Abstract
Objective
Evaluate the quality of clinical order practice patterns machine-learned from clinician cohorts stratified by patient mortality outcomes.
Materials and Methods
Inpatient electronic health records from 2010–2013 were extracted from a tertiary academic hospital. Clinicians (n=1,822) were stratified into low-mortality (21.8%, n=397) and high-mortality (6.0%, n=110) extremes using a two-sided P-value score quantifying deviation of observed vs. expected 30-day patient mortality rates. Three patient cohorts were assembled: patients seen by low-mortality clinicians, high-mortality clinicians, and an unfiltered crowd of all clinicians (n=1,046, 1,046, and 5,230 post-propensity score matching, respectively). Predicted order lists were automatically generated from recommender system algorithms trained on each patient cohort and evaluated against i) real-world practice patterns reflected in patient cases with better-than-expected mortality outcomes and ii) reference standards derived from clinical practice guidelines.
Results
Across six common admission diagnoses, order lists learned from the crowd demonstrated the greatest alignment with guideline references (AUROC range=0.86–0.91), performing on par or better than those learned from low-mortality clinicians (0.79–0.84, P<10−5) or manually-authored hospital order sets (0.65–0.77, P<10−3). The same trend was observed in evaluating model predictions against better-than-expected patient cases, with the crowd model (AUROC mean=0.91) outperforming the low-mortality model (0.87, P<10−16) and order set benchmarks (0.78, P<10−35).
Discussion
Whether machine-learning models are trained on all clinicians or a subset of experts illustrates a biasvariance tradeoff in data usage. Defining robust metrics to assess quality based on internal (e.g. practice patterns from better-than-expected patient cases) or external reference standards (e.g. clinical practice guidelines) is critical to assess decision support content.
Conclusion
Learning relevant decision support content from all clinicians is as, if not more, robust than learning from a select subgroup of clinicians favored by patient outcomes.
Keywords: data mining, machine learning, clinical decision support, electronic health records, mortality
Graphical Abstract
1. INTRODUCTION
Healthcare often falls short of recommended, evidence-based care, with overall compliance with guideline recommendations ranging from 20–80%.[1] Yet, even with recent reforms,[2] evidence-based medicine from randomized controlled trials cannot keep pace with the growing breadth of clinical questions. Only 11% of guideline recommendations are supported by high-quality evidence.[3] Variability and uncertainty in medical practice is further exacerbated by a medical knowledge base that is perpetually expanding beyond the cognitive capacity of any individual.[4] A clinician is thus left to synthesize vast streams of information in an attempt to make the best decisions for each individual patient. As such, medical practice routinely relies on anecdotal experience and individual expert opinion.
To address these issues, healthcare organizations increasingly seek clinical decision support (CDS) systems. CDS aims to reinforce best-practices by distributing knowledge-based content through order sets, templates, alerts, and prognosis scoring systems.[5] Here we focus specifically on clinical orders as concrete manifestations of point-of-care decision making. Computerized provider order entry (CPOE) typically occurs on an “a la carte” basis, where clinicians search for and select orders to trigger subsequent clinical actions (e.g., pharmacy dispensing and nurse administration of a medication, laboratory analysis of blood tests, consultation to a specialist).[5] Because clinician memory and intuition can be error-prone, health system committees manually curate order set templates to distribute standard practices for common diagnoses and procedures.[6] This top-down approach enables clinicians to draw clinical decisions from pre-constructed, human-authored order sets.
Existing approaches to CDS increase consistency and compliance with best practices,[7–8] but production of this content is limited in scale and scope by a committee-driven, manual production process that requires the collaboration of physicians, nurses, and department heads. Even once such content is produced, ongoing maintenance is required to keep it up to date with new evidence, technology, epidemiology, and culture,[9] ultimately making it infeasible to manually produce a comprehensive knowledge base.[10] One of the “grand challenges” in CDS is thus to automatically generate decision support content by data-mining clinical data sources from the bottom-up.[11] In the era of electronic health records (EHR), there is an opportunity to create data-driven CDS systems that leverage the aggregate expertise of many healthcare providers and automatically adapt to the ongoing stream of practice data.[12] This would fulfill the vision of a health system that continuously learns from real-world data and translates them into usable, point-of-care information for clinicians. Prior research into data-mining for decision support content includes association rules, Bayesian networks, and unsupervised clustering of clinical orders and diagnoses.[13–20] In our prior work, inspired by similar information retrieval problems, we developed a data-driven clinical order recommender engine[21] analogous to Netflix and Amazon.com’s “customers who bought A also bought B” system.[22] Our engine dynamically generates order recommendations based on real-world clinical practice patterns represented in EHR data, in effect enabling automatically generated order sets.
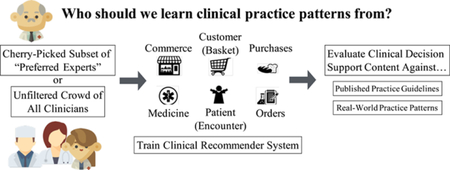
The “wisdom of the crowd” phenomenon purports that the collective assessment of a group of individuals can often be surprisingly as good as, if not better than, that of individual experts.[23] Condorcet’s Jury Theorem illustrates mathematically why one might expect collective opinion to be more likely correct than expert opinion, even when individual members of a group are less likely to be correct than individual experts.[24] In the context of data-driven CDS for medical decision making, we translate this to training machine-learning models on all available data, including patterns generated by clinicians of varying levels of experience and competence.[25] However, effective medical decision making may be compromised if patterns are learned from clinicians with systematically biased decision making that yield poorer patient outcomes. Instead, it could prove advantageous to learn from a cherry-picked subset of clinicians, such as those with notably lower observed than expected patient mortality rates, a metric commonly used to evaluate clinician and hospital performance.[26] With the emergence of data-driven CDS systems, should we learn indiscriminately from the wisdom of the entire crowd, or only from a select subset of data generated by “preferred experts?”
2. OBJECTIVE
To identify “expert” and deviant providers in a tertiary academic hospital, we develop a methodology to stratify clinician populations based on observed vs. expected patient mortality rates. To determine whether machine-learning clinical order patterns from all clinicians or a cherry-picked clinician subgroup with “better” patient outcomes yields more robust results, we evaluate association models trained on “low-mortality” clinicians, “high-mortality” clinicians, and an unfiltered clinician “crowd” against two reference standards: i) real-world practice patterns reflected in patient cases with better-than-expected patient mortality outcomes and ii) consensus order lists derived from published clinical practice guidelines for common admission diagnoses.
3. METHODS
3.1. Data Source
We extracted deidentified, structured patient data from the (Epic) EHR for inpatient hospitalizations from 2008–2013 via the Stanford University Medical Center (SUMC) Clinical Data Warehouse (Figure 1, methodology pipeline).[27] The dataset covers patient encounters from their initial (emergency room) presentation until hospital discharge, comprising >74K patients, >11 million instances of >27K distinct clinical items, and >5.3K admission diagnosis ICD9 codes. The clinical item elements include >7.8K medication, >1.6K laboratory, >1.1K imaging, and >1.0K nursing orders.
Figure 1. Methodology Pipeline to Investigate Whether Clinician Stratification by Patient Mortality Yields Better Automatically Learned Practice Patterns.
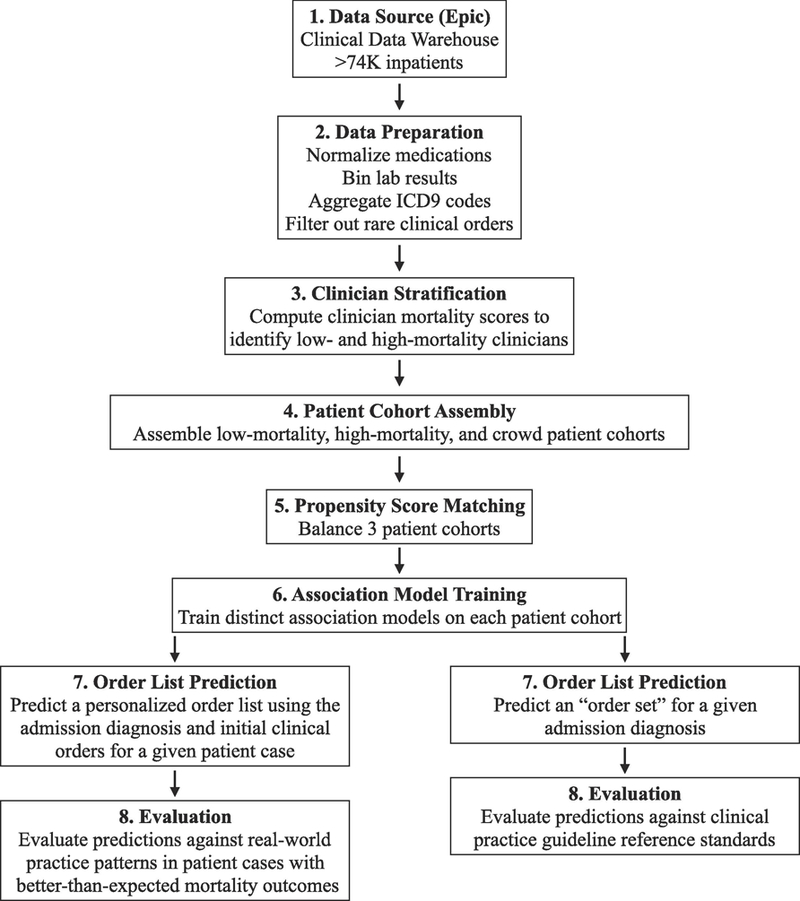
1) Data source: de-identified, structured patient data was extracted from Stanford University Medical Center’s EHR (Epic). 2) Data preparation: patient data was processed to reduce complexity across medication, lab result, and diagnosis codings. 3) Clinician stratification: to mitigate confounding factors resulting from underlying patient characteristics, a L1-regularized logistic regression model was trained on 2008–2009 clinical data to predict a patient’s probability of mortality within 30 days based on treatment team, comorbidities, demographics, severity of illness, etc. and used to predict expected 30-day mortality counts for active clinicians in 2010–2013. Using a P-value-transformation of observed versus expected mortality counts and H&P authorship to identify clinician-patient relationships, clinicians at extremes were stratified into groups with lower or higher than expected patient mortality. 4) Patient cohort assembly: low-mortality (patients seen by low-mortality clinicians and no high-mortality clinicians), high-mortality (vice versa), and crowd patient cohorts (patients seen by any clinician) were assembled. 5) Propensity score matching: to further mitigate confounding factors, the three patient cohorts were balanced across covariates including medical history, treatment specialty, and demographic data, ensuring that the patient cohorts differed primarily in which class of clinicians they saw. 6) Recommender system training: applying association rule episode mining to clinical order data extracted from each patient cohort, three distinct recommender systems were trained, each reflecting the clinical order patterns of the corresponding clinician cohort. 7) Order list prediction: each recommender system outputted order suggestions for two predictions tasks: i) given an admission diagnosis and clinical orders administered up to the usage of an order set from a real-world patient case, predict a personalized order list; ii) given an admission diagnosis, predict a general diagnosis-specific order list. For (i), we considered patient cases with better-than-expected mortality outcomes from 2010–2013 EHR data, left-out from model training. For (ii), we considered six common admission diagnoses. 8) Evaluation: predictions generated by the three association models and corresponding hospital order set benchmarks were evaluated against: i) real-world practice patterns reflected in the actual 24 hours of orders administered after the order set usage instance; ii) practice guideline reference standards curated by two board-certified internal medicine physicians based on a review of clinical practice literature. EHR: electronic health record. H&P: history & physical examination note.
3.2. Data Preparation
Medication data was normalized with RxNorm mappings[28] down to active ingredients and routes of administration. Numerical lab results were binned based on “abnormal” flags established by the clinical laboratory, or being outside two standard deviations from the population mean. ICD9 codes were aggregated up to the three digit hierarchy to compress the sparsity of diagnosis categories, but original (four-five digit) codes were retained if sufficiently prevalent. Problem list and admission diagnosis ICD9 codes were used to assign Charlson Comorbidity Indices.[29] Income levels were inferred from 2013 US census data by cross-referencing patient zip codes with the median household income in that region. Physician specialties (e.g. Otolaryngology, Plastic Surgery, Thoracic Surgery) were grouped into broader treatment team categories (e.g. Surgery Specialty). These pre-processing steps enable us to model each patient as a timeline of clinical event instances, with each event mapping a clinical item to a patient at a discrete time point.
As clinical item instances follow the 80/20 rule of a power law distribution,[30] the majority of item types may be ignored with minimal information loss. As described in our prior work,[31] ignoring rare clinical items (occurring for <1% of patients) significantly reduces the number of distinct items considered while still capturing ~98% of all clinical item event instances. Ultimately, ~2.0K candidate clinical orders were considered for prediction, including >700 medication, >350 laboratory, and >200 imaging orders.
3.3. Clinician Stratification by 30-Day Mortality Rate
Hospital clinicians are subject to performance metrics including compliance with documentation and process measures (e.g. ensuring patients suffering from a heart attack are discharged on specific medications) as well as factors like patient’s length of stay and unplanned readmission rate.[32] Here, we use 30-day patient mortality to stratify clinicians who saw patients between 2010 and 2013 (n=1,822). Mortality provides a concrete and reproducible metric prominently featured in existing quality ranking systems used to assess hospital and clinician performance (see Appendix 9).[26,32–35] We avoid process measures that do not necessarily translate into meaningful patient outcomes, and measures like length of stay that could be “gamed” by compromising other care considerations.[33]
In this step, we sought to identify “low-” and “high-mortality” clinician subgroups, defined as clinicians with lower or higher observed than expected 30-day mortality rates among the patients they treated, respectively. We attributed clinician-patient treatment relationships whenever a clinician signed a History and Physical Examination (H&P) note during a patient’s hospital encounter. After the last recorded admission, whether or not the patient died within 30 days was a binary outcome, yielding observed dead and alive patient counts for each clinician.
Observed mortality counts can be biased by variable patient sickness and clinical specialties that naturally see sicker patient populations. To address this imbalance, we computed per-clinician expected mortality counts. Given patient data commonly used to inform prognosis models and adjust for severity of illness,[36–38] we trained a L1-regularized logistic regression model using 10-fold cross validation (cross-validation AUROC=0.86) to predict patient probability of death within 30 days of their last recorded admission. Features included demographic data (age, ethnicity, gender, income level), initial vital signs (temperature, pulse, blood pressure, etc.), initial standard lab test results (white blood cell count, blood sodium level, blood urea nitrogen, etc.), Charlson Comorbidity Indices[29] to capture past medical history, treatment team designations (e.g. to distinguish medical from surgical patients), and admission diagnoses from patients seen between 2008–2009 (n=6,797). Grid-search hyperparameter tuning (systematically iterating through a manually-specified list of potential shrinkage parameters) was used to minimize the cross-validation area under the receiver operating characteristic curve (AUROC) yielding an optimal shrinkage parameter λ = 2 × 10−3.[39] We then predicted probabilities of 30-day mortality for patients seen between 2010 and 2013 (n=64,598). The expected 30-day mortality count for each clinician was computed as rounded to the nearest integer, where Nj = number of patients attributed to clinician j, and pi = patient i’s predicted probability of dying within 30 days.
Although commonly used in practice, raw observed vs. expected ratios yield unstable rate estimates for small sample sizes.[40] To address this, we integrated an assessment of numerical confidence in the estimates by calculating Pj for each clinician, the P-value for the observed vs. expected mortality contingency table (Table 1) using a two-sided Fisher Exact Test. This approach, common to genomics,[74] captures certainty of deviation from expected norms based on combined effect size and patient cohort size. We then assign each clinician j a score Sj defined as:
For clinicians with insufficient data to make statistically confident claims (or whose observed and expected rates are nearly identical), Sj converges to 0. This has the effect of assigning a large positive score to “low-mortality” clinicians (n=397) with observed mortality rates that are detectably less than expected and inversely, large negative scores to “high-mortality” clinicians (n=110).
Table 1.
Example clinician performance scores. An observed vs. expected 30-day morality contingency table was constructed for each clinician based on the set of patients they were responsible for in 2010–2013. A patient was counted as “dead” if their death occurred within 30 days of their last recorded admission order. The observed dead (DO) and alive (AO) counts can be deduced directly from admission order and mortality timestamps. The expected dead (DE) and alive (AE) counts are predicted using 30-day mortality probabilities generated by a L1-regularized logistic regression model trained on 2008–2009 patient data. In these examples, Clinician A has a lower O-to-E ratio than Clinician B, but a larger quantity of data that yields more confidence and thus an equivalent final score. Clinicians B and C have the same observed-to-expected mortality ratio, but their scores differ due to varying confidence in the estimate. Higher magnitude scores thus reflect a greater effect size or certainty of deviation from the expected mortality rate.
| Observed to Expected Ratio
= DO/DE |
Clinician Example
A O-to-E Ratio = 1.5 Score = −0.22 |
Clinician Example
B O-to-E Ratio = 3 Score = −0.22 |
Clinician Example
C O-to-E Ratio = 3 Score = −2.89 |
||||
|---|---|---|---|---|---|---|---|
| Observed Dead (DO) |
Observed Alive (AO) |
9 | 70 | 3 | 17 | 30 | 170 |
| Expected Dead (DE) |
Expected Alive (AE) |
6 | 73 | 1 | 19 | 10 | 190 |
3.4. Patient Cohort Assembly
Using clinician-patient assignments defined in Step 3.3, we assembled three patient cohorts seen from 2010–2013: 1) Low-mortality (n=8,641) comprised of patients treated by one or more low-mortality clinicians but no high-mortality clinicians, 2) high-mortality (n=1,376), comprised of patients treated by one or more high-mortality clinicians but no low-mortality clinicians, and 3) crowd (n=64,598), comprised of the unfiltered superset of all patients. Low- and high-mortality patient cohorts are mutually exclusive.
3.5. Propensity Score Matching
As we are interested in comparing the effect of different clinician decision making behaviors, we need to account for underlying differences in patient characteristics and disease severity. To minimize biases arising from confounding covariates between patient cohorts assembled in Step 3.4, we conducted common-referent 1:1:K propensity score matching.[41] In this approach, we first conduct 1:1 propensity score matching between the low- and high-mortality patient cohorts (round 1), and subsequently conduct 1:K propensity score matching between the already matched low-mortality and unmatched crowd patient cohort (round 2). Using K=5 balanced post-matching standardized mean difference (SMD) similarity against data loss.
In both rounds of matching, using demographic data, initial vital signs recorded before the onset of care, initial lab tests, and Charlson Comorbidity Indices as covariates, we applied an un-regularized logistic regression model to compute the probability p of each patient’s assignment to the low-mortality patient cohort, defined as the propensity score to match on. We then conducted caliper matching on the logit of the propensity score , using caliper widths (maximum tolerated differences between matched patients) of 0.29 and 0.11 which were chosen based on 0.2×σ where σ = the pooled standard deviation of propensity score logits across each pair of unmatched patient cohorts.[42] This ultimately yielded balanced cohorts of 1,046, 1,046, and 5,230 (1:1:5) patients.
3.6. Association Rule Episode Mining
Using patient hospitalization data from each balanced patient cohort, we conducted association rule episode mining on clinical item pairs to capture historical clinician behavior. Our previously described clinical order recommender algorithm[21,31,43–44] counts co-occurrences for all clinical item pairs occurring within 24 hours to build time-stratified item association matrices. These counts are then used to populate 2×2 contingency tables to compute association statistics such as baseline prevalence, positive predictive value (PPV), relative risk (RR), and P-value by chi-square test for each pair of clinical items. Co-occurrence counts can also be extended to identify orders associated with groups of clinical items.
For a given query item (e.g. admission diagnosis) or set of query items (e.g. admission diagnosis followed by initial clinical items administered during a given patient visit), we can then generate a list of clinical order suggestions score-ranked by a specified association statistic. Score-ranking by PPV prioritizes items that are likely to occur after the query items,[45–46] whereas score-ranking by P-value for items with odds ratio >1 prioritizes orders that are disproportionately associated with the query items.[21] We trained three distinct association models using patient encounters from the balanced low-mortality, high-mortality, and crowd patient cohorts, each reflecting clinical order patterns from the corresponding clinician population.
3.7. Hospital-Authored Order Sets
Manually-authored hospital order sets are provided as a clinician resource for addressing common diagnoses and procedures, typically curated by hospital clinical committees. Existing order sets provide us a real-world, standard-of-care benchmark for decision support content.
3.8. Evaluation Against Real-World Practice Patterns
In automatically generating order lists, we seek to inform medical decision making that results in “successful” patient cases, which we define as encounters that yield better-than-expected patient mortality outcomes. Using the patient mortality predictor defined during clinician stratification (Step 3.3), we identified patients seen between 2010–2013 with 30-day mortality probabilities >0.5 who, against expectations, survived past the 30-day threshold. We then excluded patients inputted during association model training (n=136). In our first prediction task, we sought to emulate real-world practice patterns exhibited in these “successful” patient cases.
For each patient, we isolated usage instances of manually-authored hospital order sets within the first 24 hours of a hospitalization (n=426). We then generated a personalized order list at each such moment in time by querying each association model with the patient’s admission diagnosis and clinical orders administered up to the hospital order set usage instance.[45] For each instance, we compared outputted clinical order suggestions against the “successful” set of orders that actually occurred during the patient encounter within a verification window of 24 hours post-order set usage.
3.9. Evaluation Against Practice Guidelines
In our second prediction task, we automatically generated an “order set” given an admission diagnosis. We generated order lists from each association model for six common admission diagnoses: altered mental status (ICD9: 780.97), chest pain (ICD9: 786.5), gastrointestinal (GI) hemorrhage (ICD9: 578), heart failure (ICD9: 428), pneumonia (ICD9: 486), and syncope and collapse (ICD9: 780.2). These diagnoses were selected because they have a significant quantity of clinical data examples to study and relevant published guidelines and manually-authored hospital order sets to benchmark against.
We sought to evaluate each predicted order list’s alignment with clinical practice guidelines. To develop a guideline-based reference standard for order quality, two board-certified internal medicine physicians curated reference lists of clinical orders based on clinical practice literature available from the National Guideline Clearinghouse (www.guideline.gov) and PubMed that inform common practices like hospital management of chest pain,[47–48] gastrointestinal hemorrhage,[49–51] heart failure,[52–53] and pneumonia,[54–55] as well as less well-defined standards for management of syncope,[56–57] and altered mental status.[58] The physicians were instructed to include candidate clinical orders in their reference lists if a guideline explicitly mentioned them as appropriate to consider (e.g. treating pneumonia with levofloxacin), or heavily implied them (e.g. bowel preparation and NPO diet orders are implicitly necessary to fulfill explicitly recommended endoscopy procedures for gastrointestinal bleeds). For ambiguous cases, the physicians were instructed to consider whether the order was appropriate for inclusion in a general purpose order set for the given admission diagnosis. After independently producing their lists, the two physicians resolved disagreements (items included in one list but not the other) by consensus to produce a final reference standard (Appendix 7, see reference [59]). To assess pre-consensus agreement between the two clinicians, we computed Cohen’s Kappa statistics[60] ranging from −1 to +1, with values <0 indicating poor agreement and values >0.6 indicating substantial agreement (Appendix 7).[60]
3.10. Evaluation Metrics
In both prediction tasks, we ranked predicted order lists generated from low-mortality, high-mortality, and crowd association models by PPV and evaluated predicted order lists against the corresponding guideline reference list or “successful” order history using area under the receiver operating characteristic (AUROC) and precision and recall for the top K ranked items. Comparison of such metrics sought to determine how association models differed in their i) alignment with clinical practice guidelines and ii) ability to emulate “successful” real-world patient cases.
3.11. Additional Analyses
We conducted a number of supplemental analyses to validate the robustness of our study design.
To assess the stability of our clinician stratification method relative to other potential quality measures, we compared clinician cohorts stratified by 30-day mortality against those stratified by two alternative outcome variables, 30-day readmission and joint 30-day mortality or readmission, following the same P-value based approach (Appendix 1).
To assess whether physicians assigned to low- and high-mortality cohorts held majority responsibility for patients attributed to them, we quantified clinician responsibility using a shared-attribution model based on daily H&P and progress notes (e.g. a provider who signs three out of five notes is responsible for 60% of a patient’s hospitalization).[61]
Manually-authored hospital order sets are provided as a resource for clinicians to utilize, influencing clinical order entry. As such, association models may recapitulate pre-authored order set templates in lieu of truly capturing individual clinical practice patterns. To gauge the influence of order set templates on learned patterns, we compared association models trained on clinical order data with or without the inclusion of real-world order set usage (Appendix 2).
In addition to comparing predicted order lists against reference standards derived from better-than-expected patient cases and practice guidelines, we can also directly compare similarity among predicted order lists. For this task, traditional measures of list agreement like Kendall’s τ-metric[62] are not ideal as they often require identically-sized, finite lists, and weigh all list positions equivalently. To compare scored order lists, we instead calculated agreement by Rank Biased Overlap (RBO),[63] which accounts for rank-order (see Table 3 in reference [59] for full RBO description and results). RBO values range from 0.0 (no correlation or random list order) to 1.0 (perfect agreement).
In our study design, we isolate low-mortality clinicians in an attempt to curate a high-quality clinician cohort from the bottom-up based on patient outcomes. Similarly, one could also begin with the full clinician crowd and filter out high-mortality clinicians, curating from the top-down. To compare these two approaches, we evaluated association models trained on an unfiltered crowd and top-down filtered crowd (Appendix 5).
4. RESULTS
Figure 2 shows the distribution of clinician performance scores alongside observed vs. expected (O:E) patient mortality ratios for all clinicians who saw patients between 2010 and 2013. Performance scores correlate with O:E ratios but are also dependent on patient count and effect size.
Figure 2. Distribution of clinician performance scores and corresponding patient cohort sizes among 2010–2013 clinicians.
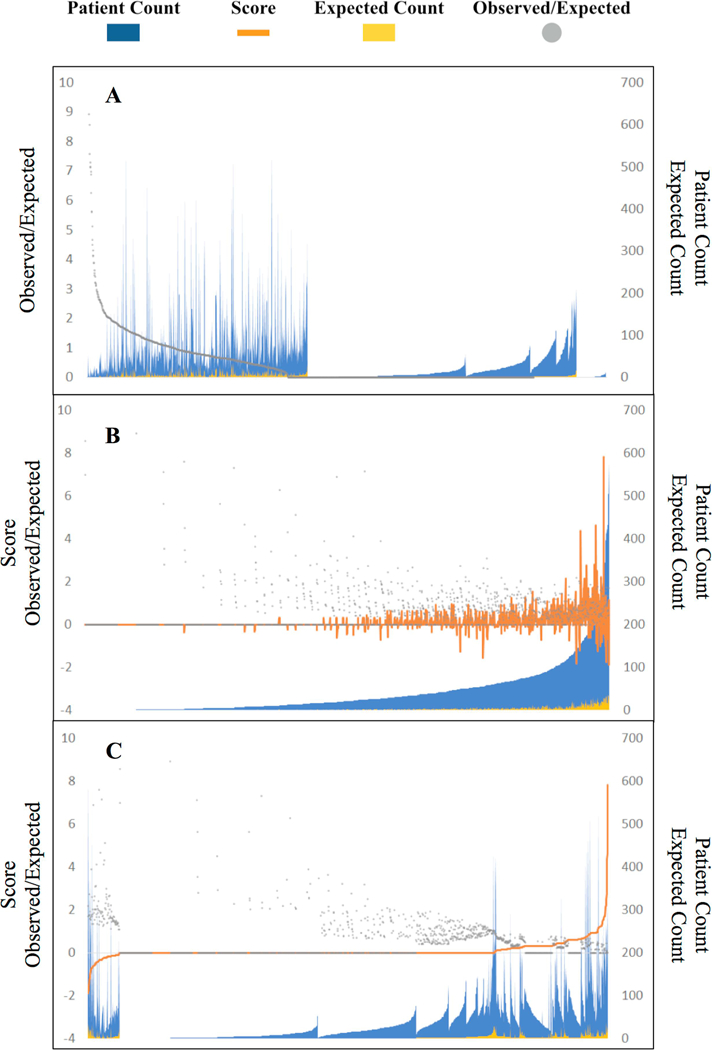
Each x-axis position represents one clinician considered with blue peaks representing the number of patients whose treatment was attributed to the clinician. Figure 1A illustrates clinicians sorted purely by observed-to-expected patient mortality ratio (gray points). The raw ratio proves unstable as a metric. Nearly half of clinicians show a “perfect” ratio of 0, mostly due to small patient cohort size, and a small cluster of clinicians on the right show an undefined ratio when the expected death denominator is zero. Figure 1B sorts clinicians by total patient count attributed to them. The clinician performance score (orange curves) accounts for both effect size of observed-to-expected patient mortality as well as certainty in those rate estimates based on the quantity of patient data available for each clinician. Figure 1C sorts clinicians by performance score, illustrating that by design the majority of clinicians (72.2%, n=1315 of 1822) are left unstratified in the middle range with an Sj score of zero, largely given patient cohort sizes too small to draw statistically detectable conclusions. Only clinicians at the extremes who demonstrate substantial deviation from expected norms are stratified into low- and high-mortality cohorts.
Appendix 4 shows covariate distributions for low-mortality, high-mortality, and crowd patient cohorts after 1:1:5 common referent matching. Post-matching standardized mean differences (SMD) were <0.2 across all covariates, indicating an insignificant difference between balanced patient cohorts.
Table 2 shows examples of clinical orders disproportionately associated with a given admission diagnosis generated by low-mortality and crowd association models for six admission diagnoses of interest (see reference [59] for extended order lists generated by all three association models). Many predicted orders are shared across cohorts, although their relative ordering and specific association statistics differ. Across all six diagnoses, the three association models exhibit substantial pairwise agreement overall as indicated by RBO values in Appendix 3 ranging from ~0.6–0.7.[59]
Table 2.
Example top ranked clinical order associations for six example admission diagnoses predicted by association models trained on low-mortality and crowd patient cohorts, sorted by P-value calculated by Yates’ chi-squared statistic. We also show positive predictive value (PPV) with 95% confidence intervals and a column denoting the presence or absence of each item in the corresponding human-authored hospital order set and guideline reference standard are included. Items with baseline prevalence <1% are excluded to avoid statistically spurious results. Although the individual item orderings and range of score metrics differ between models, the overarching order lists have sizeable overlap as computed by Rank Biased Overlap (RBO Table, Appendix 3).
| Pneumonia (ICD9: 486) | |||||||
| Low-Mortality Clinician Model Predictions | Crowd Clinician Model Prediction | ||||||
| Clinical Order |
Positive Predictive Value |
P-Value |
Order
Set/ Guideline |
Clinical Order |
Positive Predictive Value |
P-Value |
Order
Set/ Guideline |
| Levofloxacin (Intravenous) |
57% (36%,77%) |
3×10−18 | Yes/Yes | Levofloxacin (Intravenous) |
43% (34%,53%) |
4×10−29 | Yes/Yes |
| Oseltamivir (Oral) | 9% (0%,20%) |
8×10−3 | No/Yes | Blood
Culture (Aerobic & Anaerobic Bottles) |
85% (78%,92%) |
2×10−28 | Yes/Yes |
| Respiratory Sputum Induction |
13% (0%,27%) |
1×10−2 | No/Yes | Blood Culture (2 Aerobic Bottles) |
83% (76%,91%) |
1×10−27 | Yes/Yes |
| Quantiferon Test for Latent TB |
9% (0%,20%) |
2×10−2 | No/No | Azithromycin (Intravenous) |
25% (16%,33%) |
4×10−24 | Yes/Yes |
| Respiratory Culture | 26% (8%,44%) |
6×10−2 | Yes/No | Point-of-Care Venous Blood Gases and Lactate |
46% (36%,56%) |
2×10−15 | Yes/Yes |
| … | … | … | … | … | … | … | … |
| Altered Mental Status (ICD9: 780.97) | |||||||
| Low-Mortality Clinician Model Predictions | Crowd Clinician Model Prediction | ||||||
| Clinical Order |
Positive Predictive Value |
P-Value |
Order
Set/ Guideline |
Clinical Order |
Positive Predictive Value |
P-Value |
Order
Set/ Guideline |
| Point-of-Care Na, K, Glucose, Hematocrit, Hemoglobin |
14% (4%,24%) |
3×10−7 | No/No | CT Head | 59% (51%,68%) |
2×10−30 | Yes/Yes |
| Labetalol (Intravenous) |
7% (0%,15%) |
2×10−2 | No/No | Ammonia Plasma | 21% (14%,28%) |
3×10−18 | No/Yes |
| Consult to Medicine |
16% (5%,27%) |
3×10−2 | No/No | Point-of-Care Troponin I |
47% (39%,56%) |
9×10−16 | Yes/Yes |
| CT Head | 28% (15%,41%) |
4×10−2 | Yes/Yes | Point-of-Care Venous Blood Gases and Lactate |
39% (30%,47%) |
2×10−12 | No/Yes |
| Point-of-Care Creatinine |
19% (7%,30%) |
5×10−2 | No/No | Metabolic Panel Comprehensive |
89% (84%,94%) |
9×10−10 | Yes/Yes |
| … | … | … | … | … | … | … | … |
| Chest Pain (ICD9: 786.5) | |||||||
| Low-Mortality Clinician Model Predictions | Crowd Clinician Model Prediction | ||||||
| Clinical Order |
Positive Predictive Value |
P-Value |
Order
Set/ Guideline |
Clinical Order |
Positive Predictive Value |
P-Value |
Order
Set/ Guideline |
| Point-of-Care Troponin I Test |
32% (17%,46%) |
3×10−4 | Yes/Yes | Point-of-Care Troponin I Test |
69% (62%,76%) |
8×10−61 | Yes/Yes |
| Clopidogrel (Oral) | 16% (4%,27%) |
5×10−4 | Yes/Yes | Troponin Lab Test | 72% (65%,79%) |
1×10−32 | Yes/Yes |
| Nitroglycerin (Sublingual) |
8% (0%,16%) |
2×10−3 | Yes/Yes | Consult to Cardiology |
25% (18%,31%) |
7×10−31 | No/Yes |
| Creatine
Kinase MB (Mass) |
37% (22%,52%) |
1×10−2 | Yes/Yes | Nitroglycerin (Sublingual) |
13% (8%,19%) |
1×10−28 | Yes/Yes |
| Diet (Low Sodium, Low Cholesterol, Low Saturated Fat) |
16% (4%,27%) |
5×10−2 | Yes/Yes | Aspirin (Oral) | 66% (58%,73%) |
2×10−25 | Yes/Yes |
| … | … | … | … | … | … | … | … |
| Syncope and Collapse (ICD9: 780.2) | |||||||
| Low-Mortality Clinician Model Predictions | Crowd Clinician Model Prediction | ||||||
| Clinical Order |
Positive Predictive Value |
P-Value |
Order
Set/ Guideline |
Clinical Order |
Positive Predictive Value |
P-Value |
Order
Set/ Guideline |
| Point-of-Care Troponin I Test |
25% (4%,46%) |
0.2 | Yes/Yes | Point-of-Care Troponin I Test |
74% (64%,83%) |
3×10−35 | Yes/Yes |
| Aspirin (Oral) | 31% (9%,54%) |
0.3 | No/No | 12-Lead Electrocardiogram |
93% (87%,98%) |
1×10−9 | Yes/Yes |
| Thyroid- Stimulating Hormone |
25% (4%,46%) |
0.3 | No/No | Troponin Lab Test | 51% (40%,62%) |
3×10−5 | Yes/Yes |
| Carvedilol (Oral) | 13% (0%,29%) |
0.4 | No/No | Donepezil (Oral) | 6% (1%,12%) |
8×10−5 | No/No |
| Lovastatin (Oral) | 6% (0%,18%) |
0.4 | No/No | CT Head | 36% (26%,47%) |
2×10−4 | Yes/Yes |
| … | … | … | … | … | … | … | … |
| Heart Failure (ICD9: 428) | |||||||
| Low-Mortality Clinician Model Predictions | Crowd Clinician Model Prediction | ||||||
| Clinical Order |
Positive Predictive Value |
P-Value |
Order
Set/ Guideline |
Clinical Order |
Positive Predictive Value |
P-Value |
Order
Set/ Guideline |
| Isosorbide Mononitrate (Oral) |
13% (0%,29%) |
5×10−3 | Yes/Yes | N-Terminal Pro- Brain Natriuretic Peptide |
75% (66%,85%) |
6×10−64 | Yes/Yes |
| Free
Thyroxine Test |
25% (4%,46%) |
2×10−2 | No/No | Furosemide (Intravenous) |
70% (60%,80%) |
2×10−22 | Yes/Yes |
| Furosemide (Intravenous) |
44% (19%,68%) |
3×10−2 | Yes/Yes | Consult to Cardiology |
27% (17%,37%) |
9×10−19 | Yes/Yes |
| Nitroglycerin (Topical) |
13% (0%,29%) |
3×10−2 | Yes/Yes | Hydralazine (Oral) | 21% (12%,30%) |
2×10−17 | Yes/Yes |
| Abdominal Ultrasound |
13% (0%,29%) |
0.3 | No/No | Point-of-Care Troponin I Test |
57% (46%,68%) |
4×10−17 | No/Yes |
| … | … | … | … | … | … | … | … |
| Gastrointestinal Hemorrhage (ICD9: 578) | |||||||
| Low-Mortality Clinician Model Predictions | Crowd Clinician Model Prediction | ||||||
| Clinical Order |
Positive Predictive Value |
P-Value |
Order
Set/ Guideline |
Clinical Order |
Positive Predictive Value |
P-Value |
Order
Set/ Guideline |
| Bowel
Preparation Electrolyte Solution |
9% (1%,18%) |
2×10−4 | Yes/Yes | Consult to Gastroenterology |
27% (20%,35%) | 1×10−55 | Yes/Yes |
| Pantoprazole (Intravenous) |
42% (27%,57%) |
1×10−2 | Yes/Yes | Octreotide (Intravenous) |
21% (14%,28%) | 2×10−50 | Yes/Yes |
| Octreotide (Intravenous) |
5% (0%,11%) |
0.2 | Yes/Yes | Bowel
Preparation Electrolyte Solution |
19% (12%,25%) | 9×10−28 | Yes/Yes |
| Hemoglobin | 5% (0%,11%) |
0.4 | No/No | Type and Screen | 87% (82%,93%) |
3×10−23 | Yes/Yes |
| Pravastatin (Oral) | 5% (0%,11%) |
0.5 | No/No | Pantoprazole (Intravenous) |
75% (68%,83%) |
2×10−20 | Yes/Yes |
| … | … | … | … | … | … | … | … |
Figure 3 shows mean AUROC, precision, and recall values obtained by each association model in predicting personalized order lists to emulate real-world practice patterns reflected in 426 better-than-expected patient cases. In this first evaluation task, the crowd model (mean AUROC 0.91) outperformed the low-mortality model (0.87, P<10−16) and order set benchmarks (0.78, P<10−35).
Figure 3. Mean AUROC, precision, and recall metrics from evaluating predicted personalized order lists against real-world practice patterns reflected in patient cases with better-than-expected mortality outcomes (n=426 order set usage instances).
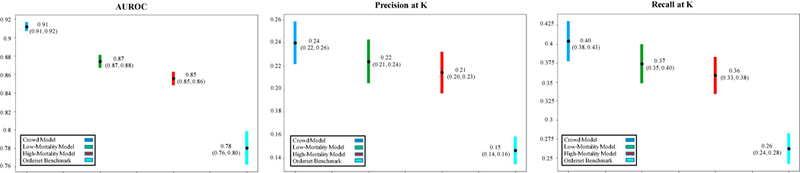
Predictions were generated each time a manually-authored hospital order set was used within 24 hours of a hospitalization. Each chart compares the performance of low-mortality, high-mortality, and crowd association models and corresponding hospital order set benchmarks in emulating the “successful” orders actually placed 24 hours post-order set usage. Mean values are plotted alongside 95% confidence interval bands empirically estimated by bootstrap resampling with replacement 1000 times. The crowd model emulates better-than-expected, real-world practice patterns as well as, or better than, the low-mortality model, high-mortality model, and order set benchmarks. K=the number of items in the corresponding hospital order set denoting the usage instance. AUROC: area under the receiver operating characteristic curve.
Figure 4 and Appendix 6 show ROC plots and precision-recall curves, respectively, evaluating alignment of automatically-learned “order sets” generated by each association model to guideline reference standards for the six admission diagnoses. In this second evaluation task, the crowd model similarly demonstrated the greatest alignment with guideline references (AUROC range 0.86–0.91), performing on par or better than the low-mortality clinician model (0.79–0.84, P<10−5) and manually-authored hospital order sets (0.65–0.77, P<10−3).
Figure 4. ROC plots evaluating predicted order lists against practice guideline reference standards for six example diagnoses.
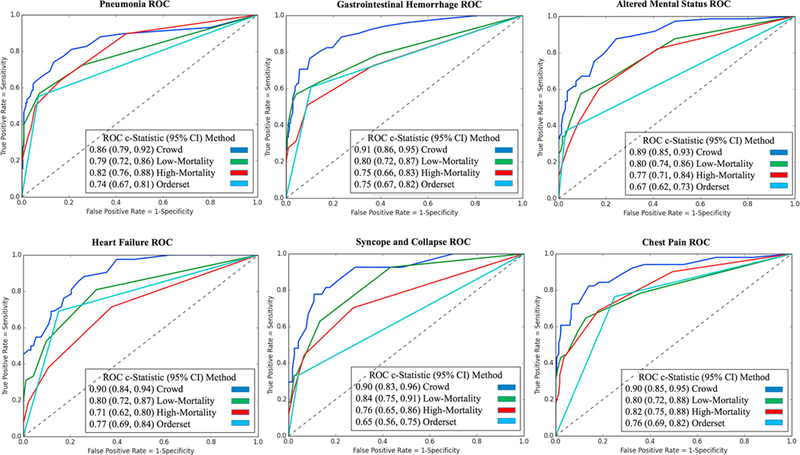
Each plot compares an order set authored by the hospital and automated predictions from low-mortality, high-mortality, and crowd association models. Pre-authored order sets have no inherent ranking or scoring system to convey relative importance and are thus depicted as a single discrete point on the ROC curve. Area-under-curve (AUROC) is reported as c-statistics with 95% confidence intervals empirically estimated by bootstrap resampling with replacement 1000 times. The unfiltered crowd of clinicians generates predictions that align with clinical practice guidelines as much as or more robustly than a cherry-picked subset of clinicians or manually-authored order sets. ROC: receiver operating characteristic.
5. DISCUSSION
In this study, we validate two trends. First, automatically learning clinical practice patterns from electronic medical records can generate decision support content that is more robust than conventional, manual methods. Second, content learned from an unfiltered crowd of all clinicians is as, or more robust, than that learned from a cherry-picked clinician subset favored by patient mortality outcomes. These findings are consistent across AUROC, precision, and recall metrics in two distinct evaluation tasks: i) emulation of real-world practice patterns represented in patient cases with better-than-expected mortality outcomes (Figure 3) and ii) alignment with clinical practice guidelines for six common admission diagnoses (Figure 4, Appendix 6).
These results may seem surprising when individual clinicians can exhibit substantial practice variability. While some clinicians will certainly make poor decisions sometimes, Condorcet’s Jury Theorem[24] posits that aggregating the non-random decisions of many converges towards correctness. This is the same argument behind the wisdom-of-the-crowd[23] and ensemble-based machine-learning algorithms that generate strong classifiers from individually weak ones.[64] Whether such models are better trained on all available cases or a cherry-picked subset of clinical decision makers illustrates a bias-variance tradeoff.[65] Intuition may suggest isolating “better” clinicians or excluding low-performing ones (Appendix 5). Instead the crowd model, which simply aggregates all available data, shows greater alignment in relation to the low-mortality model across both clinical practice guidelines (AUROC range 0.86–0.91 vs. 0.79–0.84, P<10−5) and real-world practice patterns (mean AUROC 0.91 vs. 0.87, P<10−16). Selecting a cohort of low-mortality clinicians can reduce bias in learned practice patterns towards more desirable medical decisions, but also reduces the amount of data available to the learning algorithm and can thus increase variance in model estimations.
The key risks and limitations of this study point to the importance of its contribution. Here, we focus on patient mortality as a concrete patient-centered outcome; this presumes mortality is undesirable and preventable. However, a doctor who realigns the goals of care for a terminally-ill patient towards end-of-life hospice treatment may well represent “better” treatment than one who reflexively keeps patients alive on artificial life support for prolonged periods. We do not expect this to significantly alter our usage of death as an undesirable outcome however, as less than 2% (1,262/74,880) of patients studied were ever treated with care goals purely directed towards “Comfort Care Measures.” Factoring treatment team into the expected mortality predictor was similarly important to account for different expectations of clinicians in different specialties (e.g. medical vs. surgical). Even after accounting for the aforementioned, the potential vagaries of interpretation mean we do not advise using any such scoring method to credibly distinguish care quality at the individual level. Indeed, we specifically avoid labeling individual physicians as “good, bad, correct, or incorrect” as it is difficult to reliably assign causality between individual order patterns and patient outcomes. Instead, we identify clinician populations and their practice patterns most associated with “better” patient outcomes (e.g. better-than-expected mortality). While we do not expect our system to reliably make distinctions between clinicians ranked 10 vs. 15, it is sufficient to discriminate between the top 100 vs. bottom 100. The discriminating power of the underlying mortality predictor (AUROC=0.86) is comparable with state-of-the-art mortality predictors [21,39,66]. As such, we could confidently risk-stratify a population of high-mortality clinicians from low-mortality ones. We gain further reassurance in noting that alternative clinician stratification metrics such as 30-day readmission and joint 30-day readmission or mortality produced clinician subgroups that substantially overlapped with those stratified by our 30-day mortality approach (Appendix 1). A final consideration regarding stratification methodology is physician-patient attribution in determining a clinician’s observed versus expected patient mortality rate. Multiple physicians are often responsible for a given patient’s hospitalization, distributing responsibility. In our shared-attribution analysis, we observe that attributed clinicians were responsible for the majority of patient stays, such as the high-mortality clinicians who were responsible on average for 69.2% of their patients’ hospitalization durations. Attributing mortality to the admitting attending (e.g. via H&P notes signed upon admission) rather than the discharging attending is preferable, as key diagnostic and management decisions that occur early in a patient’s care are often major drivers of patient outcomes like mortality.[67–68]
In clinical practice, the treatment regimen for an initial admission diagnosis becomes increasingly specific as the clinical investigation progresses. For heterogeneous diagnoses, a noted concern is the suitability of order set benchmarks, which may not accurately reflect real-life patient encounters. For example, the SUMC order set for altered mental status contains clinical orders used in specific manifestations of the diagnosis (e.g. naloxone to treat emergency narcotic overdose, piperacillin-tazobactam to treat sepsis) and broad spectrum orders (e.g. CT head to assess head injuries generally). However, order sets are not necessarily meant to function as explicit checklists for any one patient encounter; instead, they provide suggestions encompassing diverse possibilities to help clinicians recall potential treatment pathways. To improve upon prevailing order set curation methods, hospital order sets remain the standard-of-care benchmark although heterogeneous. A similar concern is the heterogeneity of automatically-generated order lists (Appendix 8). Automatically-curated content, like hospital order sets, provide a superset of investigations and treatments that could be the root cause for a given diagnosis. Indiscriminately following automatic suggestions could lead to over-treatment and testing. Automated decision support content is not meant to supplant complex medical decision making. Instead, such datadriven suggestions can reduce information load by enabling physicians to recognize rather than recall diverse treatment options during a patient assessment.
Notably, that there is no universal gold standard to define “good” medical decision making. Yet the impact of medical decision making on patient care makes it all the more important to develop reasonable and reproducible references to better understand these processes (see Appendix 7 and reference [59] for guideline reference standards). Here, we introduce clinical practice guidelines as an external reference standard to evaluate learned clinical order patterns, building upon previous internal statistical benchmarks (e.g. emulating existing clinical order patterns based on historical trends).[45] Although natural language processing approaches to interpreting clinical practice literature is an active field of research,[69–70] translating guidelines into reproducible and verifiable constructs (e.g. discrete clinical orders CPOE system input) still requires significant human interpretation. To mitigate variability, two physicians independently developed reference lists based on their readings of clinical practice guidelines. This yielded substantial agreement with Cohen’s Kappa values >0.6 for all diagnoses addressed (Appendix 7), offering reassurance in the stability of this reference standard. Examples of differences between the two physicians included when only one physician counted guideline references to highly conditional use of uncommon interventions (e.g. Factor IX for GI bleeds), general hospital admission orders not specifically related to the diagnosis in question (e.g. physical therapy and subcutaneous heparin orders), or intensive care unit level interventions which guidelines advised be used only if multiple other treatment modalities failed (e.g. dobutamine and nesiritide for heart failure). These were ultimately reconciled by consensus as not appropriate to include in the reference standard, based on the principle that they would not be expected for inclusion in a general purpose default “order set” for the respective admission diagnosis. Even with consistency in this reference standard, a fair question is whether clinical practice guidelines actually define “good” medical care.[71–73] To more directly identify optimal medical decision making, we introduce a second evaluation metric based on “successful” patient cases, reflected in encounters with better-than-expected mortality outcomes. Consistent trends in both evaluations provides multiple validations of our results.
In applying either evaluation methodology, we highlight the practical engineering tradeoffs that arise when deciding to cherry-pick a subset of expert clinicians or learn from an unfiltered crowd. This is a natural decision that arises when curating training data for medical informatics applications. The relative stability of patterns learned from both low-mortality and the crowd models (represented by RBO scores in Appendix 3) reflects that either approach converges towards a common basis of relevant content (e.g. established guideline-based practices). In the end, even more pointed are clinical scenarios where no robust practice guidelines exist. The recommender algorithm reviewed here is always able to produce some suggestions based on historical practice patterns. The quality of such suggestions is inherently difficult to evaluate without a proper reference standard. Here, we introduce a data-driven methodology to evaluate order predictions against “successful” real-world patient cases, even in the absence of practice guidelines. The consistent alignment of automatically-generated order lists with both practice guideline standards externally defined from the top-down and real-world practice patterns internally curated from the bottom-up gives us confidence in the utility of data-driven approaches.
6. CONCLUSIONS
A clinician scoring system based on P-values of observed vs. expected 30-day patient mortality rates can stratify clinician cohorts by taking into account combined effect size and certainty of deviation from expected norms. Automatically learning clinical practice patterns from historical EHR data can generate decision support content that aligns with clinical practice guidelines and emulates real-world practice patterns as well as, if not better than, conventional manually-authored content. Learning decision support from data generated by all clinicians using this approach is as, or more, robust than selecting a subgroup of clinicians favored by patient mortality data.
Supplementary Material
Highlights.
Patterns from clinical order entry data can yield relevant decision support content
Automatic patterns outperform manually-authored order sets by multiple metrics
Deviation in observed from expected patient outcomes can stratify clinicians
Patterns from all clinicians prove more robust than those from “preferred” clinicians
ACKNOWLEDGEMENTS
We would like to acknowledge Joyce B. Kang for technical assistance.
FUNDING
This research was supported by the NIH Big Data 2 Knowledge initiative via the National Institute of Environmental Health Sciences under Award Number K01ES026837. Patient data were extracted and de-identified by the Stanford Medicine Research Data Repository (StaRR) project with support from the Stanford NIH/National Center for Research Resources CTSA award number UL1 RR025744. A Stanford Undergraduate Advising and Research Travel Grant supported JKW in presenting this work at the 2017 American Medical Informatics Association Annual Symposium. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or Stanford Healthcare.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
COMPETING INTERESTS
The authors have no competing interests to declare.
REFERENCES
- 1.Richardson WC, Berwick DM, Bisgard JC. et al. Crossing the Quality Chasm: A New Health System for the 21st Century Washington DC: Natl Acad Press, Institute of Medicine, Committee on Quality of Health Care in America Committee on Quality of Health Care in America; 2001. [Google Scholar]
- 2.Lauer MS, Bonds D. Eliminating the ‘expensive’ adjective for clinical trials. Am Heart J 2014;167:419–20. [DOI] [PubMed] [Google Scholar]
- 3.Tricoci P, Allen JM, Kramer JM. et al. Scientific evidence underlying the ACC/AHA clinical practice guidelines. JAMA 2009;301:831–41. [DOI] [PubMed] [Google Scholar]
- 4.Durack DT. The weight of medical knowledge. N Engl J Med 1978;298:773–5. [DOI] [PubMed] [Google Scholar]
- 5.Kaushal R, Shojania KG, Bates DW. Effects of computerized physician order entry and clinical decision support systems on medication safety: a systematic review. Arch Intern Med 2003;163:1409–16. [DOI] [PubMed] [Google Scholar]
- 6.Kawamoto K, Houlihan CA, Balas EA, Lobach DF. Improving clinical practice using clinical decision support systems: a systematic review of trials to identify features critical to success. BMJ 2005;330:765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ballard DW, Kim AS, Huang J. et al. Implementation of computerized physician order entry is associated with increased thrombolytic administration for emergency department patients with acute ischemic stroke. Ann Emerg Med 2015;1–10. [DOI] [PMC free article] [PubMed]
- 8.Ballesca MA, LaGuardia JC, Lee PC. et al. An electronic order set for acute myocardial infarction is associated with improved patient outcomes through better adherence to clinical practice guidelines. J Hosp Med 2014;9:155–61. [DOI] [PubMed] [Google Scholar]
- 9.Hripcsak G, Albers DJ, Perotte A. Exploiting time in electronic health record correlations. J. Am. Med. Informatics Assoc 2011;109–115. [DOI] [PMC free article] [PubMed]
- 10.Mitchell JA, Gerdin U, Lindberg DAB, et al. 50 years of informatics research on decision support: What’s next. Methods of Information in Medicine 2011;50:525–535. [DOI] [PubMed] [Google Scholar]
- 11.Sittig DF, Wright A, Osheroff JA. et al. Grand challenges in clinical decision support. J Biomed Inform 2008;41:387–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Longhurst CA, Harrington RA, Shah NH. A ‘Green Button’ for using aggregate patient data at the point of care. Health Aff 2014;33:1229–35. [DOI] [PubMed] [Google Scholar]
- 13.Doddi S, Marathe A, Ravi SS, et al. Discovery of association rules in medical data. Med. Inform. Internet Med 2001;26:25–33. [PubMed] [Google Scholar]
- 14.Klann J, Schadow G, Downs SM. A method to compute treatment suggestions from local order entry data. AMIA Annu. Symp. Proc 2010:387–391. [PMC free article] [PubMed]
- 15.Klann J, Schadow G, McCoy JM. A recommendation algorithm for automating corollary order generation. AMIA Annu. Symp. Proc 2009:333–337. [PMC free article] [PubMed]
- 16.Wright A, Sittig DF. Automated development of order sets and corollary orders by data mining in an ambulatory computerized physician order entry system. AMIA Annu. Symp. Proc 2006: 819–823. [PMC free article] [PubMed]
- 17.Zhang Y, Padman R, Levin JE. Paving the COWpath: data-driven design of pediatric order sets. J. Am. Med. Inform. Assoc 2014;21:304–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klann JG, Szolovits P, Downs SM, et al. Decision support from local data: creating adaptive order menus from past clinician behavior. J. Biomed. Inform 2014;48:84–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wright AP, Wright AT, McCoy AB, et al. The use of sequential pattern mining to predict next prescribed medications. J. Biomed. Inform 2014;53:73–80. [DOI] [PubMed] [Google Scholar]
- 20.Wright A, Chen ES, Maloney FL. An automated technique for identifying associations between medications, laboratory results and problems. J. Biomed. Inform 2010;43:891–901. [DOI] [PubMed] [Google Scholar]
- 21.Chen JH, Altman RB. Automated physician order recommendations and outcome predictions by data-mining electronic medical records. Proc AMIA Summit Transl Sci 2014;2014:206–10. [PMC free article] [PubMed] [Google Scholar]
- 22.Linden G, Smith B, York J. Amazon.com recommendations: item-to-item collaborative filtering. IEEE Internet Comput 2003:76–80.
- 23.Surowiecki J The wisdom of crowds: why the many are smarter than the few and how collective wisdom shapes business, economies, societies, and nations Anchor: Doubleday, 2004. [Google Scholar]
- 24.Berend D, Paroush J. 1998. When Is Condorcet’s Jury Theorem Valid? Social Choice and Welfare 2015;4:481–88. [Google Scholar]
- 25.Wang JK, Schuler A, Shah NH, et al. Inpatient Clinical Order Patterns Machine-Learned From Teaching Versus Attending-Only Medical Services. AMIA Joint Summits on Translational Science Proceedings 2018;2017;226–235. [PMC free article] [PubMed] [Google Scholar]
- 26.Best WR, Cowper DC. The Ratio of Observed-to-Expected Mortality as a Quality of Care Indicator in Non-Surgical VA Patients. Medical Care 1994;32:390–400. [DOI] [PubMed] [Google Scholar]
- 27.Stanford Medicine Research IT. Stanford Medicine Research Data Repository Available at: https://med.stanford.edu/researchit.html. Accessed October 2017.
- 28.Hernandez P, Podchiyska T, Weber S, Ferris T, Lowe H. Automated mapping of pharmacy orders from two electronic health record systems to RxNorm within the STRIDE clinical data warehouse. AMIA Annu Symp Proc 2009;2009:244–8. [PMC free article] [PubMed] [Google Scholar]
- 29.Quan H, Sundararajan V, Halfon P, et al. Coding Algorithms for Defining Comorbidities in ICD-9-CM and ICD-10 Administrative Data. Medical Care 2005;43:1130–39. [DOI] [PubMed] [Google Scholar]
- 30.Wright A, Bates DW. Distribution of problems, medications and lab results in electronic health records: the pareto principle at work. Appl Clin Inform 2010;1:32–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen JH, Muthuraman A, Goldstein MK, et al. Decaying Relevance of Clinical Data towards Future Decisions in Data-Driven Inpatient Clinical Order Sets. International Journal of Medical Informatics 2017;102:71–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Medicare.gov. Hospital Compare Available at: https://www.medicare.gov/hospitalcompare/Data/Measure-groups.html. Accessed October 2017.
- 33.Agency for Healthcare Research and Quality. Mortality Measurement Available at: http://www.ahrq.gov/qual/mortality. Accessed June 2018. [DOI] [PubMed]
- 34.US News & World Reports Health. How and Why We Rank and Rate Hospitals Available at: https://health.usnews.com/health-care/best-hospitals/articles/faq-how-and-why-we-rank-and-rate-hospitals. Accessed August 2018.
- 35.DeLancey JO, Softcheck J, Chung JW, et al. Associations Between Hospital Characteristics, Measure Reporting, and the Centers for Medicare & Medicaid Services Overall Hospital Quality Star Ratings. JAMA: The Journal of the American Medical Association 2017;317(19):2015– 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.MacLean CH, Kerr EA, Qaseem A Time Out - Charting a Path for Improving Performance Measurement. The New England Journal of Medicine 2018;378(19):1757–1761. [DOI] [PubMed] [Google Scholar]
- 37.Knaus WA, Draper EA, Wagner DP, et al. APACHE II: A Severity of Disease Classification System. Critical Care Medicine 1985;13:818–29. [PubMed] [Google Scholar]
- 38.Le Gall JR, Lemeshow S, Saulnier F. A New Simplified Acute Physiology Score (SAPS II) Based on a European/North American Multicenter Study. JAMA 1993;270:2957–63. [DOI] [PubMed] [Google Scholar]
- 39.Lemeshow S, Le Gall JR. Modeling the Severity of Illness of ICU Patients. A Systems Update. JAMA 1994;272:1049–55. [PubMed] [Google Scholar]
- 40.Tibshirani R Regression Shrinkage and Selection via the Lasso. Journal of the Royal Statistical Society. Series B, Statistical Methodology 1996;58:267–288. [Google Scholar]
- 41.Sugawara E, Nikaido H. Properties of AdeABC and AdeIJK Efflux Systems of Acinetobacter Baumannii Compared with Those of the AcrAB-TolC System of Escherichia Coli. Antimicrobial Agents and Chemotherapy 2014;58:7250–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rassen JA, Shelat AA, Franklin JM, et al. Matching by Propensity Score in Cohort Studies with Three Treatment Groups. Epidemiology 2013;24:401–409. [DOI] [PubMed] [Google Scholar]
- 43.Austin PC. Optimal caliper widths for propensity-score matching when estimating differences in means and differences in proportions in observational studies. Pharm Stat 2011:150–161. [DOI] [PMC free article] [PubMed]
- 44.Chen JH, Goldstein MK, Asch SM, et al. Dynamically Evolving Clinical Practices and Implications for Predicting Medical Decisions. Pacific Symposium of Biocomputing 2016. [PMC free article] [PubMed]
- 45.Chen JH, Goldstein MK, Asch SM, et al. Predicting inpatient clinical order patterns with probabilistic topic models vs conventional order sets. JAMIA 2017;24:472–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Manrai AK, Bhatia G, Strymish J, et al. , Medicine’s uncomfortable relationship with math: calculating positive predictive value. JAMA Intern. Med 2014:991–993. [DOI] [PMC free article] [PubMed]
- 47.Institute for Clinical Systems Improvement (ICSI). Diagnosis and Treatment of Chest Pain and Acute Coronary Syndrome (ACS) 2012.
- 48.National Institute for Health and Care Excellence (NICE). Chest pain of recent onset pain or discomfort of suspected cardiac origin 2010. [PubMed]
- 49.Accounting and Corporate Regulatory Authority (ACRA). Radiologic Management of Lower Gastrointestinal Bleeding 2011:8–13.
- 50.Laine L, Jensen DM. Management of patients with ulcer bleeding. Am J Gastroenterol 2012:345–360. [DOI] [PubMed]
- 51.National Health Service (NHS). Acute upper gastrointestinal bleeding: management 2012.
- 52.National Institute for Health and Care Excellence (NICE). Acute heart failure: diagnosis and management management 2014. [PubMed]
- 53.Yancy CW, Jessup M, Bozkurt B, et al. 2013. ACCF/AHA Guideline for the Management of Heart Failure: A Report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. Circulation 2013:240–327. [DOI] [PubMed] [Google Scholar]
- 54.British Thoracic Society (BTS). Guidelines for the Management of Community Acquired Pneumonia in Adults Update 2009: A Quick Reference Guide 2009.
- 55.Mandell L, Wunderink RG, Anzueto A, et al. Infectious Diseases Society of America/American Thoracic Society consensus guidelines on the management of community-acquired pneumonia in adults. Clin Infect Dis 2007:27–72. [DOI] [PMC free article] [PubMed]
- 56.Moya A Guidelines for the diagnosis and management of syncope. he Task Force for the Diagnosis and Management of Syncope of the. Eur. Heart J 2009:2631–2671. [DOI] [PMC free article] [PubMed]
- 57.McDermott D, Quinn J. Approach to the adult patient with syncope in the emergency department Available at: https://www.uptodate.com/contents/approach-to-the-adult-patient-with-syncope-in-the-emergency-department. Accessed October 2017.
- 58.Xiao H, Wang Y, Xu T, et al. Evaluation and treatment of altered mental status patients in the emergency department: Life in the fast lane. World J. Emerg. Med 2012:270–277. [DOI] [PMC free article] [PubMed]
- 59.Wang JK, Hom J, Balasubramanian S, et al. Clinical Order Patterns Machine-Learned from Clinician Cohorts Stratified by Patient Mortality Outcomes for Six Common Admission Diagnoses. Data in Brief Submitted. [DOI] [PMC free article] [PubMed]
- 60.Landis JR, Koch GG. The Measurement of Observer Agreement for Categorical Data. Biometrics 1977:159–74. [PubMed]
- 61.Deutschendorf A, Michtalik HJ, Leung C, et al. A Method for Attributing Patient-Level Metrics to Rotating Providers in an Inpatient Setting. Journal of Hospital Medicine 2018;13(7):470–475. [DOI] [PubMed] [Google Scholar]
- 62.Kendall MG. A New Measure of Rank Correlation. Biometrika 1938:81–93.
- 63.Webber W, Moffat A, Zobel J. A similarity measure for indefinite rankings. ACM Trans. Inf. Syst 2010:1–38.
- 64.Schapire RE. The Strength of Weak Learnability. Machine Learning Hingham, MA, USA: Kluwer Academic Publishers; 1990:197–227. [Google Scholar]
- 65.James G, Witten D, Hastie T, et al. An Introduction to Statistical Learning Available at: http://link.springer.com/content/pdf/10.1007/978-1-4614-7138-7.pdf. Accessed October 2017.
- 66.Amarasingham R, Moore BJ, Tabak YP, et al. An Automated Model to Identify Heart Failure Patients at Risk for 30-Day Readmission or Death Using Electronic Medical Record Data. Medical Care 2010;48:981–88. [DOI] [PubMed] [Google Scholar]
- 67.Raghavan M, Marik PE. Management of sepsis during the early “golden hours. The Journal of Emergency Medicine 2006;31(2):185–199. [DOI] [PubMed] [Google Scholar]
- 68.Kotwal RS, Howard JT, Orman JA, et al. The Effect of a Golden Hour Policy on the Morbidity and Mortality of Combat Casualties. JAMA Surgery 2016;151(1):15–24. [DOI] [PubMed] [Google Scholar]
- 69.Zhu H, Ni Y, Cai P, et al. Automatic information extraction for computerized clinical guideline. Studies in Health Technology and Informatics 2013;192:1023. [PubMed] [Google Scholar]
- 70.Weng C, Payne PRO, Velez M, et al. Towards symbiosis in knowledge representation and natural language processing for structuring clinical practice guidelines. Studies in Health Technology and Informatics 2014;201:461–469. [PMC free article] [PubMed] [Google Scholar]
- 71.Woolf SH, Grol R, Hutchinson A, et al. Clinical Guidelines: Potential Benefits, Limitations, and Harms of Clinical Guidelines. BMJ 1999;318:527–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Woolf SH.Do Clinical Practice Guidelines Define Good Medical Care? The Need for Good Science and the Disclosure of Uncertainty When Defining ‘Best Practices. Chest 1998:166S – 171S. [DOI] [PubMed]
- 73.Kredo T, Bernhardsson S, Machingaidze S, et al. Guide to Clinical Practice Guidelines: The Current State of Play. International Journal for Quality in Health Care 2016;28:122–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rentería ME, Cortes A, & Medland SE. Using PLINK for Genome-Wide Association Studies (GWAS) and Data Analysis. In Gondro C, van der Werf J, & Hayes B (Eds.). Genome-Wide Association Studies and Genomic Prediction 2013:193–213. Totowa, NJ: Humana Press. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


