Abstract
With a unique crRNA processing capability, the CRISPR associated Cpf1 protein holds great potential for multiplex gene regulation. Unlike the well-studied Cas9 protein, however, conversion of Cpf1 to a transcription regulator and its related properties have not been systematically explored yet. In this study, we investigated the mutation schemes and crRNA requirements for the DNase deactivated Cpf1 (dCpf1). By shortening the direct repeat sequence, we obtained genetically stable crRNA co-transcripts and improved gene repression with multiplex targeting. A screen of diversity-enriched PAM library was designed to investigate the PAM-dependency of gene regulation by dCpf1 from Francisella novicida and Lachnospiraceae bacterium. We found novel PAM patterns that elicited strong or medium gene repressions. Using a computational algorithm, we predicted regulatory outputs for all possible PAM sequences, which spanned a large dynamic range that could be leveraged for regulatory purposes. These newly identified features will facilitate the efficient design of CRISPR-dCpf1 based systems for tunable multiplex gene regulation.
1. Introduction
Ever since the discovery of the Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) mechanism, its DNA-targeting strategy has been extensively characterized and masterfully adapted to a biotechnological tool for sequence-specific DNA manipulation that has rapidly revolutionized the fields of genome editing and engineering [[1], [2], [3], [4]]. This simple yet elegant system consists of the Cas9 endonuclease from Streptococcus pyogenes and a guide RNA (gRNA) that directs Cas9 to the complementary DNA target in the presence of a protospacer adjacent motif (PAM) [2,4]. The programmability, achieved through the guide sequence, has been further leveraged in variants of the system utilizing the engineered nuclease-deactivated Cas9 (dCas9) on its own or linked to diverse effector protein domains [5]. These dCas9-based CRISPR toolkits have proven extremely powerful for the systematic perturbation of single genes in regulatory and metabolic networks, advancing our knowledge in synthetic and systems biology at an unprecedented speed [6,7].
To push forward the CRISPR technology to the systems level, the ability to simultaneously manipulate multiple genes is highly demanded. Multiplex gene targeting, ideally through co-expressing multiple gRNAs in the same cell, enables the interrogation of much more complex interactions in genome-scale networks [8,9], as well as the combinatorial optimization of large heterologous pathways for metabolic engineering [5,[8], [9], [10], [11], [12]]. However, expressing gRNAs from independent plasmids suffers from a scalability issue, while encoding multiple gRNAs on the same expression cassette requires subsequent co-transcript processing, which relies on either endogenous RNase III activity, or in many systems, the introduction of sequence specific RNA endonuclease such as Csy4, the self-cleaving ribozyme sequences, or tRNA sequences that invoke the tRNA processing machinery [13,14]. For the purpose of application, these solutions either impose some level of cytotoxicity, or require lengthy additions to the gRNA sequence, causing greater genetic instability on a repeat-laden structure. This conundrum may now be solved thanks to the discovery of Cpf1, a Class II CRISPR endonuclease of Type V-A, which displays endoribonuclease activity and was shown to process CRISPR RNA (crRNA) co-transcripts into independent mature crRNAs, in addition to its DNA cleavage activity [[15], [16], [17]]. Besides functional duality, the Cpf1 system displays some enticing features – a concise crRNA, ∼40nt in its natural form, is more compatible with current DNA oligomer synthesis techniques and more resistant to homologous recombination-derived cassette disruption in a co-transcript context, and a thymine-rich PAM preference extends the targetable regions especially in AT-rich genomes. We thus believe in the great potential of a DNase deactivated Cpf1 (dCpf1) as an efficient tool for multiplex gene regulation.
Although aspects of the CRISPR-Cpf1 system as DNA endonuclease has been characterized, there have been only first attempts in using CRISPR-dCpf1 as transcriptional regulators. These studies proved its applicability in bacterial, plant, and mammalian cells [[18], [19], [20], [21]]. To harness and streamline the system for multiplex gene regulation, three specific aspects need addressing or systematic characterization: 1. a mutational scheme that abolishes Cpf1's DNase activity and yet minimally affects its DNA binding and RNase activities; 2. the requirements for pre-crRNA that contains multiple direct repeat-guide sequence units for efficient crRNA processing and DNA targeting [15,22]; and 3. the dependence of DNA binding strength on the PAM sequence [[23], [24], [25], [26]].
In this study, we designed a negative reporter assay for transcriptional repression by the CRISPR-dCpf1 system in Escherichia coli. The reporter assay was used to systematically quantify the functional effects of dCpf1 mutations and crRNA variants. We evaluated the dependence of gene repression on crRNA processing, lengths of direct repeats and guide sequences, as well as the number of target sequences tandemly located within the target gene. Most importantly, we investigated the PAM sequence preference for dCpf1 from Francisella novicida and Lachnospiraceae bacterium in a randomized 6nt PAM library. We found a broad range of repression strengths that did not conform to the previously identified PAM preferences. Therefore, we built an interpolation algorithm to predict gene repression activity for any PAM sequence based on a much limited number of sampled weak and strong PAMs. Without assuming context independency, the algorithm generated reliable estimates of PAM strengths, which could in principle lends great controllability to the CRISPR-dCpf1 system in synthetic biological applications.
2. Materials and methods
2.1. Strains and media
The E. coli DH5α was used as the host strain for all experiments. Luria-Bertani (LB) media (10 g/L tryptone, 5 g/L yeast extract, 10 g/L NaCl) was used as the growth media. Cells for flow cytometric fluorescence analysis were cultured in M9 media (12.8 g/L Na2HPO4·7H2O, 3 g/L KH2PO4, 0.5 g/L NaCl, 1.67 g/L NH4Cl, 1mM thiamine hydrochloride, 0.4% glucose, 0.2% casamino acids, 2mM MgSO4, 0.1mM CaCl2). Ampicillin, Kanamycin and Chloramphenicol concentrations for all experiments were 100 μg/ml, 50 μg/ml and 20 μg/ml, respectively.
2.2. Plasmid construction
The FnCpf1 gene were synthesized by Genscript Inc. Then it was mutated into dFnCpf1 and inserted into a vector containing a pTac-inducible promoter, an ampicillin-selectable marker, and a p15A replication origin. The crRNA plasmid backbone contained a synthetic constitutive promoter (J23119), a chloramphenicol-selectable marker, and a ColE1 replication origin. Various guide sequences were inserted by the Golden Gate method. The reporter plasmid contained sf-gfp as the reporter gene under the control of a synthetic constitutive promoter (J23100), a KanR-selectable marker, and a pSC101 replication origin. The crRNA sequences used in this study was summarized in Tables S3–S5.
2.3. Flow cytometry and analysis
Overnight culture of E. coli DH5α containing test plasmids was diluted 196 times into M9 medium with corresponding antibiotics, followed by shaking at 37 °C for 3 h. Cells were then serially diluted 1000 times into M9 medium with antibiotics and appropriate concentrations of IPTG cultured at 37 °C. The levels of fluorescence protein were analyzed by BD™ LSR II flow cytometer (Becton Dickinson, San Jose, CA, USA) with appropriate voltage settings (FSC:440, SSC:260, FITC:480) after further dilution into PBS with 20 mg/ml Kanamycin. Each sample was collected at least 50,000 events. The mean fluorescence of each sample was calculated with Flowjo software (Treestar, Inc., San Carlos, CA, USA) and analyzed with GraphPad Prism software (GraphPad Software, La Jolla, CA, USA).
2.4. PAM screen and analysis
Randomized PAM library was constructed by reverse PCR and Gibson ligation, using Random_F/Random_R consisting of six randomized nucleotides as primers and plasmid R_PAM as the backbone (Fig. S3). The PAM plasmid library was then transformed into competent E. coli DH5α harboring dFnCpf1 and crRNA plasmids. After transformation, cells were plated on LB agar supplemented with antibiotics of ampicillin, chloramphenicol and Kanamycin. After ∼16 h of growth, >107 cells were collected and pooled, diluted into fresh LB medium with antibiotics, and cultured overnight (∼16 h). The overnight culture was diluted ∼500 times into M9 medium with required antibiotics and appropriate concentrations of IPTG, followed by shaking at 37 °C for 3 h. Cultures were then diluted into PBS buffer to sort the cells with lowered fluorescence on a BD Influx Cell Sorter (Becton Dickinson, San Jose, CA, USA). From the sorted cells, random samples were collected, diluted and coated, and the remaining cells were cultured for the next round of sorting. After three rounds of sorting, colonies on the coated plates from all rounds were picked and subject to fluorescence measurements by flow cytometry and Sanger sequencing for their respective PAM sequences (Fig. S4).
2.5. PAM strength prediction and algorithm evaluation
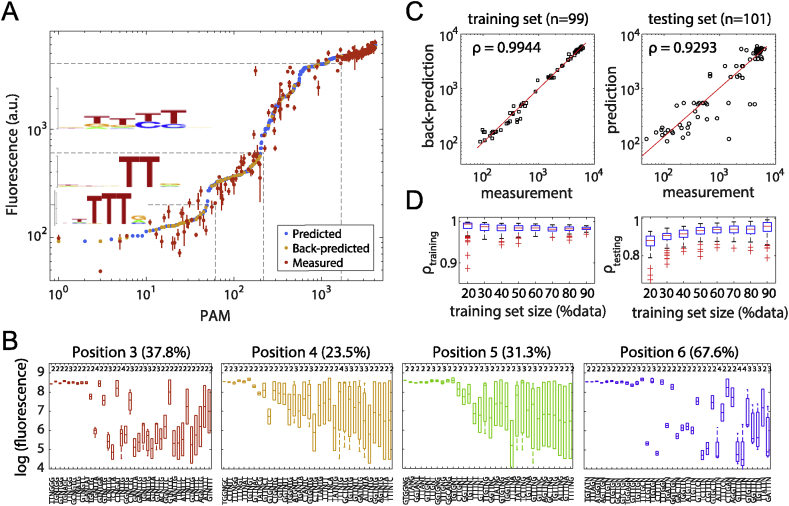
The computation algorithm used to predict PAM strength was explained in detail in Supplementary Information. The code was written in Matlab®. Cross-validation was done by randomly selecting samples from measured mean values to generate training sets. Testing was done on measured mean values for unselected PAMs (testing sets). For original selection, samples were selected randomly from the original data set. For uniform selection, samples were selected with equal numbers from equally placed bins in the entire fluorescence range of the original data set. At each training-testing set splitting ratio, 100 independent runs were conducted. Sequence logos in Fig. 6A and Fig. S7A were generated on http://weblogo.berkeley.edu/logo.cgi.
Fig. 6.
Predictions of PAM strengths for dFnCpf1. (A) Predictions of repression strengths for 4096 6-nt PAMs. PAMs are sorted by predicted values. Back predictions were made for measured PAMs in the sample pool (n = 200) from values predicted for unmeasured PAMs (n = 3896). For measured values (red dots), error bars show the standard error of mean for two to four independent measurements on different days. Sequence logos were obtained from measured PAMs with fluorescence strengths in ranges (0,200), (200, 600) and (600, 4000). (B) Site degeneracy for PAM positions 3–6. Box plots for measured PAM strengths (log fluorescence values, y-axis) of the sequence context specified on the x-axis. Percentages in parentheses indicate fractions of contexts that are degenerate by a 2-fold threshold. Numbers on top indicate the number of measured words within each sequence context. (C) Correlation between predictions and measured values in a sample run of cross-validation tests at 50% training set-testing set split. ρ: Pearson correlation between log values. (D) Summary of cross-validation tests. Distribution of correlation between predictions and measurements for the training (left) and testing (right) sets. In each run, data for training were randomly selected from and thus have the same distribution as the sample pool. 100 runs were conducted for each training set-testing set splitting ratio.
3. Results
3.1. Single mutation dCpf1 elicits stronger gene repression than double mutation dCpf1
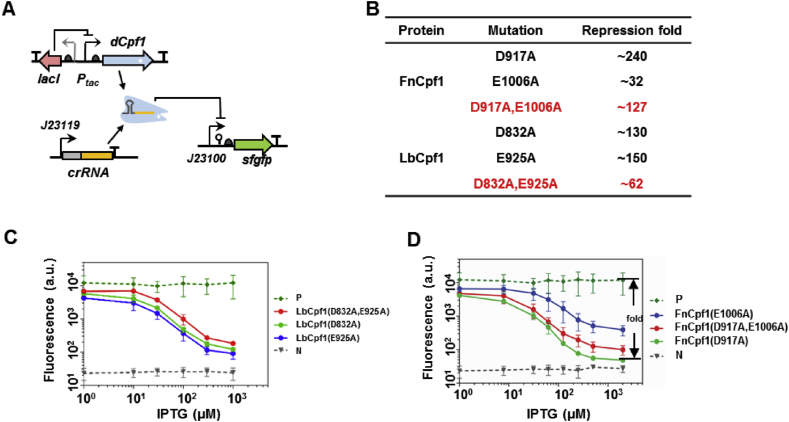
A previous study identified key amino acids in the RuvC-like domain of Cpf1 and proposed a double mutation scheme (D917A and E1006A) for deactivating the DNase activity of Cpf1 from F. novicida (FnCpf1), in much the same way as the design of dCas9 [25]. However, unlike Cas9, single mutations of either amino acid in Cpf1 was able to abolish cleavage of both DNA strands [[18], [19], [20], [21]]. As Cpf1 has a more complex domain structure than Cas9 [15,16], we suspected that double mutations may interfere with the RNA processing and DNA binding abilities of Cpf1 and thereby affect its regulatory activity. Therefore, we constructed single mutation forms of Cpf1 from F. novicida (dFnCpf1) and L. bacterium (dLbCpf1), and tested their gene repression activities against the double mutation forms. The repression activity was tested by a negative reporter assay where a constitutively expressed sf-gfp gene was targeted in its promoter region by a crRNA. Upon induction of the dCpf1 variants by IPTG, reduction in fluorescence was measured as a proxy for the binding strength of the dCpf1-crRNA duplex to the DNA target (Fig. 1A). Fig. 1C and D show the repression activity as a function of inducer concentration for dFnCpf1 and dLbCpf1, respectively. High levels of dCpf1 led to drastic reductions in gfp expression; but at all concentrations, at least one of the single mutation dCpf1s out-performed the double mutation variants. At the saturating induction level, both single mutation dLbCpf1s (D832A and E925A) showed slightly but significantly higher (>2-fold) repression activity than the double mutation dLbCpf1 (D832A+E925A). For dFnCpf1, the single mutation variant D917A elicited >200-fold gene repression, followed by the double mutation variant D917A+E1006A causing strong repression as well, whereas repression by the single mutation variant E1006A was moderate, suggesting E1006A might have destabilized DNA binding but this effect was apparently compensated by the D917A mutation in the double mutation dFnCpf1 (Fig. 1B–D). Antibiotic resistance borne on the sf-gfp plasmid was not compromised in clones carrying the single mutation dCpf1s, suggesting the enhanced repression activity was not a result of the disruption of sf-gfp gene sequence by residual DNase activities (data not shown). These data revealed a conserved D at position 917/832 responsible for the nuclease activity and its minimal interference with DNA binding ability. Thus, we adopted the single mutation dCpf1s (i.e. D917A for dFnCpf1 and D832A for dLbCpf1) in the following experiments.
Fig. 1.
Mutation variants of dCpf1 induced differential gene repression. (A) Schematic representation of the cellular circuit for evaluating performance of the dCpf1-crRNA system. In the circuit, dCpf1 and crRNA were expressed from an inducible promoter (Ptac) and a constitutive promoter (J23119), respectively, and a reporter gene (super-folded gfp, sf-gfp) is repressed by the dCpf1-crRNA complex at promoter and transcribed regions. (B) Summary of repression abilities of different dFnCpf1 and dLbCpf1 variants. Repression fold is calculated as the ratio between fluorescence of the positive control and the test systems at 103μM IPTG inducer concentration in (C) and (D). (C) Repression curves of three dLbCpf1 variants. The positive control (“P”) was of the strain with an empty crRNA plasmid, while the negative control (“N”) shows the background fluorescence of a strain with an empty gfp plasmid. (D) Repression curve of three dFnCpf1 variants. The positive and negative controls are the same as in (C). Error bars represent standard deviation of fluorescence for three independent experiments on different days. For crRNA sequences see Table S3.
3.2. Minimal crRNA length requirements for dCpf1's regulatory activity
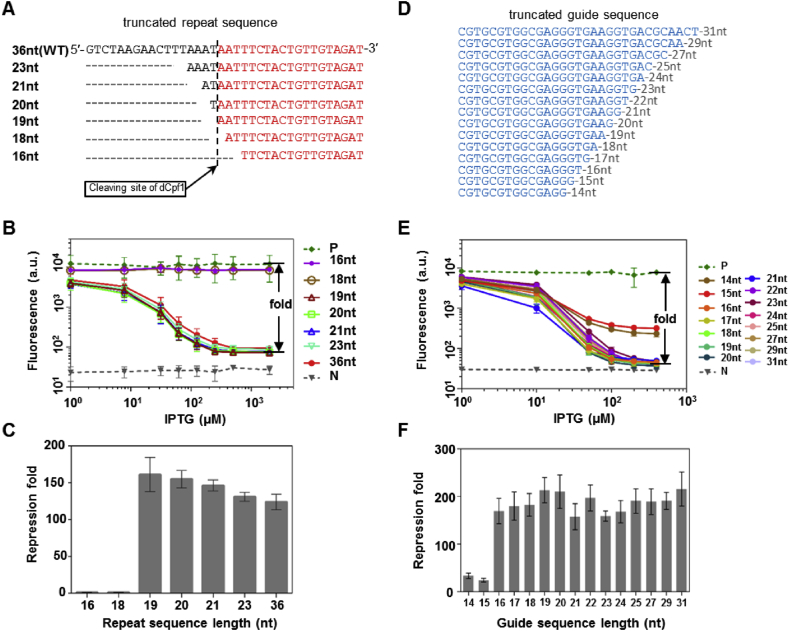
A unique function of Cpf1 is crRNA processing, where pre-crRNA containing multiple units of a 36nt direct repeat (DR) followed by gRNA is cleaved and truncated to mature crRNAs of a 19nt DR-gRNA structure [16]. In several Class I CRISPR systems, sequence- and structural-specific pre-crRNA processing by Cas6-family of endoribonucleases is a prerequisite for the subsequent assembling of a functional Cas complex on crRNA [27]. To find out if crRNA processing is essential for the gene regulatory function of dCpf1, we expressed crRNAs of various DR lengths ranging from 16nt to 36nt in the reporter system (Fig. 2A). All crRNAs with DR length >19nt showed the same repression activity as the crRNA with DR length of exactly 19nt (Fig. 2B&C). Since the latter did not undergo processing, we concluded that dCpf1 can load onto mature crRNA in the absence of extra processing signals, and thus its regulatory activity is independent of its crRNA processing activity. A previous in vitro experiment showed for Cpf1, crRNA with DR lengths of 16–18nt were still able to induce target DNA cleavage [16]. We found, however, no regulatory activity of dCpf1 with crRNAs having shorter than 19nt DRs (Fig. 2B&C). .
Fig. 2.
The effect of repeat and guide sequence lengths on gene repression by dFnCpf1-crRNA. (A) Aligned repeat sequences of different lengths used in crRNAs. The dashed line indicates the cleavage site on crRNA during crRNA processing by dFnCpf1. Red colored sequences remain in the mature crRNA, while the rest of the sequences are cleaved off. (B) Gene repression curves of dFnCpf1 with truncated repeat sequences. (C) Maximal repression folds of dFnCpf1 with the same set of truncated repeat sequence as in (B). (D) Aligned guide sequences of different lengths used in crRNAs. (E) The repression curves for different lengths of guide sequences in the dFnCpf1-crRNA system. (F) Maximal repression folds of dFnCpf1 with the same set of truncated guide sequences as in (E). Error bars represent standard deviation of fluorescence for three independent experiments on different days. Positive and negative controls are the same as in Fig. 1.
Another functional element in crRNA is the guide sequence whose length is believed to be a crucial parameter for the DNA cleaving efficiency of the Cpf1 nuclease. Cpf1 generates mature crRNAs with guide sequence of typically 24nt long. We examined how the extension and truncation of the guide sequence affect the regulatory efficiency of dCpf1 by constructing a number of guide sequences with lengths from 14nt to 31nt (Fig. 2D). The results showed a guide sequence was able to elicit 200-fold gene repression. Repression was drastically weakened with further guide sequence truncation (Fig. 2E&F). For Cpf1, previous study suggested a threshold guide sequence length of 18nt below which DNA targeting and cleavage was not observed [16]. These results together suggested a 16-18nt minimal guide sequence length required for DNA targeting, depending possibly on the specific guide sequence used.
3.3. Enhanced gene repression through multiplex targeting of dCpf1
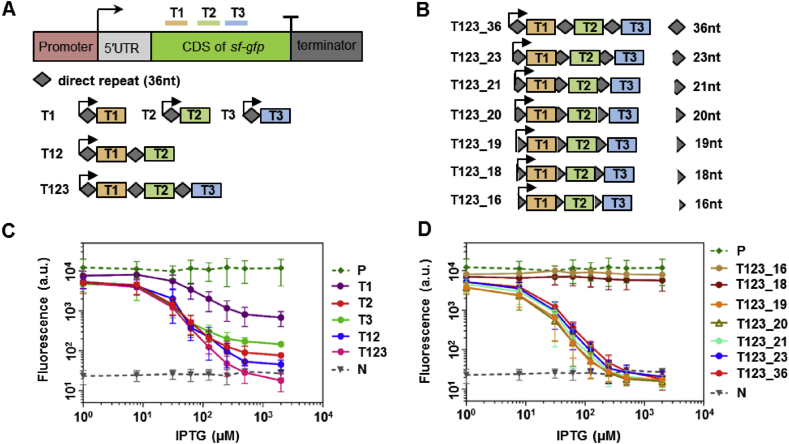
As the targeting of multiple genes has been demonstrated in several recent studies [18,19,21], and a single bound dCpf1, without dedicated inactivation domains, was not sufficient in suppressing gene expression in human HEK293T cells [21], we studied gene repression by tandemly positioned dCpf1 roadblocks. 24nt guide sequences targeting three independent segments within the coding region of the sf-gfp gene were connected by the 36nt DR sequences and co-expressed under a constitutive promoter (Fig. 3A). We found that crRNAs targeting any one of the three segments resulted in varied but significant gene repression (10–100-fold). Repression was further augmented by doubly or triply combined crRNAs, presumably through a stronger blockage of transcription elongation (Fig. 3C). Strikingly, the triply combined crRNAs completely abolished gfp expression (>300-fold reduction). The fold reduction by multiplex targeting, relative to individual targeting, was between additive and multiplicative. These results suggested that co-transcribed crRNAs targeting multiple DNA segments can be utilized by dCpf1 to combinatorially augment gene repression.
Fig. 3.
Repression by dFnCpf1 with co-transcribed crRNAs. (A) Schematic representation of target sequences for each single crRNA, as well as the design of individual and combined multiple crRNAs. (B) Different lengths of repeat sequence in the triply-combined crRNA co-transcript. (C) Repression curves of dFnCpf1 with single or multiple crRNAs. (D) Repression curves of dFnCpf1 with varied repeat sequence lengths in the same triply-combined crRNA co-transcript. Error bars represent standard deviation of fluorescence for three independent experiments on different days. Positive and negative controls are the same as in Fig. 1. For crRNA sequences see Table S3.
Co-transcription of multiple crRNAs ensures uniform expression among all gRNAs, and reduces the genetic instability associated with repeated expression cassettes. Yet, in the crRNA coding region, a repeat structure conferred by the DR sequences could also lead to genetic instability through an increased chance of homologous recombination as the length of DR increases [28]. We further optimized the system by truncating the interspersed DR sequences, and identified the minimal DR length essential for multiple DNA targeting (Fig. 3B). In consistence with the condition for single crRNAs, we found a 19nt DR is required for dCpf1-mediated multiplex repression (Fig. 3D).
3.4. dFnCpf1's regulatory activity strongly depends on the PAM sequence
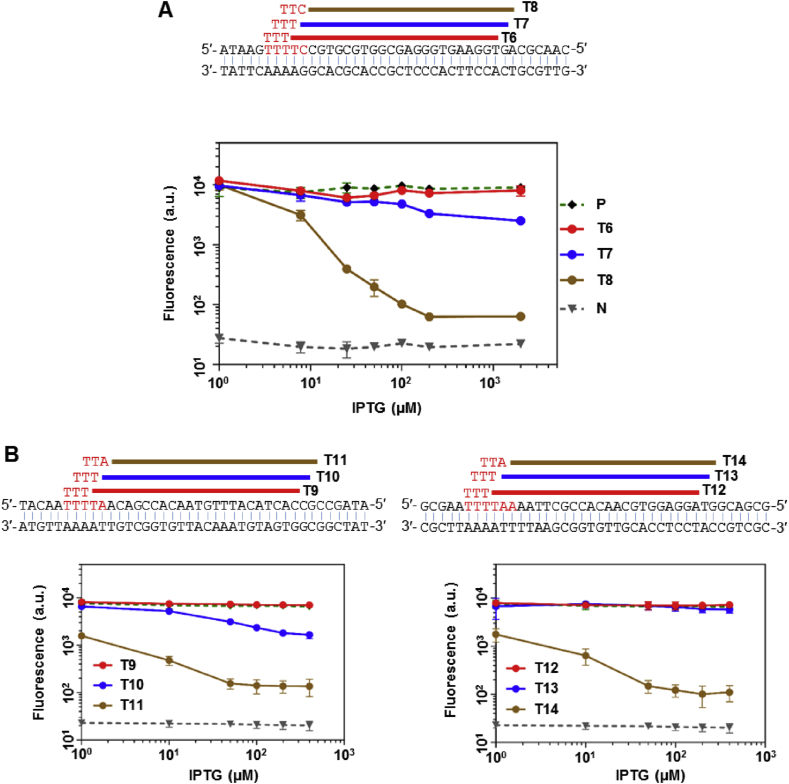
Previous studies have shown a strong dependence of CRISPR activity on the PAM sequence. For FnCpf1, CTN and TTN were identified as the preferred PAM sequences for DNA cleavage [16]. We selected two sets of targets on both the template and non-template strands of the sf-gfp gene based on these motifs, and tested the gene repression activity of dFnCpf1 (Fig. S1A). We observed that none of the non-template strand targets generated significant repression (Fig. S1B) – a strand bias also reported in other studies [21] – while the template strand targets showed a broad range of repression strengths (Fig. S1C). Unlike the case for dCas9 (Fig. S2A), for dCpf1, repression strengths were not correlated with the targets' locations within the coding region (Fig. S2B), suggesting factors other than transcript length significantly influenced dCpf1's regulatory activities. We further selected three sets of targets, each containing three targets starting from a T-rich region, but shifted by 1- or 2-nt relative to each other. Targets selected this way had similar distances from the transcription start site (TSS) and similar base/subword compositions, and all had TTN as the PAM sequence. However, within each set, repression activities were still drastically different (Fig. 4). These results were strongly indicative of TTN as an incomplete characterization of the PAM sequence preference for dFnCpf1, and we speculated that the bases adjacent to the core TTN (and perhaps CTN) motif may underlie the discrepancies in dFnCpf1's regulatory activity. For example, in Fig. 4A, the extended PAMs were GTTT, TTTT, and TTTC, respectively. While the TTTC PAM showed over 100-fold repression, the GTTT PAM was unable to repress gene expression at detectable levels.
Fig. 4.
Gene repression on sliding targets with the canonical TTN PAM motif for dFnCpf1. (A) Top panel: targets T6-T8 were selected within the gfp coding sequence by 1-nt or 2-nt shifting. Red letters show the corresponding PAM sequences. Lower panel: gene repression by dFnCpf1 targeting the respective sequences. (B) Another two sets of 1-nt or 2-nt shifted target sequences and the respective repression curves by dFnCpf1. Error bars represent the standard deviation of fluorescence for three independent experiments on different days. Positive and negative controls are the same as in Fig. 1. For crRNA sequences see Table S3.
3.5. Systematically investigating the effect of PAM sequence for dFnCpf1 and dLbCpf1
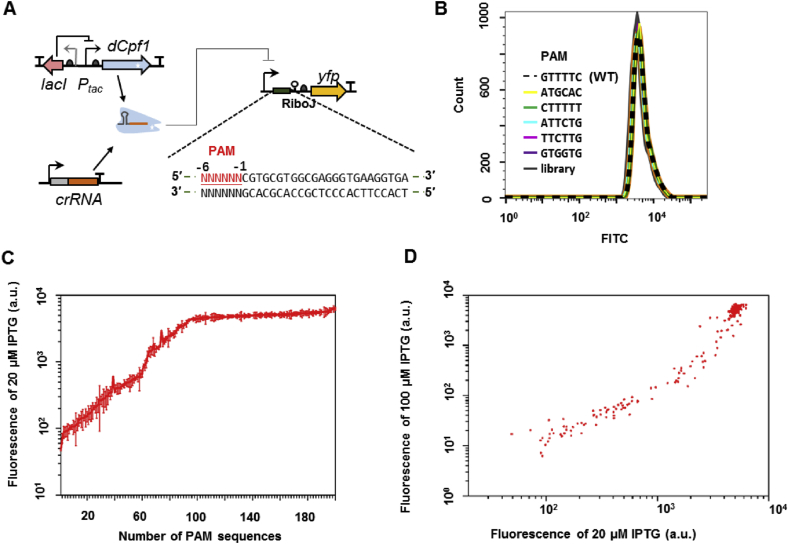
To reveal the full range of regulatory activities conveyed by PAM variation, we constructed a library of cells harboring the negative reporter system, with dCpf1 target sequence insertions varying in a randomized 6nt tract as the PAM sequence. The insertion was placed in the 5′-UTR region of the yfp gene and followed by a ribozyme-based insulator [29], such that difference in the PAM sequences would not interfere with basal transcription or translation efficiency in the absence of dCpf1 (Fig. 5A). Indeed, in Fig. 5B, under the non-induced condition, the flow cytometry measured fluorescence distributions of cells harboring the randomized PAM-library (grey line), of the construct with the previously proposed PAM (black dashed line) and of five constructs with mutated PAMs (colored lines) all collapsed onto one curve, indicating the effect of randomized PAM sequences had been successfully eliminated. The library was then subjected to three rounds of dCpf1 induction and fluorescence sorting, from which process, clones showing dramatically varied yfp expression levels were randomly picked and sequenced at the PAM locus (Fig. S4). Table S1 lists 200 and 133 non-redundant PAM sequences identified in the screens for dFnCpf1 and dLbCpf1, respectively (Fig. 5C & Fig. S5A). We further measured the fluorescence of these clones at different inducer concentrations (20μM, 50μM and 100μM IPTG, Fig. 5D and Fig. S5B). A power law scaling was observed between fluorescence at high and low inducer concentrations when PAM strength was weak or moderate, in consistence with a simple gene expression model depending on dCpf1 concentration. As PAM became strong, repression levels gradually saturated along both axes. Gene expression noise slightly increased with repression strength, but was confined within 60–80% (Fig. S6).
Fig. 5.
Negative reporter screen for the PAM dependence of dFnCpf1's regulatory activity. (A) Design of the screening circuit. A randomized 6-nt PAM sequence (red) was placed upstream of a fixed target sequence, and a ribozyme insulator (RiboJ) was inserted between the target and the reporter gene to eliminate the effect of PAM sequences on yfp translation. (B) Fluorescence distributions measured by flow cytometry for clones carrying the specified PAM sequences or cell populations carrying the randomized PAM library, under the non-induced condition. (C) Fluorescence measured under the induced condition, for n = 200 clones carrying different PAMs randomly selected from flow-cytometer sorted library cells. Error bars represent the standard deviation of two to four independent experiments on different days. (D) Fluorescence for the 200 sample clones under two different IPTG inducer concentrations (20 μM and 100 μM).
These results suggest that for the CRISPR-dCpf1 system, variations in the PAM sequences could produce a large dynamic range for gene expression regulation. In contrast to the irreversible DNA cleavage reaction for which any “good” PAM would suffice, gene regulation applications could take advantage of a more nuanced activity difference between PAMs to achieve controllable outputs. However, as our entire PAM library contained 4096 sequences, it was both impractical and uneconomical to screen and sequence all clones. Therefore, we designed an interpolation algorithm to predict PAM strengths using information gathered from a small sample pool, such as the 200 dFnCpf1 clones picked by fluorescence levels. The algorithm is based on the assumption of a semi-smooth regulatory strength landscape in the PAM sequence space, in other words, the regulatory strength of a PAM sequence of length k is computed as the average strengths of all PAMs that are different by one nucleotide at only one of the k locations, except at non-degenerate locations (see below). Strength information at location i (i = 1 … k) is weighted by the degeneracy of the location. A non-degenerate location is a location where variations in base identity have exhibited very different regulatory outputs in the sample set, and thus all information at this location is discarded for predictive purposes. Whenever possible, context dependency is considered in evaluating location degeneracy. A detailed explanation of the algorithm can be found in Supplementary Information. Unlike the conventional PWM model or sequence logo methods, the algorithm does not assume positional independence between bases, and therefore, it automatically captures all sequence patterns and features contained by the sample pool.
We predicted PAM strengths for all 6nt words based on data from 200 samples for dFnCpf1 and 133 samples for dLbCpf1 (Fig. 6A and Fig. S7A & Table S2). Conversely, we used the predicted values for unmeasured words to back-predict strengths of measured PAMs. This yielded a >0.99 correlation with measured values, indicating a minimal loss of information through the course of interpolation (data not shown). The results indicated that in general, for both dFnCpf1 and dLbCpf1, positions 1 and 2 did not had significant effects on PAM strengths. For dFnCpf1, PAM strength was most sensitive to the 4- and 5-th location, while position 3 contributed to PAM strength diversity more than position 6. For dLbCpf1, positions 3–6 all affected PAM strength strongly (Fig. 6B and Fig. S7B). When ranking samples based on repression activity, we found that for dFnCpf1, the strongest PAMs were (TT)TTTV and (T)TTV, whereas T was strongly disfavored at the last position. The other previously identified CTN motif generated only moderate repression activities (Fig. 6A). For dLbCpf1, the strongest repressions were elicited by (T) TTTV PAMs, followed by CTTV. Like dFnCpf1, there was a strong preference against T at the last position in strong and moderate PAMs. However, for dLbCpf1, TTTT was able to induce medium repression, with a 5′- T further enhancing its activity (Figs. S5A and S7A).
To evaluate the predictive power of our algorithm, cross-validation was done by splitting the sample pool for dFnCpf1 into training and testing sets, at proportions from 20% to 90% (for the training set). When using 50% randomly selected samples as the training set, the predictions for the testing set were >0.90 correlated with measured values, and back-prediction showed >0.95 correlations with data in the training sets (Fig. 6C). Even with only 20% (n = 40) of the sampled PAMs as the training data, a correlation >0.8 could be obtained with the testing set (Fig. 6D). These numbers decreased mildly for dLbCpf1, whose sample pool were smaller and less biased toward high and low repression ranges (Figs. S7C and D). When we applied a strictly uniform selection method in the low, medium, and high repression ranges, irrespective of the repression strength distribution of the original sample pools, correlation between predictions and the testing sets were around 0.75 (at 53% data as training set) for dFnCpf1 and 0.4 (at 14% data as training set) for dLbCpf1 (Fig. S8). These results underscored the importance of PAM sequences sampled at high and low repression ranges in generating sufficient information for the algorithm to successfully interpolate for any other PAM sequence. Successive shrinkage of the training set suggested a threshold sample size of n = 40 (∼1% of the sequence space), below which >50% of the prediction attempts ended up with un-predictable sequences that were not covered by available information (Fig. S9).
For the dFnCpf1 dataset, we also performed LASSO regression with a simple linear model assuming position independence (i.e. having 4 × 6 = 24 independent variables, see Supplementary Information). The best performing model came back with a (T)TT(V) preference which captures the PWM motif for dFnCpf1, but missed the fine features at positions 1–3 (Fig. S10B). The linear model had a back-prediction correlation of ∼0.86 (Fig. S10A). Cross-validation using randomly sampled training sets, as described above, generated models that predicted testing sets with 0.8–0.9 correlations (Fig. S10D). The inferiority compared to our algorithm was presumably the result of interdependency between positions which, upon close examination, had a greater impact on medium and low strength PAMs than on strong PAMs (Fig. S10A&C).
4. Discussion
In this article, we systematically investigated the key constraints and properties of the CRISPR-dCpf1 system as transcriptional repressors in E. coli cells. In comparison to the dCas9 based CRISPR systems, dCpf1 offers the unique potential of multiplex gene regulation with its ability to autonomously process crRNA co-transcripts and subsequently target multiple independent DNA sequences. This ability minimizes the uncertainty in crRNA relative dosages and genetic stabilities, as previously seen in systems with dCas9 and independently transcribed crRNAs. This is key to large scale standardized perturbation experiments such as whole transcription network engineering. There have recently been multiple reports on dCpf1's gene regulation applications in bacteria, plants, and human cells. Although repression in bacteria was attained, repression in Arabidopsis and activation in human HEK293T cells was quantitatively unstable and somewhat idiosyncratic [[19], [20], [21]]. A systematic characterization of the CRISPR-dCpf1 system with respect to its DNA binding properties is obviously in need to further enhance performance in these experimental systems.
We compared the repression activities of dCpf1 mutant forms including single and double mutations at the two previously identified catalytic residues for Cpf1's DNase activity. For both dFnCpf1 and dLbCpf1, double mutations compromised regulatory activities. Between the two single mutation variants, D917A/D832A generated consistently strong regulatory activity, whereas E1006A in dFnCpf1 was much less efficient in DNA binding than D917A. While it is possible that the single mutation variants exhibit residual DNase activities that went undetected in our growth rate measurements, based on the crystal structure of dCpf1 in complex with crRNA and DNA, we speculate that the subdued repression may reflect a genuine destabilization of dCpf1-crRNA-DNA complex, as E1006 in the RuvC-II domain of FnCpf1 is spatially close to the WED domain and the bridge helix that interact closely with the 5′ crRNA handle [30,31].
We found that for dCpf1, crRNA cleavage was not essential for subsequent DNA targeting. The wild type crRNAs adopt a 19nt DR-24nt gRNA form. In our studies, the minimal length requirements were 19nt for the direct repeat and 16nt for the guide sequence. A previous biochemical study revealed the importance of a 5′-AAU-3′ sequence at the −19 location of the processed crRNA [15]. This tri-base region may thus be crucial for RNA processing as well as stabilizing the dCpf1-crRNA complex. With shorter crRNAs, Cpf1 may still form transient complexes with DNA and produce strand breaks in vitro [16]. However, tight binding of dCpf1-crRNA to the DNA target demanded an intact 19nt direct repeat sequence according to our results.
We further demonstrated enhanced gene repression by co-transcribed crRNAs targeting DNA sequences located in tandem in the coding region. Again, a ≥19nt DR length is required in the crRNA co-transcript for crRNA processing, which sets a lower limit of 35–40nt repetitive crRNA structure for the precursor crRNA expression cassette.
We found the PAM sequence to be a major factor determining gene repression activity. The previously identified TTN and CTN motifs for FnCpf1 in DNA cleavage assays did not explained the PAM preference in terms of gene regulation by dFnCpf1. Although a T-rich PAM for Cpf1 greatly expands the genomic regions that could be targeted for cleavage, gene regulatory response was sensitively dependent on the exact PAM sequences used. On the same target sequence, a wide range of repression folds were observed when different 6nt preceding sequences were used. We designed a negative reporter screen to identify PAM sequences eliciting strong, medium and weak repressions. We further developed an interpolation algorithm based on context-dependent sequence similarities, using which, we predicted regulatory strengths for all 6nt sequences as PAMs based on measurements of 200 and 133 PAMs for dFnCpf1 and dLbCpf1, respectively. Compared to motif analysis by next-generation sequencing, the algorithm provides a fast and economic way of assessing PAM preferences, and is especially suited for revealing moderate and weak PAMs, which might be masked by biases introduced through DNA amplification. The algorithm also showed superiority over context independent linear models, revealing the significance of higher order PAM features in Cpf1-target recognition.
Our analyses suggested for both enzymes a general 4nt core sequence dependence, with T strongly disfavored in the last position, and slightly favored at the proceeding 2nt positions. Specifically, dFnCpf1 and dLbCpf1 both displayed a preference for TTTV PAMs; while for dLbCpf1, other PAMs also emerged as mediating strong regulatory responses. TTTV was previously identified for LbCpf1 [23], and recently identified in a study on genome editing by FnCpf1 in Baker's yeast [32] while we were preparing this manuscript. This suggests that differences in the strengths of extended PAMs may also be relevant when cleavage is concerned, especially for improving CRISPR DNases that did not function well in certain systems. In Ref. [32], the authors found that targets with TTTA and (CT) TTTC PAMs did not lead to genome editing, despite conforming to the TTTV motif. Our data suggest a range of 50–300 fluorescence for NNTTTA PAMs and a ∼110 fluorescence for CTTTTC. Although these are all strong repressions in the 6nt library, the six-fold difference might still significantly affect reaction outcome.
For Cas9-based CRISPR applications, the most common strategies for tuning activity include coding/non-coding strand targeting, target distance from the TSS, protein concentration, and gRNA-target sequence complementarity. While the first two do not apply to Cpf1-based systems, our results mapped out a quantitative relationship between the PAM sequence and dCpf1's regulatory activity. Modulation in cis, such as by the PAM sequence or by target complementarity, allows for the orthogonal regulation of multiple targets at a single dCpf1 induction level. This would grant much flexibility for quantitative assessment of complex transcription networks. Moreover, the screening method we developed could be utilized to introduce a control element in arbitrary genes. Compared to targets in the upstream promoter regions, insertions within the 5′UTR region followed by an insulator could minimize the interference on background gene expression levels. Compared to targets in the coding sequences, PAM sequences and target sequences in the inserted fragment can be designed separately to achieve desired repression outputs with high specificity. Besides dFnCpf1 and dLbCpf1, dCpf1s from Acidaminococcus sp. and Eubacterium eligens have also been tested in bacterial and eukaryotic cells [[18], [19], [20], [21]]. Our screening and prediction methods could serve a pipeline for rapid characterization of the natural diversity of dCpf1 proteins. When coupled with technologies already developed for dCas9 [33,34], dCpf1 may be transformed into powerful tools for sophisticated applications of multiplex gene interrogation.
Funding
National Natural Science Foundation of China (No. 31470818 and 31722002); the 973 projects of Ministry of Science and Technology of China (No. 2015CB910300); the Key Research Program of the Chinese Academy of Sciences (No. QYZDB-SSW-SMC050); and the Strategic Priority Research Program of the Chinese Academy of Sciences (No. XDB29040000).
Acknowledgements
We thank Lili Ji (Peking University), Tingting Li (Peking University) and Junying Jia (Institute of Biophysics, CAS) for technical assistance of flow cytometry.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.synbio.2018.11.002.
Contributor Information
Long Qian, Email: long.qian@pku.edu.cn.
Chunbo Lou, Email: louchunbo@im.ac.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Cho S.W., Kim S., Kim J.M., Kim J.S. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 2.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiang W., Zhao X., Gabrieli T., Lou C., Ebenstein Y., Zhu T.F. Cas9-Assisted Targeting of CHromosome segments CATCH enables one-step targeted cloning of large gene clusters. Nat Commun. 2015;6:8101. doi: 10.1038/ncomms9101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qi L.S., Larson M.H., Gilbert L.A., Doudna J.A., Weissman J.S., Arkin A.P., Lim W.A. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Didovyk A., Borek B., Hasty J., Tsimring L. Orthogonal modular gene repression in Escherichia coli using engineered CRISPR/Cas9. ACS Synth Biol. 2016;5:81–88. doi: 10.1021/acssynbio.5b00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nielsen A.A., Voigt C.A. Multi-input CRISPR/Cas genetic circuits that interface host regulatory networks. Mol Syst Biol. 2014;10:763. doi: 10.15252/msb.20145735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cress B.F., Toparlak O.D., Guleria S., Lebovich M., Stieglitz J.T., Englaender J.A., Jones J.A., Linhardt R.J., Koffas M.A. CRISPathBrick: modular combinatorial assembly of type II-a CRISPR arrays for dCas9-mediated multiplex transcriptional repression in E. coli. ACS Synth Biol. 2015;4:987–1000. doi: 10.1021/acssynbio.5b00012. [DOI] [PubMed] [Google Scholar]
- 9.Lv L., Ren Y.L., Chen J.C., Wu Q., Chen G.Q. Application of CRISPRi for prokaryotic metabolic engineering involving multiple genes, a case study: controllable P(3HB-co-4HB) biosynthesis. Metab Eng. 2015;29:160–168. doi: 10.1016/j.ymben.2015.03.013. [DOI] [PubMed] [Google Scholar]
- 10.Elhadi D., Lv L., Jiang X.R., Wu H., Chen G.Q. CRISPRi engineering E. coli for morphology diversification. Metab Eng. 2016;38:358–369. doi: 10.1016/j.ymben.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 11.Li S., Jendresen C.B., Grunberger A., Ronda C., Jensen S.I., Noack S., Nielsen A.T. Enhanced protein and biochemical production using CRISPRi-based growth switches. Metab Eng. 2016;38:274–284. doi: 10.1016/j.ymben.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 12.Zalatan J.G., Lee M.E., Almeida R., Gilbert L.A., Whitehead E.H., La Russa M., Tsai J.C., Weissman J.S., Dueber J.E., Qi L.S. Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell. 2015;160:339–350. doi: 10.1016/j.cell.2014.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nissim L., Perli S.D., Fridkin A., Perez-Pinera P., Lu T.K. Multiplexed and programmable regulation of gene networks with an integrated RNA and CRISPR/Cas toolkit in human cells. Mol Cell. 2014;54:698–710. doi: 10.1016/j.molcel.2014.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie K., Minkenberg B., Yang Y. Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc Natl Acad Sci U S A. 2015;112:3570–3575. doi: 10.1073/pnas.1420294112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fonfara I., Richter H., Bratovic M., Le Rhun A., Charpentier E. The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature. 2016;532:517–521. doi: 10.1038/nature17945. [DOI] [PubMed] [Google Scholar]
- 16.Zetsche B., Gootenberg J.S., Abudayyeh O.O., Slaymaker I.M., Makarova K.S., Essletzbichler P., Volz S.E., Joung J., van der Oost J., Regev A. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zetsche B., Heidenreich M., Mohanraju P., Fedorova I., Kneppers J., DeGennaro E.M., Winblad N., Choudhury S.R., Abudayyeh O.O., Gootenberg J.S. Multiplex gene editing by CRISPR-Cpf1 using a single crRNA array. Nat Biotechnol. 2017;35:31–34. doi: 10.1038/nbt.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim S.K., Kim H., Ahn W.C., Park K.H., Woo E.J., Lee D.H., Lee S.G. Efficient transcriptional gene repression by type V-a CRISPR-Cpf1 from Eubacterium eligens. ACS Synth Biol. 2017;6(7):1273–1282. doi: 10.1021/acssynbio.6b00368. [DOI] [PubMed] [Google Scholar]
- 19.Tak Y.E., Kleinstiver B.P., Nunez J.K., Hsu J.Y., Horng J.E., Gong J., Weissman J.S., Joung J.K. Inducible and multiplex gene regulation using CRISPR-Cpf1-based transcription factors. Nat Methods. 2017;14:1163–1166. doi: 10.1038/nmeth.4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang X., Lowder L.G., Zhang T., Malzahn A.A., Zheng X., Voytas D.F., Zhong Z., Chen Y., Ren Q., Li Q. A CRISPR-Cpf1 system for efficient genome editing and transcriptional repression in plants. Native Plants. 2017;3:17018. doi: 10.1038/nplants.2017.18. [DOI] [PubMed] [Google Scholar]
- 21.Zhang X., Wang J., Cheng Q., Zheng X., Zhao G., Wang J. Multiplex gene regulation by CRISPR-ddCpf1. Cell Discov. 2017;3:17018. doi: 10.1038/celldisc.2017.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamano T., Nishimasu H., Zetsche B., Hirano H., Slaymaker I.M., Li Y., Fedorova I., Nakane T., Makarova K.S., Koonin E.V. Crystal structure of Cpf1 in complex with guide RNA and target DNA. Cell. 2016;165:949–962. doi: 10.1016/j.cell.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim H.K., Song M., Lee J., Menon A.V., Jung S., Kang Y.M., Choi J.W., Woo E., Koh H.C., Nam J.W. In vivo high-throughput profiling of CRISPR-Cpf1 activity. Nat Methods. 2017;14:153–159. doi: 10.1038/nmeth.4104. [DOI] [PubMed] [Google Scholar]
- 24.Leenay R.T., Beisel C.L. Deciphering, Communicating, and engineering the CRISPR PAM. J Mol Biol. 2017;429:177–191. doi: 10.1016/j.jmb.2016.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leenay R.T., Maksimchuk K.R., Slotkowski R.A., Agrawal R.N., Gomaa A.A., Briner A.E., Barrangou R., Beisel C.L. Identifying and visualizing functional PAM diversity across CRISPR-Cas systems. Mol Cell. 2016;62:137–147. doi: 10.1016/j.molcel.2016.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watkins-Chow D.E., Varshney G.K., Garrett L.J., Chen Z., Jimenez E.A., Rivas C., Bishop K.S., Sood R., Harper U.L., Pavan W.J. Highly efficient Cpf1-mediated gene targeting in mice following high concentration pronuclear injection. G3 (Bethesda) 2017;7:719–722. doi: 10.1534/g3.116.038091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hochstrasser M.L., Doudna J.A. Cutting it close: CRISPR-associated endoribonuclease structure and function. Trends Biochem Sci. 2015;40:58–66. doi: 10.1016/j.tibs.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 28.Chen Y.J., Liu P., Nielsen A.A., Brophy J.A., Clancy K., Peterson T., Voigt C.A. Characterization of 582 natural and synthetic terminators and quantification of their design constraints. Nat Methods. 2013;10:659–664. doi: 10.1038/nmeth.2515. [DOI] [PubMed] [Google Scholar]
- 29.Lou C., Stanton B., Chen Y.J., Munsky B., Voigt C.A. Ribozyme-based insulator parts buffer synthetic circuits from genetic context. Nat Biotechnol. 2012;30:1137–1142. doi: 10.1038/nbt.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dong D., Ren K., Qiu X., Zheng J., Guo M., Guan X., Liu H., Li N., Zhang B., Yang D. The crystal structure of Cpf1 in complex with CRISPR RNA. Nature. 2016;532:522–526. doi: 10.1038/nature17944. [DOI] [PubMed] [Google Scholar]
- 31.Swarts D.C., van der Oost J., Jinek M. Structural basis for guide RNA processing and seed-dependent DNA targeting by CRISPR-Cas12a. Mol Cell. 2017;66:221–233 e224. doi: 10.1016/j.molcel.2017.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Swiat M.A., Dashko S., den Ridder M., Wijsman M., van der Oost J., Daran J.M., Daran-Lapujade P. FnCpf1: a novel and efficient genome editing tool for Saccharomyces cerevisiae. Nucleic Acids Res. 2017;45:12585–12598. doi: 10.1093/nar/gkx1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng A.W., Jillette N., Lee P., Plaskon D., Fujiwara Y., Wang W., Taghbalout A., Wang H. Casilio: a versatile CRISPR-Cas9-Pumilio hybrid for gene regulation and genomic labeling. Cell Res. 2016;26:254–257. doi: 10.1038/cr.2016.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Esvelt K.M., Mali P., Braff J.L., Moosburner M., Yaung S.J., Church G.M. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Methods. 2013;10:1116–1121. doi: 10.1038/nmeth.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.