Abstract
Mitochondrial dysfunction is pivotal in inducing synaptic injury and neuronal stress in Alzheimer’s disease (AD). Mitochondrial F1Fo ATP synthase deregulation is a hallmark mitochondrial defect leading to oxidative phosphorylation (OXPHOS) failure in this neurological disorder. Oligomycin sensitivity conferring protein (OSCP) is a crucial F1Fo ATP synthase subunit. Decreased OSCP levels and OSCP interaction with amyloid β (Aβ) constitute key aspects of F1Fo ATP synthase pathology in AD-related conditions. However, the detailed mechanisms promoting such AD-related OSCP changes have not been fully resolved. Here, we have found increased physical interaction of OSCP with Cyclophilin D (CypD) in AD cases as well as in an AD animal model (5×FAD mice). Genetic depletion of CypD mitigates OSCP loss via ubiquitin-dependent OSCP degradation in 5×FAD mice. Moreover, the ablation of CypD also attenuates OSCP/Aβ interaction in AD mice. The relieved OSCP changes by CypD depletion in 5×FAD mice are along with preserved F1Fo ATP synthase function, restored mitochondrial bioenergetics as well as improved mouse cognition. The simplest interpretation of our results is that CypD is a critical mediator that promotes OSCP deficits in AD-related conditions. Therefore, to block the deleterious impact of CypD on OSCP has the potential to be a promising therapeutic strategy to correct mitochondrial dysfunction for AD therapy.
Keywords: Alzheimer’s disease, mitochondrial F1Fo ATP synthase, oligomycin sensitivity conferring protein, cyclophilin D, Aβ
Introduction
Alzheimer’s disease (AD) is a devastating neurodegenerative disorder characterized by progressive cognitive decline (Cummings, 2004). Previous studies have repeatedly identified that mitochondrial dysfunction is a hallmark brain pathology underlying AD synaptic injury and neuronal death (Reddy et al., 2010; Wang et al., 2014; Zhu et al., 2013). Compromised mitochondrial oxidative phosphorylation (OXPHOS) constitutes a characteristic mitochondrial deficit in AD brains, resulting in lowered ATP production and increased oxidative stress, and eventually cell death (Du et al., 2012; Reddy, 2006; Reddy and Beal, 2008). The detailed mechanisms of OXPHOS impairment in AD remain as a long-standing scientific issue. But emerging evidence has accentuated the role of mitochondrial F1Fo ATP synthase dysfunction in AD-related mitochondrial OXPHOS failure (Beck et al., 2016).
Mitochondrial F1Fo ATP synthase is the executive site for ADP phosphorylation (He et al., 2018; Rubinstein et al., 2003). The dysfunction of this critical mitochondrial enzyme leads to disrupted OXPHOS and progressive ATP depletion. It is suggested that the AD-related F1Fo ATP synthase deregulation is strongly associated with the aberrations of its key subunit, oligomycin sensitivity conferring protein (OSCP) (Beck et al., 2016). OSCP is part of the peripheral stalk of F1Fo ATP synthase and plays a vital role in maintaining the structural stability of F1Fo complex (Carbajo et al., 2007). Decreased expression of OSCP and the physical interaction of OSCP with amyloid β (Aβ) cause the loss of OSCP function, culminating in the deregulation of F1Fo ATP synthase in AD-related conditions (Beck et al., 2016). In this regard, to determine the causative factors for such OSCP dysfunction is of paramount importance to understand mitochondrial OXPHOS defects in AD.
Intriguingly, a previous study on aging mice has shown an unexpected regulation on OSCP expression by Cyclophilin D (CypD; gene name: Ppif), which is evidenced by the observation that the brain aging-related OSCP loss is rescued by genetic depletion of CypD (Gauba et al., 2017). Given the close relationship of brain aging and AD, it is possible that decreased OSCP expression levels in AD brains are as well a result of CypD effect as increased CypD expression levels are a determined AD brain pathology (Du et al., 2008; Du et al., 2011). Furthermore, CypD has the capacity to bind both OSCP (Beutner et al., 2017; Giorgio et al., 2009) and Aβ (Du et al., 2008; Du et al., 2011). Of note, the interaction of CypD with OSCP could potentiate F1Fo complex uncoupling (Gauba et al., 2017; Giorgio et al., 2009); while the interplay of CypD with Aβ enhances CypD function (Du et al., 2008). Therefore, Aβ might serve as a key mediator that intensifies the deleterious effect of CypD on OSCP in AD-related conditions. On the other side, CypD may potentially affect the interaction of OSCP with Aβ. In this context, we thus hypothesize that CypD may play a crucial role in promoting OSCP changes in AD brains.
To this end, we examine the effect of CypD on OSCP in an AD animal model that mimics AD-like brain amyloidosis (5×FAD mice). We aim to determine whether CypD is a regulatory factor for OSCP expression and function in Aβ-rich milieus. In addition, we also examine whether ablation of CypD could be a practical strategy to protect OSCP in AD-related conditions.
Results
Aβ enhances CypD and OSCP interaction in AD subjects and 5×FAD mice
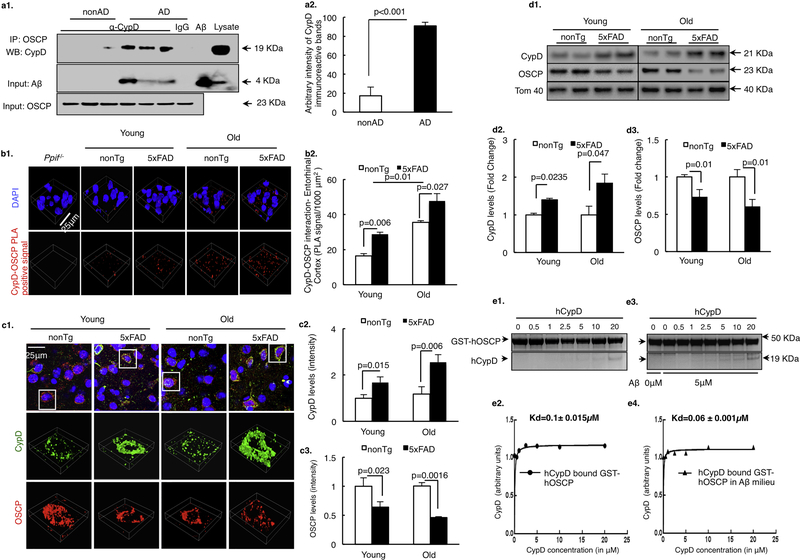
To determine the status of CypD and OSCP interplay in AD-related conditions, we subjected temporal lobe extracts from AD and nonAD subjects to perform coimmunoprecipitation for CypD and OSCP interaction. AD cases demonstrated significantly increased CypD/OSCP complexes (Fig. 1a1&2, Supplementary Table 1), suggesting enhanced CypD and OSCP interaction in AD brains. Next, we examined CypD/OSCP complexes in 5×FAD mice by antibody-based Duolink ® proximity ligation assay (PLA). PLA is a sensitive method to visualize and quantify tissue-based direct protein interactions in situ (Koos et al., 2014; Lutz et al., 2017). 5×FAD mice exhibited substantially increased density of CypD/OSCP complexes in their neocortex in an agedependent manner (Fig. 1b1&2). These results seem to implicate enhanced regulatory effect of CypD on OSCP in AD-related conditions.
Figure 1. Aβ enhances CypD and OSCP interaction.
(a) Co-immunoprecipitation of CypD and OSCP in AD patient temporal lobe extracts. Results shown are representative from three non-AD and three AD patients. AD brain lysate were used as positive controls for CypD and Aβ immunoreactive bands. Aβ peptide was used as positive control for Aβ immunoreactive bands. (b) OSCP and CypD interaction determined by in situ PLA assay in 5×FAD mouse neocortex. Scale bar, 25μm. n= 6 for nonTg and 5×FAD groups in both young and old mice. Ppif−/− mice were used as a negative control. (c) Monitoring the changes in OSCP and CypD expression levels in young and old nonTg and 5×FAD mouse neocortex. Scale bar, 25μm. n=5 per group. Quantification of staining intensity for CypD and OSCP in young and old nonTg and 5×FAD mouse brain showed inverse changes. (d) Representative immunoreactive bands for expression levels of CypD and OSCP in young and old nonTg and 5×FAD mouse synaptic mitochondria. CypD expression levels were upregulated while OSCP expression levels were downregulated in synaptic mitochondria from 5×FAD mice in an age-dependent manner. n= 5–6 per group. (e) OSCP and CypD interaction determined by an in vitro pull-down assay. The assay showed that Aβ enhances the ability of CypD to bind to OSCP. Error bars represent s.e.m.
Of note, by immunofluorescent staining as well as immuoblotting we have seen an age-dependent increase in CypD expression levels (Fig. 1c1&2, d1&2) along with decreased OSCP expression (Fig. 1c1&3, d1&3) in 5×FAD mice, which are consistent with our previous observations with AD subjects and AD mouse models (Beck et al., 2016; Du et al., 2008; Du et al., 2010). Given lowered expression levels of OSCP should disfavor the formation of CypD/OSCP complexes, we then asked whether Aβ promotes OSCP/CypD interaction. To this end, we employed in vitro pull-down assays using glutathione S-transferase (GST)-tagged OSCP as the bait protein. As shown in Fig. 1e1–4, when Aβ was added, the equilibrium dissociation constant (KD) of CypD/OSCP interaction was substantially decreased from 0.1 ± 0.015 μM (w/o Aβ) to 0.06 ± 0.001 μM (with Aβ), indicating a tighter binding of CypD with OSCP in the presence of Aβ.
CypD deficiency attenuates mitochondrial F1Fo ATP synthase deregulation in 5×FAD mice
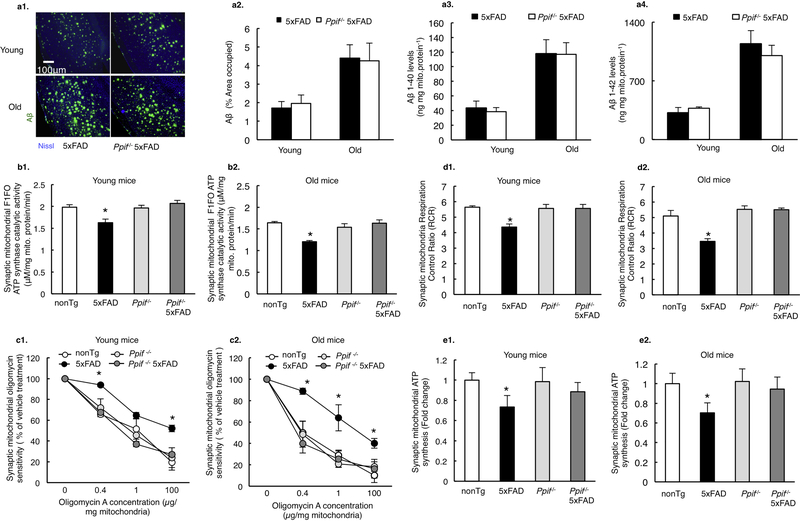
Previous studies indicate the deleterious influence of CypD and OSCP interaction on F1Fo ATP synthase function (Gauba et al., 2017; Giorgio et al., 2009). If increased CypD/OSCP complex formation as seen in 5×FAD mice underlies F1Fo ATP synthase dysfunction, we would expect a protective effect of CypD deficiency. To this end, we established CypD-knockout 5×FAD (Ppif−/−/5×FAD) mice. Of note, CypD had little impact on the levels of brain Aβ deposition in mice at both tested ages (4 and 9 months old) (Fig. 2a1&2), which is in consistence with our previous observations (Du et al., 2008). The similar levels of soluble Aβ1–40 (Fig. 2a3) and Aβ1–42 (Fig. 2a4) in 5×FAD mice with or without CypD expression were further determined by Aβ ELISA assay. To reflect F1Fo ATP synthase function, we examined F1Fo ATP synthase catalytic activity in neuron-specific synaptic mitochondria. In sharp contrast to their 5×FAD littermates, CypD-deficient 5×FAD mice exhibited substantially preserved F1Fo ATP synthase enzymatic activity at both 4 and 9 months old of age (Fig. 2b1&2). Further examination of F1Fo ATP synthase coupling assay showed a significant protection by CypD depletion (Fig. 2c1&2), indicating preserved F1Fo complex proton-flow coupling against Aβ toxicity. As a result of protected F1Fo ATP synthase function by CypD deficiency, we have observed remarkably improved synaptic mitochondrial respiratory control ratio (RCR) (Fig. 2d1&2) as well as restored ATP production (Fig. 2e1&2) in CypD deficient 5×FAD mice at both 4 and 9 months old. The above results suggest that CypD modulates F1Fo ATP synthase dysfunction in AD-related conditions.
Figure 2. CypD deficiency attenuates mitochondrial F1Fo ATP synthase deregulation in 5×FAD mice.
(a) Aβ levels in 5×FAD and CypD deficient 5×FAD mice in young and old age. n=6 for each group. Scale bar, 100μm. ELISA from isolated synaptic mitochondria showed similar levels of Aβ 1–40 and Aβ 1–42 in both AD genotype mice. (b) Synaptic mitochondria from 5×FAD mice demonstrated blunted F1Fo ATP synthase catalytic activity, which exacerbates with age. This defect was protected by CypD deficiency in both young and old Ppif−/− 5×FAD mice. n=5–7 per group. *P<0.05 vs other groups. (c) Decreased oligomycin sensitivity of synaptic mitochondria from young and old 5×FAD mice were rescued by CypD deficiency. All data are presented as percentage of the activity of the corresponding vehicle-treated mitochondrial fractions. n=5–7 per group. *P<0.05 vs other groups. (d) Synaptic mitochondria from 5×FAD mice showed an age-dependent decrease in RCR, which was protected by CypD depletion. n=5–7 mice per group. *P<0.05 vs other groups (e) Synaptic mitochondria from 5×FAD mice demonstrated an age dependent decline in ATP synthesis as compared to other groups. n=5–7 mice per group. *P<0.05 vs other groups. Error bars represent s.e.m.
CypD promotes OSCP loss via ubiquitination in 5×FAD mice
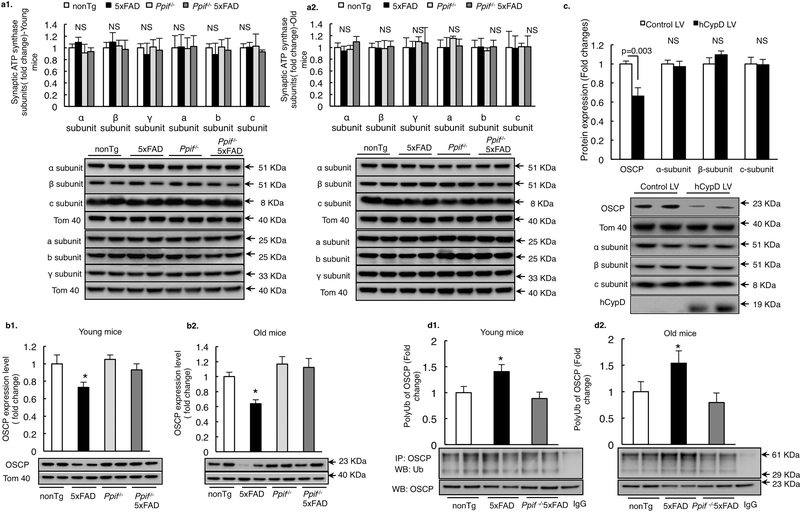
To determine whether the effect of CypD on neuronal mitochondrial F1Fo ATP synthase is associated with OSCP, we examined the influence of CypD on OSCP expression in 5×FAD mice. Purified synaptic mitochondria from nonTg, CypD deficient, 5×FAD and CypD deficient 5×FAD mice at 4 and 9 months old were subjected to immunoblotting assays for OSCP expression. In parallel, the expression levels of other major F1Fo ATP synthase subunits including α, β, γ, and a, b, c were also measured. There was little or no genotypic phenotype of CypD deficiency on the expression levels of other major subunits regardless of the presence or absence of Aβ (Fig. 3a1&2). However, CypD deficient 5×FAD mice exhibited remarkably preserved OSCP levels at both tested ages in comparison with their age- and gender-matched 5×FAD littermates (Fig. 3b1&2), implicating the effect of CypD on OSCP levels. This was further confirmed by our experiments using CypD-overexpressing neurons that showed selective decrease in OSCP expression levels (Fig. 3c). Since the proteolysis via the ubiquitin system is a critical pathway for OSCP degradation (Margineantu et al., 2007), we then examined whether CypD potentiates OSCP ubiquitination in 5×FAD mice. As expected, increased OSCP ubiquitination was detected in 5×FAD brains in both 4 and 9 months old mice, which was significantly attenuated by CypD depletion (Fig. 3d1&2). Put together, the results suggest that CypD is a mediator of OSCP loss in 5×FAD mice probably by promoting OSCP degradation via ubiquitination.
Figure 3. CypD promotes OSCP loss via ubiquitination in 5×FAD mice.
(a)Densitometric quantification of the immunoreactive bands of major F1Fo ATP synthase subunits including α, β, c, a, b, γ subunits in synaptic mitochondria of young and old nonTg, Ppif−/− mice and their AD counterparts. n=6–10 mice per group. The lower panels are representative immunoreactive bands. Tom40 was used as the loading control.*P<0.05 vs other groups. (b) Densitometric quantification of the immunoreactive bands of OSCP in synaptic mitochondria of young and old nonTg, Ppif−/− mice and their Aβ-expressing counterparts. n=6–10 mice per group. The lower panels are representative immunoreactive bands. Tom40 was used as the loading control.*P<0.05 vs other groups. (c) OSCP expression was downregulated in primary cultured mouse neurons by hCypD overexpression. Other major F1Fo ATP synthase subunits like α, β and c subunit displayed no change in expression. The lower panels are representative immunoreactive bands of indicated proteins. Tom 40 was used as the loading control. n=3 independent experiments. (d) Mouse brain mitochondria from nonTg, 5×FAD and Ppif−/− 5×FAD mice were subjected to immunoprecipitation with OSCP antibody followed by immunoblotting with ubiquitin and OSCP. Results are representative from 6 mice per group for both young and old age. *P<0.05 vs other groups. Error bars represent s.e.m.
CypD promotes OSCP/Aβ interaction in 5×FAD mice
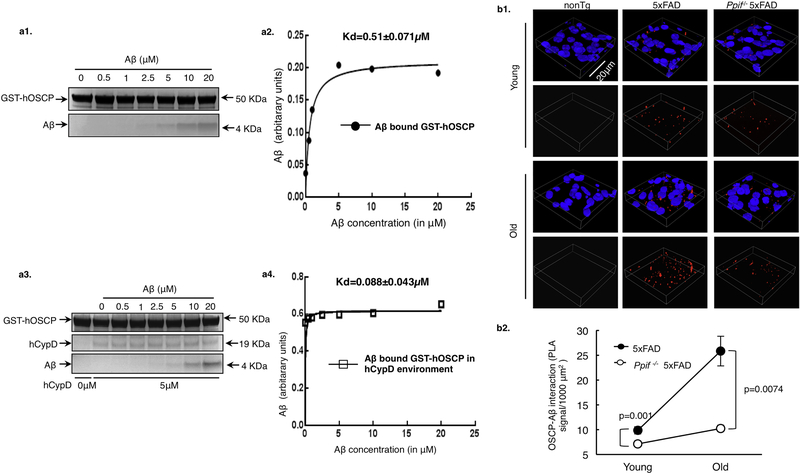
CypD, OSCP and Aβ bind to each other (Beck et al., 2016; Du et al., 2008; Gauba et al., 2017). Since Aβ could alter the binding affinity of CypD with OSCP (Fig. 1e1–4), it is thus of great interest to determine whether CypD has the similar effect on OSCP and Aβ interaction. In this regard, we adopted GST-tagged OSCP as the bait protein and performed in vitro pull down assays to examine the binding affinity of OSCP with Aβ in the absence or presence of CypD. Intriguingly, we found that the KD of OSCP/Aβ interaction was dramatically decreased from 0.51 ± 0.071 μM (w/o CypD) to 0.088 ± 0.043 μM (with CypD) (Fig. 4a1–4), indicating that CypD augments OSCP and Aβ interaction. If this in vitro observation could be extrapolated into an in vivo setting, we would expect to see less OSCP/Aβ complexes in CypD deficient 5×FAD mice. Indeed, our PLA results showed a significant decrease of OSCP/Aβ complex density in the neocortex of Ppif−/−/5×FAD mice in comparison with their age- and gender-matched 5×FAD littermates at 4 and 9 months old (Fig. 4b1&2), despite the age-dependent increase of OSCP/Aβ interaction in all the tested Aβ-producing mice (Fig. 4b1&2). Therefore, together with the observations on OSCP expression our findings indicate that CypD is a critical promoting entity underlying OSCP pathology in AD-relative pathological settings.
Figure 4. CypD promotes OSCP/Aβ interaction in 5×FAD mice.
(a) In vitro pulldown assay showed higher binding affinity of OSCP with Aβ in CypD rich environment. (b) In situ determination of OSCP and Aβ binding using proximity ligation assay in 5×FAD and Ppif−/− 5×FAD mice at young and old age. nonTg mouse brain was used as a negative control. n= 5–6 for 5×FAD and Ppif−/− 5×FAD groups at both ages. Scale bar, 20μm. Error bars represent s.e.m.
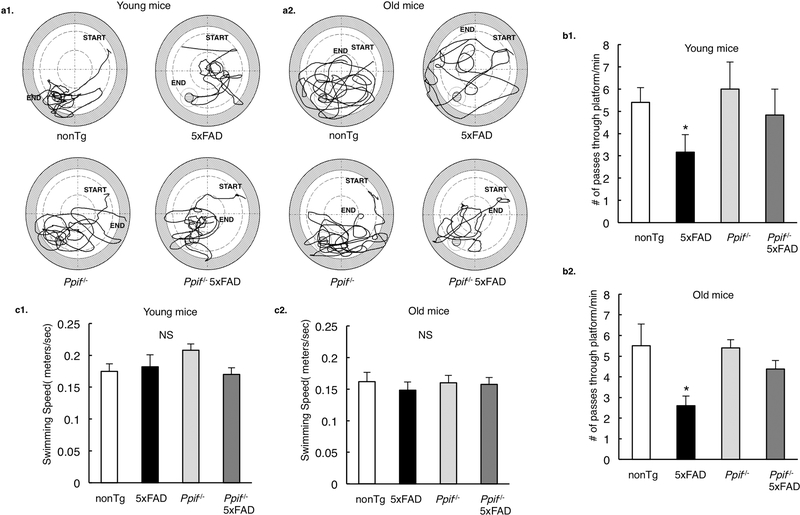
CypD deficiency improves cognitive function in 5×FAD mice
The close correlation of mitochondrial dysfunction with cognitive deficits has been repeatedly identified in AD patients as well as AD animal models. Consistent with alleviated F1Fo ATP synthase function, CypD deficient 5×FAD mice at both tested ages exhibited rescued spatial reference learning and memory (Fig. 5a1&2, b1&2) with unaltered swimming speed (Fig. 5c1&2) when comparing with their age- and gendermatched 5×FAD littermates. The results are in agreement with our previous findings on another AD mouse model (J20 line) (Du et al., 2008), further suggesting that the protection on cognition by CypD deficiency is not AD mouse model-sensitive but rather related to its protection on mitochondrial function.
Figure 5. CypD deficiency improves cognitive function in 5×FAD mice.
(a) Swimming traces for probe test after 11 day learning at Morris water maze for nonTg, Ppif−/− mice and their Aβ-expressing counterparts at young and old ages. (b) 5×FAD mice demonstrated impaired learning ability to locate the hidden platform (in an age-dependent manner), which was protected in CypD deficient AD mice. *P<0.05 vs other groups. (c) Mice in different groups did not display any difference in their swimming speeds. *P<0.05 vs other groups. n=6 for each group. Error bars represent s.e.m.
Discussion
As the major source of energy via oxidative phosphorylation (OXPHOS), mitochondria are essential organelles for neuronal physiology and survival. In recent years, mitochondrial cascade hypothesis has become a central paradigm in studying Alzheimer’s disease (AD), in particular its sporadic form (Swerdlow et al., 2014). Compromised mitochondrial OXPHOS is a determined brain pathology that induces synaptic and neuronal stress in this neurological disorder (Du et al., 2010; Manczak et al., 2004; Mastroeni et al., 2017; Onyango et al., 2016; Wang et al., 2016; Yao et al., 2009). As a result, the detailed molecular mechanisms of defected mitochondrial bioenergetics in AD have become a critical scientific question. Recent recognition of mitochondrial F1Fo ATP synthase deregulation in AD cases and AD animal models has substantially fostered our understanding of the pathogenesis of mitochondrial OXPHOS failure in AD (Beck et al., 2016). Importantly, the contribution of OSCP changes to this AD-related F1Fo ATP synthase dysfunction has indicated a potential target for AD therapy. Here, we have found that CypD plays a crucial role in mediating OSCP loss and OSCP/Aβ interaction in AD-related conditions. The ablation of CypD alleviates these ADassociated OSCP aberrations and further mitigates F1Fo ATP synthase deregulation as well as mitochondrial bioenergetics deficits and cognitive decline in 5×FAD mice. These findings have suggested a novel therapeutic strategy to protect OSCP and F1Fo ATP synthase in AD. Moreover, the current research has revealed a key role of CypD in regulating mitochondrial bioenergetics, particularly in diseases.
CypD is a mitochondrial peptidyl prolyl cis/trans isomerase (PPIase) (Baines et al., 2005). PPIases are ubiquitous enzymes that affect the structural folding of their substrate proteins by catalyzing the cis/trans isomerisation of peptides immediately preceding proline residues (Wedemeyer et al., 2002; Wu and Matthews, 2002). As a member of PPIase family in mitochondria, CypD is proposed to mediate the folding and rearrangement of mitochondrial proteins such as the adenine nucleotide translocase (ANT), mitochondrial P53 and many others (Dahout-Gonzalez et al., 2006; Lebedev et al., 2016; Woodfield et al., 1998). In recent years, the impact of CypD on mitochondrial F1Fo ATP synthase has been highlighted to be a critical mechanism regulating mitochondrial bioenergetics in health and disease (Beutner et al., 2017; Chinopoulos et al., 2011; Gauba et al., 2017; Giorgio et al., 2009; Giorgio et al., 2010). OSCP is one of the determined CypD interacting proteins in F1Fo ATP synthase (Gauba et al., 2017; Giorgio et al., 2009). The interaction of CypD with OSCP is suggested to induce uncoupling of F1Fo ATP synthase, thus reducing the activity of this pivotal mitochondrial enzyme (Gauba et al., 2017; Giorgio et al., 2009). This process might play a critical role in modulating mitochondrial bioenergetics at physiological states (Beutner et al., 2017; Giorgio et al., 2009). But in pathological settings such as brain aging (Gauba et al., 2017) as well as AD as we showed in here, increased CypD interaction with OSCP is strongly associated with F1Fo ATP synthase deregulation, suggesting its relevance to diseases. Therefore, the dual roles of CypD regulation on OSCP in physiology and pathology seem to raise a dilemma, which forms a groundwork for our future investigation to establish a delicate strategy to block excess CypD impact on OSCP while preserving its physiological function.
Of note, although it is not yet clear how CypD interaction with OSCP alters OSCP function, a possible reason is that CypD mediates the rearrangement of OSCP through their physical contact. Changes in the structural folding of a protein not only influence its function, but also potentially affect its capacity to form complexes with other entities such as proteins, peptides, and/or ions. This explains our observation that the interaction of CypD with OSCP causes an inclement of OSCP/Aβ interaction. Indeed, we cannot fully exclude the possibility that increased OSCP/Aβ interaction is at least in part a result of the formation of CypD/OSCP/Aβ complexes. Moreover, it should be noted that we have also detected a promoting effect of Aβ on OSCP/CypD interaction.This might be due to increased CypD activity by Aβ, and/or the influence of Aβ on OSCP structure which makes its binding with CypD easier. Our further study into these issues will help to address the above questions. Nevertheless, the current results suggest that blockade of CypD has the potential to relieve the deleterious impact of Aβ on OSCP through attenuated Aβ/OSCP interaction.
Another critical finding of this study that merits discussion is the role of CypD in modulating OSCP expression levels. We have found an inverse relationship between CypD and OSCP in 5×FAD mice. It should be noted that we have also seen significantly lowered OSCP levels in CypD overexpressing neurons. Importantly, our unpublished data showed that upregulation of OSCP in neurons in vivo had little influence on CypD levels. Therefore, the expression levels or the functional status of CypD seems to play a dominant role in regulating OSCP levels. Although the detailed mechanisms of CypDmediated OSCP downregulation are not yet resolved, our results have suggested OSCP degradation via ubiquitination. According to the prevailing opinions, the ubiquitindependent proteolysis is not generally accepted to be a pathway for mitochondrial protein degradation. However, increasing evidence has revealed a role of ubiquitin-mediated degradation of some mitochondrial proteins including OSCP (Heo and Rutter, 2011; Lehmann et al., 2016; Margineantu et al., 2007). It is suggested that OSCP undergoes retrograde transport to outer mitochondrial membrane (OMM), where OSCP is subsequently degraded via ubiquitination (Margineantu et al., 2007). Indeed, we have found increased ubiquitinated OSCP in 5×FAD mice, suggesting that enhanced OSCP ubiquitination is closely associated with OSCP loss in AD-related conditions. Moreover, the remarkable inhibition of OSCP ubiquitination by CypD deficiency also adds credit to the hypothesis that there is a potential link between CypD and OSCP degradation. A possible mechanism is that CypD alters the structure of OSCP and/or promotes OSCP/Aβ interaction, which mediates accelerated OSCP turnover. Since our understanding of CypD in ubiquitin-dependent proteolysis pathway is extremely limited, the detailed mechanisms of CypD-related OSCP degradation needs further study.
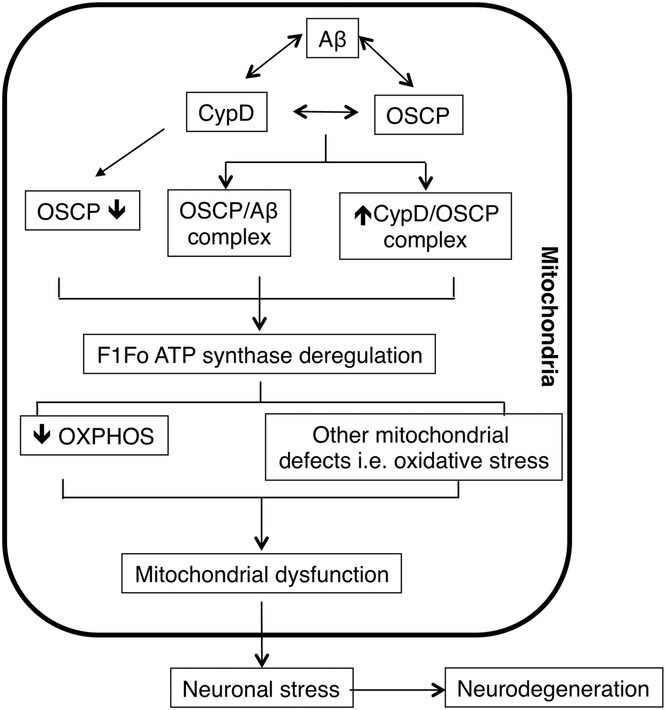
In summary, we have established a model in which CypD plays a key role in OSCP aberrations in AD-relative pathological settings, leading to F1Fo ATP synthase dysfunction and the resultant mitochondrial defects, eventually causing neuronal injury and death (Fig. 6). The ablation of CypD has demonstrated significant protection on mitochondrial F1Fo ATP synthase function as well as mitochondrial bioenergetics. Indeed, we cannot exclude the protective effects of CypD deficiency is also associated with other mechanisms such as the modulation of mitochondrial protein acetylation (Nguyen et al., 2013), the inhibition of mitochondrial permeability transition (Baines et al., 2005; Du and Yan, 2010), and the attenuation of changes of other mitochondrial proteins. Nevertheless, the results have at least suggested that CypD-mediated OSCP dysfunction is a pivotal mechanism of mitochondrial F1Fo ATP synthase deregulation and the resultant OXPHOS failure in AD-relative pathological settings. Therefore, the most parsimonious interpretation of the results is that the interplay between CypD and OSCP in Aβ-rich milieus is a primary AD change that potentially causes mitochondrial defects.
Figure 6. Schematic summary.
In response to Aβ deposition in AD brain mitochondria, CypD expression levels increase, which may promote OSCP reduction. Such changes potentiate elevated CypD/Aβ, CypD/OSCP and OSCP/Aβ interactions, leading to F1Fo ATP synthase dysfunction. The deregulation of this critical mitochondrial enzyme compromises mitochondrial OXPHOS efficacy and causes other mitochondrial defects including increased oxidative stress and calcium perturbations. Finally, the neuronal mitochondrial stress results in neuronal injury and eventually neurodegeneration.
Methods
Mice.
Animal studies were performed under the guidelines of International Animal Care and Use Committee (IACUC) at University of Texas at Dallas and National Institute of Health. Cyclophilin D deficient mice (B6;129-Ppif tm1Maf/J) and AD mice (B6SJLTg(APPSwFlLon,PSEN1*M146L*L286V)6799Vas/Mmjax) were purchased from Jackson Laboratory. We crossed these two transgenic mice to generate four genotypes of mice: nonTg, 5×FAD, Ppif−/− and Ppif−/− 5×FAD mice. Genotypes of animals were confirmed using PCR and dot blot. All the studies were performed at age of 4 months (young) and 8–9 months (old) to mimic MCI and late-stage AD respectively. Both male and female animals were used in these studies.
Human samples.
Frozen brain samples were requested from UT Southwestern Medical Center ADC Neuropathology Core, supported by ADC grant (AG12300) under a protocol approved by The UT Southwestern Medical Center with informed consent from all subjects and study adhered to Declaration of Helsinki principles.
Co-Immunoprecipitation of OSCP and CypD.
Co-Immunoprecipitation was performed as previously described (Beck et al., 2016; Du et al., 2008). Human cortical tissues were lysed in buffer (50mM Tris-HCl, 150mM NaCl, 1mM EDTA, 0.5% NP-40, 5% glycerol, and 1X Protease inhibitor (Calbiochem), pH 7.4) by keeping on ice for 30 minutes, followed by 7 freeze and thaw cycles. Lysate was pelleted at 12,500g at 4°C and supernatant was used to immunoprecipitate OSCP using anti-OSCP (Santa Cruz Biotechnologies 0.5μg IgG/100 μg protein) overnight at 4°C. Preimmuned IgG at the same concentration was used as the negative control. Prepared immuno-complex was incubated with pre-cleaned Protein agarose A/G (Pierce) for 2 hours at room temperature. Beads were washed for 5 times to remove non-specific binding of proteins. Western blot was performed and membrane was probed for anti-CypD (Calbiochem, 1:1,000). The specific bands represent CypD co-immunoprecipitated with OSCP.
Duolink® Proximity Ligation Assay.
Protein-protein interactions were detected using proximity ligation assay according to Duo-link in-situ detection (Sigma) protocol. The brain tissues were blocked for 30 minutes at 37°C and subjected to primary antibodies: anti-OSCP (Santa Cruz Biotechnologies, 1: 200), anti-Aβ (CST, 1:1000) or antiCyclophilin D (Calbiochem, 1:200) for overnight at 4°C. Following day, the slices were washed and treated with PLA probes for 1 hour at 37°C. The PLA probes were ligated at 37°C for 30 minutes and the signal was amplified using polymerase for 2 hours at 37°C. DAPI was used as the nuclear stain. Tissues were mounted and imaged under Nikon confocal microscope followed by three-dimensional reconstruction and analysis using the NIS Advanced research software.
Immunostaining for CypD and OSCP.
Brain tissues from 4 and 9 mo old nonTg and 5×FAD mice were used to perform co-staining of CypD and OSCP mitochondrial proteins. Citrate buffer was used to perform antigen retrieval followed by blocking in 5% goat serum and 0.3% triton-X for 1 hour at RT. Primary antibodies: CypD (Calbiochem, 1:500) and OSCP (Santa Cruz, 1:400) were used in blocking buffer for overnight incubation. The next day, fluorescent-tagged secondary antibodies were used at the concentrations of 1:400 and washed. DAPI was used as the nuclear stain. Tissues were mounted and imaged under Nikon confocal microscope followed by three-dimensional reconstruction and analysis using the NIS Advanced research software.
GST-pulldown assay and binding curve.
GST pulldown was performed according to manufacturer’s protocol (Pierce). Briefly, the human OSCP cDNA (Gene Name: ATP5O; NCBI Gene ID: 539) or human CypD cDNA (Gene Name: Ppif; NCBI Gene ID:10105) was transformed into BL21 (DE3) pLysS Escherichia coli (Promega) using the pGEX-4t1 plasmid (GE Healthcare). After transformation and selection a single colony was chosen for PCR to verify positive transformation. After overnight growth and induction by IPTG (Sigma-Aldrich), bacteria were pelleted and then were lysed by sonication in 1 × PBS containing 0.2 mM PMSF and 100 μg ml−1 lysozyme. After sonication bacterial debris was removed by centrifugation at 12,000g for 15 min at 4 °C. Supernatant was collected and incubated with glutathione agarose high-capacity, high-performance resin (Pierce) for 2 h. Glutathione beads were then washed and incubated overnight at 4 °C with purified hCypD protein or Aβ peptide (ApexBio). After washing, protein was eluted from the beads and separated by SDS–PAGE. Coomassie blue staining was performed to visualize results.
Purification of GST-fusion hCypD protein.
GST-hCypD protein was pulled down from E.Coli as described above. Glutathione beads (Pierce) bound to the fusion protein were subjected to thrombin proteases (GE) to cleave the protein from GST. The cleaved hCypD protein were collected, then subjected to removal of thrombin protease by using Hi-Trap Benzamidine FF (high sub) column (GE). The purified hCypD protein was concentrated into 1X PBS using Amicon Ultra filtration units (Millipore) and protein concentration was determined using Bradford assay. Coomassie staining the protein on SDS-PAGE determined the purity of the protein.
Aβ staining.
Mice brain tissues from both 4- and 9-month old 5×FAD and Ppif−/− 5×FAD mice were stained with Aβ to determine the levels of Aβ42 deposition. Briefly, the brain tissues were blocked in 5% goat serum and 0.3% triton-X for 1 hour at RT. Primary antibody Aβ (CST, 1:1000) in blocking buffer was used for overnight incubation to allow staining. The next day, fluorescent-tagged secondary antibody were used at the concentrations of 1:400 along with Nissl Blue stain (Sigma) and washed. Tissues were mounted and imaged under Nikon inverted microscope followed by analysis using the NIS Advanced research software.
ELISA for Amyloid β levels.
Synaptic amyloid β levels were measured using human Amyloid β(1–40) and Amyloid β(1–42) ELISA kit (Thermo Fisher Scientific) according to manufacturer’s instructions.
Synaptic mitochondria isolation.
Mice brain synaptic mitochondria were isolated using density gradient centrifugation as previously described (Beck et al., 2016). Firstly, cortices from mice brain were homogenized in ice-cold mitochondria isolation buffer (225 mM mannitol, 75 mM sucrose, 2 mM K2PO4, 0.1% BSA, 5 mM Hepes, 1 mM EGTA (pH 7.2)) with a Dounce homogenizer (Wheaton). The homogenate were pelleted at 1300g for 5 minutes and the supernatant was layered on a 3 × 2-ml discontinuous gradient of 15, 23 and 40% (vol/vol) Percoll (GE) and centrifuged at 34,000g for 8 min (flying time) on Beckman Coulter ultracentrifuge (Optima XPN-90 Ultracentrifuge). The band between 15 and 23% (containing synaptosomes) was collected. The fractions were then resuspended in isolation buffer containing 0.02% digitonin and incubated on ice for 5 min and then centrifuged at 16,500g for 15 min. Pellets were collected and resuspended in isolation buffer. Percoll density gradient centrifugation was performed as described above for a second time. The interface between 23 and 40% (mitochondria released from synaptosomes) was collected and resuspended in isolation buffer to centrifuge at 16,500g for 15 min. The resultant pellet was resuspended in isolation buffer followed by a centrifugation at 8,000g for 10 min. The final synaptic mitochondrial pellet was resuspended in isolation buffer and stored on ice during experiments. Protein concentration was determined using the Bio-Rad protein assay (Bio-Rad Laboratories).
F1FO ATP synthase enzyme activity assay.
F1FO ATP synthase enzymatic activity was measured spectrophotometrically using NADH-linked ATP regenerating system as previously described (Beck et al., 2016). 15 μg of synaptic mitochondria was suspended in the assay buffer [100 mM Tris-HCl (pH 7.4), 2 mM MgCl2, 50 mM KCl, 0.2 mM EDTA, 0.23 mM NADH, and 1 mM Phosphoenolpyruvate]. The reaction was triggered by the addition of 0.4 M ATP-Mg and recorded at OD340nm on a spectrophotometer (Ultraspect2100, Amersham Biosciences) for a total of 600 seconds at 10-second intervals and expressed in fold change.
F1FO ATP synthase coupling assay.
The coupling assay was performed as previously described (Beck et al., 2016). Synaptic mitochondria were treated with various concentrations of oligomycin A for 10 minutes at room temperature before conducting ATP synthase catalytic activity assay.
Mitochondrial Respiration Assay.
Freshly prepared synaptic mitochondria were used to perform this assay. Oxygen consumption was measured polarographically using temperature regulated Clark-type oxygen electrode (Oxytherm, Hansatech) as described previously (Du et al., 2008). In short, mitochondria were added to the magnetically stirred chamber and energized using 50μM of Glutamate-Malate (Sigma), followed by addition of substrate (ADP, Sigma, 300μM). State II respiration was measured as the oxygen consumption in the absence of the substrate, while addition of ADP induced state III respiration. On using up the substrate, a reduced rate of oxygen consumption was defined as the State IV respiration. Respiration Control ratio was measured as state III respiration over state IV respiration.
Mitochondrial ATP synthesis measurements.
ATP synthesis from freshly prepared synaptic mitochondria was measured using Luminescence ATP detection Assay Kit (Abcam) according to manufacturer’s instructions. 15 μg of isolated synaptic mitochondria were energized using 50μM of Glutamate/Malate, followed by addition of substrate (ADP, Sigma, 200μM). Luminescence was detected using a Microplate Reader (Synergy Mx., Biotek) with Gen5 software. Standard curve was prepared using ATP as substrate and luminescence readings were expressed in fold change.
Immunoblotting.
Immunoblotting was performed to quantify the expression levels of various proteins in isolated synaptic mitochondrial lysates. Proteins were separated in 12% Bis-Tris Gel (NuPAGE, Life Technologies) and then transferred to PVDF membrane (ImmunBlot Membrane, BioRad). Membranes were blocked with 5% non-fat dry milk (Labscientific Inc.) for 1 hour at room temperature and probed with appropriate primary antibodies overnight at 4 °C; followed by the incubation with the appropriate secondary antibody for 1 hour at room temperature. Proteins were detected using ECL (Clarity Substrate, BioRAD) and imaged using a Chemidoc system (BioRAD). The following antibodies were used in the experiments: anti-Cyclophilin D (Calbiochem, 1:5,000), anti-Tom 40 (Santa Cruz, 1: 500), and for all the major F1FO ATP synthase subunits: anti-OSCP (Santa Cruz Biotechnologies, 1: 5,000), anti-α subunit (Santa Cruz Biotechnologies, 1: 5,000), anti-β subunit (Santa Cruz Biotechnologies, 1: 5,000), anti-a subunit (ProteinTech, 1: 5000), anti-b subunit (Santa Cruz Biotechnologies, 1: 500), antiγ subunit (Santa Cruz Biotechnologies, 1: 500), anti-c subunit (abcam, 1:5000) and antiAmyloid β (CST, 1:2000) The intensities of the immunoreactive bands were analyzed using ImageJ software from NIH.
Whole brain mitochondria isolation.
Brain mitochondria were prepared as previously described (Du et al., 2008). Cortices were dissected from mouse brain and homogenized in ice cold mitochondria isolation buffer (225 mM mannitol, 75 mM sucrose, 2 mM K2PO4, 0.1% BSA, 5 mM Hepes, 1 mM EGTA (pH 7.2) with a dounce homogenizer (Wheaton). After a centrifugation at 1,300 x g for 5 minutes to remove blood and cell debris, the supernatant was layered on 15% Percoll (GE) and centrifuged at 125,00 x g for 10 minutes. The pellet was collected and resuspended in isolation buffer with 0.02% Digitonin (Sigma-Aldrich) and then subjected to the second centrifugation at 8,000 x g for another 10 minutes. The pellet was then washed by additional centrifugation step in ice-cold isolation buffer without EGTA for experiments. Protein concentrations were measured using Bradford assay for protein detection (BioRad).
Ubiquitination of OSCP.
Immunoprecipitation was performed as previously described (Beck et al., 2016; Du et al., 2008). Freshly isolated mitochondria were lysed in buffer (50mM Tris-HCl, 150mM NaCl, 1mM EDTA, 0.5% NP-40, 5% glycerol, and 1X Protease inhibitor (Calbiochem), pH 7.4) by keeping on ice for 30 minutes, followed by 7 freeze and thaw cycles. Lysate was pelleted at 12,500g at 4°C and supernatant was used to immunoprecipitate OSCP using anti-OSCP (Santa Cruz Biotechnologies 0.5μg IgG/100 μg protein) overnight at 4°C. Preimmuned IgG at the same concentration was used as the negative control. Prepared immuno-complex was incubated with pre-cleaned Protein agarose A/G (Pierce) for 2 hours at room temperature. Beads were washed for 5 times to remove non-specific binding of proteins. Western blot was performed and membrane was probed for anti-Ub (Santa Cruz, 1:2,000). The specific bands represent poly-ubiquitinated OSCP.
Primary neuron culture.
Primary neuron culture was performed using the established protocol. Briefly, brain tissues were dissected from day 0 pups and immersed in cold Hank’s balanced salt solution (HBSS, Sigma-Aldrich), dissociated with 0.05% trypsin (Sigma-Aldrich) at 37 ºC for 25–30 min followed by 10–15 times trituration. The cells were filtered through 40μm mesh cell strainer (Fisher brand) and centrifuged for 5min at 280g. The pellet was gently resuspended in neuron culture medium (Neurobasal A with 2% B27 supplement and 0.5mM L-glutamine, Gibco) and plated on poly-D-lysine (Sigma-Aldrich) coated culture plates (Corning) or chamber slides (Nunc) with an appropriate density. 10μM 5-fluoro-2′-deoxyuridine (Sigma-Aldrich) was added to the neuron cultures to inhibit non-neuronal cell proliferation. Neurons were cultured to 12 days in vitro (DIV) for experiment.
CypD overexpression in neurons.
Human CypD cDNA (Gene Name:Ppif−/− NCBI Gene ID: 10105) were inserted in to lentivirus vector with human polyubiquitin promoter-C(Addgene). Lentiviruses were packaged and applied on primary neurons. The cells were treated with lentivirus for 5 days followed by their collection in 1X LDS buffer followed by immunblotting.
Behavior studies.
Morris water maze was performed to evaluate the mice spatial learning and reference memory according to previously described protocol(Beck et al., 2016). Briefly, mice were trained to find a submerged platform in an open swimming arena. Repeated trials (n = 4) were performed each day for 11 days by starting the mice at different random start locations (NW, N, NE, E, SE) while platform was fixed at a single location (SW). Each trial lasted 60 seconds with an additional 30 seconds learning time where mice were allowed to remain on the platform. After 11 days of learning, mice were subjected to a probe test in which the platform was removed. Mice were analyzed for number of times they passed previous learning time platform location (SW). HVS Image 2015 software (HVS Image) determined swimming speeds and analyzed the behavior data.
Supplementary Material
Supplementary Table 1. The information of human brain tissues used in the experiments.
Highlights:
CypD disrupts mitochondrial oxidative phosphorylation in AD;
CypD promotes brain F1Fo ATP synthase deregulation in AD;
CypD mediates F1Fo ATP synthase dysfunction in AD via its impact on OSCP;
Loss of CypD protects F1Fo ATP synthase function in AD.
Acknowledgement:
This work was supported by research funding from NIH (R01AG053588, R01 AG059753), and Alzheimer’s Association (AARG-16–442863).
Footnotes
Conflict of interest: The authors have no conflict of interest to claim.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- Baines CP, et al. , 2005. Loss of cyclophilin D reveals a critical role for mitochondrial permeability transition in cell death. Nature. 434, 658–62. [DOI] [PubMed] [Google Scholar]
- Beck SJ, et al. , 2016. Deregulation of mitochondrial F1FO-ATP synthase via OSCP in Alzheimer’s disease. Nat Commun. 7, 11483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beutner G, et al. , 2017. Cyclophilin D regulates the dynamic assembly of mitochondrial ATP synthase into synthasomes. Sci Rep. 7, 14488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbajo RJ, et al. , 2007. How the N-terminal domain of the OSCP subunit of bovine F1Fo-ATP synthase interacts with the N-terminal region of an alpha subunit. J Mol Biol. 368, 310–8. [DOI] [PubMed] [Google Scholar]
- Chinopoulos C, et al. , 2011. Modulation of F0F1-ATP synthase activity by cyclophilin D regulates matrix adenine nucleotide levels. FEBS J. 278, 1112–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings JL, 2004. Alzheimer’s disease. N Engl J Med. 351, 56–67. [DOI] [PubMed] [Google Scholar]
- Dahout-Gonzalez C, et al. , 2006. Molecular, functional, and pathological aspects of the mitochondrial ADP/ATP carrier. Physiology (Bethesda). 21, 242–9. [DOI] [PubMed] [Google Scholar]
- Du H, et al. , 2008. Cyclophilin D deficiency attenuates mitochondrial and neuronal perturbation and ameliorates learning and memory in Alzheimer’s disease. Nat Med. 14, 1097–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, et al. , 2010. Early deficits in synaptic mitochondria in an Alzheimer’s disease mouse model. Proc Natl Acad Sci U S A. 107, 18670–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, et al. , 2012. Synaptic mitochondrial pathology in Alzheimer’s disease. Antioxid Redox Signal. 16, 1467–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, et al. , 2011. Cyclophilin D deficiency improves mitochondrial function and learning/memory in aging Alzheimer disease mouse model. Neurobiol Aging. 32, 398–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Yan SS, 2010. Mitochondrial permeability transition pore in Alzheimer’s disease: cyclophilin D and amyloid beta. Biochim Biophys Acta. 1802, 198–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauba E, et al. , 2017. Cyclophilin D Promotes Brain Mitochondrial F1FO ATP Synthase Dysfunction in Aging Mice. J Alzheimers Dis. 55, 1351–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgio V, et al. , 2009. Cyclophilin D modulates mitochondrial F0F1-ATP synthase by interacting with the lateral stalk of the complex. J Biol Chem. 284, 33982–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgio V, et al. , 2010. Cyclophilin D in mitochondrial pathophysiology. Biochim Biophys Acta. 1797, 1113–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, et al. , 2018. Assembly of the membrane domain of ATP synthase in human mitochondria. Proc Natl Acad Sci U S A. 115, 2988–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo JM, Rutter J, 2011. Ubiquitin-dependent mitochondrial protein degradation. Int J Biochem Cell Biol. 43, 1422–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koos B, et al. , 2014. Analysis of protein interactions in situ by proximity ligation assays. Curr Top Microbiol Immunol. 377, 111–26. [DOI] [PubMed] [Google Scholar]
- Lebedev I, et al. , 2016. A Novel In Vitro CypD-Mediated p53 Aggregation Assay Suggests a Model for Mitochondrial Permeability Transition by Chaperone Systems. J Mol Biol. 428, 4154–4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann G, et al. , 2016. Ubiquitination of specific mitochondrial matrix proteins. Biochem Biophys Res Commun. 475, 13–8. [DOI] [PubMed] [Google Scholar]
- Lutz MI, et al. , 2017. Novel approach for accurate tissue-based protein colocalization and proximity microscopy. Sci Rep. 7, 2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manczak M, et al. , 2004. Differential expression of oxidative phosphorylation genes in patients with Alzheimer’s disease: implications for early mitochondrial dysfunction and oxidative damage. Neuromolecular Med. 5, 147–62. [DOI] [PubMed] [Google Scholar]
- Margineantu DH, et al. , 2007. Hsp90 inhibition decreases mitochondrial protein turnover. PLoS One. 2, e1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastroeni D, et al. , 2017. Nuclear but not mitochondrial-encoded oxidative phosphorylation genes are altered in aging, mild cognitive impairment, and Alzheimer’s disease. Alzheimers Dement. 13, 510–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TT, et al. , 2013. Cyclophilin D modulates mitochondrial acetylome. Circ Res. 113, 1308–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onyango IG, et al. , 2016. Mitochondrial Dysfunction in Alzheimer’s Disease and the Rationale for Bioenergetics Based Therapies. Aging Dis. 7, 201–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy PH, 2006. Mitochondrial oxidative damage in aging and Alzheimer’s disease: implications for mitochondrially targeted antioxidant therapeutics. J Biomed Biotechnol. 2006, 31372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy PH, Beal MF, 2008. Amyloid beta, mitochondrial dysfunction and synaptic damage: implications for cognitive decline in aging and Alzheimer’s disease. Trends Mol Med. 14, 45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy PH, et al. , 2010. Amyloid-beta and mitochondria in aging and Alzheimer’s disease: implications for synaptic damage and cognitive decline. J Alzheimers Dis. 20 Suppl 2, S499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein JL, et al. , 2003. Structure of the mitochondrial ATP synthase by electron cryomicroscopy. EMBO J. 22, 6182–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swerdlow RH, et al. , 2014. The Alzheimer’s disease mitochondrial cascade hypothesis: progress and perspectives. Biochim Biophys Acta. 1842, 1219–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, et al. , 2016. Synaptosomal Mitochondrial Dysfunction in 5×FAD Mouse Model of Alzheimer’s Disease. PLoS One. 11, e0150441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, et al. , 2014. Oxidative stress and mitochondrial dysfunction in Alzheimer’s disease. Biochim Biophys Acta. 1842, 1240–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedemeyer WJ, et al. , 2002. Proline cis-trans isomerization and protein folding. Biochemistry. 41, 14637–44. [DOI] [PubMed] [Google Scholar]
- Woodfield K, et al. , 1998. Direct demonstration of a specific interaction between cyclophilin-D and the adenine nucleotide translocase confirms their role in the mitochondrial permeability transition. Biochem J. 336 ( Pt 2), 287–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Matthews CR, 2002. A cis-prolyl peptide bond isomerization dominates the folding of the alpha subunit of Trp synthase, a TIM barrel protein. J Mol Biol. 322, 7–13. [DOI] [PubMed] [Google Scholar]
- Yao J, et al. , 2009. Mitochondrial bioenergetic deficit precedes Alzheimer’s pathology in female mouse model of Alzheimer’s disease. Proc Natl Acad Sci U S A. 106, 14670–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, et al. , 2013. Abnormal mitochondrial dynamics in the pathogenesis of Alzheimer’s disease. J Alzheimers Dis. 33 Suppl 1, S253–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. The information of human brain tissues used in the experiments.