Abstract
Histone tails and their epigenetic modifications play crucial roles in gene expression regulation by altering the architecture of chromatin. However, the structural mechanisms by which histone tails influence the interconversion between active and inactive chromatin remain unknown. Given the technical challenges in obtaining detailed experimental characterizations of the structure of chromatin, multiscale computations offer a promising alternative to model the effect of histone tails on chromatin folding. Here we combine multi-microsecond atomistic molecular dynamics simulations of dinucleosomes and histone tails in explicit solvent and ions, performed with three different state-of-the-art force fields and validated by experimental NMR measurements, with coarse-grained Monte Carlo simulations of 24-nucleosome arrays to describe the conformational landscape of histone tails, their roles in chromatin compaction, and the impact of lysine acetylation, a widespread epigenetic change, on both. We find that while the wild-type tails are highly flexible and disordered, the dramatic increase of secondary-structure order by lysine acetylation unfolds chromatin by decreasing tail availability for crucial fiber-compacting internucleosome interactions. This molecular level description of the effect of histone tails and their charge modifications on chromatin folding explains the sequence sensitivity and underscores the delicate connection between local and global structural and functional effects. Our approach also opens new avenues for multiscale processes of biomolecular complexes.
Keywords: Histone tails, internucleosome interactions, lysine acetylation, chromatin compaction
Graphical Abstract
Deciphering the behaviour of histone tails within chromatin and its epigenetic modulation is essential to fully understand the detailed mechanisms that govern genome organization and influence gene expression regulation. In eukaryotic organisms, DNA and histone proteins self-assemble into nucleosomes that are connected by DNA linker segments and other architectural proteins including linker histones (LH)1. The core histone proteins (H2A, H2B, H3, and H4) contain ten positively charged protein regions, known as histone tails, that account for about a quarter of the histone core mass and extend from the nu-cleosome surface1a. It is believed that the histone tails affect chromatin compaction by mediating interactions with other proteins, nucleosomes, and linker DNAs2. Epi-genetic changes, including methylation, ubiquitination, acetylation and phosphorylation, exert their biological role by transforming chromatin structure, either directly2c, 2d, 3 or through the recruitment of other molecular species4.
The structure of the histone tails is only partially characterized crystallographycally5. Although this suggests a high degree of histone-tail structural disorder, the extent of this disorder is unknown and, despite recent advances6, the detailed mechanisms of tail-induced chromatin compaction and epigenetic-driven changes remain obscure. Experiments assessing the structure of wild type versus chemically modified histone tails within compact chro-matin are challenging because of the highly crowded environment of chromatin and the difficulty of obtaining chromatin arrays with homogeneous histone tail compositions. Circular dichroism (CD) analyses have revealed that isolated H4 tail peptides display spectra characteristic of random coils7, that the total α-helical content of histone tails attached to their nucleosomes is approximately 17%, and that hyperacetylation of all core and tail histones increases the α-helical content by 64–100%7. A previous CD nucleosome study had reported a higher (30–35%) α-helical content in the tails of H3 and H4, and suggested that such folding is conditional upon DNA binding8. More recently, an amide hydrogen exchange and multidimensional NMR study of 12-unit compact oligonu-cleosomes suggested that the H3 tail is protected from solvent exchange6b. However, this study does not clarify whether the H3 tail has a defined secondary structure or whether it adopts a disordered but compact conformation; separately, it has been suggested the histone tails in the highly condensed arrays could remain flexible yet simultaneously protected from solvent exchange on the relatively short exchange time scales investigated6a.
In contrast, a disordered histone tail behaviour has been reported in a recent magic angle spinning (MAS) NMR spectroscopy study of condensed 177-bp 17-unit oligonu-cleosomes, which showed that residues 1–38 of H3 and at least 1–21 of H4 N-terminal tails are flexible and remain available for protein binding6a. Solution NMR measurements of the nucleosome with full histone tails demonstrated that all histone-tail residues, except residues 16–22 of H4, have a disordered conformation in solution, and that a mimic of K16 acetylation (K16Q mutation) disrupts the folded region of H49. However, how well the K16Q mutation mimics K16Ac is controversial because, while the acetylated version opens chromatin, the mutation does not alter chromatin compaction2d, 10. A combined CD and solution NMR study indicates that both isolated H4-tail peptides and hyperacetylated forms adopt extended and flexible structures11.
Computational studies of ever more complex biomolecules (e.g. 12) provide detailed views, as well as mechanistic and thermodynamic information, complementing experiments that might lack the spatial resolution to provide a molecular perspective of the problem. Hardware and software advances have allowed several all-atom sim-ulations of isolated histone tails13, tails with DNA13a, 14, and even nucleosomes15 with explicit solvent and ions. All-atom molecular dynamics (MD) simulations in both implicit16 and explicit solvent13b suggested that the histone tail peptides are not fully disordered. Indeed, a recent all-atom study has shown that H4K16Ac increases the histone tail/DNA binding affinity13a. Simulations of nucleosomes with tails have shown that truncation of the H3 or H2A tails alters the structure of the nucleosome core15b. Although insightful, the longest trajectories of nucleosomes with tails this far are about 100 ns15b, and those of isolated histone tails consist of replica exchange MD (REMD) studies with 60-ns of sampling per replica (or 3-μs of accumulated simulation time)13b. Such level of sampling might not be sufficient to characterize histone tail structure and its role in internucleosome interactions (see Figure S1 showing lack of convergence at 150 ns). In fact, a multiscale approach is ideally needed to establish the conformational landscape of histone tails and relate it to changes in chromatin structure17.
Here we link the dynamics of wild-type histone tails and of lysine acetylated versions (a common epigenetic modification2c, 18) to dinucleosome and oligonucleosome structure by an unparalleled multiscale approach19 (see Fig. 1 for an overview of our study, and Tables S1 and S2 for the set of simulations performed). Our analyses indicate that while the wild-type tails are highly flexible and disordered, the dramatic increase of secondary-structure order by lysine acetylation unfolds chromatin by decreasing the tails’ availability for crucial fiber-compacting internucleosome interactions. This molecular level description of the indirect structural effect of post-translational modifications of histones on chromatin organisation explains the sequence sensitivity and underscores the delicate connection between local and global structural and functional effects. Our approach also opens new avenues for multiscale modelling of biomolecular complexes.
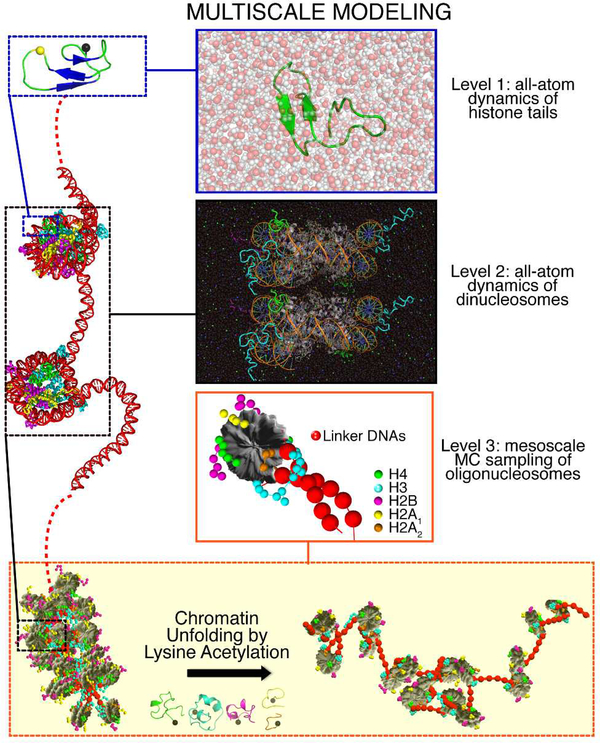
Figure 1. Overview of our multiscale computational study.
We explore the behaviour of histone tails, its epigenetic modulation, and its impact on chromatin compaction through a colossal multiscale study that combines multimicrosecond all-atom molecular dynamics simulations of dinucleosomes and histone tails with coarse-grained Monte Carlo simulations of 24-nucleosome arrays. We describe a dramatic change in histone tail conformations due to lysine acetylation and reveal a tightly orchestrated physical mechanism by which such epigenetic modifications affect DNA accessibility.
METHODS
To examine the structure and behaviour of histone tails, and their effects on chromatin organization, we developed a multiscale computational protocol that combines all-atom MD simulations of dinucleosomes in explicit solvent (800,000 atoms; 4 microseconds) and histone tails (aggregated time above 0.6 milliseconds), with three state-of-the-art force fields and validated by experimental NMR data, with Monte Carlo (MC) coarse-grained modelling of chromatin fibers (24-nucleosomes). Our all-atom simulations comprise about 1 order of magnitude more sampling than previous studies and our coarse-grained simulations study 84 different oligonu-cleosomes to extrapolate our all-atom findings into the nanoscale chromatin level. While the accuracy of atomistic models is needed for an adequate description of epigenetic marks, within chromatin their use is prohibitive due to the large number of degrees of freedom involved (e.g. a 50nucleosome array without solvent contains more than one million atoms). Coarse-graining is one of the only alternatives to model chromatin because it can dramatically reduce the system dimensionality while maintaining essential physical and chemical information. Our multiscale approach exploits the advantages of both levels of modelling to perform a unique molecular –level analysis of the effects of lysine acetylation in chromatin structure.
All-atom Studies.
All our all-atom studies were performed in Gromacs-4.520 using explicit solvent and ions. Our simulations include 27-μs (accumulated sampling time per system) REMD simulations of the four Nterminal histone tails (H4, H3, H2B, and H2A), the Cterminal H2A tail (H2AC), five acetylated H4-tail states (H4K16Ac, H4K12Ac, H4K1216Ac, H4K5,8,12Ac, and H4K5,8,12,16Ac), one acetylated H3-tail state (H3K14Ac), and two acetylated H2B-tail states (H2BK20Ac and H2B K5,12,15,20Ac). In addition, we performed 4 μs (accumulated sampling time per system) unrestrained simulations of dinucleosomes with wild-type tails and with H4K16Ac and H3K14Ac tails, and 1 μs unrestrained MD simulations of all wild-type histone tails. Periodic boundary conditions and the particle mesh Ewald method21 for long-range electrostatics were used. We modelled the shortrange repulsive and attractive dispersion interactions via a Lennard-Jones potential with a cut-off of 1.0 nm. We used the Settle algorithm22 to constrain bond lengths and angles of water molecules, and P-LINCS for all other bond lengths, with an integration time step of 2 fs. For the di-nucleosome systems, we used virtual interaction-sites20, 23 together with constraints between all bonded atoms to remove the hydrogen vibrations and allow a time step of 4 fs. The temperature was kept constant at 300 K by using the Bussi thermostat24. The pressure was kept constant and controlled by coupling the simulation box to a pressure bath of 1 atm25.
To select the most adequate force field for our study, we repeated our REMD simulations of the H4 and H4K16Ac tails with three different force fields, namely AMBER99SB*-ILDN26, AMBER99SB27, and CHARMM3628, and those of the other wild-type tails with two, AMBER99SB*-ILDN and AMBER99SB. In addition, we validated the H4 tail REMD ensemble against NMR chemical shifts (see below). We then selected the force field AMBER99SB*-ILDN to carry out the rest of the simulations. Force field parameters for acetylated lysines were taken from 29 for AMBER99SB*-ILDN and AMBER99SB (Papageorgiou’s parameters), and from 30 for CHARMM36 (Dejaegere’s parameters). To describe the nucleosomal DNA we used the AMBER99+parmBSC031 force field. We used the TIP3P model32 to describe the water molecules. All simulations were done with 150 mM of sodium and chlorine ions, which were modeled using Dang’s parameters33. Dinucleosomes were modeled in the presence of manganese with parameters taken from the Amber force field database. Table S1 lists all molecular dynamics simulations and force fields used in this work.
The initial structures for the wild-type histone tails and their acetylated versions were prepared using VMD34 based on human sequences from the UniProt Consortium35. The lengths of the N-terminal tails H3, H4, H2B, and H2A were defined as the first 38, 26, 23, and 14 residues of each histone, respectively, as done previously13b. The C-terminal H2A tail was defined as the last 9 residues of the H2A histone. Isolated histone tails were not capped in the N- or C-terminus. The tails were placed in a simulation box large enough to host the proteins completely unfolded (distance to the wall was 1.2 nm) and immersed in water. The systems were energy-minimized, then subject to 200 ps of restrained NVT simulation, and further equilibrated during 1 ns in the NPT ensemble. The systems were then simulated for 1 μs, starting with random velocities obtained from a Maxwell-Boltzmann distribution at 300 K. For the analysis of the simulations, we discarded the initial 100 ns as equilibration period and calculate the lifetime of secondary structural elements. We use the program STRIDE36 to determine the secondary structure adopted by the histone tails and measure the average time it takes for the different motifs to disappear. To define secondary structural elements, STRIDE uses empirically derived hydrogen bond energies and compares phipsi backbone torsion angles with the α-helix and β-sheet regions in a Ramachandran plot37. The parameters of STRIDE have been optimized based on visual assignments made by expert crystallographers of protein structures. α-helices start when two consecutive amino acids have i→ i+4 hydrogen bonds. The two edge residues are only labelled as belonging to the helix if they have adequate phi-psi angles, which implies that the minimum size of consecutive helical residues reported is three, not five. A similar definition is used for 310-helices but considering i→ i+3 hydrogen bonds. β-sheets are assigned if at least two hydrogen bonds occur in the sheet with adequate phi-psi angles for the residues involved. From the output of STRIDE, we obtain the maps of folding propensity versus residue number by classifying the residues as being part of an (a) α motif if they belong to a group of three consecutive residues defined as either α- or 310-helix by STRIDE, or a (b) β motif if they belong to a group of two consecutive residues that are defined as an extended conformation by STRIDE.
The initial structures for our REMD simulations were taken from our unrestrained molecular dynamics simulations described above. REMD simulations were performed using between 56 and 64 temperatures ranging from 300 K to 450K. The distribution of temperatures was chosen to guarantee an average exchange probability of 25%, with the aid of the temperature predictor of Patriksson and van der Spoel38. The exchanges between neighbouring replicas were attempted every 50 ps. Each system was simulated for 500 ns. The initial 100 ns were discarded in the subsequent analysis. We then analysed the lowest temperature replica data with the program STRIDE36 to determine if the histone tails adopt α or β structure, as define above. Following this, we reshuffled the data and divide it in 40-ns sub-ensembles to compute the average and the standard deviation of the propensity of each residue to form either α or β motifs among the sub-ensembles. In addition, for each histone tail we performed RMSD clustering as implemented in the program g_cluster in Gromacs4.520 using a total of 25,000 structures collected at 20 ps intervals from the lowest temperature replica to. For this, external translational and rotational motions were removed by minimizing the RMSD distance of the Cα atoms with respect to those of the first frame. During the clustering, we used a cutoff of 2 Å on the Cα atoms.
To probe the conformational dynamics of wild type and acetylated (H4K16Ac) histone tails in a nucleosomal environment, we performed two sets of dinucleosome simulations each with a different starting H4/H4 K16Ac setup. For the two sets, we stacked two nucleosome particles (X-ray structure with PDB code 1KX5 39) on top of each other following the positions and orientations of stacked nucleosomes in the tetranucleosome crystal (X-ray structure with PDB code 1ZBB40). For consistency with our REMD studies, the H4/H4 K16Ac tail sequences were replaced with the human ones. In the first setup, we attached an extended H4 structure from our REMD ensembles to each nucleosome and oriented it to ensure that its N-terminal region was placed near the acidic patch of its neighbouring nucleosome (Figure 4a). In the second setup, the H4 tails were oriented towards the nucleosomal DNA (Figure 4b). The dinucleosome systems were embedded in a truncated octahedron box containing 203778 water molecules, leaving 2 nm between the nucleosome atoms and the edges of the box. This separation is large enough to accommodate a fully extended H3 tail, which is the longest one. We added 895 sodium ions and 601 chlorine ions to balance the nucleosome charge and give an ionic concentration of 0.15 M NaCl. Each dinucleosome system was energy-minimized and simulated twice (two different random seeds) for 500 ns. The first 100 ns of each trajectory were discarded.
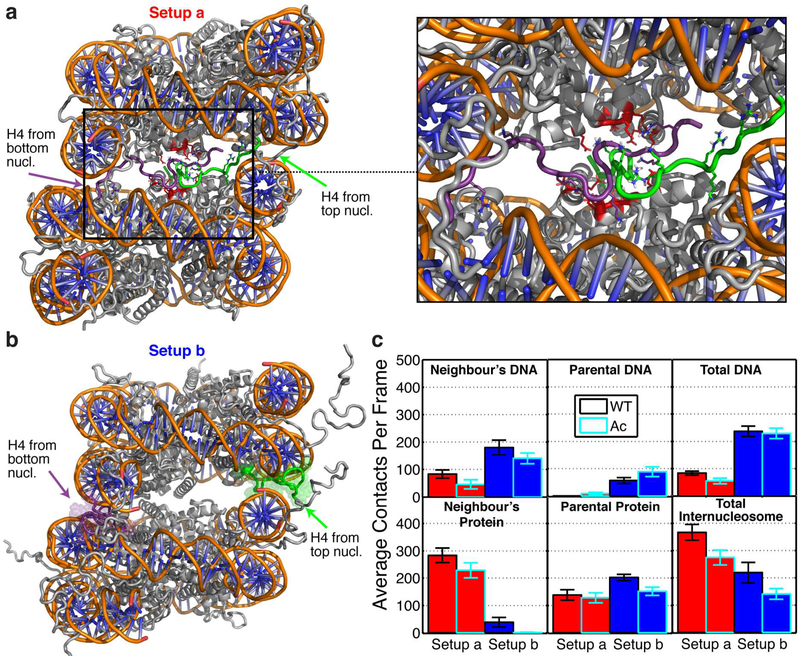
Figure 4. Internucleosome interactions within dinucleosomes mediated by the H4 tail and its mono-acetylated H4K16Ac version.
(a) Illustration of the first simulation setup (setup a). The H4 tails extend form their point of attachment in their parent nucleosome towards the acidic patch of their neighbouring nucleosome. The H4 tail of the top and bottom nucleosomes are highlighted in green and purple, respectively. At the right, we zoom in into the acidic patch region highlighting the acidic patch residues in red (H2A:E56,E61,E64,D90,E91,E92 and H2B E102, E110) and showing the positive H4-tail residues with sticks but omitting non-polar hydrogens for clarity. (b) Illustration of the second simulation setup (setup b). The H4 tails extend from their point of attachment in their parent nucleosome towards the nucleosomal DNA. (c) For both setups, total number of histone tail-DNA/protein contacts (tail and DNA atoms closer than 0.3 nm) mediated by the wild-type H4 tail versus the acetylated version (Ac) separated as: tail/DNA of neighbouring nucleosome (Neighbour’s DNA), tail/DNA of parent nucleosome (Parent DNA), total neighbouring + parent tail/DNA (Total DNA), tail/protein of neighbouring nucleosome (Neighbour’s Protein), tail/protein of parent nucleosome (Parent Protein), tail/total (neighbouring DNA + neighbouring protein; Total Internucleosome). The red bars show the results for setup a, and the blue bars the results for setup b. The bars corresponding to dinucleosomes containing wildtype or acetylated H4 versions have black and cyan borders, respectively.
Validation of all-atom studies using NMR chemical shifts.
We performed two types of validations of our simulation ensembles using experimental NMR chemical shifts of the histone tail H4 (residues 1 to 15) and the H4K16Q mutation (residues 1 to 17) attached to their nucleosomes9. The chemical shifts are available for all the Cα, and amide N and H backbone atoms, and the Cβ atoms. First, we used the experimental data to perform replica-averaged chemical shift restrained MD simulations and generate an ensemble of structures to compare against our REMD results41. Second, we used the predictor delta2D (δ2D)42 to calculate the propensity of the H4 tail to form α or β secondary structures based on the experimental chemical shifts and compared against the folding propensities resulting from our REMD ensembles.
In the replica averaging procedure, a set of chemical shifts were computed with the Camshift43 method using the coordinates of the atoms for which chemical shifts are available, and averaging over 8 replicas. The chemical shift restraints are implemented in the Gromacs engine via the Plumed44 package and the Camshift algorithm. A quadratic potential based on the differences between the reference and the computed average chemical shift is added as external penalty. The strength of the interaction was adjusted to the highest possible value that did not affect the stability of the trajectories41. Two replicas were biased using the metadynamics scheme41, 45, employing a time-dependent energy deposition that helps overcome conformational barriers. In one replica, we deposited Gaussian-shaped potential energies every 500 MD steps (1 ps) on the underlying force-field energy landscape based on the end-to-end distance of the H4 chain using a Gaussian-width of 0.1 nm and height of 0.1 kJ/mol. In another independent replica we deposited energy with the same frequency on the collective coordinate calculated as the number of hydrogen bonds between all donors and acceptors. In this case, the height of the Gaussian was also 0.1 kJ/mol, and the width was 0.5 hydrogen bonds. We used a continuous expression for the number of hydrogen bonds that involves counting the resulting product of two functions that quickly drops from 1 to 0 at a cut-off value A0 for each possible hydrogen bond. Each individual function is evaluated as (1 − A⁄A0)6⁄ (1 − A⁄A0)12. For each hydrogen bond, the distance between the hydrogen and the acceptor (A=d) has a cut-off at Ao=d0=0.3 nm, and the angle between the hydrogen-donor and hydrogenacceptor vectors (A=α) has a cut-off at A0=α0=0.66π. Finally, we estimated the secondary structure content in these restrained simulations and compared against our REMD results.
Histone tail flexibility.
To compare the relative flexibilities between wild-type histone tails and their acetylated version, we computed the tails’ persistence lengths Lp.The persistence length is a common measure of protein flexibility: the lower the persistence length, the higher the flexibility. While systems with contour lengths, L, that satisfy L ≪ Lp are rigid and maintain an average straight conformation with a preferred tangent direction, system with L ≫ Lp are flexible and bend spontaneously. Systems in which L is comparable with Lp, are considered semiflexible, and their structural rearrangements are determined by the competition of the thermal energy and the bending rigidity. For each tail, we computed the persistence length from the lowest temperature replica based on the projection of the end-to-end vector (R)with respect to its initial direction (r1) 46, as follows
| (1) |
Here is the vector joining the ith and (i+1)th Cα, l is the distance between Cα atoms (l = 0. 38 nm), and N is the total number of residues in the histone tail. To compute θ1i the angle between vectors r1 and ri, we used the program g_sgangle in Gromacs-4.520. We define the contour length of the tails as the number of amino acids in each tail multiplied by l. An illustration of the definition of the persistence is given in Fig. S2.
Chromatin coarse-grained simulations with flexible and folded tails.
To link our all-atom findings to the level of nanoscale chromatin, we carried out oligonucleosome coarse-grained simulations that explore the effects of histone tail folding. These coarse-grained studies compare the behaviour of 24-mer oligonucleosome systems without LH and two different DNA linker lengths (35 and 62 bp) with eight different concentrations (0, 5, 10, 25, 50, 75, 90, and 100%) of folded histone tails. In addition, we also include simulations in which we have reduced the charge of the H4 and H3 tails by 1e due to monoacetylation but kept the flexibility unaltered. Our coarse-grained model17a-c, 17e, 47 captures several key features of chromatin fibers, including electrostatics, DNA and nucleosome mechanics, structural irregularity, and histone tail flexibility, and averages out other effects like protein/DNA sequence effects, hydrogen bonding, atomistic fluctuations, and solvation. In essence, our model chromatin represents the nucleosome with wrapped DNA but without histone tails as an electrostatic object with Debye-Hückel charges47a, 47c; DNA linkers are described as chains of charged beads by a combined wormlike-chain (WLC) model48; histone tails are treated as flexible or almost rigid chains of charged beads with parameters that mimic their atomistic behaviour17b; and sampling of phase space is achieved through a MC algorithm. In the current version of the model, unfolded histone tails are described as flexible worm-like chains using our original model17a. In addition, transient populations of folded tails are incorporated as rigid entities with equilibrium configurations taken from the highest populated folded structure obtained for each tail in the lowest temperature replica of our REMD simulations. Specifically, to model folded tails, as done in our original model, we assign one bead per each 5 amino acids, center the bead at the Cβ atom of the middle amino acid, and assign to each bead the total charge of the 5 amino acids it represents. An image showing the histone tail beads overlaid on top of all-atom histone tails (extended and folded), and giving the sequence of amino acids grouped in each bead and the bead charge is presented in the Fig. S3. To limit tail flexibility, we increasing the stretching, bending and torsional inter-tail-bead force constants by a factor of 100. Despite being almost rigid, the tails can spontaneously fold/unfold through a new MC move (see Material) that attempts transitions between folded and flexible tails. This allows us to explore the whole conformational ensemble of oligonu-cleosomes with a given concentration of folded tails. For our additional simulations exploring the effects of charge reduction due to acetylation, we have decreased by 1e the charges of the tail-beads that contain residues H4K16 and H3K14, but have kept the flexibilities of the tail unaltered. Our 24-nucleosome simulations were performed at physiological conditions; this is, a temperature of 298 K and a high monovalent salt concentration of 0.15 M NaCl. We modelled two types of fibers characterized by uniform DNA linker lengths, either 35 or 62 bp, and no LHs. Every simulation set includes 12 trajectories that cover the mean DNA twist angle and two DNA twist deviations (±12°) from the mean twist to mimic natural variations, as done previously17c. Each simulation trajectory was run for up to 50 million MC steps. The last 5 million steps were used for statistical analysis. Convergence of these simulations is reached well before 45 million MC steps as shown elsewhere 17c. For our initial configurations, we use representative zigzag equilibrium conformations for oligonu-cleosomes with flexible tails obtained previously 17c. For more details on our coarse-grained model see 17b, 17c, 47e and the Supporting Information.
RESULTS
Histone tails are structurally highly heterogeneous.
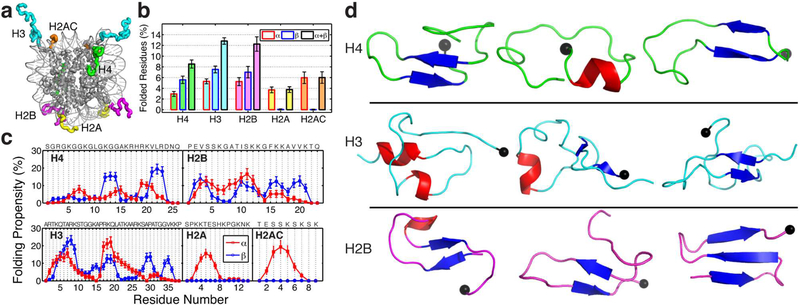
Our massive REMD simulations of the five different isolated histone tails in explicit solvent (Table S1) confirm that histone tails are mostly unstructured and exhibit only transient secondary structural elements6a, 9, 13b, 16b. The conformational ensembles obtained at physiological conditions (0.15 M monovalent salt and room temperature) contain only a small (<15%) fraction of residues with α-helical or β-strand conformations (Fig. 2a). For the H4 tail, the most abundant structure is an antiparallel β-hairpin (KGG 12–14 and VLR 21–23) connected by a sixresidue loop with four positively charged residues (15–20 AKRHRK). The H4-tail transient α-helical motifs, and the α-helical/β-motifs for the rest of the tails are short (i.e., formed by only 3–6 residues per tail) and highly diverse (i.e., many different motifs are formed by combinations involving most residues), which illustrate the overall structural disorder of histone tail conformations. Fig. S4 provides examples of the most populated folded structures with higher atomic detail. These motifs are compact and able to fit within a compact dinucleosome structure (see Fig. S5), short lived (i.e., ~48 and 88 ps for α-helical and β- strand H4 structures, respectively) and highly heterogeneous (Fig. 2b and c). Estimated persistence-tocontour-length ratios from the MD ensembles (see Table S3) measure the length scale over which histone tails remain relatively straight. Even though the tails adopt transient secondary structural elements, their persistence-tocontour-length ratios are smaller than one. Thus, histone tails are flexible and display a high degree of histone tail bending from thermal fluctuations (Table S3). The persistence lengths we obtain for the five histone tails are consistent with experimental values of flexible random coiled proteins of similar sizes (0.48—0.63 nm) and one order of magnitude smaller than that of polyproline (4.4 nm) which is considered the stiffest homo-oligopepetide49 (Table S3). The histone tail values are also much smaller than the measured persistence lengths of DNA (50 nm50) and chromatin fibers (diverging between 30–200 nm51), highlighting their high flexibility within the chromatin context.
Figure 2. Transient secondary structure populations in the wild-type histone tails.
(a) Illustration of histone tails (H4 in green, H3 in blue, H2B in magenta, H2A in yellow, and H2AC in orange) extending out of the nucleosome surface (PDB 1KX5). (b) Ensemble average and standard deviation (SD) of the percentage of residues in each of the different histone tails calculated from the last 400 ns of the 300 K REMD trajectory (SD computed by block average using 40 ns windows). The percentages of residues forming α-helices (red exterior line), β-strands (blue exterior line), and their sum (black exterior line) are shown separately. (c) Folding propensity i.e. fraction of configurations that each residue adopts α-helical (red) or β-strand (blue) structural motifs. The sequence for each tail is shown. (d) Illustration of cluster with the highest population with folded residues for H4 (first cluster with 12.54% population, ninth with 2.49%, and eleventh with 2.00%), H3 (first with 4.15%, third with 3.77%, and fourth with 3.75%), and H2B (second with 9.08%, fifth with 3.08%, and %, and sixth with 2.00%). The α-Helical motifs are coloured in red, while beta conformations are in blue. The black sphere indicates the last residue of the N-tail (point of attachment to the nucleosome).
To control for the effects of the specific force fields used in the simulations, we repeated the simulations for the H4 tail with three different state-of-the-art force fields (see Tables S1 and S2), and validated the resulting ensembles using chemical shifts of histone tails measured for reconstituted nucleosomes6b. Calculations with the different force fields reveal nearly identical results (Fig. S6). Our simulations agree well with the experimental data that shows that folding of H4 deviates from a fully random coil behaviour (Fig. S6). For further control, the atom-type-averaged chemical shifts calculated from our H4 tail ensembles also deviate from a complete random coil sequence based prediction52. Furthermore, the folding propensities obtained with our additional restrained simulations that satisfy the experimental chemical shifts and those from our REMD simulations with the AMBER99SB*-ILDN26 force field deviate from the folding propensities calculated using experimental chemical shifts almost identically.
Epigenetic modifications can change histone-tail flexibility.
REMD simulations of acetylated H4, H3, and H2B tails examine how the tails’ structure and behaviour is altered by lysine acetylation. We focus on these three tails because they have been reported to be most influential in chromatin architecture regulation53, and on lysine acetylation because this important modification for gene regulation is commonly present in transcriptionally active chromatin and is thought to directly affect chromatin structure18. For instance, hyperacetylation of histones is present in open sea urchin chromatin (with linker DNAs of 61 bp)54, and correlates with decondensed yeast chro-matin (~ 18 bp linker DNAs55)56. In addition, monoacetylation is associated with both open chromatin (H4K16Ac 2c, 2d), and with silent and compact chromatin (H4K12Ac 57).
Lysines are present in four H4-tail residues (5, 8, 12, and 16), seven H3-tail residues (4,9,14,18,23,27,36), and six H2B-tail residues (5,11,12,15,16,20). Among all the possible mono- and poly-acetylated states, we study two monoacetylated (K12Ac and K16Ac) and three poly-acetylated states (i.e., K12&16Ac, K5,8,12Ac, and K5,8,12&16Ac) for H4; for H3, we study one monoacetylation (K14Ac); and for H2B one monoacetylation (H20Ac) and one polyacety-lation (K5/12/15/20Ac) (see Table S1 for a list of all cases analysed). Comparing H4K16Ac versus H4K12Ac allows us to determine whether lysine acetylation has a sequence specific effect. We analyse H3K14Ac and H2BK20Ac because their presence is correlated to transcription activation 58. Finally, we study the poly-acetylated states to analyse the additivity or cooperativity properties of lysine acetylation.
We find that K16Ac increases the overall propensity of H4 residues to adopt both α-helical and β-strand conformations by about 50% (Fig. 3a) and increases the persistence length by 41%, thereby decreasing the flexibility of the tail (Table S4), consistent with previous theoretical suggestions13a and with circular dichroism data showing that lysine acetylation of all core and tail histones increases the α-helical content of the nucleosome by 64–100%7. An illustration of the larger space covered by H4 versus H4K16Ac during a 1 μs-long MD simulation is shown in Fig. S7.
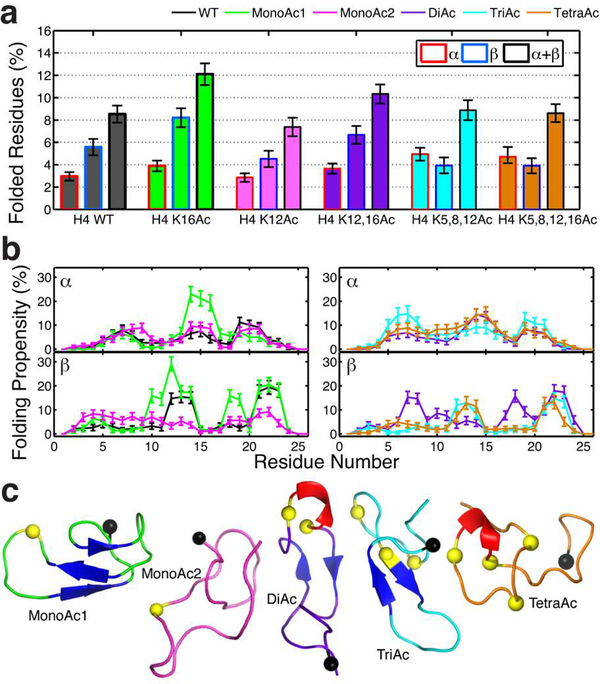
Figure 3. Effect of the acetylation of different lysine residues in the H4 tail.
(a) Percentage of residues in various lysine-acetylated tails with secondary structure motifs. (b) Effect of acetylation in the folding propensity for each residue separated by α-helical and beta strand structural motifs. (c) Illustration of highest populated clusters with folded resides. α-Helical motifs are coloured in red, while beta conformations in blue. The black sphere indicates the last residue of the N-tail (point of attachment to the nucleosome), while the yellow sphere denotes the acetylated lysine.
The greater effect of H4K16Ac over H4K12Ac as well as the polyacetylated states (Fig. 3a,b) suggests that acetylation effects are sequence-dependent, non-additive, and not explained by histone charge reduction alone. Namely, acetylation dramatically increases not only the α-helical propensity of K16 but also of the two residues preceding it (14–15), from around 7% to 21%. The stabilization of this three-residue α-helix brings residues 10–12 and 18–19 closer together, favouring a β-hairpin conformation not observed in the wild-type H4 (Fig. 3c). We also observe that acetylation increases slightly the β-strand propensity of residues 12–14 and 21–23, due to the stabilization of both the β-hairpin loop (mainly formed by positively charged residues) and the N-termini-to-C-termini partially-negative-to-partially-positive dipole in residues 12–16 (KGGAKAc). Thus, removal of the positive charge upon acetylation turns K16 into a more efficient C-terminal cap for the β-hairpin that H4 transiently samples and allows stronger dipolar interactions among the two β strands59. By contrast, K12Ac decreases formation of β-hairpins, possibly due to the destabilization of the overall β-strand dipole. Consistently, the K12,16 diacetylation increases the propensity for folding compared to the wild-type tail, but decreases it compared to K16Ac alone. The tri- and tetraacetylated cases show similar structural patterns with a preference towards α-helix formation, and a similar total folding compared to the wild type tail. For H3 and H2B, the mono-acetylations analysed do not change tail behaviour significantly, but the H2B tetra-acetylation increases notably helicity and stiffness of H2B (see Supporting Information and Fig. S8); these observations further support the sequence-dependent role of lysine acetylation.
A decrease of histone tail disorder limits internucleosome interactions in dinucleosomes.
Our 1-μs explicitsolvent all-atom dinucleosome simulations (Fig. 4) investigated the role of histone tails in dinucleosome interactions by comparing the number of histone tail-mediated nucleosome-nucleosome contacts between two stacked nucleosomes with wild-type versus H4K16Ac tails. Our dinucleosome models were built using the positions and orientations of two stacked nucleosomes in the tetranucleosome crystal. As observed in Fig. 4, the two nucleosomes are laterally displaced with respect to each other, mimicking the disposition of nucleosomes in experimentally-based chromatin models40, 60. We have explored two simulation setups, each with a different starting configuration of the H4 tails. In the first setup (setup a; Fig. 4a), the H4 tails extend from their point of attachment on their parent nucleosome to the acidic patch of their neighbouring nucleosome; this setup is expected to occur within compact chromatin. In the second setup (setup b; Fig. 4b), the H4 tails extend from their point of attachment on their parent nucleosome towards the nucleosomal DNA. Our simulations reveal that the dinucleosome with wild type tails in setup a – in which the H4 tails can interact with their neighbouring acidic patch – within the time scale of our simulations are stabilized by 40% more internucleosome interactions than that in setup b.
For both setups, the average number of H4-mediated internucleosome contacts – the sum of tail contacts with the neighbouring DNA and with the neighbouring proteins – decreases significantly in the presence of H4K16Ac (by 119 contacts in setup a, and by 77 contacts in setup b; Fig. 4c). The total number of DNA/H4 tail contacts is not notably altered upon acetylation (Fig. 4c), suggesting that acetylation decreases the ability of the H4 to extend and reach the neighbouring nucleosome but not the binding affinity of the H4 tail with the DNA. An all-atom umbrella sampling study of H4 with DNA had showed that the free energy stabilizing the DNA/H4 bound states increases upon K16 acetylation13a.
Histone tail folding triggers chromatin fiber opening.
Changes in the structure of histone tails can dramatically impact chromatin organization since the tails provide crucial bridging interactions with the charged surfaces of neighbouring nucleosomes (non-parental nucleosome) and the linker DNAs joined to other cores (non-parental linker DNA). Clearly, many factors affect chromatin structure in a cooperative way. Indeed, different internal and external factors favour different fiber organizations 61, and a fluid polymorphic structure, including irregularly folded 10-nm fibers, is more consistent with the heterogeneous conditions of in vivo chromatin 17e, 62. The length of the DNA linking consecutive nucleosomes is an important factor affecting chromatin organization, and its effects are intertwined with LH presence, ionic conditions, and others 17c, 17e, 47e, 63. The linker DNA length varies within and across species, tissues, cell-cycle states, and even within single chromatin arrays. While transcriptionally active cells are characterized by short-to-medium linker DNA lengths of ~7–42 bp, mature inactive cells have mediumto-long values of ~43–93 bp 63b. Linker DNA variations also change the way in which histone tails interact with other chromatin components and thus affect chromatin structure overall 17c, 17e, 63a.
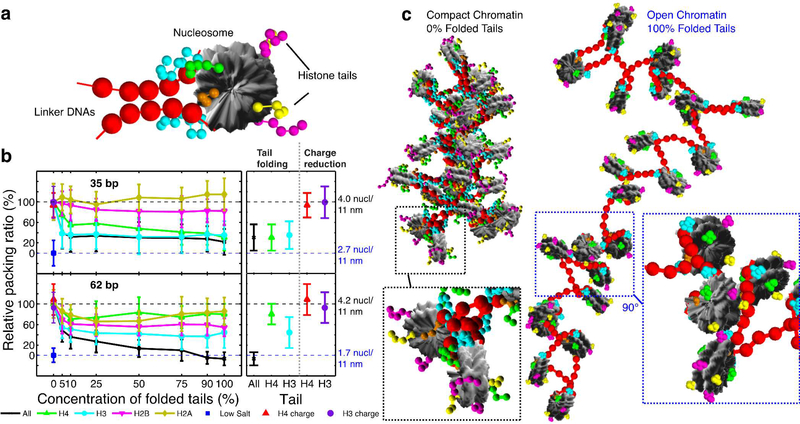
Our oligonucleosome coarse-grained model17a-c, 17e (see Methods, Fig. 5a, and Supporting Information), which has been developed and validated over the past 10 years17a-c, 17e, 47, has a DNA-DNA inter-bead separation of ~9 bp and can thus treat chromatin with linker DNA lengths of 26, 35, 44, 53, 62 bp, etc. Using this model we compare the degree of compaction of 24-nucleosome arrays without LH (more relevant to acetylation 2d) under physiological conditions of monovalent salt and two different linker DNA lengths (35 and 62 bp, corresponding to nucleosome repeat lengths of 182 and 209 bp, respectively). We analyse 80 different fibers with different distributions of folded-and-semirigid/unfolded-and-flexible tails mimicking our dinucleosome findings to help characterize how transient secondary structural elements in the different histone tails impact chromatin compaction.
Figure 5. Analysis of the effects of the folding of the histone tails on the compaction of chromatin fibers.
(a) Illustration of the coarse-grained model; nucleosome charged surfaces are shown in grey, linker DNA beads in red, and histone tail beads in green (H4), cyan (H3), magenta (H2B), yellow (H2A), and orange (H2A C-tail). (b) Effects of histone tail folding concentration on the compaction of chromatin measured through packing ratios of 24-nucleosome system with two different linker-DNA lengths (35 and 62 bp). The left section of the graph describes how the relative packing ratio (see Supporting Information) decreases as the concentration of the folded tails increases. The relative packing ratios varies between 0% for a fully unfolded fiber at low salt concentrations (10 mM NaCl), and 100% for a compact conformation at physiological salt (150 mM NaCl) and no folded tails (0% folded tails). The right section of the graph compares the effects of having 100% folded tails (black: all tails, green: H4 tail, or cyan: H3 tail) or reducing the charge by 1e to mimic the effect of acetylation but keeping the tails fully flexible. (c) Simulation snapshots for the 62-bp linker-DNA system with fully folded and flexible tails.
We find that the H3 and H4 wild-type histone tails have dominant roles in mediating internucleosome interactions (Fig. S9). Fig. 5b reveals that for the two different types of chromatin arrays analyzed (linker DNAs lengths of 35 bp and 62 bp), even a small presence of folded histone tails (>5%), induces chromatin fiber unfolding. Thus, structure-rich collapsed histone tails, for example by H4K16Ac, are consistent with weaker internucleosome contacts and accordingly poorer chromatin compaction.
We also see that chromatin decondensation due to histone-tail folding is linker-DNA-length dependent. For 62-bp DNA linkers, histone tail folding has an additive effect on chromatin decondensation; fiber opening requires the simultaneous folding of various types of histone tails, with H3- followed by H2B-tail folding being the most influential. When >50% of all histone tails are folded, the opening effect is similar to immersing the fibers in a low salt environment (Fig. 5c). Indeed, these fibers with fully folded histone tails show no salt-dependent compaction. Although the structure of 35-bp linker-DNA chromatin is more resistant overall to histone tail folding, due to higher mechanical constraints imposed by the shorter linker DNAs, folding of either H4 or H3 tails can decrease the compaction to near low-salt values. A 30% presence of H4 K16Ac has been shown to be sufficient to almost fully decompact chromatin without LH2d and a 55-bp linkerDNA.
For both linker DNAs, we verified that the crucial reduction of internucleosomal contacts we observe is not to charge reduction per se by repeating calculations for wild-type histone tails conformations, where the beads charges corresponding to H4K16Ac or H3K14Ac were reduced by 1 electron to mimic charge neutralization due to lysine monoacetylation. Charge reduction did not change the computed packing ratios (Fig. 5c), suggesting that epigenetic marks, such as lysine acetylation, do not induce fiber decompaction through charge neutralization per se, but through a cooperative multiscale mechanism involving the dramatic reduction of crucial stabilization of internucleosome interactions due to unavailability of flexible histone tails for potential contacts. Thus, we expect that other factors that enhance tail order and limit their availability, like protein binding, will have similar effects.
DISCUSSION
Using a tailored multiscale modelling approach, we show that wild type histone tails are highly flexible and mostly disordered protein regions and that these characteristics of histone tails facilitate contacts with nonparent nucleosomes because unstructured tails have larger interaction surface areas, and thus more potential binding opportunities. In addition, despite being mostly unstructured, wildtype histone tails adopt a wide range of different transient and short secondary structural motifs that might facilitate binding to a diversity of molecules (e.g. histones, DNA, and chromatin remodellers) through a conformational selection mechanism. Thus, histone tail flexibility and disorder are crucial for control of chromatin compaction via their use for recruitment of various molecules. Another important aspect of histone tail flexibility and disorder is to ensure that a larger fraction of the tail residues are exposed to the solvent, where they are more readily available to epigenetic-modifying enzymes.
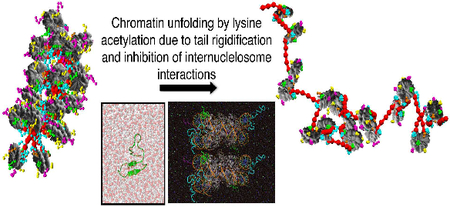
Our studies also reveal that lysine acetylation decreases the tail’s flexibility and increases their secondary structure folding propensity in a sequence-dependent manner. Indeed, although charge reduction by polyacetylation is greater, H4 polyacetylation of lysines 5,8,12 and 16 has a negligible effect in the characteristics of the tails, when compared to single K16 acetylation. Our dinucleosome simulations combined with our coarse grained modelling further reveal that the structuring effect of lysine acetylation limits the ability of the tails to extend, reach nucleosome neighbours, and establish a range of crucial proteinprotein and protein-DNA chromatin-compacting internucleosome interactions. This compromise leads to chromatin fiber opening.
Our results agree with a study that combines short (60 ns) REMD of H4 and H4K16Ac tails in explicit solvent with umbrella sampling simulations of DNA/tail systems, and demonstrates that such modification decreases the tail disorder and increases the tail/DNA bind affinity13a. The study further hypothesizes that K16Ac might induce chromatin decompaction through increase of the tail/parental DNA interactions and the sequestration of the H4 tail into its own nucleosome. Our dinucleosome simulations reveal that H4K16Ac limits the interactions of the tail with both the neighbouring protein and DNA, and our coarse-grain simulations demonstrate that enhanced rigidity of the histone tails decrease the compaction of the chromatin fiber dramatically.
In addition, we find that the effect of acetylation in tuning fiber compaction is linker DNA dependent. Our coarsegrained model suggests that H4 modifications that reduce the tail’s flexibility will have a stronger effect in chromatin with shorter linker DNAs (<62 bp) without LHs, while those on H3 tails will affect longer linker DNA arrays more strongly. This can be rationalized from the position and length of these histone tails. The H4 tail is more important for short linker DNAs because of its optimal position on the nucleosome surface, while the much longer H3 tail can reach its nonparental DNA and nucleosome neighbours more easily when they are further away in longer NRL fibers. This is consistent with experiments showing that H4K16Ac is present in decondensed yeast chromatin (linker DNA ~ 18 bp 55) 56, where 80% of H4K16 residues are acetylated56, and in active male-X Droshopila chromosomes (linker DNA ~ 43 bp 64) 65, while H4K12Ac is preferentially found in compact yeast chromatin57. Furthermore, within the H4 tail, residues 14 to 24 of H4 are essential for the condensation of 30-bp linkerDNA chromatin arrays66. H4K16Ac also opens chromatin fibers in 20-bp linker DNA 12-unit reconstituted arrays 2c and in 55-bp linker DNA 60-unit arrays in the absence of LHs 2d.
CONCLUSION
Our multiscale study sheds light into the structure and dynamics of the histone tails and identifies a mechanism by which an epigenetic modification can modulate the degree of compaction of chromatin and the corresponding accessibility of the DNA. Our results suggest that the disordered nature of wild type histone tails controls chromatin compaction and promotes the recycling of the same histones to recruit a range of different molecular partners. Structural disorder and flexibility thus allows histone tails to perform two crucial functions: (a) contact neighbouring nucleosomes and linker DNA to directly promote chromatin compaction, and (b) interact with non-histone proteins for indirect chromatin structure regulation.
That the flexibility of histone tails, which favours internucleosome interactions, can be decreased by lysine acetylation to induce different levels of chromatin deconden-sation underscores that this increased structural order of the tails upon acetylation is not merely a charge reduction phenomenon, but a subtle sequence-specific effect. Thus, the decrease of internucleosome interactions through acetylation is due to the inability of the structured histone tails to extend and reach their neighbouring nucleosomes, rather than reduced binding with DNA67. Furthermore, we propose that the effects of acetylation on chromatin compaction are linker-DNA-length dependent, with H4 modifications being most important for short-to-medium linker DNAs (<62 bp), and H3 modifications being most important for medium-to-long linker DNAs (>62 bp).
Taken together, our multiscale computational results reveal a molecular mechanism by which an epigenetic modification of the histone tails controls chromatin structure. This key role of histone tail flexibility on chromatin compaction might also help explain how molecules that remodel chromatin by binding to the tails might act to modify gene expression. These results provide a compelling illustration of the manner by which subtle modifications of the molecular structure of histone tails can profoundly impact chromatin structure and in turn gene expression. Our work also opens new avenues for multiscale computations of complex biological systems.
Supplementary Material
ACKNOWLEDGMENT
We thank the Bai group for sharing with us their NMR chemical shifts, and Dr. Annick Dejaegere for sending us CHARMM parameters for lysine acetylated residues.
Funding Sources
Computing support from the Barcelona Supercomputing Centre MareNostrum Supercomputer, and from New York University High Performance Computing Union Square cluster is acknowledged. This work was supported by the European Union Seventh Framework Programme (FP7/2007– 2013) [275096 to R.C.-G. and M.O.]; Sara Borrell Fellowships [to G.P. and M.O.]; the Spanish MINECO [BIO2012–32868 to M.O.]; the Spanish National Institute of Bioinformatics (INB) [to M.O.]; the European Research Council (ERC) [Advanced Investigator Grant to M.O.]; the National Science Foundation [MCB0316771 to T. S.]; the National Institutes of Health [R01 GM55164 to T. S.]; Philip Morris USA [to T. S.]; and Philip Morris International [to T.S.]. M.O. is an ICREA-Academia fellow.
Footnotes
ASSOCIATED CONTENT
Supporting Information. Our Supporting Information includes text describing our coarse-grained model, Monte Carlo simulation approach, calculation of chromatin packing ratios and tail mediated internucleosome interaction frequencies. It also contains four tables and eight figures.
REFERENCES
- 1. (a).Luger K; Mader AW; Richmond RK; Sargent DF; Richmond TJ, Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 1997, 389 (6648), 251–60 [DOI] [PubMed] [Google Scholar]; (b) Richmond TJ; Davey CA, The structure of DNA in the nucleosome core. Nature 2003, 423 (6936), 145–50. [DOI] [PubMed] [Google Scholar]
- 2. (a).Bannister AJ; Kouzarides T, Regulation of chromatin by histone modifications. Cell Res 2011, 21 (3), 381–395 [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Garcia-Ramirez M; Dong F; Ausio J, Role of the Histone Tails in the Folding of Oligonucleosomes Depleted of Histone-H1. J Bio Chem 1992, 267 (27), 19587–19595 [PubMed] [Google Scholar]; (c) Shogren-Knaak M; Ishii H; Sun JM; Pazin MJ; Davie JR; Peterson CL, Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 2006, 311 (5762), 844–7 [DOI] [PubMed] [Google Scholar]; (d) Robinson PJJ; An W; Routh A; Martino F; Chapman L; Roeder RG; Rhodes D, 30 nm chromatin fibre decompaction requires both H4-K16 acetylation and linker histone eviction. Journal of molecular biology 2008, 381 (4), 816–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wolffe AP; Hayes JJ, Chromatin disruption and modification. Nucleic acids research 1999, 27 (3), 711–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allis CD, Translating the histone code: A tale of tails. Abstr Pap Am Chem S 2002, 224, U45–U45. [Google Scholar]
- 5.Luger K; Richmond TJ, The histone tails of the nucleosome. Curr Opin Genet Dev 1998, 8 (2), 140–146. [DOI] [PubMed] [Google Scholar]
- 6. (a).Gao M; Nadaud PS; Bernier MW; North JA; Hammel PC; Poirier MG; Jaroniec CP, Histone H3 and H4 N-Terminal Tails in Nucleosome Arrays at Cellular Concentrations Probed by Magic Angle Spinning NMR Spectroscopy. Journal of the American Chemical Society 2013, 135 (41), 15278–15281 [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Kato H; Gruschus J; Ghirlando R; Tjandra N; Bai YW, Characterization of the N-Terminal Tail Domain of Histone H3 in Condensed Nucleosome Arrays by Hydrogen Exchange and NMR. Journal of the American Chemical Society 2009, 131 (42), 15104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang X; Moore SC; Laszckzak M; Ausio J, Acetylation increases the alpha-helical content of the histone tails of the nucleosome. The Journal of biological chemistry 2000, 275 (45), 35013–20. [DOI] [PubMed] [Google Scholar]
- 8.Baneres JL; Martin A; Parello J, The N tails of histones H3 and H4 adopt a highly structured conformation in the nucleosome. Journal of molecular biology 1997, 273 (3), 503–508. [DOI] [PubMed] [Google Scholar]
- 9.Zhou BR; Feng H; Ghirlando R; Kato H; Gruschus J; Bai Y, Histone H4 K16Q mutation, an acetylation mimic, causes structural disorder of its N-terminal basic patch in the nucleosome. Journal of molecular biology 2012, 421 (1), 30–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allahverdi A; Yang R; Korolev N; Fan Y; Davey CA; Liu C-F; Nordenskiöld L, The effects of histone H4 tail acetylations on cation-induced chromatin folding and selfassociation. Nucleic acids research 2011, 39, 1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bang E; Lee CH; Yoon JB; Lee DW; Lee W, Solution structures of the N-terminal domain of histone H4. The journal of peptide research : official journal of the American Peptide Society 2001, 58 (5), 389–98. [DOI] [PubMed] [Google Scholar]
- 12. (a).Levitt M; Warshel A, Computer simulation of protein folding. Nature 1975, 253 (5494), 694–8 [DOI] [PubMed] [Google Scholar]; (b) Su CC; Long F; Zimmermann MT; Rajashankar KR; Jernigan RL; Yu EW, Crystal structure of the CusBA heavy-metal efflux complex of Escherichia coli. Nature 2011, 470 (7335), 558–62 [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Whitford PC; Ahmed A; Yu Y; Hennelly SP; Tama F; Spahn CM; Onuchic JN; Sanbonmatsu KY, Excited states of ribosome translocation revealed through integrative molecular modeling. Proceedings of the National Academy of Sciences of the United States of America 2011, 108 (47), 18943–8 [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Liao T; Zhang Y; Kekenes-Huskey PM; Cheng Y; Michailova A; McCulloch AD; Holst M; McCammon JA, Multi-core CPU or GPU-accelerated Multiscale Modeling for Biomolecular Complexes. Molecular based mathematical biology 2013, 1 [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Zhao G; Perilla JR; Yufenyuy EL; Meng X; Chen B; Ning J; Ahn J; Gronenborn AM; Schulten K; Aiken C; Zhang P, Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics. Nature 2013, 497 (7451), 643–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. (a).Potoyan DA; Papoian GA, Regulation of the H4 tail binding and folding landscapes via Lys-16 acetylation. Proceedings of the National Academy of Sciences of the United States of America 2012, 109 (44), 17857–62 [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Potoyan DA; Papoian GA, Energy Landscape Analyses of Disordered Histone Tails Reveal Special Organization of Their Conformational Dynamics. Journal of the American Chemical Society 2011, 133 (19), 7405–7415. [DOI] [PubMed] [Google Scholar]
- 14.Korolev N; Yu H; Lyubartsev AP; Nordenskiold L, Molecular Dynamics Simulations Demonstrate the Regulation of DNA-DNA Attraction by H4 Histone Tail Acetylations and Mutations. Biopolymers 2014. [DOI] [PubMed] [Google Scholar]
- 15. (a).Bishop TC, Molecular dynamics simulations of a nucleosome and free DNA. J Biomol Struct Dyn 2005, 22 (6), 673–685 [DOI] [PubMed] [Google Scholar]; (b) Biswas M; Voltz K; Smith JC; Langowski J, Role of Histone Tails in Structural Stability of the Nucleosome. PLoS computational biology 2011, 7 (12) [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Ettig R; Kepper N; Stehr R; Wedemann G; Rippe K, Dissecting DNA-histone interactions in the nucleosome by molecular dynamics simulations of DNA unwrapping. Biophysical journal 2011, 101 (8), 1999–2008 [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Materese CK; Savelyev A; Papoian GA, Counterion atmosphere and hydration patterns near a nucleosome core particle. Journal of the American Chemical Society 2009, 131 (41), 15005–13 [DOI] [PubMed] [Google Scholar]; (e) Portella G; Battistini F; Orozco M, Understanding the connection between epigenetic DNA methylation and nucleosome positioning from computer simulations. PLoS computational biology 2013, 9 (11), e1003354. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Biswas M; Langowski J; Bishop TC, Atomistic simulations of nucleosomes. Wires Comput Mol Sci 2013, 3 (4), 378–392. [Google Scholar]
- 16. (a).Liu H; Duan Y, Effects of posttranslational modifications on the structure and dynamics of histone H3 N-terminal Peptide. Biophysical journal 2008, 94 (12), 4579–85 [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Yang D; Arya G, Structure and binding of the H4 histone tail and the effects of lysine 16 acetylation. Physical chemistry chemical physics : PCCP 2011, 13 (7), 2911–21. [DOI] [PubMed] [Google Scholar]
- 17. (a).Arya G; Schlick T, Role of histone tails in chromatin folding revealed by a mesoscopic oligonucleosome model. Proceedings of the National Academy of Sciences of the United States of America 2006, 103 (44), 16236–41 [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Arya G; Schlick T, A tale of tails: how histone tails mediate chromatin compaction in different salt and linker histone environments. The journal of physical chemistry. A 2009, 113 (16), 4045–59 [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Perisic O; Collepardo-Guevara R; Schlick T, Modeling studies of chromatin fiber structure as a function of DNA linker length. Journal of molecular biology 2010, 403 (5), 777–802 [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Kulaeva OI; Zheng G; Polikanov YS; Colasanti AV; Clauvelin N; Mukhopadhyay S; Sengupta AM; Studitsky VM; Olson WK, Internucleosomal interactions mediated by histone tails allow distant communication in chromatin. The Journal of biological chemistry 2012, 287 (24), 20248–57 [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Collepardo-Guevara R; Schlick T, Chromatin fiber polymorphism triggered by variations of DNA linker lengths. Proceedings of the National Academy of Sciences of the United States of America 2014, 111 (22), 8061–8066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turner BM, Histone acetylation and an epigenetic code. Bioessays 2000, 22 (9), 836–845. [DOI] [PubMed] [Google Scholar]
- 19.Ozer G; Luque A; Schlick T, The Chromatin Fiber: Multiscale Problems and Approaches. Curr Opin Cell Biol (Submitted) 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hess B; Kutzner C; van der Spoel D; Lindahl E, GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. J. Chem. Theory Comput 2008, 4 (3), 435–447. [DOI] [PubMed] [Google Scholar]
- 21.Darden T; York D; Pedersen L, Particle mesh Ewald: an Nlog(N) method for Ewald sums in large systems. J. Chem. Phys 1993, 98, 10089–10092. [Google Scholar]
- 22.Miyamoto S; Kollman PA, SETTLE: An Analytical Version of the SHAKE and RATTLE Algorithms for Rigid Water Models. J. Comput. Chem 1992, 13, 952–962. [Google Scholar]
- 23.Feenstra KA; Hess B; Berendsen HJC, Improving Efficiency of large time-scale of Molecular Dynamics Simulations of Hydrogen-rich Systems. J. Comput. Chem 1999, 20, 786–798. [DOI] [PubMed] [Google Scholar]
- 24.Bussi G; Donadio D; Parrinello M, Canonical sampling through velocity rescaling. J. Chem. Phys 2007, 126 (1), 014101. [DOI] [PubMed] [Google Scholar]
- 25.Berendsen HJC; Postma JPM; DiNola A; Haak JR, Molecular dynamics with coupling to an external bath. J. Chem. Phys 1984, 81, 3684–3690. [Google Scholar]
- 26.Best RB; Hummer G, Optimized molecular dynamics force fields applied to the helix-coil transition of polypeptides. The journal of physical chemistry. B 2009, 113 (26), 9004–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hornak V; Abel R; Okur A; Strockbine B; Roitberg A; Simmerling C, Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins 2006, 65 (3), 712–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Best RB; Zhu X; Shim J; Lopes PE; Mittal J; Feig M; Mackerell AD Jr., Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone phi, psi and side-chain chi(1) and chi(2) dihedral angles. Journal of chemical theory and computation 2012, 8 (9), 3257–3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Papamokos GV; Tziatzos G; Papageorgiou DG; Georgatos SD; Politou AS; Kaxiras E, Structural role of RKS motifs in chromatin interactions: a molecular dynamics study of HP1 bound to a variably modified histone tail. Biophysical journal 2012, 102 (8), 1926–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grauffel C; Stote RH; Dejaegere A, Force field parameters for the simulation of modified histone tails. Journal of computational chemistry 2010, 31 (13), 2434–51. [DOI] [PubMed] [Google Scholar]
- 31.Perez A; Marchan I; Svozil D; Sponer J; Cheatham TE 3rd; Laughton CA; Orozco M, Refinement of the AMBER force field for nucleic acids: improving the description of alpha/gamma conformers. Biophys. J 2007, 92 (11), 3817–3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jorgensen WL; Chandrasekhar J; Madura JD; Impey RW; Klein ML, Comparison of Simple Potential Functions for Simulating Liquid Water. Journal of Chemical Physics 1983, 79 (2), 926–935. [Google Scholar]
- 33.Smith DE; Dang LX, Computer simulations of NaCl association in polarizable water. J. Chem. Phys 1994, 100 (5), 3757–3766. [Google Scholar]
- 34.Humphrey W; Dalke A; Schulten K, VMD: visual molecular dynamics. Journal of molecular graphics 1996, 14 (1), 33–8, 27–8. [DOI] [PubMed] [Google Scholar]
- 35.Bateman A; Martin MJ; O’Donovan C; Magrane M; Apweiler R; Alpi E; Antunes R; Ar-Ganiska J; Bely B; Bingley M; Bonilla C; Britto R; Bursteinas B; Chavali G; Cibrian-Uhalte E; Da Silva A; De Giorgi M; Dogan T; Fazzini F; Gane P; Cas-Tro LG; Garmiri P; Hatton-Ellis E; Hieta R; Huntley R; Legge D; Liu WD; Luo J; MacDougall A; Mutowo P; Nightin-Gale A; Orchard S; Pichler K; Poggioli D; Pundir S; Pureza L; Qi GY; Rosanoff S; Saidi R; Sawford T; Shypitsyna A; Turner E; Volynkin V; Wardell T; Watkins X;Watkins; Cowley A; Figueira L; Li WZ; McWilliam H; Lopez R; Xenarios I; Bougueleret L; Bridge A; Poux S; Redaschi N; Aimo L; Argoud-Puy G; Auchincloss A; Axelsen K; Bansal P; Baratin D; Blatter MC; Boeckmann B; Bolleman J; Boutet E; Breuza L; Casal-Casas C; De Castro E; Coudert E; Cuche B; Doche M; Dornevil D; Duvaud S; Estreicher A; Famiglietti L; Feuermann M; Gasteiger E; Gehant S; Gerritsen V; Gos A; Gruaz-Gumowski N; Hinz U; Hulo C; Jungo F; Keller G; Lara V; Lemercier P; Lieberherr D; Lombardot T; Martin X; Masson P; Morgat A; Neto T; Nouspikel N; Paesano S; Pedruzzi I; Pilbout S; Pozzato M; Pruess M; Rivoire C; Roechert B; Schneider M; Sigrist C; Sonesson K; Staehli S; Stutz A; Sundaram S; Tognolli M; Verbregue L; Veuthey AL; Wu CH; Arighi CN; Arminski L; Chen CM; Chen YX; Garavelli JS; Huang HZ; Laiho KT; McGarvey P; Natale DA; Suzek BE; Vinayaka CR; Wang QH; Wang YQ; Yeh LS; Yerramalla MS; Zhang J; Consortium U, UniProt: a hub for protein information. Nucleic acids research 2015, 43 (D1), D204–D212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frishman D; Argos P, Knowledge-based protein secondary structure assignment. Proteins-Structure Function and Genetics 1995, 23 (4), 566–579. [DOI] [PubMed] [Google Scholar]
- 37.Ramachandran GN; Ramakrishnan C; Sasisekharan V, Stereochemistry of Polypeptide Chain Configurations. Journal of molecular biology 1963, 7 (1), 95-&. [DOI] [PubMed] [Google Scholar]
- 38.Patriksson A; van der Spoel D, A temperature predictor for parallel tempering simulations. Phys. Chem. Chem. Phys 2008, 10 (15), 2073–2077. [DOI] [PubMed] [Google Scholar]
- 39.Davey CA; Sargent DF; Luger K; Maeder AW; Richmond TJ, Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. Journal of molecular biology 2002, 319 (5), 1097–113. [DOI] [PubMed] [Google Scholar]
- 40.Schalch T; Duda S; Sargent DF; Richmond RK, X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature 2005, 436, 138–141. [DOI] [PubMed] [Google Scholar]
- 41.Camilloni C; Cavalli A; Vendruscolo M, Assessment of the Use of NMR Chemical Shifts as Replica-Averaged Structural Restraints in Molecular Dynamics Simulations to Characterize the Dynamics of Proteins. J. Phys. Chem. B 2013, 117 (6), 1838–1843. [DOI] [PubMed] [Google Scholar]
- 42.Camilloni C; De Simone A; Vranken WF; Vendruscolo M, Determination of secondary structure populations in disordered states of proteins using nuclear magnetic resonance chemical shifts. Biochemistry 2012, 51 (11), 2224–31. [DOI] [PubMed] [Google Scholar]
- 43.Kohlhoff KJ; Robustelli P; Cavalli A; Salvatella X; Vendruscolo M, Fast and accurate predictions of protein NMR chemical shifts from interatomic distances. J. Am. Chem. Soc 2009, 131 (39), 13894–5. [DOI] [PubMed] [Google Scholar]
- 44.Tribello GA; Bonomi M; Branduardi D; Camilloni C; Bussi G, Plumed 2: New Feathers for an Old Bird. Comput. Phys. Commun 2014, 185 (2), 604–613. [Google Scholar]
- 45.Laio A; Parrinello M, Escaping free-energy minima. Proc. Natl. Acad. Sci. U. S. A 2002, 99 (20), 12562–12566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Atkins PW; De Paula J, Atkins’ Physical chemistry. 9th ed.; Oxford University Press: Oxford; New York, 2010; p xxix, 972 p. [Google Scholar]
- 47. (a).Zhang Q; Beard DA; Schlick T, Constructing irregular surfaces to enclose macromolecular complexes for mesoscale modeling using the discrete surface charge optimization (DISCO) algorithm. Journal of computational chemistry 2003, 24 (16), 2063–74 [DOI] [PubMed] [Google Scholar]; (b) Arya G; Zhang Q; Schlick T, Flexible histone tails in a new mesoscopic oligonucleosome model. Biophysical journal 2006, 91 (1), 133–50 [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Beard DA; Schlick T, Computational modeling predicts the structure and dynamics of chromatin fiber. Structure 2001, 9 (2), 105–14 [DOI] [PubMed] [Google Scholar]; (d) Beard DA; Schlick T, Modeling salt-mediated electrostatics of macromolecules: the discrete surface charge optimization algorithm and its application to the nucleosome. Biopolymers 2001, 58 (1), 106–15 [DOI] [PubMed] [Google Scholar]; (e) Collepardo-Guevara R; Schlick T, The effect of linker histone’s nucleosome binding affinity on chromatin unfolding mechanisms. Biophysical journal 2011, 101 (7), 1670–80 [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Collepardo-Guevara R; Schlick T, Crucial role of dynamic linker histone binding and divalent ions for DNA accessibility and gene regulation revealed by mesoscale modeling of oligonucleosomes. Nucleic acids research 2012, 40 (18), 8803–17 [DOI] [PMC free article] [PubMed] [Google Scholar]; (g) Sun J; Zhang Q; Schlick T, Electrostatic mechanism of nucleosomal array folding revealed by computer simulation. Proceedings of the National Academy of Sciences of the United States of America 2005, 102 (23), 8180–5 [DOI] [PMC free article] [PubMed] [Google Scholar]; (h) Luque A; Collepardo-Guevara R; Grigoryev S; Schlick T, Dynamic condensation of linker histone C-terminal domain regulates chromatin structure. Nucleic acids research 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jian H; Vologodskii A; Schlick T, A Combined Wormlike-Chain and Bead Model for Dynamic Simulations of Long Linear DNA. Journal of Computational Physics 1997, 136, 168–179. [Google Scholar]
- 49.Schuler B; Lipman EA; Steinbach PJ; Kumke M; Eaton WA, Polyproline and the “spectroscopic ruler” revisited with single-molecule fluorescence. Proceedings of the National Academy of Sciences of the United States of America 2005, 102 (8), 2754–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu YJ; Weers B; Stellwagen NC, DNA persistence length revisited. Biopolymers 2001, 61 (4), 261–275. [DOI] [PubMed] [Google Scholar]
- 51.Lavelle C; Victor JM; Zlatanova J, Chromatin fiber dynamics under tension and torsion. International journal of molecular sciences 2010, 11 (4), 1557–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.De Simone A; Cavalli A; Hsu ST; Vranken W; Vendruscolo M, Accurate random coil chemical shifts from an analysis of loop regions in native states of proteins. J. Am. Chem. Soc 2009, 131 (45), 16332–3. [DOI] [PubMed] [Google Scholar]
- 53.Wang XD; Hayes JJ, Acetylation mimics within individual core histone tail domains indicate distinct roles in regulating the stability of higher-order chromatin structure. Mol Cell Biol 2008, 28 (1), 227–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Garciaramirez M; Rocchini C; Ausio J, Modulation of Chromatin Folding by Histone Acetylation. Journal of Biological Chemistry 1995, 270 (30), 17923–17928. [DOI] [PubMed] [Google Scholar]
- 55.Thomas JO; Furber V, Yeast Chromatin Structure. FEBS letters 1976, 66 (2), 274–280. [DOI] [PubMed] [Google Scholar]
- 56.Smith CM; Gafken PR; Zhang ZL; Gottschling DE; Smith JB; Smith DL, Mass spectrometric quantification of acetylation at specific lysines within the amino-terminal tail of histone H4. Anal Biochem 2003, 316 (1), 23–33. [DOI] [PubMed] [Google Scholar]
- 57.Braunstein M; Sobel RE; Allis CD; Turner BM; Broach JR, Efficient transcriptional silencing in Saccharomyces cerevisiae requires a heterochromatin histone acetylation pattern. Mol Cell Biol 1996, 16 (8), 4349–4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. (a).Luebben WR; Sharma N; Nyborg JK, Nucleosome eviction and activated transcription require p300 acetylation of histone H3 lysine 14. Proceedings of the National Academy of Sciences of the United States of America 2010, 107 (45), 19254–9 [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Schiltz RL; Mizzen CA; Vassilev A; Cook RG; Allis CD; Nakatani Y, Overlapping but distinct patterns of histone acetylation by the human coactivators p300 and PCAF within nucleosomal substrates. The Journal of biological chemistry 1999, 274 (3), 1189–92. [DOI] [PubMed] [Google Scholar]
- 59.FarzadFard F; Gharaei N; Pezeshk H; Marashi SA, Beta-sheet capping: Signals that initiate and terminate beta-sheet formation. J Struct Bio 2008, 161 (1), 101–110. [DOI] [PubMed] [Google Scholar]
- 60.Song F; Chen P; Sun D; Wang M; Dong L; Liang D, Cryo-EM study of the chromatin fiber reveals a double helix twisted by tetranucleosomal units. Science 2014, 344, 376–380. [DOI] [PubMed] [Google Scholar]
- 61.Schlick T; Hayes J; Grigoryev S, Toward convergence of experimental studies and theoretical modeling of the chromatin fiber. The Journal of biological chemistry 2012, 287 (8), 5183–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. (a).Maeshima K; Imai R; Tamura S; Nozaki T, Chromatin as dynamic 10-nm fibers. Chromosoma 2014, 123 (3), 225–37; [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Ozer G; Luque A; Schlick T, Current opinion in structural biology 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. (a).Routh A; Sandin S; Rhodes D, Nucleosome repeat length and linker histone stoichiometry determine chromatin fiber structure. Proceedings of the National Academy of Sciences of the United States of America 2008, 105 (26), 8872–8877 [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Correll SJ; Schubert MH; Grigoryev SA, Short nucleosome repeats impose rotational modulations on chromatin fibre folding. The EMBO journal 2012, 31 (10), 2416–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sun FL; Cuaycong MH; Elgin SCR, Long-range nucleosome ordering is associated with gene silencing in Drosophila melanogaster pericentric heterochromatin. Mol Cell Biol 2001, 21 (8), 2867–2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Akhtar A; Becker PB, Activation of transcription through histone H4 acetylation by MOF, an acetyltransferase essential for dosage compensation in Drosophila. Mol Cell 2000, 5 (2), 367–375. [DOI] [PubMed] [Google Scholar]
- 66.Dorigo B; Schalch T; Bystricky K; Richmond TJ, Chromatin fiber folding: requirement for the histone H4 Nterminal tail. Journal of molecular biology 2003, 327 (1), 85–96. [DOI] [PubMed] [Google Scholar]
- 67.Hong L; Schroth GP; Matthews HR; Yau P; Bradbury EM, Studies of the DNA-Binding Properties of Histone H4 Amino Terminus - Thermal-Denaturation Studies Reveal That Acetylation Markedly Reduces the Binding Constant of the H4 Tail to DNA. Journal of Biological Chemistry 1993, 268 (1), 305–314. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.