Abstract
The discovery of CRISPR/Cas gene editing has allowed for major advances in many biomedical disciplines and basic research. One arrangement of this biotechnology, a nuclease-based gene drive, can rapidly deliver a genetic element through a given population and studies in fungi and metazoans have demonstrated the success of such a system. This methodology has the potential to control biological populations and contribute to eradication of insect-borne diseases, agricultural pests, and invasive species. However, there remain challenges in the design, optimization, and implementation of gene drives including concerns regarding biosafety, containment, and control/inhibition. Given the numerous gene drive arrangements possible, there is a growing need for more advanced designs. In this study, we use budding yeast to develop an artificial multi-locus gene drive system. Our minimal setup requires only a single copy of S. pyogenes Cas9 and three guide RNAs to propagate three gene drives. We demonstrate how this system could be used for targeted allele replacement of native genes and to suppress NHEJ repair systems by modifying DNA Ligase IV. A multi-locus gene drive configuration provides an expanded suite of options for complex attributes including pathway redundancy, combatting evolved resistance, and safeguards for control, inhibition, or reversal of drive action.
Introduction
The discovery and implementation of the clustered regularly interspaced short palindromic repeat (CRISPR) gene editing system has revolutionized countless fields and sub-specialties across molecular biology and biotechnology to improve human health, agriculture, ecological control, and beyond. Briefly, alteration of the genetic code is accomplished using (i) a bacterial derived nuclease (typically Cas9 or Cas12a), (ii) a single-stranded fragment of “guide” RNA (sgRNA), and (iii) an optional exogenous repair fragment of DNA1–4. Priming of the nuclease with a pre-programmed guide RNA fragment targets a specific genomic sequence for a double strand break (DSB). Following DNA cleavage, eukaryotic cells activate repair systems to either fuse broken chromosomal ends together via non-homologous end joining (NHEJ) or, in the presence of donor DNA, introduce exogenous sequence via homologous recombination (HR). Moreover, the CRISPR methodology is not restricted to DSB-induced alteration of the genome—recent efforts have demonstrated that nuclease-dead variants (e.g. dCas9) can serve as delivery systems to modulate transcriptional activity5, alter epigenetic landscapes6, or introduce mutational substitutions sans any DNA cleavage event7.
One powerful biotechnological application of the CRISPR methodology is within a “gene drive” (GD) system. The basic design of a homing drive includes the expression constructs for the CRISPR nuclease and the corresponding guide RNA positioned at a desired locus of choice—the mechanism of propagation involves targeting of the homologous chromosome (within a diploid or polyploid organism) at the same genetic position (typically cleaving the wild-type gene). Creation of a DSB followed by HR-based repair (using the gene drive-containing DNA as a donor) causes the entire artificial construct (Cas9, the sgRNA, and any desired “cargo”) to be copied; in this way, a heterozygous cell is automatically converted to the homozygous state. This super-Mendelian genetic arrangement allows for the forced propagation of a genetic element within a population and has the potential to modify entire species on a global scale8,9. Some of the possible benefits of this technology include eradication of invasive species10,11, agricultural pest management12, and elimination of insect-borne diseases such as malaria9,13,14. A number of recent studies have demonstrated the potency and success of CRISPR-based gene drives in fungi15–18, and metazoans19–22. While ongoing technical challenges remain in the design, optimization, and field testing of gene drive-harboring organisms, there are also serious biosafety and ethical concerns regarding use of this biotechnology as even current drive systems are expected to be highly invasive within native populations23. There is an immediate need for further study (in silico and in vivo) of gene drive systems that focus on issues of safety15,24,25, control and reversal26,27, and optimal design11.
There are many types of gene drive designs including “daisy-chain drives,” “underdominance drives,” and “anti-drives,” each with a distinct arrangement of the basic CRISPR components that is predicted to sweep through native populations at varying levels/rates9,28,29. Moreover, the need for additional drive components (more than one guide RNA construct), genetic safeguards, and built-in redundancy, calls for a new level of complexity within drive architecture. Here, we demonstrate use of multiple gene drives across three chromosomal loci within an artificial budding yeast system. Our “minimal” multi-locus gene drive arrangement requires only a single copy of the S. pyogenes Cas9 gene (installed at one position), along with three distinct guide RNAs to multiplex the nuclease throughout the genome. We demonstrate that this technique could be used to perform targeted replacement of a native gene (under its endogenous promoter) in trans from the Cas9-harboring locus. Finally, reducing or modulating NHEJ by targeting the highly conserved DNA Ligase IV may provide a means to further bias HR-dependent repair and action of gene drives across eukaryotic systems. Our method includes multiple layers of genetic safeguards as well as recommendations for future designs of multi-locus drive systems.
Results
Rationale and design of a multi-locus CRISPR gene drive
To date, a number of studies in fungi, insects, and now vertebrates, have demonstrated that CRISPR-based gene drive systems are effective in both single-celled and multicellular eukaryotes15–17,19–22. One of the benefits of homing systems is the ability to install additional genetic “cargo” proximal to the gene drive (consisting of a nuclease gene and an expression cassette for the guide RNA). Current strategies use the gene drive cassette itself to delete and replace an endogenous gene, and/or include exogenous material as a desired cargo. However, there are a number of limitations to the use of a single locus harboring the entirety of the gene drive. First, addition of entire genetic pathways or large numbers of gene expression systems may be less efficient at HR-based copying of the drive. Second, introduction of additional endogenous gene(s) or modified alleles may require the native promoter system and/or epigenetic landscape to provide accurate and timely expression—this would not be possible at a single generic drive-containing locus. Third, given the observation of both natural (e.g. single nucleotide polymorphisms) and evolved resistance to gene drives through insertions or deletions (indel) resulting from NHEJ within insect populations20,30–34, mechanisms for fortifying drive systems are still being elucidated. The proposal to increase the number of targeted double strand breaks (and corresponding sgRNAs) by the single nuclease of choice (e.g. S. pyogenes Cas9) would greatly aid in combatting resistance35–37. However, an independent means to both minimize or escape resistance and ensure the intended biological outcome (deletion of the intended gene or introduction of the exogenous cargo) would involve a redundant delivery system. In this way, multiple gene drives (with multiple guide RNAs) within the same organism could target independent genetic loci either from the same, distinct, or parallel genetic pathways to achieve the desired outcome(s).
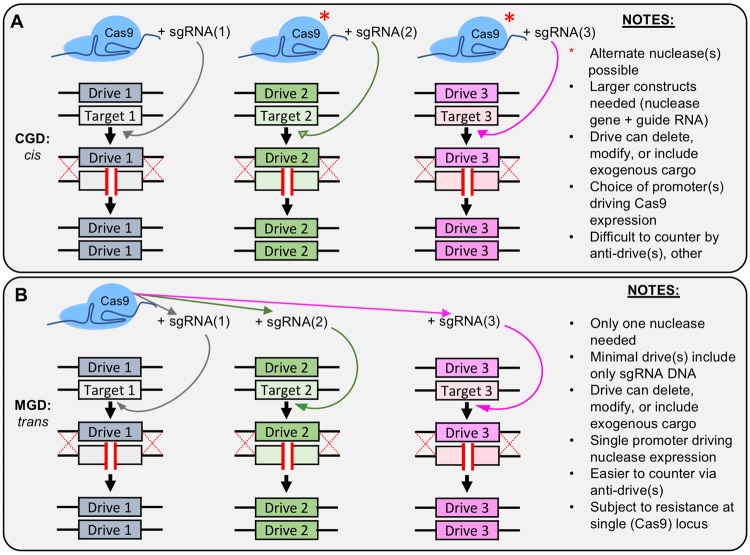
We envisioned two general strategies for the development of a gene drive system across distinct chromosomal positions: (i) each multi-locus “Complete” Gene Drive (CGD) would contain both a nuclease and corresponding guide RNA or (ii) a multi-locus “Minimal” Gene Drive (MGD) would include a nuclease and sgRNA, and all other genetic loci would only contain additional guide RNA cassettes (Fig. 1). We chose to focus on the latter strategy for a number of reasons, but we recognize that both would have distinct challenges and advantages. For one, a possible technical hurdle to development of a modified organism with multiple CGDs would be the generation of distinct “large” expression system consisting of the entire nuclease gene, flanking untranslated region (UTR), the guide expression cassette(s), and any optional cargo compared to the MGD which removes the bulk of the drive system (nuclease expression) at additional loci.
Figure 1.
Models for multi-locus CRISPR gene drive systems. (A) A proposed gene drive arrangement in cis. Each locus to be modified contains a “complete” system (nuclease and guide RNA cassette). These may be identical nuclease genes, altered variants, or sourced from separate species (e.g. Cas9 versus Cas12a). The action of each drive is fully independent from other drive-containing loci. (B) A single nuclease functions in trans across multiple loci with separate guide RNAs. This “minimal” design allows for greater safety and security (easily countered by a single anti-drive system or other means) but may be more susceptible to resistance at the primary (Cas9-harboring) locus.
Along these lines, the issue of appropriate expression of each of the nuclease gene(s) (whether identical or distinct) would need to be addressed using identical or modified promoter elements; this issue does not exist for a MGD with only a single copy of Cas9. Second, the issue of biosecurity and safeguarding against accidental or malicious release was taken into consideration. Given that a MGD would only harbor one copy of the nuclease, it would provide far less hurdles to counter and inactivate—either through use of an anti-drive system15,26, by induced self-excision17, or by removal of the Cas9-containing drive guide RNA17. Therefore, we have chosen to focus our study on design and testing of a three-locus MGD in budding yeast using the S. pyogenes Cas9 nuclease.
An efficient triple gene drive system functions independently at each locus
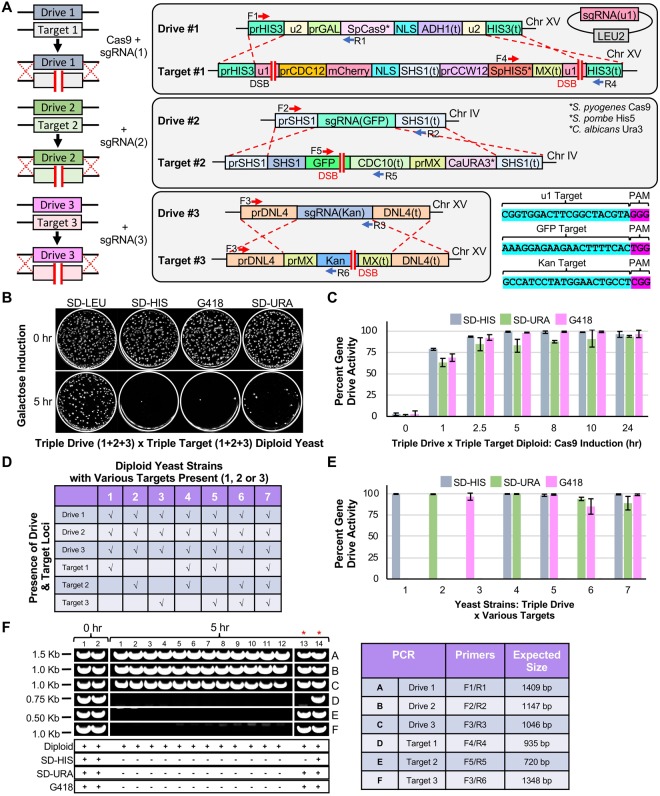
Our novel system includes the most potent genetic safeguard known to date used within a gene drive: artificial and non-native sequences used as targets. In this way, we have not only generated a haploid yeast strain harboring the MGD system at three genetic loci (HIS3, SHS1, and DNL4), but have also created a corresponding haploid strain with three distinct artificial targets at the same three loci (Fig. 2A). The “primary” drive at the HIS3 locus includes (i) Cas9 under an inducible promoter (GAL1/10) commonly used for overexpression, (ii) flanking (u2) artificial sequences to be used for self-excision as a safeguard, and (iii) the absence of any selectable marker. The corresponding guide RNA cassette was installed on a high-copy plasmid for security reasons, but could have also been integrated proximal to the drive itself. The “secondary” and “tertiary” drive systems (SHS1 and DNL4) are both non-essential genes and contain the minimum required components in the MGD design; in both cases, the native gene was deleted and fully replaced by the guide expression cassette (455 bp, although this could be reduced further) with no selectable marker. Construction of this complex haploid yeast strain used a combination of traditional HR-based integrations (with selectable markers), universal Cas9-targeting systems (CRISPR-UnLOCK)18, and novel “self-editing” integration events (Supplementary Fig. 1). To test the efficacy of the MGD, a three-locus “target” strain was generated: the HIS3 locus was flanked by two (u1) sequences and included the S.p.HIS5 selectable marker, the SHS1 gene was fused with GFP and contained the C.a.URA3 marker, and finally, the DNL4 locus was deleted and replaced with the KanR drug cassette (Fig. 2A).
Figure 2.
Design of a CRISPR/Cas9-based gene drive system in S. cerevisiae across three loci. (A) Left, An artificial gene drive was installed at three loci in haploid yeast. Each drive system (Drive 1–3) contained a guide RNA cassette targeting an artificial target (Target 1–3) at the same locus. Only Drive 1 contained the cassette for S. pyogenes Cas9. Right, Artificial (u1) and (u2) sites63 were used flanking the gene drive at the HIS3 locus (Chromosome XV) and the S.p.HIS5 selectable marker. The SHS1 locus (Chromosome IV) included a C-terminal GFP and C.a.URA3. DNL4 (Chromosome XV) was deleted with the KanR cassette. All sgRNAs were targeted to non-native sequences. The sgRNA(u1) cassette was on a high-copy plasmid (LEU2 marker). S.p.Cas9 was under control of the inducible GAL1/10 promoter. (B) Haploid yeast harboring the triple drive (GFY-3675) were mated to the triple target strain (GFY-3596) to form diploids. Cas9 expression was induced by galactose (0 or 5 hr). Cultures were diluted to 100–500 cells per plate, grown for 2 days, and transferred to SD-LEU, SD-HIS, SD-URA, and G418 plates. (C) A time course of galactose activation using the [GFY-3675 x GFY-3596] diploid in triplicate. Error, SD. (D) Seven haploid strains (GFY-3206, 3593, 3264b, 3578, 3594, 3623, and 3596) were tested as in (B) against the triple drive strain (GFY-3675). (E) Each of the diploids from (D) were cultured for 5 hr and quantified for drive success. Error, SD. (F) Clonal isolates were obtained from diploids generated in (B) at either 0 hr (2 isolates) or 5 hr activation of Cas9 (14 isolates). All yeast were confirmed as diploids and assayed on each media type (below). Diagnostic PCRs were performed on genomic DNA to detect the presence (or absence) of each locus; oligonucleotide (Supplementary Table 5) positions can be found in (A) and the expected sizes are illustrated (right). Two isolates (13,14) were chosen for their incomplete growth profile (red asterisks). Images were cropped from separate portions of larger gels or from independent DNA gels and are separated by white lines. The unedited images can be found in Supplementary Fig. S8.
The triple MGD strain was first mated with the triple target strain to form a diploid, and Cas9 was activated by culturing in medium containing galactose (Fig. 2B). In the absence of nuclease expression (Fig. 2B, top), nearly all yeast colonies contained the (u1) guide plasmid (LEU2), and three selectable markers (HIS5, URA3, and KanR—providing resistance to G418). However, following a 5 hr incubation in galactose, >95% of all colonies were sensitive to all three growth conditions indicating a loss of all three selectable markers and replacement via the MGD (Fig. 2B, bottom). A time course of galactose induction illustrated highly efficient drive activity for all three loci by five hours; we noticed a slight lag in efficiency for the loss of the URA3 marker (SHS1 locus) until the 24 hr mark (Fig. 2C). This observation may be due to the HIS3 and DNL4 loci both being present on chromosome XV whereas SHS1 was located on chromosome IV. Alternatively, differences in available guide RNAs (plasmid-borne versus integrated) or local epigenetic effects could cause this slight reduction in editing. Next, to ensure that action of the MGD at each locus was not dependent on the presence or absence of one or more of the intended targets (simulating “resistance” at one or more loci), we retested the triple drive strain against six additional strains, each lacking one or two of the proper targets and instead, contained the native yeast sequence: his3∆1, SHS1, or DNL4 (Fig. 2D). We obtained similar results for each combination as the triple MGD strain (#7) indicating that each gene drive functioned independent of the presence of additional target(s) (Fig. 2E). We also observed that drive success at the SHS1 locus slightly increased when fewer targets were presented. Finally, to ensure that the loss of the selectable marker was coupled to replacement of the target locus by the drive locus, we isolated clonal yeast from the MGD triple cross (Fig. 2B) and confirmed both the growth profile and ploidy status of random samples (Fig. 2F, bottom). Diagnostic PCRs were performed on all six distinct loci to assay for the presence or absence of each engineered drive and target (Fig. 2F, Supplementary Fig. 3). Oligonucleotides (Supplementary Table 5) unique to specific drive/target elements were chosen; prior to Cas9 activation (0 hr), diploids contain all six distinct loci (two isolates). However, following activation of the nuclease, diploids maintained all three drive loci (PCRs A,B, and C), but lost all three target loci (PCRs D, E, and F) (twelve independent isolates).
We recognized that following the 5 hr drive activation, a small number of yeast colonies (<5% in most cases) still contained one or more selectable marker(s). We reasoned that these rare colonies likely arose from either complete or partial failure of the gene drive system for various possible reasons (poor expression, loss of guide RNA plasmid, NHEJ, alterations in ploidy, etc.). Therefore, we isolated and tested additional clones that displayed incomplete growth profiles across the three selection plates (isolates 13,14) (Fig. 2F). One isolate (13) had lost the (u1)-flanked target at the HIS3 locus yet still contained the SHS1 and DNL4 markers. The second isolate (14) appeared to have lost the LEU2-based plasmid and all three target loci were still present (Fig. 2F). Following transformation with the (u1) guide vector, we examined a second round of drive activation from these two isolates and obtained a similar growth profile with a loss of the remaining loci indicating that at least some of the “failed” drive occurrences resulted from improper activation and/or targeting (Supplementary Fig. 4). Of note, our gene drive system was activated in the absence of any selection—diploids were grown in rich medium containing galactose, and grown on SD-LEU plates prior to testing of the drive status on various medium. In this way, the action of the gene drive was performed in the absence of any selection or challenge.
DNA Ligase IV as a target for gene drives
Our choice of the yeast DNL4 gene as one of the MGD targets was intended to highlight the ability of a drive itself to modify or eliminate non-homologous end joining (NHEJ)—the DNA repair process that directly counteracts the action of gene drives. Following DSB formation by Cas9, the function of the homing drive requires repair of the broken chromosome via homology directed repair using the homologous chromosome (and drive itself) as the source of the donor DNA. However, should NHEJ repair systems ligate the broken chromosome ends prior to HR-based copying, the drive will fail to copy; in fact, imprecise repair by NHEJ may even generate alleles of the target that would be resistant to further rounds of editing. Therefore, this competing DNA repair system remains one major technical hurdle to optimal gene drive design in higher eukaryotes. Of note, interest in modulating, tuning, or inhibiting NHEJ-based repair pathways is not unique to CRISPR gene drives as this mode of repair still competes with the introduction of exogenous DNA via HR38–43.
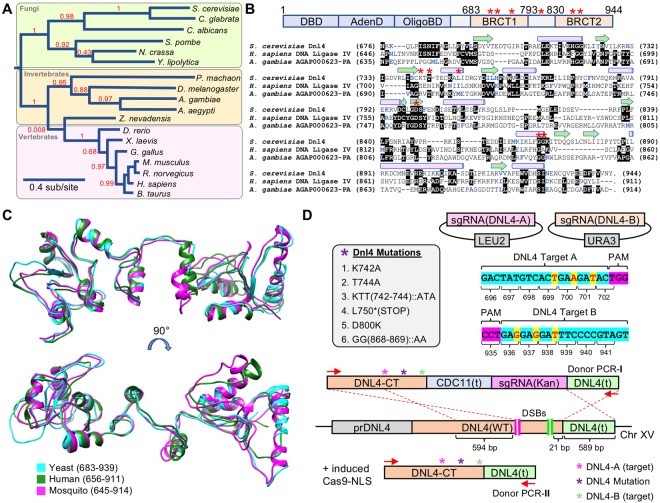
The NHEJ pathway is highly conserved from yeast to humans and functions to directly fuse exposed DNA ends44,45. DNA Ligase IV (Dnl4 in yeast, Lig4 in humans) is required for the final step of DNA ligation along with other conserved binding partners46. We examined the genomes of other fungi and metazoans using the yeast Dnl4 protein sequence as a query and a phylogenetic history of this enzyme illustrated the evolution of this enzyme through deep time (Fig. 3A). Note, the branching of Z. nevadensis (termite) was poorly supported and has been previously shown to be included within the Insecta class47. The DNA Ligase IV enzyme is divided into multiple subdomains including DNA binding, adenylation, oligonucleotide binding, and a C-terminal BRCA1 C-terminal domain (BRCT) that interacts with binding partner Lif1 (XRCC4 in human). A previous study identified a number of mutational substitutions within the C-terminus of yeast Dnl4 that resulted in a partial loss of function of NHEJ48. Examination of protein sequence alignments between yeast, mosquito, and human DNA Ligase IV C-terminal domains revealed only a minor conservation of sequence identity (Fig. 3B). However, several of the identified yeast residues were conserved by either insects and/or humans (yeast T744, D800, G868, and G869). Using the crystal structure of the C-terminus of yeast Dnl4 as a template, we generated models (I-TASSER) for the corresponding domains of mosquito and human Lig4—both displayed a much higher conservation of structure as opposed to primary sequence (Fig. 3C, Supplementary Table 4). The N-terminal region also displayed strong structural homology using the human Lig4 crystal structure as a template (Supplementary Fig. 5).
Figure 3.
DNA Ligase IV, critical for NHEJ and conserved across eukaryotes, provides a unique candidate for gene drives. (A) Phylogenic analysis of Ligase IV candidates (Supplementary Table 3) across fungi and metazoans by Phylogeny.fr65,66. Branch lengths correspond to the number of substitutions per site and the confidence of most branches is illustrated as a decimal (red text). (B) Top, Illustration of the domain structure of yeast Dnl4. The catalytic N-terminal portion includes a DNA binding domain, adenylation domain, and oligonucleotide domain (blue). The C-terminal portion includes tandem BRCA1 C-Terminal domains (BRCT). Bottom, A multiple sequence alignment was performed using Clustal Omega67 of the yeast, mosquito, and human Ligase IV protein C-termini. Identical residues are shown against a black background and similar residues are colored in blue. Secondary structures (pink cylinder, α-helix; green arrow, β-strand) for the yeast Dnl4 C-terminal as determined by the crystal structure are illustrated70. The position of six alleles (K742, T744, L750, D800, G868, and G869) are also illustrated (red asterisk) that were identified from a previous study48. (C) The protein sequences of the A. gambiae (645–914) and H. sapiens (656–911) Ligase IV were modeled against the crystal structure of the S. cerevisiae (683–939) Dnl4 (PDB:1Z56) using I-TASSER68 (Supplementary Table 4) and illustrated using Chimera64. (D) Cas9-based genomic integration methodology for introduction of mutational substitutions to the native DNL4 locus in yeast. Two sgRNA-expressing cassettes were cloned onto high-copy plasmids (marked with LEU2 and URA3) to induce two DSBs within the C-terminus of DNL4. Silent substitutions were generated within the intended repair DNA to prevent re-targeting of Cas9 (silent alterations in yellow). Two repair strategies were used to include either a non-native terminator coupled with a sgRNA(Kan) cassette, or the native DNL4 terminator; the included amount of homology (bp) is illustrated.
While total loss of NHEJ (e.g. dnl4∆) is tolerated in yeast, it is unclear whether a DNA Ligase IV null allele would be viable in higher eukaryotes. Along these lines, truncations or mutations of Lig4 in humans can lead to the rare DNA Ligase IV syndrome49,50. However, given that reduction in transcript or replacement by a partially functioning allele could reduce, but not eliminate NHEJ repair, it could be utilized in other systems to maximize gene drive efficiency, even at the (potential) expense of overall fitness. Therefore, we utilized a “self-editing” methodology to integrate six dnl4 alleles—five partial loss of function substitutions, one truncation, and a WT control (Fig. 3D). In a strain harboring integrated Cas9 at the HIS3 locus17, we introduced two DSBs within the C-terminus of native DNL4 and integrated two different constructs: (i) a modified dnl4 allele with a sgRNA(Kan) cassette and (ii) a modified dnl4 locus using the native terminator sequence. Both Cas9 target sites were also mutated within the repair (donor) DNA to prevent subsequent rounds of unintended editing.
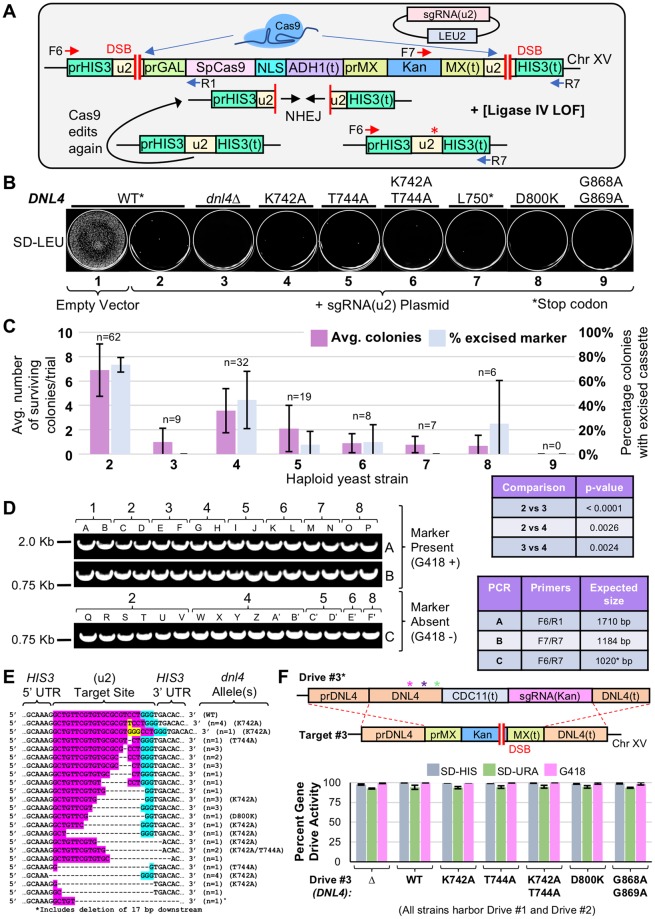
We utilized these eight haploid strains to quantify the level of NHEJ repair (Fig. 4). Our system of DSB formation followed by DNA repair utilized the dual programmed (u2) sites flanking the Cas9 expression cassette (Fig. 4A). With only a single guide construct, Cas9 would be multiplexed to both identical sites, causing complete excision of the nuclease gene and KanR marker. Following transformation of the sgRNA(u2) plasmid, yeast were analyzed for the number of surviving colonies on SD-LEU medium (Fig. 4B). Editing by Cas9 at both (u2) sites followed by precise DNA ligation of the broken ends would generate a “new” (u2) site, and would be subject to a second round of Cas9-dependent cleavage—continual DSB formation followed by exacting repair causes inviability in yeast17.
Figure 4.
Partial loss of function alleles of yeast DNA Ligase IV reduce NHEJ. (A) Design of a self-excising Cas9-based assay for NHEJ. Strain GFY-2383 included an inducible Cas9 cassette paired with the KanR marker. Transformation of the sgRNA(u2) plasmid would result in multiplexing to two flanking (u2) sites. Repair via NHEJ would result in the formation of the original (u2) site, and would be subject to further rounds of editing; introduction of an indel (red asterisk) would cause destruction of the target. (B) Strains GFY-3850 through GFY-3856 and GFY-3864 (Supplementary Table 1, Conditions 1–9) were transformed with the sgRNA(u2) plasmid (pGF-V809) or empty vector control (pRS425) and plated onto SD-LEU for three days. The DNL4 (WT) gene contained six silent substitutions (asterisk). (C) The average number of surviving colonies was quantified for all trials—labeled as in (B); the number of colonies (n) obtained across all experiments is displayed. Error, SD. For the pRS425 vector, 2897+/−357 colonies were obtained. The percentage of isolates that excised the cassette at the HIS3 locus (by sensitivity to G418) is displayed. Error, SD. Statistical analyses of strain comparisons (colonies per trial) were performed using an unpaired t-test. (D) Diagnostic PCRs were performed on chromosomal DNA from isolates from (B) to illustrate presence (2 isolates each) or loss (between 2–6 shown) of the Cas9 cassette. Conditions 2–8 correspond to (B). Oligonucleotides (Supplementary Table 5) are in (A) and the expected sizes are illustrated (right). Images of independent DNA gels are separated by white lines; unedited gel images can be found in Supplemental Fig. S8. (E) DNA sequencing of the HIS3 locus following NHEJ (on isolates sensitive to G418). For each insertion or deletion, the number of identical clones is displayed. All sequences were obtained from WT yeast unless otherwise noted. Target, pink. PAM, blue. Insertions, yellow. (F) Triple-drive containing strains were constructed with a modified DNL4 and sgRNA(Kan) cassette. Haploid strains (GFY-3675, 3865–3867, 3871, 3872, and 3875) containing the sgRNA(u1) plasmid were mated with GFY-3596, diploids selected, and drives activated. Percentage of colonies sensitive to each condition, gene drive activity. Error, SD.
However, introduction of an insertion, deletion, or substitution within the target sequence would render the site immune from subsequent rounds of editing. Furthermore, loss of the KanR marker provided a growth phenotype associated with targeting of the (u2) sites and excision of the entire cassette at the HIS3 locus. Both the total number of surviving colonies as well as the percentage of isolates with an excised marker were quantified in triplicate (Fig. 4C). In our assay, the presence of WT DNL4 allowed for approximately 7 colonies/experimental trial, whereas dnl4∆ yeast resulted in 0–1 colonies on average. Importantly, of the WT DNL4 isolates, 73% had properly excised the entire cassette whereas this was found to be 0% for dnl4∆ yeast across numerous independent trials (Fig. 4C). The partial loss of function dnl4 alleles provided a range of NHEJ efficiencies: the K742A mutant averaged 4 colonies/trial with an excision rate of nearly 50% and other substitutions displayed excision rates of between 0–25%. As expected, the C-terminal dnl4 truncation at L750 phenocopied the null allele. Diagnostic PCRs confirmed the presence or absence of the Cas9-KanR expression cassette for clonal isolates from each of the aforementioned haploid strains tested (Fig. 4D). For strains that had undergone editing and marker excision, the HIS3 locus was amplified and sequenced; NHEJ followed by imprecise ligation introduced either insertions or deletions at the site of Cas9 cleavage 3 bp upstream of the 5′ end of the PAM sequence (Fig. 4E). Finally, each of the dnl4 alleles was tested within our MGD system as a native cargo-based delivery system (Fig. 4F, top). Given that our artificial DNL4 target was the dnl4∆KanR null allele, we recognize that in the context of a [gene drive x WT] diploid genome, further modifications would be required to bias the HR-based repair of the intended dnl4 allele. This could include recoding (silent substitutions) of the DNL4 C-terminal domain sequence to prevent promiscuous cross-over downstream of the intended mutation(s). Following expression of Cas9 and activation of the MGD, the growth profiles of 7 diploid strains were assessed in triplicate and demonstrated efficient drive activity at all three loci (Fig. 4F, bottom). Moreover, PCRs from clonal isolates confirmed the presence or absence of each drive and target locus (Supplementary Fig. 6). These data demonstrate that the MGD strategy can be used as a knock-out or allele replacement strategy (at a native locus) with only minimal added sequence (782 bp).
Discussion
In this study, we have developed a multi-locus CRISPR gene drive with a minimal design (MGD) that allows for multiplexing of Cas9 in trans across three distinct chromosomal locations (Fig. 1). An alternative strategy could also be employed to create more than one gene drive system within a single genome—a CGD where each locus of interest contains the full complement of genetic information (nuclease, UTR, sgRNA, and optional cargo). In this way, each drive would be completely independent from all other drive(s). While this design clearly provides a maximum level of potential redundancy, there are other technical and safety issues inherent to this multi-nuclease arrangement. For one, countering or inhibiting a CGD with more than one active nuclease would require more sophisticated anti-drive systems, the discovery of additional anti-CRISPR proteins, or complex regulatory systems to ensure inactivation or destruction of each drive. In contrast, our minimal GD design can be inhibited by the AcrIIA2/A4 proteins27, self-excised by our flanking (u2) sites17, or targeted by an anti-drive system no different than a traditional single-locus gene drive. We argue that this type of design provides a higher level of biosecurity and can still accomplish the same task as n-number of “full” gene drives. Moreover, the issue of tightly regulated control of the nuclease transcript may pose additional challenges if the same promoter elements are positioned across multiple chromosomes and epigenetic landscapes in the CGD design.
One potential issue facing our MGD design (or any gene drive design, for that matter) is that of natural or evolved resistance to the action of the drive. Since our multi-locus arrangement includes only a single nuclease gene powering all drives, any resistance or escaped action to the Cas9-containing locus would render all three gene drives inactivated in subsequent generations. However, evidence now exists both in silico11,35,37 and in vivo51 that the addition of multiple guide RNAs (to the same genetic target) can reduce (or potentially eliminate) resistance to the gene drive. Using current estimations for a given target, five separate guide RNAs may provide a sufficiently rare or improbable event requiring mismatch or mutation to occur at all five target DNA sites35,37. This would provide greater than 99% confidence in eliminating an A. gambiae population on a continent-wide scale35. Therefore, our recommendation would be to greatly bias multiplexing (via multiple guide RNAs) to the gene drive locus harboring the sole copy of Cas9 in the MGD design (in our system, Cas9 creates two DSBs flanking the entire locus). To ensure even higher fidelity of the nuclease and to combat resistance, one could combine the two strategies (CGD and MGD) to have a secondary copy of the nuclease (of the same variant or a different species) positioned at a second locus—additional multiplexing across numerous other loci could include the minimal design (guide RNA cassette only). Finally, while our MGD methodology includes a gene drive consisting of only 455 nucleotides (sgRNA expression cassette), this could be reduced even further to be only a few bases or the absence of any base pairs. Additional sgRNA cassettes could be installed at one or more loci to allow for targeting of chromosomal positions where the “drive” is nothing more than a single base substitution or deletion. Provided few bases separate the DSB site and the intended mutation(s), HR-based repair would allow for propagation of the few bases no different than a “full” gene drive (consisting of many thousands or tens of thousands of bases) into the homozygous condition. The only requirement would involve the sgRNA expression cassette(s) to also be installed within a drive-containing locus in trans.
Finally, we chose one of the chromosomal targets within our minimal gene drive system to include both truncations and substitution alleles of DNL4—one of the essential components of the NHEJ repair pathway. While loss of DNA Ligase IV is non-lethal in yeast52 and flies53, it is embryonic lethal in mouse54 and not tolerated in mosquito55. However, suppression or inhibition of this enzyme or the NHEJ repair pathway has been shown to increase rates of recombination and genomic integration of exogenous DNA using CRISPR systems in vivo38,40–43,55,56. We demonstrate that this critical factor could be an additional target for a multi-locus gene drive system—suppression of NHEJ, whether by mutated alleles, regulation of transcript, or direct inhibition of the enzyme—would aid in successful HR-based copying of the drive and further reduce the possibility for drive resistance, especially when coupled with multiple guide RNAs. Numerous strategies might be employed to accomplish targeted suppression of NHEJ including testing of additional Ligase IV loss of function alleles that may be widely conserved across eukaryotes; our study focused on the C-terminal BRCT-domain containing portion of Dnl4, but other substitutions have been also been characterized within the N-terminal catalytic domain57.
A multi-locus CRISPR gene drive system should help advance current designs and provide additional options for (i) biosecurity, (ii) drive redundancy, (iii) combatting of evolved resistance, (iv) native gene replacement, (v) multiple gene cargo/genetic pathway delivery, (vi) suppression of NHEJ or activation of HR-promoting repair pathways, and (vii) multiple phenotypic outcomes. Advanced drive arrangements28 could accomplish multiple outcomes within a single-genome system—the additional of exogenous cargo could also be paired with (native) allele introduction and modulating of organism fitness by perturbing numerous other genetic pathways in a single step. As the design and application of CRISPR gene drives continues to advance, we continue to stress the need for multiple levels of control, tunability, inhibition, and drive reversal.
Methods
Yeast Strains and Plasmids
Standard molecular biology protocols were used to engineer all S. cerevisiae strains (Supplementary Table 1) used in this study58. The overall methodology for construction of the triple gene drive strain utilized both standard HR-based chromosomal integrations (sans any DSB) and Cas9-based editing (Supplementary Fig. 1). Briefly, DNA constructs were first assembled onto CEN-based plasmids (typically pRS315) using in vivo assembly in yeast59. If necessary, point mutations were introduced using PCR mutagenesis60. Next, the engineered cassette was amplified with a high-fidelity polymerase (KOD Hot Start, EMD Millipore), transformed into yeast using a modified lithium acetate method61, and integrated at the desired genomic locus. PCR was used to diagnose proper chromosomal position for each integration event followed by DNA sequencing. The DNA maps for manipulated yeast loci are included in Supplementary Fig. 2. DNA plasmids used in this study can be found in Supplementary Table 2. Expression cassettes for sgRNA were based on a previous study62, purchased as synthetic genes (Genscript), and sub-cloned to high-copy plasmids using unique flanking restriction sites. All vectors were confirmed by Sanger sequencing.
Culture Conditions
Budding yeast were cultured in liquid or solid medium. YPD-based medium included 2% peptone, 1% yeast extract, and 2% dextrose. Synthetic (drop-out) medium included yeast nitrogen base, ammonium sulfate, and amino acid supplements. The supplement mixture included adenine, arginine, tyrosine, isoleucine, phenylalanine, glutamic acid, aspartic acid, threonine, serine, valine, lysine, and methionine. For specific drop-out combinations, one or more of the following were removed: leucine, uracil, and/or histidine. Tryptophan (filter sterilized solution) was also added to media before final plating. A raffinose/sucrose mixture (2%/0.2%) was used to pre-induce cultures prior to treatment with galactose (2%). Yeast cultures were all grown in a 30 °C incubator with shaking. All media was autoclaved or filter sterilized (sugars). For agar plates containing G418 sulfate, the final concentration was 240 µg/mL.
Cas9-based editing in vivo
Editing of haploid S. cerevisiae strains was performed as previously described17. Briefly, an integrated copy of S. pyogenes Cas9 was designed with two flanking “unique” (u2) sites—23 base pairs artificially introduced into the genome. This sequence contains a maximum mismatch to the native yeast genome and is used in order to (i) multiplex at two separate sites using a single guide RNA construct, (ii) minimize (or likely eliminate) potential off-target effects, and (iii) allow for increased biosecurity in testing of active CRISPR gene drive systems63. Haploid yeast were pre-induced overnight in a raffinose/sucrose mixture to saturation, back-diluted to an OD600 of approximately 0.35 in rich medium containing galactose, and cultured for 4.5 hr at 30 °C. Equimolar amounts (1,000 ng) of high-copy plasmid (sgRNA) were transformed into yeast followed by recovery overnight in galactose and a final plating onto SD-LEU medium. Colonies were imaged and quantified after 3–4 days of growth. Haploid yeast editing experiments included three replicates in triplicate—all as separate transformation events—for each strain (n = 9).
Gene drive activation and containment
Haploid yeast strains harboring the gene drive (Cas9) system were first transformed with the sgRNA-containing plasmid (LEU2-marked). Next, drive strains were mated to target strains of the opposite mating type on rich medium for 24 hr. Third, yeast were velvet-transferred to synthetic drop-out medium to select diploids (e.g. SD-URA-LEU or SD-URA-LEU-HIS); each haploid genome contained at least one unique selectable marker. Diploids were selected three consecutive rounds with 1–2 days incubation at each step. Fourth, yeast were cultured in pre-induction medium (raffinose/sucrose) lacking leucine overnight, back-diluted into rich medium containing galactose, and grown for 5 hr (or appropriate time intervals). Strains were diluted to approximately 100–500 cells per mL and plated onto SD-LEU for 2 days. Finally, colonies were transferred to the appropriate selection plates (e.g. SD-HIS, G418, SD-URA, and a fresh SD-LEU plate) for 1 additional day of growth before being imaged (Supplemental Fig. S7). The number of surviving colonies on each media type was quantified; experiments were performed in at least triplicate.
A number of safeguards were included in the design of all gene drive systems. First, the genomic targets for all guide RNAs included only non-yeast sequences (u1, GFP, and KanR)18,63. Second, the primary guide RNA cassette (u1) for targeting of the HIS3 locus which included Cas9 was maintained on an unstable high-copy (2μ) plasmid; previous work has demonstrated loss of this vector type in the absence of active selection15,17. Third, the S. cerevisiae BY4741/BY4742 genetic background does not readily undergo sporulation, even under optimal conditions. Fourth, Cas9 expression was repressed by growth on dextrose until gene drives were activated. And finally, all diploid strains, plates, and consumables were autoclaved and inactivated.
Images, Graphics, Data and Evolutionary Analysis
Images (DNA gels, agar plates) were processed using ImageJ (National Institute of Health). For PCR reactions demonstrating the absence of a gene target (following gene drive activation), the original, unedited raw images were also included in Supplementary Figs S8–S9 for comparison.
Data analysis (Fig. 4C) included error illustrated as the standard deviation of multiple independent trials and statistical comparisons were performed using an unpaired t-test.
Molecular graphics were generated using the Chimera software package from the Univ. of California, San Francisco64. Homologous sequences to the yeast DNA Ligase IV (Dnl4) protein were obtained using multiple BLAST (NCBI) searches within either the fungal or metazoan clade (Supplementary Table 3). The phylogenetic tree of DNA Ligase IV was created using the Phylogeny.fr software65,66. Multiple sequence alignments were performed using Clustal Omega67. The predicted structures of the human, yeast, and mosquito Ligase IV enzyme were generated using I-TASSER68. The template structures included the human Lig4 N-terminus (PDB:3W1B)69 and the yeast Dnl4 C-terminus (PDB:1Z56)70. Predicted models were individually aligned against the crystal structures using MatchMaker in Chimera. Metrics for the predicted structures are included in Supplementary Table 4.
Animal and Human Subject Statement
This study does not use any animals or human subjects.
Electronic supplementary material
Acknowledgements
We thank Emily Wedeman (Kansas State University) and Muriel Eaton (Purdue University) for useful comments and Megan Halloran (Kansas State University) for laboratory assistance. This project was supported by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant number P20 GM103418 to G.C.F. This work was also supported by the USDA National Institute of Food and Agriculture, Hatch Project 1013520 to G.C.F. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institute of Health.
Author Contributions
Y.Y. and G.C.F. built all reagents (plasmids and yeast), performed all experiments, and performed data analyses and figure preparation. G.C.F. wrote the manuscript.
Data Availability
The datasets generated during and analyzed during the current study are available from the corresponding author on reasonable request.
Competing Interests
G.C.F. (Kansas State University) has filed for a provisional patent entitled “Multi-Locus Gene Drive System,” U.S. Serial No. 62/697,855 on July 13, 2018 for the intellectual property and technology described within this work. G.C.F declares no potential non-financial conflict of interest. Y.Y. declares no potential conflict of interest of any kind.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-34909-3.
References
- 1.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science (New York, N.Y.) 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jinek M, et al. RNA-programmed genome editing in human cells. eLife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science (New York, N.Y.) 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 4.Zetsche B, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qi LS, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thakore PI, et al. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nature methods. 2015;12:1143–1149. doi: 10.1038/nmeth.3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaudelli NM, et al. Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature. 2017;551:464–471. doi: 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bull JJ, Barrick JE. Arresting Evolution. Trends in genetics: TIG. 2017;33:910–920. doi: 10.1016/j.tig.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Godfray HCJ, North A, Burt A. How driving endonuclease genes can be used to combat pests and disease vectors. BMC biology. 2017;15:81. doi: 10.1186/s12915-017-0420-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Esvelt KM, Gemmell NJ. Conservation demands safe gene drive. PLoS biology. 2017;15:e2003850. doi: 10.1371/journal.pbio.2003850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prowse Thomas A. A., Cassey Phillip, Ross Joshua V., Pfitzner Chandran, Wittmann Talia A., Thomas Paul. Dodging silver bullets: good CRISPR gene-drive design is critical for eradicating exotic vertebrates. Proceedings of the Royal Society B: Biological Sciences. 2017;284(1860):20170799. doi: 10.1098/rspb.2017.0799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Courtier-Orgogozo V, Morizot B, Boete C. Using CRISPR-based gene drive for agriculture pest control. EMBO reports. 2017 doi: 10.15252/embr.201744822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lambert B, North A, Burt A, Godfray HCJ. The use of driving endonuclease genes to suppress mosquito vectors of malaria in temporally variable environments. Malaria journal. 2018;17:154. doi: 10.1186/s12936-018-2259-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hammond AM, Galizi R. Gene drives to fight malaria: current state and future directions. Pathogens and global health. 2017;111:412–423. doi: 10.1080/20477724.2018.1438880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DiCarlo JE, Chavez A, Dietz SL, Esvelt KM, Church GM. Safeguarding CRISPR-Cas9 gene drives in yeast. Nature biotechnology. 2015;33:1250–1255. doi: 10.1038/nbt.3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shapiro RS, et al. A CRISPR-Cas9-based gene drive platform for genetic interaction analysis in Candida albicans. Nature microbiology. 2018;3:73–82. doi: 10.1038/s41564-017-0043-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roggenkamp E, et al. Tuning CRISPR-Cas9 Gene Drives in Saccharomyces cerevisiae. G3 (Bethesda, Md.) 2018;8:999–1018. doi: 10.1534/g3.117.300557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roggenkamp E, et al. CRISPR-UnLOCK: multipurpose Cas9-based strategies for Conversion of yeast libraries and strains. Frontiers in microbiology. 2017;8:1773. doi: 10.3389/fmicb.2017.01773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gantz VM, et al. Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E6736–6743. doi: 10.1073/pnas.1521077112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Champer J, et al. Novel CRISPR/Cas9 gene drive constructs reveal insights into mechanisms of resistance allele formation and drive efficiency in genetically diverse populations. PLoS genetics. 2017;13:e1006796. doi: 10.1371/journal.pgen.1006796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hammond A, et al. A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae. Nature biotechnology. 2016;34:78–83. doi: 10.1038/nbt.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grunwald, H. A. et al. Super-Mendelian inheritance mediated by CRISPR/Cas9 in the female mouse germline. bioRxiv, 10.1101/362558 (2018).
- 23.Noble, C., Adlam, B., Church, G. M., Esvelt, K. M. & Nowak, M. A. Current CRISPR gene drive systems are likely to be highly invasive in wild populations. eLife7, 10.7554/eLife.33423 (2018). [DOI] [PMC free article] [PubMed]
- 24.James S, et al. Pathway to Deployment of Gene Drive Mosquitoes as a Potential Biocontrol Tool for Elimination of Malaria in Sub-Saharan Africa: Recommendations of a Scientific Working Group(dagger) The American journal of tropical medicine and hygiene. 2018;98:1–49. doi: 10.4269/ajtmh.18-0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Najjar DA, Normandin AM, Strait EA, Esvelt KM. Driving towards ecotechnologies. Pathogens and global health. 2017;111:448–458. doi: 10.1080/20477724.2018.1452844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vella MR, Gunning CE, Lloyd AL, Gould F. Evaluating strategies for reversing CRISPR-Cas9 gene drives. Scientific reports. 2017;7:11038. doi: 10.1038/s41598-017-10633-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Basgall EM, et al. Gene drive inhibition by the anti-CRISPR proteins AcrIIA2 and AcrIIA4 in Saccharomyces cerevisiae. Microbiology (Reading, England) 2018;164:464–474. doi: 10.1099/mic.0.000635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dhole S, Vella MR, Lloyd AL, Gould F. Invasion and migration of spatially self-limiting gene drives: A comparative analysis. Evolutionary applications. 2018;11:794–808. doi: 10.1111/eva.12583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burt A, Crisanti A. Gene Drive: Evolved and Synthetic. ACS chemical biology. 2018;13:343–346. doi: 10.1021/acschembio.7b01031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Drury DW, Dapper AL, Siniard DJ, Zentner GE, Wade MJ. CRISPR/Cas9 gene drives in genetically variable and nonrandomly mating wild populations. Science advances. 2017;3:e1601910. doi: 10.1126/sciadv.1601910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hammond AM, et al. The creation and selection of mutations resistant to a gene drive over multiple generations in the malaria mosquito. PLoS genetics. 2017;13:e1007039. doi: 10.1371/journal.pgen.1007039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buchman A, Marshall JM, Ostrovski D, Yang T, Akbari OS. Synthetically engineered Medea gene drive system in the worldwide crop pest Drosophila suzukii. Proceedings of the National Academy of Sciences of the United States of America. 2018;115:4725–4730. doi: 10.1073/pnas.1713139115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Unckless RL, Clark AG, Messer PW. Evolution of Resistance Against CRISPR/Cas9 Gene Drive. Genetics. 2017;205:827–841. doi: 10.1534/genetics.116.197285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.KaramiNejadRanjbar M, et al. Consequences of resistance evolution in a Cas9-based sex conversion-suppression gene drive for insect pest management. Proceedings of the National Academy of Sciences of the United States of America. 2018;115:6189–6194. doi: 10.1073/pnas.1713825115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marshall JM, Buchman A, Sanchez CH, Akbari OS. Overcoming evolved resistance to population-suppressing homing-based gene drives. Scientific reports. 2017;7:3776. doi: 10.1038/s41598-017-02744-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bull JJ, Malik HS. The gene drive bubble: New realities. PLoS genetics. 2017;13:e1006850. doi: 10.1371/journal.pgen.1006850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Noble C, Olejarz J, Esvelt KM, Church GM, Nowak MA. Evolutionary dynamics of CRISPR gene drives. Science advances. 2017;3:e1601964. doi: 10.1126/sciadv.1601964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vartak SV, Raghavan SC. Inhibition of nonhomologous end joining to increase the specificity of CRISPR/Cas9 genome editing. The FEBS journal. 2015;282:4289–4294. doi: 10.1111/febs.13416. [DOI] [PubMed] [Google Scholar]
- 39.Schwartz C, Frogue K, Ramesh A, Misa J, Wheeldon I. CRISPRi repression of nonhomologous end-joining for enhanced genome engineering via homologous recombination in Yarrowia lipolytica. Biotechnology and bioengineering. 2017;114:2896–2906. doi: 10.1002/bit.26404. [DOI] [PubMed] [Google Scholar]
- 40.Robert F, Barbeau M, Ethier S, Dostie J, Pelletier J. Pharmacological inhibition of DNA-PK stimulates Cas9-mediated genome editing. Genome medicine. 2015;7:93. doi: 10.1186/s13073-015-0215-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Canny MD, et al. Inhibition of 53BP1 favors homology-dependent DNA repair and increases CRISPR-Cas9 genome-editing efficiency. Nature biotechnology. 2018;36:95–102. doi: 10.1038/nbt.4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maruyama T, et al. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nature biotechnology. 2015;33:538–542. doi: 10.1038/nbt.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chu VT, et al. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nature biotechnology. 2015;33:543–548. doi: 10.1038/nbt.3198. [DOI] [PubMed] [Google Scholar]
- 44.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annual review of biochemistry. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chiruvella KK, Liang Z, Wilson TE. Repair of double-strand breaks by end joining. Cold Spring Harbor perspectives in biology. 2013;5:a012757. doi: 10.1101/cshperspect.a012757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ellenberger T, Tomkinson AE. Eukaryotic DNA ligases: structural and functional insights. Annual review of biochemistry. 2008;77:313–338. doi: 10.1146/annurev.biochem.77.061306.123941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Misof B, et al. Phylogenomics resolves the timing and pattern of insect evolution. Science (New York, N.Y.) 2014;346:763–767. doi: 10.1126/science.1257570. [DOI] [PubMed] [Google Scholar]
- 48.Chiruvella KK, et al. Yeast DNA ligase IV mutations reveal a nonhomologous end joining function of BRCT1 distinct from XRCC4/Lif1 binding. DNA repair. 2014;24:37–45. doi: 10.1016/j.dnarep.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Altmann T, Gennery AR. DNA ligase IV syndrome; a review. Orphanet journal of rare diseases. 2016;11:137. doi: 10.1186/s13023-016-0520-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chistiakov DA. L IV Syndrome. Advances in experimental medicine and biology. 2010;685:175–185. doi: 10.1007/978-1-4419-6448-9_16. [DOI] [PubMed] [Google Scholar]
- 51.Champer Jackson, Liu Jingxian, Oh Suh Yeon, Reeves Riona, Luthra Anisha, Oakes Nathan, Clark Andrew G., Messer Philipp W. Reducing resistance allele formation in CRISPR gene drive. Proceedings of the National Academy of Sciences. 2018;115(21):5522–5527. doi: 10.1073/pnas.1720354115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilson TE, Grawunder U, Lieber MR. Yeast DNA ligase IV mediates non-homologous DNA end joining. Nature. 1997;388:495–498. doi: 10.1038/41365. [DOI] [PubMed] [Google Scholar]
- 53.Gorski MM, et al. The Drosophila melanogaster DNA Ligase IV gene plays a crucial role in the repair of radiation-induced DNA double-strand breaks and acts synergistically with Rad54. Genetics. 2003;165:1929–1941. doi: 10.1093/genetics/165.4.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barnes DE, Stamp G, Rosewell I, Denzel A, Lindahl T. Targeted disruption of the gene encoding DNA ligase IV leads to lethality in embryonic mice. Current biology: CB. 1998;8:1395–1398. doi: 10.1016/S0960-9822(98)00021-9. [DOI] [PubMed] [Google Scholar]
- 55.Basu S, et al. Silencing of end-joining repair for efficient site-specific gene insertion after TALEN/CRISPR mutagenesis in Aedes aegypti. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:4038–4043. doi: 10.1073/pnas.1502370112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cen Y, Timmermans B, Souffriau B, Thevelein JM, Van Dijck P. Comparison of genome engineering using the CRISPR-Cas9 system in C. glabrata wild-type and lig4 strains. Fungal genetics and biology: FG & B. 2017;107:44–50. doi: 10.1016/j.fgb.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 57.Chiruvella KK, Liang Z, Birkeland SR, Basrur V, Wilson TE. Saccharomyces cerevisiae DNA ligase IV supports imprecise end joining independently of its catalytic activity. PLoS genetics. 2013;9:e1003599. doi: 10.1371/journal.pgen.1003599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sambrook, J. & Russell, D. W. Molecular Cloning: A Laboratory Manual. 3rd edn, (Cold Spring Harbor Laboratory Press, 2001).
- 59.Finnigan GC, Thorner J. Complex in vivo ligation using homologous recombination and high-efficiency plasmid rescue from Saccharomyces cerevisiae. Bio-protocol. 2015;5:e1521. doi: 10.21769/BioProtoc.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zheng L, Baumann U, Reymond JL. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic acids research. 2004;32:e115. doi: 10.1093/nar/gnh110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Eckert-Boulet N, Pedersen ML, Krogh BO, Lisby M. Optimization of ordered plasmid assembly by gap repair in Saccharomyces cerevisiae. Yeast (Chichester, England) 2012;29:323–334. doi: 10.1002/yea.2912. [DOI] [PubMed] [Google Scholar]
- 62.DiCarlo JE, et al. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic acids research. 2013;41:4336–4343. doi: 10.1093/nar/gkt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Finnigan GC, Thorner J. mCAL: a new approach for versatile multiplex action of Cas9 using one sgRNA and loci flanked by a programmed target sequence. G3 (Bethesda, Md.) 2016;6:2147–2156. doi: 10.1534/g3.116.029801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pettersen EF, et al. UCSF Chimera–a visualization system for exploratory research and analysis. Journal of computational chemistry. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 65.Dereeper A, Audic S, Claverie JM, Blanc G. BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC evolutionary biology. 2010;10:8. doi: 10.1186/1471-2148-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dereeper A, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic acids research. 2008;36:W465–469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li W, et al. The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic acids research. 2015;43:W580–584. doi: 10.1093/nar/gkv279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nature protocols. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ochi T, Gu X, Blundell TL. Structure of the catalytic region of DNA ligase IV in complex with an Artemis fragment sheds light on double-strand break repair. Structure (London, England: 1993) 2013;21:672–679. doi: 10.1016/j.str.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dore AS, et al. Structure of an Xrcc4-DNA ligase IV yeast ortholog complex reveals a novel BRCT interaction mode. DNA repair. 2006;5:362–368. doi: 10.1016/j.dnarep.2005.11.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and analyzed during the current study are available from the corresponding author on reasonable request.