Abstract
Rhabdomyosarcoma (RMS) is the most common soft tissue sarcoma of childhood with presumed origins of skeletal muscle because of its myogenic phenotype. RMS is composed of two main subtypes, embryonal RMS (eRMS) and alveolar (aRMS). While eRMS histologically resembles embryonic skeletal muscle, the aRMS subtype is more aggressive and exhibits a poorer prognosis. In addition, whereas the genetic profile of eRMS is less established, aRMS is commonly associated with distinct chromosome translocations that fuse domains of transcription factors, Pax3 or Pax7 to FKHR (Foxo1A). Both eRMS and aRMS tumor cells express myogenic markers such as MyoD, but their ability to complete differentiation is impaired. How this impairment occurs is the subject of this review, which will focus on several themes that include signaling pathways that converge on Pax-FKHR gene targets, alterations in MyoD function, epigenetic modifications of myogenic promoters, and microRNAs whose expression patterns in RMS alter key regulatory circuits to help maintain tumor cells in an opportunistically less differentiated state.
Introduction
RMS is considered a relatively rare cancer, yet among children and young adults is the most common soft-tissue sarcoma. The annual incidence of RMS is approximately 350 cases in the US (1). These tumors are classified into two main subtypes. The embryonal form (eRMS) occurs more often in children ages 10 years or younger, and accounts for approximately 67% of all RMS cases and have a favorable prognosis. In contrast, the alveolar form (aRMS) prototypically occurs in adolescents in 30% of RMS cases, and has a higher rate of metastasis upon initial diagnosis (2). Therefore, aRMS patients experience a poorer clinical outcome (3–5). eRMS and aRMS tumors are diagnosed by expression of skeletal markers, such as transcription factors, MyoD and myogenin, as well as structural proteins, myosin heavy chain, skeletal α-actin, and desmin (6–8). These markers link RMS to a skeletal muscle lineage, yet whether the tumor cell of origin is sometimes a non-myogenic cell is distinctly possible (9). Furthermore, although RMS tumors commonly originate from within skeletal muscle, they can also develop from non-muscle sites such as the salivary glands, skull base (parameninges), biliary tree, and genitourinary tract (bladder/prostate)(9, 10).
In contrast to eRMS, where the cytogenetic characterization of these tumors can be complex, aRMS exhibit common chromosomal translocations that occurs in 80% or more of cases between chromosomes 2 and 13, t(2;13)(q35;q14) or else chromosomes 1 and 13, t(1;13)(q36;q14) (11–13). These genetic imbalances lead to the fusion of two transcription factor families. The first on chromosomes 1 or 2 involve members of the paired box transcription factor family, Pax7 and Pax3, respectively. Pax genes contain an N-terminal DNA binding domain composed of paired box and homeobox motifs, and a C-terminal transactivation domain. The second class of transcription factors involves the fork head (FKHR) family member, FOXO1A. Like Pax genes, FKHR also possess N-terminal DNA binding and C-terminal transactivation domains. The breakpoint occurs in intron 7 for Pax genes, and intron 1 for FKHR, which upon fusion encodes chimeric proteins, Pax3-FKHR and Pax7-FKHR that consist of the 5′ DNA binding domain of Pax and the 3′ transactivation domain of FKHR.
Several studies support the transforming properties of Pax-FKHR proteins. In cultured fibroblasts, addition of Pax3-FKHR, but not Pax3 alone, promoted cells to become transformed (14, 15). Similar methods expressing Pax3-FKHR in eRMS cell lines caused growth rates to increase, and accelerated tumor formation in immune compromised mice (16). In comparison, mice genetically engineered to conditionally express Pax3-FKHR in maturing muscle progenitor cells (Myf6+ myoblasts) developed RMS with low frequency (17), but tumor latency could be significantly reduced when Pax3-FKHR was expressed in a p53 deficient background. These results suggested that the Pax3-FKHR translocation is required but not sufficient to induce aRMS.
A considerable number of studies have also examined the mechanisms by which Pax-FKHR proteins contribute to oncogenesis. The dose and cellular localization of the chimeric protein appear critical for its transformation activity. Both Pax3-FKHR and Pax7-FKHR exhibit up to 100 fold more transcriptional activity compared to wild type Pax3 and Pax7 proteins (18, 19). The fusion proteins themselves are expressed at a higher level compared to their wild type counterparts. For Pax3-FKHR, over-expression results from a copy number independent increase in transcription (20). In contrast, the elevated expression of Pax7-FKHR associates with gene amplification (21). In addition, unlike the rapid proteolytic turnover of Pax3 during myogenic differentiation, Pax3-FKHR is significantly more stable (22). With regards to cellular localization, Akt tightly controls the cytoplasmic to nuclear shuttling of wild type FKHR (23). Upon stimulation, Akt activity phosphorylates FKHR, which causes its cytoplasmic retention. However, in aRMS, Pax-FKHR fusion proteins can be resistant to Akt activity and therefore predominantly reside in the nucleus (24). A means by which Pax-FKHR proteins contribute to RMS is by protecting tumor cells from apoptosis (25), which is mediated through the expression of anti-apoptotic genes, such as BCL-XL (26). In addition, Pax-FKHR proteins are thought to promote RMS by fractionally suppressing terminal differentiation. Gene expression profiling revealed that Pax3-FKHR positively regulates a wide array of myogenic genes that under physiological conditions are necessary to promote terminal muscle differentiation (27–32). Importantly, similar defects in differentiation occur in RMS tumors lacking the Pax-FKHR translocation, or in aRMS when Pax3:FKHR is knocked down, which highlights a common phenotype presented among both aRMS and eRMS subtypes (33). In drosophila, Pax7:FKHR expression leads to disorganized muscle (34), reminiscent of preneoplastic lesions, which recapitulates the findings in mouse models (35).
The intent of this review is to summarize the mechanisms that underlie the inability of RMS cells to achieve terminal differentiation. Seemingly centric to these mechanisms is the role of the master switch, skeletal muscle specific, transcription factor MyoD that controls terminal differentiation. Studies support that the deregulation of MyoD activity and not its expression level is likely to account for the block in differentiation of RMS. How this deregulation occurs, and what signaling pathways crosstalk with, or independently of, Pax-FKHR to mediate the dysregulated activity on MyoD will be discussed.
Mechanisms of MyoD dysregulation in RMS
MyoD is a basic helix-loop-helix (bHLH) transcription factor specifically expressed in skeletal muscle that binds DNA as a heterodimer with E-proteins, E2A, E2-2, or HEB (36). MyoD, along with other skeletal muscle specific bHLH members, Myf5, myogenin, and MRF4/Myf6, are responsible for coordinating the myogenic program as proliferating myoblasts transition towards terminal differentiation. Early efforts to investigate the mechanisms of impaired differentiation in RMS focused on MyoD. Those findings revealed that MyoD was competent to bind as a heterodimer to its consensus DNA binding site, but in RMS cells exhibited poor transactivation potential (37). It was later discovered by using an eRMS cell line named RD (38) that inhibition of MyoD activity occurs from the binding with bHLH repressor proteins. Musculin, otherwise known as MyoR, or a spliced form of E2A, repress MyoD-induced gene expression by competing for binding to E-box proteins (39). Importantly, this inhibitory activity is reversible, as exogenous addition of a forced dimer linking MyoD and an E protein can overcome the effects from bHLH repressors, and rescue terminal differentiation in RD cells.
Another mechanism proposed to interfere with MyoD activity in RMS tumors relates to enzymes that modify histone proteins in chromatin to influence gene expression. MyoD is well known to interact in a genome wide fashion with histone acetyltransferases (HATs) such as p300/CBP, as well as histone deacetylases (HDACs) to control the temporal expression of myogenic genes (40, 41). In addition, in RMS, the histone methyltransferase KMT1A (Suv39) was shown to associate with MyoD in Rh28 and Rh30 aRMS cell lines (42). Through this interaction, KMT1A suppresses transcription of myogenin by inducing the trimethylation on lysine 9 of histone H3 (H3K9me3). Under a physiological setting, this suppression occurs in undifferentiated myoblasts and is relieved as KMT1A levels decrease during differentiation. This in turn allows MyoD to stimulate myogenin transcription, which is needed to complete myogenic differentiation (42). In aRMS, but not eRMS, KMT1A expression and H3K9me3 activity are maintained, even in cells exposed to differentiation conditions (43). This results in sustained suppression of myogenin promoter activity. Experiments to test the direct involvement of KMT1A in aRMS showed that KMT1A knockdown by targeted shRNA reduced the growth rate and tumor volume of Rh28 and Rh30 cells and tumor xenografts, respectively (43). Furthermore, p53 has a tight interplay with KMT1A by decreasing H3K9me3 repressive marks (44), but since the Arf-Mdm2-p53 axis is often lost in aRMS (45, 46), epigenetic suppression on myogenin and other terminally differentiation gene promoters would be expected to be maintained. Thus, targeting KMT1A in aRMS allows the reactivation of MyoD function to promote terminal differentiation, irrespective of, or in response to, p53 status.
Downstream effects of MyoD dysregulation in RMS
As a master switch transcription factor in skeletal muscle, MyoD has been implicated in the transcriptional regulation of a myriad of genes, most of which are vital in ensuring that the differentiation program is efficiently completed. However, these genes may or may not be directly involved in the development or progression of RMS. One exception seems to be the non-coding microRNA gene, miR-206, whose involvement has been strongly linked to RMS (47). Mir-206 is referred to as a MyomiR, due to its ability to control myogenic cell fate (48). Included in this family is miRs-1 and -133. The miR-1/miR-133 and miR-206 family is encoded on 3 bicistronic microRNA gene clusters on 3 separate chromosomes. While miR-206 is specifically expressed in skeletal muscle, miR-1-1/miR-133a-2 and miR-1-2/miR-133a-1 are expressed in both skeletal and cardiac muscles. MiRs1-1 and 1-2 are identical in sequence while miR-206 differs by only 4 nucleotides, which lie outside the seed sequence. The seed forms part of an 18–22 mature nucleotide sequence that dictates the mRNA target that a miR will bind to and typically suppress by inhibiting protein translation or inducing mRNA cleavage (49). Because of their similarity, miR-1 and miR-206 share common mRNA targets, although some unique target genes have also been described (47).
Skeletal muscle expression of miR-206 is due in large part to direct transcriptional regulation by MyoD (50, 51). In cultured myoblasts, miR-206 is prominently induced during differentiation and functions to promote myogenesis by inhibiting proliferation and signaling factors who themselves serve as negative regulators of skeletal muscle differentiation (52–56). In primary RMS tumors and established cell lines, both aRMS and eRMS subtypes are firmly associated with silencing miR-1/miR-206 expression (57–60). The clinical relevance of downregulating miR-206 was recently shown using a large cohort of RMS patients, where overall patient survival was found to inversely correlate with miR-206 expression levels (61). Interestingly, this correlation occurred in patients that lacked Pax-FKHR translocations, and in addition no correlation was found with miR-1.
Recent evidence in RD cells has shed some light as to what controls the suppression of miR-206 in RMS. Chromatin immunoprecipitation (ChIP) experiments showed that while MyoD occupies two E box binding sites in the miR-206 promoter in RD cells, the adjacent E box site is occupied by Musculin (62). Occupation by Musculin is sufficient to impede MyoD DNA binding and activation of miR-206. This co-occupancy model was further supported by genome wide ChIP sequencing in undifferentiated RD cells where mapping concluded that MyoD and Musculin bindings sites existed on the miR-206 promoter. A similar mapping analysis revealed a highly comparable MyoD binding pattern between primary myotubes and RD cells (63), suggesting that the lack of proper differentiation in RMS tumors may relate to a subset of myogenic genes unable to properly be activated by MyoD due to the interference by Musculin or other altered E box transcription factors.
Direct evidence that the downregulation of miR-206 might be relevant in rhabdomyosarcomagenesis derives from the demonstration that reconstituting this microRNA in RMS cell lines is capable of re-establishing differentiation by reducing cell growth and promoting myotube formation (59, 62). Consistent with these morphological changes, gene expression profiling showed that exogenous addition of miR-206 caused both the suppression of cell cycle promoting factors while stimulating expression of terminally differentiated genes (61, 62). Analogous reconstitution of miR-206 in RMS xenografts led to the inhibition of tumor growth in mice, thus supporting the role of miR-206 as a tumor suppressor (59, 60). Mechanistically, the pro-differentiation/tumor suppressor activity of miR-206 was associated with its regulated suppression of c-Met (59, 60). The c-Met receptor is expressed in myogenic precursor cells, and upon signaling stimulation by its ligand, Hepatocyte Growth Factor/Scatter Factor (HGF/SF), induces cell proliferation and migration (64). Another target of miR-206 is the deactelyase enzyme, HDAC4, which inhibits differentiation by suppressing the activity of the myogenic transcription factor, MEF2c (65). More recently, miR-206 (as well as miR-1) was found to target Pax3. Interestingly, this regulation was limited to the JR1 eRMS cell line, as similar repression of Pax3 was not observed in the more commonly studied Rh30 aRMS cells (57). Other miR-206 targets that interfere with myogenic differentiation by regulating myoblast proliferation and survival were also recently described (66). Previous studies have shown that Pax3 is expressed in myogenic precursors, and analogous to Pax7, functions as an inhibitor of terminal differentiation (67). Therefore, by targeting Pax3, miR-206 may be capable of over ridding a block in the differentiation program.
Pax-FKHR mechanisms that impair RMS differentiation
Given the high prevalence of Pax-FKHR fusions in aRMS, significant efforts have been placed on determining the mechanisms by which Pax-FKHR proteins are capable of impairing differentiation. Our studies of Pax-FKHR expression in the embryo suggest that Pax3-FKHR suppresses many, but not all Pax3 target genes as well as Pax3 itself (35). Interestingly, Pax3-FKHR suppresses Pax7 effectors but leaves Pax7 expression itself unaffected (35). These regulations lead to Pax-FKHR promoting cell growth, as any event limiting myoblasts to properly exit cell cycle after receiving their differentiation cue would predictably act to block terminal differentiation. This notion is supported by microarray expression analysis of primary aRMS tumors, which found that Pax-FKHR fusions exhibited a distinct gene signature whose functional outcome was related to promoting proliferation and suppressing differentiation (68). More refined genome wide ChIP-Seq technology was used to identify direct transcriptional targets of Pax3-FKHR that potentially mediate the transforming activity of this oncogenic protein. Such analysis revealed Fibroblast Growth Factor Receptor 4 (FGFR4) as a target gene (69). Reporter assays were used to confirm the functional relevance of Pax3-FKHR binding elements at the FGFR4 promoter, and shRNA knockdown further demonstrated the dependence of Pax3-FKHR in regulating FGFR4 expression (69). FGFR4 has also been shown to associate with enhanced proliferation and cell survival in aRMS and eRMS cell lines and xenograft tumors (70). Since Pax3 was previously shown to regulate FGF signaling, which stimulates myoblast growth (71), Pax3-FKHR likely functions to promote proliferation through FGFR4 and its tyrosine receptor kinase activity. Another tyrosine receptor kinase gene identified as a Pax3-FKHR target is the Insulin Growth Factor Receptor (IGF1R). IGFR expression is elevated in Pax3-FKHR expressing cells and similar to FGF signaling, IGF activity stimulates myoblast growth (72). However, since IGF is also important for promoting myoblast differentiation, it is possible that Pax3-FKHR acts through IGF1R to maintain RMS cells in a proliferative state while at the same time promoting expression of differentiation markers. Yet another growth promoting gene whose expression is elevated in fusion positive aRMS cells is the c-Met receptor (73–75), also mentioned above. There is strong likelihood that c-Met is a direct target of Pax3-FKHR as Pax3 binding sites are present in the c-Met promoter, and Pax3 deficient mice exhibit reduced levels of c-Met during embryonic development (73, 76). c-Met is activated by HGF/SF (64, 77), which functions to stimulate quiescent muscle precursor cells to re-enter cell cycle during post-natal muscle injury. Direct evidence that c-Met is relevant in aRMS was demonstrated with an shRNA knockdown approach in Rh30 cells, which led to reduced tumor burden in immune compromised mice (78). Histologically, tumors with lower c-Met expression were characterized as more mature and differentiated, suggesting that c-Met expression in aRMS functions as a suppressor of differentiation. This notion is consistent with findings described above that miR-206 induced differentiation of aRMS cells is mediated through the targeting of c-Met (59, 60).
Recently, the histone demethylase protein, JARID2, belonging to the Jumonji family that demethylates mono-, di-, and trimethyl marks, was found to be widely elevated in RMS, and particularly high in fusion positive tumors tightly linked with metastasis (79). JARID2 associates with the PRC2 polycomb complex that functions to represses transcription via its Ezh2 subunit that causes trimethylation on H3K27, but JARID2 itself lacks active site residues to regulate demethylase activity in this complex (80, 81). Significantly, JARID2 was identified as a direct Pax3-FKHR transcriptional target, and a functional binding site was located in the JARID2 proximal promoter (79). JARID2 knockdown in Rh30 cells reduced cell growth and also promoted myogenic differentiation. Researchers determined that JARID2 inhibits differentiation through its association with the PRC2 complex that promoted H3K27me3 activity and transcription silencing of the myogenin and myosin light chain promoters. Taken together, studies support that the ability of Pax-FKHR fusion proteins to inhibit differentiation in aRMS occurs through multiple gene targets.
Aside from direct targets, recent developments indicate that Pax-FKHR proteins can also suppress differentiation by strongly inhibiting MyoD transcriptional activity. This presumably would impact the expression of MyoD target genes, such as the microRNAs discussed above. Findings showed that exogenous expression of Pax3-FKHR and Pax7-FKHR in myoblasts significantly reduced expression of MyoD regulated genes, myogenin, p21, and muscle creatine kinase, which was associated with their concomitant activity to block differentiation (82). Mechanistically, Pax-FKHR proteins are unable to disrupt MyoD expression, localization, phosphorylation, interactions with E box proteins, or binding to the myogenin promoter, but these fusion proteins are capable of preventing chromatin remodeling on MyoD target genes, such as myogenin. Specifically on the myogenin locus, Pax3-FKHR expression diminishes the acetylation of histone H4 and the occupancy of RNA polymerase II. Since MyoD transactivation function depends on interactions with HATs, it is likely that such interactions or their resulting activities become perturbed in the presence of Pax-FKHR proteins. Interestingly, Pax-FKHR products are themselves not bound to MyoD target promoters (82), which suggests that their ability to impede MyoD is indirect, or at least is independent of MyoD DNA binding.
Anti-differentiation signaling pathway in RMS
In addition to Pax-FKHR fusions in aRMS, and the general inactivation of MyoD, several signaling pathways have been identified that contribute to impairing myogenic differentiation in RMS. In most cases the deregulation of these pathways are not specific to RMS subtypes. Such is the case for the IL-4 receptor (IL-4R) whose gene expression significantly increases in both human and mouse RMS tumors, as well as in primary and metastatic tissue (83, 84). Given that IL-4R signaling is mediated through JAK/STAT, PI3K/AKT, and MAPK pathways, investigators looked for differences in these signaling mediators in eRMS and aRMS cells treated with IL-4. Their findings showed robust induction of STAT6 and AKT. Treatment with IL-13, which similar to IL-4 signals through the IL-4Rα component of the IL-4 receptor, exhibited analogous activation profiles, with the exception that ERK1/2 could also be stimulated. Functionally, these cytokines promote proliferation and inhibit expression of differentiation markers in both eRMS and aRMS cells (83). Similar conclusions were reached with IL-4 signaling in RD cells (84).
RD cells can also be induced to differentiate in response to the compound, O-Tetradecanoylphorbol-13-acetate (TPA), an activity that depends in part on protein kinase C (PKCα) (85). What lies downstream of PKCα has not yet been resolved, but evidence indicates that several MAPK proteins are involved, including p38, ERK1/2, and JNK. It will be interesting in future studies to determine whether direct links exist between these MAPK proteins and pro-myogenic transcription factors. P38 is a strong candidate, given its ability to partially restore RMS differentiation in response to MKK6 (86), as well as promote differentiation in myoblasts by inhibiting the anti-myogenic function of Pax7 (87) and stimulating the myogenic activity of MyoD (88). Furthermore, TNF-alpha has been shown to be upstream of p38 and EZH2, and to lay down H3K27 repressive marks at the Pax7 promoter (87). One might speculate that a similar approach may apply to silencing Pax3-FKHR – especially given that isolated limb perfusion with TNF-alpha has been shown in pilot studies to exhibit a clinical benefit as a limb saving approach (89).
An additional signaling mediator, whose dysfunction is widely associated with a multitude of skeletal muscle disorders, is myostatin (90, 91). Myostatin belongs to the TGFβ superfamily and signals through the activin receptor type IIb to activate Smad proteins and repress myogenic differentiation. Significantly, with regards to RMS, suppression of myostatin in RD cells with a dominant negative form of the activin receptor type IIb promotes differentiation by inhibiting SMAD2/3 and activating p38 (92). Similar pro-myogenic effects resulting from myostatin inhibition were observed in eRMS cells (93). How myostatin inhibits differentiation in RMS cells is not clear, but evidence indicates that similar to Pax-FKHR proteins, this might occur by negatively targeting the transactivation function of MyoD (94). Whether such overlapping activities occur due to crosstalk between Pax-FKHR fusion proteins and myostatin signaling remains to be determined.
Interestingly, both AKT and myostatin have been shown to mediate their signaling activities in part through the activation of NF-κB (95–98), which itself functions as a negative regulator of myogenic differentiation (99–101). In RMS, the p65/RelA subunit of NF-κB is elevated in aRMS and eRMS tumors, and functions by epigenetically suppressing the expression of miR-29 through Ying Yang 1 and the PRC2 Polycomb complex (102). In myoblasts, miR-29 promotes differentiation (103), so the epigenetic silencing of miR-29 in RMS is speculated to maintain these tumors in a less differentiated state. Consistent with this notion, re-expression of miR-29 in Rh30 cells or xenografts tumors reduced cell growth and restored expression of myogenic terminally differentiated genes (102). Whether IL-4 induced AKT activity or myostatin signaling in RMS cells requires NF-κB to limit differentiation of tumor cells remains to be investigated.
Tumor suppressors such as PTEN and p53 negatively regulate the transcriptional activity of NF-κB (104, 105). Studies indicate that NF-κB and p53 antagonize each other’s activities to regulate cell growth and cell survival properties by competing for the transcriptional co-activator, CREB Binding Protein, CBP (105). This may be relevant in controlling the differentiated state of RMS tumors since p53, and its structural homologs, p63 and p73, have been shown to promote myogenic differentiation (106). In addition, the transdominant inhibitor of the p53 family members, Np73, is able to block myoblasts from exiting the cell cycle in response to a differentiation cue, and significantly, similar expression of Np73 correlates highly with human RMS tumors. Np73 is thought to function in RMS by providing cells with a growth advantage. However, this activity alone is not sufficient to impair differentiation and promote RMS oncogenesis, suggesting a role for additional oncogenic signals. Such a notion was supported by results showing that addition of IGF or Pax3:FKHR to Np73 expressing myoblasts could successfully induce RMS tumors in mice (106). Consistent with the tumor suppressor, pro-myogenic role of p53 family members, the negative regulator of p53, MDM2, is elevated in RMS (107) and was shown to be capable of inhibiting C2C12 myoblasts (108). Although it is possible that MDM2 controls myogenesis in RMS tumors via p53, genetic evidence indicates MDM2-mediated sarcomagenesis occurs irrespective of p53 function (109), and may instead result from a complex competition between MDM2 and pRB for SP-1 (110). This is in line with other findings showing that MDM2 inhibits myogenic differentiation by impairing the transcriptional activity of MyoD. Since genetics studies have yet to be confirmed in mouse models of RMS, a role for p53 still remains. This is supported by evidence from RMS cell lines where alternative spliced forms of MDM2 were identified to mediate p53 activity in association with defective cell growth and differentiation (111).
Recent studies implicate the cell of origin having influence over the degree of myogenic differentiation of tumors derived thereof (112). Rb1 becomes a natural candidate for determining tumor phenotype and the differentiation potential given its requisite role in embryonic and postnatal muscle differentiation (113, 114). A high frequency of retinoblastoma (Rb1) gene mutation has been reported in a subset of human eRMS (115), and we previously reported that Rb1 nullizygosity in combination with other mutations might lead to loss of differentiation in eRMS and spindle cell sarcomas (112). However, the role of Rb1 loss in aRMS remains controversial (115, 116). Studies employing conditional mouse genetics to define the role of Rb1 in the initiation and progression of aRMS are highly anticipated.
Discussion
Although histological and genetic features of eRMS and aRMS tumors are distinct, they share a common phenotype of a defective differentiation program that contributes to oncogenesis. Mechanistically, numerous signaling pathways have been described, but in many cases their anti-differentiation activities act through a central hub of the myogenic transcription factor MyoD. From a multitude of studies, we can now fully appreciate that the expression of MyoD protein is useful as a reliable immunological marker to clinical diagnosis RMS, but MyoD expression does not reflect its function, as aRMS and eRMS tumors typically contain MyoD whose transactivation activity has been impaired either directly or indirectly. For Pax3-FKHR translocations, this impairment might occur indirectly by negatively influencing the epigenetic landscape of MyoD regulated genes such as myogenin. Since myogenin is absolutely essential for myogenic differentiation, any event blocking myogenin transcription would be predicted to maintain RMS cells in a less differentiated state. Epigenetic control occurs via the Pax-FKHR gene targets, JARID2, which stimulates PRC2 activity to suppress myogenin transcription, or through KMT1A that directly binds MyoD and silences myogenin transcription by methylating H3K9. Another example whereby MyoD transcriptional activity, but not expression, is affected is through the E box protein, Musculin. In this case, enhanced expression of Musculin in eRMS tumor cells competes with MyoD for binding to E box proteins, thus lowering the concentration of MyoD heterodimers available to transcribe MyoD target genes to promote a differentiated phenotype. As these mechanisms become more refined, additional effort will be needed to test whether MyoD can serve as a bona fide therapeutic target in RMS. If such strategies are to be successful, they will need to focus on increasing MyoD transactivation function, without compromising its expression. Appealing avenues worth exploring might be the generation of anti-Musculin compounds or the use of existing small molecules to inhibit the PRC2 suppressor complex. However, such strategies will nevertheless remain challenging, as correcting one pathway might not necessarily diminish other pathways from their sustained impedance on MyoD activity (Figure 1).
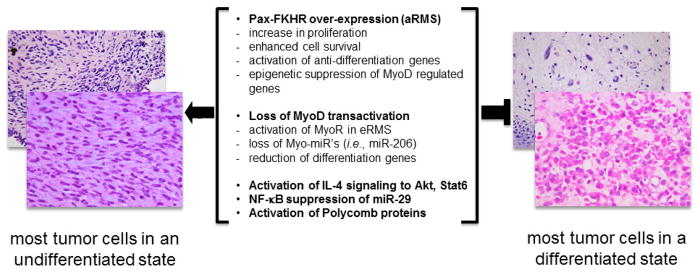
Fig. 1.
Mechanisms of differentiation suppression in rhabdomyosarcoma. Middle text represents transcriptional, epigenetic and cytokine signaling converge on the myogenic differentiation state of this sarcoma. Shown on left side are examples of embryonal rhabdomyosarcoma with less differentiation (as seen at diagnosis), and on the right are examples of embryonal rhabdomyosarcoma with mature rhabdomyoblasts whose eosinophilic cytoplasm contains myofibrillary proteins (as often seen at completion of therapy). Photomicrographs kindly provided by Drs. David Parham and Atiya Mansoor.
Another potential therapeutic avenue to explore is the ability to restore downstream genes whose expression is lost as a result of impaired MyoD function. Such genes would certainly include microRNAs that have surfaced as relevant factors in rhabdomyosarcomagenesis. Particularly, loss of miR-206 appears to be a strong prognostic marker of both aRMS and eRMS, and importantly, in both subtypes, restoration of miR-206 expression is capable of re-establishing terminal differentiation that in vivo associates with delayed tumor progression. Although other myomiRs and non-muscle miRs are capable of possessing similar pro-differentiation activities, additional studies will be needed to determine whether such miRs share similar prognostic features as miR-206, and are capable of exhibiting similar pro-differentiation activities when reconstituted in RMS tumors. The delivery of miRs as a therapeutic for RMS is an alluring option, as their overexpression would predictably reduce tumor cell proliferation and promote cellular differentiation. This may in turn help differentiated tumor cells overcome their radiation or chemotherapy resistance (Figure 1). However, translating miR therapy will require additional pre-clinical data to demonstrate that anti-tumor efficacy can be maintained when miRs are delivered systemically by a route of administration suitable for the clinic, and using genetic models of RMS that better recapitulate the onset and progression of aRMS and eRMS tumors.
Finally, unexpected frontiers in myodifferentiation are emerging as the field goes beyond a cell-autonomous view of rhabdomyosarcoma biology and gives consideration to the tumor cell heterogeneity that has been observed for decades in human tumors. New evidence suggests that more differentiated tumor cells march in advance of the stem-like tumor cells to prepare a receptive ‘bed’ for tumors at a new site (117). This more sophisticated view of cellular control is likely to bring improved, if not a fascinating understanding to the concept of differentiation therapy.
References
- 1.Ries LAG, Smith MA, Gurney JG, et al. Cancer incidence and survival among children and adolescents: United States SEER Program 1975–1995. NIH Pub. NCI SEER Program. 1999;99:4649. [Google Scholar]
- 2.Perez EA, et al. Rhabdomyosarcoma in children: a SEER population based study. J Surg Res. 2011;170:e243–251. doi: 10.1016/j.jss.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 3.Punyko JA, et al. Long-term survival probabilities for childhood rhabdomyosarcoma. A population-based evaluation. Cancer. 2005;103:1475–1483. doi: 10.1002/cncr.20929. [DOI] [PubMed] [Google Scholar]
- 4.Punyko JA, et al. Long-term medical effects of childhood and adolescent rhabdomyosarcoma: a report from the childhood cancer survivor study. Pediatr Blood Cancer. 2005;44:643–653. doi: 10.1002/pbc.20310. [DOI] [PubMed] [Google Scholar]
- 5.Xia SJ, Pressey JG, Barr FG. Molecular pathogenesis of rhabdomyosarcoma. Cancer Biol Ther. 2002;1:97–104. doi: 10.4161/cbt.51. [DOI] [PubMed] [Google Scholar]
- 6.Dias P, et al. Strong immunostaining for myogenin in rhabdomyosarcoma is significantly associated with tumors of the alveolar subclass. Am J Pathol. 2000;156:399–408. doi: 10.1016/S0002-9440(10)64743-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sebire NJ, Malone M. Myogenin and MyoD1 expression in paediatric rhabdomyosarcomas. J Clin Pathol. 2003;56:412–416. doi: 10.1136/jcp.56.6.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tonin PN, Scrable H, Shimada H, Cavenee WK. Muscle-specific gene expression in rhabdomyosarcomas and stages of human fetal skeletal muscle development. Cancer Res. 1991;51:5100–5106. [PubMed] [Google Scholar]
- 9.Hatley ME, et al. A mouse model of rhabdomyosarcoma originating from the adipocyte lineage. Cancer Cell. 2012;22:536–546. doi: 10.1016/j.ccr.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gurney JG. Topical topics: Brain cancer incidence in children: time to look beyond the trends. Med Pediatr Oncol. 1999;33:110–112. doi: 10.1002/(sici)1096-911x(199908)33:2<110::aid-mpo9>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 11.Barr FG. Molecular genetics and pathogenesis of rhabdomyosarcoma. J Pediatr Hematol Oncol. 1997;19:483–491. doi: 10.1097/00043426-199711000-00001. [DOI] [PubMed] [Google Scholar]
- 12.Davis RJ, D’Cruz CM, Lovell MA, Biegel JA, Barr FG. Fusion of PAX7 to FKHR by the variant t(1;13)(p36;q14) translocation in alveolar rhabdomyosarcoma. Cancer Res. 1994;54:2869–2872. [PubMed] [Google Scholar]
- 13.Galili N, et al. Fusion of a fork head domain gene to PAX3 in the solid tumour alveolar rhabdomyosarcoma. Nat Genet. 1993;5:230–235. doi: 10.1038/ng1193-230. [DOI] [PubMed] [Google Scholar]
- 14.Lam PY, Sublett JE, Hollenbach AD, Roussel MF. The oncogenic potential of the Pax3-FKHR fusion protein requires the Pax3 homeodomain recognition helix but not the Pax3 paired-box DNA binding domain. Mol Cell Biol. 1999;19:594–601. doi: 10.1128/mcb.19.1.594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scheidler S, Fredericks WJ, Rauscher FJ, 3rd, Barr FG, Vogt PK. The hybrid PAX3-FKHR fusion protein of alveolar rhabdomyosarcoma transforms fibroblasts in culture. Proc Natl Acad Sci U S A. 1996;93:9805–9809. doi: 10.1073/pnas.93.18.9805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anderson J, Ramsay A, Gould S, Pritchard-Jones K. PAX3-FKHR induces morphological change and enhances cellular proliferation and invasion in rhabdomyosarcoma. Am J Pathol. 2001;159:1089–1096. doi: 10.1016/S0002-9440(10)61784-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keller C, et al. Alveolar rhabdomyosarcomas in conditional Pax3:Fkhr mice: cooperativity of Ink4a/ARF and Trp53 loss of function. Genes Dev. 2004;18:2614–2626. doi: 10.1101/gad.1244004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bennicelli JL, Advani S, Schafer BW, Barr FG. PAX3 and PAX7 exhibit conserved cis-acting transcription repression domains and utilize a common gain of function mechanism in alveolar rhabdomyosarcoma. Oncogene. 1999;18:4348–4356. doi: 10.1038/sj.onc.1202812. [DOI] [PubMed] [Google Scholar]
- 19.Fredericks WJ, et al. The PAX3-FKHR fusion protein created by the t(2;13) translocation in alveolar rhabdomyosarcomas is a more potent transcriptional activator than PAX3. Mol Cell Biol. 1995;15:1522–1535. doi: 10.1128/mcb.15.3.1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davis RJ, Barr FG. Fusion genes resulting from alternative chromosomal translocations are overexpressed by gene-specific mechanisms in alveolar rhabdomyosarcoma. Proc Natl Acad Sci U S A. 1997;94:8047–8051. doi: 10.1073/pnas.94.15.8047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barr FG, et al. In vivo amplification of the PAX3-FKHR and PAX7-FKHR fusion genes in alveolar rhabdomyosarcoma. Hum Mol Genet. 1996;5:15–21. doi: 10.1093/hmg/5.1.15. [DOI] [PubMed] [Google Scholar]
- 22.Miller PJ, Hollenbach AD. The oncogenic fusion protein Pax3-FKHR has a greater post-translational stability relative to Pax3 during early myogenesis. Biochim Biophys Acta. 2007;1770:1450–1458. doi: 10.1016/j.bbagen.2007.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brunet A, et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 24.del Peso L, Gonzalez VM, Hernandez R, Barr FG, Nunez G. Regulation of the forkhead transcription factor FKHR, but not the PAX3-FKHR fusion protein, by the serine/threonine kinase Akt. Oncogene. 1999;18:7328–7333. doi: 10.1038/sj.onc.1203159. [DOI] [PubMed] [Google Scholar]
- 25.Bernasconi M, Remppis A, Fredericks WJ, Rauscher FJ, 3rd, Schafer BW. Induction of apoptosis in rhabdomyosarcoma cells through down-regulation of PAX proteins. Proc Natl Acad Sci U S A. 1996;93:13164–13169. doi: 10.1073/pnas.93.23.13164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Margue CM, Bernasconi M, Barr FG, Schafer BW. Transcriptional modulation of the anti-apoptotic protein BCL-XL by the paired box transcription factors PAX3 and PAX3/FKHR. Oncogene. 2000;19:2921–2929. doi: 10.1038/sj.onc.1203607. [DOI] [PubMed] [Google Scholar]
- 27.Armeanu-Ebinger S, et al. Differential expression of invasion promoting genes in childhood rhabdomyosarcoma. Int J Oncol. 2011;38:993–1000. doi: 10.3892/ijo.2011.921. [DOI] [PubMed] [Google Scholar]
- 28.Khan J, et al. cDNA microarrays detect activation of a myogenic transcription program by the PAX3-FKHR fusion oncogene. Proc Natl Acad Sci U S A. 1999;96:13264–13269. doi: 10.1073/pnas.96.23.13264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khan J, et al. Gene expression profiling of alveolar rhabdomyosarcoma with cDNA microarrays. Cancer Res. 1998;58:5009–5013. [PubMed] [Google Scholar]
- 30.Mercado GE, et al. Identification of PAX3-FKHR-regulated genes differentially expressed between alveolar and embryonal rhabdomyosarcoma: focus on MYCN as a biologically relevant target. Genes Chromosomes Cancer. 2008;47:510–520. doi: 10.1002/gcc.20554. [DOI] [PubMed] [Google Scholar]
- 31.Schaaf GJ, et al. Full transcriptome analysis of rhabdomyosarcoma, normal, and fetal skeletal muscle: statistical comparison of multiple SAGE libraries. FASEB J. 2005;19:404–406. doi: 10.1096/fj.04-2104fje. [DOI] [PubMed] [Google Scholar]
- 32.Wachtel M, et al. Gene expression signatures identify rhabdomyosarcoma subtypes and detect a novel t(2;2)(q35;p23) translocation fusing PAX3 to NCOA1. Cancer Res. 2004;64:5539–5545. doi: 10.1158/0008-5472.CAN-04-0844. [DOI] [PubMed] [Google Scholar]
- 33.Kikuchi K, et al. Effects of PAX3-FKHR on malignant phenotypes in alveolar rhabdomyosarcoma. Biochem Biophys Res Commun. 2008;365:568–574. doi: 10.1016/j.bbrc.2007.11.017. [DOI] [PubMed] [Google Scholar]
- 34.Galindo RL, Allport JA, Olson EN. A Drosophila model of the rhabdomyosarcoma initiator PAX7-FKHR. Proc Natl Acad Sci U S A. 2006;103:13439–13444. doi: 10.1073/pnas.0605926103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keller C, Hansen MS, Coffin CM, Capecchi MR. Pax3:Fkhr interferes with embryonic Pax3 and Pax7 function: implications for alveolar rhabdomyosarcoma cell of origin. Genes Dev. 2004;18:2608–2613. doi: 10.1101/gad.1243904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lassar AB, et al. Functional activity of myogenic HLH proteins requires hetero-oligomerization with E12/E47-like proteins in vivo. Cell. 1991;66:305–315. doi: 10.1016/0092-8674(91)90620-e. [DOI] [PubMed] [Google Scholar]
- 37.Tapscott SJ, Thayer MJ, Weintraub H. Deficiency in rhabdomyosarcomas of a factor required for MyoD activity and myogenesis. Science. 1993;259:1450–1453. doi: 10.1126/science.8383879. [DOI] [PubMed] [Google Scholar]
- 38.McAllister RM, Melnyk J, Finkelstein JZ, Adams EC, Jr, Gardner MB. Cultivation in vitro of cells derived from a human rhabdomyosarcoma. Cancer. 1969;24:520–526. doi: 10.1002/1097-0142(196909)24:3<520::aid-cncr2820240313>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 39.Yang Z, et al. MyoD and E-protein heterodimers switch rhabdomyosarcoma cells from an arrested myoblast phase to a differentiated state. Genes Dev. 2009;23:694–707. doi: 10.1101/gad.1765109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Puri PL, Sartorelli V. Regulation of muscle regulatory factors by DNA-binding, interacting proteins, and post-transcriptional modifications. J Cell Physiol. 2000;185:155–173. doi: 10.1002/1097-4652(200011)185:2<155::AID-JCP1>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 41.Sartorelli V, Puri PL. The link between chromatin structure, protein acetylation and cellular differentiation. Front Biosci. 2001;6:D1024–1047. doi: 10.2741/sartorel. [DOI] [PubMed] [Google Scholar]
- 42.Mal AK. Histone methyltransferase Suv39h1 represses MyoD-stimulated myogenic differentiation. EMBO J. 2006;25:3323–3334. doi: 10.1038/sj.emboj.7601229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee MH, Jothi M, Gudkov AV, Mal AK. Histone methyltransferase KMT1A restrains entry of alveolar rhabdomyosarcoma cells into a myogenic differentiated state. Cancer Res. 2011;71:3921–3931. doi: 10.1158/0008-5472.CAN-10-3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mungamuri SK, et al. p53-mediated heterochromatin reorganization regulates its cell fate decisions. Nat Struct Mol Biol. 2012;19:478–484. doi: 10.1038/nsmb.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Felix CA, et al. Frequency and diversity of p53 mutations in childhood rhabdomyosarcoma. Cancer Res. 1992;52:2243–2247. [PubMed] [Google Scholar]
- 46.Takahashi Y, et al. Altered expression and molecular abnormalities of cell-cycle-regulatory proteins in rhabdomyosarcoma. Mod Pathol. 2004;17:660–669. doi: 10.1038/modpathol.3800101. [DOI] [PubMed] [Google Scholar]
- 47.Rota R, Ciarapica R, Giordano A, Miele L, Locatelli F. MicroRNAs in rhabdomyosarcoma: pathogenetic implications and translational potentiality. Mol Cancer. 2011;10:120–135. doi: 10.1186/1476-4598-10-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McCarthy JJ. MicroRNA-206: the skeletal muscle-specific myomiR. Biochim Biophys Acta. 2008;1779:682–691. doi: 10.1016/j.bbagrm.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Elbashir SM, et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 50.Rao PK, Kumar RM, Farkhondeh M, Baskerville S, Lodish HF. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc Natl Acad Sci U S A. 2006;103:8721–8726. doi: 10.1073/pnas.0602831103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sweetman D, et al. Specific requirements of MRFs for the expression of muscle specific microRNAs, miR-1, miR-206 and miR-133. Dev Biol. 2008;321:491–499. doi: 10.1016/j.ydbio.2008.06.019. [DOI] [PubMed] [Google Scholar]
- 52.Chen JF, et al. microRNA-1 and microRNA-206 regulate skeletal muscle satellite cell proliferation and differentiation by repressing Pax7. J Cell Biol. 2010;190:867–879. doi: 10.1083/jcb.200911036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dey BK, Gagan J, Dutta A. miR-206 and -486 induce myoblast differentiation by downregulating Pax7. Mol Cell Biol. 2011;31:203–214. doi: 10.1128/MCB.01009-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gagan J, Dey BK, Layer R, Yan Z, Dutta A. Notch3 and Mef2c proteins are mutually antagonistic via Mkp1 protein and miR-1/206 microRNAs in differentiating myoblasts. J Biol Chem. 2012;287:40360–40370. doi: 10.1074/jbc.M112.378414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim HK, Lee YS, Sivaprasad U, Malhotra A, Dutta A. Muscle-specific microRNA miR-206 promotes muscle differentiation. J Cell Biol. 2006;174:677–687. doi: 10.1083/jcb.200603008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rosenberg MI, Georges SA, Asawachaicharn A, Analau E, Tapscott SJ. MyoD inhibits Fstl1 and Utrn expression by inducing transcription of miR-206. J Cell Biol. 2006;175:77–785. doi: 10.1083/jcb.200603039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li L, Sarver AL, Alamgir S, Subramanian S. Downregulation of microRNAs miR-1, -206 and -29 stabilizes PAX3 and CCND2 expression in rhabdomyosarcoma. Lab Invest. 2012;92:571–583. doi: 10.1038/labinvest.2012.10. [DOI] [PubMed] [Google Scholar]
- 58.Rao PK, et al. Distinct roles for miR-1 and miR-133a in the proliferation and differentiation of rhabdomyosarcoma cells. FASEB J. 2010;24:3427–3437. doi: 10.1096/fj.09-150698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Taulli R, et al. The muscle-specific microRNA miR-206 blocks human rhabdomyosarcoma growth in xenotransplanted mice by promoting myogenic differentiation. J Clin Invest. 2009;119:2366–2378. doi: 10.1172/JCI38075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yan D, et al. MicroRNA-1/206 targets c-Met and inhibits rhabdomyosarcoma development. J Biol Chem. 2009;284:29596–29604. doi: 10.1074/jbc.M109.020511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Missiaglia E, et al. MicroRNA-206 expression levels correlate with clinical behaviour of rhabdomyosarcomas. Br J Cancer. 2010;102:1769–1777. doi: 10.1038/sj.bjc.6605684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Macquarrie KL, Yao Z, Young JM, Cao Y, Tapscott SJ. miR-206 integrates multiple components of differentiation pathways to control the transition from growth to differentiation in rhabdomyosarcoma cells. Skelet Muscle. 2012;2:7. doi: 10.1186/2044-5040-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Macquarrie KL, et al. Comparison of Genome-Wide Binding of MyoD in Normal Human Myogenic Cells and Rhabdomyosarcomas Identifies Regional and Local Suppression of Promyogenic Transcription Factors. Mol Cell Biol. 2013;33:773–784. doi: 10.1128/MCB.00916-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Allen RE, Sheehan SM, Taylor RG, Kendall TL, Rice GM. Hepatocyte growth factor activates quiescent skeletal muscle satellite cells in vitro. J Cell Physiol. 1995;165:307–312. doi: 10.1002/jcp.1041650211. [DOI] [PubMed] [Google Scholar]
- 65.McKinsey TA, Zhang CL, Lu J, Olson EN. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature. 2000;408:106–111. doi: 10.1038/35040593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Goljanek-Whysall K, et al. Regulation of multiple target genes by miR-1 and miR-206 is pivotal for C2C12 myoblast differentiation. J Cell Sci. 2012;125:3590–3600. doi: 10.1242/jcs.101758. [DOI] [PubMed] [Google Scholar]
- 67.Epstein JA, Lam P, Jepeal L, Maas RL, Shapiro DN. Pax3 inhibits myogenic differentiation of cultured myoblast cells. J Biol Chem. 1995;270:11719–11722. doi: 10.1074/jbc.270.20.11719. [DOI] [PubMed] [Google Scholar]
- 68.Davicioni E, et al. Identification of a PAX-FKHR gene expression signature that defines molecular classes and determines the prognosis of alveolar rhabdomyosarcomas. Cancer Res. 2006;66:6936–6946. doi: 10.1158/0008-5472.CAN-05-4578. [DOI] [PubMed] [Google Scholar]
- 69.Cao L, et al. Genome-wide identification of PAX3-FKHR binding sites in rhabdomyosarcoma reveals candidate target genes important for development and cancer. Cancer Res. 2010;70:6497–6508. doi: 10.1158/0008-5472.CAN-10-0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Crose LE, et al. FGFR4 blockade exerts distinct antitumorigenic effects in human embryonal versus alveolar rhabdomyosarcoma. Clin Cancer Res. 2012;18:3780–3790. doi: 10.1158/1078-0432.CCR-10-3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lagha M, et al. Pax3 regulation of FGF signaling affects the progression of embryonic progenitor cells into the myogenic program. Genes Dev. 2008;22:1828–1837. doi: 10.1101/gad.477908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Engert JC, Berglund EB, Rosenthal N. Proliferation precedes differentiation in IGF-I-stimulated myogenesis. J Cell Biol. 1996;135:431–440. doi: 10.1083/jcb.135.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Epstein JA, Shapiro DN, Cheng J, Lam PY, Maas RL. Pax3 modulates expression of the c-Met receptor during limb muscle development. Proc Natl Acad Sci U S A. 1996;93:4213–4218. doi: 10.1073/pnas.93.9.4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ferracini R, et al. Retrogenic expression of the MET proto-oncogene correlates with the invasive phenotype of human rhabdomyosarcomas. Oncogene. 1996;12:1697–1705. [PubMed] [Google Scholar]
- 75.Ginsberg JP, Davis RJ, Bennicelli JL, Nauta LE, Barr FG. Up-regulation of MET but not neural cell adhesion molecule expression by the PAX3-FKHR fusion protein in alveolar rhabdomyosarcoma. Cancer Res. 1998;58:3542–3546. [PubMed] [Google Scholar]
- 76.Phelan SA, Loeken MR. Identification of a new binding motif for the paired domain of Pax-3 and unusual characteristics of spacing of bipartite recognition elements on binding and transcription activation. J Biol Chem. 1998;273:19153–19159. doi: 10.1074/jbc.273.30.19153. [DOI] [PubMed] [Google Scholar]
- 77.Cornelison DD, Wold BJ. Single-cell analysis of regulatory gene expression in quiescent and activated mouse skeletal muscle satellite cells. Dev Biol. 1997;191:270–283. doi: 10.1006/dbio.1997.8721. [DOI] [PubMed] [Google Scholar]
- 78.Miekus K, et al. The decreased metastatic potential of rhabdomyosarcoma cells obtained through MET receptor downregulation and the induction of differentiation. Cell Death Dise. 2013;4:e459. doi: 10.1038/cddis.2012.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Walters ZS, et al. JARID2 is a direct target of the PAX3-FOXO1 fusion protein and inhibits myogenic differentiation of rhabdomyosarcoma cells. Oncogene. 2013 Feb 25; doi: 10.1038/onc.2013.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet. 2006;7:715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- 81.Shen X, et al. Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell. 2009;139:1303–1314. doi: 10.1016/j.cell.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Calhabeu F, Hayashi S, Morgan JE, Relaix F, Zammit PS. Alveolar rhabdomyosarcoma-associated proteins PAX3/FOXO1A and PAX7/FOXO1A suppress the transcriptional activity of MyoD-target genes in muscle stem cells. Oncogene. 2013;32:651–662. doi: 10.1038/onc.2012.73. [DOI] [PubMed] [Google Scholar]
- 83.Hosoyama T, et al. IL-4R drives dedifferentiation, mitogenesis, and metastasis in rhabdomyosarcoma. Clin Cancer Res. 2011;17:2757–2766. doi: 10.1158/1078-0432.CCR-10-3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nanni P, et al. Opposing control of rhabdomyosarcoma growth and differentiation by myogenin and interleukin 4. Mol Cancer Ther. 2009;8:754–761. doi: 10.1158/1535-7163.MCT-08-0678. [DOI] [PubMed] [Google Scholar]
- 85.Mauro A, et al. PKCalpha-mediated ERK, JNK and p38 activation regulates the myogenic program in human rhabdomyosarcoma cells. J Cell Sci. 2002;115:3587–3599. doi: 10.1242/jcs.00037. [DOI] [PubMed] [Google Scholar]
- 86.Puri PL, et al. Induction of terminal differentiation by constitutive activation of p38 MAP kinase in human rhabdomyosarcoma cells. Genes Dev. 2000;14:574–584. [PMC free article] [PubMed] [Google Scholar]
- 87.Palacios D, et al. TNF/p38alpha/polycomb signaling to Pax7 locus in satellite cells links inflammation to the epigenetic control of muscle regeneration. Cell Stem Cell. 2010;7:455–469. doi: 10.1016/j.stem.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Serra C, et al. Functional interdependence at the chromatin level between the MKK6/p38 and IGF1/PI3K/AKT pathways during muscle differentiation. Mol Cell. 2007;28:200–213. doi: 10.1016/j.molcel.2007.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lejeune FJ, et al. Limb salvage by neoadjuvant isolated perfusion with TNFalpha and melphalan for non-resectable soft tissue sarcoma of the extremities. Eur J Surg Oncol. 2000;26:669–678. doi: 10.1053/ejso.2000.0979. [DOI] [PubMed] [Google Scholar]
- 90.Han HQ, Mitch WE. Targeting the myostatin signaling pathway to treat muscle wasting diseases. Curr Opin Support Palliat Care. 2011;5:334–341. doi: 10.1097/SPC.0b013e32834bddf9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lee SJ. Regulation of muscle mass by myostatin. Annu Rev Cell Dev Biol. 2004;20:61–86. doi: 10.1146/annurev.cellbio.20.012103.135836. [DOI] [PubMed] [Google Scholar]
- 92.Rossi S, Stoppani E, Puri PL, Fanzani A. Differentiation of human rhabdomyosarcoma RD cells is regulated by reciprocal, functional interactions between myostatin, p38 and extracellular regulated kinase signalling pathways. Eur J Cancer. 2011;47:1095–1105. doi: 10.1016/j.ejca.2010.12.010. [DOI] [PubMed] [Google Scholar]
- 93.Langley B, et al. Myostatin inhibits rhabdomyosarcoma cell proliferation through an Rb-independent pathway. Oncogene. 2004;23:524–534. doi: 10.1038/sj.onc.1207144. [DOI] [PubMed] [Google Scholar]
- 94.Ricaud S, et al. Inhibition of autocrine secretion of myostatin enhances terminal differentiation in human rhabdomyosarcoma cells. Oncogene. 2003;22:8221–8232. doi: 10.1038/sj.onc.1207177. [DOI] [PubMed] [Google Scholar]
- 95.Ozes ON, et al. NF-kappaB activation by tumour necrosis factor requires the Akt serine-threonine kinase. Nature. 1999;401:82–85. doi: 10.1038/43466. [DOI] [PubMed] [Google Scholar]
- 96.Romashkova JA, Makarov SS. NF-kappaB is a target of AKT in anti-apoptotic PDGF signalling. Nature. 1999;401:86–90. doi: 10.1038/43474. [DOI] [PubMed] [Google Scholar]
- 97.Sizemore N, Leung S, Stark GR. Activation of phosphatidylinositol 3-kinase in response to interleukin-1 leads to phosphorylation and activation of the NF-kappaB p65/RelA subunit. Mol Cell Biol. 1999;19:4798–4805. doi: 10.1128/mcb.19.7.4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Trendelenburg AU, Meyer A, Jacobi C, Feige JN, Glass DJ. TAK-1/p38/nNFkappaB signaling inhibits myoblast differentiation by increasing levels of Activin A. Skelet Muscle. 2012;2:3. doi: 10.1186/2044-5040-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Guttridge DC, Albanese C, Reuther JY, Pestell RG, Baldwin AS., Jr NF-kappaB controls cell growth and differentiation through transcriptional regulation of cyclin D1. Mol Cell Biol. 1999;19:5785–5799. doi: 10.1128/mcb.19.8.5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Guttridge DC, Mayo MW, Madrid LV, Wang C-Y, Baldwin AS., Jr NF-kB-Induced Loss of MyoD Messenger RNA: Possible Role in Muscle Decay and Cachexia. Science. 2000;289:2363–2366. doi: 10.1126/science.289.5488.2363. [DOI] [PubMed] [Google Scholar]
- 101.Peterson JM, Bakkar N, Guttridge DC. NF-kappaB signaling in skeletal muscle health and disease. Curr Top Dev Biol. 2011;96:85–119. doi: 10.1016/B978-0-12-385940-2.00004-8. [DOI] [PubMed] [Google Scholar]
- 102.Wang H, et al. NF-kappaB-YY1-miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell. 2008;14:369–381. doi: 10.1016/j.ccr.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang H, et al. NF-kappaB regulation of YY1 inhibits skeletal myogenesis through transcriptional silencing of myofibrillar genes. Mol Cell Biol. 2007;27:4374–4387. doi: 10.1128/MCB.02020-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mayo MW, et al. PTEN blocks tumor necrosis factor-induced NF-kappa B-dependent transcription by inhibiting the transactivation potential of the p65 subunit. J Biol Chem. 2002;277:11116–11125. doi: 10.1074/jbc.M108670200. [DOI] [PubMed] [Google Scholar]
- 105.Webster GA, Perkins ND. Transcriptional cross talk between NF-kappaB and p53. Mol Cell Biol. 1999;19:3485–3495. doi: 10.1128/mcb.19.5.3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cam H, et al. p53 family members in myogenic differentiation and rhabdomyosarcoma development. Cancer Cell. 2006;10:281–293. doi: 10.1016/j.ccr.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 107.Meddeb M, et al. MDM2 amplification in a primary alveolar rhabdomyosarcoma displaying a t(2;13)(q35;q14) Cytogenet Cell Genet. 1996;73:325–330. doi: 10.1159/000134368. [DOI] [PubMed] [Google Scholar]
- 108.Fiddler TA, Smith L, Tapscott SJ, Thayer MJ. Amplification of MDM2 inhibits MyoD-mediated myogenesis. Mol Cell Biol. 1996;16:5048–5057. doi: 10.1128/mcb.16.9.5048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jones SN, Hancock AR, Vogel H, Donehower LA, Bradley A. Overexpression of Mdm2 in mice reveals a p53-independent role for Mdm2 in tumorigenesis. Proc Natl Acad Sci U S A. 1998;95:15608–15612. doi: 10.1073/pnas.95.26.15608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Guo CS, Degnin C, Fiddler TA, Stauffer D, Thayer MJ. Regulation of MyoD activity and muscle cell differentiation by MDM2, pRb, and Sp1. J Biol Chem. 2003;278:22615–22622. doi: 10.1074/jbc.M301943200. [DOI] [PubMed] [Google Scholar]
- 111.Singh RK, Tapia-Santos A, Bebee TW, Chandler DS. Conserved sequences in the final intron of MDM2 are essential for the regulation of alternative splicing of MDM2 in response to stress. Exp Cell Res. 2009;315:3419–3432. doi: 10.1016/j.yexcr.2009.07.017. [DOI] [PubMed] [Google Scholar]
- 112.Rubin BP, et al. Evidence for an unanticipated relationship between undifferentiated pleomorphic sarcoma and embryonal rhabdomyosarcoma. Cancer Cell. 2011;19:177–191. doi: 10.1016/j.ccr.2010.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hosoyama T, Nishijo K, Prajapati SI, Li G, Keller C. Rb1 gene inactivation expands satellite cell and postnatal myoblast pools. J Biol Chem. 2011;286:19556–19564. doi: 10.1074/jbc.M111.229542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Huh MS, Parker MH, Scime A, Parks R, Rudnicki MA. Rb is required for progression through myogenic differentiation but not maintenance of terminal differentiation. J Cell Biol. 2004;166:865–876. doi: 10.1083/jcb.200403004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kohashi K, et al. Alterations of RB1 gene in embryonal and alveolar rhabdomyosarcoma: special reference to utility of pRB immunoreactivity in differential diagnosis of rhabdomyosarcoma subtype. J Cancer Res Clin Oncol. 2008;134:1097–1103. doi: 10.1007/s00432-008-0385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.De Chiara A, T’Ang A, Triche TJ. Expression of the retinoblastoma susceptibility gene in childhood rhabdomyosarcomas. J Natl Cancer Inst. 1993;85:152–157. doi: 10.1093/jnci/85.2.152. [DOI] [PubMed] [Google Scholar]
- 117.Ignatius MS, et al. In vivo imaging of tumor-propagating cells, regional tumor heterogeneity, and dynamic cell movements in embryonal rhabdomyosarcoma. Cancer Cell. 2012;21:680–693. doi: 10.1016/j.ccr.2012.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]