Abstract
Ultrafast affinity extraction was evaluated and used with microcolumns containing human serum albumin (HSA) to measure the global affinity constants and dissociation rate constants for several second- and third-generation sulfonylurea drugs with solution-phase normal HSA or glycated HSA. Glibenclamide, glimepiride and glipizide were used as model drugs for this work. Both single- and two-column systems were considered for the analysis of global affinities for the model drugs. These methods were optimized with respect to the flow rates, column sizes and sample residence times that were employed with each drug for ultrafast affinity extraction. Data acquired with single-column systems were further utilized to estimate the dissociation rate constants for normal HSA and glycated HSA with the given drugs. The binding constants obtained by the single- and two-column systems showed good agreement with each other and with values obtained from the literature. Use of a single-column system indicated that levels of glycation found in controlled or advanced diabetes resulted in a 18-44% decrease in the overall binding strength of the model drugs with HSA. Although the two-column system allowed work with smaller free drug fractions and clinically-relevant drug/protein concentrations, the single-column system required less protein, provided better precision, and was easier to use in binding studies.
Keywords: Ultrafast affinity extraction, Drug-protein binding, Sulfonylurea drugs, Human serum albumin, Glycation, Diabetes
Introduction
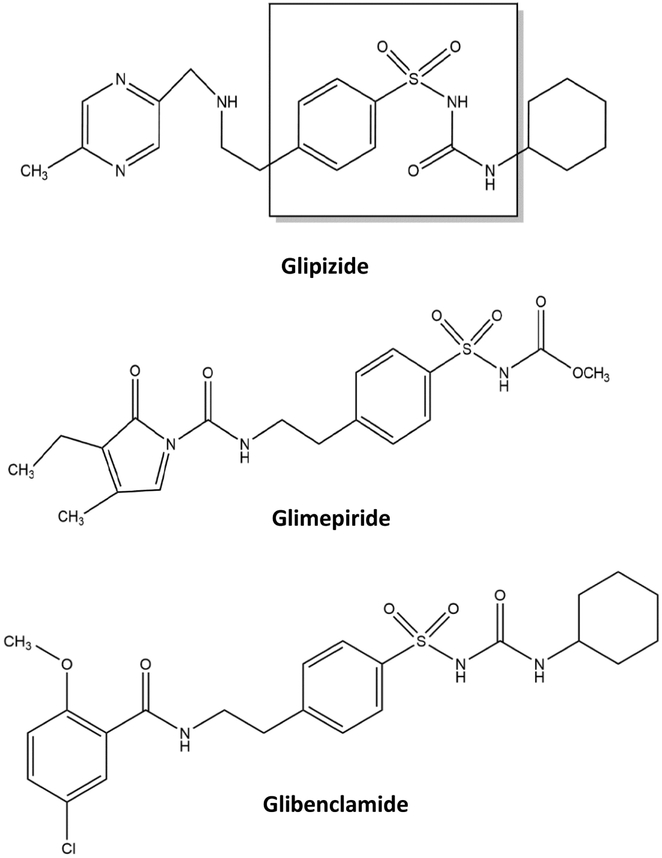
It is estimated that over 360 million people in the world have diabetes [1-3], with type 2 diabetes accounting for 90% or more of these cases [3-5]. In type 2 diabetes, the body exhibits insulin resistance and does not use insulin properly or produce enough insulin for proper glucose control [5]. This disease can be treated by stimulating the secretion of insulin from β cells in the pancreas, such as by using sulfonylurea drugs (see Figure 1) [2,6,7]. Sulfonylurea drugs are usually categorized as being first-, second- or third-generation and vary in terms of their dosages and effectiveness in treating type 2 diabetes [8,9]. For example, the second-generation sulfonylurea drugs glibenclamide and glipizide have therapeutic ranges in serum of only 0.08-0.4 μM and 0.22-2.24 μM, respectively, while the first-generation sulfonylurea drug tolbutamide has a therapeutic range of 185-370 μM [11].
Figure 1.
Structures of several second- or third-generation sulfonylurea drugs (Note: glibenclamide is also known as glyburide). The portion in the dashed box shows the core structure of a sulfonylurea drug.
All categories of sulfonylurea drugs have been found to bind human serum albumin (HSA) in the circulatory system [9,10,12-21]. HSA, which has a molar mass of 66.5 kDa, is the most abundant protein in blood and acts as a solubilizing and/or transporting agent for a variety of drugs and other small solutes [22]. There are several sites on HSA that can interact with sulfonylurea drugs. Two regions that have been noted to bind many sulfonylurea drugs are Sudlow sites I and II, which are located in subdomains IIA and IIIA of HSA [9,10,12-15,17,18,21-24]. The digitoxin site of HSA has also been found to bind some sulfonylurea drugs [17]. These sites are all relatively well-defined and often result in multi-site saturable interactions between sulfonylurea drugs and this protein [9,10,12-19,22-24].
HSA and other serum proteins can be modified by glucose during diabetes through a process known as glycation [25-29]. Glycation is a non-enzymatic reaction in which an amine group on a protein undergoes reversible coupling with glucose to form a Schiff base, which may then rearrange to form a stable ketoamine [25,26]. Healthy individuals can have 11-16% of their HSA present in a glycated form, an amount that can increase up to two- to five-fold in diabetic patients [25,29]. This type of modification has been of recent interest in that it has been shown that glycation can alter the interactions between HSA and sulfonylureas [9,10,12-20,25,27,28], which in turn can alter the non-bound and bioavailable forms of these drugs in blood [16,25]. Glycation has also been found to alter the binding of other classes of compounds with HSA (e.g., polyphenols and L-tryptophan) [25,30-32].
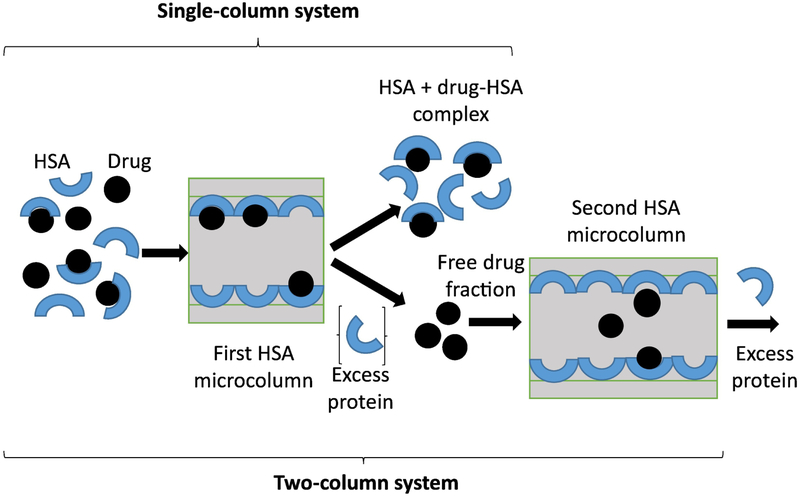
A variety of spectroscopic or separation-based methods have been used to study the changes glycation has on the binding of HSA to sulfonylureas and other pharmaceutical agents [20,21,25,27,28]. This has included a number of methods that have used covalently immobilized, adsorbed or entrapped samples of normal HSA or glycated HSA in high-performance affinity chromatography (HPAC) [9,10,12-15,17-19,33,34]. An alternative approach that has been recently considered for such work is ultrafast affinity extraction [16,35-37]. This latter method makes use of small HPAC columns to isolate a free drug fraction from a sample on the millisecond time scale (e.g., see possible schemes in Figure 2, as described later in more detail) [16,37]. This method has recently been shown to be a convenient technique for quickly studying drug- or solute-protein interactions in solution, and without the need for labeling either the drug/solute or protein [16,35-37].
Figure 2.
General process in ultrafast affinity extraction for the separation of a free drug fraction from a protein and drug-protein complex by using either a single- or two-column system. This example is for a drug that binds to soluble HSA and that uses an HSA microcolumn to isolate and measure the captured free drug fraction. The second HSA microcolumn is used to further isolate the captured free drug fraction from any co-eluting excess protein or non-retained sample components, as can be employed during the measurement of small free fractions or in work with samples that contain high protein concentrations.
This study will evaluate and use various schemes based on ultrafast affinity extraction to examine the binding strength and interaction rates of normal or glycated HSA with various second- or third-generation sulfonylurea drugs. The drugs that will be used as models in this study (see Figure 1) will include glipizide and glibenclamide (i.e., second-generation sulfonylureas), as well as glimepiride (a third-generation sulfonylurea) [9,16-18]. The binding of these drugs with immobilized samples of HSA has previously been examined by traditional HPAC methods such as frontal analysis and zonal elution competition studies [9,10,17,18]. In addition, preliminary work in using ultrafast affinity extraction for free fraction measurements has been reported for glibenclamide [16], which makes this drug useful as a reference compound in examining the general robustness and reproducibility of the experimental conditions required for this approach.
In this report, both single- and two-column systems will be developed and optimized to measure the global affinities for these drugs with normal HSA and glycated HSA. The single-column systems will also be used to estimate the dissociation rate constants for the given drugs with these protein preparations. These results will make it possible to directly compare the relative advantages or limitations for the single- and two-column methods in examining the interactions of various drugs with soluble HSA, as well as on the experimental conditions that are needed for such studies. In addition, the information acquired with these techniques will provide further data on how the process of glycation may alter the strength and rates of these interactions during diabetes [9,12-14].
2. Experimental
2.1. Reagents
The glibenclamide (≥ 99% pure), glimepiride (≥ 99%), glipizide (≥ 96%), and HSA (Cohn fraction V, essentially fatty acid free, ≥ 96%) were from Sigma (St. Louis, MO, USA). Nucleosil Si-300 silica (7 μm particle diameter, 300 Å pore size) was acquired from Macherey Nagel (Dűren, Germany). The components for the bicinchoninic acid (BCA) protein assay were purchased from Pierce (Rockford, IL, USA). All aqueous solutions and buffers were prepared using water that was generated by a NANOpure system (Barnstead, Dubuque, IA, USA) and were passed through Osmonics 0.22 μM nylon filters from Fisher Scientific (Pittsburgh, PA, USA).
2.2. Apparatus
The microcolumns were packed by utilizing a Prep 24 pump from ChromTech (Apple Valley, MN, USA). The HPLC system consisted of an AS-2057 autosampler, a PU-2080 Plus pump, and a UV-2075 absorbance detector from Jasco (Easton, MD, USA), and included a six-port LabPro valve (Rheodyne, Cotati, CA, USA). A Jasco X-LC 3167CO column oven was used to maintain a temperature of 37.0 (± 0.1) °C during all the chromatographic experiments. The HPLC system was controlled by using ChromNAV v1.18.04 and LCNet software from Jasco. The chromatographic data were analyzed by using PeakFit v4.12 software (Jandel Scientific, San Rafael, CA, USA).
2.3. Protein glycation
The glycated HSA samples were made in vitro, as described in previous papers [13,14,32]. The HSA was incubated in two separate batches with 15 mM or 30 mM glucose (i.e., giving final samples which will be referred to as “gHSA1” and “gHSA2”, respectively) under sterile conditions for four weeks at a physiological concentration of HSA and at pH 7.4 and 37°C (i.e., conditions that mimic the glycation of HSA in the circulation). After the incubation was finished, the glycated HSA was purified according to prior methods [13,14,38] and stored at 4°C. The level of modification was measured by using a glycated serum protein assay from Diazyme (San Diego, CA, USA). This assay gave a measured glycation level of 1.40 (± 0.06) mol hexose/mol HSA for gHSA1 and 3.24 (± 0.07) mol hexose/mol HSA for gHSA2 (Note: the values in parentheses represent ± 1 S.D.). These glycation levels for gHSA1 and gHSA2 were representative of values that would be seen for patients with mild or advanced diabetes, respectively [13,25,38].
2.3. Microcolumn preparation
The microcolumns used in this study were prepared by using Nucleosil Si-300 silica as the starting material. This support was converted into a diol-bonded form and then used in the Schiff base method for the immobilization of HSA [35,38]. A control support was made in the same manner but with no protein being added during the immobilization step. The protein content of these supports was measured by using a BCA protein assay, using the control support as the blank and soluble HSA as the standard. The final support used for studies involving drug interactions with normal HSA had a measured protein content of 53.6 (± 2.2) mg HSA/g silica, and the support used for work that examined drug binding with glycated HSA contained 58.7 (± 3.5) mg HSA/g silica.
The microcolumns that were used in this report were based on the column designs described in Ref. [16] and had lengths of 1.0 mm, 5.0 mm or 10.0 mm and an inner diameter of 2.1 mm. The microcolumns were packed at 27.6 MPa (4000 psi) by using pH 7.4, 0.067 M potassium phosphate buffer as the packing solution. The same buffer was placed within these columns for their storage at 4°C when not in use.
2.3. Ultrafast affinity extraction
In both the single-column and two-column systems that were employed for ultrafast affinity extraction (see Figure 2), the first HSA microcolumn was used to extract a free drug fraction from a sample. In the two-column system, a longer second HSA column was later placed on-line with the first to further isolate the extracted free fraction from other sample components (e.g., any remaining non-retained HSA from samples with high protein contents) [16]. The samples used on the single-column system contained 10 or 20 μM HSA and 10 μM of the drug of interest in pH 7.4, 0.067 M phosphate buffer, with the same buffer also being used as the mobile phase. In studies with the two-column system, the samples contained the following therapeutic drug concentrations: glibenclamide, 0.08 μM; glimepiride, 0.4 μM; or glipizide, 2.24 μM [11,39]. The concentration of HSA in these latter samples was 500 or 600 μM, as chosen to represent a physiological level for this protein (496-782 μM) [40]. All drug/HSA mixtures were mixed and incubated at 37°C for at least 30 min before injection to reach equilibrium between the drug and its drug-protein complex in the sample [16]. A 1 or 5 μL injection volume was used, and all samples were analyzed in quadruplicate.
The HSA microcolumn used in a single-column system or as the first column in a two-column system had the following dimensions: 5.0 mm × 2.1 mm i.d. for glipizide or glimepiride; and 1.0 mm × 2.1 mm i.d. for glibenclamide. The size of second HSA column in the two-column system was 10.0 mm × 2.1 mm i.d. for glipizide or glimepiride, and 5.0 mm × 2.1 mm i.d. for glibenclamide. The final optimized flow rates for this system are provided in Section 3. The wavelengths used for absorbance detection were as follows: 242 nm for glibenclamide, 275 nm for glipizide, and 245 nm for glimepiride. The peaks for the retained free drug fractions were processed by using the autofit and subtract baseline functions of PeakFit 4.12 and were fit to exponentially-modified Gaussian curves [16].
3. Results and discussion
3.1. Optimization of single-column systems for ultrafast affinity extraction
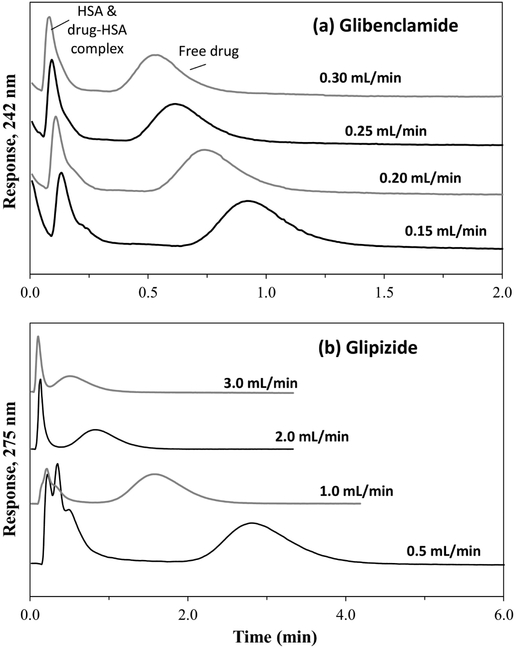
Before ultrafast affinity extraction could be used to measure free drug fractions, there were a number of conditions that had to be considered [16,35]. First, the residence time for the sample in the affinity microcolumn had to be sufficiently small to avoid significant dissociation of the protein-bound drug fraction in the sample. This factor was varied by adjusting the microcolumn size and flow rate [16,35-37]. Changing the size of the microcolumn also affected the degree of retention and separation of the retained free drug fractions from the non-retained sample components (see Figure 3). A drug such as glibenclamide, which has strong binding to HSA (global affinity constant, ~2 × 106 M−1) [16,17], was found to have good retention and resolution for its free drug fraction at low-to-moderate flow rates on a 1.0 mm × 2.1 mm I.D. microcolumn. A longer 5.0 mm × 2.1 mm microcolumn and higher flow rates were needed for glipizide, which has a moderate binding strength for HSA (global affinity constant, ~4-5 × 105 M−1) [18]. As is illustrated in Figure 3, these column sizes allowed the captured free drug fractions to be eluted and measured within 1.0 min at the higher flow rates that were used in this study and within 1.5-4.0 min at the lower end of each flow rate range that was employed.
Figure 3.
Typical chromatograms obtained at pH 7.4 and 37°C on (a) a 1.0 mm × 2.1 mm i.d. HSA microcolumn for 5 μL injections of 10 μM glibenclamide plus 10 μM HSA or (b) a 5.0 mm × 2.1 mm i.d. HSA microcolumn for 1 μL injections of 10 μM glipizide plus 20 μM HSA.
Along with retention, another factor that was considered when selecting the conditions for these experiments was the variation of back pressure with column size [16]. The microcolumns that were used in this report for ultrafast affinity extraction had sizes ranging from 1.0 to 5.0 mm × 2.1 mm i.d., with the longer columns showing proportionally higher back pressures at any given flow rate. These microcolumns had back pressures up to only 4.0 MPa (580 psi) even at flow rates as high as 4.0 mL/min, which were well within the typical usable range of 28-41 MPa (4000-6000 psi) for a standard HPLC system.
Studies were next conducted with each drug and an appropriately-sized microcolumn to find the optimum flow rate range that could be used to isolate and retain the drug’s free fraction in the presence of HSA. These experiments were carried out by injecting samples containing 10 μM of only the desired drug or 10 μM of this drug combined with 10 or 20 μM of normal HSA. The following sizes were employed in this work for the HSA microcolumns: 5.0 mm × 2.1 mm i.d. for glipizide and glimepiride, and 1.0 mm × 2.1 mm i.d. for glibenclamide. The injection of each drug alone gave quantitative extraction (i.e., >95%) by these microcolumns at the flow rates that were tested; this was in agreement with results noted previously with other drugs/solutes and similar microcolumns [16,35,36]. In each case, the addition of soluble HSA to the sample resulted in a lower peak for the retained drug because part of this drug was now bound within a non-retained drug-protein complex [16].
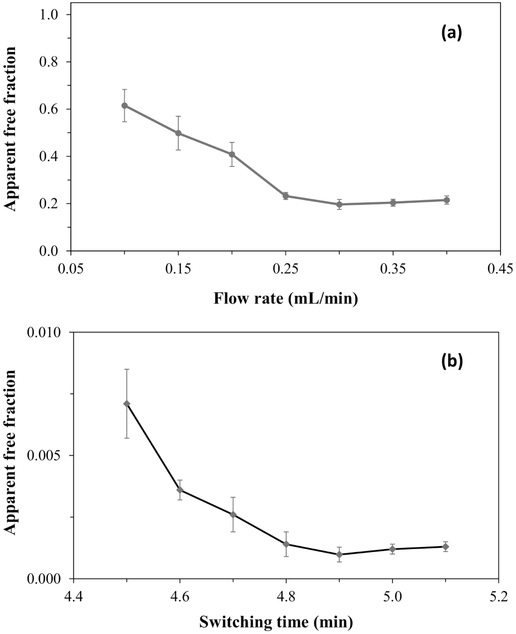
The size of the apparent free drug fraction in the drug/HSA sample mixtures decreased as the injection flow rate was increased (i.e., as the residence time in the microcolumn decreased) but reached a steady value at higher flow rates. This behavior was seen for each drug, as is illustrated for glibenclamide in Figure 4(a) (see Supplemental Material for additional examples). Such an effect has been noted for other drugs and solutes during ultrafast affinity extraction [16,35,36] and is due to dissociation of these drugs/solutes from their complexes with proteins in the sample during passage through a column at low-to-moderate flow rates.
Figure 4.
Effects in ultrafast affinity extraction on the measurement of free drug fractions when (a) changing the injection flow rate on a single-column system or (b) altering the switching time for placing the second column on-line in a two-column system. These effects are illustrated by using data for samples containing a mixture of glibenclamide and soluble normal HSA. The single-column system used a 1.0 mm × 2.1 mm i.d. HSA microcolumn; the two-column system used the same first column, operated at 0.3 mL/min, followed by the addition of a 5.0 mm × 2.1 mm I.D. HSA microcolumn that was operated at 0.15 mL/min. The samples in (a) contained 10 μM glibenclamide +10 μM HSA, while the samples in (b) contained 0.08 μM glibenclamide + 500 μM HSA. The error bars represent a range of ± 1 S.D.
It has been shown in prior work that this dissociation effect can be minimized by raising the flow rate (i.e., decreasing the residence time in the microcolumn) to a given threshold level [16,35,36]. For instance, glibenclamide gave a consistent free fraction in the presence of normal HSA when using an injection flow rate of 0.30 mL/min or greater on a 1.0 mm × 2.1 mm i.d. HSA microcolumn. These conditions were in agreement with those that have been utilized in prior work for the measurement of free glibenclamide fractions with a two-column system [16]. Glimepiride gave a consistent free fraction at a flow rate of 2.5-3.0 mL/min or greater when using a 5.0 mm × 2.1 mm i.d. HSA microcolumn, and glipizide gave a consistent free fraction at an injection flow rate of 3.0-3.5 mL/min or greater on a 5.0 mm × 2.1 mm i.d. HSA microcolumn. The maximum column residence times for the non-retained components under these conditions were as follows: glibenclamide, ~550-560 ms; glimepiride, ~280-330 ms; and glipizide, ~240-280 ms. These column residence times agreed with the general range of a few-to-several hundred milliseconds that have been found to work for other drug/serum transport protein systems that have been examined by ultrafast affinity extraction [16,35,37,41]. In addition, this range of residence times was consistent with what has been reported in the use of a two-column system for examining the original free fractions of other sulfonylurea drugs in the presence of normal HSA (e.g., ~330-670 ms for acetohexamide and gliclazide, which have global affinities for HSA of around 1.7 × 105 M−1 and 0.7-0.8 × 105 M−1, respectively) [16].
3.2. Analysis of sulfonylurea binding to HSA by single-column ultrafast affinity extraction
The free fractions measured by single-column ultrafast affinity extraction at high flow rates, as optimized in Section 3.1, were next employed to estimate the global affinity constants for each drug in the presence of normal HSA. This was accomplished by using Eq. (1) [16,37,41].
| (1) |
This equation relates the original free drug fraction (F0) in a drug/protein mixture that was at equilibrium to the association equilibrium constant (Ka) for a drug-protein interaction that has saturable binding at a single site or to the global affinity constant (nKa’) for a drug that has multiple and independent saturable binding sites on the protein [16,35-37,41]. The other terms that appear in Eq. (1) are [A]0 and [P]0, which represent the total and original sample concentrations of the drug/analyte and soluble protein, respectively, where [A]0 ≤ [P]0. Binding constants that have been previously determined for various solute/protein systems by using ultrafast affinity extraction and Eq. (1) have spanned from approximately 103 to 109 M−1 [37].
Table 1 shows the binding constants that were obtained by using Eq. (1) and the free drug fractions that were determined by a single-column system for the model sulfonylurea drugs with normal HSA. These binding constants are all reported as global affinity constants because it is known that they each have two or three saturable and independent binding sites on HSA [9,17,18]. This approach gave a nKa’ value of 20.9 (± 2.3) × 105 M−1 for glibenclamide with normal HSA at pH 7.4 and 37°C, along with nKa’ values of 9.1 (± 1.4) × 105 M−1 for glimepiride and 5.4 (± 0.5) × 105 M−1 for glipizide at the same temperature and pH. These binding constants were based on the measurement of free drug fractions spanning from 14-28% and covering free drug concentrations of 1.4 to 2.8 μM. The precisions for these binding constants, as represented by the relative standard deviation of the mean, ranged from ± 4.7 to ± 7.7% (n = 4).
Table 1.
Global affinity constants for second- and third-generation sulfonylurea drugs with normal HSAa
| Drug | Method & conditions | Measured free drug fraction, F0b |
Global affinity constant, nKa’(× 105 M−1)c |
|---|---|---|---|
| Glibenclamide | Ultrafast affinity extraction, single-column system: 10 μM drug + 10 μM HSA |
19.6 (±1.5)% | 20.9 (± 2.3) |
| Ultrafast affinity extraction, two-column system: 0.08 μM drug + 500 μM HSA |
0.10 (± 0.03)% | 20.0 (± 6.0) | |
| Ultrafast affinity extraction, two-column system: 0.4 μM drug + 526 μM HSA [16] |
0.09 (± 0.02)% | 21.1 (± 4.7) | |
| Zonal competition + frontal analysis studies using immobilized HSA [17] |
N/A | 21.6 (± 8.0) | |
| Glimepiride | Ultrafast affinity extraction, single-column system: 10 μM drug + 10 μM HSA |
28.1 (±2.9)% | 9.1 (± 1.4) |
| Ultrafast affinity extraction, two-column system: 0.4 μM drug + 500 μM HSA |
0.22 (± 0.11)% | 9.1 (±4.5) | |
| Zonal competition + frontal analysis studies using immobilized HSA [9] |
N/A | 9.7 (± 0.7) | |
| Glipizide | Single-column system Drug (10 μM) + HSA (20 μM) |
13.9 (±1.2)% | 5.4 (± 0.5) |
| Two-column system Drug (2.24 μM) + HSA (600 μM) |
0.37 (± 0.12)% | 4.5 (± 1.5) | |
| Ref. [18] - Zonal competition ± frontal analysis studies using immobilized HSA |
N/A | 4.4 (± 0.2) |
The free fractions and reference values for nKa’ were all determined in aqueous buffers at pH 7.4 and 37°C, as well as using fatty acid free HSA. The values in parentheses represent a range of ± 1 S.D.
The free fractions listed for this study were measured at the following flow rates: glibenclamide, 0.30 mL/min; glimepiride, 3.0 mL/min; and glipizide, 3.5 mL/min.
The overall binding of normal HSA to glimepiride and glipizide was also examined in Ref. [20] by using fluorescence quenching. Binding affinities of 1.41 × 105 M−1 and 3.74 × 105 M−1, respectively, were reported at pH 7.4 and 37°C. However, dimethyl sulfoxide was also present in these solutions (i.e., to dissolve the drugs), which may had led to decreased drug-protein binding [22].
A comparison of these binding constants with literature values is provided in Table 1 [9,16-18,42]. The literature results were obtained by various approaches involving both soluble HSA [16] and immobilized HSA [9,17,18] and were also acquired in aqueous solutions at pH 7.4 and 37°C. It was found that the results of single-column ultrafast affinity extraction overlapped within ± 2 S.D. with all of the listed literature values and/or ranges that have been obtained by other methods for the same drugs with normal HSA [9,16-18]. It was further noted that the global affinity constants of 4.5 to 21 × 106 M−1 that were measured in this report for second- and third-generation sulfonylurea drugs were 2.6- to 19-fold higher than values that have been previously determined for normal HSA with the first-generation sulfonylurea drugs acetohexamide and tolbutamide [13-16].
3.3. Analysis of sulfonylurea binding to HSA by two-column ultrafast affinity extraction
Although the binding constants determined in Section 3.2 showed good agreement with values based on the literature, the drug and protein concentrations used with the single-column system (i.e., 10 or 20 μM) were not typical therapeutic or physiological levels for these agents. Work at such levels required samples with total concentrations of only a few μM or less for the drugs that were used in this study [11,39] and concentrations for HSA in the range of roughly 500-780 μM [40]. It was found in preliminary studies that the single-column systems used in Sections 3.1-3.2 did not have sufficient resolving power to adequately separate the retained free drug fractions from the large amount of HSA that was present in such samples (i.e., a 270- to 6250-fold mol excess of HSA versus the drug). Thus, a two-column system was instead used in which a second HSA microcolumn was placed on-line after capture of a free drug fraction by the first microcolumn, with the second column acting to provide further separation of the free fraction from any remaining non-retained sample components. Such an approach has been shown in prior work to be useful in measuring the free fractions of glibenclamide and other sulfonylurea drugs at therapeutically-relevant concentrations [16]. This method has also been employed in simultaneously examining the protein binding of several solutes (e.g., warfarin enantiomers) in drug/protein mixtures or serum [40].
One factor that had to be considered in the use of a two-column system for ultrafast affinity extraction was the time at which the second microcolumn was switched on-line with the first [16]. Figure 4(b) shows how the apparent free drug fraction for glibenclamide changed when using various times for this event. In this case, the apparent free drug fraction reached a minimum and steady value for glibenclamide when using a valve switching time of 4.9 min or longer to place the first HSA microcolumn (i.e., as optimized in Section 3.1) on-line with a 5.0 mm × 2.1 mm i.d. HSA microcolumn (Note: the flow rate was simultaneously changed to 0.15 mL/min to aid in peak resolution). This switching time was consistent with that reported for glibenclamide in Ref. [16] for free drug fraction measurements in a similar two-column system.
The increase in Figure 4(b) for the apparent free fraction at small switching times reflects the greater amount of non-retained sample components that co-eluted with the free drug fraction at these shorter times. Using an intermediate switching time, such as 4.9 min in Figure 4(b), had the advantage of minimizing the amount of these co-eluting agents while still allowing a reasonable amount of the retained free fraction to transfer to the second column. Although longer times could be used for this switching event, a further increase in this parameter will also decrease the amount of free drug that enters the second column for analysis and lead to lower precision for the measurement of this fraction [16].
Similar behavior to that in Figure 4(b) was seen for glipizide and glimepiride (see Supplemental Material). For glipizide, the final two-column system used a switching time of 1.3-1.4 min or more with a second HSA microcolumn that had a size of 10.0 mm × 2.1 mm i.d. and that was used at a flow rate of 1.5 mL/min. Glimepiride gave a consistent free fraction when using a switching time of 1.4 min or greater and a second HSA column that had a size of 10.0 mm × 2.1 mm i.d. and that was operated at 1.0 mL/min.
Table 1 shows the free drug fractions that were measured for drug/protein samples at therapeutically-relevant levels under these optimized conditions. These free fractions now spanned from 0.10% for glibenclamide to 0.37% for glipizide, demonstrating the ability of this approach to work even at low free drug levels. Although the two-column approach did allow much lower free drug fractions to be analyzed than was possible with a single-column system, the two-column method also gave less precise free fractions (i.e., with 3.8- to 4.8-fold larger variations in their values). In addition, the amount of HSA that was used within the samples examined by the two-column method at physiological levels was much higher (i.e., 30- to 50-fold greater) than was used in Sections 3.1-3.2 for samples that were analyzed by the single-column method.
The global affinity constants that were acquired by using Eq. (1) and the results of the two-column system are included in Table 1. The nKa’ values obtained for the drug/protein samples at therapeutically-relevant levels were statistically identical, at the 95% confidence level, to the values determined by the single-column method for samples that contained both the drug and HSA at concentrations in the 10-20 μM range. This consistency in the observed global affinity over a broad range of concentrations fits with previous observations made with other drugs and HSA samples that have been examined by ultrafast affinity extraction [16,35-37].
These results indicated that for these drug/protein systems, which are known to follow saturable interactions [9,17,18], the binding constants measured at higher concentrations with the single-column system could be used to model behavior of the same drugs with HSA at therapeutic levels. The consistency of the binding data in Table 1 also indicated that the global affinity constants obtained at the higher concentrations by the single-column system could be used, with a rearranged form of Eq. (1), to estimate F0 for the same drugs at their therapeutic levels [43]. Given the agreement of these two approaches for ultrafast affinity extraction, along with the greater ease-of-use and better precision of the single-column method, the remainder of this study (i.e., looking at sulfonylurea interactions with glycated HSA) focused on using the single-column method with drug/protein samples in the 10-20 μM range.
3.4. Analysis of sulfonylurea binding to glycated HSA by ultrafast affinity extraction
Ultrafast affinity extraction and single-column systems were next used to examine the binding by second- and third-generation sulfonylurea drugs with glycated forms of HSA. The conditions for this approach were optimized in the same manner as described in Section 3.1. It was found that the same types of microcolumns and flow rates as were used for normal HSA could be used for samples with glycated HSA (see Supplemental Material). This consistency in the conditions for different protein samples can be explained by the fact that the same types of HSA microcolumns were used to capture the free drug fractions. In addition, the forms of modified HSA that were present in the samples were later found to have similar, although not identical, levels of binding and dissociation rates for the model drugs when compared to normal HSA (see following discussion and Section 3.5).
Table 2 shows the free drug fractions that were measured for mixtures of the second- and third-generation sulfonylurea drugs with gHSA1 and gHSA2. It was observed that all of these sulfonylurea drugs gave a net increase in their free fractions in going from normal HSA to gHSA2 (i.e., a change that was significant at the 90% confidence level). The corresponding values of nKa’ that were obtained by using Eq. (1) are provided in Table 2. There was an overall decrease seen in the global affinity constants in going from normal HSA to either gHSA1 or gHSA2. Most of the observed changes in nKa’ were significant at the 95% confidence level. The only exception was for glimepiride with gHSA1; however, this result did differ at the 90% confidence level from that measured for normal HSA.
Table 2.
Global affinity constants measured with a single-column system for second- and third-generation sulfonylurea drugs with normal versus glycated HSAa
| Drug and sample | Measured free drug fraction, F0 |
Global affinity constant, nKa’ (× 10s M−1) |
% Change vs. normal HSA |
|---|---|---|---|
| Glibenclamideb | |||
| Drug (10 μM) + HSA (10 μM) | 19.6 (±1.5)% | 20.9 (± 2.3) | N/A |
| Drug (10 μM) + gHSA1 (10 μM) | 21.4 (±1.4)% | 17.2 (± 1.6) | −17.7 (± 2.6)% |
| Drug (10 μM) + gHSA2 (10 μM) | 22.9 (±2.8)% | 14.7 (± 2.6) | −29.7 (± 6.2)% |
| Glimepiride | |||
| Drug (10 μM) + HSA (10 μM) | 28.1 (±2.9)% | 9.1 (± 1.4) | N/A |
| Drug (10 μM) + gHSA1 (10 μM) | 30.9 (±2.2)% | 7.2 (± 0.8) | −20.9 (± 4.0)% |
| Drug (10 μM) + gHSA2 (10 μM) | 35.5 (±6.2)% | 5.1 (± 1.4) | −44.0 (± 13.8)% |
| Glipizide | |||
| Drug (10 μM) + HSA (20 μM) | 13.9 (±1.2)% | 5.4 (± 0.5) | N/A |
| Drug (10 μM) + gHSA1 (20 μM) | 17.3 (±2.7)% | 4.1 (± 0.7) | −24.1 (± 4.7)% |
| Drug (10 μM) + gHSA2 (20 μM) | 18.2 (±3.8)% | 3.8 (± 0.8) | −29.6 (± 6.8)% |
The values in parentheses represent a range of ± 1 S.D. (n = 4).
The values for normal HSA are the same as shown in Table 1 for the single-column systems and are provided for reference. The free fractions listed for glycated HSA were measured at the following flow rates: glibenclamide, 0.35 mL/min (gHSA1) and 0.40 mL/min (gHSA2); glimepiride, 3.5 mL/min (gHSA1 and gHSA2); and glipizide, 3.0 mL/min (gHSA1) and 3.5 mL/min (gHSA2).
These results indicated that glycation can alter the binding strength of these sulfonylurea drugs for HSA. Glimepiride showed the largest overall change (−44.0%) in affinity in going from normal HSA to gHSA2 (i.e., the more highly modified of the two glycated samples), while glibenclamide and glipizide had similar changes of −29.7% and −29.6% in their relative binding for this protein preparation vs. normal HSA. These three drugs were more consistent in their change in binding strength when going from normal HSA to gHSA1, giving variations that ranged from −17.7% to −24.1%.
These changes in affinity will lead to higher free drug fractions for gHSA1 and gHSA2 when compared to normal HSA, as observed in Table 2. Based on Eq. (1) and the values of nKa’ that are provided in Table 2, it is possible to estimate the relative changes in the free fraction F0 (and effective dosages of these drugs) that would be expected at therapeutic levels and in the presence of physiological levels of HSA. These calculations indicate that the free fraction will increase by up to 1.4-fold for glibenclamide or glipizide and by almost 1.8-fold for glimepiride under these conditions when going from normal to glycated HSA.
The general trends in Table 2 gave good agreement with previous reports that have examined the site-specific or overall changes in drug interactions with glycated HSA that had similar levels of modification to those present in this study [9,17,18]. For instance, the decrease in global affinity with glycation that was noted for glibenclamide agrees with changes due to glycation that have been observed in the binding of this drug with the digitoxin site of HSA (i.e., the highest affinity site found for this drug, with a site-specific Ka of 2.1 × 106 M−1 for normal HSA versus Ka values of 2.4-3.9 × 104 M−1 for Sudlow sites I and II) [17]. The decrease in global affinity for glipizide in going from normal HSA to gHSA1 and gHSA2 fits with prior changes that have been noted in the overall binding of this drug with glycated HSA [16], as well as with an observed decrease with glycation in the site-specific binding constant for this drug at its major site, Sudlow site I (Ka equal to 3.9 × 105 M−1 for normal HSA, with a Ka of 1.1 × 104 M−1 for Sudlow site II) [18]. The binding of glimepiride with HSA is slightly more complex and is known to occur at both Sudlow sites I and II with roughly equal affinities (i.e., Ka values of 4.2-5.5 × 105 M−1) [9]. For this drug, the affinity at Sudlow site II has been found to decrease at levels of glycation like those in gHSA2 (i.e., following the trend seen in Table 2 for glimepiride), while the binding strength at Sudlow site I may increase or follow a more complex interaction model [9].
3.5. Determination of dissociation rate constants by ultrafast affinity extraction
The information generated by the single-column system was further used to estimate the dissociation rate constants for the second- and third-generation sulfonylurea drugs in the presence of HSA. This was done by analyzing the data obtained at low-to-moderate flow rates through the use of Eq. (2) [35-37],
| (2) |
Dissociation rate constants that have recently been measured by using Eq. (2) and single-column ultrafast affinity extraction have ranged from roughly 10−2 to 101 s−1 for solute/protein systems with binding constants of 104 to 109 M−1 [37].
In Eq. (2), F0 is the free fraction of the drug in the original sample at equilibrium, and Ft is the apparent free fraction that is measured when the drug-protein complex has been allowed to dissociate in the column for time t. The value of t is equal to the sample residence time, which depends on the column void volume and the flow rate that was used for sample injection. Eq. (2) indicates that a linear relationship should be obtained in a plot of ln[1/(1 – Ft)] versus t, with a slope of kd and an intercept that is related to the value of F0 [35-37].
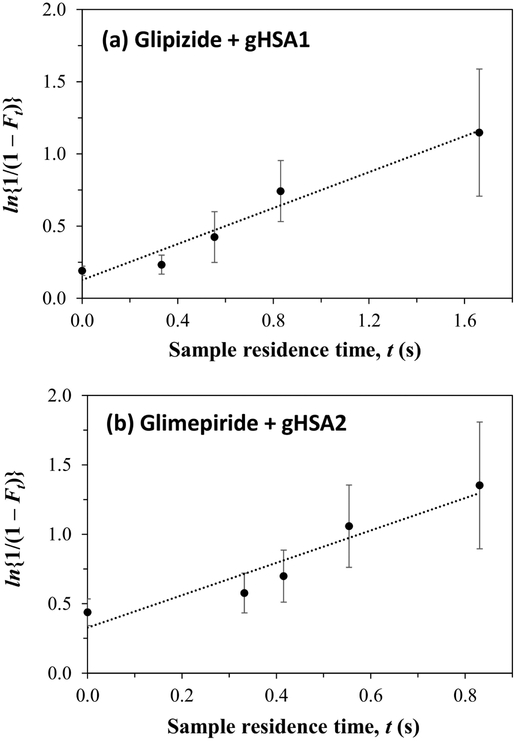
Some typical plots that were obtained when using Eq. (2) are provided in Figure 5. These plots gave an approximately linear response for all of the tested drugs and types of HSA, with correlation coefficients ranging from 0.9145 to 0.9955 (n = 4 to 6) over dissociation times that allowed measurable changes to be made in the apparent free fractions (e.g., see time scales used in Figure 5). Table 3 shows the kd values that were obtained from plots that were prepared according to Eq. (2). These dissociation rate constants ranged from 0.44-0.78 s−1 for the given sulfonlyurea drugs and normal HSA, which gave good agreement with results that have been reported for other sulfonylrea drugs [35]. For instance, the first-generation sulfonlyurea drugs tolbutamide and acetohexamide and the second-generation sulfonlyurea gliclazide (all of which bind to Sudlow sites I and II and which have global affinities for HSA spanning 0.69-1.8 × 105 M−1) have been found to have kd values for normal HSA in the range of 0.58-0.67 s−1 under the same pH and temperature conditions as used in this study [35].
Figure 5.
Determination of the dissociation rate constant (kd) for (a) glipizide in the presence of soluble gHSA1 or (b) glimepiride in the presence of soluble gHSA2 at pH 7.4 and 37°C by using ultrafast affinity extraction and a single-column system. The results were analyzed by using Eq. (2), with measured equilibrium values of F0 being utilized to obtain the points shown at t = 0. The correlation coefficients for these plots were 0.9792 and 0.9498 (n = 5), respectively. The error bars represent a range of ± 1 S.D., as based on error propagation and the measured precision of the Ft values.
Table 3.
Dissociation rate constants and apparent association rate constants for second- and third-generation sulfonylurea drugs with normal or glycated HSA
| Dissociation rate constant, kd (s−1)a | |||
|---|---|---|---|
| Drug | Normal HSA | gHSA1 | gHSA2 |
| Glibenclamide | 0.44 (± 0.03) | 0.33 (± 0.06) | 0.42 (± 0.07) |
| Glimepiride | 0.78 (± 0.12) | 0.87 (± 0.18) | 1.17 (± 0.22) |
| Glipizide | 0.65 (± 0.14) | 0.62 (± 0.07) | 0.93 (± 0.14) |
| Apparent association rate constant, ka (M−1 s−1)b | |||
|---|---|---|---|
| Drug | Normal HSA | gHSA1 | gHSA2 |
| Glibenclamide | 9.2 (± 1.2) × 105 | 5.7 (± 1.2) × 105 | 6.2 (± 1.5) × 105 |
| Glimepiride | 7.1 (± 1.5) × 105 | 6.3 (± 1.5) × 105 | 6.0 (± 2.0) × 105 |
| Glipizide | 3.5 (± 0.8) × 105 | 2.5 (± 0.5) × 105 | 3.5 (± 0.9) × 105 |
The kd values were determined at pH 7.4 and at 37 °C. The values in the parentheses represent a range of ± 1 S.D., as determined from the slopes of the best-fit lines according to Eq. (2).
The ka values were calculated by using the relationship ka = kd nKa’. The numbers in the parentheses for the ka values represent a range of ± 1 S.D., as determined from error propagation and the precisions provided for kd and nKa’.
A slightly larger range of 0.33-1.17 s−1 for the dissociation rate constants was obtained for glibenclamide, glimepiride and glipizide in the experiments that were conducted with glycated HSA. When compared to the results for normal HSA, each drug gave one kd value for glycated HSA that was significantly different at the 95% confidence level (glipizide, in going from normal HSA to gHSA2) or 90% confidence level (glibenclamide, in going from normal HSA to gHSA1; or glimepiride, when comparing normal HSA and gHSA2). These changes suggested that alterations in the dissociation rate accounted for at least part of the changes in global affinities that were seen in Table 2 for these drugs in the normal versus glycated forms of HSA.
The dissociation rate constants and global affinities that were measured earlier in this report were also used to estimate the apparent association rate constants (ka) for these drug-protein interactions. These ka values were calculated from kd and nKa’ by using the relationship ka = kd nKa’. Apparent association rate constants of 5.7 to 9.2 × 105 M−1 s−1 were obtained for glibenclamide with normal HSA or glycated HSA. A similar range of 6.0 to 7.1 × 105 M−1 s−1 occurred for glimepiride with these protein samples. A slightly lower set of values spanning from 2.5 to 3.5 × 105 M−1 s−1 was noted for glipizide. Some of the ka values had significant differences in going from normal to glycated HSA for a particular drug. This situation occurred for glibenclamide when comparing normal HSA and gHSA1 (95% confidence level) or gHSA2 (90% confidence level) and for glipizide in going from normal HSA to gHSA1 (~90% confidence level). These differences indicated that changes in the net association rate might also have led to some of the alterations in global affinities that were seen when comparing normal and glycated HSA.
4. Conclusions
This report developed and used ultrafast affinity extraction to determine the global affinity constants and dissociation rate constants for several second- and third-generation sulfonylurea drugs with normal HSA and glycated HSA. Affinity microcolumns containing immobilized HSA were utilized to capture and retain the free fractions of glibenclamide, glimepiride and glipizide in mixtures of these drugs with soluble HSA.
Both single- and two-column systems for ultrafast affinity extraction were employed and compared in these studies. Factors that were optimized for these methods included the flow rates, column sizes and sample residence times that were employed with each drug for ultrafast affinity extraction. These conditions were consistent with those identified in a separate study using ultrafast affinity extraction in free fraction measurements for one of the tested drugs (i.e., glibenclamide) [16], indicating that such systems can be made and used in a robust and reproducible manner. In addition, the final optimized conditions that were identified for glibenclamide and the other drugs in this study should be valuable in the future for the selection of a single set of analysis conditions that can be employed for a larger set of compounds.
Of the two methods that were examined based on ultrafast affinity extraction, it was found that the single-column approach was simpler to operate. This single-column technique allowed free fraction measurements and global affinity constants with good precisions to be obtained within only 1.0 min for samples containing comparable levels of the drugs and HSA. Another advantage of the single-column method was it could be modified to estimate dissociation rate constants for drug-protein interactions. The two-column approach had more factors to consider in its design, making it more complicated to utilize. However, this second method also allowed the measurement of binding constants in samples that contained much lower free drug fractions, as occurred when therapeutic levels of these drugs were in the presence of physiological levels of HSA.
The global affinity constants that were obtained by both of these methods showed good agreement with each other and with reference values. The results indicated that the levels of glycation seen in diabetes can alter the overall binding of second- and third-generation sulfonylurea drugs with HSA. The drugs examined in this report had a net decrease in affinity in going from normal to glycated HSA, which would correspond to an increase in the free fractions and biologically-active forms of these drugs in the circulation. It was further found that these changes in affinity could be the result of alterations in either the dissociation or association rates for the drugs with HSA. The observations made in this report produced greater insight as to how glycation can alter drug interactions with HSA and provided fundamental information that can be used in the future for the adjustment of drug dosages for patients with diabetes. In addition, the methods that were employed in this report could be modified for use with other pharmaceuticals and binding agents and to study the effects of other diseases on drug-protein interactions. This includes the use of this approach for the analysis of these processes with proteins obtained from, or present in, clinical samples [12,35-37,43].
Supplementary Material
Highlights.
Ultrafast affinity extraction was used to study binding by sulfonylureas with albumin
Both single- and two-column systems were optimized and compared for this work
Normal and glycated human serum albumin were examined in these studies
The global affinity constants obtained showed good agreement with the literature
The dissociation rate constants of these interactions were also measured
Acknowledgements
This work was supported by the National Institutes of Health under grant R01 DK069629.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Wild S, Roglic G, Green A, Sicree R, King H, Global prevalence of diabetes: estimates for the year 2000 and projections for 2030, Diabetes Care 27 (2004) 1047–1053. [DOI] [PubMed] [Google Scholar]
- [2].Olokoba AB, Obateru OA, Olokoba LB, Type 2 diabetes mellitus: a review of current trends, Oman Med. J 27 (2012) 269–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Gan D (Ed.), Diabetes Atlas, 2nd ed., International Diabetes Federation, Brussels, 2003. [Google Scholar]
- [4].National diabetes fact sheet: general information and national estimates on diabetes in the United States, 2011, U.S. Centers of Disease Control, Atlanta, GA, 2011. [Google Scholar]
- [5].Pratley RE, The early treatment of type 2 diabetes, Am. J. Med 126 (2013) S2–S9. [DOI] [PubMed] [Google Scholar]
- [6].Ashcroft FM, Gribble FM, ATP-sensitive K+ channels and insulin secretion: their role in health and disease, Diabetologia 42 (1999) 903–919. [DOI] [PubMed] [Google Scholar]
- [7].Harrigan RA, Nathan MS, Beattie P, Oral agents for the treatment of type 2 diabetes mellitus: pharmacology, toxicity, and treatment, Ann. Emerg. Med 38 (2001) 68–71. [DOI] [PubMed] [Google Scholar]
- [8].Sola D, Rossi L, Schianca GPC, Maffioli P, Bigliocca M, Mella R, Corlianò F, Fra GP, Bartoli E, Derosa G, Sulfonylureas and their use in clinical practice, Arch. Med. Sci 11 (2015) 840–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Matsuda R, Li Z, Zheng X, Hage DS, Analysis of multi-site drug-protein interactions by high-performance affinity chromatography: binding by glimepiride to normal or glycated human serum albumin, J. Chromatogr. A 1408 (2015) 133–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Matsuda R, Li Z, Zheng X, Hage DS, Corrigendum to ‘Analysis of multi-site drug-protein interactions by high-performance affinity chromatography: binding by glimepiride to normal or glycated human serum albumin’ [J. Chromatogr. A, 1408 (2015) 133-144], J. Chromatogr. A, 1423 (2015) 190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Regenthal R, Krueger M, Koeppel C, Preiss R, Drug levels: therapeutic and toxic serum/plasma concentrations of common drugs, J. Clin. Monit. Comput 15 (1999) 529–544. [DOI] [PubMed] [Google Scholar]
- [12].Anguizola J, Joseph KS, Barnaby OS, Matsuda R, Alvarado G, Clarke W, Cerny RL, Hage DS, Development of affinity microcolumns for drug-protein binding studies in personalized medicine: interactions of sulfonylurea drugs with in vivo glycated human serum albumin, Anal. Chem 85 (2013) 4453–4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Joseph KS, Anguizola J, Jackson AJ, Hage DS, Chromatographic analysis of acetohexamide binding to glycated human serum albumin, J. Chromatogr. B 878 (2010) 2775–2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Joseph KS, Anguizola J, Hage DS, Binding of tolbutamide to glycated human serum albumin, J. Pharmaceut. Biomed. Anal 54 (2011) 426–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Joseph KS, Hage DS, Characterization of the binding of sulfonylurea drugs to HSA by high-performance affinity chromatography, J. Chromatogr. B 878 (2010) 1590–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zheng X, Matsuda R, Hage DS, Analysis of free drug fractions by ultrafast affinity extraction: interactions of sulfonylurea drugs with normal or glycated human serum albumin, J. Chromatogr. A 1371 (2014) 82–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Matsuda R, Anguizola J, Joseph KS, Hage DS, Analysis of drug interactions with modified proteins by high-performance affinity chromatography: binding of glibenclamide to normal and glycated human serum albumin, J. Chromatogr. A 1265 (2012) 114–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Matsuda R, Li Z, Zheng X, Hage DS, Analysis of glipizide binding to normal or glycated human serum albumin by high-performance affinity chromatography, Anal. Bioanal. Chem 407 (2015) 5309–5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Jackson AJ, Anguizola J, Pfaunmiller EL, Hage DS, Use of entrapment and high-performance affinity chromatography to compare the binding of drugs and site-specific probes with normal and glycated human serum albumin, Anal. Bioanal. Chem 405 (2013) 5833–5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Seeder N, Kanojia M, Mechanism of interaction of hypoglycemic agents glimepiride and glipizide with human serum albumin, Cent. Eur. J. Chem 7 (2009) 96–104. [Google Scholar]
- [21].Ascenzi P, Bocedi A, Notari S, Menegatti E, Fasano M, Heme impairs allosterically drug binding to human serum albumin Sudlow’s site I, Biochem. Biophys. Res. Commun 334 (2005)481–486. [DOI] [PubMed] [Google Scholar]
- [22].Peters T Jr., All About Albumin: Biochemistry, Genetics and Medical Applications, Academic Press, San Diego, 1996. [Google Scholar]
- [23].Sudlow G, Birkett DJ, Wade DN, Further characterization of specific drug binding sites on human serum albumin, Mol. Pharmacol 12 (1976) 1052–1061. [PubMed] [Google Scholar]
- [24].Ascoli GA, Domenic E, Bertucci D, Drug binding to human serum albumin: abridged review of results obtained with high-performance liquid chromatography and circular dichroism, Chirality 18 (2006) 667–679. [DOI] [PubMed] [Google Scholar]
- [25].Anguizola J, Matsuda R, Barnaby OS, Hoy KS, Wa C, Debolt E, Koke M, Hage DS, Review: glycation of human serum albumin, Clin. Chim. Acta 425 (2013) 64–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Garlick RL, Mazer JS, The principal site of nonenzymatic glycosylation of human serum albumin in vivo, J. Biol. Chem 258 (1983) 6142–6146. [PubMed] [Google Scholar]
- [27].Tsuchiya S, Sakurai T, Sekiguchi SI, Nonenzymatic glucosylation of human serum albumin and its influence on binding capacity of sulfonylureas, Biochem. Pharmacol 33 (1984) 2967–2971. [DOI] [PubMed] [Google Scholar]
- [28].Koyama H, Sugioka N, Lino A, Mori S, Nakajima K, Effects of glycosylation of hypoglycaemic drug binding to serum albumin, Biopharm. Drug Dispos 18 (1997) 791–801. [DOI] [PubMed] [Google Scholar]
- [29].Roohk HV, Zaidi AR, A review of glycated albumin as an intermediate glycation index for controlling diabetes, J. Diabetes Sci. Technol 2 (2008) 1114–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Xiao JB, Högger P, Influence of diabetes on the pharmacokinetic behavior of natural polyphenols, Curr. Drug Metabol 15 (2014) 23–29. [DOI] [PubMed] [Google Scholar]
- [31].Y X. Xie, Xiao JB, Kai GY, Chen XQ, Glycation of plasma proteins in type II diabetes lowers the non-covalent interaction affinities for dietary polyphenols, Integrative Biol. 4 (2012) 502–507. [DOI] [PubMed] [Google Scholar]
- [32].Joseph KS, Hage DS, The effects of glycation on the binding of human serum albumin to warfarin and L-tryptophan, J. Pharm. Biomed. Anal 53 (2010) 811–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Matsuda R, Jobe D, Beyersdorf J, Hage DS, Analysis of drug-protein binding using on-line immunoextraction and high-performance affinity microcolumns: studies with normal and glycated human serum albumin, J. Chromatogr. A, 1416 (2015) 112–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Cao H, Liu XJ, Ulrih NP, Sengupta PK, Xiao JB, Plasma protein binding of dietary polyphenols to human serum albumin: a high performance affinity chromatography approach, Food Chem. 270 (2019) 257–263. [DOI] [PubMed] [Google Scholar]
- [35].Zheng X, Li Z, Podariu M, Hage DS, Determination of rate constants and equilibrium constants for solution-phase drug-protein interactions by ultrafast affinity extraction, Anal. Chem 86 (2014) 6454–6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Zheng X, Brooks M, Hage DS, Analysis of hormone-protein binding in solution by ultrafast affinity extraction: interactions of testosterone with human serum albumin and sex hormone binding globulin, Anal. Chem 87 (2015) 11187–11194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Beeram SR, Zheng X, Suh K, Hage DS, Characterization of solution-phase drug-protein interactions by ultrafast affinity extraction, Methods 146 (2018) 46–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Matsuda R, Kye S-H, Anguizola J, Hage DS, Studies of drug interactions with glycated human serum albumin by high-performance affinity chromatography, Rev. Anal. Chem 33 (2014) 79–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Launiainen T, Ojanpera I, Drug concentrations in post-morten femoral blood compared with therapeutic concentratiosn in plasma, Drug Test. Analysis 6 (2014) 308–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Rifai N, Horvath AR, Witter CT (eds.), Tietz Textbook of Clinical Chemistry and Molecular Diagnostics, 6th ed., Elsevier, Amsterdam, 2018. [Google Scholar]
- [41].Mallik R, Yoo MJ, Briscoe CJ, Hage DS, Analysis of drug-protein binding by ultrafast affinity chromatography using immobilized human serum albumin, J. Chromatogr. A 1217 (2010) 2796–2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Patel S, Wainer IW, Lough WJ, Affinity-based chiral stationary phases, in: Hage DS (ed.), Handbook of Affinity Chromatography, 2nd ed., CRC Press, Boca Raton, 2006, pp. 571–594. [Google Scholar]
- [43].Zheng X, Yoo MJ, Hage DS, Analysis of free fractions for chiral drugs using ultrafast extraction and multi-dimensional high-performance affinity chromatography, Analyst, 138 (2013) 6262–6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.