Summary
Myelin allows for fast and efficient axonal conduction, but much remains to be determined about the mechanisms that regulate myelin formation. To investigate the genetic basis of myelination, we carried out a genetic screen using zebrafish. Here we show that the lysosomal Gprotein RagA is essential for CNS myelination. In rraga−/− mutant oligodendrocytes, target genes of the lysosomal transcription factor Tfeb are upregulated, consistent with previous evidence that RagA represses Tfeb activity. Loss of Tfeb function is sufficient to restore myelination in RagA mutants, indicating that hyperactive Tfeb represses myelination. Conversely, tfeb−/− single mutants exhibit ectopic myelin, further indicating that Tfeb represses myelination during development. In a mouse model of de- and remyelination, TFEB expression is increased in oligodendrocytes, but the protein is localized to the cytoplasm, and hence inactive, especially during remyelination. These results define essential regulators of myelination and may advance approaches to therapeutic remyelination.
Graph Abstract
eTOC
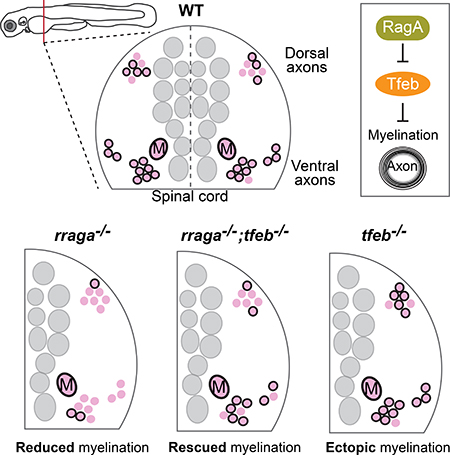
Using genetic approaches in zebrafish, Meireles, Shen et al. shows that the lysosomal regulators RagA and Tfeb have opposing roles to regulate myelin formation. Tfeb represses myelination, and RagA represses Tfeb. In RagA mutants, Tfeb is hyperactive and myelin is nearly eliminated.
Introduction
Myelin, the membranous sheath that insulates axons in vertebrates, is essential for both rapid conduction of action potentials and for metabolic and trophic support of axons (Funfschilling et al., 2012; Sherman and Brophy, 2005; Simons and Nave, 2016). Oligodendrocytes mature from oligodendrocyte precursor cells (OPCs), some of which differentiate during development, while others persist into adulthood (Emery, 2010). Adult OPCs can differentiate into myelinating oligodendrocytes to form new myelin in the adult brain, which occurs in response to motor learning in the healthy brain and also in diseases such as Multiple Sclerosis (MS) (Almeida and Lyons, 2017; Bengtsson et al., 2005; McKenzie et al., 2014; Münzel et al., 2013). MS results from inflammatory disruption of myelin in the CNS. Demyelination and irreversible axon loss in MS lead to impaired vision, loss of coordination, muscle weakness, fatigue, and cognitive impairment (Browne et al., 2014; Dutta and Trapp, 2011; Franklin and Ffrench-Constant, 2008; Münzel et al., 2013). Despite the importance of myelination in the healthy and diseased CNS, the molecular mechanisms that control oligodendrocyte development are only partly understood.
Lysosomes, long recognized as degradative organelles in the cell, are emerging as important signaling hubs that integrate nutrient availability with specialized cellular functions (Appelqvist et al., 2013; Ferguson, 2015; Saftig and Haas, 2016; Settembre and Ballabio, 2014; Settembre et al., 2013). Biochemical studies have defined the roles of several essential lysosomal proteins in the context of nutrient sensing and regulation of metabolic pathways (Efeyan et al., 2015). For example, in the presence of amino acids, GTP-bound heterodimeric Rag-GTPases (RagA or B bound to Rag C or D) recruit mTORC1 to the lysosomal membrane, where its then activated to induce protein synthesis and cell growth (Kim et al., 2008; Sancak et al., 2008; Shaw, 2008). This process requires the guanine nucleotide exchange factor (GEF) activity of the Ragulator complex, encoded by the lamtor genes (Bar-Peled et al., 2012; Sancak et al., 2010). Additionally, RagA regulates the activity of Transcription Factor EB (TFEB), which controls lysosomal biogenesis and autophagy (Sardiello, 2016; Sardiello et al., 2009; Settembre et al., 2011). When nutrients are available and lysosomal activity is sufficient, RagA recruits TFEB to the lysosome, where it is phosphorylated and inactivated. When the cell is starved or when lysosomal activity is disrupted or insufficient, TFEB is dephosphorylated, allowing it to enter the nucleus and activate target genes that control lysosome biogenesis, autophagy, and lipid catabolism (Martina and Puertollano, 2013; Napolitano and Ballabio, 2016). In addition to controlling lysosomal activity in phagocytic cells (Shen et al., 2016), RagA and RagB also function in cardiomyocytes, underscoring the important roles of the lysosomes in diverse cell types (Kim et al., 2014). Moreover, TFEB activation promotes clearing of intracellular debris in laboratory models of neurodegenerative diseases such as Huntington’s disease (Appelqvist et al., 2013; Martini-Stoica et al., 2016; Williams et al., 2008), but the roles of RagA and TFEB in other aspects of CNS development and disease remain unexplored.
Starting with a forward genetic screen in zebrafish, we define essential functions for several key lysosomal signaling molecules in myelination. We show that mutations in rraga or the Ragulator component lamtor4 result in reduced CNS myelination. We find that TFEB target genes are significantly upregulated in myelinating glia of rraga−/− mutants. Consistent with the hypothesis that increased TFEB activity blocks myelination in rraga−/− mutants, elimination of tfeb activity rescues myelination in rraga−/− mutants. Additionally, in tfeb−/− mutants we observe ectopic myelination in the dorsal spinal cord and ectopic expression of myelin basic protein (MBP) in the hindbrain. Moreover, transient overexpression of a nonphosphorylatable Tfeb construct represses mbp mRNA expression, further demonstrating that tfeb represses myelination in the developing CNS. We also explored the possibility that TFEB may be regulated during remyelination after injury. We find that localization of TFEB to the cytoplasm of oligodendrocytes (where it is inactive as a transcription factor) increases during remyelination in a mouse model of focal demyelination. Together, our results reveal essential roles for RagA and TFEB in regulating myelination, and suggest that manipulation of TFEB or its downstream effectors may represent a new avenue for improving therapeutic remyelination in MS.
Results
The Rag-Ragulator complex is essential for CNS myelination
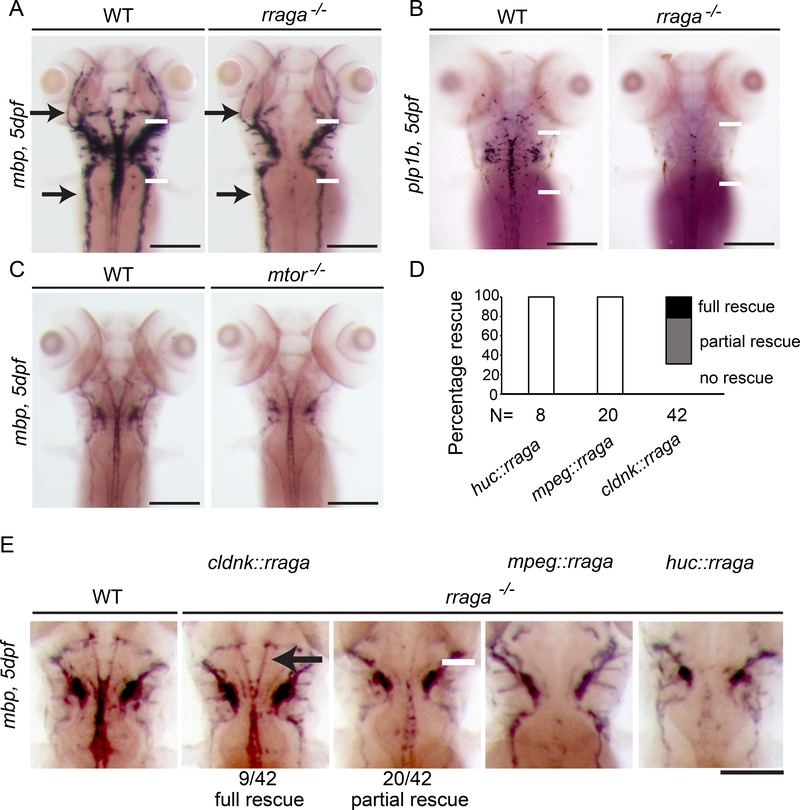
In a forward genetic screen using zebrafish, we identified rraga and lamtor4, which encode RagA and the Ragulator component Lamtor4, respectively, as genes essential for microglia development (Shen et al., 2016). To examine myelination in zebrafish carrying mutations in these genes, we analyzed the expression of the myelin markers myelin basic protein (mbp) and proteolipid protein (plp1b) by in situ hybridization (ISH). Homozygous rraga−/− mutants exhibited reduced mbp mRNA expression in the central nervous system (CNS) (Figure 1A, white arrows), whereas mbp expression in the peripheral nervous system (PNS) was normal (Figure 1A, black arrows). Similarly, expression of plp1b mRNA was reduced in rraga−/− homozygous mutants (Figure 1B). As described previously (Shen et al., 2016), lamtor4−/− homozygous mutants and rraga−/− mutants have very similar phenotypes (Figure S1).
Figure 1. rraga is essential for CNS myelination and acts autonomously in oligodendrocytes.
(A-E) Analysis of mbp or plp1b mRNA expression at 5 dpf by whole mount in situ hybridization. (A) Compared to their wildtype siblings, rraga−/− mutants show reduced mbp expression in the CNS (white arrows), whereas mbp expression in the PNS is normal (black arrows). (B) Expression of plp1b is also reduced in rraga−/− mutants. See also Figure S1. (C) mtor−/− mutants are developmentally delayed and have a small reduction in mbp mRNA expression levels in the CNS and PNS. See also Figure S2. (D) Quantification of rescue of mbp expression in rraga-/mutants following expression of wildtype rraga under the control of the claudink, mpeg (expressed in macrophages) or huc promoter (expressed in neurons). (E) Whole mount in situ hybridization analysis of mbp mRNA expression in 5 dpf rraga−/− mutants following expression of wild type rraga under the control of different tissue-specific promoters. mbp mRNA expression is partially or fully rescued only when wild type rraga is expressed in oligodendrocytes (claudinK promoter). All panels show dorsal views, with anterior to the top. Genotypes of all animals shown were determined by PCR after imaging. Scale bar = 50 μm.
Previous biochemical studies have provided evidence that the Rag-Ragulator complex recruits mTORC1 to lysosomes, where it is activated in response to amino acids (Napolitano and Ballabio, 2016; Sancak et al., 2010). To determine if reduced mTORC1 activity might cause the reduction of myelin in rraga−/− mutants, we compared the phenotypes of rraga−/− and mtor−/− mutants using the previously identified mtorxu015 insertional allele (Ding et al., 2011). This analysis revealed important differences in the rraga−/− and mtor−/− mutant phenotypes. mtor -/mutants exhibited a slight delay in overall development and partial reduction in mbp expression both in the CNS and PNS (Figure 1C), whereas rraga−/− mutants exhibit a strong, CNS-specific reduction of mbp expression (Figure 1A). In addition, treatment of wildtype zebrafish with Torin1, a potent and specific inhibitor of mTOR signaling (Thoreen et al., 2012), led to an overall developmental delay and slight reduction of mbp expression in both CNS and PNS, similar to mtor−/− mutants (Figure S2). Collectively, these experiments show that mtor−/− mutants express nearly normal levels of mbp in the CNS, in contrast to the strong and specific reduction of mbp in the CNS that is evident in rraga−/− mutants. These phenotypic studies indicate that the RagRagulator complex has a role in oligodendrocyte myelination that is independent of mTOR.
RagA acts autonomously in oligodendrocytes
To determine the cellular basis of RagA function in CNS myelination, we transiently expressed full-length wildtype rraga mRNA in rraga−/− mutants under the control of different cell typespecific regulatory sequences. Expression of mbp was rescued in rraga−/− mutants when the wildtype rraga gene was expressed in oligodendrocytes (claudinK promoter), but not when expressed in neurons or microglia (huC or mpeg promoter, respectively) (Figure 1D,E). Previous transcriptomic studies in mammals show that Rraga expression increases in myelinating oligodendrocytes, consistent with its cell autonomous function in myelination (Marques et al., 2016). Combined with our previous analysis of microglia (Shen et al., 2016), these results indicate that RagA has independent and cell autonomous functions in oligodendrocytes and microglia.
Myelin is reduced but oligodendrocytes are present in rraga−/− mutants
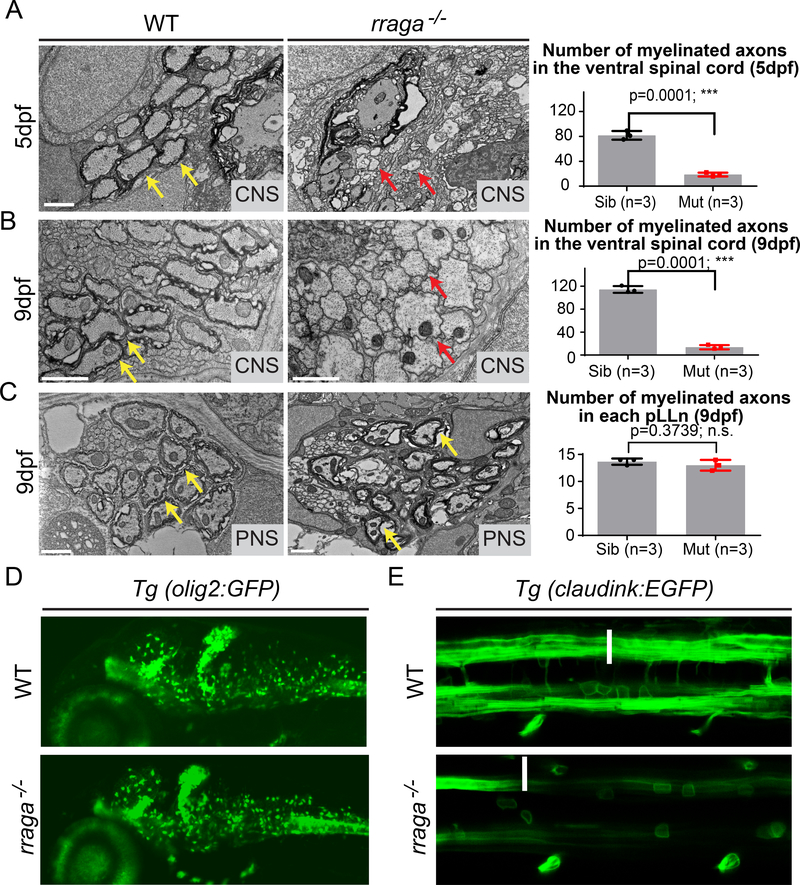
To determine if myelin ultrastructure is disrupted in rraga−/− mutants, we performed transmission electron microscopy on transverse sections of the ventral spinal cord in wildtype and rraga−/− mutant zebrafish. At 5 dpf, wildtype siblings had approximately 4-fold more myelinated axons than rraga−/− mutants (Figure 2A). At 9 dpf, the number of myelinated axons increased in wildtype siblings, whereas no additional myelinated axons were detected in rraga−/− mutants (Figure 2B). rraga−/− mutants and their wildtype siblings had similar numbers of myelinated axons in the posterior lateral line nerve, which is a component of the PNS (Figure 2C). This ultrastructural analysis of rraga−/− mutants confirms that RagA is essential for myelination in the developing CNS, but not the PNS.
Figure 2. CNS myelination is severely reduced in rraga mutants although oligodendrocytes are present.
(A,B) TEM images of transverse sections of the ventral spinal cord (CNS) at 5 dpf (A) and 9 dpf (B) show fewer myelinated axons in rraga−/− mutants (middle panel) compared to wildtype siblings (left panel). Quantification of the number of myelinated axons in the ventral spinal cord at 5 dpf and 9 dpf is shown on the right; bar graph depicts average values and standard error; individual measurements are also shown. (C) TEM images of transverse sections of the posterior lateral line nerve (pLLn) at 9 dpf show normal myelination in rraga−/− mutants (middle panel) compared to the wildtype sibling (left panel). Quantification of the number of myelinated axons in the pLLn at 9 dpf is shown on the right; bar graph depicts average values and standard error; individual measurements are also shown. Red arrows indicate unmyelinated axons and yellow arrows indicate myelinated axons. 3 wildtype and 3 rraga−/− mutants were analyzed. Scale bar = 1μm. (*** p <0.001, Student t-test, two-tailed). (D) Lateral view of 4 dpf Tg(olig2:EGFP) larvae showing comparable numbers of olig2-positive cells in both wildtype (top panel) and rraga−/− mutant (lower panel); panel shows anterior to the left and dorsal up. See also Figure S2. (E) Dorsal view of 4 dpf Tg(cldnk:EGFP) embryos showing presence of oligodendrocytes (arrows) in both wildtype (top panel) and rraga−/− mutants (lower panel), but reduced expression of GFP along myelinated axonal tracts (arrowheads). Panel shows anterior to the left and dorsal up. Genotypes of all animals shown were determined by PCR after imaging.
To determine if CNS myelin is reduced because rraga−/− mutants have fewer oligodendrocytes than wildtype, we examined the expression of olig2. Expression of olig2 mRNA, which marks motor neurons and cells of the oligodendrocyte lineage (Ravanelli and Appel, 2015; Zhou and Anderson, 2002), was normal in rraga−/− mutants at 3 dpf, as detected by in situ hybridization (Figure S3A). Similarly, analysis of the transgenic reporter Tg(olig2:GFP) (Shin et al., 2003) at 4 dpf showed that the number and distribution of olig2-GFP expressing cells were comparable between rraga−/− mutants and their wildtype siblings (Figure 2D).
To examine later stages of oligodendrocyte development, we used the transgenic reporter line Tg(claudink:EGFP ), which expresses GFP in mature oligodendrocytes and Schwann cells (myelinating glia of the PNS) (Münzel et al., 2012). The claudink:EGFP reporter is expressed in the CNS of rraga−/− mutants, but at lower levels than in wildtype siblings (Figure 2E, Figure S3B). In agreement with the olig2 marker analysis, oligodendrocyte cell bodies are visible in both rraga−/− mutants and their wildtype siblings (arrows), but in rraga−/− mutants the myelinating processes that normally extend along the axonal tracts are severely reduced (Figure 2E, arrowheads). These studies indicate that cells of the oligodendrocyte lineage are present in rraga−/− mutants, and that the onset of myelination is disrupted.
Transcriptomic analysis reveals upregulation of lysosomal genes and TFEB targets in myelinating glia of rraga−/− mutants
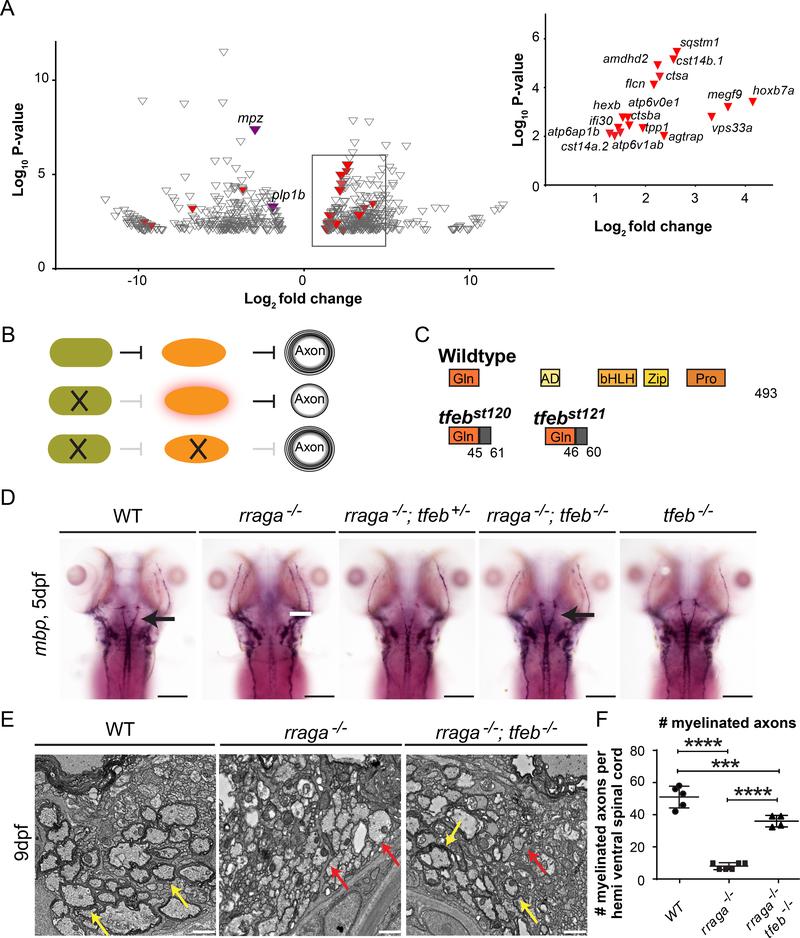
To investigate the pathways that are disrupted in myelinating glia of RagA mutants, we conducted RNA-sequencing of FACS-sorted Tg(claudinK:GFP) expressing cells from wildtype and rraga−/− mutant larvae at 5 dpf. claudink is expressed in oligodendrocytes and Schwann cells (Münzel et al., 2012), but because myelin appears to be normal in the PNS of rraga−/− mutants (Figure 1A, 2E), differences detected in Tg(claudink:GFP) expressing cells are likely to reflect changes in the oligodendrocyte transcriptome. Comparison of wildtype and rraga−/− mutant samples revealed 343 genes significantly upregulated in Tg(claudink:GFP) expressing cells of rraga−/− mutants (>2.5X, p<0.01) and 298 significantly downregulated genes (<2.5X, p<0.01) (Figure 3A, Tables S1, S2 and S3). Consistent with the reduction of myelination in the CNS of rraga−/− mutants, expression of myelin genes including plp1b and myelin protein zero (mpz) was significantly reduced (Figure 3A). Also consistent with our observation that PNS myelin is normal in rraga−/− mutants, the key regulators of Schwann cell myelination egr2b and pou3f1 are expressed at similar levels in wildtype and rraga−/− mutants.
Figure 3. RagA promotes CNS myelination by repressing TFEB.
(A) Volcano plot depicting the genes differentially expressed (p<0.01) in rraga−/− mutants relative to wildtype animals. Among the significantly downregulated genes (<2.5X) are mpz and plp1b (purple), which at this stage are characteristic of CNS myelin. Expression levels of 17 previously defined targets of TFEB (red) are upregulated (>2.5) in rraga−/− mutants. Inset graph depicts previously known TFEB targets (red) upregulated in rraga−/− mutants. See also Table S1-S3 (B) Diagrammatic representation of the hypothesis that RagA promotes myelination by repressing TFEB, which inhibits CNS myelination. (C) Diagrammatic representation of TFEB protein and of predicted truncated proteins encoded by tfebst120 and tfebst121 mutant alleles. (D) Expression of mbp mRNA, as detected by whole mount in situ hybridization, is reduced in CNS of rraga−/− mutants, but is restored in rraga−/−;tfeb−/− double mutants. rraga−/−;tfeb−/− double mutants and tfeb−/− mutants are indistinguishable from wildtype larvae. See also Figure S5. Scale bar = 50 μm. (E) TEM images of transverse sections of the ventral spinal cord (CNS) at 9 dpf show that myelination is restored in rraga−/−;tfeb−/− double mutants. Red arrows indicate unmyelinated axons and yellow arrows indicate myelinated axons. Scale bar = 1μm. (F) Quantification of the number of myelinated axons per hemi ventral spinal cord at 9 dpf is shown on the right; graph depicts average values and standard deviation; individual measurements are also shown. 3 individuals of each genotype were analyzed. (Statistical analysis: Pairwise comparisons using one-way ANOVA, Tukey post-hoc test significant interaction p<0.0001 ****, p <0.001***).
Functional annotation with PANTHER (Mi et al., 2013, 2017) revealed that genes associated with lysosomal activities were significantly enriched within the upregulated genes in rraga−/− mutant myelinating glia (GO term: cellular component, Table S1). TFEB directly activates the expression of more than 400 genes involved in lysosomal biogenesis, autophagy, mitophagy, lipid catabolism and lysosomal biogenesis (Medina et al., 2011; Napolitano and Ballabio, 2016; Nezich et al., 2015; Palmieri et al., 2011; Settembre et al., 2011). We compared our lists of genes differentially expressed in myelinating glia of rraga−/− mutants (upregulated and downregulated) to a previously compiled list of TFEB target genes (Palmieri et al., 2011). 17 previously defined TFEB target genes were upregulated in myelinating glia of rraga−/− mutants, including ctsa, ctsba, atp6ap1b, agtrap, hexb and sqstm1 (Figure 3A and Table S3). Similar analysis identified 4 TFEB target genes in the downregulated dataset, namely hoxc13a, prkag2b, naglu and comtd (Figure 3A and TableS3).
Confirming and extending our analysis of TFEB target gene expression in myelinating glia, we performed quantitative RT-PCR on total RNA from wildtype and rraga−/− mutants. Our analysis of 21 well established TFEB target genes (Palmieri et al., 2011) confirmed that ctsba, ctsa and sqstm1 are significantly upregulated in rraga−/− mutants, and identified 9 more TFEB target genes upregulated in whole-animal RNA samples, including gla, hexa, lamp1a, psap, clcn7, gba and neu (Figure S4A). Taken together these experiments provide evidence that TFEB target genes are upregulated in oligodendrocytes of rraga−/− mutants, consistent with previous reports demonstrating that RagA represses TFEB in other cell types (Martina and Puertollano, 2013).
The Rag-Ragulator complex promotes CNS myelination by repressing TFEB
Recent work has shown that active Rag GTPases repress TFEB by binding and recruiting it to lysosomes, where TFEB is phosphorylated and inactivated (Martina and Puertollano, 2013). Previous studies show that both Rraga and Tfeb are expressed in oligodendrocytes and that their expression increases as myelination progresses, consistent with the possibility that RagA antagonizes TFEB activity in oligodendrocytes (Lister et al., 2011; Marques et al., 2016). These previous studies, together with the upregulation of TFEB target genes in myelinating glia of rraga−/− mutants, led us to investigate whether aberrantly increased TFEB activity blocks myelination in rraga−/− mutant oligodendrocytes. This hypothesis predicts that TFEB represses myelination and that inactivation of TFEB would rescue CNS myelination in rraga−/− mutants (Figure 3B).
We used Crispr–Cas9 to generate mutations near the 5’end of the tfeb coding sequence and establish two new alleles of tfeb: tfebst120 and tfebst121 (Figure 3C). As expected for a loss of function allele, quantitative RT-PCR analysis showed that some known TFEB target genes are indeed downregulated in tfeb transheterozygous mutants (tfeb st120/st121, henceforth indicated as tfeb−/− mutant), including hexa, gba, gla, neu1, psap and sqstm1 (Figure S4B). Strikingly, expression of mbp and plp1b mRNA was partially restored in rraga−/− mutants heterozygous for tfeb+/− mutations, and mbp was restored to wildtype levels in rraga−/−;tfeb−/− double mutants (Figure 3D, Figure S5). Furthermore, in the ventral spinal cord, electron microscopy analysis revealed that rraga−/−; tfeb−/− double mutants have many more myelinated axons than rraga−/− single mutants (36 and 8, respectively) (Figure 3E,F). Our analysis of myelin gene expression and myelin ultrastructure in these mutants indicates that TFEB is hyperactive in oligodendrocytes of rraga−/− mutants and that inhibiting TFEB activity can rescue myelination in rraga−/− mutants.
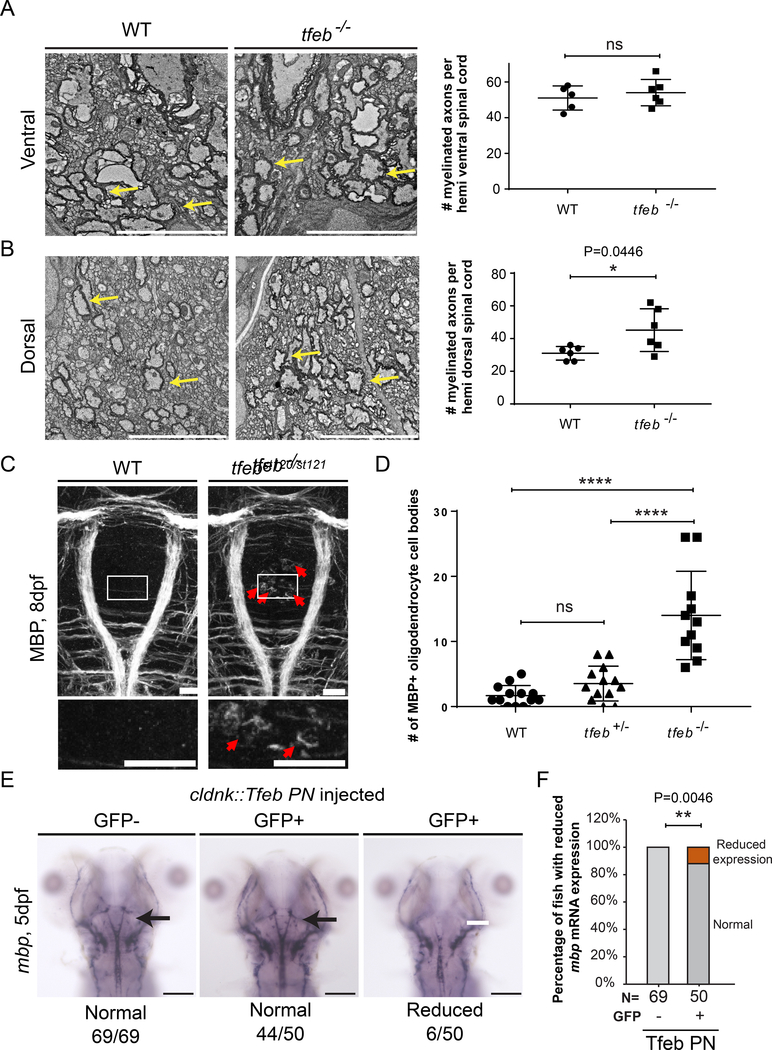
To test the hypothesis that TFEB represses myelination in the developing CNS, we examined expression of mbp mRNA, myelin ultrastructure, and MBP protein in tfeb−/− mutant larvae. At 5 dpf mbp and plp1b mRNA levels are indistinguishable in tfeb−/− mutants and their wildtype siblings (Figure 3D and Figure S5). To determine if tfeb−/− mutants have normal myelin, we next examined CNS ultrastructure by transmission electron microscopy. While our analysis revealed no statistical difference in the number of myelinated axons in the ventral spinal cord of tfeb−/− mutants and their wildtype siblings (ventral: 54 vs. 51, Figure 4A), there was a significant increase in the number of myelinated axons in the dorsal spinal cord of tfeb−/− mutants (dorsal: 48 vs. 32, Figure 4B). To determine if oligodendrocytes are altered in the brain of tfeb−/− mutants, we examined the expression and localization of MBP protein in the hindbrain of 8 dpf tfeb−/− mutants and siblings. MBP protein is normally localized in oligodendrocyte processes and largely excluded from the cell body (Figure 4C). In the hindbrain of 8 dpf tfeb−/− mutants, however, we observed MBP ectopically expressed in a region of the hindbrain that normally contains oligodendrocyte cell bodies with little or no MBP (Figure 4 C,D, red arrows). Our analysis of tfeb−/− and rraga−/−;tfeb−/− double mutants indicates that TFEB represses myelination in CNS, and that RagA is essential to prevent hyperactivity of TFEB in oligodendrocytes.
Figure 4. TFEB represses myelination in the CNS.
(A,B) TEM images of transverse sections of the spinal cord at 9 dpf show an increased number of myelinated axons in the dorsal spinal cord of tfeb mutants. Quantification of the number of myelinated axons per ventral (A) or dorsal (B) hemi spinal cord is shown on the right. Graph depicts average values and standard deviation; individual measurements are also shown. Yellow arrows indicate myelinated axons. Scale bar =5 μm. (Statistical analysis: Two-tailed unpaired t-test with Welch’s correction). 3 wildtype and 4 tfeb−/− mutant animals were analyzed. (C) Dorsal view of the hindbrain of 8 dpf larvae shows robust expression of MBP protein in myelinating oligodendrocyte processes in wild type and tfeb−/− mutants, and ectopic expression of MBP protein in tfeb−/− mutant cell bodies (red arrows). Scale bar = 50 μm. (D) Quantification of the number of ectopic MBP positive cells in tfeb−/− mutants (tfeb−/−, n=11) and siblings (n=13 for wildtype and tfeb+/− heterozygotes) (Pairwise comparisons using one-way ANOVA, Tukey posthoc test significant interaction p<0.0001 ****); bars represent standard deviation values. Individual values are also depicted. Genotypes of all animals shown were determined by PCR after imaging. (E) Whole mount in situ hybridization analysis of mbp mRNA expression in 5 dpf fish following transgenic expression of phosphorylation-null Tfeb (Tfeb NP) under the control of claudink promoter. mbp mRNA expression is reduced in some fish overexpressing the construct. (F) Quantification of the percentage of fish with reduced mbp mRNA expression. (Statistical analysis: Fisher’s exact test, two tailed, p< 0.01 **).
Our hypothesis that TFEB activity represses myelination in the CNS predicts that overexpression of a constitutively active (i.e. non-phosphorylatable) form of TFEB will repress myelination (Puertollano et al., 2018; Sardiello et al., 2009; Settembre et al., 2011). To determine if overexpression of constitutively active Tfeb represses myelination we transiently expressed a phosphorylation-null Tfeb mutant (Tfeb PN: S3A+S149A+S221A+S484A+S486A), under the control of the claudink promoter. Transient transgenesis results in high variability of transgene expression both within and between individuals. To select individuals with a higher rate of transgenesis, the transgenesis vector contained a cmlc2:GFP reporter, which allowed transgenic animals to be identified by GFP expression in the heart. At 4 dpf, injected fish were sorted into GFP+ or GFP- groups (transgenic and weakly or non transgenic, respectively), and mbp expression was monitored by in situ hybridization. As predicted, transient overexpression of phosphorylation-null Tfeb resulted in a decrease in mbp mRNA expression levels in some of the injected fish (Phospho-null 12%, 6/50 fish) (Figure 4E). Together, our data provide evidence that RagA functions to promote myelination by repressing TFEB, which in turn inhibits myelination in the CNS.
Levels of cytoplasmic TFEB increase in remyelinating lesions in mouse
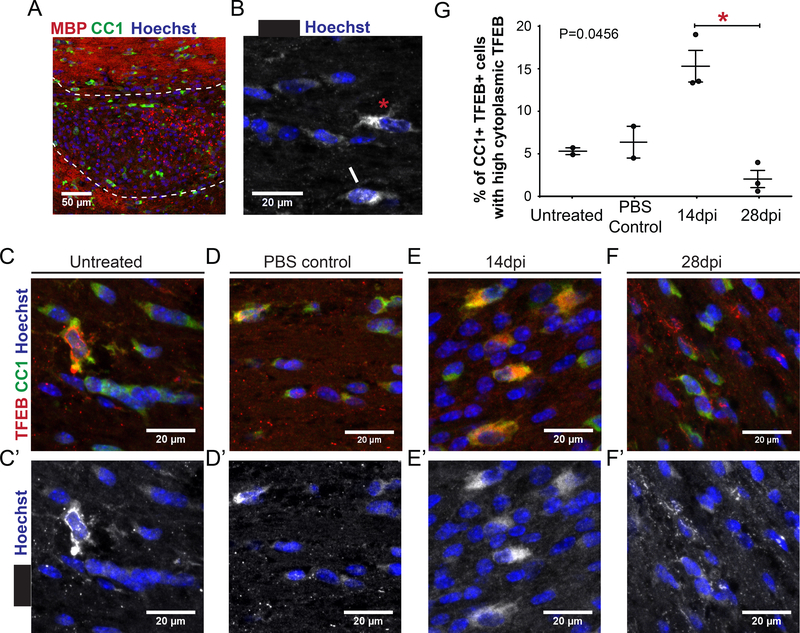
Our data indicate that Tfeb represses myelination during development. We next explored whether TFEB is altered after injury during the process of remyelination. We used an adult mouse model of demyelination and remyelination, where focal demyelination is induced by stereotactic injection of the demyelinating agent L-α-lysophosphatidylcholine (LPC) into the corpus callosum (Boyd et al., 2013) (Figure 5A). Remyelination relies on the recruitment of oligodendrocyte precursor cells (OPCs) to demyelinated lesions and their subsequent differentiation into myelin-forming oligodendrocytes. In this model, oligodendrocyte differentiation begins at 10 days post injection (dpi), early remyelination is present at 14 days dpi and is complete by 28 days dpi (Boyd et al., 2013). Immunohistochemical analysis of control adult mouse corpus callosum (either uninjected or injected with PBS) showed that the majority of CC1+ oligodendrocytes have low levels of cytoplasmic TFEB expression, but there were a small number of oligodendrocytes with high TFEB expression (Figure 5B, C, D). TFEB expression was almost exclusively localized to the cytoplasm of CC1+ oligodendrocytes, with nuclear TFEB localization rarely observed (approximately 1/cell per optical frame) (Figure 5B, C, D). During the early stages of remyelination (14 dpi), we observed a significant increase in the number of oligodendrocytes with high cytoplasmic TFEB expression in the lesion and perilesion areas (Figure 5E and E’). The number of oligodendrocytes with high cytoplasmic expression of TFEB dropped significantly at the late remyelination stage (28 dpi), when the lesion is mostly remyelinated (Figure 5F and F’).
Figure 5. Remyelinating lesions in mouse show increased levels of cytoplasmic TFEB.
(A) Low power image of LPC injected corpus callosum, 14 dpi stained for MBP (red), CC1 (green) and Hoechst (blue). The area of the lesion is outlined with dashed white line, and the perilesion area is marked. (B) TFEB is mostly found in the cytoplasm in mature oligodendrocytes of the rodent corpus callosum, both in those expressing low levels (white asterisk) and high levels (red asterisk). Rarely both cytoplasmic and nuclear TFEB expression is detected (white arrow). (C) A few mature, CC1 (green) expressing oligodendrocytes of the adult, normal, rodent corpus callosum show high level expression of TFEB (red) in the cytoplasm. Most express low levels of TFEB in the cytoplasm (C’, TFEB white). (D) PBSinjected control corpus callosum 14 dpi. CC1+ oligodendrocytes display a similar TFEB expression profile to untreated controls (C,C’) (E) LPC-injected tissue 14 dpi (early remyelination time point). An increased number of CC1+ oligodendrocytes with high cytoplasmic TFEB expression appear in the corpus callosum (E,E’). (F) LPC-injected tissue 28 dpi (late remyelination time point). Very few CC1+ oligodendrocytes have high cytoplasmic TFEB expression (F,F’). (G) Quantification of the percentage of CC1+TFEB+ cells with high cytoplasmic TFEB levels in the corpus callosum after LPC focal lesion induction. Graph depicts average value and SEM. Individuals values are also represented. A marked increase is observed during early remyelination in lesion and perilesion areas (14 dpi) that significantly drops at later remyelination stages. Statistical analysis: non parametric ANOVA (Kruskal-Wallis test) with Dunn’s post hoc test. Total P value 0.0456 and significance between 14dpi and 28 dpi.
Discussion
TFEB represses myelination
Our experiments demonstrate that TFEB inhibits myelination. TFEB loss-of-function mutations cause ectopic myelination in the dorsal spinal cord and ectopic expression of Myelin Basic Protein in the hindbrain. Conversely, rraga−/− mutants have increased TFEB activity, which in turn blocks myelination. Furthermore, expression of constitutively active TFEB also inhibits mbp expression. Consistent with previous biochemical studies (Martina and Puertollano, 2013), our analyses showed that RagA and TFEB have opposing functions in CNS myelination in vivo. In striking contrast to rraga−/− single mutants, rraga−/−; tfeb−/− double mutants have apparently normal levels of mbp expression. Expression of mbp is also partially rescued in rraga−/−; tfeb+/− mutants, indicating that myelination is very sensitive to the level of tfeb function. Thus, the analysis of both loss and gain of TFEB activity defines TFEB as a repressor of myelination.
Oligodendrocytes are present in rraga−/− mutants and are competent to myelinate if tfeb activity is partially (haploinsufficiency) or fully eliminated, indicating that TFEB blocks mbp expression in pre-myelinating oligodendrocytes. Previous studies have defined a brief developmental window (~5 hours in zebrafish) during which an oligodendrocyte can initiate new myelin segments (Czopka et al., 2013). After this critical period, new myelin segments are not initiated, although previously established segments are maintained and expanded. It is possible that TFEB closes the critical period, such that myelination is greatly curtailed by hyperactive TFEB in rraga−/− mutants.
There are many possible mechanisms by which TFEB might repress myelination. TFEB is a transcriptional regulatory protein, raising the possibility that TFEB might directly repress myelin gene expression in oligodendrocytes. There are, however, no reported TFEB binding sites in the vicinity of the myelin basic protein gene, suggesting that TFEB represses myelination indirectly. In other cell types, TFEB activates hundreds of target genes that regulate diverse lysosome-related processes, including lysosome biogenesis and exocytosis, autophagy, and lipid catabolism (Napolitano and Ballabio, 2016). Our analysis indicates that TFEB activates at least some of these previously known target genes in oligodendrocytes as well. It is likely that inappropriately increased activity of one or more TFEB-regulated processes disrupts myelination in oligodendrocytes. For example, TFEB hyperactivity might disrupt the trafficking of endolysosomal organelles to the membrane or synthesis of lipids critical for the formation of the membranous myelin sheath, which could in turn impede myelination.
RagA and other regulators of TFEB in myelination
Our mutational analysis demonstrates that the lysosomal genes rraga and lamtor4 are essential for myelination in the CNS, but not the PNS. RagA and Lamtor4 are components of the RagRagulator complex, which recruits TFEB to the lysosome, where it is phosphorylated and inactivated (Efeyan et al., 2015). mTOR, which is recruited to the lysosome by RagAdependent and -independent mechanisms, is one kinase that can phosphorylate and inactivate TFEB (Martina et al., 2012; Roczniak-Ferguson et al., 2012; Settembre et al., 2011, 2012). If mTOR inactivated TFEB in the context of myelination, we would expect mtor−/− mutants to have hyperactive TFEB and a phenotype similar to rraga−/− mutants. Our data, however, suggest that mTOR is not the only kinase that can repress TFEB activity in oligodendrocytes, because mtor−/− mutants have much more mbp expression in oligodendrocytes than rraga−/− mutants. Previous studies show that the kinases GSKβ3, ERK2, AKT, and PKCβ can also phosphorylate TFEB, and some combination of these likely controls TFEB activity during myelination (Li et al., 2016; Martina and Puertollano, 2013; Medina et al., 2015; Palmieri et al., 2017; Puertollano et al., 2018; Roczniak-Ferguson et al., 2012; Settembre et al., 2011).
TFEB during remyelination
Our analysis indicates that TFEB represses myelination during development, raising the question of its role during remyelination. In control mouse white matter, TFEB is mainly detected at low levels in the cytoplasm of mature oligodendrocytes and is rarely localized in the nucleus. TFEB protein levels increase in the cytoplasm of mature oligodendrocytes during remyelination, suggesting that it is predominantly inactive and thus permitting the remyelination process. After remyelination is complete, TFEB expression declined to near normal levels. It may seem somewhat paradoxical that levels of TFEB - a repressor of myelination - increase in remyelinating cells. One possibility is that TFEB, as a component of the integrated stress response (Martina et al., 2016), is more highly expressed due to the stress of the injury, and is subsequently inactivated by cytoplasmic localization when remyelination commences. RagA and kinases that phosphorylate TFEB (GSKβ3, ERK2, AKT, PKCβ, mTORC) may be essential to retain TFEB in the cytoplasm during remyelination. Animals with increased TFEB function, e.g rraga−/− mutants or Tfeb phosphorylation-null transgenic animals, will be valuable models in which to better characterize the role of TFEB and its regulators in the context of remyelination. Understanding the nature of the signals that regulate TFEB expression and localization in oligodendrocytes of the healthy and diseased brain will be critical to understand its function in oligodendrocyte development, myelination, and remyelination. Elucidating the target genes and processes downstream of TFEB that are inhibitory to myelination may define additional regulators of myelination and myelin repair, which in turn may suggest strategies for therapeutic remyelination.
STAR Methods
Contact for reagent and resource sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, William S Talbot (william.talbot@stanford.edu).
Experimental model and subject details
Zebrafish
Zebrafish embryos, larvae and adults were produced, grown and maintained according to standard protocols approved by the Stanford University Institutional Animal Care and Use Committee. Ethical approval was obtained from the Stanford University Institutional Animal Care and Use Committee. To obtain embryos and larvae used in experiments, adults 3–18 months were crossed. Adult density was maintained at 5–10 fish/L, with a 14hr light/ 10nr dark cycle and fish were fed twice daily. Water temperature was maintained at 28ºC. Embryos and larvae were treated with 0.003% 1-phenyl-2-thiourea (PTU) to inhibit pigmentation, and they were anesthetized with 0.016% (w/v) Tricaine prior to experimental procedures. Published strains used in this study include: wildtype TL, mtorxu015 (Ding et al., 2011), Tg(olig2:GFP) (Shin et al., 2003), Tg(cldnk:GFP) (Münzel et al., 2012), rragast77 and lamtor4 st99/st99 (Shen et al., 2016). Details of the construction of the new strains generated in this study are described below.
Mouse
Mice were housed and used according to standard UK Home Office regulations, under project license 60/ 4524 awarded to AW. 12–14 week old C57BL/6 male mice were used for all experiments. Experimental details are described below.
Method details
In situ hybridization
In situ hybridization on embryos and larvae was performed using standard methods (Thisse et al., 2004). Briefly, embryos were fixed overnight in 4% paraformaldehyde, dehydrated for at least 2 hr in 100% methanol, rehydrated in PBS, permeabilized with proteinase K, and incubated overnight with antisense riboprobes at 65°C. The probe was detected with an antidigoxigenin antibody conjugated to alkaline phosphatase (11093274910, Sigma-Aldrich). Images were captured using the Zeiss AxioCam HRc camera with the AxioVision software. Probes for mbp and olig2 were previously described (Lyons et al., 2009; Park et al., 2002). A 700 bp fragment of plp1b gene was amplified from 5 dpf whole larvae cDNA using the following primers: 5’- AACAGCCGTGGTTGTGTAAAGC-3’ and 5’-TATGTTGGCCAGTGCATTTCCAC-3’. The resulting fragment was cloned into the pCRII vector using Dual promoter TA cloning Kit (K207020, Invitrogen) to generate a riboprobe for plp1b. The construct was linearized with SpeI and antisense probe was transcribed with T7 RNA polymerase (AM2085, Ambion). For each genetic cross, at least 3 independent clutches were analyzed. For each genotype a minimum of 10 total fish were analyzed.
Whole-mount immunofluorescence
For MBP immunofluorescence 8 dpf zebrafish larvae were fixed in 4%PFA/PBS, overnight at 4ºC. Larval brains were dissected and incubated with rabbit anti-MBP (Lyons et al., 2005). Anti- rabbit Alexa Fluor568 was used at 1/1000 (A-11011, ThermoFisher). Embryos were mounted in DAPI Fluoromount-G (SouthernBiotech). Transgenic zebrafish embryos were mounted in 1.5% low melting point agarose in distilled water. Images were captured using a Zeiss LSM 5 Pascal or Zeiss LSM 7 microscope. Objectives used were Plan-Neofluar 10× (numerical aperture 0.30) and 20× (numerical aperture 0.75). All fish were genotyped after imaging and analysis.
Crispr/Cas9 targeting
sgRNAs were designed using CHOPCHOP (https://chopchop.rc.fas.harvard.edu/, (Labun et al., 2016; Montague et al., 2014), transcribed with T7 polymerase (E2040S, New England Biolabs) and purified using mirVana miRNA isolation kit (AM1560, Ambion). Cas9 protein (Macrolab, Berkeley, http://qb3.berkeley.edu/macrolab/cas9-nls-purified-protein/) was injected together with 300 ng sgRNA into 1-cell stage embryos, and the embryos were genotyped after analysis to detect lesions. sgRNAs used were: TFEB (5’-GGTGCACTGATGGCTGGCGT-3’).
tfebst120 and tfebst121 mutant alleles
tfeb mutants were generated by raising embryos injected with sgRNA 5’-GGTGCACTGATGGCTGGCGT-3’. Two mutants were isolated and tested. tfebst120 contains a 5 bp deletion which results in a change in reading frame after amino acid 45 and a premature stop codon after amino acid 61 compared to 491 amino acids for the WT protein. tfebst121 contains a 8 bp deletion which results in a change in reading frame after amino acid 46 and a premature stop codon after amino acid 60. tfeb mutants were genotyped using the primers 5′- GCTCATGCGGGACCAAATGC −3′ and 5′- GGTCACACTAACAAATGTGG −3′. PCR products were digested with Cac8I (R0579, New England Biolabs). Mutant (233+83 bp) and WT (213bp + 83 +25 bp) bands were distinguished by running the digested PCR product on a 3% agarose gel. tfebst120/st120, tfebst121/st121 and tfebst120/st121 transheterozygotes all displayed equal phenotypes. All experiments were performed in transheterozygous fish.
FAC sorting and RNA sequencing
WT and rraga−/− mutant Tg(cldnk:GFP) larvae at 5 dpf were distinguished by neutral red staining and pooled for RNA isolation (Shen et al., 2016), in two independent experiments. Dissociation of embryos was carried out as previously described (Manoli and Driever, 2012) and GFPpositive cells were isolated at the Stanford Shared FACS Facility. RNA was isolated using the Qiagen RNeasy micro kit (74004, Qiagen) and provided to the Stanford Functional Genomics Facility. Libraries were prepared and sequenced using the Illumina NextSeq Mid platform.
qRT-PCR
Total RNA was extracted from larvae at 5 dpf with the RNAeasy kit (QIAGEN). cDNA was synthesized using iScript supermix (Biorad). qPCR was performed with SsoAdvancedTM Universal SYBR Green Supermix (Bio-Rad) on the Bio-Rad CFX384 Real-Time PCR Detection System. All experiments were done in biological and technical triplicates. Transcript levels were normalized to ef1-alpha (primers were previously reported (Shen et al., 2016; Shiau et al., 2013). Relative mRNA levels were calculated using ΔΔCT.
RNA Seq analysis
Over 7.5M paired end reads were obtained for each sample. These reads were mapped to the zebrafish reference genome (NCBI) using CLC Genomics Workbench 11.0 (http://www.clcbio.com/products/clc-genomics-workbench/) and over 90% of the sequenced fragments mapped to annotated genes. Differential expression analysis was performed by comparing the mapped WT and rraga−/− mutant reads, while controlling for the two independent experiments. Annotation was performed using the Danio rerio annotation file downloaded from Gene Ontology Consortium (http://www.geneontology.org/page/download-annotations). Raw and analyzed data were deposited with GEO, GSE119332.
Gene Ontology Enrichment
The list of 343 genes upregulated (p<0.01, >2,5X) in myelinating glia of rraga−/− was queried in PANTHER11 using gene ontology term cellular component (Mi et al., 2013, 2017).
Identification of Tfeb targets in genes upregulated in rraga−/− mutants
Significantly upregulated (p<0.01, >2.5X) and downregulated genes (p<0.01, <2.5X) in rraga−/− mutants were mapped to human orthologues using the Ensemble Biomart feature and compared to the list of TFEB targets (471 genes, (Palmieri et al., 2011)). 15 zebrafish genes orthologous to human Tfeb target genes were identified in the upregulated dataset, and 4 were identified in the downregulated dataset. Manual curation identified two additional upregulated genes with human TFEB target orthologues. Processing and analysis of gene lists was performed using Python.
Expression constructs and transient injections
The coding sequence of rraga was cloned from 5 dpf embryonic cDNA pool and directionally inserted into pCR8/GW/TOPO vector (Invitrogen). Synthetic DNA encoding sequence of Tfeb (ENSDART00000182200.1) and Tfeb phosphorylation-null mutant was obtained from Integrated DNA Technologies (IDT) and subcloned into pCR8/GW by Gibson assembly (New England Biolabs). For transient transgenesis, rraga, tfeb and tfeb phospho-null sequences were subcloned into pDestTol2CG2 by multisite gateway. The tissue-specific regulatory sequences used have been reported elsewhere (Shen et al., 2016; Shiau et al., 2013). Tissue-specific transgenes were transiently expressed by co-injecting 12–25 pg of Tol2 plasmids described above and 100–200 pg of Tol2 transposase mRNA in 1 cell embryos.
Transmission Electron Microscopy
TEM was performed as described previously (Lyons et al., 2008). Briefly, decapitated embryos were fixed in 2% glutaraldehyde and 4% paraformaldehyde in 0.1 M sodium cacodylate buffer (pH 7.4). The posterior portion of the larvae was used to isolate DNA for genotyping. For secondary fixation, samples were fixed in 2% osmium tetraoxide, 0.1 M imidazole in 0.1 M sodium cacodylate (pH7.4), stained with saturated uranyl acetate, and dehydrated in ethanol and acetone. Fixation and dehydration were accelerated using the PELCO 3470 Multirange Laboratory Microwave System (Pelco) at 15°C. Sample s were then incubated in 50% Epon/50% acetone overnight, followed by 100% Epon for 4 hr at room temperature. Samples were then embedded in 100% Epon and baked for 48 hr at 60°C. Blocks were sectioned using a Leica Ultramicrotome. Thick sections (500–1,000 μm) for toluidine blue staining were collected on glass slides, stained at 60°C for 5 s, and imaged with the Leica DM 2000 microscope using the Leica DFC290 HD camera and Leica Application Suite software. After the desired region of the spinal cord was reached, we collected ultrathin sections for TEM analysis on copper grids, and we stained them with uranyl acetate and Sato’s lead stain (1% lead citrate, 1% lead acetate, and 1% lead nitrate). Sections were imaged on a JEOL JEM-1400 transmission electron microscope.
Drug Treatment
For Torin1 treatment, WT fish were incubated in 1 μM Torin1 (4247, Tocris Bioscience) in embryo water with PTU from 24 hpf to 5 dpf; fresh embryo medium, PTU and Torin1 were replaced daily.
LPC-induced demyelinating lesions in rodents
Focal demyelination was induced via stereotactic injection of L-α-Lysophosphatidylcholine (LPC, L4129, Sigma-Aldrich) into the corpus callosum of 12–14 week old C57BL/6 male mice (n=2–3 per time point). Anesthetized animals received 2μL of 1% LPC through a burr hole in the skull and at stereotactic coordinates 1.2 mm posterior, 0.5 mm lateral, 1.4 mm deep to the bregma over 4 minutes using a 30 gauge needle attached to a Hamilton syringe and driven by a Nano pump (KD Scientific Inc., Holliston, MA). The needle was additionally left on site for additional 4 minutes to avoid backflow. Similar stereotactic injection of PBS was followed to create a surgical control (PBS control). LPC causes reversible focal demyelination without axonal loss. The time course of de- and remyelination is well established and reproducible with demyelinated lesions appearing as early as 3 days post injection (3dpi). Two time points with early (14 dpi) and late (28 dpi) remyelination were analyzed. Mice were subsequently perfused with 4% PFA, the brain tissue was harvested, cryoprotected in 30% sucrose and frozen embedded in OCT compound.
Immunohistochemistry of mouse tissue and quantification.
10 μm thick cryosections were briefly washed in PBS and microwaved for 10 minutes in Vector Unmasking Solution for antigen retrieval (H-3300, Vector) before blocking with 10% normal horse serum, 0.3% Triton-X in 1×PBS for 1 hr at RT. Sections were then incubated with primary antibodies in the same solution overnight at 4oC in a humidified chamber. Following washes in PBS, the cryosections were incubated with Alexa Fluor secondary antibodies (Thermo Fischer Scientific, 1:1000) for 1 ½ hrs at room temperature and counterstained with Hoechst for the visualization of the nuclei. Primary antibodies used: rabbit polyclonal anti-TFEB (Bethyl laboratories inc, A303–673A, 1:1000), mouse IgG anti-APC (CC1, OP80, Merck-Millipore, 1:500) and rat IgG2a anti-Myelin Basic Protein (clone 12, AbD Serotec, 1:300). All slides were mounted using mowiol mounting medium (475904, MERCK- Millipore) and imaged using Leica TCS SP8 confocal microscopy. From each section 3–5 random fields (145.31μm x 145.31μm each) were obtained. 2 sections were analyzed per animal. Total number of CC1/TFEB double positive oligodendrocytes was counted in lesion and perilesion areas (CC1+TFEB+/mm2). For the same area, the number of CC1/TFEB double positive cells with high cytoplasmic TFEB levels was also counted (CC1+ TFEB+ cytoplasmic high/mm2). Values for each sample were averaged, and data are depicted as mean percentage of CC1+ TFEB+ cytoplasmic high/CC1+TFEB+ cells.
Quantification and Statistical analysis
Sample sizes were chosen based on previous publications and are indicated in each figure and/or figure legend. No animal or sample was excluded from the analysis unless the animal died during the procedure. Except for electron microscopy samples, zebrafish larvae were only genotyped after image acquisition and analysis. EM samples were blinded before counts of myelinated axons numbers were performed. The experiments were not randomized. Sample sizes, statistical test and P values are indicated in the figures or figure legends. Statistical significance was assigned at P < 0.05. Statistical tests were performed using GraphPad Prism 6 or Prism 7 software.
Supplementary Material
Table S1. Related to Figure 3. Genes significantly upregulated in FAC-sorted cldnk:GFP positive cells of rraga−/− mutants
Table S2. Related to Figure 3. Genes significantly downregulated in FAC-sorted cldnk:GFP positive cells of rraga−/− mutants
RagA is required autonomously in oligodendrocytes for CNS myelination in zebrafish.
Inactivation of TFEB rescues the myelin defect in RagA mutants.
Tfeb mutants exhibit ectopic myelination, showing that TFEB represses myelination.
During remyelination in mammals, TFEB localizes to the cytoplasm, where it is inactive.
Acknowledgements
We thank Talbot laboratory members for helpful discussions and technical advice and Tuky K. Reyes and Chenelle Hill for fish maintenance. Transmission electron microscopy was performed with assistance from John J. Perrino in the Stanford Cell Sciences Imaging Facility; this work was supported in part by ARRA Award number 1S10RR026780–01 from the National Center for Research Resources (NCRR). Cell sorting for this project was performed with assistance from Brandon J. Carter and Qianyi Lee, using instruments in the Stanford Shared FACS Facility. RNA sequencing was performed with assistance from the Stanford Functional Genomics Facility. Mouse tissue was kindly generated by Amanda Boyd. K.S was supported by a fellowship from A*STAR Singapore, and E.L.B. was supported by a fellowship from the National Science Foundation. W.S.T. is a Catherine R. Kennedy and Daniel L. Grossman Fellow in Human Biology. LZ and AW are supported by a MS Society UK Centre Grant. This work was supported by NIH grant R01NS050223 and NMSS grant RG-1707–28694 to W.S.T.
Footnotes
Declaration of Interests
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Almeida RG, and Lyons DA (2017). On Myelinated Axon Plasticity and Neuronal Circuit Formation and Function. J. Neurosci 37, 10023–10034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appelqvist H, Wäster P, Kågedal K, and Öllinger K (2013). The lysosome: from waste bag to potential therapeutic target. J. Mol. Cell Biol 5, 214–226. [DOI] [PubMed] [Google Scholar]
- Bar-Peled L, Schweitzer LD, Zoncu R, and Sabatini DM (2012). Ragulator Is a GEF for the Rag GTPases that Signal Amino Acid Levels to mTORC1. Cell 150, 1196–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtsson SL, Nagy Z, Skare S, Forsman L, Forssberg H, and Ullén F (2005). Extensive piano practicing has regionally specific effects on white matter development. Nat. Neurosci. 8, 1148–1150. [DOI] [PubMed] [Google Scholar]
- Boyd A, Zhang H, and Williams A (2013). Insufficient OPC migration into demyelinated lesions is a cause of poor remyelination in MS and mouse models. Acta Neuropathol. (Berl.) 125, 841–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne P, Chandraratna D, Angood C, Tremlett H, Baker C, Taylor BV, and Thompson AJ (2014). Atlas of Multiple Sclerosis 2013: A growing global problem with widespread inequity. Neurology 83, 1022–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czopka T, ffrench-Constant C, and Lyons DA (2013). Individual Oligodendrocytes Have Only a Few Hours in which to Generate New Myelin Sheaths In Vivo. Dev. Cell 25, 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Sun X, Huang W, Hoage T, Redfield M, Kushwaha S, Sivasubbu S, Lin X, Ekker S, and Xu X (2011). Haploinsufficiency of Target of Rapamycin Attenuates Cardiomyopathies in Adult ZebrafishNovelty and Significance. Circ. Res 109, 658–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta R, and Trapp BD (2011). Mechanisms of neuronal dysfunction and degeneration in multiple sclerosis. Prog. Neurobiol 93, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efeyan A, Comb WC, and Sabatini DM (2015). Nutrient-sensing mechanisms and pathways. Nature 517, 302–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery B (2010). Regulation of oligodendrocyte differentiation and myelination. Science 330, 779–782. [DOI] [PubMed] [Google Scholar]
- Ferguson SM (2015). Beyond indigestion: emerging roles for lysosome-based signaling in human disease. Curr. Opin. Cell Biol 35, 59–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin RJM, and Ffrench-Constant C (2008). Remyelination in the CNS: from biology to therapy. Nat. Rev. Neurosci 9, 839–855. [DOI] [PubMed] [Google Scholar]
- Funfschilling U, Supplie LM, Mahad D, Boretius S, Saab AS, Edgar J, Brinkmann BG, Kassmann CM, Tzvetanova ID, Mobius W, et al. (2012). Glycolytic oligodendrocytes maintain myelin and long-term axonal integrity. Nature 485, 517–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Goraksha-Hicks P, Li L, Neufeld TP, and Guan K-L (2008). Regulation of TORC1 by Rag GTPases in nutrient response. Nat. Cell Biol 10, 935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YC, Park HW, Sciarretta S, Mo J-S, Jewell JL, Russell RC, Wu X, Sadoshima J, and Guan K-L (2014). Rag GTPases are cardioprotective by regulating lysosomal function. Nat. Commun 5, 4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labun K, Montague TG, Gagnon JA, Thyme SB, and Valen E (2016). CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res. 44, W272–W276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Xu M, Ding X, Yan C, Song Z, Chen L, Huang X, Wang X, Jian Y, Tang G, et al. (2016). Protein kinase C controls lysosome biogenesis independently of mTORC1. Nat. Cell Biol 18, 1065–1077. [DOI] [PubMed] [Google Scholar]
- Lister JA, Lane BM, Nguyen A, and Lunney K (2011). Embryonic expression of zebrafish MiT family genes tfe3b, tfeb, and tfec. Dev. Dyn. Off. Publ. Am. Assoc. Anat 240, 2529–2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons DA, Pogoda H-M, Voas MG, Woods IG, Diamond B, Nix R, Arana N, Jacobs J, and Talbot WS (2005). erbb3 and erbb2 are essential for schwann cell migration and myelination in zebrafish. Curr. Biol. CB 15, 513–524. [DOI] [PubMed] [Google Scholar]
- Lyons DA, Naylor SG, Mercurio S, Dominguez C, and Talbot WS (2008). KBP is essential for axonal structure, outgrowth and maintenance in zebrafish, providing insight into the cellular basis of Goldberg-Shprintzen syndrome. Development 135, 599–608. [DOI] [PubMed] [Google Scholar]
- Lyons DA, Naylor SG, Scholze A, and Talbot WS (2009). Kif1b is essential for mRNA localization in oligodendrocytes and development of myelinated axons. Nat. Genet. 41, 854–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoli M, and Driever W (2012). Fluorescence-Activated Cell Sorting (FACS) of Fluorescently Tagged Cells from Zebrafish Larvae for RNA Isolation. Cold Spring Harb. Protoc 2012, pdb.prot069633. [DOI] [PubMed] [Google Scholar]
- Marques S, Zeisel A, Codeluppi S, van Bruggen D, Mendanha Falcão A, Xiao L, Li H, Häring M, Hochgerner H, Romanov RA, et al. (2016). Oligodendrocyte heterogeneity in the mouse juvenile and adult central nervous system. Science 352, 1326–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina JA, and Puertollano R (2013). Rag GTPases mediate amino acid-dependent recruitment of TFEB and MITF to lysosomes. J. Cell Biol 200, 475–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina JA, Chen Y, Gucek M, and Puertollano R (2012). MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy 8, 903–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina JA, Diab HI, Brady OA, and Puertollano R (2016). TFEB and TFE3 are novel components of the integrated stress response. EMBO J. 35, 479–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martini-Stoica H, Xu Y, Ballabio A, and Zheng H (2016). The Autophagy–Lysosomal Pathway in Neurodegeneration: A TFEB Perspective. Trends Neurosci. 39, 221–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie IA, Ohayon D, Li H, Faria J.P. de, Emery B, Tohyama K, and Richardson WD (2014). Motor skill learning requires active central myelination. Science 346, 318–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina DL, Fraldi A, Bouche V, Annunziata F, Mansueto G, Spampanato C, Puri C, Pignata A, Martina JA, Sardiello M, et al. (2011). Transcriptional Activation of Lysosomal Exocytosis Promotes Cellular Clearance. Dev. Cell 21, 421–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina DL, Di Paola S, Peluso I, Armani A, De Stefani D, Venditti R, Montefusco S, Scotto-Rosato A, Prezioso C, Forrester A, et al. (2015). Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat. Cell Biol 17, 288–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Casagrande JT, and Thomas PD (2013). Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc 8, 1551–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Huang X, Muruganujan A, Tang H, Mills C, Kang D, and Thomas PD (2017). PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 45, D183–D189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montague TG, Cruz JM, Gagnon JA, Church GM, and Valen E (2014). CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res. 42, W401–W407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münzel EJ, Schaefer K, Obirei B, Kremmer E, Burton EA, Kuscha V, Becker CG, Brösamle C, Williams A, and Becker T (2012). Claudin k is specifically expressed in cells that form myelin during development of the nervous system and regeneration of the optic nerve in adult zebrafish. Glia 60, 253–270. [DOI] [PubMed] [Google Scholar]
- Münzel EJ, Jolanda Münzel E, and Williams A (2013). Promoting remyelination in multiple sclerosis-recent advances. Drugs 73, 2017–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napolitano G, and Ballabio A (2016). TFEB at a glance. J. Cell Sci 129, 2475–2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nezich CL, Wang C, Fogel AI, and Youle RJ (2015). MiT/TFE transcription factors are activated during mitophagy downstream of Parkin and Atg5. J. Cell Biol 210, 435–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri M, Impey S, Kang H, di Ronza A, Pelz C, Sardiello M, and Ballabio A (2011). Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum. Mol. Genet 20, 3852–3866. [DOI] [PubMed] [Google Scholar]
- Palmieri M, Pal R, Nelvagal HR, Lotfi P, Stinnett GR, Seymour ML, Chaudhury A, Bajaj L, Bondar VV, Bremner L, et al. (2017). mTORC1-independent TFEB activation via Akt inhibition promotes cellular clearance in neurodegenerative storage diseases. Nat. Commun 8, 14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H-C, Mehta A, Richardson JS, and Appel B (2002). olig2 Is Required for Zebrafish Primary Motor Neuron and Oligodendrocyte Development. Dev. Biol 248, 356–368. [DOI] [PubMed] [Google Scholar]
- Puertollano R, Ferguson SM, Brugarolas J, and Ballabio A (2018). The complex relationship between TFEB transcription factor phosphorylation and subcellular localization. EMBO J. 37, e98804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravanelli AM, and Appel B (2015). Motor neurons and oligodendrocytes arise from distinct cell lineages by progenitor recruitment. Genes Dev. 29, 2504–2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roczniak-Ferguson A, Petit CS, Froehlich F, Qian S, Ky J, Angarola B, Walther TC, and Ferguson SM (2012). The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci. Signal 5, ra42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saftig P, and Haas A (2016). Turn up the lysosome. Nat. Cell Biol 18, 1025–1027. [DOI] [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, and Sabatini DM (2008). The Rag GTPases Bind Raptor and Mediate Amino Acid Signaling to mTORC1. Science 320, 1496–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, and Sabatini DM (2010). Ragulator-Rag Complex Targets mTORC1 to the Lysosomal Surface and Is Necessary for Its Activation by Amino Acids. Cell 141, 290–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardiello M (2016). Transcription factor EB: from master coordinator of lysosomal pathways to candidate therapeutic target in degenerative storage diseases. Ann. N. Y. Acad. Sci. 1371, 3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardiello M, Palmieri M, Ronza A.di, Medina DL, Valenza M, Gennarino VA, Malta CD, Donaudy F, Embrione V, Polishchuk RS, et al. (2009). A Gene Network Regulating Lysosomal Biogenesis and Function. Science 325, 473–477. [DOI] [PubMed] [Google Scholar]
- Settembre C, and Ballabio A (2014). Lysosomal adaptation: how the lysosome responds to external cues. Cold Spring Harb. Perspect. Biol 6, a016907–a016907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, et al. (2011). TFEB links autophagy to lysosomal biogenesis. Science 332, 1429–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S, Huynh T, Ferron M, Karsenty G, Vellard MC, et al. (2012). A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 31, 1095–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, Fraldi A, Medina DL, and Ballabio A (2013). Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat. Rev. Mol. Cell Biol 14, 283–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw RJ (2008). mTOR signaling: RAG GTPases transmit the amino acid signal. Trends Biochem. Sci 33, 565–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen K, Sidik H, and Talbot WS (2016). The Rag-Ragulator Complex Regulates Lysosome Function and Phagocytic Flux in Microglia. Cell Rep. 14, 547–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman DL, and Brophy PJ (2005). Mechanisms of axon ensheathment and myelin growth. Nat. Rev. Neurosci 6, 683–690. [DOI] [PubMed] [Google Scholar]
- Shiau CE, Monk KR, Joo W, and Talbot WS (2013). An anti-inflammatory NOD-like receptor is required for microglia development. Cell Rep. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Park H-C, Topczewska JM, Mawdsley DJ, and Appel B (2003). Neural cell fate analysis in zebrafish using <Emphasis Type=“Italic”>olig2</Emphasis> BAC transgenics. Methods Cell Sci. 25, 7–14. [DOI] [PubMed] [Google Scholar]
- Simons M, and Nave K-A (2016). Oligodendrocytes: Myelination and Axonal Support. Cold Spring Harb. Perspect. Biol 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse B, Heyer V, Lux A, Alunni V, Degrave A, Seiliez I, Kirchner J, Parkhill J-P, and Thisse C (2004). Spatial and temporal expression of the zebrafish genome by large-scale in situ hybridization screening. Methods Cell Biol. 77, 505–519. [DOI] [PubMed] [Google Scholar]
- Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, and Sabatini DM (2012). A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 485, 109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams A, Sarkar S, Cuddon P, Ttofi EK, Saiki S, Siddiqi FH, Jahreiss L, Fleming A, Pask D, Goldsmith P, et al. (2008). Novel targets for Huntington’s disease in an mTORindependent autophagy pathway. Nat. Chem. Biol 4, 295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, and Anderson DJ (2002). The bHLH Transcription Factors OLIG2 and OLIG1 Couple Neuronal and Glial Subtype Specification. Cell 109, 61–73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Related to Figure 3. Genes significantly upregulated in FAC-sorted cldnk:GFP positive cells of rraga−/− mutants
Table S2. Related to Figure 3. Genes significantly downregulated in FAC-sorted cldnk:GFP positive cells of rraga−/− mutants