Abstract
Background & Aims
The tumor-suppressor sterile α motif– and Src-homology 3–domain containing 1 (SASH1) has clinical relevance in colorectal carcinoma and is associated specifically with metachronous metastasis. We sought to identify the molecular mechanisms linking decreased SASH1 expression with distant metastasis formation.
Methods
SASH1-deficient, SASH1-depleted, or SASH1-overexpressing HCT116 colon cancer cells were generated by the Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR-associated 9-method, RNA interference, and transient plasmid transfection, respectively. Epithelial-mesenchymal transition (EMT) was analyzed by quantitative reverse-transcription polymerase chain reaction, immunoblotting, immunofluorescence microscopy, migration/invasion assays, and 3-dimensional cell culture. Yeast 2-hybrid assays and co-immunoprecipitation/mass-spectrometry showed V-Crk avian sarcoma virus CT10 oncogene homolog-like (CRKL) as a novel interaction partner of SASH1, further confirmed by domain mapping, site-directed mutagenesis, co-immunoprecipitation, and dynamic mass redistribution assays. CRKL-deficient cells were generated in parental or SASH1-deficient cells. Metastatic capacity was analyzed with an orthotopic mouse model. Expression and significance of SASH1 and CRKL for survival and response to chemotherapy was assessed in patient samples from our department and The Cancer Genome Atlas data set.
Results
SASH1 expression is down-regulated during cytokine-induced EMT in cell lines from colorectal, pancreatic, or hepatocellular cancer, mediated by the putative SASH1 promoter. Deficiency or knock-down of SASH1 induces EMT, leading to an aggressive, invasive phenotype with increased chemoresistance. SASH1 counteracts EMT through interaction with the oncoprotein CRKL, inhibiting CRKL-mediated activation of SRC kinase, which is crucially required for EMT. SASH1-deficient cells form significantly more metastases in vivo, depending entirely on CRKL. Patient tumor samples show significantly decreased SASH1 and increased CRKL expression, associated with significantly decreased overall survival. Patients with increased CRKL expression show significantly worse response to adjuvant chemotherapy.
Conclusions
We propose SASH1 as an inhibitor of CRKL-mediated SRC signaling, introducing a potentially druggable mechanism counteracting chemoresistance and metastasis formation.
Keywords: EMT, Tumor Suppressor, SRC-Kinase, Chemoresistance
Abbreviations used in this paper: BSA, bovine serum albumin; cDNA, complementary DNA; CRISPR/Cas9, Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR-associated 9; CRKL, V-Crk avian sarcoma virus CT10 oncogene homolog-like; DMEM, Dulbecco's modified Eagle medium; EMT, epithelial-mesenchymal transition; GFP, green fluorescent protein; gRNA, guide RNA; GTPase, guanosine triphosphatase; mRNA, messenger RNA; MS, mass spectrometry; NLS, nuclear localization signal; PBS, phosphate-buffered saline; qRT-PCR, quantitative reverse-transcription polymerase chain reaction; SASH1, sterile α motif– and Src-homology 3–domain containing 1; SH2, Src-homology 2 domain; SH3, Src-homology 3 domain; SH3N, N-terminal Src-homology 3 domain; TGF, transforming growth factor; TNF, tumor necrosis factor; ZEB, zinc-finger δEF1 family
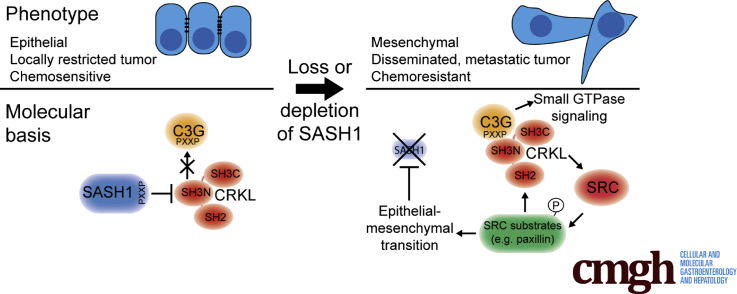
Graphical abstract
Summary.
This study showed a metastasis-suppressive function of sterile α motif– and Src-homology 3–domain containing 1 (SASH1) in vivo. Furthermore, SASH1 antagonizes epithelial–mesenchymal transition, tumor aggressiveness, and chemoresistance in colon cancer. Mechanistically, SASH1 directly inhibits the oncoprotein V-Crk avian sarcoma virus CT10 oncogene homolog-like, introduced as its new interaction partner.
Most cancer-related deaths can be attributed to metastasis from epithelial-derived carcinomas, while local, undisseminated tumors only rarely are lethal.1 Therefore, it is of major clinical importance to understand the molecular mechanisms that regulate the metastatic cascade. The transdifferentiation process referred to as epithelial-mesenchymal transition (EMT) is believed to be crucial for the formation of metastases because it converts sessile epithelial cells into motile and invasive mesenchymal-like cells. EMT is physiologically required for embryonic development and wound healing, but can be hijacked by cancer cells (reviewed by Kalluri and Weinberg2 and Lamouille et al3). During EMT, key epithelial characteristics are suppressed, notably the epithelial adherens junction protein E-cadherin. Thus, intercellular adhesions are weakened or completely lost, promoting cell individualization.3 In addition, typical mesenchymal properties are induced, increasing cell survival, motility, and invasiveness. These processes are mediated primarily by master transcription factors such as the 2-handed zinc-finger δEF1 family (ZEB1/2) factors.3, 4, 5, 6 Because cells that underwent EMT have gained an invasive behavior, they are believed to drive metastasis by penetrating tissue adjacent to the primary tumor. These cells then can enter the blood and lymph system, disseminate systemically, and may finally settle down at distant sites to spawn metastases.2 The scaffold protein sterile α motif– and Src-homology 3 (SH3)-domain containing 1 (SASH1) is a candidate tumor suppressor with clinical relevance in breast and colon cancer, as well as in other solid cancer entities.7, 8, 9, 10, 11, 12, 13 In addition, SASH1 is involved in atherosclerosis, innate immune responses, and skin pigmentation defects.14, 15, 16, 17, 18 Our group previously showed that down-regulation of SASH1 specifically correlates with poor prognosis and formation of metachronous distant metastases in patients with colorectal cancer.8, 9 Although these data confirm a clinical implication of reduced or absent intratumoral SASH1 expression, it is still unknown how its loss or decrease mechanistically aggravates tumor progression and induces formation of distant metastases. Considering that SASH1 is thought to be a multitissue tumor suppressor, better understanding of this process could provide a basis for therapeutic strategies in a wide variety of cancer entities.
Results
Loss of SASH1 Induces Epithelial-Mesenchymal Transition
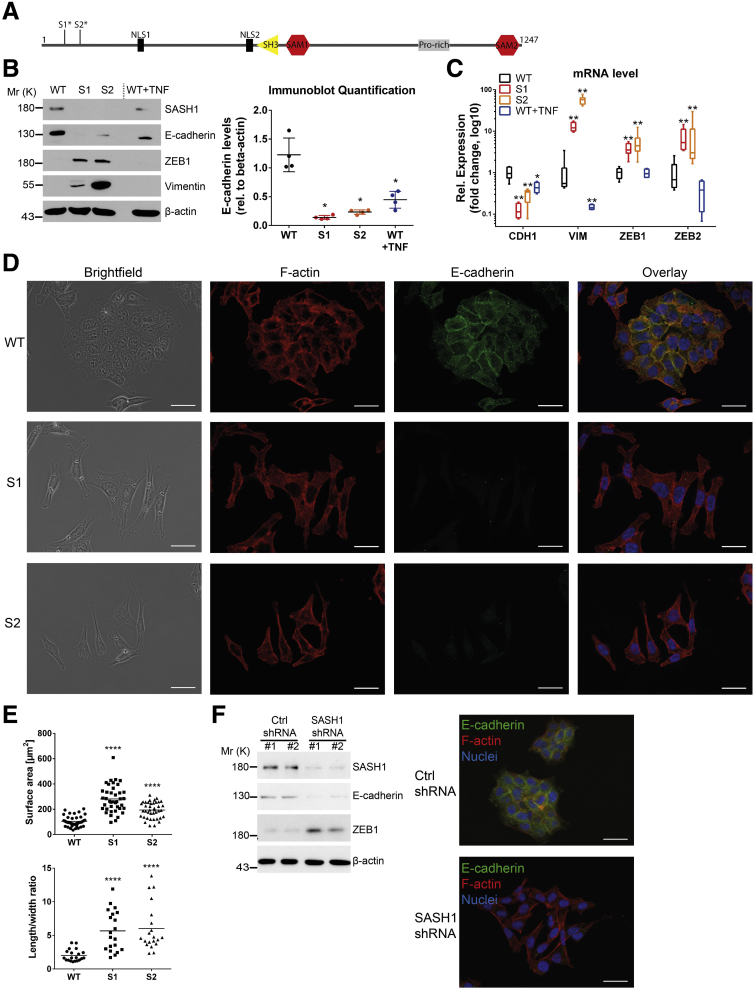
We sought to identify the mechanistic role of SASH1 in cancer progression and metastasis formation. Because SASH1 frequently is lost or down-regulated in colorectal cancer,7, 8 its deficiency was induced by CRISPR/Cas9-editing in human HCT116 colon cancer cells. Clones were derived from 2 independent guide RNAs to minimize the risk of confounding off-target effects. Deficiency of SASH1 was confirmed by immunoblot analysis and next-generation sequencing of the genomic target area, revealing single base pair insertions in the coding sequence of exon 1 (clone S1) or exon 2 (clone S2), respectively, leading to premature stop codons and absent protein expression (Figure 1A and B). Proliferation was essentially unaffected by loss of SASH1 (data not shown), however, cellular morphology was altered remarkably. Because a fibroblast-like morphology is indicative of EMT, expression of EMT-specific markers was investigated, using treatment with the cytokine tumor necrosis factor (TNF), which is known to induce EMT in HCT116 cells, as positive control for EMT19 (Figure 1B). The epithelial marker E-cadherin was down-regulated significantly upon SASH1 deficiency, while the mesenchymal marker vimentin and the EMT-promoting transcription factor ZEB1 were up-regulated. Compared with SASH1-deficient cells, TNF-treated parental cells showed a minor reduction of E-cadherin and no increase in ZEB1 or vimentin levels. Interestingly, SASH1 also was reduced upon TNF treatment. In accordance with these results, EMT marker expression was altered significantly at the messenger RNA (mRNA) level (Figure 1C). Whereas parental cells (wild type) formed dense epithelial islets, SASH1-deficient cells (S1 and S2) acquired a fibroblast-like morphology with concomitant cell individualization (Figure 1D). In accordance, immunofluorescence microscopy showed barely detectable E-cadherin staining of SASH1-deficient cells, whereas E-cadherin localized to cell–cell adhesion sites in parental cells with primarily cortical distribution of F-actin (Figure 1D). Loss of SASH1 induced the formation of actin stress fibers and a fibroblast morphology (Figure 1D). This observation also was reflected by a significant increase in cell size and length-to-width ratio (Figure 1E). As a complementary approach, stable short hairpin RNA–mediated depletion of SASH1 was performed. SASH1 down-regulation led to reduced E-cadherin levels, while ZEB1 was increased (Figure 1F). E-cadherin was absent from cell–cell interfaces, and cells also acquired a fibroblast-like morphology (Figure 1F). Therefore, both down-regulation and loss of SASH1 induced EMT in HCT116 cells.
Figure 1.
CRISPR/Cas9-mediated SASH1 deficiency in HCT116 induces a mesenchymal phenotype. (A) Domain architecture of SASH1, and premature stop codons of SASH1-deficient clones (S1*, S2*), which were generated by independent guide RNAs. (B) Immunoblot analysis of SASH1-deficient (S1, S2) and parental (wild-type [WT]) HCT116 cells. As control, parental cells were treated with 20 ng/mL TNF for 48 hours to induce EMT. Immunoblot quantification of E-cadherin levels (Mann–Whitney test; n = 4 independent experiments; P = .0286) is also shown. (C) qRT-PCR was performed to determine expression of CDH1, ZEB1, ZEB2, and VIM (Mann–Whitney test; n = 4–6 independent experiments; P ≤ .019). (D) Immunofluorescence staining (40× objective; scale bar: 20 μm) for E-cadherin (green), F-actin (red), and 4′,6-diamidino-2-phenylindole for nuclei (blue). (E) Quantification of cell size (unpaired t test; n = 40 cells; P < .0001) and length-to-width ratio (unpaired t test; n = 20 cells; P < .0001). (F) Immunoblot and immunofluorescence microscopic analysis of parental HCT116 stably transfected with vector control (Ctrl) or short hairpin RNA (shRNA) vector targeting SASH1. Mr (K), molecular range (kilodalton); Pro-rich, Proline-rich region; Rel, relative; SAM, sterile α motif. ∗P ≤ .05; ∗∗P ≤ .01; P ≤ .001; ∗∗∗∗P ≤ .0001.
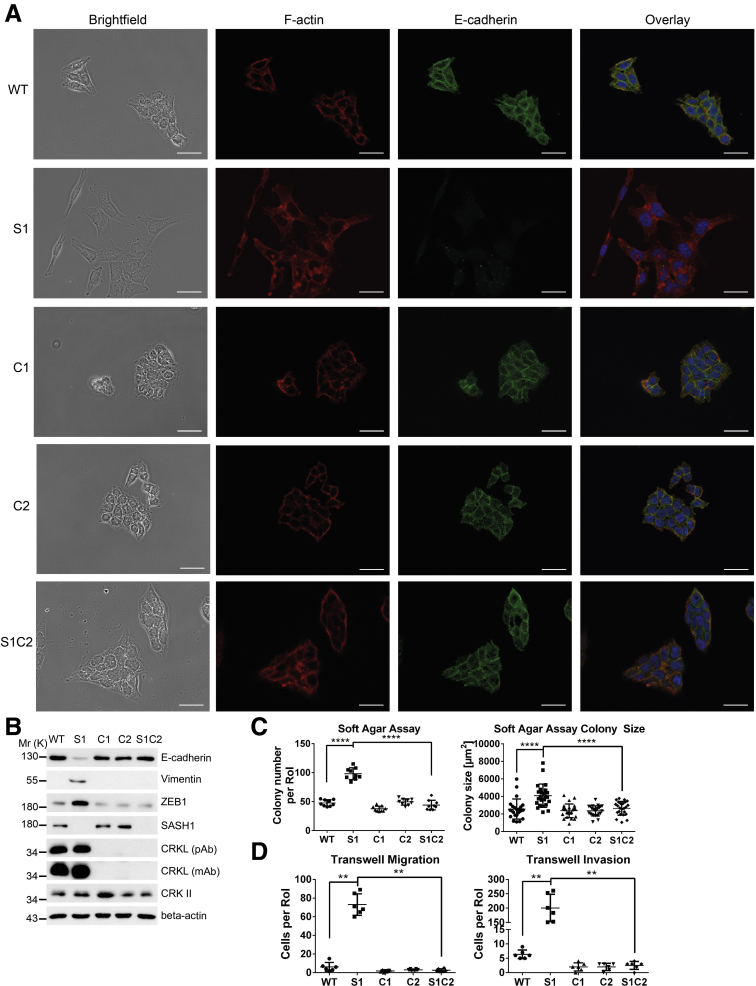
SASH1 Is a Negative Regulator of EMT-Associated Aggressiveness
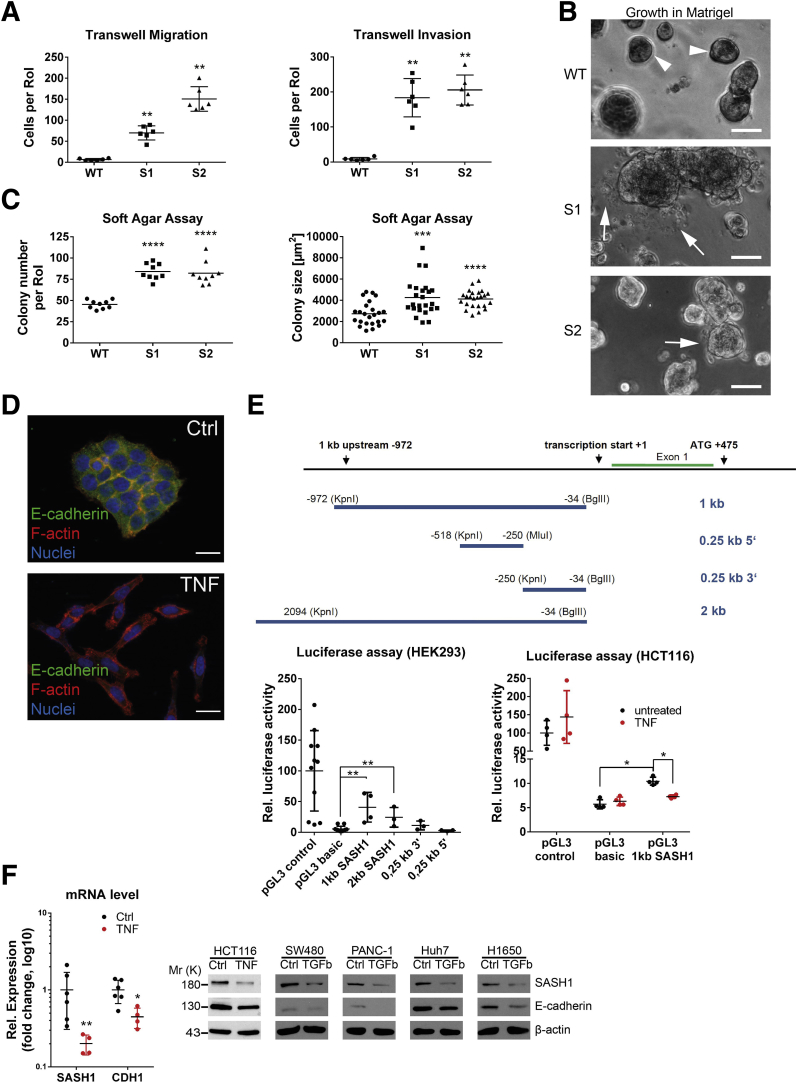
To verify whether loss of SASH1 induces a bona fide EMT that generates aggressive cancer cells, its impact was functionally analyzed by migration and invasion assays. SASH1-deficient clones showed a highly significant increase in transmigrated and Matrigel (Sigma Aldrich, Steinheim, Germany)-invading cells (Figure 2A). Furthermore, cells were cultured in Matrigel to analyze colony morphology in a 3-dimensional matrix. Parental cells formed rounded colonies with a smooth surface (Figure 2B, arrowheads). Consistent with the EMT-like phenotype in 2-dimensional cultures, SASH1-deficient cells formed morphologically differing cell clusters, as single cells disseminated from the colony and invaded the extracellular matrix (Figure 2B, arrows). Next, soft agar assays were used to probe anchorage-independent growth and survival. The number and size of colonies were increased significantly upon loss of SASH1 (Figure 2C). Thus, SASH1 deficiency induces a bona fide EMT with increased migration and invasion, and increased survival in soft agar. Based on our observation of reduced SASH1 levels upon TNF treatment, we focused on the hitherto unknown transcriptional regulation of SASH1. TNF indeed induced EMT within 48 hours, as judged by loss of E-cadherin–mediated intercellular adhesions and fibroblast-like morphology (Figure 2D). HEK293 cells were transfected with luciferase reporters under the control of various fragments of the putative regulatory region of the SASH1 gene (Figure 2E, upper panel). Of note, a robust and significant constitutive reporter activity was detected for a 1-kb region immediately upstream of the transcription start (Figure 2E, lower left panel). HCT116 cells were transfected with the 1-kb reporter construct, and stimulation with TNF caused a significant decrease of SASH1 reporter activity, while controls showed no alterations (Figure 2E, lower right panel). In accordance, expression of both CDH1 and SASH1 were reduced at the mRNA level (Figure 2F). Importantly, SASH1 expression also was down-regulated in cancer cell lines derived from further entities, including SW480 (colorectal cancer), PANC-1 (pancreatic ductal adenocarcinoma), Huh7 (hepatocellular carcinoma), and H1650 (non–small-cell lung cancer) after treatment with transforming growth factor (TGF)-β1 for 48 hours, indicating a general role of SASH1 in EMT, independent from the inducing agent and cancer entity (Figure 2F).
Figure 2.
Loss of SASH1 activates a bona fide EMT program in HCT116 cells. (A) Transwell assays were performed with SASH1-deficient (S1, S2) and parental (wild-type [WT]) HCT116 cells (Mann–Whitney test; n = 6 independent experiments; P = .0022). (B) In contrast to parental cells, which form dense colonies (arrowheads), SASH1-deficient cells grown in a 3-dimensional Matrigel culture for 5 days frequently detach from the colony and invade the extracellular matrix as single cells (arrows; scale bar: 50 μm). (C) Cells were grown in soft agar for 14 days, before the number of colonies (unpaired t test; n = 9 independent experiments; P < .0001) and colony size (unpaired t test; n = 24 colonies; S1: P = .0007; S2: P < .0001) was assessed. (D) Parental cells treated with 20 ng/mL TNF for 48 hours undergo EMT, as judged by staining for immunofluorescence microscopy (40× objective, scale bar: 20 μm) for E-cadherin (green), F-actin (red), and 4′,6-diamidino-2-phenylindole (DAPI) for nuclei (blue). (E) Shown is the proximal promoter of SASH1 and corresponding pGL3 reporter constructs. Luciferase assays were performed to compare the activity of SASH1 promoter sequences with the promoter-less pGL3-basic vector, as well as the SV40 promoter containing positive pGL3-control plasmid in HEK293 cells (lower left panel; Mann–Whitney test; n ≥ 3 independent experiments; P = .0286). Luciferase assays (lower right panel) also were performed with HCT116 cells treated with 20 ng/mL TNF for 24 hours (Mann–Whitney test; n = 4 independent experiments; P = .0286). (F) SASH1 and CDH1 expression levels after induction of EMT by 20 ng/mL TNF for 48 hours (Mann–Whitney test; n = 4–6 independent experiments; P ≤ .019), as determined by qRT-PCR (left panel). Immunoblot analysis of SASH1 and E-cadherin levels upon treatment of a panel of cell lines with 20 ng/mL TNF (HCT116), or 10 ng/mL TGF-β1 (SW480, PANC-1, Huh7, H1650) for 48 hours. Ctrl, control; Rel, relative; RoI, region of interest.
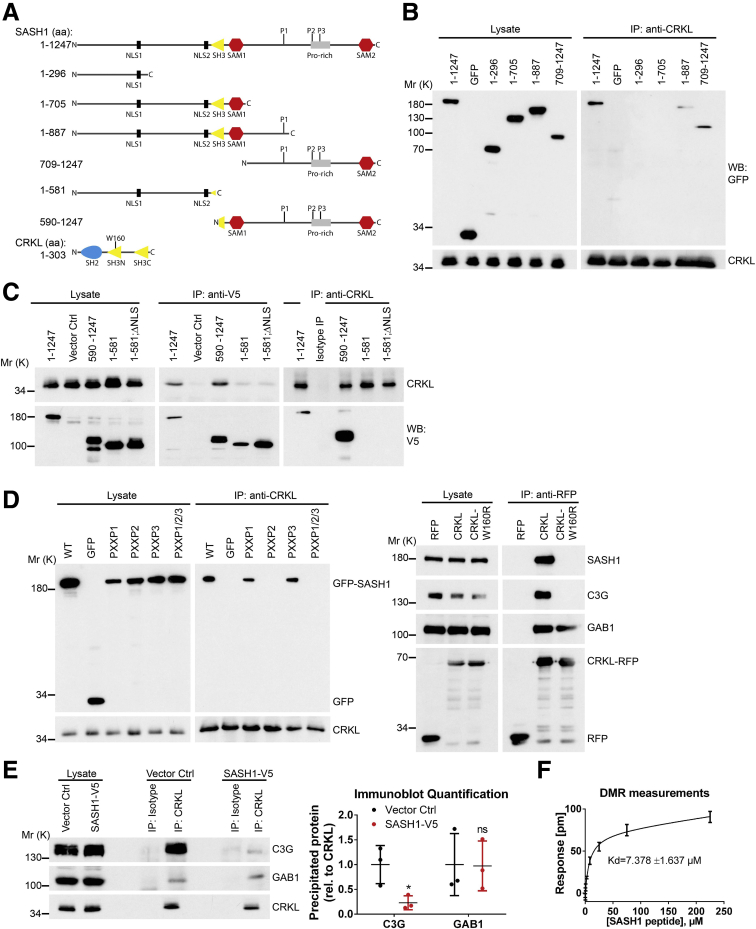
SASH1 Interacts With the Oncoprotein and Signal Adaptor V-Crk Avian Sarcoma Virus CT10 Oncogene Homolog-Like
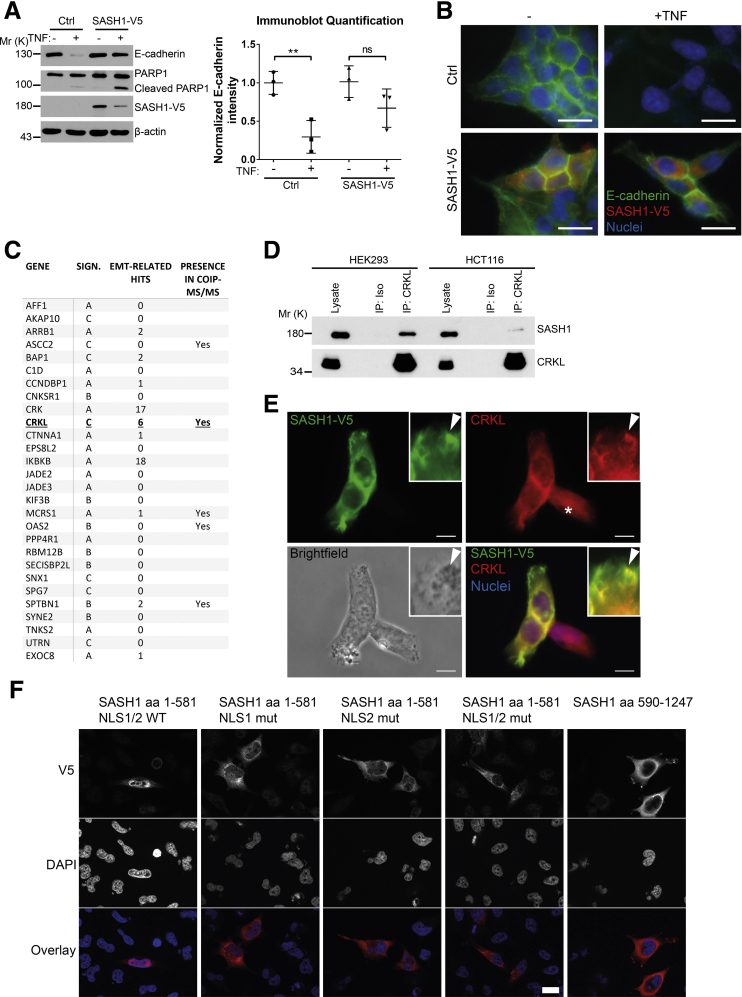
To determine if forced expression of SASH1 can counteract TNF-induced EMT, SASH1 with a C-terminal V5-epitope tag was expressed transiently in parental HCT116. In contrast to control transfected cells, TNF treatment induced only a minor reduction of E-cadherin levels in SASH1-V5–expressing cells. Of note, overexpression of SASH1 was associated with apoptosis, as indicated by Poly(ADP-Ribose)-Polymerase 1 cleavage and reduced SASH1-V5 levels (Figure 3A). The epithelial state of SASH1-V5–expressing cells after TNF administration was highlighted further by a cobblestone-like morphology and E-cadherin strongly localizing to intercellular adhesions (Figure 3B). Next, to investigate the molecular mechanism of the negative regulatory role of SASH1 in EMT, a yeast 2-hybrid screen was performed in a human placental complementary DNA (cDNA) bank with SASH1 as bait. Complementary to the yeast 2-hybrid screen, co-immunoprecipitations were performed with SASH1 with an N-terminal green fluorescent protein (GFP) tag expressed in HEK293 cells, and analyzed by mass spectrometry, using GFP-only transfected cells as control. As potential binding partners, the yeast 2-hybrid screen showed 28 protein coding hits (Figure 3C). Among these candidates, the V-Crk avian sarcoma virus CT10 oncogene homolog family of adaptor proteins (the V-Crk avian sarcoma virus CT10 oncogene homolog [CRK] and V-Crk avian sarcoma virus CT10 oncogene homolog-like [CRKL]) were published to be EMT-related. However, only CRKL was detected independently by co-immunoprecipitation. For these reasons, we focused on CRKL as a potential EMT-related interaction partner of SASH1. First, the putative interaction between SASH1 and CRKL was verified by co-immunoprecipitation of endogenous proteins (Figure 3D). Furthermore, SASH1 with a V5-epitope tag and endogenous CRKL co-localized in the cytoplasm, and at phase-dense structures, partly identified as focal adhesions by paxillin staining (data not shown) (Figure 3E). Because of relatively low SASH1-V5 levels in the nuclei, as well as increased nuclear CRKL staining in cells lacking SASH1-V5 (Figure 3E, asterisk), the predicted nuclear localization signals (NLS)1 and NLS2 within SASH1 were disabled by site-directed mutagenesis, either alone or combined, efficiently blocking nuclear import of the N-terminal fragment (Figure 3F). Therefore, nuclear shuttling of SASH1 is mediated by both amino-terminal NLS signals, and SASH1 likely interacts with CRKL both in cytoplasm and nucleus.
Figure 3.
Identification of the signal adaptor CRKL as an EMT-related interaction partner of SASH1. (A) Immunoblot analysis of parental HCT116 cells transiently transfected with control vector or a vector to express SASH1 with a C-terminal V5 tag. Cells were stimulated with 20 ng/mL TNF or vehicle control for 48 hours. Immunoblot quantifications also are shown (unpaired t test; n = 3 independent experiments; P = .0094). (B) Immunofluorescence microscopy (scale bar: 20 μm) of control or SASH1-V5–transfected cells, either untreated or stimulated with 20 ng/mL TNF for 48 hours. Fixed cells were stained with an anti-V5 antibody (red), anti–E-cadherin antibody (green), and 4′,6-diamidino-2-phenylindole for nuclei (DAPI; blue). (C) To screen for potential interaction partners of SASH1, a yeast 2-hybrid screen was performed with SASH1 as bait protein and a human placental cDNA bank. Shown are protein coding hits of good and high confidence (significance A–C), and the corresponding number of articles from “<gene name> and EMT” searches in the PubMed database. In addition, SASH1 with an N-terminal GFP tag was transiently expressed in HEK293 cells and precipitated to analyze co-precipitating proteins via mass spectrometry. Yeast 2-hybrid hits that also were found to interact with SASH1 via co-precipitation/mass spectrometry are indicated. (D) Endogenous CRKL co-precipitates endogenous SASH1 in HCT116 and HEK293 cell lysates, while the isotype control antibody shows no signal. (E) HEK293 cells transiently expressing SASH1 with a C-terminal V5 epitope were stained for immunofluorescence microscopy (scale bar: 10 μm) with an anti-V5 and anti-CRKL antibody. SASH1-V5 and CRKL showed a pronounced co-localization at phase-dense structures in peripheral regions of the cell (arrowheads). (F) Immunofluorescence microscopy (scale bar: 20 μm) of HeLa cells expressing SASH1-V5 deletion constructs with wild-type (WT) or mutated (mut) nuclear localization signals. Cells were fixed and stained with an anti-V5 antibody (red) and DAPI for nuclei (blue). COIP, co-immunoprecipitation; IP, immunoprecipitation; ISO, isotype antibody; Mr (K), molecular range (kilodalton); PARP, Poly(ADP-Ribose)-Polymerase 1; SIGN, significance.
The N-Terminal SH3 Domain of CRKL Interacts With a PXXPXK Motif of SASH1
Next, the binding mode between SASH1 and CRKL was mapped using full-length and truncated SASH1 constructs with N-terminal GFP or C-terminal V5 epitopes (Figure 4A) expressed in HEK293 cells, as described before.20 Immunoprecipitation showed that only constructs containing the C-terminal part of SASH1 bind to endogenous CRKL (Figure 4B and C). However, the NLS showed no impact on CRKL binding (Figure 4C). CRKL is known to bind specific PXXPXK/R motifs of its downstream effector proteins (eg, C3G), mediated by its N-terminal SH3 domain (SH3N).21, 22 Analysis of the SASH1 primary structure (Research Collaboratory for Structural Bioinformatics Protein Data Bank-entry: O94885) showed 3 PXXPXK motifs within the C-terminal part as putative CRKL binding sites (aa865-870, 984-989, and 1016-1021). Site-directed mutagenesis of SASH1 was performed by alanine substitution of the 3 PXXPXK motifs to AXXAXK. Immunoprecipitations of endogenous CRKL showed that only mutation of the second PXXPXK motif (aa984-989), or of all PXXPXK motifs together, disrupted CRKL binding (Figure 4D, left panel). Furthermore, mutation of a tryptophan residue to arginine (W160R) within the CRKL SH3N domain, reported to be important for domain function,23 abolished the interaction with C3G, as well as with SASH1 (Figure 4D, right panel). As a control, the CRKL interaction partner GAB1,24 which binds to the Src-homology 2 (SH2) domain of CRKL, still interacted with CRKLW160R. Thus, the interaction between SASH1 and CRKL is mediated by the CRKL SH3N domain, binding the second PXXPXK motif of SASH1. Importantly, recombinant expression of SASH1 significantly inhibited CRKL binding to its effector C3G, while the interaction with GAB1 remained unaffected (Figure 4E). The apparent affinity between a peptide corresponding to the second PXXPXK motif of SASH1 (QPPPVPAKKS) and the recombinant CRKL SH3N domain was determined by a dynamic mass redistribution-based assay (dissociation constant = 7.4 ± 1.637 μmol/L) (Figure 4F).
Figure 4.
The N-terminal SH3 domain of CRKL binds to a specific PXXPXK motif of SASH1. (A) Shown are several SASH1 deletion constructs tagged with an N-terminal GFP- or C-terminal V5-epitope, which were used to map the CRKL binding site. Length of SASH1 constructs as well as the CRKL domain architecture also are indicated. (B) Endogenous CRKL was precipitated with an anti-CRKL antibody in HEK293 cells expressing GFP or different SASH1 deletion constructs with N-terminal GFP tags. Full-length GFP-SASH1 (aa1–1247) and deletion constructs containing the C-terminal part of SASH1 (aa1–887 and 709–1247) co-precipitated with CRKL. (C) Reciprocal immunoprecipitations were performed with SASH1 constructs carrying a C-terminal V5-epitope tag. In accordance to GFP-SASH1 constructs, precipitation with anti-V5 and anti-CRKL antibodies showed that endogenous CRKL binds to the C-terminal part of SASH1 (aa590–1247). (D) SASH1 contains 3 distinct PXXPXK motifs (P1–3) in the C-terminal region. Mutation of the second PXXPXK motif (P2, aa984–989) of GFP-SASH1 is sufficient to disrupt co-precipitation with endogenous CRKL (left panel). Anti-RFP immunoprecipitations show that the interaction of CRKL (fused to RFP at the C-terminus) with C3G and SASH1, but not GAB1, is abolished by the W160R mutation that inactivates the N-terminal SH3 domain of CRKL (right panel). (E) Recombinant expression of SASH1 with a C-terminal V5 tag in HEK293 cells abrogates co-precipitation of C3G by endogenous CRKL (unpaired t test; n = 3 independent experiments; P = .0310), while the interaction with GAB1 is not disturbed. (F) The affinity between SASH1 and CRKL was determined by dynamic mass redistribution (DMR) experiments using a peptide containing the second PXXPXK motif of SASH1 (aa982–991) and the N-terminal SH3 domain of CRKL purified from E coli. SASH1 binds CRKL with an apparent dissociation constant of dissociation constant (±SD) of 7.378 ± 1.637 μmol/L. Ctrl, control; IP, immunoprecipitation; Mr (K), molecular range (kilodalton); P1–3, PXXPXK motifs; Pro-rich, proline-rich region; rel, relative; RFP, red fluorescent protein; SAM, sterile α motif; WB, Western blot.
SASH1 Negatively Regulates EMT Through CRKL
As a working hypothesis, SASH1 physically associates with CRKL and counteracts its EMT-promoting signaling. CRKL deficiency thus was induced in parental and SASH1-deficient cells, using 2 independent guide RNAs (clones C1 and C2 in parental cells). CRKL deficiency induced a more pronounced epithelial morphology with densely packed, cobblestone-like islets (Figure 5A). Strikingly, loss of CRKL in SASH1-deficient cells (S1C2) completely reverted their fibroblast-like morphology into a clear epithelial phenotype (Figure 5A). Essentially the same phenotype was observed for an independent SASH1/CRKL-deficient clone (S2C2, data not shown). Loss of CRKL in SASH1-deficient cells restored the expression of EMT markers to the parental levels (Figure 5B). CRKL deficiency did not affect anchorage-independent growth in parental cells, but highly significantly reduced colony number and size of SASH1-deficient cells (Figure 5C). Furthermore, increased migration and invasion of SASH1-deficient cells was completely reverted by loss of CRKL (Figure 5D). Thus, EMT induced by loss of SASH1 completely depends on the presence of its interaction partner CRKL.
Figure 5.
Loss of CRKL rescues the EMT phenotype of SASH1-deficient cells. (A) Parental (wild-type [WT]), SASH1-deficient (S1), CRKL-deficient (C1, C2), and compound SASH1/CRKL-deficient (S1C2) HCT116 cells were subjected to immunofluorescence staining (40× objective, scale bar: 20 μm) for E-cadherin (green), F-actin (red), and 4′,6-diamidino-2-phenylindole for nuclei (blue). (B) Immunoblot analysis of EMT markers and CRKL, using 2 different CRKL-specific antibodies. (C) Cells were grown for 14 days in soft agar before quantification of colony number (unpaired t test; n = 9 independent experiments; P < .0001) and colony size (unpaired t test; n = 24 colonies; P < .0001). (D) Transwell assays to quantify migration and invasion (Mann–Whitney test; n = 6 independent experiments; P = .0022). mAb, monoclonal antibody; Mr (K), molecular range (kilodalton); pAb, polyclonal antibody; RoI, region of interest. ∗P ≤ .05; ∗∗P ≤ .01; P ≤ 0.001; ∗∗∗∗P ≤ .0001.
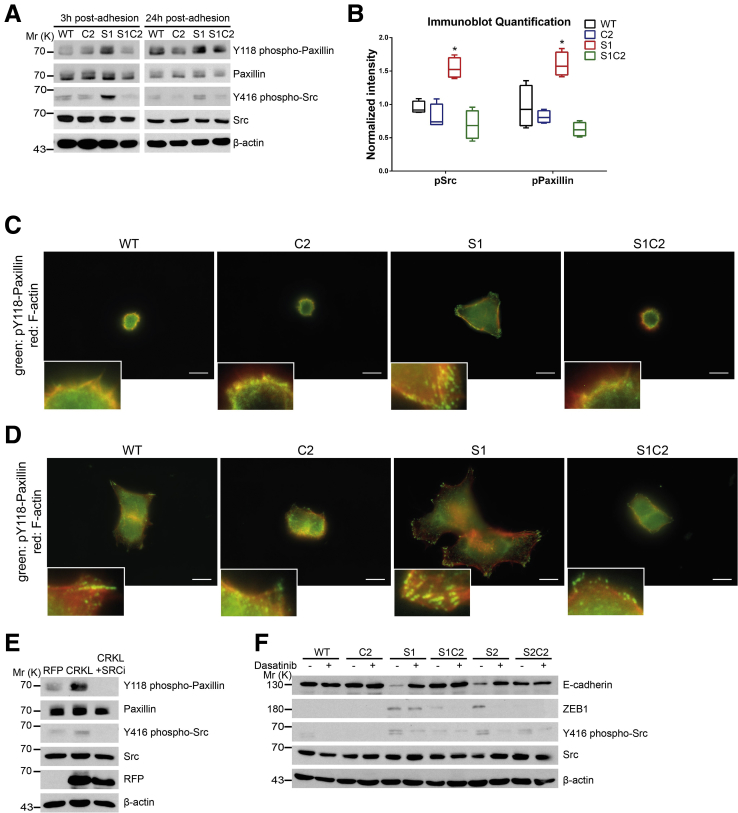
EMT Induced by Loss of SASH1 Depends on CRKL-Mediated SRC Signaling
CRKL was reported to play unique roles in integrin signaling at focal adhesions.25, 26, 27 To investigate if SASH1 negatively regulates EMT through these signaling pathways in a CRKL-dependent manner, cells were cultured on fibronectin-coated plates. SASH1-deficient cells showed significantly increased tyrosine-phosphorylation levels of SRC family kinases at the activating residue Y416, as well as of paxillin at Y118, which was abolished by loss of CRKL (Figure 6A and B). SASH1-deficient cells formed focal adhesions after 3 hours of adhesion, while parental or CRKL-deficient cells poorly spread and rarely formed phospho-paxillin–positive foci (Figure 6C). After 24 hours, SASH1-deficient cells showed multipolar lamellipodia with abundant phospho-paxillin–positive focal adhesions, while parental cells showed few adhesive structures (Figure 6D). CRKL deficiency strongly reduced the development of focal adhesions in parental and SASH1-deficient cells.
Figure 6.
SASH1 inhibits CRKL-mediated SRC signaling, which is required for the mesenchymal phenotype. (A) SASH1- and/or CRKL-deficient cells that adhered to fibronectin-coated plates for 3 hours (left panel) or for 24 hours (right panel) were subjected to immunoblot analysis. (B) Immunoblot quantification of SRC (Y416) and paxillin (Y118) phosphorylation after 3 hours of adhesion (Mann–Whitney test; n = 4 independent experiments; P = .0286). ∗P ≤ .05. (C) Cells that adhered for 3 hours to fibronectin-coated plates were subjected to immunofluorescence staining (100× objective, scale bar: 10 μm) for Y118 phospho-paxillin (green), F-actin (red), and 4′,6-diamidino-2-phenylindole for nuclei (blue). (D) The same analysis was performed after 24-hour adhesion to fibronectin-coated plates. (E) Immunoblot analysis of parental HCT116 cells recombinantly expressing RFP as control or CRKL with a C-terminal RFP tag. In addition, CRKL-RFP–expressing cells were treated with 100 nmol/L dasatinib for 24 hours. (F) Parental (wild-type [WT]), CRKL-deficient (C2), SASH1-deficient (S1, S2), and compound SASH1/CRKL-deficient (S1C2, S2C2) cells were treated with 100 nmol/L dasatinib for 72 hours before they were subjected to immunoblot analysis. Mr (K), molecular range (kilodalton); pPaxillin, phospho-Paxillin; pSrc, phospho-Src; RFP, red fluorescent protein.
Next, CRKL was recombinantly expressed in parental HCT116, which led to tyrosine phosphorylation of SRC and paxillin, phenocopying a loss of SASH1. This effect was inhibited by the Abl/SRC-inhibitor dasatinib (Figure 6E). Importantly, pharmacologic inhibition of SRC via dasatinib counteracted the mesenchymal phenotype of SASH1-deficient cells (S1 and S2), as E-cadherin was increased to the levels of compound SASH1/CRKL-deficient cells (S1C2 and S2C2) (Figure 6F). ZEB1 was slightly decreased (S1) or undetectable (S2) after treatment with dasatinib. Thus, EMT induced by loss of SASH1 directly depends on CRKL-dependent SRC signaling.
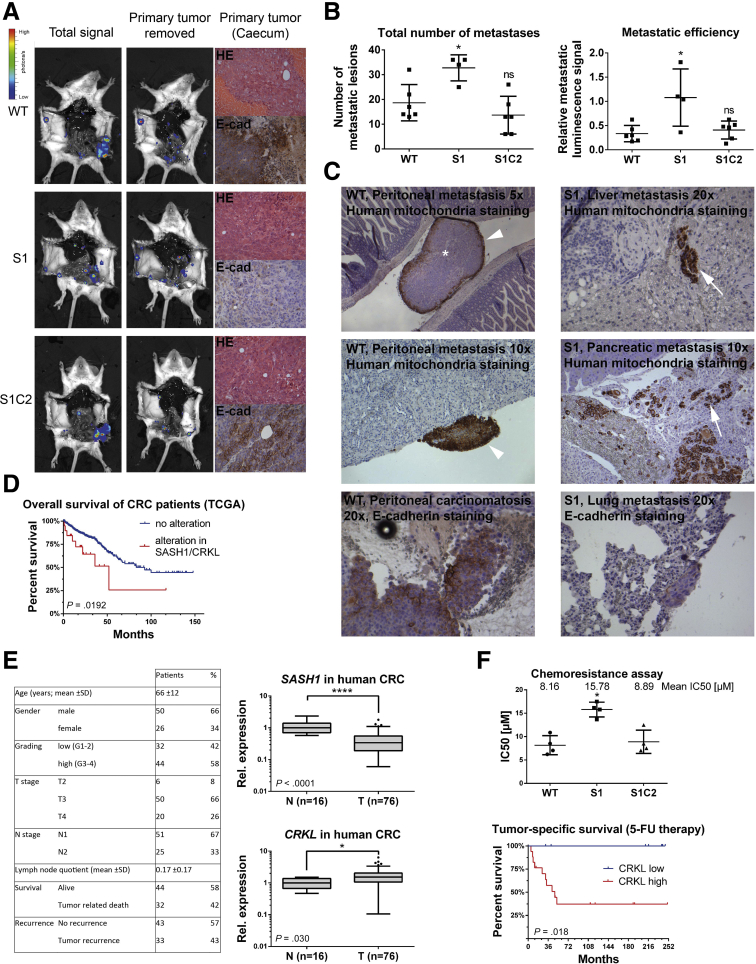
SASH1 Inhibits Metastasis Formation In Vivo in a CRKL-Dependent Manner
An orthotopic xenograft model was applied to analyze the metastatic capability of SASH1-deficient (S1), compound SASH1/CRKL-deficient (S1C2), and parental (wild-type) HCT116 cells. Cells were retrovirally transduced to express luciferase and injected in immune-deficient mice into the tip of the cecum, previously enclosed by absorbable surgical suture. This model allowed tumors to form upon local invasion of the cecum mucosa at the site of implantation. After 34 days, animals were killed and analyzed by bioluminescence imaging (Figure 7A). Mice harbored palpable primary tumors, histologically confirmed as poorly differentiated colon carcinoma with similar overall tissue architecture for all 3 cell lines (Figure 7A). Importantly, tumors derived from SASH1-deficient cells featured a strongly reduced E-cadherin staining, in accordance with our in vitro findings (Figure 7A).
Figure 7.
SASH1 suppresses metastasis formation in a CRKL-dependent manner. (A) Immunodeficient (severe combined immunodeficiency/beige) mice were injected orthotopically with luciferase expressing parental (wild-type [WT]), SASH1-deficient (S1), or compound SASH1/CRKL-deficient (S1C2) HCT116 cells into the cecum (n = 6 mice per group). Metastasis was assessed by bioluminescence imaging before and after removal of the primary tumor. Primary tumors were subjected to H&E and immunohistochemistry staining for human E-cadherin. (B) Metastasis was quantified by automatically counting the number of lesions by their luminescence signal after removal of the primary tumor (left panel; Mann–Whitney test; n = 4–6 mice; P = .019). In addition, metastatic efficiency was assessed by the ratio of total luminescence intensity before and after removal of primary tumors (right panel; Mann–Whitney test; n = 4–6 mice; P = .0381). (C) Organ sections were analyzed for the presence of tumor cells (brown staining) by immunohistochemistry with human-specific antibodies against mitochondria or E-cadherin. In parental and compound SASH1/CRKL-deficient cells, peritoneal carcinomatosis frequently occurred (arrowheads), with metastatic lesions showing central necrosis (asterisk). SASH1-deficient cells spawned metastases enclosed by normal tissue (arrows), which showed low E-cadherin staining compared with the parental line (lower panels). (D) Genomic deletion of SASH1 and/or amplification of CRKL expression was observed in 10.8% of colorectal cancers (23 of 213 patients) and was associated significantly with poor overall survival in Kaplan–Meier analysis (log-rank test; n = 213 patients; P = .0266), based on the TCGA data set. (E) Clinicopathologic characteristics of the Technical University of Munich patient cohort (left panel). qRT-PCR analysis of the patient cohort with Union for International Cancer Control/American Joint Committee on Cancer stage III colorectal cancer (T, n = 76 patient samples) showed a highly significant reduction of SASH1 expression (upper right panel; Mann–Whitney U test; P < .0001) compared with nondiseased colon mucosa (N, n = 16 patient samples). The expression of CRKL was increased significantly in tumor tissue (lower right panel; Mann–Whitney U test; P = .03). (F) Chemoresistance of parental (WT), SASH1-deficient (S1), and compound SASH1/CRKL-deficient (S1C2) cells was analyzed by XTT (Roche) assays after treatment with varying concentrations of 5-fluorouracil for 24 hours to determine the median inhibitory concentration (IC50) values (upper panel; Mann–Whitney test; n = 4 independent experiments; P = .0286). Tumor-specific survival for stage III patients who received adjuvant 5-fluorouracil (5-FU) was strongly reduced in the group with high intratumoral CRKL expression (lower panel; log-rank test; n = 25 patients; P = .018). CRC, colorectal cancer; E-cad, E-cadherin; Rel, relative. ∗P ≤ .05; ∗∗P ≤ .01; ∗∗∗P ≤ .001; ∗∗∗∗ P ≤ .0001.
To quantitatively assess the metastatic capability, bioluminescence was measured post-mortem before and after resection of primary tumors. Automated quantification after removal of the primary tumor showed a significant increase in the total number of metastatic lesions generated by SASH1-deficient cells compared with parental cells (Figure 7B, left panel). This increase was fully blocked by CRKL deficiency as compound SASH1/CRKL-deficient cells were essentially identical to the parental line. To quantify the relative distribution of tumor cells, the signal ratio of distant lesions after removal of the primary tumor was calculated relative to the total signal before removal of the primary tumor (Figure 7B, right panel). In contrast to parental and compound SASH1/CRKL-deficient lines, SASH1-deficient cells showed a significantly higher relative metastatic luminescence signal. Thus, although a similar tumor burden was induced by the different cell lines, a drastic difference was apparent in tumor dissemination. Parental and compound SASH1/CRKL-deficient cells showed locoregional spread, while SASH1-deficient cells showed massive distant dissemination.
Tissue sections were analyzed with an antibody specifically staining human cells.28 Peritoneal carcinomatosis occurred in essentially all mice injected with parental or compound SASH1/CRKL-deficient cells as lesions were attached to the outer surface of host organs within the peritoneum (Figure 7C, left panels). In contrast, secondary lesions from SASH1-deficient cells were completely enclosed by host tissue, and tumor cells often were dispersed extensively as small clusters or single cells (Figure 7C, right panels). In addition, only SASH1-deficient cells were detected within intrahepatic blood vessels (data not shown). SASH1-deficient cells were capable of hematogenic and lymphogenic systemic dissemination, as evidenced by detection of human cancer cells in lungs of the host (Figure 7C, lower right panel). Human E-cadherin was strongly expressed in distant lesions from parental cells, whereas SASH1-deficient secondary lesions showed only low staining (Figure 7C, lower panels).
To address the question of whether SASH1 and CRKL play a clinical role and are involved in survival of patients with colorectal cancer, genetic alterations and aberrant expression of both genes were assessed in the publicly available TCGA data set consisting of 233 colorectal cancer patients, with full sequence information and available follow-up data in the cBioPortal (Memorial Sloan Kettering Cancer Center, New York, NY) platform.29, 30 SASH1 was down-regulated or homodeleted, and CRKL was increased or genomically amplified in 10.8% of cases (23 of 213). There was a trend toward mutual exclusivity of both alterations in individual patients. Kaplan–Meier analysis showed a significantly reduced overall survival in the subgroup with up-regulated CRKL and/or down-regulated SASH1 (Figure 7D). Therefore, SASH1 and its interaction partner CRKL have prognostic significance in colorectal cancer. In addition, SASH1 and CRKL expression was analyzed in an independent patient cohort with locally advanced colorectal cancer (Union for International Cancer Control/American Joint Committee on Cancer stage III, n = 76) by quantitative reverse-transcription polymerase chain reaction (qRT-PCR) (Figure 7E). In accordance with earlier findings, SASH1 expression was highly significantly down-regulated in tumor tissue (Figure 7E, upper right panel), whereas expression of CRKL was increased significantly compared with normal colon tissue (Figure 7E, lower right panel). Because of the relevance of EMT in chemoresistance,31, 32 the resistance to 5-fluorouracil was probed in vitro (Figure 7F, upper panel). SASH1-deficient cells showed an increased resistance to 5-fluorouracil, which was dependent on the presence of CRKL, because loss of CRKL either in SASH1-deficient (Figure 7F, upper panel) or parental cells (not shown) significantly increased the cytotoxic effects. In accordance, survival of stage III colorectal cancer patients receiving 5-fluorouracil as an adjuvant chemotherapeutic agent was reduced significantly in the subgroup with high intratumoral CRKL expression (Figure 7F, lower panel). Intratumoral SASH1 expression was not correlated significantly with response to 5-fluorouracil therapy (data not shown).
Discussion
Metastases account for the majority of cancer-related deaths,1 in the case of colorectal cancer most frequently spreading to the liver or lungs. Thus, it is of huge clinical importance to understand metastasis formation and to find druggable targets. The differentiation process of EMT turns epithelial cells into highly invasive mesenchymal-like cells, proposed as an essential prerequisite for the formation of metastases.2, 3, 33 The present study shows that loss or down-regulation of the tumor-suppressor SASH1 is sufficient to induce EMT. Furthermore, cytokine-induced EMT resulted in a strong reduction of SASH1 expression in a panel of carcinoma cell lines. Forced expression of SASH1 in turn counteracted EMT induced by TNF. Therefore, multiple lines of evidence have indicated a reciprocal relation between EMT and SASH1, and established SASH1 as a key negative regulator of EMT. In support, EMT induced by hypoxia or TGF-β1 was shown independently to down-regulate SASH1 in a pancreatic and gastric cell line, respectively.34, 35 Down-regulation of SASH1 expression during EMT thus fosters the conversion toward a mesenchymal phenotype, representing a feed-forward loop.
Tumor cells acquire an invasive phenotype during EMT, enabling them to detach from the primary tumor and invade adjacent tissue, ultimately spawning metastases.2 Loss of SASH1 was sufficient to induce a drastic increase in migration and invasion, even in the absence of cytokines. These data are in accordance with our previous data because forced expression of SASH1 reduced cell migration and increased cell-matrix adhesion,20 as well as with our clinical results showing a significant association between distant metastasis and reduced SASH1 expression in colon cancer.9 Indeed, orthotopic xenograft models showed a significant increase of metastatic lesions upon loss of SASH1. This report confirms that SASH1 is a tumor suppressor in vivo, counteracting metastasis. Our data also support the relevance of EMT during the metastatic process, which currently is discussed controversially.31, 32, 36, 37 Furthermore, the mechanistic involvement of SASH1 in these processes could be clarified because the phenotype of SASH1-deficient cells depended exclusively on CRKL, which we introduce as a novel interaction partner of SASH1.
CRKL and CRK were identified as putative new interaction partners of SASH1 by yeast 2-hybrid screening. Although both proteins are involved in EMT,38, 39, 40, 41, 42, 43 only CRKL was confirmed independently by co-immunoprecipitation. This can be explained by domain organization differences between CRK and CRKL since the N-terminal SH3 domain of CRK is regulated negatively by phosphorylation. CRK therefore might not be absolutely excluded as an interaction partner.44 CRKL is a SH2- and SH3-domain-containing oncogenic adaptor protein involved in signaling pathways downstream of receptor tyrosine kinases and integrins.21, 24, 25, 39, 45 It binds phosphorylated, membrane-associated proteins (eg, GAB1) via its SH2 domain.24, 27 Thereby, CRKL recruits PXXPX(K/R)-motif containing effectors such as C3G through the SH3N domain to the membrane, leading to activation of small guanosine triphosphatases (GTPases), and thus inducing downstream signaling.21, 24, 45 Our data indicate that SASH1 binds to the SH3N domain of CRKL primarily through a PXXPXK motif (aa984–989) with an affinity in the low micromolar range. The interaction may be supported through other PXXPXK motifs of SASH1 because a deletion construct containing another motif (aa1–887) also co-precipitated with CRKL. Importantly, SASH1 inhibited interactions between CRKL and its effector protein C3G, elucidating a potential inhibitory mode of action for SASH1: by tightly binding the SH3N domain of CRKL, SASH1 abolishes interactions between CRKL, C3G, and further effector proteins, which are essential for CRKL signaling.21 In the absence of SASH1, C3G, and other factors thus can act as a guanine nucleotide exchange factor for small GTPases to induce cell migration. Because CRKL is well established to promote EMT,39, 41, 42 we assume that SASH1 inhibits EMT directly through the competitive inhibition of CRKL downstream signaling (eg, by preventing activation of small GTPases required for cytoskeletal rearrangement and efficient cell migration, see the graphic abstract). A recent study on lung carcinoma suggested that knockdown of CRKL affects migration and invasion, but not EMT-marker expression in mesenchymal-like carcinoma cells.25 In the present study, however, loss of CRKL in SASH1-deficient cells not only rescued the aggressive phenotype, but also reverted the expression of EMT markers. Furthermore, genetic deletion of CRKL in SASH1-deficient cells significantly decreased their aggressive prometastatic phenotype toward the parental state in vivo. Therefore, EMT induced by SASH1 deficiency clearly depends on CRKL-mediated signaling. CRKL is involved in signaling downstream of integrins at focal adhesions, which potentially could promote EMT.22, 25, 27 Focal adhesions also are important players during metastasis.46 Here, we establish SASH1 as a negative regulator of CRKL-mediated activation of SRC family kinases. In accordance, pharmacologic inhibition of SRC in SASH1-deficient cells phenocopied loss of CRKL, indicating that SRC activation upon loss of SASH1 is required for the EMT phenotype. CRK has been shown to activate SRC by association with its negative regulatory kinase C-terminal SRC kinase,47, 48 which also may apply to CRKL. In the absence of SASH1, SRC signaling also could be activated directly by binding of effector proteins (eg, C3G in this study) to the CRKL SH3N domain, leading to activation of small GTPases.
According to our working model, loss of SASH1 induces EMT and formation of metastases owing to a lack of CRKL inhibition. Our in vitro and in vivo findings are supported by data obtained from clinical samples that show a striking decrease of SASH1 and an increase of CRKL expression in colorectal cancer compared with normal colon mucosa. Moreover, analysis of the TCGA data set on an independent patient cohort showed that genetic loss of SASH1, and/or amplification of the CRKL gene locus, occur in approximately 11% of patients, and are associated significantly with worse overall survival. This is in good accordance with our earlier findings on SASH1 as a positive prognostic factor in colorectal cancer.8, 9
Importantly, chemoresistance, which has been linked to EMT,31, 32 was increased significantly in SASH1-deficient cells, whereas loss of CRKL reverted this phenotype. The decreased resistance in vitro against the cytotoxic compound 5-fluorouracil upon loss of CRKL was in accordance with clinical findings. Patients with stage III colorectal cancer who received adjuvant 5-fluorouracil therapy had significantly worse survival when intratumoral CRKL expression was increased. These findings highlight CRKL as a promising therapeutic target. In contrast, intratumoral SASH1 expression was not associated significantly with prognosis in 5-fluorouracil–treated patients. Even though decreased SASH1 expression has been reported as a negative prognostic factor,7, 8, 9, 10, 11, 12, 13 it recently was shown that increased SASH1 expression is associated with a reduced response to adjuvant therapy in a subgroup of breast cancer patients.13 Therefore, independent patient cohorts are required to make conclusive statements about the clinical implications of our findings. However, it is tempting to speculate that patients might be stratified by expression of SASH1 and CRKL to identify subgroups that benefit from pharmacologic inhibition of SRC kinases, which remarkably reduced the malignant properties of cancer cells in our analyses. Furthermore, the inhibition of CRKL, based on the interaction with its natural inhibitor SASH1, could be a suitable new strategy for therapeutic intervention, especially in combination with conventional chemotherapeutic agents. Such treatments potentially could inhibit EMT-induced tumor progression, metastasis, and chemoresistance, thereby preventing minimal residual disease.
Patients and Methods
Cell Culture and Constructs
Human cell lines HCT116 and HEK293 (ATCC, Rockville, MD) were cultured in Dulbecco’s modified Eagle medium (DMEM; Invitrogen, Karlsruhe, Germany) containing 10% fetal bovine serum (Biochrom, Berlin, Germany). HEK293 cells were transfected by precipitation with calcium phosphate as described earlier.20 HCT116 cells were transfected using FuGENE HD (Promega, Madison, WI) according to the manufacturer's protocol. Human CRKL was cloned from cDNA into pmRFP-N2 with primers containing KpnI and XmaI sites. SASH1 deletion constructs were described earlier.20 Mutagenesis of GFP–SASH1 and CRKL–RFP was performed according to published protocols,49 using 1 pair of primers per PXXPXK motif. For short hairpin RNA–mediated knockdown of SASH1, pSUPER.neo+GFP (Oligoengine, Seattle, WA), was used as described.20 The luciferase reporter construct was generated by cloning the proximal 1 kb of the SASH1 gene regulatory region into the pGL3 vector. For induction of epithelial–mesenchymal transition, cells were stimulated with 10 ng/mL TGF-β1 (Stemcell Technologies, Cologne, Germany) or 20 ng/mL TNF (Gibco, Waltham, MA) for 48 hours in normal growth medium. Proliferation and response to 5-fluorouracil (24-hour treatment) was probed using the Cell Proliferation Kit II (XTT) from Roche (Mannheim, Germany). The following antibodies were used: E-cadherin (3195; CST), β-actin (3700; CST), SASH1 (A302-265A; Bethyl, Montgomery, TX), GAB1 (A303-288A; Bethyl), C3G (Bethyl A301-965A), RFP (5F8; Chromotek, Martinsried, Germany), V5 epitope tag (MA5-15253; Thermo Scientific, Waltham, MA), Poly(ADP-Ribose)-Polymerase 1 (ab137653; Abcam, Cambridge, UK), GFP (3H9; Chromotek), CRK-II (sc-289; SCBT, Heidelberg, Germany), CRKL (05-414; EMD Millipore, Darmstadt, Germany), CRKL (sc-319; SCBT), ZEB1 (HPA027524; Sigma, Steinheim, Germany), vimentin (ab92547; Abcam), Y118 phospho-paxillin (2541; CST, Danvers, MA), paxillin (12065; CST), Y416 phospho-Src family (6943; CST), Src (2123; CST), human mitochondria (ab92824; Abcam), human E-cadherin (ab40772; Abcam), RFP-trap (rta-10; Chromotek), and GFP trap (gta-10; Chromotek). The following primers were used: cloning of CRKL-RFP: 5′-CAAGGTACCATGTCCTCCGCCAGG-3′ and 5′-GCCCGGGACTCGTTTTCATCTGGGTTTTG-3′; mutagenesis of PXXP1: 5′-GATTGTAGCAGAAGTGGCACAGAAGACGACCGCC-3′ and 5′-CTGTGCCACTTCTGCTACAATCTGAGGGGGCGG-3′; PXXP2: 5′-CAGCCTGCACCTGTTGCAGCCAAAAAGAGCAGAGAACG-3′ and 5′-CTTTTTGGCTGCAACAGGTGCAGGCTGTGATGGAATTTTGG-3′; PXXP3: 5′-GATGCGGCATGCCTGGCAGTGAAAAGGGGCAGCCCCGC-3′ and 5′-CTTTTCACTGCCAGGCATGCCGCATCGGGACTGGGGAG-3′; CRKL W160R: 5′-GAACAGCGGTGGAGTGCCCGGAACAAGG-3′ and 5′-GCACTCCACCGCTGTTCTTCAGGCTTCTC-3′; amplification of guide RNA (gRNA) target locus gRNA1: 5′-CACATCCGAGGCGTTCTC-3′ and 5′-TGTGTGTATTTAGCCCCCTAAG-3′; gRNA2: 5′-GCGCAGCAGTATGCAGATTA-3′ and 5′-GTGCAGCAGCCTACAGATTG-3′; cloning of luciferase reporter constructs with SASH1_1 kb forward (-972): 5′-CCGGTACCGACATGCTCTTATCCCCTTTCAAG-3′; SASH1_0.25 kb-3′ forward (-250): 5′-CCGGTACCGCATTGTAGCGACACGGACTACTTG-3′; SASH1_0.25 kb-3′ reverse (-250): 5′-GAACGCGTAAGTAGTCCGTGTCGCTACAATGC-3′; SASH1_0.25 kb-5′ forward (-518): 5′-CCGGTACCCTGTGTCCCTAATGTAAAGG-3′; SASH1_2 kb forward (-2094): 5′-CCGGTACCTTGAAAACCCATCTGGCCTAACTC-3′; SASH1_1 kb reverse (-34): 5′-GGAGATCTCTCTCCCTCGAGGCTAAAGAA automatic gain control-3′.
Generation of SASH1- and/or CRKL-Deficient Cells
SASH1- and CRKL-deficient cells were generated with the CRISPR/Cas9 system. The vector pSpCas9(BB)-2A-Puro (PX459) V2.0 was a gift from Feng Zhang (plasmid 62988; Addgene, Cambridge, MA).50 Two different guide RNAs per gene were cloned into the PX459 vector according to published protocols,50 with the target genes SASH1 (gRNA1: 5′-CGACTCTGGACCGACGTGAT-3′; gRNA2: 5′-GTTTCTCCGACGTGTGCGAG-3′) and CRKL (gRNA1: 5′-GTCCGAGGAGTCGAACCTGG-3′; gRNA2: 5′-CGAGGAGTCGAACCTGGCGG-3′). HCT116 cells were transfected using FuGENE HD (Promega) according to the manufacturer’s protocol. Two days later, cells were selected with 5 μg/mL puromycin for 72 hours. Single colonies were isolated and analyzed for SASH1 or CRKL protein levels by immunoblotting during several passages to ensure a complete loss of the respective protein. Specific primers were used for amplification of the genomic CRISPR/Cas9 target area. These amplicons then were sequenced via NGS CRISPR amplicon sequencing (CCIB DNA Core, Massachusetts General Hospital, Boston, MA).
Co-immunoprecipitation
Co-immunoprecipitation was performed as described previously.20 Briefly, cells were lysed in RIPA buffer (50 mmol/L Tris-HCl [pH 7.4], 150 mmol/L NaCl, 1% NP-40, 0.25% sodium deoxycholate, 1 mmol/L EDTA [pH 8.0], 1 mmol/L Na3VO4, 1 mmol/L NaF, 1 mmol/L Pefabloc, 2× protease inhibitor cocktail [Roche], 1 mmol/L β-glycerophosphate, 1 mmol/L benzamidine, 1 mmol/L phenylmethylsulfonyl fluoride, 1 μg/mL Pepstatin), total protein was quantified with the Pierce BCA Protein Quantification Kit (Thermo Fisher Scientific), and equal concentrations of cleared lysates were incubated with 2 μg antibody for 2 hours at 4°C with end-over-end rotation before protein A– or protein G–Sepharose (Sigma-Aldrich) was added. After further rotation for 1 hour at 4°C, beads were washed 4 times with RIPA buffer. Bound proteins were eluted by addition of Laemmli buffer and subjected to immunoblotting.
Reverse Transcription and qRT-PCR
RNA isolation and reverse transcription from cell lines and tissue was performed with the RNeasy Kit (Qiagen, Hilden, Germany) as described before.8, 9 Gene expression at the mRNA level was determined by qRT-PCR using the LightCycler 480 II system (Roche) and specific primers. Hypoxanthine-guanine phosphoribosyltransferase was used as an internal reference transcript and a pool of human mucosal cDNA for normalization. The following primers were used: CDH1 (universal probe library [UPL] #84): 5′-TGGAGGAATTCTTGCTTTGC-3′ and 5′-CGCTCTCCTCCGAAGAAAC-3′; CRKL (UPL #77): 5′-GGCCCTGCTGGAGTTTTAC-3′ and 5′-TTGGTGGGCTTGGATACCT-3′; SASH1 (UPL #25): 5′-CTTGGCACAGGACTGAGGA-3′ and 5′-GGTCAAAGAGAACCGCACTAA-3′; VIM (UPL #39): 5′-GACCAGCTAACCAACGACAAA-3′ and 5′-GAAGCATCTCCTCCTGCAAT-3′; ZEB1 (UPL #34): 5′-TTTTTCCTGAGGCACCTGAA-3′ and 5′-TGAAAATGCATCTGGTGTTCC-3′; ZEB2 (UPL #6): 5′-GCTCGCACTACAATGCATCA-3′ and 5′-GGGAAATTGATGAATAGCGAAA-3′.
Immunofluorescence Microscopy
Immunofluorescence microscopy was performed as described previously.20 Briefly, cells were fixed with 3% paraformaldehyde in phosphate-buffered saline (PBS) for 20 minutes, permeabilized with 0.1% Triton X-100 (Carl Roth, Karlsruhe, Germany) in PBS for 3 minutes, blocked with 2% (wt/vol) bovine serum albumin (BSA; Sigma-Aldrich) in PBS for 1 hour at room temperature, and incubated with the appropriate primary antibody in blocking buffer overnight at 4°C. Afterward, cells were incubated with Alexa Fluor 488– or Cy3-conjugated secondary antibodies (Thermo Scientific) in blocking buffer for 1 hour at room temperature, before they were mounted on glass slides with Prolong Gold antifade (Life Technologies, Waltham, MA). Images were acquired with a Zeiss AxioObserver Z1 microscope (Carl Zeiss, Jena, Germany).
Transwell Migration and Invasion Assays
Transwell migration (8.0-μm Transwell Permeable Supports, 3422; Costar, Washington DC) and invasion assays (8.0-μm Matrigel Invasion Chamber, 354480; Corning, Corning, NY) were performed according to the manufacturer’s protocol. Briefly, inserts were coated for 2 hours at 37°C with DMEM containing 10% fetal bovine serum. For Transwell assays, 1 × 105 cells were placed on the upper layer of the insert in DMEM without supplements and allowed to migrate toward DMEM containing 10% fetal bovine serum for 20 hours. The same gradient was used for the invasion assay, but with 2 × 105 cells in the upper chamber and a 40-hour incubation time. Afterwards, cells were fixed with 3% paraformaldehyde in PBS for 20 minutes, permeabilized with 0.5% Triton X-100 for 10 minutes, and stained with 4′,6-diamidino-2-phenylindole. Cells at the upper layer of the insert were carefully scratched off. Images of fluorescent nuclei were acquired with a 10× objective (Zeiss AxioObserver Z1) to quantify the number of cells per region with the image analysis software ImageJ v1.51 (National Institutes of Health, Bethesda, MD).
Soft Agar Assay and 3-Dimensional Culture in Matrigel
Soft agar assays for anchorage-independent growth were performed in 12-well plates, in which a bottom layer of 0.5 mL DMEM containing 10% fetal bovine serum and 0.6% agarose was allowed to solidify. Cells (2500 per well) were seeded in 0.5 mL DMEM with 10% fetal bovine serum and 0.4% agarose on top of the bottom layer. After solidification, 0.5 mL DMEM containing 10% fetal bovine serum was added on top of the well. Cells were grown for 2 weeks with medium being replenished every 3 days. The colony number then was counted manually with a 5× objective and an inverted phase-contrast microscope (Zeiss Axiovert 100). In addition, image acquisition of single colonies was performed with a 20× objective in a Zeiss AxioObserver Z1. Colony size then was determined by measuring the area using ImageJ software v1.51 (National Institutes of Health). To culture cells in a 3-dimensional matrix, Matrigel was diluted 1:1 with DMEM containing 10% fetal bovine serum and allowed to solidify in a 96-well plate to generate a bottom layer. On top of this layer, 100 cells were seeded in the same extracellular matrix gel composition. Cell growth was monitored daily and images were acquired after 7 days of incubation (Zeiss AxioObserver Z1).
Luciferase Reporter Assay
The plasmids pGL3 control (Promega), pGL3 basic (Promega), and pGL3 SASH1 (containing the -972 to -34 bp upstream of the SASH1 transcription start site) were co-transfected with the pCMVR renilla reporter plasmid (Promega) in a ratio of 1:1. Cells were grown for 24 hours before media was changed. After 6 hours, cells were incubated with 10 ng/mL TNF (Gibco) for a further 24 hours. Cells were washed with PBS before being lysed in dual-glow luciferase assay lysis buffer. Enzymatic activity was elucidated following the Promega dual-glow luciferase assay kit manual, using a Fluoroskan 2300 device (Thermo Fisher). Relative luciferase activity was calculated as the quotient of firefly-to-renilla luciferase signals.
Mass Spectrometry
Precipitations were performed as described earlier against GFP-only or GFP–SASH1 transiently expressed in HEK293 cells. The samples were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis and gels were stained with Sypro Ruby (Thermo Scientific). Specific gel bands were excised and submitted to reduction (10 mmol/L dithiothreitol in 50 mmol/L ammonium bicarbonate for 30 min at 56°C) and alkylation (50 mmol/L iodoacetamide in 50 mmol/L ammonium bicarbonate for 30 min, room temperature). In-gel digestion was performed using 250 ng trypsin overnight at 37°C, then tryptic peptides were concentrated in a speed vacuum dryer and resuspended in 20 μL of 5% ACN with 0.1% formic acid. Liquid chromatography–mass spectrometry(MS)/MS analyses were performed using an Ultimate 3000 Rapid Separation liquid chromatographic system coupled to an Orbitrap Fusion Tribrid mass spectrometer (Thermo Fisher Scientific). Mass spectrometer settings were as follows: full MS (automatic gain control target, 2E5; resolution, 60K; m/z range, 350–1500; maximum ion injection time, 60 ms); MS/MS (normalized collision energy, 30% ± 5%; detector, ion trap; isolation window, 1.6 m/z; dynamic exclusion time setting, 30 s; automatic gain control target, 2E4; maximum injection time, 100 ms). Fragmentation was permitted for a precursor with a charge state of 2 to 7. Protein identification was performed using Mascot 2.2 (Matrix Science, Boston, MA) with a concatenated forward and reverse SwissProt (Swiss Institute of Bioinformatics, Lausanne, Switzerland) Human database (April 13, 2016, 40,422 entries) generated with DBToolKit. ProteinScape 2.1 (Bruker, Billerica, MA) was used to run Mascot with the following parameters: 1 missed cleavage, carbamidomethylation of Cys as a fixed modification, acetylation of protein N-ter and oxidation of Met as variable modifications, MS tolerance at 5 ppm and MS/MS tolerance at 0.5 daltons, and a protein list compilation by Protein Extractor (University of Alberta, Alberta, Canada). At least 2 peptides with a P value less than .05 (score, 20) were required for protein validation. With these parameters, the false-discovery rate was less than 1%.
Yeast Two-Hybrid Screen
Yeast 2-hybrid screening was performed with the ULTImate Y2HTM platform (Hybrigenics, Paris, France) on a human placenta cDNA library as prey, with human full-length SASH1 (aa1–1247) as bait, cloned in a pB27 plasmid (N-LexA-bait-C fusion; Hybrigenics), according to published protocols.51 The number of processed clones was 447, and 65 million interactions were analyzed. A confidence score previously shown to correlate with the biological significance of the interactions (predicted biological score) was attributed to each interaction, essentially as described elsewhere.52, 53 This score (the predicted biological score) is based first on a local score, considering the redundancy and independency of prey fragments, as well as the distribution of reading frames and stop codons in overlapping fragments. Second, a global score measures the interactions found in all the screens performed at Hybrigenics using the same library. The global score represents the probability of an interaction being nonspecific. Only hits from the highest confidence categories were selected for further analysis, from A (highest confidence, 15 hits), B (high confidence, 9 hits), to C (good confidence, 7 hits), and all other categories (D to F, lowest confidence, 52 hits in total) were discarded from further analysis. Hits from categories A–C were verified independently for their presence in co-immunoprecipitation/mass spectrometry experiments, and curated manually for the number of EMT-related articles by searching PubMed for “<gene name> and EMT.”
Dynamic Mass Redistribution–Based Affinity Measurement
The affinity between SASH1 and CRKL was assessed using a peptide corresponding to the CRKL binding sequence within SASH1 (aa982–991, QPPPVPAKKS, synthesized by JPT Peptide Technologies, Berlin, Germany), and the recombinant SH3N of CRKL. The SH3N domain was fused to a hexahistidine-tag at the C-terminus and purified from Escherichia coli BL21 Star DE3 (Thermo Fisher Scientific). Assay plates (Enspire-LFB, 384-well High Sensitivity, User-Activated Biochemical Plates, Corning Epic System; Corning) were activated with N-hydroxysulfosuccinimide and 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide for 30 minutes at room temperature. After washing 4 times with 25 μL ddH2O, 15 μL of the purified domain at a concentration of 25 μg/mL in 20 mmol/L sodium acetate (pH 5.0) was added to each well and allowed to immobilize overnight at 4°C. The plate then was washed 4 times with 15 μL PBS (pH 7.4), containing 0.005% Tween-20, and equilibrated to room temperature for 3 hours, before the baseline was measured using the Enspire Multimode Plate Reader (PerkinElmer, Waltham, MA). Volumes of 15 μL peptide solution at different concentrations then were added and the final read was performed. Response was calculated by subtracting the baseline from the final peak wavelength.
Orthotopic Mouse Model
HCT116 cells (parental, and clones S1 and S1C2) were transduced with retroviral particles carrying a luciferase encoding vector (LPP-hLUC-Lv201-025-C; GeneCopoeia, Rockville, MD). Cells were selected with 1 μg/mL puromycin for 3 weeks. Bioluminescence of cultured cells was measured in a luminometer (FLUOstar OPTIMA; BMG Labtech GMBH, Ortenberg, Germany). Female SCID/beige mice (Barcelona Biomedical Research Park, Barcelona, Spain) were used at 8 weeks of age. All mice were housed under specific pathogen-free conditions in mixed groups and in accordance with institutional guidelines approved by the Use Committee for Animal Care of the University of Santiago de Compostela (Santiago de Compostela, Spain). For surgery, mice were anesthetized under constant isoflurane application. The cecum was exposed with an incision through the abdominal wall. The luminal content was carefully pushed away from the tip of the cecum and approximately the first 2 mm of the tip of the cecum then was tied up using absorbable surgical sutures. Cells resuspended in ice-cold Matrigel were injected into the enclosed cecal lumen (5 × 106 cells in 25 μL Matrigel). The cell/Matrigel suspension was allowed to rigidify before the surgical wound was closed. Mice were killed upon signs of distress or weight loss greater than 10%, otherwise tumors were allowed to grow for 34 days with regular daily follow-up evaluation. Mice then were injected intraperitoneally with 10 μL/g D-luciferine substrate (P/N 122796; PerkinElmer) before death. Spatially resolved luminescence measurements were performed on whole dissected mice with an IVIS Spectrum In Vivo Imaging System (Xenogen Corp, Alameda, CA; Caliper Life Sciences, Inc, Hopkinton, MA; PerkinElmer, Waltham, MA), and data were analyzed using Living Image 3.1. In 2 of 6 mice injected with SASH1-deficient cells, primary tumors were barely developed, obviously failing to attach to the mucosa. These mice were excluded from further analysis.
Immunohistochemistry
Formalin-fixed tissue samples from the orthotopic xenograft experiments were embedded in paraffin, from which 2-μm sections were generated. These sections were deparaffinized in Roticlear (Carl Roth) 3 times for 10 minutes. Rehydration was performed with an ethanol gradient before samples were boiled in citrate buffer (10 mmol/L, pH 6.0) at 94°C for 20 minutes. After washing with PBS containing 0.1% BSA, peroxidase activity was blocked by incubating the samples in 3% H2O2 in methanol for 5 minutes at room temperature. Samples were washed 3 times with PBS, 0.1% BSA, and then treated with 0.1% Triton X-100 in PBS, 0.1% BSA for 5 minutes at room temperature. The tissue sections then were blocked with 10% goat serum in PBS, 0.1% BSA for 1 hour at room temperature, before they were incubated with the primary antibody in blocking solution overnight at 4°C. As primary antibodies, either anti–E-cadherin (ab40772; Abcam) or anti-mitochondria (ab92824; Abcam) were used. The next day, samples were washed 3 times with PBS, 0.1% BSA, before they were incubated with the appropriate secondary anti-mouse or anti-rabbit antibody–horseradish-peroxidase conjugate (EnVision+ System; DAKO, Agilent, Leuven, Belgium) for 1 hour at room temperature. Samples were washed 3 times with PBS, and chromogen-substrate mixture (DAKO) was added for 8 minutes at room temperature. After washing in ddH2O, sections were counterstained with hematoxylin for 5 minutes at room temperature, washed again with H2O, and mounted with gelatin. Images were acquired with a Zeiss Axiolab microscope (Carl Zeiss).
Analysis of Human Tissue Samples
Seventy-six patients with stage III primary colon cancer were analyzed, who underwent curative surgery (R0) and provided informed consent for analysis of their tissue and data before surgery at the Department of Surgery (Technical University of Munich, Munich, Germany). The tissue was shock-frozen immediately after resection. Clinicopathologic characteristics of the patients are summarized in Figure 7E. Patients with documented local recurrence were not included in this study to circumvent putative bias by poor surgical technique. Ethical and data protection standards of this study were ensured by supervision of the Ethics Committee of the Faculty of Medicine (Technical University of Munich), which approved the study (#1926/07, #5428/12). Tumor-specific overall survival and recurrence-free survival (ie, distant metastasis-free survival) was considered as the primary end point for risk prediction. Statistical evaluation was performed using IBM SPSS Statistics software version 20.0 (SPSS, Inc, Chicago, IL). To derive optimal cut-off values of gene expression levels (SASH1, 0.29; CRKL, 0.44), maximally selected log-rank statistics performed by R Software version 2.13.0 (R Foundation for Statistical Computing, Vienna, Austria) were used. To consider multiple test issues within these analyses, the R-function maxstat.test was used. Survival analysis was performed using Kaplan–Meier estimates. All statistical tests were performed 2-sided, and the significance level was set at 0.05. No correction of P values was applied to adjust for multiple test issues. However, results of all statistical tests being conducted were thoroughly reported so that an informal adjustment of P values could be performed while reviewing the data. The Cancer Genome Atlas TCGA data set was analyzed for prognostic association of SASH1 and CRKL genes with the cBioPortal platform, consisting of 213 fully sequenced colorectal cancer samples with available follow-up documentation.29, 30
Statistical Evaluation
Statistical analysis was performed with GraphPad Prism 6.01 (GraphPad Software, San Diego, CA). All error bars represent the SD. Statistical tests, P values, and sample size are indicated in figure legends.
Acknowledgments
The authors wish to thank Alexandra Gnann and Widya Johannes for excellent technical assistance, Dr Julia Slotta-Huspenina for histopathologic evaluation of tissue samples, and Maximilian Ehrenfeld, Dr Bernhard Holzmann, and Dr Melanie Laschinger for critical discussion. The authors thank the 3P5 proteomic facility (Institut Cochin, Paris, France) for liquid chromatography–MS/MS data acquisition.
Footnotes
Current address of H.W.: Institute of Clinical Chemistry and Laboratory Medicine, University Medical Center Hamburg-Eppendorf, Hamburg, Germany. Current address of J.M.: Max Planck Institute of Biochemistry, Research Dept. Proteomics and Signal Transduction, Martinsried, Germany.
Author contributions Fabian Christoph Franke and Klaus-Peter Janssen were responsible for the study concept and design; Fabian Christoph Franke, Johannes Müller, Eduardo Domínguez Medina, Henri Weidmann, and Solenne Chardonnet acquired data; Fabian Christoph Franke, Johannes Müller, Miguel Abal, Eduardo Domínguez Medina, Ulrich Nitsche, Henri Weidmann, Solenne Chardonnet, Ewa Ninio, and Klaus-Peter Janssen analyzed and interpreted data; Fabian Christoph Franke, Johannes Müller, Solenne Chardonnet, and Klaus-Peter Jansse collected and assembled data; Fabian Christoph Franke and Klaus-Peter Janssen drafted the manuscript; Klaus-Peter Janssen critically revised the manuscript for important intellectual content; Ulrich Nitsche performed the statistical analysis; Ewa Ninio and Klaus-Peter Janssen obtained funding; Miguel Abal provided technical and material support; and Klaus-Peter Janssen supervised the study.
Conflicts of interest The authors disclose no conflicts.
Funding Supported by the Institut National de la Santé et de la Recherche Médicale and Fondation de France (201300038584 for project R13072DD-Bigot) (E.N.). This study was initiated based on findings obtained by grant 111822 from the Deutsche Krebshilfe e.V. (K.-P.J.).
References
- 1.Mehlen P., Puisieux A. Metastasis: a question of life or death. Nat Rev Cancer. 2006;6:449–458. doi: 10.1038/nrc1886. [DOI] [PubMed] [Google Scholar]
- 2.Kalluri R., Weinberg R.A. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lamouille S., Xu J., Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peinado H., Olmeda D., Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7:415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 5.Shirakihara T., Saitoh M., Miyazono K. Differential regulation of epithelial and mesenchymal markers by deltaEF1 proteins in epithelial mesenchymal transition induced by TGF-beta. Mol Biol Cell. 2007;18:3533–3544. doi: 10.1091/mbc.E07-03-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krebs A.M., Mitschke J., Lasierra Losada M., Schmalhofer O., Boerries M., Busch H., Boettcher M., Mougiakakos D., Reichardt W., Bronsert P., Brunton V.G., Pilarsky C., Winkler T.H., Brabletz S., Stemmler M.P., Brabletz T. The EMT-activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nat Cell Biol. 2017;19:518–529. doi: 10.1038/ncb3513. [DOI] [PubMed] [Google Scholar]
- 7.Zeller C., Hinzmann B., Seitz S., Prokoph H., Burkhard-Goettges E., Fischer J., Jandrig B., Schwarz L.E., Rosenthal A., Scherneck S. SASH1: a candidate tumor suppressor gene on chromosome 6q24.3 is downregulated in breast cancer. Oncogene. 2003;22:2972–2983. doi: 10.1038/sj.onc.1206474. [DOI] [PubMed] [Google Scholar]
- 8.Rimkus C., Martini M., Friederichs J., Rosenberg R., Doll D., Siewert J.R., Holzmann B., Janssen K.P. Prognostic significance of downregulated expression of the candidate tumour suppressor gene SASH1 in colon cancer. Br J Cancer. 2006;95:1419–1423. doi: 10.1038/sj.bjc.6603452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nitsche U., Rosenberg R., Balmert A., Schuster T., Slotta-Huspenina J., Herrmann P., Bader F.G., Friess H., Schlag P.M., Stein U., Janssen K.P. Integrative marker analysis allows risk assessment for metastasis in stage II colon cancer. Ann Surg. 2012;256:763–771. doi: 10.1097/SLA.0b013e318272de87. discussion 771. [DOI] [PubMed] [Google Scholar]
- 10.Burgess J.T., Bolderson E., Saunus J.M., Zhang S.D., Reid L.E., McNicol A.M., Lakhani S.R., Cuff K., Richard K., Richard D.J., O'Byrne K.J. SASH1 mediates sensitivity of breast cancer cells to chloropyramine and is associated with prognosis in breast cancer. Oncotarget. 2016;7:72807–72818. doi: 10.18632/oncotarget.12020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang L., Zhang H., Yao Q., Yan Y., Wu R., Liu M. Clinical significance of SASH1 expression in glioma. Dis Markers. 2015;2015:383046. doi: 10.1155/2015/383046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meng Q., Zheng M., Liu H., Song C., Zhang W., Yan J., Qin L., Liu X. SASH1 regulates proliferation, apoptosis, and invasion of osteosarcoma cell. Mol Cell Biochem. 2013;373:201–210. doi: 10.1007/s11010-012-1491-8. [DOI] [PubMed] [Google Scholar]
- 13.Peng L., Wei H., Liren L. Promoter methylation assay of SASH1 gene in hepatocellular carcinoma. J BUON. 2014;19:1041–1047. [PubMed] [Google Scholar]
- 14.Shellman Y.G., Lambert K.A., Brauweiler A., Fain P., Spritz R.A., Martini M., Janssen K.P., Box N.F., Terzian T., Rewers M., Horvath A., Stratakis C.A., Robinson W.A., Robinson S.E., Norris D.A., Artinger K.B., Pacheco T.R. SASH1 is involved in an autosomal dominant lentiginous phenotype. J Invest Dermatol. 2015;135:3192–3194. doi: 10.1038/jid.2015.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weidmann H., Touat-Hamici Z., Durand H., Mueller C., Chardonnet S., Pionneau C., Charlotte F., Janssen K.P., Verdugo R., Cambien F., Blankenberg S., Tiret L., Zeller T., Ninio E. SASH1, a new potential link between smoking and atherosclerosis. Atherosclerosis. 2015;242:571–579. doi: 10.1016/j.atherosclerosis.2015.08.013. [DOI] [PubMed] [Google Scholar]
- 16.Wang J., Zhang J., Li X., Wang Z., Lei D., Wang G., Li J., Zhang S., Li Z., Li M. A novel de novo mutation of the SASH1 gene in a Chinese family with multiple lentigines. Acta Derm Venereol. 2017;97:530–531. doi: 10.2340/00015555-2575. [DOI] [PubMed] [Google Scholar]
- 17.Courcet J.B., Elalaoui S.C., Duplomb L., Tajir M., Riviere J.B., Thevenon J., Gigot N., Marle N., Aral B., Duffourd Y., Sarasin A., Naim V., Courcet-Degrolard E., Aubriot-Lorton M.H., Martin L., Abrid J.E., Thauvin C., Sefiani A., Vabres P., Faivre L. Autosomal-recessive SASH1 variants associated with a new genodermatosis with pigmentation defects, palmoplantar keratoderma and skin carcinoma. Eur J Hum Genet. 2015;23:957–962. doi: 10.1038/ejhg.2014.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dauphinee S.M., Clayton A., Hussainkhel A., Yang C., Park Y.J., Fuller M.E., Blonder J., Veenstra T.D., Karsan A. SASH1 is a scaffold molecule in endothelial TLR4 signaling. J Immunol. 2013;191:892–901. doi: 10.4049/jimmunol.1200583. [DOI] [PubMed] [Google Scholar]
- 19.Wang H., Wang H.S., Zhou B.H., Li C.L., Zhang F., Wang X.F., Zhang G., Bu X.Z., Cai S.H., Du J. Epithelial-mesenchymal transition (EMT) induced by TNF-alpha requires AKT/GSK-3beta-mediated stabilization of snail in colorectal cancer. PLoS One. 2013;8:e56664. doi: 10.1371/journal.pone.0056664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martini M., Gnann A., Scheikl D., Holzmann B., Janssen K.P. The candidate tumor suppressor SASH1 interacts with the actin cytoskeleton and stimulates cell-matrix adhesion. Int J Biochem Cell Biol. 2011;43:1630–1640. doi: 10.1016/j.biocel.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 21.Bell E.S., Park M. Models of crk adaptor proteins in cancer. Genes Cancer. 2012;3:341–352. doi: 10.1177/1947601912459951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arai A., Nosaka Y., Kohsaka H., Miyasaka N., Miura O. CrkL activates integrin-mediated hematopoietic cell adhesion through the guanine nucleotide exchange factor C3G. Blood. 1999;93:3713–3722. [PubMed] [Google Scholar]
- 23.Senechal K., Heaney C., Druker B., Sawyers C.L. Structural requirements for function of the Crkl adapter protein in fibroblasts and hematopoietic cells. Mol Cell Biol. 1998;18:5082–5090. doi: 10.1128/mcb.18.9.5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sakkab D., Lewitzky M., Posern G., Schaeper U., Sachs M., Birchmeier W., Feller S.M. Signaling of hepatocyte growth factor/scatter factor (HGF) to the small GTPase Rap1 via the large docking protein Gab1 and the adapter protein CRKL. J Biol Chem. 2000;275:10772–10778. doi: 10.1074/jbc.275.15.10772. [DOI] [PubMed] [Google Scholar]
- 25.Ungewiss C., Rizvi Z.H., Roybal J.D., Peng D.H., Gold K.A., Shin D.H., Creighton C.J., Gibbons D.L. The microRNA-200/Zeb1 axis regulates ECM-dependent beta1-integrin/FAK signaling, cancer cell invasion and metastasis through CRKL. Sci Rep. 2016;6:18652. doi: 10.1038/srep18652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lamorte L., Rodrigues S., Sangwan V., Turner C.E., Park M. Crk associates with a multimolecular Paxillin/GIT2/beta-PIX complex and promotes Rac-dependent relocalization of Paxillin to focal contacts. Mol Biol Cell. 2003;14:2818–2831. doi: 10.1091/mbc.E02-08-0497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li L., Guris D.L., Okura M., Imamoto A. Translocation of CrkL to focal adhesions mediates integrin-induced migration downstream of Src family kinases. Mol Cell Biol. 2003;23:2883–2892. doi: 10.1128/MCB.23.8.2883-2892.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thibaudeau L., Taubenberger A.V., Holzapfel B.M., Quent V.M., Fuehrmann T., Hesami P., Brown T.D., Dalton P.D., Power C.A., Hollier B.G., Hutmacher D.W. A tissue-engineered humanized xenograft model of human breast cancer metastasis to bone. Dis Model Mech. 2014;7:299–309. doi: 10.1242/dmm.014076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E., Antipin Y., Reva B., Goldberg A.P., Sander C., Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E., Cerami E., Sander C., Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fischer K.R., Durrans A., Lee S., Sheng J., Li F., Wong S.T., Choi H., El Rayes T., Ryu S., Troeger J., Schwabe R.F., Vahdat L.T., Altorki N.K., Mittal V., Gao D. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature. 2015;527:472–476. doi: 10.1038/nature15748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng X., Carstens J.L., Kim J., Scheible M., Kaye J., Sugimoto H., Wu C.C., LeBleu V.S., Kalluri R. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature. 2015;527:525–530. doi: 10.1038/nature16064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thiery J.P. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 34.Pan J., Liu Y. SASH1 inhibits hypoxia-induced epithelial-to-mesenchymal transition via suppression of the PI3K/Akt pathway in human pancreatic cancer cells. Int J Clin Exp Pathol. 2016;9:750–757. [Google Scholar]
- 35.Zong W., Yu C., Wang P., Dong L. Overexpression of SASH1 inhibits TGF-beta1-induced EMT in gastric cancer cells. Oncol Res. 2016;24:17–23. doi: 10.3727/096504016X14570992647203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aiello N.M., Brabletz T., Kang Y., Nieto M.A., Weinberg R.A., Stanger B.Z. Upholding a role for EMT in pancreatic cancer metastasis. Nature. 2017;547:E7–E8. doi: 10.1038/nature22963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ye X., Brabletz T., Kang Y., Longmore G.D., Nieto M.A., Stanger B.Z., Yang J., Weinberg R.A. Upholding a role for EMT in breast cancer metastasis. Nature. 2017;547:E1–E3. doi: 10.1038/nature22816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsumoto R., Tsuda M., Wang L., Maishi N., Abe T., Kimura T., Tanino M., Nishihara H., Hida K., Ohba Y., Shinohara N., Nonomura K., Tanaka S. Adaptor protein CRK induces epithelial-mesenchymal transition and metastasis of bladder cancer cells through HGF/c-Met feedback loop. Cancer Sci. 2015;106:709–717. doi: 10.1111/cas.12662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lamorte L., Royal I., Naujokas M., Park M. Crk adapter proteins promote an epithelial-mesenchymal-like transition and are required for HGF-mediated cell spreading and breakdown of epithelial adherens junctions. Mol Biol Cell. 2002;13:1449–1461. doi: 10.1091/mbc.01-10-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamada S., Yanamoto S., Kawasaki G., Rokutanda S., Yonezawa H., Kawakita A., Nemoto T.K. Overexpression of CRKII increases migration and invasive potential in oral squamous cell carcinoma. Cancer Lett. 2011;303:84–91. doi: 10.1016/j.canlet.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 41.Han G., Wu D., Yang Y., Li Z., Zhang J., Li C. CrkL meditates CCL20/CCR6-induced EMT in gastric cancer. Cytokine. 2015;76:163–169. doi: 10.1016/j.cyto.2015.05.009. [DOI] [PubMed] [Google Scholar]
- 42.Cheng S., Guo J., Yang Q., Yang X. Crk-like adapter protein regulates CCL19/CCR7-mediated epithelial-to-mesenchymal transition via ERK signaling pathway in epithelial ovarian carcinomas. Med Oncol. 2015;32:47. doi: 10.1007/s12032-015-0494-1. [DOI] [PubMed] [Google Scholar]
- 43.Elmansuri A.Z., Tanino M.A., Mahabir R., Wang L., Kimura T., Nishihara H., Kinoshita I., Dosaka-Akita H., Tsuda M., Tanaka S. Novel signaling collaboration between TGF-beta and adaptor protein Crk facilitates EMT in human lung cancer. Oncotarget. 2016;7:27094–27107. doi: 10.18632/oncotarget.8314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jankowski W., Saleh T., Pai M.T., Sriram G., Birge R.B., Kalodimos C.G. Domain organization differences explain Bcr-Abl's preference for CrkL over CrkII. Nat Chem Biol. 2012;8:590–596. doi: 10.1038/nchembio.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Birge R.B., Kalodimos C., Inagaki F., Tanaka S. Crk and CrkL adaptor proteins: networks for physiological and pathological signaling. Cell Commun Signal. 2009;7:13. doi: 10.1186/1478-811X-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barbazan J., Alonso-Alconada L., Elkhatib N., Geraldo S., Gurchenkov V., Glentis A., van Niel G., Palmulli R., Fernandez B., Viano P., Garcia-Caballero T., Lopez-Lopez R., Abal M., Vignjevic D.M. Liver metastasis is facilitated by the adherence of circulating tumor cells to vascular fibronectin deposits. Cancer Res. 2017;77:3431–3441. doi: 10.1158/0008-5472.CAN-16-1917. [DOI] [PubMed] [Google Scholar]
- 47.Kumar S., Lu B., Davra V., Hornbeck P., Machida K., Birge R.B. Crk tyrosine phosphorylation regulates PDGF-BB-inducible Src activation and breast tumorigenicity and metastasis. Mol Cancer Res. 2018;16:173–183. doi: 10.1158/1541-7786.MCR-17-0242. [DOI] [PubMed] [Google Scholar]
- 48.Watanabe T., Tsuda M., Makino Y., Konstantinou T., Nishihara H., Majima T., Minami A., Feller S.M., Tanaka S. Crk adaptor protein-induced phosphorylation of Gab1 on tyrosine 307 via Src is important for organization of focal adhesions and enhanced cell migration. Cell Res. 2009;19:638–650. doi: 10.1038/cr.2009.40. [DOI] [PubMed] [Google Scholar]
- 49.Zheng L., Baumann U., Reymond J.L. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 2004;32:e115. doi: 10.1093/nar/gnh110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rain J.C., Cribier A., Gerard A., Emiliani S., Benarous R. Yeast two-hybrid detection of integrase-host factor interactions. Methods. 2009;47:291–297. doi: 10.1016/j.ymeth.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 52.Rain J.C., Selig L., De Reuse H., Battaglia V., Reverdy C., Simon S., Lenzen G., Petel F., Wojcik J., Schachter V., Chemama Y., Labigne A., Legrain P. The protein-protein interaction map of Helicobacter pylori. Nature. 2001;409:211–215. doi: 10.1038/35051615. [DOI] [PubMed] [Google Scholar]
- 53.Formstecher E., Aresta S., Collura V., Hamburger A., Meil A., Trehin A., Reverdy C., Betin V., Maire S., Brun C., Jacq B., Arpin M., Bellaiche Y., Bellusci S., Benaroch P., Bornens M., Chanet R., Chavrier P., Delattre O., Doye V., Fehon R., Faye G., Galli T., Girault J.A., Goud B., de Gunzburg J., Johannes L., Junier M.P., Mirouse V., Mukherjee A., Papadopoulo D., Perez F., Plessis A., Rosse C., Saule S., Stoppa-Lyonnet D., Vincent A., White M., Legrain P., Wojcik J., Camonis J., Daviet L. Protein interaction mapping: a Drosophila case study. Genome Res. 2005;15:376–384. doi: 10.1101/gr.2659105. [DOI] [PMC free article] [PubMed] [Google Scholar]