Abstract
MucR is a member of the Ros/MucR family of prokaryotic zinc-finger proteins found in the α-proteobacteria which regulate the expression of genes required for the successful pathogenic and symbiotic interactions of these bacteria with the eukaryotic hosts. The structure and function of their distinctive zinc-finger domain has been well-studied, but only recently the quaternary structure of the full length proteins was investigated demonstrating their ability to form higher-order oligomers. The aim of this study was to identify the region of MucR involved in higher-order oligomer formation by analysing deletion and point mutants of this protein by Light Scattering, and to determine the role that MucR oligomerization plays in the regulatory function of this protein. Here we demonstrate that a conserved hydrophobic region at the N-terminus of MucR is responsible for higher-order oligomer formation and that MucR oligomerization is essential for its regulatory function in Brucella. All these features of MucR are shared by the histone-like nucleoid structuring protein, (H-NS), leading us to propose that the prokaryotic zinc-finger proteins in the MucR/Ros family control gene expression employing a mechanism similar to that used by the H-NS proteins, rather than working as classical transcriptional regulators.
Introduction
The Ros/MucR protein family1,2 includes prokaryotic zinc-finger proteins such as Ros from Agrobacterium tumefaciens3 and MucR from Brucella spp.4–6, both of which regulate genes required for the virulence of these strains in their respective plant and animal hosts2,5–7. Also included in this family are MucR from Sinorhizobium meliloti7,8, and from Sinorhizobium fredii9; RosR from Rhizobium etli10 and from Rhizobium leguminosarum11 which regulate genes required for the successful symbiosis of these bacteria with plants. Additionally, the structural homologs MucR1 and MucR2 play important roles in coordinating the orderly expression of the cell cycle genes in Caulobacter crescentus12. Many structural features related to the DNA-binding domains of Ros from Agrobacerium tumefaciens and Mls from Mesorhizobium loti have been described13–21. One of the interesting features of these and other Ros/MucR homologs is that direct binding studies suggest that these proteins recognize A-T rich regions in and around bacterial promoters that have little sequence consensus12,22–25. Recently, we demonstrated that the AT-rich DNA targets sites for the Mesorhizobium Mls and Brucella MucR contain T-A steps, and that these proteins contact DNA mostly in the minor groove and are able to form higher-order oligomers26. Furthermore, we have shown that MucR from Brucella abortus is able to recognize multiple AT-rich sites in the promoter of its own gene and that it is a heat-stable protein with a Tm of 63 °C27. The ability to bind AT-rich sites containing T-A steps in the minor groove, the capacity to oligomerize and the heat-stability are also features of another prokaryotic protein family, the histone-like nucleoid structuring proteins, H-NS28–34. H-NS proteins not only play important roles in nucleoid compaction, but they also serve as gene silencers, preventing the potentially toxic expression of bacterial genes acquired by horizontal gene transfer35–37 and repressing the gratuitous expression of virulence genes in bacterial pathogens38,39. One of the important features of H-NS proteins with regard to their ability to serve as gene silencers is their capacity to recognize AT-rich DNA-target sites containing T-A steps in and around promoters. They use these sequences as nucleation sites to form higher-order oligomers that prevent RNA polymerase access to these promoters40,41.
Here, we identify a hydrophobic region as responsible for the higher-order oligomer formation at the N-terminus of the prokaryotic zinc-finger protein MucR and definitively demonstrate the importance of MucR oligomerization for its regulatory function in Brucella. Based on the results presented here, together with previously published findings26,27, we propose that the prokaryotic zinc-finger proteins in the Ros/MucR family control gene expression by employing a mechanism similar to that used by the H-NS proteins, rather than working as classical transcriptional regulators.
Results
A conserved hydrophobic region located at the N-terminus of MucR is responsible for higher-order oligomer formation
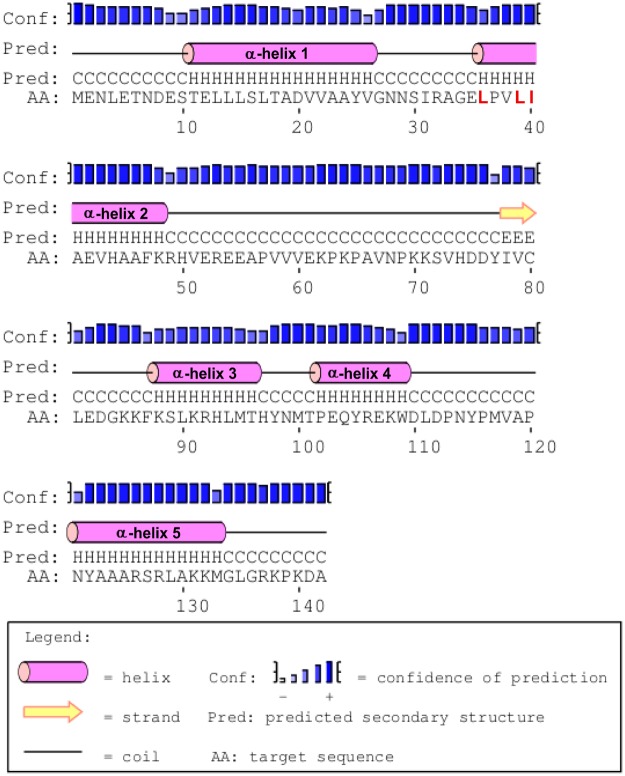
As previously reported26, the N-terminal region of MucR and of the homologous Ml proteins is responsible for higher-order oligomer formation. Using the PSIPRED tool42, we obtained a secondary structure prediction of the Brucella MucR (Fig. 1). In the N-terminal region, two α-helices were predicted, one extending from the threonine in position 11 to the valine in position 27 and the other from the leucine in position 36 to the lysine in position 48. In an attempt to identify the amino acids in the N-terminal region of MucR involved in oligomerization, we designed the deletion mutant MucR33–142, in which the first 32 amino acids including the first putative α-helix, were deleted, and the deletion mutant MucR45–142, in which the deletion was extended to the amino acid in position 44 breaking down the second predicted α-helix (Fig. 1). To investigate the oligomeric state of MucR33–142 and MucR45–142, we performed a static Light Scattering (LS) analysis (Table 1). The results obtained under the conditions tested show that the deletion mutant MucR33–142 is still able to form higher-order oligomers showing a decameric state, whereas the deletion mutant MucR45–142 turns out to be in a monomeric state. The same result was obtained with the deletion mutant MucR57–142 (Table 1) lacking the N-terminal region and comprising only the region corresponding to the Ros DNA-binding domain whose structure was solved by NMR15. These results identify the region spanning from the alanine in position 33 to that in position 44 as responsible for oligomerization. This amino acid sequence contains two leucines in position 36 and 39, and an isoleucine in position 40 that constitute a hydrophobic region highly conserved in MucR homologs (Suppl. Fig. 1). We thus designed a mutant version of MucR33–142, named MucR33-142mut, in which these three conserved amino acids were mutated into alanines. Analysed by LS, MucR33-142mut turned out to be a monomer under the conditions tested, indicating that the conserved residues leucine 36, leucine 39 and isoleucine 40 are involved in higher-order oligomer formation (Table 1). To investigate the role of this highly conserved hydrophobic region in the full-length MucR, we expressed and purified the MucRL36L39I40A mutant, a version of MucR in which the leucines 36 and 39, and the isoleucine 40 are mutated into alanines. Performing LS analysis, we found that MucRL36L39I40A turns out to be a monomer losing its quaternary decameric structure (Fig. 2) indicating that the here identified highly conserved hydrophobic region in the second putative α-helix at the N-terminus of MucR is responsible for higher-order oligomer formation.
Figure 1.
Secondary structure prediction of MucR by PSIPRED tool. The predicted α-helices are indicated and numbered in the barrels; the amino acids mutated in the MucRL36L39I40A are reported in red.
Table 1.
Light Scattering (LS) analysis of the MucR deletion mutants.
| Protein | Theoretical molecular weight monomer | Experimental Molecular weight by Static LS | ExMw/ThMw |
|---|---|---|---|
| MucR | 16024 Da | 167000 (±1, 0%) Da | 10.40 |
| MucR33–142 | 12559 Da | 132600 (±2, 0%) | 10.50 |
| MucR45–142 | 11330 Da | 11510 (±0, 6%) | 1.01 |
| MucR57–142 | 9905 Da | 10740 (±3%) | 1.08 |
| MucR33–142 mut | 12334 Da | 12690 (±0, 7%) | 1.02 |
The theoretical molecular weight of the monomer calculated by ProtParam tool (http://web.expasy.org/protparam/) is reported in the second column; the results by LS in the third column; the ratio between the experimental weights found and the monomer weights are reported in the fourth column.
Figure 2.
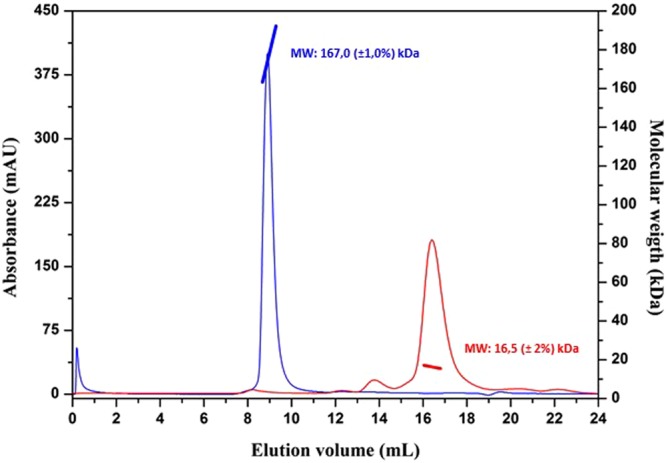
Gel-filtration light-scattering analysis of MucR (blue) and MucRL36L39I40A (red). The solid lines represent the signals of the eluted proteins at 280 nm. The molecular weight of each peak calculated by LS of MucR (reported in blue) and MucRL36L39I40A (reported in red) is also reported.
NMR analysis of MucR structure and oligomerization
We performed a structural analysis of the studied proteins by means of NMR spectroscopy. The NMR study started with the analysis of the DOSY experiment recorded for the MucR57–142 deletion mutant. This experiment gave a diffusion coefficient (Dt) of 1.22 (±0.12) *10−10 m2 sec−1, which is very similar to the value measured for Ros87, thus confirming the monomeric form of MucR57-142 also at the NMR concentration used43. Figure 3, panel a, reports the 1H-15N HSQC spectrum of the same protein. The spectrum is consistent with a well-defined native structure with extensive tertiary interactions. It shows a large number of resonances well dispersed over a chemical shift range of about 3 ppm in the proton dimension and 25 ppm in the 15N dimension. This feature clearly indicates the presence of β-strands, as this kind of secondary structure is typically associated with a good dispersion of the NMR signals. The more crowded region in the centre of the spectrum is likely to contain resonances from helical structures which lead to a minor degree of dispersion.
Figure 3.
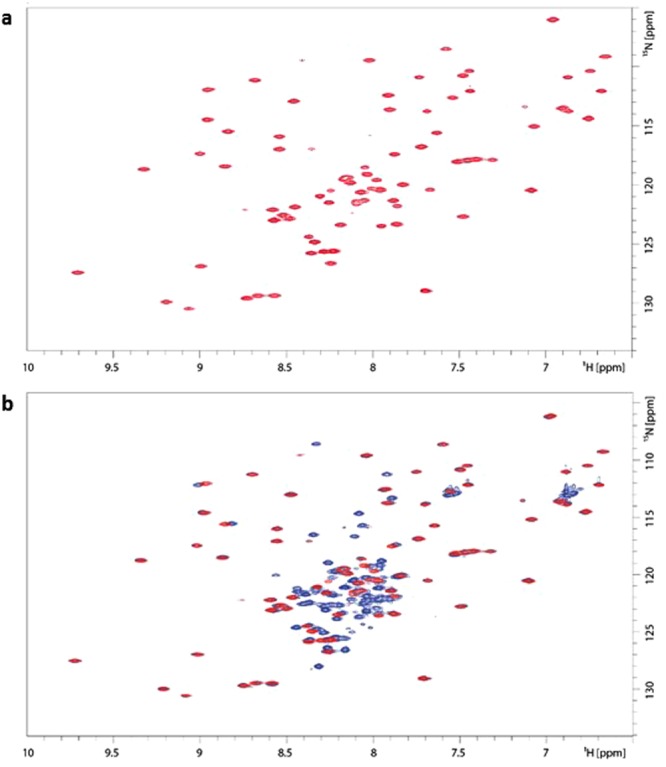
(a) 1H-15N HSQC spectrum of MucR57–142 acquired at 600 MHz and at 298 K; (b) superposition of MucR57–142 and MucRL36L39I40A 1H−15N HSQC spectra acquired at 600 MHz and at 298 K. The spectrum of MucR57–142 is in red whereas that of MucRL36L39I40A is in blue.
The same spectra were then recorded for the wild-type full-length protein MucR and for the full-length mutant MucRL36L39I40A. Consistent with the data acquired using other techniques, the wild-type full-length protein gave very low quality spectra bearing only a few broad signals suggesting the presence of high-order oligomers. The behaviour of the protein remained the same also when lowering the concentration, indicating a concentration independent tendency of this protein to form oligomers. Quite opposite is the behaviour of MucRL36L39I40A. This triple point mutant gave good quality spectra (Fig. 3b) which are consistent with a well-defined monomeric tertiary structure (Dt = 1.18 (±0.13) * 10−10 m2 sec−1). Figure 3, panel b, reports the superposition of the spectra recorded for the MucR57–142 mutant with those of MucRL36L39I40A and shows how the DNA-binding domain, apart from minor local differences, is essentially contained within the structure of MucRL36L39I40A.
MucRL36L39I40A fails to form the slow mobility protein-DNA complex observed in EMSA of MucR
To investigate the ability of the mutant protein MucRL36L39I40A to bind DNA, we performed electrophoretic mobility shift assays (EMSAs) using one of the MucR DNA-binding site, named Site1, previously identified in the mucR gene promoter and located at −174 bp from the ATG start codon27. Comparing the protein/DNA complexes formed by MucR and MucRL36L39I40A, it is evident that the mutant protein is unable to form the slow mobility MucR/DNA complex indicated by the arrow in Fig. 4 and in Suppl. Fig. 2. It is likely that the mutant MucRL36L39I40A cannot form higher-order oligomer bound to DNA, but only the DNA-protein complexes with lower molecular masses which turn to be faster in their electrophoretic mobility. The results observed by EMSAs are in line with NMR data, which show that the DNA-binding domain is not altered by the mutation at the N-terminus of the protein and with those obtained by LS which demonstrates that MucRL36L39I40A is not able to form higher-order oligomers.
Figure 4.
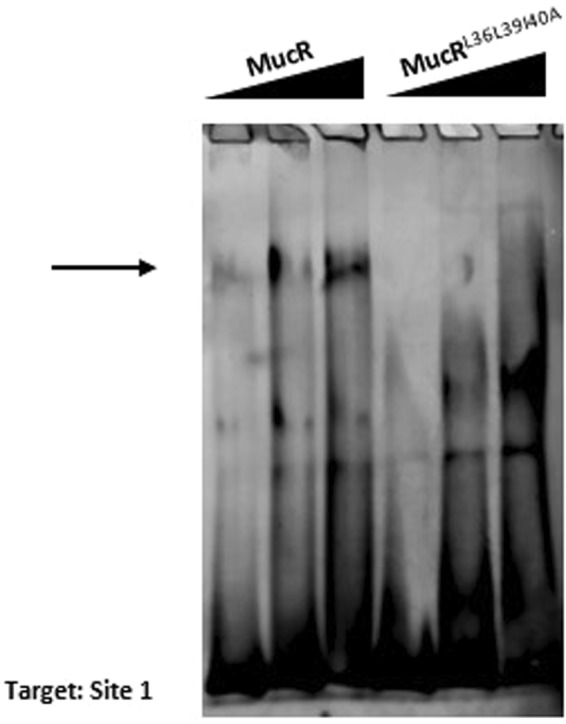
MucR and MucRL36L39I40A binding to Site 1 by EMSA. The increasing amount of the protein used is indicated on the top of the lanes as well as the name of the proteins. The target sequence Site1 was previously identified in the promoter of mucR gene at −174 bp from the ATG start codon27. The arrow indicate the protein/DNA complex formed by MucR with Site1 which does not appear in the EMSAs of MucRL36L39I40A.
The region of MucR responsible for oligomerization is necessary for its wild-type regulatory function in B. abortus 2308
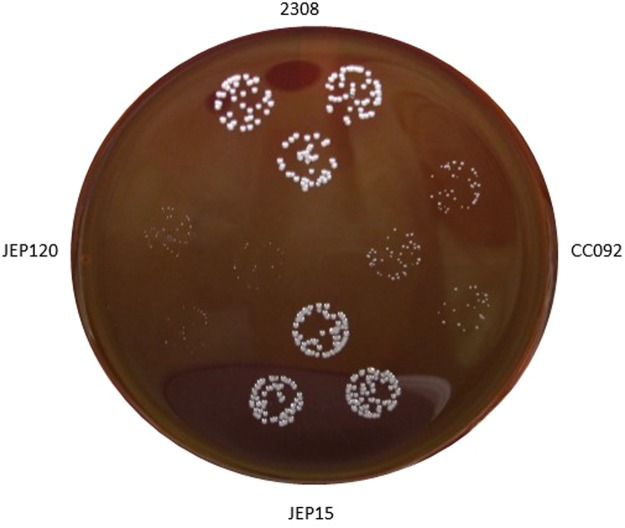
The B. abortus mucR mutant CC092 displays a delay in its ability to form colonies on agar plates compared to its parent strain B. abortus 23084, and rescue of this growth defect by genetic complementation with plasmid-borne copies of mutated mucR alleles has proven to be a useful tool for structure/function analysis of the MucR protein. To investigate whether or not MucR requires the region responsible for oligomerization to rescue the growth defect exhibited by B. abortus CC092, we performed a genetic complementation experiment transforming this mucR mutant with the plasmids containing the wild-type gene mucR (pJep011) and the mutated mucRL36L39I40A gene (pJep120). Transcription of the mucR wild-type and mucRL36L39I40A in the B. abortus transformants was verified by q-RT PCR, and colony formation by these strains was monitored during cultivation on Schaedler blood agar supplemented with bovine blood4. Unlike the native mucR gene, a plasmid-borne copy of the mucRL36L39I40A allele did not restore the capacity of the B. abortus CC092 mucR mutant to form the same sized colonies as the parental 2308 strain following 72 h growth on agar plates (Fig. 5). These results strongly suggest that oligomerization is required for the normal physiologic function of MucR.
Figure 5.
Colony formation by B. abortus 2308, an isogenic mucR mutant CC092 and derivatives of CC092 carrying plasmid-borne copies of mucR (JEP15) or mucRL36L39I40A (JEP120) following 72 h incubation at 37 °C on Schaedler agar supplemented with 5% defibrinated bovine blood.
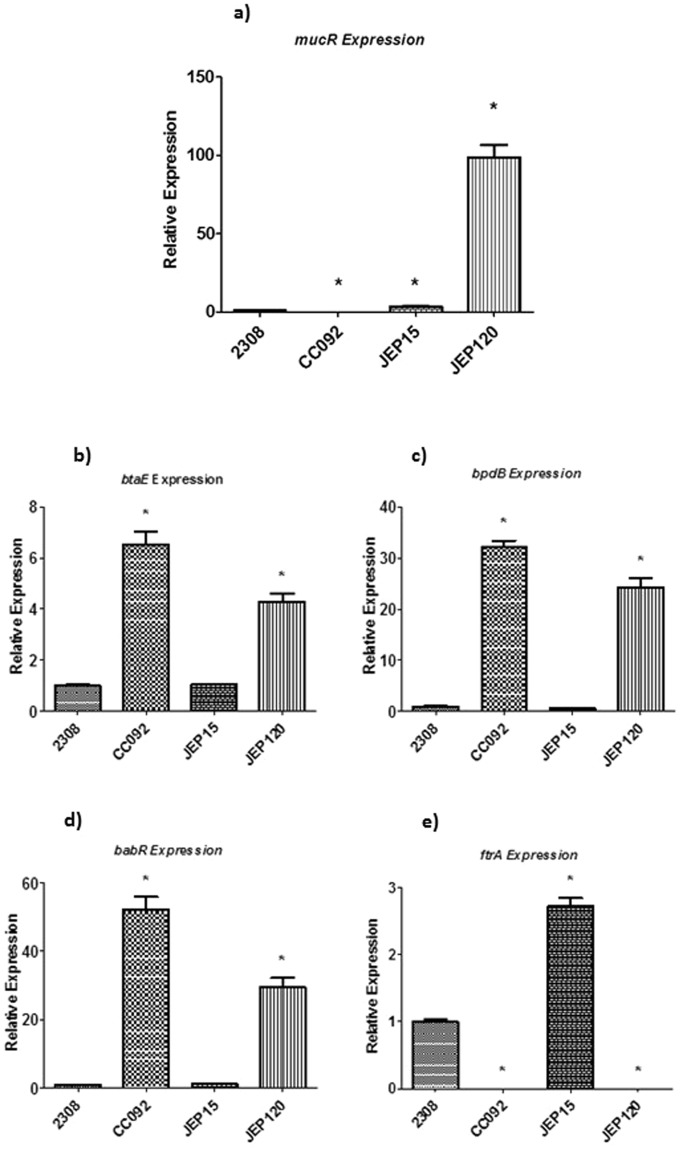
Microarray analysis indicates that MucR controls the expression of more than 90 genes in B. abortus 23084. Genes that have been found to be direct targets of MucR repression by EMSA analysis (J. Pitzer, unpublished data) include the gene encoding the LuxR-type quorum sensing regulator BabR44, the gene encoding the c-diGMP degrading phosphodiesterase BpdB45, the gene encoding the polar adhesin BtaE46 and the mucR gene itself4. EMSA analysis (J. Pitzer, unpublished data) also indicates that the operon encoding the Fe2+ transporter FtrABCD47 is a direct target of MucR activation in Brucella. To investigate whether or not the MucRL36L39I40A mutant is able to regulate these genes, we used q-RT PCR analysis to compare the transcription patterns of babR, bpdB, btaE, mucR and ftrA in B. abortus 2308, the isogenic mucR mutant CC092, and derivatives of the mucR mutant carrying plasmid-borne copies of the native mucR gene (JEP15) or the mutant mucRL36L39I40A gene (JEP120). The results clearly show that the mutant MucRL36L39I40A has lost its ability to repress btaE, bpdB, babR and mucR expression, or activate ftrA expression, with the same efficiency as the wild-type MucR (Fig. 6). These experimental findings strongly suggest that MucR oligomerization is essential for its regulatory function in Brucella.
Figure 6.
Relative expression of (a) mucR, (b) btaE, (c) bpdB, (d) babR and (e) ftrA in B. abortus 2308, an isogenic mucR mutant (CC092), the mucR mutant carrying a plasmid-borne copy of mucR (JEP15), and the mucR mutant carrying a plasmid-borne copy of the mucRL36L39I40A (JEP120) determined by q-RT PCR. RNA was obtained from mid-log phase bacterial cultures grown in brucella broth. The results presented are from a single representative experiment with three technical replicates for each experimental condition. The experiment was repeated three times with equivalent results. *P < 0.05 for comparisons of 2308 vs. the other three strains using the Student t-test.
Discussion
The Ros/MucR prokaryotic zinc-finger proteins are global transcriptional regulators in the α-proteobacteria, where they work in concert with other transcriptional regulators to coordinate the expression of genes required for the symbiotic8–10,48,49 and pathogenic3–6,50 interactions of these bacteria with their eukaryotic hosts, and the orderly progression of the aquatic bacterium Caulobacter crescentus through its well-described developmental cycle12. Studies employing transposon mutagenesis51, transcriptomic4,6,8,9,52 and proteomic analyses53, and chromatin immunoprecipitation coupled with high throughput DNA sequencing (ChIPseq)12 have shown that many genes involved in basic metabolic and physiologic processes and virulence and symbiotic properties in a variety of different α-proteobacteria are regulated by Ros/MucR homologs, and in most cases these genes appear to be targets of Ros/MucR repression. The zinc-finger domains of these proteins have been studied in detail both structurally and functionally1,13–20,54. Previous studies have shown that these proteins bind AT-rich DNA sequences12,13,22–25. We recently demonstrated that AT-rich DNA sequences containing T-A steps are necessary and sufficient for DNA-binding of the Ros/MucR proteins and that these proteins contact DNA mainly in the minor groove26.
From a structural standpoint, the prokaryotic and eukaryotic C2H2 zinc-finger domains share similarities, including their ability to bind DNA, the tetrahedral coordination of a structural zinc ion and the presence of ββα secondary structures15. On the other hand, the prokaryotic C2H2 zinc-finger shows some peculiarities such as the second α-helix, the flexibility of the zinc-coordination sphere and the larger hydrophobic core that together with the zinc-finger motif constitutes the DNA-binding domain1,14–18. Our recent studies with full-length Ros/MucR proteins has determined that these proteins form higher-order oligomers26, are stable at high temperature and are able to bind multiple sites present in the promoters of their target genes26,27. These combined experimental findings have led us to propose that the Ros/MucR proteins regulate gene expression in the same manner as the DNA structuring protein H-NS, which is found in many other Gram-negative bacteria55.
In the present study, we define a hydrophobic region including residues L36, L39and I40 as being responsible for higher-order oligomer formation by the Brucella MucR, and show that a targeted mutation of this region is sufficient to get the protein to switch from a decameric state to a monomeric one. This finding together with the predicted structure of the MucR N-terminus suggests the possibility that the second α-helix comprising the hydrophobic region identified here at the N-terminus of MucR could be involved in the formation of higher-order oligomers. Our data clearly demonstrate that the absence of the first α-helix at the N-terminus of MucR is not sufficient to change the oligomeric state of the protein. In fact, the deletion mutant 33–142 containing this first α-helix is still able to form higher-order oligomers, whereas the 45–142 deletion mutant of MucR lacking the first α-helix and the hydrophobic region L36L39I40, presents only a monomeric state. The MucRL36L39I40A mutant demonstrates that this hydrophobic region included in the second α-helix at the N-terminus of MucR has an essential role in oligomerization. More structural studies are necessary to understand how the interaction between MucR monomers might occur and whether or not other secondary structure elements have a role in stabilizing the higher-order oligomers. In H-NS more than one region is involved in higher-order oligomer formation56–58 whereas in the case of MucR, the mutation of three residues at the N-terminus is sufficient to get a switch from higher-order oligomers to a monomer. The presence of H-NS lower-order oligomers in solution was thoroughly investigated29,57–63 showing many different degrees of H-NS oligomers depending on both different experimental conditions and/or portions of the protein analysed. All the studies agreed in pointing out the crucial role of higher-order oligomer formation for proper H-NS function. Our findings with MucR cannot exclude the existence of lower-order oligomers which could be present at very low concentration in solution and contribute to the formation of higher-order oligomers similar to the H-NS. However, we can state that the main region forming higher-order oligomers is located at the N-terminus of MucR and that the hydrophobic region that this protein shares with all of the MucR/Ros homologs plays a major role in building the higher-order oligomers. Our preliminary results indicate that there are about 5.5 × 104 MucR molecules for B. abortus cells, or about 3.6% of the total cellular protein (J. Pitzer, unpublished data). This is consistent with the number of H-NS and other nucleoid-associated proteins that have been reported in other bacteria which ranges from 104 to 105 molecules per cell corresponding to μM concentration56,64. This high intracellular level of MucR also indicates that the in vitro experimental conditions used here to observe its oligomerization are likely to be physiologically relevant.
The observation that the mucRL36L39I40A allele cannot repress the btaE, bpdB, babR or mucR expression and cannot activate the ftrA expression in B. abortus indicates that MucR must be able to form oligomers to retain its wild-type regulatory function. These experimental findings also provide further support for our hypothesis that MucR plays a similar role to that proposed for H-NS in terms of its ability to serve as a transcriptional regulator. Its capacity to form extensive oligomers allows H-NS to compact DNA, and accordingly, it plays an important role in establishing and maintaining nucleoid structure in bacteria59–66. But the members of H-NS protein family also play an important role in regulating prokaryotic gene expression. The capacity of these proteins to bind to AT-rich regions in and around bacterial promoters, oligomerize and make these promoters inaccessible to RNA polymerase allows H-NS proteins to serve as global gene ‘silencers’38,39,41. Experimental evidence suggests that the gene silencing capacity of H-NS proteins is important for both protecting bacteria from the uncontrolled expression of xenogeneic genes acquired by horizontal gene transfer30,32–39,56 and preventing the gratuitous expression of genes that only provide bacteria with a fitness benefit in a particular environment (e.g. genes required for virulence in a mammalian host)30–39. Notably, previous genetic and biochemical studies suggest that MucR and its homologs function predominately as transcriptional repressors in the α-proteobacteria4,6,8,12,51 and that they work in concert with antagonistic transcriptional activators in these bacteria to ensure that virulence, symbiosis and cell cycle genes are only expressed when they are needed by these bacteria67. However, it is important to note that the inability of the mucRL36L39I40A allele to activate ftrA transcription in B. abortus gene is also compatible with the proposed role of MucR as an H-NS-like regulator. Specifically, in addition to its negative effects, the nucleoid structuring role of H-NS proteins can also have positive effects on the transcription of some genes because this structuring provides easier access of RNA polymerase and transcriptional activators to their promoter regions33.
Previous studies have provided evidence that Ros/MucR proteins in other α-proteobacteria form oligomeric complexes11,68, and the data presented here suggest that the ability to form these complexes plays an important role in the capacity of these proteins to function as transcriptional regulators. The capacity to bind to low consensus AT-rich regions of DNA in and around the promoters of the genes they regulate also seems to be a shared feature of the Ros/MucR proteins that have been characterized7,12,13,22–25,69. Further biochemical and genetic studies will be necessary to fully understand how MucR homologs function as transcriptional regulators, but a model in which they recognize AT-rich sequences containing T-A steps in DNA as nucleation sites, bind other AT-rich regions, form high-order oligomers and prevent access to bacterial promoters by RNA polymerase is certainly consistent with the data presented here and with the information available in the literature with regard to the capacity of these proteins to serve as transcription repressors. This suggests that from a functional standpoint, perhaps the Ros/MucR family of zinc-finger prokaryotic proteins should be considered to be new members of the family of so-called H-NS-‘like’ gene silencers30,32,35–37. Although the role of H-NS as a global gene silencer is well established, not all bacteria, including the majority of the α-proteobacteria, possess an H-NS homolog55. However, other small nucleoid binding proteins that have no significant amino acid identity with H-NS have been found to perform a similar regulatory role. The MvaT and MvaU proteins in Pseudomonas29,32,34 and the Lsr2 protein in Mycobacterium30, for instance, are small nucleoid binding proteins that also recognize AT-rich regions containing T-A steps in DNA, bind to these regions and form oligomers, and serve as global repressors of gene expression.
Conclusion
The experimental findings presented here show that the hydrophobic region defined by the amino acids L36L39I40 at the N-terminus of the Brucella MucR is required for this protein to be able to form higher-order oligomers and to perform its normal function as a transcriptional regulator. Based on these and earlier findings26,27, a new functional model is arising for the prokaryotic zinc-finger proteins in the Ros/MucR family. Specifically, these proteins appear to be binding to low consensus AT-rich regions in DNA and functioning as H-NS-‘like’ gene silencers, unlike their counterparts in eukaryotes which function mainly as DNA sequence-specific transcriptional regulators70.
Methods
Cloning, protein expression and purification
The DNA fragments encoding for MucRL36L39I40A, MucR33–142, MucR33–142mut, MucR45–142, MucR57–142 were generated by PCR. Primers were designed on the basis of the wild-type mucR gene sequence (GenBank: SHO30402.1) to get the deletions or the point mutations of the wild-type gene. The sequence of the primers are shown in the Suppl. Table 1; the template used for PCRs was the pET-22b(+) plasmid containing the wild-type mucR gene previously published26. The PCR products encoding for MucR33–142, MucR33–142mut, MucR45–142, MucR57–142 were digested with NdeI/EcoRI restriction enzymes and cloned into pET-22b(+) expression vector digested with the same enzymes; the PCR product encoding for MucRL36L39I40A was digested with NcoI/EcoRI restriction enzymes and cloned into the pET-11D expression vector digested with NcoI/EcoRI as well. All the proteins were expressed as previously reported1 in E. coli host strain BL21(DE3) grown in Luria Bertani medium (for proteins to be analysed by Light Scattering) or grown in a minimal medium1 containing 0.5 g/L 15NH4Cl as the only nitrogen source (for proteins to be analysed by NMR). The expression was induced for 1 h at 28 °C with 1 mM IPTG. Protein purification was carried out as previously reported1. The proteins were eluted from a Mono S HR 5/5 cation exchange chromatography column in the 0.3–0.6 M NaCl concentration range.
Light Scattering
For molecular weight measurements, a MiniDAWN Treos spectrometer (Wyatt Instrument Technology Corp.) equipped with a laser operating at 658 nm was used connected on-line to a size-exclusion chromatography column. Samples at 1 mg/ml were loaded onto a Superdex 200 column (10 × 30 cm, GEHealthcare) equilibrated in the same buffer used for the final purification procedure and connected to a triple-angle light scattering detector equipped with a QELS (Quasi-Elastic Light Scattering) module71. A constant flow rate of 0.5 ml/min was applied. Elution profiles were detected by a Shodex interferometric refractometer and a mini Dawn TREOS light scattering system. Data were analyzed using the Astra 5.3.4.14 software (Wyatt Technology). The experiments were carried out in duplicate.
NMR spectroscopy
The NMR spectra of the proteins purified as described above were recorded at 298 K on a Bruker Avance III HD 600 MHz equipped with cryoprobe at the Department of Environmental, Biological and Pharmaceutical Science and Technology, University of Campania - Luigi Vanvitelli (Caserta, Italy). The NMR samples of MucR, MucRL36L39I40A and MucR57–142 contained 200 μM or 500 μM of purified 15N labelled proteins in a 20 mM phosphate buffer, 0.3 M NaCl at pH = 6.8.
The 1H-15N HSQC spectra of MucR, MucR57–142 and MucRL36L39I40A were acquired at 298 K using 256 complex points for 15N (F1) and 2048 for 1H (F2). The translational diffusion coefficient (Dtrans)72 was obtained by using the PFG diffusion measurements with the PG-SLED (pulse gradient-stimulated echo longitudinal encode-decode) sequence73.
Data were processed using the TopSpin 3.5 software (Bruker) and analyzed with the CARA software as previously reported16 (downloaded from cara.nmr.ch+).
Electrophoretic Mobility Shift Assay (EMSA)
The EMSA experiments were performed as previously described74,75. In detail, 0.4, 0.6 or 0.8 μg of each protein were incubated 10 min on ice in binding buffer (25 mM HEPES pH 7.9, 50 mM KCl, 6.25 mM MgCl2, 5% glycerol) with 5 pmol of double-stranded oligonucleotides Site 1(5′-GTTGCCTATTATTAATGTAATATGGTTTGA-3′) previously published as a target site of MucR and located at −174 bp from the ATG start codon of mucR gene27. The protein/DNA ratio for each sample was 5, 8, 10 when 0.4, 0.6, 0.8 μg of protein were used respectively. To obtain a negative control, 0.8 μg of each protein were incubated with 5 pmol of the double-stranded oligonucleotide NC (5′-CGCGGCACGACCGCAGCGGTCGGGTGGCAC-3′) in the same binding buffer whose composition has been already described above. The total volume of each reaction mixture was 20 μl. After incubation on ice, the samples were loaded onto a 5% polyacrylamide gel in 0.5X TBE and run at room temperature for 70 min at 200 V. Gels were stained 20 minutes using Diamond™ Nucleic Acid Dye (Promega) following the manufacturer’s instructions and imaged by Typhoon Trio+ scanner (GE Healthcare). The results by EMSAs shown in this study are representative of more than 10 replicates.
Genetic complementation of a Brucella mucR mutant
Genetic complementation of the B. abortus mucR mutant CC0924 with a wild-type version of the corresponding gene was performed as previously described26. The Q5 Site Directed Mutagenesis kit (NEB) was used to make mucRL36L39I40A gene using the mutagenesis primers mucRL36AL39AI40A SDMF and R (Suppl. Table 1) and plasmid pJep011 which encodes the wild-type Brucella mucR26 as a template. The mutagenesis primers were designed using the web-based NEB Base Changer Program available at www.neb.com. The nucleotide sequence of the mucRL36L39I40A gene confirmed by DNA sequence analysis.
The B. abortus mucR mutant CC092 was transformed by electroporation with either the pJep01126 or pJep120 plasmid and selected on Schaedlar agar supplemented with 5% defibrinated bovine blood (SBA)4 containing 45 μg/ml kanamcyin. The B. abortus strains were grown to mid log phase in brucella broth, and diluted in this medium to a cell density of approximately 104 colony forming units (CFU) per ml. Twenty-five microliters (25 μ) of each bacterial cell suspension was spotted onto SBA plates, the plates incubated at 37 °C under 5% CO2, and the bacterial colonies produced by the individual strains observed and photographed after 72 h cultivation on this medium.
q-RT PCR
Total RNA was isolated from B. abortus cells following growth to mid log phase in brucella broth (BD) using previously described procedures4. cDNA was generated from the final RNA preparation using the SuperScript III First Strand kit (Invitrogen) following the manufacturer’s protocol. The cDNA preparations were then used as the templates for real-time RT-PCR with SsoAdvanced Sybr Green Supermix (Bio-Rad) to evaluate the relative levels of gene-specific mRNA transcripts in the total cellular RNA preparations. The gene-specific oligonucleotide primers used for amplification of the experimental (babR [BAB1_0190], bpdB [BAB1_0512], ftrA [BAB2_0840], btaE [BAB1_0069], mucR [BAB1_0594] and control (GAP [BAB1_1741] and 16 S rRNA [BAB1_2223]) transcripts are listed in Supp. Table 1. The parameters used for the PCR reaction were a single denaturing step for 30 sec at 95 °C, followed by 40 cycles (denature for 15 sec at 95 °C, anneal/extend for 30 sec at 60 °C) of amplification. Fluorescence from SYBR green incorporation into double-stranded DNA was measured with a Bio-Rad CFX96 Thermocycler and analyzed on Bio-Rad Maestro Software. The results were confirmed by three biological and three technical replications. Data were analysed by the 2−ΔΔCt method. Student T-test was used to evaluate the statistical significance of the results.
Electronic supplementary material
Acknowledgements
This work was supported by Ministero della Salute funding CUP C75E17000050001 granted to I.B. Work in the Roop laboratory was funded by a grant (AI112745) from the National Institute of Allergy and Infectious Diseases. This work was also supported by VALERE project of University of Campania “Luigi Vanvitelli”.
Author Contributions
L.P.: performed static Light Scattering. J.E.P.: performed genetic complementation; q-RT-PCRs. G.D. and G.M.: performed NMR experiments. R.F. and G.M.: analysed NMR spectra and data. E.M.P. and L.P.: analysed Light Scattering data; contributed to write the manuscript. P.V.P. and R.M.R. II: analysed data, contributed to conceive the project and to write the manuscript. I.B.: conceived the project, designed the experiments; wrote the manuscript; performed cloning; expression and protein purifications; EMSAs. All the authors revised the manuscript.
Data Availability
Data generated or analysed during this study are included in this published article (and its Supplementary Information files). The unpublished data are available from the corresponding author on reasonable request.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Luciano Pirone and Joshua Edison Pitzer contributed equally.
Contributor Information
Paolo Vincenzo Pedone, Email: paolov.pedone@unicampania.it.
Roy Martin Roop, II, Email: ROOPR@ecu.edu.
Ilaria Baglivo, Email: ilaria.baglivo@unicampania.it.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-35432-1.
References
- 1.Baglivo I, et al. The structural role of the zinc ion can be dispensable in prokaryotic zinc-finger domains. Proc. Natl. Acad. Sci. USA. 2009;106:6933–6938. doi: 10.1073/pnas.0810003106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Netti F, et al. An experimentally tested scenario for the structural evolution of eukaryotic Cys2His2 zinc fingers from eubacterial ros homologs. Mol. Biol. Evol. 2013;30:1504–1513. doi: 10.1093/molbev/mst068. [DOI] [PubMed] [Google Scholar]
- 3.Close TJ, Tait RC, Kado CI. Regulation of Ti plasmid virulence genes by a chromosomal locus of Agrobacterium tumefaciens. J. Bacteriol. 1985;164:774–781. doi: 10.1128/jb.164.2.774-781.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caswell CC, et al. Diverse genetic regulon of the virulence-associated transcriptional regulator MucR in Brucella abortus. Infection and Immunity. 2013;81:1040–1051. doi: 10.1128/IAI.01097-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mirabella A, et al. Brucella melitensis MucR, an orthologue of Sinorhizobium meliloti MucR, is involved in resistance to oxidative, detergent, and saline stresses and cell envelope modifications. J. Bacteriol. 2013;195:453–465. doi: 10.1128/JB.01336-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dong H, et al. The effects of MucR on expression of type IV secretion system, quorum sensing system and stress responses in Brucella melitensis. Vet. Microbiol. 2013;166:535–542. doi: 10.1016/j.vetmic.2013.06.023. [DOI] [PubMed] [Google Scholar]
- 7.Keller M, et al. Molecular analysis of the Rhizobium meliloti mucR gene regulating the biosynthesis of the exopolysaccharides succinoglycan and galactoglucan. Mol. Plant. Microbe Interact. 1995;8:267–277. doi: 10.1094/MPMI-8-0267. [DOI] [PubMed] [Google Scholar]
- 8.Mueller K, González JE. Complex regulation of symbiotic functions is coordinated by MucR and quorum sensing in Sinorhizobium meliloti. J. Bacteriol. 2011;193:485–496. doi: 10.1128/JB.01129-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiao J, et al. MucR is required for transcriptional activation of conserved ion transporters to support nitrogen fixation of Sinorhizobium fredii in soybean nodules. Mol. Plant Microbe. Interact. 2016;29:352–361. doi: 10.1094/MPMI-01-16-0019-R. [DOI] [PubMed] [Google Scholar]
- 10.Bittinger MA, et al. rosR, a determinant of nodulation competitiveness in Rhizobium etli. Mol. Plant Microbe Interact. 1997;10:180–186. doi: 10.1094/MPMI.1997.10.2.180. [DOI] [PubMed] [Google Scholar]
- 11.Janczarek M, Skorupska A. The Rhizobium leguminosarum bv. trifolii RosR: transcriptional regulator involved in exopolysaccharide production. Mol. Plant Microbe Interact. 2007;20:867–881. doi: 10.1094/MPMI-20-7-0867. [DOI] [PubMed] [Google Scholar]
- 12.Fumeaux C, et al. Cell cycle transition from S-phase to G1 in Caulobacter is mediated by ancestral virulence regulators. Nat. Commun. 2014;5:4081. doi: 10.1038/ncomms5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esposito S, et al. A novel type of zinc finger DNA binding domain in the Agrobacterium tumefaciens transcriptional regulator Ros. Biochemistry. 2006;45:10394–10405. doi: 10.1021/bi060697m. [DOI] [PubMed] [Google Scholar]
- 14.Baglivo I, et al. Molecular strategies to replace the structural metal site in the prokaryotic zinc finger domain. Biochim. Biophys. Acta. 2014;1844:497–504. doi: 10.1016/j.bbapap.2013.12.019. [DOI] [PubMed] [Google Scholar]
- 15.Malgieri G, et al. The prokaryotic Cys2His2 zinc-finger adopts a novel fold as revealed by the NMR structure of Agrobacterium tumefaciens Ros DNA-binding domain. Proc. Natl. Acad. Sci. USA. 2007;104:17341–6. doi: 10.1073/pnas.0706659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.D’Abrosca G, et al. The (unusual) aspartic acid in the metal coordination sphere of the prokaryotic zinc finger domain. J. Inorg. Biochem. 2016;161:91–98. doi: 10.1016/j.jinorgbio.2016.05.006. [DOI] [PubMed] [Google Scholar]
- 17.Palmieri M, et al. Structural Zn(II) implies a switch from fully cooperative to partly downhill folding in highly homologous proteins. J. Am. Chem. Soc. 2013;135:5220–5228. doi: 10.1021/ja4009562. [DOI] [PubMed] [Google Scholar]
- 18.Palmieri M, et al. Deciphering the zinc coordination properties of the prokaryotic zinc finger domain: The solution structure characterization of Ros87 H42A functional mutant. J. Inorg. Biochem. 2014;131:30–36. doi: 10.1016/j.jinorgbio.2013.10.016. [DOI] [PubMed] [Google Scholar]
- 19.Malgieri G, et al. Zinc to cadmium replacement in the prokaryotic zinc-finger domain. Metallomics. 2014;6:96–104. doi: 10.1039/C3MT00208J. [DOI] [PubMed] [Google Scholar]
- 20.Russo L, et al. NMR assignments of the DNA binding domain of Ml4 protein from Mesorhizobium loti. Biomol. NMR Assign. 2010;4:55–57. doi: 10.1007/s12104-009-9206-0. [DOI] [PubMed] [Google Scholar]
- 21.Russo L, et al. Towards understanding the molecular recognition process in prokaryotic zinc-finger domain. Eur. J. Med. Chem. 2015;91:100–108. doi: 10.1016/j.ejmech.2014.09.040. [DOI] [PubMed] [Google Scholar]
- 22.D’Souza-Ault MR, et al. Analysis of the Ros repressor of Agrobacterium virC and virD operons: molecular intercommunication between plasmid and chromosomal genes. J. Bacteriol. 1993;175:3486–3490. doi: 10.1128/jb.175.11.3486-3490.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bertram-Drogatz PA, et al. The regulatory protein MucR binds to a short DNA region located upstream of the mucR coding region in Rhizobium meliloti. Mol. Gen. Genet. 1997;254:529–538. doi: 10.1007/s004380050448. [DOI] [PubMed] [Google Scholar]
- 24.Bertram-Drogatz PA, et al. The Sinorhizobium meliloti MucR protein, which is essential for the production of high-molecular-weight succinoglycan exopolysaccharie, binds short DNA regions upstream of exoH and exoY. Mol. Gen. Genet. 1998;257:433–441. doi: 10.1007/s004380050667. [DOI] [PubMed] [Google Scholar]
- 25.Bahlawane C, et al. Sinorhizobium meliloti regulator MucR couples exopolysaccharide synthesis and motility. Mol. Plant Microbe Interact. 2008;21:1498–1509. doi: 10.1094/MPMI-21-11-1498. [DOI] [PubMed] [Google Scholar]
- 26.Baglivo I, et al. Ml proteins from Mesorhizobium loti and MucR from Brucella abortus: an AT-rich core DNA-target site and oligomerization ability. Sci Rep. 2017;7:15805. doi: 10.1038/s41598-017-16127-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baglivo Ilaria, Pirone Luciano, Malgieri Gaetano, Fattorusso Roberto, Roop II Roy Martin, Pedone Emilia Maria, Pedone Paolo Vincenzo. MucR binds multiple target sites in the promoter of its own gene and is a heat-stable protein: Is MucR a H-NS-like protein? FEBS Open Bio. 2018;8(4):711–718. doi: 10.1002/2211-5463.12411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lang B, et al. High-affinity DNA binding sites for H-NS provide a molecular basis for selective silencing within proteobacterial genomes. Nucleic Acids Res. 2007;35:6330–6337. doi: 10.1093/nar/gkm712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Castang S, Dove SL. High-order oligomerization is required for the function of the H-NS family member MvaT in Pseudomonas aeruginosa. Mol Microbiol. 2010;78:916–931. doi: 10.1111/j.1365-2958.2010.07378.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gordon BR, et al. Structural basis for recognition of AT-rich DNA by unrelated xeogeneic silencing proteins. Proc. Natl. Acad. Sci. US A. 2011;108:10690–10695. doi: 10.1073/pnas.1102544108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cordeiro TN, et al. Indirect DNA Readout by H-NS Related protein: Structure of the DNA Complex of the C-terminal Domain of Ler. Plos Pathogen. 2011;7:e1002380. doi: 10.1371/journal.ppat.1002380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ding P, et al. A Novel AT-Rich DNA Recognition Mechanism for Bacterial Xenogeneic Silencer MvaT. Plos Pathogen. 2015;11:e1004967. doi: 10.1371/journal.ppat.1004967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dorman CJ. H-NS: A Universal Regulator For a Dynamic genome. Nature Rev Microbiol. 2004;2:391–399. doi: 10.1038/nrmicro883. [DOI] [PubMed] [Google Scholar]
- 34.Castang S, McManus HR, Turner KH, Dove SL. H-NS family members function coordinately in an opportunistic pathogen. Proc. Natl. Acad. Sci. USA. 2008;105:18947–18952. doi: 10.1073/pnas.0808215105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lucchini S, et al. H-NS Mediates the Silencing of Laterally Acquired Genes in Bacteria. Plos Pathogen. 2006;2:e81. doi: 10.1371/journal.ppat.0020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Navarre WW, et al. Selective Silencing of Foreign DNA with Low GC Content by the H-NS Protein in Salmonella. Science. 2006;313:236–238. doi: 10.1126/science.1128794. [DOI] [PubMed] [Google Scholar]
- 37.Navarre WW. Silencing of xenogeneic DNA by H-NS – facilitation of lateral gene transfer in bacteria by a defense system that recognizes foreign DNA. Genes dev. 2007;21:1456–1471. doi: 10.1101/gad.1543107. [DOI] [PubMed] [Google Scholar]
- 38.Dorman CJ. H-NS, the genome sentinel. Nat. Rev. Microbiol. 2007;5:157–161. doi: 10.1038/nrmicro1598. [DOI] [PubMed] [Google Scholar]
- 39.Will WR, Navarre WW, Fang FC. Integrated Circuits: How Transcriptional Silencing and Counter-silencing Facilitate Bacterial Evolution. Curr Opin Microbiol. 2015;0:8–13. doi: 10.1016/j.mib.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ulissi U, Fabbretti A, Sette M, Giuliodori AM, Spurio R. Time-resolved assembly of a nucleoprotein complex between Shigella Flexneri virF promoter and its transcriptional repressor H-NS. Nucleic Acids Res. 2014;42:13039–13050. doi: 10.1093/nar/gku1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ayala JC, Silva AJ, Benitez JAH-NS. An overarching regulator of the Vibrio cholera life cycle. Res. Microbiol. 2017;168:16–25. doi: 10.1016/j.resmic.2016.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 43.Arena Giuseppe, Fattorusso Roberto, Grasso Giuseppe, Grasso Giuseppa Ida, Isernia Carla, Malgieri Gaetano, Milardi Danilo, Rizzarelli Enrico. Zinc(II) Complexes of Ubiquitin: Speciation, Affinity and Binding Features. Chemistry - A European Journal. 2011;17(41):11596–11603. doi: 10.1002/chem.201101364. [DOI] [PubMed] [Google Scholar]
- 44.Rambow-Larsen A. A., Rajashekara G., Petersen E., Splitter G. Putative Quorum-Sensing Regulator BlxR of Brucella melitensis Regulates Virulence Factors Including the Type IV Secretion System and Flagella. Journal of Bacteriology. 2008;190(9):3274–3282. doi: 10.1128/JB.01915-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Petersen E, et al. Brucella melitensis cyclic di-GMP phosphodiesterase BpdA controls expression of flagellar genes. J. Bacteriol. 2011;193:5683–5691. doi: 10.1128/JB.00428-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ruiz-Ranwez V, et al. BtaE, an adhesin that belongs to the trimeric autotransporter family, is required for full virulence and defines a specific adhesive pole of Brucella suis. Infect. Immun. 2013;81:996–1007. doi: 10.1128/IAI.01241-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Elhassanny AE, et al. The ferrous iron transporter FtrABCD is required for the virulence of Brucella abortus 2308 in mice. Mol. Microbiol. 2013;88:1070–1082. doi: 10.1111/mmi.12242. [DOI] [PubMed] [Google Scholar]
- 48.Janczarek J, et al. Rhizobium leguminosarum bv. trifolii rosR is required for interaction with clover, biofilm formation and adaptation to the environment. BMC Microbiol. 2010;10:e284. doi: 10.1186/1471-2180-10-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Acosta-Jurado S, et al. The Sinorhizobium fredii HH103 MucR1 global regulator is connected with the nod regulon and is required for efficient symbiosis with Lotus burttii and Glycine max cv. Williams. Mol. Plant Microbe Interact. 2016;29:700–712. doi: 10.1094/MPMI-06-16-0116-R. [DOI] [PubMed] [Google Scholar]
- 50.Wu W, et al. Mariner mutagenesis of Brucella melitensis reveals genes with previously uncharacterized role in virulence and survival. BMC Microbiol. 2006;6:e102. doi: 10.1186/1471-2180-6-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bittinger MA, Handelsman J. Identification of genes in the RosR regulon of Rhizobium etli. J. Bacteriol. 2000;182:1706–1713. doi: 10.1128/JB.182.6.1706-1713.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rachwel K, et al. Transcriptome profiling of a Rhizobium leguminosarum bv. trifolii rosR mutant reveals the role of the transcriptional regulator RosR in motility, synthesis of cell-surface components, and other cellular processes. BMC Genomics. 2015;16:e1111. doi: 10.1186/s12864-015-2332-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rachwal K, et al. The regulatory protein RosR affects Rhizobium leguminosarum bv. trifolii protein profiles, cell surface properties, and symbiosis with clover. Front. Microbiol. 2016;7:e1302. doi: 10.3389/fmicb.2016.01302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chou AY, et al. Agrobacterium transcriptional regulator Ros is a prokaryotic zinc finger protein that regulates the plant oncogene ipt. Proc. Natl. Acad. USA. 1998;95:5293–5298. doi: 10.1073/pnas.95.9.5293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tendeng C, Bertin PN. H-NS in Gram-negative bacteria: a family of multifaceted proteins. Trends in Microbiol. 2003;11:511–518. doi: 10.1016/j.tim.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 56.Esposito D, et al. H-NS oligomerization domain structure reveals the mechanism for high order self-association of the intact protein. J Mol Biol. 2002;324:841–850. doi: 10.1016/S0022-2836(02)01141-5. [DOI] [PubMed] [Google Scholar]
- 57.Bloch V, et al. The H-NS dimerization domain defines a new fold contributing to DNA recognition. Nat Struct Biol. 2003;10:212–218. doi: 10.1038/nsb904. [DOI] [PubMed] [Google Scholar]
- 58.Arold ST, Leonard PG, Parkinson GN, Ladbury JE. H-NS forms a superhelical protein scaffold for DNA condensation. Proc. Natl. Acad. Sci. US A. 2010;107:15728–15732. doi: 10.1073/pnas.1006966107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Winardhi RS, Yan J, Kenney LJ. H-NS Regulates Gene Expression and Compacts the Nucleoid: Insights from Single-Molecule Experiments. Biophys J. 2015;109:1321–1329. doi: 10.1016/j.bpj.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Renault MR, Garcia JCTN, Baldus M, Pons M. Protein oligomers studied by solid-state NMR – the case of the full-length nucleoid-associated protein histone-like nucleoid structuring protein. FEBS J. 2013;280:2916–2928. doi: 10.1111/febs.12297. [DOI] [PubMed] [Google Scholar]
- 61.Stella S, Spurio R, Falconi M, Pon CL, Gualerzi CO. Nature and mechanism of the in vivo oligomerization of nucleoid protein H-NS. EMBO J. 2005;24:2896–2905. doi: 10.1038/sj.emboj.7600754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smyth CP, et al. Oligomerization of the chromatin-structuring protein H-NS. Mol Microbiol. 2000;36:962–972. doi: 10.1046/j.1365-2958.2000.01917.x. [DOI] [PubMed] [Google Scholar]
- 63.Ceschini S, et al. Multimeric self-assembly equilibria involving the histone-like protein H-NS. A thermodynamic study. J Biol Chem. 2000;275:729–734. doi: 10.1074/jbc.275.2.729. [DOI] [PubMed] [Google Scholar]
- 64.Tupper AE, et al. The chromatin-associated protein H-NS alters DNA topology in vitro. EMBO J. 1994;13:258–268. doi: 10.1002/j.1460-2075.1994.tb06256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ueguchi C, Suzuki T, Tanaka TYK, Mizuno T. Systematic Mutational Analysis Revealing the Functional Domain Organization of Escherichia coli Nucleoid Protein H-NS. J. Mol. Biol. 1996;263:149–162. doi: 10.1006/jmbi.1996.0566. [DOI] [PubMed] [Google Scholar]
- 66.Dorman CJ, Kane KA. DNA bridging and antibridging: a role for bacterial nucleoid-associated proteins in regulating the expression of laterally acquired genes. FEMS Microbiol Rev. 2009;33:587–592. doi: 10.1111/j.1574-6976.2008.00155.x. [DOI] [PubMed] [Google Scholar]
- 67.Panis G, Murray SR, Viollier PH. Versatility of global transcriptional regulators in alpha-Proteobacteria: from essential cell cycle control ancillary function. FEMS Microbiol. Rev. 2015;39:120–133. doi: 10.1093/femsre/fuu002. [DOI] [PubMed] [Google Scholar]
- 68.Bahlawane C, et al. Fine-tuning of galactoglucan biosynthesis in Sinorhizobium meliloti by differential WggR (ExpG)-, PhoB-, and MucR-dependent regulation of two promoters. J. Bacteriol. 2008;190:3456–3466. doi: 10.1128/JB.00062-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cooley MB, D’Souza MR, Kado CI. The virC and virD Operons of the Agrobacterium Ti Plasmid Are Regulated by the ros Chromosomal Gene: Analysis of the Cloned ros Gene. J. Bacteriol. 1991;173:2608–2616. doi: 10.1128/jb.173.8.2608-2616.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Malgieri G. The prokaryotic zinc-finger: structure, function and comparison with the eukaryotic counterpart. FEBS J. 2015;282:4480–4496. doi: 10.1111/febs.13503. [DOI] [PubMed] [Google Scholar]
- 71.Contursi P, et al. C68 from the Sulfolobus islandicus plasmid-virus pSSVx is a novel member of the AbrB-like transcription factor family. Biochem. J. 2011;435:157–66. doi: 10.1042/BJ20101334. [DOI] [PubMed] [Google Scholar]
- 72.Wilkins DK, et al. Hydrodynamic radii of native and denatured proteins measured by pulse field gradient NMR techniques. Biochemistry. 1999;38:16424–16431. doi: 10.1021/bi991765q. [DOI] [PubMed] [Google Scholar]
- 73.Gibbs SJ, Johnson CS., Jr. A PFG-NMR experiment for accurate diffusion and flow studies in the presence of eddy currents. J Magn Reson. 1991;93:395–402. [Google Scholar]
- 74.Baglivo I, et al. Genetic and epigenetic mutations affect the DNA binding capability of human ZFP57 in transient neonatal diabetes type 1. FEBS Lett. 2013;587:1474–1481. doi: 10.1016/j.febslet.2013.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Anvar Z, et al. ZFP57 recognizes multiple and closely spaced sequence motif variants to maintain repressive epigenetic marks in mouse embryonic stem cells. Nucleic Acids Res. 2016;44:1118–1132. doi: 10.1093/nar/gkv1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data generated or analysed during this study are included in this published article (and its Supplementary Information files). The unpublished data are available from the corresponding author on reasonable request.