Abstract
Most genetic transformation protocols for the model diatom Phaeodactylum tricornutum rely on one of two available antibiotics as selection markers: Zeocin (a formulation of phleomycin D1) or nourseothricin. This limits the number of possible consecutive genetic transformations that can be performed. In order to expand the biotechnological possibilities for P. tricornutum, we searched for additional antibiotics and corresponding resistance genes that might be suitable for use with this diatom. Among the three different antibiotics tested in this study, blasticidin-S and tunicamycin turned out to be lethal to wild-type cells at low concentrations, while voriconazole had no detectable effect on P. tricornutum. Testing the respective resistance genes, we found that the blasticidin-S deaminase gene (bsr) effectively conferred resistance against blasticidin-S to P. tricornutum. Furthermore, we could show that expression of bsr did not lead to cross-resistances against Zeocin or nourseothricin, and that genetically transformed cell lines with resistance against Zeocin or nourseothricin were not resistant against blasticidin-S. In a proof of concept, we also successfully generated double resistant (against blasticidin-S and nourseothricin) P. tricornutum cell lines by co-delivering the bsr vector with a vector conferring nourseothricin resistance to wild-type cells.
Keywords: Phaeodactylum tricornutum, Voriconazole, Genetic transformation, Genome editing, Tunicamycin, Resistance gene, Blasticidin-S, Selection marker, Antibiotics
Introduction
Phaeodactylum tricornutum is one of the most widely used models in diatom research. The genome of P. tricornutum is completely sequenced (Bowler et al., 2008) and different methods for the introduction of transgenes are available, for example, particle bombardment (Apt, Grossman & Kroth-Pancic, 1996), conjugation with bacteria (Karas et al., 2015), or electroporation (Miyahara et al., 2013; Niu et al., 2012; Zhang & Hu, 2014). In addition, genome editing methods like TALEN or CRISPR have been introduced recently, allowing the direct knockout of specific genes (Daboussi et al., 2014; Gruber & Kroth, 2017; Nymark et al., 2016; Serif et al., 2017; Slattery et al., 2018). Despite these important achievements, the availability of antibiotics and the number of selection markers for diatoms is very low (compiled in Huang & Daboussi (2017)). Essentially two antibiotics are widely used for the selection of transformed diatom cells, both of which were introduced more than two decades ago: phleomycin (e.g., Zeocin/phleomycin D1), together with the respective ShBle resistance gene, as well as nourseothricin and the nat/sat-1 genes (Apt, Grossman & Kroth-Pancic, 1996; Falciatore et al., 1999; Huang & Daboussi, 2017; Sakaguchi, Nakajima & Matsuda, 2011; Zaslavskaia et al., 2000). There are two more pairs of antibiotics and resistance genes that have been reported, but that are not widely used: The antibiotic G418/neomycin and the resistance gene nptII have been used by Zaslavskaia et al. (2000), however, the antibiotic is only effective at low salt concentrations (Apt, Grossman & Kroth-Pancic, 1996; Zaslavskaia et al., 2000), and strains expressing nptII reach only limited resistance to G418 (Zaslavskaia et al., 2000). Furthermore, the chloramphenicol acetyltransferase was previously published as a reporter gene by Apt, Grossman & Kroth-Pancic (1996), and more recently used by Niu et al. (2012) as a resistance marker for the selection of genetically transformed cell lines on chloramphenicol. Other antibiotics commonly used in protists, like hygromycin and puromycin, were tested in P. tricornutum, but either proved ineffective (hygromycin) or only effective at very high concentrations (e.g. puromycin) (Apt, Grossman & Kroth-Pancic, 1996).
The lack of selection markers currently limits genetic modifications of diatoms to a maximum of two independent (or consecutive) screening steps. This is a limiting factor in biotechnological applications, because already the knockout of a single gene via the TALEN system requires two antibiotic resistance markers to select for the presence of the two (left and right) required TALEN constructs (Serif et al., 2017). Further steps like complementation or introduction of a second gene knockout therefore are difficult to achieve. While transient genetic transformations and protein delivery might offer workarounds to this limitation in the future (Karas et al., 2015; Serif et al., 2018; Slattery et al., 2018), additional selection markers will nonetheless be needed whenever stable genome integration of transgenes is required.
Phleomycins target and cleave the DNA of eukaryotic as well as prokaryotic organisms, ultimately resulting in cell death (Berdy, 1980; Maeda et al., 1956). In human cell lines, the expression of the resistance gene ShBle does not completely inhibit DNA cleavage and unpredictable mutations still occur (Trastoy, Defais & Larminat, 2005). Such phenomena have not yet been observed in P. tricornutum, however, particularly for genome editing, it appears essential to use antibiotics that do not affect non-targeted DNA.
In this study, we tested the toxicity of three antibiotics, tunicamycin, blasticidin-S and voriconazole to P. tricornutum cells. Only the N-glycosylation inhibitor tunicamycin and the protein synthesis inhibitor blasticidin-S effectively caused cell death of P. tricornutum. For blasticidin-S, we were able to establish the appropriate resistance gene blasticidin-S deaminase (bsr) as selection marker in P. tricornutum. We further showed that the bsr confers resistance to blasticidin-S only and does not alter the effectiveness of Zeocin or nourseothricin.
Materials and Methods
Phaeodactylum tricornutum culture media
One liter of modified f/2 medium (Guillard, 1975) was prepared by dissolving 16.6 or 8.3 g of “Tropic-Marin CLASSIC” artificial sea salt (Dr. Biener GmbH, Wartenberg, Germany) in demineralized water (resulting in one half or one quarter of the salinity of natural sea water). After autoclaving the standard f/2 supplements (Guillard, 1975) and additionally one ml of 2M Tris–HCl pH 8.0 were added. For preparation of solid media, 12 g bacto agar (Becton; Dickinson and Company, Le Pont de Claix, France) per liter of salt water were added prior to autoclaving. After addition of the supplements, plates each containing 25 ml of solid medium were prepared in Petri dishes (633181; Greiner Bio-One, Kremsmünster, Austria).
Phaeodactylum tricornutum selection media
Zeocin (Thermo Fisher, Waltham, MA, USA): 750 μl of a 100 mg/ml solution of Zeocin was added to one liter of the f/2 medium to a final concentration of 75 μg/ml. Nourseothricin (clonNAT; WERNER BioAgents, Jena, Germany) was prepared as a stock solution of 100 mg/ml in water and added to the f/2 medium a final concentration of 150 μg/ml. Blasticidin-S (R21001; Thermo Fisher, Waltham, MA, USA) and tunicamycin (Abcam, Cambridge, UK) were prepared as stock solutions of 10 mg/ml in water. Zeocin, nourseothricin, blasticidin-S and tunicamycin stock solutions were added in the desired quantities to the media after autoclaving and prior to preparing the culture plates, or prior to the inoculation in case of liquid selection media. Voriconazole (Sigma-Aldrich, St. Louis, MO, USA) was dissolved in dimethyl sulfoxide as stock solution of 20 mg/ml. Each voriconazole plate was prepared by plating the appropriate amount of the stock solution onto culture plates prepared as described above.
Transformation of P. tricornutum by particle bombardment
Genetic transformation of P. tricornutum Pt4 (single colony derived from UTEX646 which was previously used by Apt, Grossman & Kroth-Pancic (1996); UTEX culture collection of algae, Austin, TX, USA) was performed as previously described (Apt, Grossman & Kroth-Pancic, 1996; Kroth, 2007) with three single ballistic shots for one attempt. Selection on blasticidin-S plates was performed at 70 μE (Biolux L 36/965; Osram, Munich, Germany) continuous light conditions and at 22 °C for 3–5 weeks.
Construction of the vectors carrying the resistance genes
Amino acid sequences of the tunicamycin resistance protein (tmrB, GenBank P12921.4; Harada et al., 1988) and the bsr (GenBank P33967.1; Kobayashi et al., 1991) were retrieved from GenBank. The amino acid sequences were reverse translated with the online-tool “Reverse Translate” of the “Sequence Manipulation Suite” (SMS, http://www.bioinformatics.org/sms2/rev_trans.html) (Stothard, 2000). Recognition sites of popular commercially available restriction enzymes were then manually eliminated (the final gene sequences are shown in Data S1). We then checked the codon efficiency of the genes with a custom generated codon usage table for P. tricornutum (Table S1), prepared in the following way: The coding sequences (cds) of the P. tricornutum gene catalog Phatr3 (Bowler et al., 2008; Rastogi et al., 2018) were downloaded from ftp://ftp.ensemblgenomes.org/pub/protists/release-39/fasta/phaeodactylum_tricornutum/cds/ (file name Phaeodactylum_tricornutum.ASM15095v2.cds.all_.fa). The 12178 cds were then screened for sequence lengths that are divisible by three. If necessary, sequences were processed to a sequence length that is divisible by three with help of the custom perl script “divisible_by_three.pl” (https://bitbucket.org/grubio-/divisible_by_three). 12177 cds had lengths that were either divisible by three, or could be truncated to complete codons in reference to a start or a stop codon, resulting in a total of 18112998 base pairs (compared to 18113425 base pairs in the complete Phatr3 cds catalog). Sequences were then concatenated into a single sequence with the “Combine FASTA” tool of a locally installed version of SMS (Stothard, 2000), and the codon table was generated using the tool “Codon Usage” of SMS (Stothard, 2000).
The reverse translated sequences were synthesized and cloned into a standard plasmid by Eurofins Genomics (Ebersberg, Germany). The genes were amplified by PCR (primers 01 and 02 for bsr and 03 and 04 for tmrB, Table S2) using KapaHifi polymerase (Roche, Basel, Switzerland).
The plasmid pPha-T1 (Zaslavskaia et al., 2000) was used as a template for a deletion PCR (primers 05 and 06, Table S2) to remove the ShBle gene and to introduce recognition sites for the restriction enzymes MssI and EcoRI. The resulting PCR product was ligated and delivered to E. coli XL1 Blue by electroporation, followed by selection on ampicillin plates. Plasmid preparation was done according to the manufacturer’s protocol using “QIAprep Spin Miniprep Kit” (QIAGEN, Hilden, Germany). The plasmid DNA was treated with the restriction enzyme MssI (Thermo Fisher, Waltham, MA, USA) and ligated with the amplified resistance genes using T4 DNA ligase (Thermo Fisher, Waltham, MA, USA). These constructs were genetically transformed to E. coli XL1 Blue by electroporation. Resulting colonies were screened by PCR for correct orientation of the insert. Extracted plasmid DNAs of single colonies growing on ampicillin plates were sequenced (Microsynth Seqlab, Göttingen, Germany) and used for the experiments with P. tricornutum.
Vector sequences and maps
Vector maps were generated with Geneious version 9 (Kearse et al., 2012). The sequences of the reverse translated resistance genes and of the whole plasmids can be found in Supplemental Information S1. The sequence of the pPTbsr vector is additionally available at GenBank (accession number MH541819) and the vector will be available at Addgene with the deposit number 117696.
PCR test for presence of bsr gene
A small amount of blasticidin-S resistant colonies (transformed with pPTbsr) was scraped off the plates, dissolved in 100 μl of water and heated to 99 °C for 10 min to serve as template. The PCR mixture contained five μl of this template, 0.1 μl of Taq DNA Polymerase E (Genaxxon bioscience, Ulm, Germany), 2.5 μl of 10× buffer, 0.25 μl of each primer (01 and 02 for the whole gene, 07 and 08 for a shorter fragment; 10 mM, Table S2), 0.25 μl of dNTPs (10 mM), 17 μl of H2O. The PCRs were performed in a PCR cycler (ThermoCycler; Eppendorf, Hamburg, Germany) as described by the manufacturer of the DNA polymerase with annealing temperature of 54 °C and an extension time of 40 s.
Cross activities of marker genes
Nine of the blasticidin-S-resistant P. tricornutum colonies, transformed with the plasmid “pPTbsr” and nine nourseothricin-resistant colonies created by transformation of wild-type P. tricornutum with the plasmid “pNat” (Zaslavskaia et al., 2000), as well as five of the Zeocin resistant “lhcf1” cell lines generated by Chu et al. (2016) as well as two TGS1 and two TGS2 cell lines generated by Huang, Río Bártulos & Kroth, (2016) were used for the detection of possible cross-resistances against the antibiotics conferred by one of the tested marker genes.
Results and Discussion
To identify new antibiotics suitable for selection of P. tricornutum, we first established a workflow consisting of three main steps. In the first step, the adequate amount of an antibiotic was determined that leads to cell death in P. tricornutum. In the second step, potential resistance genes that conferred resistance against the respective antibiotics were tested in the target organism. Finally, the respective resistance proteins were checked for any cross activity with the two other antibiotics in use (Zeocin and nourseothricin), which is required for independent selection with combinations of the antibiotics.
Determination of effective concentrations of the antibiotics
A classical way to determine the lethal concentration of an antibiotic is a dose-response experiment, in which cells are subjected to increasing amounts of an antibiotic to determine the minimal efficient antibiotic concentration that is needed to inhibit cell growth. Accordingly, we tested the efficiency of the antibiotics by cultivating P. tricornutum cells on plates containing different concentrations of antibiotics. According to previous publications, a lethal dose of blasticidin-S of about two μg/ml was reported for mammalian cells (Izumi et al., 1991), for tunicamycin the LD50 was reported between 0.1 and five μg/ml (Elbein, 1981), while voriconazole at 0.35 μg/ml impaired plant growth (Rozhon et al., 2013). For our experiments, we used concentrations ranges from higher to lower than these values (Table 1). We plated 2.5 × 107 wild-type cells, and the growth of cells in each plate was determined after 3 and 5 weeks, classifying the results into “no growth”, “scattered growth”, or “diatom lawns” (Fig. 1; Table 1). Some antibiotics like G418 and nourseothricin are inhibited by high salt concentrations in marine media (Zaslavskaia et al., 2000). Blasticidin-S has been described by the manufacturer to be affected by the presence of salts, we therefore additionally tested a lower salt concentration in the medium (50% and 25% sea salt concentration). P. tricornutum is known to occur in places with highly variable growth conditions (Martino et al., 2007) and to tolerate a wide range of salt concentrations (Kräbs & Büchel, 2011).
Table 1. Determination of effective antibiotic concentrations.
| Antibiotic | Salt concentration | Antibiotic concentration (μg/ml) | Results | Ind. Repl. | |
|---|---|---|---|---|---|
| 3 Weeks | 5 Weeks | ||||
| Blasticidin-S | 1/2 | 2 | Lawn | Lawn | 6 |
| 1/2 | 4, 6 | Scattered | Scattered | 6 | |
| 1/2 | 8, 10 | No | No | 6 | |
| 1/4 | 0.5, 1, 1.5, 2, 2.5 | Lawn | Lawn | 10 | |
| 1/4 | 3 | Scattered | Scattered | 10 | |
| 1/4 | 3.5 | No | No | 4 | |
| 1/4 | 4 | No | No | 24 | |
| 1/4 | 6 | No | No | 2 | |
| Tunicamycin | 1/2 | 0.075, 0.15, 0.3, 0.6 | Lawn | Lawn | 1 |
| 1/2 | 1.25, 2.5 | Scattered | Scattered | 1 | |
| 1/2 | 5, 10 | No | Scattered | 6 | |
| 1/4 | 5 | No | No | 6 | |
| 1/4 | 10 | No | No | 2 | |
| Voriconazole | 1/2 | 0.4, 0.7, 1.1, 3.5, 7, 10.5, 35, 70, 105 | Lawn | Lawn | 1 |
Note:
2.5 × 107 P. tricornutum cells were plated on each agar plate containing different concentrations of antibiotics. Tunicamycin and blasticidin-S were tested both on 50% and 25% sea salinity plates. Growth was checked after 3 and 5 weeks of incubation. “Ind. repl.” = independent replicates, “no” = no growth, “scattered” = growth in single colonies, “lawn” = lawn growth.
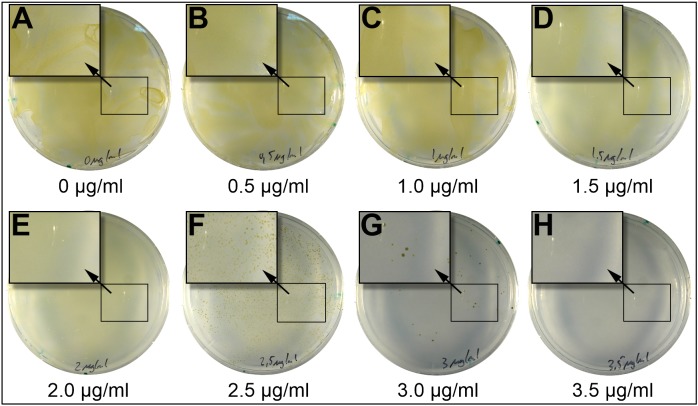
Figure 1. Incubation of P. tricornutum on plates containing different concentrations of blasticidin-S.
2.5 × 107 P. tricornutum cells were spread on plates (25% salinity of seawater) and containing different concentrations of blasticidin-S. At concentrations between 0 and two μg/ml of blasticidin-S (A–E) we observed a lawn of cells. At 2.5 μg/ml (F) and 3.0 μg/ml (G) we observed scattered growth, and above 3.5 μg/ml (H) no growth was observed. Magnified areas in the picture are indicated by an arrow.
Voriconazole, an inhibitor of P450 in plants (Rozhon et al., 2013), did not show any effect on P. tricornutum growth even at 105 μg/ml, which is about 300-fold the concentration of effective doses for most land plants (Rozhon et al., 2013). Therefore, this antibiotic was not investigated further. Blasticidin-S is an inhibitor of protein synthesis binding to the P-site of the 60S ribosomal subunit (Garreau De Loubresse et al., 2014). Blasticidin-S inhibited the growth of P. tricornutum completely at a concentration of 3.5 μg/ml on plates with 25% salt and eight μg/ml on 50% salt plates (Table 1). In liquid 50% salt medium, blasticidin-S inhibited growth at a concentration of three μg/ml (Fig. S1). Tunicamycin, which acts as inhibitor of biosynthesis of peptidoglycan structures in prokaryotes and of N-glycosylation in eukaryotes (Takatsuki, Arima & Tamura, 1971; Wyszynski et al., 2012), inhibited the growth at concentrations of five μg/ml on 25% salt plates, and 10 μg/ml on 50% salt plates. However, in contrast to blasticidin-S, tunicamycin was not able to stop growth completely, as single colonies appeared after 5 weeks on the 50% salt plates.
Functionality of the respective resistance genes
In order to confer resistance against tunicamycin and blasticidin-S in P. tricornutum, we generated DNA fragments encoding the tunicamycin resistance protein (tmrB) (Harada et al., 1988) and the bsr (Endo et al., 1987), with codons suitable for P. tricornutum and manually eliminated recognition sites of popular commercially available restriction enzymes. We inserted the synthetic genes into the plasmid pPha-T1 (Zaslavskaia et al., 2000), replacing the Zeocin resistance cassette ShBle between the fcpB-promoter and the fcpA-terminator (Fig. 2, example for the bsr gene). P. tricornutum was genetically transformed with the constructs by particle bombardment. To test the antibiotic resistance, we plated the cells on selection plates containing four μg/ml (25% salt) or 10 μg/ml (50% salt) of blasticidin-S, and 10 μg/ml (50% salt) tunicamycin, respectively. For unknown reasons, we only obtained blasticidin-S-resistant colonies for the cells transformed with the bsr gene, while no tunicamycin-resistant colonies were obtained when we tested the tmrB gene using two independent attempts.
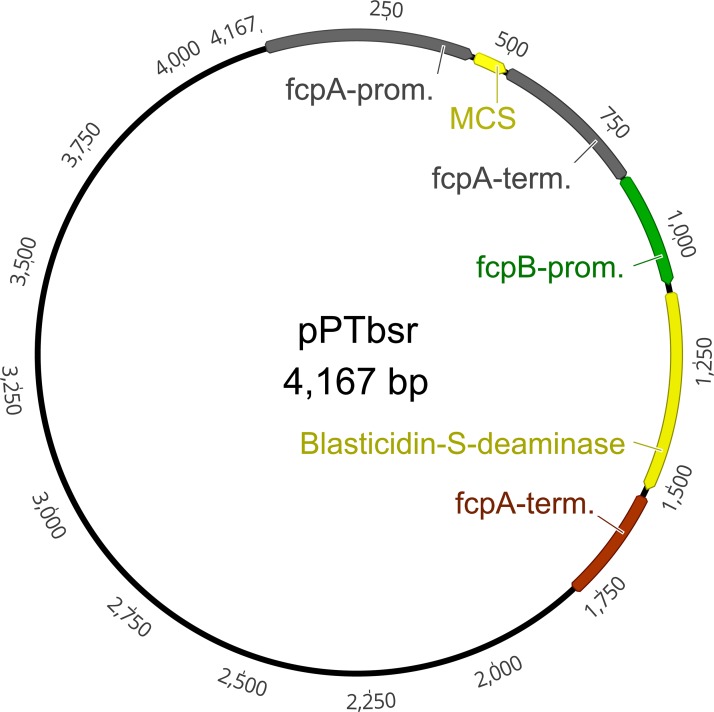
Figure 2. Plasmid map of the vector pPTbsr.
The vector is based on pPha-T1 (derived from Zaslavskaia et al. (2000)) but includes the resistance gene blasticidin-S deaminase (bsr) instead of the Zeocin resistance cassette ShBle. MCS, multiple cloning site; fcpA/B, fucoxanthin-chlorophyll-binding protein A/B; prom, promoter; term, terminator.
In total, we obtained 78 blasticidin-S-resistant colonies on four μg/ml, 25% salt medium in two independent transformations, plus 22 colonies on 10 μg/ml, 50% salt medium in one transformation approach. We re-plated each of the obtained colonies on fresh blasticidin-S plates, on which all of them continued to grow stably and were able to survive in liquid medium under selection pressure (Fig. S1). In more than one year that has passed since we obtained the first blasticidin-S cell lines, we did not observe a loss of resistance in any of the colonies. For further experiments we focused only on colonies that were selected on the 25% salt plates. These 78 colonies were checked by PCR for the presence of the bsr gene. We could amplify a PCR product in the expected size range from 75 of the 78 analyzed templates derived from the colonies, indicating the presence of the resistance gene (Fig. S2).
Cross reactivity test
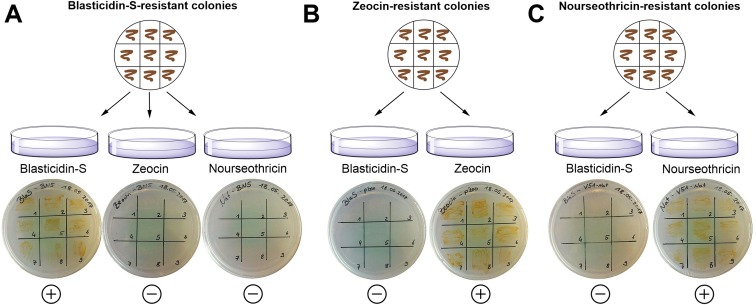
In order to perform successive transformations with different selection markers, we tested whether blasticidin-S might interfere with the usage of Zeocin- or nourseothricin-based screening protocols. We therefore plated nine of the blasticidin-S-resistant colonies on agar plates containing either Zeocin or nourseothricin (Fig. 3). Conversely, nine nourseothricin- or Zeocin-resistant colonies were plated on plates with four μg/ml blasticidin-S. As a control, the same colonies were inoculated on culture plates containing the antibiotic to which they show resistance. After 11 days of growth, colonies were detected only on plates on which cells were plated that had the respective antibiotic resistance (Fig. 3). This indicates that blasticidin-S selection can be used in combination with Zeocin- or nourseothricin-resistant cells. Such double resistant strains were already reported for the combination of Zeocin and nourseothricin (Serif et al., 2017). Especially the combination of blasticidin-S- and nourseothricin might be important for future applications, for instance to replace Zeocin in the TALEN vectors or for complementation experiments following gene knockouts. We co-transformed the plasmids pPTbsr and pPha-NR (without BsaI) (Serif et al., 2017), which mediates nourseothricin resistance, leading to 29 resistant colonies on the 50% salt selection plates containing 10 μg/ml blasticidin-S and 150 μg/ml nourseothricin.
Figure 3. Exclusion of potential interference between different resistance genes using strains showing either Zeocin, nourseothricin or blasticidin-S resistance.
(A) Nine blasticidin-S-resistant P. tricornutum colonies were spread on plates containing Zeocin or nourseothricin and on a control plate containing blasticidin-S. (B) Nine Zeocin-resistant colonies were spread on blasticidin-S plates and on control plates with Zeocin. (C) Nine nourseothricin-resistant colonies were spread on plates with blasticidin-S and on control plates with nourseothricin. The cells were able to survive only on the appropriate antibiotics. (+) = growth of the cells; (−) = no growth.
Transferability of the results to other strains and diatom species
In this study we used P. tricornutum strain UTEX 646 (Pt4 in the nomenclature by Martino et al. (2007)) in all our experiments. Experience with other antibiotics like Zeocin/phleomycin shows that there is apparently no difference between different strains regarding their sensitivity (Apt, Grossman & Kroth-Pancic, 1996; Falciatore et al., 1999; Siaut et al., 2007; Tanaka et al., 2005). Furthermore, Martino et al. (2007) used the ShBle marker gene for ten different strains of P. tricornutum. We therefore expect that our results regarding blasticidin-S are directly transferable to other P. tricornutum strains.
In contrast to this, the sensitivity of different diatom species to the currently used antibiotics is quite variable (Huang & Daboussi, 2017) and although some antibiotics, after dose adjustment, can be used in several species of diatoms, marker genes are not generally transferrable between diatom species (Huang & Daboussi, 2017). It therefore remains to be shown if blasticidin-S is suitable for the selection of genetically transformed cell lines of other diatom species. For this, most likely adjustments of the promoters and antibiotic doses would be needed.
Conclusions
Here we report a new and easy to handle system of antibiotic selection with blasticidin-S and the resistance gene bsr in P. tricornutum. Blasticidin-S works by blocking protein translation and therefore it is not expected to affect the genomic DNA of transformed cells. Especially for reverse genetics studies via genome editing, when phenotypes should be determined based on cell-linages with as less off-target genetic changes as possible, utilization of this antibiotic is a major improvement. Additionally, blasticidin-S selection can be used in combination with nourseothricin- and potentially also with Zeocin-resistant cells. This extension of the genetic toolbox will be very useful for further molecular characterization or biotechnological application of the model diatom P. tricornutum and possibly for other species of diatoms.
Supplemental Information
The table shows the codon frequency for each codon in P. tricornutum. “Number” is the total count for each codon within all protein coding genes (see text for details), “/1000” is the frequency of a codon (per 1000 total codons), “Fraction” is the relative proportion of a single codon within the synonymous codons.
P. tricornutum was inoculated with 105 cells/ml in liquid 50% salt medium with different concentrations of blasticidin-S. The growth was determined six days after inoculation. (A) Wild type cells in 1 μg/ml blasticidin-S (B) Wild type cells in 2 μg/ml blasticidin-S (C) wild type cells in 3 μg/ml blasticidin-S (D) Wild type cells in 4 μg/ml blasticidin-S (E) Wild type cells in 5 μg/ml blasticidin-S (F) Resistant cells in 2 μg/ml blasticidin-S (G) Resistant cells in 3 μg/ml blasticidin-S (H) Resistant cells in 4 μg/ml blasticidin-S. Growth of the wild-type cells was inhibited at a concentration of 3 μg/ml or higher, while the pPTbsr transformed cell line survived 4 μg/ml.
(A) Primers for amplification of the whole bsr gene (primers 01 and 02; expected length of 423 base pairs) or (B) for an internal part (primers 07 and 08; expected lenght of 216 base pairs) were used. As negative control, wild-type cells were used as template. “1kb” = O&GeneRuler 1 kb DNA-ladder (Thermo Fisher, Waltham, MA, USA); “50bp” = O&GeneRuler 50 bp DNA-ladder (Thermo Fisher, Waltham, MA, USA); “P” = positive control (positive colonies as PCR Template); “C” = Control (plasmid-DNA (pPTbsr) as PCR template); “bp” = base pairs.
Acknowledgments
The authors are grateful to Doris Ballert for help with the cultivation and transformation of P. tricornutum, Nandor Hegyi and Olga Klat for experimental support and to Kalen Robeson for proofreading this manuscript.
Funding Statement
This work was financed by the University of Konstanz and the Gordon and Betty Moore Foundation GBMF 4966 (grant DiaEdit to Peter G. Kroth). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
The authors declare that they have no competing interests.
Author Contributions
Jochen M. Buck conceived and designed the experiments, performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft.
Carolina Río Bártulos conceived and designed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft.
Ansgar Gruber conceived and designed the experiments, analyzed the data, authored or reviewed drafts of the paper, approved the final draft.
Peter G. Kroth analyzed the data, authored or reviewed drafts of the paper, approved the final draft.
DNA Deposition
The following information was supplied regarding the deposition of DNA sequences:
Sequences were uploaded to GenBank (accession number MH541819) and can also be found in Supplemental Dataset 1.
Data Availability
The following information was supplied regarding data availability:
Data can be found at GenBank, accession number GenBank: MH541819
References
- Apt, Grossman & Kroth-Pancic (1996).Apt KE, Grossman AR, Kroth-Pancic PG. Stable nuclear transformation of the diatom Phaeodactylum tricornutum. MGG Molecular & General Genetics. 1996;252(5):572–579. doi: 10.1007/bf02172403. [DOI] [PubMed] [Google Scholar]
- Berdy (1980).Berdy J. Bleomycin-Type Antibiotics. Boca Raton: CRC Press; 1980. pp. 459–497. [Google Scholar]
- Bowler et al. (2008).Bowler C, Allen AE, Badger JH, Grimwood J, Jabbari K, Kuo A, Maheswari U, Martens C, Maumus F, Otillar RP, Rayko E, Salamov A, Vandepoele K, Beszteri B, Gruber A, Heijde M, Katinka M, Mock T, Valentin K, Verret F, Berges JA, Brownlee C, Cadoret J-P, Chiovitti A, Choi CJ, Coesel S, De Martino A, Detter JC, Durkin C, Falciatore A, Fournet J, Haruta M, Huysman MJJ, Jenkins BD, Jiroutova K, Jorgensen RE, Joubert Y, Kaplan A, Kröger N, Kroth PG, La Roche J, Lindquist E, Lommer M, Martin–Jézéquel V, Lopez PJ, Lucas S, Mangogna M, McGinnis K, Medlin LK, Montsant A, Secq M-POL, Napoli C, Obornik M, Parker MS, Petit J-L, Porcel BM, Poulsen N, Robison M, Rychlewski L, Rynearson TA, Schmutz J, Shapiro H, Siaut M, Stanley M, Sussman MR, Taylor AR, Vardi A, Von Dassow P, Vyverman W, Willis A, Wyrwicz LS, Rokhsar DS, Weissenbach J, Armbrust EV, Green BR, Van De Peer Y, Grigoriev IV. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature. 2008;456(7219):239–244. doi: 10.1038/nature07410. [DOI] [PubMed] [Google Scholar]
- Chu et al. (2016).Chu L, Ewe D, Rio Bartulos C, Kroth PG, Gruber A. Rapid induction of GFP expression by the nitrate reductase promoter in the diatom Phaeodactylum tricornutum. PeerJ. 2016;4:e2344. doi: 10.7717/peerj.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daboussi et al. (2014).Daboussi F, Leduc S, Maréchal A, Dubois G, Guyot V, Perez-Michaut C, Amato A, Falciatore A, Juillerat A, Beurdeley M, Voytas DF, Cavarec L, Duchateau P. Genome engineering empowers the diatom Phaeodactylum tricornutum for biotechnology. Nature Communications. 2014;5(1):3831. doi: 10.1038/ncomms4831. [DOI] [PubMed] [Google Scholar]
- Elbein (1981).Elbein AD. The tunicamycins—useful tools for studies on glycoproteins. Trends in Biochemical Sciences. 1981;6:219–221. doi: 10.1016/0968-0004(81)90080-3. [DOI] [Google Scholar]
- Endo et al. (1987).Endo T, Furuta K, Kaneko A, Katsuki T, Kobayashi K, Azuma A, Watanabe A, Shimazu A. Inactivation of blasticidin S by Bacillus cereus. I. Inactivation mechanism. Journal of Antibiot. 1987;40(12):1791–1793. doi: 10.7164/antibiotics.40.1791. [DOI] [PubMed] [Google Scholar]
- Falciatore et al. (1999).Falciatore A, Casotti R, Leblanc C, Abrescia C, Bowler C. Transformation of nonselectable reporter genes in marine diatoms. Marine Biotechnology. 1999;1(3):239–251. doi: 10.1007/pl00011773. [DOI] [PubMed] [Google Scholar]
- Garreau De Loubresse et al. (2014).Garreau De Loubresse N, Prokhorova I, Holtkamp W, Rodnina MV, Yusupova G, Yusupov M. Structural basis for the inhibition of the eukaryotic ribosome. Nature. 2014;513(7519):517–522. doi: 10.1038/nature13737. [DOI] [PubMed] [Google Scholar]
- Gruber & Kroth (2017).Gruber A, Kroth PG. Intracellular metabolic pathway distribution in diatoms and tools for genome-enabled experimental diatom research. Philosophical Transactions of the Royal Society B: Biological Sciences. 2017;372(1728):20160402. doi: 10.1098/rstb.2016.0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillard (1975).Guillard RRL. Culture of phytoplankton for feeding marine invertebrates. In: Smith WL, Chanley MH, editors. Culture of Marine Invertebrate Animals: Proceedings—1st Conference on Culture of Marine Invertebrate Animals Greenport. Boston: Springer; 1975. pp. 29–60. [Google Scholar]
- Harada et al. (1988).Harada S, Yoda K, Mori M, Tanimoto A, Furusato T, Yamane K, Takatsuki A, Yamasaki M, Tamura G. The gene responsible for tunicamycin resistance, tmr B, in Bacillus subtilis. Agricultural and Biological Chemistry. 1988;52(7):1785–1789. doi: 10.1271/bbb1961.52.1785. [DOI] [Google Scholar]
- Huang & Daboussi (2017).Huang W, Daboussi F. Genetic and metabolic engineering in diatoms. Philosophical Transactions of the Royal Society B: Biological Sciences. 2017;372(1728):20160411. doi: 10.1098/rstb.2016.0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Río Bártulos & Kroth (2016).Huang W, Río Bártulos C, Kroth PG. Diatom vacuolar 1, 6-β-Transglycosylases can functionally complement the respective yeast mutants. Journal of Eukaryotic Microbiology. 2016;63(4):536–546. doi: 10.1111/jeu.12298. [DOI] [PubMed] [Google Scholar]
- Izumi et al. (1991).Izumi M, Miyazawa H, Kamakura T, Yamaguchi I, Endo T, Hanaoka F. Blasticidin S-resistance gene (bsr): a novel selectable marker for mammalian cells. Experimental Cell Research. 1991;197(2):229–233. doi: 10.1016/0014-4827(91)90427-V. [DOI] [PubMed] [Google Scholar]
- Karas et al. (2015).Karas BJ, Diner RE, Lefebvre SC, McQuaid J, Phillips APR, Noddings CM, Brunson JK, Valas RE, Deerinck TJ, Jablanovic J, Gillard JTF, Beeri K, Ellisman MH, Glass JI, Hutchison CA, III, Smith HO, Venter JC, Allen AE, Dupont CL, Weyman PD. Designer diatom episomes delivered by bacterial conjugation. Nature Communications. 2015;6(1):6925. doi: 10.1038/ncomms7925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse et al. (2012).Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28(12):1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi et al. (1991).Kobayashi K, Kamakura T, Tanaka T, Yamaguchi I, Endō T. Nucleotide sequence of the bsr gene and N-Terminal amino acid sequence of Blasticidin S Deaminase from Blasticidin S resistant Escherichia coli TK 121. Agricultural and Biological Chemistry. 1991;55(12):3155–3157. doi: 10.1080/00021369.1991.10859777. [DOI] [PubMed] [Google Scholar]
- Kräbs & Büchel (2011).Kräbs G, Büchel C. Temperature and salinity tolerances of geographically separated Phaeodactylum tricornutum Böhlin strains: maximum quantum yield of primary photochemistry, pigmentation, proline content and growth. Botanica Marina. 2011;54(3):231–241. doi: 10.1515/bot.2011.037. [DOI] [Google Scholar]
- Kroth (2007).Kroth PG. Genetic transformation. In: Van Der Giezen M, editor. Protein Targeting Protocols. Totowa: Humana Press; 2007. pp. 257–267. [Google Scholar]
- Maeda et al. (1956).Maeda K, Kosaka H, Yagishita K, Umezawa H. A new antibiotic, phleomycin. Journal of Antibiotics. 1956;9(2):82–85. [PubMed] [Google Scholar]
- Martino et al. (2007).Martino AD, Meichenin A, Shi J, Pan K, Bowler C. Genetic and phenotypic characterization of Phaeodactylum tricornutum (Bacillariophyceae) accessions. Journal of Phycology. 2007;43:992–1009. doi: 10.1111/j.1529-8817.2007.00384.x. [DOI] [Google Scholar]
- Miyahara et al. (2013).Miyahara M, Aoi M, Inoue-Kashino N, Kashino Y, Ifuku K. Highly efficient transformation of the diatom Phaeodactylum tricornutum by multi-pulse electroporation. Bioscience, Biotechnology, and Biochemistry. 2013;77(4):874–876. doi: 10.1271/bbb.120936. [DOI] [PubMed] [Google Scholar]
- Niu et al. (2012).Niu Y-F, Yang Z-K, Zhang M-H, Zhu C-C, Yang W-D, Liu J-S, Li H-Y. Transformation of diatom Phaeodactylum tricornutum by electroporation and establishment of inducible selection marker. Biotechniques. 2012;52:1–3. doi: 10.2144/000113881. [DOI] [PubMed] [Google Scholar]
- Nymark et al. (2016).Nymark M, Sharma AK, Sparstad T, Bones AM, Winge P. A CRISPR/Cas9 system adapted for gene editing in marine algae. Scientific Reports. 2016;6(1):24951. doi: 10.1038/srep24951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastogi et al. (2018).Rastogi A, Maheswari U, Dorrell RG, Vieira FRJ, Maumus F, Kustka A, McCarthy J, Allen AE, Kersey P, Bowler C, Tirichine L. Integrative analysis of large scale transcriptome data draws a comprehensive landscape of Phaeodactylum tricornutum genome and evolutionary origin of diatoms. Scientific Reports. 2018;8(1):4834. doi: 10.1038/s41598-018-23106-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozhon et al. (2013).Rozhon W, Husar S, Kalaivanan F, Khan M, Idlhammer M, Shumilina D, Lange T, Hoffmann T, Schwab W, Fujioka S, Poppenberger B. Genetic variation in plant CYP51s confers resistance against voriconazole, a novel inhibitor of brassinosteroid-dependent sterol biosynthesis. PLOS ONE. 2013;8(1):e53650. doi: 10.1371/journal.pone.0053650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi, Nakajima & Matsuda (2011).Sakaguchi T, Nakajima K, Matsuda Y. Identification of the UMP synthase gene by establishment of uracil auxotrophic mutants and the phenotypic complementation system in the marine diatom Phaeodactylum tricornutum. Plant Physiology. 2011;156(1):78–89. doi: 10.1104/pp.110.169631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serif et al. (2018).Serif M, Dubois G, Finoux A-L, Teste M-A, Jallet D, Daboussi F. One-step generation of multiple gene knock-outs in the diatom Phaeodactylum tricornutum by DNA-free genome editing. Nature Communications. 2018;9(1):3924. doi: 10.1038/s41467-018-06378-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serif et al. (2017).Serif M, Lepetit B, Weißert K, Kroth PG, Rio Bartulos C. A fast and reliable strategy to generate TALEN-mediated gene knockouts in the diatom Phaeodactylum tricornutum. Algal Research. 2017;23:186–195. doi: 10.1016/j.algal.2017.02.005. [DOI] [Google Scholar]
- Siaut et al. (2007).Siaut M, Heijde M, Mangogna M, Montsant A, Coesel S, Allen A, Manfredonia A, Falciatore A, Bowler C. Molecular toolbox for studying diatom biology in Phaeodactylum tricornutum. Gene. 2007;406(1–2):23–35. doi: 10.1016/j.gene.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Slattery et al. (2018).Slattery SS, Diamond A, Wang H, Therrien JA, Lant JT, Jazey T, Lee K, Klassen Z, Desgagné-Penix I, Karas BJ, Edgell DR. An expanded plasmid-based genetic toolbox enables Cas9 genome editing and stable maintenance of synthetic pathways in Phaeodactylum tricornutum. ACS Synthetic Biology. 2018;7(2):328–338. doi: 10.1021/acssynbio.7b00191. [DOI] [PubMed] [Google Scholar]
- Stothard (2000).Stothard P. The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. BioTechniques. 2000;28(6):1102–1104. doi: 10.2144/00286ir01. [DOI] [PubMed] [Google Scholar]
- Takatsuki, Arima & Tamura (1971).Takatsuki A, Arima K, Tamura G. Tunicamycin, a new antibiotic. I. Journal of Antibiotics. 1971;24(4):215–223. doi: 10.7164/antibiotics.24.215. [DOI] [PubMed] [Google Scholar]
- Tanaka et al. (2005).Tanaka Y, Nakatsuma D, Harada H, Ishida M, Matsuda Y. Localization of soluble β-carbonic anhydrase in the marine diatom Phaeodactylum tricornutum. Sorting to the chloroplast and cluster formation on the girdle lamellae. Plant Physiology. 2005;138(1):207–217. doi: 10.1104/pp.104.058982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trastoy, Defais & Larminat (2005).Trastoy MO, Defais M, Larminat F. Resistance to the antibiotic Zeocin by stable expression of the Sh ble gene does not fully suppress Zeocin-induced DNA cleavage in human cells. Mutagenesis. 2005;20(2):111–114. doi: 10.1093/mutage/gei016. [DOI] [PubMed] [Google Scholar]
- Wyszynski et al. (2012).Wyszynski FJ, Lee SS, Yabe T, Wang H, Gomez-Escribano JP, Bibb MJ, Lee SJ, Davies GJ, Davis BG. Biosynthesis of the tunicamycin antibiotics proceeds via unique exo-glycal intermediates. Nature Chemistry. 2012;4(7):539–546. doi: 10.1038/nchem.1351. [DOI] [PubMed] [Google Scholar]
- Zaslavskaia et al. (2000).Zaslavskaia LA, Lippmeier JC, Kroth PG, Grossman AR, Apt KE. Transformation of the diatom Phaeodactylum tricornutum (Bacillariophyceae) with a variety of selectable marker and reporter genes. Journal of Phycology. 2000;36(2):379–386. doi: 10.1046/j.1529-8817.2000.99164.x. [DOI] [Google Scholar]
- Zhang & Hu (2014).Zhang C, Hu H. High-efficiency nuclear transformation of the diatom Phaeodactylum tricornutum by electroporation. Marine Genomics. 2014;16:63–66. doi: 10.1016/j.margen.2013.10.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The table shows the codon frequency for each codon in P. tricornutum. “Number” is the total count for each codon within all protein coding genes (see text for details), “/1000” is the frequency of a codon (per 1000 total codons), “Fraction” is the relative proportion of a single codon within the synonymous codons.
P. tricornutum was inoculated with 105 cells/ml in liquid 50% salt medium with different concentrations of blasticidin-S. The growth was determined six days after inoculation. (A) Wild type cells in 1 μg/ml blasticidin-S (B) Wild type cells in 2 μg/ml blasticidin-S (C) wild type cells in 3 μg/ml blasticidin-S (D) Wild type cells in 4 μg/ml blasticidin-S (E) Wild type cells in 5 μg/ml blasticidin-S (F) Resistant cells in 2 μg/ml blasticidin-S (G) Resistant cells in 3 μg/ml blasticidin-S (H) Resistant cells in 4 μg/ml blasticidin-S. Growth of the wild-type cells was inhibited at a concentration of 3 μg/ml or higher, while the pPTbsr transformed cell line survived 4 μg/ml.
(A) Primers for amplification of the whole bsr gene (primers 01 and 02; expected length of 423 base pairs) or (B) for an internal part (primers 07 and 08; expected lenght of 216 base pairs) were used. As negative control, wild-type cells were used as template. “1kb” = O&GeneRuler 1 kb DNA-ladder (Thermo Fisher, Waltham, MA, USA); “50bp” = O&GeneRuler 50 bp DNA-ladder (Thermo Fisher, Waltham, MA, USA); “P” = positive control (positive colonies as PCR Template); “C” = Control (plasmid-DNA (pPTbsr) as PCR template); “bp” = base pairs.
Data Availability Statement
The following information was supplied regarding data availability:
Data can be found at GenBank, accession number GenBank: MH541819