Khaldi N., Joardar V.S. Genomic islands in the pathogenic filamentous fungus Aspergillus fumigatus. PLoS Genetics. 2008;4 doi: 10.1371/journal.pgen.1000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fremlin L., Piggott A., Lacey E. Cottoquinazoline A and cotteslosins A and B, metabolites from an Australian marine-derived strain of Aspergillus versicolor. Journal of Natural Products. 2009;72:666–670. doi: 10.1021/np800777f. [DOI] [PubMed] [Google Scholar]
- Galagan J.E., Calvo S.E., Cuomo C. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature. 2005;438:1105–1115. doi: 10.1038/nature04341. [DOI] [PubMed] [Google Scholar]
- Gruben B.S., Zhou M., de Vries R.P. GalX regulates the D-galactose oxido-reductive pathway in Aspergillus niger. FEBS Letters. 2012;586:3980–3985. doi: 10.1016/j.febslet.2012.09.029. [DOI] [PubMed] [Google Scholar]
- Gruben B.S., Zhou M., Wiebenga A. Aspergillus niger RhaR, a regulator involved in L-rhamnose release and catabolism. Applied Microbiology and Biotechnology. 2014;98:5531–5540. doi: 10.1007/s00253-014-5607-9. [DOI] [PubMed] [Google Scholar]
- Harris P., Stone B. Chemistry and molecular organization of plant cell walls. In: Himmel M., editor. Biomass Recalcitrance. Blackwell Publishing Ltd.; Oxford, UK: 2008. pp. 61–93. [Google Scholar]
- Hasegawa S., Takizawa M., Suyama H. Characterization and expression analysis of a maltose-utilizing (MAL) cluster in Aspergillus oryzae. Fungal Genetics and Biology. 2010;47:1–9. doi: 10.1016/j.fgb.2009.10.005. [DOI] [PubMed] [Google Scholar]
- Hasper A.A., Dekkers E., van Mil M. EglC, a new endoglucanase from Aspergillus niger with major activity towards xyloglucan. Applied and Environmental Microbiology. 2002;68:1556–1560. doi: 10.1128/AEM.68.4.1556-1560.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Vivas Duarte A., van den Brink J. Enhancing saccharification of wheat straw by mixing enzymes from two genetically modified strains Trichoderma reesei and Aspergillus niger. Biotechnology Letters. 2016;38:65–70. doi: 10.1007/s10529-015-1951-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaubauf S., Narang H.M., Post H. Similar is not the same: differences in the function of the (hemi-)cellulolytic regulator XlnR (Xlr1/Xyr1) in filamentous fungi. Fungal Genetics and Biology. 2014;72:73–81. doi: 10.1016/j.fgb.2014.07.007. [DOI] [PubMed] [Google Scholar]
- Kowalczyk J., Benoit I., de Vries R.P. Regulation of plant biomass utilization in Aspergillus. Advances in Applied Microbiology. 2014;88:31–56. doi: 10.1016/B978-0-12-800260-5.00002-4. [DOI] [PubMed] [Google Scholar]
- Kowalczyk J.E., Lubbers R.J.M., Peng M. Combinatorial control of gene expression in Aspergillus niger grown on sugar beet pectin. Scientific Reports. 2017;7:12356. doi: 10.1038/s41598-017-12362-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühnel S., Hinz S.W.A., Pouvreau L. Chrysosporium lucknowense arabinohydrolases effectively degrade sugar beet arabinan. Bioresource Technology. 2010;101:8300–8307. doi: 10.1016/j.biortech.2010.05.070. [DOI] [PubMed] [Google Scholar]
- Kunitake E., Tani S., Sumitani J., Kawaguchi T. A novel transcriptional regulator, ClbR, controls the cellobiose- and cellulose-responsive induction of cellulase and xylanase genes regulated by two distinct signaling pathways in Aspergillus aculeatus. Applied Microbiology and Biotechnology. 2013;97:2017–2028. doi: 10.1007/s00253-012-4305-8. [DOI] [PubMed] [Google Scholar]
- Li Q., Loman A.A., Coffman A.M. Soybean hull induced production of carbohydrases and protease among Aspergillus and their effectiveness in soy flour carbohydrate and protein separation. Journal of Biotechnology. 2017;248:35–42. doi: 10.1016/j.jbiotec.2017.03.013. [DOI] [PubMed] [Google Scholar]
- Lo Leggio L., Simmons T.J., Poulsen J.-C.N. Structure and boosting activity of a starch-degrading lytic polysaccharide monooxygenase. Nature Communications. 2015;6:5961. doi: 10.1038/ncomms6961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard V., Ramulu H.G., Drula E. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Research. 2014;42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonial S., Williams L., Carrum G. Neosartorya fischeri: an invasive fungal pathogen in an allogeneic bone marrow transplant patient. Bone Marrow Transplantation. 1997;19:753–755. doi: 10.1038/sj.bmt.1700715. [DOI] [PubMed] [Google Scholar]
- Machida M., Asai K., Sano M. Genome sequencing and analysis of Aspergillus oryzae. Nature. 2005;438:1157–1161. doi: 10.1038/nature04300. [DOI] [PubMed] [Google Scholar]
- Mardones W., Callegari E., Eyzaguirre J. Heterologous expression of a Penicillium purpurogenum exo-arabinanase in Pichia pastoris and its biochemical characterization. Fungal Biology. 2015;119:1267–1278. doi: 10.1016/j.funbio.2015.09.009. [DOI] [PubMed] [Google Scholar]
- Martens-Uzunova E.S., Schaap P.J. An evolutionary conserved D-galacturonic acid metabolic pathway operates across filamentous fungi capable of pectin degradation. Fungal Genetics and Biology. 2008;45:1449–1457. doi: 10.1016/j.fgb.2008.08.002. [DOI] [PubMed] [Google Scholar]
- Murphy C., Powlowski J., Wu M. Curation of characterized glycoside hydrolases of fungal origin. Database (Oxford) 2011;2011:bar020. doi: 10.1093/database/bar020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäkelä M.R., Bredeweg E.L., Magnuson J.K. Fungal ligninolytic enzymes and their applications. Microbiology Spectrum. 2016;4:1–13. doi: 10.1128/microbiolspec.FUNK-0017-2016. [DOI] [PubMed] [Google Scholar]
- Nierman W.C., Pain A., Anderson M.J. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature. 2005;438:1151–1156. doi: 10.1038/nature04332. [DOI] [PubMed] [Google Scholar]
- Niu J., Alazi E., Reid I.D. An evolutionarily conserved transcriptional activator-repressor module controls expression of genes for D-galacturonic acid utilization in Aspergillus niger. Genetics. 2017;205:169–183. doi: 10.1534/genetics.116.194050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi Y., Sano M., Kanamaru K. Genes regulated by AoXlnR, the xylanolytic and cellulolytic transcriptional regulator, in Aspergillus oryzae. Applied Microbiology and Biotechnology. 2009;85:141–154. doi: 10.1007/s00253-009-2236-9. [DOI] [PubMed] [Google Scholar]
- Ogawa M., Kobayashi T., Koyama Y. ManR, a novel Zn(II)2Cys6 transcriptional activator, controls the β-mannan utilization system in Aspergillus oryzae. Bioscience, Biotechnology and Biochemistry. 2012;77:426–429. doi: 10.1271/bbb.120795. [DOI] [PubMed] [Google Scholar]
- Ogawa M., Kobayashi T., Koyama Y. ManR, a transcriptional regulator of the β-mannan utilization system, controls the cellulose utilization system in Aspergillus oryzae. Bioscience, Biotechnology and Biochemistry. 2013;77:426–429. doi: 10.1271/bbb.120795. [DOI] [PubMed] [Google Scholar]
- Okabe M., Lies D., Kanamasa S. Biotechnological production of itaconic acid and its biosynthesis in Aspergillus terreus. Applied Microbiology and Biotechnology. 2009;84:597–606. doi: 10.1007/s00253-009-2132-3. [DOI] [PubMed] [Google Scholar]
- Payne G.A., Nierman W.C., Wortman J.R. Whole genome comparison of Aspergillus flavus and A. oryzae. Medical Mycology. 2006;44:S9–S11. doi: 10.1080/13693780600835716. [DOI] [PubMed] [Google Scholar]
- Petersen K.L., Lehmbeck J., Christensen T. A new transcriptional activator for amylase genes in Aspergillus. Molecular and General Genetics. 1999;262:668–676. doi: 10.1007/s004380051129. [DOI] [PubMed] [Google Scholar]
- Pel H.J., de Winde J.H., Archer D.B. Genome sequence of Aspergillus niger strain CBS 513.88: a versatile cell factory. Nature Biotechnology. 2007;25:221–231. doi: 10.1038/nbt1282. [DOI] [PubMed] [Google Scholar]
- Raulo R., Kokolski M., Archer D.B. The roles of the zinc finger transcription factors XlnR, ClrA and ClrB in the breakdown of lignocellulose by Aspergillus niger. AMB Express. 2016;6:5. doi: 10.1186/s13568-016-0177-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polizeli M.L.T.M., Rizzatti A.C.S., Monti R. Xylanases from fungi: properties and industrial applications. Applied Microbiology and Biotechnology. 2005;67:577–591. doi: 10.1007/s00253-005-1904-7. [DOI] [PubMed] [Google Scholar]
- Pollet A., Delcour J.A., Courtin C.M. Structural determinants of the substrate specificities of xylanases from different glycoside hydrolase families. Critical Reviews in Biotechnology. 2010;30:176–191. doi: 10.3109/07388551003645599. [DOI] [PubMed] [Google Scholar]
- Price M.N., Dehal P.S., Arkin A.P. FastTree 2 - approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5 doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T., Ihara H., Shibano A. Molecular characterization of a Penicillium chrysogenum exo-1,5-α-L-arabinanase that is structurally distinct from other arabinan-degrading enzymes. FEBS Letters. 2004;560:199–204. doi: 10.1016/S0014-5793(04)00106-1. [DOI] [PubMed] [Google Scholar]
- Sakamoto T., Thibault J.F. Exo-arabinanase of Penicillium chrysogenum able to release arabinobiose from α-1,5-L-arabinan. Applied and Environmental Microbiology. 2001;67:3319–3321. doi: 10.1128/AEM.67.7.3319-3321.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saykhedkar S., Ray A., Ayoubi-Canaan P. A time course analysis of the extracellular proteome of Aspergillus nidulans growing on sorghum stover. Biotechnology for Biofuels. 2012;5:52. doi: 10.1186/1754-6834-5-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmoll M., Dattenböck C., Carreras-Villaseñor N. The genomes of three uneven siblings: footprints of the lifestyles of three Trichoderma species. Microbiology and Molecular Biology Reviews. 2016;80:205–327. doi: 10.1128/MMBR.00040-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloothaak J., Schilders M., Schaap P. Overexpression of the Aspergillus niger GatA transporter leads to preferential use of D-galacturonic acid over D-xylose. AMB Express. 2014;4:66. doi: 10.1186/s13568-014-0066-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser K., McDonnell E., Nyaga C. mycoCLAP, the database for characterized lignocellulose-active proteins of fungal origin: resource and text mining curation support. Database (Oxford) 2015;2015:bav008. doi: 10.1093/database/bav008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukagoshi N., Kobayashi T., Kato M. Regulation of the amylolytic and (hemi-)cellulolytic genes in aspergilli. Journal of General and Applied Microbiology. 2001;47:1–19. doi: 10.2323/jgam.47.1. [DOI] [PubMed] [Google Scholar]
- van Peij N.N.M.E., Visser J., de Graaff L.H. Isolation and analysis of xlnR, encoding a transcriptional activator co-ordinating xylanolytic expression in Aspergillus niger. Molecular Microbiology. 1998;27:131–142. doi: 10.1046/j.1365-2958.1998.00666.x. [DOI] [PubMed] [Google Scholar]
- van Peij N.N.M.E., Gielkens M.M.C., de Vries R.P. The transcriptional activator XlnR regulates both xylanolytic and endoglucanase gene expression in Aspergillus niger. Applied and Environmental Microbiology. 1998;64:3615–3619. doi: 10.1128/aem.64.10.3615-3619.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardakou M., Katapodis P., Samiotaki M. Mode of action of family 10 and 11 endoxylanases on water-unextractable arabinoxylan. International Journal of Biological Macromolecules. 2003;33:129–134. doi: 10.1016/s0141-8130(03)00077-1. [DOI] [PubMed] [Google Scholar]
- Vizcaíno J.A., Csordas A., Del-Toro N. 2016 update of the PRIDE database and its related tools. Nucleic Acids Research. 2016;44:D447–D456. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voragen A.G.J., Coenen G.J., Verhoef R.P. Pectin, a versatile polysaccharide present in plant cell walls. Structural Chemistry. 2009;20:263–275. [Google Scholar]
- Wortman J.R., Gilsenan J.M., Joardar V. The 2008 update of the Aspergillus nidulans genome annotation: a community effort. Fungal Genetics and Biology. 2009;46:S2–S13. doi: 10.1016/j.fgb.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y., Mao X., Yang J. dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Research. 2012;40:W445–W451. doi: 10.1093/nar/gks479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X.L., Roubos J.A., van den Hondel C.A. Identification of InuR, a new Zn(II)2Cys6 transcriptional activator involved in the regulation of inulinolytic genes in Aspergillus niger. Molecular Genetics and Genomics. 2008;279:11–26. doi: 10.1007/s00438-007-0290-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng F., Lian X., Zhang G. A comparative genome analysis of Cercospora sojina with other members of the pathogen genus Mycosphaerella on different plant hosts. Genomics Data. 2017;8:54–63. doi: 10.1016/j.gdata.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.