Abstract
Background
Tumour budding is an important prognostic factor in colorectal cancer (CRC). Molecular profiling of tumour buds suggests (partial) epithelial–mesenchymal transition and cancer stem-cell phenotype, similarly described in the “mesenchymal” Consensus Molecular Subtype 4 (CMS4), which identifies a particularly poor prognostic subgroup. Here, we determine the association of tumour budding with CMS classification, prognosis, and response to therapy.
Methods
AMC-AJCCII-90 cohort (n = 76, stage II) was evaluated for peritumoural budding on H&E slides. LUMC (n = 270, stage I–IV), CAIRO (n = 504, metastatic CRC) and CAIRO2 (n = 472, metastatic CRC) cohorts were investigated for intratumoural budding using pan-cytokeratin-stained tissue microarrays. Budding was scored as count/area, then classified as <5 or ≥5 buds. For all cohorts, CMS classifications were available (gene-expression/immunohistochemistry-based classifiers).
Results
High (≥5) budding predicted a worse outcome in multivariate analysis in AMC-AJCCII-90 (p = 0.018), LUMC (p < 0.0001), and CAIRO (p = 0.03), and in CAIRO2 (continuous variable, p = 0.02). Tumour budding counts were higher in CMS4 compared to epithelial CMS2/3 cancers (p < 0.01, all), and associated with KRAS/BRAF mutations (p < 0.01, AMC-AJCCII-90, CAIRO, CAIRO2).
Conclusion
Tumour budding is an adverse prognostic factor across all CRC stages and is associated with the mesenchymal CMS4 phenotype. KRAS/BRAF mutations are strongly correlated with tumour budding suggesting their involvement in the regulation of this process.
Subject terms: Biomarkers, Cancer
Introduction
Tumour budding in colorectal cancer is now established as a strong, independent prognostic factor.1 It is associated with advanced stage of disease as well as shorter disease-free survival (DFS) and overall survival (OS).2 The International Tumour Budding Consensus Conference (ITBCC) underlined the importance of this prognostic parameter and, as a consequence, this histologic feature has recently been added to both the 8th edition of the TNM staging manual and CAP guidelines.
Tumour budding is described as the presence of single tumour cells or small tumour cell clusters dissociated from the main tumour body. The frequency of tumour budding is approximately 40% but varies significantly in the reported literature due to methodological and scoring differences.2 Although typically described as occurring at the invasive front (peritumoural budding, PTB) and detected in surgical resections of colorectal cancer, the presence of tumour budding within the main tumour body (intratumoural budding, ITB) has also been described.1 This latter observation has highlighted a clinically important role for tumour budding in the pre-operative management of patients with rectal cancers since the presence of ITB can also be detected in pre-treatment biopsies.
Tumour buds are thought to have the properties of cells undergoing an epithelial–mesenchymal transition (EMT), or a partial-EMT state.3 In fact, laser capture microdissection studies in both colorectal cancers and oral squamous cell carcinomas show that tumour buds over-express EMT-related genes such as ZEB1, ZEB2, DES, TGFB3, and VIM in comparison to the main tumour mass.4 At the protein level, frequent loss of E-cadherin and β-catenin from the membrane can be observed, not only in colorectal cancer but also in pancreatic ductal adenocarcinomas and oral squamous cell carcinoma, indicating that activation of both WNT and TGF-β signalling may be occurring.4–6 In addition to reduced proliferation and apoptosis in these cells, tumour buds are reported as having an overexpression of genes involved in extracellular matrix degradation, invasion, and migration and may co-express both vimentin (a mesenchymal marker) and cytokeratin (an epithelial marker), again in line with a transitory EMT-like phenotype.3,7
In 2015, Guinney, following a large collaborative effort to which we have contributed, published a widely adapted comprehensive classification of colorectal cancer and identified four distinct subtypes.8 The first subtype, CMS1, included predominantly hypermethylated cancers with microsatellite instability (MSI), BRAF mutation, and a rich immune cell infiltrate. The second subtype, CMS2, was identified as the canonical colorectal cancer type with WNT pathway activation and chromosomal instability, while CMS3 encompassed tumours with profound metabolic deregulation and frequent KRAS mutations. The fourth group called “mesenchymal”, is found in 23% of all cases and characterised by stromal infiltration, overexpression of genes involved in EMT and TGF-β activation, matrix remodeling, and angiogenesis among others. Prognostically, the CMS4 group exhibited the worst OS and DFS demonstrating an urgent clinical need to develop therapies that would be effective for this subtype.
In addition, we recently established an immunohistochemistry-based tissue microarray classifier based on five protein markers (CDX2, FRMD6, HTR2B, ZEB1, and pan-cytokeratin) to determine patients’ CMS in cohorts where gene-expression profiling is unavailable.9 The accuracy of the classifier was found to be 87% in discriminating between the epithelial (CMS2/3) and mesenchymal (CMS4) groups. Moreover, this classifier distinguished patients with the worst OS and DFS, validating the CMS4 subtype.
Taken together, the molecular and prognostic profile of CMS4 patients is reminiscent of a high-grade tumour budding phenotype, and we speculated an association between tumour cell budding and the mesenchymal colorectal cancer subtype. To test this hypothesis, we investigated the association between tumour budding, CMS groups, and prognosis in four cohorts including patients from a series of stage II patients (AMC-AJCCII-90), a stage I–IV cohort (LUMC), and two randomised clinical trials of metastatic colorectal cancer (CAIRO and CAIRO2).
Material and methods
Patient cohorts
Four independent patient cohorts were used in this study: the AMC-AJCCII-90 series collected between 1997 and 2006,10 LUMC collected between 1991 and 2001 following surgical intervention,11 the CAIRO trial enrolling patients between 2003 and 2004 (Trial Registration ID: NCT00312000),12 and the CAIRO2 trial enrolling patients between 2005 and 2006 (Trial Registration ID: NCT00208546)13 series. The CAIRO trial assesses the efficacy of sequential compared to combination treatment with capecitabine, irinotecan, and oxaliplatin. The CAIRO2 trial assesses the added benefit of the anti-EGFR agent cetuximab to a regimen of capecitabine, oxaliplatin, and bevacizumab.
Patient sample specimens and clinical information were obtained and processed using IRB-approved protocols. The gold standard CMS classification based on microarray (Affymetrix) gene expression data was available for the AMC-AJCCII-90 cohort, while LUMC, CAIRO, and CAIRO2 were classified into CMS groups using an IHC-based tissue microarray classifier described in ref. 9
Patients were excluded from this study due to the following criteria: (i) insufficient patient material for tissue microarray or H&E slide construction; (ii) poor quality or missing tissue microarray cores; and/or (iii) incomplete clinical records, as summarised in Supplementary Figure 1.
Tissue microarray construction
Three 0.6 mm cores were included in the construction of the tissue microarray for the LUMC cohort whereas for CAIRO and CAIRO2 cohorts, tissues were mounted onto single-punch tissue microarrays of 2.0 mm in diameter. Tissue microarray construction is described in detail in Trinh et al.9 Slides were sectioned at 4 µm and immunohistochemistry for cytokeratin AE1/AE3 (1:500; cat M351529-2 Aligent) was performed. After a secondary incubation with anti-rabbit-HRP or anti-mouse-HRP (Powervision), staining was developed using DAB+ Chromogen (Dako) and counterstained with haematoxylin. For the AMC-AJCCII-90 cohort, whole slide tumour sections were cut at 4 µm and after preparation and antigen retrieval (sodium citrate buffer, pH 6.0) of the formalin fixed paraffin embedded (FFPE) sections they were stained with haematoxylin (Mayer) and eosin Y (Sigma).
KRAS/BRAF mutation detection
KRAS and BRAF mutation status was determined in the AMC-AJCCII-90 series and CAIRO2 cohort by PCR of the V600E site in BRAF and codons 12 and 13 of KRAS; and validated by Sanger sequencing of exon 2 of KRAS and exon 15 in BRAF as previously reported.13–15 BRAF status in the CAIRO cohort was determined by high resolution melting sequencing analysis16 and KRAS status was determined by next-generation sequencing using the TruSeq Amplicon Cancer Panel.17 KRAS and BRAF mutation data were not available for the LUMC cohort.
BRAF and KRAS mutation testing was performed in the CAIRO, CAIRO2 cohorts only after the completion of these trials for scientific purposes, for example following the discovery that patients with mutations in the KRAS/BRAF axis are refractory to anti-EGFR therapy, and for comparison to other putative (genetic) biomarkers of therapy response.18
Quantifying tumour budding
Peritumoural budding (PTB) using ITBCC criteria
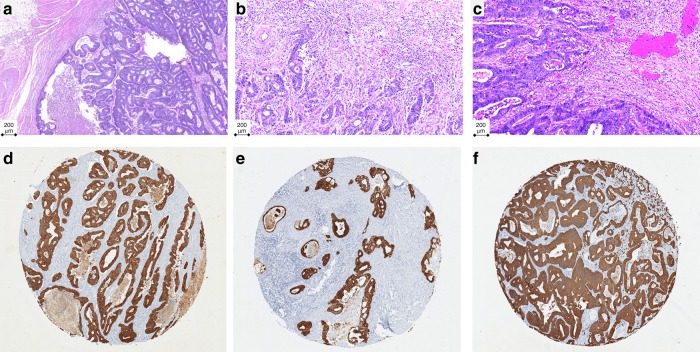
Tumour budding was assessed in the AMC-AJCCII-90 cohort according to recommendations of the ITBCC on scanned haematoxylin and eosin (H&E) stained slides.1 One representative slide of tumour per case was available for scoring. Tumour buds were counted in a single circular hotspot measuring 0.785 mm2 at the invasive front. Both tumour budding count and tumour budding category, namely BD1 (0–4 buds/0.785 mm2), BD2 (5–9 buds/0.785 mm2), and BD3 (≥10 buds/0.785 mm2) were recorded. In three cases, tumour budding could not be assessed as the selected slide contained only mucinous and/or signet ring cell formations. Representative areas of the three ITBCC budding categories are shown in Fig. 1a–c.
Fig. 1.
Representative images of tumour budding on H&E slides depicting ITBCC classifications. a BD1 (0–4 buds/0.785 mm2), b BD2 (5–9 buds/0.785 mm2), and c BD3 (≥10 buds/0.785 mm2), and pan-cytokeratin-stained slides representing (d) absence, e low-grade budding (<5 buds), and f high-grade budding (≥5 buds)
Intratumoural budding (ITB)
Pan-cytokeratin-stained tissue microarrays were used to assess tumour budding in the LUMC, CAIRO, and CAIRO2 cohorts by means of ITB. ITB was defined as one single cell or a cell cluster of up to five tumour cells each containing a distinct nucleus and surrounded by tumour stroma.2,19 Areas of necrosis were avoided. Punches with more than 40% missing area were removed from the analysis. For patients with multiple cores, the average number of buds across all viable samples was used. Representative pan-cytokeratin stained tissue microarray punches showing low- and high-grade tumour budding are shown in Fig. 1d–f.
Statistics
Descriptive statistics were performed for tumour budding on all cohorts. ITB was assessed both as a categorical variable (using a threshold of 5) and a continuous variable represented in the log10 scale. Differences in the number of tumour buds by CMS groups or KRAS/BRAF mutation were analysed using the Wilcoxon’s Rank Sum Test. OS and DFS analyses were performed using univariate and multivariate Cox proportional hazards models accounting for age, stage, and sex with 60 months (5-year) follow-up, after which samples were right censored. Differences in survival were expressed as hazard ratios (HR) with 95% confidence intervals. Significance was tested using the log-rank test. Survival curves were constructed using the Kaplan–Meier method. All p-values < 0.05 were considered statistically significant.
Results
Summary of patient cohorts and tumour budding counts
The association of tumour budding with prognosis and CMS subtypes was performed using four different patient cohorts of various TNM stages and using different methods.
The AMC-AJCCII-90 series (n = 76) included only patients with stage II disease. PTB was evaluated on H&E slides according to the ITBCC guidelines (number of buds followed by categorisation into BD1 < 5 buds versus BD2 + BD3 ≥ 5 buds), in a hotspot of 0.785 mm2. The average budding count was six buds (std ±8), with a median of three buds per tumour. Each tumour in the AMC-AJCCII-90 cohort was previously classified into one of the four CMS subtypes based on gene expression profiles.8
The LUMC cohort included patients (n = 268) with TNM stages I–IV. Tumours were mounted onto 0.6 mm diameter, multi-core tissue microarrays. ITB was assessed after pan-cytokeratin immunohistochemistry. The average ITB count was four (std ±9) buds per punch, with a median of zero buds per tumour. Results from the IHC-based tissue microarray classifier were used to determine the CMS of each case.
In the CAIRO (n = 504) and CAIRO2 (n = 472) cohorts, all patients had metastatic cancers (stage IV). Colorectal cancers from both trials were included onto 2.0 mm diameter tissue microarrays stained for pan-cytokeratin. Average ITB budding counts were 30 (std ±40) and 44 buds (std ±72) in CAIRO and CAIRO2, respectively, with median counts of 15 and 18. The IHC-based tissue microarray classifier results were used to assign CMS groups.9
Patient characteristics of all four cohorts are provided in Table 1.
Table 1.
Summary of patient characteristics
| Feature | CAIRO | LUMC | CAIRO2 | AMC-AJCCII-90 | |
|---|---|---|---|---|---|
| No. of patients | 504 | 270 | 472 | 76 | |
| Sample information | TMA or slide | TMA | TMA | TMA | Whole slide |
| Core diameter | 2 mm | 0.6 mm | 2 mm | – | |
| Staining | CK | CK | CK | H&E | |
| Age (years) | 62.8 (41–78) | 65.4 (39.3–86.4) | 61.3 (40.3–76) | – | |
| Sex | F | 181 | 135 | 197 | 40 |
| M | 322 | 135 | 275 | 36 | |
| Stage | 1 | 0 | 43 | 0 | 0 |
| 2 | 0 | 99 | 0 | 76 | |
| 3 | 0 | 75 | 0 | 0 | |
| 4 | 504 | 50 | 472 | 0 | |
| Event status | None | 61 | 91 | 104 | 60 |
| Yes | 443 | 179 | 368 | 16 | |
| Treatment arma | A | 252 | – | 243 | – |
| B | 252 | – | 229 | – | |
| Mutational information | |||||
| MSI | MSI-H | 12 | 29 | 8 | 23 |
| MSS/MSI-L | 423 | 189 | 464 | 53 | |
| KRAS/BRAF | mut | 174 | – | 218 | 31 |
| wt | 139 | – | 227 | 45 | |
| CMS classification | |||||
| CMS classifier | IHC | IHC | IHC | GeneExp | |
| CMS available | 401 | 214 | 340 | 76 | |
| Subtype | CMS2/3 | 247 | 96 | 162 | 35 |
| CMS4 | 142 | 89 | 170 | 15 | |
TMA tissue microarray, CK pan-cytokeratin, H&E haematoxylin and eosin, MSI microsatellite instability, CMS Consensus Molecular Subtype
aTreatment arms: CAIRO—sequential (Arm A) versus combination (Arm B) capecitabine, irinotecan, and oxaliplatin. CAIRO2—capecitabine, oxaliplatin, and bevacizumab (Arm A) with cetuximab (Arm B)
Prognostic effect of tumour budding
The association of tumour budding with DFS (LUMC, AMC-AJCCII-90) or OS (CAIRO, CAIRO2) was investigated in two ways. The first analysis was based on dichotomizing budding counts into <5 and ≥5 buds for PTB in the AMC-AJCCII-90 cohort and a proposed cut-off from the literature for ITB for the remaining tissue microarray cohorts,19 while the second was based on the raw tumour budding counts in each cohort.
High tumour budding (>5 buds) is associated with poor disease-free and overall survival
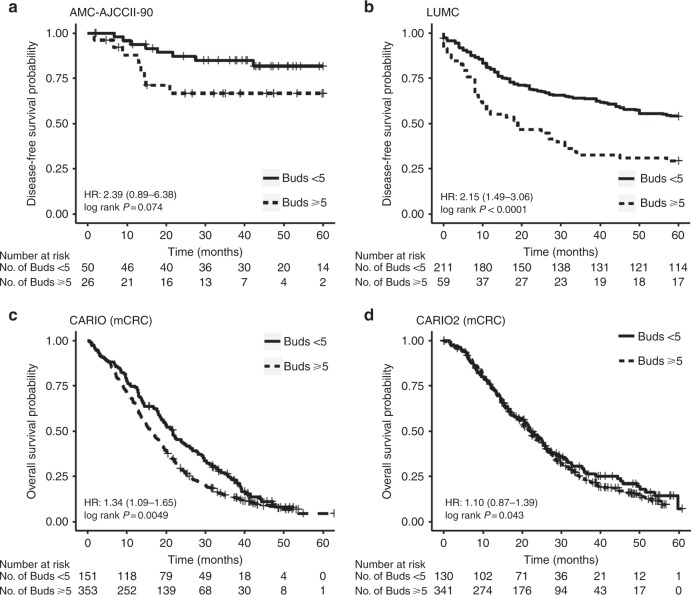
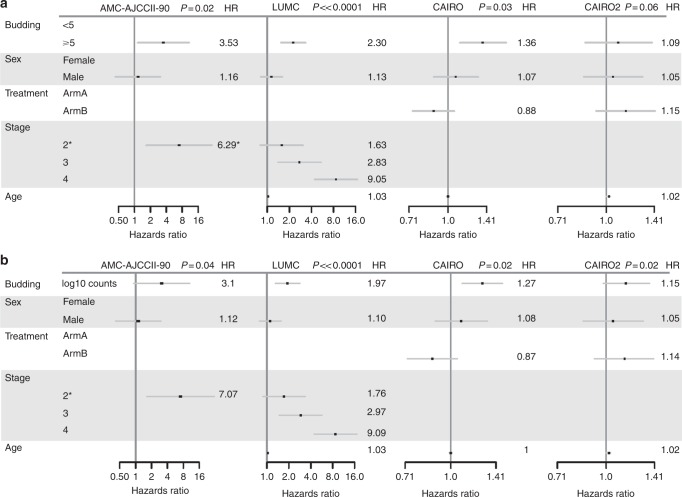
Since the survival time of patients with BD2 and BD3 cancers was similar and due to the small number of patients in each BD group, we combined BD2 + BD3 into a single category for analysis. High tumour budding was associated with poor DFS in both the AMC-AJCCII-90 and LUMC cohorts. PTB in the AMC-AJCCII-90 cohort shows a worse DFS in patients with high-grade budding although the difference is not statistically significant (HR: 2.39 (0.89–6.38), p = 0.074; Fig. 2a). However, adjusting for age, sex, and T stage, a significant worse and independent effect on outcome in this cohort for patients with high-grade budding was observed (3.53 (1.15–10.83), p = 0.02, Fig. 3a).
Fig. 2.
Kaplan–Meier curves representing survival time differences across the different cohorts with a cut-off of five tumour buds for low- or high-grade budding. a AMC-AJCCII-90, b LUMC, c CAIRO, and d CAIRO2. Differences in survival are reported as univariate hazard ratios with 95% confidence intervals, and p values were calculated using the log-rank method
Fig. 3.
Forest plots showing hazard ratios (HR) and 95% confidence intervals from multivariable Cox regression analysis across all cohorts using tumour budding as a a threshold of 5 buds and b as a continuous variable. Note the AMC-AJCCII-90 analysis compares the effects for stage 2A (reference level) and 2B
Similarly, univariate analysis of high- versus low-grade ITB in the LUMC cohort demonstrated a significantly worse DFS (HR: 2.15 (1.49–3.09); p < 0.0001; Fig. 2b), which again was maintained in multivariable analysis adjusting for sex, age, and TNM stage (HR: 2.30 (1.56–3.38), p ≪ 0.0001) (Fig. 3a). In fact, patients with high-grade budding exhibited a 2.3 times greater relative risk of recurrence as compared to patients with low-grade budding, indicating that tumour budding is an independent prognostic factor in this cohort.
However, the relation between ITB and disease outcome was variable in the metastatic setting. In the CAIRO cohort, treatment arms were combined and the association between budding OS time was analysed. OS time was significantly worse in patients with high-grade budding within the primary tumour in univariate analysis (HR: 1.34 (1.09–1.65); p = 0.0049; Fig. 2c), with a median time to death decreasing from 21.7 (18.6–25.9) months to 16.2 (14.6–18.2) months. In a multivariate analysis that included age, sex, and treatment arm, tumour budding maintained its effect on prognosis, namely, patients with high-grade budding had a worse outcome compared to patients with low-grade budding tumours (HR: 1.36 (1.11–1.68), p = 0.03 (Fig. 3a)). In CAIRO2, no association between tumour budding and survival could be found (Figs. 2d, 3a).
Modeling tumour budding as a continuous variable
Evaluating the number of tumour buds and DFS or OS, a significant association was found between a higher number of tumour buds and worse outcome in all four cohorts in Cox models adjusted for age, sex, and TNM stage (Fig. 3b).
The AMC-AJCCII-90 cohort showed a trend of higher budding counts associated with worse DFS in a multivariate model adjusted for age, sex, and T stage (HR: 3.1 (0.92–10.52), p = 0.04). Similarly, for the LUMC cohort, a higher number of tumour buds was found to have prognostic significance when analysed in a multivariable model adjusting for sex and TNM stage (HR: 1.97 (1.35–2.87); p ≪ 0.0001).
A similar association between budding and OS in metastatic colorectal cancer was observed in the CAIRO cohort in multivariate analysis (HR: 1.27 (1.09–1.48); p = 0.02) adjusting for sex and age. In CAIRO2, a significant impact of tumour budding on OS was found in multivariate analysis taking into account age and sex (HR: 1.15 (0.98–1.36), p = 0.02) (Fig. 3b).
Predictive effect of tumour budding to different treatment
We sought to determine whether ITB had any predictive value in the metastatic setting through the retrospective analysis of the CAIRO and CAIRO2 clinical trials.
The CAIRO clinical trial assessed the efficacy of sequential versus combination chemotherapy involving capecitabine, irinotecan, and oxaliplatin. Stratifying patients by treatment regimen in the CAIRO cohort showed that a high-grade tumour budding phenotype was found to correlate with worse prognosis both in the sequential treatment arm (Arm A) and the combination arm (Arm B) in univariate analysis using a cut-off of 5 (Arm A: HR = 1.4 (1.06–1.87), p = 0.02; Arm B: HR: 1.34 (0.99–1.81), p = 0.06) (Supplemental Figure 2A). However, modeling budding as a continuous variable shows a prognostic effect only in Arm A (Arm A: HR = 1.38 (1.1–1.72), p = 0.04; Arm B: HR = 1.20 (0.97–1.47), p = 0.23) (Supplemental Figure 2B). These data suggest that ITB is an unfavourable prognostic factor in both treatment arms, but does not have predictive value.
Similarly, we stratified the CAIRO2 cohort according to whether patients were treated with the anti-EGFR agent cetuximab in addition to a baseline regimen of capecitabine, oxaliplatin, and bevacizumab. Patients with cetuximab therapy did not differ in prognosis when tumours were stratified by budding status using a cut-off of 5 (Supplemental Figure 2C), but showed a trend of increased budding counts associated with poorer prognosis in the cetuximab arm in multivariate analysis including sex and age (HR: 1.25 (0.98–1.58), p = 0.1) (Supplemental Figure 2D). This association was however lost when taking into account KRAS and BRAF status (HR budding: 1.21 (0.83–1.75), p = 0.15). These results indicate that tumour budding is not a predictive factor of response to anti-EGFR therapy in RAS/RAF wild-type patients.
Association with CMS and molecular features
We next determined the association of tumour budding within CMS subgroups, focusing on the difference in tumour buds between the epithelial-like CMS2/3 subtypes versus the mesenchymal-like CMS4 group.
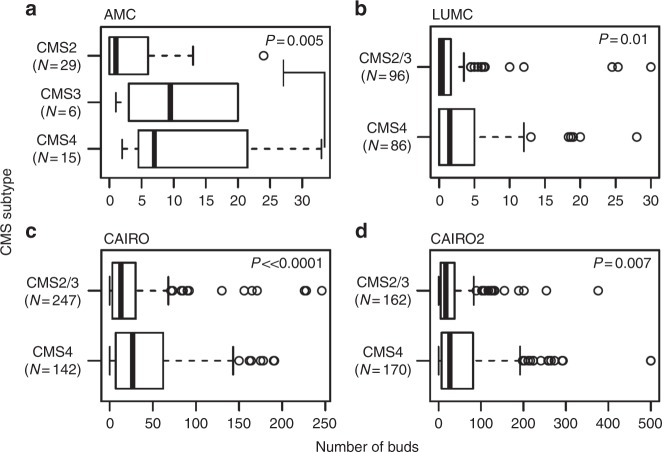
In all four cohorts, there was a significantly greater number of tumour buds identified in the CMS4 subtype compared to CMS2/3, namely in AMC-AJCCII-90 (mean: 11.5 versus 4.4 buds; p = 0.005), LUMC (mean: 3.84 versus 2.1 buds; p = 0.01), CAIRO (mean: 41.4 versus 23.9 buds; p « 0.0001), and CAIRO2 (mean: 59.6 versus 32.5 buds; p = 0.007) (Fig. 4).
Fig. 4.
Boxplots showing the distribution of tumour budding counts by CMS subtype in the a AMC-AJCCII-90, b LUMC, c CAIRO, and d CAIRO2 cohort. p-Values from Wilcoxon Rank Sum Test indicate a significant association between higher budding counts and the CMS4 subtype in all cohorts
Given that budding was not shown to be predictive for cetuximab therapy, we also sought to determine whether this could be due to an association between budding and mutational status in the KRAS/BRAF axis.
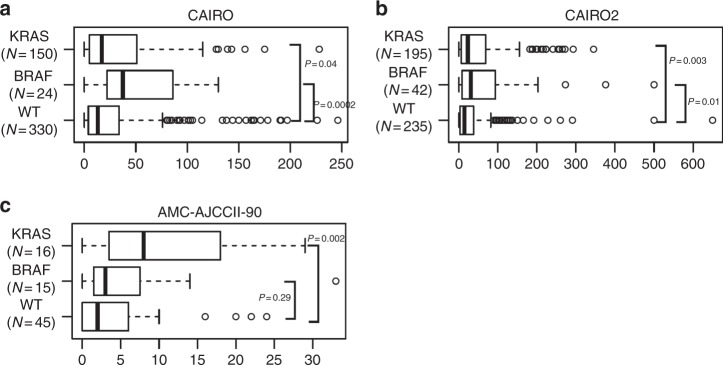
Interestingly, a significant correlation was found between a higher number of tumour buds and KRAS mutation in all three cohorts (AMC-AJCCII-90 p = 0.002; CAIRO2 p = 0.003; CAIRO p = 0.04, Wilcoxon Rank Sum Test). Additionally, higher ITB was observed in BRAF mutant metastatic samples (CAIRO p = 0.0002; CAIRO2 p = 0.01, Wilcoxon Rank Sum Test) (Fig. 5).
Fig. 5.
Boxplots showing the distribution of tumour budding counts by mutational status in the a CAIRO, b CAIRO2, and c AMC-AJCCII-90 cohort. p-Values from Wilcoxon Rank Sum Test indicate a significant association between higher budding counts and the KRAS/BRAF mutation in all cohorts
Discussion
In this study, we investigated the prognostic effect of tumour budding on 1320 patients with colorectal cancers across various cohorts, including two randomised clinical trials for metastatic patients. In addition, we evaluated the association of tumour budding with the CMS in order to test the hypothesis that the “mesenchymal-like” subtype (CMS4), which shares similarities in molecular profile with tumour buds and their microenvironment, would be associated with a high-grade tumour budding phenotype.
We validated the prognostic effect of tumour budding independent of age, stage, and sex in three of the four cohorts. It is known that a higher number of tumour buds is correlated with worse survival in patients with colorectal cancer. Evidence suggests that regardless of scoring method, the presence of tumour budding is associated with advanced disease stage and worse clinical outcome.2,20,21 There are, however, several new elements in this study. Firstly, we can show using a randomised clinical trial of metastatic patients (CAIRO) that tumour budding is indeed prognostic in this setting. Secondly, we could validate the newly established ITBCC criteria for the scoring of tumour budding on H&E slides in a stage II cohort with DFS as the endpoint. The cutoff proposed by the ITBCC (BD1, BD2, BD3) is based on the scoring method already integrated into the Japanese Guidelines for the reporting of colorectal cancer. A recent retrospective study by Oh and colleagues pooled results from more than 4000 Japanese patients from all stages and validated the effect of high-grade budding on worse OS and DFS.22 However, in contrast to this study, Oh and colleagues evaluated budding using a three-tiered system (equivalent to BD1, BD2, and BD3) and only found an association between BD1/2 versus BD3 and outcome. Thirdly, this study underlines the value of ITB as a prognostic factor in both the metastatic and mixed stage cohorts, since counts were evaluated on tissue microarrays constructed without specific regard to the localisation (centre/invasion front) within the tumour. Although ITB rather than PTB could be evaluated in three of the four cohorts here, both ITB and PTB counts are highly correlated when ITB is present.19 It should be mentioned that differences in methodology regarding ITB counts have been described. Lugli et al. used CK22 staining and counted buds in multi-punch tissue microarrays, taking the maximum number of buds as the final count, whereas in this study AE1/AE3 staining was performed and the average number of buds was taken. Unfortunately, false-negatives, namely cases which exhibit PTB but no ITB, may occur and this could be a part of the explanation for the lack of correlation between tumour budding and outcome in the CAIRO2 cohort in multivariable analysis. In addition, the distribution of budding counts in this cohort suggests that a cutoff of five buds is not optimal for this setting, as we demonstrated a prognostic effect when ITB was assessed on a continuous scale. Despite the different composition of cohorts and the various methods used for assessing tumour budding including tissue microarrays, whole slides as well as H&E or pan-cytokeratin, tumour budding is validated here as an independent prognostic factor.
Additionally, we evaluated the association between tumour budding and the CMS taxonomy. For the AMC-AJCCII-90 series, assignment of CMS was performed using gene expression,8 while the IHC-based classifier was used to assign CMS classes to the tumours from the CAIRO, CAIRO2, and LUMC cohorts.9 Indeed, we found in all four cohorts a significantly greater number of tumour buds in the CMS4 compared to CMS2/3. The CMS4 group represents a set of tumours with upregulated expression of mesenchymal and EMT-related genes, as well as those involved with TGF-β signalling, matrix remodeling, stemness, wound healing, and angiogenesis.8 These tumours have both low content of immune cells and low-expression of immune cell activation markers. These findings are in line with the described profiles of tumour buds, or more precisely the microenvironment of tumour buds.3 RNA- or DNA-based studies to date have attempted to analyse molecular profiles of tumour buds.4,6,23 These profiles include mesenchymal, EMT, and cancer stem-like genes consistent with the patterns of the CMS4 group. As they appear as single cells or small cell clusters, it is challenging to perform experiments directly on tumour buds, hence one criticism of this approach is that the expression patterns observed are not strictly from the buds themselves but very likely also from the adjacent stromal microenvironment. It is also known that proteins such as TWIST1 and SNAI1 are only rarely expressed in tumour buds, but are profusely found in cancer-associated fibroblasts in regions of high-grade budding.5,24 We believe that the CMS4 group contains high-grade tumour budding cancers and that the molecular profile of these CMS4 tumours reflects not only the tumour cells themselves, but also the stromal microenvironment in which they reside.
Finally, we observed that the extent of tumour budding was significantly greater in cancers with KRAS and BRAF mutations. The relationship between KRAS mutation and tumour budding has previously been reported.25–27 Jang and colleagues found that 21/34 colorectal cancers with high-grade tumour budding also had mutations in KRAS.28 They encourage the assessment of tumour budding for patients receiving anti-EGFR therapy. Our group has previously investigated progression-free survival in patients treated with anti-EGFR therapy based on tumour budding status.29 We showed not only that tumour budding led to worse clinical outcome in cetuximab-treated patients but that there was an added benefit to KRAS mutation status in terms of identifying possible non-responders. In this study however, we could not show a predictive effect of tumour budding specific to anti-EGFR therapy or chemotherapy regimen, but patients with high-grade budding had worse OS in both sequential or combined chemotherapy groups. Furthermore, Cho et al. present RAS destabilisation as a potential approach for preventing metastatic progression.30 The RAS protein was shown to be significantly more abundant in tumour buds than in other cells in human colorectal cancers. In vitro treatment of KRAS mutated cell lines with KY011, a compound that destabilises β-catenin and RAS proteins, prevented spindle cell morphology as well as E-cadherin loss and vimentin over-expression. Taken together, these results all point toward KRAS mutation as an element regulating the tumour budding phenotype, which deserves exploration in a further study. Of note, KRAS mutations are not acquired in tumour buds as shown in a next-generation sequencing study of driver mutations comparing tumour buds with other regions of the tumour.31
Our study is limited by the use of tissue microarrays for the assessment of tumour budding, since areas selected for tissue microarray construction were selected for epithelial-rich regions rather than targeting regions containing buds. Nonetheless, we could evaluate ITB in these punches and could still show an association of tumour budding with outcome, using single punch 2.0 mm cores or 3 × 0.6 mm cores. Secondly, an IHC-based tissue microarray classifier trained on the AMC-AJCCII-90 cohort had an accuracy of 87%, which itself introduces a misclassification rate of 13% in discriminating CMS groups in the LUMC, CAIRO, and CAIRO2 cohorts. On the other hand, our study was complemented by the AMC-AJCCII-90 cohort, for which gold-standard RNA-based CMS group assignment was available. Moreover, results from all cohorts showed an association between higher number of tumour buds and the mesenchymal subtype.
To conclude, tumour budding is not only an independent and adverse prognostic factor in all stages of colorectal cancer, but is associated with the poor “mesenchymal-like” prognostic group of the consensus molecular subtypes. We believe that the CMS4 group is enriched for high-grade budding cancers, which is also underlined by the similar molecular profiles associated with both tumour budding and CMS4 tumours. In addition, KRAS/BRAF mutation is strongly correlated with tumour budding suggesting that this molecular change may play a role in regulating the budding phenotype.
Electronic supplementary material
Acknowledgements
L.V. was supported by grants from the New York Stem Cell Foundation, the Dutch Cancer Society (KWF; UVA2014-7245 and 0651/2016-2), het Innovatiefonds Zorgverzekeraars, the European Research Council (ERG-StG 638193), and ZonMw (Vidi 016.156.308). L.V. is a New York Stem Cell Foundation—Robertson Investigator. I.Z. was supported by grants from the Johanna-Dürmüller Bol and Mach-Gaensslen Foundation.
Author contributions
I.Z., L.V., A.T., C.L.: study design, C.L., H.D.: carried out experiments, A.T.: performed statistical analysis, I.Z., A.T.: wrote the manuscript, S.T.H., P.J.K.K., M.S.R., M.K., C.J.A.P., A.L.: gave critical inputs, all authors reviewed and approved the final version of the paper.
Competing interests
The authors declare no competing interests.
Data availability:
Analyses were performed using R and code for the analysis can be found at the following github repository (github.com/trinhan/crc-ihc-classification).
Ethics approval and consent to participate
Informed consent was obtained from all patients; and the collection, processing, and use of tissue specimens and clinical information were approved by the ethics committees/institutional review boards at the respective institutions in accordance with the declaration of Helsinki (Leiden University Medical Centre, Committee on research involving human subjects Arnhem-Nijmegen, Academic Medical Center in Amsterdam, Central Committee of Human-related Research).
Footnotes
These authors contributed equally: Anne Trinh, Claudia Lädrach, Louis Vermeulen, Inti Zlobec
Electronic supplementary material
Supplementary information is available for this paper at 10.1038/s41416-018-0230-7.
References
- 1.Lugli A, et al. Recommendations for reporting tumour budding in colorectal cancer based on the International Tumour Budding Consensus Conference (ITBCC) 2016. Mod. Pathol. 2017;30:1299–1311. doi: 10.1038/modpathol.2017.46. [DOI] [PubMed] [Google Scholar]
- 2.Koelzer VH, Zlobec I, Lugli A. Tumour budding in colorectal cancer—ready for diagnostic practice? Hum. Pathol. 2016;47:4–19. doi: 10.1016/j.humpath.2015.08.007. [DOI] [PubMed] [Google Scholar]
- 3.Grigore, A. D., Jolly, M. K., Jia, D., Farach-Carson, M. C. & Levine, H. Tumour budding: the name is EMT. Partial EMT. J. Clin. Med.5, E51 (2016). [DOI] [PMC free article] [PubMed]
- 4.De Smedt L, et al. Expression profiling of budding cells in colorectal cancer reveals an EMT-like phenotype and molecular subtype switching. Br. J. Cancer. 2017;116:58–65. doi: 10.1038/bjc.2016.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galvan JA, et al. Expression of E-cadherin repressors SNAIL, ZEB1 and ZEB2 by tumour and stromal cells influences tumour-budding phenotype and suggests heterogeneity of stromal cells in pancreatic cancer. Br. J. Cancer. 2015;112:1944–1950. doi: 10.1038/bjc.2015.177. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6.Jensen DH, et al. Molecular profiling of tumour budding implicates TGFβ-mediated epithelial–mesenchymal transition as a therapeutic target in oral squamous cell carcinoma. J. Pathol. 2015;236:505–516. doi: 10.1002/path.4550. [DOI] [PubMed] [Google Scholar]
- 7.Kohler I, et al. Detailed analysis of epithelial–mesenchymal transition and tumour budding identifies predictors of long-term survival in pancreatic ductal adenocarcinoma. J. Gastroenterol. Hepatol. 2015;30(Suppl. 1):78–84. doi: 10.1111/jgh.12752. [DOI] [PubMed] [Google Scholar]
- 8.Guinney J, et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 2015;21:1350–1356. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trinh A, et al. Practical and robust identification of molecular subtypes in colorectal cancer by immunohistochemistry. Clin. Cancer Res. 2017;23:387–398. doi: 10.1158/1078-0432.CCR-16-0680. [DOI] [PubMed] [Google Scholar]
- 10.De Sousa EMF, et al. Poor-prognosis colon cancer is defined by a molecularly distinct subtype and develops from serrated precursor lesions. Nat. Med. 2013;19:614–618. doi: 10.1038/nm.3174. [DOI] [PubMed] [Google Scholar]
- 11.Goossens-Beumer IJ, et al. Clinical prognostic value of combined analysis of Aldh1, Survivin, and EpCAM expression in colorectal cancer. Br. J. Cancer. 2014;110:2935–2944. doi: 10.1038/bjc.2014.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koopman M, et al. Sequential versus combination chemotherapy with capecitabine, irinotecan, and oxaliplatin in advanced colorectal cancer (CAIRO): a phase III randomised controlled trial. Lancet. 2007;370:135–142. doi: 10.1016/S0140-6736(07)61086-1. [DOI] [PubMed] [Google Scholar]
- 13.Tol J, et al. Chemotherapy, bevacizumab, and cetuximab in metastatic colorectal cancer. N. Engl. J. Med. 2009;360:563–572. doi: 10.1056/NEJMoa0808268. [DOI] [PubMed] [Google Scholar]
- 14.de Sousa EMF, et al. Methylation of cancer-stem-cell-associated Wnt target genes predicts poor prognosis in colorectal cancer patients. Cell Stem Cell. 2011;9:476–485. doi: 10.1016/j.stem.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 15.Tol J, et al. Markers for EGFR pathway activation as predictor of outcome in metastatic colorectal cancer patients treated with or without cetuximab. Eur. J. Cancer. 2010;46:1997–2009. doi: 10.1016/j.ejca.2010.03.036. [DOI] [PubMed] [Google Scholar]
- 16.Venderbosch S, et al. Mismatch repair status and BRAF mutation status in metastatic colorectal cancer patients: a pooled analysis of the CAIRO, CAIRO2, COIN, and FOCUS studies. Clin. Cancer Res. 2014;20:5322–5330. doi: 10.1158/1078-0432.CCR-14-0332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van den Broek E, et al. High prevalence and clinical relevance of genes affected by chromosomal breaks in colorectal cancer. PLoS One. 2015;10:e0138141. doi: 10.1371/journal.pone.0138141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Dijk E, et al. Loss of chromosome 18q11.2-q12.1 is predictive for survival in patients with metastatic colorectal cancer treated with Bevacizumab. J. Clin. Oncol. 2018;36:2052–2060. doi: 10.1200/JCO.2017.77.1782. [DOI] [PubMed] [Google Scholar]
- 19.Lugli A, et al. Intratumoural budding as a potential parameter of tumour progression in mismatch repair-proficient and mismatch repair-deficient colorectal cancer patients. Hum. Pathol. 2011;42:1833–1840. doi: 10.1016/j.humpath.2011.02.010. [DOI] [PubMed] [Google Scholar]
- 20.Cappellesso R, et al. Tumour budding as a risk factor for nodal metastasis in pT1 colorectal cancers: a meta-analysis. Hum. Pathol. 2017;65:62–70. doi: 10.1016/j.humpath.2017.04.013. [DOI] [PubMed] [Google Scholar]
- 21.Rogers AC, et al. Systematic review and meta-analysis of the impact of tumour budding in colorectal cancer. Br. J. Cancer. 2016;115:831–840. doi: 10.1038/bjc.2016.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oh BY, et al. Prognostic impact of tumour-budding grade in stages 1–3 colon cancer: a retrospective cohort study. Ann. Surg. Oncol. 2017;25:204–211. doi: 10.1245/s10434-017-6135-5. [DOI] [PubMed] [Google Scholar]
- 23.Knudsen KN, Lindebjerg J, Nielsen BS, Hansen TF, Sorensen FB. MicroRNA-200b is downregulated in colon cancer budding cells. PLoS One. 2017;12:e0178564. doi: 10.1371/journal.pone.0178564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Galvan JA, et al. TWIST1 and TWIST2 promoter methylation and protein expression in tumour stroma influence the epithelial–mesenchymal transition-like tumour budding phenotype in colorectal cancer. Oncotarget. 2015;6:874–885. doi: 10.18632/oncotarget.2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barresi V, Bonetti LR, Bettelli S. KRAS, NRAS, BRAF mutations and high counts of poorly differentiated clusters of neoplastic cells in colorectal cancer: observational analysis of 175 cases. Pathology. 2015;47:551–556. doi: 10.1097/PAT.0000000000000300. [DOI] [PubMed] [Google Scholar]
- 26.Graham RP, et al. Tumour budding in colorectal carcinoma: confirmation of prognostic significance and histologic cutoff in a population-based cohort. Am. J. Surg. Pathol. 2015;39:1340–1346. doi: 10.1097/PAS.0000000000000504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prall F, Ostwald C. High-degree tumour budding and podia-formation in sporadic colorectal carcinomas with K-ras gene mutations. Hum. Pathol. 2007;38:1696–1702. doi: 10.1016/j.humpath.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 28.Jang S, et al. KRAS and PIK3CA mutations in colorectal adenocarcinomas correlate with aggressive histological features and behavior. Hum. Pathol. 2017;65:21–30. doi: 10.1016/j.humpath.2017.01.010. [DOI] [PubMed] [Google Scholar]
- 29.Zlobec I, et al. Tumour budding predicts response to anti-EGFR therapies in metastatic colorectal cancer patients. World J. Gastroenterol. 2010;16:4823–4831. doi: 10.3748/wjg.v16.i38.4823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cho YH, et al. KY1022, a small molecule destabilizing Ras via targeting the Wnt/beta-catenin pathway, inhibits development of metastatic colorectal cancer. Oncotarget. 2016;7:81727–81740. doi: 10.18632/oncotarget.13172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Centeno I, et al. DNA profiling of tumour buds in colorectal cancer indicates that they have the same mutation profile as the tumour from which they derive. Virchows Arch. 2017;470:341–346. doi: 10.1007/s00428-017-2071-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Analyses were performed using R and code for the analysis can be found at the following github repository (github.com/trinhan/crc-ihc-classification).