Abstract
Loss-of-function (LOF) mutations in CC2D1A cause a spectrum of neurodevelopmental disorders, including intellectual disability, autism spectrum disorder, and seizures, identifying a critical role for this gene in cognitive and social development. CC2D1A regulates intracellular signaling processes that are critical for neuronal function, but previous attempts to model the human LOF phenotypes have been prevented by perinatal lethality in Cc2d1a-deficient mice. To overcome this challenge, we generated a floxed Cc2d1a allele for conditional removal of Cc2d1a in the brain using Cre recombinase. While removal of Cc2d1a in neuronal progenitors using Cre expressed from the Nestin promoter still causes death at birth, conditional postnatal removal of Cc2d1a in the forebrain via calcium/calmodulin-dependent protein kinase II-alpha (CamKIIa) promoter-driven Cre generates animals that are viable and fertile with grossly normal anatomy. Analysis of neuronal morphology identified abnormal cortical dendrite organization and a reduction in dendritic spine density. These animals display deficits in neuronal plasticity and in spatial learning and memory that are accompanied by reduced sociability, hyperactivity, anxiety, and excessive grooming. Cc2d1a conditional knockout mice therefore recapitulate features of both cognitive and social impairment caused by human CC2D1A mutation, and represent a model that could provide much needed insights into the developmental mechanisms underlying nonsyndromic neurodevelopmental disorders.
Keywords: autism, cognitive development, dendritic spines, intellectual disability, long-term plasticity
Introduction
Genes involved in the etiology of intellectual disability (ID) and autism spectrum disorder (ASD) are being identified at a steadily increasing rate, and recent predictions suggest that there may be hundreds or even thousands of genes that when mutated confer susceptibility to one or both of these disorders (Hoischen et al. 2014). A portion of ASD cases have been shown to result from the cumulative effects of common variants (Gaugler et al. 2014); however, the overlapping genetic architecture of ID and ASD contains many rare variants of large effect, where disruption of a single gene is sufficient to cause the disease (Gratten et al. 2014). These findings highlight the extreme etiologic complexity of ASD and ID, and in order to identify the molecular pathways and developmental processes that are targeted by these genetic changes, we must understand the role of each gene in brain development and how different genes relate to each other (Hoischen et al. 2014).
We have recently shown that null mutations in “coiled-coil and C2-domain containing 1A,” CC2D1A, cause both ID and ASD with no other clinical findings, suggesting that the loss of this gene primarily affects brain function (Manzini et al. 2014). Studies on CC2D1A and its orthologues in several model organisms have shown that this protein is involved in the regulation of endosomal trafficking and signaling (Gallagher and Knoblich 2006; Jaekel and Klein 2006; Troost et al. 2012). CC2D1A acts as a molecular scaffold for the assembly of signaling protein complexes (Nakamura et al. 2008; Chang et al. 2011; Al-Tawashi et al. 2012), and disruption of CC2D1A affects a remarkable range of biochemical pathways, including nuclear factor-κB (NF-κB; Zhao et al. 2010; Manzini et al. 2014), protein kinase A (PKA), and cAMP responsive element-binding protein (CREB; Al-Tawashi et al. 2012), protein kinase B (PKB/AKT; Nakamura et al. 2008), Notch (Gallagher and Knoblich 2006; Troost et al. 2012), and bone morphogenetic protein (BMP; Morawa et al. 2015). In addition, CC2D1A has been implicated in transcriptional regulation of both serotonin and dopamine receptors in the murine and human brain (Ou et al. 2003; Rogaeva et al. 2007; Szewczyk et al. 2010). These studies show that CC2D1A regulates multiple pathways that are critical for neuronal differentiation and cognitive function, but appropriate animal models to understand how Cc2d1a loss of function (LOF) affected neuronal circuits in the developing and adult brain have been lacking.
While most genetic changes causing ASD cause partial LOF as heterozygous de novo variants or recessive hypomorphic mutations, CC2D1A stands out since complete LOF causes ID in all cases and ASD with reduced penetrance. However, multiple Cc2d1a-knockout mouse lines generated to date have been limited in their impact, because Cc2d1a null mice suffer from early postnatal lethality (Zhao et al. 2011; Al-Tawashi et al. 2012; Chen et al. 2012). Cc2d1a-knockout pups are anatomically normal and indistinguishable from their wild-type (WT) littermates, indicating that loss of Cc2d1a does not affect gross body and brain development, but pups die in the first few hours following birth (Zhao et al. 2011). In vitro studies of Cc2d1a-knockout or knockdown hippocampal neurons have shown a reduction in the size and complexity of the dendritic arbor (Al-Tawashi et al. 2012; Manzini et al. 2014). Moreover, a delay in synaptic maturation was observed in cortical neurons in a Cc2d1a-knockout line (Zhao et al. 2011).
To bypass early postnatal lethality and study how loss of Cc2d1a affects the developing and adult brain, we generated a Cc2d1a-deficient line with a conditional gene-trap, conditional knockout (cKO) construction. Since the global gene-trap animals die at birth, similar to the other 3 Cc2d1a-knockout lines, we removed the gene-trap cassette and used the floxed allele, Cc2d1aflx, to remove Cc2d1a only in the brain. Animals where Cc2d1a is removed in the forebrain postnatally using the Cre recombinase expressed under a CamKIIa promoter are viable and fertile, and display an array of anatomical, physiological, and behavioral defects reflecting the cognitive and social deficits observed in the patients, including defects in dendritic spine density, long-term potentiation (LTP), learning and memory, and sociability. Taken together, these findings show that conditional postnatal removal of Cc2d1a from forebrain neurons is sufficient to disrupt function at the cellular and circuit level in mice, and that these Cc2d1a-deficient animals model the nonsyndromic neurodevelopmental deficits caused by CC2D1A mutation in human patients.
Materials and Methods
Animals
All animal care and use was in accordance with the institutional guidance and approved by the Institutional Animal Care and Use Committee of The George Washington University and Boston Children's Hospital. A Cc2d1a null mouse line (KO) was generated by the Knockout Mouse Project Repository (Project Design ID 49663) at the University of California, Davis, from a C57BL/6 embryonic stem cell clone (Chen et al. 2012). The Cc2d1a targeting constructs contains an “engrailed 2” splice acceptor (En2SA) gene-trap allele with bicistronic expression of β-galactosidase. In addition, a neomycin resistance cassette is expressed under the human β-actin (Actb) promoter. These 2 elements are flanked by flippase recombinase target (FRT) recombination sites, in intron 11 of Cc2d1a. Finally, the construct carries LoxP sites flanking exons 12–14 (see the diagram in Fig. 1A). Complete description of the targeting construct can be found at https://www.mousephenotype.org/imits/targ_rep/targeted_alleles/13317.
Figure 1.
Cc2d1a KO mice exhibit early postnatal lethality. (A) Gene trap containing En2SA sequence, and a separate neomycin resistance cassette is flanked by FRT sites and produces a transcript truncated between exons 11 and 12. LoxP sites for Cre recombinase targeting flank exons 12–14. (B) Domain organization of full length human and murine proteins (4 DM14 domains and 1 C2 domain) compared with a predicted protein product from a disease-causing CC2D1A mutation (Basel-Vanagaite et al. 2006) and the truncated proteins produced in our gene-trap mouse and a previously reported Cc2d1a null mouse (Al-Tawashi et al. 2012). (C) Immunoblot analysis of Cc2d1a expression in WT (+/+), heterozygous (+/−), and null mice (−/−) shows the complete loss of full length (fl) Cc2d1a protein. (D) Brains dissected from pups at embryonic day (E) 18.5 show now difference in size and organization is preserved in E18.5 brain tissue sections stained with H&E (E). (F) The weight of pups following Cesarean delivery at E18.5 is the same across genotypes. Results expressed as mean ± SEM of indicated number of pups from 4 litters. (G) Cc2d1a null mice are born in reduced numbers relative to predicted Mendelian ratios and do not survive beyond postnatal day (P) 0 (data from 5 to 10 litters per age).
The Cc2d1a KO line was backcrossed to at least 6 generations on 3 independent strains: 129/SvEvTac (Stock 129SVE, Taconic), C57BL/6, and Swiss Webster (the background used for each experiment will be listed in the Results section). To generate cKO lines, Cc2d1a KO 129/SvEvTac mice were first crossed with the FLPeR line (Stock 003946; 129Sv background; Jackson Laboratories; Farley et al. 2000), to remove the gene-trap allele while leaving LoxP sites in introns 11 and 14 (Cc2d1aflx). Conditional removal of Cc2d1a exons 12, 13, and 14 was achieved by crossing Cc2d1aflx mice with Nestin-Cre (Stock 003771; C57BL/6 background; Jackson Laboratories; Tronche et al. 1999) or CamKIIa-Cre mouse lines (Stock 005359; C57BL/6 background; Jackson Laboratories; Tsien et al. 1996), with expression of Cre recombinase driven by either the Nestin (Nestincre) or CamKIIa (CamKIIacre) promoter. To visualize dendrites and spines, Cc2d1flx/flx-CamKIIacre (cKO) mice were crossed with the Thy1-YFP-H reporter line (Stock 003782; C57BL/6 background; Jackson Laboratories; Feng et al. 2000). Morphological, electrophysiological, and behavioral analyses of Cc2d1a cKO mice were performed on fully backcrossed C57BL/6 male mice at the indicated ages.
For genotyping, polymerase chain reaction (PCR) amplifications were performed on 1 µL of proteinase K (New England Biolabs) digested tail DNA samples. PCRs (50 µL) consisted of GoTaq Flexi buffer (Promega), 100 µM dNTPs, 50 µM each of forward and reverse primers (sequence available upon request), 1 mM MgCl2 (Promega), and 1.25 U GoTaq Flexi DNA polymerase (Promega), and were run with optimized reaction profiles determined for each genotype. A 25 µL aliquot from each reaction was analyzed by gel electrophoresis on a 1.0–1.5% agarose gel for the presence of the indicated bands.
Neonatal Breathing Assessment
Timed pregnant Cc2d1a heterozygote dams from heterozygous crosses on a 129/SvEvTac or Swiss Webster background were sacrificed and E18.5/E19 embryos were removed as rapidly as possible from the uterus. Pups were placed on a warm pad, cleaned, and their abdomen was gently massaged until amniotic fluid was regurgitated. Each pup was numbered and given the same amount of attention to determine the lag between birth and the first breath. After the initial diaphragm, contraction was observed and the pup was able to breathe on its own (usually within 5 min for WTs), it was placed back on the warm pad. Poor breathers were pups that never fully perfused, remain cyanotic, and showed irregular rhythm of diaphragm contraction. Nonbreathers were pups that never attempted to breathe even 30–40 min after birth.
Histological Preparation
To prepare tissue for histological analysis, deeply anesthetized mice were transcardially perfused with phosphate-buffered saline followed by 4% paraformaldehyde (PFA). Brains were removed, postfixed in PFA, and embedded as indicated. Paraffin sections from mouse fetuses were prepared and stained for hematoxylin and eosin (H&E) at the Histology Core at the Children's Hospital Intellectual and Developmental Disabilities Research Center in Boston. Cryosections from adult mouse brains were prepared by mounting in Neg-50 (Thermo Scientific) and cut at 20 µm on a Cryostar NX50 cryostat (Thermo Scientific) in the Biomarker Core at the George Washington University, then stained with H&E to visualize tissue architecture (see Supplementary Material for additional details). For dendrite morphology and dendritic spine analysis, brains were embedded in 2% agarose and sectioned at 150 µm on a Pelco 101 Series 1000 vibratome. Floating sections were immunostained with a green fluorescent protein (GFP) antibody (see Supplementary Table 1) that also recognizes YFP to enhance visualization of layer V cortical neurons and CA1 pyramidal neurons of the hippocampus in Thy1-YFP-expressing WT and cKO mice. For tracing of dendritic arbors, freshly dissected brain tissue was stained using the FD Rapid GolgiStain kit (FD NeuroTechnologies) according to the manufacturer's instructions. Impregnated brains were frozen in dry ice-cooled isopentane, sectioned at 80 µm on a Cryostar NX50 cryostat (Thermo Scientific), mounted on gelatin-coated slides (Southern Biotech), and developed according to the kit manufacturer's instructions.
Microscopy and Image Analysis
Imaging of H&E-stained sections was performed on a Leica M165 FC. Apical and basal dendrite arbors of Golgi-stained layer V cortical were imaged with a Plan-Apochromat 10× objective on a Zeiss Axio Imager M2. Confocal imaging was performed within the George Washington University Center for Microscopy and Image Analysis (CMIA). For dendrite morphology and spine analysis, GFP-stained brain sections were imaged with Plan-Apochromat 10× or Plan-Apochromat 100× objectives using a Zeiss LSM 710 laser scanning confocal microscope equipped with an argon laser producing excitation at 488 nm.
Dendrite Arbor Analysis
Layer V cortical neurons that were well isolated and undamaged were imaged as 100–150 µm Z-stacks to capture the full arbor of the cell of interest. Captured images encompassing the entire cell (6–8 cells per animal) were imported into Neurolucida 360 (MBF Bioscience) for interactive tracing of the apical and basal arbors. Means were derived from 4 animals per genotype.
Cortical Dendrite Branching Analysis
Randomly selected fields (4–6 per animal) containing GFP-stained layer V cortical neurons were imaged as 100 µm Z-stacks to capture the entire depth of the section in each field. Primary apical dendrites (30–50 per field) were traced from the soma to the first bifurcation point using the Simple Neurite Tracer plugin (Longair et al. 2011) in Fiji (Schindelin et al. 2012). The branch depth relative to the cortical surface was calculated as the difference between the branch depth (db) and the soma depth (ds), presented as a percentage of the total soma depth (100 × [db− ds/ds]). Means were derived from 5 animals per genotype.
Dendritic Spine Analysis
Secondary branches of the apical dendrites of GFP-stained layer V cortical neurons and pyramidal cells from CA1 of the hippocampus were imaged as Z-stacks containing the dendrite's entire depth (5–25 µm), including the branch point. Selected dendrites (7–10 per animal) were well stained and unobscured by neighboring cells to a distance of at least 80 µm from the branch point. Image stacks were imported into Imaris (Bitplane) and were traced using a semi-automated Filament procedure to mark spine locations and to determine dendritic spine density.
Quantitative PCR
cDNA samples from mouse brain tissue that was dissected at the gestational day indicated from embryos collected from timed pregnant dams. mRNA was extracted using the ReliaPrep RNA Miniprep System (Promega), and cDNA was synthetized using iScript Reverse Transcription Supermix (Bio-Rad). Quantitative PCR (qPCR) was performed using SYBR green reagents (SsoFast EvaGreen Supermix from Bio-Rad) on a Bio-Rad platform (CFX384). Expression of Cc2d1a was normalized to Actb; relative expression levels are indicated as ratio to the cDNA from the earliest time available. Primer sequences are available upon request.
Immunoblots
Protein was prepared from fresh, frozen mouse cortical tissue that was homogenized in a buffer containing Tris–HCl (10 mM), NaCl (100 mM), ethylenediaminetetraacetic acid (1 mM), and sucrose (250 mM), and extracted for 30 min with Triton X-100 (1%), sodium deoxycholate (0.5%), and sodium dodecyl sulfate (0.1%). Lysates were cleared by centrifugation at 15 000 × g for 20 min, then combined with one volume of Laemmli sample buffer (Bio-Rad) containing 5% beta-mercaptoethanol, and denatured by heating at 95 °C for 5 min. Protein was separated by SDS–PAGE on 4–12% Bis-Tris gels (Life Technologies) and transferred to Immobilon-FL (Millipore) polyvinylidine fluoride membranes. Immunoblots were probed with primary antibodies at optimized concentrations (see Supplementary Table 1) that were detected with fluorophore-conjugated secondary antibodies (LI-COR Biosciences) and imaged on an Odyssey Imager (LI-COR Biosciences). Tubulin or actin was used as a control for equal protein loading.
Slice Preparation and Electrophysiology
Transverse hippocampal slices (400 µm) were prepared from mice 2–5 months of age using a vibratome as previously described (Hoeffer et al. 2013). Slices were maintained at room temperature in a submersion chamber with artificial cerebral spinal fluid (aCSF) containing the following (in mM): 125 NaCl, 2.5 KCl, 2 CaCl2, 1 MgCl2, 1.25 NaH2PO4, 24 NaHCO3, and 15 glucose bubbled with 95% O2–5% CO2. Slices were incubated in a recording chamber (preheated to 32 °C), where they were superfused with oxygenated aCSF for at least 1 h before starting the recordings. Recordings were performed by placing a bipolar stimulating electrodes (92 : 8 Pt : Y) at the border of area CA3 and area CA1 along the Schaffer-collateral pathway. ACSF-filled glass recording electrodes (1–3 MΩ) were positioned in the stratum radiatum of area CA1. Stable baseline synaptic transmission was established for at least 30 min. Early-phase LTP (E-LTP) and late-phase LTP (L-LTP) were induced with 1 train (100 Hz for 1 s) and 2 trains (100 Hz for 1 s, with 20 s interval) of high-frequency stimulation, respectively. Measurements of field excitatory postsynaptic potential (fEPSP) were acquired and analyzed using pCLAMP 10 (Molecular Devices).
Behavioral Tests
All behavioral experiments were performed on Cc2d1aflx/flx-CamKIIacre male mice of at least 12 weeks of age. Cohorts of littermates included roughly equal numbers of WT (WT or CamKIIacre or Cc2d1aflx/flx), heterozygotes (Cc2d1a+/flx-CamKIIacre), and cKO mice (Cc2d1aflx/flx-CamKIIacre) were used (see Supplementary Table 2). Initial characterization included an analysis of basic sensory and motor function as described by Rogers et al. (2001). Briefly, adult males of each genotype were tested for weight, righting reflex, wire hang, gait, tail pinch, visual reach, and the Preyer reflex (acoustic startle). Testing of cognitive and social behaviors was performed in a dedicated behavior analysis suite in the George Washington University Animal Research Facility. A battery of behavioral tests was initially applied to a cohort of animals at 2–3 months of age, including the open field test, the novel object recognition test (NORT; Bevins and Besheer 2006), the Morris water maze (MWM; Vorhees and Williams 2006), and the marble burying test (Deacon 2006). A subsequent cohort of animals underwent the three-chamber social interaction test (Moy et al. 2004; Kaidanovich-Beilin et al. 2011), an analysis of social approach between males and females (Kazdoba et al. 2015), and a test for ultrasonic vocalization (USV) during male–female interactions (Grimsley et al. 2011; Scattoni et al. 2011). Additional details of the behavioral methods may be found in Supplementary Material.
Results
Cc2d1a KO Mice Die at Birth Following Breathing and Swallowing Defect
Cc2d1a KO mice were generated using a targeted gene-trap construct inserted between exons 11 and 12 of the murine gene (Fig. 1A). This strategy closely replicates one of the truncating mutations found in humans where a 3.5-kb deletion interrupts the gene at exon 13 (Basel-Vanagaite et al. 2006) and leaves an N-terminal fragment that includes the first 3 DM14 domains of the protein (Fig. 1B). The gene-trap is highly effective as immunoblots show that the full length 100-kDa Cc2d1a band is completely absent in homozygous animals, leaving only a 50-kDa truncated band which is expressed at lower levels than the full length protein (Fig. 1C). In previously reported Cc2d1a-deficient lines where a deletion generated a 263 amino acid fragment (Al-Tawashi et al. 2012) or where Cc2d1a was completely removed starting from exon 1 (Zhao et al. 2011), no homozygous Cc2d1a gene-trap pups were found past postnatal day (P)0, suggesting that all of these alleles lead to Cc2d1a LOF and early postnatal lethality. At birth, homozygous Cc2d1a KO pups (initially crossed on a 129/SvEvTac background) are indistinguishable from WT littermates, with normal brain anatomy and size (Fig. 1D,E) and normal weight (Fig. 1F). The analysis of the internal organs observed on paraffin sections stained with H&E conducted by a pathologist at the Rodent Histopathology Core at Harvard Medical School reported no differences from WT littermates (data not shown). Mendelian ratios are preserved in the Cc2d1a-deficient litters up to embryonic day 18 (E18); however, homozygous pups die at or shortly after birth (Fig. 1G). Fostering the newborns with an experienced mother following Cesarean recovery from the heterozygous dam still resulted in early postnatal death, indicating that maternal care was not involved in reduced survival. As early lethality can sometimes be rescued by breeding a mutation onto a different genetic background (Sibilia and Wagner 1995; Threadgill et al. 1995), we fully backcrossed the Cc2d1a gene trap on a C57BL/6 background and a more outbred Swiss Webster background and found no difference in survival.
While breathing defects have been suggested as the cause of lethality in a different KO line, diaphragm innervation was found to be normal (Zhao et al. 2011) and the cause of sudden perinatal death in Cc2d1a KO mice has not been determined. To better understand the cause of early postnatal death caused by Cc2d1a LOF, we examined the behavior of the pups in the first hour out of the womb using dams from both 129/SvEvTac and Swiss Webster backgrounds. Following Cesarean delivery at E19, 25% of Cc2d1a KO pups never took a first breath and around 38% had difficulties in breathing and did not perfuse, remaining fully or mildly cyanotic (Fig. 2A). We analyzed the anatomy of the respiratory centers in the brain stem and identified no obvious abnormalities (see Supplementary Fig. 1A,B). Further examination of brain stem nuclei function will be required to identify the specific defects in Cc2d1a KO mice. Since breathing difficulties accounted for only a portion of the early perinatal death, we then asked whether swallowing might be affected. A few minutes after the pups started breathing they were fed a drop of infant formula labeled with green fluorescent microbeads. Pups from all genotypes had milk in their stomach showing that swallowing occurred (Fig. 2B). However, Cc2d1a KO pups also aspirated milk into the lungs, which were labeled brightly by the fluorescent beads (Fig. 2C). Thus, Cc2d1a KO pups have abnormalities in both breathing and swallowing. These are critical functions that are localized to neuronal populations of the brain stem, although these deficiencies have not been linked to a specific anatomical locus in Cc2d1a KO animals.
Figure 2.
Cc2d1a KO mice show breathing and swallowing deficits. (A) Ratio of pups breathing at birth following Cesarean delivery. (B) Ratio of pups that could be induced to swallow at birth following Cesarean delivery. Results expressed as the percent of total of pups from 4 litters (A and B). (C) Cc2d1a KO pups show accumulation of fluorescently labeled milk (in green; right panels) in the lungs (L) and stomach (S), while milk can only be found in the stomach in WTs.
A β-Gal reporter included in the Cc2d1a targeting construct showed that Cc2d1a expression is highly enriched in developing neural structures at least as early as E10.5 (see Supplementary Fig. 2A). Previous work has shown that Cc2d1a is expressed in the ventricular zone, cortical plate, and ganglionic eminence at E12 (Basel-Vanagaite et al. 2006), and by E14.5 it is detectible throughout the developing mouse brain (Richardson et al. 2014). At E16 and E18.5, Cc2d1a is expressed in all regions of the brain, with the strongest expression in the gray matter, including that of the hippocampus and cortex (Basel-Vanagaite et al. 2006; Zhao et al. 2011), where expression peaks around birth (Manzini et al. 2014) and remains enriched relative to other brain regions into adulthood (see Supplementary Fig. 2B). The early expression of Cc2d1a in the tissues from which neural progenitors originate suggests that the death of Cc2d1a KO pups is due to a neuron-specific effect. Indeed, in primary cortical cultures, we have observed that Cc2d1a is primarily expressed in neurons relative to astrocytes (see Supplementary Fig. 2C), a finding which has recently been replicated in proteomic screen exploring expression differences in murine neurons and glial subtypes, showing that Cc2d1a is expressed exclusively in neurons (Sharma et al. 2015).
To determine whether early death was, in fact, caused by neuronal dysfunction, we took advantage of the conditional gene-trap/cKO design of the Cc2d1a targeting construct to generate a conditional Cc2d1a allele. The gene-trap/Neo cassette was removed by recombination using the FLPeR mouse, which expresses the FLPe recombinase in the germline (Farley et al. 2000), leaving intact the floxed Cc2d1a allele (Cc2d1aflx) with exons 12–14 flanked by LoxP sites (Fig. 3A). We then crossed the Cc2d1aflx allele with a line expressing the Cre recombinase under the Nestin promoter (Nestincre), which targets the nervous tissue starting at E11 (Tronche et al. 1999). Cc2d1aflx/flx-Nestincre pups still died immediately after birth (Fig. 3B) and looked indistinguishable from WT littermates, indicating that loss of Cc2d1a in neurons at an early stage of development is responsible for the perinatal lethality of this line. The existence of a functional deficit at the level of individual brain stem nuclei or the involved circuits is an attractive explanation for perinatal failure of the respiratory and swallowing reflexes in Cc2d1a null mice and future work should address in detail the cellular origin of these phenotypes.
Figure 3.
Postnatal forebrain removal of Cc2d1a rescues lethality. (A) Removal of gene-trap allele by FRT following cross of Cc2d1a heterozygous mice with the FLPeR mouse line, leaving LoxP sites flanking exons 12–14 in Cc2d1aflx mice. (B) Cre recombinase expression under the CamKIIa promoter rescues early postnatal lethality (data from 16 litters), while crossing with Nestincre does not (data from 6 litters). (C) Cc2d1a is almost completely absent from the cortex of adult cKO mice, even though CamKIIcre is only expressed in excitatory neurons. (D) Analysis of steady-state Cc2d1a mRNA expression in Cc2d1aflx/flx (+/+) and cKO brain tissue also shows almost complete ablation of mRNA both upstream (5′) and downstream (3′) of the predicted truncation. Levels in cKO cortex (ctx) and hippocampus (hip) are expressed as fold change relative to respective WT tissues. Results expressed as mean from 4 mice per genotype, *P < 0.05, ***P < 0.001 (two-tailed t-tests in D). (E) The size and organization of the adult cKO brain is indistinguishable from WT littermates Cc2d1aflx/flx stained with H&E.
Forebrain-Specific Postnatal Removal of Cc2d1a Rescues Lethality
Since our goal was to study the impact of Cc2d1a LOF in circuit function and behavior in the adult brain, we sought to remove Cc2d1a only in the forebrain after birth to bypass respiratory control centers in the brain stem. We used a line expressing Cre under the CamKIIa promoter (CamKIIacre; Tsien et al. 1996), which recombines during the first 2 postnatal weeks and we confirmed this pattern using a Cre-reporter mouse where the red fluorescent marker TdTomato is expressed whenever Cre recombinase is active (Madisen et al. 2010). The reporter shows an initial presence of Cre in the frontal lobe and hippocampus at P10 (see Supplementary Fig. 3A) with widespread expression throughout the cortex and the hippocampus by P21 (see Supplementary Fig. 3B). The Cc2d1aflx/flx-CamKIIacre (cKO) mice were born in the expected Mendelian ratios (Fig. 3B), developed normally, had normal somatosensory and motor responses (see Supplementary Table 3), and were fertile. Although inactivation of Cc2d1a in cKO mice is predicted to be restricted to excitatory neurons in the cortex and hippocampus, we observed a significant reduction in Cc2d1a expression in adult animals both by immunoblot (Fig. 3C) and by qPCR, suggesting that the truncated mRNA is largely degraded (Fig. 3D). Brain histology in adult mice is grossly indistinguishable from WT (Fig. 3E). Thus, we have generated a forebrain-specific Cc2d1a LOF line, which can be studied to model how mutations in CC2D1A cause cognitive deficits.
Cc2d1a LOF Alters Cortical Dendrite Morphology and Spine Density in the Adult Brain
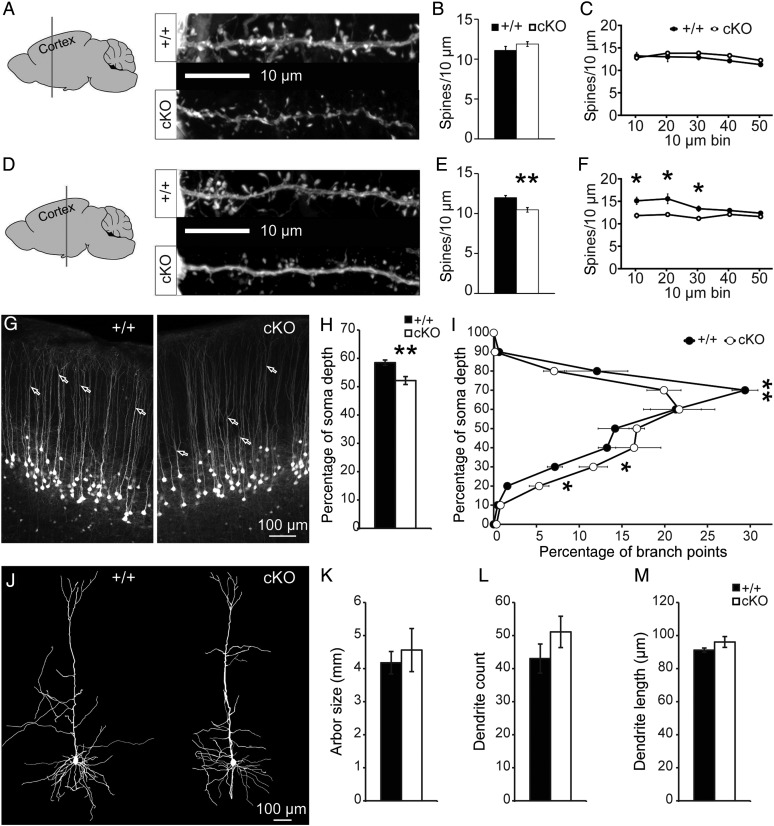
Several reports have shown that Cc2d1a LOF disrupts dendrite morphogenesis in vitro, with significant reduction in dendrite complexity reported in cultured cortical neurons from Cc2d1a KO mice (Al-Tawashi et al. 2012) as well as following shRNA-mediated depletion of Cc2d1a from hippocampal neurons (Manzini et al. 2014). Both synaptic number (Al-Tawashi et al. 2012) and spine density (Manzini et al. 2014) were reported to be reduced after depletion of Cc2d1a in neurons and synaptic maturation was delayed (Zhao et al. 2011). In addition, we have previously observed that early knockdown of Cc2d1a by in utero electroporation of shRNA at E15.5 reduces dendrite complexity at P11 in the intact brain (Manzini et al. 2014). Since dendritic complexity and spine number are often reduced in the cortex of patients with ID (Huttenlocher 1974; Purpura 1974; Penzes et al. 2011), we studied neuronal morphology in the brains of adult Cc2d1a cKO mice. To analyze dendrite morphology and dendritic spine density, Cc2d1a cKO mice were crossed with the Thy1-YFP-H reporter line (cKO-YFP), which expresses YFP in layer V neurons of the cortex (Feng et al. 2000). While the overall density and morphological features of layer V neurons were similar between cKO-YFP and WT-YFP mice, we examined dendritic spine density on the secondary branches of the apical dendrites in the primary somatosensory cortex to determine whether structural changes that could affect cortical connectivity were present, and identified remarkable regional differences. Layer V cells in frontal areas corresponding to the hindlimb and forelimb regions were unaffected (Fig. 4A), with density (Fig. 4B) and distribution (Fig. 4C) of dendritic spines that were similar to WT-YFP mice. Spine length, surface area, and volume were likewise unaffected (see Supplementary Fig. 4A–C). In contrast, the distribution of dendritic spines on layer V cells of the primary somatosensory cortex corresponding to the trunk and barrel fields (Fig. 4D) was significantly affected. The shift in spine distribution, which was of sufficient magnitude to produce a significant reduction in the average spine density in cKO-YFP mice (Fig. 4E), was restricted to the initial 30 µm of secondary apical dendrites (Fig. 4F). Although spine density was reduced, no effect on spine length, area, or volume was detected (see Supplementary Fig. 4D–F).
Figure 4.
Dendrite and spine morphology is altered in the cortex of cKO mice. (A) No changes in dendritic spine (B) density and (C) distribution are observed on secondary apical dendrites on layer V cells of the anterior somatosensory cortex (Bregma −0.23 mm) in WT (+/+) and Cc2d1a cKO mice. (D) Dendritic spine density is reduced (E), especially in the initial segment of secondary dendrites (F) in more posterior somatosensory cortex (Bregma −1.55 mm). Results expressed as mean ± SEM of n = 5 mice per genotype (7–10 dendrites per animal), *P < 0.05, **P < 0.01 (two-tailed t-tests in C and F). (G) The branching point of the apical tuft in layer V neurons of the posterior somatosensory cortex (bifurcations marked by arrows) was found to be lower in cKO brains, as indicated by (H) population average and (I) distribution of branch depths relative to soma depth (0% = closest to soma; 100% = furthest from soma). Results expressed as mean ± SEM of n = 5 mice per genotype (100–200 branch points per animal), *P < 0.05, **P < 0.01 (two-tailed t-tests in H and I). (J) No difference was observed in dendrite (K) size, (L) number, and (M) average length in the dendritic arbor of Golgi-stained layer V neurons of the posterior somatosensory cortex in WT (+/+) and Cc2d1a cKO mice. Results expressed as mean ± SEM of n = 4 mice per genotype (6–8 arbor traces per animal; two-tailed t-tests in K, L, and M).
Since we observed a reduction in spine density in a subset of layer V cells, we decided to investigate further the morphology of these neurons. It was evident from micrographs that the primary branch point of the apical tuft was shifted in cKO-YFP mice (Fig. 4G). We quantified the average depth of these apical bifurcations and found that it was deeper in cKO-YFP mice (Fig. 4H), with significantly more layer V neurons branching in the first third of their apical dendrites, around the depth of layer IV (Fig. 4I). Since arbor organization was affected, we also asked whether dendritic complexity would also be altered, but neurons in these dorsal locations are too densely packed in the Thy1-YFP-H mice to measure reliably the number and length of dendritic processes. Therefore, we completed this analysis in a separate set of cKO mice using a modified Golgi technique and found no significant differences between WT and cKO mice in layer V neurons in the somatosensory cortex (Fig. 4J–M). The observed changes in spine density and dendritic organization show that Cc2d1a LOF disrupts the morphological development of neurons in a manner that is likely to have significant impacts on the function of individual neurons as well as the circuits they comprise.
Learning Deficits and Hippocampal Dysfunction in Cc2d1a cKO Mice
CC2D1A LOF leads to fully penetrant ID in humans (Basel-Vanagaite et al. 2006; Manzini et al. 2014). We tested cognitive function on a cohort of Cc2d1a cKO male mice using the novel object recognition test (NORT) and Morris water maze (MWM). As mice are innately curious of novel objects, the NORT tests the ability of a mouse to discriminate between a previously encountered object and an unfamiliar object (see Supplementary Fig. 5A), as measured by the relative preference to explore each object (Bevins and Besheer 2006). The mice were exposed to 2 identical objects (Fig. 5A; baseline) and after 15 min tested with a familiar and a novel object (Fig. 5A; novel/familiar). While WT mice showed a significant preference for the novel object, cKO mice did not spend significantly more time with the novel object, showing that they could not remember the training (Fig. 5A).
Figure 5.
Hippocampus-dependent memory formation is impaired in the Cc2d1a cKO mice. (A) Short-term memory was examined by the NORT test, with a novel object replacing a familiar object after a 15-min intersession interval. In contrast to WT (+/+; n = 11), Cc2d1a cKO mice (n = 15) showed no preference for the novel object relative to a familiar object. Results expressed as mean ± SEM, *P < 0.05 (two-tailed t-tests in A). (B) Spatial learning was measured as latency to escape in 3 stages of the MWM with a visible platform (V; 2 days), hidden platform (H; 5 days), or the reversal (R; 2 days) of the hidden platform location. Cc2d1a cKO (n = 15) mice were significantly impaired in learning the location of the hidden platforms compared with WT mice (+/+; n = 14) over 4 trials per day per stage. Results expressed as mean ± SEM of mice from each genotype, *P < 0.05 (Mann–Whitney U-tests or two-tailed t-tests for stages with unequal or equal variance, respectively, in B, following detection of significant interaction by two-way ANOVA). (C and D) Dendritic spine density on secondary apical dendrites of CA1 pyramidal neurons was slightly reduced. Results expressed as mean ± SEM of n = 5 mice per genotype (8–10 dendrites per animal), ***P < 0.001 (effect of genotype by two-way ANOVA in D). (E and F) fEPSP recordings show a defect in L-LTP (F; 100 Hz for 1 s with 20 s interval; 10 WT and 11 Cc2c1a cKO hippocampal preparations), but not in E-LTP (E; 100 Hz for 1 s; 8 hippocampal preparations per genotype). Representative traces recorded (1) before and (2) after stimulus protocols are displayed (insets). (G) Averaging of the last 10 min of fEPSPs recorded in the L-LTP indicates a significant reduction in synaptic strength. Results expressed as mean ± SEM of 10 WT and 11 Cc2c1a cKO hippocampal preparations, **P < 0.01 (two-tailed t-test in H).
Cc2d1a cKO mice were also significantly impaired in spatial learning in the MWM. In this test, mice are trained to use spatial cues to learn the locations of an underwater platform in a large water tub. Mice are tested in the MWM over a period of 10 days (see Supplementary Material for detailed methods) for their latency to exit the water onto a visible (V) or hidden (H) platform. After a series of tests with the hidden platform in an initial location, reversal (R) learning was also tested by moving the platform to a new location. While the identification of the visible platform was not different between genotypes (Fig. 5B; V1–V2), cKO mice demonstrated a significant delay in learning the location of the hidden platform as their latencies were significantly higher on the second and third days of testing (Fig. 5B; H2–H3). cKO animals eventually improved their latency with successive trials reaching WT levels by day 5 (Fig. 5B; H4–H5). Performance of cKO and WT mice was similar in the reversal trials (Fig. 5B; R1–R2), and no differences were detected in a probe trial (see Supplementary Fig. 5B).
As both the NORT and MWM rely strongly on hippocampal function (Vorhees and Williams 2006), we also analyzed dendritic spines in hippocampal CA1 pyramidal neurons using in WT-YFP and cKO-YFP mice and found a mild but significant reduction in spine density on secondary apical dendrites (Fig. 5C,D). The behavioral impairments suggested a defect in synaptic plasticity; therefore, we also studied LTP in hippocampal slices from Cc2d1a cKO mice. We examined changes in synaptic strength induced by 1 train of tetanic stimulation (100 Hz for 1 s) and 2 such trains, which generally results in short-lasting (E-LTP) and long-lasting (L-LTP) LTP, respectively. In slices from Cc2d1a cKO mice, E-LTP was unchanged (Fig. 5E), whereas there was a significant impairment in L-LTP (Fig. 5F,G). This effect is not likely to be due to defective basal synaptic transmission, since neither input/output (data not shown) nor paired-pulse facilitation (see Supplementary Fig. 5C) was impaired in the cKO mice. These results suggest that Cc2d1a LOF prevents the conversion of E-LTP into L-LTP. Thus, the disruption of long-lasting synaptic plasticity in the hippocampus is associated with changes in dendrite morphology and learning deficits in adult Cc2d1a cKO mice.
Social Impairments in Cc2d1a cKO Mice
In addition to cognitive deficits, CC2D1A LOF causes a variable constellation of behavioral phenotypes including ASD or autistic traits in some of the cases (Manzini et al. 2014). We examined social behavior in cKO mice using the three-chamber test (see Supplementary Fig. 6A; see Supplementary Material for detailed methods) to assess social preference as indicated by the time spent exploring chambers containing a novel object or an unfamiliar mouse of the same sex. While WT mice show a clear social preference, spending more time in the chamber containing the unfamiliar mouse, cKO mice present the opposite pattern of behavior, spending more time exploring the object chamber (Fig. 6A). We also analyzed the social approach and interaction behavior of these animals. Compared with WT, the cKO made significantly fewer approaches to the mouse-containing cup (Fig. 6B), and spent significantly less time in close proximity to the unfamiliar mouse (Fig. 6C). Thus, cKO mice engage less frequently in social approach and spend less time in social interactions with unfamiliar male mice. To further examine the social function of Cc2d1a cKO mice, we also analyzed a social approach between males and females and USV during male–female interactions. During a 5-min social approach test between virgin Cc2d1a cKO male mice and virgin WT females, cKO male mice performed significantly less social sniffing than WT males (Fig. 6D), with a significant reduction in nose-to-body contact with females (Fig. 6E), while other forms of social sniffing were unchanged (Fig. 6F,G). This may reflect a specific reduction in the number of copulatory attempts by Cc2d1a cKO males, while other male–female interactions are not affected. Exploration time during this test was increased in cKO mice relative to WT (see Supplementary Fig. 6B). In addition, USVs were recorded in a multi-day testing paradigm as the males were attempting to mate with the females (Grimsley et al. 2011; Scattoni et al. 2011). Although the syllable distribution (see Supplementary Fig. 6C) and sonic characteristics (Fig. 6H and Supplementary Fig. 6D) of USVs are similar to WT, the vocalization rate of Cc2d1a cKO mice during these encounters is significantly reduced (Fig. 6I). Taken together, these data show that Cc2d1a LOF in cKO mice is associated with deficits across many aspects of social interaction.
Figure 6.
Social interaction deficits in Cc2d1a cKO mice. (A–C) Sociability is reduced for cKO mice in all analyses. Social interaction was assessed in the three-chamber test (see Supplementary Fig. 6A) and is presented as (A) time spent in each chamber, (B) entries to the perimeter around each cup, and (C) time spent interacting with each cup. Results expressed as mean ± SEM of 10 mice per genotype; *P < 0.05, **P < 0.01, ***P < 0.001 (two-tailed t-test within the genotype); #P < 0.05, ##P < 0.01, #P < 0.001 (two-tailed t-test between genotypes). Male–female social interaction was assessed during interaction between virgin WT (+/+) or cKO males and virgin WT females as (D) total social sniffing time which is the sum of (E) nose-to-body, (F) nose-to-genital, and (G) nose-to-nose sniffing. Values calculated as a percentage of the total testing duration (300 s) are expressed as mean ± SEM of 8 mice per genotype; **P < 0.01 (two-tailed t-tests in D–G). (H) Representative sonograms of USVs recorded during interaction between virgin WT (+/+) or cKO males and virgin WT females. (I) Average number of vocalizations per pairing recorded across 5 pairings. Results expressed as mean ± SEM of 8 mice per genotype; *P < 0.05 (Mann–Whitney U-test in I).
Anxiety, Hyperactivity, and Compulsivity in Cc2d1a cKO Mice
In addition to social impairments, ASD often includes repetitive behaviors or a restricted pattern of interests. As a large percentage of Cc2d1a cKO animals (∼30%) developed severe ulcerative dermatitis between 4 and 8 months of age, due to obsessive grooming (see Supplementary Fig. 7A), we also tested these mice for repetitive and obsessive behaviors, as well as anxiety. Initial exploration of the open field is a measure of the baseline motor activity level. The analysis of track plots of WT (Fig. 7A) and cKO mice (Fig. 7B) showed that cKO cover significantly more distance in the open field, suggesting that Cc2d1a cKO mice are hyperactive (Fig. 7C). Although cKO mice covered more distance in the open field, they spent only half as much time in the center zone of the arena, showing an increase in anxiety-like behavior (Fig. 7D). In the marble burying test of repetitive behavior, although the number of marbles buried was unchanged (see Supplementary Fig. 7B), cKO mice showed a decreased tendency to dig compared with WT mice (Fig. 7E), distributing more time during the testing period across other types of behavior. Taken together, these abnormalities in activity, anxiety-like behavior, and compulsivity show that cKO mice have a multifaceted cognitive–behavioral phenotype that recapitulates additional common features of neurodevelopmental disorders.
Figure 7.
cKO mice display behavioral phenotypes consistent with ID/ASD. (A–D) Exploratory activity in a novel environment was assessed on the open field test. Representative paths from (A) WT (+/+; n = 14) and (B) cKO mice (n = 15) mice show a substantial increase in locomotion in cKO animals. (C) Total path length is significantly increased, while (D) time spent in the center zone is reduced showing anxiety. Results expressed as mean ± SEM of mice from each genotype, **P < 0.01, ***P < 0.001 (two-tailed t-tests in C and D). (E) In the marble burying test, digging activity is significantly reduced in cKO mice, while other activities, such as exploring, sitting, and grooming (+/+, n = 12; cKO, n = 18), are unaffected. Results expressed as mean ± SEM of mice from each genotype, *P < 0.05 (two-tailed t-tests in E).
Discussion
Here, we report a novel mouse model of nonsyndromic ID and ASD that recapitulates key features of the human presentation of CC2D1A LOF. Null mutations in CC2D1A cause severe cognitive deficits, with a variable presentation of ID, ASD, and seizures (Manzini et al. 2014). This report shows that conditional postnatal inactivation of Cc2d1a in forebrain neurons in cKO mice results in a robust behavioral phenotype that includes learning and memory impairments, social deficits, hyperactivity, anxiety, and obsessive grooming. This is the first successful effort to model the effects of CC2D1A LOF in adult animals, as previously reported null mutants suffer from early postnatal lethality (Zhao et al. 2011; Al-Tawashi et al. 2012; Chen et al. 2012). The behaviors we report here are also associated with disrupted LTP and a reduction in dendritic spine density.
Modeling CC2D1A to Study the Mechanisms of ID/ASD
Recent next-generation sequencing studies have identified thousands of mutations in hundreds of genes that may be involved in neurodevelopmental disorders (Hoischen et al. 2014). This work has generated long lists of candidate genes for ID (Gilissen et al. 2014) and ASD (Betancur 2011; Abrahams et al. 2013), with only a limited growth in the repertoire of well-characterized animal models (Crawley 2012). Many of these disease mutations can be difficult to model because they are hypomorphic or de novo alleles, or because the disease is caused by large deletions of multiple genes that may not be contiguous in the mouse genome. Recessive LOF mutations have the advantage of requiring the homozygous removal of a single gene in the mouse to generate a representative animal model. Rare X-linked single gene mutations account for around 10% of ID cases and autosomal recessive genes could represent even more, but are more difficult to identify (Chelly et al. 2006). Complete gene knockouts are also enriched in ASD cases relative to controls, and autosomal and X-linked null mutations are predicted to contribute to up to 5% of ASD cases (Lim et al. 2013; Yu et al. 2013), but only a handful of these genes have been identified to date. While variably penetrant ASD presentations are often found in conjunction with ID in disorders such as Fragile X syndrome (Koukoui and Chaudhuri 2007) and tuberous sclerosis complex (Curatolo et al. 2015), CC2D1A mutations are among the few to identify similar variability of presentation in autosomal recessive nonsyndromic disorders. An appropriate model to study Cc2d1a LOF in mice will not only provide insights into how loss of Cc2d1a affects neuronal differentiation and cognitive development, but may also be used to explore the mechanisms leading to phenotypic variability.
Murine Cc2d1a in Breathing and Swallowing Regulation
The primary hurdle in studying how Cc2d1a LOF affects cognitive development has been the early neonatal mortality of Cc2d1a KO pups. Three independent KO lines have been generated, all of which die a few hours after birth (Zhao et al. 2011; Al-Tawashi et al. 2012; Chen et al. 2012). Nonetheless, KO pups are grossly indistinguishable from WT littermates at the time of birth, with normal weight and anatomy, indicating that loss of Cc2d1a does not significantly affect embryonic morphogenesis. Lethality is extremely robust, as we backcrossed our line on 2 inbred (129/SvEvTac and C57BL/6) and 1 outbred (Swiss Webster) genetic backgrounds and found no differences in survival. In addition, transferring the pups to an expert foster mother had no effect, indicating that maternal care is not a contributing factor. Respiratory failure had been suggested as the cause of death in another Cc2d1a KO line (Zhao et al. 2011), and after careful behavioral analysis of newborns following Cesarean birth at E19, we identified a combination of breathing and swallowing defects. Two-thirds of pups never breathe or breathe poorly leading to a cyanosis and death. The remaining third breathe and show normal tissue perfusion, but swallowing is impaired with substantial amounts of milk aspirated in the lungs leading to asphyxiation. In another Cc2d1a KO mouse line, diaphragm structure and innervation were previously shown to be normal (Zhao et al. 2011), as is the anatomy of breathing centers in the brain stem in our animals. The fact that neuron-specific removal of Cc2d1a during embryonic development using Nestincre produces the same perinatal lethality indicates that these deficits originate in the brain, possibly in central pattern generators in the brain stem. The neural regulation of breathing and swallowing are believed to be linked, and many of the same muscles are recruited by both processes (Martin-Harris 2008). Thus, whether a particular Cc2d1a KO animal dies of respiratory failure or dysphagia may depend on the severity of effects on the same neural structure, or may represent some interindividual variability in the anatomical extent of those effects.
There are no reports of sudden infant death or respiratory deficits in patients with CC2D1A mutations, and it is not known whether loss of CC2D1A contributes to respiratory failure in humans. It is possible that respiratory deficits are limited to the mouse for evolutionary reasons, such as differences in the breathing regulation and compensation from other genes. Alternatively, respiratory deficits could be variably penetrant in human patients with loss of CC2D1A, as are ASD and epilepsy, and therefore may have escaped detection because of early lethality in a limited number of cases. The fact that no other phenotype is present in this animal model indicates that Cc2d1a has an essential, neuron-specific function that is disrupted in KO mice.
Cc2d1a LOF Affects Neuronal Morphology and Plasticity
Mouse models are often heterogeneous in their presentation of morphological, physiological, and behavioral phenotypes, and one current goal in the field is to use them to yield generalizable principles of how development is affected in neurodevelopmental disorders. To understand how Cc2d1a cKO mice fit in the landscape of neurodevelopmental disease models and how they can help us understand the mechanisms of disease, we studied a variety of morphological, physiological, and behavioral features. Morphological parameters such as dendritic arbor complexity and dendritic spine number and shape are often increased or decreased in size, length, and number in patients and different models of neurodevelopmental disease (Huttenlocher 1974; Hutsler and Zhang 2010; Penzes et al. 2011). However, the magnitude and directionality of these changes vary widely across even well-studied models. Cc2d1a LOF has led to a reduction in dendritic complexity and spine density in cultured neurons, which is present in some models (Al-Tawashi et al. 2012; Manzini et al. 2014) and not in others (Zhao et al. 2011). In vivo we have found a reduction in spine density in the adult brain in the cortex and hippocampus, but no change in dendrite number or length. Interestingly, spine changes are regionally distributed in the cortex, where layer V pyramidal neurons in more posterior somatosensory areas are affected, while more frontal somatosensory areas are not. Morphological features have been shown to be dynamic in neurodevelopmental disorders (Takashima et al. 1981; Hutsler and Zhang 2010), but developmental and anatomical patterns of overgrowth and/or progressive loss of dendrites and spines are still poorly characterized. It is possible that these morphological changes can point to specific circuits that are more severely disrupted, but it is unclear how morphological and anatomical changes relate to functional deficits. A recent neuroanatomical analysis by magnetic resonance imaging of the brain of a collection of 26 mouse models of neurodevelopmental disorders, ranging from single gene mutations to large deletions, has shown that anatomical parameters vary widely (Ellegood et al. 2014). The mouse models in this study were clustered in groups where expansion or contraction of individual brain regions was correlated, but genes with similar functions and presentation were found in different groups, suggesting that different anatomical deficits can lead to similar outcomes.
The morphological changes reported in postmortem analyses (Penzes et al. 2011) have been associated with functional deficits at the cellular or local circuit level in experimental models (He and Portera-Cailliau 2013), but substantial deficits in synaptic physiology can still be found in model systems where morphological changes are very subtle or not present at all (Boda et al. 2004; Meng et al. 2005). Despite very mild alterations in spine density in the hippocampus, Cc2d1a cKO mice showed significant impairment in learning and memory skills that are associated with hippocampal function. When we examined synaptic function and plasticity in the hippocampus, we found no defects in short-term plasticity, but a complete suppression of L-LTP, a sustained increase in synaptic strength that is often associated with memory consolidation (i.e., the formation of long-term memory). Recently, it was shown that L-LTP and long-term memory are dependent on structural plasticity and actin dynamics (Huang et al. 2013), which are impaired in the hippocampus of the cKO mice. Thus, genetic disruption of Cc2d1a results in a series of behavioral, structural, and synaptic defects, consistent with the human disorder.
Behavioral Deficits Parallel Models of Synaptic and Signaling Proteins
As observed in patients, animal models of neurodevelopmental disorders show very heterogeneous behavioral presentations with different combinations of deficits. Cc2d1a cKO mice display learning and memory deficits in the MWM and the NORT. In addition, these animals have a broad array of social impairments that are detected across multiple testing paradigms. In a three-chamber test of male–male interactions, Cc2d1a cKO mice show an antisocial preference and reduced sociability. Male–female interactions are also impacted, and are associated with the reduced production of USVs by cKO males encountering virgin females. These cognitive and social phenotypes are associated with a distinctive combination of hyperactivity, anxiety-like behaviors, and repetitive behaviors. Increased locomotor activity in the open field and obsessive grooming are accompanied by a reduction in digging activity in the marble burying test. While this association is somewhat counterintuitive, it corresponds with a report showing that marble burying in this task may be incidental, and that the test is largely a measure of repetitive digging behavior that does not necessarily correlate with other measures of either anxiety or motor activity (Thomas et al. 2009). In addition, as repetitive behaviors have been linked in striatal defects and CamKII-cre is not expressed in the striatum, additional tissue-specific deletions may be necessary to fully understand how Cc2d1a deficiency affects these behaviors.
A similar combination of behavioral phenotypes has been described in mice with mutations of Shank2 (Schmeisser et al. 2012; Won et al. 2012), Shank3 (Wang et al. 2011; Yang et al. 2012), or “phosphatase and tensin homolog on chromosome ten” (Pten; Kwon et al. 2006; Napoli et al. 2012). The ProSAP/Shank gene family has been linked to ID and ASD, most prominently in the case of Phelan-McDermid syndrome, which is caused by haploinsufficiency of the SHANK3-containing locus 22q13.3 (Phelan and McDermid 2012). Other mutations of SHANK3, as well as the related genes SHANK1 and SHANK2, have also been linked to ID and ASD (Jiang and Ehlers 2013). The SHANK genes encode large molecular scaffolds that are present at synapses and organize multiple proteins at the postsynaptic density. Learning impairments, social deficits, and repetitive grooming behavior are found with both Shank2 (Won et al. 2012) and Shank3 (Wang et al. 2011; Yang et al. 2012), but while Shank2 KOs display hyperactivity like Cc2d1a cKO, Shank3 KOs tend to be less active than controls. Social deficits in SHANK models have in some cases been associated with changes in vocalization, although results are not consistent across all reports (Wöhr 2014). In Shank3 heterozygotes, reduced social interaction is accompanied by a reduced number of USVs elicited by exposure of males to a female in estrus (Bozdagi et al. 2010), similar to what we find with Cc2d1a cKO mice, and reduced female–female vocalizations have been observed in Shank2 KO mice (Schmeisser et al. 2012) with some changes in the organization of USVs also reported in this model (Ey et al. 2013).
Conditional removal of Pten in the brain is also consistent with the Cc2d1a behavioral phenotypes. Pten is a critical negative regulator of PI3K and mTor signaling, a pathway involved in the growth and proliferation of many cell types, including neurons (Kwon et al. 2003; Jaworski et al. 2005). LOF PTEN mutations lead to overgrowth syndromes in different tissues (Waite and Eng 2002), while de novo truncating mutations have been identified in several cases of ASD (O'Roak et al. 2012). Homozygous inactivation of Pten in the mouse is embryonically lethal, and several lines with conditional inactivation of Pten in the brain have been generated to study how this gene affects neural development. Cre-mediated recombination in the brain at embryonic (Lugo et al. 2014) or postnatal (Kwon et al. 2006; Napoli et al. 2012) time points shows impaired spatial learning, hyperactivity, reduced social preference, increased anxiety-like behaviors, repetitive grooming, and reduced digging as we observe for Cc2d1a LOF.
Despite producing similar behavioral phenotypes, Shank2 and Pten deficiency affects the morphological development of neurons in different ways. Shank2 LOF is more similar to Cc2d1a LOF since spine density is mildly reduced in the hippocampus (Schmeisser et al. 2012), while disruption of Pten leads to neuronal hypertrophy and an increase in dendritic spine formation (Kwon et al. 2006; Fraser et al. 2008), again suggesting that morphological deficits may not be predictive of the behavioral outcome. A common thread among all these models is LTP, and in particular, L-LTP impairment (Fraser et al. 2008; Schmeisser et al. 2012).
In relation to gene function, the parallel between Cc2d1a and Pten is not surprising as Cc2d1a is also a regulator of Akt signaling (Nakamura et al. 2008). It is therefore plausible that deficiency of either Cc2d1a or Pten could be modulating the PI3K/Akt/mTor pathway leading to similar physiological and behavioral outcomes. The similarity to Shank is also interesting as mild deficits in postsynaptic maturation have been described in one of the Cc2d1a global KOs (Zhao et al. 2011), but not much work has been devoted to the role of this protein at synapses. Cc2d1a is a regulator of endosomal trafficking and signaling, and these mechanisms are critical for synaptic maintenance and spine function (Cosker and Segal 2014). The Cc2d1a cKO would be an ideal model to further explore whether Cc2d1a LOF leads to localized trafficking and signaling disruptions at the synapse.
Conclusions
In summary, our findings have demonstrated that Cc2d1a is involved in many aspects of neurodevelopment, and point to a previously unknown role of Cc2d1a in activity-dependent functional plasticity in mature neurons. We have shown for the first time that Cc2d1a deficiency disrupts early postnatal brain development, which is when cognitive, social, and behavioral deficits are thought to originate in human patients. The behavioral, morphological, and functional deficits we have identified in Cc2d1a cKO mice parallel those found in ID and ASD, and indicate that Cc2d1a-dependent neuronal development is a critical determinant of cognitive and social function. Given the many signaling pathways known to be modulated by Cc2d1a, this animal model presents an excellent opportunity to identify new therapeutic targets in the treatment of neurodevelopmental disorders.
Future work on this model should directly address the mechanisms involved in the development and plasticity of Cc2d1a-deficient neurons. Answering this question may clarify the reasons for both the similarities and the differences between our model and others. It remains unknown which, if any, of the intracellular pathways known to be regulated by Cc2d1a is involved in its effects on neuronal plasticity or whether a more general dysregulation of endosomal trafficking is involved. Further investigation of the endosomal trafficking of signaling molecules and neurotransmitter receptors recruited by the induction of plasticity is a clear priority as work on both in vivo and in vitro models of Cc2d1a LOF proceeds.
Supplementary Material
Supplementary material can be found at: http://www.cercor.oxfordjournals.org/.
Funding
This work was supported by the NIH grants R00HD067379 to M.C.M., R01s MH083565 and NS032457 to C.A.W., K99NS087112 to E.S., and R01s NS034007 and NS047384 to E.K. The GWU Center for Microscopy and Image Analysis is supported by the Intellectual and Developmental Disabilities Research Center (IDDRC) at Children's Research Institute (P30HD040677) and by a grant from the NIH National Center for Research Resources (1S10RR025565).
Supplementary Material
Notes
The authors thank Anthony LaMantia and Thomas Maynard for discussion and for sharing the milk swallowing protocol developed in their laboratory, Matthew Fralish for help with the td-Tomato line, and Grzergorz Gorski at the Boston Children's Cellular Neuroscience Core for histology. We are particularly indebted to Anastas Popratiloff at the GWU CMIA for guidance in setting up multiple imaging and analytical approaches. The Cc2d1a KO mouse strain used for this research project was generated by the trans-NIH KnockOut Mouse Project (KOMP) and obtained from the KOMP Repository (www.komp.org). NIH grants to Velocigene at Regeneron, Inc. (U01HG004085) and the CSD Consortium (U01HG004080) funded the generation of gene-targeted ES cells for 8500 genes in the KOMP Program and archived and distributed by the KOMP Repository at UC Davis and CHORI (U42RR024244). For more information or to obtain KOMP products go to www.komp.org or email service@komp.org. Conflict of Interest: None declared.
References
- Abrahams BS, Arking DE, Campbell DB, Mefford HC, Morrow EM, Weiss LA, Menashe I, Wadkins T, Banerjee-Basu S, Packer A. 2013. SFARI Gene 2.0: a community-driven knowledgebase for the autism spectrum disorders (ASDs). Mol Autism. 4:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Tawashi A, Jung S, Liu D, Su B, Qin J. 2012. Protein implicated in nonsyndromic mental retardation regulates protein kinase A (PKA) activity. J Biol Chem. 287:14644–14658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basel-Vanagaite L, Attia R, Yahav M, Ferland RJ, Anteki L, Walsh CA, Olender T, Straussberg R, Magal N, Taub E et al. 2006. The CC2D1A, a member of a new gene family with C2 domains, is involved in autosomal recessive non-syndromic mental retardation. J Med Genet. 43:203–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancur C. 2011. Etiological heterogeneity in autism spectrum disorders: more than 100 genetic and genomic disorders and still counting. Brain Res. 1380:42–77. [DOI] [PubMed] [Google Scholar]
- Bevins R, Besheer J. 2006. Object recognition in rats and mice: a one-trial non-matching-to-sample learning task to study “recognition memory”. Nat Protoc. 1:1306–1311. [DOI] [PubMed] [Google Scholar]
- Boda B, Alberi S, Nikonenko I, Node-Langlois R, Jourdain P, Moosmayer M, Parisi-Jourdain L, Muller D. 2004. The mental retardation protein PAK3 contributes to synapse formation and plasticity in hippocampus. J Neurosci. 24:10816–10825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozdagi O, Sakurai T, Papapetrou D, Wang X, Dickstein DL, Takahashi N, Kajiwara Y, Yang M, Katz AM, Scattoni ML et al. 2010. Haploinsufficiency of the autism-associated Shank3 gene leads to deficits in synaptic function, social interaction, and social communication. Mol Autism. 1:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Lai L, Cheng H, Chen K, Syue Y, Lu H, Lin W, Chen S, Huang H, Shiau A et al. 2011. TBK1-associated protein in endolysosomes (TAPE) is an innate immune regulator modulating the TLR3 and TLR4 signaling pathways. J Biol Chem. 286:7043–7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelly J, Khelfaoui M, Francis F, Chérif B, Bienvenu T. 2006. Genetics and pathophysiology of mental retardation. Eur J Hum Genet. 14:701–713. [DOI] [PubMed] [Google Scholar]
- Chen K, Chang C, Huang C, Lin C, Lin W, Lo Y, Yang C, Hsing E, Chen L, Shih S et al. 2012. TBK1-associated protein in endolysosomes (TAPE)/CC2D1A is a key regulator linking RIG-I-like receptors to antiviral immunity. J Biol Chem. 287:32216–32221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosker K, Segal R. 2014. Neuronal signaling through endocytosis. Cold Spring Harb Perspect Biol. 6:a020669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawley J. 2012. Translational animal models of autism and neurodevelopmental disorders. Dialog Clin Neurosci. 14:293–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curatolo P, Moavero R, de Vries PJ. 2015. Neurological and neuropsychiatric aspects of tuberous sclerosis complex. Lancet Neurol. 14:733–745. [DOI] [PubMed] [Google Scholar]
- Deacon RMJ. 2006. Digging and marble burying in mice: simple methods for in vivo identification of biological impacts. Nat Protoc. 1:122–124. [DOI] [PubMed] [Google Scholar]
- Ellegood J, Anagnostou E, Babineau BA, Crawley JN, Lin L, Genestine M, DiCicco-Bloom E, Lai JKY, Foster JA, Peñagarikano O et al. 2014. Clustering autism: using neuroanatomical differences in 26 mouse models to gain insight into the heterogeneity. Mol Psychiatry. 20:118–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ey E, Torquet N, Le Sourd A-M, Leblond C, Boeckers T, Faure P, Bourgeron T. 2013. The Autism ProSAP1/Shank2 mouse model displays quantitative and structural abnormalities in ultrasonic vocalisations. Behav Brain Res. 256:677–689. [DOI] [PubMed] [Google Scholar]
- Farley F, Soriano P, Steffen L, Dymecki S. 2000. Widespread recombinase expression using FLPeR (flipper) mice. Genesis. 28:106–110. [PubMed] [Google Scholar]
- Feng G, Mellor RH, Bernstein M, Keller-Peck C, Nguyen QT, Wallace M, Nerbonne JM, Lichtman JW, Sanes JR. 2000. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 28:41–51. [DOI] [PubMed] [Google Scholar]
- Fraser M, Bayazitov I, Zakharenko S, Baker S. 2008. Phosphatase and tensin homolog, deleted on chromosome 10 deficiency in brain causes defects in synaptic structure, transmission and plasticity, and myelination abnormalities. Neuroscience. 151:476–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher C, Knoblich J. 2006. The conserved c2 domain protein lethal (2) giant discs regulates protein trafficking in Drosophila. Dev Cell. 11:641–653. [DOI] [PubMed] [Google Scholar]
- Gaugler T, Klei L, Sanders SJ, Bodea CA, Goldberg AP, Lee AB, Mahajan M, Manaa D, Pawitan Y, Reichert J et al. 2014. Most genetic risk for autism resides with common variation. Nat Genet. 46:881–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilissen C, Hehir-Kwa JY, Thung DT, Vorst M, Bon BWM, Willemsen MH, Kwint M, Janssen IM, Hoischen A, Schenck A et al. 2014. Genome sequencing identifies major causes of severe intellectual disability. Nature. 511:344–347. [DOI] [PubMed] [Google Scholar]
- Gratten J, Wray NR, Keller MC, Visscher PM. 2014. Large-scale genomics unveils the genetic architecture of psychiatric disorders. Nat Neurosci. 17:782–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimsley JMS, Monaghan JJM, Wenstrup JJ. 2011. Development of social vocalizations in mice. PLoS One. 6:e17460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He CX, Portera-Cailliau C. 2013. The trouble with spines in fragile X syndrome: density, maturity and plasticity. Neuroscience. 251:120–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeffer CA, Santini E, Ma T, Arnold EC, Whelan AM, Wong H, Pierre P, Pelletier J, Klann E. 2013. Multiple components of eIF4F are required for protein synthesis-dependent hippocampal long-term potentiation. J Neurophysiol. 109:68–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoischen A, Krumm N, Eichler EE. 2014. Prioritization of neurodevelopmental disease genes by discovery of new mutations. Nat Neurosci. 17:764–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Zhu PJ, Zhang S, Zhou H, Stoica L, Galiano M, Krnjević K, Roman G, Costa-Mattioli M. 2013. mTORC2 controls actin polymerization required for consolidation of long-term memory. Nat Neurosci. 16:441–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutsler JJ, Zhang H. 2010. Increased dendritic spine densities on cortical projection neurons in autism spectrum disorders. Brain Res. 1309:83–94. [DOI] [PubMed] [Google Scholar]
- Huttenlocher PR. 1974. Dendritic development in neocortex of children with mental defect and infantile spasms. Neurology. 24:203–210. [DOI] [PubMed] [Google Scholar]
- Jaekel R, Klein T. 2006. The Drosophila Notch inhibitor and tumor suppressor gene lethal (2) giant discs encodes a conserved regulator of endosomal trafficking. Dev Cell. 11:655–669. [DOI] [PubMed] [Google Scholar]
- Jaworski J, Spangler S, Seeburg DP, Hoogenraad CC, Sheng M. 2005. Control of dendritic arborization by the phosphoinositide-3′-kinase-Akt-mammalian target of rapamycin pathway. J Neurosci. 25:11300–11312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Ehlers MD. 2013. Modeling autism by SHANK gene mutations in mice. Neuron. 78:8–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaidanovich-Beilin O, Lipina T, Vukobradovic I, Roder J, Woodgett JR. 2011. Assessment of social interaction behaviors. J Vis Exp. (48):e2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazdoba TM, Hagerman RJ, Zolkowska D, Rogawski MA, Crawley JN. 2015. Evaluation of the neuroactive steroid ganaxolone on social and repetitive behaviors in the BTBR mouse model of autism. Psychopharmacology (Berl). 233:309–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koukoui SD, Chaudhuri A. 2007. Neuroanatomical, molecular genetic, and behavioral correlates of Fragile X syndrome. Brain Res Rev. 53:27–38. [DOI] [PubMed] [Google Scholar]
- Kwon CH, Luikart BW, Powell CM, Zhou J, Matheny SA, Zhang W, Li Y, Baker SJ, Parada LF. 2006. Pten regulates neuronal arborization and social interaction in mice. Neuron. 50:377–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon C-H, Zhu X, Zhang J, Baker SJ. 2003. mTor is required for hypertrophy of Pten-deficient neuronal soma in vivo. Proc Natl Acad Sci USA. 100:12923–12928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim ET, Raychaudhuri S, Sanders SJ, Stevens C, Sabo A, MacArthur DG, Neale BM, Kirby A, Ruderfer DM, Fromer M et al. 2013. Rare complete knockouts in humans: population distribution and significant role in autism spectrum disorders. Neuron. 77:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longair M, Baker D, Armstrong J. 2011. Simple Neurite Tracer: open source software for reconstruction, visualization and analysis of neuronal processes. Bioinformatics. 27:2453–2454. [DOI] [PubMed] [Google Scholar]
- Lugo JN, Smith GD, Arbuckle EP, White J, Holley AJ, Floruta CM, Ahmed N, Gomez MC, Okonkwo O. 2014. Deletion of PTEN produces autism-like behavioral deficits and alterations in synaptic proteins. Front Mol Neurosci. 7:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin S, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR et al. 2010. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 13:133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzini M, Xiong L, Shaheen R, Tambunan D, Costanzo S, Mitisalis V, Tischfield D, Cinquino A, Ghaziuddin M, Christian M et al. 2014. CC2D1A regulates human intellectual and social function as well as NF-κB signaling homeostasis. Cell Rep. 8:647–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Harris B. 2008. Clinical implications of respiratory-swallowing interactions. Curr Opin Otolaryngol Head Neck Surg. 16:194–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng J, Meng Y, Hanna A, Janus C, Jia Z. 2005. Abnormal long-lasting synaptic plasticity and cognition in mice lacking the mental retardation gene Pak3. J Neurosci. 25:6641–6650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morawa KS, Schneider M, Klein T. 2015. Lgd regulates the activity of the BMP/Dpp signalling pathway during Drosophila oogenesis. Development. 142:1325–1335. [DOI] [PubMed] [Google Scholar]
- Moy SS, Nadler JJ, Perez A, Barbaro RP, Johns JM, Magnuson TR, Piven J, Crawley JN. 2004. Sociability and preference for social novelty in five inbred strains: an approach to assess autistic-like behavior in mice. Genes Brain Behav. 3:287–302. [DOI] [PubMed] [Google Scholar]
- Nakamura A, Naito M, Tsuruo T, Fujita N. 2008. Freud-1/Aki1, a novel PDK1-interacting protein, functions as a scaffold to activate the PDK1/Akt pathway in epidermal growth factor signaling. Mol Cell Biol. 28:5996–6009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli E, Ross-Inta C, Wong S, Hung C, Fujisawa Y, Sakaguchi D, Angelastro J, Omanska-Klusek A, Schoenfeld R, Giulivi C. 2012. Mitochondrial dysfunction in Pten haplo-insufficient mice with social deficits and repetitive behavior: Interplay between Pten and p53. PLoS One. 7:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Roak B, Vives L, Fu W, Egertson J, Stanaway I, Phelps I, Carvill G, Kumar A, Lee C, Ankenman K et al. 2012. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 338:1619–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X-M, Lemonde S, Jafar-Nejad H, Bown CD, Goto A, Rogaeva A, Albert PR. 2003. Freud-1: a neuronal calcium-regulated repressor of the 5-HT1A receptor gene. J Neurosci. 23:7415–7425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzes P, Cahill ME, Jones KA, VanLeeuwen JE, Woolfrey KM. 2011. Dendritic spine pathology in neuropsychiatric disorders. Nat Neurosci. 14:285–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan K, McDermid HE. 2012. The 22q13.3 deletion syndrome (Phelan-McDermid Syndrome). Mol Syndromol. 2:186–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purpura DP. 1974. Dendritic spine “dysgenesis” and mental retardation. Science. 186:1126–1128. [DOI] [PubMed] [Google Scholar]
- Richardson L, Venkataraman S, Stevenson P, Yang Y, Moss J, Graham L, Burton N, Hill B, Rao J, Baldock RA et al. 2014. EMAGE mouse embryo spatial gene expression database: 2014 update. Nucleic Acids Res. 42:D835–D844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogaeva A, Ou XM, Jafar-Nejad H, Lemonde S, Albert PR. 2007. Differential repression by Freud-1/CC2D1A at a polymorphic site in the dopamine-D2 receptor gene. J Biol Chem. 282:20897–20905. [DOI] [PubMed] [Google Scholar]
- Rogers DC, Peters J, Martin JE, Ball S, Nicholson SJ, Witherden AS, Hafezparast M, Latcham J, Robinson TL, Quilter CA et al. 2001. SHIRPA, a protocol for behavioral assessment: validation for longitudinal study of neurological dysfunction in mice. Neurosci Lett. 306:89–92. [DOI] [PubMed] [Google Scholar]
- Scattoni ML, Ricceri L, Crawley JN. 2011. Unusual repertoire of vocalizations in adult BTBR T+tf/J mice during three types of social encounters. Genes Brain Behav. 10:44–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods. 9:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeisser MJ, Ey E, Wegener S, Bockmann J, Stempel AV, Kuebler A, Janssen A-L, Udvardi PT, Shiban E, Spilker C et al. 2012. Autistic-like behaviours and hyperactivity in mice lacking ProSAP1/Shank2. Nature. 486:256–260. [DOI] [PubMed] [Google Scholar]
- Sharma K, Schmitt S, Bergner CG, Tyanova S, Kannaiyan N, Manrique-Hoyos N, Kongi K, Cantuti L, Hanisch U-K, Philips M-A et al. 2015. Cell type– and brain region–resolved mouse brain proteome. Nat Neurosci. 18:1819–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibilia M, Wagner EF. 1995. Strain-dependent epithelial defects in mice lacking the EGF receptor. Science. 269:234–238. [DOI] [PubMed] [Google Scholar]
- Szewczyk B, Albert PR, Rogaeva A, Fitzgibbon H, May WL, Rajkowska G, Miguel-Hidalgo JJ, Stockmeier CA, Woolverton WL, Kyle PB et al. 2010. Decreased expression of Freud-1/CC2D1A, a transcriptional repressor of the 5-HT1A receptor, in the prefrontal cortex of subjects with major depression. Int J Neuropsychopharmacol. 13:1089–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashima S, Becker L, Armstrong D, Chan F. 1981. Abnormal neuronal development in the visual cortex of the human fetus and infant with Down's syndrome. A quantitative and qualitative Golgi study. Brain Res. 225:1–21. [DOI] [PubMed] [Google Scholar]
- Thomas A, Burant A, Bui N, Graham D, Yuva-Paylor LA, Paylor R. 2009. Marble burying reflects a repetitive and perseverative behavior more than novelty-induced anxiety. Psychopharmacology (Berl). 204:361–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Threadgill DW, Dlugosz AA, Hansen LA, Tennenbaum T, Lichti U, Yee D, LaMantia C, Mourton T, Herrup K, Harris RC. 1995. Targeted disruption of mouse EGF receptor: effect of genetic background on mutant phenotype. Science. 269:230–234. [DOI] [PubMed] [Google Scholar]
- Tronche F, Kellendonk C, Kretz O, Gass P, Anlag K, Orban PC, Bock R, Klein R, Schütz G. 1999. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat Genet. 23:99–103. [DOI] [PubMed] [Google Scholar]
- Troost T, Jaeckel S, Ohlenhard N, Klein T. 2012. The tumour suppressor Lethal (2) giant discs is required for the function of the ESCRT-III component Shrub/CHMP4. J Cell Sci. 125:763–776. [DOI] [PubMed] [Google Scholar]
- Tsien J, Chen D, Gerber D, Tom C, Mercer E, Anderson D, Mayford M, Kandel ER, Kandel E, Tonegawa S. 1996. Subregion- and cell type-restricted gene knockout in mouse brain. Cell. 87:1317–1326. [DOI] [PubMed] [Google Scholar]
- Vorhees C, Williams M. 2006. Morris water maze: procedures for assessing spatial and related forms of learning and memory. Nat Protoc. 1:848–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waite KA, Eng C. 2002. Protean PTEN: form and function. Am J Hum Genet. 70:829–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, McCoy P, Rodriguiz R, Pan Y, Je H, Roberts AC, Kim C, Berrios J, Colvin J, Bousquet-Moore D et al. 2011. Synaptic dysfunction and abnormal behaviors in mice lacking major isoforms of Shank3. Hum Mol Genet. 20:3093–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wöhr M. 2014. Ultrasonic vocalizations in Shank mouse models for autism spectrum disorders: detailed spectrographic analyses and developmental profiles. Neurosci Biobehav Rev. 43:199–212. [DOI] [PubMed] [Google Scholar]
- Won H, Lee H-R, Gee HY, Mah W, Kim J-I, Lee J, Ha S, Chung C, Jung ES, Cho YS et al. 2012. Autistic-like social behaviour in Shank2-mutant mice improved by restoring NMDA receptor function. Nature. 486:261–265. [DOI] [PubMed] [Google Scholar]
- Yang M, Bozdagi O, Scattoni M, Wohr M, Roullet F, Katz A, Abrams D, Kalikhman D, Simon H, Woldeyohannes L et al. 2012. Reduced excitatory neurotransmission and mild autism-relevant phenotypes in adolescent Shank3 null mutant mice. J Neurosci. 32:6525–6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu TW, Chahrour MH, Coulter ME, Jiralerspong S, Okamura-Ikeda K, Ataman B, Schmitz-Abe K, Harmin DA, Adli M, Malik AN et al. 2013. Using whole-exome sequencing to identify inherited causes of autism. Neuron. 77:259–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Li X-D, Chen Z. 2010. CC2D1A, a DM14 and C2 domain protein, activates NF-kappaB through the canonical pathway. J Biol Chem. 285:24372–24380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Raingo J, Chen Z, Kavalali E. 2011. Cc2d1a, a C2 domain containing protein linked to nonsyndromic mental retardation, controls functional maturation of central synapses. J Neurophysiol. 105:1506–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.