Abstract
Background
Glycosylation plays a critical role in the aggressiveness of pancreatic cancer (PC). Emerging evidences indicate significant involvement of cancer stem cells (CSCs) in PC aggressiveness. However, the importance of glycosylation in pancreatic cancer stem cells (PCSCs) is yet to be addressed. Hence, we evaluated the potential role of glycosylation in maintenance of stemness of PCSCs.
Methods
Effect of glycosylation specific inhibitors on growth and PCSCs of PC cells was assessed by MTT assay and Side Population (SP) analysis. Isolated PCSCs/SP were characterized using molecular and functional assays. Expression of tumor-associated carbohydrate antigens (TACAs) was analyzed in PCSCs by western blotting. Effect of tunicamycin on PCSCs was analyzed by tumorsphere, clonogenicity, migration assay and immunoblotting for CSCs markers. The differential expression of glycogenes in PCSCs compared to non-CSCs were determined by RT-qPCR, immunoblotting and immunofluorescence. Co-expression of GALNT3 and B3GNT3 with CD44v6 was assessed in progression stages of KrasG12D; Pdx-1-Cre (KC) and KrasG12D; p53R172H; Pdx-1-Cre (KPC) tumors by immunofluorescence. Transient and CRISPR/Cas9 silencing of GALNT3 and B3GNT3 was performed to examine their effect on CSCs maintenance.
Results
Inhibition of glycosylation decreased growth and CSCs/SP in PC cells. PCSCs overexpressed CSC markers (CD44v6, ESA, SOX2, SOX9 and ABCG2), exhibited global expressional variation of TACAs and showed higher self-renewal potential. Specifically, N-glycosylation inhibition, significantly decreased tumorsphere formation, migration, and clonogenicity of PCSCs, as well as hypo-glycosylated CD44v6 and ESA. Of note, glycosyltransferases (GFs), GALNT3 and B3GNT3, were significantly overexpressed in PCSCs and co-expressed with CD44v6 at advanced PDAC stages in KC and KPC tumors. Further, GALNT3 and B3GNT3 knockdown led to a decrease in the expression of cell surface markers (CD44v6 and ESA) and self-renewal markers (SOX2 and OCT3/4) in PCSCs. Interestingly, CD44v6 was modified with sialyl Lewis a in PCSCs. Finally, CRISPR/Cas9-mediated GALNT3 KO significantly decreased self-renewal, clonogenicity, and migratory capacity in PCSCs.
Conclusions
Taken together, for the first time, our study showed the importance of glycosylation in mediating growth, stemness, and maintenance of PCSCs. These results indicate that elevated GALNT3 and B3GNT3 expression in PCSCs regulate stemness through modulating CSC markers.
Electronic supplementary material
The online version of this article (10.1186/s12885-018-5074-2) contains supplementary material, which is available to authorized users.
Keywords: Pancreatic cancer stem cells, Glycosylation, Tunicamycin, GALNT3, B3GNT3, CD44v6
Background
Pancreatic cancer is a highly lethal malignancy, estimated to become the second-leading cause of cancer related deaths by 2030 [1]. Early metastasis and increased chemoresistance make this cancer extremely difficult to treat and results in worse than typical prognosis [2, 3]. Emerging evidences support the involvement of CSCs in metastasis and resistance to chemotherapy [4, 5]. CSCs are a rare, small subset of cells with the capacity to give rise to full tumor mass. They have a self-renewal capacity and undergo asymmetric or symmetric cell division producing a heterogeneous cell population [5–7]. The presence of CSCs has been observed in many solid tumors, including PC [8–10]. PCSCs have been identified by expression of markers such as CD44+CD24+ESA+, CD133+, ALDH1+ and Side population (SP, Hoechst dye exclusion) [8]. CD133+CXCR4+ was identified as a subpopulation of PCSCs that mediates tumor metastasis in PC [8, 10, 11].
In recent years, various reports have shown the role of glycosylation in tumorigenesis. Glycosylation is a co- or post-translational modification of proteins and lipids with glycans (Glycoproteins; N- or O-linked, O-GlcNAc and Glycolipids). Glycosylation regulates various biological and cellular signaling, including embryogenesis, pluripotency, cell-to-cell & cell-to-environment interaction, signal transduction, protein folding, defense against microbial infection, and immune modulation [12–14]. Aberrant glycosylation is associated with tumor initiation, development, and metastasis. The expression of TACAs like Tn, sTn, T, sT (truncated) and sialyl Lewis a (sLea), sialyl Lewis x (sLex) (de novo; neo-synthesis) are especially implicated in tumor formation and metastasis in many cancers, including PC [15–17].
Several studies have explored the role of O-linked glycosylation in the regulation of tumorigenesis in PC. Expression of truncated O-glycans induced oncogenic signaling in PC; however, expression of core-3 derived O-glycans (extended structures) was shown to inhibit tumor growth and metastasis [18, 19]. In our recent study, we demonstrated that the crossing of C1galt1 floxed mice with KrasG12D; p53R172H; Pdx-1-Cre mice resulted in elevated synthesis of truncated O-linked glycans promoting development of aggressive PDAC with increased metastasis [17]. De-regulated mucin expression (MUC1, MUC4, MUC5AC, MUC16; majorly O-linked glycoprotein) has been implicated in PC development, metastasis, and chemoresistance [16, 20–24]. However, there are limited studies on the role of glycosylation in stemness of PCSCs [25]. One study, for example, demonstrated the enrichment of fucosylation in gemcitabine-resistant and CD44+CD24+ CSC-like populations in PC [26]. In another, the glycosyltransferase ST6Gal-1 was shown to regulate stem cell transcription factors, confer CSC phenotype, and promote gemcitabine resistance in pancreatic and ovarian cancer [27]. However, no detailed study has been performed to understand the role of glycosylation in stemness of PCSCs.
Given that both CSC and altered glycosylation have been implicated in tumorigenesis, metastasis, and chemoresistance, we tested the hypothesis that glycosylation maintains the stemness in PCSCs. The present study was undertaken to elucidate the significance of glycosylation and to identify the glycosyltransferases involved in the maintenance of stemness of PCSCs, and our results demonstrated significant importance of glycosylation in stemness. We also defined the role of specific GFs in the maintenance of stemness in PCSCs.
Methods
Cell culture
SW1990 (Cat. No. CRL-2172) and Capan1 (Cat. No. HTB-79) cells were procured from American type Culture Collection (Manassas, VA, USA) and cultured in Dulbecco’s Modified Eagles Medium (DMEM) (HyClone Laboratories, Logan, UT, USA) supplemented with 10% fetal bovine serum, 2 mM glutamine and 1% penicillin-streptomycin solution (Sigma-Aldrich, St Louis, MO, USA). These cell lines were recently authenticated by STR confirmation (Nebraska Medical Center HDI Lab, UNMC, Omaha, NE) and checked for mycoplasma contamination by using Roche BM cyclin kit (Cat. No. 10799050001) according to manufacturer’s instruction (Roche, Peznberg, Germany).
Growth inhibitory studies
Cells seeded at 4 × 103 cells/per well density in 96 well plate and grown in complete DMEM for 24 h. Cells were treated with different concentration of tunicamycin ™ (5–20 μg/ml) and benzyl-2-acetamido-2-deoxy-α-D-galactopyranoside (BAG) (0.5–2 mM) for different time points (24-72 h). At specified time points, 10 μl of MTT (5 mg/ml) in plain DMEM (90 μl) added to the each well and incubated for 3–4 h. Finally, cells were lysed by adding 200 μl DMSO to each well and absorbance was measured at 570 nm, with reference wavelength of 640 nm using ELISA plate reader.
SP assay
We performed SP analysis to determine the effect of TM (1.2 μM) and BAG (0.5 mM) on PCSCs. SP analysis was carried out by staining cells with Hoechst 33342 dye (AnaSpec Inc., Fremont, CA, USA) as described earlier [28]. Verapamil/Reserpine, a calcium channel blocker, was used as a control for gating SP cells.
Isolation of SP/CSC population from PC cells and culture
SP cells or putative CSCs and non-side population were isolated from SW1990 and Capan1 cells by FACS (fluorescent activated cell sorting) using standard method [28]. Verapamil/Reserpine was used to gate and sort SP cells. Isolated SP from SW1990 and Capan1 were grown in standardized stem cell-specific media as described previously [29]. SP cells were grown in 0.1 μM gemcitabine to obtain an enriched CSC population.
Immunoblot and lectin blot assay
SW1990, Capan1, SW1990 SP and Capan1 SP processed for protein extraction and western blotting using standard procedures [30]. For protein detection, the following antibodies were used; anti-OCT3/4, anti-SOX2, anti-ESA, anti-CD24, anti-β-Actin; anti-GAPDH (Sanatacruz Biotechnology, TX, USA); anti-CD44S (Cell signaling technology, MA, USA); anti-CD44v6 (eBiosciences, CA, USA); anti-CD133 (Abnova, CA, USA); anti-SOX9, anti-GALNT3, anti-B3GNT3, anti-MGAT4A (Abcam, MA, USA). Anti-CC49, Anti-sLex (Additional file 1: Table S1); Anti-sLea (Thermo Fischer Scientific, USA). β-Actin /GAPDH was used as a loading control. Blots were developed using chemiluminescent HRP kit (Bio-Rad, CA, USA). For lectin blotting, blots were blocked by 3% BSA (Jackson Immunoresearch Labs, Inc.) and probed for biotinylated lectins including VVA (binds Tn antigen) and PNA (binds T antigen) (Vector Labs. CA, USA). Streptavidin HRP was applied and bands were visualized using a chemiluminescent HRP kit.
Tumorsphere assay
NSP and SP cells of SW1990 and Capan1 were seeded in a 24/96 well ultra-low attachment plate (Corning Inc., New York, USA) in CSC-specific media at a concentration of 500/2500 cells per well in triplicate. Tumorsphere formation was observed under the microscope after 7–14 days. The number of tumor spheres were counted and plotted. A similar procedure was carried out for TM-treated (0.6 and 1.2 μM) and CRISPR knockout (KO) cells to study tumorsphere formation with respective controls.
RNA isolation and real time-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from cells using RNAeasy kit (Qiagen, CA, USA). 1–2 μg of RNA from each sample was converted to cDNA and used for RT-qPCR analysis [31]. Expression of CSC genes and glycogenes was profiled using gene-specific primers (Additional file 1: Table S2) and relative expression calculated using 2−ΔΔCT method. Data expressed as mean ± s.e.m. and statistical significance was set at P ≤ 0.05.
Human glycosylation polymerase chain reaction (PCR) array
Total RNA isolated from NSP and SP cells of SW1990 were reverse-transcribed using RT2 SYBR qPCR master mix (330,401, Qiagen). 25 μl aliquot of mix of both samples was added in separate 96-well PCR array kit containing lyophilized gene-specific primer set (PAHS-046ZF-12, Qiagen). Threshold cycles were used to calculate fold change using free online web server of RT2 profiler PCR array data analysis [31].
Confocal immunofluorescence (IF) microscopy
Cells were grown on sterilized cover slips for 24 h and primary antibodies specific for mouse-CD44v6 (1:500), rabbit-ESA (1:200), rabbit-SOX2 (1:50), rabbit-GALNT3 (1:200), mouse-GALNT3 (1:20), rabbit-B3GNT3 (1:200) were used to stain cells overnight at 4 °C. After incubation, cells were processed using standard procedure [29]. We used KC and KPC spontaneous mouse models and their control littermate tissue sections to examine GFs expression in CSCs. KC and KPC tissue sections were stained with mouse-CD44v6 (1:100), rabbit-GALNT3 (1:50), and rabbit-B3GNT3 (1:200) overnight at 4 °C. After incubation, tissues were processed as described earlier [29, 32].
Colony forming assay
Cells treated with or without TM (0.6 and 1.2 μM) were seeded in triplicate in six well plates at a concentration of 500/1000 cells per well in CSC-specific media. After 10–14 days of seeding, cells were fixed with ice-cold methanol for 5 min and stained with crystal violet stain solution (0.1%, w/v in 20 nM 4-morpholinepropanesulfonic acid; Sigma, MO, USA). Colonies were counted manually or with ImageJ software. Similar procedure was carried out for CRISPR knockout cells to study colony formation with controls.
Trans well migration assay
SP cells treated with or without TM (0.6 and 1.2 μM) were seeded at a concentration of 0.1 million cells per well in 100 μl of plain medium into the upper well of Boyden chamber with 8-μm pore, 6.5 mm polycarbonate trans well filters (Corning Costar Corp., MA, USA). The lower chamber contained 600 μl of CSC media. After 24 h, cells that had migrated at the lower surface of the membrane were fixed, stained, and counted under microscope. Similar procedure followed to study migration upon GALNT3KO clones.
Knockdown (KD) of GALNT3 and B3GNT3
Transient KD of GALNT3 (Cat. No. SR301729) and B3GNT3 (Cat. No. SR307003) was performed in SP cells of SW1990 and Capan1 cells using gene-specific siRNA (Origene, MD, USA). SP cells of SW1990 and Capan1 were plated 0.2 million cells/well in a six-well plate. Next day, cells were serum-starved for 3–4 h and transfected with gene-specific SiRNA or control siRNA at a concentration of 80 nM/well by using lipofectamine 2000 reagent (Invitrogen, Life Technologies Inc., NY, USA) in plain DMEM. After 4–6 h of transfection, serum containing media was added and cell lysate collected at 48–72 h post transfection.
Immunoprecipitation (IP) analysis
Cells were lysed with IP buffer (20 mM Tris, pH 7.5, 200 mM NACL, 1% NP-40, 10% Glycerol, 1 mM DTT, Protease inhibitor cocktail). IP was performed with anti-CD44v6, anti-CD44S, and anti-SLea (Thermo Fischer, MA, USA) antibodies as described earlier [30]. Immunoprecipitated samples were resolved on 10% SDS-PAGE (sodium dodecyl sulfate polyacrylamide gel electrophoresis) and transferred on PVDF (Polyvinylidene difluoride) membrane. Blots were incubated with protein-specific primary antibody, followed by species-specific secondary antibody and developed with chemiluminescent kit.
CRISPR/Cas9-mediated KO
KO of GALNT3 was performed by using CRISPR/Cas9 system in Capan1 SP cells. Briefly, cells were transfected with GALNT3 guide RNA (5’ATTTCTTTGCACCGAGATCT3’, GenScript, NJ, USA) in CRISPR/Cas9 vector (pSpCas9 BB-2A-GFP (PX458), GenScript, NJ, USA) by using lipofectamine 2000 reagent. GFP positive single cells were sorted in 96 well plate after 48 h of transfection by FACS. Single cells were allowed to grow into colonies in CSC-specific media and later used for further analysis.
Statistical analysis
Different statistical analysis including student t-test, one-way, and two-way Annova were used for different experiments to determine statistical significance. Error bars were set calculate standard error values. P value of ≥0.05 was considered statistically significant. * P ≤ 0.05 ** P ≤ 0.01 *** P ≤ 0.001 **** P ≤ 0.0001.
Results
Inhibition of glycosylation reduces the SP of PC cells
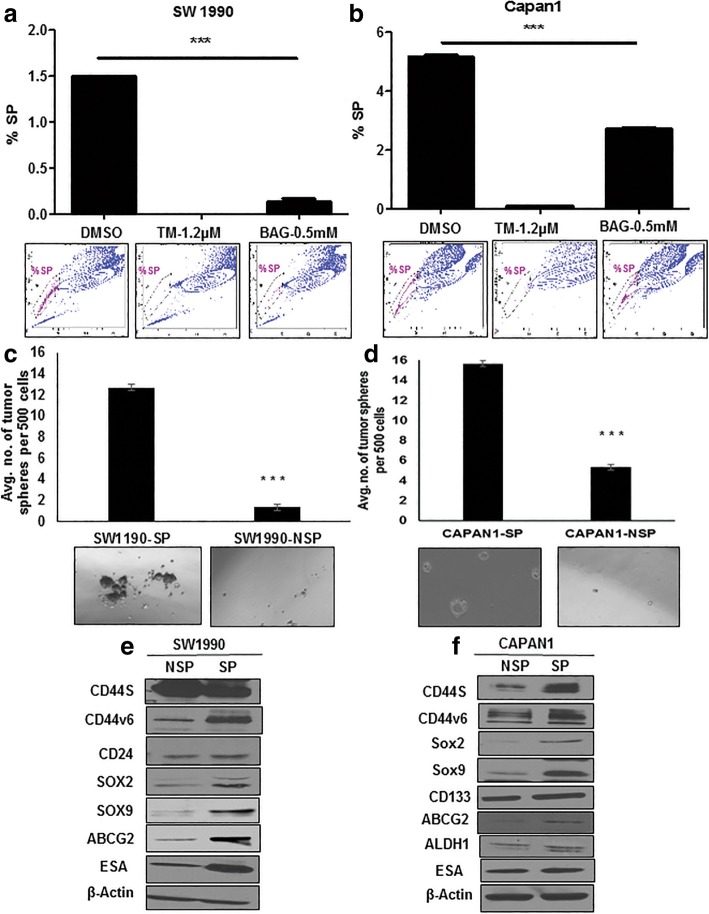
To investigate the functional importance of glycosylation in the stemness of PC cells, we used glycan inhibitors and analyzed a CSC population. TM and BAG were used to inhibit N-linked and O-linked glycosylation, respectively. Both TM and BAG showed dose- and time-dependent growth inhibition of SW1990 and Capan1 cells (Additional file 2: Figure S1a-S1d). We further analyzed the SP/CSCs and altered glycosylation with TM and BAG treated SW1990 and Capan1 cells. Both TM and BAG significantly reduced the SP/CSC population in SW1990 and Capan1 cells, as determined by SP analysis (Fig. 1a and b). TM treatment resulted in altered N-linked glycosylation of stem cell markers (ESA and CDDv6) and BAG treatment resulted in global variation of Tn antigen, O-linked glycosylation, as detected by VVA staining in PC cells (Additional file 2: Figure S1e and S1f).
Fig. 1.
Inhibition of global glycosylation reduces the SP cells of PC. a and b SP analysis in SW1990 and Capan 1 cells after treatment with TM and BAG for 48 h, respectively. Reserpine used as control to gate the SP cells. c and d Tumorsphere formation assay from NSP and SP cells of SW1990 and Capan1, respectively. e and f Immunoblotting analysis of CSC marker expression from NSP and SP cells of SW1990 and Capan1, respectively
Cancer stem cell-like phenotype of isolated SP cells
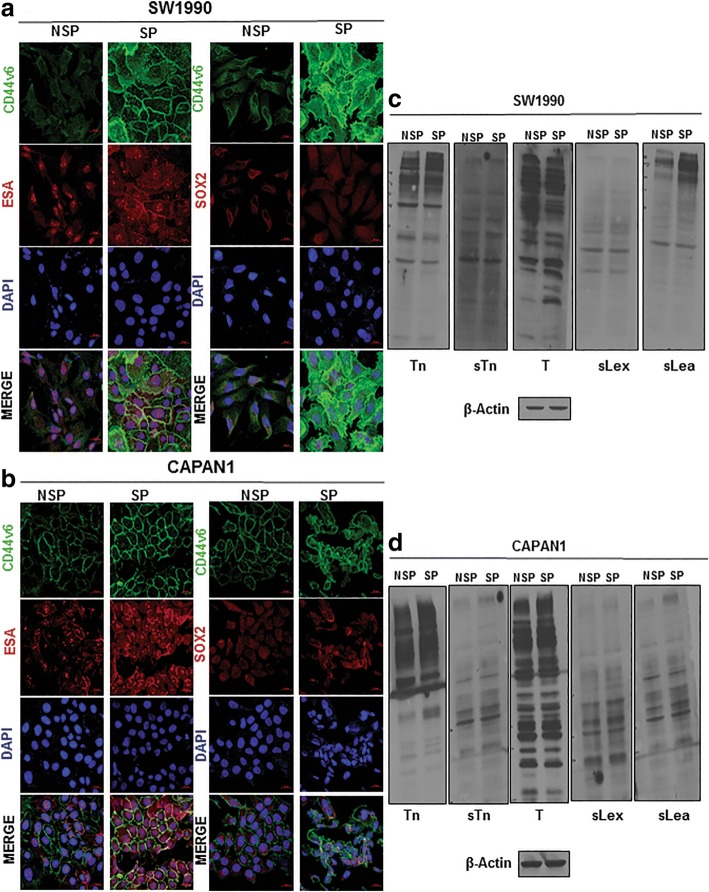
Because PCSCs display elevated expression of CSC markers and pose a marked self-renewal property, we characterized the SP of SW1990 and Capan1 cells [28]. We isolated SP cells and non-side population (NSP) cells from SW1990 and Capan1 cells (Additional file 3: Figure S2a), and found that the cellular morphology of SP cells was distinct from that of NSP cells derived from either SW1990 or Capan1 (Additional file 3: Figure S2b). Furthermore, the CSC phenotype of SP cells were characterized by tumorsphere assay and CSC markers expression. Tumorsphere assay determines the self-renewal capacity and in vitro tumorigenic potential of stem cells [33]. SP cells from SW1900 and Capan1 formed a significantly higher number of tumorspheres compared to NSP cells (Fig. 1c and d). Further, SP cells showed increased expression of cell surface markers (viz CD44v6, CD44s, ESA, and ABCG2) and self-renewal markers (viz SOX2 and SOX9) compared to NSP as determined by RT-qPCR (Additional file 3: Figure S2c and S2d) and western blotting (Fig. 1e and f). Increased expression of CSCs markers in SP cells (CD44v6 with SOX2/ESA) was also confirmed by IF analysis (Fig. 2a and b).
Fig. 2.
Global variation in expression of TACAs in PCSCs. a and b IF analysis of CSC marker expression from NSP and SP cells of SW1990 and Capan1, respectively. c and d Immunoblotting analysis of TACAs from NSP and SP cells of SW1990 and Capan1, respectively
TACAs differentially expressed in PCSCs
Aberrant glycosylation in cancer results in the expression of TACAs that are implicated in early tumor progression and metastasis [15]. Because CSCs have been implicated in tumor initiation and metastasis, we next examined if expression of TACAs, such as Tn, T, sTn, sLex, and sLea, is altered in CSCs/SP cells of SW1990 and Capan1 PC cells. SP cells of SW1990 and Capan1 showed differential expression of TACAs compared to NSP (Fig. 2c and d). Few higher and low molecular bands of Tn and sLea exhibited greater expression in SP cells of SW1990 and Capan1 compared to NSP cells (Fig. 2c and d).
N-linked glycosylation regulates stemness, clonogenicity, migration of PCSCs
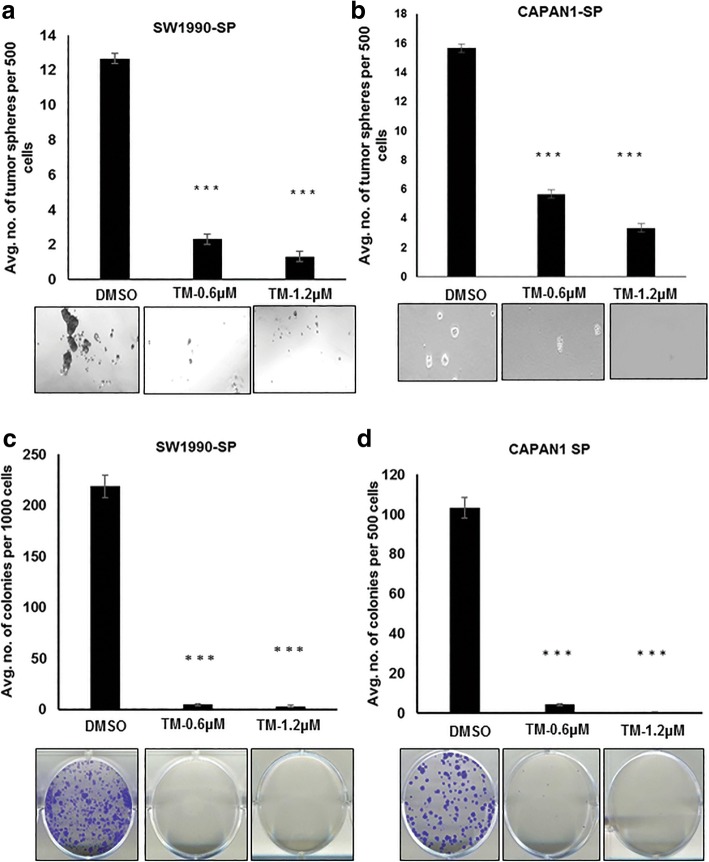
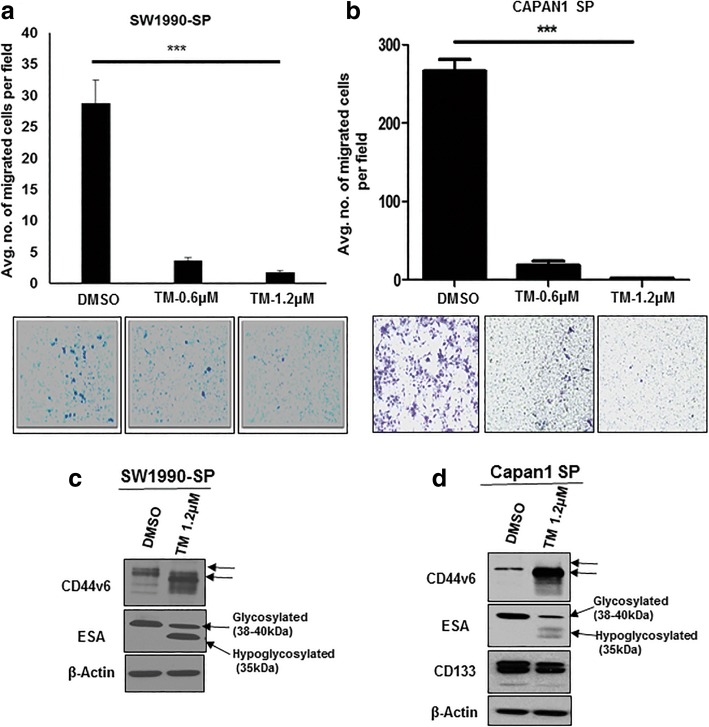
Because many of the CSC markers, such as CD44 and ESA, are N-glycosylated, we evaluated the role of N-linked glycosylation in the stemness of PCSCs by treating them with TM [34, 35]. TM treatment significantly inhibited the tumorsphere formation of SP cells of SW1990 and Capan1 compared to control cells treated with dimethyl sulfoxide (DMSO) (Fig. 3a and b). Further, experiments were performed to understand the role of N-linked glycosylation in growth and migration of PCSCs. Of note, TM treatment resulted in a complete loss of ability of SP cells from SW1990 and Capan1 to form visible colonies, compared to DMSO-treated cells (Fig. 3c and d). In addition, TM treatment significantly inhibited the migratory capacity of SW1990 and Capan1 SP cells compared to DMSO treatment (Fig. 4a and b). These results suggest the importance of N-linked glycans in stemness, clonogenicity and migration. We further examined TM effect on CSC marker expression, and were interested to find that TM treatment resulted in a shift in molecular weight of CD44v6 and ESA in SP cells of SW1990 and Capan1 compared to DMSO treatment, and resulted in a reduction of expression of CD133 in Capan1 SP cells (Fig. 4c and d). This observation indicates the possible involvement of N-linked glycan modification of CD44v6 and ESA in PCSC stemness, tumorigencity, and metastasis.
Fig. 3.
TM inhibits tumorsphere formation and clonogenicity of PCSCs. a and b Tumorsphere formation assay from SP cells of SW1990 and Capan1, respectively, after pre-treatment with TM for 48 h. c and d Colony formation assay from SP cells of SW1990 and Capan1, respectively, after pre-treatment with TM for 48 h
Fig. 4.
TM inhibits migration of PCSCs and hypo-glycosylates CSC markers. a and b Trans-well migration assay from SP cells of SW1990 and Capan1, respectively, after treatment with TM. c and d Immunoblotting analysis of CSC marker expression from SP cells of SW1990 and Capan1, respectively, after treatment with TM for 48 h
GALNT3 and B3GNT3 overexpressed in PCSCs
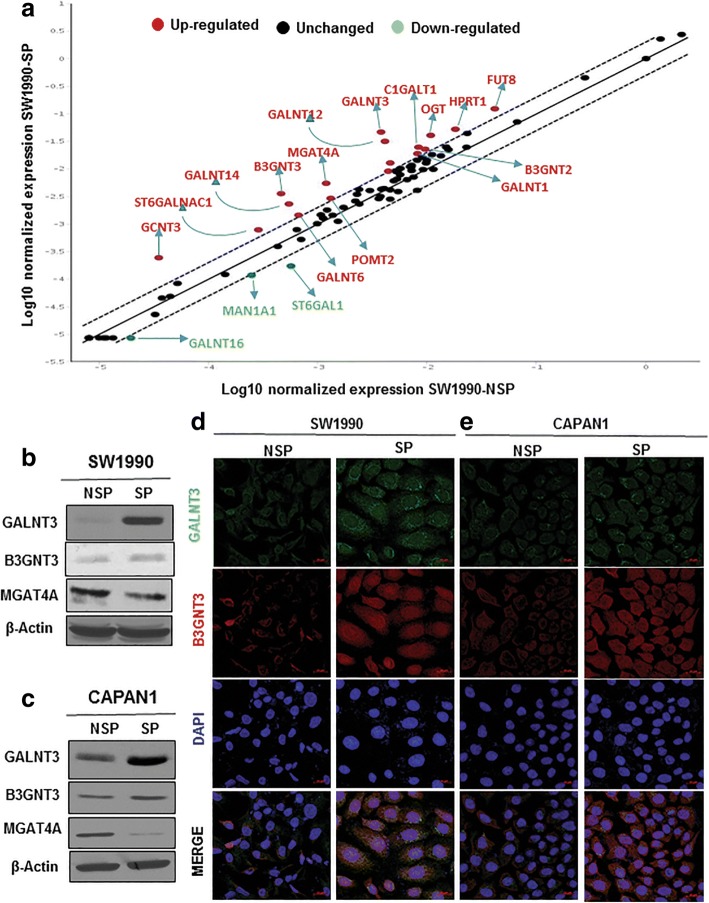
Since aberrant glycosylation is attributed to altered expression of GFs, we next performed a PCR array for human glycosylation genes in SW1990 NSP and SP cells. Twenty glycogenes were differentially expressed from PCR array for human glycosylation between NSP and SP cells of SW1990 (Fig. 5a, Additional file 1: Table S3). Differentially expressed glycogenes (DEGs) were further validated using RT-qPCR, revealing that only GALNT3, GALNT12, GALNT16, B3GNT2, B3GNT3, and MGAT4A were differentially expressed (Additional file 4: Figure S3a, Additional file 1: Table S3). These DEGs were also validated in NSP and SP of Capan1 by RT-qPCR (Additional file 4: Figure S3b). In particular, increased expression of GALNT3 and B3GNT3 and decreased expression of MGAT4A proteins was observed in SP cells from SW1990 and Capan1 cells (Fig. 5b and c). Increased expression of GALNT3 and B3GNT3 was seen in both the mRNA and protein levels, whereas MGAT4A expression alone was less in protein level. The IF expression of GALNT3 and B3GNT3 confirmed overexpression of these GFs in PCSCs (Fig. 5d and e).
Fig. 5.
GALNT3 and B3GNT3 are overexpressed in PCSCs. a PCR array for human glycosylation from NSP and SP cells of SW1990. b and c Immunoblotting analysis of DEGs between NSP and SP cells of SW1990 and Capan 1, respectively. d and e IF analysis of GALNT3 and B3GNT3 expression between NSP and SP cells of SW1990 and Capan 1, respectively
GALNT3 and B3GNT3 specifically over-expressed in advanced stages of KC and KPC autochthonous tumors along with CD44v6
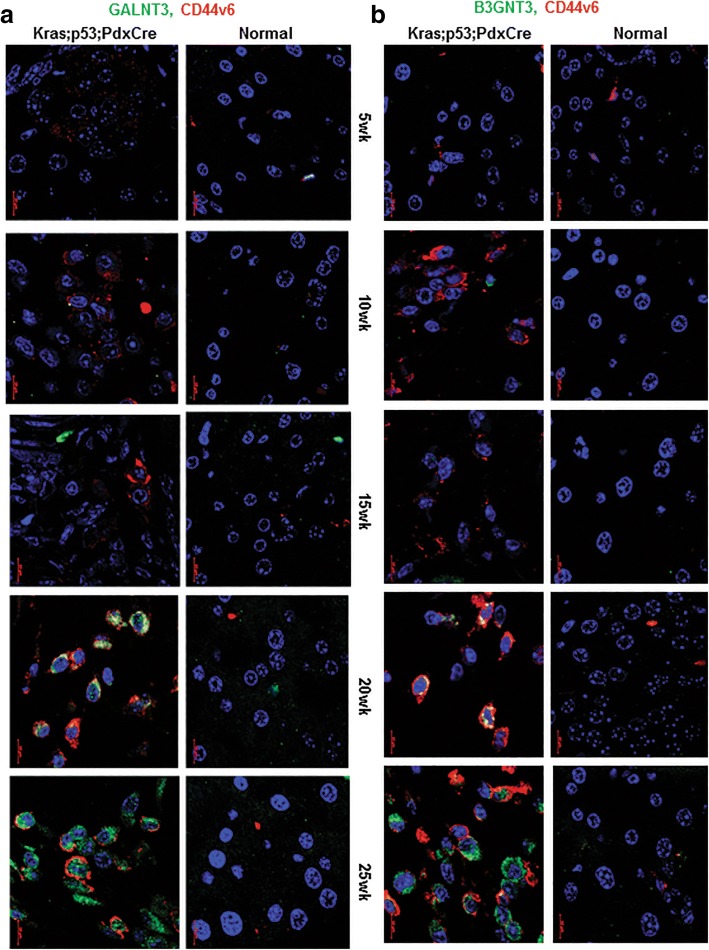
To understand the involvement of GALNT3 and B3GNT3 in the pathogenesis of PC progression, the expression of these GFs, along with CSC marker CD44v6, were analyzed in different stages of KC and KPC mouse tumor tissues by IF. In KC tissues, GALNT3 and B3GNT3 co-expression with CD44v6 were observed at fully advanced PDAC stage at the 50th week (Additional file 5: Figure S4a and S4b). Similarly, in KPC tissues, co-expression of GALNT3 and B3GNT3 with CD44v6 observed at PDAC stages at the 20th and 25th weeks (Fig. 6a and b). These results indicate that both GALNT3 and B3GNT3 are specifically overexpressed in advanced stages of PC progression, along with CSC marker CD44v6 expression.
Fig. 6.
GALNT3 and B3GNT3 expresses at PDAC stage in PCSCs of KPC. a and b IF analysis for co-expression of GALNT3 and B3GNT3 with CD44v6, respectively, at different stages of KPC
KD of GALNT3 and B3GNT3 decreases PCSC markers
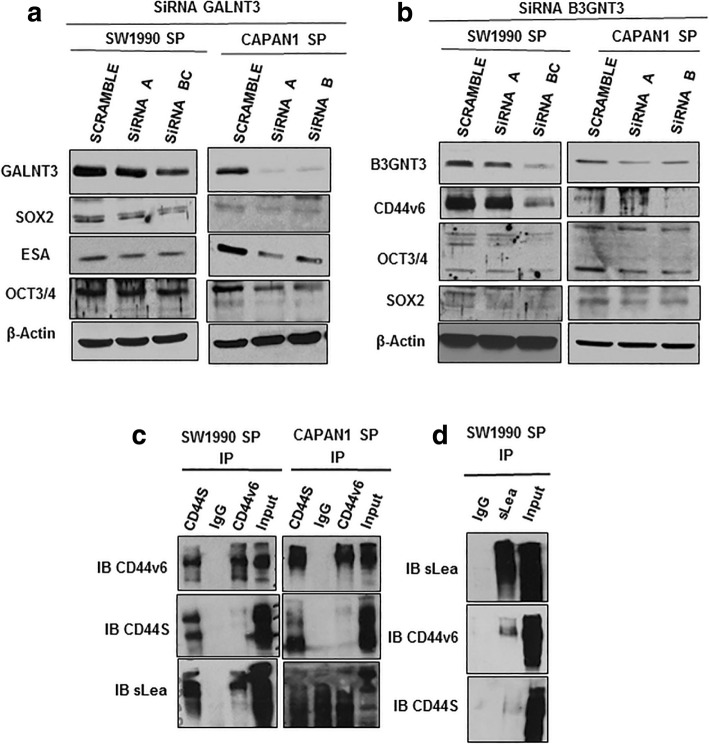
To identify the mechanism by which GALNT3 and B3GNT3 impact CSC stemness, we performed transient KD of GALNT3 and B3GNT3 in PCSCs. Silencing of GALNT3 and B3GNT3 showed altered expression of the cell surface, and self-renewal stem cell markers in PCSCs. Specifically, GALNT3 KD reduced expression levels of SOX2, OCT3/4 and ESA (Fig. 7a). KD of B3GNT3 reduced CD44v6, SOX2 and OCT3/4 expression (Fig. 7b). Our results suggest that both GALNT3 and B3GNT3 maintain stemness by altering expression of PCSC markers.
Fig. 7.
GALNT3 and B3GNT3 regulates expression of PCSC markers. a siRNA mediated knockdown of GALNT3 and immunoblotting analysis of CSC markers expression from SP cells of SW1990 and Capan1, respectively. b siRNA mediated knockdown of B3GNT3 and immunoblotting analysis of CSC markers expression from SP cells of SW1990 and Capan1, respectively. c IP of CD44s and CD44v6 from SP cells of SW1990 and Capan1, and immunoblotting with CD44s, CD44v6 and sLea. d IP of sLea from SP cells of SW1990, and immunoblotting with sLea, CD44v6 and CD44s
CD44v6 carries sLea glycans in PCSCs
In SP cells of SW1990 and Capan1, increased expression of sLea glycans at higher molecular weight (100-250 kDa) was observed. Since CD44v6 molecular weight falls in that size range, we analyzed for its potential modification with sLea. CD44S and CD44v6 were immunoprecipitated and blotted with sLea expression in PCSCs. We were interested to find that only CD44v6 carries sLea modification in significant amounts, but not CD44S in SW1990 and Capan1 SP cells (Fig. 7c). We also performed reciprocal IP with sLea and probed for CD44v6 and CD44S, and found that CD44v6 underwent sLea modification (Fig. 7d). Our results indicate that the modification of sLea on CD44v6 of PCSC, which may be involved in tumor metastasis.
GALNT3 regulates stemness in PCSCs
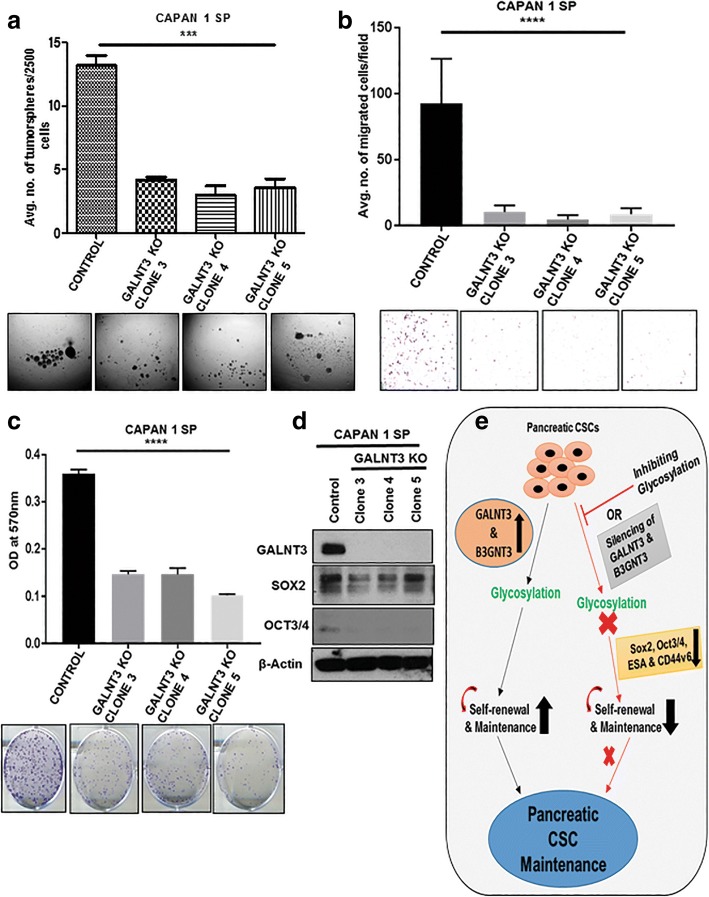
Transient KD of GALNT3 alluded to its role in regulating the expression of PCSCs markers. We further used CRISPR/Cas9 system to KO GALNT3 expression to explore its functional importance in PCSCs, with three complete GALNT3 KO Capan1 SP clones developed (Fig. 8d, Additional file 6: Figure S5). GALNT3 KO clones formed a lesser number of tumorspheres compared to control SP cells (Fig. 8a). KO of GALNT3 resulted in loss of self-renewal and the in-vitro tumorigenic potential of SP cells. We next evaluated the effect of GALNT3 KO on clonogenicity and migration of SP cells. GALNT3 KO clones formed very few colonies (Fig. 8c), and migrated much less compared to control SP cells (Fig. 8b). Furthermore, GALNT3 KO clones showed reduced expression of self-renewal markers, such as SOX2 and OCT3/4 (Fig. 8d). These results imply that GALNT3 regulates growth, stemness, and migration of PCSCs.
Fig. 8.
GALNT3 regulates stemness, clonogenicity and migration in PCSCs. a Tumorsphere sphere formation assay in GALNT3 KO clones compared to control Capan 1 SP cells. b and c Transwell migration and colony formation assay in GALNT3 KO clones compared to control SP cells. d Immunoblotting analysis of CSC markers expression compared to control SP cells. e Schematic representation describing the importance of glycosylation in the maintenance of stemness of PCSCs. Silencing of GALNT3 and B3GNT3 specifically involved in decreasing the self-renewal markers and stemness in PCSCs
Discussion
Cancer stem cells have been correlated to metastasis to distant organs and to chemo- and radio- resistance including pancreatic cancer [11, 36]. Although there is ongoing work to target CSCs, mainly at the genomic and proteomic levels, CSC nevertheless remains a challenge that must be addressed. It is crucial, therefore, to understand CSC maintenance from a unique angle that may pave the way for better management of PC. To that end, in the present study, we investigated the importance of glycosylation in maintenance of PCSC populations, identifying and defining the role of GALNT3 and B3GNT3 in maintenance of stemness of PCSCs.
CSCs or tumor initiating cells (TICs) are responsible for tumor initiation, metastasis, chemo/radio-resistance, and recurrence in many cancers including PC [4, 5, 11, 36]. Aberrant glycosylation is also implicated for tumor development, metastasis and chemoresistance in PC and several other malignancies [15–17]. Our results indicate that glycosylation plays an important role in the maintenance of stemness of PCSCs. Inhibition of N- and O-linked glycosylation decreased SP/CSCs, in PC cells suggesting that both glycan modifications are important for PCSCs maintenance. Further, inhibition of N-linked glycosylation (TM treatment) reduced tumorsphere formation, clonogenicity, migration, and hypo-glycosylated CD44v6 and ESA of PCSCs. TM, a mixture of homologous nucleoside antibiotics, blocks N-linked glycosylation by inhibiting GlcNAc phophotransferase. TM has been reported to induce unfolded protein response (UPR) and endoplasmic reticulum (ER)-stress mediated apoptosis in many cancers [37, 38]. Similar to our observation, a previous study shows TM treatment reduces the in vitro subpopulation and invasion of breast CSCs through ER stress [39]. In another study, TM treatment increased hypo-glycosylated ESA by decreasing its glycosylated form in head and neck cancer [35]. Our results with these studies suggest that N-linked glycosylation is involved in the maintenance of PCSCs by altering the glycosylation of CSC markers.
Based on the importance of glycosylation in PCSCs, we further identified the glycogenes involved in stemness. Our results indicated upregulation of GALNT3 and B3GNT3 among 84 GFs in isolated CSCs/SP cells. Both GALNT3 and B3GNT3 are involved in the synthesis of mucin-type O-linked glycosylation. For example, GALNT3 is member of GalNAc-transferases that catalyze the addition of GalNAc on the serine or threonine residue of polypeptide to from the Tn structure [40]. B3GNT3 is member of the B3GlcNAc family that catalyzes the extended core 1 structure (6-sulfo sLex), synthesizes the poly-N-acetyllactoseamine structures and dimeric sLea antigens, and is involved in L-selectin ligand biosynthesis, lymphocyte homing, and trafficking [41–43]. Since GALNT3 is linked to biosynthesis of the Tn carbohydrate antigen and B3GNT3 is correlated to the sLea antigen, increased expression of GALNT3 and B3GNT3 in CSCs could be the plausible mechanism of observed increased expression of Tn and sLea in PCSCs.
The expression of GALNT3 and B3GNT3 along with CSC marker CD44v6 were analyzed in the progression of KC and KPC mouse tissues. In the KC model, PanIN (Pancreatic Intraepithelial neoplasia) I lesions start at 10 weeks of age and develop to fully blown PDAC (Pancreatic ductal adeno carcinoma) at 50th weeks of age [32]. In KPC model, PanINs observed at 5 weeks of age that progress to PDAC stage by 20th and 25th weeks of age [29]. Our results showed co-expression of GALNT3 and B3GNT3 along with CD44v6 at an advanced stage of PDAC in KC (50th week) and KPC (20th and 25th week). These results suggest the possible role of GALNT3 and B3GNT3 in the maintenance of CSCs in advanced stages of PC progression.
GALNT3 and B3GNT3 have been implicated in the regulation of tumorigenesis in many cancers. GALNT3 is overexpressed in some cancers including PC, and is known for its tumor-promoting role in pancreatic and ovarian cancers [44, 45]. In our previous study, by contrast, we demonstrated that down-regulation of GALNT3 leads to aggressiveness in poorly differentiated PC cells [46]. In the present study, significantly increased expression of GALNT3 was shown in PCSCs, which is an undifferentiated population. These studies and our observation suggest that GALNT3 is differentially expressed based on the differentiation potential of cancer cells, and may play a different functional role based on its level of expression. Similarly, we demonstrated the multifunctional role of PD2/PAF1, a stem cell marker, based on the expressional variation in different cell types [29, 47–51]. Expression of B3GNT3 is also involved in the development and progression of many cancers, including non-hodgkin lymphoma (NHL), colon, pancreatic, esophageal squamous cell, hepatocellular, and cervical [41, 43, 52, 53]. In contrast, over expression of B3GNT3 increases the migration and invasion in neuroblastoma cells [54]. These reports indicate opposite roles of these GFs in the tumorigenesis of different cancers.
Our next goal was to investigate the GALNT3 and B3GNT3 mediated mechanism for maintenance of PCSCs. We observed that KD of GALNT3 reduced expression of SOX2, OCT3/4, and ESA, and that B3GNT3 KD reduced CD44v6, SOX2 and OCT3/4 expression. These results confirm that O-GFs GALNT3 and B3GNT3 are involved in the maintenance of stemness in PCSCs by altering the CSC markers. Similar to our observation, numerous studies have shown the significant involvement of GFs in the regulation of stemness of many cancers [25]. GALNT1 has been reported to regulate stemness in bladder CSCs through activating SHH signaling and inducing gli1 expression [55]. The two GFs, B4GALNT3 and MGAT5, are involved in synthesis of N-linked glycosylation and have been implicated in regulating stemness in colon CSCs. B4GALNT3 and MGAT5 have been shown to regulate stemness by modifying EGFR and WNT glycosylation, respectively [56, 57]. In another study, ST6Gal-1 has been reported to regulate stemness and gemcitabine resistance in pancreatic and ovarian cancer by modulating SOX9 and SLUG expression [27]. Our study supports the concept that specific GFs are involved in the maintenance of stemness in PC.
Expression of TACAs are implicated in tumorigenesis in numerous cancers [15–17]. Therefore, we analyzed the expression of TACAs in PCSC, given that they are implicated in tumor development and metastasis. Our results showed a global change in expression of TACAs, and increased expression of Tn and sLea at higher molecular weight proteins. Further, we noticed the reduction in expression of CD44v6 in PCSCs by KD of B3GNT3. Because both CD44v6 and sLea are implicated in the tumor metastasis process, B3GNT3 may be involved in the possible modification of CD44 for sLea in PCSCs. Of interest, our IP results showed the modification of sLea on CD44v6 but not on CD44S in PCSCs. There are many reports in the literature relating the expression of CD44v6 and sLea to tumor metastasis. CD44v6 expression promotes tumor growth and metastasis in many cancer including PC and colorectal CSCs [58–62]. In one study, sLea modification on CD44v6 was shown to mediate detachment of ploymorphonuclear leukocytes during trans-epithelial migration from the intestinal epithelium [63]. The H type glycan on CD44v6 was also shown to augment motility of tumor cells and tumorigencity in rat colon carcinoma [64, 65]. Further, CD44v6 is known to be modified with T and sTn antigens in colon cancer and is implicated in tumor metastasis [66]. Our results and those of previous studies indicate the possible involvement of CD44v6 modified with sLea, possibly by B3GNT3, which may facilitate tumor metastasis. However, this insight warrants further detailed investigation.
CRISPR/Cas9-mediated GALNT3 KO in PCSCs showed decreased tumorspheres formation, colony formation and migration potential, along with reduced expression of the stem cell transcription factors, SOX2 and OCT3/4. These results suggest that GALNT3 regulates stemness, growth and migration by regulating SOX2 and OCT3/4 expression in PCSCs. Similar to our observation, stable knockdown of GALNT1 was shown to inhibit oncosphere formation and soft agar colony formation in bladder CSCs by regulating gli1 expression [55]. Our results and the literature further support the involvement of GFs in regulating stemness of CSCs.
Conclusions
In summary, our present work thus delineates the importance of glycosylation in the maintenance of stemness in PCSCs. We have identified for the first time the role of GALNT3 and B3GNT3 in regulating the stemness of PCSCs by altering CSC markers (Fig. 8e). Specifically, modification of sLea on CD44v6 occurs in PCSCs, which may be involved in the tumor metastasis. We conclude that O-GFs GALNT3 and B3GNT3 are involved in the maintenance of PCSCs and the observed findings opens new avenues to target CSC populations in PC.
Additional files
Table S1. Antibodies used for the study. Table S2. Primers used for the study. A. Cancer stem cell markers and B. Glycogenes. Table S3. Fold change values of differentially expressed genes in SP when compared to NSP of SW1990. (DOCX 21 kb)
Figure S1. TM and BAG inhibits growth and alters glycosylation of PC cells. (a) and (b), Effect of TM on growth of SW1990 and Capan1 cells. (c) and (d), Effect of BAG on growth of SW1990 and Capan1 cells. (e) Effect of TM on N-linked glycosylation of PC cells. (f) Effect of BAG treatment on O-linked glycosylation of PC cells. (TIF 2768 kb)
Figure S2. Isolation and Characterization of PCSCs. Isolation and Characterization of PCSCs. (a) Sorting of SP and NSP cells from SW1990 and Capan 1 by Hoechst staining. Reserpine/Verapamil used as control to gate the SP cells. (b) Morphology of NSP and SP cells of SW1990 and Capan1. (c) and (d), RT-qPCR analysis of CSC markers expression between NSP and SP cells of SW1990 and Capan1, respectively. β-Actin is used to normalize the fold change values. (TIF 4154 kb)
Figure S3. RT-qPCR analysis of DEGs in PCSCs. (a) and (b), RT-qPCR analysis for validation of DEGs identified by PCR array between NSP and SP cells of SW1990 and Capan1. (TIF 2283 kb)
Figure S4. GALNT3 and B3GNT3 expresses at PDAC stage in PCSCs of KC. (a) and (b), IF analysis for co-expression of GALNT3 and B3GNT3 with CD44v6, respectively, at different stages of KC. (TIF 8121 kb)
Figure S5. KO of GALNT3 in Capan 1 SP cells. IF analysis for expression of GALNT3 in control and GALNT3 KO clones of Capan 1 SP. (TIF 2895 kb)
Acknowledgements
We thank Craig Semerad, Victoria B. Smith and Samantha Wall of the Flow Cytometry Research Facility, University of Nebraska Medical Center, for providing assistance with flow cytometry. We also thank confocal core facility at UNMC. We also thank Mrs. Kavita Mallya, for her invaluable technical support. We also thank Dr. Adrian Koesters, Research Editor at UNMC, for her substantial editorial contribution to the manuscript.
Funding
No specific funding is received for this study. The authors on this manuscript are, in parts, supported by grants from the National Institutes of Health RO1 CA210637, R01CA183459, PO1 CA217798, UO1 CA185148, UO1 CA200466, P50 CA127297 and P30 CA036727) the Nebraska Department of Health and Human Services LB595.
Abbreviations
- BAG
Benzyl-2-acetamido-2-deoxy-α-D-galactopyranoside
- CSCs
Cancer stem cells
- DEGs
Differentially expressed glycogenes
- DMEM
Dulbecco’s Modified Eagles Medium
- DMSO
Dimethyl sulfoxide
- ER
Endoplasmic reticulum
- FACS
Fluorescence activated cell sorting
- GFs
Glycosyltransferases
- IF
Immunofluorescence
- IP
Immunoprecipitation
- KC
Kras G12D ; Pdx-1-Cre
- KD
Knockdown
- KO
Knockout
- KPC
Kras G12D ; p53 R172H ; Pdx-1-Cre
- NHL
Non-hodgkin lymphoma
- NSP
Non side population
- PanIN
Pancreatic Intraepithelial neoplasia
- PC
Pancreatic cancer
- PCR
Polymerase chain reaction
- PCSCs
Pancreatic cancer stem cells
- PDAC
Pancreatic ductal adenocarcinoma
- PVDF
Polyvinylidene difluoride
- RT-qPCR
Real time quantitative polymerase chain reaction
- SDS-PAGE
Sodium dodecyl sulfate polyacrylamide gel electrophoresis
- sLea
Sialyl Lewis a
- sLex
Sialyl Lewis x
- SP
Side population
- TACAs
Tumor associated carbohydrate antigens
- TICs
Tumor initiating cells
- TM
Tunicamycin
- UPR
Unfolded protein response
Authors’ contributions
SB, SKB and MPP designed and executed the study. SB performed the experiments, compiled the data and wrote the manuscript. SC, SK, GK, SR involved in the execution of experiments and edited the manuscript. SNR performed the animal model experiments. MPP and SKB interpreted the data and edited the manuscript. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
We have received Institutional Animal Care and Use Committee (IACUC) approval to perform the animal experiments. There is no animal sacrificed for this study, we have used previously available paraffin tissue sections. We did not use human samples for this study.
Consent for publication
Not applicable.
Competing interests
SKB is one of co-founders of Sanguine Diagnostics and Therapeutics, Inc. No potential conflicts of interest were disclosed by the other authors.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Srikanth Barkeer, Email: barkeer.srikanth@gmail.com.
Seema Chugh, Email: seemac@med.umich.edu.
Saswati Karmakar, Email: saswati.karmakar@unmc.edu.
Garima Kaushik, Email: garima.kaushik@unmc.edu.
Sanchita Rauth, Email: sanchita.rauth@unmc.edu.
Satyanarayana Rachagani, Email: srachagani@unmc.edu.
Surinder K. Batra, Email: sbatra@unmc.edu
Moorthy P. Ponnusamy, Phone: 402-559-1170, Email: mpalanim@unmc.edu
References
- 1.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74(11):2913–2921. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 2.Jiang JH, Liu C, Cheng H, Lu Y, Qin Y, Xu YF, Xu J, Long J, Liu L, Ni QX, et al. Epithelial-mesenchymal transition in pancreatic cancer: is it a clinically significant factor? Biochim Biophys Acta. 2015;1855(1):43–49. doi: 10.1016/j.bbcan.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 3.Hezel AF, Kimmelman AC, Stanger BZ, Bardeesy N, Depinho RA. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 2006;20(10):1218–1249. doi: 10.1101/gad.1415606. [DOI] [PubMed] [Google Scholar]
- 4.Medema JP. Cancer stem cells: the challenges ahead. Nat Cell Biol. 2013;15(4):338–344. doi: 10.1038/ncb2717. [DOI] [PubMed] [Google Scholar]
- 5.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414(6859):105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 6.Lobo NA, Shimono Y, Qian D, Clarke MF. The biology of cancer stem cells. Annu Rev Cell Dev Biol. 2007;23:675–699. doi: 10.1146/annurev.cellbio.22.010305.104154. [DOI] [PubMed] [Google Scholar]
- 7.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100(7):3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vaz AP, Ponnusamy MP, Batra SK. Cancer stem cells and therapeutic targets: an emerging field for cancer treatment. Drug Deliv Transl Res. 2013;3(2):113–120. doi: 10.1007/s13346-012-0095-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mimeault M, Hauke R, Mehta PP, Batra SK. Recent advances in cancer stem/progenitor cell research: therapeutic implications for overcoming resistance to the most aggressive cancers. J Cell Mol Med. 2007;11(5):981–1011. doi: 10.1111/j.1582-4934.2007.00088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hermann PC, Huber SL, Herrler T, Aicher A, Ellwart JW, Guba M, Bruns CJ, Heeschen C. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1(3):313–323. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 11.Zhan HX, Xu JW, Wu D, Zhang TP, Hu SY. Pancreatic cancer stem cells: new insight into a stubborn disease. Cancer Lett. 2015;357(2):429–437. doi: 10.1016/j.canlet.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 12.Wang YC, Lin V, Loring JF, Peterson SE. The ‘sweet’ spot of cellular pluripotency: protein glycosylation in human pluripotent stem cells and its applications in regenerative medicine. Expert Opin Biol Ther. 2015;15(5):679–687. doi: 10.1517/14712598.2015.1021329. [DOI] [PubMed] [Google Scholar]
- 13.Jang H, Kim TW, Yoon S, Choi SY, Kang TW, Kim SY, Kwon YW, Cho EJ, Youn HD. O-GlcNAc regulates pluripotency and reprogramming by directly acting on core components of the pluripotency network. Cell Stem Cell. 2012;11(1):62–74. doi: 10.1016/j.stem.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 14.Varki A, Lowe JB. Biological Roles of Glycans. In: Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME, editors. Essentials of Glycobiology. 2nd ed. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2009. [PubMed]
- 15.Pinho SS, Reis CA. Glycosylation in cancer: mechanisms and clinical implications. Nat Rev Cancer. 2015;15(9):540–555. doi: 10.1038/nrc3982. [DOI] [PubMed] [Google Scholar]
- 16.Chugh S, Gnanapragassam VS, Jain M, Rachagani S, Ponnusamy MP, Batra SK. Pathobiological implications of mucin glycans in cancer: sweet poison and novel targets. Biochim Biophys Acta. 2015;1856(2):211–225. doi: 10.1016/j.bbcan.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chugh Seema, Barkeer Srikanth, Rachagani Satyanarayana, Nimmakayala Rama Krishna, Perumal Naveenkumar, Pothuraju Ramesh, Atri Pranita, Mahapatra Sidharth, Thapa Ishwor, Talmon Geoffrey A., Smith Lynette M., Yu Xinheng, Neelamegham Sriram, Fu Jianxin, Xia Lijun, Ponnusamy Moorthy P., Batra Surinder K. Disruption of C1galt1 Gene Promotes Development and Metastasis of Pancreatic Adenocarcinomas in Mice. Gastroenterology. 2018;155(5):1608–1624. doi: 10.1053/j.gastro.2018.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Radhakrishnan P, Dabelsteen S, Madsen FB, Francavilla C, Kopp KL, Steentoft C, Vakhrushev SY, Olsen JV, Hansen L, Bennett EP, et al. Immature truncated O-glycophenotype of cancer directly induces oncogenic features. Proc Natl Acad Sci U S A. 2014;111(39):E4066–E4075. doi: 10.1073/pnas.1406619111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Radhakrishnan P, Grandgenett PM, Mohr AM, Bunt SK, Yu F, Chowdhury S, Hollingsworth MA. Expression of core 3 synthase in human pancreatic cancer cells suppresses tumor growth and metastasis. Int J Cancer. 2013;133(12):2824–2833. doi: 10.1002/ijc.28322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gautam SK, Kumar S, Cannon A, Hall B, Bhatia R, Nasser MW, Mahapatra S, Batra SK, Jain M. MUC4 mucin- a therapeutic target for pancreatic ductal adenocarcinoma. Expert Opin Ther Targets. 2017;21(7):657–669. doi: 10.1080/14728222.2017.1323880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Das S, Batra SK. Understanding the unique attributes of MUC16 (CA125): potential implications in targeted therapy. Cancer Res. 2015;75(22):4669–4674. doi: 10.1158/0008-5472.CAN-15-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nath S, Mukherjee P. MUC1: a multifaceted oncoprotein with a key role in cancer progression. Trends Mol Med. 2014;20(6):332–342. doi: 10.1016/j.molmed.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Joshi S, Kumar S, Choudhury A, Ponnusamy MP, Batra SK. Altered mucins (MUC) trafficking in benign and malignant conditions. Oncotarget. 2014;5(17):7272–7284. doi: 10.18632/oncotarget.2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaur S, Kumar S, Momi N, Sasson AR, Batra SK. Mucins in pancreatic cancer and its microenvironment. Nat Rev Gastroenterol Hepatol. 2013;10(10):607–620. doi: 10.1038/nrgastro.2013.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barkeer S, Chugh S, Batra SK, Ponnusamy MP. Glycosylation of Cancer stem cells: function in Stemness, tumorigenesis, and Metastasis. Neoplasia. 2018;20(8):813–825. doi: 10.1016/j.neo.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Terao N, Takamatsu S, Minehira T, Sobajima T, Nakayama K, Kamada Y, Miyoshi E. Fucosylation is a common glycosylation type in pancreatic cancer stem cell-like phenotypes. World J Gastroenterol. 2015;21(13):3876–3887. doi: 10.3748/wjg.v21.i13.3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schultz MJ, Holdbrooks AT, Chakraborty A, Grizzle WE, Landen CN, Buchsbaum DJ, Conner MG, Arend RC, Yoon KJ, Klug CA, et al. The tumor-associated glycosyltransferase ST6Gal-I regulates stem cell transcription factors and confers a Cancer stem cell phenotype. Cancer Res. 2016;76(13):3978–3988. doi: 10.1158/0008-5472.CAN-15-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li D, Su D, Xue L, Liu Y, Pang W. Establishment of pancreatic cancer stem cells by flow cytometry and their biological characteristics. Int J Clin Exp Pathol. 2015;8(9):11218–11223. [PMC free article] [PubMed] [Google Scholar]
- 29.Vaz AP, Ponnusamy MP, Rachagani S, Dey P, Ganti AK, Batra SK. Novel role of pancreatic differentiation 2 in facilitating self-renewal and drug resistance of pancreatic cancer stem cells. Br J Cancer. 2014;111(3):486–496. doi: 10.1038/bjc.2014.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lakshmanan I, Rachagani S, Hauke R, Krishn SR, Paknikar S, Seshacharyulu P, Karmakar S, Nimmakayala RK, Kaushik G, Johansson SL, et al. MUC5AC interactions with integrin beta4 enhances the migration of lung cancer cells through FAK signaling. Oncogene. 2016;35(31):4112–4121. doi: 10.1038/onc.2015.478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar S, Das S, Rachagani S, Kaur S, Joshi S, Johansson SL, Ponnusamy MP, Jain M, Batra SK. NCOA3-mediated upregulation of mucin expression via transcriptional and post-translational changes during the development of pancreatic cancer. Oncogene. 2015;34(37):4879–4889. doi: 10.1038/onc.2014.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rachagani S, Torres MP, Kumar S, Haridas D, Baine M, Macha MA, Kaur S, Ponnusamy MP, Dey P, Seshacharyulu P, et al. Mucin (Muc) expression during pancreatic cancer progression in spontaneous mouse model: potential implications for diagnosis and therapy. J Hematol Oncol. 2012;5:68. doi: 10.1186/1756-8722-5-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hirschhaeuser F, Menne H, Dittfeld C, West J, Mueller-Klieser W, Kunz-Schughart LA. Multicellular tumor spheroids: an underestimated tool is catching up again. J Biotechnol. 2010;148(1):3–15. doi: 10.1016/j.jbiotec.2010.01.012. [DOI] [PubMed] [Google Scholar]
- 34.Mallard BW, Tiralongo J. Cancer stem cell marker glycosylation: nature, function and significance. Glycoconj J. 2017;34(4):441–452. doi: 10.1007/s10719-017-9780-9. [DOI] [PubMed] [Google Scholar]
- 35.Pauli C, Munz M, Kieu C, Mack B, Breinl P, Wollenberg B, Lang S, Zeidler R, Gires O. Tumor-specific glycosylation of the carcinoma-associated epithelial cell adhesion molecule EpCAM in head and neck carcinomas. Cancer Lett. 2003;193(1):25–32. doi: 10.1016/S0304-3835(03)00003-X. [DOI] [PubMed] [Google Scholar]
- 36.Fitzgerald TL, McCubrey JA. Pancreatic cancer stem cells: association with cell surface markers, prognosis, resistance, metastasis and treatment. Adv Biol Regul. 2014;56:45–50. doi: 10.1016/j.jbior.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 37.Lei Y, Wang S, Ren B, Wang J, Chen J, Lu J, Zhan S, Fu Y, Huang L, Tan J. CHOP favors endoplasmic reticulum stress-induced apoptosis in hepatocellular carcinoma cells via inhibition of autophagy. PLoS One. 2017;12(8):e0183680. doi: 10.1371/journal.pone.0183680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schonthal AH. Endoplasmic reticulum stress: its role in disease and novel prospects for therapy. Scientifica (Cairo) 2012;2012:857516. doi: 10.6064/2012/857516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nami B, Donmez H, Kocak N. Tunicamycin-induced endoplasmic reticulum stress reduces in vitro subpopulation and invasion of CD44+/CD24- phenotype breast cancer stem cells. Exp Toxicol Pathol. 2016;68(7):419–426. doi: 10.1016/j.etp.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 40.Bennett EP, Mandel U, Clausen H, Gerken TA, Fritz TA, Tabak LA. Control of mucin-type O-glycosylation: a classification of the polypeptide GalNAc-transferase gene family. Glycobiology. 2012;22(6):736–756. doi: 10.1093/glycob/cwr182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.He M, Wu C, Xu J, Guo H, Yang H, Zhang X, Sun J, Yu D, Zhou L, Peng T, et al. A genome wide association study of genetic loci that influence tumour biomarkers cancer antigen 19-9, carcinoembryonic antigen and alpha fetoprotein and their associations with cancer risk. Gut. 2014;63(1):143–151. doi: 10.1136/gutjnl-2012-303434. [DOI] [PubMed] [Google Scholar]
- 42.Yeh JC, Hiraoka N, Petryniak B, Nakayama J, Ellies LG, Rabuka D, Hindsgaul O, Marth JD, Lowe JB, Fukuda M. Novel sulfated lymphocyte homing receptors and their control by a Core1 extension beta 1,3-N-acetylglucosaminyltransferase. Cell. 2001;105(7):957–969. doi: 10.1016/S0092-8674(01)00394-4. [DOI] [PubMed] [Google Scholar]
- 43.Shiraishi N, Natsume A, Togayachi A, Endo T, Akashima T, Yamada Y, Imai N, Nakagawa S, Koizumi S, Sekine S, et al. Identification and characterization of three novel beta 1,3-N-acetylglucosaminyltransferases structurally related to the beta 1,3-galactosyltransferase family. J Biol Chem. 2001;276(5):3498–3507. doi: 10.1074/jbc.M004800200. [DOI] [PubMed] [Google Scholar]
- 44.Taniuchi K, Cerny RL, Tanouchi A, Kohno K, Kotani N, Honke K, Saibara T, Hollingsworth MA. Overexpression of GalNAc-transferase GalNAc-T3 promotes pancreatic cancer cell growth. Oncogene. 2011;30(49):4843–4854. doi: 10.1038/onc.2011.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang ZQ, Bachvarova M, Morin C, Plante M, Gregoire J, Renaud MC, Sebastianelli A, Bachvarov D. Role of the polypeptide N-acetylgalactosaminyltransferase 3 in ovarian cancer progression: possible implications in abnormal mucin O-glycosylation. Oncotarget. 2014;5(2):544–560. doi: 10.18632/oncotarget.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chugh S, Meza J, Sheinin YM, Ponnusamy MP, Batra SK. Loss of N-acetylgalactosaminyltransferase 3 in poorly differentiated pancreatic cancer: augmented aggressiveness and aberrant ErbB family glycosylation. Br J Cancer. 2016;114(12):1376–1386. doi: 10.1038/bjc.2016.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ponnusamy MP, Deb S, Dey P, Chakraborty S, Rachagani S, Senapati S, Batra SK. RNA polymerase II associated factor 1/PD2 maintains self-renewal by its interaction with Oct3/4 in mouse embryonic stem cells. Stem Cells. 2009;27(12):3001–3011. doi: 10.1002/stem.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dey P, Ponnusamy MP, Deb S, Batra SK. Human RNA polymerase II-association factor 1 (hPaf1/PD2) regulates histone methylation and chromatin remodeling in pancreatic cancer. PLoS One. 2011;6(10):e26926. doi: 10.1371/journal.pone.0026926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dey P, Rachagani S, Vaz AP, Ponnusamy MP, Batra SK. PD2/Paf1 depletion in pancreatic acinar cells promotes acinar-to-ductal metaplasia. Oncotarget. 2014;5(12):4480–4491. doi: 10.18632/oncotarget.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vaz AP, Deb S, Rachagani S, Dey P, Muniyan S, Lakshmanan I, Karmakar S, Smith L, Johansson S, Lele S, et al. Overexpression of PD2 leads to increased tumorigenicity and metastasis in pancreatic ductal adenocarcinoma. Oncotarget. 2016;7(3):3317–3331. doi: 10.18632/oncotarget.6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Karmakar S, Seshacharyulu P, Lakshmanan I, Vaz AP, Chugh S, Sheinin YM, Mahapatra S, Batra SK, Ponnusamy MP. hPaf1/PD2 interacts with OCT3/4 to promote self-renewal of ovarian cancer stem cells. Oncotarget. 2017;8(9):14806–14820. doi: 10.18632/oncotarget.14775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang W, Hou T, Niu C, Song L, Zhang Y. B3GNT3 expression is a novel marker correlated with pelvic lymph node metastasis and poor clinical outcome in early-stage cervical Cancer. PLoS One. 2015;10(12):e0144360. doi: 10.1371/journal.pone.0144360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cerhan JR, Ansell SM, Fredericksen ZS, Kay NE, Liebow M, Call TG, Dogan A, Cunningham JM, Wang AH, Liu-Mares W, et al. Genetic variation in 1253 immune and inflammation genes and risk of non-Hodgkin lymphoma. Blood. 2007;110(13):4455–4463. doi: 10.1182/blood-2007-05-088682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ho WL, Che MI, Chou CH, Chang HH, Jeng YM, Hsu WM, Lin KH, Huang MC. B3GNT3 expression suppresses cell migration and invasion and predicts favorable outcomes in neuroblastoma. Cancer Sci. 2013;104(12):1600–1608. doi: 10.1111/cas.12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li C, Du Y, Yang Z, He L, Wang Y, Hao L, Ding M, Yan R, Wang J, Fan Z. GALNT1-mediated glycosylation and activation of sonic hedgehog signaling maintains the self-renewal and tumor-initiating capacity of bladder Cancer stem cells. Cancer Res. 2016;76(5):1273–1283. doi: 10.1158/0008-5472.CAN-15-2309. [DOI] [PubMed] [Google Scholar]
- 56.Guo H, Nagy T, Pierce M. Post-translational glycoprotein modifications regulate colon cancer stem cells and colon adenoma progression in Apc(min/+) mice through altered Wnt receptor signaling. J Biol Chem. 2014;289(45):31534–31549. doi: 10.1074/jbc.M114.602680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Che MI, Huang J, Hung JS, Lin YC, Huang MJ, Lai HS, Hsu WM, Liang JT, Huang MC. beta1, 4-N-acetylgalactosaminyltransferase III modulates cancer stemness through EGFR signaling pathway in colon cancer cells. Oncotarget. 2014;5(11):3673–3684. doi: 10.18632/oncotarget.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Matzke-Ogi A, Jannasch K, Shatirishvili M, Fuchs B, Chiblak S, Morton J, Tawk B, Lindner T, Sansom O, Alves F, et al. Inhibition of tumor growth and metastasis in pancreatic Cancer models by interference with CD44v6 signaling. Gastroenterology. 2016;150(2):513–525. doi: 10.1053/j.gastro.2015.10.020. [DOI] [PubMed] [Google Scholar]
- 59.Eom BW, Joo J, Park B, Jo MJ, Choi SH, Cho SJ, Ryu KW, Kim YW, Kook MC. Nomogram Incorporating CD44v6 and Clinicopathological Factors to Predict Lymph Node Metastasis for Early Gastric Cancer. PLoS One. 2016;11(8):e0159424. doi: 10.1371/journal.pone.0159424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hu B, Luo W, Hu RT, Zhou Y, Qin SY, Jiang HX. Meta-analysis of prognostic and clinical significance of CD44v6 in esophageal Cancer. Medicine (Baltimore) 2015;94(31):e1238. doi: 10.1097/MD.0000000000001238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Todaro M, Gaggianesi M, Catalano V, Benfante A, Iovino F, Biffoni M, Apuzzo T, Sperduti I, Volpe S, Cocorullo G, et al. CD44v6 is a marker of constitutive and reprogrammed cancer stem cells driving colon cancer metastasis. Cell Stem Cell. 2014;14(3):342–356. doi: 10.1016/j.stem.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 62.Ni J, Cozzi PJ, Hao JL, Beretov J, Chang L, Duan W, Shigdar S, Delprado WJ, Graham PH, Bucci J, et al. CD44 variant 6 is associated with prostate cancer metastasis and chemo−/radioresistance. Prostate. 2014;74(6):602–617. doi: 10.1002/pros.22775. [DOI] [PubMed] [Google Scholar]
- 63.Brazil JC, Liu R, Sumagin R, Kolegraff KN, Nusrat A, Cummings RD, Parkos CA, Louis NA. alpha3/4 Fucosyltransferase 3-dependent synthesis of Sialyl Lewis A on CD44 variant containing exon 6 mediates polymorphonuclear leukocyte detachment from intestinal epithelium during transepithelial migration. J Immunol. 2013;191(9):4804–4817. doi: 10.4049/jimmunol.1301307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hallouin F, Goupille C, Bureau V, Meflah K, Le Pendu J. Increased tumorigenicity of rat colon carcinoma cells after alpha1,2-fucosyltransferase FTA anti-sense cDNA transfection. Int J Cancer. 1999;80(4):606–611. doi: 10.1002/(SICI)1097-0215(19990209)80:4<606::AID-IJC20>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 65.Goupille C, Hallouin F, Meflah K, Le Pendu J. Increase of rat colon carcinoma cells tumorigenicity by alpha(1-2) fucosyltransferase gene transfection. Glycobiology. 1997;7(2):221–229. doi: 10.1093/glycob/7.2.221. [DOI] [PubMed] [Google Scholar]
- 66.Singh R, Campbell BJ, Yu LG, Fernig DG, Milton JD, Goodlad RA, FitzGerald AJ, Rhodes JM. Cell surface-expressed Thomsen-Friedenreich antigen in colon cancer is predominantly carried on high molecular weight splice variants of CD44. Glycobiology. 2001;11(7):587–592. doi: 10.1093/glycob/11.7.587. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Antibodies used for the study. Table S2. Primers used for the study. A. Cancer stem cell markers and B. Glycogenes. Table S3. Fold change values of differentially expressed genes in SP when compared to NSP of SW1990. (DOCX 21 kb)
Figure S1. TM and BAG inhibits growth and alters glycosylation of PC cells. (a) and (b), Effect of TM on growth of SW1990 and Capan1 cells. (c) and (d), Effect of BAG on growth of SW1990 and Capan1 cells. (e) Effect of TM on N-linked glycosylation of PC cells. (f) Effect of BAG treatment on O-linked glycosylation of PC cells. (TIF 2768 kb)
Figure S2. Isolation and Characterization of PCSCs. Isolation and Characterization of PCSCs. (a) Sorting of SP and NSP cells from SW1990 and Capan 1 by Hoechst staining. Reserpine/Verapamil used as control to gate the SP cells. (b) Morphology of NSP and SP cells of SW1990 and Capan1. (c) and (d), RT-qPCR analysis of CSC markers expression between NSP and SP cells of SW1990 and Capan1, respectively. β-Actin is used to normalize the fold change values. (TIF 4154 kb)
Figure S3. RT-qPCR analysis of DEGs in PCSCs. (a) and (b), RT-qPCR analysis for validation of DEGs identified by PCR array between NSP and SP cells of SW1990 and Capan1. (TIF 2283 kb)
Figure S4. GALNT3 and B3GNT3 expresses at PDAC stage in PCSCs of KC. (a) and (b), IF analysis for co-expression of GALNT3 and B3GNT3 with CD44v6, respectively, at different stages of KC. (TIF 8121 kb)
Figure S5. KO of GALNT3 in Capan 1 SP cells. IF analysis for expression of GALNT3 in control and GALNT3 KO clones of Capan 1 SP. (TIF 2895 kb)