Abstract
Background:
Autophagy is an extensive self-degradation process for the disposition of cytosolic aggregated or misfolded proteins and defective organelles which executes the functions of pro-survival and pro-death to maintain cellular homeostasis. The pathway plays essential roles in several neurological disorders. Subarachnoid Hemorrhage (SAH) is a devastating subtype of hemorrhagic stroke with high risk of neurological deficit and high mortality. Early brain injury (EBI) plays a role in the poor clinical course and outcome after SAH. Recent studies have paid attention on the role of the autopha-gy pathway in the development of EBI after SAH. We aim to update the multifaceted roles of autophagy pathway in the pathogenesis of SAH, especially in the phase of EBI.
Methods:
We reviewed early researches related to autophagy and SAH. The following three aspects of contents will be mainly discussed: the process of the autophagy pathway, the role of the autophagy in SAH and the interaction between organelle dysfunction and autophagy pathway after SAH.
Results:
Accumulating evidence shows an increased autophagy reaction in response to early stages of SAH. However, oth-ers suggest inadequate or excessive autophagy activation can result in cell injury and death. In addition to autophagy, apopto-sis and necrosis can occur in neurons simultaneously after SAH, leading to mixed features of cell death morphologies. And it is also known that there is extensive crosstalk between autophagy and apoptosis pathway. Subcellular organelles of neural cells generally participate in the formation and functional parts of autophagy process.
Conclusion:
Autophagy plays an important role in the SAH-induced brain injury. A better understanding of the interrelationship among autophagy, apoptosis, and necrosis might provide us better therapeutic targets for the treatment of SAH
Keywords: Autophagy, subarachnoid hemorrhage, early brain injury, pathogenesis, subcellular organelles, treatment
1. Introduction
Autophagy, a cell self-digestive and lysosomal degradation pathway, widely exists in yeast and mammal’s cells [1]. It sequesters the cytosolic components into autophagosomes, and joins with lysosomes to form autolysosomes [2]. Under the process of this pathway many cargos are degraded, such as aggregated or misfolded proteins, damaged organelles, and intracellular pathogens [3]. Autophagy also executes a crucial role in balancing sources of energy at critical times in cellular growth and in stress-related responses [4]. It had been shown that autophagy pathway was the essential part of several central nervous system (CNS) diseases such as neurodegenerative diseases, brain ischemia [5, 6], myelin diseases [7], and traumatic brain injury [8].
Subarachnoid hemorrhage (SAH) is a devastating subtype of stroke with high risk of neurological deficit and even death [9, 10]. Research has determined that the complications after SAH was due to vasospasm, and subsequently research focused on vasospasm as an important mechanism in SAH [11]. However, the mortality and clinical outcomes were not improved by decreasing vasospasm rate in the clinical trials [12]. Recently, studies have drawn their attention on the early brain injury (EBI) after SAH, as it is currently being considered to contribute to the poor outcome of SAH patients [13, 14]. The possible mechanisms of EBI include: raised intracranial pressure (ICP), decreased cerebral blood flow (CBF) and cerebral perfusion pressure (CPP), blood-brain barrier (BBB) disruption, brain edema, acute vasospasm, and dysfunction of autoregulation [14-17]. In addition, pathological events such as inflammation response, excitotoxicity events, oxidative stress, and ionic homeostasis, have also been proposed in the development of EBI [18]. Nowadays, subcellular organelles dysfunction, such as endoplasmic reticulum (ER) stress, mitochondrial damage, and lysosome activation has become the hot spot of EBI after SAH [19].
Recently, accumulating evidence has shown that autophagy pathway plays a multifaceted role in the development of EBI after SAH [20, 21]. Considering the interconnections between the autophagy pathway and EBI development, the common process of the autophagy pathway and the role of autophagy in EBI development after SAH were summarized in this review. The interaction between organelle dysfunction and autophagy pathway in the pathogenesis of EBI after SAH was also reviewed. A circumstantial recognition of autophagy may provide us a novel therapeutic potential for SAH treatment.
2. The process of autophagy pathway
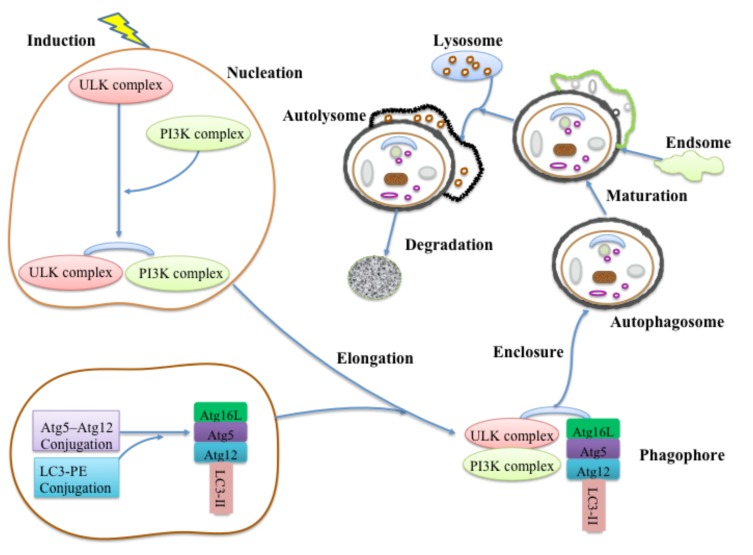
Autophagy is a cellular degradation process conserved in the eukaryotic cell [1]. It recycles futile contents by sequestering long-lived proteins and dysfunctional organelles into a double-membrane vesicle and delivers them to the lysosome for digestion [22]. The subtypes of autophagy can be divided according to the cargo degraded. Generally, three forms of autophagy are described, which include macroautophagy, microautophagy, and chaperone-mediated autophagy (CMA) [23]. Among them, the macroautophagy is the best-characterized type. Under normal circumstances in the brain, autophagy works at a lower basal level and executes lots of physiological functions in most cells, such as intracellular quality control, cell development and death, immunity inanition, and anti-aging mechanism [24]. Meanwhile, autophagy plays important roles in mediating the invaluable cellular contents during starvation and participating in physiological processes of replacing futile organisms and toxic protein after stress response [22, 25]. The initial steps of autophagy pathway, as phagophores form, include nucleation and elongation of isolated membranes. The edges of the phagophore fuse together to constitute a sealed double-membraned vesicle (autophagosome), which gathers and sequesters the cytoplasmic material (enclosure). In the end, the endosomes and lysosomes continuously integrate with the autophagosomes to form autolysosomes (maturation and fusion) for degradation [3, 26]. The degraded products include amino acid and fatty acid building blocks [26] (Fig. 1).
Fig. (1).
Molecular mechanism of autophagy pathway. The autophagy pathway formation involves several steps: nucleation, elongation, enclosure, maturation, fusion and degradation. Under the induction of physiological signals, the autophagy cascade initiated from the assembly of unc-51-like kinase (ULK) complex and followed class III phosphatidylinositol 3-kinase (PI3K) formation, which integrate the isolated membranes. With the assistance of autophagy-related genes (Atgs), two ubiquitin like conjugation systems, Atg5-Atg12 conjugation and protein light chain 3 (LC3) -phosphatidyl ethanolamine (PE) conjugation, involved in the expansion of membranes to form the phagophores. After edges of the phagophore fusing together, the autophagosomes continuously combining with endosomes and lysosomes to form the autolysomes for degradation.
The molecular mechanism of autophagy pathway is closely related with the autophagy-related genes (Atgs) and their encoded proteins [27]. It has been shown in yeast that Atg1 forms a complex with the product of the Atg13 and Atgs including Atg17, Atg29 and Atg31, which join the Atg13:Atg1 complex [28, 29]. The mammalian counterpart of this complex has recently been found, named unc-51-like kinase (ULK):Atg13:FIP200; a 200-kDa focal adhesion kinase family-interacting protein [30]. The assembly of this complex completes the beginning step of the autophagy cascade. In the mammalian cell, ULK1 and ULK2 are the two homologues of Atg1; while the FIP200 is the homologue of Atg17. After induction signals, the ULK and FIP200 connect to the Atg13 to form a stable complex [30]. This complex is regulated by nutrition conditions [31]. Meanwhile, a new mammalian Atg13 binding protein, Atg101, also plays a crucial role in initiating autophagy as well as protecting Atg13 from proteasomal degradation [32, 33].
After the formation of the ULK complex, the class III phosphatidylinositol 3-kinase (PI3K) complex is the essential part of vesicle nucleation [34]. In mammals, PI3K complex is composed of vacuolar protein sorting-associated protein 34 (Vps34), Beclin-1 (a homolog of Atg6), p150 (a homolog of Vps15) and Barkor (Beclin-1-associated autophagy-related key regulator) [34, 35]. This complex generates phosphatidylinositol-3 phosphate (PtdIns) and recruits additional Atg proteins to the formation phagophore membrane [36]. Beclin-1, a molecular platform, contacts several special factors, such as UV irradiation resistance-associated tumor suppressor gene (UVRAG), Atg14L, Ambra1, and Bif-1 to promote Vps34 activation. Thus, these key regulators act as Barkor for the formation of autophagosome [34, 37].
Elongation of autophagosome membrane involves two ubiquitin like conjugation systems, including Atg5–Atg12 conjugation and protein light chain 3 (LC3)-phosphatidyl ethanolamine (PE) conjugation [38]. The two ubiquitin-like reactions share the same activating enzyme, Atg7. In the Atg5-Atg12 conjugation, with the assistance of E1 ubiquitin activating enzyme (Atg7) and E2 ubiquitin activating enzyme (Atg10), Atg5 and Atg12 are combined into a covalent complex [39, 40]. Next, the Atg12-Atg5 conjugation recruits Atg16L (mammalian homologue of Atg16) to form the Atg12-Atg5:Atg16L complex of approximately 800 kDa which plays an important role in the elongation of phagophores. It also acts as the ligase of the LC3 ubiquitin-like reaction [41]. The Apg12-Apg5:Apg16L complex is primarily needed for the expansion of the isolated membranes, but not for the initial step of isolated membranes generation [42]. The LC3-PE conjugation involves the evolution of LC3. The mammalian homologue of yeast Atg8, LC3, is a common marker of autophagosome formation in mammalian systems [43]. With the assistance of Atg4, the C-terminus of LC3 is cleaved to form LC3-I, and LC3-I is subsequently activated by Atg7 for transferring to the Atg3 (E2 ubiquitin activating enzyme) to form the LC3-II. In addition, this step also involves the participation of lipid PE [44]. Normally, LC3 seemed to be a critical signal and mediator for membrane prolonging during the autophagosome formation [45]. Finally, LC3 connected with Atg12-Atg5:Atg16L and subsequently join to ULK-PI3K membranes. Meanwhile, Atg12-Atg5 and Atg16L get separated from the membrane after completion of autophagosome formation, while LC3-II remains on the autophagosome membranes [42].
The autophagosomes maturate by sequentially fusing with endosomes and lysosomes, acidifying the autophagic cargos, and recycling metabolites from the lysosomal degradation process [46]. During the autophagosome fusion, some lysosomal membrane proteins are required, such as GTPase Rab7, lysosomal-associated membrane protein 1 (LAMP-1), and LAMP-2; however, the mechanism is vague [47, 48]. After fusion of autophagosomes, some lysosomal/vacuolar acid hydrolases are also required for autophagosomes degradation including cathepsins B, D and L. Lastly, the products of autophagosomes degradation are released back into the cytosol [49].
3. The regulators of autophagy pathway
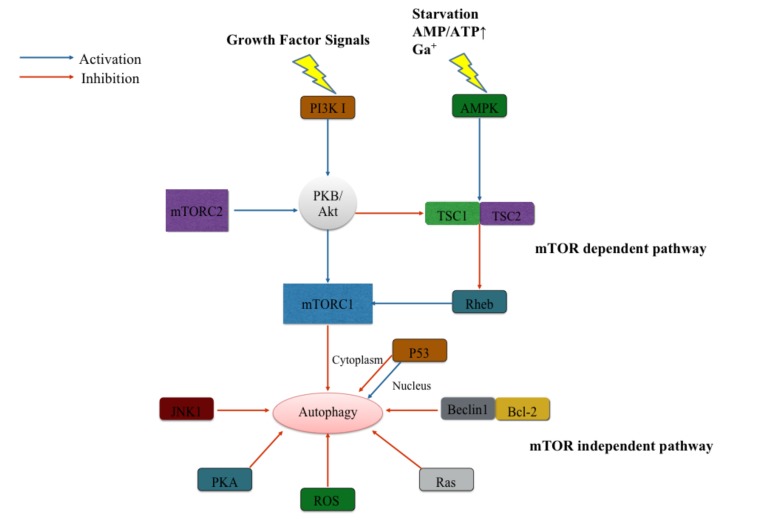
There are several signaling pathways in autophagy regulation of mammalian cells. According to the mammalian target of rapamycin (mTOR), the signaling pathways can be divided into mTOR dependent and mTOR independent pathways [50] (Fig. 2).
Fig. (2).
Regulation of the autophagy pathway. Regulation of the autophagy pathway can be divided in mammalian target of rapamycin (mTOR) dependent pathway and independent pathway. Depending on different composition and structure, there are two mTOR complexes, mTORC1 and mTORC2. mTORC1 is regulated upon growth factors or starvation or other stress signals. While mTORC2 works via PI3K-protein kinase B (PKB/Akt) pathway in skeletal muscle cells only when in response to the emergency circumstance; The mTOR independent regulators involve P53, c-Jun N-terminal protein kinase 1 (JNK1), protein kinase A (PKA), reactive oxygen species (ROS), Ras and Beclin1-Bcl-2 interaction.
In the mTOR dependent pathways, there is induction of nutrients (amino-acids or glucose), energy or several growth factors. Then, mTOR inhibits autophagy activity through several upstream signaling pathways, such as PI3K I-Akt pathway and AMP-activated protein kinase (AMPK) pathway mTOR inhibits autophagy activity through several upstream signaling pathways, such as PI3K I-Akt pathway and AMP-activated protein kinase (AMPK) pathway [50]. Normally, mTOR binds several different proteins to form two distinct mTOR complexes (mTORC), mTORC1 and mTORC2, which are different in their sensitivity to rapamycin [51]. Under the stimulation of growth factors, PI3K I promotes protein kinase B (PKB, also named Akt) and leads to the phosphorylation of plasma membrane lipids [52]. This mechanism finally activates mTORC1 signaling and inhibits autophagy. However, the overexpression of PKB indirectly promotes the phosphorylation of tuberous sclerosis complex (TSC)2 and prevents interaction between TSC2 and TSC1 [52, 53]. The complex of TSC1/2 can facilitate the small GTPase Rheb to inhibit the mTORC1 [54]. Another upstream regulator of mTORC1, AMPK pathway, suppresses mTORC1 activity while the cell undergoes induction of starvation or calcium signals. It regulates autophagy pathway by sensing levels of cellular bioenergetics. In low energy states, the increasing ratio of AMP/ATP leads to the activation of AMPK and subsequently inhibits mTORC1 through the assistance of TSC1/TSC2 and Rheb protein [55]. mTORC2 regulates autophagy pathway only in the skeletal muscle and works through Akt pathway when it responds to the emergency circumstance [56].
The mTOR independent pathway may include several regulators. The cancer suppressor and transcription factor, p53, plays different roles in the regulation of autophagy pathway depending on the function location at the subcellular level [57]. In the cytoplasm, p53 suppresses autophagy pathway by directly interacting with FIP200 [58]. In the nucleus, p53 promotes transcriptional activation of autophagy-associated genes to facilitate the autophagy process [59, 60]. p53 also activates AMPK to inhibit mTOR activity and stimulates the initiation of autophagy [61]. Beclin 1 is a Bcl-2 interacting protein via its BH3 domain. The interaction between Beclin 1 and Bcl-2 indirectly suppresses the Beclin 1-dependent autophagy activation through replacement of association between Beclin 1 and PI3K, hVps34, and p150 [62]. Starvation-induced activation of c-Jun N-terminal protein kinase 1 (JNK1) may also promote autophagy by dissociating Bcl-2 from the autophagy inhibitory Beclin 1–Bcl-2 complex to abolish their inhibitory effects on autophagy while indirectly promoting the formation of the Beclin-1–Vps34 complex [63].
The reactive oxygen species (ROS) can modulate the autophagy process in the mTOR-independent autophagy pathways. By ameliorating the ROS level, antioxidant treatment can effectively weaken the autophagy activation [64, 65]. Under oxidative stress, the oxidized Atg4 loses action on LC3, inhibiting the LC3-PE conjugation [66]. Ras signals also evolve in the mTOR-independent autophagy pathways by sensing growth factors [67]. Other studies show that mammalian protein kinase A (PKA) inhibits autophagy either by directly phosphorylating LC3 [68] or by activating mTORC1. Additionally, in the downstream of the mTORC, there are several regulators of autophagy, such as positive regulator p70S6 kinase or S6K (homologue of Sch9), and negative regulator death-associated protein 1 (DAP1) [69, 70].
4. Autophagy in Subarachnoid hemorrhage
SAH is a disastrous and multisystem disorder of acute cerebral diseases, which owns complex pathological processes [71, 72]. It has been recognized that EBI represents the major cause of morbidity and mortality in SAH progress [73]. The sudden raised ICP, the decrease of CBF and CPP, BBB disruption, and brain edema may play critical roles in the development of EBI [10, 74, 75]. However, the exact underlying molecular mechanisms during this early period following SAH are not well understood. Meanwhile, as we discussed above, the autophagy pathway has been reportedly activated in many neurological diseases [76-78]. In the SAH process, the effect of autophagy activation is still controversial (Fig. 3).
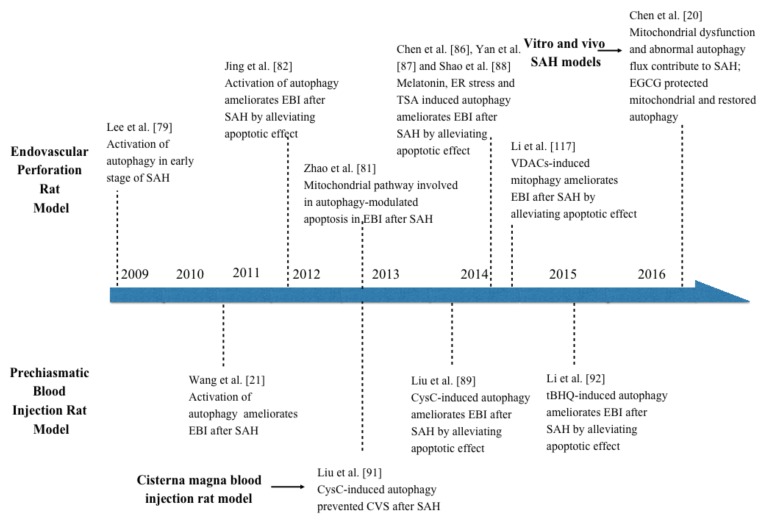
Fig. (3).
Recent findings of autophagy pathway after subarachnoid hemorrhage. This figure summarized the studies on autophagy in subarachnoid hemorrhage.
The activation of autophagy pathway in neurons in the acute phase after SAH was presented by Lee and his partners [79]. They determined that there is a significant increase in autophagosomes and autolysosomes in the neurons during the entire phase of EBI (up to 3 days, peak at 24h) in a modified endovascular perforation rat model of SAH. Additionally, the conversion of LC3-I to LC3-II and the expression of beclin-1 and Cathepsin-D, an important hydrolytic enzyme of lysosomes, were also increased along with autophagosomes and autolysosomes. However, the role of autophagy pathway in SAH-induced EBI was not mentioned in their study. By injuring blood vessels and causing direct hemorrhagic brain lesions due to arterial blood pressure, the endovascular perforation model is more likely to bring human SAH pathophysiological and histological changes in the brain. These changes include higher ICP, increased hemoglobin content, lower CBF, neuronal cell edema, inflammation, and acute vasospasm [75]. However, it provided little control over the amount of blood released in the subarachnoid space, resulting in an experimental model with variable bleeding. In addition, the mortality after inducing SAH of this model was almost 50%. In the prechiasmatic blood injection SAH rat model, which has a controllable amount of blood, the autophagy pathway was also activated in the early stages after SAH in rats [21, 80]. The time–course study showed how autophagy reached at peak at 24 h and recovered at 48 h following SAH [21]. Application of Rapamycin (an autophagy inducer targeting at mTOR) and 3-Methyladenine (3-MA) (an autophagy inhibitor targeting at PI3K complex) to manipulate the autophagy activity, revealed a preferable course with ameliorated brain edema, higher BBB permeability, and improved clinical behavior function in the EBI [21]. The activation of autophagy pathway may play a potential role against the development of EBI in the rat SAH model.
The endovascular perforation rat model demonstrated how autophagy activation reduces translocation of Bax from cytosol to the mitochondrial membrane to work against the apoptosis effect [81]. The Mitochondrial pathway was considered to be involved in autophagy-modulated apoptosis in EBI after SAH [82]. Following the apoptotic stimuli after SAH, the outer mitochondrial membrane became permeabilized, and Bax, a Bcl-2 family member, was increased in the outer mitochondrial membrane, forming a channel to release cytochrome c into cytosol [83]. Cytoplasmic cytochrome c caused apoptosome formation and activated caspase-9, which subsequently resulted in caspase-3 function and finally led to the activation of cellular apoptosis [84]. Additionally, melatonin, a powerful pharmacological antioxidant has been proven to avert brain injury after ischemia and reperfusion via autophagy induction [85]. In endovascular perforation rat model, melatonin stimulated autophagy to ameliorate apoptotic cell death subjected to SAH [86]. This process also prevents mitochondrial release of cytochrome c to mediate apoptotic cascades. Similarly, ER stress inducer tunicamycin (Tm) and the inhibitor tauroursodeoxycholic acid (TUDCA) manipulate ER stress and demonstrate that enhanced ER stress can also improve neurological deficits and attenuate the expression of apoptosis [87]. In contrast, the ER stress inhibitor TUDCA aggravated neurological deficits and apoptotic cell death. Meanwhile, Histone deacetylase inhibitor Trichostatin A (TSA) performs the same effect as melatonin, ameliorating apoptosis and promoting autophagy activation in endovascular perforation rat model [88]. However, the mechanism of autophagy activation by TSA still remains unclear. It may be through promoting the Beclin 1 transcription and expression to increase the activation of autophagy. Other pathways and mechanisms they did not find may also be involved, and the underlying molecular mechanism of suppressing neuronal apoptosis is not clarified in their study [88].
Cystatin C (CysC) is a cysteine protease inhibitor which has been proven to have therapeutic implications on brain ischemia and neurodegenerative disorders [89]. In accordance with the results of autophagy function in prechiasmatic blood injection models, the effects of exogenous CysC on EBI were investigated after the SAH burst [89]. The group under treatment of CysC with low and medial concentrations had a lower degree of EBI compared with other groups. Autophagosomes, autolysosomes, LC3, and beclin-1 significantly increased in the cortex 48h after SAH, while the moderate CysC concentration effectively up-regulated these autophagic factors. However, the high-dose therapy may unfortunately result in a toxic outcome in the brain of SAH rats [89]. A recent study demonstrated the protective role of CysC under neuronal stress by inhibiting mTOR to promote autophagy, and how the neuroprotective effects can be prevented by inhibiting autophagy through beclin-1 siRNA or 3-MA [90]. Interestingly, another study [91] found that CysC also ameliorates a degree of vasospasm by promoting autophagy activation in cisterna magna blood injection rat model. Despite these findings, the exact anti-vasospasm mechanisms of autophagy remain unknown. Furthermore, a recent study showed that Tert-butylhydroquinone (tBHQ), a commonly used nuclear factor erythroid 2-related factor 2 (Nrf2) activator, significantly enhanced autophagy activation, decreased neuronal degeneration by apoptotic and improved the neurological score after SAH in rats. Enhanced autophagy and restoration of the apoptotic regulatory proteins (Bcl-2, Bax and cleaved caspase-3) were explored at 24 h after tBHQ treatment [92].
It should be noted that the autophagy may play a potential role in endothelial cells of the brain vessels following SAH. Brain endothelial cells injury after stress induction is the initial phase of BBB disruption and brain edema [93]. Recently, some evidences suggested that the moderate autophagy acts a beneficial role on endothelial protection after stress induction [94-96]. However, the autophagy effect on the endothelial cells after SAH needs to be further studied in the future.
5. Subcellular Organelles changes of autophagy in subarachnoid hemorrhage
The primary subcellular organelles of neural cells include the nucleus, lysosomes, ER, mitochondria, ribosomes, and Golgi body. The functional alterations of subcellular organelles in the CNS cells may lead to pathogenesis of the SAH. In the past, several experimental studies have demonstrated that the nucleus, ER, mitochondria, and lysosomes interact with autophagy pathway in the development of SAH [19] (Fig. 4).
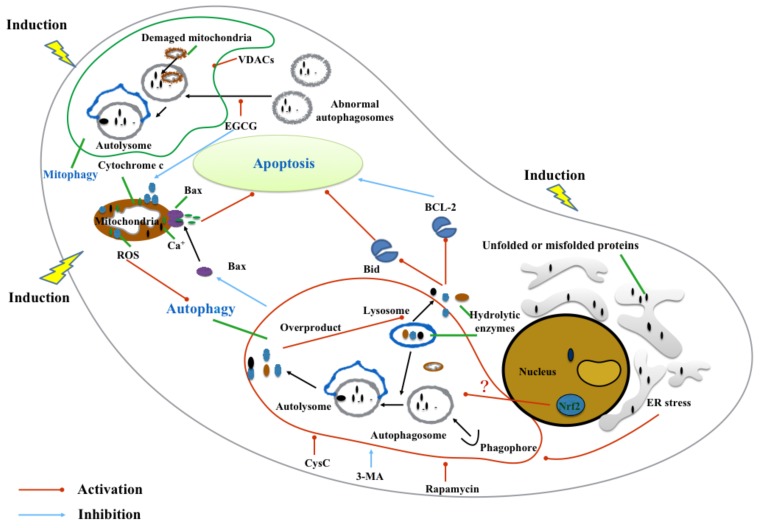
Fig. (4).
Subcellular organelles alteration with autophagy pathway in pathophysiology of subarachnoid hemorrhage. After subarachnoid hemorrhage (SAH) burst, the interaction between subcellular organelles alterations (nucleus, lysosomes, endoplasmic reticulum and mitochondria) and autophagy pathway contributes to pathogenesis of the SAH. These subcellular organelles interact with autophagy pathway via autophagy regulation, autophagy formation, signal transmission and subcellular organelles quality control.
The nucleus, a membrane-enclosed organelle functions as the control center of neural cells. It maintains the integrity of genes and controls the activities of the cell by regulating gene expression [97]. Three key regulators were activated in the nucleus after the SAH, including Nrf2-antioxidant response element (ARE) pathway, NF-κB signaling, and Hypoxia-Inducible Factor (HIF)-1. The Nrf2-ARE pathway maintains cellular homeostasis via antioxidant defense which alleviates EBI and vasospasm after SAH [98, 99]. It was documented in cancer biology that Nrf2 was observed to take apart in Nrf2/Keap-1-mediated oxidative autophagy [100], but its role in the brain was poorly studied [101]. However, a recent study revealed that tBHQ treatment ameliorated EBI after SAH in Nrf2-deficient mice by enhancing autophagy activation [92]. The mechanisms of Nrf2 involved autophagy after SAH require further clarification for complete understanding.
As for the lysosome, a cytoplasmic membrane-enclosed organelle, it works as the terminal proteolytic compartment in the cells. With the assistance of different hydrolytic enzymes, it degrades macromolecules by endocytosis, phagocytosis, and autophagy [102]. In the last link of autophagy pathway, the lyosomes fuse with autophaosomes to form autolyosomes to degrade all the cargo that autophaosomes captured [103]. It is important to note that the permeabilization of lysosomes would be unstable and lead to the releasing of hydrolytic enzymes into the cytoplasm while under the situation of oxidative stress after SAH [104]. This pathological change would trigger the lysosomal pathway of apoptosis through cleavage of the pro-apoptotic Bcl-2 family member Bid and the degradation of the anti-apoptotic Bcl-2 members such as Bcl-2, Bcl-xL, and Mcl-1 [104]. Overactivation of autophagy may also lead to this apoptosis pathway through lysosomal destabilization by accumulation of enlarged and unstable acidic vesicles [105, 106]. However, to find whether there are other mediators in SAH-induced autophagy that can monitor lysosomal pathway of apoptosis or lysosomal degradation processes deserves further investigation.
The ER is involved in the autophagy pathway by way of activation and formation, as autophagosomes are generated or in close proximity to the ER [107]. Under pathophysiological insults, dysfunction of ER results in the accumulation of unfolded or misfolded proteins in the lumen of ER, which activates ER stress [108]. ER stress stimulates autophagy by removing detrimental components of cytoplasm and serving a beneficial role for cell survival [109]. As mentioned above, ER stress can also alleviate early brain injury following SAH via the inhibition of apoptosis [87]. However, the mechanism of how ER stress activates autophagy pathway and promotes neuroprotection remains to be explored in the future. Despite moderate levels of autophagy, ER stress predominantly acts as a pro-survival pathway in the cell [110, 111]. Other studies argued that excessive ER stress or autophagy might promote cell injury and death [112, 113].
The mitochondria is a double-membrane organelle highly correlated with energy generating in most eukaryotic cells, and is also critical for the functioning of autophagy [5]. Outer stress leading to mitochondrial dysfunction may bring a series of awful consequences, such as collapse of the mitochondrial membrane, disorder of transmembrane product transport, outflow of matrix calcium. It results to overproduction of ROS and releasing of apoptogenic proteins [114, 115]. As noted, the mitochondrial pathway is considered to be involved in autophagy-modulated apoptosis in EBI after SAH [82]. Autophagy activation has been shown to reduce Bax transport to the mitochondrial membrane which is able to work against cytochrome c leaving the mitochondria to activate apoptosis [81]. In addition, this pathologic alteration can induce mitophagy, a special type of macroautophagy, to regulate mitochondrial number to match metabolic demand and is responsible for quality control and removing damaged mitochondria [116]. Voltage-dependent anion channel 1 (VDAC1) induced mitophagy following SAH injury may take part in neuroprotection by attenuation of the apoptotic and necrositic molecular pathways [117]. ROS produced by damaged mitochondria could directly regulate the formation of autophagosomes [118]. By applying VDCA1, mitochondria actively contributed to the autophagic response in the prechiasmatic blood injection SAH rats model through providing proautophagic ROS, which contributed to autophagic formation and elongation products (LC3) and avoided autophagic degradation (mitophagy). In another study about mitochondria dysfunction after SAH, abnormal autophagy flux synergistically took part in SAH pathogenesis [20]. They found that a new pharmacological agent, Epigallocatechin-3-gallate (EGCG) can act on the mitochondria and alert the autophagy pathway in SAH therapy. EGCG is the main biological activator of tea catechin, a beneficial treatment for CNS diseases [20, 119, 120]. EGCG protects mitochondrial function after SAH and restores the abnormal autophagy flux which can then eliminate dysfunctional mitochondria and prevent EBI from progressing. After EBI in SAH, the accumulation of damaged mitochondria leads to autophagic flux, but excessive autophagosomes might not be parallel with normal autophagy activity, and overwhelmed and dysfunctional mitochondria continuously lead to autophagosome accumulation with the loss of autophagosomes maturation (autolysosomes) [20]. EGCG can up-regulate the Atg5, Becn-1, and LC3-II expression and eliminate autophagysomes from autolysosomes in a timely and complete manner [20, 121]. Taken together, considering the participation of mitochondria, mitochondrial autophagy pathway should be also studied in the future to help us better understand SAH pathogenesis.
Future Directions and Conclusion
SAH is a complicated and multifaceted disease with high mortality and morbidity. In conjunction with loose translational research, without knowing details of the SAH pathological process, the path to finding effective and clinically applicable targeted therapies remains slow for this devastating disease. The autophagy pathway has shown to play a crucial role in the pathogenesis of many CNS diseases [7]. Also, the significance of autophagy pathophysiology of SAH has been highlighted. Accumulating evidences show an increased autophagy reaction in response to early stages of SAH. However, others suggest that inadequate or excessive activation of autophagy can result in cell injury and death [113]. In addition to autophagy, apoptosis and necrosis can occur in neurons simultaneously after SAH, leading to mixed features of cell death morphologies. Moreover, it is also know that there is extensive crosstalk between autophagy and apoptosis pathway [122]. Subcellular organelles of neural cells generally participate in the formation and functional parts of autophagy process [19]. Thus, future investigation should be focused on integration of both the pathways rather than examining them separately. Also, a better understanding of the interrelationship among autophagy, apoptosis, and necrosis might provide us better therapeutic targets for the treatment of SAH.
ACKNOWLEDGEMENTS
This study was supported by the National Natural Science Foundation of China (81500992), Natural Science Foundation of Zhejiang (LQ16H090002), Medical and health key project of Zhejiang Province (2016RCA015).
Consent for Publication
Not applicable.
Conflict of Interest
The authors report no conflict of interest concerning the materials used in this study or findings specified in this paper.
References
- 1.Liu J., Fan L., Wang H., Sun G. Autophagy, a double-edged sword in anti-angiogenesis therapy. Med. Oncol. 2016;33(1):10. doi: 10.1007/s12032-015-0721-9. [http://dx.doi.org/10.1007/s12032-015-0721-9]. [PMID: 26715036]. [DOI] [PubMed] [Google Scholar]
- 2.Zeki A.A., Yeganeh B., Kenyon N.J., Post M., Ghavami S. Autophagy in airway diseases: a new frontier in human asthma? Allergy. 2016;71(1):5–14. doi: 10.1111/all.12761. [http://dx.doi.org/10.1111/all.12761]. [PMID: 26335713]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee Y.J., Jang B.K. The role of autophagy in hepatocellular carcinoma. Int. J. Mol. Sci. 2015;16(11):26629–26643. doi: 10.3390/ijms161125984. [http:// dx.doi.org/10.3390/ijms161125984]. [PMID: 26561802]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glick D., Barth S., Macleod K.F. Autophagy: cellular and molecular mechanisms. J. Pathol. 2010;221(1):3–12. doi: 10.1002/path.2697. [http://dx.doi. org/10.1002/path.2697]. [PMID: 20225336]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qi Z., Dong W., Shi W., Wang R., Zhang C., Zhao Y., Ji X., Liu K.J., Luo Y. Bcl-2 phosphorylation triggers autophagy switch and reduces mitochondrial damage in limb remote ischemic conditioned rats after ischemic stroke. Transl. Stroke Res. 2015;6(3):198–206. doi: 10.1007/s12975-015-0393-y. [http://dx.doi.org/10.1007/s12975-015-0393-y]. [PMID: 25744447]. [DOI] [PubMed] [Google Scholar]
- 6.Wei H., Li Y., Han S., Liu S., Zhang N., Zhao L., Li S., Li J. cPKCγ-modulated autophagy in neurons alleviates ischemic injury in Brain of Mice with Ischemic stroke through Akt-mTOR pathway. Transl. Stroke Res. 2016;7(6):497–511. doi: 10.1007/s12975-016-0484-4. [http://dx.doi. org/10.1007/s12975-016-0484-4]. [PMID: 27510769]. [DOI] [PubMed] [Google Scholar]
- 7.Hu Z., Yang B., Mo X., Xiao H. Mechanism and regulation of autophagy and is role in neuronal diseases. Mol. Neurobiol. 2015;52(3):1190–1209. doi: 10.1007/s12035-014-8921-4. [http://dx.doi.org/10.1007/s12035-014-8921-4]. [PMID: 25316381]. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y.B., Li S.X., Chen X.P., Yang L., Zhang Y.G., Liu R., Tao L.Y. Autophagy is activated and might protect neurons from degeneration after traumatic brain injury. Neurosci. Bull. 2008;24(3):143–149. doi: 10.1007/s12264-008-1108-0. [http://dx.doi.org/10.1007/s12264-008-1108-0]. [PMID: 18500386]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Etminan N. Aneurysmal subarachnoid hemorrhage-status quo and perspective. Transl. Stroke Res. 2015;6(3):167–170. doi: 10.1007/s12975-015-0398-6. [http://dx.doi. org/10.1007/s12975-015-0398-6]. [PMID: 25860440]. [DOI] [PubMed] [Google Scholar]
- 10.Cheng C., Jiang L., Yang Y., Wu H., Huang Z., Sun X. Effect of APOE gene polymorphism on early cerebral perfusion after aneurysmal subarachnoid hemorrhage. Transl. Stroke Res. 2015;6(6):446–450. doi: 10.1007/s12975-015-0426-6. [http://dx.doi.org/10.1007/s12975-015-0426-6]. [PMID: 26370543]. [DOI] [PubMed] [Google Scholar]
- 11.Inagawa T. Risk factors for cerebral vasospasm following aneurysmal subarachnoid hemorrhage: A review of the literature. World Neurosurg. 2016;85:56–76. doi: 10.1016/j.wneu.2015.08.052. [http://dx.doi.org/10.1016/ j.wneu.2015.08.052]. [PMID: 26342775]. [DOI] [PubMed] [Google Scholar]
- 12.Macdonald R.L., Kassell N.F., Mayer S., Ruefenacht D., Schmiedek P., Weidauer S., Frey A., Roux S., Pasqualin A., Investigators C. Clazosentan to overcome neurological Ischemia and infarction occurring after subarachnoid hemorrhage (CONSCIOUS-1): randomized, double-blind, placebo-controlled phase 2 dose-finding trial. Stroke. 2008;39(11):3015–3021. doi: 10.1161/STROKEAHA.108.519942. [http://dx.doi.org/10. 1161/STROKEAHA.108.519942]. [PMID: 18688013]. [DOI] [PubMed] [Google Scholar]
- 13.Yuksel S., Tosun Y.B., Cahill J., Solaroglu I. Early brain injury following aneurysmal subarachnoid hemorrhage: emphasis on cellular apoptosis. Turk Neurosurg. 2012;22(5):529–533. doi: 10.5137/1019-5149.JTN.5731-12.1. [PMID: 23015327]. [DOI] [PubMed] [Google Scholar]
- 14.Pang J., Chen Y., Kuai L., Yang P., Peng J., Wu Y., Chen Y., Vitek M.P., Chen L., Sun X., Jiang Y. Inhibition of blood-brain barrier disruption by an apolipoprotein E-Mimetic peptide ameliorates early brain injury in experimental subarachnoid hemorrhage. Transl. Stroke Res. 2016 doi: 10.1007/s12975-016-0507-1. [PMID: 27796945]. [DOI] [PubMed] [Google Scholar]
- 15.Fujii M., Yan J., Rolland W.B., Soejima Y., Caner B. ; Zhang J.H. Early brain injury, an evolving frontier in subarachnoid hemorrhage research. Transl. Stroke Res. 2013;4(4):432–446. doi: 10.1007/s12975-013-0257-2. [http://dx.doi.org/10.1007/s12975-013-0257-2]. [PMID: 23894255]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo D., Wilkinson D.A., Thompson B.G., Pandey A.S., Keep R.F., Xi G., Hua Y. MRI characterization in the acute phase of experimental subarachnoid hemorrhage. Transl. Stroke Res. 2017;8(3):234–243. doi: 10.1007/s12975-016-0511-5. [PMID: 27896625]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi Y., Leak R.K., Keep R.F., Chen J. Translational stroke research on blood-brain barrier damage: challenges, perspectives, and Goals. Transl. Stroke Res. 2016;7(2):89–92. doi: 10.1007/s12975-016-0447-9. [http://dx. doi.org/10.1007/s12975-016-0447-9]. [PMID: 26757714]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sehba F.A., Pluta R.M., Zhang J.H. Metamorphosis of subarachnoid hemorrhage research: from delayed vasospasm to early brain injury. Mol. Neurobiol. 2011;43(1):27–40. doi: 10.1007/s12035-010-8155-z. [http://dx.doi.org/10. 1007/s12035-010-8155-z]. [PMID: 21161614]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen S., Wu H., Tang J., Zhang J., Zhang J.H. Neurovascular events after subarachnoid hemorrhage: focusing on subcellular organelles. Acta Neurochir. Suppl. (Wien) 2015;120:39–46. doi: 10.1007/978-3-319-04981-6_7. [PMID: 25366597]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Y., Huang L., Zhang H., Diao X., Zhao S., Zhou W. Reduction in Autophagy by (-)-Epigallocatechin-3-Gallate (EGCG): a potential mechanism of prevention of mitochondrial dysfunction after subarachnoid hemorrhage. Mol. Neurobiol. 2017;54(1):392–405. doi: 10.1007/s12035-015-9629-9. [PMID: 26742518]. [DOI] [PubMed] [Google Scholar]
- 21.Wang Z., Shi X.Y., Yin J., Zuo G., Zhang J., Chen G. Role of autophagy in early brain injury after experimental subarachnoid hemorrhage. J. Mol. Neurosci. 2012;46(1):192–202. doi: 10.1007/s12031-011-9575-6. [http://dx. doi.org/10.1007/s12031-011-9575-6]. [PMID: 21728063]. [DOI] [PubMed] [Google Scholar]
- 22.Mizushima N. Physiological functions of autophagy. Curr. Top. Microbiol. Immunol. 2009;335:71–84. doi: 10.1007/978-3-642-00302-8_3. [http://dx.doi.org/10.1007/ 978-3-642-00302-8_3]. [PMID: 19802560]. [DOI] [PubMed] [Google Scholar]
- 23.Mizushima N., Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147(4):728–741. doi: 10.1016/j.cell.2011.10.026. [http://dx.doi.org/10.1016/ j.cell.2011.10.026]. [PMID: 22078875]. [DOI] [PubMed] [Google Scholar]
- 24.Levine B., Klionsky D.J. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell. 2004;6(4):463–477. doi: 10.1016/s1534-5807(04)00099-1. [http://dx.doi.org/10.1016/S1534-5807(04) 00099-1]. [PMID: 15068787]. [DOI] [PubMed] [Google Scholar]
- 25.Klionsky D.J. Autophagy in mammalian systems, Part B. Preface. Methods Enzymol. 2009;452:xxi–xxii. doi: 10.1016/S0076-6879(08)03631-8. [http://dx.doi.org/10. 1016/S0076-6879(08)03631-8]. [PMID: 19200871]. [DOI] [PubMed] [Google Scholar]
- 26.Levine B., Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132(1):27–42. doi: 10.1016/j.cell.2007.12.018. [http://dx.doi.org/10.1016/j.cell.2007. 12.018]. [PMID: 18191218]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klionsky D.J., Cregg J.M., Dunn W.A., Jr, Emr S.D., Sakai Y., Sandoval I.V., Sibirny A., Subramani S., Thumm M., Veenhuis M., Ohsumi Y. A unified nomenclature for yeast autophagy-related genes. Dev. Cell. 2003;5(4):539–545. doi: 10.1016/s1534-5807(03)00296-x. [http://dx.doi.org/10.1016/ S1534-5807(03)00296-X]. [PMID: 14536056]. [DOI] [PubMed] [Google Scholar]
- 28.Funakoshi T., Matsuura A., Noda T., Ohsumi Y. Analyses of APG13 gene involved in autophagy in yeast, Saccharomyces cerevisiae. Gene. 1997;192(2):207–213. doi: 10.1016/s0378-1119(97)00031-0. [http://dx.doi.org/10.1016/ S0378-1119(97)00031-0]. [PMID: 9224892]. [DOI] [PubMed] [Google Scholar]
- 29.Ohsumi Y. Historical landmarks of autophagy research. Cell Res. 2014;24(1):9–23. doi: 10.1038/cr.2013.169. [http://dx.doi.org/10.1038/cr.2013.169]. [PMID: 24366340]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hosokawa N., Hara T., Kaizuka T., Kishi C., Takamura A., Miura Y., Iemura S., Natsume T., Takehana K., Yamada N., Guan J.L., Oshiro N., Mizushima N. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol. Biol. Cell. 2009;20(7):1981–1991. doi: 10.1091/mbc.E08-12-1248. [http://dx.doi.org/10.1091/mbc.e08-12-1248]. [PMID: 19211835]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jung C.H., Jun C.B., Ro S.H., Kim Y.M., Otto N.M., Cao J., Kundu M., Kim D.H. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol. Biol. Cell. 2009;20(7):1992–2003. doi: 10.1091/mbc.E08-12-1249. [http://dx.doi.org/10.1091/mbc.e08-12-1249]. [PMID: 19225151]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mercer C.A., Kaliappan A., Dennis P.B. A novel, human Atg13 binding protein, Atg101, interacts with ULK1 and is essential for macroautophagy. Autophagy. 2009;5(5):649–662. doi: 10.4161/auto.5.5.8249. [http://dx.doi. org/10.4161/auto.5.5.8249]. [PMID: 19287211]. [DOI] [PubMed] [Google Scholar]
- 33.Hosokawa N., Sasaki T., Iemura S., Natsume T., Hara T., Mizushima N. Atg101, a novel mammalian autophagy protein interacting with Atg13. Autophagy. 2009;5(7):973–979. doi: 10.4161/auto.5.7.9296. [http://dx.doi.org/10.4161/auto.5.7.9296]. [PMID: 19597335]. [DOI] [PubMed] [Google Scholar]
- 34.Itakura E., Kishi C., Inoue K., Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol. Biol. Cell. 2008;19(12):5360–5372. doi: 10.1091/mbc.E08-01-0080. [http://dx.doi.org/10.1091/mbc.e08-01-0080]. [PMID: 18843052]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun Q., Fan W., Chen K., Ding X., Chen S., Zhong Q. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc. Natl. Acad. Sci. USA. 2008;105(49):19211–19216. doi: 10.1073/pnas.0810452105. [http://dx.doi.org/10.1073/ pnas.0810452105]. [PMID: 19050071]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Decuypere J.P., Parys J.B., Bultynck G. Regulation of the autophagic bcl-2/beclin 1 interaction. Cells. 2012;1(3):284–312. doi: 10.3390/cells1030284. [http://dx.doi.org/10.3390/cells1030284]. [PMID: 24710477]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Furuya N., Yu J., Byfield M., Pattingre S., Levine B. The evolutionarily conserved domain of Beclin 1 is required for Vps34 binding, autophagy and tumor suppressor function. Autophagy. 2005;1(1):46–52. doi: 10.4161/auto.1.1.1542. [http://dx.doi.org/10.4161/auto.1.1.1542]. [PMID: 16874027]. [DOI] [PubMed] [Google Scholar]
- 38.Mehrpour M., Esclatine A., Beau I., Codogno P. Overview of macroautophagy regulation in mammalian cells. Cell Res. 2010;20(7):748–762. doi: 10.1038/cr.2010.82. [http://dx.doi.org/10.1038/cr.2010.82]. [PMID: 20548331]. [DOI] [PubMed] [Google Scholar]
- 39.Ohsumi Y., Mizushima N. Two ubiquitin-like conjugation systems essential for autophagy. Semin. Cell Dev. Biol. 2004;15(2):231–236. doi: 10.1016/j.semcdb.2003.12.004. [http://dx.doi.org/10.1016/j.semcdb.2003.12.004]. [PMID: 15209383]. [DOI] [PubMed] [Google Scholar]
- 40.Yamaguchi M., Noda N.N., Yamamoto H., Shima T., Kumeta H., Kobashigawa Y., Akada R., Ohsumi Y., Inagaki F. Structural insights into Atg10-mediated formation of the autophagy-essential Atg12-Atg5 conjugate. Structure. 2012;20(7):1244–1254. doi: 10.1016/j.str.2012.04.018. [http:// dx.doi.org/10.1016/j.str.2012.04.018]. [PMID: 22682742]. [DOI] [PubMed] [Google Scholar]
- 41.Mizushima N., Noda T., Ohsumi Y. Apg16p is required for the function of the Apg12p-Apg5p conjugate in the yeast autophagy pathway. EMBO J. 1999;18(14):3888–3896. doi: 10.1093/emboj/18.14.3888. [http://dx.doi.org/ 10.1093/emboj/18.14.3888]. [PMID: 10406794]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burman C., Ktistakis N.T. Autophagosome formation in mammalian cells. Semin. Immunopathol. 2010;32(4):397–413. doi: 10.1007/s00281-010-0222-z. [http://dx. doi.org/10.1007/s00281-010-0222-z]. [PMID: 20740284]. [DOI] [PubMed] [Google Scholar]
- 43.Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19(21):5720–5728. doi: 10.1093/emboj/19.21.5720. [http://dx.doi.org/10.1093/emboj/19.21.5720]. [PMID: 11060023]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ichimura Y., Kirisako T., Takao T., Satomi Y., Shimonishi Y., Ishihara N., Mizushima N., Tanida I., Kominami E., Ohsumi M., Noda T., Ohsumi Y. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408(6811):488–492. doi: 10.1038/35044114. [http://dx.doi.org/ 10.1038/35044114]. [PMID: 11100732]. [DOI] [PubMed] [Google Scholar]
- 45.Nakatogawa H., Ichimura Y., Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007;130(1):165–178. doi: 10.1016/j.cell.2007.05.021. [http://dx. doi.org/10.1016/j.cell.2007.05.021]. [PMID: 17632063]. [DOI] [PubMed] [Google Scholar]
- 46.Codogno P., Meijer A.J. Autophagy and signaling: their role in cell survival and cell death. Cell Death Differ. 2005;12(Suppl. 2):1509–1518. doi: 10.1038/sj.cdd.4401751. [http://dx.doi.org/10.1038/sj.cdd.4401751]. [PMID: 16247498]. [DOI] [PubMed] [Google Scholar]
- 47.Jäger S., Bucci C., Tanida I., Ueno T., Kominami E., Saftig P., Eskelinen E.L. Role for Rab7 in maturation of late autophagic vacuoles. J. Cell Sci. 2004;117(Pt 20):4837–4848. doi: 10.1242/jcs.01370. [http://dx.doi. org/10.1242/jcs.01370]. [PMID: 15340014]. [DOI] [PubMed] [Google Scholar]
- 48.Tanaka Y., Guhde G., Suter A., Eskelinen E.L., Hartmann D., Lüllmann-Rauch R., Janssen P.M., Blanz J., von Figura K., Saftig P. Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2-deficient mice. Nature. 2000;406(6798):902–906. doi: 10.1038/35022595. [http://dx.doi.org/10.1038/35022595]. [PMID: 10972293]. [DOI] [PubMed] [Google Scholar]
- 49.Xu F., Gu J.H., Qin Z.H. Neuronal autophagy in cerebral ischemia. Neurosci. Bull. 2012;28(5):658–666. doi: 10.1007/s12264-012-1268-9. [http://dx.doi.org/10. 1007/s12264-012-1268-9]. [PMID: 22968594]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schmelzle T., Hall M.N. TOR, a central controller of cell growth. Cell. 2000;103(2):253–262. doi: 10.1016/s0092-8674(00)00117-3. [http://dx.doi.org/10.1016/S0092-8674(00)00117-3]. [PMID: 11057898]. [DOI] [PubMed] [Google Scholar]
- 51.Guertin D.A., Sabatini D.M. The pharmacology of mTOR inhibition. Sci. Signal. 2009;2(67):pe24. doi: 10.1126/scisignal.267pe24. [http://dx.doi.org/10.1126/ scisignal.267pe24]. [PMID: 19383975]. [DOI] [PubMed] [Google Scholar]
- 52.Alessi D.R., James S.R., Downes C.P., Holmes A.B., Gaffney P.R., Reese C.B., Cohen P. Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Balpha. Curr. Biol. 1997;7(4):261–269. doi: 10.1016/s0960-9822(06)00122-9. [http://dx.doi. org/10.1016/S0960-9822(06)00122-9]. [PMID: 9094314]. [DOI] [PubMed] [Google Scholar]
- 53.Long X., Lin Y., Ortiz-Vega S., Yonezawa K., Avruch J. Rheb binds and regulates the mTOR kinase. Curr. Biol. 2005;15(8):702–713. doi: 10.1016/j.cub.2005.02.053. [http://dx.doi.org/10.1016/j.cub.2005.02.053]. [PMID: 15854902]. [DOI] [PubMed] [Google Scholar]
- 54.Inoki K., Li Y., Zhu T., Wu J., Guan K.L. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat. Cell Biol. 2002;4(9):648–657. doi: 10.1038/ncb839. [http://dx.doi.org/10.1038/ncb839]. [PMID: 12172553]. [DOI] [PubMed] [Google Scholar]
- 55.Gwinn D.M., Shackelford D.B., Egan D.F., Mihaylova M.M., Mery A., Vasquez D.S., Turk B.E., Shaw R.J. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol. Cell. 2008;30(2):214–226. doi: 10.1016/j.molcel.2008.03.003. [http://dx.doi.org/10.1016/j.molcel.2008.03. 003]. [PMID: 18439900]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jung C.H., Ro S.H., Cao J., Otto N.M., Kim D.H. mTOR regulation of autophagy. FEBS Lett. 2010;584(7):1287–1295. doi: 10.1016/j.febslet.2010.01.017. [http:// dx.doi.org/10.1016/j.febslet.2010.01.017]. [PMID: 20083114]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maiuri M.C., Galluzzi L., Morselli E., Kepp O., Malik S.A., Kroemer G. Autophagy regulation by p53. Curr. Opin. Cell Biol. 2010;22(2):181–185. doi: 10.1016/j.ceb.2009.12.001. [http://dx.doi.org/10.1016/j.ceb.2009.12.001]. [PMID: 20044243]. [DOI] [PubMed] [Google Scholar]
- 58.Morselli E., Shen S., Ruckenstuhl C., Bauer M.A., Mariño G., Galluzzi L., Criollo A., Michaud M., Maiuri M.C., Chano T., Madeo F., Kroemer G. p53 inhibits autophagy by interacting with the human ortholog of yeast Atg17, RB1CC1/FIP200. Cell Cycle. 2011;10(16):2763–2769. doi: 10.4161/cc.10.16.16868. [http://dx.doi.org/10.4161/cc.10.16.16868]. [PMID: 21775823]. [DOI] [PubMed] [Google Scholar]
- 59.Mah L.Y., O’Prey J., Baudot A.D., Hoekstra A., Ryan K.M. DRAM-1 encodes multiple isoforms that regulate autophagy. Autophagy. 2012;8(1):18–28. doi: 10.4161/auto.8.1.18077. [http://dx.doi.org/10.4161/auto.8.1.18077]. [PMID: 22082963]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gao W., Shen Z., Shang L., Wang X. Upregulation of human autophagy-initiation kinase ULK1 by tumor suppressor p53 contributes to DNA-damage-induced cell death. Cell Death Differ. 2011;18(10):1598–1607. doi: 10.1038/cdd.2011.33. [http://dx.doi.org/10.1038/cdd.2011.33]. [PMID: 21475306]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Feng Z., Zhang H., Levine A.J., Jin S. The coordinate regulation of the p53 and mTOR pathways in cells. Proc. Natl. Acad. Sci. USA. 2005;102(23):8204–8209. doi: 10.1073/pnas.0502857102. [http://dx.doi.org/10.1073/pnas. 0502857102]. [PMID: 15928081]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pattingre S., Tassa A., Qu X., Garuti R., Liang X.H., Mizushima N., Packer M., Schneider M.D., Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122(6):927–939. doi: 10.1016/j.cell.2005.07.002. [http://dx.doi.org/10.1016/j.cell.2005.07.002]. [PMID: 16179260]. [DOI] [PubMed] [Google Scholar]
- 63.Wei Y., Pattingre S., Sinha S., Bassik M., Levine B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol. Cell. 2008;30(6):678–688. doi: 10.1016/j.molcel.2008.06.001. [http://dx.doi.org/10. 1016/j.molcel.2008.06.001]. [PMID: 18570871]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sarkar S., Davies J.E., Huang Z., Tunnacliffe A., Rubinsztein D.C. Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and alpha-synuclein. J. Biol. Chem. 2007;282(8):5641–5652. doi: 10.1074/jbc.M609532200. [http://dx.doi.org/10. 1074/jbc.M609532200]. [PMID: 17182613]. [DOI] [PubMed] [Google Scholar]
- 65.Yang Y., Chen S., Zhang J. The updated role of oxidative stress in subarachnoid hemorrhage. Curr. Drug Deliv. 2016;14(6):832–842. doi: 10.2174/1567201813666161025115531. [PMID: 27784210]. [DOI] [PubMed] [Google Scholar]
- 66.Scherz-Shouval R., Shvets E., Fass E., Shorer H., Gil L., Elazar Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 2007;26(7):1749–1760. doi: 10.1038/sj.emboj.7601623. [http://dx.doi.org/10.1038/sj.emboj.7601623]. [PMID: 17347651]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.He C., Klionsky D.J. Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [http:// dx.doi.org/10.1146/annurev-genet-102808-114910]. [PMID: 19653858]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Azad M.B., Chen Y., Gibson S.B. Regulation of autophagy by reactive oxygen species (ROS): implications for cancer progression and treatment. Antioxid. Redox Signal. 2009;11(4):777–790. doi: 10.1089/ars.2008.2270. [http://dx.doi.org/10.1089/ars.2008.2270]. [PMID: 18828708]. [DOI] [PubMed] [Google Scholar]
- 69.Armour S.M., Baur J.A., Hsieh S.N., Land-Bracha A., Thomas S.M., Sinclair D.A. Inhibition of mammalian S6 kinase by resveratrol suppresses autophagy. Aging (Albany N.Y.) 2009;1(6):515–528. doi: 10.18632/aging.100056. [http://dx.doi.org/10.18632/aging.100056]. [PMID: 20157535]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Koren I., Reem E., Kimchi A. DAP1, a novel substrate of mTOR, negatively regulates autophagy. Curr. Biol. 2010;20(12):1093–1098. doi: 10.1016/j.cub.2010.04.041. [http://dx.doi.org/10.1016/j.cub.2010.04.041]. [PMID: 20537536]. [DOI] [PubMed] [Google Scholar]
- 71.Munakata A., Naraoka M., Katagai T., Shimamura N., Ohkuma H. Role of cyclooxygenase-2 in relation to nitric oxide and endothelin-1 on pathogenesis of cerebral vasospasm after subarachnoid hemorrhage in rabbit. Transl. Stroke Res. 2016;7(3):220–227. doi: 10.1007/s12975-016-0466-6. [http://dx.doi.org/10.1007/s12975-016-0466-6]. [PMID: 27044361]. [DOI] [PubMed] [Google Scholar]
- 72.Marbacher S. Animal models for the study of subarachnoid hemorrhage: Are we moving towards increased standardization? Transl. Stroke Res. 2016;7(1):1–2. doi: 10.1007/s12975-015-0442-6. [http://dx.doi.org/10.1007/s12975-015-0442-6]. [PMID: 26754973]. [DOI] [PubMed] [Google Scholar]
- 73.Caner B., Hou J., Altay O., Fujii M., Zhang J.H. Transition of research focus from vasospasm to early brain injury after subarachnoid hemorrhage. J. Neurochem. 2012;123(Suppl. 2):12–21. doi: 10.1111/j.1471-4159.2012.07939.x. [http:// dx.doi.org/10.1111/j.1471-4159.2012.07939.x]. [PMID: 23050638]. [DOI] [PubMed] [Google Scholar]
- 74.Cahill J., Calvert J.W., Zhang J.H. Mechanisms of early brain injury after subarachnoid hemorrhage. J. Cereb. Blood Flow Metab. 2006;26(11):1341–1353. doi: 10.1038/sj.jcbfm.9600283. [http://dx.doi.org/10.1038/sj.jcbfm. 9600283]. [PMID: 16482081]. [DOI] [PubMed] [Google Scholar]
- 75.Atangana E., Schneider U.C., Blecharz K., Magrini S., Wagner J., Nieminen-Kelha M., Kremenetskaia I., Heppner F.L., Engelhardt B., Vajkoczy P. Intravascular inflammation triggers intracerebral activated microglia and contributes to secondary brain injury after experimental subarachnoid hemorrhage (eSAH). Transl. Stroke Res. 2017;8(2):144–156. doi: 10.1007/s12975-016-0485-3. [PMID: 27477569]. [DOI] [PubMed] [Google Scholar]
- 76.He Y., Wan S., Hua Y., Keep R.F., Xi G. Autophagy after experimental intracerebral hemorrhage. J. Cereb. Blood Flow Metab. 2008;28(5):897–905. doi: 10.1038/sj.jcbfm.9600578. [http://dx.doi.org/10.1038/sj.jcbfm.9600578]. [PMID: 17987045]. [DOI] [PubMed] [Google Scholar]
- 77.Wen Y.D., Sheng R., Zhang L.S., Han R., Zhang X., Zhang X.D., Han F., Fukunaga K., Qin Z.H. Neuronal injury in rat model of permanent focal cerebral ischemia is associated with activation of autophagic and lysosomal pathways. Autophagy. 2008;4(6):762–769. doi: 10.4161/auto.6412. [http://dx.doi.org/10.4161/auto.6412]. [PMID: 18567942]. [DOI] [PubMed] [Google Scholar]
- 78.Smith C.M., Chen Y., Sullivan M.L., Kochanek P.M., Clark R.S. Autophagy in acute brain injury: feast, famine, or folly? Neurobiol. Dis. 2011;43(1):52–59. doi: 10.1016/j.nbd.2010.09.014. [http://dx.doi.org/10.1016/j.nbd. 2010.09.014]. [PMID: 20883784]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lee J.Y., He Y., Sagher O., Keep R., Hua Y., Xi G. Activated autophagy pathway in experimental subarachnoid hemorrhage. Brain Res. 2009;1287:126–135. doi: 10.1016/j.brainres.2009.06.028. [http://dx.doi.org/10.1016/j. brainres.2009.06.028]. [PMID: 19538949]. [DOI] [PubMed] [Google Scholar]
- 80.Prunell G.F., Mathiesen T., Svendgaard N.A. Experimental subarachnoid hemorrhage: cerebral blood flow and brain metabolism during the acute phase in three different models in the rat. 2004. [DOI] [PubMed]
- 81.Zhao H., Ji Z., Tang D., Yan C., Zhao W., Gao C. Role of autophagy in early brain injury after subarachnoid hemorrhage in rats. Mol. Biol. Rep. 2013;40(2):819–827. doi: 10.1007/s11033-012-2120-z. [http://dx.doi.org/10. 1007/s11033-012-2120-z]. [PMID: 23054025]. [DOI] [PubMed] [Google Scholar]
- 82.Jing C.H., Wang L., Liu P.P., Wu C., Ruan D., Chen G. Autophagy activation is associated with neuroprotection against apoptosis via a mitochondrial pathway in a rat model of subarachnoid hemorrhage. Neuroscience. 2012;213:144–153. doi: 10.1016/j.neuroscience.2012.03.055. [http://dx.doi.org/ 10.1016/j.neuroscience.2012.03.055]. [PMID: 22521819]. [DOI] [PubMed] [Google Scholar]
- 83.Cahill J., Calvert J.W., Marcantonio S., Zhang J.H. p53 may play an orchestrating role in apoptotic cell death after experimental subarachnoid hemorrhage. Neurosurgery. 2007;60(3):531–545. doi: 10.1227/01.NEU.0000249287.99878.9B. [http://dx.doi.org/10.1227/01.NEU.0000249287.99878.9B]. [PMID: 17327799]. [DOI] [PubMed] [Google Scholar]
- 84.Li P., Nijhawan D., Budihardjo I., Srinivasula S.M., Ahmad M., Alnemri E.S., Wang X. Cytochrome C and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell. 1997;91(4):479–489. doi: 10.1016/s0092-8674(00)80434-1. [http://dx.doi.org/10.1016/ S0092-8674(00)80434-1]. [PMID: 9390557]. [DOI] [PubMed] [Google Scholar]
- 85.Guo Y., Wang J., Wang Z., Yang Y., Wang X., Duan Q. Melatonin protects N2a against ischemia/reperfusion injury through autophagy enhancement. J. Huazhong Univ. Sci. Technolog. Med. Sci. 2010;30(1):1–7. doi: 10.1007/s11596-010-0101-9. [http://dx.doi.org/10.1007/s11596-010-0101-9]. [PMID: 20155447]. [DOI] [PubMed] [Google Scholar]
- 86.Chen J., Wang L., Wu C., Hu Q., Gu C., Yan F., Li J., Yan W., Chen G. Melatonin-enhanced autophagy protects against neural apoptosis via a mitochondrial pathway in early brain injury following a subarachnoid hemorrhage. J. Pineal Res. 2014;56(1):12–19. doi: 10.1111/jpi.12086. [http://dx.doi.org/10.1111/jpi.12086]. [PMID: 24033352]. [DOI] [PubMed] [Google Scholar]
- 87.Yan F., Li J., Chen J., Hu Q., Gu C., Lin W., Chen G. Endoplasmic reticulum stress is associated with neuroprotection against apoptosis via autophagy activation in a rat model of subarachnoid hemorrhage. Neurosci. Lett. 2014;563:160–165. doi: 10.1016/j.neulet.2014.01.058. [http://dx. doi.org/10.1016/j.neulet.2014.01.058]. [PMID: 24513235]. [DOI] [PubMed] [Google Scholar]
- 88.Shao A., Wang Z., Wu H., Dong X., Li Y., Tu S., Tang J., Zhao M., Zhang J., Hong Y. Enhancement of autophagy by histone deacetylase inhibitor trichostatin a ameliorates neuronal apoptosis after subarachnoid hemorrhage in rats. Mol. Neurobiol. 2016;53(1):18–27. doi: 10.1007/s12035-014-8986-0. [http://dx.doi.org/10.1007/s12035-014-8986-0]. [PMID: 25399954]. [DOI] [PubMed] [Google Scholar]
- 89.Liu Y., Li J., Wang Z., Yu Z., Chen G. Attenuation of early brain injury and learning deficits following experimental subarachnoid hemorrhage secondary to Cystatin C: possible involvement of the autophagy pathway. Mol. Neurobiol. 2014;49(2):1043–1054. doi: 10.1007/s12035-013-8579-3. [http://dx.doi.org/10.1007/s12035-013-8579-3]. [PMID: 24203677]. [DOI] [PubMed] [Google Scholar]
- 90.Tizon B., Sahoo S., Yu H., Gauthier S., Kumar A.R., Mohan P., Figliola M., Pawlik M., Grubb A., Uchiyama Y., Bandyopadhyay U., Cuervo A.M., Nixon R.A., Levy E. Induction of autophagy by cystatin C: a mechanism that protects murine primary cortical neurons and neuronal cell lines. PLoS One. 2010;5(3):e9819. doi: 10.1371/journal.pone.0009819. [http://dx.doi.org/10.1371/journal.pone.0009819]. [PMID: 20352108]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liu Y., Cai H., Wang Z., Li J., Wang K., Yu Z., Chen G. Induction of autophagy by cystatin C: a potential mechanism for prevention of cerebral vasospasm after experimental subarachnoid hemorrhage. Eur. J. Med. Res. 2013;18:21. doi: 10.1186/2047-783X-18-21. [http://dx.doi.org/ 10.1186/2047-783X-18-21]. [PMID: 23816364]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li T., Sun K.J., Wang H.D., Zhou M.L., Ding K., Lu X.Y., Wei W.T., Wang C.X., Zhou X.M. Tert-butylhydroquinone ameliorates early brain injury after experimental subarachnoid hemorrhage in mice by enhancing NRF2-independent autophagy. Neurochem. Res. 2015;40(9):1829–1838. doi: 10.1007/s11064-015-1672-4. [http://dx.doi.org/10. 1007/s11064-015-1672-4]. [PMID: 26260377]. [DOI] [PubMed] [Google Scholar]
- 93.Chen Y., Tachibana O., Hasegawa M., Xu R., Hamada J. ; Yamashita J., Hashimoto N., Takahashi J.A. Absence of tight junctions between microvascular endothelial cells in human cerebellar hemangioblastomas. Neurosurgery. 2006;59(3):660–670. doi: 10.1227/01.NEU.0000223372.18607.D7. [http://dx.doi.org/10.1227/01.NEU.0000223372.18607.D7]. [PMID: 16955048]. [DOI] [PubMed] [Google Scholar]
- 94.Liu X., Zhao J., Xu J., Zhao B., Zhang Y., Zhang S., Miao J. Protective effects of a benzoxazine derivative against oxidized LDL-induced apoptosis and the increases of integrin beta4, ROS, NF-kappaB and P53 in human umbilical vein endothelial cells. Bioorg. Med. Chem. Lett. 2009;19(10):2896–2900. doi: 10.1016/j.bmcl.2009.03.070. [http://dx.doi. org/10.1016/j.bmcl.2009.03.070]. [PMID: 19362839]. [DOI] [PubMed] [Google Scholar]
- 95.Xie Y., You S.J., Zhang Y.L., Han Q., Cao Y.J., Xu X.S., Yang Y.P., Li J., Liu C.F. Protective role of autophagy in AGE-induced early injury of human vascular endothelial cells. Mol. Med. Rep. 2011;4(3):459–464. doi: 10.3892/mmr.2011.460. [PMID: 21468592]. [DOI] [PubMed] [Google Scholar]
- 96.Li H., Gao A., Feng D., Wang Y., Zhang L., Cui Y., Li B., Wang Z., Chen G. Evaluation of the protective potential of brain microvascular endothelial cell autophagy on blood-brain barrier integrity during experimental cerebral ischemia-reperfusion injury. Transl. Stroke Res. 2014;5(5):618–626. doi: 10.1007/s12975-014-0354-x. [http://dx.doi.org/10. 1007/s12975-014-0354-x]. [PMID: 25070048]. [DOI] [PubMed] [Google Scholar]
- 97.Wieslander L. The cell nucleus. Exp. Cell Res. 2004;296(1):1–3. doi: 10.1016/j.yexcr.2004.03.003. [http://dx.doi.org/10.1016/j.yexcr.2004.03.003]. [PMID: 15120986]. [DOI] [PubMed] [Google Scholar]
- 98.Wang Z., Ma C., Meng C.J., Zhu G.Q., Sun X.B., Huo L., Zhang J., Liu H.X., He W.C., Shen X.M., Shu Z., Chen G. Melatonin activates the Nrf2-ARE pathway when it protects against early brain injury in a subarachnoid hemorrhage model. J. Pineal Res. 2012;53(2):129–137. doi: 10.1111/j.1600-079X.2012.00978.x. [http://dx.doi.org/10.1111/j.1600-079X. 2012.00978.x]. [PMID: 22304528]. [DOI] [PubMed] [Google Scholar]
- 99.Zhang J., Zhu Y., Zhou D., Wang Z., Chen G. Recombinant human erythropoietin (rhEPO) alleviates early brain injury following subarachnoid hemorrhage in rats: possible involvement of Nrf2-ARE pathway. Cytokine. 2010;52(3):252–257. doi: 10.1016/j.cyto.2010.08.011. [http://dx.doi. org/10.1016/j.cyto.2010.08.011]. [PMID: 20864352]. [DOI] [PubMed] [Google Scholar]
- 100.Nezis I.P., Stenmark H. p62 at the interface of autophagy, oxidative stress signaling, and cancer. Antioxid. Redox Signal. 2012;17(5):786–793. doi: 10.1089/ars.2011.4394. [http://dx.doi.org/10.1089/ars.2011.4394]. [PMID: 22074114]. [DOI] [PubMed] [Google Scholar]
- 101.Hensley K., Harris-White M.E. Redox regulation of autophagy in healthy brain and neurodegeneration. Neurobiol. Dis. 2015;84:50–59. doi: 10.1016/j.nbd.2015.03.002. [http://dx.doi.org/10.1016/j.nbd.2015.03.002]. [PMID: 25771170]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Repnik U., Stoka V., Turk V., Turk B. Lysosomes and lysosomal cathepsins in cell death. Biochim. Biophys. Acta. 2012;1824(1):22–33. doi: 10.1016/j.bbapap.2011.08.016. [http://dx.doi.org/10.1016/j.bbapap.2011.08.016]. [PMID: 21914490]. [DOI] [PubMed] [Google Scholar]
- 103.Wu H., Niu H., Wu C., Li Y., Wang K., Zhang J., Wang Y., Yang S. The autophagy-lysosomal system in subarachnoid haemorrhage. J. Cell. Mol. Med. 2016;20(9):1770–1778. doi: 10.1111/jcmm.12855. [http://dx.doi. org/10.1111/jcmm.12855]. [PMID: 27027405]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Droga-Mazovec G., Bojic L., Petelin A., Ivanova S., Romih R., Repnik U., Salvesen G.S., Stoka V., Turk V., Turk B. Cysteine cathepsins trigger caspase-dependent cell death through cleavage of bid and antiapoptotic Bcl-2 homologues. J. Biol. Chem. 2008;283(27):19140–19150. doi: 10.1074/jbc.M802513200. [http://dx.doi.org/10.1074/jbc.M802513200]. [PMID: 18469004]. [DOI] [PubMed] [Google Scholar]
- 105.Degtyarev M., De Mazière A., Orr C., Lin J., Lee B.B., Tien J.Y., Prior W.W., van Dijk S., Wu H., Gray D.C., Davis D.P., Stern H.M., Murray L.J., Hoeflich K.P., Klumperman J., Friedman L.S., Lin K. AKT inhibition promotes autophagy and sensitizes PTEN-null tumors to lysosomotropic agents. J. Cell Biol. 2008;183(1):101–116. doi: 10.1083/jcb.200801099. [http://dx.doi.org/10.1083/jcb.200801099]. [PMID: 18838554]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gonzalez P., Mader I., Tchoghandjian A., Enzenmüller S., Cristofanon S., Basit F., Debatin K.M., Fulda S. Impairment of lysosomal integrity by B10, a glycosylated derivative of betulinic acid, leads to lysosomal cell death and converts autophagy into a detrimental process. Cell Death Differ. 2012;19(8):1337–1346. doi: 10.1038/cdd.2012.10. [http://dx.doi.org/10.1038/cdd.2012.10]. [PMID: 22343715]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mizushima N., Levine B. Autophagy in mammalian development and differentiation. Nat. Cell Biol. 2010;12(9):823–830. doi: 10.1038/ncb0910-823. [http://dx.doi.org/10.1038/ncb0910-823]. [PMID: 20811354]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kaufman R.J. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 1999;13(10):1211–1233. doi: 10.1101/gad.13.10.1211. [http://dx.doi.org/ 10.1101/gad.13.10.1211]. [PMID: 10346810]. [DOI] [PubMed] [Google Scholar]
- 109.Zou X., Xu J., Yao S., Li J., Yang Y., Yang L. Endoplasmic reticulum stress-mediated autophagy protects against lipopolysaccharide-induced apoptosis in HL-1 cardiomyocytes. Exp. Physiol. 2014;99(10):1348–1358. doi: 10.1113/expphysiol.2014.079012. [http://dx.doi.org/10.1113/expphysiol. 2014.079012]. [PMID: 24951501]. [DOI] [PubMed] [Google Scholar]
- 110.Fouillet A., Levet C., Virgone A., Robin M., Dourlen P., Rieusset J., Belaidi E., Ovize M., Touret M., Nataf S., Mollereau B. ER stress inhibits neuronal death by promoting autophagy. Autophagy. 2012;8(6):915–926. doi: 10.4161/auto.19716. [http://dx.doi.org/10.4161/auto.19716]. [PMID: 22660271]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Levine B., Yuan J. Autophagy in cell death: an innocent convict? J. Clin. Invest. 2005;115(10):2679–2688. doi: 10.1172/JCI26390. [http://dx.doi.org/10. 1172/JCI26390]. [PMID: 16200202]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mizushima N., Levine B., Cuervo A.M., Klionsky D.J. Autophagy fights disease through cellular self-digestion. Nature. 2008;451(7182):1069–1075. doi: 10.1038/nature06639. [http://dx.doi.org/10.1038/nature06639]. [PMID: 18305538]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Salazar M., Carracedo A., Salanueva I.J., Hernández-Tiedra S., Lorente M., Egia A., Vázquez P., Blázquez C., Torres S., García S., Nowak J., Fimia G.M., Piacentini M., Cecconi F., Pandolfi P.P., González-Feria L., Iovanna J.L., Guzmán M., Boya P., Velasco G. Cannabinoid action induces autophagy-mediated cell death through stimulation of ER stress in human glioma cells. J. Clin. Invest. 2009;119(5):1359–1372. doi: 10.1172/JCI37948. [http://dx.doi.org/10. 1172/JCI37948]. [PMID: 19425170]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chan D.C. Mitochondria: dynamic organelles in disease, aging, and development. Cell. 2006;125(7):1241–1252. doi: 10.1016/j.cell.2006.06.010. [http://dx.doi.org/ 10.1016/j.cell.2006.06.010]. [PMID: 16814712]. [DOI] [PubMed] [Google Scholar]
- 115.Kroemer G., Dallaporta B., Resche-Rigon M. The mitochondrial death/life regulator in apoptosis and necrosis. Annu. Rev. Physiol. 1998;60:619–642. doi: 10.1146/annurev.physiol.60.1.619. [http://dx.doi.org/10.1146/annurev.physiol.60. 1.619]. [PMID: 9558479]. [DOI] [PubMed] [Google Scholar]
- 116.Youle R.J., Narendra D.P. Mechanisms of mitophagy. Nat. Rev. Mol. Cell Biol. 2011;12(1):9–14. doi: 10.1038/nrm3028. [http://dx.doi.org/10.1038/ nrm3028]. [PMID: 21179058]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Li J., Lu J., Mi Y., Shi Z., Chen C., Riley J., Zhou C. Voltage-dependent anion channels (VDACs) promote mitophagy to protect neuron from death in an early brain injury following a subarachnoid hemorrhage in rats. Brain Res. 2014;1573:74–83. doi: 10.1016/j.brainres.2014.05.021. [http://dx.doi.org/10.1016/j.brainres.2014.05.021]. [PMID: 24880016]. [DOI] [PubMed] [Google Scholar]
- 118.Kubli D.A., Gustafsson A.B. Mitochondria and mitophagy: the yin and yang of cell death control. Circ. Res. 2012;111(9):1208–1221. doi: 10.1161/CIRCRESAHA.112.265819. [http://dx.doi.org/10.1161/CIRCRESAHA.112.265819]. [PMID: 23065344]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Itoh T., Tabuchi M., Mizuguchi N., Imano M., Tsubaki M., Nishida S., Hashimoto S., Matsuo K., Nakayama T., Ito A., Munakata H., Satou T. Neuroprotective effect of (-)-epigallocatechin-3-gallate in rats when administered pre- or post-traumatic brain injury. J. Neural Transm. (Vienna) 2013;120(5):767–783. doi: 10.1007/s00702-012-0918-4. [http://dx.doi.org/10.1007/s00702-012-0918-4]. [PMID: 23180302]. [DOI] [PubMed] [Google Scholar]
- 120.Hyung S.J., DeToma A.S., Brender J.R., Lee S., Vivekanandan S., Kochi A., Choi J.S., Ramamoorthy A., Ruotolo B.T., Lim M.H. Insights into antiamyloidogenic properties of the green tea extract (-)-epigallocatechin-3-gallate toward metal-associated amyloid-β species. Proc. Natl. Acad. Sci. USA. 2013;110(10):3743–3748. doi: 10.1073/pnas.1220326110. [http://dx.doi.org/10.1073/pnas.1220326110]. [PMID: 23426629]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wang Y., Gao A., Xu X., Dang B., You W., Li H., Yu Z., Chen G. The neuroprotection of lysosomotropic agents in experimental subarachnoid hemorrhage Probably involving the apoptosis pathway triggering by Cathepsins via chelating intralysosomal Iron. Mol. Neurobiol. 2015;52(1):64–77. doi: 10.1007/s12035-014-8846-y. [http://dx.doi.org/10.1007/ s12035-014-8846-y]. [PMID: 25112680]. [DOI] [PubMed] [Google Scholar]
- 122.Wu H.J., Pu J.L., Krafft P.R., Zhang J.M., Chen S. The molecular mechanisms between autophagy and apoptosis: potential role in central nervous system disorders. Cell. Mol. Neurobiol. 2015;35(1):85–99. doi: 10.1007/s10571-014-0116-z. [http://dx.doi.org/10.1007/s10571-014-0116-z]. [PMID: 25257832]. [DOI] [PMC free article] [PubMed] [Google Scholar]