Abstract
Background
A variety of treatment strategies have been developed for clear cell kidney carcinoma (KIRC); however, there is still a need for effective therapeutic targets and prognostic molecular biomarkers. Given that long noncoding RNAs (lncRNAs) has been emerging as an important regulator in tumorigenesis, we explored potential functional lncRNAs in KIRC by comprehensively analyzing the lncRNA–miRNA–mRNA regulatory network with bioinformatics processing tools.
Material/Methods
RNA-seq/miRNA-seq data of KIRC in The Cancer Genome Atlas (TCGA) were obtained and analyzed. The “edgeR” package in R software was used to identify differentially expressed lncRNAs (DElncRNAs, differentially expressed long noncoding RNAs), miRNAs (DEmiRNAs, differentially expressed micro RNAs), and mRNAs (DEmRNAs, differentially expressed messenger RNAs) in KIRC and normal samples. A global triple network was conducted based on the competing endogenous RNA (ceRNA) theory, and survival analysis was conducted by “survival” package in R software.
Results
A total of 4246 DElncRNAs, 179 DEmiRNAs, and 5758 DEmRNAs were identified, among which a subset of them (321 lncRNAs, 26 miRNAs, and 1068 mRNAs) were found to constitute a global ceRNA network in KIRC. Four lncRNAs (ENTPD3-AS1, FGD5-AS1, LIFR-AS1, and UBAC2-AS1) were revealed to be potential therapeutic targets as well as prognostic biomarkers of KIRC by our extensive functional analysis.
Conclusions
We reported here the identification of functional lncRNAs in KIRC via a TCGA data-based bioinformatics analysis. We believe that this study might contribute to improving the comprehension of the lncRNA-mediated ceRNA regulatory mechanisms in the tumorigenesis of KIRC. Meanwhile, our results suggested that 4 lncRNAs might act as potential therapeutic targets or candidate prognostic biomarkers in KIRC.
MeSH Keywords: Carcinoma, Renal Cell; MicroRNAs; RNA, Long Noncoding
Background
Renal cancer, which ranks the top 10 most frequent cancer in humans, is a matter of great public health concern in the world. In the United States, the estimated new cases and deaths of renal cancer in both genders will be 65 349 and 14 970 in 2018, respectively [1]. Being the most common subtype, clear cell kidney carcinoma (KIRC) accounts for about 75% of all renal cancers. Histologically, the KIRC cells have clear cytoplasm with nested clusters of cells surrounded by a delicate vascular network [2,3]. Currently, multiple clinical managements, including surgery and radiation therapy, are available for patients with KIRC in an early stage; the 5-year survival rate after diagnosis has shown some improvement in recent years. However, the overall prognosis remains poor especially for patients with advanced disease, mostly due to lacking more effective therapeutics [4]. Thus, a more comprehensive understanding of the underlyingly molecular mechanism of KIRC is in urgent need for further development of novel treatment strategy against this disease.
In addition to oncoprotein as tumor suppressor, non-protein coding RNAs such as microRNAs (miRNAs) and long noncoding RNAs (lncRNAs) are emerging as important regulators in tumor biology. Actually, the detection of extensive RNAs transcribed from non-protein coding regions of the genome is one of the most inconceivable findings of the genomics era of biology [5,6]. The lncRNAs, functionally defined as an RNA transcript longer than 200 nucleotides in length that cannot be translated into a protein, widely exist in metazoans including humans [7]. In general, the exons of lncRNAs are more divergent compared with their promoters. In addition, lncRNAs are less evolutionarily conserved compared to protein-coding genes. During the past decade, thousands of lncRNAs have been identified, and many of which have been shown to be uniquely expressed at specific times in different tissues or specific cancer types, however, most of them have not been functionally characterized [8–10]. Fortunately, with the rapid progress of technologies, such as high-throughput RNA sequencing, it has becoming more feasible to unravel the precise function of lncRNAs in tumor biology [11].
Recent studies suggested that lncRNAs might be involved in epigenetic regulation of the expression of proteins and play an integral part in a series of physiological and pathological processes such as tumorigenesis of various human cancers [12,13]. According to the competing endogenous RNA (ceRNA) hypothesis, lncRNAs, mRNAs (messenger RNAs), and other RNAs, all of which serve as natural miRNA sponges to restrain the function of intracellular miRNA via sharing one or more miRNA response elements (MREs) [14]. This hypothesis has been verified by accumulated experimental evidence, and the critical roles of lncRNAs have been identified gradually [15–18].
The Cancer Genome Atlas (TCGA), a publicly available database with information of the clinical pathology of 11 000 patients with over 30 cancers, has help to improve the diagnostic accuracy and the effectiveness of treatment in a wide range of human malignant diseases [19]. These immense data provide an ideal opportunity for investigators to comprehensively explore the molecular mechanisms of tumorigenesis of various cancers as well as identify novel therapeutic targets and prognostic molecular markers. In an attempt to unravel the lncRNA–miRNA–mRNA regulatory network in KIRC, the RNA-seq/miRNA-seq data from 530 KIRC cases in TCGA were downloaded and subjected to comprehensive analysis with bioinformatics tools. Our study reconstructed a global lncRNA-miRNA-mRNA ceRNA network in KIRC. Moreover, relevant survival and location analyses of lncRNAs were performed to determine the potential therapeutic targets or candidate prognostic biomarkers in KIRC. The verification of their exact biological function awaits further investigation.
Material and Methods
Collection of raw TCGA data
The RNA-seq/miRNA-seq data and clinical information were downloaded from GDC Data Portal (https://portal.gdc.cancer.gov/) for comprehensive integrated analysis with Data Transfer Tool (provided by GDC Apps) according to the published guidelines provided by TCGA (http://cancergenome.nih.gov/publications/publicationguidelines). All TCGA data are now available without restrictions on their use in publications or presentations according to the posted statement from the TCGA website. The TCGA website lists “clear cell kidney carcinoma (KIRC)” as a cancer in the database with “no restrictions; all data available without limitations”. Further analysis was approved by the Ethics Committee of Fuzhou Dongfang Hospital, Xiamen University.
Exploring of differentially expressed lncRNAs (DElncRNAs), DEmiRNAs, and DEmRNAs in KIRC
The KIRC RNA-Seq data were derived from 539 KIRC samples and 72 matched normal samples of 530 cases for analysis of mRNAs and lncRNAs. Simultaneously, the miRNA-seq data of 545 KIRC samples and 71 matched normal samples of 516 cases were obtained for analysis of miRNAs. Meanwhile, expression data closing to zero were excluded and data from tumor samples and normal samples were merged. To explore the DEmRNAs, DEmiRNAs, and DElncRNAs, we used R software (https://www.r-project.org/) with the “edgeR” package to compare the KIRC with the normal samples. The adjust P-values were used to reduce the false-positive rate using Benjamini-Hochberg (false discovery rate) method by default. The threshold was set as |log2 (fold change [FC]) | >1.0 and adjusted P-value <0.01.
Correlation analysis of lncRNAs, miRNAs, and mRNAs and construction of lncRNA-miRNA-mRNA network
Perl software (https://www.perl.org/) was used to explore the DEmiRNAs interacting with DElncRNAs in the mircode database (http://www.mircode.org/). The obtained DEmiRNAs were standardized in the starBase database (http://starbase.sysu.edu.cn/), while their target genes were found in 3 databases of miRNAs (miRDB_http://www.mirdb.org/, miRTarBase_http://mirtarbase.mbc.nctu.edu.tw/php/index.php, and TargetScan_http://www.targetscan.org/vert_71/) using Perl software with the criteria that each target gene appears at least in 2 different databases. Nonconforming target genes were excluded. Predicted target genes of miRNAs were intersected with DEmRNAs by “VennDiagrams” package in R software. Then, the lncRNA–miRNA–mRNA network was reconstructed based on the ceRNA theory as follows: 1) for a given co-expressed lncRNA–mRNA pair, both mRNA and lncRNA in this pair were targeted by and co-expressed inversely with a certain common miRNA, and this lncRNA–miRNA–mRNA was identified as the co-expression competing triplet. 2) The lncRNA–miRNA–mRNA network was reconstructed by assembling all identified co-expression competing triplets and was visualized using Cytoscape software (http://www.cytoscape.org/).
Functional enrichment analysis
To interactively analyze the Kyoto Encyclopedia of Genes and Genomes (KEGG, http://www.genome.jp/kegg/) pathways of the DEmRNAs in the network, the plug-in ClueGo in Cytoscape was conducted and the P value <0.05 was set as the screening condition. Gene Ontology (GO, http://www.geneontology.org) function enrichment analysis was performed based on GO database in FunRich (http://www.funrich.org/) with 3 main categories including molecular function (MF), biological process (BP), and cellular component (CC).
Survival analysis
To reveal the potential prognostic miRNAs and lncRNAs biomarker, the KIRC patients’ clinical data from TCGA were downloaded and the Kaplan-Meier method survival analysis of DEmiRNAs and DElncRNAs in the network was carried out using R software with “survival” package with a threshold of P-value <0.05.
Reconstruction of the lncRNA–miRNA–mRNA subnetwork
The lncRNAs’ location information was searched in lncATLAS (http://lncatlas.crg.eu/) based on relative concentration index (RCI), and the obtained lncRNAs located in the cytoplasm were retained for subsequence analysis. For the interest lncRNAs, their linked miRNAs and mRNAs in the global triple network were extracted and used to reconstruct the new subnetwork using Cytoscape software. To enhance the data’s reliability, only lncRNAs and miRNAs that have meaningful survival curve (P<0.05) were retained. Thereafter, GO-KEGG intersection networks were reconstructed using Metascape (http://metascape.org).Terms with P-value <0.01, minimum count 3, and enrichment factor >1.5 were collected according to the Metascape’s default parameters, and grouped into clusters based on their membership similarities.
Statistical analysis
For functional enrichment analysis conducted by Cytoscape plug-in ClueGo, the P-value was calculated with Fisher’s exact test. For overall survival (OS) analysis, we calculated the survival rate with the Kaplan-Meier method, compared the survival curves in Log rank test. A P-value less than 0.05 was considered as statistically significance.
Results
Identifying DEmRNAs, DEmiRNAs, and DElncRNAs in KIRC
To explore DEmRNAs, DEmiRNAs, and DElncRNAs in KIRC, R software with the “edgeR” package was used to compare the expression levels of mRNAs, miRNAs, and lncRNAs in KIRC with that in the normal tissue group. As a result, a total of 5758 DEmRNAs, 179 DEmiRNAs, and 4246 DElncRNAs were identified in KIRC, among which 3895 DEmRNAs, 117 DEmiRNAs, and 3116 DElncRNAs were upregulated, while 1863 DEmRNAs, 62 DEmiRNAs, and 1130 DElncRNAs were downregulated. The top 10 DEmRNAs with |log2FC| >3.0, DEmiRNAs with |log2FC| >3.0, and DElncRNAs with |log2FC| >3.0 are shown in Table 1.
Table 1.
Lists of top 10 DEmRNAs, top 10 DEmiRNAs, and top 10 DElncRNAs.
| Gene symbol | logFC | logCPM | Pvalue | FDR | Stage |
|---|---|---|---|---|---|
| Top 10 DEmRNAs | |||||
| GSG1L2 | 10.13932235 | 2.609234989 | 1.75E-26 | 9.44E-26 | Up |
| PAEP | 9.296582129 | 3.215953219 | 3.56E-22 | 1.55E-21 | Up |
| MUC17 | 8.897464861 | 1.38316872 | 2.68E-24 | 1.29E-23 | Up |
| SLC18A3 | 8.731543336 | 1.621173476 | 1.20E-20 | 4.85E-20 | Up |
| CFHR5 | 8.337594048 | −0.148783213 | 2.63E-12 | 6.78E-12 | Up |
| AQP2 | −8.921997403 | 8.615757188 | 2.03E-135 | 3.47E-133 | Down |
| UMOD | −8.538154717 | 10.10423007 | 8.06E-129 | 1.18E-126 | Down |
| SLC12A1 | −8.248671314 | 8.546152794 | 1.71E-237 | 1.82E-234 | Down |
| TMEM207 | −8.117681211 | 0.883434713 | 3.88E-124 | 5.16E-122 | Down |
| ELF5 | −7.828424697 | 2.601269721 | 2.84E-278 | 6.42E-275 | Down |
| Top 10 DEmiRNAs | |||||
| hsa-mir-122 | 6.469661662 | 4.643895842 | 1.23E-80 | 4.01E-79 | Up |
| hsa-mir-875 | 4.339977994 | 0.858647482 | 6.01E-15 | 2.20E-14 | Up |
| hsa-mir-1293 | 4.02677539 | 0.570800834 | 1.44E-18 | 6.19E-18 | Up |
| hsa-mir-891a | 3.888011524 | 9.67019917 | 2.99E-07 | 6.40E-07 | Up |
| hsa-mir-4773-1 | 3.849423442 | 0.157334403 | 6.36E-29 | 4.94E-28 | Up |
| hsa-mir-514b | −6.0169279 | 0.356895636 | 3.42E-154 | 3.35E-152 | Down |
| hsa-mir-934 | −5.77369524 | 0.448266948 | 4.56E-135 | 3.19E-133 | Down |
| hsa-mir-506 | −5.604961065 | 0.738941469 | 4.48E-169 | 1.10E-166 | Down |
| hsa-mir-514a-3 | −4.352581963 | 3.475332738 | 9.52E-155 | 1.17E-152 | Down |
| hsa-mir-514a-1 | −4.349319616 | 3.501254002 | 5.14E-155 | 8.39E-153 | Down |
| Top 10 DElncRNAs | |||||
| OSTM1-AS1 | 8.424888513 | 9.217351016 | 3.58E-58 | 1.13E-56 | Up |
| TTC21B-AS1 | 8.213393051 | 11.47965363 | 2.90E-89 | 2.11E-87 | Up |
| AC113410.2 | 7.418820525 | 5.251043499 | 7.53E-12 | 2.21E-11 | Up |
| AC008060.4 | 7.188134257 | 5.571997961 | 1.07E-16 | 4.49E-16 | Up |
| AL590644.1 | 6.951085884 | 8.89791889 | 2.80E-59 | 9.41E-58 | Up |
| AC079310.1 | −8.444248085 | 4.601017824 | 4.95E-229 | 2.97E-226 | Down |
| LINC02437 | −8.12004929 | 6.534142577 | 9.71E-217 | 4.38E-214 | Down |
| LINC02121 | −8.115309639 | 6.068605258 | 5.76E-126 | 7.42E-124 | Down |
| AC073336.1 | −8.045973803 | 3.717169509 | 1.99E-161 | 4.26E-159 | Down |
| AC090709.1 | −7.615037785 | 4.427023078 | 3.86E-301 | 6.95E-298 | Down |
FC – fold change; CPM – counts per million; FDR – false discovery rate; DEmRNAs – differentially expressed messenger RNAs; DEmiRNAs – differentially expressed micro RNAs; DElncRNAs – differentially expressed long noncoding RNAs.
Construction of lncRNA-miRNA-mRNA network in KIRC
Next, in an effort to construct lncRNA-miRNA-mRNA network in KIRC, the MREs of DElncRNAs were first explored, and then 399 of 4246 DElncRNAs were predicted by miRcode to share MREs mediating the binding of 26 DEmiRNAs. Importantly, several miRNAs such as hsa-miR-21, hsa-miR-155, hsa-miR-204, and hsa-miR-221, and lncRNAs such as MALAT1 and TCL6 have been previously reported to be highly associated with KIRC [20–25]. Then, the possible mRNA targets of all 26 DEmiRNAs were searched using Perl software based on 3 databases of miRNAs (miRDB, miRTarBase, and TargetScan). According to the searching criteria described, a total of 7233 potential mRNA targets were found with each appearing at least in 2 different miRNAs databases. These mRNA targets were then crossed with 5758 identified DEmRNAs using VennDiagrams (http://bioinformatics.psb.ugent.be/webtools/Venn/) to sort out 1675 intersection genes in KIRC. Interestingly, some of intersection genes such as FOXO1 have been demonstrated to have a significant correlation with KIRC, such as oncogenesis [26], which indicated that those data we obtained are valuable for further investigation.
Cytoscape, as an open source software platform, is widely employed to integrate any type of attribute data for visualizing complex networks. Thus, based on current information we received, the lncRNA-miRNA-mRNA network in KIRC was constructed and visualized using Cytoscape (Figure 1). The constructed lncRNA-miRNA-mRNA network was composed of 321 lncRNA nodes, 26 miRNA nodes, 1068 mRNA nodes, and 2313 edges. These nodes include 221 upregulated lncRNAs and 100 downregulated lncRNAs, 13 upregulated miRNAs and 13 downregulated miRNAs, and 599 upregulated mRNAs and 469 downregulated mRNAs.
Figure 1.
The holistic view of the lncRNA–miRNA–mRNA network. The diamond represents lncRNAs, the round rectangle represents miRNAs, and the ellipse represents mRNAs. The red color represents upregulated and the blue color represents downregulated, respectively. Each edge represents interaction between 2 nodes. There were 321 lncRNAs, 26 miRNAs, 1068 mRNAs, and 2313 edges in the network. LncRNAs – long noncoding RNAs; miRNAs – microRNAs; mRNAs – messenger RNAs.
Functional pathways revealed by GO enrichment analysis
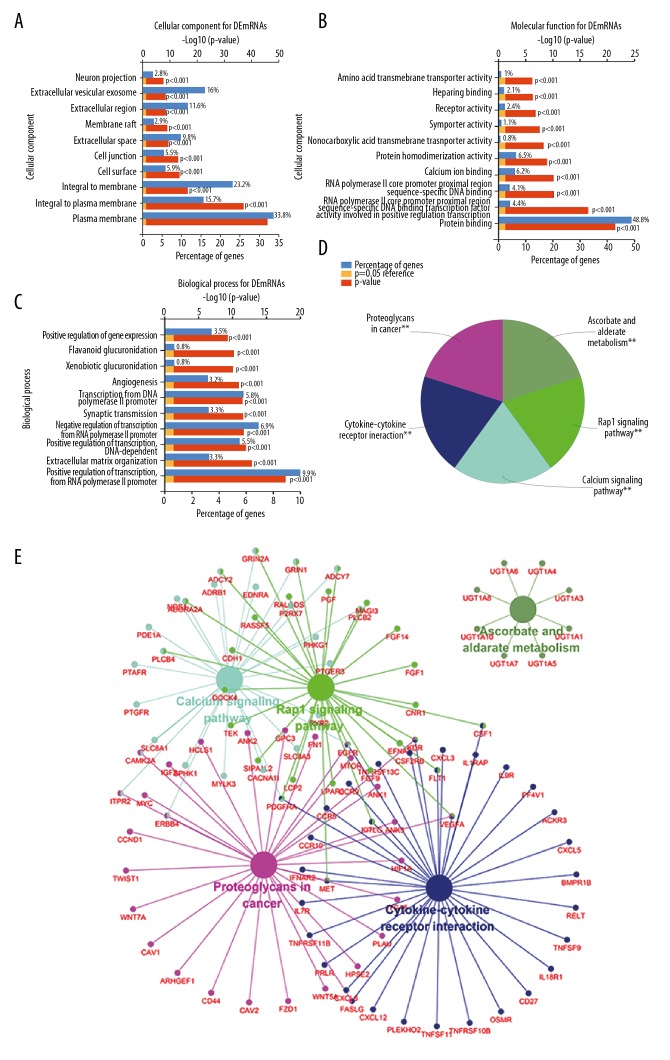
To further unravel the functional pathways that the constructed lncRNA–miRNA–mRNA network in KIRC might involve, GO function enrichment analysis was carried out with FunRich software. Cellular component analysis indicated that proteins encoded by those DEmRNAs are mainly located in plasma membrane, integral to plasma membrane, or extracellular vesicular exosome (Figure 2A). Their molecular functions involved in protein binding, protein homodimerization activity, receptor activity, etc. (Figure 2B), which mainly participate in biological processes such as positive regulation of transcription from RNA polymerase II promoter, transcription from RNA polymerase II promoter, angiogenesis, etc. (Figure 2C). Since as a functional plug-in in Cytoscape, ClueGO can functionally annotate gene ontology as well as visualize pathway. Thus, the KEGG pathway enrichment analysis was conducted by ClueGO (kappa score threshold=0.4). As shown in Figure 2D and 2E, the top 5 pathways revealed by KEGG pathway enrichment analysis are mainly involved in the proteoglycans in cancer, cytokine-cytokine receptor interaction, Rap1 signaling pathway, calcium signaling pathway, and ascorbate and aldarate metabolism.
Figure 2.
GO enrichment and KEGG analysis of DEmRNAs in the network. (A–C) Cellular component, molecular function, and biological process enrichments for DEmRNAs in the network. (D, E) KEGG pathway analysis of DEmRNAs in the network. Statistical significance was accepted at the P<0.05 level (** represents P<0.05). GO – Gene Ontology; KEGG – Kyoto Encyclopedia of Genes and Genomes; DE – differentially expressed; mRNA – messenger RNA.
Identification of prognostic miRNAs and lncRNAs in KIRC
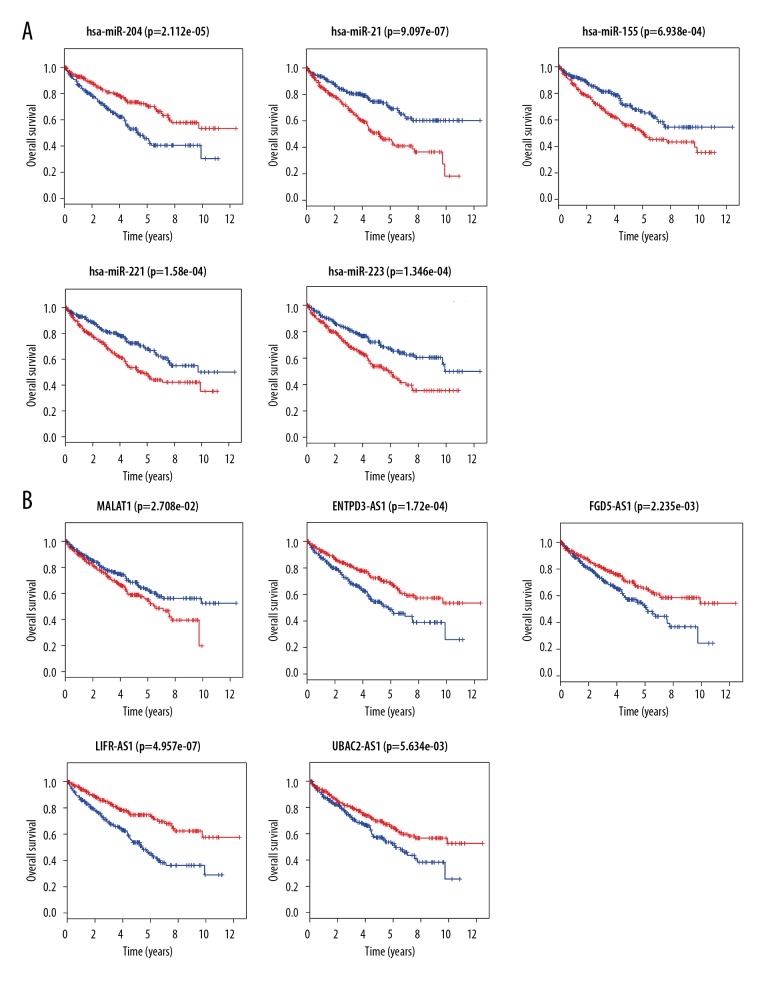
After a successful construction of lncRNA-miRNA-mRNA network in KIRC and recognition of functional pathways that the aforementioned network might involve, to further investigate the lncRNAs and miRNAs participating in the regulation of the development and progression of KIRC, the Kaplan-Meier survival analysis of DEmiRNAs and DElncRNAs in the network was carried out using R software with “survival” package. In a total of 26 DEmiRNAs analyzed, 5 of them were shown to significantly associate with patients’ overall survival (OS, P<0.05). While higher expression of miR-204 was shown to correlate with favorable outcome of patients with KIRC; on the contrary, the expression levels of miR-21, miR-155, miR-221, and miR-223 were inversely correlated with patients’ OS (Figure 3A). In terms of 321 DElncRNAs analyzed, there were 49 DElncRNAs which associated with not only the aforementioned 5 DEmiRNAs but also patients’ OS (P<0.05). For example, as shown in Figure 3B, while the expression of some DElncRNAs such as MALAT1 exhibits inversely correlation with patients’ OS, the expression of others such as ENTPD3-AS1, FGD5-AS1, LIFR-AS1, and UBAC2-AS1 exhibits positively correlation with patients’ OS. Taken together, our results indicated that 49 DElncRNAs and their associated 5 DEmiRNAs might serve as prognostic biomarkers in KIRC
Figure 3.
Survival analysis of miRNAs and lncRNAs. (A) Survival analysis of miRNAs. (B) Survival analysis of lncRNAs. Low expression samples are in blue while high expression samples are in red. Survival years are shown along the x-axis. Overall survival rates are shown along the y-axis. LncRNAs – long noncoding RNAs; miRNAs – microRNAs.
Reconstruction of the functional lncRNA–miRNA–mRNA subnetwork in KIRC
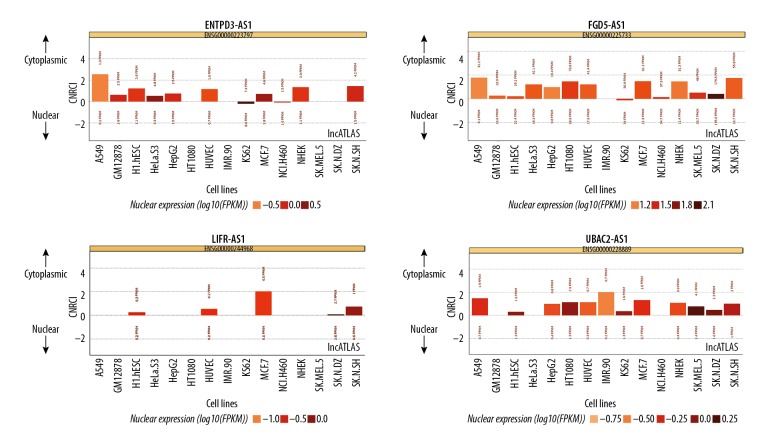
Since based on the ceRNA hypothesis, the competitive binding of target miRNAs by lncRNAs might mainly happen in cellular compartment of cytoplasm; thus, to reconstruct a more specific lncRNA–miRNA–mRNA subnetwork in KIRC, those identified prognostic DElncRNAs located in cytoplasm were sorted out first via analysis with lncATLAS. Among 49 DElncRNAs analyzed, 17 of them were excluded because they lacked localization data. In addition, 19 DElncRNAs located in nuclear and 5 DElncRNAs whose localization had no significant difference between the cytoplasm and nuclear were also excluded. Although 8 remaining DElncRNAs were all shown to locate in the cytoplasm, 4 of them (DIAP2-AS1, HULC, LINC00443, and PHEX-AS1) were still excluded because the localization could only be validated in one cell line. Ultimately, 4 DElncRNAs (ENTPD3-AS1, FGD5-AS1, LIFR-AS1, and UBAC2-AS1) whose cytoplasma location was verified in 9, 13, 5, and 12 different cell lines, respectively, were chosen for further investigation (Figure 4). Interestingly, the expression of all 4 DElncRNAs was shown to be positively correlated with OS of patients with KIRC (Figure 3B).
Figure 4.
LncRNAs’ cytoplasmic/nuclear localization: RCI and expression values (15 cell types). LncRNAs – long noncoding RNAs; RCI – relative concentration index.
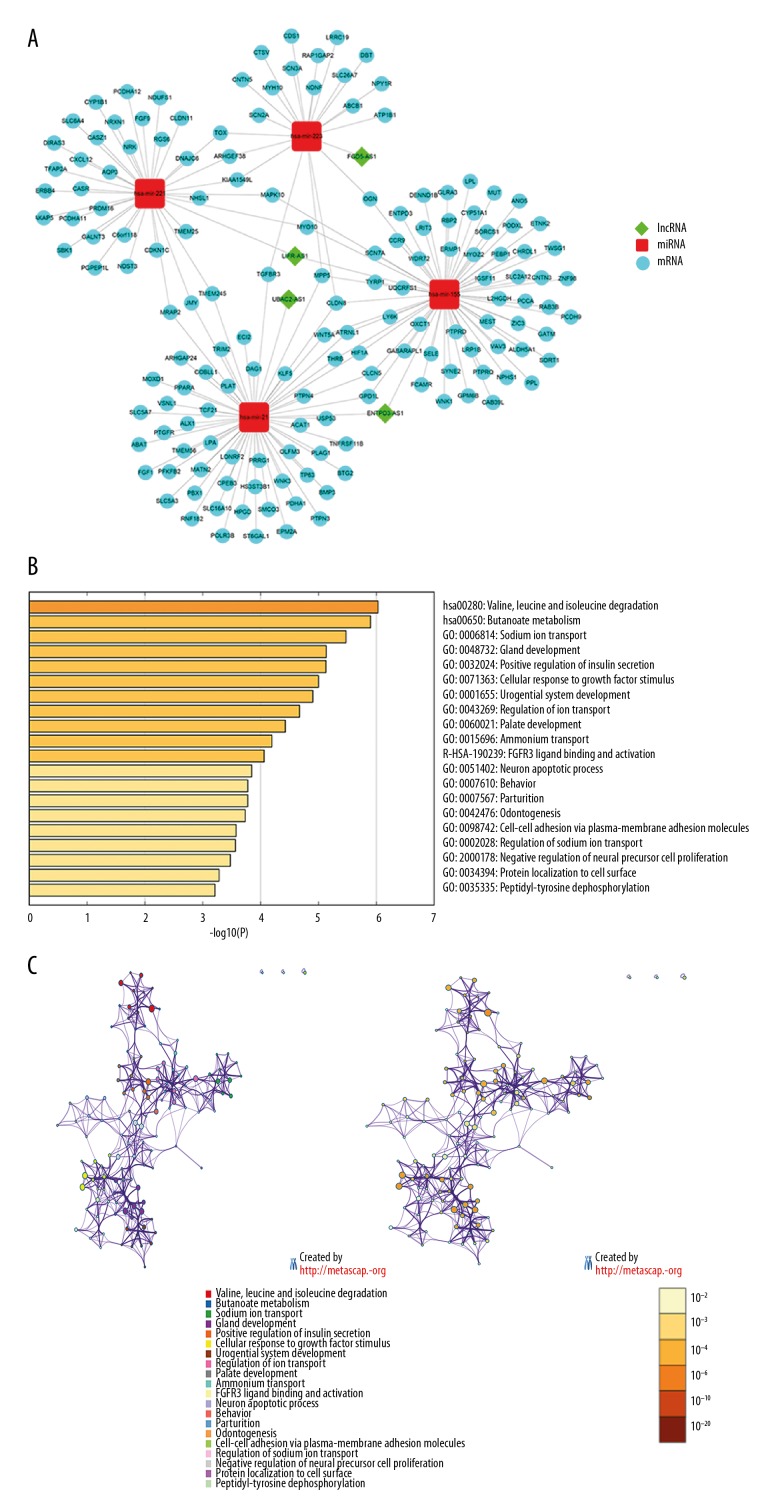
Again, a more specific functional lncRNA-miRNA-mRNA subnetwork in KIRC was reconstructed based on 4 DElncRNAs located in the cytoplasm with the use of Cytoscape (Figure 5A). This newly reconstructed subnetwork was comprised of 4 lncRNA nodes, 4 miRNA nodes, 155 mRNA nodes, and 181 edges. Further analysis of these 155 mRNAs with Metascape revealed that the top 3 enriched terms are hsa00280 (valine, leucine and isoleucine degradation), hsa00650 (butanoate metabolism), and GO: 0006814 (sodium ion transport) with most significant log10 P-values being −6.03, −5.90, and −5.48, respectively (Figure 5B). Additionally, all other identified biological processes with a significant P-value were closely interrelated and largely reflected a change in the cytokine regulation and tissue development related pathways (Figure 5C). Top 20 clusters with their representative enriched terms were shown in Table 2.
Figure 5.
The sub-network of lncRNA-miRNA-mRNA and GO-KEGG intersection networks. (A) The sub-network of 4 lncRNAs. The diamond represents lncRNAs, the round rectangle represents miRNAs, and the ellipse represents mRNAs. Each edge represents interaction between 2 nodes. (B) Heatmap of the top 20 enriched terms, colored by P-values. (C) Network of enriched terms, colored by cluster ID and P-value, respectively. Nodes share the same cluster are typically close to each other and terms containing more genes tend to have a more significant P-value. LncRNAs – long noncoding RNAs miRNAs – microRNAs; mRNAs – messenger RNAs; GO – Gene Ontology; KEGG – Kyoto Encyclopedia of Genes and Genomes.
Table 2.
Top 20 clusters with their representative enriched term.
| GO | Category | Description | Count | % | Log10(P) | Log10(q) |
|---|---|---|---|---|---|---|
| hsa00280 | KEGG pathway | Valine, leucine and isoleucine degradation | 6 | 3.87 | −6.03 | −2.2 |
| hsa00650 | KEGG pathway | Butanoate metabolism | 5 | 3.23 | −5.9 | −2.2 |
| GO: 0006814 | GO biological processes | Sodium ion transport | 10 | 6.45 | −5.48 | −2.07 |
| GO: 0048732 | GO biological processes | Gland development | 13 | 8.39 | −5.14 | −1.86 |
| GO: 0032024 | GO biological processes | Positive regulation of insulin secretion | 6 | 3.87 | −5.13 | −1.86 |
| GO: 0071363 | GO biological processes | Cellular response to growth factor stimulus | 16 | 10.32 | −5 | −1.82 |
| GO: 0001655 | GO biological processes | Urogenital system development | 11 | 7.1 | −4.9 | −1.82 |
| GO: 0043269 | GO biological processes | Regulation of ion transport | 15 | 9.68 | −4.67 | −1.69 |
| GO: 0060021 | GO biological processes | Palate development | 6 | 3.87 | −4.43 | −1.6 |
| GO: 0015696 | GO biological processes | Ammonium transport | 6 | 3.87 | −4.2 | −1.51 |
| R-HSA-190239 | Reactome gene sets | FGFR3 ligand binding and activation | 3 | 1.94 | −4.06 | −1.45 |
| GO: 0051402 | GO biological processes | Neuron apoptotic process | 8 | 5.16 | −3.85 | −1.27 |
| GO: 0007610 | GO biological processes | Behavior | 13 | 8.39 | −3.78 | −1.22 |
| GO: 0007567 | GO biological processes | Parturition | 3 | 1.94 | −3.78 | −1.22 |
| GO: 0042476 | GO biological processes | Odontogenesis | 6 | 3.87 | −3.73 | −1.2 |
| GO: 0098742 | GO biological processes | Cell–cell adhesion via plasma-membrane adhesion molecules | 8 | 5.16 | −3.58 | −1.11 |
| GO: 0002028 | GO biological processes | Regulation of sodium ion transport | 5 | 3.23 | −3.56 | −1.11 |
| GO: 2000178 | GO biological processes | Negative regulation of neural precursor cell proliferation | 3 | 1.94 | −3.48 | −1.06 |
| GO: 0034394 | GO biological processes | Protein localization to cell surface | 4 | 2.58 | −3.28 | −0.96 |
| GO: 0035335 | GO biological processes | Peptidyl-tyrosine dephosphorylation | 5 | 3.23 | −3.21 | −0.91 |
GO – Gene Ontology; KEGG – Kyoto Encyclopedia of Genes and Genomes; Count – the number of input genes with membership in the given ontology term; % – the percentage of total input genes that are found in the given ontology term; Log10(P) – the P-value in log base 10; Log10(q) – the multi-test adjusted P-value in log base 10.
Discussion
Over the past few years, great efforts have been made to explore the molecular mechanisms of KIRC and the focus of previous studies has been centered on protein-coding genes or miRNAs. However, accumulated data have shown that lncRNAs participate in a variety of biological processes in KIRC recently. Despite that, to our knowledge, limited studies have been conducted to predict the prognosis of KIRC, and no reliable specific lncRNAs have been identified as biomarkers for the detection and risk stratification of KIRC. Therefore, the identification of lncRNA biomarkers in KIRC is still poor characterized and poses a great challenge to clarify the functions of them.
According to the latest studies, researchers found an efficient and effective way to investigate the potential functions of lncRNAs by establishing relationship between interested lncRNAs and function annotated miRNAs/mRNAs. For instance, some methodical analysis of the ceRNA network has been performed in many cancers, such as pancreatic cancer, breast cancer, and lung cancer [27–29]. Thus, in an effort to find out the potential implications of lncRNAs for the diagnosis and prognosis of KIRC, investigate and examine the possible regulatory mechanisms and functional roles of lncRNAs as ceRNAs in the progression of KIRC are going to be crucial. In this study, we used the interactions data of KIRC from TCGA database to generate the co-expression competing triplet for the first time, which the lncRNA and mRNA sharing a common miRNA according to the ceRNA theory. This novel lncRNA–miRNA–mRNA ceRNA network of KIRC, which was comprised of 321 lncRNA nodes, 26 miRNA nodes, 1068 mRNA nodes, and 2313 edges.
Over the past decade, the enrichment of GO functional analysis and biological pathways (KEGG pathways) analysis were widely used to estimate and evaluate biological functions enriched among multiple coding genes [30]. In our present study, the results of some significant GO and pathway analysis on the 1068 mRNAs are involved in KIRC. GO terms such as angiogenesis, positive/negative regulation of transcription from RNA polymerase II promoter, and extracellular matrix organization, all of them have been reported in KIRC [31–33]. KEGG pathways such as proteoglycans in cancer, calcium signaling pathway, and rap1 signaling pathway, all of which have been shown to play important roles in KIRC [34–36].
With increasing attention to the function roles of lncRNAs, several studies indicated that lncRNAs can improve the diagnosis and prognosis of some diseases, such as pancreatic ductal adenocarcinoma, acute myocardial infarction, and thyroid cancer [37–39]. However, the prognostic role of lncRNAs in KIRC has not been fully investigated. For the sake of discovering the appropriate lncRNAs, which can be served as potential novel biomarkers for clinical prognostic and treatment targets of KIRC, the OS analysis and location of lncRNAs were utilized. In the nucleus, lncRNAs are mainly served as a regulator which affects chromosomal spatial conformation, transcription factor activity, and alternative splicing. In the cytoplasm, lncRNAs are predominantly affected mRNAs stability, translation regulation, and conducted ceRNA mechanism by adsorbing miRNAs [40]. In general, a lncRNA that located in the cytoplasm indicates that the lncRNA has very likely participated in ceRNA interaction. Additionally, OS analysis demonstrated a lncRNA which related to prognosis to a certain disease indicate that this specific lncRNA plays a significant role in this disease. In this study, 4 lncRNAs (ENTPD3-AS1, FGD5-AS1, LIFR-AS1, UBAC2-AS1) were selected as potential prognostic biomarkers for whose location was in the cytoplasm and closely related to OS of KIRC.
Recently, a similar study which committed discovering potential pathogenic biomarkers in KIRC based on Gene Expression Omnibus (GEO) database has been conducted by Wang et al. [41]. They used the “preprocessCore” package and found that HAPLN1, hsa-miR-204, and hsa-miR-218 might serve as potential biomarkers in KIRC. The sample size they used, however, was small in the profiles and verification, which might lead to the identified genes and miRNAs have greater specificity and less universality. Moreover, this study only made use of the mRNASeq and miRNASeq data, and there were no combinations with lncRNAs or other types of biological molecules. Conversely, our research has identified DElncRNAs, DEmiRNAs, and DEmRNAs based on TCGA database with larger sample size, and the lncRNA-miRNA-mRNA network in KIRC was constructed successfully. Furthermore, our ceRNA network findings and lncRNA location analysis pointed out that 4 lncRNAs (ENTPD3-AS1, FGD5-AS1, LIFR-AS1, and UBAC2-AS1) could serve as potential biomarker in KIRC. Due to the differences in data sources and methods used for data processing, there was no overlap between our research and Wang et al’s study.
Although these 4 lncRNAs have been seldom reported before, miRNAs and their target mRNAs, which interacted with these lncRNAs, have been known to be involved and play a vital role in KIRC. These findings indicated that these 4 lncRNAs might have a critical role in the development of KIRC, despite the contribution of these 4 lncRNAs to the progression of KIRC has not been fully clarified from the present studies. According to the lncRNA-miRNA-mRNA sub network, we speculated that these 4 lncRNAs might have a decisive role in altering the expression of the downstream KIRC related disease mRNAs through competitive interactions with miRNAs (miR-21, miR-155, miR-221, and miR-223). As support of our speculation, recent studies have demonstrated that these 4 miRNAs play crucial roles in the development of KIRC. For example, Vergho et al. reported that the expression of miR-21 was significantly upregulated and involved in KIRC tumorigenesis [42]. In addition, Nijhuis et al. demonstrated that miR-21 expression was upregulated in colorectal cancer and involved in amino acid metabolism, which has been reported have a momentous influence on cell growth and drug resistance [43]. Another study illustrated that overexpression of miR-221 promotes proliferation, migration and invasion by targeting TIMP2 in KIRC [22]. Moreover, recent studies have demonstrated that tumor cells have a reprogrammed metabolism compared with normal cells, and the nutrients consumed by cancer cells such as amino acid, which is required in various cancer subtypes and plays an important role in the whole process of cancer metabolism reprogram [44]. Based on the Metascape analysis, 2 metabolic changes (butanoate metabolism, valine, leucine, and isoleucine degradation) were observed in the top 20 clusters, all of them are involved in various cancers including kidney cancer. Other GO terms, such as cell-cell adhesion via plasma-membrane adhesion molecules and cellular response to growth factor stimulus, are related to KIRC as well. For instance, Nagata et al. reported that the expression of cell adhesion molecules CADM4 suppresses the tumor invasion both in vitro and in vivo, which might be through the regulation of invasion [45]. Additionally, further studies have demonstrated that cell-cell adhesion promotes the proliferation of tumor cells and enhance the expression of gene products related to tumor invasion, which is closely related to the tumor and metastasis [46–48].
Conclusions
In this study, we re-established a lncRNA–miRNA–mRNA network, which for the first time enables a holistic view and comprehensive analysis of the lncRNA-associated ceRNA mediated genes in the development and progression of KIRC at a system-wide level. Our findings revealed that lncRNAs play indispensable roles in the development of KIRC. Our study further pointed out that 4 lncRNAs (ENTPD3-AS1, FGD5-AS1, LIFR-AS1, and UBAC2-AS1) could possibly be selected as key lncRNAs. This study will further our understanding of the pathogenesis of KIRC from the perspective of lncRNAs and highlight several novel lncRNAs as candidate prognostic biomarkers or potential therapeutic targets. Despite the results obtained in this study, there were certain limitations to this study with no verification experiments based on cells or tissues to confirm our results. Therefore, the biological functions and molecular mechanisms of these 4 specific lncRNAs in KIRC needed to be explored in the further studies.
Acknowledgements
This study was completed based on publicly available data; we would like to thank TCGA for providing RNA-seq/miRNA-seq data and high-quality clinical data on clear cell renal cell carcinoma. Moreover, the authors are grateful to those open source initiatives, such as the R Project, Perl, Cytoscape, Funrich, Metascape, etc.
Abbreviations
- RCC
renal cell carcinoma
- WHO
World Health Organization
- KIRC
clear cell kidney carcinoma
- PRCC
papillary renal cell carcinoma
- chRCC
chromophobe renal cell carcinoma
- miRNAs
microRNAs
- lncRNAs
long noncoding RNAs
- mRNAs
messenger RNA
- ceRNA
competing endogenous RNA
- TCGA
The Cancer Genome Atlas
- NCI
National Cancer Institute
- NHGRI
National Human Genome Research Institute
- DEmRNAs
differentially expressed messenger RNAs
- DEmiRNAs
differentially expressed micro RNAs
- DElncRNAs
differentially expressed long noncoding RNAs
- MREs
miRNA response elements
- GO
Gene Ontology
- CC
cellular component
- MF
molecular function
- BP
biological process
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- OS
overall survival
- RCI
relative concentration index
- ARF6
ADP-ribosylation factor 6
Footnotes
Source of support: This work was supported by National Nature Science Foundation of China (No.81570748), Foundation for Distinguished Young Scholars of Fuzhou General Hospital (No.2017Q05), and the Natural Science Foundation of Fujian Province of China (No.2016J01577)
Conflict of interest
None.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Jonasch E, Gao J, Rathmell WK. Renal cell carcinoma. BMJ. 2014;349:g4797. doi: 10.1136/bmj.g4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moch H, Cubilla AL, Humphrey PA, et al. The 2016 WHO classification of tumours of the urinary system and male genital organs – part A: Renal, penile, and testicular tumours. Eur Urol. 2016;70(1):93–105. doi: 10.1016/j.eururo.2016.02.029. [DOI] [PubMed] [Google Scholar]
- 4.Ho TH, Kapur P, Eckel-Passow JE, et al. Multicenter validation of enhancer of zeste homolog 2 expression as an independent prognostic marker in localized clear cell renal cell carcinoma. J Clin Oncol. 2017;35(32):3706–13. doi: 10.1200/JCO.2017.73.3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morris KV, Mattick JS. The rise of regulatory RNA. Nat Rev Genet. 2014;15(6):423–437. doi: 10.1038/nrg3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Ann Rev Biochem. 2012;81:145–66. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iyer MK, Niknafs YS, Malik R, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47(3):199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo S, Lu JY, Liu L, et al. Divergent lncRNAs regulate gene expression and lineage differentiation in pluripotent cells. Cell Stem Cell. 2016;18(5):637–52. doi: 10.1016/j.stem.2016.01.024. [DOI] [PubMed] [Google Scholar]
- 9.Sun M, Kraus WL. From discovery to function: The expanding roles of long noncoding RNAs in physiology and disease. Endocr Rev. 2015;36(1):25–64. doi: 10.1210/er.2014-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ward M, McEwan C, Mills JD, Janitz M. Conservation and tissue-specific transcription patterns of long noncoding RNAs. J Hum Transcr. 2015;1(1):2–9. doi: 10.3109/23324015.2015.1077591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang C, Yuan N, Wu L, et al. An integrated analysis for long noncoding RNAs and microRNAs with the mediated competing endogenous RNA network in papillary renal cell carcinoma. OncoTargets Ther. 2017;10:4037–50. doi: 10.2147/OTT.S141951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hosseini ES, Meryet-Figuiere M, Sabzalipoor H, et al. Dysregulated expression of long noncoding RNAs in gynecologic cancers. Mol Cancer. 2017;16(1):107. doi: 10.1186/s12943-017-0671-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan Y, Xu Z, Li Z, et al. An insight into the increasing role of LncRNAs in the pathogenesis of gliomas. Front Mol Neurosci. 2017;10:53. doi: 10.3389/fnmol.2017.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–58. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang WC, Fu WM, Wong CW, et al. The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget. 2015;6(26):22513–25. doi: 10.18632/oncotarget.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi X, Sun M, Liu H, et al. Long non-coding RNAs: A new frontier in the study of human diseases. Cancer Lett. 2013;339(2):159–66. doi: 10.1016/j.canlet.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 17.Song X, Cao G, Jing L, et al. Analysing the relationship between lncRNA and protein-coding gene and the role of lncRNA as ceRNA in pulmonary fibrosis. J Cell Mol Med. 2014;18(6):991–1003. doi: 10.1111/jcmm.12243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang WT, Ye H, Wei PP, et al. LncRNAs H19 and HULC, activated by oxidative stress, promote cell migration and invasion in cholangiocarcinoma through a ceRNA manner. J Hematol Oncol. 2016;9(1):117. doi: 10.1186/s13045-016-0348-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robertson AG, Kim J, Al-Ahmadie H, et al. Comprehensive molecular characterization of muscle-invasive bladder cancer. Cell. 2017;171(3):540–56. doi: 10.1016/j.cell.2017.09.007. e525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fritz HK, Lindgren D, Ljungberg B, et al. The miR(21/10b) ratio as a prognostic marker in clear cell renal cell carcinoma. Eur J Cancer. 2014;50(10):1758–65. doi: 10.1016/j.ejca.2014.03.281. [DOI] [PubMed] [Google Scholar]
- 21.Hall DP, Cost NG, Hegde S, et al. TRPM3 and miR-204 establish a regulatory circuit that controls oncogenic autophagy in clear cell renal cell carcinoma. Cancer Cell. 2014;26(5):738–53. doi: 10.1016/j.ccell.2014.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu GJ, Dong YQ, Zhang QM, et al. miRNA-221 promotes proliferation, migration and invasion by targeting TIMP2 in renal cell carcinoma. Int J Clin Exp Pathol. 2015;8(5):5224–29. [PMC free article] [PubMed] [Google Scholar]
- 23.Su H, Sun T, Wang H, et al. Decreased TCL6 expression is associated with poor prognosis in patients with clear cell renal cell carcinoma. Oncotarget. 2017;8(4):5789–99. doi: 10.18632/oncotarget.11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.White NM, Bao TT, Grigull J, et al. miRNA profiling for clear cell renal cell carcinoma: Biomarker discovery and identification of potential controls and consequences of miRNA dysregulation. J Urol. 2011;186(3):1077–83. doi: 10.1016/j.juro.2011.04.110. [DOI] [PubMed] [Google Scholar]
- 25.Xiao H, Tang K, Liu P, et al. LncRNA MALAT1 functions as a competing endogenous RNA to regulate ZEB2 expression by sponging miR-200s in clear cell kidney carcinoma. Oncotarget. 2015;6(35):38005–15. doi: 10.18632/oncotarget.5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kojima T, Shimazui T, Horie R, et al. FOXO1 and TCF7L2 genes involved in metastasis and poor prognosis in clear cell renal cell carcinoma. Genes Chromosomes Cancer. 2010;49(4):379–89. doi: 10.1002/gcc.20750. [DOI] [PubMed] [Google Scholar]
- 27.Li X, Zheng L, Zhang F, et al. STARD13-correlated ceRNA network inhibits EMT and metastasis of breast cancer. Oncotarget. 2016;7(17):23197–211. doi: 10.18632/oncotarget.8099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tay Y, Karreth FA, Pandolfi PP. Aberrant ceRNA activity drives lung cancer. Cell Res. 2014;24(3):259–60. doi: 10.1038/cr.2014.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou M, Diao Z, Yue X, et al. Construction and analysis of dysregulated lncRNA-associated ceRNA network identified novel lncRNA biomarkers for early diagnosis of human pancreatic cancer. Oncotarget. 2016;7(35):56383–94. doi: 10.18632/oncotarget.10891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao Z, Bai J, Wu A, et al. Co-LncRNA: Investigating the lncRNA combinatorial effects in GO annotations and KEGG pathways based on human RNA-Seq data. Database. 2015;2015 doi: 10.1093/database/bav082. pii: bav082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brugarolas J. Molecular genetics of clear-cell renal cell carcinoma. J Clin Oncol. 2014;32(18):1968–76. doi: 10.1200/JCO.2012.45.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Na X, Duan HO, Messing EM, et al. Identification of the RNA polymerase II subunit hsRPB7 as a novel target of the von Hippel-Lindau protein. EMBO J. 2003;22(16):4249–59. doi: 10.1093/emboj/cdg410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y, Guo X, Bray MJ, et al. An integrative genomics approach for identifying novel functional consequences of PBRM1 truncated mutations in clear cell renal cell carcinoma (ccRCC) BMC Genom. 2016;17(Suppl 7):515. doi: 10.1186/s12864-016-2906-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dormoy V, Beraud C, Lindner V, et al. Vitamin D3 triggers antitumor activity through targeting hedgehog signaling in human renal cell carcinoma. Carcinogenesis. 2012;33(11):2084–93. doi: 10.1093/carcin/bgs255. [DOI] [PubMed] [Google Scholar]
- 35.Kim WJ, Gersey Z, Daaka Y. Rap1GAP regulates renal cell carcinoma invasion. Cancer Lett. 2012;320(1):65–71. doi: 10.1016/j.canlet.2012.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Valsechi MC, Oliveira AB, Conceicao AL, et al. GPC3 reduces cell proliferation in renal carcinoma cell lines. BMC Cancer. 2014;14:631. doi: 10.1186/1471-2407-14-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li L, Cong Y, Gao X, et al. Differential expression profiles of long non-coding RNAs as potential biomarkers for the early diagnosis of acute myocardial infarction. Oncotarget. 2017;8(51):88613–21. doi: 10.18632/oncotarget.20101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murugan AK, Munirajan AK, Alzahrani AS. Long noncoding RNAs: Emerging players in thyroid cancer pathogenesis. Endocr Relat Cancer. 2018;25(2):R59–82. doi: 10.1530/ERC-17-0188. [DOI] [PubMed] [Google Scholar]
- 39.Song J, Yan Q, Zhang H, et al. Five key lncRNAs considered as prognostic targets for predicting Pancreatic Ductal Adenocarcinoma. J Cell Biochem. 2018;119(6):4559–69. doi: 10.1002/jcb.26598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang K, Shi ZM, Chang YN, et al. The ways of action of long non-coding RNAs in cytoplasm and nucleus. Gene. 2014;547(1):1–9. [Google Scholar]
- 41.Wang Z, Zhang Z, Zhang C, Xu Y. Identification of potential pathogenic biomarkers in clear cell renal cell carcinoma. Oncol Lett. 2018;15(6):8491–99. doi: 10.3892/ol.2018.8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vergho D, Kneitz S, Rosenwald A, et al. Combination of expression levels of miR-21 and miR-126 is associated with cancer-specific survival in clear-cell renal cell carcinoma. BMC Cancer. 2014;14:25. doi: 10.1186/1471-2407-14-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nijhuis A, Thompson H, Adam J, et al. Remodelling of microRNAs in colorectal cancer by hypoxia alters metabolism profiles and 5-fluorouracil resistance. Hum Mol Genet. 2017;26(8):1552–64. doi: 10.1093/hmg/ddx059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsun ZY, Possemato R. Amino acid management in cancer. Semin Cell Dev Biol. 2015;43:22–32. doi: 10.1016/j.semcdb.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nagata M, Sakurai-Yageta M, Yamada D, et al. Aberrations of a cell adhesion molecule CADM4 in renal clear cell carcinoma. Int J Cancer. 2012;130(6):1329–37. doi: 10.1002/ijc.26160. [DOI] [PubMed] [Google Scholar]
- 46.Heinzelmann J, Unrein A, Wickmann U, et al. MicroRNAs with prognostic potential for metastasis in clear cell renal cell carcinoma: A comparison of primary tumors and distant metastases. Ann Surg Oncol. 2014;21(3):1046–54. doi: 10.1245/s10434-013-3361-3. [DOI] [PubMed] [Google Scholar]
- 47.Salama MF, Carroll B, Adada M, et al. A novel role of sphingosine kinase-1 in the invasion and angiogenesis of VHL mutant clear cell renal cell carcinoma. FASEB J. 2015;29(7):2803–13. doi: 10.1096/fj.15-270413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weygant N, Qu D, May R, et al. DCLK1 is a broadly dysregulated target against epithelial-mesenchymal transition, focal adhesion, and stemness in clear cell renal carcinoma. Oncotarget. 2015;6(4):2193–205. doi: 10.18632/oncotarget.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]