Abstract
The genus Nitrospira is considered to be the most widespread and abundant group of nitrite-oxidizing bacteria in many natural and man-made ecosystems. However, the ecophysiological versatility within this phylogenetic group remains highly understudied, mainly due to the lack of pure cultures and genomic data. To further expand our understanding of this biotechnologically important genus, we analyzed the high quality draft genome of “Nitrospira lenta” strain BS10, a sublineage II Nitrospira that was isolated from a municipal wastewater treatment plant in Hamburg, Germany. The genome of “N. lenta” has a size of 3,756,190 bp and contains 3968 genomic objects, of which 3907 are predicted protein-coding sequences. Thorough genome annotation allowed the reconstruction of the “N. lenta” core metabolism for energy conservation and carbon fixation. Comparative analyses indicated that most metabolic features are shared with N. moscoviensis and “N. defluvii”, despite their ecological niche differentiation and phylogenetic distance. In conclusion, the genome of “N. lenta” provides important insights into the genomic diversity of the genus Nitrospira and provides a foundation for future comparative genomic studies that will generate a better understanding of the nitrification process.
Keywords: “Nitrospira lenta”, Nitrite oxidation, Nitrospira, Wastewater treatment
Introduction
Nitrification, the two-step oxidation of ammonia to nitrate via nitrite, is a key process of the biogeochemical nitrogen cycle. Nitrite-oxidizing bacteria (NOB) are chemolithoautotrophic microorganisms that catalyze the oxidation of nitrite to nitrate, the second step of the nitrification process. For decades NOB where considered as metabolically restricted microorganisms solely associated with nitrification. However, experimental findings contradict this opinion, indicating a versatile ecophysiology of many NOB [1–4] and highlighting their important role in and possibly outside of the nitrogen cycle [5].
The genus Nitrospira is the most diverse known NOB genus and is divided in six different phylogenetic sublineages [6–8]. Members of the genus are ubiquitously present in different natural and engineered ecosystems [5, 9–11]. Despite their high abundance, only eleven representative species, distributed within the six Nitrospira sublineages, have been obtained in enrichment or pure culture so far [7, 8, 12–15]. Sublineage I and II Nitrospira are considered to be the dominant NOB in most wastewater treatment plants (WWTPs), playing a key role in the efficient removal of nitrogen via nitrification [6, 16]. Besides their widespread distribution and crucial role, the physiology of Nitrospira species is highly understudied, mainly due to the lack of pure cultures and genomic data [3, 17, 18]. The recent identification of complete ammonia-oxidizing (comammox) Nitrospira [15, 19] not only redefined the nitrification process, but also further indicated the importance of the genus and emphasized our poor understanding of the metabolic versatility present within this phylogenetic group.
“Nitrospira lenta” strain BS10 was isolated from a municipal WWTP [13] and it is the fourth isolate belonging to the sublineage II Nitrospira, besides N. moscoviensis [20], “N. japonica” [14], and the comammox organism “N. inopinata” [12]. Thus, insights into the “N. lenta” genome will shed light onto the genomic flexibility and metabolic diversity of the genus Nitrospira and aid further comparative studies between Nitrospira species.
Organism information
Classification and features
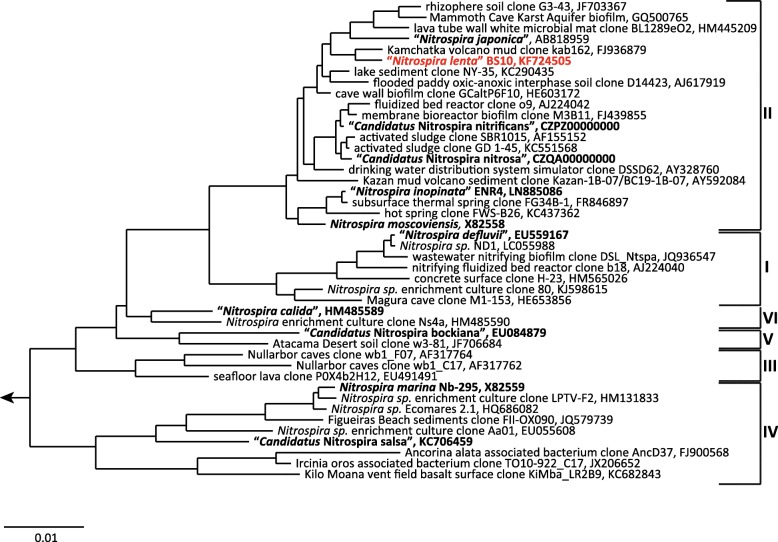
“N. lenta” strain BS10 is a Gram negative, aerobic NOB isolated from activated sludge of a municipal WWTP in Hamburg, Germany (basic properties are summarized in Table 1) [13]. Based on 16S rRNA gene-based phylogentic analysis, “N. lenta” is affiliated with Nitrospira sublineage II but is only distantly related to the sublineage II type strain, N. moscoviensis (Fig. 1).
Table 1.
Classification and general features of “Nitrospira lenta” strain BS10 [34]
| MIGS ID | Property | Term | Evidence codea |
|---|---|---|---|
| Classification | Domain Bacteria | TAS [35] | |
| Phylum Nitrospirae | TAS [36] | ||
| Class “Nitrospira” | TAS [36] | ||
| Order “Nitrospirales” | TAS [36] | ||
| Family “Nitrospiraceae” | TAS [36] | ||
| Genus Nitrospira | TAS [32] | ||
| Species Nitrospira lenta | TAS [13] | ||
| Strain: BS10 | TAS [13] | ||
| Gram stain | Negative | TAS [13] | |
| Cell shape | Spiral-shaped rods | TAS [13] | |
| Motility | Non-motileb | TAS [13] | |
| Sporulation | Not reported | NAS | |
| Temperature range | 10–32 °C | TAS [13] | |
| Optimum temperature | 28 °C | TAS [13] | |
| pH range; Optimum | 7.4–8.0; 7.4–7.6 | TAS [13] | |
| Carbon source | Carbon dioxide | TAS [13] | |
| MIGS-6 | Habitat | Wastewater treatment plant | TAS [13] |
| MIGS-6.3 | Salinity | 0,5% w/v | TAS [13] |
| MIGS-22 | Oxygen requirement | Aerobic | TAS [13] |
| MIGS-15 | Biotic relationship | Free-living | TAS [13] |
| MIGS-14 | Pathogenicity | Non-pathogen | NAS |
| MIGS-4 | Geographic location | Germany/Hamburg | TAS [13] |
| MIGS-5 | Sample collection | 12/12/2006 | TAS [13] |
| MIGS-4.1 | Latitude | 53° 31′ 8″ N | TAS [13] |
| MIGS-4.2 | Longitude | 9° 54′ 53″E | TAS [13] |
| MIGS-4.4 | Altitude | – | NAS |
aEvidence codes – IDA Inferred from Direct Assay, TAS Traceable Author Statement (i.e., a direct report exists in the literature), NAS Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from the Gene Ontology project [37]
bGenes encoding for a flagellum were identified in “N. lenta” BS10 genome
Fig. 1.
Phylogenetic analysis of selected representatives from the genus Nitrospira. A 16S rRNA gene-based maximum likelihood tree is shown. Sequences of cultured representatives are printed in bold, “N. lenta” in red. The tree was constructed using sequences ≥1165 bp and a 50% conservation filter, resulting in 1463 valid alignment positions. Leptospirillum ferriphilum (AF356829) and L. ferrooxidans (X86776) were used as outgroup, which is indicated by the arrow. Scale bar indicates 1% estimated sequence divergence
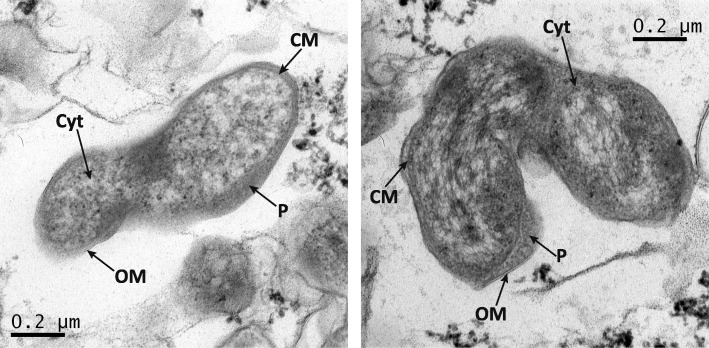
A pure culture of “N. lenta” was obtained after applying a combination of standard isolation methods (density gradient centrifugation and serial dilutions) with an optical tweezer system for the sorting of single cells [13]. “N. lenta” grows mainly planktonic and forms helical-shaped cells. Cells are 1.0–2.3 μm in length and 0.2–0.3 μm in diameter (Table 1, Fig. 2). As shown by Nowka et al. [13], “N. lenta” is able to grow at lower temperatures (10 °C) than N. moscoviensis. Interestingly, while “N. lenta” is not able to tolerate high concentrations of nitrite, it exhibits a lower affinity for nitrite compared to N. moscoviensis and “N. defluvii”, which indicates a clear niche differentiation among these Nitrospira species [21].
Fig. 2.
Electron micrographs of “N. lenta” strain BS10. The cells, in accordance to most of the bacteria belonging to the genus Nitrospira, feature a characteristic spiral shape and an enlarged periplasm. The individual cell components are indicated. OM, outer membrane; P, periplasm; CM, cytoplasmic membrane; Cyt, cytoplasm
Genome sequencing information
Genome project history
“N. lenta” was selected for whole genome sequencing on the basis of its relevance within the nitrogen cycle as well as due to the general lack of genomic information for Nitrospira species. Furthermore, because “N. lenta” was isolated from activated sludge, its genome was expected to yield insights that allow optimization and stabilization of the nitrification process in wastewater treatment. The draft genome sequence of “N. lenta” BS10 was completed on 27/07/2013. The high-quality draft genome is available in the European Nucleotide Archive (ENA) under study accession number PRJEB26290. An overview of the genome sequencing project is given in Table 2.
Table 2.
Project information
| MIGS ID | Property | Term |
|---|---|---|
| MIGS 31 | Finishing quality | High quality draft |
| MIGS-28 | Libraries used | 1 |
| MIGS 29 | Sequencing platforms | Roche GS FLX |
| MIGS 31.2 | Fold coverage | 40 |
| MIGS 30 | Assemblers | GS De Novo Assembler |
| MIGS 32 | Gene calling method | AMIGene [24] |
| Locus Tag | NITLEN | |
| GenBank ID | OUNR00000000.1 | |
| GenBankDate of Release | 4 June 2018 | |
| GOLD ID | – | |
| BioProject | PRJEB26290 | |
| MIGS 13 | Source Material Identifier | BS10 |
| Project relevance | Microbiology, Biotechnology |
Growth conditions and genomic DNA extraction
“N. lenta” was cultivated as described by Nowka et al. [13] in mineral salt medium amended with 0.02 g L− 1 NaNO2− as energy source. The cultures were incubated in the dark at 28 °C, with moderate stirring (100–300 rpm). The genomic DNA was extracted following the hexadecyltrimethylammoniumbromide (CTAB) protocol provided by the DOE Joint Genome Institute (JGI, https://jgi.doe.gov/user-program-info/pmo-overview/protocols-sample-preparation-information/) as described elsewhere [22].
Genome sequencing and assembly
High-throughput sequencing was performed at GATC Biotech (Constance, Germany) using Roche GS FLX technology. The final draft genome of “N. lenta” was obtained using the GS De Novo Assembler (Newbler) and comprised 3.8 Mb on 22 contigs. Genome completeness was evaluated with CheckM [23]. Similarly to the complete genomes of “N. defluvii” (98% completeness, 2.3% contamination) and N. moscoviensis (96% completeness, 6.6% contamination), the “N. lenta” genome was estimated to be 96% complete with 3.2% contamination.
Genome annotation
The draft genome of “N. lenta” was annotated using the MicroScope platform [24] as described in detail elsewhere [17]. The automatic annotation was manually checked and curated using the MicroScope Web interface MaGe [25]. The genomic features of “N. lenta” were compared to N. moscoviensis and “N. defluvii”, the type strains of the Nitrospira sublineages II and I, respectively, using the OrthoVenn web service [26] for the identification and comparison of orthologous gene groups. Sequence similarities were calculated using an E-value of 1e-05. An inflation value of 1.5 was applied to generate the orthologous clusters.
Genome properties
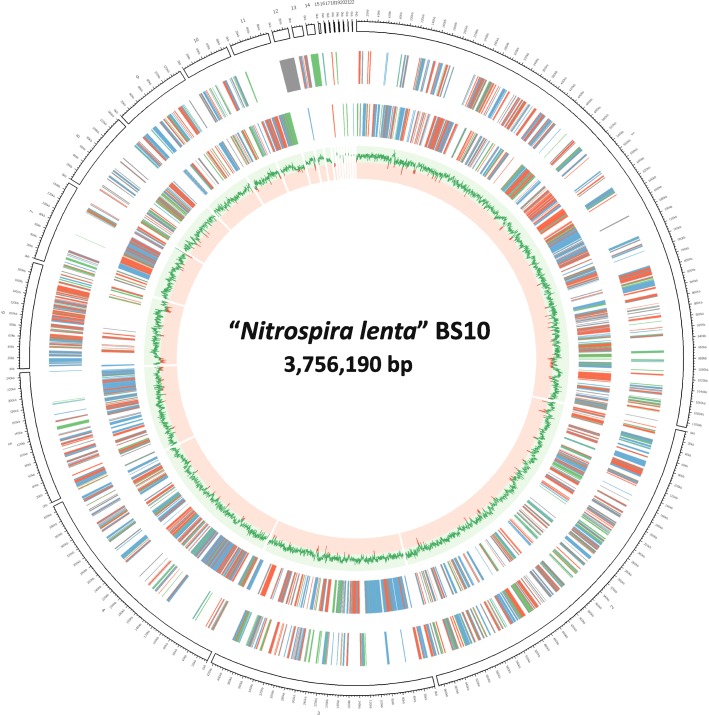
The “N. lenta” draft genome consists of 22 contigs and has a total size of 3,756,190 bp with an overall G + C content of 57.9% (Fig. 3, Table 3). From a total of 3968 predicted genes, 3907 (98.5%) and 61 (1.5%) are protein and RNA coding sequences, respectively. The genome encodes for 1 complete rRNA operon and 46 tRNAs, with 1 to 5 copies for each tRNA type. Moreover, 66.8% of the predicted genes were assigned into to Clusters of Orthologous Groups (COG) functional categories (Table 4).
Fig. 3.
Circular representation of the “N. lenta” genome. From inside out the circles display: (1) G + C content (red < 50%, green > 50%), (2) CDS on reverse strand, (3) CDS on forward strand, (4) contig organization. The tick marks correspond to 20 kbp. CDS colors indicate COG classification (blue, cellular processes and signaling; green, information storage and processing; red, metabolism, grey; poorly characterized). Contigs are ordered by size, and their arrangement in the figure may not reflect the positions of the respective regions on the chromosome. The graphical circular map of “N. lenta” chromosome was generated using the CIRCOS software [33]
Table 3.
Genome statistics
| Attribute | Value |
|---|---|
| Genome size (bp) | 3,756,190 |
| DNA coding (bp) | 3,487,097 |
| DNA G + C (%) | 57.9 |
| DNA scaffolds | 22 |
| Total genes | 3968 |
| Protein coding genes | 3907 |
| RNA genes | 61 |
| rRNA genes | 3 |
| tRNA genes | 46 |
| Pseudo genes | 0 |
| Genes in internal clusters | – |
| Genes with function prediction | 1990 |
| Genes assigned to COGs | 2609 |
| Genes with Pfam domains | 3868 |
| Genes with signal peptidesa | 196 |
| Genes with transmembrane helices | 965 |
| CRISPR repeats | 0 |
aOnly signal peptides with a prediction probability greater 70% were taken into consideration
Table 4.
Number of genes associated with general COG functional category prediction
| Code | Value | %agea | Description |
|---|---|---|---|
| J | 163 | 4.17 | Translation, ribosomal structure and biogenesis |
| A | 0 | 0 | RNA processing and modification |
| K | 148 | 3.79 | Transcription |
| L | 142 | 3.64 | Replication, recombination and repair |
| B | 1 | 0.03 | Chromatin structure and dynamics |
| D | 48 | 1.23 | Cell cycle control, Cell division, chromosome partitioning |
| V | 44 | 1.13 | Defense mechanisms |
| T | 256 | 6.55 | Signal transduction mechanisms |
| M | 303 | 7.76 | Cell wall/membrane biogenesis |
| N | 114 | 2.92 | Cell motility |
| U | 119 | 3.05 | Intracellular trafficking and secretion |
| O | 151 | 3.87 | Posttranslational modification, protein turnover, chaperones |
| C | 205 | 5.25 | Energy production and conversion |
| G | 133 | 3.40 | Carbohydrate transport and metabolism |
| E | 198 | 5.07 | Amino acid transport and metabolism |
| F | 59 | 1.51 | Nucleotide transport and metabolism |
| H | 136 | 3.48 | Coenzyme transport and metabolism |
| I | 76 | 1.95 | Lipid transport and metabolism |
| P | 156 | 3.99 | Inorganic ion transport and metabolism |
| Q | 77 | 1.97 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 389 | 9.96 | General function prediction only |
| S | 221 | 5.66 | Function unknown |
| – | 1298 | 33.22 | Not in COGs |
aThe total is based on the total number of protein coding genes in the genome
Insights from the genome sequence
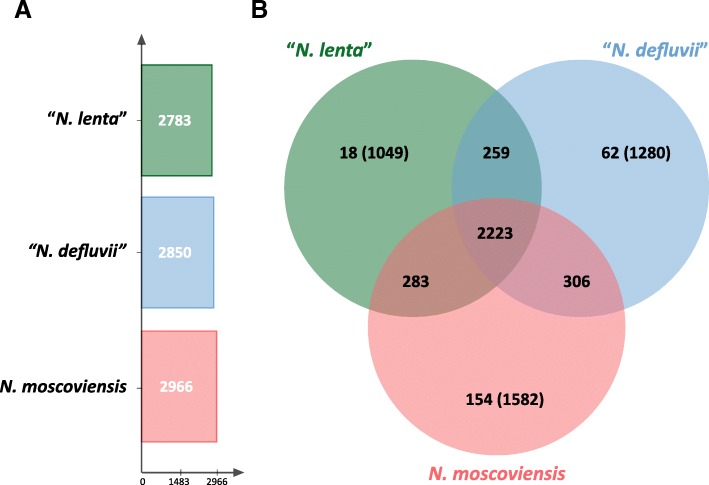
Nitrospira species belonging to sublineages I and II are the most abundant NOB in many environments and play a key role in N-cycling in engineered ecosystems [6, 10]. Recent experimental data indicates a clear niche differentiation between sublineage I and II Nitrospira [21, 27]. More specifically, “N. lenta”, like other members of sublineage II, exhibits a lower maximum activity [21, 27] and could be outcompeted by sublineage I Nitrospira at higher nitrite concentrations [28]. Despite their ecophysiological differences, sublineage I and II Nitrospira exhibit substantial genomic similarities. More specifically, “N. lenta” shares a core genome including 2223 orthologous protein clusters with N. moscoviensis and “N. defluvii”. This corresponds to 67.3% of the pan-genome of the Nitrospira species included in this analysis (Fig. 4). Moreover, “N. lenta” features the lowest number of unique genes (1100, of which 51 are grouped in 18 paralogous protein clusters). Most of these unique genes lack any function prediction (Fig. 4).
Fig. 4.
Genome comparison of “N. lenta” with representatives of Nitrospira sublineages I and II. (a) Total number of inferred orthologous protein clusters for each species. (b) Venn diagram depicting the distribution of orthologous clusters among the genomes of “N. lenta”, “N. defluvii” and N. moscoviensis. The numbers indicate orthologous protein clusters; numbers in brackets depict the unique, unclustered proteins of each genome
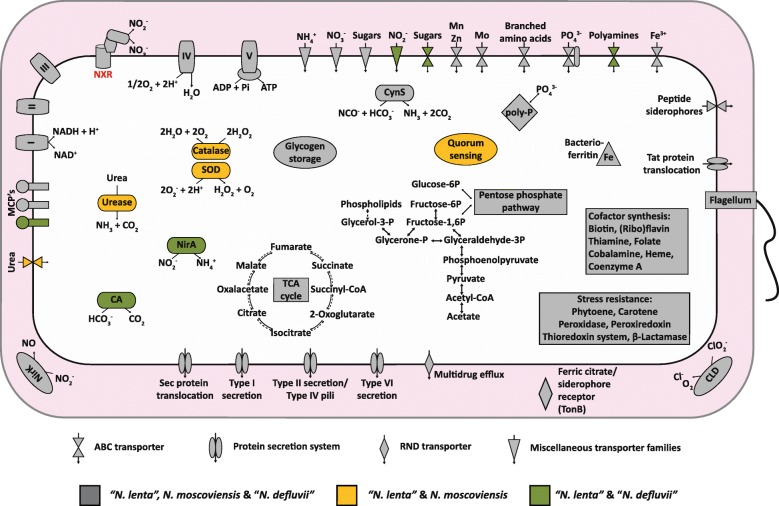
In accordance with its ability to oxidize nitrite to nitrate [13], “N. lenta” encodes all proteins required for nitrite oxidation (Fig. 5), for which the key enzyme is a membrane-associated nitrite oxidoreductase (NXR) [29]. This protein complex belongs to the type II dimethyl sulfoxide reductase family of molybdopterin-binding enzymes and consists of three subunits [17, 29]. The “N. lenta” genome contains two paralogous copies of nxrA and nxrB, encoding the NXR α and β subunits, respectively, and two copies of nxrC for the candidate γ subunit. Like all Nitrospira genomes analyzed to date, the “N. lenta” genome contains nirK, encoding the copper-dependent NO-forming nitrite reductase. While the function of NirK in Nitrospira is still unclear, a role in dissimilatory nitrite reduction is unlikely as no other genes involved in denitrification were identified in “N. lenta” or any other Nitrospira. One cannot exclude the possibility that NO plays a regulatory role in Nitrospira, for example in the regulation of forward versus reverse electron flow as proposed for Nitrobacter [30]. Moreover, “N. lenta” exhibits the genetic capacity for nitrogen assimilation from nitrite as its genome features nirA encoding the ferredoxin-dependent nitrite reductase. Interestingly, NirA is conserved in “N. defluvii”, but not the other genome-sequenced sublineage II Nitrospira, which either encode an octaheme nitrite reductase [3, 18], or, in the case of the comammox Nitrospira, lack assimilatory nitrite reduction pathways [15, 19, 31]. Interestingly, the “N. lenta” BS10 genome also features an ure operon encoding a functional urease (UreC), as well as a complete gene set (urtABCDE) for a high affinity urea ABC transporter. This implies that “N. lenta” is able to hydrolyse urea to ammonium and CO2, facilitating nitrogen and carbon assimilation from urea and reciprocal feeding between “N. lenta” and urease-negative ammonia-oxidizing bacteria [3, 4]. The “N. lenta” urease is closely related to the enzyme of N. moscoviensis, but significantly differs from known ureases of ammonia-oxidizing bacteria [3].
Fig. 5.
Cell metabolic cartoon of “N. lenta” in comparison to N. moscoviensis and “N. defluvii”. CA, carbonic anhydrase; SOD, superoxide dismutase; CLD, chlorite dismutase; CynS, cyanate hydratase (cyanase); MCPs, methyl-accepting chemotaxis proteins; NirA, ferredoxin nitrite reductase; NirK, dissimilatory nitrite reductase. Enzyme complexes of the electron transport chain are labeled by Roman numerals
“N. lenta” conserves energy by nitrite oxidation with oxygen as terminal electron acceptor. During nitrite oxidation catalyzed by NXR, two electrons are shuttled (putatively via cytochrome c) towards a putative novel bd-like terminal oxidase [17]. The proton motive force established through active proton pumping by this novel complex IV and/or the release of scalar protons during nitrite oxidation drives ATP synthesis by a FOF1-type ATPase (complex V). The other respiratory complexes (complexes I to III) will not contribute to energy conservation during lithoautotrophic growth on nitrite, but will operate in reverse to provide reducing equivalents for carbon fixation [17]. Moreover, the complete gene repertoire for the oxidative and reductive tricarboxylic acid (TCA) cycle is present in “N. lenta” for pyruvate oxidation via acetyl-CoA and CO2 fixation, respectively. Moreover, the complete glycolysis/gluconeogenesis and pentose phosphate pathways were identified. The observed presence of glycolysis and the oxidative TCA cycle might indicate that “N. lenta” can benefit from a mixotrophic lifestyle in the presence of organic carbon, as has been reported for other Nitrospira representatives [6, 16, 32].
Finally, the “N. lenta” genome contains various defense mechanisms against heavy metals, antibiotics, and the antibacterial agent acriflavine. “N. lenta” encodes a superoxide dismutase for defense mechanisms against oxidative stress, as well as several bacterioferritins, which can participate in oxidative stress resistance mechanisms [17].
Conclusions
Together with N. moscoviensis and “N. japonica”, “N. lenta” represents only the third cultured species of canonical nitrite-oxidizing Nitrospira from sublineage II. In this study, the genome of “N. lenta” was analyzed, demonstrating that “N. lenta” shares a significant amount of genomic features with other representatives of the genus. However, physiological differences observed by Nowka et al. regarding growth conditions and nitrite affinities [13, 21] clearly suggest a niche differentiation between different species. The “N. lenta” genome will facilitate a better understanding of the metabolic versatility of the genus Nitrospira and will be useful for future comparative studies, especially those with a focus on species obtained from engineered systems.
Acknowledgements
We are grateful to Lianna Poghosyan, Jeroen Frank and Maartje van Kessel for helpful discussions. The LABGeM (CEA/IG/Genoscope & CNRS UMR8030) and the France Génomique National infrastructure (funded as part of Investissement d’avenir program managed by Agence Nationale pour la Recherche, contract ANR-10-INBS-09) are acknowledged for support within the MicroScope annotation platform.
Funding
This work was supported by the European Research Council (ERC AG 339880 ECOMOM), the Netherlands Organisation for Scientific Research (NWO; VENI grant 863.14.019), the Vienna Science and Technology Fund (WWTF; grant LS09–40), the Austrian Science Fund (FWF; grants P24101-B22 and P25231-B21), and the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation grant SP 667/10–1 + 2).
Abbreviations
- NOB
Nitrite-oxidizing bacteria
- NXR
Nitrite oxidoreductase
- WWTP
Wastewater treatment plant
Authors’ contributions
ES, HD, and SL designed the study, DS, BN, and SL performed research, DS and SL analyzed data, DS, ES, HD and SL wrote the paper, all authors reviewed and agreed with the final version of the manuscript.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Füssel J, Lücker S, Yilmaz P, Nowka B, van Kessel MAHJ, Bourceau P, et al. Adaptability as the key to success for the ubiquitous marine nitrite oxidizer Nitrococcus. Sci Adv. 2017;3(11):e1700807. doi: 10.1126/sciadv.1700807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koch H, Galushko A, Albertsen M, Schintlmeister A, Gruber-Dorninger C, Lücker S, et al. Growth of nitrite-oxidizing bacteria by aerobic hydrogen oxidation. Science. 2014;345(6200):1052–1054. doi: 10.1126/science.1256985. [DOI] [PubMed] [Google Scholar]
- 3.Koch H, Lücker S, Albertsen M, Kitzinger K, Herbold C, Spieck E, et al. Expanded metabolic versatility of ubiquitous nitrite-oxidizing bacteria from the genus Nitrospira. Proc Natl Acad Sci U S A. 2015;112(36):11371–11376. doi: 10.1073/pnas.1506533112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Palatinszky M, Herbold C, Jehmlich N, Pogoda M, Han P, von Bergen M, et al. Cyanate as an energy source for nitrifiers. Nature. 2015;524(7563):105–108. doi: 10.1038/nature14856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daims H, Lücker S, Wagner M. A new perspective on microbes formerly known as nitrite-oxidizing bacteria. Trends Microbiol. 2016;24(9):699–712. doi: 10.1016/j.tim.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daims H, Nielsen JL, Nielsen PH, Schleifer KH, Wagner M. In situ characterization of Nitrospira-like nitrite-oxidizing bacteria active in wastewater treatment plants. Appl Environ Microbiol. 2001;67(11):5273–5284. doi: 10.1128/AEM.67.11.5273-5284.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lebedeva EV, Alawi M, Maixner F, Jozsa PG, Daims H, Spieck E. Physiological and phylogenetic characterization of a novel lithoautotrophic nitrite-oxidizing bacterium, ‘Candidatus Nitrospira bockiana’. Int J Syst Evol Microbiol. 2008;58(Pt 1):242–250. doi: 10.1099/ijs.0.65379-0. [DOI] [PubMed] [Google Scholar]
- 8.Lebedeva EV, Off S, Zumbragel S, Kruse M, Shagzhina A, Lucker S, et al. Isolation and characterization of a moderately thermophilic nitrite-oxidizing bacterium from a geothermal spring. FEMS Microbiol Ecol. 2011;75(2):195–204. doi: 10.1111/j.1574-6941.2010.01006.x. [DOI] [PubMed] [Google Scholar]
- 9.Freitag TE, Chang L, Clegg CD, Prosser JI. Influence of inorganic nitrogen management regime on the diversity of nitrite-oxidizing bacteria in agricultural grassland soils. Appl Environ Microbiol. 2005;71(12):8323–8334. doi: 10.1128/AEM.71.12.8323-8334.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Juretschko S, Timmermann G, Schmid M, Schleifer K-H, Pommerening-Röser A, Koops H-P, et al. Combined molecular and conventional analyses of nitrifying bacterium diversity in activated sludge: Nitrosococcus mobilis and Nitrospira-like bacteria as dominant populations. Appl Environ Microbiol. 1998;64(8):3042–3051. doi: 10.1128/aem.64.8.3042-3051.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lebedeva EV, Alawi M, Fiencke C, Namsaraev B, Bock E, Spieck E. Moderately thermophilic nitrifying bacteria from a hot spring of the Baikal rift zone. FEMS Microbiol Ecol. 2005;54(2):297–306. doi: 10.1016/j.femsec.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 12.Kits KD, Sedlacek CJ, Lebedeva EV, Han P, Bulaev A, Pjevac P, et al. Kinetic analysis of a complete nitrifier reveals an oligotrophic lifestyle. Nature. 2017;549:269–272. doi: 10.1038/nature23679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nowka B, Off S, Daims H, Spieck E. Improved isolation strategies allowed the phenotypic differentiation of two Nitrospira strains from widespread phylogenetic lineages. FEMS Microbiol Ecol. 2015;91(3):fiu031. doi: 10.1093/femsec/fiu031. [DOI] [PubMed] [Google Scholar]
- 14.Ushiki N, Fujitani H, Aoi Y, Tsuneda S. Isolation of Nitrospira belonging to sublineage II from a wastewater treatment plant. Microbes Environ. 2013;28(3):346–353. doi: 10.1264/jsme2.ME13042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Kessel MA, Speth DR, Albertsen M, Nielsen PH, Op den Camp HJ, Kartal B, et al. Complete nitrification by a single microorganism. Nature. 2015;528(7583):555–559. doi: 10.1038/nature16459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gruber-Dorninger C, Pester M, Kitzinger K, Savio DF, Loy A, Rattei T, et al. Functionally relevant diversity of closely related Nitrospira in activated sludge. ISME J. 2015;9(3):643–655. doi: 10.1038/ismej.2014.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lücker S, Wagner M, Maixner F, Pelletier E, Koch H, Vacherie B, et al. A Nitrospira metagenome illuminates the physiology and evolution of globally important nitrite-oxidizing bacteria. Proc Natl Acad Sci U S A. 2010;107(30):13479–13484. doi: 10.1073/pnas.1003860107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ushiki N, Fujitani H, Shimada Y, Morohoshi T, Sekiguchi Y, Tsuneda S. Genomic Analysis of Two Phylogenetically Distinct Nitrospira Species Reveals Their Genomic Plasticity and Functional Diversity. Front Microbiol. 2018;8:2637. doi: 10.3389/fmicb.2017.02637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daims H, Lebedeva EV, Pjevac P, Han P, Herbold C, Albertsen M, et al. Complete nitrification by Nitrospira bacteria. Nature. 2015;528(7583):504–509. doi: 10.1038/nature16461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ehrich S, Behrens D, Lebedeva E, Ludwig W, Bock E. A new obligately chemolithoautotrophic, nitrite-oxidizing bacterium, Nitrospira moscoviensis sp. nov. and its phylogenetic relationship. Arch Microbiol. 1995;164(1):16–23. doi: 10.1007/BF02568729. [DOI] [PubMed] [Google Scholar]
- 21.Nowka B, Daims H, Spieck E. Comparison of oxidation kinetics of nitrite-oxidizing bacteria: nitrite availability as a key factor in niche differentiation. Appl Environ Microbiol. 2015;81(2):745–753. doi: 10.1128/AEM.02734-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lücker S, Nowka B, Rattei T, Spieck E, Daims H. The genome of Nitrospina gracilis illuminates the metabolism and evolution of the major marine nitrite oxidizer. Front Microbiol. 2013;4:27. doi: 10.3389/fmicb.2013.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25(7):1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vallenet D, Belda E, Calteau A, Cruveiller S, Engelen S, Lajus A, et al. MicroScope - an integrated microbial resource for the curation and comparative analysis of genomic and metabolic data. Nucleic Acids Res. 2013;41(D1):D636–D647. doi: 10.1093/nar/gks1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vallenet D, Labarre L, Rouy Z, Barbe V, Bocs S, Cruveiller S, et al. MaGe: a microbial genome annotation system supported by synteny results. Nucleic Acids Res. 2006;34(1):53–65. doi: 10.1093/nar/gkj406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, Coleman-Derr D, Chen G, Gu YQ. OrthoVenn: a web server for genome wide comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res. 2015;43(W1):W78–W84. doi: 10.1093/nar/gkv487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ushiki N, Jinno M, Fujitani H, Suenaga T, Terada A, Tsuneda S. Nitrite oxidation kinetics of two Nitrospira strains: the quest for competition and ecological niche differentiation. J Biosci Bioeng. 2017;123(5):581–589. doi: 10.1016/j.jbiosc.2016.12.016. [DOI] [PubMed] [Google Scholar]
- 28.Maixner F, Noguera DR, Anneser B, Stoecker K, Wegl G, Wagner M, et al. Nitrite concentration influences the population structure of Nitrospira-like bacteria. Environ Microbiol. 2006;8(8):1487–1495. doi: 10.1111/j.1462-2920.2006.01033.x. [DOI] [PubMed] [Google Scholar]
- 29.Spieck E, Ehrich S, Aamand J, Bock E. Isolation and immunocytochemical location of the nitrite-oxidizing system in Nitrospira moscoviensis. Arch Microbiol. 1998;169(3):225–230. doi: 10.1007/s002030050565. [DOI] [PubMed] [Google Scholar]
- 30.Starkenburg SR, Arp DJ, Bottomley PJ. Expression of a putative nitrite reductase and the reversible inhibition of nitrite-dependent respiration by nitric oxide in Nitrobacter winogradskyi Nb-255. Environ Microbiol. 2008;10(11):3036–3042. doi: 10.1111/j.1462-2920.2008.01763.x. [DOI] [PubMed] [Google Scholar]
- 31.Lawson CE, Lücker S. Complete ammonia oxidation: an important control on nitrification in engineered ecosystems? Curr Opin Biotechnol. 2018;50:158–165. doi: 10.1016/j.copbio.2018.01.015. [DOI] [PubMed] [Google Scholar]
- 32.Watson SW, Bock E, Valois FW, Waterbury JB, Schlosser U. Nitrospira marina gen. Nov. sp. nov.: a chemolithotrophic nitrite-oxidizing bacterium. Arch Microbiol. 1986;144(1):1–7. doi: 10.1007/BF00454947. [DOI] [Google Scholar]
- 33.Krzywinski MI, Schein JE, Birol I, Connors J, Gascoyne R, Horsman D, et al. Circos: An information aesthetic for comparative genomics. Genome Research. 2009;19(9):1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol. 2008;26(5):541–547. doi: 10.1038/nbt1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains archaea, Bacteria, and Eucarya. Proc Natl Acad Sci U S A. 1990;87(12):4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garrity GM, Holt JG, Spieck E, Bock E, Johnson DB, Spring S, et al. Phylum BVIII. Nitrospirae phy. nov. In: Boone DR, Castenholz RW, Garrity GM, et al., editors. Bergey’s Manual® of Systematic Bacteriology: Volume One : The Archaea and the Deeply Branching and Phototrophic Bacteria. New York: Springer New York; 2001. pp. 451–464. [Google Scholar]
- 37.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. Nat Genet. 2000;25:25. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]