SUMMARY
An understanding of how heterozygous loss-of-function mutations in ASD risk genes, such as TBR1, contribute to ASD remains elusive. Conditional Tbr1 deletion during late mouse gestation in cortical layer 6 neurons (Tbr1layer6 mutants) provides novel insights into its function, including dendritic patterning, synaptogenesis, and cell intrinsic physiology. These phenotypes occur in heterozygotes, providing insights into mechanisms that may underlie ASD pathophysiology. Restoring expression of Wnt7b, largely rescues the synaptic deficit in Tbr1layer6 mutant neurons. Furthermore, Tbr1layer6 heterozygotes have increased anxiety-like behavior, a phenotype seen ASD. Integrating TBR1 ChIP-Seq and RNA-Seq data from layer 6 neurons, and activity of TBR1 bound candidate enhancers, provides evidence for how TBR1 regulates layer 6 properties. Moreover, several putative TBR1 targets are ASD risk genes, placing TBR1 in a central position both for ASD risk and for regulating transcriptional circuits that control multiple steps in layer 6 development essential for the assembly of neural circuits.
eTOC blurb:
TBR1 directly regulates transcriptional circuits in heterozygous mutant mice that specify layer 6 identity and synapse number. As TBR1 is an ASD risk gene, our results provide insights into mechanisms that underlie ASD pathophysiology.
INTRODUCTION
Autism Spectrum Disorder (ASD) is defined by impairments in reciprocal social interaction, often accompanied by abnormalities in language development as well as repetitive behaviors and/or restricted interests. Recent progress in detection and analysis of rare variants in ASD has led to reliable and systematic gene discovery and revealed a group of 28 genes with the strongest statistical evidence for association with ASD risk (defined by FDR <0.01) (Sanders et al., 2015). These highest confidence (hcASD) genes encode various groups of proteins, including transcription factors (Tbr1, Tcf7l2; mouse orthologs listed), synaptic genes (Scn2a1, Syngap1), and chromatin remodelers (Chd8, Arid1b) (Sanders et al., 2015). Furthermore, amongst the top 65 ASD genes (FDR <0.1) (Sanders et al., 2015), Tbr1, Bcl11a and Foxp1 transcription factors (TF) are implicated in mouse cortical development. Systems analyses of ASD genes revealed that there is a convergence of ASD-risk gene expression in mid-fetal prefrontal cortex; concentrated in the excitatory neurons of deep cortical layers 5 and 6 (Willsey et al., 2013). However, an actionable understanding of how large-effect, heterozygous loss-of-function mutations in risk genes are contributing to the pathophysiology of ASD remains elusive. Thus, we explored how conditional deletion of mouse Tbr1 in cortical layer 6, at a developmental interval roughly equivalent to human mid-fetal stages, alters neuronal identity and function in homozygous and heterozygous mutants.
T-brain-1 (Tbr1), a T-box TF, has a central role in early cortical development. During neurodevelopment and in adulthood Tbr1 is expressed in the excitatory neurons of the neocortex (subplate, layer 6, rostral layer 5, layers 2/3), hippocampus, entorhinal cortex, pallial amygdala, piriform cortex, olfactory bulb and Cajal Retzius (CR) neurons (Hevner et al., 2003, Hevner et al., 2001). The encoded protein regulates development of early-born pallial projection neurons, including CR cells, subplate and layer 6 projection neurons (Bedogni et al., 2010, Hevner et al., 2001, Bulfone et al., 1995). Tbr1 constitutive null (Tbr1constitutive null) mouse showed defects in layer 6 corticothalamic neurons (Bedogni et al., 2010, Bulfone et al., 1998). Tbr1 promotes layer 6 identity by repressing Fezf2 and Bcl11b, TFs that control layer 5 fate (McKenna et al., 2011, Han et al., 2011). Moreover, Tbr1constitutive heterozygous mice have abnormal inter- and intraamygdalar axonal projections (Chuang et al., 2015, Huang et al., 2014). Lastly, TBR1 binds to the Grin2b promoter and promotes Grin2b expression upon neuronal activation (Chuang et al., 2014). However, many important aspects of Tbr1 function have yet to be clarified, including elucidating its role in early post-natal mouse brain development, characterizing the composition of Tbr1-regulated gene networks and their cis-regulatory elements; and understanding the consequence of Tbr1 heterozygosity on neocortical development and function.
Here, we have used conditional mutagenesis to define early postnatal functions of Tbr1 in layer 6 cortical projection neurons by creating a viable Tbr1layer6 mutant. The Tbr1layer6 homozygous mutant neurons take on a hybrid layer 5/layer 6 identity based on their gene expression profile, dendritic pattern, and physiology. Tbr1 promotes expression of layer 6 markers (Foxp2, Nr4a2, Tle4, Wnt7b) and represses expression of layer 5 identity regulators (Fezf2, Bcl11b). Tbr1layer6 homozygous mutants also have altered RNA levels of Scn2a1 and Grin2b (orthologs of hcASD genes), as well as of Bcl11a, Foxp1, Nuak1 and Wnt7b (orthologs of probable ASD genes (FDR < 0.3)) (Sanders et al., 2015).
TBR1 and other ASD genes have been identified based on heterozygous rare variants observed in cases, and therefore, it is of interest to characterize the phenotype of Tbr1layer6 heterozygotes. In these animals, we observe that Wnt7b and Bcl11a expression are reduced in layer 6, whereas Fezf2 is ectopically expressed in layer 6, providing insight into perturbations that may occur in ASD patients. Furthermore, neurons from both the Tbr1layer6 heterozygous and homozygous mice have reduced excitatory and inhibitory synaptic density as well as spontaneous EPSCs and IPSCs. Restoring expression of Wnt7b (a direct TBR1 target), largely rescues the synaptic deficit phenotype in Tbr1layer6 heterozygous and homozygous neurons in vitro and in vivo. Collectively, we propose that these phenotypes, and the Tbr1-regulated gene regulatory networks, shed light on how Tbr1 loss-of-function mutation disrupt neural function and connectivity. Importantly, we find that Tbr1layer6 heterozygous mutants, have reduced synapse numbers and functions; mechanisms that have been strongly implicated in ASD pathogenesis (De Rubeis et al., 2014). Thus, our analysis adds fundamentally to understanding of how a single TF regulates a temporal sequence of steps in cortical development that have implications for understanding complex human social behaviors.
RESULTS
Tbr1 conditional mutant allele.
To investigate the function(s) of Tbr1 in specific subtypes of cortical neurons at later stages of development, we generated a Tbr1 conditional mutant (Tbr1flox) allele by inserting LoxP sites into introns 1 and 3 (Fig. S1A). We validated that recombination using β−actin-Cre eliminated expression of TBR1 protein and RNA encoded by the deleted exons (Fig. S1C, S1E). Thus, it is likely to be a null allele, even though a truncated RNA continues to be expressed (Fig. S1D). Upon recombination with Ntsr1-cre, TBR1 protein levels were reduced by approximately 90% in layer 6 and subplate at P0 (Fig. S1F).
Tbr1 maintains layer 6 identity in postnatal cortex.
To restrict Tbr1 perturbation to layer 6 and subplate, we deleted Tbr1 5–6 days after Tbr1 expression begins using Neurotensin receptor 1-Cre mice (Ntsr1-cre). We refer to these mice as Tbr1layer6 mutants. Ntsr1-cre expression begins in layer 6 at ~E16.5 (data not shown). To identify putative Tbr1 regulatory targets in layer 6 and subplate we compared gene expression profiles in FAC-sorted layer 6 neurons from mutant and wildtype mice. More specifically, we generated RNA-Seq data from layer 6 neurons isolated from postnatal day 5 (P5) Tbr1wildtype, Tbr1layer6 heterozygous and homozygous mutant somatosensory cortex (SSCx). We identified 178 differentially expressed (DEX) genes in Tbr1layer6 homozygous mutants (false discovery rate ≤ 0.05) (Fig. 1A; Table S1 and S2). However, transcriptomic analysis of Tbr1layer6 heterozygous mutants did not reveal conclusive evidence for changes in RNA levels (data not shown).
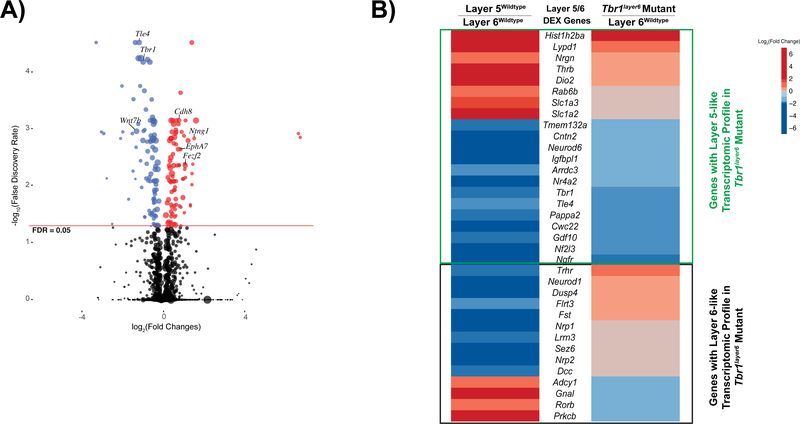
Figure 1. Tbr1 transcriptional regulation in the neonatal cortex.
(A) Volcano plot of genes up-regulated (red) and downregulated (blue) in FACS purified layer 6 neurons from P5 Tbr1layer6 homozygous mutant SSCx. Black dots represent the genes that did not reach statistical significance (adjusted p value > 0.05). The size of each point represents the difference in the median gene expression between Tbr1wildtype and Tbr1layer6 mutant (large dots mean large differences). (B) P5 transcriptomic comparison of DEX genes between layer 5wildtype vs. layer 6wildtype and Tbr1layer6 homozygotes vs. layer6wildtype. With respect to genes that mark wildtype layer 5 (red genes), we observed eight genes with increased expression in the Tbr1layer6 mutants (p < 0.05). With respect to genes that mark wildtype layer 6 (blue genes), we observed 13 with reduced expression in the Tbr1layer6 mutants (p < 0.05). However, there were 14 of the layer 5 and layer 6 marker genes whose expression did not significantly change in the Tbr1layer6 mutants (Fig. 1B). Genes with layer 5-like transcriptome profile (green box) and layer 6-like expression profile (black box) are indicated. See also Figure S1 and Tables S1, S2 and S3.
We observed increased expression of several regulators of layer 5 identity in layer 6 neurons isolated from Tbr1layer6 homozygous mutants, including Fezf2 and Bcl11b. Additionally we observed decreased expression of RNAs encoding regulators/markers of layer 6 identity, including Foxp2, Nr4a2, Tle4, and Wnt7b. Together, this suggests that layer 6 neurons from P5 Tbr1layer6 mutants have changed fate to a layer 5-like identity.
To better understand the consequence of these transcriptomic changes on the identity of the layer 6 mutant neurons, we identified genes that distinguish layer 5 and layer 6 pyramidal neurons in P5 wildtype mice. To accomplish this, we generated RNA-Seq data from FACS purified layer 5 neurons and compared with RNA-Seq data from FACS purified layer 6 neurons. We identified 35 DEX genes that distinguish layer 6 and layer 5 wildtype neurons (denoted as Layer 5/6 DEX genes; Fig. 1B). Next, we compared the Layer 5/6 DEX genes with the genes dysregulated in layer 6 neurons from Tbr1layer6 null mice. With respect to genes that mark wildtype layer 5 (red genes; Fig. 1B), we observed eight genes with increased expression in the Tbr1layer6 mutants (p < 0.05). Moreover, with respect to genes that mark wildtype layer 6 (blue genes in Fig. 1B), we observed 13 with reduced expression in the Tbr1layer6 mutants (p < 0.05; Fig. 1B; Table S3). On the contrary, the analysis identified 14 of the layer 5 and layer 6 marker genes, whose expression did not significantly change in the Tbr1layer6 mutants (Fig. 1B; Table S3). This suggests that Tbr1layer6 mutant neurons have a hybrid identity, with transcriptomic properties of both layer 5 and layer 6 pyramidal neurons.
In situ hybridization confirmed some of these results at P3 (Fig. 2, Table S4). Regulators/markers of layer 6 identity were downregulated in Tbr1layer6 homozygous (e.g. Foxp2, Tle4, Nr4a2, Wnt7b) and Tbr1layer6 heterozygous mutants (e.g. Wnt7b, Bcl11a; Fig. 2); whereas, the expression of Fezf2, a regulator of layer 5 identity, was upregulated in layer 6 mutant neurons (Fig. 2). Overall, our RNA expression data (RNA-Seq and ISH) demonstrates that late gestational/neonatal Tbr1 expression is essential to maintain layer 6 identity.
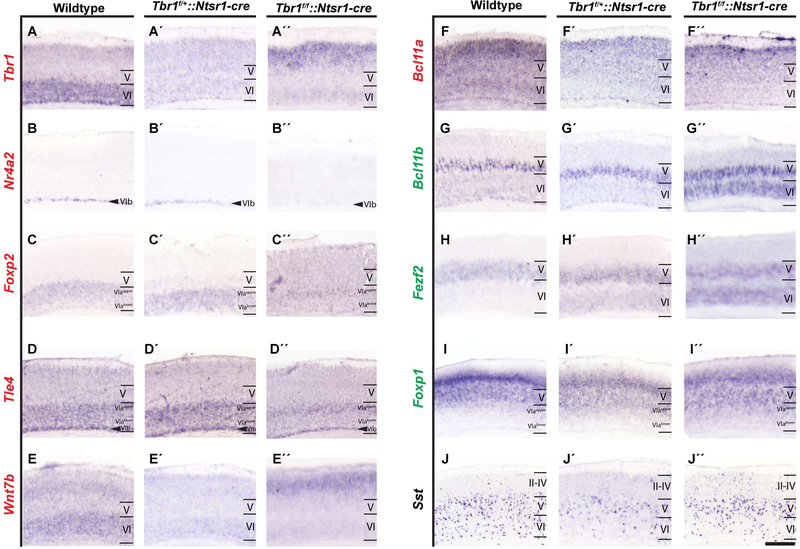
Figure 2. Tbr1 is required to maintain layer 6 identity in postnatal cortex.
In situ hybridization on SSCx coronal sections of Tbr1wildtype (A-J), Tbr1layer6 heterozygous (Tbr1f/+::Ntrs1-cre) (A´-J´), and Tbr1layer6 homozygous mutants (Tbr1f/f::Ntrs1-cre) (A´´- J´´) at P3 (n=2). In Tbr1layer6 heterozygotes and homozygotes, Tbr1 (A-A´´), Nr4a2 (B-B´´), Wnt7b (EE´´), and Bcl11a (F-F´´) expressions are reduced in layer 6 and subplate. The expression of Tbr1 and Wnt7b are increased in the superficial layers (A-A´´, E-E´´). The expression of Foxp2 (C, C´´) and Tle4 (D, D´´) are reduced in layer 6alower of Tbr1layer6 homozygous mutants. Tbr1layer6 heterozygotes and homozygotes exhibit an ectopic expression of Fezf2 (H-H´´) in layer 6. In Tbr1layer6 homozygous mutants, Bcl11b (G, G´´) and Foxp1 (I-I´´) are ectopically expressed in layer 6. Furthermore, Tbr1layer6 mutants exhibit changes in the number and laminar distribution of Sst+ CINs (J-J´´). Transcriptome levels of each gene is reflected as downregulated (red), upregulated (green) and unchanged (black) in Tbr1layer6 mutants. II-IV = layers 2–4, V = layer 5, VI = layer 6. VIb = Subplate. Scale bar = 50μm. See also Figure S2, Table S4.
Tbr1 directly regulates the transcription of genes that control layer 6 identity.
To determine the regions of the genome that TBR1 interacts with, and whether the changes in gene expression in Tbr1layer6 mutants are due to direct regulation by TBR1, we performed ChIP-Seq using P2 wildtype cortex. TBR1 binds to 68,218 regions genome-wide, (Fig. 3A). No enrichment was found at TBR1 ChIP-Seq peaks in two different control datasets: input and negative (TBR1 blocking peptide and IgG) (Fig. 3A, Fig. S3A). Approximately 20% of peaks (13,973 peaks) overlap with transcriptional start site (TSS), 31% (21,189 peaks) are on gene body, 43% (29,010 peaks) are located intergenically, with 3% on exons (2,060 peaks), 2% on 3′ UTR (1,356 peaks) and 1% on 5′ UTR (630 peaks; Fig. 3B). De novo motif discovery identified the canonical TBR1 binding motif in 17% of peaks (Fig 3B).
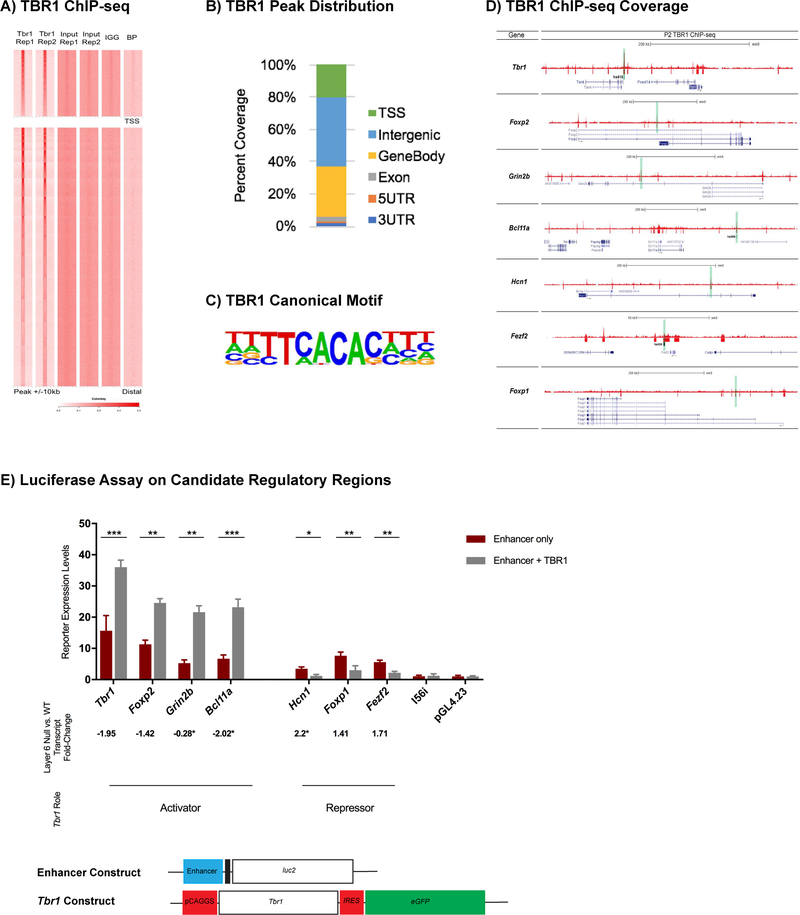
Figure 3. Genome-wide analysis of TBR1 binding and transcriptional regulation of candidate enhancer regions in loci adjacent to Tbr1-regulated genes.
(A) Heatmap of TBR1 ChIP-seq replicates compared to the controls. (B) Summary of the genomic distribution of TBR1 ChIP-Seq peaks at P2. (C) TBR1 canonical motif. (D) TBR1 ChIP-Seq on wildtype whole cortex at P2 (red tracks). Red boxes represent the TBR1 binding that reached statistical significance. Genes are shown in blue. Candidate REs that were tested in the luciferase transcription assay are highlighted in green. Black boxes indicate REs that have proven enhancer activity in E11.5 cortex corresponding to hs416 (Tbr1 locus), hs434 (Fezf2 locus) and hs399 (Bcl11a locus). Black arrow indicates the direction of transcription. Genomic scale (in kb) are shown for each locus. (E) Luciferase transcription assay was utilized to measure activity of Tbr1, Foxp2, Grin2b, Bcl11a, Foxp1, Fezf2, Hcn1 candidate enhancers in P0 primary cortical cultures. The reporter activity was measured under enhancer alone (red) and enhancer co-transfected with TBR1 (grey). TBR1 activates candidate REs of Tbr1 (FC= 2.3, p= 0.0007), Foxp2 (FC= 2.17, p= 0.0023), Grin2b (FC= 4.11, p= 0.0015), and Bcl11a (FC= 3.46, p= 0.0002), whereas it represses candidate REs of Foxp1 (FC= −2.52, p= 0.0087), Fezf2 (FC= −2.55, p= 0.0015) and Hcn1 (FC= −2.9, p=0.0248). I56i enhancer and pGL4.23 empty vectors were used as negative controls. The error bars represent the standard error of the mean. (*) represents the transcript fold-change using qPCR. T-test with Welch’s correction was used for the statistical analysis. (*p<0.05) (**p< 0.01) (***p<0.001). Rep1=Replicate 1, Rep2=Replicate 2, BP=blocking peptide, TSS=Transcriptional Start Site, FC=Fold Change. See also Figure S4, Tables S5, S6.
To gain further evidence that these peaks represent candidate regulatory elements (REs) influenced by TBR1, we assessed the number of genes with putative TBR1 regulatory loci that are dysregulated in the Tbr1layer6 mutants (Fig. 3D, Table S5). TBR1 binds to the 89% of the promoter regions and 77% of the candidate REs near the genes dysregulated in the Tbr1layer6 mutants. Genes with TBR1 binding at the promoter, or with binding within 100kb, exhibit higher overall expression levels relative to genes with no TBR1 binding (Fig S3B), indicating a general positive relationship between local TBR1 binding and gene expression. In contrast, there was no significant relationship between TBR1 promoter or distal binding and differential gene expression in the Tbr1layer6 mutants (Fig S3C), likely due in part to the relatively high number of TBR1 binding sites. While we did not observe overall enrichment for TBR1 binding at DEX genes, regions that have both a TBR1 ChIP signal and a canonical TBR1 motif were enriched at promoters for genes downregulated in the Tbr1layer6 mutants (Fig. S3C).
We compared TBR1-bound regions to regions of open chromatin identified in fetal human cerebral cortex (germinal zone and cortical plate) (de la Torre-Ubieta et al., 2018), finding overlap between fetal human cortex ATAC-seq with 80% of mouse TBR1-bound promoters and 23% of TBR1-bound distal regions. Compared to a control cardiac mesoderm ATAC-seq dataset (Koh et al., 2016), we found significant enrichment for TBR1-binding at open chromatin at both promoters and distal elements in fetal human cerebral cortex (Fisher’s exact test, P-value < 0.001), with no consistent differences in enrichment between ATAC-seq peaks specific to germinal zone or cortical plate (Fig S3D). Many DEX loci exhibited overlapping mouse TBR1 binding with human fetal cortex ATAC-seq regions (Fig S3E). This comparison demonstrates that the regulatory targets of TBR1 at P2 in the mouse cortex overlap with regulatory elements active in mid-fetal human cortical development.
We assessed the function of nine TBR1-bound REs containing a canonical TBR1 motif (T-box motif; Fig. 3C, 3D and Table S6). RE expression vectors were transfected into P0 wildtype cortical cultures and assayed for luciferase activity 3 days later. In parallel, we cotransfected the RE vectors with a Tbr1 expression vector. We tested 3 classes of putative REs that were candidates for regulating: (1) downregulated genes (Tbr1, Foxp2, Grin2b and Bcl11a), (2) upregulated genes (Hcn1, Fezf2 and Foxp1), and (3) unchanged genes (Dlx5/6) (Table S1, S2). Luciferase activity was driven by all of the REs (Fig. 3E), except the negative control Dlx5/6, I56i enhancer (active in forebrain GABAergic neurons) (Zerucha et al., 2000). Tbr1 cotransfection only activated the Tbr1, Foxp2, Grin2b and Bcl11a candidate REs, consistent with down-regulation of these cognate genes in the Tbr1layer6 mutants (Table S1, Fig. 1A). Moreover, Tbr1 co-transfection reduced luciferase expression with the Hcn1, Fezf2 and Foxp1 REs, which corresponded to the upregulated genes in the Tbr1layer6 mutants (Table S1, Fig. 1A).
Thus, using TBR1 ChIP-Seq and a RE functional assay, we have identified REs that function either as activators or repressors in the presence of TBR1. This data solidifies our evidence that TBR1 directly controls the molecular properties of layer 6 pyramidal neurons, and that TBR1 functions as an activator or a repressor depending on the nature of the RE.
Tbr1 specifies a program that patterns apical dendritic lamination of layer 6 neurons.
A key feature of a neuron’s identity is its dendritic patterning (Lefebvre et al., 2015). Normally, the apical dendrites of layer 6 pyramidal neurons grow into layer 4 where they elaborate their branches (Ledergerber and Larkum, 2010). We used tdTomato expression driven by Ntsr1-cre in layer 6, to compare the dendritic patterning of wildtype and Tbr1layer6 heterozygous and homozygous mutants (Fig. 4). In the wildtype control, the apical dendrites of the layer 6 neurons extended to layer 4 at P3, P21 and P60 (Fig. 4A-C). On the contrary, the changes in the dendritic morphology of the Tbr1layer6 heterozygotes were not detected at P3 and began to emerge later in development at P21 and persisted into adulthood (P56) (Fig. 4A′-C′).
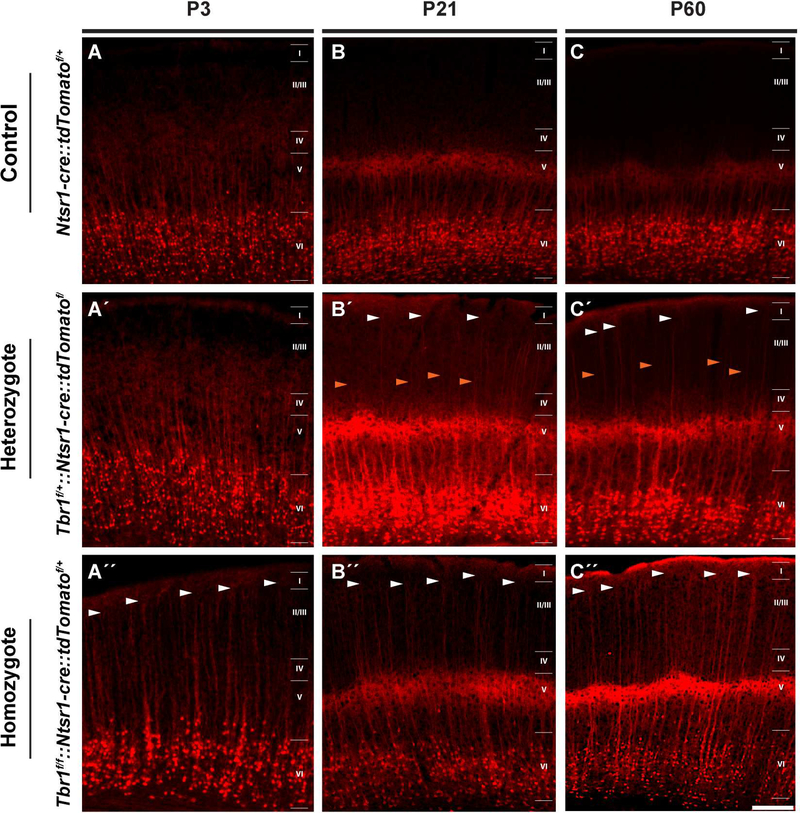
Figure 4: Ectopic growth of layer 6 apical dendrites into superficial layer 1 in Tbr1layer6 mutants.
The endogenous tdTomato fluorescence (red) in the SSCx of Control (Ntsr1-cre::tdTomatof/+) (A-C), Tbr1layer6 heterozygous (Tbr1f/+::Ntsr1-cre::tdTomatof/+) (A´-C´), and Tbr1layer6 homozygous mutants (Tbr1f/f::Ntsr1-cre::tdTomatof/+) (A´´-C´´). These mice had the Ntsr1cre::tdTomatof/+ alleles to label the layer 6 cell bodies and their dendrites. Changes in the dendritic patterning of layer 6 neurons were examined at P3 (A-A´´), P21 (B-B´´) and P56 (CC´´). White arrowheads indicate some of the apical dendrites extending through layers 2/3 to layer 1 in Tbr1layer6 mutants. Orange arrowheads indicates a group of apical dendrites that only extend to layers 2/3. Cortical layers are labelled. Scale bar: 50μm. See also Figure S4.
Interestingly, Tbr1layer6 heterozygotes had two different subtypes of apical dendrites; first group extended to layer1 (white arrowheads) similar to those in the null (Fig. 4A″-C″), and a second group that extended to layers 2/3 (orange arrowheads) (Fig. 4A′-C′). On the other hand, in the Tbr1layer6 homozygotes, the apical dendrites extended to layer 1 as early as P3, and these persisted into adulthood (P56) (white arrowheads, Fig. 4A″-C″). The change in dendritic morphology further supports the hypothesis that the mutant layer 6 neurons have layer 5-like properties. It is noteworthy that Tbr1 also regulates dendritic patterning of retinal ganglion cells (Liu et al., 2018).
Tbr1 is required after E17.5 for corticothalamic projections into the anteromedial thalamus.
Layer 6 and subplate neurons extend their axons through the basal ganglia to the thalamus, where they form a stereotypic topographic map between cortical areas and specific thalamic nuclei (Deck et al., 2013). Tbr1constitutive null axons fail to grow to the thalamus (Hevner et al., 2002). Here, we investigated corticothalamic projections in Tbr1layer6 mutants (Fig. S4). Despite the evidence that Tbr1layer6 mutant neurons have molecular and dendritic properties of layer 5 neurons (Figs. 1, 2 and 4), the mutant layer 6 neurons, like in the wildtype, have corticothalamic projections that enter the thalamus at P3 and P21 (Fig. S4A, S4B). The quantification of the corticothalamic projections in Tbr1layer6 mutants demonstrates that this reduction was most strongly seen in the anterior and anteromedial thalamus of rostral coronal sections at P21 (regions 4 and 5; Fig. S4C, S4D). However, Tbr1layer6 heterozygotes did not exhibit such deficit in their corticothalamic projections (data not shown).
To evaluate whether the phenotype was due to a failure to maintain the projections, or a failure to establish them, we studied neonatal Tbr1layer6 mutants at P3 (Fig. S4A). The P3 and P21 phenotypes were very similar. Thus, while Tbr1 is required prior to E17.5 for corticothalamic projections to emerge from the subpallium and enter the diencephalon (Hevner et al., 2002, Hevner et al., 2001), these processes take place in the Tbr1layer6 mutants, consistent with the presence the functional TBR1 protein in the corticothalamic neurons when these cells are initially specified and grow their axons to the thalamus. However, the Tbr1layer6 mutants show that after ~E17.5 Tbr1 is required for the maturation of corticothalamic connectivity preferentially in the anterior and anteromedial thalamus.
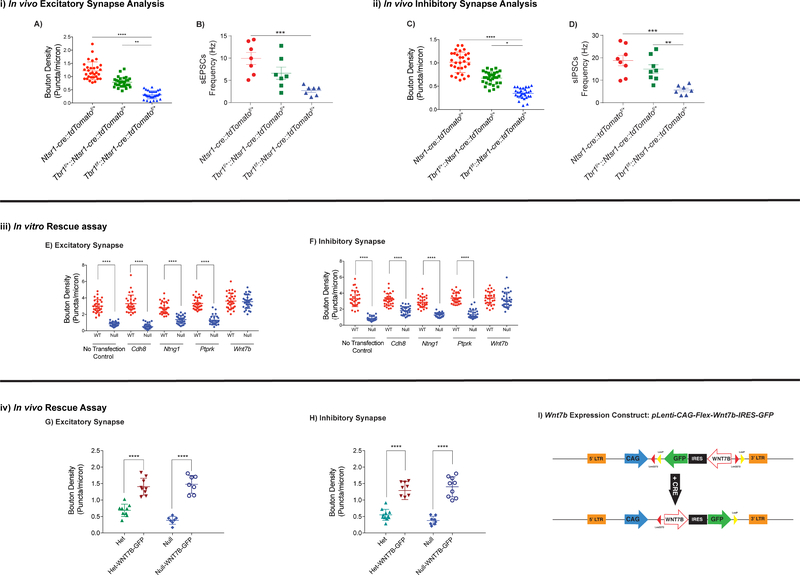
Excitatory synapse numbers are reduced in Tbr1layer6 mutants at P21 and P56.
We used immunofluorescence to label and analyze the excitatory presynaptic terminals (VGlut1+) that are apposed to dendritic postsynaptic zones (PSD95+) in SSCx of Tbr1wildtype, Tbr1layer6 heterozygous and homozygous mutants on the apical spines of layers 6 neurons (n=30) at P56 (Fig. 5i) and P21 (Fig. S5i). As depicted in Fig. S5A, we examined the excitatory synapses in layer 5 of SSCx of Tbr1wildtype (Ntsr1-cre::tdTomatof/+), Tbr1layer6 heterozygous (Tbr1f/+::Ntsr1-cre::tdTomatof/+) and Tbr1layer6 homozygous mutants (Tbr1f/f::Ntsr1cre::tdTomatof/+; Fig. S5A). Confocal fluorescent microscopy analysis of the synapse numbers showed a 30% decrease in Tbr1layer6 heterozygous (BD=0.772, p<0.0001) and 60% in Tbr1layer6 homozygous mutants at P56 (BD=0.415, p<0.0001; Fig. 5A). The synaptic deficit phenotype was also present at P21, where excitatory synapse numbers were reduced by 34% in Tbr1layer6 heterozygous (BD=0.501, p<0.0001) and 64% in Tbr1layer6 homozygous mutants at P21 (BD= 0.273, p<0.0001; Fig. S5G).
Figure 5: Tbr1 is required for the excitatory and inhibitory synaptic development of layer 6 pyramidal neurons at P56.
(i) Excitatory synapses were analyzed via synaptic bouton staining onto apical dendrites of layer 6 neurons (n=30) and spontaneous EPSC (sEPSC) recordings from the soma of Tbr1 wildtype, Tbr1layer6 heterozygous, and Tbr1layer6 homozygous mutants at P56. Ntsr1-cre::tdTomatof/+ allele was used to label the layer 6 neurons. ImageJ software was used to process confocal images for quantification. Excitatory synapses were analyzed by VGlut1+ boutons and PSD95+ clusters colocalizing onto the dendrites of layer 6 neurons. (A) Quantification of excitatory synaptic density at P56. (B) Quantification of the sEPSC frequency in layer 6 neurons at P56.
(ii) Inhibitory synapses were examined by synaptic bouton staining onto apical dendrites of layer 6 neurons and spontaneous IPSC (sIPSC) recordings from the soma of the layer 6 neurons of Tbr1 wildtype, Tbr1layer6 heterozygous, and Tbr1layer6 homozygous mutants at P56. Inhibitory synaptic input was measured by VGat+ boutons and Gephyrin+ clusters co-localizing onto the dendrites of layer 6 neurons. (C) Quantification of inhibitory synaptic density at P56. (D) Quantification of the sIPSC frequency in layer 6 neurons at P56.
(iii) In vitro rescue assay was conducted using Cdh8, Ntng1, Ptprk and Wnt7b expression vectors in cultured P0 cells from Tbr1wildtype (red) and Tbr1layer6 mutant (blue) (n=2). (E, F) Quantification of excitatory and inhibitory synaptic density in vitro.
(iv) In vivo rescue assay was conducted by injecting Wnt7b-IRES-GFP lentivirus into the layer 6 of SSCx of Tbr1layer6 heterozygous and Tbr1layer6 homozygous mutants at P1. (G, H) Quantification of excitatory and inhibitory synapse numbers onto the layer 6 neurons of Tbr1layer6 heterozygous (Het) and Tbr1layer6 homozygous mutants (Null) expressing GFP at P21.
(I) Schematic representation of the lentiviral CAG-Flex-Wnt7b-IRES-GFP (Wnt7b-IRES-GFP expressing) construct. CRE inverts the Wnt7b coding region enabling its expression. Two-way ANOVA was used for the statistical analysis of the control, heterozygote and null. Two-tailed Ttest with tukey correction was used for pairwise comparisons. (*p<0.05) (**p< 0.01) (***p<0.001) (****p<0.0001). See also Figure S5.
To study the physiological ramification of reduced excitatory synaptic density in Tbr1layer6 mutants, we measured spontaneous Excitatory Post-Synaptic Current (sEPSCs) using whole-cell patch clamp at P21 and P56. We recorded from neurons expressing Ntsr1-cre, identified using the fluorescent tdTomato Cre-dependent reporter, in SSCx from coronal slices of Tbr1wildtype, Tbr1layer6 heterozygous, and Tbr1layer6 homozygous mutant (Fig. S5A-C). The frequency of sEPSCs was reduced in Tbr1layer6 homozygous mutants as compared to cells from Tbr1wildtype mice at P56 (Fig. 5B: n = 7/7/7, wildtype/ heterozygous/ homozygous (n=number of patched cells); One-way ANOVA, F(2,18) = 10.17, p = 0.0011; t-test, Tukey correction, wildtype v. homozygous: q(18) = 6.371, p = 0.0008). The decreased sEPSC frequency was also present at P21 (Fig. S5I: n = 7/7/7, wildtype/ heterozygous/ homozygous; One-way ANOVA, F(2,18) = 6.625, p = 0.007; t-test, Tukey correction, wildtype v. homozygous: q(18) = 5.123, p = 0.0053).
Tbr1layer6 mutants exhibit altered cortical interneuron lamination and reduced inhibitory synaptic density.
The pattern of Sst+ cortical interneurons (CINs) and their lamination is abnormal in the SSCx of P3 Tbr1layer6 mutants (Fig. 2J-J″). Tbr1layer6 heterozygotes exhibited a decrease in the Sst+ CINs in layers 5 and 6 (Fig. 2J, J′; Fig. S3A); whereas, in Tbr1layer6 homozygotes, the Sst+ CINs were reduced in layer 6, unchanged in layer 5 and increased in layers 2–4 (Fig. 2J, J″; Fig. S3A). The reduction in Sst+ CINs was persistent at P21 (data not shown). However, there was no changes in PV+ CINs at P21 (Fig. S3B).
We suggest that the Tbr1layer6 mutation disrupts the laminar distribution of Sst+ CINs by altering the signals coming from the dendrites of the miss-specified layer 6 pyramidal neurons. This result, in conjunction with the reduction in excitatory synapses, led us to measure inhibitory synapse numbers in the Tbr1layer6 mutants. From confocal images, we counted the numbers of inhibitory terminals (VGat+ presynaptic structures) apposed to dendritic postsynaptic zones (Gephyrin+) onto the apical dendrites of layers 6 neurons (n=30) of Tbr1wildtype, Tbr1layer6 heterozygous and homozygous mutants at P56 (Fig. 5ii) and P21 (Fig. S5ii). Analysis of inhibitory synapse numbers showed a 33% decrease in Tbr1layer6 heterozygous (BD= 0.673, p<0.0001) and 66% in Tbr1layer6 homozygous mutants at P56 (BD= 0.346, p<0.0001; Fig. 5C). This phenotype was also detectable at P21, where the inhibitory synapse numbers were reduced 37% in Tbr1layer6 heterozygous mutants (BD= 0.574, p<0.0001) and 72% decrease in Tbr1layer6 homozygous mutants (BD= 0.252, p<0.0001) at P21 (Fig. S5M).
To test whether the reduced inhibitory synaptic density in Tbr1layer6 mutants had physiological ramifications, we measured spontaneous Inhibitory Post-Synaptic Current (sIPSCs) using whole-cell patch clamp on brain slices at P21 and P56. The frequency of sIPSCs were reduced in Tbr1layer6 homozygotes at P56 (Fig. 5D: n = 8/8/7, wildtype/ heterozygous/ homozygous; One-way ANOVA, F(2,20) = 12.44, p = 0.0003; t-test, Tukey correction, wildtype v. homozygous: q(20) = 6.907, p = 0.0003, heterozygous v. homozygous: q(20) = 4.901, p = 0.0066). The reduction in sIPSC frequency was also present at P21 (Fig. S5O: n = 7/6/7, wildtype/ heterozygous/ homozygous; One-way ANOVA, F(2,17) = 4.738, p = 0.023; t-test, Tukey correction, wildtype v. homozygous: q(17) = 3.847, p = 0.037, heterozygous v. homozygous: q(17) = 3.635, p = 0.0495). Lastly, we did not observe any changes in the amplitude of sEPSCs and sIPSCs at P21 and P56 (data not shown).
Restoring Wnt7b expression rescues the decreased synaptic phenotype of Tbr1layer6 mutant neurons in vitro and in vivo.
The decrease in the excitatory and inhibitory synaptic density in the layer 6 mutant neurons, in conjunction with the transcriptome changes in Tbr1layer6 FACS purified neurons, prompted us to identify a subset of genes that are dysregulated in Tbr1layer6 mutant neurons that may contribute to the synaptic deficit. These genes, including Cdh8 (Friedman et al., 2015, Liu et al., 2018), Ntng1 (Zhang et al., 2016), Ptprk (Lim et al., 2009) and Wnt7b (Budnik and Salinas, 2011) have been shown to contribute to synaptic development, maintenance and/or plasticity. Thus, we examined the impact of transfecting Cdh8, Ntng1, Ptprk and Wnt7b expression vectors in primary cortical cultures derived from Tbr1wildtype and Tbr1layer6 mutant neurons at P0 (n=2).
After 14 days in vitro, we analyzed the number of excitatory (VGlut+ presynaptic and PSD95+ postsynaptic) and inhibitory (VGat+ presynaptic and Gephyrin+ postsynaptic) terminals of Tbr1wildtype and Tbr1layer6 homozygous mutant neurons (Fig. 5iii). The reduced excitatory and inhibitory synaptic density was recapitulated in vitro, where excitatory and inhibitory synapse numbers were reduced by 71% (BD = 0.945, p<0.0001) and 78% (BD = 0.836, p<0.0001), respectively (Fig. 5E, 5F).
Amongst the four tested genes, only Wnt7b rescued the reduction in both excitatory (Fig. 5E) and inhibitory (Fig. 5F) synapse numbers. Therefore, we further investigated the impact of Wnt7b expression on rescuing synapse numbers in vivo (Fig. 5iv). We generated a flex lentiviral constructs that would express WNT7B and GFP upon Cre-recombination (Fig. 5I). We injected the Wnt7b-IRES-GFP lentivirus into layer 6 of SSCx of Tbr1layer6 heterozygotes and homozygotes at P1. The virus was only injected in the right hemisphere; the left hemisphere was used as a control. We analyzed excitatory and inhibitory synapse numbers on the apical dendrites of layer 6 neurons at P21 (Fig. 5G, 5H). The regions expressing GFP in layer 6 cells, showed an increase in excitatory and inhibitory synapse numbers (Fig. 5G, 5H); thus providing in vivo evidence that restoring Wnt7b expression can rescue the synaptic deficits of Tbr1layer6 mutants.
Tbr1 mutants have increased hyperpolarization-activated cation currents (Ih).
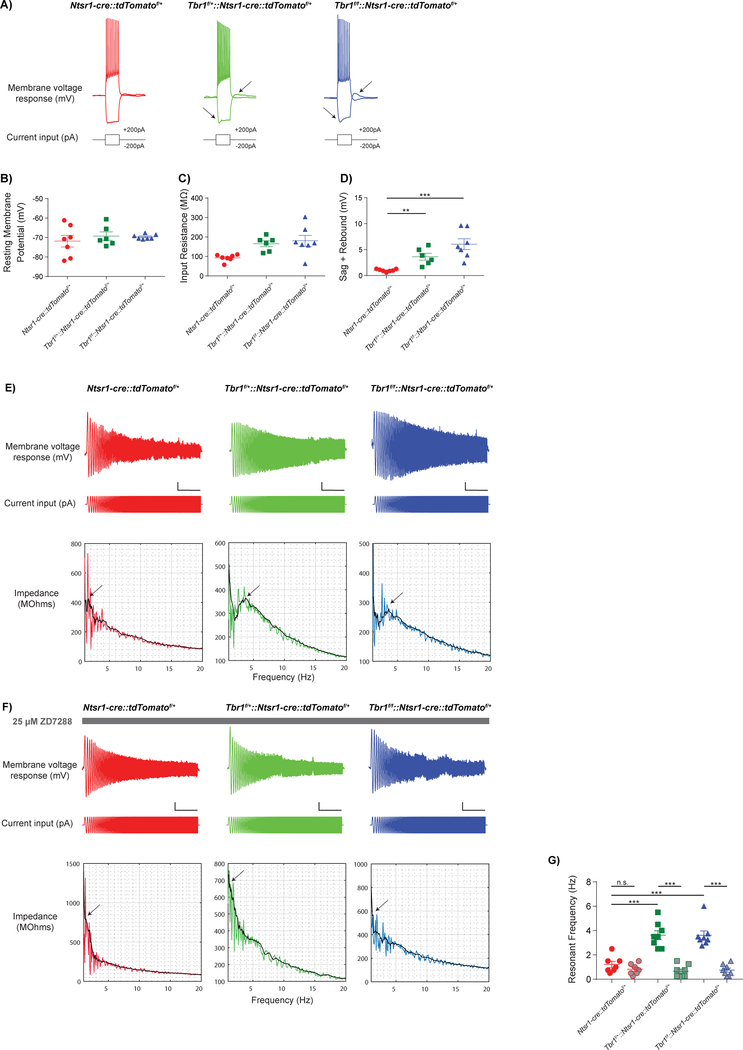
We next examined the intrinsic properties of layer 6 neurons in Tbr1layer6 mutants using whole-cell patch clamp to measure intrinsic physiological properties of Ntsr1-cre::tdTomato+ neurons of layer 6 in SSCx (Fig. 6). Resting membrane potential (Fig. 6B, Fig. S6B) and input resistance (Fig. 6C, Fig. S6C) were not different between Tbr1wildtype, Tbr1layer6 heterozygotes, and Tbr1layer6 homozygotes (n=8) at P56 (Fig. 6) and P21 (Fig. S6).
Figure 6: Loss of Tbr1 in layer 6 somatosensory cortex results in an increase in hyperpolarization-activated cation currents (Ih).
Whole-cell patch clamp recordings from layer 6 SSCx at P56 (A-D) show that many intrinsic electrophysiological properties were unaffected by loss of Tbr1, including resting membrane potential (B), input resistance (C), and action potential half-width (data not shown). (E) Neurons were held in current clamp at −70mV. The resonant frequency was measured as the frequency at which the impedance profile reached its peak (arrows). Scale bar = 5 mV, 5 s. (F) ZD7288, an HCN channel blocker, decreased resonance frequency by over 50% in Tbr1layer6 heterozygous (green), and Tbr1layer6 homozygous mutants (blue). (G) Quantification of changes in resonant frequency of Tbr1wildtype (red), Tbr1layer6 heterozygous (green) and Tbr1layer6 homozygous mutants (blue) after ZD7288 treatment. (**p< 0.01) (***p<0.001). See also Figure S6.
A prominent feature of many layer 5 pyramidal neurons that is largely absent from layer 6, is a hyperpolarization-activated cation current (Ih or h-current) mediated by HCN channels (Shepherd, 2013). Ih causes a characteristic “sag” and “rebound” in current clamp recordings of responses to steps of hyperpolarizing current. We examined responses to a −200 pA step, and found that SSCx layer 6 pyramidal neurons from P56 Tbr1layer6 heterozygotes and homozygotes exhibited significantly increased “sag + rebound” compared to Tbr1wildtype controls, suggesting increased Ih, while other intrinsic electrophysiological properties were largely unaltered (Fig. 6D: n = 7/6/7, wildtype/ heterozygous/ homozygous; One-way ANOVA, F(2,17) = 13.18, p = 0.0003; t-test, Tukey correction, wildtype v. heterozygous: q(17) = 3.693, p = 0.0457; wildtype v. homozygous: q(17) = 7.258, p = 0.0002). Likewise, the neurons from Tbr1layer6 homozygotes at P21 also exhibited an increased Ih compared to Tbr1wildtype controls (Fig. S6D: n = 8/8/8, wildtype/ heterozygous/ homozygous; One-way ANOVA, F(2,21) = 17.68, p < 0.001; t-test, Tukey correction, wildtype v. homozygous: q(21) = 8.331, p < 0.0001; heterozygous v. homozygous: q(21) = 5.16, p = 0.0041). Furthermore, HCN1 protein levels were increased ~5-fold in Tbr1layer6 homozygotes compared to Tbr1wildtype controls (Fig. S6E, S6F), suggesting that upregulation of HCN1 could be contributing to the changes in the Ih of Tbr1layer6 mutant neurons.
In deep layer neocortical pyramidal neurons, the presence of Ih shifts the resonant frequency towards higher frequencies (Dembrow et al., 2010). Therefore, to further characterize potential increases in Ih in Tbr1layer6 mutants, we estimated the resonant frequency. For this, we injected constant current to hold Ntsr1-cre+ neurons in current clamp near −70mV, then introduced a sinusoidal current stimulus with constant amplitude (100 pA peak-to-peak) and a frequency that increased linearly from 0 to 20 Hz over 20 seconds (Fig. 6E). We used the ratio of the fast Fourier transform of the voltage response (Fig. 6E top) to the fast Fourier transform of the sinusoidal current stimulus (Fig. 6E middle) to calculate the impedance amplitude profile (Fig. 6E bottom). We defined the resonant frequency as the frequency at which the impedance profile reached its peak. Tbr1layer6 heterozygous and Tbr1layer6 homozygous mutants exhibited an increase in their resonant frequency compared to Tbr1wildtype controls at P56 (Fig. 6G: n = 7/8/8, wildtype/ heterozygous/ homozygous; One-way ANOVA, F(2,20) = 16.24, p < 0.0001; t-test, Tukey correction, wildtype v. heterozygous: q(20) = 7.075, p = 0.0002; wildtype v. homozygous: q(20) = 7.038, p = 0.0002).
Finally, we blocked Ih by bath applying the specific HCN channel antagonist ZD7288 (25 μM; Fig. 6F). The resonant frequency was reduced by over 50% in the Tbr1layer6 heterozygous (Fig. 6G: n = 8; paired T-test, t(7) = 7.723, p < 0.0001) and Tbr1layer6 homozygous mutants (Fig. 6G: n = 8; paired T-test, t(7) = 8.194, p < 0.0001). However, the resonant frequency was not significantly altered by ZD7288 in the neurons from the Tbr1wildtype mice, indicating that Ih contributes to intrinsic resonance in mutant, but not wildtype, layer 6 pyramidal neurons. Thus, both Tbr1layer6 heterozygotes and homozygotes have an increased Ih resembling that of layer 5 neurons.
Tbr1 mutants exhibit increased aggressive and anxiety-like behaviors.
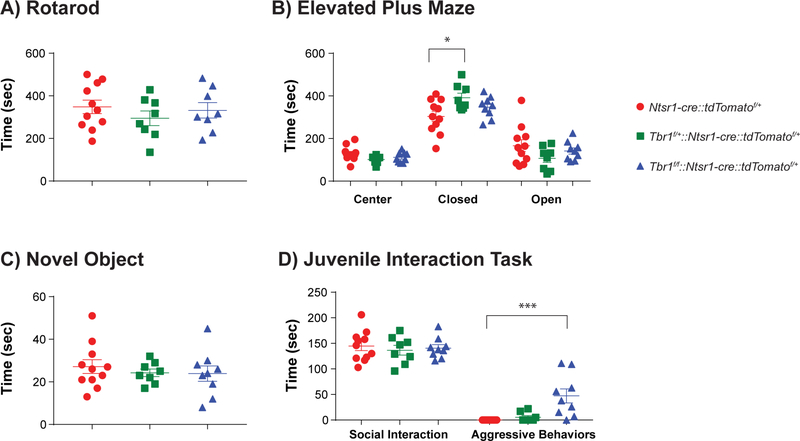
To identify behavioral abnormalities linked to loss of Tbr1 function in layer 6 neurons, we performed assays in littermate cohorts of Tbr1wildtype, Tbr1layer6 heterozygous and homozygous mutant male and female mice between P56–80. Tbr1layer6 heterozygous and Tbr1layer6 homozygous mutants did not show impairments in locomotion as measured by speed in the open field (data not shown) or in motor coordination as measured by performance on a rotarod (Fig. 7A) compared to wildtype controls. Tbr1layer6 mutants did not differ in the amount of time spent in the center of the open field (data not shown). However, in the elevated plus maze, Tbr1layer6 heterozygous mutants spent more time in the closed arms, suggesting an increase in anxiety-like behavior (Fig. 7B: n = 11/8/9, wildtype/ heterozygous/ homozygous; One-way ANOVA, F(2,25) = 4.155, p = 0.028; t-test, Tukey correction, wildtype v. heterozygous: q(25) = 4.065, p = 0.022).
Figure 7: Tbr1layer6 mutants exhibit increased aggressive and anxiety-like behavior at P56P80.
Behavioral analysis of Tbr1wildtype (red), Tbr1layer6 heterozygous (green), and Tbr1layer6 homozygous mutants (blue). (A) Rotarod assay did not demonstrate any impaired movement or motor coordination in Tbr1layer6 mutants. (B) Tbr1layer6 heterozygous (green) mutants spent more time in the closed arm of the elevated plus maze (an anxiety-like phenotype) compared to their Tbr1wildtype littermates. (C) Loss of Tbr1 did not affect the time spent engaged in novel object exploration or (D) social interactions. (D) Tbr1layer6 homozygous mutants (blue) exhibited aggressive behaviors when interacting with a novel juvenile mouse. Two-tailed T-test with tukey correction was used for pairwise comparisons (*p<0.05) (***p<0.001).
To assay mouse social behavior, we measured the time each experimental animal spent exploring a novel object and a novel juvenile wildtype mouse of the same sex introduced to its home cage. There were no differences in the time Tbr1layer6 mutants spent exploring a novel object (Fig. 7C). However, the Tbr1layer6 homozygous mutants spent more time with the novel juvenile mouse (Fig. 7D: n = 11/8/9, wildtype/ heterozygous/ homozygous; One-way ANOVA, F(2,25) = 4.534, p = 0.021; t-test, Tukey correction, wildtype v. homozygous: q(25) = 3.731, p = 0.0364). To further assess the type of interaction, we divided this time into “social interaction”, defined as time spent in sniffing, close following or allo-grooming, vs. “aggressive” behavior, defined as biting, fighting, or close following associated with periods of active fighting. Tbr1wildtype, Tbr1layer6 heterozygous and homozygous mutants spent similar time in social interaction (Fig. 7D). However, Tbr1layer6 homozygous mutants exhibited a marked increase in aggressive interactions with the juvenile (Fig. 7D: n = 11/8/9, wildtype/ heterozygous/ homozygous; One-way ANOVA, F(2,25) = 10.97, p = 0.0004; t-test, Tukey correction, wildtype v. homozygous: q(25) = 6.237, p = 0.0005). These data indicate that neonatal Tbr1 deletion in layer 6 neurons in adult mice leads to increased anxiety-like behavior in heterozygotes and increased aggressive behavior in homozygous mutants.
DISCUSSION
Neonatal Tbr1 specifies properties of a sublamina of neocortical layer 6 (corticothalamic) and represses layer 5 (corticofugal) identity.
Tbr1 is expressed in the excitatory neurons of the neocortex (subplate, layer 6, rostral layer 5, layers 2/3), hippocampus, entorhinal cortex, pallial amygdala, piriform cortex, olfactory bulb and CR neurons (Hevner et al., 2001, Hevner et al., 2003). Analysis of Tbr1constitutive null mice demonstrated its function in the differentiation of the first waves of pallial glutamatergic neurons including CR cells, olfactory bulb mitral cells, subplate cells and layer 6 cells (Bedogni et al., 2010). Further analyses of Tbr1constitutive null mice revealed that Tbr1 promotes the identity of layer 6 neurons by repressing layer 5 molecular properties in layer 6 (McKenna et al., 2011, Han et al., 2011).
Here, by deleting Tbr1 late in gestation, we have demonstrated that Tbr1 is required in maintaining subplate and layer 6 identity. The impaired differentiation of subplate and layer 6 neurons is indicated by molecular (Figs. 1, 2) and dendritic defects (Fig. 4). Tbr1layer6 mutant neurons have reduced expression of layer 6 markers including Wnt7b, Foxp2 and Tle4 and have ectopic layer 6 expression of genes controlling layer 5 molecular properties including Bcl11b, Fezf2 and Foxp1. Strikingly, the expression of Tle4 and Foxp2 are most strongly reduced in the deep part of layer 6 but is maintained in the subplate and the superficial part of layer 6. This result suggests the existence of layer 6 sublamina, which is consistent with the previously reported study (Chevée et al., 2018), that we refer to as layer 6aupper and layer 6alower. Subplate is considered layer 6b (Hoerder-Suabedissen et al., 2018). The ectopic expression of layer 5 markers in the Tbr1layer6 mutants suggest that layer 6 mutant neurons have an altered fate that is a hybrid of layer 5 and layer 6 pyramidal neurons. This conclusion is supported by our computational transcriptomic analysis of DEX genes between layers 5 and 6 in wildtype at P5 and comparing those to the group of DEX genes from Tbr1layer6 mutant neurons (Fig 1B). We discovered that 60% of the common genes between the two datasets changed in the same direction, demonstrating that Tbr1layer6 mutant neurons share transcriptomic properties of layer 5 neurons. On the contrary, the remaining 40% of genes showed no significant change in the Tbr1layer6 mutant neurons, showing that Tbr1layer6 mutant neurons possess a hybrid fate of layer 5 and layer 6 neurons.
Despite their hybrid molecular characteristic, some phenotypes suggested a transformation towards layer 5 identity. For instance, dendritic patterning of layer 6 neurons resembled that of layer 5 neurons, as their apical dendrites extended superficially into the marginal zone (Lefebvre et al., 2015). In addition to changes in the transcriptome and in dendritic patterning, Tbr1layer6 mutant neurons have increased Ih, similar to that of layer 5 neurons. There are at least two subclasses of pyramidal neurons within layer 5. Type B intratelencephalic (IT) cells lack prominent Ih, whereas Type A corticofugal (CF) cells have a prominent Ih (Shepherd, 2013). In layer 6 of the Tbr1layer6 mutants, the increase in Ih and levels of HCN1 protein suggests that the mutant layer 6 neurons have properties similar to Type A layer 5 pyramidal neurons. Thus, Tbr1 persistent function is required to initiate, orchestrate and maintain a layer 6 specific program, and while repressing the layer 5 (specifically Type A/CF) molecular, dendritic and physiological program.
Contrary to the Tbr1constitutive null mice where corticothalamic axons fail to grow into the thalamus (Hevner et al., 2001, Hevner et al., 2002), Tbr1layer6 mutants have corticothalamic projections that enter the thalamus. However, their intrathalamic ramifications are abnormal, with decreased projections in the anterior and anteromedial thalamus. Thus, even though Tbr1 is required to initiate the corticothalamic pathway, it is not required to maintain these axons through P56 in the Tbr1layer6 mutants. Furthermore, no ectopic subcortical projections or corpus callosum projections are generated by Tbr1layer6 mutant neurons. Thus, despite taking on many layer 5 properties, the mutant layer 6 neurons do not grow layer 5-like axonal projections. Therefore, once the layer 6 axonal pathway choice program is established by Tbr1, it is irreversibly maintained in the absence of Tbr1. On the other hand, Tbr1 dependent programs for promoting layer 6 gene expression, repression of layer 5 gene expression, layer 6-specific dendritic patterning and physiological properties (Ih) remain plastic and are dependent upon Tbr1 function during later stages of development and adulthood.
Overall, these results support our hypothesis that deleting Tbr1 late in mouse gestation, induces a hybrid fate in layer 6 and subplate neurons. Tbr1layer6 mutant neurons transform to have many properties of layer 5 pyramidal neurons including ectopic expression of regulators of layer 5 identity, dendritic patterning, and cell intrinsic physiology, while also maintaining some aspects of layer 6 identity, such as their axonal projections to the thalamus.
Tbr1 directly regulates the transcription of genes that control layer 6 identity.
Towards elucidating TBR1-regulated transcriptional pathways that control layer 6 properties, we combined transcriptomic analysis of FACS purified neonatal wildtype and Tbr1layer6 mutant, with whole genome neonatal TBR1 ChIP-Seq. These genomic analyses show that TBR1 directly regulates the transcriptional program driving layer 6 identity via genomic binding to gene promoters and distal enhancers. Our data further suggest TBR1 interaction is mediated both by direct binding of TBR1 to the canonical or degenerate TBR1 motif, as well as via secondary interaction where the TBR1 motif is absent but TBR1 still interacts. Differential expression changes, especially for genes that control layer 6 identity were more strongly associated with direct TBR1 binding to its cognate motif, evidenced by ChIP-Seq signal and putative motif presence at regulatory DNA elements associated with down- and up-regulated genes. Despite these associations, TBR1 interaction, as identified via ChIP-Seq alone, is not strongly indicative of an activating or repressive function. Furthermore, in general, TBR1 appears to be present at regulatory sequences of highly expressed genes in the P2 cortex. Investigation of the impact of Tbr1 loss-of-function on chromatin state and transcriptional activation and repression, as has been done for other key transcriptional regulators (Sandberg et al., 2016), could further elucidate the regulatory function of TBR1 binding.
Transcription assays performed herein, using neonatal primary cortical cultures demonstrated that TBR1 functions as an activator or repressor of specific REs adjacent to genes whose expression changes in Tbr1layer6 mutant neurons. Importantly, TBR1 activated REs near to genes whose expression was reduced in layer 6, and repressed REs near to genes whose expression was increased in layer 5. Future studies are needed to identify the nuclear co-factors that determine whether TBR1 acts as a transcriptional activator or repressor, although it is conceivable that the DNA sequence of the REs modifies TBR1’s confirmation to control its activity. The discovery of these TBR1 regulated REs opens the possibility that these elements will show in vivo layer-specific activity which could be elucidated using transgenic experiments. These REs also serve as essential nodes for establishing the transcriptional circuits that drive TBR1 mediated gene expression, as well as sites where mutations may contribute risk for human neurodevelopmental disorders.
Tbr1 is required for proper synaptic development in layer 6 neurons.
A reduced density of excitatory and inhibitory dendritic synapses is a central phenotype of the Tbr1layer6 heterozygous and homozygous mutants as seen in tissue sections from P21 and P56. A similar reduction was also seen in primary cultures grown from P0 cortex. These findings were substantiated using slice physiology, where Tbr1layer6 mutant neurons exhibit reduced sEPSCs and sIPSCs at P21 and P56. It is pertinent that Tbr1layer6 heterozygotes have reduced synapse numbers and reduced sEPSCs and sIPSCs, since de novo Tbr1 loss-of-function mutations are found in the heterozygous state. Genes annotated with having synaptic function are enriched among ASD genes, suggesting this is a relevant pathway, although more work is needed to clarify this (Sanders et al., 2015). Therefore, the observed synaptic deficit in Tbr1layer6 mutants may be relevant to ASD. We further examined the impact of Tbr1 loss-of-function on synaptogenesis and synaptic function by identifying a subset of Tbr1 regulated genes (66/178) that are linked to biological processes that could affect synapse development. These genes encode proteins implicated in signaling through G-protein coupled receptors, WNTs and retinoids, and that regulate cell adhesion (Table 3) (Yee and Chen, 2016).
WNT signaling is well-known to control synapse development (Davis et al., 2008). WNTs promote synaptic assembly by signaling to the developing pre and postsynaptic compartments (Budnik and Salinas, 2011, Salinas and Zou, 2008). Importantly, WNTs also are implicated in synaptic changes induced by neuronal activity in mature neurons (Budnik and Salinas, 2011). Here we showed that restoring Wnt7b expression in Tbr1layer6 mutant neurons rescued the decrease in excitatory and inhibitory synapse numbers in vitro and in vivo. This provides evidence that downregulation of Wnt7b may contribute to the synaptic deficits in Tbr1layer6 mutants.
Currently, there are no effective somatic treatments for most core deficits of ASD. More broadly there are no current treatments for neuropsychiatric illness in humans that restore normal biology. The ability to successfully restore synapse numbers by expressing Wnt7b may provide a possible avenue to restoring synapse numbers in humans with TBR1 mutations using small molecule WNT7B agonists. In short, these observations provide an important initial step in conceptualizing rational therapies for ASD patients – though, of course, critically important hurdles remain, including demonstrating that the observed biology is truly relevant for pathology in humans and, if this is the case, determining at what developmental stages interventions may have an impact on core components of the ASD phenotype.
Tbr1layer6 mutants exhibit increased aggression and anxiety-like behaviors.
Tbr1layer6 mutants are viable allowing us to interrogate their behavior, which was remarkably normal in many assays including assays of their motor functions (rotarod and open field) and interest in novel objects. On the other hand, Tbr1layer6 heterozygous mutants spent more time in the closed arms of the elevated plus maze, reflecting an increase in innate anxietylike behaviors. Furthermore, homozygotes Tbr1layer6 mutants exhibited prolonged periods of aggression towards juvenile mice.
Ntsr1-Cre recombination of Tbr1 does not extend into many cortical regions, including the olfactory bulb, dorsomedial neocortex (cingulate and retrosplenial), hippocampus and parahippocampus, piriform cortex, and pallial amygdala, ruling out the possibility that defects in Tbr1+ neurons in these structures contributes to the behavioral phenotypes. This is pertinent, as Tbr1constitutive heterozygotes have abnormal amygdala connectivity that has been associated with deficits in social interaction, cognitive flexibility and associative memory (Huang et al., 2014). Thus, the highly specific molecular and physiological defects in the early born pyramidal neurons of the neocortical subplate and layer 6 can be implicated in the mutants’ increased aggression and anxiety-like behavior(s). While these phenotypes have similarities to common comorbidities of ASD, the relevance is unclear. However, it is relevant that loss of a single Tbr1 copy in mouse leads to alterations in complex behaviors reflecting the type of circuit-based dysfunctions that likely underlies ASD.
Insights into how Tbr1 loss-of-function mutations contribute risk for ASDs.
Genetic analyses of ASD patients have identified TBR1 as a high confidence risk factor for ASD (Sanders et al., 2015). Analyses of co-expression networks of ASD risk genes provides evidence that reduced dosage of genes, such as Tbr1, may underlie ASD by disrupting processes in immature projection neurons of deep cortical layers during human mid-fetal development (Willsey et al., 2013). Here, by deleting Tbr1 at a stage similar to the mid-fetal human, we have identified several novel Tbr1 functions in mouse that provide hypotheses about how a reduction in Tbr1 dosage may contribute to ASD pathophysiology.
As ASD loss-of-function mutations have their effect in the heterozygous state, a key discovery is that Tbr1layer6 heterozygous mice have a reduced density of excitatory and inhibitory dendritic synapses and reduced sEPSCs and sIPSCs. This supports a hypothesis that reduced TBR1 dosage increases ASD risk by reducing synaptic input onto layer 6 cortico-thalamic neurons. This model converges with the observation that many hcASD genes encode proteins that regulate synapse development and function (Sanders et al., 2015). Tbr1layer6 heterozygotes also had defects in dendritic patterning, an increased Ih, and anxiety-like behaviors in the elevated plus maze assay. While some of the other phenotypes detected in Tbr1layer6 mutants were only present in the homozygotes, these observations could have relevance for ASD as they signify biological pathways that could be altered in Tbr1 heterozygotes; including corticothalamic projections, and aggression.
Furthermore, the transcriptome analysis revealed that Tbr1 regulates other ASD genes, including Scn2a1, Foxp1, Wnt7b, Nuak1 (Sanders et al., 2015) and Foxp2 (Gong et al., 2004). In situ hybridization showed that Bcl11a expression, a probable ASD gene (Sanders et al., 2015) is also reduced in Tbr1layer6 mutants. Additionally, previous work provided evidence that the Tbr1constitutive null regulates expression of ASD risk genes during cortical development (Notwell et al., 2016). Of the five reported TBR1 de novo mutations associated with ASD, two generate truncated proteins that lack the DNA-binding T-box domain. These TBR1 mutant proteins lose their ability to regulate transcription, have an altered intracellular distribution and fail to interact with CASK (Huang and Hsueh, 2017) and FOXP2 (Deriziotis et al., 2014). Additional evidence of Tbr1 involvement in molecular pathways relevant to ASD includes its regulation of Grin2b, Bcl11a, Foxp1, Foxp2, and Wnt7b, a subset of ASD genes that also regulate cortical development (Sanders et al., 2015). Furthermore, CASK phosphorylation of TBR1, by protein kinase A, enhances TBR1’s direct activation of Grin2b expression (Chuang et al., 2014).
In summary, here we have elucidated core elements of a Tbr1-driven transcriptional circuit operating in neonatal mouse cortical layer 6 neurons (Fig. 8). This program represses layer 5 transcriptomic properties, layer 5 dendritic pattern and layer 5-like Ih. It promotes and maintains cortical axons entering thalamus and promotes excitatory and inhibitory synapse formation onto layer 6 pyramidal neurons. Tbr1 is a hcASD gene and drives a transcriptional network that includes a subset of ASD genes, including Scn2a1, Grin2b, Bcl11a, Foxp1, Nuak1 and Wnt7b. Considering that Tbr1layer6 heterozygotes, studied at a developmental stage that roughly approximates a point of convergent vulnerability in human pathology, have reduced synapse numbers, sEPSCs and sIPSCs, and increased Ih, we propose that these phenotypes offer important new insights into how Tbr1 loss-of-function mutations may contribute to ASD pathology in humans.
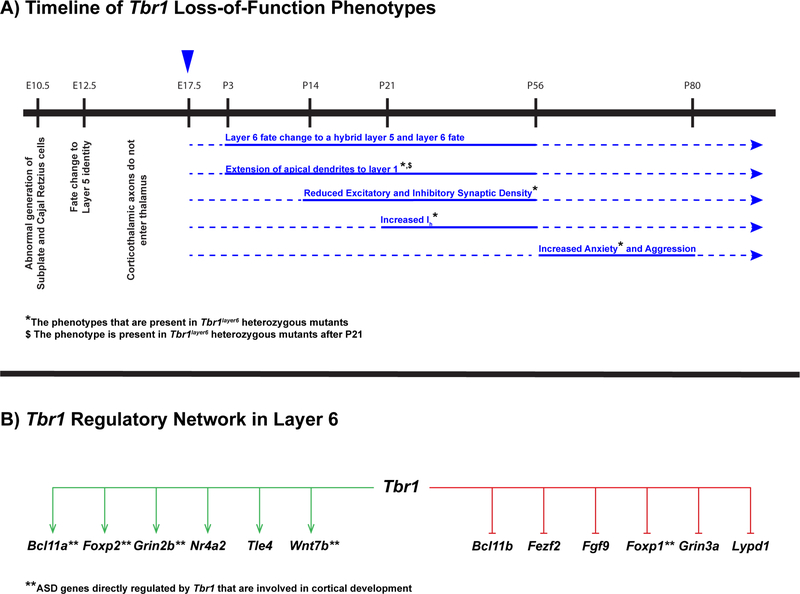
Figure 8:
(A) Schematic representation of a timeline of Tbr1 loss-of-function phenotypes from embryonic stages until adulthood in mouse. The blue arrowhead at E17.5 corresponds to the timing of knocking out Tbr1 in layer 6 using conditional mutagenesis. Postnatal phenotypes associated with Tbr1 loss-of-function are shown in blue. Solid lines correspond to the developmental window in which we have provided evidence for the reported phenotypes. Dotted line represents the presumed duration of the reported phenotypes. (*) Indicates the phenotypes that are observed in Tbr1layer6 heterozygotes and homozygotes. (B) Schematic representation of regulatory network of Tbr1 in cortical layer 6. Tbr1 is a repressor (red) of determinants of layer 5 identity including Bcl11b, Fezf2, Fgf9, Foxp1, Grin3a and Lypd1. Conversely, Tbr1 dictates layer 6 identity through activation (green) of layer 6 markers including Bcl11a, Foxp2, Grin2b, Nr4a2, Tle4 and Wnt7b. (**) Indicates ASD genes directly regulated by Tbr1 (TBR1 genomic binding and expression changes in the mutant) that are involved in cortical development.
STAR Methods:
CONTACT FOR REAGENT AND RESOURCE SHARING:
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. John L. Rubenstein (john.rubenstein@ucsf.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS:
Animals:
All procedures and animal care were approved and performed in accordance with the University of California San Francisco Laboratory Animal Research Center (LARC) guidelines. All strains were maintained on a C57Bl/6 background. Animals were housed in a vivarium with a 12hr light, 12hr dark cycle. Postnatally, experimental animals were kept with their littermates. For timed pregnancies, noon on the day of the vaginal plug was counted as embryonic day 0.5.
The Tbr1flox allele was generated by inGenious Targeting Laboratory (Ronkonkoma, NY). LoxP sites were inserted into introns 1 and 3, flanking Tbr1 exons 2 and 3 (Fig. 1A). To enable selection of homologous recombinants, the LoxP site in intron 3 was embedded in a neo cassette that was flanked by Flp sites. The neo cassette was removed by mating to a Flp-expressing mouse to generate the Tbr1flox allele. Cre excision removes exons 2 and 3, including the T-box DNA binding region, similar to the constitutive null allele (Bulfone et al., 1998). Ntsr1-cre mice (Gensat 220) were used to delete Tbr1 in layer 6 projection neurons. tdTomatofl/+ (Ai14) mice were crossed with Tbr1f/f mice and used as an endogenous reporter. Tbr1 layer 6 knockout mice (Tbr1layer6 mutant) were generated by crossing Tbr1f/f::tdTomatof/+ mice with Tbr1f/+::Ntsr1cre+. The specific gender and age of experimental animals can be found in the Results section and corresponding figure legends.
TRANSGENIC ANIMAL MODELS:
Information about the generation and genotyping of the transgenic lines used in this study can be found in the corresponding original studies: Ntsr1-Cre (Gong et al., 2007), lox-STOP-loxtdTomato (Ai14; Madisen et al., 2010). Mice were maintained on C57BL/6J background.
METHOD DETAILS:
Genomic DNA extraction and genotyping:
Tissue samples were digested in a solution containing 1 mg/mL of proteinase K, 50 mM Tris-HCl pH 8.0, 100 mM EDTA, 100 mM NaCl and 1% SDS. Genomic DNA was extracted using a standard ethanol precipitation protocol. Genotyping was performed with PCR-based assays using purified genomic DNA, and primer-pair combinations flanking the deleted region and detecting Cre and tdTomato alleles.
RNA extraction and cDNA synthesis:
Total RNA was extracted from the cortices of wildtype and Tbr1 constitutive null mice at E15.5 and P0 using RNeasy Plus® Mini Kit (QIAGEN) following the manufacturer’s protocol. First strand cDNA was synthesized using Superscript reverse transcriptase II following manufacturer’s protocol (Thermofisher).
Quantitative real time PCR (qPCR):
Quantitative RT-PCR was performed to measure RNA levels using SYBR Green (BioRad) and 7900HT Fast Real-Time PCR System. Gene-specific primers for Tbr1 exons 1, 2 and 4, Bcl11a, Grin2b and Hcn1 as well as ef1α housekeeping genes (HKG) were designed using the Primer 3 program. The expression levels of the genes in both wildtype and Tbr1 mutant mice were normalized to the expression levels of ef1α. Subsequently, the gene expression levels in Tbr1 mutant mice were measured relative to the wildtype littermates.
Western blot (WB):
Cortices of 2 Tbr1 constitutive null and 2 wildtype brains were dissected at E15.5 and P0 in ice-cold PBS. For assessing HCN1 protein levels, the somatosensory cortex was dissected in ice-cold HBSS from P7 mice. Cortices were dissociated using a Papain Dissociation System (Worthington Biochemical Corporation) following manufacturer’s protocol. tdTomato+ cells were sorted using BD FACS Aria II Cell Sorter at Center for Advanced Technology (UCSF).
Tissues were homogenized in 300 μL ice-cold RIPA lysis buffer. Following an incubation at 4°C for 2 hrs with agitation, the samples were centrifuged at 13,500 rpm for 20 min at 4°C. 20–30 μg total protein was combined with Laemmli buffer supplemented with 1:20 βmercaptoethanol and was heat to 95°C for 5 min. The protein lysate was electrophoresed using Mini-PROTEIN®TGX 4–20% precasted gels (Bio-Rad) and ran for 1–2 hrs at 100V. The franctionated proteins were transferred to a nitrocellulose membrane (GE Amersham Protran). The membrane was blocked with 7.5% nonfat dried milk, washed 3X with 1X PBS with 0.1% Tween-20, and then was incubated for 12 hrs with the primary antibody at 4°C. The following day, the membrane was washed 3X with 1X PBS with 0.1% Tween-20, incubated with the Goat Anti-Rabbit-HRP secondary for 1 hr. Signals were detected using a DAB system (Vector Laboratories) following manufacturer’s protocol.
RNA-Seq on FAC-Sorted Cells:
Layer specific transcriptome profiling was conducted by using RNA-seq on FAC-Sorted cells from somatosensory cortex of Tbr1wildtype and Tbr1layer6 mutants. The somatosensory cortex was dissected in HBSS from P5 mice (Thermofisher). Cortices were dissociated using a Papain Dissociation System (Worthington Biochemical Corporation) following manufacturer’s protocol. tdTomato+ cells were sorted using BD FACS Aria II Cell Sorter at Center for Advanced Technology (UCSF). Approximately 25,000 cells were collected from each sample and immediately proceeded with RNA extraction using RNeasy®Plus Micro Kit (QIAGEN) following manufacturer’s protocol. using Agilent RNA 6000 Nano Kit (Agilent Technologies) and ran on Bioanalyzer 2100 (Agilent Technologies) and samples that had RIN scores of 8.5–9.5 were used to generate libraries. Library preparation and amplification was performed by TruSeq® Stranded Total RNA Library Prep Kit with Ribo-Zero Gold Set A (Illumina). The amplification of adapter-ligated fragments was carried out for 12 cycles during which individual index sequences were added to each distinct sample. Library concentration was assessed with Qubit (INFO) and library fragment size distribution was assessed on the Agilent Bioanalyzer 2100 (Agilent Technologies) and Agilent High Sensitivity DNA Kit (Agilent Technologies) following manufacturer’s protocol. Libraries were validated using qPCR. Pooled, indexed RNA-seq libraries were sequenced on Hiseq 4000 at Center for Advanced Technology (UCSF) to produce 100 bp paired-end reads.
Bioinformatics analysis of FAC-Sorted layer 6 RNA-Seq:
Collectively, we analyzed 8 RNA-Seq libraries, which comprised of 4 Tbr1wildtype layer 6, 5 Tbr1wildtype layer 5, and 4 Tbr1layer6 layer 6 mutant RNA-Seq libraries. Sequencing was conducted on HiSeq 4000 using Paired-End 100 (PE100) with the Library fragment size of approximately 300 bp.
RNA-Seq alignment, and quality control:
The RNA-Seq reads were aligned to the mm9 mouse genome reference using STAR in gene annotation mode. Picard was utilized to generate alignment quality control (QC) metrics for every RNA-Seq samples. Principal component analysis (PCA) of the quality control matrices was employed to determine the presence of RNASeq sample outliers (The outliner is defined as a sample whose QC metrics are at least three standard deviations away from the mean in any of the first three principal components). The analysis did not indicate any outliers in layer 6 samples. We found and removed one outlier in layer 5 WT control samples. After quality control, we had 4 Tbr1wildtype layer 5, 4 Tbr1wildtype layer 6, and 4 Tbr1layer6 layer 6 mutant samples.
Gene expression estimation and normalization:
Gene expression was quantified with HTSeq in intersection-strict mode. We created two subsets of samples after obtaining raw gene expression counts. The first subset contained 4 layer 6 Tbr1wildtype and 4 layer 6 Tbr1layer6 mutant samples. The second subset had 4 layer 5 Tbr1wildtype and 4 layer 6 Tbr1wildtype samples. We employed the same gene filtering and normalization approach to process both subsets of samples. We removed genes have less than or equal to one read in more than 50% of the samples. Filtered genes were normalized for gene length, GC content, and sample library size using CQN R-package. Gene length is obtained directly from the gene annotation file (.GTF) of mouse mm9 genome build reference. BedTools is used to compute the gene GC content. After normalization, the genes whose expression value don’t change across all samples are removed. PCA is applied to identify any sample outliners with those filtered and normalized gene expression. The expression values were scaled and centered before PCA. PCA over gene expression shows that there are not outliers in the datasets.
Differential gene expression analysis (DEX analysis) with layer 6 Tbr1wildtype and Tbr1layer6 mutant samples:
To identify differentially expressed genes (DEX genes), we identified all possible confounding variables including ribosomal bases in the mapped reads, percentage of bases in intronic region, RIN, Sex and RNA concentration to produce a reliable conclusion. Thousands of negative binomial regression models are built to model expressions of each genes. The best model is formed using Bayesian information criterion (BIC) and forward stepwise algorithm. The DEX analysis was performed with edgeR. Genes that pass a 0.05 significant threshold are considered as significantly differentially expressed genes.
Differential gene expression analysis (DEX analysis) with layer 5 Tbr1wildtype and layer 6 Tbr1wildtype samples:
Mice were of varying sex (4 males, 1 females in layer 5 wildtype versus 2 males and 2 females in layer 6 wildtype) and therefore we controlled for sex in DEX analyses. The DEX analysis was performed with edgeR. Significantly differentially expressed genes are genes that pass a 0.05 significant threshold and have a log2fold change large than or equal to 1.5 or less than or equal to 1.5 (log2fold change ≥ 1.5 or log2fold change ≤ −1.5).
Comparison between DEX genes identified in two DEX analyses:
We utilized three different analyses to examine the relationship between DEX gene lists identified by two comparisons. We used hypergeometric test to see what is the likelihood of observing an enrichment of “layer 5 Tbr1wildtype versus layer 6 Tbr1wildtype DEX genes” in the “Tbr1layer6 mutant versus layer 6 Tbr1wildtype DEX gene list”. We conducted permutation test to further study this relationship without having the hypergeometric distribution assumption. The permutation test was run for 500,000 times to ensure we obtained a highly accurate underline distribution. P-value of the permutation test was defined as proportion of randomly generated gene lists that have at least the same number of overlapped gene as what we observed in our dataset. We determined the background for these calculations as the number of overlapped genes in two DEX analyses. In additional to the enrichment tests, we studied gene expression pattern of our data. Genes with same effect direction are genes that are 1) up in “layer 6 Tbr1layer6 mutant versus layer 6 Tbr1wildtype” and “layer 5 Tbr1wildtype versus layer 6 Tbr1wildtype”. 2) down in “layer 6 Tbr1layer6 mutant versus Tbr1wildtype” and “layer 5 Tbr1wildtype versus layer 6 Tbr1wildtype”. Genes with opposite effect direction are genes that are 1) up in “layer 6 Tbr1layer6 mutant versus layer 6 Tbr1wildtype” and down in “layer 5 Tbr1wildtype versus layer 6 Tbr1wildtype” or 2) down in “layer 6 Tbr1layer6 mutant versus layer 6 Tbr1wildtype” and up in “layer 5 Tbr1wildtype versus layer 6 Tbr1wildtype”.
Layer specific gene set enrichment analysis:
We conducted hypergeometric test to examine the relationship between the significant differentially expressed genes identified in “layer 6 Tbr1wildtype versus Tbr1layer6 mutant” comparison and layer specific genes reported by Willsey et al (Willsey et al., 2013). Willsey et al provided a list of layer specific genes in Table S5. The mouse samples used in our experiments are estimated to be most similar to P4 and P5 developing cells. Therefore, we restricted our analysis to genes layer specific at P4 and P5. We used a hypergeometric test (one-sided) to assess enrichment. We determined the background for these calculations as the number of filtered genes in our dataset.
TBR1 Chromatin immunoprecipitation (ChIP-Seq):
Transcription factor ChIP was performed as previously published with a few modifications (McKenna et al., 2011, Sandberg et al., 2016). P2 somatosensory cortices were dissected and dissociated by pipetting in cold PBS. Dissociated cells were fixed in 1% formaldehyde for 10 min at RT and neutralized with 1 mL 2.5M glycine. Fixed chromatin was lysed and sheared into 200 – 1,000 bp fragments using a Covaris S2 (14 cycles of duty cycle = 5%, intensity = 3 and cycles per burst = 200). Immunoprecipitation (IP) reactions of two biological replicates at P2 were performed using 5 μg TBR1 polyclonal antibody (Santa Cruz Biotech, SC48816 X (M-200)). 20X molar excess TBR1 blocking peptide was used as negative control. Protein/antibody complexes were collected using Dynabeads (20 mL protein A + 20 mL protein G). ChIP-seq libraries were generated using Ovation Ultralow System V2 (NuGEN) following manufacturer’s protocol. The resulting libraries were size selected (180–350 bp) and sequenced at the Center for Advanced Technology at UCSF (Illumina HiSeq 4000; http://cat.ucsf.edu/) using a single read 50-bp strategy.
ChIP-Seq Computational Analysis:
Clustering, base calling, and quality metrics were performed using standard Illumina software. Sequenced libraries were analyzed for overall quality and were filtered to remove artifacts and low-quality sequences using Trim Galore version 0.4.2
(https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/), and reads were mapped to the mouse genome (mm9) using BWA version 0.7.16a:
bwa aln -t 12 mm9 sample.trimmed.fastq.gz).
Significant binding peaks were called on individual replicates using MACS version 2.1.0 against matched input control and blocking peptide negative control samples, with both the model-based peak identification and local significance testing disabled:
macs2callpeak -t chip.bam -c input.bam -n chip_vs.input -f BAM -g mm --call-summ its -B -q 0.01 --nolambda --nomodel --extsize=350 --outdir output_directory
Downstream analyses were conducted on merged peaks across replicates, filtered to remove ENCODE blacklisted regions and annotated using custom scripts. Coverage plots and heatmap diagrams were generated using ngs.plot version 2.61. We performed de novo motif discovery and enrichment analysis of significant known motifs using HOMER version 4.9 with default settings and genomic background. Plots of motif distribution around peaks and heatmaps were generated using custom R scripts (data not shown). The data used in this publication have been deposited in NCBI’s Gene Expression Omnibus (GEO) under accession number GSE119362 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE119362); ChIP-seq experiments can be visualized in the UCSC genome browser via track hubs that are hosted at https://github.com/NordNeurogenomicsLab/Publications/tree/master/Darbandi_Cell_2018.
TBR1 ChIP-seq peaks from P2 mouse cortex defined here were compared to regions of open chromatin identified via ATAC-seq analysis of micro-dissected human fetal cerebral cortex (germinal zone and cortical plate, (de la Torre-Ubieta et al., 2018) and to a human embryonic stem cell derived cardiac mesoderm dataset (Koh et al., 2016) as an unrelated tissue. Called peaks from the human ATAC-seq datasets were annotated to the mouse genome via the UCSC liftover tool and overlap was compared between the human fetal cortex and control peaks for promoter and distal regions. The proportion of TBR1-bound peaks from each ATAC-seq dataset was compared via Fisher’s exact test.
Primary Cell Culture and Luciferase assay:
Plasmids:
To generate luciferase constructs candidate regulatory elements of mouse Tbr1 (hs416, chr2: 61494203–61494886, 683bp), Foxp2 (chr6: 15097241–15098146, 905 bp), Grin2b (chr6: 135813640–135814770, 1,130 bp), Bcl11a (chr11: 24270818–24271924, 1,383 bp), Hcn1 (chr13: 118669041–118670541, 1,500 bp), Fezf2 (hs434, chr14: 13170235–13171693, 1,458bp), Foxp1 (chr6: 99325484–99327361, 1,877 bp), and DlxI5/6i enhancer (chr6: 6819420–6819819, 400 bp) were amplified by PCR, and cloned into the pGL4.23 vector (Promega). The vectors were transformed with DH5α E. coli cells at 42°C.
Luciferase assay:
Primary cortical neurons were harvested from P0 wildtype cortex and transfected using Lipofectamine 2000 (Invitrogen) and one of the regulatory element luciferase vectors that were generated as described above. To test whether TBR1 modified the regulatory elements activity, pCAG-Tbr1-IRES-eGFP was co-transfected together with one of the aforementioned regulatory element vectors. A renilla luciferase plasmid (pRL, Promega) was cotransfected to control for transfection efficiency. The luciferase assay was performed 48hrs after transfection using the dual-luciferase kit (Promega) according to manufacturer’s instructions. Reporter activity was measured using Veritas™ Microplate Luminometer (Turner BioSystems, Model# 9100–001).
Primary Cell Culture:
Cortex was dissected from P0 Tbr1wildtype and Tbr1layer6 homozygous mutants and dissociated using papain dissociation kit following manufacturer’s protocol (Worthington). A total of 100,000 cells were seeded into tissue culture slides pre-coated with poly-L-lysine (10 mg/ml, Sigma) and then laminin (5 mg/ml, Sigma), and grown in vitro with media containing DMEM-H21 with 5% fetal bovine serum for 2 hrs. After the cells recovered, DMEM-H21 media was replaced by Neurobasal medium containing B27 supplement, 25% glucose, and glutamax overnight. Tbr1layer6 mutant cells were transfected with Cdh8, Ntng1, Ptprk and Wnt7b expression vectors and Tbr1wildtype were transfected with mock empty vector using Lipofectamine 2000 (Invitrogen) for 6 hrs. Following incubation, the media was replaced by Neurobasal medium containing B27 supplement, Penicillin/Streptomycin, 25% glucose, and glutamax. Cultures were grown for 14 days in vitro. Cultures were fixed with 4% PFA for 10 min and processed for immunohistochemistry. Briefly, they were washed in PBS, quenched 2 times for 15 min with 2 mg/ml sodium borohydrate solution, blocked in PBS containing 10% Normal Serum, 0.1% Triton X- 100 and 2% BSA, incubated in primary antibody overnight (4°C), washed in PBS, incubated in secondary antibody for 1–2 hrs (room temperature), washed in PBS, and mounted. This experiment was repeated twice (n=2).
Lentiviral Injection and in vivo Rescue Assay:
In vivo rescue assay was carried out by cloning Wnt7b into a Cre-dependent lentiviral backbone (pLenti-CAG-Flex-Wnt7b-IRES-GFP). HEK293T cells were transfected with pLentiCAG-Flex-Wnt7b-IRES-GFP and pLenti-BG-GFP-T2a-Cre using Polyplus jetPRIME® transfection reagent following manufacturer’s protocol. WNT7B levels were examined by performing western blot against WNT7B on HEK293T cell lysates that were transfected previously. Upon validation, the Wnt7b-IRES-GFP expressing lentivirus (pLenti-CAG-FlexWnt7b-IRES-GFP) was generated in HEK293T cells as previously reported (Vogt et al., 2015) using Polyplus jetPRIME® transfection reagent following manufacturer’s protocol.
Lentivirus expressing Wnt7b-IRES-GFP was injected in the SSCx of Tbr1layer6 heterozygous and homozygous mutants pups at P1. For injections, a glass micropipette of 50 μm diameter (with a beveled tip) was preloaded with sterile mineral oil and viral suspension was front-loaded into the tip of the needle using a plunger connected to a hydraulic drive (Narishige) that was mounted to a stereotaxic frame. P1 pups from Tbr1layer6 wildtype and Tbr1layer6 heterozygous and homozygous mutants were anesthetized on ice for 1–2 min before injections. Each pup received 3–5 viral injections (70 nl per site) in the right hemisphere. These sites were about 1 mm apart along the rostral to caudal axis. Viral suspensions were injected into layer 6 of the neonatal SSCx. After injections, pups were put back with the mother to recover after they began to move around on their own. Mice were sacrificed 21 days after injection and transcardially perfused with PBS followed by 4% PFA.
Histology:
At the time of experiment, for P0 and P3 experiments, animals were anesthetized on ice while postnatal (P21 and P56) animals were anesthetized with intraperitoneal avertin (0.015 ml/g of a 2.5% solution) injection. Animals were perfused transcardially with cold PBS and then with 4% PFA in PBS, followed by brain isolation, 1–2 hr post-fixation, cryoprotected in 30% sucrose in PBS, and cut frozen (coronally or sagittally) on a sliding microtome at 40μm for immunohistochemistry or in situ hybridization. All primary and secondary antibodies were diluted in PBS containing 10% Normal Serum, 0.25% Triton X-100 and 2% BSA. The following primary antibodies were used: Chicken anti-GFP (1:2000, Aves), mouse anti-Vglut1 (1:200, Synaptic Systems), rabbit anti-Vgat (1:500, Synaptic Systems), rabbit anti-PSD95 (1:200, Cell Signaling), mouse anti-gephyrin (1:200, Synaptic Systems). The secondary antibodies for immunofluorescence were Alexa Fluor-conjugated and purchased from Thermofisher. For synapse immunohistochemistry, a total of n=30 apical dendrites were counted from each of Tbr1wildtype, Tbr1layer6 heterozygous and Tbr1layer6 homozygous mutants. The coronal sections were pre-treated with pepsin to enhance the staining. Immunofluorescence specimens were counterstained with 1% DAPI to assist the delineation of cortical layers. For in situ hybridization a rostro-caudal coronal series of at least ten sections from n=2 brains from Tbr1wildtype and Tbr1layer6 homozygous mutants were examined. Anti-sense riboprobes for Tbr1, Nr4a2, Foxp2, Tle4, Wnt7b, Cntn2, Ptprk, Bcl11a, Bcl11b, Fezf2, Foxp1 and Sst were prepared as previously described (Cobos et al., 2005, Long et al., 2003). ISH was performed using digoxigenin-labeled riboprobes.
Image Acquisition and Analysis:
Fluorescent and bright-field images were taken using a Coolsnap camera (Photometrics) mounted on a Nikon Eclipse 80i microscope using NIS Elements acquisition software (Nikon). Confocal imaging experiments were conducted at the Cancer Research Laboratory (CRL) Molecular Imaging Center, supported by Helen Wills Neuroscience Institute at UC Berkeley. Confocal images were acquired using Zeiss LSM 880 with Airyscan with a 63X objective at 1,024×1,024 pixels resolution using ZEN 2.0 software. Brightness and contrast were adjusted, and images merged using Photoshop or ImageJ software. ImageJ software was used for image processing. For synapse counting (presynaptic and postsynaptic boutons), confocal image stacks (0.4μm step size) were processed with ImageJ software. In brief, background subtraction and smooth filter were applied to each stack. Using a threshold function, each stack was converted into a ‘masks’ image. Furthermore, the channels were co-localized with the Image Calculator plugging. Lastly, the number of co-localizations were counted, and the length of each dendrite was measured in each of the focal plane. Staining for control and mutant were done in parallel as well as the image capturing.
Electrophysiology:
Coronal brain slices (250 μm) including somatosensory cortex were made from three mice (n=3) at age p21–28 and at p56-p80. Slicing solution was chilled to 4°C and contained (in mM): 234 sucrose, 26 NaHCO3, 11 glucose, 10 MgSO4, 2.5 KCl, 1.25 NaH2PO4, 0.5 CaCl2, bubbled with 5% CO2/ 95% O2. Slices were incubated in artificial cerebrospinal fluid (aCSF) at 32°C for 30 minutes and then at room temperature until recording. aCSF contained (in mM): 123 NaCl, 26 NaHCO3, 11 glucose, 3 KCl, 2 CaCl2, 1.25 NaH2PO4, 1 MgCl2, also bubbled with 5% CO2/ 95% O2. Neurons were visualized using differential interference contrast or DODT contrast microscopy on an upright microscope (Olympus). Ntsr1-cre positive neurons were identified by fluorescent visualization of cre-dependent tdTomato. We obtained somatic whole-cell patch clamp recordings using a Multiclamp 700B (Molecular Devices) amplifier and acquired with pClamp. Patch pipettes (2–5 MΩ tip resistance) were filled with the following (in mM): 130 KGluconate, 10 KCl, 10 HEPES, 10 EGTA, 2 MgCl2, 2 MgATP, 0.3 Na3GTP. All recordings were made at 32–34°C. Series resistance was compensated in all current clamp experiments and monitored throughout recordings. Recordings were discarded if Rs changed by >25%. For spontaneous EPSC and IPSC recordings cells were held in voltage clamp at −70 mV and +10mV, respectively. In both cases patch pipettes were filled with the following (in mM): 135 Cesium Methanesulfonate, 8 NaCl, 10 HEPES, 0.3 EGTA, 5 QX314, 4 MgATP, 0.3 Na3GTP.
Behavioral Assays:
Experiments were conducted during the light cycle (8am to 8pm). Mice were habituated to investigator handling for 1–2min on three consecutive days. On the testing day, mice were transferred to experimental room and allowed to habituate for at least 45 minutes prior to testing. All behavior assays were performed on mice age P56 to P80. We were blind to the genotypes during scoring of videos.
Open-field test:
An individual mouse was placed near the wall-side of 50 × 50 cm open-field arena, and the movement of the mouse was recorded by a video camera for 10 min. The recorded video file was analyzed with Any-Maze software (San Diego Instruments). Time in the center of the field (a 25 × 25 cm square) was measured. The open field arena was cleaned with 70% ethanol and wiped with paper towels between each trial.
Elevated plus maze test:
An individual mouse was placed at the junction of the open and closed arms, facing the arm opposite to the experimenter, of an apparatus with two open arms without walls (30 × 5 × 0.5 cm) across from each other and perpendicular to two closed arms with walls (30 × 5 × 15 cm) with a center platform (5 × 5 cm), and at a height of 40 cm above the floor. The movement of the mouse was recorded by a video camera for 10 min. The recorded video file was analyzed with Any-Maze software and time in the open arms of the apparatus was measured. The arms of the elevated plus maze apparatus was cleaned with 70% ethanol and wiped with paper towels between each trial.
Rotarod test:
The assay consisted of four trials per day over the course of 2 days with the rotarod set to accelerate from 4rpm to 45rpm over 5 minutes. The trial started once five mice were placed on the rotarod rotating at 4rpm in separate partitioned compartments. Each trial ended when a mouse fell off, made three complete revolutions while hanging on, or reached 300 s. Digital videos of the mice on the rotarod were recorded from behind. The rotarod apparatus was cleaned with 70% ethanol and wiped with paper towels between each trial.
Social interaction and novel object task:
An individual mouse was allowed to habituate for 5 minutes in their home cage prior to starting the trial. A juvenile (3–4 weeks old) mouse of the same strain and sex was introduced to the home cage. After 5 minutes, the juvenile was removed from the home cage. After a 5 min break a novel object (typically a plastic test tube cap) was introduced into the home cage for five minutes. We scored videos offline, blind to genotype. We measured the number of seconds the mouse spent with its nose in direct contact with the novel object or engaged in social interaction with the juvenile (defined as sniffing, close following, or allo-grooming) in the 300 seconds following the time the juvenile or object was introduced into the cage. In addition, we noted any aggressive-appearing behaviors toward the juvenile, freezing, and grooming behaviors.
Quantification and Statistical Analysis:
All individual data points are shown as well as mean ± SEM. All statistical analyses were performed using GraphPad Prism 7.0 software. Statistical significance was accepted at the level p < 0.05. We used student’s t-test to compare pairs of groups if data were normally distributed (verified using Lillie test). If more than two groups were compared, we used one-way ANOVA with post-hoc tests between groups corrected for multiple comparisons (Holm-Sidak or Tukey). For the ISH experiments reported in this paper n=2 represents two biological replicates for each of the reported genes. We examined the changes in synapse numbers of n=30 different dendrites from n=2 animals for each genotype. Whole-cell patch clamp experiments at P21 and P56 were conducted from n=3 different animals for each age and genotype. Lastly, behavioral analysis was conducted from n = 11/8/9, wildtype/ heterozygous/ homozygous animals. The specific n for each experiment as well as the post-hoc test, exact F and corrected p values can be found in the Results section.
Data and Software Availability:
Data and MATLAB analysis scripts are available upon request from the Lead Contact.
Supplementary Material
RNA-Seq analysis of FACS purified Layer 6 neurons at P5 identified different classes of genes that are dysregulated in the Tbr1layer6 homozygous mutant compared to Tbr1wildtype at P5. Related to Figure 1A.
Complete list of differentially expressed genes in the Tbr1layer6 mutant compared to Tbr1wildtype at P5. Related to Figure 1A.
Highlights:
Tbr1 specifies layer 6 dendritic patterning and cell intrinsic physiology.
Tbr1 promotes synapse numbers through Wnt7b.
Tbr1 heterozygotes provide insight into ASD pathophysiology.
TBR1 directly regulates transcriptional circuits that controls ASD risk genes.
Acknowledgments
This work was supported by the research grants to J.L.R.R. from: Nina Ireland, NIMH R37 MH049428 and NINDS R01 NS34661, grants to V.S.S. NIMH R01MH100292 and R01MH106507, grants to B.C. NIH R01MH094589. A.N. and R.C.-P. were supported by NIGMS R35 GM119831and R.C.-P. was supported via a Science Without Borders Fellowship from CNPq (Brazil).
Footnotes
Declaration of Interests
J.L.R.R. is cofounder, stockholder, and currently on the scientific board of Neurona, a company studying the potential therapeutic use of interneuron transplantation.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- BEDOGNI F, HODGE RD, ELSEN GE, NELSON BR, DAZA RAM, BEYER RP, BAMMLER TK, RUBENSTEIN JLR & HEVNER RF (2010). Tbr1 regulates regional and laminar identity of postmitotic neurons in developing neocortex. Proceedings of the National Academy of Sciences, 107, 13129–13134. https://doi.org/10.1073/pnas.1002285107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- BUDNIK V & SALINAS PC (2011). Wnt signaling during synaptic development and plasticity. Current Opinion in Neurobiology, 21, 151–159. https://doi.org/10.1016/j.conb.2010.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- BULFONE A, SMIGA SM, SHIMAMURA K, PETERSON A, PUELLES L & RUBENSTEIN JLR (1995). T-Brain-1: A homolog of Brachyury whose expression defines molecularly distinct domains within the cerebral cortex. Neuron, 15, 63–78. https://doi.org/10.1016/0896-6273(95)90065-9 [DOI] [PubMed] [Google Scholar]
- BULFONE A, WANG F, HEVNER R, ANDERSON S, CUTFORTH T, CHEN S, MENESES J, PEDERSEN R, AXEL R & RUBENSTEIN JLR (1998). An Olfactory Sensory Map Develops in the Absence of Normal Projection Neurons or GABAergic Interneurons. Neuron, 21, 1273–1282. https://doi.org/10.1016/S08966273(00)80647-9 [DOI] [PubMed] [Google Scholar]
- CHEVÉE M, ROBERTSON JDJ, CANNON GH, BROWN SP & GOFF LA (2018). Variation in Activity State, Axonal Projection, and Position Define the Transcriptional Identity of Individual Neocortical Projection Neurons. Cell Reports, 22, 441–455. https://doi.org/10.1016/j.celrep.2017.12.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHUANG H-C, HUANG T-N & HSUEH Y-P (2014). Neuronal excitation upregulates Tbr1, a high-confidence risk gene of autism, mediating Grin2b expression in the adult brain. Frontiers in Cellular Neuroscience, 8, 280 https://doi.org/10.3389/fncel.2014.00280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHUANG H-C, HUANG T-N & HSUEH Y-P (2015). T-Brain-1 – A Potential Master Regulator in Autism Spectrum Disorders. Autism Research, 1–13. https://doi.org/10.1002/aur.1456 [DOI] [PubMed] [Google Scholar]
- COBOS I, CALCAGNOTTO ME, VILAYTHONG AJ, THWIN MT, NOEBELS JL, BARABAN SC, & RUBENSTEIN J (2005). Mice lacking Dlx1 show subtypespecific loss of interneurons, reduced inhibition and epilepsy. Nature Neuroscience, 8, 1059–1068. https://doi.org/10.1038/nn1499 [DOI] [PubMed] [Google Scholar]
- DAVIS EK, ZOU Y & GHOSH A (2008). Wnts acting through canonical and noncanonical signaling pathways exert opposite effects on hippocampal synapse formation. Neural Development, 3, 32–32. https://doi.org/10.1186/1749-8104-3-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DE LA TORRE-UBIETA L, STEIN JL, WON H, OPLAND CK, LIANG D, LU D & GESCHWIND DH (2018). The Dynamic Landscape of Open Chromatin during Human Cortical Neurogenesis. Cell, 172, 289–304.e18. https://doi.org/10.1016/j.cell.2017.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DE RUBEIS S, HE X, GOLDBERG AP, POULTNEY CS, SAMOCHA K, ERCUMENT CICEK A, KOU Y, LIU L, FROMER M, WALKER S, et al. (2014). Synaptic, transcriptional and chromatin genes disrupted in autism. Nature, 515, 209–215. https://doi.org/10.1038/nature13772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DECK M, LOKMANE L, CHAUVET S, MAILHES C, KEITA M, NIQUILLE M, YOSHIDA M, YOSHIDA Y, LEBRAND C, MANN F et al. (2013). Pathfinding of Corticothalamic Axons Relies on a Rendezvous with Thalamic Projections. Neuron, 77, 472–484. http://dx.doi.org/10.1016/j.neuron.2012.11.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DEMBROW NC, CHITWOOD RA & JOHNSTON D (2010). Projection-Specific Neuromodulation of Medial Prefrontal Cortex Neurons. The Journal of Neuroscience, 30, 16922 https://doi.org/10.1523/JNEUROSCI.3644-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DERIZIOTIS P, O’ROAK BJ, GRAHAM SA, ESTRUCH SB, DIMITROPOULOU D, BERNIER RA, GERDTS J, SHENDURE J, EICHLER EE & FISHER SE (2014). De novo TBR1 mutations in sporadic autism disrupt protein functions. Nat Commun, 5 https://doi.org/10.1038/ncomms5954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- FRIEDMAN LG, RIEMSLAGH FW, SULLIVAN JM, MESIAS R, WILLIAMS FM, HUNTLEY GW & BENSON DL (2015). Cadherin-8 expression, synaptic localization and molecular control of neuronal form in prefrontal cortico-striatal circuits. The Journal of comparative neurology, 523, 75–92. https://doi.org/10.1002/cne.23666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- GONG S, DOUGHTY M, HARBAUGH CR, CUMMINS A, HATTEN ME, HEINTZ N & GERFEN CR (2007). Targeting Cre Recombinase to Specific Neuron Populations with Bacterial Artificial Chromosome Constructs. The Journal of Neuroscience, 27, 9817 https://doi.org/10.1523/JNEUROSCI.2707-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- GONG X, JIA M, RUAN Y, SHUANG M, LIU J, WU S, GUO Y, YANG J, LING Y, YANG X & ZHANG D (2004). Association between the FOXP2 gene and autistic disorder in Chinese population. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics, 127B, 113–116. https://doi.org/10.1002/ajmg.b.20162 [DOI] [PubMed] [Google Scholar]
- HAN W, KWAN KY, SHIM S, LAM MMS, SHIN Y, XU X, ZHU Y, LI M & ŠESTAN N (2011). TBR1 directly represses Fezf2 to control the laminar origin and development of the corticospinal tract. Proceedings of the National Academy of Sciences, 108, 3041–3046. https://doi.org/10.1073/pnas.1016723108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- HEVNER RF, MIYASHITA-LIN E & RUBENSTEIN JLR (2002). Cortical and thalamic axon pathfinding defects in Tbr1, Gbx2, and Pax6 mutant mice: Evidence that cortical and thalamic axons interact and guide each other. The Journal of Comparative Neurology, 447, 8–17. https://doi.org/10.1002/cne.10219 [DOI] [PubMed] [Google Scholar]
- HEVNER RF, NEOGI T, ENGLUND C, DAZA RAM & FINK A (2003). Cajal– Retzius cells in the mouse: transcription factors, neurotransmitters, and birthdays suggest a pallial origin. Developmental Brain Research, 141, 39–53. http://dx.doi.org/10.1016/S0165-3806(02)00641-7 [DOI] [PubMed] [Google Scholar]
- HEVNER RF, SHI L, JUSTICE N, HSUEH Y-P, SHENG M, SMIGA S, BULFONE A, GOFFINET AM, CAMPAGNONI AT & RUBENSTEIN JLR (2001). Tbr1 Regulates Differentiation of the Preplate and Layer 6. Neuron, 29, 353–366. http://dx.doi.org/10.1016/S0896-6273(01)00211-2 [DOI] [PubMed] [Google Scholar]
- HOERDER-SUABEDISSEN A, HAYASHI S, UPTON L, NOLAN Z, CASASTORREMOCHA D, GRANT E, VISWANATHAN S, KANOLD PO, CLASCA F, KIM Y & MOLNÁR Z (2018). Subset of Cortical Layer 6b Neurons Selectively Innervates Higher Order Thalamic Nuclei in Mice. Cerebral Cortex, 28, 1882–1897. https://doi.org/10.1093/cercor/bhy036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUANG T-N, CHUANG H-C, CHOU W-H, CHEN C-Y, WANG H-F, CHOU S-J & HSUEH Y-P (2014). Tbr1 haploinsufficiency impairs amygdalar axonal projections and results in cognitive abnormality. Nat Neurosci, 17, 240–247. https://doi.org/ 10.1038/nn.3626 [DOI] [PubMed] [Google Scholar]
- HUANG T-N & HSUEH Y-P (2017). Calcium/calmodulin-dependent serine protein kinase (CASK), a protein implicated in mental retardation and autism-spectrum disorders, interacts with T-Brain-1 (TBR1) to control extinction of associative memory in male mice. Journal of Psychiatry & Neuroscience : JPN, 42, 37–47. https://doi.org/10.1503/jpn.150359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- KOH PW, SINHA R, BARKAL AA, MORGANTI RM, CHEN A, WEISSMAN IL, ANG LT, KUNDAJE A & LOH KM (2016). An atlas of transcriptional, chromatin accessibility, and surface marker changes in human mesoderm development. Scientific Data, 3, 160109 https://doi.org/10.1038/sdata.2016.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAI M-C, LOMBARDO MV & BARON-COHEN S (2014). Autism. The Lancet, 383, 896–910. http://dx.doi.org/10.1016/S0140-6736(13)61539-1 [DOI] [PubMed] [Google Scholar]
- LEDERGERBER D & LARKUM ME (2010). Properties of Layer 6 Pyramidal Neuron Apical Dendrites. The Journal of Neuroscience, 30, 13031 https://doi.org/10.1523/JNEUROSCI.2254-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEFEBVRE JL, SANES JR & KAY JN (2015). Development of Dendritic Form and Function. Annual Review of Cell and Developmental Biology, 31, 741–777. https://doi.org/10.1146/annurev-cellbio-100913-013020 [DOI] [PubMed] [Google Scholar]
- LIM SH, KWON SK, LEE MK, MOON J, JEONG DG, PARK E, KIM SJ, PARK BC, LEE SC, RYU SE et al. (2009). Synapse formation regulated by protein tyrosine phosphatase receptor T through interaction with cell adhesion molecules and Fyn. The EMBO Journal, 28, 3564 https://doi.org/10.1038/emboj.2009.289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIU J, REGGIANI JDS, LABOULAYE MA, PANDEY S, CHEN B, RUBENSTEIN JLR, KRISHNASWAMY A & SANES JR (2018). Tbr1 instructs laminar patterning of retinal ganglion cell dendrites. Nature Neuroscience, 21, 659–670. https://doi.org/10.1038/s41593-018-0127-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- LONG JE, GAREL S, DEPEW MJ, TOBET S & RUBENSTEIN J (2003). DLX5 Regulates Development of Peripheral and Central Components of the Olfactory System. The Journal of Neuroscience, 23, 568–578. https://doi.org/10.1523/JNEUROSCI.23-0200568.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MADISEN L, ZWINGMAN TA, SUNKIN SM, OH SW, ZARIWALA HA, GU H, NG LL, PALMITER RD, HAWRYLYCZ MJ, JONES AR et al. (2010). A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nature neuroscience, 13, 133–140. https://doi.org/10.1038/nn.2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MCKENNA WL, BETANCOURT J, LARKIN KA, ABRAMS B, GUO C, RUBENSTEIN JLR & CHEN B (2011). Tbr1 and Fezf2 Regulate Alternate Corticofugal Neuronal Identities during Neocortical Development. The Journal of Neuroscience, 31, 549–564. https://doi.org/ 10.1523/JNEUROSCI.4131-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- NOTWELL JH, HEAVNER WE, DARBANDI SF, KATZMAN S, MCKENNA WL, ORTIZ-LONDONO CF, TASTAD D, ECKLER MJ, RUBENSTEIN JLR, MCCONNELL SK et al. (2016). TBR1 regulates autism risk genes in the developing neocortex. Genome Research, 26, 1013–1022. https://doi.org/10.1101/gr.203612.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- SALINAS PC & ZOU Y (2008). Wnt Signaling in Neural Circuit Assembly. Annual Review of Neuroscience, 31, 339–358. https://doi.org/10.1146/annurev.neuro.31.060407.125649 [DOI] [PubMed] [Google Scholar]
- SANDBERG M, FLANDIN P, SILBERBERG S, SU-FEHER L, PRICE JD, HU JS, KIM C, VISEL A, NORD AS & RUBENSTEIN JLR (2016). Transcriptional Networks Controlled by NKX2–1 in the Development of Forebrain GABAergic Neurons. Neuron, 91, 1260–1275. https://doi.org/10.1016/j.neuron.2016.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- SANDERS SJ, HE X, WILLSEY AJ, ERCAN-SENCICEK AG, SAMOCHA KAITLINE, CICEK AE, MURTHA MT, BAL VH, BISHOP SL et al. (2015). Insights into Autism Spectrum Disorder Genomic Architecture and Biology from 71 Risk Loci. Neuron, 87, 1215–1233. http://dx.doi.org/10.1016/j.neuron.2015.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHEPHERD GMG (2013). Corticostriatal connectivity and its role in disease. Nature reviews. Neuroscience, 14, 278–291. https://doi.org/10.1038/nrn3469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VOGT D, WU P-R, SORRELLS SF, ARNOLD C, ALVAREZ-BUYLLA A & RUBENSTEIN JLR (2015). Viral-mediated Labeling and Transplantation of Medial Ganglionic Eminence (MGE) Cells for In Vivo Studies. Journal of Visualized Experiments : JoVE, 52740 https://doi.org/10.3791/52740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- WILLSEY AJ, SANDERS SJ, LI M, DONG S, TEBBENKAMP AT, MUHLE RA, REILLY SK, LIN L, FERTUZINHOS S, MILLER JA et al. (2013). Coexpression Networks Implicate Human Midfetal Deep Cortical Projection Neurons in the Pathogenesis of Autism. Cell, 155, 997–1007. https://doi.org/10.1016/j.cell.2013.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- YEE A & CHEN L (2016). Differential regulation of spontaneous and evoked inhibitory synaptic transmission in somatosensory cortex by retinoic acid. Synapse (New York, N.Y.), 70, 445–452. https://doi.org/10.1002/syn.21921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZERUCHA T, STUHMER T, GAMBAROTTA A, HATCH G, SCHULTZ J, PARK B, LONG Q, RUBENSTEIN J, YU G & EKKER M (2000). A Highly Conserved Enhancer in the Dlx5/Dlx6 Intergenic Region is the Site of Cross-Regulatory Interactions between Dlx Genes in the Embryonic Forebrain. The Journal of Neuroscience, 20, 709–721. https://doi.org/10.1523/JNEUROSCI.20-02-00709.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG Q, GOTO H, AKIYOSHI-NISHIMURA S, PROSSELKOV P, SANO C, MATSUKAWA H, YAGUCHI K, NAKASHIBA T & ITOHARA S (2016). Diversification of behavior and postsynaptic properties by netrin-G presynaptic adhesion family proteins. Molecular Brain, 9, 1–23. https://doi.org/10.1186/s13041-016-0187-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
RNA-Seq analysis of FACS purified Layer 6 neurons at P5 identified different classes of genes that are dysregulated in the Tbr1layer6 homozygous mutant compared to Tbr1wildtype at P5. Related to Figure 1A.
Complete list of differentially expressed genes in the Tbr1layer6 mutant compared to Tbr1wildtype at P5. Related to Figure 1A.